Abstract
WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) is a multifunction protein containing an N-terminal C2 domain, four tandem WW domains for substrate binding, and a C-terminal catalytic HECT domain for ubiquitin transferring. WWP1 has been suggested to function as the E3 ligase for several PY motif-containing proteins, such as Smad2, KLF5, p63, ErbB4/HER4, RUNX2, JunB, RNF11, SPG20, and Gag, as well as several non-PY motif containing proteins, such as TβR1, Smad4, KLF2, and EPS15. WWP1 regulates a variety of cellular biological processes including protein trafficking and degradation, signaling, transcription, and viral budding. WWP1 has been implicated in several diseases, such as cancers, infectious diseases, neurological diseases, and aging. In this review article, we extensively summarize the current knowledge of WWP1 with special emphasis on the roles and action of mechanism of WWP1 in signaling and human diseases.
Keywords: WWP1, WW domain, PY motif, Ubiquitin E3 ligase, Ubiquitination, Cancer
WWP1 (WW domain-containing E3 ubiquitin protein ligase 1), also known as TIUL1 (TGIF-interacting ubiquitin ligase 1) [1] or AIP5 (Atropin-1-interacting protein 5) [2], is a C2-WW-HECT (homologous to E6-AP COOH terminus)-type ubiquitin E3 ligase. The C2-WW-HECT E3 ligase family contains nine members, i.e. NEDD4-1 NEDD4-2/NEDD4L, WWP1, WWP2/AIP2, Itch/AIP4, SMURF1, SMURF2, NEDL1, and NEDL2. They all share three common functional domains: an N-terminal Ca2+/lipid-binding C2 domain, 2-4 substrate-binding WW domains, and a C-terminal catalytic HECT domain [3]. The NEDD4-like family is involved in a diverse variety of cellular processes, such as membrane protein trafficking, protein degradation, signaling, transcription, and apoptosis [3–6]. Several family members have been implicated in cancers, hypertension, neurological diseases, bone metabolism, and immune diseases [3]. Accumulating evidence indicates that WWP1 plays important roles in cancers, infectious diseases, and neurological diseases. We attempt to comprehensively summarize the recent progresses of WWP1 investigations including the protein structure and functions, gene expression, regulation, substrates, and animal models.
Biochemistry of WWP1
WWP1 protein structure
The human WWP1 protein contains 922 amino acid residues with the molecular weight of ~110 kDa. It contains a C2 domain, four WW domains, and a HECT domain (Fig. 1a). The N-terminal C2 domain may mediate the protein–protein interaction, Ca2+ dependent membrane binding, and enzyme activation according to the studies on other WWP1 family members, such as NEDD4 [7–9] and SMURF2 [10]. Consistently, WWP1 is predominately localized to the endosome [11].
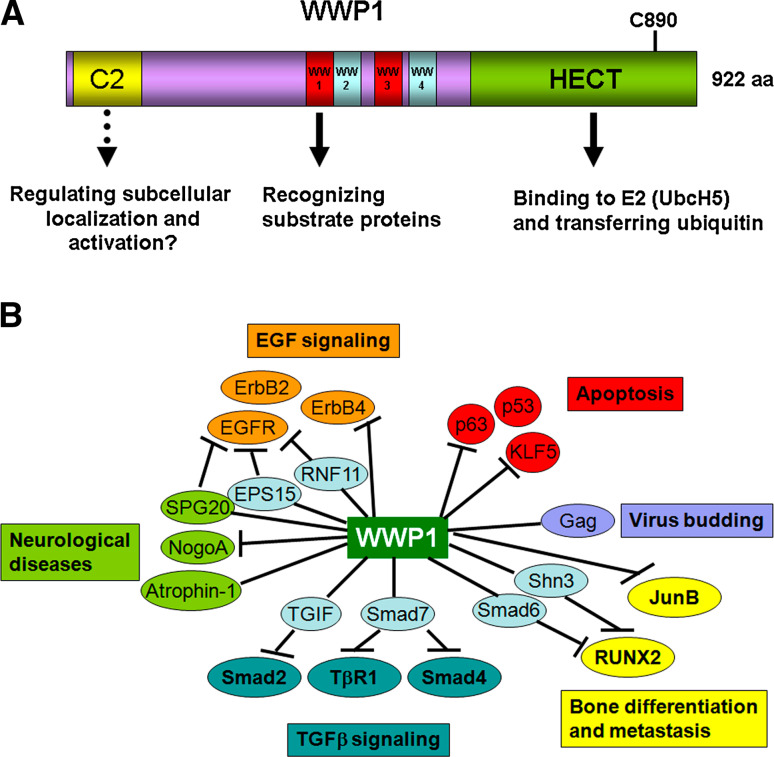
Fig. 1.
a The diagram of the WWP1 protein domains. The WWP1 protein contains 922 amino acid residues. The C2 domain at N-terminus is responsible for membrane and protein binding. The four WW domains in the central region are responsible for the interaction with substrate proteins. The WW1 and WW3 are type I WW domains that recognize PY motifs. The HECT domain at the C-terminus is responsible for the ubiquitin–protein ligase activity. The Cystein-890 is the catalytic activation site. b WWP1 regulates different substrates in different cellular processes
WWP1 has four tandem WW domains in its central part. The WW domain is composed of 38–40 semi-conserved residues with two conserved tryptophans (W) that are spaced 20–22 residues apart and has been shown to mediate protein–protein interaction [12]. The WW domain binds to Pro-rich polypeptide ligands, such as PPXY (PY motif), PPLP, PPR, or p(S/T)P [13]. WW#1 and #3 of WWP1 are type I WW domains that bind to PY motifs with higher affinities as compared to WW#2 and #4 [14–16]. The C-terminus of WWP1 possesses a HECT domain that interacts with a ubiquitin (Ub) conjugation enzyme (i.e. UbcH5 or UbcH7) and is responsible for its ubiquitin E3 ligase enzyme activity [17]. E3 ligases are key enzymes for protein ubiquitination because they determine substrate specificity. Unlike the RING finger type E3 ligases, the HECT type E3 ligases contain a catalytic cystein (C) that can form a covalent isopeptide bond with ubiquitin. To date, 28 of over 600 ubiquitin E3 ligases have been identified to be HECT type E3s in mammalian cells. The catalytic C890 of human WWP1 is responsible for ubiquitin transferring from the WWP1 protein to a substrate protein [1].
WWP1 gene expression
The WWP1 gene is highly conserved among different animals including C. elegans, chicken, mouse, and human. The C. elegans wwp-1 gene encodes 792 amino acid residues [18]. The human WWP1 gene is localized on chromosome 8q21 and encompasses 26 exons, which span at least 142 kb DNA [19]. The 8q21 chromosome region is frequently amplified in several human cancers, including prostate and breast cancers. We reported that gain of gene copy number for WWP1 was detected in 31–51% of prostate and breast cancers [20, 21]. Furthermore, the WWP1 gene is mutated in human cancers. In 30 prostate cancer samples, two sequence alterations that change the WWP1 protein sequence were detected [20]. However, the functions of these mutations have not been characterized.
The WWP1 mRNA is ubiquitously expressed in multiple tissues. Mosser et al. [14] investigated the WWP1 mRNA expression by northern blot and found that WWP1 is highly expressed in the heart and skeletal muscle. This result was confirmed by an independent northern blot study [2]. Quantitative PCR results indicated that WWP1 is highly expressed in the liver, heart, and testis [22]. Several studies showed that the WWP1 mRNA is normally expressed in the prostate and breast at low levels [22, 23]; however, it is elevated in a subset of prostate and breast tumors [20, 21, 24].
The WWP1 mRNA undergoes alternative splicing. WWP1 shows two splice variants in different human tissues by northern blot [2, 14]. Flasza et al. [25] identified six WWP1 splice variants by RT-PCR in the T47D human breast cancer cell line. By northern blot, Huang et al. [18] demonstrated that mouse Wwp1 mRNA has four different splice variants that are highly expressed in the heart, kidney, liver, and testis. The relative ratios of different splice products appear to be tissue-specific. The WWP1 mRNA alternative splicing may be functional relevant. Some isoforms without the C2 domain may have distinct or dominant negative functions [25]. We found an extra, smaller WWP1 protein band below the wild-type WWP1 band in two prostate cancer cell lines PC-3 and LAPC-4 [20].
WWP1 protein subcellular localization: membrane, cytoplasm, or nucleus?
The WWP1 protein is predominantly localized in the early endosome, in the cytoplasm, and occasionally in the nucleus. By immunofluorescence (IF) analysis, Seo et al. [1] demonstrated that exogenous WWP1/Tiul1 protein is expressed in both the nucleus and cytoplasm of MDCK canine kidney epithelial cells. Consistently, we found that the exogenous WWP1 protein is expressed in both the nucleus and cytoplasm of the 22Rv1 prostate cancer cell line although WWP1 has no nuclear localization signal [11, 23]. It is worth pointing out that the exogenous WWP1 protein is predominantly localized in cytoplasmic punctate membrane structures and occasionally localized in the nucleus [11]. Flasza et al. [26] demonstrated that the exogenous WWP1 protein is localized to the early endosome in C2C12 murine skeletal muscle-derived myoblast cells. By immunohistochemistry (IHC), Huu et al. [24] detected both nuclear and cytoplasmic endogenous WWP1 protein expression in breast tumors. However, we only detected the cytoplasmic WWP1 protein expression in breast tumors using a mouse monoclonal anti-WWP1 antibody [27]. Thus, all studies support the cytoplasmic localization of WWP1 while the nuclear localization of WWP1 is context dependent. In agreement with different WWP1 subcellular localizations, WWP1 ubiquitinates not only cytoplasmic substrates, such as ErbB4 and TβR1, but also nuclear substrates, such as KLF5 and p63.
Additionally, the cellular localization of WWP1 is regulated by the Notch signaling. Notch could deplete WWP1 out of the nucleus, and co-localize with WWP1 in early endosomes. In non-differentiated C2C12 cells, WWP1 is distributed in both the nucleus and cytoplasm. When the cells were induced to differentiate, the localization of WWP1 was exclusively in the cytoplasm [26].
Regulation of WWP1
WWP1 has been shown to be regulated under the physiological processes. WWP1 mRNA levels are increased during osteoblast differentiation [28]. Zhao et al. [29] recently reported that tumor necrosis factor α (TNFα) induced the WWP1 mRNA expression in mesenchymal stem cells. We found that transforming growth factor β (TGFβ) induced WWP1 mRNA expression in HaCaT and PC-3 cells [3]. It is possible that the induction of WWP1 by TGFβ is a negative feedback mechanism for controlling the TGFβ signaling pathway since WWP1 suppresses TGFβ signaling [1, 22]. During the cellular senescence, WWP1 has been shown to be downregulated [30].
Under the pathological conditions, DNA damage chemotherapeutic drugs (Doxorubicin and Cisplatin) induced WWP1 in a p53-dependent manner in MCF10A and HCT116 cells [15]. However, Wwp1 mRNA levels decreased after exposure to UV- or γ-irradiation in p53 wild-type mouse embryo fibroblast cells [31]. Thus, DNA damage may increase the p53 protein level that regulates the WWP1 expression in a cell line-dependent manner. Nevertheless, these results imply that WWP1 is a downstream target gene of p53. Interestingly, WWP1 negatively regulates the activities of p53 and its closely related family member p63 [15, 31]. Further investigations are necessary to elucidate the mechanism by which WWP1 is regulated by p53.
In addition, WWP1 has been suggested to be induced by androgen in the LAPC9 prostate cancer cell line [32]. We reported that the WWP1 protein expression correlates with the estrogen receptor (ERα) positive status in breast tumors [27]. Most recently, estrogen was shown to induce the formation of complexes containing ERβ, KLF5, and WWP1, resulting in the ubiquitination and degradation of KLF5 in prostate cancer [33]. Whether the WWP1 expression is regulated by other hormones is unclear.
Autoubiquitination and proteasomal degradation are features common to most E3 ligases. We previously showed that the WWP1 protein is degraded by the 26S proteasome [3]. The half-life of WWP1 protein is about 3 h in 22Rv1 cells. The proteasome inhibitor MG132 could dramatically increase the WWP1 protein levels in MCF10A and 22Rv1 cells [3].
Whether WWP1 is regulated by other posttranslational modifications, such as phosphorylation is currently unknown. Understanding the regulatory mechanisms of WWP1 will help us design effective strategies for targeting the WWP1 pathway in diseases.
WWP1-ubiquitinated substrate proteins
As an ubiquitin E3 ligase, WWP1 interacts with a variety of substrate proteins and regulates their expression levels and activities. To date, WWP1 has been shown to function as the E3 ligase for TβR1, Smad2, ErbB4/HER4, RNF11, SPG20, KLF5, p63, RUNX2, JunB, p27, and others (Fig. 1b; Table 1). How WWP1 specifically binds to different substrates is not completely understood at present. The subcellular localization, the expression level in a specific cell type, and posttranslational modifications of substrates may contribute to the specificity.
Table 1.
The list of WWP1 interacting proteins
| WWP1 interacting proteins | Regulation | Functions | References |
|---|---|---|---|
| PY motif containing substrates | |||
| ErbB4/HER4 | Degradation | Differentiation and cancer | [16, 41] |
| p63 | Degradation | Apoptosis and cancer | [15] |
| KLF5 | Degradation | Proliferation, apoptosis, and cancer | [23, 51] |
| RNF11 | Ubiquitination, no degradation | EGFR and ErbB2 degradation | [11] |
| Spartin (SPG20) | mUb, no degradation | Troyer syndrome | [46, 47] |
| Nogo-A | Degradation | Central nervous system regeneration | [71] |
| Smad2 | Degradation | TGFβ signaling | [1, 20] |
| Smad6 | No degradation | Adaptor for RUNX2 | [28] |
| Smad7 | No degradation | TGFβ signaling | [1, 22] |
| RUNX2 | Degradation | Osteoblast differentiation | [28, 58, 59] |
| JunB | Degradation | Osteoblast differentiation | [29] |
| Gag | mUb, no degradation | Virus budding | [63, 64, 67, 75] |
| ARRDC1 | Ubiquitination, no degradation | Virus budding | [68] |
| Atrophin-1 | Muscular dystrophy | [2] | |
| WBP-1/2 | Co-activator of PR and ER | [75, 76] | |
| RasGAP | Apoptosis and migration | [75] | |
| p53-BP2 | [75] | ||
| IL6Rα | Inflammatory response | [75] | |
| NF-E2 (p45) | Globin gene transcription | [14] | |
| Non-PY motif containing substrates | |||
| TβR1 | Degradation | TGFβ signaling | [1, 22] |
| Smad4 | Degradation | TGFβ signaling | [20, 34] |
| TGIF | No degradation | Adaptor for Smad2 | [1] |
| Shn3 | No degradation | Adaptor for RUNX2 | [58] |
| p53 | Subcellular localization | Apoptosis | [31] |
| p27 | Degradation | Senescence | [30] |
| KLF2 | Degradation | Transcription | [52] |
| EPS15 | mUb, no degradation | Trafficking | [3] |
TGFβ signaling pathway
The TGFβ superfamily regulates cell proliferation, differentiation, apoptosis, and migration in numerous cell types. TGFβ ligands bind to a type II receptor (TβR2), which recruits and phosphorylates TβR1. Then, TβR1 recruits and phosphorylates receptor-regulated Smads (R-Smads), Smad2 and Smad3. The phosphorylated R-Smads bind to the common Smad4, enter the nucleus, and form transcription complexes to regulate gene expression.
Accumulating evidence suggests that WWP1 negatively regulates the TGFβ signaling pathway [1, 20, 22]. WWP1 inhibits TGFβ-induced transcriptional activities as well as PAI-1 and JunB expression [1, 22]. In addition, overexpression of WWP1 in MDCK cells reduces the TGFβ-induced growth inhibition [1]. Among eight Smads, WWP1 can strongly interact with Smad2, 3, 6, 7 proteins, weakly with Smad1 and 5, but not with Smad4 and Smad8, which do not contain a PY motif [1, 22]. WWP1 directly targets Smad2 for ubiquitination and degradation in the presence of TGIF [1, 22]. Two independent studies suggest that WWP1 targets TβR1 for ubiquitin-mediated degradation through the Smad7 adaptor [1, 22]. Morén et al. [34] reported that WWP1 uses Smad7 to induce Smad4 ubiquitination and degradation. Thus, WWP1 inhibits the TGF-β signaling pathway directly or indirectly by targeting the TβR1, Smad2, and/or Smad4 proteins for degradation.
Epithelial growth factor (EGF) signaling pathway
The EGF receptor (EGFR) subfamily of receptor tyrosine kinases are cell-surface receptors, consisting of 4 homologous members: EGFR, ErbB2/HER2, ErbB3, and ErbB4/HER4 [35]. EGFRs are overexpressed in multiple tumors, including breast cancer and lung cancer [36].
ErbB4 has three PY motifs that are the targets of several WW domain-containing proteins, such as WWOX [37], YAP [38, 39] and AIP4/Itch [40]. Recently, two independent studies suggest that WWP1 promotes ErbB4 protein ubiquitination and degradation [16, 41]. The protein–protein interaction between WWP1 and ErbB4 is through the first and third WW domains of WWP1 and the second PY motif (PPAY) of ErbB4-CYT1 that overlaps with the docking site (YTPM) of PI3K [16]. The phosphorylated tyrosine will activate the PI3K/AKT survival signaling pathway while the non-phosphorylated PY motif may facilitate WWP1 and Itch binding. Thus, the tyrosine phosphorylation of ErbB4 may act as a binary switch for ErbB4’s activity and stability. A similar effect of the tyrosine phosphorylation has been observed in RNA Polymerase II [13].
ErbB4 is well known to undergo a two-step proteolytic cleavage, during which a membrane-anchored fragment of 80 kDa is first produced and a soluble 80-kDa fragment is subsequently liberated [42]. WWP1-mediated degradation is more efficient for the full-length ErbB4 protein and for the membrane-anchored fragment as compared to the soluble fragment [41]. Unlike other EGFR subfamily members, the ErbB4/HER4 expression correlates with positive ERα, lower tumor grade, and a better prognosis [43, 44]. Intriguingly, the expression levels of WWP1 and ErbB4 seem to be correlated in breast cancer cell lines [16].
Additionally, our previous study showed that WWP1 overexpression also upregulated the EGFR and ErbB2 expression levels in MCF10A and MDA-MB-231 breast cell lines in a manner independent of E3 ligase activity [11]. WWP1 depletion decreased the cell surface EGFR and ErbB2 levels. The regulation of EGFR and ErbB2 by WWP1 may partially rely on RNF11, a PY motif containing RING finger type E3 ligase promoting EGFR and ErbB2 protein degradation. The interaction between WWP1 and RNF11 is through the first and third WW domains of WWP1 and the PY motif of RNF11 [11].
EGFR is well known to undergo rapid endocytosis upon ligand stimulation. SPG20/Saprtin is necessary for efficient degradation of EGFR [45]. Recently, two independent studies suggested that WWP1 interacts with SPG20 through WW/PY motifs [46, 47]. SPG20 can be recruited to the endosome and to lipid droplets [46, 47]. While WWP1 overexpression can promote SPG20 mono-ubiquitination, the mono-ubiquitination of endogenous SPG20 is not affected by WWP1 depletion [46, 47]. Interestingly, WWP1 ubiquitinates another endocytosis protein, EPS15, that interacts with SPG20 [3, 46]. Further investigation will be required to clarify the role of WWP1 in regulating SPG20, EPS15, and endocytosis.
Taken together, WWP1 may regulate the EGF signaling pathway by not only directly targeting ErbB4 for degradation but also indirectly inhibiting EGFR and ErbB2 endocytosis through other proteins, including RNF11, SPG20, and EPS15.
Krüppel-like factor (KLF) family
The KLF family members are well-established zinc finger transcription factors that play critical roles in cell proliferation, survival, differentiation, and stemness [48]. Both KLF5 and KLF2 have been implicated in cancers and cardiovascular diseases and reported to be WWP1 substrates.
KLF5 promotes cell proliferation, survival, and angiogenesis [49]. KLF5 turns over rapidly through the ubiquitin–proteasome pathway [50]. The KLF5 transactivation domain contains a PY motif that interacts with the WW domains of WWP1 [23]. The E3 ligase activity of WWP1 is essential for WWP1 to target KLF5 for ubiquitination and degradation [23]. WWP1 depletion increases the endogenous KLF5 protein levels in MCF10A, MCF7, and PC-3 [23, 51]. As expected, the expression of WWP1 is negatively correlated with that of KLF5 in human breast cancer cell lines [51]. Most recently, Yuka Nakajima et al. [33] reported that estrogen induced the formation of complexes containing ERβ, KLF5, and WWP1, resulting in the ubiquitination and degradation of KLF5.
In contrast to KLF5, KLF2 does not have a PY motif. Zhang et al. [52] reported that WWP1 interacted with the inhibitory domain of KLF2, ubiquitinated KLF2, and targeted it for proteasomal degradation in an E3 ligase activity-independent manner. These findings suggest that WWP1 may facilitate KLF2 ubiquitination and degradation as an adaptor protein. Whether endogenous WWP1 regulates KLF2 has not been investigated.
p53 family
The p53 transcription factor family contains three members, p53, p63, and p73. p53 is a well-known tumor suppressor that responds to diverse cellular stresses and induces cell cycle arrest, apoptosis, and senescence [53]. p53 has been reported to be ubiquitinated by several E3 ligases, such as MDM2 [54], Pirh2 [55], and WWP1 [31].
Laine et al. [31] reported that WWP1 interacts with the p53 DNA-binding domain. The proline-rich domain of p53 does not have a PY motif but increases the efficiency of protein–protein interaction. WWP1 ubiquitinated p53 in an E3 ligase-dependent manner and increased the nuclear export of p53 [31]. We demonstrated that WWP1 targeted p63 for ubiquitin-mediated protein degradation [15]. Using different promoters, p63 can be expressed as ΔNp63 and TAp63 that have an opposite function in terms of apoptosis. The pro-survival ΔNp63 protein is the predominant p63 isotype in the prostate and breast [15]. There are three isoforms (α, β, γ) for both ΔNp63 and TAp63. WWP1 interacts with p63α through WW-PY motifs and targets p63α for ubiquitin-mediated proteasomal degradation [15]. In ΔNp63-positive breast cells, WWP1 depletion confers apoptosis resistance [15]. Paradoxically, Peschiaroli et al. [56] found that WWP1 did not trigger ΔNp63 proteasome-dependent degradation. WWP1 modified ΔNp63 with K63-linked polyubiquitin chains, increased the ΔNp63-mediated transcription, and induced cell cycle arrest in human primary keratinocytes [56]. Thus, WWP1 may regulate p63 in a cell type-dependent manner. It is likely that WWP1 uses different E2s to ubiquitinate ΔNp63 in different cell types. Whether WWP1 targets the PY motif containing p73 for degradation has not been tested. Interestingly, WWP1 is induced by DNA damage agents in a p53-dependent manner [15]. Thus, WWP1 is p53 downstream gene that can inhibit p53 family member activities through a feedback mechanism.
RUNX2 and JunB
RUNX2 and JunB are principal transcription factors for osteoblast differentiation and associate with tumor growth in the bone [57]. RUNX2 is frequently overexpressed in invasive breast and prostate cancers [57]. WWP1 uses Shn3 or Smad6 as adaptors to target RUNX2 for ubiquitination and degradation although RUNX2 itself has a PY motif [58, 59]. Shn3 increases the protein interaction between WWP1 and RUNX2 [58]. Mice lacking Shn3 display increased bone mass [58]. Surprisingly, the expression levels of Wwp1/Shn3 and Runx2 increased during osteoblast differentiation [28]. It was speculated that WWP1 only functions at the late stage of osteoblast differentiation, as RUNX2 must be downregulated in fully matured osteoblasts. Most recently, WWP1 was reported to promote the ubiquitin-mediated degradation of JunB, which is another transcription factor critical for osteoblast formation in mesenchymal stem cells [29].
WWP1 has been reported to modify substrates with mono-ubiquitin (SPG20 and EPS15), K48-linked polyubiquitin chains (KLF5 and p27), and K63-linked polyubiquitin chains (ΔNp63). Although it is well known that different types of ubiquitination have different physiological functions, the determination of the ubiquitin conjugate types catalyzed by WWP1 requires further study.
WWP1 and diseases
WWP1 is ubiquitously expressed in human tissues and participates in multiple physiological and pathological processes, such as cancers, infectious diseases, neurological diseases, and even aging.
Cancers
WWP1 appears to function in a context-dependent manner in cancers. The WWP1 mRNA and protein levels are frequently up-regulated in a subset of prostate and breast cancers [20, 21]. Interestingly, the expression of WWP1 in breast tumors correlates with positive ERα and insulin-like growth factor 1 receptor (IGF-1R) statuses, which are good prognosis biomarkers in breast tumors. Indeed, Huu et al. [24] found that tumors with low/absent WWP1 expression have a worse prognosis than tumors with WWP1 expression. Similar results were observed in head and neck squamous cell carcinoma [60]. In MCF10A breast cells, when WWP1 was knocked down by siRNA, the cells became more resistant to doxorubicin-induced apoptosis [15]. This result suggests that WWP1 has a pro-apoptotic function in MCF10A. The WWP1 pro-apoptotic function can be attributed to targeting several pro-survival proteins, such as ΔNp63 [15] and KLF5 [23], for ubiquitin-mediated degradation. Consistently, the expression of WWP1 is negatively correlated with the expression of ΔNp63 [15] and KLF5 [51] in breast cancers.
In contrast to the WWP1 pro-apoptotic function in ERα-negative breast cells, WWP1 depletion in ERα-positive MCF7 and HCC1500 breast cancer cell lines suppressed cell proliferation and induced apoptosis [11, 21, 24, 27]. Depletion of WWP1 activated caspase-8 and sensitized these breast cancer cells to TRAIL-induced apoptosis [61]. Similar results were obtained in MDCK [1], PC-3 [20], and HCT116 cells [15]. In addition, WWP1 overexpression promoted breast epithelial cell growth and anchorage-independent growth [24]. In 2BS and WI38 fibroblast cell lines, overexpression of WWP1 increased cell proliferation while depletion of WWP1 induced senescence [30]. WWP1 enhances cell proliferation and survival likely through both ubiquitin ligase-dependent and -independent activities. In MDCK cells, WWP1 suppressed the expression of TβR1, Smad2, and TGFβ-induced PAI1 and JunB [1]. In PC-3, WWP1 suppressed the expression of TβR1, Smad4, and the cell cycle-dependent kinase inhibitor p15 [20]. In HCT116, WWP1 targeted the TAp63 for degradation [15]. In addition, WWP1 enhances MAPK signaling through decreasing the ErbB2 and EGFR turnover [11]. Thus, WWP1 may have a context-dependent role in cancer development. To conclusively sort this out, the physiological role of WWP1 needs to be elucidated in animal models.
Infectious diseases
Available evidence suggests that WWP1 may participate in viral internalization and budding. Galinier et al. [62] first reported that WWP1 interacted with the adenovirus penton base protein that has two PY motifs and is involved in viral internalization. It is becoming obvious that WWP1 promotes retrovirus particle assembly and release through binding to Gag. The Gag precursor protein is well known to drive the assembly and release of enveloped retroviruses. Many Gag late assembly function domains contain a PY motif [63]. Heidecker et al. [64] reported that mutation of the PY motif severely decreased human T cell leukemia virus type 1 (HTLV-1) virus budding. Chen et al. [65] suspected that Gag of avian retroviruses and LMP2 of the Epstein-Barr virus (EBV) may interact with WW domain-containing proteins through their PY motifs. Consistently, cis expression of a WW domain in the Gag protein can block budding of Rous sarcoma virus (RSV) [66]. These studies suggest that the Gag PY motif is essential for budding through recruiting the WW domain proteins.
WWP1 has been demonstrated to mono-ubiquitinate Gag proteins at a single lysine [46, 64], and mutation of the Gag ubiquitination site dramatically inhibited viral budding [67], suggesting that the Gag ubiquitination by WWP1 may play a role for viral budding. This notion is supported by another independent study [63]. Martin-Serrano et al. [63] reported that the E3 ligase activity was required for WWP1 functions in this context. The HECT domains from WWP1, WWP2, or Itch recruited class E vacuolar protein-sorting (VPS) factors and ubiquitinated them for viral budding [63]. Importantly, trans expression of dominant negative WWP1 (C2-WW or WW) proteins inhibited budding of HTLV-1, murine leukemia virus (MLV), and Ebola viruses [63, 64]. Most recently, WWP1 was proposed to recruit the ESCRT (endosomal sorting complex required for transport)-III to the site of viral budding through interacting with ARRDC1 (arrestin domain-containing protein 1) [68]. These studies suggest that the WW domain polypeptide may be a potential therapeutic intervention for viral infection.
WWP1 may also be involved in innate immunity. Wwp-1 deficient C. elegans are hypersensitive to killing by pore-forming toxins and the bacterial pathogen Pseudomonas aeruginosa [69]. Wwp-1 may regulate innate immunity through antagonizing the DAF-2 insulin-like signaling pathway in C. elegans. Whether WWP1 plays a role in mammalian immunity is currently unknown.
Neurological diseases
Ubiquitination is well documented to play important roles in neurological diseases. WWP1 interacts with several proteins implicated in neurological diseases. Firstly, WWP1 interacts with Atrophin-1, a nuclear receptor corepressor containing PY motifs and a polyglutamine repeat [2]. Atrophin-1 was identified as a neurodegenerative disease gene because it is mutated in dentatorubral–pallidoluysian atrophy. Disease-causing mutations are expansions of a DNA triplet repeat leading to the expression of a protein with an extended stretch of glutamine residues that causes neuronal death [70]. The functional consequences of the WWP1/Atrophin-1 interaction have not yet been explored. Secondly, WWP1 interacts with Nogo-A, a key inhibitor for central nervous system (CNS) regeneration [71]. The Nogo-A protein contains a PY motif and localizes to the endoplasmic reticulum. The protein solution structure of WWP1 WW4 domain with the Nogo-A PY motif peptide has been determined by NMR [71]. Qin et al. [71] claimed that WWP1 ubiquitinated Nogo-A and regulated the Nogo-A protein level. It would be interesting to investigate whether WWP1 regulates CNS regeneration. Finally, WWP1 interacts with Saprtin/SPG20, a protein mutated in Troyer syndrome [46, 47, 72]. The aforementioned studies suggest that WWP1 mono-ubiquitinates SPG20 and regulates its protein levels and subcellular localization [46, 47].
Aging and other diseases
In response to dietary restriction, wwp-1 acts as a positive regulator of lifespan in an E3 ubiquitin ligase-dependent manner in C. elegans [73]. In contrast to three WWP ligases (WWP1, WWP2, and AIP4) in mammalians, there is only one WWP E3 ligase in C. elegans. AIP4 is the wwp-1 most related human WWP ligase according to the conserved WW and HECT domain sequences [18]. Depletion of wwp-1 leads to abnormal embryogenesis and a lethal phenotype [18]. Carrano et al. [73] demonstrated that loss of wwp-1 functions by RNAi or mutation decreases lifespan. The wwp-1 transgenic lines lived up to 20% longer than the controls. Chen et al. [69] confirmed that wwp-1 mutations decreased lifespan in C. elegans. The possible molecular mechanism by which wwp-1 participates in longevity regulation is that wwp-1 is inhibited by the PDK-1 kinase downstream of the DAF-2 insulin/IGF-1 signaling pathway in C. elegans [69]. It would be interesting to elucidate the role and mechanism of action of WWP1 in mammalian longevity. Most recently, Cao et al. [30] reported that the expression level of WWP1 in young human fibroblasts is significantly higher than that in aged fibroblasts with senescence. Further investigation revealed that WWP1 targets the cell cycle-dependent kinase inhibitor p27kip protein for ubiquitin-mediated degradation in human fibroblasts [30].
In addition, WWP1 may be involved in bone diseases because it targets RUNX2 and JunB for degradation [29, 58, 59]. Patients with chronic inflammation, such as rheumatoid arthritis, often have severe bone loss and increased incidence of bone fractures. One possible molecular mechanism is that TNFα induces the expression of WWP1, which in turn promotes JunB for ubiquitination and degradation in mesenchymal stem cells [29]. Inhibition of WWP1 may provide a new therapy for inflammatory osteoporosis.
WWP1 has also been reported to be responsible for chicken muscular dystrophy [74]. A WWP1 missense mutation was identified in dystrophic chickens [74].
Conclusions and perspectives
In summary, WWP1 is a WW domain-containing HECT-type ubiquitin E3 ligase. WWP1 targets a number of PY motif containing substrate proteins, including Smad2, KLF5, p63, ErbB4, RUNX2, RNF11, and SPG20 for ubiquitination. In addition, WWP1 may indirectly regulate other substrates without a PY motif through adaptors. WWP1 plays important roles in a variety of diseases, such as cancers, infectious diseases, neurological diseases, and aging.
In the future, Wwp1 knock-out and tissue-specific transgenic mouse models will be urgently required to elucidate the physiological and pathological functions of WWP1 in human diseases. Xenograft mouse models will also be useful to determine the role of WWP1 in cancer growth and metastasis. Whether the genetic and expression alterations of WWP1 can be used as biomarkers in cancers and other diseases should be studied in large cohorts of patients. Additionally, it is very important to dissect the distinct and redundant functions of the WWP1 E3 family members in order to design the most effective interventions. Furthermore, the mechanism of WWP1 regulation is still poorly understood. Finally, if further studies clarify the role of WWP1 and other family members in disease processes, it would be interesting to develop small molecular inhibitors or peptides for treating WWP1-related diseases.
Acknowledgments
This work was supported in part by grants from National Nature Science Foundation of China (81072162 and 81120108019) and a grant from Yunnan Province High-Profile Scientists Program (云南省高端科技人才计划 2010CI114).
References
- 1.Seo SR, Lallemand F, Ferrand N, Pessah M, L’Hoste S, Camonis J, Atfi A. The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J. 2004;23(19):3780–3792. doi: 10.1038/sj.emboj.7600398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood JD, Yuan J, Margolis RL, Colomer V, Duan K, Kushi J, Kaminsky Z, Kleiderlein JJ, Sharp AH, Ross CA. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol Cell Neurosci. 1998;11(3):149–160. doi: 10.1006/mcne.1998.0677. [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26(3–4):587–604. doi: 10.1007/s10555-007-9091-x. [DOI] [PubMed] [Google Scholar]
- 4.Harvey KF, Kumar S. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999;9(5):166–169. doi: 10.1016/S0962-8924(99)01541-X. [DOI] [PubMed] [Google Scholar]
- 5.Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23(11):1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- 6.Shearwin-Whyatt L, Dalton HE, Foot N, Kumar S. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays. 2006;28(6):617–628. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
- 7.Plant PJ, Yeger H, Staub O, Howard P, Rotin D. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J Biol Chem. 1997;272(51):32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- 8.Plant PJ, Lafont F, Lecat S, Verkade P, Simons K, Rotin D. Apical membrane targeting of Nedd4 is mediated by an association of its C2 domain with annexin XIIIb. J Cell Biol. 2000;149(7):1473–1484. doi: 10.1083/jcb.149.7.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Peng Q, Lin Q, Childress C, Carey D, Yang W. Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. J Biol Chem. 2010;285(16):12279–12288. doi: 10.1074/jbc.M109.086405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130(4):651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, Zhou Z, Liu R, Li Y, Azmi PB, Seth AK. The WW domain containing E3 ubiquitin protein ligase 1 upregulates ErbB2 and EGFR through RING finger protein 11. Oncogene. 2008;27(54):6845–6855. doi: 10.1038/onc.2008.288. [DOI] [PubMed] [Google Scholar]
- 12.Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module–the WW domain. FEBS Lett. 1995;369(1):67–71. doi: 10.1016/0014-5793(95)00550-S. [DOI] [PubMed] [Google Scholar]
- 13.Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103(7):1001–1004. doi: 10.1016/S0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 14.Mosser EA, Kasanov JD, Forsberg EC, Kay BK, Ney PA, Bresnick EH. Physical and functional interactions between the transactivation domain of the hematopoietic transcription factor NF-E2 and WW domains. Biochemistry. 1998;37(39):13686–13695. doi: 10.1021/bi981310l. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008;15(12):1941–1951. doi: 10.1038/cdd.2008.134. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Zhou Z, Alimandi M, Chen C. WW domain containing E3 ubiquitin protein ligase 1 targets the full-length ErbB4 for ubiquitin-mediated degradation in breast cancer. Oncogene. 2009;28(33):2948–2958. doi: 10.1038/onc.2009.162. [DOI] [PubMed] [Google Scholar]
- 17.Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11(1):249–259. doi: 10.1016/S1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 18.Huang K, Johnson KD, Petcherski AG, Vandergon T, Mosser EA, Copeland NG, Jenkins NA, Kimble J, Bresnick EH. A HECT domain ubiquitin ligase closely related to the mammalian protein WWP1 is essential for Caenorhabditis elegans embryogenesis. Gene. 2000;252(1–2):137–145. doi: 10.1016/S0378-1119(00)00216-X. [DOI] [PubMed] [Google Scholar]
- 19.Malbert-Colas L, Fay M, Cluzeaud F, Blot-Chabaud M, Farman N, Dhermy D, Lecomte MC. Differential expression and localisation of WWP1, a Nedd4-like protein, in epithelia. Pflugers Arch. 2003;447(1):35–43. doi: 10.1007/s00424-003-1152-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Sun X, Guo P, Dong XY, Sethi P, Zhou W, Zhou Z, Petros J, Frierson HF, Jr, Vessella RL, Atfi A, Dong JT. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene. 2007;26(16):2386–2394. doi: 10.1038/sj.onc.1210021. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Zhou Z, Ross JS, Zhou W, Dong JT. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer. 2007;121(1):2834–2841. doi: 10.1002/ijc.22653. [DOI] [PubMed] [Google Scholar]
- 22.Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, Miyazawa K. Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1) Oncogene. 2004;23(41):6914–6923. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280(50):41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen Huu NS, Ryder WD, Zeps N, Flasza M, Chiu M, Hanby AM, Poulsom R, Clarke RB, Baron M. Tumour-promoting activity of altered WWP1 expression in breast cancer and its utility as a prognostic indicator. J Pathol. 2008;216(1):93–102. doi: 10.1002/path.2385. [DOI] [PubMed] [Google Scholar]
- 25.Flasza M, Gorman P, Roylance R, Canfield AE, Baron M. Alternative splicing determines the domain structure of WWP1, a Nedd4 family protein. Biochem Biophys Res Commun. 2002;290(1):431–437. doi: 10.1006/bbrc.2001.6206. [DOI] [PubMed] [Google Scholar]
- 26.Flasza M, Nguyen Huu NS, Mazaleyrat S, Clemence S, Villemant C, Clarke R, Baron M. Regulation of the nuclear localization of the human Nedd4-related WWP1 protein by Notch. Mol Membr Biol. 2006;23(3):269–276. doi: 10.1080/09687860600665010. [DOI] [PubMed] [Google Scholar]
- 27.Chen C, Zhou Z, Sheehan CE, Slodkowska E, Sheehan CB, Boguniewicz A, Ross JS. Overexpression of WWP1 is associated with the estrogen receptor and insulin-like growth factor receptor 1 in breast carcinoma. Int J Cancer. 2009;124(12):2829–2836. doi: 10.1002/ijc.24266. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Huang M, Zheng H, Wang Y, Ren F, Shang Y, Zhai Y, Irwin DM, Shi Y, Chen D, Chang Z. CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J Cell Biol. 2008;181(6):959–972. doi: 10.1083/jcb.200711044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L, Huang J, Zhang H, Wang Y, Matesic LE, Takahata M, Awad H, Chen D, Xing L. Tumor necrosis factor inhibits mesenchymal stem cell differentiation into osteoblasts via the ubiquitin E3 ligase Wwp1. Stem Cells. 2011;29(10):1601–1610. doi: 10.1002/stem.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao X, Xue L, Han L, Ma L, Chen T, Tong T. WW Domain-containing E3 ubiquitin protein ligase 1 (WWP1) Delays cellular senescence by promoting p27Kip1 degradation in human diploid fibroblasts. J Biol Chem. 2011;286(38):33447–33456. doi: 10.1074/jbc.M111.225565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laine A, Ronai Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene. 2007;26(10):1477–1483. doi: 10.1038/sj.onc.1209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu Z, Rubin MA, Yang Y, Deprimo SE, Zhao H, Horvath S, Brooks JD, Loda M, Reiter RE. Reg IV: a promising marker of hormone refractory metastatic prostate cancer. Clin Cancer Res. 2005;11(6):2237–2243. doi: 10.1158/1078-0432.CCR-04-0356. [DOI] [PubMed] [Google Scholar]
- 33.Nakajima Y, Akaogi K, Suzuki T, Osakabe A, Yamaguchi C, Sunahara N, Ishida J, Kako K, Ogawa S, Fujimura T, Homma Y, Fukamizu A, Murayama A, Kimura K, Inoue S, Yanagisawa J. Estrogen regulates tumor growth through a nonclassical pathway that includes the transcription factors ERbeta and KLF5. Sci Signal. 2011;4(168):ra22. doi: 10.1126/scisignal.2001551. [DOI] [PubMed] [Google Scholar]
- 34.Moren A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280(23):22115–22123. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- 35.Herbst A, Salghetti SE, Kim SY, Tansey WP. Multiple cell-type-specific elements regulate Myc protein stability. Oncogene. 2004;23(21):3863–3871. doi: 10.1038/sj.onc.1207492. [DOI] [PubMed] [Google Scholar]
- 36.Yarden Y. The EGFR family and its ligands in human cancer: signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(Suppl 4):S3–S8. doi: 10.1016/S0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 37.Aqeilan RI, Donati V, Gaudio E, Nicoloso MS, Sundvall M, Korhonen A, Lundin J, Isola J, Sudol M, Joensuu H, Croce CM, Elenius K. Association of Wwox with ErbB4 in breast cancer. Cancer Res. 2007;67(19):9330–9336. doi: 10.1158/0008-5472.CAN-07-2147. [DOI] [PubMed] [Google Scholar]
- 38.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278(35):33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 39.Omerovic J, Puggioni EM, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp Cell Res. 2004;294(2):469–479. doi: 10.1016/j.yexcr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Omerovic J, Santangelo L, Puggioni EM, Marrocco J, Dall’Armi C, Palumbo C, Belleudi F, Di Marcotullio L, Frati L, Torrisi MR, Cesareni G, Gulino A, Alimandi M. The E3 ligase Aip4/Itch ubiquitinates and targets ErbB-4 for degradation. Faseb J. 2007;21(11):2849–2862. doi: 10.1096/fj.06-7925com. [DOI] [PubMed] [Google Scholar]
- 41.Feng SM, Muraoka-Cook RS, Hunter D, Sandahl MA, Caskey LS, Miyazawa K, Atfi A, Earp HS., 3rd The E3 ubiquitin ligase WWP1 selectively targets HER4 and its proteolytically derived signaling isoforms for degradation. Mol Cell Biol. 2009;29(3):892–906. doi: 10.1128/MCB.00595-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vecchi M, Carpenter G. Constitutive proteolysis of the ErbB-4 receptor tyrosine kinase by a unique, sequential mechanism. J Cell Biol. 1997;139(4):995–1003. doi: 10.1083/jcb.139.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes NL, Khavari S, Boland GP, Cramer A, Knox WF, Bundred NJ. Absence of HER4 expression predicts recurrence of ductal carcinoma in situ of the breast. Clin Cancer Res. 2005;11(6):2163–2168. doi: 10.1158/1078-0432.CCR-04-1633. [DOI] [PubMed] [Google Scholar]
- 44.Naresh A, Long W, Vidal GA, Wimley WC, Marrero L, Sartor CI, Tovey S, Cooke TG, Bartlett JM, Jones FE. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;66(12):6412–6420. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]
- 45.Bakowska JC, Jupille H, Fatheddin P, Puertollano R, Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol Biol Cell. 2007;18(5):1683–1692. doi: 10.1091/mbc.E06-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eastman SW, Yassaee M, Bieniasz PD. A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J Cell Biol. 2009;184(6):881–894. doi: 10.1083/jcb.200808041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards TL, Clowes VE, Tsang HT, Connell JW, Sanderson CM, Luzio JP, Reid E. Endogenous spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases AIP4 and AIP5. Biochem J. 2009;423(1):31–39. doi: 10.1042/BJ20082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188(2):143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 49.Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66(16):2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW, Dong JT. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24(20):3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 51.Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70(11):4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Kruppel-like factor, KLF2. Biochem Biophys Res Commun. 2004;316(1):139–148. doi: 10.1016/j.bbrc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 53.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/S0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 54.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–1245. doi: 10.1016/0092-8674(92)90644-R. [DOI] [PubMed] [Google Scholar]
- 55.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112(6):779–791. doi: 10.1016/S0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 56.Peschiaroli A, Scialpi F, Bernassola F, El Sherbini el S, Melino G. The E3 ubiquitin ligase WWP1 regulates DeltaNp63-dependent transcription through Lys63 linkages. Biochem Biophys Res Commun. 2010;402(2):425–430. doi: 10.1016/j.bbrc.2010.10.050. [DOI] [PubMed] [Google Scholar]
- 57.Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25(4):589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- 58.Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312(5777):1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- 59.Shen R, Chen M, Wang YJ, Kaneki H, Xing L, O’Keefe RJ, Chen D. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J Biol Chem. 2006;281(6):3569–3576. doi: 10.1074/jbc.M506761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung CH, Parker JS, Ely K, Carter J, Yi Y, Murphy BA, Ang KK, El-Naggar AK, Zanation AM, Cmelak AJ, Levy S, Slebos RJ, Yarbrough WG. Gene expression profiles identify epithelial-to-mesenchymal transition and activation of nuclear factor-kappaB signaling as characteristics of a high-risk head and neck squamous cell carcinoma. Cancer Res. 2006;66(16):8210–8218. doi: 10.1158/0008-5472.CAN-06-1213. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Z, Liu R, Chen C (2011) The WWP1 ubiquitin E3 ligase increases TRAIL resistance in breast cancer. Int J Cancer. doi:10.1002/ijc.26122 [DOI] [PubMed]
- 62.Galinier R, Gout E, Lortat-Jacob H, Wood J, Chroboczek J. Adenovirus protein involved in virus internalization recruits ubiquitin-protein ligases. Biochemistry. 2002;41(48):14299–14305. doi: 10.1021/bi020125b. [DOI] [PubMed] [Google Scholar]
- 63.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168(1):89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heidecker G, Lloyd PA, Fox K, Nagashima K, Derse D. Late assembly motifs of human T cell leukemia virus type 1 and their relative roles in particle release. J Virol. 2004;78(12):6636–6648. doi: 10.1128/JVI.78.12.6636-6648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92(17):7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patnaik A, Wills JW. In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J Virol. 2002;76(6):2789–2795. doi: 10.1128/JVI.76.6.2789-2795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heidecker G, Lloyd PA, Soheilian F, Nagashima K, Derse D. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T cell leukemia virus type 1. J Virol. 2007;81(18):9769–9777. doi: 10.1128/JVI.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rauch S, Martin-Serrano J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J Virol. 2011;85(7):3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen CS, Bellier A, Kao CY, Yang YL, Chen HD, Los FC, Aroian RV. WWP-1 is a novel modulator of the DAF-2 insulin-like signaling network involved in pore-forming toxin cellular defenses in Caenorhabditis elegans. PLoS One. 2010;5(3):e9494. doi: 10.1371/journal.pone.0009494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanazawa I. Molecular pathology of dentatorubral-pallidoluysian atrophy. Philos Trans R Soc Lond B. 1999;354(1386):1069–1074. doi: 10.1098/rstb.1999.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qin H, Pu HX, Li M, Ahmed S, Song J. Identification and structural mechanism for a novel interaction between a ubiquitin ligase WWP1 and Nogo-A, a key inhibitor for central nervous system regeneration. Biochemistry. 2008;47(51):13647–13658. doi: 10.1021/bi8017976. [DOI] [PubMed] [Google Scholar]
- 72.Patel H, Cross H, Proukakis C, Hershberger R, Bork P, Ciccarelli FD, Patton MA, McKusick VA, Crosby AH. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat Genet. 2002;31(4):347–348. doi: 10.1038/ng937. [DOI] [PubMed] [Google Scholar]
- 73.Carrano AC, Liu Z, Dillin A, Hunter T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature. 2009;460(7253):396–399. doi: 10.1038/nature08130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsumoto H, Maruse H, Inaba Y, Yoshizawa K, Sasazaki S, Fujiwara A, Nishibori M, Nakamura A, Takeda S, Ichihara N, Kikuchi T, Mukai F, Mannen H. The ubiquitin ligase gene (WWP1) is responsible for the chicken muscular dystrophy. FEBS Lett. 2008;582(15):2212–2218. doi: 10.1016/j.febslet.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 75.Pirozzi G, McConnell SJ, Uveges AJ, Carter JM, Sparks AB, Kay BK, Fowlkes DM. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272(23):14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 76.Dhananjayan SC, Ramamoorthy S, Khan OY, Ismail A, Sun J, Slingerland J, O’Malley BW, Nawaz Z. WW domain binding protein-2, an E6-associated protein interacting protein, acts as a coactivator of estrogen and progesterone receptors. Mol Endocrinol. 2006;20(10):2343–2354. doi: 10.1210/me.2005-0533. [DOI] [PubMed] [Google Scholar]