Abstract
Degradation of dysfunctional intracellular components in the lysosome system can occur through three different pathways, i.e., macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). In this review, we focus on CMA, a type of autophagy distinct from the other two autophagic pathways owing to its selectivity, saturability and competitivity by which a subset of long-lived cytosolic soluble proteins are directly delivered into the lysosomal lumen via specific receptors. CMA participates in quality control to maintain normal cell functions by clearing “old” proteins and provides energy to cells under nutritional stress. Deregulation of CMA has recently been shown to underlie some diseases, especially neurodegenerative disorders for which the decline with age in the activity of CMA may become a major aggravating factor. Therefore, targeting aberrant alteration in CMA under pathological conditions could serve as a potential therapeutic strategy for treating related diseases.
Keywords: CMA, Lysosome, Hsc70, Lamp2a, Autophagy, Unibiquitin
Introduction
Maintaining a balance between protein synthesis and degradation is absolutely essential for proper cellular function, homeostasis and survival in a changing extracellular environment. The two major protein degradation mechanisms in eukaryotes are the ubiquitin-proteasome system (UPS) and the autophagy-lysosome system [1, 2]. Collectively, they (1) exert quality control by clearing damaged or incorrectly synthesized proteins, (2) recycle proteins that are no longer needed into constitutive amino acid components, (3) mount the cellular defense by breaking down components of various invading pathogens, and (4) regulate cellular response to stress or the alteration in extracellular micro-environments, assisting cells in adapting to new changes [3, 4].
The UPS is a multi-subunit protease complex in the cytosol that permits entry and subsequent degradation of proteins tagged with one or more covalently bound ubiquitin molecules [5]. Ubiquitin is a small protein highly conserved from yeast to man. It was first described in the context of protein degradation, but later shown to participate in regulation of other cellular processes, such as endocytosis, signal transduction and DNA repair. Conjugation of ubiquitin is a complex reaction that requires E1, E2 and E3 enzymes, and leads to the formation of an isopeptide bond between the C-terminal glycine of ubiquitin and the 3-amino group of a lysine residue of the substrate protein [6, 7]. Subunits of the regulatory complex of the proteasome recognize the ubiquitin tag, remove it and mediate the unfolding of the substrates required to gain access to the catalytic region of the proteasome barrel. With certain exceptions, most proteasome substrates have short half-lives [6, 8]. Interestingly, recent studies suggest that ubiquitin and lysosomal pathways are linked. For example, new evidence suggests that attachment of ubiquitin to various cellular cargos not only constitutes a degradation signal for proteasome, but also serves to signal for cargo removal by lysosomal system via autophagy [2, 9].
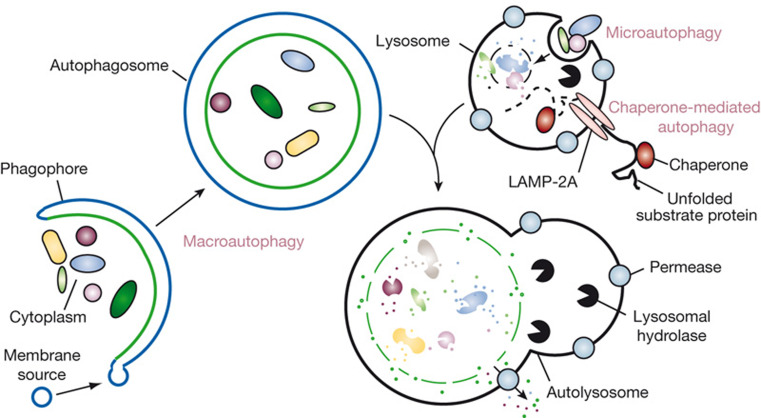
Autophagy is a conserved cellular “self-eating” process that involves sequestration and delivery of cytosolic components to the lysosome for degradation and recycling [10]. This evolutionarily conserved process can be categorized into three classes depending on their respective sequestration and delivery mechanism (Fig. 1) [11]. In macroautophagy, a double-membrane vesicle termed the autophagosome is formed to engulf long-lived proteins and organelles. The autophagosome is subsequently fused with a lysosome, releasing its cargo for degradation by lysosomal hydrolases. The resultant nucleotides, amino acids and fatty acids are eventually recycled back into the cytosol for reuse. In microautophagy, the sequestration of cytosolic content is facilitated by direct invagination or exvagination of lysosomal membrane, and subsequent budding of the invaginated vesicles into the lysosomal lumen releases the sequestered cytosolic material. In contrast to the vesicle-mediated substrate delivery of macro and microautophagy, chaperone-mediated autophagy (CMA) targets and delivers substrate proteins directly across the lysosomal membrane via the specific receptor [9, 12]. Only proteins containing a consensus peptide sequence are recognized by a chaperone complex. This CMA substrate-chaperone complex locates to the lysosome through interaction with the lysosome receptor and translocates the substrate across the lysosomal membrane with the assistance of lysosomal chaperones on the lumenal side [12, 13]. In this review, we first describe the machinery of CMA, summarize the regulation mechanisms involved in activation of CMA, and finally discuss the biological consequences of CMA under various physiological and pathological conditions, and relevant therapeutic strategies via regulating CMA.
Fig. 1.
Autophagy refers to the conserved degradation of intracellular components by lysosomes. In mammals, three types of autophagy have been described: macroautophagy, microautophagy and CMA. Adapted from [11] with permission
History of CMA
In the early 1980s, Professor Dice’s group first reported that radiolabeled RNase A introduced into the cytoplasm of human fibroblasts by using erythrocyte-mediated microinjection or osmotic lysis of pinosomes was degraded with a half-life of approximately 90 h in the presence of serum, whereas in response to serum deprivation its rate of degradation was enhanced 1.6-fold [14]. This enhanced breakdown following serum withdrawal was highly selective and based on a feature present within the N-terminal 20 amino acids of RNase A [15]. During subsequent years, the essential motifs related to KFERQ were identified in proteins serving as substrates of this selective degradation pathway, which is referred to as the selective pathway for degradation of cytosolic proteins by lysosomes [16, 17]. In 1989, a 73-kDa heat shock cognate protein (Hsc73, now commonly referred to as Hsc70) was found to bind to KFERQ-like regions in intracellular proteins that are targeted for lysosomal degradation in response to serum deprivation [18]. In 1994, isolated rat liver lysosomes were used to probe the selective binding and uptake of RNase A and glyceraldehyde-3-phosphate dehydrogenase. Their uptake and degradation by lysosomes were progressively activated in rat liver by starvation [19]. In 1996, Lamp2a, also named Lgp96 (lysosomal glycoprotein of 96 kDa), was identified as a receptor for the selective import and degradation of proteins within lysosomes [20]. In 1997, an intralysosomal Hsc73 was determined to be required for the selective pathway of lysosome-mediated protein degradation [21]. Since 2000, this pathway has been formally named chaperone-mediated autophagy (CMA) [22, 23]. Throughout the subsequent decade, new substrates, more detailed machinery, new components, physiological roles and associated diseases for CMA have been extensively investigated and elucidated [3].
Machinery of CMA
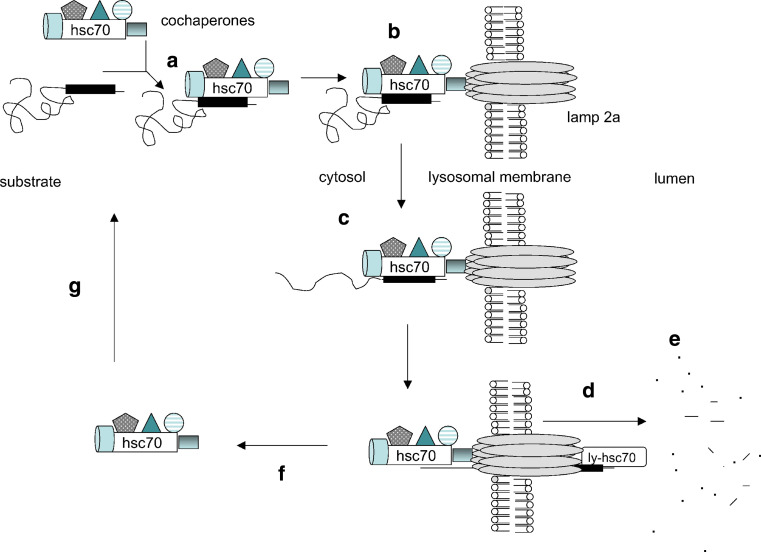
CMA uses the unique and distinctive machinery from the other two autophagy pathways to carry out this process [3, 24]. The basic machinery consists of at least three types of proteins, including (1) chaperone proteins, which are responsible for recognizing substrates based on their specific motifs and delivering them to lysosomes, (2) receptor proteins, which bind and transport/pull substrates into lysosome lumens, and (3) substrate proteins, which are a subset of soluble cytosolic proteins containing specific motifs related to KFERQ. Following activation of CMA, these three subsets of proteins collaborate and complete the process (Fig. 2) [3, 25].
Fig. 2.
The diagram of proposed mechanisms of CMA: a Hsc70 with co-chaperones recognizes a KFERQ-related peptide in cytosolic substrate proteins; b the complex binds to the Lamp2a receptor on the lysosomal membrane; c the substrate protein is unfolded before traversing the lysosomal membrane; d lys-Hsc70 pulls the substrate into the lysosome matrix; e the substrate protein is degraded by lysosomal proteases; f the Hsc70-cochaperone complex is released from the lysosomal membrane; g Hsc70 is available to bind to another CMA substrate. Adapted from [25] with permission
Workshop—lysosome
Lysosomes are the primary catabolic compartment of eukaryotic cells. The name lysosome derives from the Greek words lysis, which means dissolution or destruction, and soma, which means body [24]. Lysosomes were discovered by the Belgian cytologist Christian de Duve in 1949 [26]. They are created by the addition of hydrolytic enzymes to early endosomes from the Golgi apparatus [24]. The membrane around a lysosome allows the digestive enzymes to work at pH 5.1–5.5, which is optimal to these acidic hydrolases. Lysosomes fuse with vacuoles and dispense their enzymes within digesting the contents [27].
A healthy cell is dependent on the proper targeting of newly synthesized lysosomal proteins. Two classes of proteins are essential for the function of lysosomes: soluble lysosomal hydrolases (i.e., acid hydrolases) and integral lysosomal membrane proteins (LMPs) [24]. Lysosomal acidic pH and the large variety of hydrolases present in the lysosomal lumen (including proteases, lipases, glycosidases and nucleases) confer upon this organelle its high capacity of degradation and mediate complete breakdown of all types of molecules. In addition to bulk degradation, lysosomal hydrolases are involved in antigen processing, degradation of the extracellular matrix and initiation of apoptosis [24, 28]. LMPs reside mainly in the lysosomal limiting membrane and have diverse functions, including acidification of the lysosomal lumen, protein import from the cytosol, membrane fusion and transport of degradation products to the cytoplasm [29]. The main LMPs are lysosome-associated membrane protein 1 and 2 (Lamp1 and 2), lysosomal integral membrane protein 2 and tetraspanin CD63 [24]. Substrates can reach lysosomes via heterophagy, in which the cargo to be degraded originates at the plasma membrane or extracellularly, or via autophagy, for cargo located in the cytosol [3, 24, 30].
Chaperones—Hsc70 and partners
Hsc70, a 73-kDa protein, is the constitutive member of the heat shock protein 70 family of chaperone [19, 25]. Hsc70s have been found to be involved in many cellular processes including dissociation of clathrin and assembly proteins [25, 31]. Binding of Hsc70 to substrate proteins is regulated by ATP binding and hydrolysis, and the ADP-bound form of Hsc70 has the highest affinity for protein substrates for CMA [19, 32]. Hsc70 is located in the cytosol or in the lumen of lysosomes. Cytosolic Hsc70 (cyt-Hsc70) can recognize a peptide sequence including the KFERQ motif in CMA substrate proteins and aid in their transport to the lysosomal receptor [3, 19]. Cyt-Hsc70 docked to the lysosomal membrane helps to unfold substrate proteins, a necessary step for their entry into lysosomes [33].
Other co-chaperones interact with Hsc70 and regulate its activities. Hsp40 may activate the ATPase activity of Hsc70 to facilitate substrate binding; and the Hsp70-interacting protein (Hip) stimulates the assembly of Hsc70 with Hsp40 and the protein substrate [25]. Cell division cycle 48 (Cdc48) can also enhance the activity of Hsc70-Hsp40 complexes [32, 34]. Hsc70-Hsp90 organizing protein acts as an adapter between Hsc70 and Hsp90, which recognize unfolded regions within proteins and prevent substrate protein aggregation [25, 32]. There are co-chaperones of Hsp90 that may be in the molecular chaperone complex. Activator of Hsp90 ATPase is a family of heat shock proteins that activates the ATPase activity of Hsp90 and thereby stimulates both protein binding and release [32, 35]. The Bcl2-associated athanogene 1 protein (Bag-1) was initially described as a co-chaperone of Hsc70 that uncouples the ATPase cycle from substrate binding [36]. But several subsequent studies showed that it functions as a nucleotide exchange factor that stimulates substrate release [37]. The carboxyl terminus of Hsc70-interacting protein (Chip), which can regulate protein refolding [38], acts mainly as a chaperone-associated ubiquitin ligase to stimulate the degradation of Hsc70 client proteins [39]. The chaperone complex present on the cytosolic face of lysosomal membranes is linked to Lamp2a by the substrate protein via a site that is different from sites of interaction for the molecular chaperone complex. Each of the components in the molecular chaperone complex is required for transport of substrates into the lysosome lumen [25, 32].
Both Hsc70 and Hsp90 have been found to be also present in the lysosomal lumen. Lysosomal-Hsc70 (lys-Hsc70) is required for the CMA pathway [21]. The more active population of lysosomes contains abundant lys-Hsc70, whereas the less active ones contain little Hsc70. The latter group of lysosomes can be made more active for CMA if they are allowed to take up Hsc70. A role for lys-Hsc70 in the uptake of substrate proteins has been demonstrated in cultured confluent human fibroblasts. It has been speculated that lys-Hsc70 may be required to pull proteins into the lysosomal lumen because of analogous roles of Hsp70s in the translocation of proteins into the endoplasmic reticulum, mitochondria and chloroplasts [21, 25]. Nearly half of the lysosomal Hsp90s associate with the lumenal side of the lysosomal membrane where this chaperone may contribute to the stabilization of essential components of the translocation complex when it is organized into a multimeric structure [3, 40].
Receptor—Lamp2a
In 1996, a lysosomal membrane glycoprotein Lgp96 (called Lamp2a in human) was identified as a receptor for binding and uptake of lysosome substrates [20]. Lamp2a is one of the three splice variants of the Lamp2 gene that gives rise to three single-span membrane proteins, Lamp2a, b and c. These variants all have a common highly N-glycosylated lumenal region, but possess different transmembrane and C-terminal cytosolic tail regions. Lamp2a has a short cytosolic tail (GLKRHHTGYEQF) to which CMA substrate proteins bind [25]. The positively charged residues in the Lamp2a cytosolic tail are important for binding of substrate proteins. However, the specific amino acids on the CMA substrate proteins required for receptor binding remain elusive [41]. Binding of substrate proteins to Lamp2a is a rate-limiting step for the CMA process, and overexpression of Lamp2a in Chinese hamster ovary cells can increase CMA activity. Lamp2a is not limited to acting as a classic receptor. Increasing evidence shows that Lamp2a is involved in many other aspects of the CMA process such as substrate translocation [3, 25, 40]. Lamp2 deficiency in mice causes extensive accumulation of autophagic vacuoles in many tissues [42]. Contrary to the proposed receptor function of Lamp2a in CMA, confluent mouse embryonic fibroblasts deficient in lamp2 appear to have normal levels of CMA-associated lysosomal proteolysis after prolonged serum withdrawal, suggesting that Lamp2a may not be the only receptor for CMA [29, 43].
Substrates—KFERQ motif
Substrates for CMA are defined by an amino acid sequence motif related to KFERQ [3, 16]. This pentapeptide targeting motif was first identified in microinjected RNase A [44]. It consists of a glutamine (Q) preceded or followed by a combination of four amino acids that are basic (R, K), acidic (D, E), or bulky and hydrophobic (F, I, L, V) residues. In some cases, Q may be substituted by the related N [45]. Antibodies raised against KFERQ can immunoprecipitate 30% of cytosolic proteins in mammalian cells [46].
Without denaturation, more than 80% of cytosolic proteins that contain KFERQ motifs can be recognized by the antibody to this motif, indicating that most KFERQ motifs in these proteins are exposed. However, certain proteins such as aldolase B have hidden KFERQ motifs due to multimeric formation. The ubiquitination may dissociate the aldolase tetramer and result in the exposure of KFERQ motif [25, 47]. In addition, certain monomers may have hidden KFERQ sequences that may be exposed following partial unfolding. Therefore, the presence of a KFERQ sequence alone in the primary structure of a protein is not sufficient for determining them as substrates of CMA [48]. Such identification requires rigorous experimental proof. Recently, we have identified MEF2D, a protein known to promote neuronal survival as a CMA substrate. Interestingly, MEF2D uses a set of over-lapping imperfect KFERQ motifs to mediate its interaction with Hsc70 [49].
Characteristics of CMA
Compared to macro- and microautophagy, which occurs in a wide range of eukaryotes including mammals, plants and fungi, CMA has only been described in mammals. This process has some distinctive features.
Selectivity
Lysosome-mediated degradation had traditionally been perceived as a process performed in bulk with poor selectivity. The initial idea of selective autophagy originated from the observation that starvation in animals or serum removal in cultured cells accelerates the degradation of particular cytosolic proteins in lysosomes but not others [50]. Selectivity thus became one of the hallmarks of CMA [51]. CMA mediates selective targeting of non-essential proteins for degradation to obtain the amino acids required for the synthesis of essential proteins. The intrinsic selectivity of CMA is also well suited for the removal of specific proteins damaged during stress without interfering with nearby normally functioning forms of the same protein [51, 52]. This selectivity is achieved by making the KFERQ motifs in the altered protein accessible to the chaperone but inaccessible when it is properly folded or hidden. Compare to CMA, macroautophagy has traditionally been considered as a nonselective degradation process. However, recent evidence suggests that this view needs modification. New experimental data demonstrate that some macroautophagic processes, termed chaperone-assisted selective autophagy, are assisted by chaperone proteins and can be selective in targeting protein complexes, organelles and microbes [2]. In this process, macroautophagy has been proposed to be initiated by a selective ubiquitylation of cellular targets and followed by recognition via autophagic ubiquitin adaptors such as p62, NBR1 and HDAC6. These molecules can mediate docking of ubiquitinated proteins or damaged organelles to autophagosomes and lysosomes, thereby ensuring their selective degradation [2, 53, 54].
Saturability
The unusual characteristics of CMA are not limited to its selectivity. As the mechanism for cargo delivery was revealed, it became evident that, in contrast to the other forms of autophagy, vesicle formation was not required in CMA. Instead, the substrate proteins were translocated across the lysosomal membrane [55]. The receptor Lamp2a is mainly responsible for this translocation. Because of the requirement for receptor binding before translocation can occur, this delivery process becomes saturable. In contrast to Hsc70, which is often in excess in the cytosol, levels of Lamp2a are limiting for CMA and hence subjected to tight regulation [23].
Competitivity
The selectivity and saturability of CMA directly lead to the competitive binding of CMA substrates. During the process of CMA, different substrates compete for binding Hsc70 and limited pool of Lamp2a [23, 56]. Several studies have shown that degradation of some substrates is slowed down by overexpression of other substrates. This has been proposed to underlie the pathogenesis of some diseases [49, 57]. For example, both the wild-type α-synuclein and neuronal transcription factor MEF2D are substrates of CMA. Elevated levels of wild-type α-synuclein may reduce the degradation of MEF2D via CMA [49].
Regulation of CMA
The signal transduction pathways involved in the regulation of CMA from cell membranes to cytosolic chaperone proteins and lysosomes remain largely elusive [3]. The p38 MAPK inhibitor can partly prevent the activation of CMA, implicating this pathway in CMA [45]. On the other hand, the extensively investigated local regulation of CMA activity in the lysosomes has precise, fine-tuned mechanisms [3, 25].
Regulation of CMA via Lamp2a
The level of Lamp2a at the lysosomal membrane is proportional to the activity of CMA. Therefore, changes in the Lamp2a level at the lysosomal membrane can quickly regulate the activity of CMA. The level of Lamp2a at the lysosomal membrane may be changed by synthesis, degradation and redistribution [41, 58]. De novo synthesis of Lamp2a or its inhibition by protein synthesis inhibitors can directly change the level of Lamp2a in cells, but the physiological or pathological relevance remains unclear [3, 59]. The distribution of Lamp2a at the lysosomal membrane is regulated by its dynamic association with discrete membrane lipid microdomains [60]. Under basal conditions, sequestration of Lamp2a in cholesterol-enriched regions favors its cleavage by two proteases, including an unidentified metalloprotease at the membrane and a serine protease—cathepsin A, which associates dynamically with the lumenal side of the lysosomal membrane [60, 61]. Exclusion of Lamp2a from these regions allows its multimerization (see below), a step required for the uptake of CMA substrate by lysosomes. Interestingly, the degradation of Lamp2a can be reduced by the presence of substrates or stimuli that induce the activation of CMA. This blockage in degradation, rather than the increase of synthesis, to increase Lamp2a at the lysosomal membrane is particularly advantageous to cells with limited access to amino acids such as during nutrient deficit [3, 62].
The levels of Lamp2a can be further increased at the lysosomal membrane through the mobilization of the pool normally resident in the lysosomal lumen. The exact nature of this luminal pool of Lamp2a remains unclear, but intact molecules of this protein exist inside lysosomes, and a gradual decrease in the percentage of Lamp2 in this compartment occurs as activation of CMA persists beyond 1 day [3, 60]. Membrane chaperones and an intact membrane potential are needed for the mobilization of Lamp2a from the lumen to membrane [3]. Fractionation studies have shown that luminal Lamp2a associates with lipid, indicating the possible existence of luminal Lamp2a-containing micelles. It is thought that these micelles fuse or integrate into the lysosomal membrane under specific stress, resulting in the incorporation of Lamp2a in the membrane and exposure of its C terminus to the cytosol [60, 63].
Lamp2a can undergo cycles of rapid assembly into a 700-kDa protein complex at the lysosomal membrane. Monomers of Lamp2a at the lysosomal membrane can accept substrate proteins, and this interaction drives the organization of Lamp2a into multimeric complexes needed for substrate translocation into lysosomes. Once the substrate protein reaches the lysosomal lumen, Lamp2a will disassemble from the multimeric complex to enable subsequent rounds of substrate binding [3, 40]. This continuous assembly and disassembly of Lamp2a from the multimeric translocation complex highlights the importance of the lateral mobility of this protein in the lysosomal membrane [40]. Chaperone proteins located at both sides of the lysosomal membrane may regulate the lateral mobility of Lamp2a. Lys-Hsc70 induces disassembly of Lamp2a from the 700-kDa complex once the substrate has crossed the membrane. A lysosome-associated form of the glial fibrillary acidic protein (GFAP), a component of the intermediate filament network, associates to Lamp2a once it is organized into multimers and contributes to stabilizing the CMA translocation complex against the disassembling activity of Hsc70, whereas GTP-mediated release of elongation factor-1α from the lysosomal membrane promotes self-association of GFAP, disassembly of the CMA translocation complex and the consequent decrease in CMA [12, 64]. In addition, Hsp90 at the lumenal side is also required to preserve the stability of Lamp2a during the transition [40].
Regulation of CMA via Hsc70
The other limiting lysosomal component is lys-Hsc70. As indicated in the previous section, the presence of this chaperone at the luminal side of the membrane is necessary for substrate translocation [18]. Levels of lys-Hsc70 increase gradually with increasing CMA activity, although the mechanisms modulating this increase are still poorly understood. Although Hsc70 contains two KFERQ sequences and is a putative substrate of CMA [21, 25], it appears that neither CMA nor macroautophagy is involved in delivering lys-Hsc70 to lysosomes [3, 65]. It is possible that this chaperone reaches lysosomes through maturation of late endosomes, a compartment in which high levels of luminal Hsc70 have been detected, thus highlighting a possible relationship between CMA and endocytosis [3, 65, 66].
Regulation of CMA via macroautophagy
The activity of CMA is also directly modulated by changes in other autophagic and proteolytic systems inside the cell. Cells in culture respond to CMA blockage by upregulating macroautophagy. Similarly, blockage of macroautophagy results in constitutive activation of CMA [65]. These pathways are clearly not redundant, as CMA is, for example, unable to degrade organelles normally turned over by macroautophagy, whereas macroautophagy lacks the selectivity of CMA in the degradation of individual soluble cytosolic proteins [67]. Nevertheless, the compensatory activation of one form of autophagy when the other is compromised allows cells to preserve homeostasis, at least under basal conditions. Additionally, blockage of either form of autophagy also has a direct impact on proteasomal activity [3]. The molecular mechanisms that regulate crosstalk between these two different pathways are currently under investigation. In the case of the interrelationship between macroautophagy and CMA, continuous fusion of autophagosomes to lysosomes when macroautophagy is upregulated results in transient dissipation of the lumenal lysosomal pH, which negatively affects lys-Hsc70 stability. In fact, although lys-Hsc70 is normally stable at pH ranges of 5.2–5.4, changes in pH values above 5.6 in the lysosomal lumen result in its rapid degradation in this compartment. The reduced levels of lys-Hsc70 in those lysosomes decrease their capability to perform CMA [68].
Regulation of CMA via UPS
UPS and autophagy were long viewed as independent, parallel degradation systems with no point of intersection. Increasing evidence shows that the UPS and autophagy are functionally interrelated catabolic processes. Specifically, these degradation systems share certain substrates and regulatory molecules, and show coordinated and, in some contexts, compensatory function. For example, the neuronal protein α-synuclein can be degraded by the UPS, macroautophagy and CMA. Under conditions in which the UPS is compromised, enhanced degradation by CMA and macroautophagy may become critical to maintaining pools of amino acids for protein synthesis and may protect against the accumulation of a toxic species [1, 3]. On the other hand, during the acute stages of CMA blockage, there is an accumulation of polyubiquitinated proteins, often in the form of protein aggregates, attributable to the observed reduction in their removal through the proteasome system, and the underlying mechanisms remain to be clarified [69]. Interestingly, recent evidence suggests that ubiquitin may play a key role in the crosstalk between proteasome-mediated degradation and selective autophagy [2, 70]. Furthermore, ubiquilin, a ubiquitin-like protein, functions to regulate macroautophagy by facilitating maturation of LC3 protein, and is also a substrate of CMA, indicating that ubiquilin may also be at a crossroad between protein degradation pathways [71, 72]. However, detailed mechanisms are under investigation.
Physiological relevance
Selective degradation of cytosolic proteins via CMA contributes to both quality control (housekeeping) and response to stress [2, 3]. CMA was initially identified as an inducible pathway in response to stress. However, increasing evidence shows that there is a certain level of CMA activity under basal conditions. Most cell types analyzed to date display some level of continuous CMA activity detectable in the absence of typical CMA-inducing conditions. Basal CMA requires participation of the same effectors at the lysosomal membrane—the membrane chaperones and the protein translocation complex. It has been speculated that there is a difference between basal and inducible CMA. Whether the regulation of basal and inducible CMA occurs through different signaling mechanisms is under investigation [65]. However, both basal and inducible CMA may have physiological relevance.
Recycling and quality control
The delivery of intracellular substrates such as misfolded proteins and damaged organelles from the cytosol to the lysosomes for degradation is crucial for cell survival. Under physiological conditions, renewal of cytosolic proteins is needed to maintain their normal function through recycling their old versions [4]. Besides bulk autophagy (microautophagy and macroautophagy), CMA efficiently facilitates the transit of specific proteins from the cytoplasm to lysosomes and is responsible for their recycling. If this process is inhibited, excessive old and dysfunctional proteins will accumulate in the cytosol and disturb the physiological functions of cells [3, 49].
Immune response
Durable adaptive immunity is dependent on CD4+ T cell recognition of the major histocompatibility complex (MHC) class II molecules that display peptides from exogenous and endogenous antigens. Specialized antigen-presenting cells use the endosomal/lysosomal systems to internalize exogenous antigens, which can then be presented on MHC to CD4+ T cells. Beside the proteasome and macroautophagy in processing and MHC loading with endogenous and exogenous antigens, CMA has also been recently investigated in antigen processing/presentation. Cells with reduced levels of Lamp2a or Hsc70 exhibit decreased presentation of cytoplasmic epitopes on class II molecules [73]. Conversely, an increase in cytoplasmic autoantigen presentation is observed upon overexpression of either Lamp2a or Hsc70. Furthermore, there is cross-talk between autophagy pathways in the expression of MHC class II molecules. Macroautophagy can deliver antigens into autophagosomes for processing by acidic proteases before MHC class II presentation. However, other endogenous antigens are processed by cytoplasmic proteases, yielding fragments that translocate via CMA into the endosomal network to intersect MHC class II. This cross-talk, particularly in response to stress, appears to balance the relative efficiency of each pathway, limits redundancy and gives MHC class II broader access to antigens within different intracellular compartments. Whether alteration in CMA activity such as its reduction with age could contribute to the altered immune response requires further investigation [74].
Starvation
The first stimulus identified to activate CMA was nutritional starvation. In most mammalian cells, macroautophagy is activated during the first hours of serum removal. After prolonged starvation in cells and animals, CMA activity increases progressively, reaching maximal activation at about 10 h after the removal of serum in the cultured cells, or at 3 days in animals, and remains at this level for the duration of the starvation period [25, 75]. These findings suggest that starvation-induced activation of CMA coincides with the decay in macroautophagy. Cells may benefit from switching to a more selective degradation, such that particular essential proteins could be maintained in the cytosol while other less critical ones can be specifically targeted for degradation [3, 75]. It has been found that many glycolytic enzymes undergo degradation through CMA during prolonged starvation as they may be unnecessary in the cells in the absence of nutrients [55]. In addition, CMA activation during nutrient deprivation is both tissue- and cell type-specific. While starvation is a potent CMA stimulus in liver, spleen, kidney, and heart, neurons display higher basal levels of CMA activity and do not appear to greatly augment their CMA in response to starvation [3].
Oxidative stress
Antioxidants can at least partially prevent the increased degradation of some specific proteins by the CMA pathway in response to starvation, indicating that the oxidation of proteins may accelerate their degradation via CMA [59, 76]. This speculation is supported by a more direct study comparing lysosomal binding and uptake of different CMA substrates, either unmodified or upon exposure to pro-oxidizing conditions. More importantly, the upregulation of CMA during oxidative stress has been observed using lysosomes isolated from cultured cells and livers of rodents treated with pro-oxidant compounds [59]. Interestingly, these lysosomes not only have endogenous cytosolic oxidized proteins in their lumens, but also have higher CMA activity even for non-modified substrate proteins. Transcriptional upregulation of Lamp2a seems to account for the increased CMA activity under such conditions. Oxidative stress also increases the levels of other key CMA components, such as lysosomal Hsc70 and Hsp90 [24, 59]. Decline of CMA with age in whole organs may contribute to the accumulation of oxidized proteins characteristic of most tissues in old organisms, and manipulations to prevent the age-dependent decrease of CMA in liver reduces the amount of oxidized proteins in this organ even at advanced age [77, 78].
Age-related decline in CMA
Phenomena
Total rates of protein degradation decrease in almost all tissues and organisms with age [3, 23]. In cultured senescent fibroblasts, CMA degradation of cytosolic proteins is slower than in early passage cells. Furthermore, the decline of this degradation is no longer upregulated even upon serum starvation [14]. This decrease in CMA activity has been confirmed by comparing the ability of lysosomes isolated from livers of young and aged rats to take up substrates. Lysosomes from aging rats show lower rates of CMA, and both substrate binding to the lysosomal membrane and transport into lysosomes seem to decline with age [23]. These findings strongly suggest that the decline of CMA activity is an index of aging.
Mechanisms
The analysis of the different steps involved in the CMA pathway has revealed that CMA substrate proteins are still recognized by Hsc70 and targeted to the membrane of old lysosomes with age. Furthermore, if substrates are presented directly to the lysosomal hydrolases, the rate of their degradation in the old lysosome is similar to that in the young lysosome. Consistently, there is a decrease in the levels of Lamp2a at the lysosomal membrane [23]. Interestingly, the decline in Lamp2a levels with age is not due to transcriptional downregulation or changes in rates of synthesis and delivery of this receptor to the lysosomal compartment. It is the dynamics and stability of Lamp2a at the lysosomal membrane that appear to be severely compromised with age. The reason for this alteration is probably related to the changes in the composition of the lipid microdomains of the lysosomal membrane, which regulates the association of Lamp2a with cathepsin A [60]. Rapid degradation of Lamp2a in the lysosomal lumen likely contributes to the lower levels of this protein in aging lysosomes. This role of reduced levels of Lamp2a in age is supported by findings in a double-transgenic mouse model in which enhancing of Lamp2a expression preserves normal CMA activity in the livers of the old transgenic mice [77].
Aberrance of CMA in diseases
Besides being involved in different aspects of cellular physiology, CMA has been implicated in the pathogenesis of some diseases, especially age-associated disorders, when CMA is deregulated by both genetic and environmental factors. In some of these diseases, the CMA defect is likely to underlie the pathogenesis, whereas in others, it may be secondary to the main defect but contribute to the pathology and course of the respective diseases [3].
Lysosomal storage diseases
LSD is the common name for disorders that may be caused by improper degradation of certain substrates in lysosomes [78]. Altered CMA in LSD is often secondary to the primary impairment in the lysosomal compartment. The global lysosomal dysfunction caused by mutations in specific enzymes or disrupted trafficking of these enzymes to lysosomes may disrupt CMA activity along with other autophagic pathways. However, several LSDs have defective CMA activity, which may occur before global failure of the lysosomal system. Abnormal CMA upregulation has been observed in galactosialidosis, a disorder resulting from defective function of protective protein/cathepsin A. Cathepsin A is one of the two proteases required for the regulated degradation of Lamp2a. The abnormally high levels of Lamp2a observed in galactosialidosis and the consequent increase in CMA activity can be normalized after cathepsin A levels are restored [61]. Mucolipidosis type IV disease may be associated with a primary defect in CMA. The transient receptor potential mucolipin-1 mutated in this disorder associates with Hsc70 and Hsp40 at the lysosomal membrane and may be a novel regulator of the CMA translocation pathway. Fibroblasts from mucolipidosis type IV patients have lower CMA activity compared with those from normal controls, leading to the increase of oxidized proteins in the cells [79]. Aberrant alteration of CMA also occurs in Danon disease, a lysosomal glycogen storage disorder with normal acid maltase activity resulting from a mutation in the Lamp2 gene. However, due to the multiplicity of functions of the different Lamp2 protein variants and the defects in lysosomal biogenesis caused by altered intracellular trafficking of essential lysosomal enzymes observed in this disease, more studies are needed to dissect whether and how CMA activity is altered in Danon patients [80].
Neurodegenerative diseases
The hallmark of most neurodegenerative disorders is the accumulation of pathogenic proteins. Alterations in the cellular quality control mechanisms have been proposed to contribute to the neuronal inability to clear the pathogenic proteins [81, 82]. Besides dysfunction of macroautophagy, dysfunction of CMA has been found to cause the accumulation of some pathogenic proteins involved in neurodegenerative disorders, such as α-synuclein, UCH-L1 and MEF2D in Parkinson’s (PD), APP and tau in Alzheimer’s (AD) and huntingtin in Huntington’s (HD) disease. These proteins contain KFERQ motifs in their amino acid sequences and have been proved to be CMA substrates [49, 83, 84]. In the following sections, we will discuss the recent findings regarding the involvement of CMA in the pathogenesis of these disorders.
PD
PD, the most common neurodegenerative movement disorder, is a degenerative disorder of the central nervous system characterized by the progressive loss of dopaminergic neurons in substantia nigra and their terminals, leading to impairments of the victim’s motor skills, speech and other functions. The molecular mechanisms of PD are not fully understood, and no cure is presently available [85]. However, an increase in the amount of α-synuclein protein may constitute a cause of PD. α-Synuclein is degraded at least in part by CMA [83, 86]. α-Synuclein in its mutant forms can cause dysfunction of CMA. Conversely, aberration in CMA can lead to the accumulation of α-synuclein protein.
Mutant α-synuclein and CMA
The first pathogenic protein related to CMA was α-synuclein, the main component of Lewy bodies in PD patients. Although mutations in α-synuclein have only been described in a small percentage of PD patients, this protein is aggregated in the form of Lewy bodies in sporadic PD. Wild-type α-synuclein is a CMA substrate, whereas pathogenic A53T and A30P α-synuclein mutants bind to the receptor Lamp2a, but act as an uptake blocker [83, 87]. Therefore, the presence at the lysosomal membrane of the tightly bound forms of mutant α-synuclein not only inhibits their own degradation, but also interferes with the degradation of other cytosolic proteins through the CMA pathway [85].
Modified forms of α-synuclein and CMA
Similar to mutants of α-synuclein, some post-translationally modified forms of wild-type α-synuclein resulting from the reaction with dopamine also block translocation in CMA [3, 86]. In mouse neurons, SH-SY5Y cells and isolated mouse lysosomes, most posttranslational modifications of α-synuclein impair degradation of this protein by CMA but do not affect degradation of other substrates. However, dopamine-modified α-synuclein not only is poorly degraded by CMA, but also blocks degradation of other CMA substrates. Cellular viability improves if lysosomal targeting of the pathogenic forms of α-synuclein is prevented by eliminating its CMA-targeting motif. Since blockage of CMA increases cellular vulnerability to stressors, dopamine-induced autophagic inhibition may explain the selective degeneration of PD dopaminergic neurons. However, the exact mechanisms by which the pathogenic forms of α-synuclein block translocation of CMA substrates remain to be clarified [86].
UCH-L1 and CMA
Ubiquitin C-terminal hydrolase L1 (UCH-L1), a neuronal de-ubiquitin enzyme, is expressed abundantly in neurons and has been reported to be a major target of oxidative/carbonyl damage associated with sporadic PD. Mutant forms of UCH-L1 are associated with familial PD [88]. UCH-L1 interacts with three of the main components of CMA, i.e., Lamp2a, Hsc70 and Hsp90. These interactions are abnormally enhanced by the I93M mutation and independent of the monoubiquitin binding of UCH-L1. Expression of I93M UCH-L1 in cells can induce an increase in the amount of α-synuclein that is associated with CMA inhibition. The findings may provide novel insights into the molecular links between α-synuclein and UCH-L1. Aberrant interaction of mutant UCH-L1 with CMA machinery may underlie the pathogenesis of PD associated with I93M UCH-L1 [89].
MEF2D and CMA
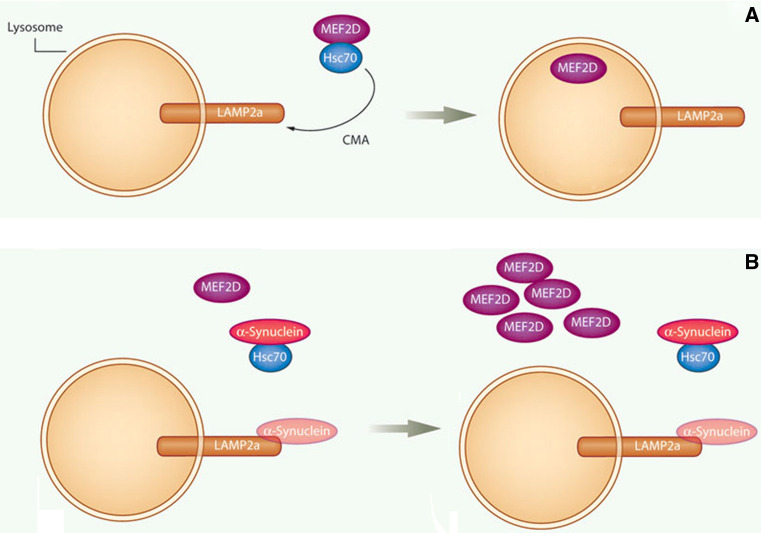
Transcription factor MEF2D plays an important role in neuronal survival [90–92]. Our recent studies identify MEF2D as a direct substrate of CMA. In a neuronal cell line, MEF2D continuously shuttles to the cytoplasm under basal conditions, interacts with Hsc70 and undergoes CMA-mediated degradation (Fig. 3). Inhibition of CMA leads to MEF2D accumulation in the cytoplasm. But the accumulated MEF2D has apparently lost its DNA binding capacity and therefore is non-functional. Importantly, both wild-type and PD-associated mutant α-synuclein disrupts the MEF2D-Hsc70 binding, which leads to neuronal death. Consistently, MEF2D levels are increased in the brains of α-synuclein transgenic mice and PD patients. Thus, CMA modulates the neuronal survival factor MEF2D, and dysregulation of this pathway may contribute to the pathogenesis of PD [49, 93].
Fig. 3.
Regulation of neuronal survival factor MEF2D by CMA: a Hsc70 binds to and accompanies MEF2D to Lamp2a for uptake and digestion by the lysosome under physiological conditions; b in PD, the CMA process is deregulated, preventing the normal uptake and removal of MEF2D and resulting in its accumulation. Modified from [93] with permission
AD
AD is the most common form of dementia. The disease is characterized neuropathologically by the presence of amyloid plaques, neurofibrillary tangles, and synaptic and neuronal loss with severe atrophy of cortical and subcortical regions [94]. The role for CMA in AD has been proposed based on the aberrant interaction of major components of the CMA machinery with particular mutant forms of cytoskeleton-associated protein tau. These tau mutants undergo proteolytic cleavage, which generates highly amyloidogenic fragments. Tau mutants in the cytosol are targeted for CMA and cleaved by lysosomal enzymes [95]. Compared to regular CMA substrates, the truncated forms of tau undergo only partial insertion into the CMA translocon complex, enough to present their C terminus to the lysosomal hydrolases resulting in their cleavage into amyloidogenic peptides. The pathogenic forms of tau also organize into irreversible oligomeric structures at the lysosomal membrane, which interferes with normal CMA, and leads to destabilization of the lysosomal membrane and leakage of lysosomal enzymes into the cytosol [95, 96].
Regulator of calcineurin 1 (RCAN1), a gene identified from the critical region of Down syndrome, has been implicated in the pathogenesis of AD. Besides being regulated by the ubiquitin proteasome pathway, RCAN1 protein can also be degraded through the lysosomal pathway. Inhibition of macroautophagy leads to the reduction of RCAN1 expression; conversely, inhibition of CMA increases RCAN1 level. Two CMA recognition motifs are identified in RCAN1 protein and function to mediate its degradation through CMA. Furthermore, promoter assay demonstrates that inhibition of RCAN1 degradation in cells reduces calcineurin-NFAT activity. This finding suggests additional connections between possible effectors involved in AD and CMA [57].
HD
HD is a dominantly inherited pathology caused by the accumulation of mutant huntingtin protein (HTT). Mutant huntingtin contains an expanded polyglutamine tract contributing to its accumulation as nuclear and cytosolic aggregates in HD patients [97]. Huntingtin has three putative CMA-targeting motifs. They are outside exon 1 where the abnormally extended stretch of glutamines localizes in the mutant pathogenic form. It has been found that regulated phosphorylation of exon 1 by the inflammatory kinase IKK enhances the normal clearance of this product by both the proteasome and lysosomes. Interestingly, the lysosomal clearance is dependent on Lamp2a and Hsc70, indicating that CMA may contribute to the removal of the pathogenic protein before aggregation. Therefore, decline in CMA activity may be one of the aggravating factors in the progression of HD [3, 98].
Kidney pathology
The first association of CMA with disease was established in kidney, an organ where CMA activity is markedly high. Study of a form of chemically induced hyaline droplet nephropathy reveals that CMA activity increases in the early stages of the disease to prevent the accumulation of α-2-microglobulin. α-2-Microglobulin is altered by the various chemical compounds that trigger this kidney condition and is itself a bona fide CMA substrate. However, as the disease progresses, CMA is not sufficient to accommodate the high levels of altered α-2-microglobulin, leading to its accumulation in kidney cells in the form of hyaline droplets. This kidney lesion often evolves to chronic progressive nephropathy and carcinogenesis [3, 99].
Another kidney pathology where alterations in the degradation of selective proteins by CMA may play a role is renal hypertrophy. Hypertrophy is characteristic of the diabetic kidney, a condition where reduced levels of Lamp2a has been observed. Hypertrophic cellular growth can result from imbalance between protein synthesis and protein degradation. Decreased CMA activity under these conditions leads to high levels of the nuclear transcription factor Pax2, a factor essential in regulation of cellular growth in the kidney. The increased levels of the transcription factor Pax2 may contribute to maintenance of the hypertrophic state through upregulation of tubular cell growth [100].
Other pathologies
Alteration in macroautophagy has been described in infectious diseases and cancer. In light of the contribution of CMA to antigen presentation, CMA dysfunction may underlie some immunoreactive and autoimmune disorders, but this aspect remains to be explored [73, 74]. Another area of investigation is the possible role of CMA in oncogenesis. Epidermal growth factor receptor pathway substrate 8 (Eps8) promotes the growth of various solid malignancies. A novel association of Eps8 with the late endosomal/lysosomal compartment specifically occurs in cancer cells. In metastatic pancreatic cancer cell lines, endogenous Eps8 is localized to large vesicular lysosomal structures. Structure-function analysis reveals that amino acids 184–535 of Eps8 were sufficient to mediate its lysosomal recruitment. Notably, this fragment harbors two KFERQ-like motifs for CMA, and Eps8 itself has been identified as a CMA substrate. These findings serve as an example for the possible role of CMA and oncogenesis [56]. In addition to CMA, the newly recognized selective macroautophagy, chaperone-assisted selective autophagy, seems to be an essential proteostasis mechanism and may have important implications for understanding some diseases and age-dependent pathologies [30]. For example, it has been found that impaired chaperone-assisted selective autophagy results in Z disk disintegration and progressive muscle weakness in several species [9].
A potential therapeutic target
Increasing evidence suggests that CMA acts as a cell protector and its dysfunction is correlated with diverse pathologies [3, 12]. A better understanding of CMA, ranging from detailed processes to its role in physiology and pathology, may be needed to allow its manipulation for therapeutic purposes. New insights into the molecular mechanisms of CMA are now leading to the discovery of exciting new potential drug targets.
Upregulation of CMA acts as an adaptive response that protects cells from stresses. Therefore, activation of CMA and/or enhancement of CMA activity by agents and genetic methods may prevent the process of aging and some CMA-associated diseases. Indeed, a recent study has demonstrated that restoration of CMA in aging liver improves cellular maintenance and hepatic function, and reduces intracellular accumulation of oxidized and aggregated proteins [77]. These findings strongly underscore the therapeutic potential of maintenance of proper CMA activity. The polyglutamine binding peptide 1 (QBP1) binds an expanded polyQ tract in mutated but not normal HTT. Expression of a fusion molecule comprising two copies of QBP1 and copies of two different Hsc70-binding motifs has been employed to selectively target mutant HTT for degradation in cellular and mouse models of HD. This strategy results in specific degradation of mutant HTT in both cultured cells expressing the construct and the R6/2 mouse model of HD. Therefore, similar adaptor molecules comprising Hsc70-binding motifs fused to an appropriate structure-specific binding agent may have broader therapeutic potential for treating diseases caused by misfolded proteins [97].
Both calorie restriction and the ketogenic diet possess broad therapeutic potential in various clinical settings and in various animal models of neurological diseases [98]. Following calorie restriction or consumption of a ketogenic diet, there is notable improvement in mitochondrial function, a decrease in the expression of apoptotic and inflammatory mediators and an increase in the activity of neurotrophic factors. Recent research aimed at identifying compounds that can reproduce, at least partially, the neuroprotective effects of the diets with less demanding changes to food intake suggests that ketone bodies may represent an appropriate candidate, protecting neurons against multiple types of neuronal injury [101, 102]. As mentioned above, CMA is activated in higher organisms under conditions of prolonged starvation. Interestingly, it has been reported that ketone bodies stimulate CMA. Ketone bodies are comprised of β-hydroxybutyrate, acetoacetate and acetone, which circulate during starvation. Physiological concentrations of β-hydroxybutyrate and acetoacetate induce proteolysis in cells. Lysosomes isolated from β-hydroxybutyrate-treated cells display an increased ability to degrade CMA substrates. Pretreatment of CMA substrates with β-hydroxybutyrate increases their rate of degradation by isolated lysosomes [103]. Given these, we speculate that CMA may, at least in part, be involved in the neuroprotective effects of calorie restriction and ketogenic diet. The induction of CMA by ketone bodies may provide an important physiological mechanism for the activation of CMA during prolonged starvation.
Concluding remarks
Cargo delivery into lysosomes is not always “in bulk” and instead can occur in the selective manner, i.e., CMA and other selective autophagy. Distinct from the other two autophagy pathways, CMA possesses unique characteristics, i.e., selectivity, saturability and competitivity, by which substrate proteins are directly delivered into the lysosomal lumen through Lamp2a receptor associated with the assistance of Hsc70 chaperone complex. These characteristics are not only implicated in mechanisms and regulation of CMA, but also involved in alteration of CMA under pathological conditions.
The machinery, mechanisms and regulation of CMA have been extensively analyzed. Despite the considerable advance in the molecular dissection of this pathway, there remain many questions: (1) what pathways are involved in signal transduction from cell membranes to lysosomes to regulate CMA following various stresses? (2) In addition to Hsc70 chaperone complex and the receptor Lamp2a, are there other chaperone complexes and receptors involved in the CMA process? (3) Are there additional and specific interactions between CMA and other autophagic pathways or the ubiquitin–proteasome system? (4) What new substrates are important for various physiological and pathological conditions? (5) Can one develop better and more specific small molecules to modulate CMA in an acute manner?
Acknowledgments
We thank Brian Ciliax and Gary Miller for their critical comments. This work was supported by NIH grants (AG023695, NS048254, ES015317 and ES016731-0002) and a Michael J. Fox Foundation grant to Z. Mao.
References
- 1.Nedelsky NB, Todd PK, Taylor JP. Autophagy and the ubiquitin–proteasome system: collaborators in neuroprotection. Biochim Biophys Acta. 2008;782:691–699. doi: 10.1016/j.bbadis.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 3.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubota H. Quality control against misfolded proteins in the cytosol: a network for cell survival. J Biochem. 2009;146:609–616. doi: 10.1093/jb/mvp139. [DOI] [PubMed] [Google Scholar]
- 5.Sato Y, Sakamoto K, Sei M, Ewis AA, Nakahori Y. Proteasome subunits are regulated and expressed in comparable concentrations as a functional cluster. Biochem Biophys Res Commun. 2009;378:795–798. doi: 10.1016/j.bbrc.2008.11.125. [DOI] [PubMed] [Google Scholar]
- 6.Nagy V, Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. J Biol Chem. 2010;391:163–169. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- 7.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 8.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin–proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584:1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 9.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK, Hoch M, Höhfeld J. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20(2):143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mul JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koga H, Cuervo AM. 17 Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiol Dis. 2010 doi: 10.1016/j.nbd.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dice JF. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624–14627. [PubMed] [Google Scholar]
- 15.Backer JM, Bourret L, Dice JF. Regulation of catabolism of microinjected ribonuclease A requires the amino-terminal 20 amino acids. Proc Natl Acad Sci USA. 1983;80:2166–2170. doi: 10.1073/pnas.80.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dice JF, Chiang HL, Spencer EP, Backer JM. Regulation of catabolism of microinjected ribonuclease A. Identification of residues 7–11 as the essential pentapeptide. J Biol Chem. 1986;261:6853–6859. [PubMed] [Google Scholar]
- 17.Dice JF. Molecular determinants of protein half-lives in eukaryotic cells. FASEB J. 1987;1:349–357. doi: 10.1096/fasebj.1.5.2824267. [DOI] [PubMed] [Google Scholar]
- 18.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-KDa heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 19.Cuervo AM, Terlecky SR, Dice JF, Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 20.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 21.Agarraberes FA, Terlecky SR, Dice JF. An intralysosomal Hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dice JF (2000) Lysosomal pathways of protein degradation, Molecular Biology Intelligence Unit, Landes Bioscience, Austin, TX
- 23.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 24.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meeting function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 25.Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Murphy RF. Maturation models for endosome and lysosome biogenesis. Trends Cell Biol. 1991;1:77–82. doi: 10.1016/0962-8924(91)90022-2. [DOI] [PubMed] [Google Scholar]
- 27.Holtzman E. Lysosomes (p. 439) New York: Plenum Press; 1989. [Google Scholar]
- 28.Conus S, Simon HU. Cathepsins: key modulators of cell death and inflammatory responses. Biochem Pharmacol. 2008;76:1374–1382. doi: 10.1016/j.bcp.2008.07.041. [DOI] [PubMed] [Google Scholar]
- 29.Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 30.Kettern N, Dreiseidler M, Tawo R, Höhfeld J. Chaperone-assisted degradation: multiple paths to destruction. Biol Chem. 2010;391:481–489. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- 31.Newmyer S, Christensen A, Sever S. Auxilindynamin interactions link the uncoating ATPase chaperone machinery with vesicle formation. Developmental Cell. 2003;4:929–940. doi: 10.1016/s1534-5807(03)00157-6. [DOI] [PubMed] [Google Scholar]
- 32.Agarraberes F, Dice J. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–2499. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 33.Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem. 2000;275:27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 34.Thoms S. Cdc48 can distinguish between native and nonnative proteins in the absence of cofactors. FEBS Lett. 2002;520:107–110. doi: 10.1016/s0014-5793(02)02777-1. [DOI] [PubMed] [Google Scholar]
- 35.Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan J, Singh S, Millson S, Clarke P, Naaby-Hansen S, Stein R, Cramer R, Mollapour M, Workman P, Piper P, Pearl L, Prodromou C. Activation of the ATPase activity of Hsp90 by the stress-regulated cochaperone aha1. Molecular Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 36.Bimston D, Song J, Winchester D, Takayama S, Reed JC, Morimoto RI. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alberti S, Esser C, Höhfeld J. BAG-1–a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones. 2003;8:225–231. doi: 10.1379/1466-1268(2003)008<0225:bnefoh>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 39.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Höhfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276(46):42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 40.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperonemediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuervo A, Dice J. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 43.Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lüllmann-Rauch R, Hartmann D, Heeren J, von Figura K, Knecht E, Saftig P. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell. 2004;15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dice J. Microinjected ribonuclease A as a probe for lysosomal pathways of intracellular protein degradation. J Protein Chem. 1988;7:115–127. doi: 10.1007/BF01025241. [DOI] [PubMed] [Google Scholar]
- 45.Finn PF, Mesires NT, Vine M, Dice JF. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 46.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 47.Chiang H, Dice J. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem. 1988;262:6797–6805. [PubMed] [Google Scholar]
- 48.Cuervo A, Gomes J, Barnes A, Dice J. Selective degradation of annexins by chaperone-mediated autophagy. J Biol Chem. 2000;275:33329–33335. doi: 10.1074/jbc.M005655200. [DOI] [PubMed] [Google Scholar]
- 49.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuervo AM, Dice JF. Lysosomes, a meeting point of proteins, chaperones, and proteases. J Mol Med. 2009;76:6–12. doi: 10.1007/s001090050185. [DOI] [PubMed] [Google Scholar]
- 51.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2010;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bandyopadhyay U, Cuervo AM. Chaperone-mediated autophagy in aging and neurodegeneration: lessons from alpha-synuclein. Exp Gerontol. 2007;42:120–128. doi: 10.1016/j.exger.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 53.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8(13):1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 54.Kirkin V, Lamark T, Johansen T, Dikic I. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 55.Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 56.Welsch T, Younsi A, Disanza A, Rodriguez JA, Cuervo AM, Scita G, Schmidt J. Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp Cell Res. 2010;316:1914–1924. doi: 10.1016/j.yexcr.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H, Wang P, Song W, Sun X. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J. 2009;23:3383–3392. doi: 10.1096/fj.09-134296. [DOI] [PubMed] [Google Scholar]
- 58.Bandyopadhyay U, Cuervo AM. Entering the lysosome through a transient gate by chaperone-mediated autophagy. Autophagy. 2008;4:1101–1103. doi: 10.4161/auto.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cuervo AM, Mann L, Bonten EJ, d′Azzo A, Dice JF. Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 2003;22:47–59. doi: 10.1093/emboj/cdg002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cuervo AM, Dice JF. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 63.Kaushik S, Kiffin R, Cuervo AM. Chaperone-mediated autophagy and aging: a novel regulatory role of lipids revealed. Autophagy. 2007;3:387–389. doi: 10.4161/auto.4246. [DOI] [PubMed] [Google Scholar]
- 64.Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010;39:535–547. doi: 10.1016/j.molcel.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh R, Czaja MJ. Compensatory mechanisms and the type of injury determine the fate of cells with impaired macroautophagy. Autophagy. 2008;4:516–518. doi: 10.4161/auto.5800. [DOI] [PubMed] [Google Scholar]
- 68.Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 69.Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 2008;4:442–4456. doi: 10.4161/auto.5654. [DOI] [PubMed] [Google Scholar]
- 70.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 71.Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, Monteiro MJ. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–3232. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rothenberg C, Monteiro MJ. Ubiquilin at a crossroads in protein degradation pathways. Autophagy. 2010;6:101–102. doi: 10.4161/auto.6.7.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou D, Li P, Lin Y, Lott JM, Hislop AD, Canaday DH, Brutkiewicz RR, Blum JS. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 74.Strawbridge AB, Blum JS. Autophagy in MHC class II antigen processing. Curr Opin Immunol. 2007;19:87–92. doi: 10.1016/j.coi.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Wing S, Chiang H, Goldberg A, Dice J. Proteins containing peptide sequences related to KFERQ are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem J. 1991;275:165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cuervo AM, Hu W, Lim B, Dice JF. IκB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bejarano E, Cuervo AM. Chaperone-mediated autophagy. Proc Am Thorac Soc. 2010;7:29–39. doi: 10.1513/pats.200909-102JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Venugopal B, Mesires NT, Kennedy JC, Curcio-Morelli C, Laplante JM, Dice JF, Slaugenhaupt SA. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009;219:344–353. doi: 10.1002/jcp.21676. [DOI] [PubMed] [Google Scholar]
- 80.Fidzianska A, Walczak E, Walski M. Abnormal chaperone-mediated autophagy (CMA) in cardiomyocytes of a boy with Danon disease. Folia Neuropathol. 2007;45:133–139. [PubMed] [Google Scholar]
- 81.Shacka JJ, Roth KA, Zhang J. The autophagy-lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front Biosci. 2008;13:718–736. doi: 10.2741/2714. [DOI] [PubMed] [Google Scholar]
- 82.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 83.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 84.Kon Maria, Cuervo AnaMaria. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2010;584:1399–1404. doi: 10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Veazey C, Aki SO, Cook KF, Lai EC, Kunik ME. Prevalence and treatment of depression in Parkinson’s disease. J Neuropsychiatry Clin Neurosci. 2005;17:310–323. doi: 10.1176/jnp.17.3.310. [DOI] [PubMed] [Google Scholar]
- 86.Martinez-Vicente M, Talloczy Z, Kaushik S, Massey AC, Mazzulli J, Mosharov EV, Hodara R, Fredenburg R, Wu DC, Follenzi A, Dauer W, Przedborski S, Ischiropoulos H, Lansbury PT, Sulzer D, Cuervo AM. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, Di Monte DA. Lysosomal degradation of alpha-synuclein in vivo. J Biol Chem. 2010;285:13621–13629. doi: 10.1074/jbc.M109.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 89.Kabuta T, Furuta A, Aoki S, Furuta K, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–23738. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J, Mao Z. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron. 2003;38:33–46. doi: 10.1016/s0896-6273(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 91.Wang X, Tang X, Li M, Marshall J, Mao Z. Regulation of neuroprotective activity of myocyte-enhancer factor 2 by cAMP-protein kinase A signaling pathway in neuronal survival. J Biol Chem. 2005;280:16705–16713. doi: 10.1074/jbc.M501819200. [DOI] [PubMed] [Google Scholar]
- 92.Tang X, Wang X, Gong X, Tong M, Park D, Xia Z, Mao Z. Cyclin-dependent kinase 5 mediates neurotoxin-induced degradation of the transcription factor myocyte enhancer factor 2. J Neurosci. 2005;25:4823–4834. doi: 10.1523/JNEUROSCI.1331-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Irrcher I, Park DS. Parkinson’s disease: to live or die by autophagy. Sci Signal. 2009;2:pe21. doi: 10.1126/scisignal.265pe21. [DOI] [PubMed] [Google Scholar]
- 94.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y, Martinez-Vicente M, Krüger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E. Synergy and antagonism of macroautophagy and chaperone-mediated autophagy in a cell model of pathological tau aggregation. Autophagy. 2010;6:182–183. doi: 10.4161/auto.6.1.10815. [DOI] [PubMed] [Google Scholar]
- 97.Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, Miyazaki H, Matsumoto G, Kino Y, Nagai Y, Nukina N. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 98.Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cuervo A, Hildebrand H, Bomhard E, Dice J. Direct lysosomal uptake of 2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 1999;55:529–545. doi: 10.1046/j.1523-1755.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- 100.Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 101.Maalouf M, Rhob J, Mattson M. The neuroprotective properties of calorie restriction, the ketogenic diet, and ketone bodies. Brain Res Rev. 2009;59:293–315. doi: 10.1016/j.brainresrev.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mattson M. Dietary factors, hormesis and health. Ageing Res Rev. 2008;7:43–48. doi: 10.1016/j.arr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]