Abstract
Maintenance of ploidy in sexually reproducing organisms requires a specialized form of cell division called meiosis that generates genetically diverse haploid gametes from diploid germ cells. Meiotic cells halve their ploidy by undergoing two rounds of nuclear division (meiosis I and II) after a single round of DNA replication. Research in Saccharomyces cerevisiae (budding yeast) has shown that four major deviations from the mitotic cell cycle during meiosis are essential for halving ploidy. The deviations are (1) formation of a link between homologous chromosomes by crossover, (2) monopolar attachment of sister kinetochores during meiosis I, (3) protection of centromeric cohesion during meiosis I, and (4) suppression of DNA replication following exit from meiosis I. In this review we present the current understanding of the above four processes in budding yeast and examine the possible conservation of molecular mechanisms from yeast to humans.
Keywords: Meiosis, Gametogenesis, Ploidy, Cell cycle, Shugoshin, Monopolin, FEAR
Introduction
Sex entails mixing of paternal and maternal genomes which allows union of two or more favorable mutations of different genotypes into a single genotype in the progeny. This is in contrast to the less plausible scenario in asexually reproducing organisms where favorable mutations need to occur in the same cell lineage. Sex therefore enables rapid evolution of a population in comparison to an otherwise equivalent population that reproduces asexually. However, the requirement of two parents in the sexual mode of reproduction introduces a problem about maintenance of ploidy (chromosome number). The ploidy of a sexually reproducing species would double every generation if diploid (somatic) cells were used as gametes. In order to maintain ploidy, sexually reproducing organisms have evolved a specialized way of cell division called meiosis that produces haploid gametes from diploid germ cells.
How do meiotic cells halve their ploidy? Meiotic cells undergo two nuclear divisions after one round of DNA replication which is in contrast to somatic cells where ordinarily a round of nuclear division is followed by a round of DNA replication. Research on meiosis in various model systems including Saccharomyces cerevisiae (budding yeast), Schizosaccharomyces pombe (fission yeast), Drosophila melanogaster (flies), Caenorhabditis elegans (worms) and Mus musculus (mice) have given valuable insights into how this might be accomplished. In this review we discuss how budding yeast research has led to understanding of meiotic cell cycle events including entry into meiosis, DNA recombination, protection of centromeric cohesion, monopolar attachment of sister kinetochores and exit from meiosis I. At the end we discuss whether the above mechanisms are conserved in higher eukaryotes. An understanding of meiosis is relevant to human health as errors in regulation of the meiotic cell cycle could result in fetal aneuploidy and spontaneous abortions in humans [1].
Overview of mitotic cell cycle in budding yeast
To fully appreciate the logic of meiosis, it is necessary to review how the mitotic cell cycle works as several elements of the mitotic cell cycle machinery are used during meiosis either with no changes or with subtle improvizations. The cell cycle can be divided into G1, S (synthesis), G2 and M (mitotic) phases. The DNA in cells replicates during S phase, and chromosomes segregate and the cytoplasm divides (cytokinesis) during M phase. These phases are separated by two gaps namely, G1 and G2 phases. Progression through the cell cycle is regulated by the highly conserved family of proteins called cyclin-dependent kinases (Cdk). The kinase activity of Cdk on its own is very low but is stimulated by binding to its regulatory subunit called cyclin. Budding yeast has a single Cdk which is encoded by the CDC28 gene. Cdc28 associates with distinct cyclins during different phases of the cell cycle and controls the activity of downstream effector proteins by phosphorylation to orchestrate cell cycle transitions. Entry into S phase is promoted by Cdk bound to cyclins Cln1 and Cln2. Cdk bound to cyclins Clb5 and Clb6 is involved in DNA replication. Cdk bound to cyclins Clbs1–4 regulates progression through mitosis. Oscillating levels of cyclins leads to periodic activation of Cdk which ensures the correct sequence of events such as budding, DNA replication, chromosome segregation and cytokinesis during the cell cycle. When errors occur during the cell cycle, control mechanisms referred to as checkpoints delay or prevent cell cycle progression until the previous step has been successfully completed.
The cell cycle is coordinated with chromosome segregation. Replication of DNA during S phase generates sister chromatids which are held together by a highly conserved protein complex called cohesin. Cohesin is composed of four subunits Smc1, Smc3, Scc1 and Scc3 and forms a 50-nm ring that holds sister chromatids together by trapping them topologically within it [2–4]. The spindle pole bodies (SPBs, functional equivalent of centrosomes in mammals) duplicate during G1/S phase. As cells enter mitosis, the SPBs separate and a bipolar spindle is formed. A specialized structure composed of several multiprotein subcomplexes called the kinetochore is assembled at the centromere on every chromosome. The kinetochore has the ability to bind to growing ends of microtubules. When sister kinetochores bind to microtubules emanating from opposite spindle poles, their pole-ward movement is opposed by sister chromatid cohesion. The ensuing tug-of-war generates tension that stabilizes microtubule–kinetochore connections. When all the sister kinetochores are bioriented on the mitotic spindle (metaphase), the spindle checkpoint (a surveillance mechanism that delays cell cycle progression in response to kinetochore–microtubule binding defects) is inactivated. This results in activation of the ubiquitin ligase called anaphase-promoting complex (APC) bound to the substrate specificity factor Cdc20 (APC/Cdc20). APC/Cdc20 activates a cysteine protease separase (Esp1) by targeting its inhibitor securin (Pds1) for proteasomal degradation by ubiquitination. Separase cleaves the cohesin subunit Scc1 which destroys sister chromatid cohesion and triggers the movement of sister chromatids towards the opposite poles of the cell (anaphase). Following anaphase, cells exit from mitosis by abrogating M-Cdk (mitotic Cdk) activity. This is achieved by destruction of Clbs and dephosphorylation of Cdk targets that result in spindle disassembly, cytokinesis (separation of cytoplasm) and preparation for the next round of DNA replication.
Entry into meiosis
Budding yeast cells can exists as either haploids or diploids. Haploid cells could be one of two possible mating types ‘a’ or ‘α’ (alpha). The mating type is specified by a single locus called MAT and is determined by whether the MAT locus contains the MAT a (encoding gene a1) or the MATα allele (encoding genes α1 and α2). When haploid cells of opposite mating type are in close juxtaposition they mate using a pheromone-based signaling pathway. The cytoplasmic fusion of the two cells is followed by fusion of their nuclei resulting in the formation of a diploid cell. When subjected to nutritional starvation, only diploid cells (MAT a/MATα) can enter meiosis which is a key developmental decision in budding yeast. Meiosis is coupled to sporulation as the four haploid products are packaged as spores in a sac referred to as the ascus. The fact that it is possible to analyze all the four products of an individual meiosis in budding yeast has been invaluable in dissecting the mechanism of meiotic recombination and chromosome segregation.
Yeast cells enter meiosis on the basis of two kinds of cues:
Genetic: cells have to express both MAT a and MATα genes. This ensures that only diploid and not haploid cells enter meiosis.
Environmental: the absence of glucose and nitrogen and the presence of a nonfermentable carbon source in the growth medium. The requirement for a nonfermentable carbon source ensures that only respiration-competent cells can enter meiosis. The coupling of nutrient starvation to sporulation enhances the chance of survival of yeast cells under adverse conditions as the spores are genetically distinct and have the ability to stay dormant for long periods of time.
Integration of these two cues occurs at the promoter of the master transcription factor Ime1 (Initiator of meiosis) which is required for expression of several early-meiosis genes involved in entry into meiosis, DNA replication and homologous recombination [5, 6]. During vegetative growth multiple mechanisms ensure that IME1 is not expressed. In haploid cells the zinc-finger protein Rme1 (repressor of meiosis) binds to the IME1 promoter and prevents its transcription [7]. Rme1 also binds to the CLN2 promoter, but this activates CLN2 expression [8]. Cln2 interferes with entry into meiosis (see below). In diploid cells, the MATa1 and MATα2 gene products form a complex that represses transcription of RME1 [7]. In addition, the MATa1/MATα2 complex activates expression of IME4, a putative RNA methyltransferase which in turn enhances IME1 expression through an unknown mechanism [9, 10].
How does the absence of glucose and nitrogen trigger entry into meiosis? Ime1 is recruited to the promoters upstream of early-meiosis genes and activates their transcription via its interaction with a DNA-binding protein Ume6 [11]. During vegetative growth Ume6 binds to promoters of early-meiosis genes and represses their expression. This requires association of Ume6 with the histone deacetylase Sin3/Rpd3 complex and chromatin remodeling complex Isw2 [12, 13]. The absence of glucose and nitrogen results in the replacement of Sin3/Rpd3 and Isw2 by Ime1 and this switch activates the transcription of early-meiosis genes [14]. Rim11, the homolog of human glycogen synthase kinase (GSK-3) phosphorylates Ime1 which enhances its interaction with Ume6 and thereby activates the expression of early-meiosis genes [15]. Glucose also prevents transcription of IME1 by inhibiting a kinase called Rim15 which promotes interaction of Ime1 with Ume6 [16]. Amongst various targets of Ime1 are two crucial genes IME2 and NDT80 which encode a Cdk-related kinase and a transcription factor for middle-meiosis genes, respectively.
G1/S transition
Entry into premitotic S phase requires the activity of Cdk bound to cyclins Cln1 and Cln2 which target the Clb-Cdk inhibitor Sic1 for degradation. During entry into premeiotic S phase, Sic1 degradation is promoted by Ime2 instead of Cdk-Cln1/2 [17]. Mechanisms driving entry into premitotic and premeiotic S phases might be different as conditions that trigger entry into the two phases are contrastingly different (presence vs. absence of nutrients). In addition, the Cln-Cdks inhibit IME1 expression making entry into mitosis and meiosis mutually exclusive [18]. Cln3-Cdk induces expression of CLN1 and CLN2 during premitotic G1 phase. CLN3 expression is inhibited by Whi3 during meiosis which binds to CLN3 mRNA and localizes it into discrete cytoplasmic foci [19].
Premeiotic S phase
It is generally thought that DNA replication during mitotic and meiotic cell cycles requires the same set of proteins. Consistent with this idea, the replication fork progression rates and choice of replication origins during premitotic and premeiotic DNA replication are quite similar [20]. However, the premeiotic S phase is two to three times longer than the premitotic S phase [21], and this is true in several organisms besides budding yeast. Preparation for recombination and homolog pairing could account for this difference. The length of the premeiotic S phase is negatively regulated by the noncatalytic activity of Spo11 (the endonuclease that initiates recombination by introducing double-strand breaks (DSBs) in DNA) and positively regulated by the meiotic cohesin subunit Rec8 [22]. Clb5 and Clb6 cyclins are not essential for premitotic DNA replication but are essential for premeiotic DNA replication. While Clb1, Clb3 and Clb4 can compensate for the absence of Clb5 and Clb6 during vegetative growth [23], they cannot support premeiotic DNA replication for unknown reasons [24]. Premeiotic, but not premitotic, DNA replication requires the protein Mum2 [25]. However, the precise function of Mum2 in premeiotic DNA replication is unknown.
The four crucial innovations during meiosis
Following premitotic DNA replication, sister chromatids are bioriented on the mitotic spindle and segregated towards opposite poles during anaphase. However, the fate of chromosomes after premeiotic S phase is quite different. Four key differences between mitosis and meiosis I allow cells to halve the chromosome number. They are:
Reciprocal recombination between homologous chromosomes leads to formation of crossovers or chiasmata which is essential for bi-orientation of recombined homologs or bivalents on the metaphase I spindle.
While sister kinetochores bi-orient during mitosis, they mono-orient during meiosis I, i.e. they bind to microtubules emanating from the same spindle pole. This is essential for setting up the reductional mode of meiosis I chromosome segregation.
Cohesin at the centromeric regions is protected from cleavage by separase. Destruction of arm cohesion resolves chiasmata that link homologs together. Centromeric cohesion that persists after meiosis I is required for bi-orientation of sister centromeres on the meiosis II spindle.
S phase is suppressed between meiosis I and II.
The crux of understanding how meiotic cells halve ploidy lies in deciphering how the above four processes occur.
Recombination and formation of chiasmata
After premeiotic DNA replication the sister chromatids are connected by cohesin, but there is no linkage between homologous chromosomes. The result of meiotic recombination is creation of a link between the homologs so that they can be pulled towards opposite spindle poles during meiosis I. Although recombination contributes to genetic diversity, it is important to remember that its principal function is to facilitate chromosome segregation during meiosis I. In the absence of recombination, homologous chromosomes are randomly segregated during meiosis [26, 27].
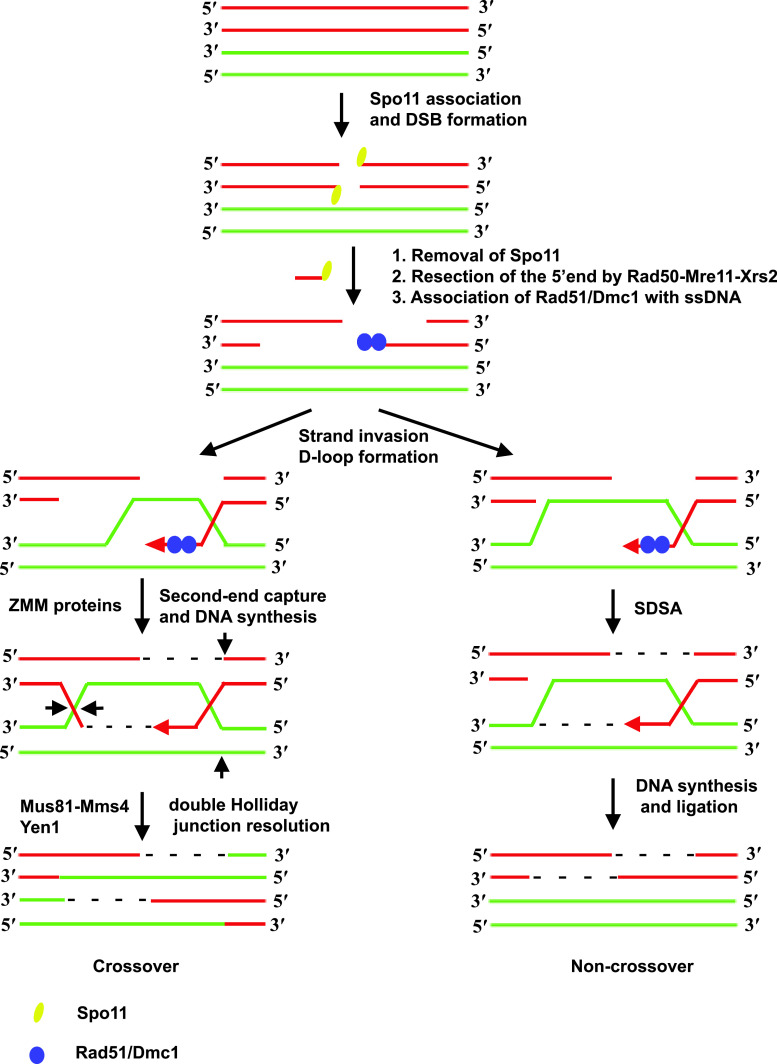
Recombination in budding yeast is initiated by introduction of DSBs in DNA by the topoisomerase-related protein Spo11 [26] with the help of several accessory proteins. Following this, a complex series of steps culminates in the formation of covalent connections between homologs called crossovers (Fig. 1). The DSBs are generated by a trans-esterification reaction and results in the covalent linkage of Spo11 to the 5′ end of DNA at the DSB. The 5′ ends linked to Spo11 are recognized by the RMX complex (Rad50/Mre11/Xrs2). The RMX complex eliminates Spo11 from the 5′ ends and, in collaboration with Sae2/Com1 and Exo1, resects the 5′ ends [28]. This results in the formation of single-stranded 3′ overhangs which are bound by the bacterial RecA-related recombinase proteins Rad51 and Dmc1. The resulting ssDNA-Rad51/Dmc1 nucleoprotein filaments, along with proteins Rad52 and Rad54, are recombinogenic and can scan the genome for homologous sequences. When the nucleoprotein filaments find a match the ssDNA base pairs with a complementary strand from the homologous chromosome displacing the other strand to form a displacement loop (D-loop). Regulatory mechanisms ensure that a homolog and not a sister chromatid is used to repair the DSB [29]. At this point there are two possibilities. The invading 3′ strand could dissociate from its complementary strand and return to its original chromatid. This pathway, referred to as synthesis-dependent strand annealing (SDSA), does not result in a covalent connection between the two homologs (noncrossovers). Alternatively, the displaced D-loop could base-pair with the second strand from the chromatid where the DSB was made. Extension of the 3′ ends and ligation of the gaps result in a structure called a double Holliday junction (dHJ). These dHJs are then resolved by HJ resolvases Mus81/Mms4 and Yen1 which make a pair of nicks and ligations to create a crossover [30].
Fig. 1.
Pathway for meiotic recombination in budding yeast. Recombination is initiated by creation of DSBs in DNA by Spo11. The 5′ ends are resected by the Rad50-Mre11-Xrs2 (RMX) complex acting together with Com2/Sae1 and Exo1. Nucleoprotein filaments formed by association of recombinases Rad51 and meiosis-specific Dmc1 with single-stranded DNA scan the genome for homologous sequences. When a match is found the 3′ end base pairs with the complementary strand displacing the other strand to form a D-loop. At this stage there are two possible routes. In the SDSA pathway the invading strand dissociates from the homologous strand and re-anneals with its complementary strand leading to a non-crossover. A crossover is formed when the displaced strand base-pairs with the strand complementary to the invading strand to form a dHJ which is subsequently resolved by a pair of nick and ligation reactions by resolvases Mus81/Mms4 and Yen1
The formation of crossovers requires pairing of homologs, and this is brought about by synapsis which involves assembly of a highly conserved proteinaceous structure called the synaptonemal complex (SC) that aligns homologs together. Recombination and SC assembly have to be tightly coordinated, and this is achieved by the ZMM (Zip1, Msh4, Msh5) group of proteins [31]. The ZMM group comprises at least seven proteins Zip1–4, Msh4, Msh5 and Mer3. The transverse filament protein Zip1 establishes stable homolog juxtaposition by polymerizing as an integral component of the SC. Zip2, Zip3, and Zip4 likely mediate protein–protein interactions, while Mer3, Msh4, and Msh5 directly promote steps in DNA recombination. Poorly understood mechanisms ensure that recombination generates at least one crossover between a pair of homologs, which is essential for their bi-orientation on the meiosis I spindle. For a more detailed understanding of recombination, readers are directed to a couple of reviews [31, 32].
Linking premeiotic S phase and DNA recombination
Multiple lines of evidence suggest that premeiotic DNA replication is coupled to recombination and homolog pairing. Firstly, there is a direct correlation between the amount of replication observed in clb5clb6 mutants and the number of DSBs they produce [33]. Secondly, deleting all replication origins on the left arm of chromosome III causes similar delays in replication and initiation of recombination in the same chromosomal region, suggesting that replication is directly coupled to recombination [34]. The precise mechanism by which this coupling is achieved is unknown. The presence of a replication fork is required for coupling replication to recombination as mutants that fail to initiate replication (therefore will not produce replication forks) produce DSBs normally and try to repair them [35]. Clb5-Cdk and Dbf4-dependent Cdc7 kinase are required for initiation of replication and for activating Mer2 (a meiosis-specific DSB protein) by phosphorylation [36, 37]. By using the same set of proteins to activate initiation of premeiotic DNA replication and DSB formation, cells might be able to coordinate the two processes.
Recombination checkpoint
The recombination checkpoint ensures that entry into meiosis I occurs after completion of DSB repair. Errors in DNA recombination and synapsis activate this checkpoint which causes an arrest in a stage of meiotic prophase called pachytene. Pachytene is characterized by the presence of unresolved dHJs and persistent SC. Although Cdk activity is very low at this stage of meiosis, the term prophase is used for cytological reasons (due to the appearance of condensed chromosomes with SC).
The recombination checkpoint requires kinases (Mec1 and Mek1), mitotic DNA damage checkpoint components (Rad17 and Rad24), SC components (Hop1 and Red1), Sir2 (a rDNA silencing protein), Pch2 (a AAA-based ATPase) and a histone methyl transferase (Dot1). A recent comprehensive review provides a detailed understanding of how this checkpoint senses errors in recombination [38]. In this review we focus only on how the checkpoint influences the meiotic cell cycle.
The recombination checkpoint primarily operates by controlling Cdk activity and expression of the transcription factor Ndt80. Cdk is regulated by phosphorylation and availability of its regulatory subunit Clb1. Errors in recombination activate Swe1 which inhibits Cdk by phosphorylating Cdc28 at tyrosine-19 [39]. Expression of Clb1 is regulated by Ndt80 which is required for exit from pachytene [40]. Ndt80 binds to middle sporulation elements within the promoters of middle-sporulation genes and activates their transcription. In vegetative cells, the middle sporulation elements are bound by Sum1 which acts as a transcriptional repressor of middle-sporulation genes [41]. Sum1 levels remain high during checkpoint activation and decline during exit from pachytene. Relative amounts of Sum1 and Ndt80 and their competitive binding to middle sporulation elements determines whether cells exit from pachytene [42–44]. Cdk and Ime2 inactivate Sum1 by phosphorylating it, and this leads to activation of Ndt80 [45]. The activity of Ndt80 might also be controlled by its nuclear localization [46] and phosphorylation by Ime2 [47].
Activation of Ndt80 results in the expression of more than 200 middle-meiosis genes which drive exit from pachytene. The middle-meiosis genes are also required for meiotic divisions and spore formation, and include cyclins Clb1, Clb3 and Clb4, polo-like kinase Cdc5 and Ndt80 itself [48, 49]. Activation of polo-like kinase Cdc5 results in the resolution of dHJs and disassembly of SC [50]. Increased M-Cdk activity caused by expression of cyclins results in the separation of SPBs and the formation of a metaphase I spindle.
Monopolar attachment of sister kinetochores
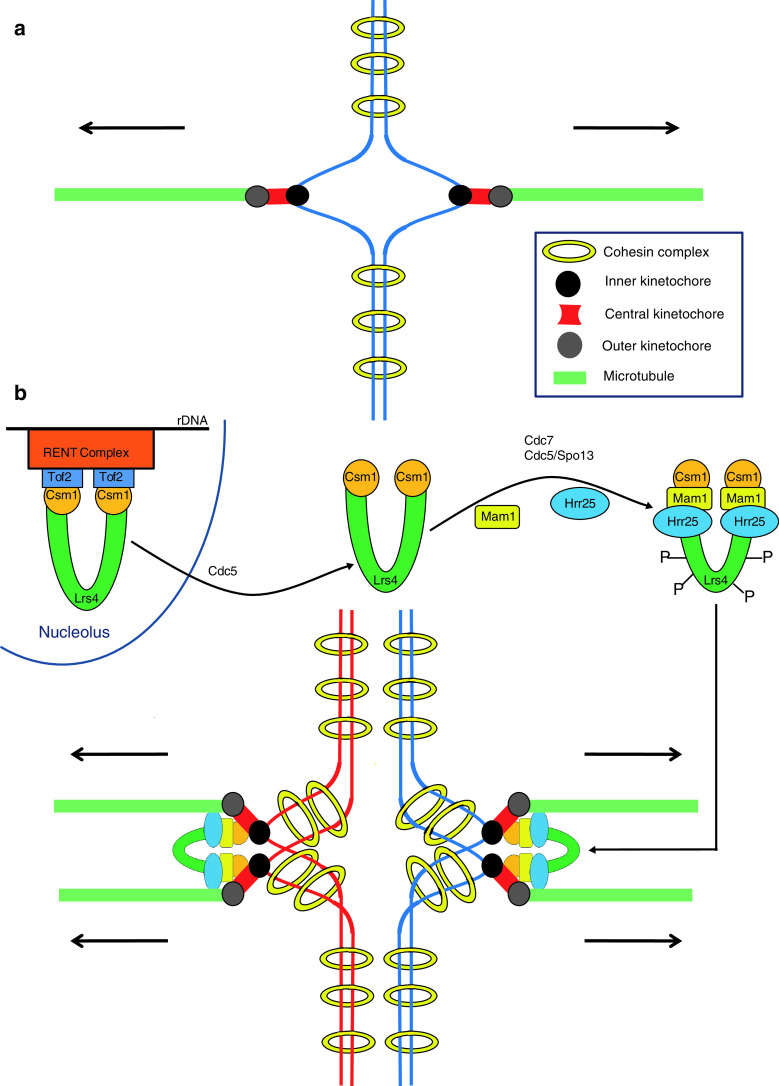
Homologs connected by chiasmata bi-orient on the meiosis I spindle. Tension created by sister chromatid cohesion distal to the chiasmata stabilizes the bi-oriented state. For homologs to go towards opposite spindle poles, it is essential that sister kinetochores are mono-oriented, i.e. they bind to microtubules originating from the same spindle pole. This is in contrast to the scenario during mitosis where sister kinetochores biorient, i.e. they bind to microtubules from opposite spindle poles (Fig. 2a). So how is bi-orientation of sister kinetochores suppressed during meiosis I? Research over the last 10 years has shown that this is accomplished by the ‘monopolin’ complex which is composed of Csm1, Lrs4, Mam1 and Hrr25 [51–53]. Monopolin mutants attempt to biorient sister centromeres on the meiosis I spindle which results in a massive chromosome segregation defect.
Fig. 2.
Monopolar attachment of sister kinetochores during meiosis I. a Cohesion between sister chromatids (blue) is mediated by cohesin (yellow rings). A specialized structure called the kinetochore is formed at the centromere on each chromatid. The kinetochore is composed of three distinct layers the inner (black), the central (red) and the outer (gray) kinetochores. Sister kinetochores bind to microtubules (green rods) emanating from opposite spindle poles during mitosis (bi-orientation). b Csm1 and Lrs4 proteins localize to the nucleolus via their binding to Tof2 which is bound to rDNA via its association with the RENT (regulator of nucleolar silencing and telophase) complex. After exit from pachytene, Cdc5 triggers release of Csm1 and Lrs4 from the nucleolus. The Csm1/Lrs4 complex associates with meiosis-specific Mam1 and casein kinase-1 (Hrr25) subunits to form the monopolin complex. After nucleolar release, Lrs4 is phosphorylated by Dbf4-dependent kinase Cdc7 and Cdc5 acting together with meiosis-specific protein Spo13. The monopolin complex associates with sister kinetochores and crosslinks them such that they bind to microtubules from the same spindle pole. For the sake of simplicity chiasmata are not shown
Csm1 and Lrs4 are nucleolar proteins expressed during the mitotic cell cycle and are required for rDNA silencing and preventing unequal sister chromatid exchange at the rDNA repeats [54, 55]. They interact with Tof2 which binds to rDNA via its interaction with the RENT (regulator of nucleolar silencing and telophase) complex composed of Net1, Cdc14 and Sir2. As cells exit from pachytene, Csm1 and Lrs4 are released from the nucleolus (Fig. 2b) and this requires the activity of polo-like kinase Cdc5 [56, 57]. Csm1 and Lrs4 associate with meiosis-specific protein Mam1 and casein kinase-1 Hrr25 to form the monopolin complex which associates with kinetochores. After nucleolar release Lrs4 is hyperphosphorylated by Dbf4-dependent kinase Cdc7 and Cdc5 acting together with a meiosis-specific protein called Spo13. Lrs4 hyperphosphorylation is thought to assist monopolin binding to kinetochores [56–58]. The precise mechanism of how monopolin co-orients sister kinetochores is unknown. Crystal structure and electron microscopic analysis of Csm1/Lrs4 complexes has indicated that they form a V-shaped structure with two kinetochore-binding globular domains separated by 10 nm [59]. It was therefore suggested that monopolins could work by crosslinking two sister kinetochores via the two globular kinetochore-binding domains such that they face the same spindle pole. However, the mechanism is likely to be more complex as the kinase activity of Hrr25 is required for monopolar attachment but not for monopolin binding to kinetochores [53]. Identifying substrates of Hrr25 at the kinetochores and monopolin binding sites on the kinetochore is crucial for understanding the mechanism of monopolar attachment.
Pcs1 and Mde4, which are fission yeast homologs of Csm1 and Lrs4 respectively, are required to prevent merotelic attachments (where a single kinetochore binds to microtubules from both spindle poles) during mitosis and meiosis II, but are not required for monopolar attachment during meiosis I [52, 60, 61]. An attractive hypothesis has been proposed which might reconcile the contrasting phenotypes of monopolin mutants in budding and fission yeasts [52]. The fission yeast kinetochore has three MT binding sites compared to just one site for the budding yeast kinetochore. Monopolins are proposed to be a nearest-neighbor clamp for MT binding sites. In budding yeast, monopolins clamp microtubule binding sites from two sister kinetochores. In fission yeast, monopolins crosslink adjacent MT binding sites from the same sister kinetochore. Interestingly, it has been recently reported that Pcs1 and Mde4 prevent merotelic attachment in fission yeast by targeting the condensin complex (which is related to the cohesin complex) to the kinetochores [62]. It is therefore possible that the budding yeast monopolin complex targets condensin to the kinetochores to effect monopolar attachment during meiosis I. Consistent with this possibility, the condensin subunits Brn1 and Ycs4 are required for efficient monopolar attachment during meiosis I [63].
Protection of centromeric cohesion
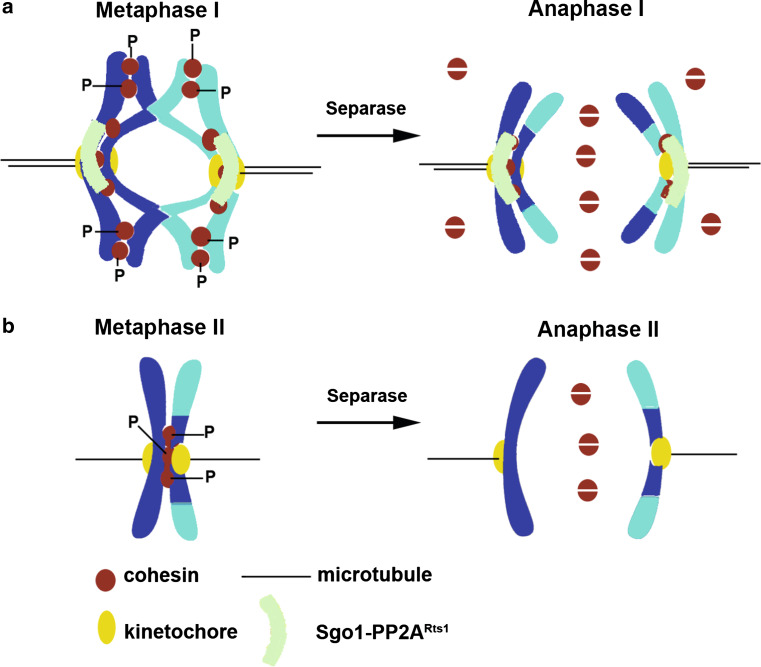
During meiosis there are two nuclear divisions after one round of DNA replication. Since sister chromatid cohesion is established during DNA replication, complete loss of cohesion during meiosis I will result in separation of sister chromatids and random chromosome segregation during meiosis II. Meiotic cells have solved this problem by destroying cohesion in two steps. Cohesion along chromosome arms but not at centromeres is destroyed during meiosis I. Cohesion at centromeres is dissolved during anaphase II. Since separase becomes active during anaphase I, centromeric cohesion has to be protected from separase cleavage. Protected centromeric cohesin is essential for biorientation of sister centromeres during meiosis II. Meiotic cells which fail to protect centromeric cohesion during meiosis I segregate dyad chromosomes randomly during meiosis II.
Protection of centromeric cohesion during meiosis I in budding yeast requires two crucial changes. Firstly, the cohesin subunit Scc1 is replaced by the meiosis-specific Rec8. In meiotic cells expressing Scc1 instead of Rec8, protection of centromeric cohesion during anaphase I fails [51]. Secondly, meiotic cells target Sgo1 to the kinetochore to protect centromeric Rec8 from separase cleavage during meiosis I. Sgo1 belongs to a highly conserved family of coiled-coil proteins called ‘shugoshins’ (guardian spirit in Japanese) and are required for protection of centromeric cohesion during meiosis I in budding yeast, fission yeast, flies, plants and mice [64–70].
How does Sgo1 protect centromeric cohesion? Work in the last 5 years has uncovered an elegant mechanism for protection of centromeric cohesion. Sgo1 recruits the protein phosphatase PP2ARts1 [71, 72] to the kinetochores. The PP2A phosphatases consist of three subunits—a catalytic subunit (Pph21/Pph22), a scaffold subunit (Tpd3) and a regulatory subunit (Rts1/Cdc55). PP2ARts1 refers to a form that contains Pph21/22, Tpd3 and Rts1. Recruitment of PP2ARts1 to the kinetochores is achieved by its physical interaction with Sgo1 and a mutant form of Sgo1 that fails to bind to PP2ARts1 is defective in protection of centromeric cohesion [73]. PP2ARts1 opposes phosphorylation of the meiotic cohesin subunit Rec8 at the centromeres by Hrr25, a casein kinase-1 and the Dbf4-dependent kinase Cdc7 [74]. This results in phosphorylated cohesin along the chromosome arms and dephosphorylated cohesin at the centromeres (Fig. 3). It has already been established that phosphorylation of mitotic cohesin subunit Scc1 by polo-like kinase Cdc5 sensitizes its cleavage by separase [75]. If a similar relationship between phosphorylation and sensitivity to separase extends to Rec8 then arm cohesin (phosphorylated) but not centromeric cohesin (dephosphorylated) would be susceptible to separase cleavage during meiosis I. Consistent with this idea, replacement of 24 phosphorylated serine/threonine residues in Rec8 by alanine (rec8-24A) has been shown to inhibit its cleavage by separase [74]. Conversely phosphomimetic substitution of 14 phosphorylated serine/threonine residues with aspartate (rec8-14D) causes loss of centromeric cohesion during meiosis I [74]. The above mechanism for protection of centromeric cohesion is conserved in fission yeast in which it has been shown that the shugoshin–PP2A complex opposes casein kinase-1-dependent separase cleavage of Rec8 during meiosis I [76, 77].
Fig. 3.
Protection of centromeric cohesion during meiosis I. a During metaphase I, Sgo1 targets the protein phosphatase PP2ARts1 to the kinetochore which shields centromeric cohesin from phosphorylation by casein kinase-1 (Hrr25) and Dbf4-dependent kinase (DDK). Separase cleaves arm cohesin (phosphorylated) to resolve chiasmata during anaphase I, but does not cleave centromeric cohesin (dephosphorylated). b Loss of Sgo1/PP2ARts1 function after meiosis I results in phosphorylation of centromeric cohesin. Separase gets reactivated during meiosis II and cleaves centromeric cohesin to separate dyad chromosomes
The polo-like kinase Cdc5 is required for phosphorylation of Rec8 and for its cleavage by separase during meiosis I [56, 57]. However, alanine replacement of serine/threonine residues in Rec8 that were phosphorylated by Cdc5 has only a minor effect on the kinetics of Rec8 cleavage during meiosis I [78]. Given the overwhelming evidence for casein kinase-1 and the Dbf4-dependent kinase, Cdc5 might play an indirect role in Rec8 phosphorylation. For instance, Cdc5 might be required for full activation of Cdc7 during meiosis. Consistent with this idea, Cdc5 physically interacts with Cdc7 during meiosis I [58].
Exit from meiosis I
For halving ploidy, meiotic cells also have to prevent an occurrence of S phase between meiosis I and II. Since high Cdk activity prevents licensing of replication origins [79], it is envisaged that meiotic cells might prevent a second round of DNA replication by maintaining relatively high levels of Cdk activity between meiosis I and II. Meiotic cells have to perform a careful balancing act in terms of Cdk activity during exit from meiosis I. The Cdk activity has to be reduced sufficiently to disassemble the meiosis I spindle but should be high enough to prevent licensing of replication origins. It is suggested that budding yeast cells have solved this tricky problem by employing the FEAR (Cdc fourteen early anaphase release) network instead of the MEN (mitotic exit network) to downregulate Cdk activity during exit from meiosis I [80, 81].
Exit from mitosis requires inactivation of Clb–Cdk. This is achieved in budding yeast by the activation of the Cdk-antagonizing phosphatase Cdc14 during anaphase. Activated Cdc14 dephosphorylates Cdk substrates and targets the mitotic cyclins for degradation. Both FEAR and MEN signaling cascades activate Cdc14 during anaphase. However, MEN but not FEAR is essential for mitotic exit as it causes a more complete inactivation of Cdk activity during anaphase.
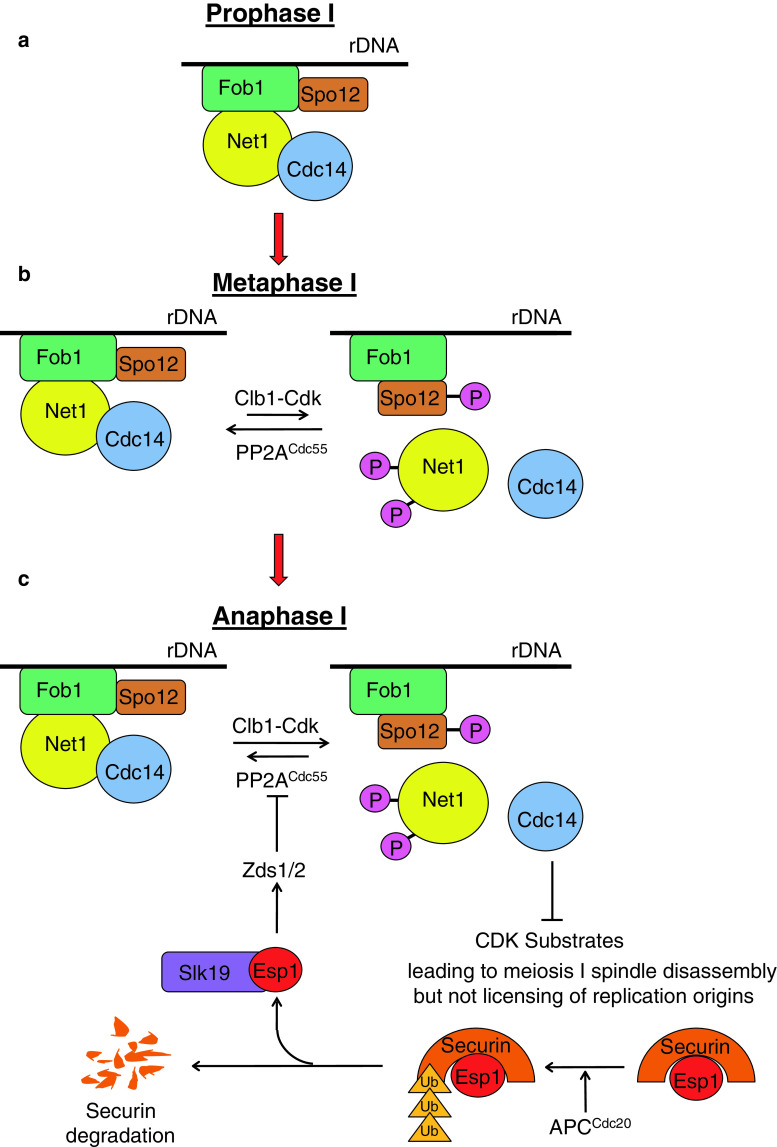
How does FEAR work? Cdc14 is kept in an inactive state bound to a nucleolar anchor protein Net1 for much of the cell cycle (Fig. 4a). Phosphorylation of Net1 by Clb–Cdk causes dissociation of Net1–Cdc14 complexes and Cdc14 release from the nucleolus during anaphase [82, 83]. Premature release of Cdc14 from the nucleolus is prevented by the protein phosphatase PP2ACdc55 which opposes Net1 phosphorylation by Cdk until early anaphase (Fig. 4b) [84, 85]. Inhibition of PP2ACdc55 by separase (which triggers the metaphase–anaphase transition by destroying sister chromatid cohesion) results in Net1 phosphorylation and Cdc14 release from the nucleolus (Fig. 4c). The precise mechanism by which separase inhibits PP2ACdc55 is not known, but it requires assistance from two fungal-specific proteins Zds1 and Zds2 [86] and the kinetochore protein Slk19. Interestingly the proteolytic activity of separase is not required for its FEAR function. In a parallel branch of the FEAR pathway a nucleolar protein Spo12 promotes Cdc14 release from the nucleolus during anaphase. Net1–Cdc14 interaction is promoted by the nucleolar protein Fob1. Activation of Spo12 by Cdk-mediated phosphorylation during anaphase weakens Net1–Cdc14 interaction by inhibiting Fob1 [87, 88].
Fig. 4.
The FEAR network regulates exit from meiosis I. a Until prophase I, Cdc14 is bound to the nucleolus via its physical interaction with its competitive inhibitor Net1 which is enhanced by the replication fork barrier protein Fob1. b Increase in Cdk activity following exit from pachytene promotes phosphorylation of Net1, but this is opposed by the protein phosphatase PP2ACdc55 restraining Cdc14 release from the nucleolus. This state persists until metaphase I. c During anaphase I, separase is activated by destruction of its inhibitor securin which is targeted for proteasomal degradation by APCCdc20. Activated separase in association with Slk19 inhibits PP2ACdc55 via Zds1/2 proteins which results in phosphorylation of Net1 and Cdc14 release from the nucleolus. In addition, activation of Spo12 by Cdk-mediated phosphorylation weakens the effect of Fob1 in stabilizing Net1–Cdc14 interaction. The Cdc14 released antagonizes Cdk activity which is sufficient to cause spindle disassembly but not licensing of DNA replication origins
The FEAR network is required for exit from meiosis I
The Cdc14 release initiated by the FEAR network is transient. This is possibly because the declining Cdk activity is unable to sustain Net1 phosphorylation, thus leading to return of Cdc14 to the nucleolus. FEAR-induced Cdc14 release activates MEN, a G-protein mediated signaling cascade, which causes a complete release of Cdc14 from the nucleolus and is essential for mitotic exit. Although the transience of FEAR-induced Cdc14 release limits its ability to control mitotic exit, it is however appropriate for exit from meiosis I which requires an incomplete loss of Cdk activity. Indeed, the FEAR network has been shown to be essential for exit from meiosis I. Mutations in SPO12, SLK19 and ESP1 genes delay Cdc14 release from the nucleolus and disassembly of anaphase I spindles [80, 81]. Although these mutant cells are arrested in anaphase I the meiosis II events go unperturbed and they perform two nuclear divisions on the same spindle resulting in the formation of dyads (two-spored asci). Thus FEAR is essential for coupling the chromosome segregation cycle to spindle assembly and disassembly during meiosis. Conversely, premature activation of FEAR during meiosis caused by loss of PP2ACdc55 activity results in a failure to assemble bipolar spindles and undergo nuclear divisions [89, 90]. These results highlight the importance of FEAR and its timing of activation in regulating exit from meiosis I.
Is FEAR upregulated during meiosis I?
Although much of the knowledge about FEAR has come out of mitotic experiments, it is quite likely to operate in the same way during meiosis I. However, action of FEAR during meiosis I, but not during mitosis, achieves disassembly of anaphase spindles. MEN is inactive during meiosis I as a few essential MEN components like Cdc15 and Tem1 are not expressed during meiosis I [91]. Therefore, the question that arises is: how does FEAR cause disassembly of anaphase I spindles? One possibility is that meiotic cells have intrinsically low levels of Cdk activity compared to mitotic cells, thus leading to increased sensitivity to FEAR. An alternative possibility is that FEAR is upregulated during meiosis to enable disassembly of anaphase I spindles. A strong candidate for this role is the Cdk-related kinase Ime2 which has two peaks of expression during meiosis [92]. The first peak is during early meiosis which would be consistent with its role in initiating premeiotic DNA replication. The second peak of expression coincides with meiosis I, but its functional significance is yet to be determined. Although Ime2 has overlapping substrate specificity with Cdk, the Ime2 phosphorylation sites are resistant to dephosphorylation by Cdc14 [93]. Therefore, it has been suggested that Ime2 limits the activity of Cdc14 during exit from meiosis I and prevents re-replication. Just as Ime2 and Cdk collaborate during exit from pachytene by phosphorylating Sum1, they could also phosphorylate Net1 during exit from meiosis I. Since Ime2 phosphorylation sites in Net1 could be resistant to dephosphorylation by Cdc14, the Cdc14 release could be stronger than during mitotic FEAR activation and might suffice to disassemble anaphase I spindles. Consistent with this possibility, ime2 mutants accentuate the phenotype of FEAR mutants [94]. Identifying substrates of Ime2 during meiosis I is key to understanding how exit from meiosis I is regulated in budding yeast.
Meiosis II
After exit from meiosis I, the SPBs duplicate and separate to form two sets of bipolar spindles (metaphase II). While chromosome segregation during meiosis I is reductional, it is equational during meiosis II and similar to mitosis. Mechanisms supporting monopolar attachment of sister kinetochores and protection of centromeric cohesion would have to be disabled to accomplish this switch. Centromeric cohesion is required for bi-orientation of sister centromeres during metaphase II. Separase becomes active again during anaphase II and cleaves centromeric cohesin, triggering the metaphase II–anaphase II transition. Although the levels of Sgo1 decline after meiosis I, it is still bound to chromatin until anaphase II [67]. Centromeric cohesin is no longer protected during meiosis II, and this suggests that either Sgo1 bound to chromosomes after meiosis I is not functional or activated separase is able to cleave centromeric cohesin as a result of a high enzyme/substrate ratio. How monopolins are inactivated after meiosis I is completely unknown.
How do meiotic cells ensure that there is adequate Cdk activity for meiosis II? A clue to how this might be achieved came from an elegant study investigating the kinetics of cyclin expression during synchronous meiosis [95]. While Clb1 was expressed during meiosis I, Clb3 was expressed only during meiosis II. The fact that CLB1 and CLB3 are both transcribed at the same time following Ndt80 activation implies that the lag in Clb3 expression is due to a delay in CLB3 mRNA translation. A 153-base sequence at the 5′ untranslated region of CLB3 mRNA was necessary for meiosis II-specific accumulation of Clb3. The same sequence was also sufficient to prevent protein accumulation during meiosis I. It is not known how 5′ untranslated region of CLB3 mRNA prevents translation during meiosis I. It might recruit a meiosis I-specific RNA-binding protein which either prevents its access to the translational machinery or delays export of CLB3 RNA from the nucleus to the cytoplasm. By engineering a delay in translation of CLB3 mRNA meiotic cells might ensure that Cdk activity builds up quickly after meiosis I.
Following meiosis II, a forespore membrane is synthesized within the cytoplasm of the mother cell which eventually becomes the plasma membrane of ascospores. The growth of the forespore membrane begins at the outer plaque of the SPBs and then proceeds outwards, encapsulating the four haploid nuclei.
Features of meiotic cell cycle regulation
A study of budding yeast meiosis has helped us uncover general mechanisms by which ploidy is halved. Firstly, the transcription of genes that are specifically required during meiosis is linked to nutrient starvation. Secondly, meiotic cells make subtle improvizations to the mitotic cell cycle machinery to effect dramatic changes that result in halving of ploidy. The use of the meiosis-specific cohesin complex subunit Rec8 is an outstanding example. Rec8 is required for recombination, SC assembly, and protection of centromeric cohesion. Thirdly, as in many processes in biology, the use of phosphorylation and dephosphorylation as a molecular switch is also true for the meiotic program in yeast. Fourthly, meiotic cells use master kinases to regulate multiple key processes during meiosis. The Cdk-related Ime2 kinase is required for DNA replication and exit from pachytene. The Dbf4-dependent kinase Cdc7 is required for initiating DNA replication during the mitotic cell cycle. However, during meiosis Cdc7 is also required for recombination, monopolar attachment of sister kinetochores and sensitizing arm cohesin cleavage during meiosis I. The casein kinase-1 is required for monopolar attachment of sister kinetochores and for sensitizing cohesin cleavage along chromosome arms during meiosis I. Cdc5 is required for chiasmata formation, monopolar attachment and for cohesin cleavage during meiosis I. The presence of master kinases might allow cells to better coordinate the relative timing of landmark events during meiosis.
Are meiotic regulatory mechanisms conserved from budding yeast to humans?
Since meiosis is fundamental for all sexually reproducing organisms, one would imagine the ploidy-halving mechanisms to be evolutionarily conserved. While mechanisms controlling recombination and possibly protection of centromeric cohesion are conserved, those regulating monopolar attachment and exit from meiosis I may have diverged significantly.
Multiple elements of the recombination pathway that leads to crossover between homologs are conserved from yeast to humans. Paralogs of genes encoding Spo11 (endonuclease that creates DSBs), the Rad50–Mre11–Xrs2 complex (processing of DSBs), Dmc1 (recombinase), Mus81–Mms4 and Yen1 (DHJ resolvases) are present in mammalian genomes. Inactivation of some of the above genes results in phenotypes that are expected from their deduced function in yeast [96]. Defects in recombination and synapsis also trigger an arrest in pachytene in mice, suggesting the existence of a recombination checkpoint. However, none of the genes required for this checkpoint have been identified. Interestingly, the murine homolog of the yeast checkpoint protein Pch2 is not required for checkpoint signaling in mice but for recombination [97].
Members of the shugoshin family of proteins are required for protection of centromeric cohesion during meiosis I in mice. Inactivation of murine Sgo2 by RNAi in oocytes results in loss of protein phosphatase PP2A-B56 (counterpart of budding yeast PP2ARts1) binding to centromeres and premature sister chromatid separation during anaphase I [70]. Since separase is required for cleavage of arm cohesin during anaphase I in murine oocytes [98], it is quite likely that Sgo2 protects cohesin from separase cleavage by recruiting PP2A-B56 to the centromeric regions during meiosis I.
Since homologs of Lrs4 and Csm1 are found only amongst closely related yeasts and the fact that the fission yeast counterpart (Pcs1/Mde4 complex) is not required for monopolar attachment, the mechanism of monopolar attachment in mammals is bound to be different. In fission yeast, cohesion at the core centromeric region mediated by cohesin containing meiosis-specific subunit Rec8 is required for monopolar attachment [99, 100]. By juxtaposing the sister kinetochores side-by-side, centromeric cohesin constrains the sister kinetochores to face the same spindle pole during meiosis I [101]. The function of Rec8 in monopolar attachment appears to be conserved, as inactivation of Rec8 results in bi-orientation of sister kinetochores during meiosis I in plants [102, 103]. It remains to be tested whether Rec8 is also required for monopolar attachment during mammalian meiosis I.
The mechanism regulating exit from meiosis I in mammals is likely to be quite different from that in budding yeast. Although homologs of Cdc14 exist in humans, they do not appear to be required for cell cycle progression [104]. In addition, the orthologs of FEAR components in fission yeast are not required for nucleolar release of the Cdc14 homolog Clp1 [105]. In fission yeast, a meiosis-specific protein Mes1 inhibits APC, which results in a partial destruction of cyclin (Cdc13) during exit from meiosis I [106]. This ensures that there is adequate Cdk activity for meiosis II. Inactivation of Mes1 results in complete destruction of cyclins and a failure to undergo meiosis II. In Xenopus oocytes, the presence of a meiosis-specific S phase inhibitor Mos and the absence of Cdk-inactivating Wee1 kinase are important for preventing an extra round of DNA replication between the two divisions [107, 108]. How mammalian cells exit from meiosis I is not known.
Conclusion and future perspective
Although most of the genes regulating the key meiotic events in budding yeast have been identified, the precise molecular mechanisms by which the encoded proteins work remain poorly understood. Structural biochemistry and in vitro reconstitution of physiological reactions with purified protein complexes will be crucial for advancing our knowledge of meiosis. For a complete understanding of recombination, each step would need to be reconstituted in the proposed pathway with defined components. Poor synchrony of meiotic yeast cultures limits interpretation of mutant phenotypes. Live imaging of meiosis and synchronous meiotic cultures are two powerful tools that will play an increasingly important role in analyzing the phenotypes of mutants. The translation of CLB3 mRNA during meiosis II was discovered because of a new protocol for synchronous meiosis. Since phosphorylation is used as a molecular switch in multiple meiotic processes, quantitative phosphoproteomics of synchronous meiotic cultures might be illuminating. Ultimately, it is crucial to test whether the mechanisms discovered in yeast are conserved in mammals. A more detailed understanding of mammalian meiosis might shed light on the basis of infertility and aneuploidy-related disorders in humans.
Acknowledgments
We apologize to numerous colleagues if, due to space constraints, their work is not cited in this review. P.A. and S.S. are supported by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC, BB/G00353X/1). G.W.K. is supported by a BBSRC-funded studentship.
Abbreviations
- APC
Anaphase promoting complex
- Cdk
Cyclin-dependent kinase
- dHJ
Double Holliday junction
- DSB
Double-strand break
- FEAR
Cdc fourteen early anaphase release
- MEN
Mitotic exit network
- MT
Microtubule
- SC
Synaptonemal complex
- SPB
Spindle pole body
References
- 1.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 2.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 3.Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- 4.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 5.Kassir Y, Adir N, Boger-Nadjar E, Raviv NG, Rubin-Bejerano I, Sagee S, Shenhar G. Transcriptional regulation of meiosis in budding yeast. Int Rev Cytol. 2003;224:111–171. doi: 10.1016/s0074-7696(05)24004-4. [DOI] [PubMed] [Google Scholar]
- 6.Honigberg SM, Purnapatre K. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J Cell Sci. 2003;116:2137–2147. doi: 10.1242/jcs.00460. [DOI] [PubMed] [Google Scholar]
- 7.Covitz PA, Herskowitz I, Mitchell AP. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1-alpha 2. Genes Dev. 1991;5:1982–1989. doi: 10.1101/gad.5.11.1982. [DOI] [PubMed] [Google Scholar]
- 8.Toone WM, Johnson AL, Banks GR, Toyn JH, Stuart D, Wittenberg C, Johnston LH. Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J. 1995;14:5824–5832. doi: 10.1002/j.1460-2075.1995.tb00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah JC, Clancy MJ. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1078–1086. doi: 10.1128/mcb.12.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 13.Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 14.Pnueli L, Edry I, Cohen M, Kassir Y. Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast. Mol Cell Biol. 2004;24:5197–5208. doi: 10.1128/MCB.24.12.5197-5208.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malathi K, Xiao Y, Mitchell AP. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidan S, Mitchell AP. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol. 1997;17:2688–2697. doi: 10.1128/mcb.17.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirick L, Goetsch L, Ammerer G, Byers B. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science. 1998;281:1854–1857. doi: 10.1126/science.281.5384.1854. [DOI] [PubMed] [Google Scholar]
- 18.Colomina N, Garí E, Gallego C, Herrero E, Aldea M. G1 cyclins block the Ime1 pathway to make mitosis and meiosis incompatible in budding yeast. EMBO J. 1999;18:320–329. doi: 10.1093/emboj/18.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gari E, Volpe T, Wang H, Gallego C, Futcher B, Aldea M. Whi3 binds the mRNA of the G1 cyclin CLN3 to modulate cell fate in budding yeast. Genes Dev. 2001;15:2803–2808. doi: 10.1101/gad.203501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins I, Newlon CS. Chromosomal DNA replication initiates at the same origins in meiosis and mitosis. Mol Cell Biol. 1994;14:3524–3534. doi: 10.1128/mcb.14.5.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson DH, Johnston LH, Fennell DJ, Simchen G. The timing of the S phase and other nuclear events in yeast meiosis. Exp Cell Res. 1983;145:209–217. doi: 10.1016/s0014-4827(83)80022-6. [DOI] [PubMed] [Google Scholar]
- 22.Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- 23.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 24.Stuart D, Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis L, Barbera M, McDonnell A, McIntyre K, Sternglanz R, Jin Q, Loidl J, Engebrecht J. The Saccharomyces cerevisiae MUM2 gene interacts with the DNA replication machinery and is required for meiotic levels of double strand breaks. Genetics. 2001;157:1179–1189. doi: 10.1093/genetics/157.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 27.Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 28.Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lao JP, Hunter N. Trying to avoid your sister. PLoS Biol. 2010;8:e1000519. doi: 10.1371/journal.pbio.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 32.Whitby MC. Making crossovers during meiosis. Biochem Soc Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 33.Smith KN, Penkner A, Ohta K, Klein F, Nicolas A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- 34.Borde V, Goldman AS, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 35.Hochwagen A, Tham WH, Brar GA, Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005;122:861–873. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 36.Sasanuma H, Ohta K. Formation of DNA double-strand breaks in meiosis. Tanpakushitsu Kakusan Koso. 2009;54:459–465. [PubMed] [Google Scholar]
- 37.Wan L, Niu H, Futcher B, Zhang C, Shokat KM, Boulton SJ, Hollingsworth NM. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 2008;22:386–397. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Leu JY, Roeder GS. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol Cell. 1999;4:805–814. doi: 10.1016/s1097-2765(00)80390-1. [DOI] [PubMed] [Google Scholar]
- 40.Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindgren A, Bungard D, Pierce M, Xie J, Vershon A, Winter E. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 2000;19:6489–6497. doi: 10.1093/emboj/19.23.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pak J, Segall J. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:6430–6440. doi: 10.1128/MCB.22.18.6430-6440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierce M, Benjamin KR, Montano SP, Georgiadis MM, Winter E, Vershon AK. Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol Cell Biol. 2003;23:4814–4825. doi: 10.1128/MCB.23.14.4814-4825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin ME, Skokotas A, Winter E. The Cdk1 and Ime2 protein kinases trigger exit from meiotic prophase in Saccharomyces cerevisiae by inhibiting the Sum1 transcriptional repressor. Mol Cell Biol. 2010;30:2996–3003. doi: 10.1128/MCB.01682-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Chang CY, Wu JF, Tung KS. Nuclear localization of the meiosis-specific transcription factor Ndt80 is regulated by the pachytene checkpoint. Mol Biol Cell. 2011;22:1878–1886. doi: 10.1091/mbc.E10-12-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sopko R, Raithatha S, Stuart D. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol Cell Biol. 2002;22:7024–7040. doi: 10.1128/MCB.22.20.7024-7040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- 49.Hepworth SR, Friesen H, Segall J. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sourirajan A, Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008;22:2627–2632. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- 52.Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- 53.Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 54.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waples WG, Chahwan C, Ciechonska M, Lavoie BD. Putting the brake on FEAR: Tof2 promotes the biphasic release of Cdc14 phosphatase during mitotic exit. Mol Biol Cell. 2009;20:245–255. doi: 10.1091/mbc.E08-08-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- 57.Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- 58.Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135:662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 59.Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, Kearsey SE, Shirahige K, Allshire RC, Nasmyth K. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol. 2007;17:1190–1200. doi: 10.1016/j.cub.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rumpf C, Cipak L, Schleiffer A, Pidoux A, Mechtler K, Tolic-Norrelykke IM, Gregan J. Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle. 2010;9:3997–4004. doi: 10.4161/cc.9.19.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- 63.Brito IL, Yu HG, Amon A. Condensins promote coorientation of sister chromatids during meiosis I in budding yeast. Genetics. 2010;185:55–64. doi: 10.1534/genetics.110.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 65.Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- 66.Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenhaber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 67.Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Tang TT, Bickel SE, Young LM, Orr-Weaver TL. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamant O, Golubovskaya I, Meeley R, Fiume E, Timofejeva L, Schleiffer A, Nasmyth K, Cande WZ. A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr Biol. 2005;15:948–954. doi: 10.1016/j.cub.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 70.Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- 71.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Gálová M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 72.Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 73.Xu Z, Cetin B, Anger M, Cho US, Helmhart W, Nasmyth K, Xu W. Structure and function of the PP2A-shugoshin interaction. Mol Cell. 2009;35:426–441. doi: 10.1016/j.molcel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell. 2010;18:397–409. doi: 10.1016/j.devcel.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by polo/cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 76.Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol. 2010;12:500–506. doi: 10.1038/ncb2052. [DOI] [PubMed] [Google Scholar]
- 77.Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, Ammerer G, Mechtler K, Gregan J. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle. 2010;9:2657–2662. doi: 10.4161/cc.9.13.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- 79.Dahmann C, Diffley JF, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 80.Marston AL, Lee BH, Amon A. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev Cell. 2003;4:711–726. doi: 10.1016/s1534-5807(03)00130-8. [DOI] [PubMed] [Google Scholar]
- 81.Buonomo SB, Rabitsch KP, Fuchs J, Gruber S, Sullivan M, Uhlmann F, Petronczki M, Tóth A, Nasmyth K. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev Cell. 2003;4:727–739. doi: 10.1016/s1534-5807(03)00129-1. [DOI] [PubMed] [Google Scholar]
- 82.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by inhibiting and sequestering the Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 83.Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 84.Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A (Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 85.Yellman CM, Burke DJ. The role of Cdc55 in the spindle checkpoint is through regulation of mitotic exit in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:658–666. doi: 10.1091/mbc.E05-04-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Queralt E, Uhlmann F. Separase cooperates with Zds1 and Zds2 to activate Cdc14 phosphatase in early anaphase. J Cell Biol. 2008;182:873–883. doi: 10.1083/jcb.200801054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tomson BN, Rahal R, Reiser V, Monje-Casas F, Mekhail K, Moazed D, Amon A. Regulation of Spo12 phosphorylation and its essential role in the FEAR network. Curr Biol. 2009;19:449–460. doi: 10.1016/j.cub.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stegmeier F, Huang J, Rahal R, Zmolik J, Moazed D, Amon A. The replication fork block protein Fob1 functions as a negative regulator of the FEAR network. Curr Biol. 2004;14:467–480. doi: 10.1016/j.cub.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 89.Kerr GW, Sarkar S, Tibbles KL, Petronczki M, Millar JB, Arumugam P. Meiotic nuclear divisions in budding yeast require PP2A(Cdc55)-mediated antagonism of Net1 phosphorylation by Cdk. J Cell Biol. 2011;193:1157–1166. doi: 10.1083/jcb.201103019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bizzari F, Marston AL. Cdc55 coordinates spindle assembly and chromosome disjunction during meiosis. J Cell Biol. 2011;193:1213–1228. doi: 10.1083/jcb.201103076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kamieniecki RJ, Liu L, Dawson DS. FEAR but not MEN genes are required for exit from meiosis I. Cell Cycle. 2005;4:1093–1098. [PubMed] [Google Scholar]
- 92.Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holt LJ, Hutti JE, Cantley LC, Morgan DO. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol Cell. 2007;25:689–702. doi: 10.1016/j.molcel.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schindler K, Winter E. Phosphorylation of Ime2 regulates meiotic progression in Saccharomyces cerevisiae. J Biol Chem. 2006;281:18307–18316. doi: 10.1074/jbc.M602349200. [DOI] [PubMed] [Google Scholar]
- 95.Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 97.Li XC, Schimenti JC. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet. 2007;3:e130. doi: 10.1371/journal.pgen.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kudo NR, Wassmann K, Anger M, Schuh M, Wirth KG, Xu H, Helmhart W, Kudo H, McKay M, Maro B, Ellenberg J, de Boer P, Nasmyth K. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 2006;126:135–146. doi: 10.1016/j.cell.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 99.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 100.Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- 102.Shao T, Tang D, Wang K, Wang M, Che J, Qin B, Yu H, Li M, Gu M, Cheng Z. OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol. 2011;156:1386–1396. doi: 10.1104/pp.111.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Márquez-Lema A, Bhatt AM, Horlow C, Mercier R, Mézard C, Grelon M. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118:4621–4632. doi: 10.1242/jcs.02583. [DOI] [PubMed] [Google Scholar]
- 104.Mocciaro A, Schiebel E. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci. 2010;123:2867–2876. doi: 10.1242/jcs.074815. [DOI] [PubMed] [Google Scholar]
- 105.Chen CT, Peli-Gulli MP, Simanis V, McCollum D. S. pombe FEAR protein orthologs are not required for release of Clp1/Flp1 phosphatase from the nucleolus during mitosis. J Cell Sci. 2006;119:4462–4466. doi: 10.1242/jcs.03220. [DOI] [PubMed] [Google Scholar]
- 106.Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature. 2005;434:529–533. doi: 10.1038/nature03406. [DOI] [PubMed] [Google Scholar]
- 107.Furuno N, Nishizawa M, Okazaki K, Tanaka H, Iwashita J, Nakajo N, Ogawa Y, Sagata N. Suppression of DNA replication via Mos function during meiotic divisions in Xenopus oocytes. EMBO J. 1994;13:2399–2410. doi: 10.1002/j.1460-2075.1994.tb06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nakajo N, Yoshitome S, Iwashita J, Iida M, Uto K, Ueno S, Okamoto K, Sagata N. Absence of Wee1 ensures the meiotic cell cycle in Xenopus oocytes. Genes Dev. 2000;14:328–338. [PMC free article] [PubMed] [Google Scholar]