Abstract
In eukaryotes, binding of the six-subunit origin recognition complex (ORC) to DNA provides an interactive platform for the sequential assembly of pre-replicative complexes. This process licenses replication origins competent for the subsequent initiation step. Here, we analyze the contribution of human Orc6, the smallest subunit of ORC, to DNA binding and pre-replicative complex formation. We show that Orc6 not only interacts with Orc1–Orc5 but also with the initiation factor Cdc6. Biochemical and imaging experiments reveal that this interaction is required for licensing DNA replication competent. Furthermore, we demonstrate that Orc6 contributes to the interaction of ORC with the chaperone protein HMGA1a (high mobility group protein A1a). Binding of human ORC to replication origins is not specified at the level of DNA sequence and the functional organization of origins is poorly understood. We have identified HMGA1a as one factor that might direct ORC to AT-rich heterochromatic regions. The systematic analysis of the interaction between ORC and HMGA1a revealed that Orc6 interacts with the acidic C-terminus of HMGA1a and also with its AT-hooks. Both domains support autonomous replication if targeted to DNA templates. As such, Orc6 functions at different stages of the replication initiation process. Orc6 can interact with ORC chaperone proteins such as HMGA1a to facilitate chromatin binding of ORC and is also an essential factor for pre-RC formation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0675-9) contains supplementary material, which is available to authorized users.
Keywords: Origin recognition complex, Orc6, Chromatin, HMGA1a, Replication initiation
Introduction
The origin recognition complex (ORC) plays a central role in the initiation of DNA replication in all eukaryotic systems. However, the different eukaryotes have apparently developed multiple mechanisms by which ORC interacts with DNA and promotes replication initiation. In S. cerevisiae, replication origins are genetically defined. ScORC binds sequence-specifically to a consensus motif within the autonomous replicating sequences (ARS) [1]. In contrast, replication origins in higher eukaryotes are poorly defined. Besides epigenetic features, a permissive local chromatin structure, DNA topology, and DNA sequence motifs contribute to origin formation. ORC interacting proteins like Ku80, AlF-C, c-Myc, Trf2, EBNA1, and the architectural chromatin protein HMGA1a were shown to function as ORC-chaperones targeting ORC to chromatin regions and contributing to origin formation [2, 15, 16, 27, 37, 40, 44]. A coherent picture of a replication origin in metazoans has not yet emerged. HMGA1a is the first example that chromatin constituents can contribute to ORC DNA-binding. The HMGA-family is characterized by AT-hook motifs, which are required for chromatin association [20]. Co-immunoprecipitation and bimolecular fluorescence complementation (BiFC) experiments indicated that HMGA1a interacts with the hexameric ORC including Orc6 via direct protein–protein interactions [44]. Moreover, HMGA1a can recruit ORC to AT-rich heterochromatin and promote DNA replication in a site-specific manner if it is specifically tethered to DNA. Altering the molecular ratio between ORC and HMGA1a after overexpression of HMGA1a causes relocalization of ORC to AT-rich chromatin regions [44]. Conversely, displacement of HMGA1a from heterochromatin results in relocation of ORC (R. Hock, personal communication; [44]). HMGA1a is a very mobile chromatin component. Fluorescence recovery after photobleaching (FRAP) studies displayed recovery times of less than 1 s for euchromatin and 3.3 s for heterochromatin in interphase cells [20]. Similarly, ORC is a highly dynamic chromatin binding complex with FRAP kinetics comparable to those of the family of HMGA proteins [26]. These very high motilities hamper ChIP-experiments to map chromosomal origins characterized by the co-localization of HMGA1a and ORC [36].
Orc6 is the smallest subunit of ORC and the evolutionarily least conserved between budding yeast and metazoans [14, 18]. S. cerevisiae Orc6 (ScOrc6) is not essential for DNA binding of ORC [14, 18, 24, 25], but the interaction between ScOrc6 and ScCdt1 is required for Mcm2–7 loading and for the maintenance of pre-replicative complexes (pre-RC) [7, 39]. In contrast to ScOrc6, Drosophila Orc6 plays an important role in DNA recognition [3]. The unique position of Orc6 is also becoming evident in siRNA depletion experiments. A very recent study demonstrates that protein levels of Orc2–Orc5 are reduced after RNAi depletion of individual Orc2–5 subunits, whereas Orc6 levels are not affected [31]. Furthermore, it is still under debate whether a stoichiometric human Orc1–Orc6 complex exists in vivo [41] and how Orc6 contributes to the function of human ORC, because the physical association of Orc6 with the Orc1–5 complex is weak and biochemically less reproducible in vitro [3, 13, 32, 41, 46]. Orc6 RNAi-mediated depletion in human cells indicated a vital role of Orc6 in cytokinesis and caused reduced BrdU incorporation into newly replicated DNA indicating a smaller number of active replication origins [30]. This finding provides evidence for a replicative function for human Orc6, but its exact contribution to replication initiation has yet to be elucidated.
Our previous findings suggested that HMGA1a might play a role in defining origins rather than replication initiation. We build this model on our observation that HMGA1a functions as ORC chaperone and mediates replication competence [44]. Our experiments aim at deciphering the domains of HMGA1a that contribute to DNA replication. By mutational analysis of HMGA1a, we show that ORC interacts with two different domains of HMGA1a. The bipartite binding surface consists of the acidic C-terminus, which interacts with the hexameric complex, and the AT-hook domains of HMGA1a, which might preferentially interact with Orc6. Both domains support the extrachromosomal replication of plasmids independently from each other. Replication competence strongly correlates with the mutant forms’ potential to cooperate with human ORC, arguing for an active role of human Orc6 in origin activation. Our experiments suggest that Orc6, which is abundantly expressed in relation to Orc2, also contributes to the replication initiation process. Gel shift experiments indicate that human Orc1–5 and Orc6 can bind independently to DNA, but Orc6 is an integral part of ORC. Employing a binding assay with immobilized plasmid DNA and replication competent nuclear extracts as previously reported [38, 47, 48], we demonstrate that Orc6 contributes to pre-RC formation. Biochemical and imaging data indicate that Orc6 and Cdc6 interact. We conclude from these data that human Orc6 is essential for the recruitment of Cdc6 to origins and therefore crucial for pre-RC formation to provide replication competence.
Materials and methods
DNA transfection, HIRT extract and Southern blotting
Plasmid DNAs were transfected with Polyfect into HEK293 cells stably expressing EBNA1 and scTetR:HMGA1a derivatives, which were selected with 80 μg/ml hygromycin, 250 μg/ml puromycin, or 200 μg/ml neomycin. For Southern blotting, 6 μg of Hirt extracted and purified DNA was digested with DpnI and HindIII, separated on agarose gels, transferred onto nylon membranes (Amersham) and probed with a radiolabeled prokaryotic probe. To control the integrity of the extrachromosomal plasmids, 500 ng of Hirt-extracted DNA was transfected into electrocompetent DH10B (Invitrogen) and a representative number of plasmids recovered from the transformants were analyzed by restriction enzyme analysis.
Chromatin-immunoprecipitation and real-time PCR analysis
ChIP experiments were performed as described [37]. For each sample, 1 × 107 nuclei were isolated, cross-linked with formaldehyde for 10 min at 37°C, washed, and lysed by adding N-Laurylsarcosine (2% final concentration). Chromatin was washed with PBS, centrifuged, resuspended in 2 ml TE and sonicated. For immunoprecipitation, 500 μg nucleoprotein and 10 μg polyclonal (Orc2) or monoclonal (Orc6) antibodies were incubated in NET (50 mM Tris, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP40). Co-precipitated DNA was isolated, purified and quantitative real-time PCR was performed as described [37]. Primer pairs are listed in Supplementary Table 1.
Immunofluorescence and live cell microscopy
Bimolecular fluorescence complementation was analyzed with a Leica TCS-SP2/AOBS instrument 22–26 h post transfection of 1 μg of the indicated expression plasmids as described [44].
Co-immunoprecipitation assays and western blot
Chromatin-bound proteins were isolated by 450 mM salt extraction from 2.5 × 107 cells and, after dialysis to 125 mM salt, incubated with 5–10 μg antibodies coupled to protein A or G Sepharose. Co-precipitated proteins were eluted, separated on SDS-PAGE, blotted, and detected with the indicated antibodies.
Plasmid binding assay
pEPI-UPR was biotinylated by UV-cross-linking at a molar ratio of 1:18 according to the manufacturers instructions [Photoprobe (S-S Biotin); Vector Laboratories]. 10 μg of biotinylated plasmid were bound to 500 μg of Streptavidin beads using the Dynabeads kilobaseBINDER kit (Dynal). For each binding reaction, 32 μg of HeLa high salt nuclear extract [4] were incubated with an equivalent of 90 ng bead-bound plasmid DNA as described [47]. Where indicated, 2 mM ATP and an ATP regenerating system (20 mM creatine phosphate, 6.3 μg/ml creatine phosphokinase, 2 mM ATP) were added to the binding reaction. Analysis of plasmid-bound proteins was performed by immunoblotting using the indicated antibodies.
Recombinant protein expression
Recombinant human Orc1–5 expression was performed as described [32]. Semi-confluent 15-cm dishes of High Five cells were co-infected with human Orc1–Orc5 baculoviruses coding for the different human ORC subunits. The Orc1 subunit was C-terminal His-tagged to facilitate affinity purification of the complex using Ni–NTA–Agarose beads (Qiagen). Wild-type and mutant N-terminal His6-tagged Orc6 proteins were expressed and purified from BL21 [Rosetta (DE3) pLysS] and purified via Ni–NTA–Agarose beads (Qiagen). GST-tagged Cdc6 was expressed using a baculovirus system and affinity purified using Glutathione–Sepharose 4 FAST flow beads according to the manufacturer’s instructions (GE Healthcare) [21].
Results
Orc6 is part of the ORC holocomplex and mediates replication competence
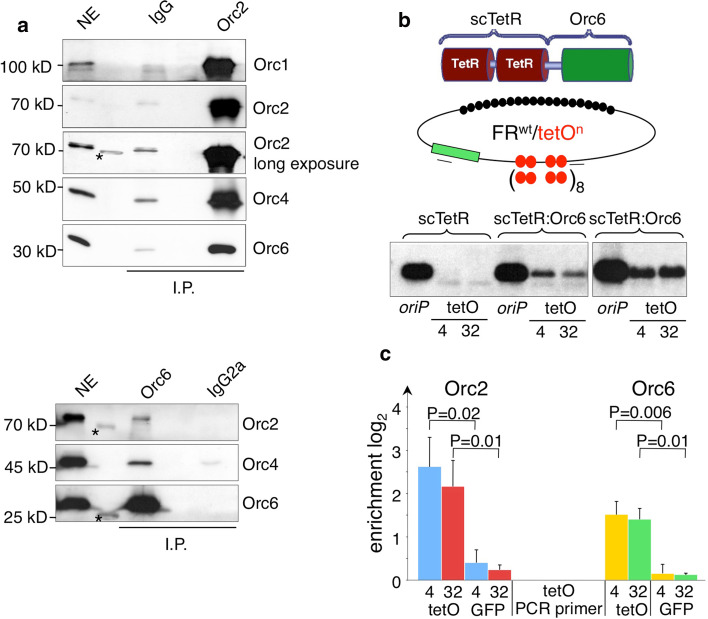
The molecular basis of the replicative function of human Orc6 is still unknown and its contribution to DNA replication differs between species [18]. This led us to study different aspects of the replicative functions of human Orc6 in detail. First, we aimed at confirming the integrity of human ORC as a six-subunit complex. Bimolecular fluorescence complementation (BiFC) experiments visualized the Orc1–Orc6 complex in vivo [44]. The holocomplex was difficult to reconstitute in vitro with recombinant proteins because Orc6 dynamically associates with the ORC core complex resulting in a biochemically weak interaction [32, 41]. We started with co-immunoprecipitation experiments from HeLaS3 nuclear extracts with a polyclonal Orc2 and a monoclonal Orc6-specific antibody to further confirm the existence of the heterohexameric complex Orc1–6 [37]. Rabbit IgG and a rat monoclonal antibody of the same isotype the Orc6 antibody (IgG2a) were used as controls to identify non-specific interactions. Co-immunoprecipitation experiments were performed from a 450-mM potassium acetate nuclear extract that was dialyzed against 100 mM potassium acetate constantly measuring the conductivity. The extract preparation procedure was crucial for the detection of the heterohexameric complex, because prolonged dialysis resulted in the precipitation of ORC-subunits and in the disintegration of the holocomplex. Also, a high protein concentration of the nuclear extracts is crucial to maintain the Orc1–Orc6 interaction. Our co-immunoprecipitation with a polyclonal Orc2 antibody revealed the efficient co-precipitation of Orc1, Orc4 and Orc6 with Orc2 (Fig. 1a, upper panel). Conversely, the Orc6 antibody co-precipitated Orc2 and Orc4 with lower efficiency (Fig. 1a, lower panel). Orc6 being abundantly expressed relative to other ORC subunits could explain this result and therefore only a fraction of Orc6 is constantly associated with Orc1–Orc5 [9]. Alternatively, the binding of the monoclonal antibody to Orc6 might interfere with its association with other ORC subunits. Quantitative western blot experiments from whole cell extracts indicate that Orc6 is in different cell lines abundantly expressed in comparison to Orc2 (Supplementary Fig. 1A).
Fig. 1.
Orc6 is part of the ORC holocomplex and mediates replication competence. a Co-immunoprecipitation experiments with HeLa nuclear extracts were performed with the indicated antibodies. Immunoblot analysis confirmed the co-precipitation of ORC subunits. For comparison 0.5% of the input material (NE) is shown. The asterisks mark an unspecific interaction of the antibody with the protein marker. b Schematics of the single-chain tetracycline repressor Orc6 fusion protein (scTetR:Orc6) and the reporter plasmids. The four or 32 tet-operator (tetO) sites are illustrated as red circles and the EBNA1 binding sites within the nuclear retention element FR as black circles. An oriP control plasmid and the reporter plasmids were transfected into HEK293 EBNA1+/scTetR:Orc6+ cells, selected and assessed for their replication competence by Southern blotting. Two independent experiments are shown. c ChIP analysis of HEK293 EBNA1+/scTetR:Orc6+ carrying either FRwttetO4 or FRwttetO32 plasmid DNAs indicated the site-specific recruitment of the Orc2 subunit via scTetR:Orc6. The locations of the primer pairs are indicated by black bars in (b). The GFP gene (green rectangle) served as a reference site and is located 3.5 kbp from the tetO cluster. The histogram indicates the relative enrichment values and standard deviations of three independent experiments on a logarithmic scale expressed as difference between PCR values (Cp) obtained with the Orc6 and Orc2 specific antibodies versus controls obtained with the corresponding IgG (Orc6) and pre-immune serum (Orc2). Student’s t test was used to analyze the statistical significance
To assess whether Orc6 can mediate replication competence, we made use of a plasmid reporter assay. To target Orc6 to plasmid DNA, we constructed a fusion of Orc6 and the single-chain tetracycline repressor to generate a scTetR:Orc6 fusion protein (Fig. 1b). The reporter system is based on the extrachromosomal maintenance of plasmids containing a modified plasmid origin of Epstein–Barr virus, oriP. The family of repeats of oriP (FR; Fig. 1b) is bound by the EBV-protein EBNA1, which tethers oriP to chromatin and confers long-term nuclear retention and segregation. The replicator element of oriP, the EBNA1-bound dyad symmetry (DS) element, was replaced by four (FRwttetO4; Fig. 1b) or 32 tetO-sites (FRwttetO32), respectively. This strategy separates the plasmid maintenance and replication functions of oriP and allowed us to determine the long-term replication competence of the protein of interest [44].
scTetR:Orc6 was stably integrated into HEK293/EBNA1+ cells and single cell clones were identified that co-express EBNA1 and scTetR:Orc6 (Supplemntary Fig. 1B). HEK293/EBNA1+/scTetR:Orc6+ cells were transfected with three different reporter plasmids: oriP as a positive control as well as FRwttetO4 and FRwttetO32 as two reporter plasmids to analyze their replication competence. Two weeks after transfection and selection, low molecular weight DNA was isolated and digested with DpnI to eliminate non-replicated plasmid DNA. After restriction with HindIII, replicated DNA was detected by Southern blot hybridization with a plasmid-specific prokaryotic probe. The results indicated that scTetR:Orc6 efficiently supports extrachromosomal replication of FRwttetOn plasmids (Fig. 1b). The negative control scTetR alone did not allow replication of plasmids FRwttetO4 and FRwttetO32 (Fig. 1b).
Next, we asked whether scTetR:Orc6 mediates replication by associating additional ORC-subunits to plasmid DNA. As expected, ChIP with an Orc6-specific antibody showed a significant enrichment at the tetO-plasmid locus as compared to a reference site located in the GFP gene 3.5 kbp apart (P ≤ 0.01; Fig. 1c). A similar specific binding was observed with an Orc2-specific antibody representative for the ORC holocomplex (P = 0.01; Fig. 1c). This finding indicates the site-specific recruitment of the entire ORC complex via Orc6 to DNA, which is in agreement with the co-immunoprecipitation experiments (Fig. 1a). Reporter plasmids bearing 4 or 32 tetO-sites did not differ in this respect. Similar to a previous report that analyzed targeted Orc2- and Cdc6-proteins, our results clearly indicated that site-specific targeting of Orc6 to plasmid DNA generated a functional origin of replication and mediated replication competence, probably by co-associating the other ORC-subunits [42].
Human Orc6 and Orc1–Orc5 bind independently to DNA
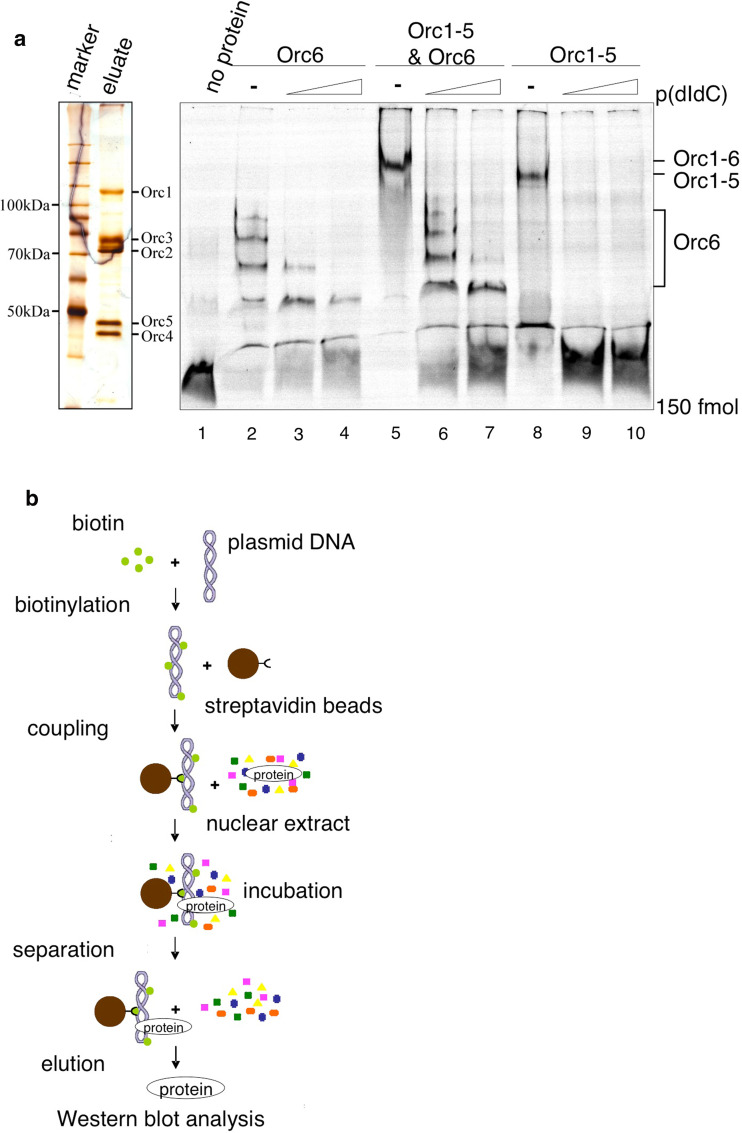
The molecular contribution of Orc6 to DNA binding of ORC and to replication initiation is unclear and differs between species. S. cerevisiae, Orc6 is dispensable for origin recognition and for progression through mitosis and cytokinesis, but it is essential for viability, formation and maintenance of pre-RCs and cell cycle progression [24, 25, 39]. In contrast, DmOrc6 is required for DNA binding of DmORC, and HsOrc6 depletion results in severe mitotic defects [3, 30]. To determine the role of human Orc6 in DNA replication in more detail, we first studied the DNA binding characteristics of Orc1–Orc5 and Orc6 alone as well as in combination. To this end, we used recombinant proteins in in vitro DNA binding assays. High Five™ cells (BTI-TN-5B1-4) were co-infected with baculoviruses expressing Orc1 C-terminally tagged with 3xHA-His; Orc2, Orc3, Orc4, and Orc5 [32]. Nuclear extracts from these cells were subjected to affinity-purification using a Nickel-resin (Supplementary Fig. 2A). After extensive washes, the complex was eluted and separated on a denaturing SDS-PAGE and visualized using silver staining (Fig. 2a, left). Recombinant His6-Orc6 was expressed in E. coli and affinity purified (Supplemntary Fig. 2B). We determined the DNA binding abilities of human Orc6 and Orc1–Orc5 in gel shift experiments in 4.5% native polyacrylamide gels (EMSA). For our EMSA experiments, we chose an AT-rich sequence, because HsORC preferentially binds to synthetic AT-rich DNA [45]. Furthermore, DmOrc6, which has DNA binding activity, preferentially binds to poly(dA) sequences [3]. Recombinant wild-type Orc6 formed distinct complexes, which competed with increasing amounts of poly(dI-dC) (Fig. 2a right, lanes 2–4). HsOrc1–5 formed one distinct complex. Orc1–Orc5 was competed with excess of poly(dI-dC) (lanes 8–10). Previous experiments suggested that ORC contacts the minor grooves of AT-rich DNA and poly (dI-dC), which are structurally similar between these two sequences [45]. Co-incubation of Orc6 and Orc1–Orc5 resulted in one complex that migrated slightly slower than the Orc1–Orc5-DNA complex. The small size of the probe does not allow the binding of multiple separately associated origin recognition complexes as suggested by recent structural information on metazoan ORC [11, 12]. This implies that the observed slower migrating species reflects an Orc1–Orc6 holocomplex that bound to the probe. In accordance with our quantification experiments, we used a fivefold molar excess of Orc6 relative to Orc1–Orc5. Orc1–Orc6 was competed with low amounts of poly(dI-dC), whereas the competition of excess Orc6 required larger concentrations of competitor DNA (lanes 6 and 7). This observation might indicate that poly(dI-dC) is a less efficient competitor for Orc6, which has higher affinity to AT-rich DNA by analogy with DmOrc6 [3]. In summary, the EMSA experiments demonstrate that Orc1–Orc5 and Orc6 are able to bind DNA independently from each other, but also form a DNA-bound heterohexameric complex if co-incubated with DNA.
Fig. 2.
Human Orc6 is required for pre-RC formation but not for ORC DNA-binding. a Left Silver-stained SDS-PAGE of affinity-purified human Orc1–5 after baculovirus mediated overexpression. Right HsOrc1–HsOrc5 and bacterially expressed HsOrc6 were employed to monitor DNA binding activities in EMSA experiments; 150 fmol of a Cy5-labeled double-stranded AT-rich oligonucleotide [41] was incubated with 5 pmol Orc6 (lanes 2–4), 1 pmol Orc1–Orc5 (lanes 8–10), and Orc1–Orc6 (lanes 5–7). Increasing amounts of poly(dI-dC) were used as competitor (lanes 3, 6, and 9 50 ng, lanes 4, 7, and 10 250 ng). Control lane 1 no protein. b Outline of the cell-free plasmid-binding assay to study the assembly of pre-RCs in vitro. Biotinylated supercoiled pEPI-UPR was immobilized to paramagnetic beads and incubated with nuclear extract [4]. Proteins bound to the beads were detected by immunoblotting. c ATP is required for Mcm2–Mcm7 protein loading and induces phosphorylation of DNA-bound Cdc6. Lane 1 beads only control. The binding reaction was performed with DNA without adding further ATP (lane 2) or in the presence of an ATP-regenerating system (lane 3). The putative phosphorylation of Cdc6 was addressed in an independent experiment (lanes 4–6) by λ-phosphatase treatment (PPase, lane 6) of the bead-bound material before the SDS-PAGE. A λ-PPase-untreated reaction is shown for comparison (lane 5). Lane 4 beads only control. d Depletion of ORC subunits reduces pre-RC formation. Orc2- but not Orc6-specific antibodies co-deplete Orc1–Orc5 proteins (lanes 1–3). Immobilized plasmid DNA was incubated with nuclear extract either depleted with control (α-IgG, lanes 6, 10), Orc2- (lane 7) or Orc6-specific antibodies (lane 11). For complementation, 120 ng of recombinant Orc6 protein was added to the Orc6-depleted nuclear extract (lane 12). Lanes 5–7 DNA binding of Orc1, 2, 4, 6 and Cdc6 was quantified in relation to the amount of protein bound to DNA (lane 5). Lanes 9–12 Plasmid binding of Orc1, -2, -4 and -6 proteins, as well as Mcm7 and Cdc6 was monitored and quantified in relation to lane 9. All experiments were performed in the presence of 1 mM ATP and an ATP regenerating system
Orc6 is essential for pre-RC formation
The data presented above demonstrated that human Orc6 is part of the heterohexameric ORC, and that its specific targeting to DNA can recruit ORC to support efficient replication initiation. Even though we observed DNA binding ability for the smallest ORC subunit, Orc6 does not seem to be essential for DNA binding of the human origin recognition complex. We therefore wanted to investigate whether Orc6 contributes to pre-RC formation. To address this question, we made use of immobilized plasmid DNA that is incubated with in vitro replication competent HeLaS3 nuclear extracts (Supplementary Fig. 3A; [4]). We took advantage of a pEPI-1-based plasmid, pEPI-UPR, that replicates as an extrachromosomal plasmid faithfully in human cells in an ORC-dependent manner similar to experiments published earlier [35]. Plasmid DNA-bound proteins were analyzed by Western blotting [38, 47, 48]. This cell-free experimental system allowed us to investigate individual steps of pre-RC formation in vitro (Fig. 2b).
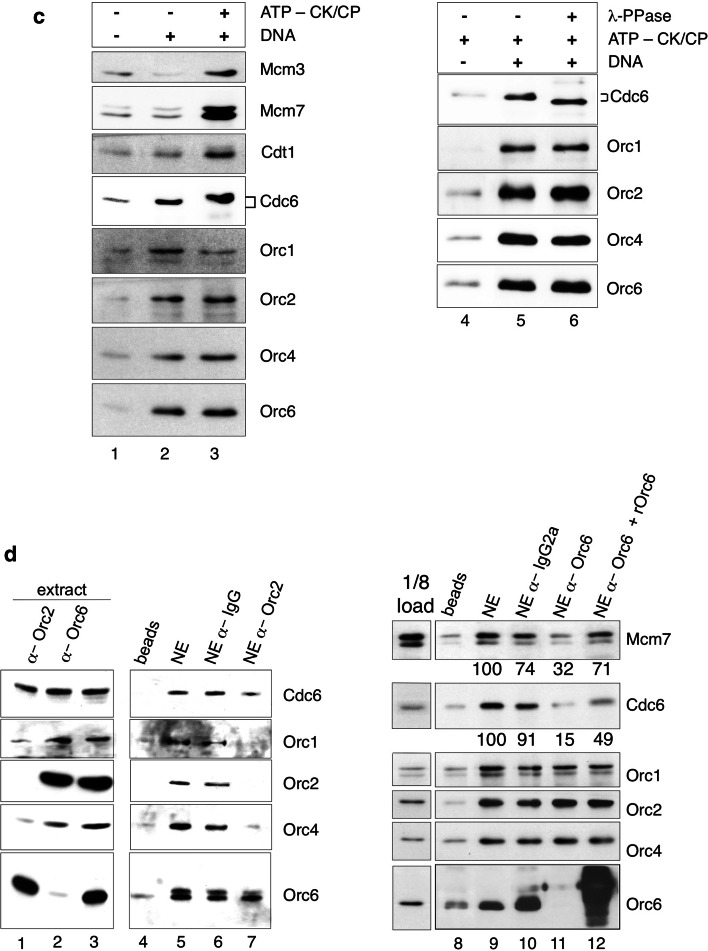
Replication competent nuclear extracts of HeLaS3-cells were generated in the presence of 1 mM ATP according to a protocol that allowed plasmid replication in vitro in a cell-free system [4]. We had previously used the same extracts for the co-immunoprecipitation experiments with Orc2- and Orc6-specific antibodies (Fig. 1a). Binding experiments without supplementing additional ATP revealed the loading of only ORC-components to immobilized circular plasmid DNA (Fig. 2c, lane 2). Cdc6 showed some unspecific bead-binding that varied between experiments (lane 1). With an ATP-regenerating system, the entire pre-RC assembled to the immobilized DNA as indicated by the specific association of Cdt1, Mcm3 and Mcm7 proteins (lane 3). Quantification of these proteins in relation to lane 2 indicates an at least threefold increase of Mcm2–Mcm7 and Cdt1 protein binding. We also observed that Cdc6 migrated slightly slower in the presence of creatine phosphokinase/creatine phosphate, probably due to a DNA-dependent phosphorylation (lanes 3 and 5). Treatment of the plasmid-bound proteins with λ-phosphatase resulted in a faster migrating form of Cdc6 (lane 6), confirming that this modification is indeed caused by phosphorylation of Cdc6. Our findings indicated that pre-RCs are successfully assembled on plasmid DNA. Pre-RC assembly requires ATP for efficient Cdc6 recruitment, whose DNA-bound form is phosphorylated during the assembly. These findings are in line with reports of the S. cerevisiae in vitro assembly system [38], but phosphorylation of Cdc6 does not seem to have a direct functional relevance for pre-RC assembly [33].
To investigate the contribution of Orc6 to pre-RC assembly, we depleted Orc6 with a specific monoclonal antibody and, for comparison, Orc2 [34]. Orc2-depletion of the nuclear extract co-depleted Orc1 and 4, whereas depletion of Orc6 does not co-deplete Orc1–Orc5 confirming our results that Orc1–Orc5 is able to bind DNA independently from Orc6 (Figs. 1 and 2d, lanes 1–3). Consequently, after Orc2 depletion, Orc1–Orc5 plasmid-binding was drastically reduced in comparison to non-depleted nuclear extracts and to extracts incubated with unspecific control antibodies (lanes 5–7). Plasmid-binding of Cdc6 was slightly reduced in Orc2-depleted extracts (lane 7). Complementation experiments with recombinant Orc1–Orc5 proteins have been unsuccessful in in vitro replication studies and have not been tested in this context [4]. Binding of Orc1, -2 and -4 proteins to immobilized plasmid DNA was unaffected by Orc6 depletion as their abundance was comparable to mock depleted extracts (Fig. 2d, lanes 9 and 10), which is consistent with our EMSA experiments (Fig. 2a). Interestingly, Orc6 depletion impaired Cdc6 and Mcm7 association with the immobilized plasmid (Fig. 2d, lane 11). Complementation of the Orc6-depleted extract with bacterially expressed Orc6 partially restored Cdc6 and Mcm7 binding (Fig. 2d, lane 12) as judged by the quantification of the western blot signals. These data suggest that human Orc6 is important for pre-RC assembly.
Orc6 and Cdc6 interact
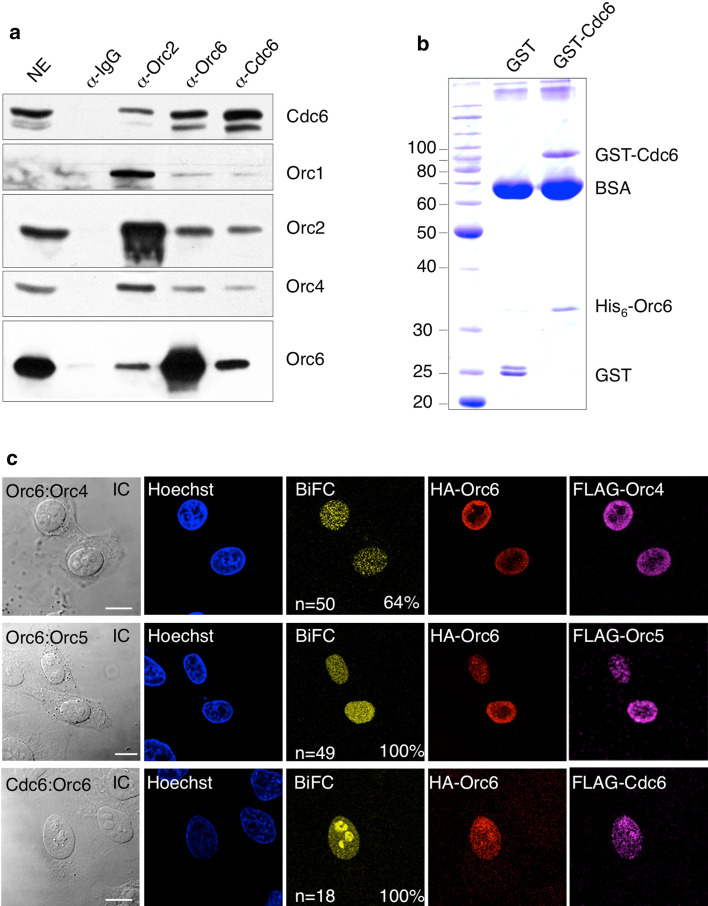
Our EMSA and plasmid binding data indicated that human Orc6 and Orc1–Orc5 bind DNA independently, which is different to DmORC [3]. The plasmid binding experiments further demonstrated that human Orc6 is essential for pre-RC formation. To elaborate whether human Orc6 is directly involved in pre-RC formation, we performed co-immunoprecipitation experiments from nuclear extracts with an Orc6-specific antibody. Cdc6 co-precipitated with the Orc6-specific antibody, but not with a control IgG (Fig. 3a). The Orc2-specific antibody efficiently co-precipitated Orc1, Orc4 and Cdc6. However, Cdc6 co-precipitation was more efficient using an Orc6-specific antibody. This finding is confirmed in Cdc6 co-immunoprecipitation experiments, which indicate that Orc6 co-precipitates more efficiently than the other ORC-subunits.
Fig. 3.
Cdc6 and Orc6 interact to support pre-RC formation. a Co-immunoprecipitation experiments with HeLa nuclear extracts were performed with the indicated antibodies. Immunoblot analysis confirmed the co-precipitation of Cdc6 and Orc subunits. 0.5% of the input material is shown for comparison (NE). b Purified GST-tagged Cdc6 or GST were co-precipitated with affinity-purified His-tagged Orc6. Glutathione–Sepharose beads were blocked with BSA to reduce unspecific binding of Orc6. The complexes were analyzed by SDS-PAGE followed by Coomassie staining. An immunoblot with Cdc6 and Orc6 specific antibodies confirmed the identity of the recombinant proteins (Supplementary Fig. 3). c Cdc6 associates with Orc6 in vivo. BiFC analysis of Orc6 and Orc4 (top row), Orc6 and Orc5 (middle row) and Orc6 and Cdc6 confirm that Orc6 is part of the heterohexameric ORC and of the pre-RC in vivo (lower row). Immunofluorescence with HA- and FLAG-specific antibodies monitored the transfection of the individual constructs as indicated. BIFC efficiencies were calculated from double transfected HepG2 cells only. Differential interference contrast images of analyzed cells are shown (left). (Scale bars 10 μm)
This intrinsic DNA-binding activity of Cdc6 might mask smaller effects in the plasmid-binding assay, which might explain, why DNA-binding of Cdc6 is less efficient in Orc6-depleted extracts than in Orc2-depleted.
The co-immunoprecipitation of Cdc6 suggested that Orc6 and Cdc6 form a stable complex in nuclear cell extracts. However, this observation does not reveal whether this interaction is direct or indirect or whether Orc6 is directly involved in Cdc6 association. To test this hypothesis, we performed pull-down experiments with purified proteins. Recombinant GST-Cdc6 but not GST co-precipitated His6-Orc6, which points to a direct interaction that is independent of DNA (Fig. 3b and Supplementary Fig. 3B). To verify the existence of a Cdc6:Orc6 complex in cells, we again employed BiFC that visualizes protein–protein interactions in vivo [22]. As positive controls, we monitored the localization of ORC via BiFC between Orc6/Orc4 and Orc6/Orc5 (Fig. 3c). The diffuse nuclear distribution is characteristic for interphase cells as previously shown [44]. To corroborate the interaction between Orc6 and Cdc6, we fused Cdc6 to the C-terminal part of YFP and Orc6 to the N-terminal fragment. Fluorescence complementation was also observed for Cdc6 and Orc6 in all cells that co-expressed both proteins (Fig. 3c). The HA- and FLAG-tags allow the detection of the individual fusion-proteins. Both, the co-immunoprecipitation and the BiFC experiments clearly argue for the existence of a Cdc6:Orc6 complex in the nucleus of human cells.
Taken together, the experiments presented so far indicate that Orc6 might be involved in several steps of replication initiation. First, Orc6, as an integral part of the ORC holocomplex, mediates replication competence. Second, Orc6 interacts with Cdc6 and is essential for pre-RC formation.
Orc6 interacts with the AT-hooks of HMGA1a
In a previous study, we have shown that different subunits of ORC including Orc6 interact with the ORC chaperone HMGA1a suggesting that this interaction might specify replication origins in HMGA1a-rich chromatin regions [44]. The family of HMGA proteins consists of an acidic C-terminal domain and an unstructured domain containing three AT-hook motifs, which bind dynamically to minor grooves of AT-rich DNA [6, 20]. While this manuscript was under revision, Eilebrecht et al. [19] reported that the non-coding 7SK RNA can compete with DNA for the binding to AT-hooks and thereby influences the regulatory functions of this chromatin factor. Here we addressed the question whether a specific domain of HMGA1a physically interacts with ORC and also mediates replication competence. In particular, we wanted to know whether the Orc6:HMGA1a interaction is relevant for HMGA1a’s ability to support replication.
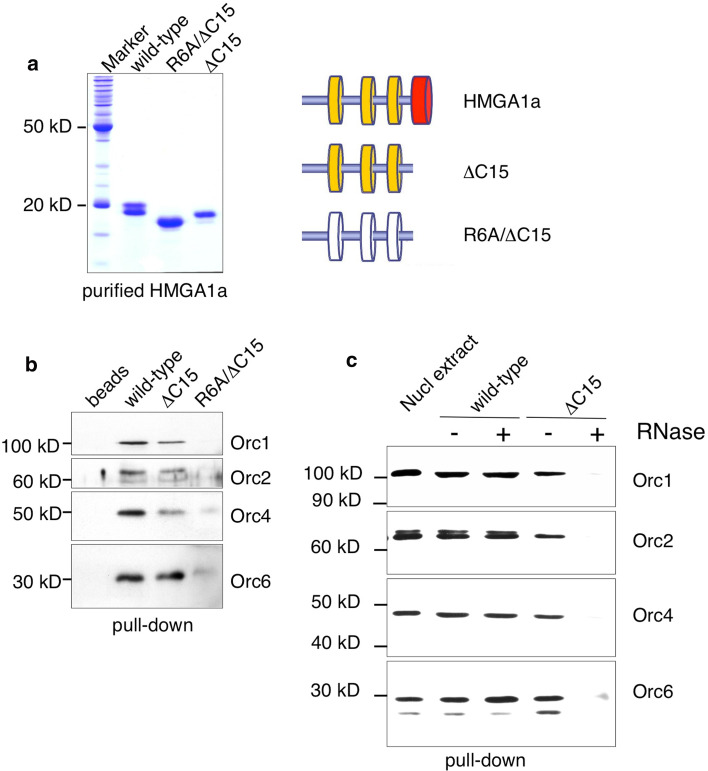
As HMGA1a interacts with ORC in pull-down and imaging experiments, it most likely mediates replication competence through this interaction setting the stage for pre-RC assembly [44]. To determine whether the AT-hooks and the acidic C-terminal patch of HMGA1a interact with the same subunits of ORC, we performed pull-down experiments with different affinity-purified recombinant HMGA1a mutant proteins (Fig. 4a) and HeLaS3-derived nuclear extracts (Figs 1, 3). Full-length HMGA1a co-precipitated the entire ORC as indicated by the detection of Orc1, -2 and -4 including Orc6 (Fig. 4b). A C-terminal deletion mutant of HMGA1a (ΔC15) interacted with Orc6 with similar efficiency as wild-type HMGA1a, whereas Orc1, Orc2 and Orc4 co-precipitated to a slightly lower efficiency indicating that the C-terminus stabilizes the interaction between Orc1–Orc5 (Fig. 4b). To test whether the AT-hook function mediates ORC binding, we constructed a mutant called R6A/ΔC15. In this mutant, all six arginines in the AT-hook RGRP motifs were replaced by alanines. The acidic C-terminus has been truncated to study the contribution of the AT-hooks only. We could not detect any interaction of this mutant HMGA1a protein with any of the ORC subunits tested (Fig. 4b).
Fig. 4.
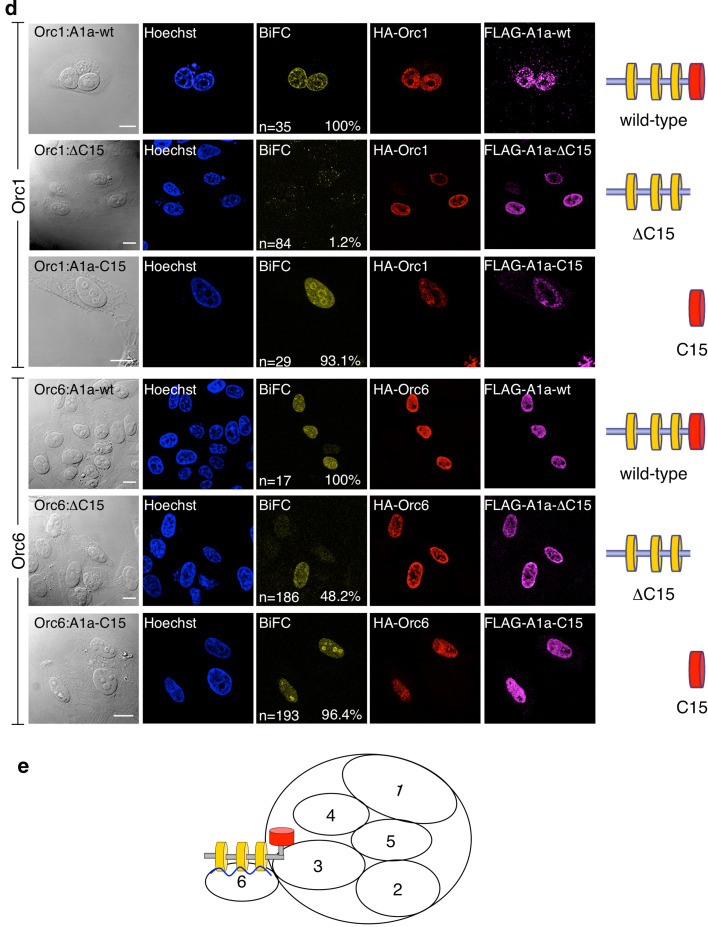
Replicative domains of HMGA1a interact with ORC. a Recombinant STREP-tagged full-length HMGA1a (wild-type), the mutant lacking the acidic C-terminal 15 amino acids (ΔC15) and the R6A/ΔC15 mutant were expressed in E. coli and affinity-purified. The Coomassie gel shows the purified proteins. The schemes of the mutants are depicted on the right. b Pull-down experiments were performed with HeLa nuclear extracts and 7 nmol of each recombinant HMGA1a protein. Co-precipitated ORC-proteins were detected by immunoblot analysis. Orc6 also interacted with the ΔC15 mutant but this interaction was abrogated with the AT-hook mutant R6A/ΔC15 (right). Left lane beads only control. c Affinity-purified HMGA1a and HMGA1aΔC15 incubated with HeLaS3 nuclear extracts and then treated with 75U RNase A and buffer control. Co-precipitated ORC components were monitored by immunoblotting with anti-ORC1, Orc2, Orc4 and Orc6 antibodies. d BiFC analysis of Orc1 or Orc6 with HMGA1a mutant proteins. Orc1 and Orc6 were fused to the HA-tagged N-terminal fragment of YFP. Full-length HMGA1a (top row), HMGA1a containing amino acid (aa) residues 1–92 (ΔC15, middle row), or only the C-terminal acidic domain aa93–107 (C15, lower row) of HMGA1a were linked to the Flag-tagged C-terminal fragment of YFP. Single HMGA1a variants and an ORC BiFC construct were co-transfected into HepG2 cells and fluorescence images were acquired 24 h post-transfection. Expression of ORC-fusion proteins was controlled using monoclonal rat antibodies directed against HA-tag; expression of HMGA1a-fusion proteins was controlled using mouse antibodies directed against the Flag-tag. The frequencies of BIFC signals in cells that co-expressed both YFP-fusion proteins are indicated (%). Scale bars 10 μm. e Model of the HMGA1a interaction. The acidic C-terminal domain of HMGA1a and the AT-hook motifs interact with ORC. The interaction between ORC and the AT-hooks is mediated by RNA (blue line)
The co-precipitation experiments indicate that two domains of HMGA1a might interact independently with ORC. Previously, we have shown that the ORC interaction of HMGA1a is partially disrupted by RNase and that the complex between the AT-hook motifs and ORC might be RNA-dependent [29]. To assess whether the AT-hook mediated interaction is indeed RNA-dependent, recombinant full-length HMGA1a (Fig. 4c, lanes 2 and 3) and a mutant HMGA1a lacking the acidic C-terminus (HMGA1aΔC15; lanes 4 and 5), respectively, were incubated with HeLa nuclear extract as above and then treated with a control buffer (lanes 2 and 4) or RNase A (lanes 3 and 5). In the absence of RNase, both HMGA1a proteins efficiently co-precipitated Orc1, -2, -4 and -6 as above. RNase A treatment of the deletion mutant abolished the ORC interaction, indicating that the AT-hook requires RNA for their interaction with ORC. The observation that the full-length protein does require RNA for the ORC-interaction suggests that the C-terminal part interacts in an RNA-independent manner. The importance of this RNA-dependent interaction in the context of the full-length protein is unclear and requires further investigation (Fig. 4c, lane 3).
Our findings suggest that the AT-hooks as well as the acidic C-terminus of HMGA1a interact with ORC. To verify these HMGA1a:ORC interactions in cells, we again used bimolecular fluorescent complementation (Fig. 4d). To this end, different HMGA1a mutants were fused to the Flag-tagged C-terminal fragment of YFP, whereas Orc1 and Orc6 were fused to YFP’s HA-tagged N-terminal part. As in previous experiments, fluorescence complementation indicated the interaction between HMGA1a and both ORC subunits (Fig. 4c; [44]). The AT-hook domains of HMGA1a localized the HMGA1a:ORC complex to AT-rich heterochromatic domains [44]. BiFC assays with only the last 15 amino acids of HMGA1a (C15) or an HMGA1a mutant lacking its acidic C-terminus (ΔC15) confirmed that this small domain is crucial for the interaction with ORC. Only 1.2% of cells transfected with ΔC15 and Orc1 displayed BiFC signals, whereas 93.1% scored positively when Orc1 was co-expressed with C15 of HMGA1a. BiFC assays with Orc6 and HMGA1a confirmed our in vitro data indicating that Orc6 interacts with different domains of HMGA1a but Orc6 interaction with the AT-hook motifs might be less efficient (48.2%) than the interaction with the C-terminus of HMGA1a (96.4%). However, the BiFC analysis might be interpreted as such that the AT-hooks of HMGA1a mainly confer Orc6 binding and that Orc1–Orc5 association with the ΔC15 mutant in pull-downs is indirect via Orc6.
The model in Fig. 4e summarizes the results of the pull-down experiments with recombinant HMGA1a-mutants and HeLaS3-nuclear extracts combined with the BiFC. Our results suggest two modes of HMGA1a binding to ORC. On the one hand, the ORC interaction is mediated via the acidic C-terminal 15 amino acids of HMGA1a that associate with Orc1 and Orc6. On the other hand, an RNA-dependent association with ORC is mediated by HMGA1a’s AT-hook motifs that contact Orc6 (Fig. 4b lower panel).
Acidic domain and AT-hooks of HMGA1a support replication of plasmid DNA
The biochemical and imaging experiments presented above indicate that HMGA1a provides a dual interface for its Interaction with ORC. Next, we addressed whether these interactions have functional relevance with respect to HMGA1a’s replicative function.
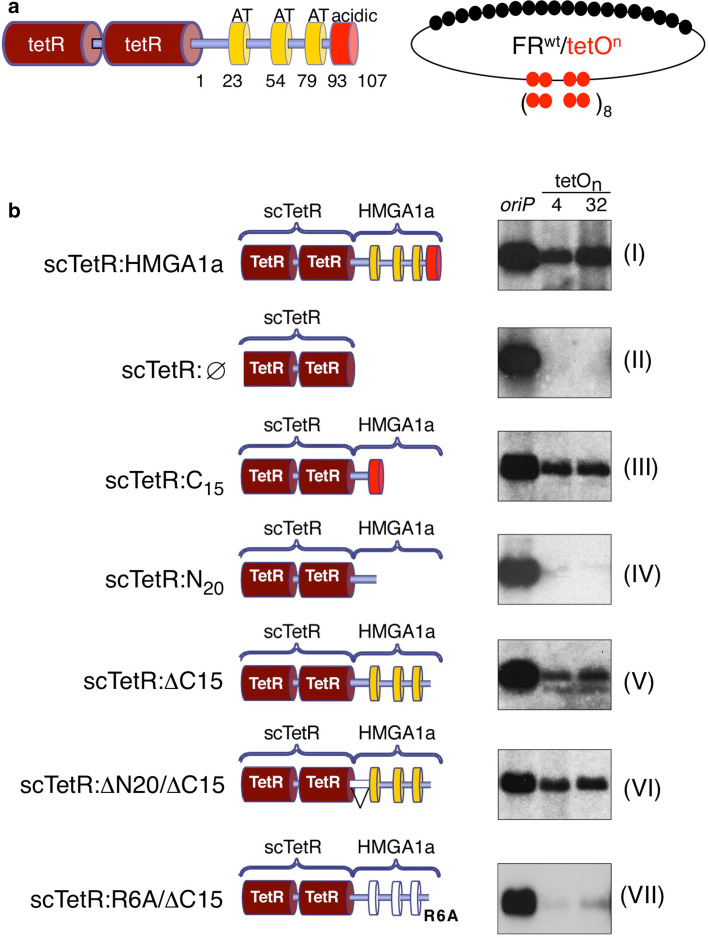
To resolve whether both the AT-hook domains and the acidic C-Terminal domain support replication, we made use of the plasmid assay as described above [44]. By fusing HMGA1a to the single-chain tetracycline repressor (scTetR:HMGA1a) we demonstrated that scTetR:HMGA1a can target ORC to cognate tetracycline-operator (tetO) sites generating functional origins of DNA replication [44]. We introduced mutations into the N-terminus and the C-terminal acidic patch of scTetR:HMGA1a as shown in Fig. 5. We constructed four deletion mutants and also tested the 6RA/ΔC15 mutant, which did not interact with ORC in pull-down assays (Fig. 5b). The different HMGA1a mutants were stably introduced into HEK293/EBNA1+ cells (Supplemntary Fig. 4). Again, replication competence of the HMGA1a mutants was determined in the plasmid propagation system that is based on the bipartite structure of EBV’s oriP (Fig. 5a). The replicator element of the test plasmids consists of four (FRwttetO4) or 32 tetO sites (FRwttetO32, Fig. 5a), respectively [44].
Fig. 5.
AT-hooks and C-terminal domain of HMGA1a contribute to replication competence of HMGA1a. a Schematics of the single-chain tet-repressor HMGA1a fusion protein (scTetR:HMGA1a, left). Indicated are the single-chain dimer (dark red), the AT-hook domains (yellow boxes) and the acidic domain (red box). A scheme of the reporter plasmids FRwttetO4 and FRwttetO32 is shown for clarity. The 6RA/ΔC15 mutant lacks the last 15 amino acid residues and carries point mutations changing the six arginines of the three AT-hook RGRP-motifs to alanines. b Both reporter constructs were transfected into HEK293 cells expressing EBNA1 and scTetR:HMGA1a (HEK293/EBNA1+/scTetR:HMGA1a+), the indicated mutants of HMGA1a (III–VII), or scTetR only (II). Southern blot analysis was used to visualize extrachromosomally maintained and replicated plasmid DNA using the prokaryotic backbone as plasmid-specific probe. The oriP plasmid was used as a positive experimental control for EBNA1, scTetR:HMGA1a (I) and scTetR:Ø (II) as positive and negative controls, respectively. One representative result out of three or more independent experiments is shown
Transient replication assays with the two FRwttetOn reporter plasmids and a wild-type oriP control plasmid were performed in HEK293/EBNA1+ cell lines expressing the different scTetR:HMGA1a-fusion proteins. The cells were drug-selected for 2 weeks and extrachromosomally replicating plasmids were visualized by Southern blot hybridization using a plasmid-specific probe. Full-length HMGA1a supported plasmid replication as reported previously [44] (Fig. 5b, I). Neither scTetR alone nor scTetR fused to the HMGA1a’s 20 amino-terminal residues were functional in this assay (Fig. 5b, II and IV). Interestingly, the acidic C-terminal domain and the AT-hook motifs of HMGA1a supported replication independently from each other (Fig. 5b, III, V and VI). This result is consistent with our findings from biochemical and BiFC analysis, showing that these domains can bind ORC and thereby may generate replication origins on the test plasmids. In agreement with this hypothesis, the 6RA/ΔC15 mutant that did not interact with ORC in pull-down assays was drastically impaired in its ability to support replication of extrachromosomal plasmids (Fig. 5b, VII). This result confirms the functional relevance of the AT-hooks for DNA replication. Assuming that the ΔC15 mutant mainly confers Orc6 binding this assay implies that recruiting Orc6 is sufficient to create a replication origin. This model is supported by the fact that a single-chain TetR:Orc6 fusion protein also mediates replication competence (Fig. 1b). In conclusion, the plasmid replication experiments demonstrate the functional relevance of the identified dual ORC binding interface of HMGA1a.
In summary, we demonstrate that the human Orc6 protein fulfils different functions as replication factor. It may contribute to ORC targeting through its interaction with chaperone proteins like HMGA1a and, although Orc6 is not required for DNA binding of ORC, this subunit is essential for pre-RC assembly.
Discussion
In this study, we focused on the role of the human Orc6 protein in DNA replication. Orc6 is the most divergent and evolutionarily least conserved subunit among all ORC proteins. As other ORC subunits, Orc6 is involved in different cellular processes and our data argue for different functional roles of the smallest subunit of human ORC in DNA replication. On the one hand, the Orc6 interaction with chaperone proteins such as HMGA1a might contribute to the targeting of ORC to specific chromatin regions. On the other hand, Orc6 also functions in the assembly of pre-RCs.
In previous studies, we have shown that RNA is required for the interaction between ORC and the conserved AT-hook motifs of EBNA1 and that RNase treatment also partially interferes with the HMGA1a–ORC interaction [29]. Here, we provide evidence for a dual ORC binding interface within HMGA1a. ORC interacts independently of RNA with the acidic C-terminus of HMGA1a and, in addition, with the AT-hook domains of HMGA1a in an RNA-dependent manner (Fig. 4e). Our bimolecular fluorescence complementation experiments indicate that the latter interaction occurs via the Orc6 subunit. Interestingly, both domains of HMGA1a that interact with ORC provide replication competence in plasmid replication experiments. This implies that Orc6 recruitment should be sufficient for specifying replication start sites. In agreement with this model, we find that artificial targeting of Orc6 to plasmids provides replication competence in replication assays, which extends a previous report by Takeda et al. [43] (Fig. 1). Takeda et al. created functional artificial origins by tethering Gal4 DNA binding domain-HsOrc2 and Gal4-HsCdc6 fusion proteins to plasmid containing Gal4-binding sites. However, these experiments failed when fusing human Orc6 to Gal4. The authors argue that this negative result might also be due to sterical problems with the fusion proteins. Gal4 needs to dimerize in order to bind to its cognate DNA sequences, which could interfere with origin recognition complex formation. Our tethering system is based on the single-chain TetR system, which recruits a single fusion protein per binding site. Furthermore, Takeda et al. [43] used transient transfections and short-term experiments to test the replicative potential of the factors. We used an oriP-background that allows separation of plasmid retention and replication functions. Nuclear retention of plasmid is supported by EBNA1 binding to the family of repeats of EBV. With respect to its cytokinetic function that is reflected in Orc6 localization to the mid-body, Orc6 is very likely less potent in conferring nuclear retention than Orc2 and therefore Orc6-tethered plasmids might get lost during mitosis [30]. In our experimental setup, the single-chain tet-repressor:Orc6 protein promoted efficient plasmid replication that seemed independent of the number of tetO-binding sites (Fig. 1b). This dose-independency indicates that a single or few Orc6 molecules are sufficient to create a functional replication origin that supports replication for at least 2 weeks.
Whether or not human Orc6 is an integral part of the human origin recognition complex was controversial for a long time as purifications of recombinant ORC expressed via the baculovirus system lack the Orc6 subunit [46]. In this respect, human Orc6 differs from the corresponding Drosophila melanogaster and S. cerevisiae ORC subunits, which are tightly bound to the ORC core complex [5, 8, 18]. Our in vitro as well as in vivo data demonstrated here support previous reports that Orc6 is an integral ORC component. We do find that the extract concentration and presumably also the salt concentration are critical determinants for co-immunoprecipitations of Orc6 with the other ORC subunits (Fig. 1). In cells, Orc6 efficiently associates with ORC1–Orc5 as judged from our bimolecular fluorescence-complementation data (Fig. 4c). In addition, as demonstrated by our ChIP experiments, artificial targeting of Orc6 also leads to the recruitment of other ORC subunits confirming that Orc6 forms a complex with Orc1–Orc5 (Fig. 1b). It is likely that the Orc6 interaction with Orc1–Orc5 is in vivo not only stabilized by the relative abundance of Orc6 but also by additional factors such as pre-RC components or proteins like HMGA1a that interact with Orc6 and the other ORC subunits. At least HMGA1a stabilizes the chromatin association of ORC, probably in a cell-cycle dependent manner from late mitosis and during G1, suggesting that pre-RC formation might be facilitated in heterochromatic regions by the presence of HMGA1a (R. Hock, personal communication).
Several lines of evidence suggest that RNAs might stabilize the heterohexameric human ORC or contribute to origin recognition. Tetrahymena ORC was shown to contain a ribosomal RNA fragment that participates in rDNA origin recognition [28]. In BiFC experiments, we also observe that human Orc6 interacts with the Pescadillo protein, a factor that is involved in rRNA processing (data not shown). A preferred nucleolar localization of a HMGA1a/Orc6 complex was also observed in Hoechst 33342 competition experiments and with an HMGA1a(R3xG)-mutant with impaired chromatin binding activities [44]. This provides another link between the ribosome biogenesis pathway and DNA replication as has already been postulated for S. cerevisiae [17, 23]. Given that the interaction between Orc6 and the HMGA1a AT-hooks is RNA dependent, it is tempting to speculate that Orc6 not only binds DNA but also RNA, and that this may either contribute to origin recognition or to the stabilization of the heterohexameric complex. Eilebrecht et al. recently proposed a molecular switch model for HMGA1a, which proposes that the HMGA1a bound by 7SK snRNA has reduced DNA binding activity and, therefore, reduced oncogenic potential [19]. The 7SK RNA-bound form functions as regulator of RNA Pol II transcription elongation. As such, the equilibrium between 7SK and HMGA1a affects the chromatin function and RNA Pol II-regulator function. We suggest that snRNAs other than 7SK might bind to HMGA1a, which specifies the interaction with other proteins. However, such RNAs still need to be identified.
Our EMSA experiments with recombinant proteins demonstrate that human Orc6 is dispensable for DNA binding of Orc1–Orc5. In this respect, HsOrc6 differs from DmOrc6, whose Orc6 subunit is essential for ORC-DNA binding [10]. Our data provide evidence that HsOrc6 has DNA binding activity on its own. In DmOrc6, this activity maps to a helix-turn-helix motif that is shared by metazoan Orc6 proteins and also mediates DNA binding of TFIIB [3] (Supplementary Fig. 2B–D). Future experiments will have to clarify the functional importance of the HsOrc6 DNA-binding domain, for example, whether it is crucial for pre-RC formation.
We used a plasmid binding system to analyze the role of Orc6 in pre-RC formation. This system is based on 450 mM salt nuclear extracts that contain all proteins required for replication initiation to support plasmid replication in vitro (Supplementary Fig. 3) [4]. Our biochemical and imaging analyses indicated that Orc6 is essential for pre-RC assembly (Figs. 2 and 3). Depletion of Orc6 impairs pre-RC formation in vitro, which can be partially rescued by adding recombinant Orc6 (Fig. 2). Currently, we cannot rule out that the HsOrc6 DNA-binding activity contributes to pre-RC formation. We find that Cdc6 is phosphorylated when associated with DNA; however, the functional significance of this modification is unclear. A recent study by Remus et al. [33] in S. cerevisiae suggested that ScCdc6 phosphorylation is not required for Mcm2–Mcm7 loading, and that regulation of Cdc6 function depended on SCFCDC4 function. Co-immunoprecipitation experiments show that Cdc6 and ORC interact, but the precipitation efficiency was higher between Orc6 and Cdc6 compared to Orc1–Orc5 and Cdc6. In our experiments, the direct interaction between Orc6 and Cdc6 further suggests that Cdc6 might also stabilize the dynamic interaction between Orc1–Orc5 and Orc6, possibly in a cell-cycle dependent manner. Complex formation of Cdc6 and ORC specifically stabilizes the ORC:origin interaction to facilitate pre-RC formation [42]. In S. cerevisiae, Orc6 interacts with Cdt1 and is required for pre-RC assembly and its maintenance [7, 39]. These and our observations provide a mechanistic explanation for a replication defect after siRNA depletion of human Orc6 [30] and argue for a conserved replicative function of Orc6 in recruiting Cdc6. However, further experiments are required to elucidate whether or not human Orc6 is also essential for the maintenance of pre-RCs.
In conclusion, the human Orc6 protein qualifies as a regulatory subunit of ORC that seems to fulfil integrating functions at different stages of the replication initiation process and the cell cycle. It supports DNA association of ORC with the help of chaperone proteins before licensing DNA, it is essential for the formation of pre-RCs, and vital for executing mitosis. Our co-immunoprecipitation experiments suggested that Orc6 is part of the Orc1–Orc6 complex. The interaction between Orc6 and Orc1–Orc5 might be regulated by additional factors such as Cdc6 and HMGA1a that could stabilize ORC or by post-translational modifications. This evolutionary adaption to a dynamic association of Orc6 with the Orc1–Orc5 core complex might play a role in regulating and specifying protein:protein interactions of Orc6 allowing a flexible regulation of the association of this protein with different partners to conduct its diverse cellular functions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Christoph-Erik Mayer and Stefanie Fülöp for discussions and suggestions and J.F.X. Diffley for the protocol of the plasmid-binding system. We gratefully acknowledge R. Knippers for ideas and advice. This work was supported by the following institutional grants: Deutsche Forschungsgemeinschaft grants SFB646 (A.S.), SFB/TR05 (A.S.), SPP1230 (A.S. and W.H.), and NIH grant CA70723 (W.H.).
Abbreviations
- BiFC
Bimolecular fluorescent complementation
- ChIP
Chromatin immunoprecipitation
- DS
Dyad symmetry element
- EBV
Epstein–Barr virus
- FR
Family of repeats
- HMG
High mobility group
- ORC
Origin recognition complex
- pre-RC
Prereplicative complex
- tetR
Tetracyclin repressor
References
- 1.Aladjem MI, Falaschi A, Kowalski D. Eukaryotic DNA replication origins. Cold Spring Harbor: Cold Spring Harbor Press; 2006. [Google Scholar]
- 2.Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 2006;7:716–721. doi: 10.1038/sj.embor.7400730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster . Mol Cell Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltin J, Leist S, Odronitz F, Wollscheid HP, Baack M, Kapitza T, Schaarschmidt D, Knippers R. DNA replication in protein extracts from human cells requires ORC and Mcm proteins. J Biol Chem. 2006;281:12428–12435. doi: 10.1074/jbc.M510758200. [DOI] [PubMed] [Google Scholar]
- 5.Bell SP, Stillman B. Nucleotide dependent recognition of chromosomal origins of DNA replication by a multi-protein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 6.Catez F, Hock R. Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim Biophys Acta. 2010;1799:15–27. doi: 10.1016/j.bbagrm.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, de Vries MA, Bell SP. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2–7 loading. Genes Dev. 2007;21:2897–2907. doi: 10.1101/gad.1596807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesnokov I, Gossen M, Remus D, Botchan M. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 1999;13:1289–1296. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesnokov I, Remus D, Botchan M. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc Natl Acad Sci USA. 2001;98:11997–12002. doi: 10.1073/pnas.211342798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chesnokov IN. Multiple functions of the origin recognition complex. Int Rev Cytol. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- 11.Clarey MG, Botchan M, Nogales E. Single particle EM studies of the Drosophila melanogaster origin recognition complex and evidence for DNA wrapping. J Struct Biol. 2008;164:241–249. doi: 10.1016/j.jsb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat Struct Mol Biol. 2006;13:684–690. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- 13.Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J Biol Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- 14.Dhar SK, Dutta A. Identification and characterization of the human ORC6 homolog. J Biol Chem. 2000;275:34983–34988. doi: 10.1074/jbc.M006069200. [DOI] [PubMed] [Google Scholar]
- 15.Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A. Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–296. doi: 10.1016/S0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- 16.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 17.Du YC, Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/S0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- 18.Duncker BP, Chesnokov IN, McConkey BJ. The origin recognition complex protein family. Genome Biol. 2009;10:214. doi: 10.1186/gb-2009-10-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eilebrecht S, Brysbaert G, Wegert T, Urlaub H, Benecke BJ, Benecke A. 7SK small nuclear RNA directly affects HMGA1 function in transcription regulation. Nucleic Acids Res. 2010;39:2057–2072. doi: 10.1093/nar/gkq1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrer M, Luhrs H, Bustin M, Scheer U, Hock R. Dynamic interaction of HMGA1a proteins with chromatin. J Cell Sci. 2004;117:3459–3471. doi: 10.1242/jcs.01160. [DOI] [PubMed] [Google Scholar]
- 21.Herbig U, Marlar CA, Fanning E. The Cdc6 nucleotide-binding site regulates its activity in DNA replication in human cells. Mol Biol Cell. 1999;10:2631–2645. doi: 10.1091/mbc.10.8.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 23.Killian A, Le Meur N, Sesboue R, Bourguignon J, Bougeard G, Gautherot J, Bastard C, Frebourg T, Flaman JM. Inactivation of the RRB1-Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability. Oncogene. 2004;23:8597–8602. doi: 10.1038/sj.onc.1207845. [DOI] [PubMed] [Google Scholar]
- 24.Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 26.McNairn AJ, Okuno Y, Misteli T, Gilbert DM. Chinese hamster ORC subunits dynamically associate with chromatin throughout the cell-cycle. Exp Cell Res. 2005;308:345–356. doi: 10.1016/j.yexcr.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minami H, Takahashi J, Suto A, Saitoh Y, Tsutsumi K. Binding of AlF-C, an Orc1-binding transcriptional regulator, enhances replicator activity of the rat aldolase B origin. Mol Cell Biol. 2006;26:8770–8780. doi: 10.1128/MCB.00949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammad MM, Donti TR, Sebastian Yakisich J, Smith AG, Kapler GM. Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J. 2007;26:5048–5060. doi: 10.1038/sj.emboj.7601919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norseen J, Thomae A, Sridharan V, Aiyar A, Schepers A, Lieberman PM. RNA-dependent recruitment of the origin recognition complex. EMBO J. 2008;27:3024–3035. doi: 10.1038/emboj.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 31.Prasanth SG, Shenb Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci USA. 2010;107:15093–15098. doi: 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranjan A, Gossen M. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc Natl Acad Sci USA. 2006;103:4864–4869. doi: 10.1073/pnas.0510305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remus D, Beuron F, Toulon G, Griffith JD, Morris EP, Diffley JFX. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A. Complex protein–DNA dynamics at the latent origin of DNA replication of Epstein–Barr virus. J Cell Sci. 2003;116:3971–3984. doi: 10.1242/jcs.00708. [DOI] [PubMed] [Google Scholar]
- 35.Schaarschmidt D, Baltin J, Stehle IM, Lipps HJ, Knippers R. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO J. 2004;23:191–201. doi: 10.1038/sj.emboj.7600029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schepers A, Papior P. Why are we where we are? Understanding replication origins and initiation sites in eukaryotes using ChIP-approaches. Chromosome Res. 2010;18:63–77. doi: 10.1007/s10577-009-9087-1. [DOI] [PubMed] [Google Scholar]
- 37.Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J. 2001;20:4588–4602. doi: 10.1093/emboj/20.16.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seki T, Diffley JF. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc Natl Acad Sci USA. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semple JW, Da-Silva LF, Jervis EJ, Ah-Kee J, Al-Attar H, Kummer L, Heikkila JJ, Pasero P, Duncker BP. An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. EMBO J. 2006;25:5150–5158. doi: 10.1038/sj.emboj.7601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sibani S, Price GB, Zannis-Hadjopoulos M. Ku80 binds to human replication origins prior to the assembly of the ORC complex. Biochemistry. 2005;44:7885–7896. doi: 10.1021/bi047327n. [DOI] [PubMed] [Google Scholar]
- 41.Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J Biol Chem. 2007;282:32370–32383. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- 42.Speck C, Stillman B. Cdc6 ATPase activity regulates ORC x Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem. 2007;282:11705–11714. doi: 10.1074/jbc.M700399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomae A, Pich D, Spindler MP, Berens C, Hammerschmidt W, Schepers A. Interaction between HMGA1a and ORC creates site-specific origins. Proc Natl Acad Sci USA. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vashee S, Simancek P, Challberg MD, Kelly TJ. Assembly of the human origin recognition complex. J Biol Chem. 2001;276:26666–26673. doi: 10.1074/jbc.M102493200. [DOI] [PubMed] [Google Scholar]
- 47.Waga S, Zembutsu A. Dynamics of DNA binding of replication initiation proteins during de novo formation of pre-replicative complexes in Xenopus egg extracts. J Biol Chem. 2006;281:10926–10934. doi: 10.1074/jbc.M600299200. [DOI] [PubMed] [Google Scholar]
- 48.Zembutsu A, Waga S. De novo assembly of genuine replication forks on an immobilized circular plasmid in Xenopus egg extracts. Nucleic Acids Res. 2006;34:e91. doi: 10.1093/nar/gkl512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.