Abstract
AGC kinases are important regulators of cell growth, metabolism, division, and survival in mammalian systems. Mutation or deregulation of members of this family of protein kinases contribute to the pathogenesis of many human diseases, including cancer and diabetes. Although AGC kinases are conserved in the plant kingdom, little is known about their molecular functions and targets. Some of the best-studied plant AGC kinases mediate auxin signaling and are thereby involved in the regulation of growth and morphogenesis. Furthermore, certain members are regulated by lipid-derived signals via the 3-phosphoinositide-dependent kinase 1 (PDK1) and the kinase target of rapamycin (TOR), similar to its animal counterparts. In this review, we discuss recent findings on plant AGC kinases that unravel important roles in the regulation of plant growth, immunity and cell death, and connections to stress-induced mitogen-activated protein kinase signaling cascades.
Keywords: AGC kinases, PDK1, TOR, Plant immunity, Cell death
Introduction
Protein phosphorylation is a conserved strategy used to regulate various cellular functions in prokaryotes and eukaryotes. In animals, AGC kinases regulate signaling events that affect cell size, cell number, and cell death, and thereby influence growth and morphogenesis, and dysfunction of members of this kinase group cause various important diseases, including cancer and diabetes [1]. The AGC family of protein kinases defines a group of serine/threonine protein kinases that share sequence similarity in their catalytic kinase domains with cAMP-dependent protein kinase A (PKA), cGMP-dependent protein kinase G (PKG) and phospholipid-dependent protein kinase C (PKC) [1]. AGC kinases were found to have important functions in the model plant Arabidopsis (Arabidopsis thaliana) [2] and members of this family of kinases have also been studied in other plant species such as rice (Oryza sativa), tomato (Solanum lycopersicum), and bean (Phaseolus vulgaris), but functions are only known for a few plant AGC kinases [3–5].
The genome of Arabidopsis encodes 39 AGC kinases that are classified into five subfamilies: AGCVI, AGCVII, AGCVIII, AGC other, and homologs of PDK1 [2]. In Arabidopsis, the largest subfamily is AGCVIII, which comprises 23 AGC kinases subdivided into the groups AGC1, AGC2, AGC3, and AGC4 [6, 7]. Among the best-studied AGC kinases in Arabidopsis, there is the AGC3 kinase PINOID, which phosphorylates PIN-FORMED (PIN) auxin transporters and is involved in determining polar auxin distribution, and the AGC4 kinases PHOTOTROPIN1 (PHOT1) and PHOT2 that mediate blue light signals [8, 9]. This shows that plant AGC kinases are also involved in growth and morphogenesis like its animal counterparts. Furthermore, various reports show that plant AGC kinases participate in the responses to environmental stresses, especially in the regulation of plant immunity [3, 10–12].
Plants have an acute ability to sense and respond to environmental stimuli. Some of the most destructive environmental stresses come from microbial pathogens and the plant has evolved a multilayered innate immune system to resist infection by the majority of pathogenic microorganisms. An early layer in plant immunity relies on pattern recognition receptors that recognize conserved pathogen or microbe-associated molecular patterns (PAMPs or MAMPs), such as bacterial flagellin [13]. Characterized plant PAMP receptors belong to the family of plasma membrane receptor kinases (RKs) and their activation leads to various defense responses such as an oxidative burst, activation of mitogen-activated protein kinases (MAPKs), transcriptional reprogramming, and finally to PAMP-triggered immunity (PTI). For example, the Arabidopsis RK FLS2 recognizes bacterial flagellin through binding of the peptide flg22 and initiates defense signaling [14, 15]. PTI is generally efficient in stopping microbial colonization and therefore successful pathogens have evolved effectors that interfere with PTI and allow infection. Recognition of effectors by primarily intracellular Resistance (R) proteins activates the second branch of plant immunity known as effector-triggered immunity (ETI) [16]. ETI is typically accompanied by a hypersensitive response (HR) that leads to localized programmed cell death and stops further colonization by biotrophic pathogens. Furthermore, various plant hormones are involved in balancing defense responses and pathogens modulate hormone signaling to their benefit. In a simplistic view, salicylic acid (SA) is necessary to combat biotrophic pathogens, whereas jasmonic acid and ethylene are required for defenses against necrotrophs that feed on dead tissue. Nevertheless, recent reports show that most plant hormones such as auxin, gibberellins, abscisic acid, cytokinins, and brassinosteroids play important roles in shaping the outcomes of plant–microbe interactions [17].
In this review, we will focus on recent advances on the role of plant AGC kinases in modulating the outcome of plant–microbe interactions.
Plant AGC kinases are activated by lipid and stress signals via PDK1 and TOR
AGC kinases possess certain common domain architecture that includes a central kinase catalytic domain, a C-terminal regulatory region with a consensus hydrophobic sequence and an N-terminal phospholipid-binding domain, such as pleckstrin homology (PH), conserved region 1 (C1) or conserved region 2 (C2) domains [1]. For many AGC kinases, activation is achieved by phosphorylation of two conserved regulatory motifs: the activation loop in the catalytic domain and the C-terminal hydrophobic sequence [1]. With these domain modules, various AGC kinases function to transduce phospholipid signals to the phosphorylation of downstream targets. Even if most membrane phospholipids have structural roles, a small percentage is induced by environmental and endogenous stimuli and performs important cellular signaling functions. Some key conserved signaling lipids in eukaryotes are phosphatidylinositol (PtdIns) lipids, diacylglycerol (DAG) and phosphatidic acid (PA) [18, 19]. The structural lipid PtdIns is used to produce diverse polyphosphoinositides through phosphorylation and dephosphorylation of the inositol head group by specific phosphoinositide kinases and phosphoinositide phosphatases. Phospholipase C (PLC) can cleave the inositol head group of the polyphosphoinositides to produce membrane-bound DAG and water-soluble inositol polyphosphates. The DAG produced through PLC-mediated hydrolysis can be further phosphorylated to PA via DAG kinase. PA can alternatively be produced through direct hydrolysis of structural phospholipids by phospholipase D (PLD) [18, 19].
In animals, the activation of receptor kinases at the cell surface by growth factors leads to phosphoinositide 3-kinase (PI3K)-mediated phosphorylation of PtdIns(4,5)P2 to generate the lipid messenger PtdIns(3,4,5)P3. Animal AGC kinases are activated downstream of PI3K by PDK1 and mediate diverse cellular responses to these important stimuli [1]. AGC kinases bind to PDK1 through a PDK1-interacting fragment (PIF) domain in their C-terminal hydrophobic sequence that docks to the PIF-binding pocket in PDK1. Subsequently, PDK1-binding to the PIF domain results in the phosphorylation of the activation loop of AGC kinases [20]. PDK1 is itself an AGC kinase with a C-terminal PH domain that can bind to PtdIns(3,4,5)P3 at the plasma membrane or to cytosolic inositol phosphates, which thereby allows the regulation of AGC kinases with different subcellular distribution, such as membrane-bound protein kinase B (PKB, also known as Akt) and soluble S6 kinase [21, 22]. PDK1 shows constitutive as well as lipid-inducible activity and the regulation of PDK1-mediated activation of AGC kinases is thought to be achieved through a previous substrate “priming” through changes in conformation and/or phosphorylation of the AGC kinase hydrophobic segment [20, 23].
PDK1 is conserved in higher and lower plants [5, 24–27]. Similarly to the severe defects displayed by animal organisms or cell lines with reduced PDK1 activity [20], silencing of both tomato PDK1 genes is lethal [3] and PDK1 gene disruption in the moss Physcomitrella patens impairs normal growth and resistance to abiotic stresses [25]. In rice, mutation of the only PDK1 encoding gene leads to a mild dwarf phenotype and its overexpression leads to enhanced disease resistance [5]. Interestingly, Arabidopsis plants mutated in the two PDK1-encoding genes look like wild-type plants and they are only impaired in the growth promotion conferred by the mutualistic fungi Piriformospora indica [28]. In sum, plant PDK1s regulate signaling pathways necessary for proper growth in normal and stress conditions similar to what happens in animals. Plants do not produce PtdIns(3,4,5)P3 and Arabidopsis PDK1 was shown to bind and be activated by a wider array of lipid molecules including PA and PtdIns(4,5)P2 [24, 29]. Whereas PINOID was found to display phospholipid binding capability [26], PDK1 is the only AGC kinase in plants with a known lipid-binding domain, suggesting that other plant AGC kinases may normally integrate lipid signals via PDK1 or other upstream regulators [2]. PDK1 has been shown to phosphorylate various AGC kinases in plants [5, 24, 26, 27, 30, 31]. Similar to animals, most plant AGC kinases possess a PIF domain at the C-terminal hydrophobic region that is involved in the interaction with and activation by PDK1 [24, 26]. Nevertheless, the different subgroups of AGC kinases in Arabidopsis display differences in their PIF domains and in their regulation by PDK1 [2, 24, 27].
In mammals, the protein kinase TOR (target of rapamycin) regulates some AGC kinases through phosphorylation of the C-terminal hydrophobic segment [1, 32]. TOR is a serine/threonine kinase that is highly conserved among eukaryotes and controls cell growth and metabolism in response to nutrients, growth factors, and stress [32, 33]. TOR forms distinct complexes with proteins that are conserved across kingdoms, such as KOG1 (kontroller of growth 1)/RAPTOR1 (regulatory-associated protein of TOR), which is thought to serve as a scaffold that recruits substrates for TOR, and LST8 that may function to stimulate TOR kinase activity [32]. TOR, RAPTOR, and LST8 exist in plants and the corresponding Arabidopsis mutants show severe growth defects suggesting that the TOR pathway plays an essential role also in plant development [34–37]. Furthermore, TOR seems important for plant responses to stress, as reduced and enhanced expression of the single Arabidopsis TOR-encoding gene correlate with reduced and enhanced growth and stress resistance, respectively [38]. In line with a function of TOR complexes in stress resistance, RAPTOR1 was found to regulate the activity of the AGCVI kinase S6K1 in response to osmotic stress [30]. Interestingly, overexpression of PLDα3 in Arabidopsis, which leads to increased root growth and hyperosmotic tolerance, results in enhanced accumulation of TOR and OXI1 transcripts under hyperosmotic stress [39]. All these data suggest a potential stress-induced pathway including phospholipid signaling to TOR and AGC kinases similar to animals.
Downstream targets of AGC kinases
Animal AGC kinases phosphorylate a wide range of proteins and the targets of these kinases are involved in the regulation of phosphorylation, transcription, translation, apoptosis, cell cycle, and metabolism, indicating that a large amount of cellular processes are affected by AGC kinases [1, 40]. These kinases have similar consensus sequences for substrate phosphorylation and as a consequence, certain proteins are substrates of more than one AGC kinase allowing one cellular process to be regulated by various stimuli. The preferred consensus sequences for animal AGC kinases normally include basic residues (lysine/arginine) N-terminal of the phosphorylatable serine or threonine [1]. For example, the animal AGC kinases Akt, ribosomal S6 kinase (RSK) and p70 S6 kinase (S6K) phosphorylate substrates in the RxRxxS/T motif (R, arginine; S, serine; T, threonine and x, any amino acid) [40]. Plant AGC kinases were shown to have similar substrate preferences as mammalian enzymes [24, 41]. For instance, Arabidopsis AGC1-1 and OXI1 can use the kemptide peptide as substrate and are inhibited by a kinase inhibitor peptide derived from PKI [24], thereby resembling animal PKA mode of function. Furthermore, similar to mammalian cells, the Arabidopsis S6K1 was found to phosphorylate the C-terminal segment of the ribosomal protein S6 [30]. Therefore, although not many kinase substrates are known for plant AGC kinases, substrate preference seems to be conserved between plant and animal AGC kinases.
Using yeast-two-hybrid screens, different groups have identified homologues of tomato Pti1 (Pto-interacting protein 1) as interactors and downstream targets of the OXI1 kinase in Arabidopsis and rice [11, 42–44]. Tomato Pti1 is a serine/threonine protein kinase that is activated by the tomato kinase Pto (resistance to Pseudomonas syringae pv. tomato) through phosphorylation on a conserved threonine (T233) in the kinase activation loop [45]. Substituting the corresponding threonine with an alanine in Arabidopsis Pti1-2 (T238) or in rice OsPti1a (T233) compromises OXI1-mediated phosphorylation, suggesting that the Pti1 phosphorylation site is conserved across species even if the upstream kinases may differ [11, 42]. As compared with the animal AGC kinase substrates, the phosphorylated threonine lies within a highly conserved motif that includes only one basic residue, which is an arginine at position-4 [42, 46], but the importance of this residue for OXI1-mediated phosphorylation has not been demonstrated.
The phosphoproteome of Arabidopsis oxi1 plants was recently analyzed using two approaches. One used two-dimensional PAGE coupled with Pro-Q diamond staining of phosphoproteins and the other stable isotope labeling of whole plants followed by phosphopeptide identification by mass spectrometry [47]. These approaches allowed the identification of proteins that were differentially phosphorylated in oxi1 as compared with wild-type Ws-2, and therefore putative OXI1 kinase substrates, upon pathway activation by cellulase treatment. Interestingly, among the proteins identified, phosphopeptides corresponding to PDK1.2 and PLDγ showed decreased and increased abundance, respectively pointing to some possible feedback regulation of the pathway. Phosphoproteome analyses of plants carrying loss-of-function and/or gain-of-function mutations in AGC kinases could be a good strategy to find their potential phosphorylation targets. Recently, an ATP analog-sensitive version of the tomato Adi3 AGC kinase was identified, through the mutation of a “gatekeeper” residue in the ATP-binding pocket to a less bulky amino acid that thereby allows the use of bulky ATP analogs [48]. The advantage of this method is that only the modified kinase can use the bulky ATP analogs and thus can be used to identify their in vivo phosphorylation substrates from plant extracts. The recent expansion of technologies for the identification of plant kinase targets will surely prove useful for the identification of substrates of plant AGC kinases.
AGC kinases in plant immunity and modulation of MAPK cascades
MAPK cascades are an important component of the signaling events induced during PTI [49]. They are minimally composed of a MAPKKK (MAPKK kinase), a MAPKK (MAPK kinase) and a MAPK, which activate each other by phosphorylation and link perception of external cues by upstream receptors with the activation of downstream targets. In Arabidopsis, the FLS2-mediated recognition of the PAMP flg22 activates two MAPK cascades [50, 51]. One cascade includes the MAPKKK MEKK1, the partially redundant MAPKKs MKK1 and MKK2, and the MAPK MPK4 and negatively regulates plant immunity. Arabidopsis mekk1, mkk1/mkk2 double and mpk4 mutants display similar phenotypes including spontaneous cell death, accumulation of SA and constitutive disease resistance against biotrophic pathogens [51, 52]. MKK4/MKK5 and MPK3/MPK6 constitute another MAPK cascade that acts positively on PTI [50].
Plant AGC protein kinases have been shown to participate in immune responses and regulate pathogen-induced MAPK cascades. The tomato MAPKKKα functions in the resistance to the bacterial speck disease produced by P. syringae pv. tomato (Pst) infection, downstream of the Pto kinase [53, 54]. Pto is a serine/threonine protein kinase that recognizes the Pst effectors AvrPto and AvrPtoB and initiates ETI [54, 55]. MAPKKKα, which belongs to the same MAPKKK family as MEKK1, positively regulates the Pto-mediated cell death response against Pst AvrPto infection and activates at least two MAPK cascades [53]. The AGC kinase Adi3 (AvrPto-dependent Pto-interacting protein 3) was identified in a search for proteins that interact with the Pto kinase in an AvrPto-dependent manner and was found to be a direct phosphorylation target of Pto [3]. Adi3 functions to suppress MAPKKKα-dependent cell death, as virus-induced gene silencing of Adi3 in tomato led to the appearance of spontaneous cell death lesions and co-silencing MAPKKKα abolished this response [3]. Furthermore, Adi3 gene silencing led to reduced growth and enhanced resistance, suggesting that Adi3 is a broader negative regulator of plant immune responses. Tomato PDK1 regulates Adi3 activity, and consequently cell death, via phosphorylation of serine 539 (S539) of the Adi3 activation loop, which is necessary for Adi3 nuclear accumulation and cell death suppression activity [3, 56]. Therefore, it was proposed that during ETI, the Pto/AvrPto complex sequesters Adi3 and impairs its nuclear accumulation, which allows the HR-associated cell death to occur [57] (Fig. 1a). Interestingly, the tomato PLC-encoding gene PLC6 is required for resistance to Pst AvrPto [58], further implicating phospholipid signaling in Pto-mediated resistance. The Pti1 kinase previously mentioned to be a direct target of Pto, was also found to positively regulate Pto-mediated cell death when over-expressed in tobacco (Nicotiana tabacum) [44], although no loss-of-function phenotype has been found for this kinase likely due to redundancy [57]. Therefore the Pto kinase triggers various parallel kinase cascades to modulate cell death and resistance in response to P. syringae infection.
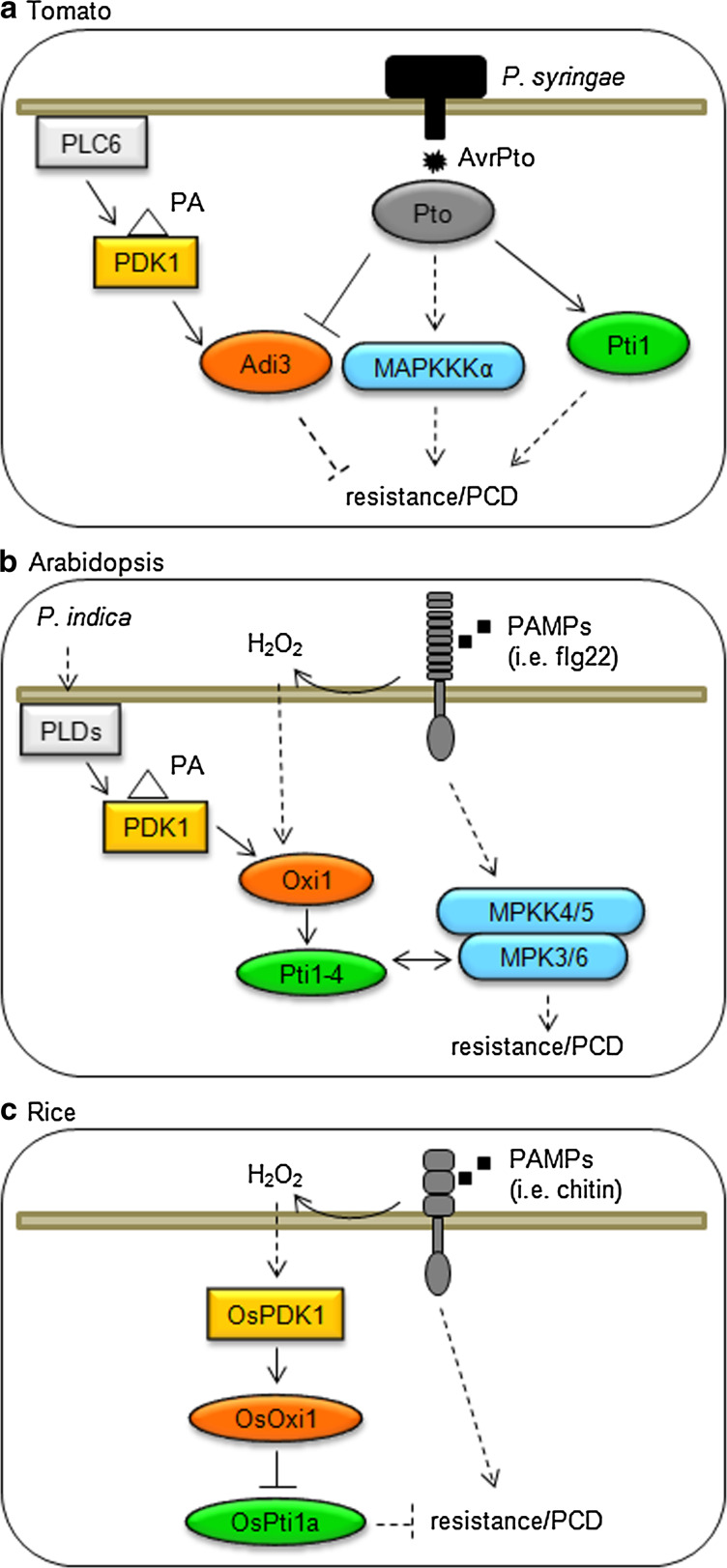
Fig. 1.
Schematic representation of AGC kinase signaling cascades in plant immune responses. a The tomato AGC kinase Adi3 is regulated by the PDK1 and Pto kinases to negatively modulate disease resistance and programmed cell death (PCD). Two parallel kinase pathways mediated by MAPKKKα and Pti1 kinases are activated downstream of Pto and also participate in the modulation of resistance and PCD. b The Arabidopsis AGC kinase OXI1 is activated by PDK1 and H2O2 and is required for disease resistance. OXI1 modulates the stress-induced activation of the MAPKs MPK3 and MPK6, putatively through its downstream kinase Pti1-4. c In rice, a phosphorylation cascade including OsPDK1, the AGC kinase OsOXI1 and the downstream kinase OsPti1a was shown to positively regulate disease resistance and cell death in response to H2O2 and pathogen recognition. Filled arrows represent direct regulation. Dashed arrows represent processes comprising various or unknown steps. Double-headed arrows represent protein association. PA phosphatidic acid
In Arabidopsis, OXI1 is required for complete activation of MPK3 and MPK6 by H2O2 and cellulase treatments and for resistance against the biotrophic pathogens Hyaloperonospora arabidopsidis and P. syringae [10, 12] (Fig. 1b). OXI1 gene expression and kinase activity are rapidly induced by PAMPs and danger signals such as treatment with Flg22, cellulase, and H2O2 in Arabidopsis leaves and roots [10, 59]. Interestingly, OXI1 transcriptional induction is reduced in Arabidopsis mutant plants in the NADPH oxidase AtrbohD or by treatment with the NADPH oxidase inhibitor DPI (diphenylene iodinium) [12]. AtrbohD is responsible of most H2O2 produced upon treatment with PAMPs and biotrophic pathogens [60, 61], indicating that the production of reactive oxygen species by NADPH oxidases mediates OXI1 induction during plant–pathogen interactions. As mentioned before, OXI1 regulates a group of kinases named Pti1-1 to Pti1-4 due to their similarity with tomato Pti1 [42, 43]. Interestingly, the stress-inducible Pti1-4 was found to interact with MPK6 in vivo and MPK3 and MPK6 were found to phosphorylate Pti1-4 and Oxi1 in in vitro kinase assays [43]. These results suggest a complex picture where the OXI1-Pti1-4 signaling pathway could regulate MAPKs and/or be regulated by MAPKs. As is characteristic for AGC kinases, OXI1 activity is regulated by PDK1 and PA was reported to activate OXI1 in a PDK1-dependent manner [24]. Interestingly, H2O2 and flg22 lead to PDK1-independent, but OXI1-dependent activation of Pti1 kinases [42], showing that other stress-induced kinases function upstream of OXI1. Recently, a rice OXI1 signaling cascade composed by OsPDK1, OsOXI1, and OsPti1a was shown to regulate defenses against the causative agent of the rice blast fungus Magnaporthe oryzae [5, 11]. In rice, perception of the fungal PAMP chitin by a receptor kinase complex at the plasma membrane triggers various PTI hallmarks, such as an oxidative burst [62]. OsPDK1 and OsOXI1 are activated by chitin and H2O2 treatments [5, 11], and therefore seem to be activated by PAMP signaling similar to OXI1 in Arabidopsis. Different to the observations done in tomato, the downstream kinase OsPti1a appears to negatively regulate ETI-mediated resistance and cell death [44]. OsOXI1 signaling increases disease resistance by phosphorylating OsPti1a and thereby releasing OsPti1a-mediated inhibition of disease resistance [11] (Fig. 1c).
Other AGC kinases have also been involved in plant–pathogen interactions. The protein kinase PvPK-1 is induced in bean cell cultures treated with the fungal pathogen Colletotrichum lindemuthianum and the viroid-induced protein kinase PKV is transcriptionally induced in tomato leaves following Potato spindle tuber viroid (PSTVd) infection [4, 63]. Similarly to PSTVd infection symptoms, transgenic tobacco plants overexpressing tomato PKV exhibited dwarfism and a reduced root system, whereas antisense suppression of PKV resulted in enhanced growth and root development [63]. Interestingly, PKV overexpression symptoms could be rescued by exogenous application of the hormone gibberellic acid, suggesting that PSTVd regulates plant growth via PKV modulation of GA levels. These data shows that AGC kinases have evolved in various plant species to regulate plant immunity.
What is the mechanism of cell death regulation by AGC kinases?
Similar to animals, the present data suggest that a common theme in the function of plant AGC kinases is the regulation of programmed cell death, although how this is accomplished is not clear. In animals, AGC kinases regulate apoptotic cell death and mediate autophagy induction by TOR [1, 64, 65]. Autophagy is a degradation process found in fungi and higher eukaryotes that is characterized by the formation of double-membrane vesicles, called autophagosomes, which engulf and target cytoplasmic contents to the vacuole and lysosome for degradation. Autophagy-related proteins (ATGs) execute autophagy and Arabidopsis ATG loss-of-function mutants show that autophagy plays an important role in development and responses to biotic and abiotic stresses [66–68]. Defects in autophagy leading to reduced or enhanced autophagic flux have been shown to result in inappropriate activation of cell death programs in animals and plants, such as senescence and apoptosis [69]. The TOR kinase complex was recently shown to negatively regulate autophagy in plants [70, 71]. Furthermore, Adi3 was found to interact with the tomato autophagy protein ATG8h in yeast-two-hybrid [72]. Recent reports demonstrate that both apoptotic and autophagy-related cell death programs play an important role in plant immunity through regulation of HR development [66, 73]. Therefore, it will be of interest to assess whether the role of AGC kinases in the regulation of plant cell death programs is related to a function in the modulation of autophagy.
AGC kinases are required for the mutualistic interaction between Arabidopsis and Piriformospora indica
Recently, Arabidopsis OXI1 and its closest homologue AGC2-2 were shown to participate in the symbiotic interaction between Arabidopsis and the basidiomycete P. indica [28]. P. indica colonizes the roots of an exceptionally large range of plants and confers beneficial traits to its hosts such as increased growth and resistance to abiotic and biotic stresses [74]. The colonization of Arabidopsis roots starts with a biotrophic growth phase that is followed by a cell death-associated colonization phase [59]. The broad host range of P. indica suggests the beneficial interaction relies on general and conserved recognition and signaling systems. Accordingly, a fungal cell wall extract can induce growth promotion and mimics the presence of the fungus at early stages [75] and two Arabidopsis leucine-rich repeat proteins found at the plasma membrane play a role in the interaction [76]. These results suggest that the interaction may be based on the recognition of a conserved PAMP at the cell surface. Using Arabidopsis and barley (Hordeum vulgare) mutant plants, it was established that the symbiosis requires and alters various hormone signaling pathways. Whereas P. indica-induced systemic resistance depends on a functional jasmonate signaling pathway [77], growth promotion requires gibberellic acid and cytokinins [78, 79], and ethylene seems crucial for the beneficial outcome of the interaction [80]. Furthermore, Arabidopsis mutants compromised in SA accumulation allowed increased fungal colonization and P. indica suppresses various hallmarks of PTI showing that suppression rather than evasion of plant immunity is required for successful colonization [59].
A recent study used genetic and physiological analyses to show an important role for phospholipid and OXI1 signaling in the establishment of the beneficial interaction. At low beneficial doses, P. indica inoculation induces PA accumulation via the phospholipases PLDα1 and PLDγ, which in turn leads to PDK1 signaling to OXI1 and AGC2-2 [28]. Arabidopsis plants carrying mutations in the two PDK1-encoding genes present in Arabidopsis PDK1.1 and PDK1.2, as well as single mutant plants in OXI1 and AGC2-2 are impaired in the interaction and do not show growth promotion upon co-cultivation with P. indica [28]. Furthermore, Arabidopsis MPK3 and MPK6 are rapidly activated in roots by treatment with the P. indica cell wall extract and mpk6 mutant plants do not show P. indica-induced growth promotion [75]. It is not clear what molecular function OXI1 and AGC2-2 play in the signaling cascade induced by P. indica. P. indica changes the oxidative status of the host [74] and suppresses basal and PAMP-induced H2O2 production in roots [28, 59]. Thus it seems possible that OXI1 (and AGC2-2) could play a role in modulating the oxidative stress and/or MAPK activities upon P. indica perception. Interestingly, Arabidopsis oxi1 agc2-2 double-knockout mutant plants could not be isolated, indicating some redundant and essential roles for these kinases [28].
Conclusions
The data gathered so far indicate that, similar to its animal counterparts, plant AGC kinases play important roles in plant immunity and cell death regulation. Furthermore, the involvement of AGC kinases in the relationship with growth-promoting endophytic fungi as well as in auxin and light signaling position AGC kinases at the intersection between environmental stress responses and internal growth programs. Unfortunately, compared to the amount of information on the molecular function of AGC kinases in animals, research on plant AGC kinases is much less developed. For instance, only few upstream regulators and kinase substrates of AGC kinases are known in plants. At least in Arabidopsis, it seems that a likely source of complexity in the study of AGC kinases comes from functional redundancies between close homologues [28, 81–83]. Recent technical advances in mass spectrometry have enabled the development of semi-quantitative methods to analyze protein complex constituents and its post-translational modifications [84]. As a consequence, it now seems feasible to study the composition and post-translational modifications of AGC kinase protein complexes in vivo, and thereby to identify protein partners and amino acid residues that are important upon activation of a certain cellular process. Furthermore, mass spectrometry analysis of the phosphoproteome of plants carrying loss-of-function and gain-of-function mutations in AGC kinases will help in finding potential targets and signaling partners in planta. The recent development of new technologies for the study of kinases in plants will surely contribute to advance our knowledge on AGC kinases and the identification of their targets.
Acknowledgments
The project is supported by funding from the following projects: ANR MAPK, EU ADYSARC, and Systems Biology SHIPREC. Due to space limitations, we apologize to all colleagues whose work has not been included in the review.
References
- 1.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11(1):9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 2.Bogre L, Okresz L, Henriques R, Anthony RG. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 2003;8(9):424–431. doi: 10.1016/S1360-1385(03)00188-2. [DOI] [PubMed] [Google Scholar]
- 3.Devarenne TP, Ekengren SK, Pedley KF, Martin GB. Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. EMBO J. 2006;25(1):255–265. doi: 10.1038/sj.emboj.7600910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawton MA, Yamamoto RT, Hanks SK, Lamb CJ. Molecular cloning of plant transcripts encoding protein kinase homologs. Proc Natl Acad Sci USA. 1989;86(9):3140–3144. doi: 10.1073/pnas.86.9.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsui H, Miyao A, Takahashi A, Hirochika H. Pdk1 kinase regulates basal disease resistance through the OsOxi1-OsPti1a phosphorylation cascade in rice. Plant Cell Physiol. 2010;51(12):2082–2091. doi: 10.1093/pcp/pcq167. [DOI] [PubMed] [Google Scholar]
- 6.Galvan-Ampudia CS, Offringa R. Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 2007;12(12):541–547. doi: 10.1016/j.tplants.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, McCormick S. AGCVIII kinases: at the crossroads of cellular signaling. Trends Plant Sci. 2009;14(12):689–695. doi: 10.1016/j.tplants.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Ding Z, Galvan-Ampudia CS, Demarsy E, Langowski L, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, Friml J. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis . Nat Cell Biol. 2011;13(4):447–452. doi: 10.1038/ncb2208. [DOI] [PubMed] [Google Scholar]
- 9.Robert HS, Offringa R. Regulation of auxin transport polarity by AGC kinases. Curr Opin Plant Biol. 2008;11(5):495–502. doi: 10.1016/j.pbi.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Rentel MC, Lecourieux D, Ouaked F, Usher SL, Petersen L, Okamoto H, Knight H, Peck SC, Grierson CS, Hirt H, Knight MR. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis . Nature. 2004;427(6977):858–861. doi: 10.1038/nature02353. [DOI] [PubMed] [Google Scholar]
- 11.Matsui H, Yamazaki M, Kishi-Kaboshi M, Takahashi A, Hirochika H. AGC kinase OsOxi1 positively regulates basal resistance through suppression of OsPti1a-mediated negative regulation. Plant Cell Physiol. 2010;51(10):1731–1744. doi: 10.1093/pcp/pcq132. [DOI] [PubMed] [Google Scholar]
- 12.Petersen LN, Ingle RA, Knight MR, Denby KJ. OXI1 protein kinase is required for plant immunity against Pseudomonas syringae in Arabidopsis . J Exp Bot. 2009;60(13):3727–3735. doi: 10.1093/jxb/erp219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009;150(4):1638–1647. doi: 10.1104/pp.109.139709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18(2):465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428(6984):764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 16.Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 2011;12(9):817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- 17.Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 18.Munnik T, Nielsen E. Green light for polyphosphoinositide signals in plants. Curr Opin Plant Biol. 2011;14(5):489–497. doi: 10.1016/j.pbi.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Testerink C, Munnik T. Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J Exp Bot. 2011;62(7):2349–2361. doi: 10.1093/jxb/err079. [DOI] [PubMed] [Google Scholar]
- 20.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15(2):161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 21.Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Peter Downes C, Safrany ST, Alessi DR, van Aalten DM. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 2004;23(20):3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McManus EJ, Collins BJ, Ashby PR, Prescott AR, Murray-Tait V, Armit LJ, Arthur JS, Alessi DR. The in vivo role of PtdIns(3,4,5)P3 binding to PDK1 PH domain defined by knocking mutation. EMBO J. 2004;23(10):2071–2082. doi: 10.1038/sj.emboj.7600218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biondi RM. Phosphoinositide-dependent protein kinase 1, a sensor of protein conformation. Trends Biochem Sci. 2004;29(3):136–142. doi: 10.1016/j.tibs.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Anthony RG, Henriques R, Helfer A, Meszaros T, Rios G, Testerink C, Munnik T, Deak M, Koncz C, Bogre L. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis . EMBO J. 2004;23(3):572–581. doi: 10.1038/sj.emboj.7600068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dittrich AC, Devarenne TP. Characterization of a PDK1 homologue from the moss Physcomitrella patens . Plant Physiol. 2012;158(2):1018–1033. doi: 10.1104/pp.111.184572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zegzouti H, Anthony RG, Jahchan N, Bogre L, Christensen SK. Phosphorylation and activation of PINOID by the phospholipid signaling kinase 3-phosphoinositide-dependent protein kinase 1 (PDK1) in Arabidopsis . Proc Natl Acad Sci USA. 2006;103(16):6404–6409. doi: 10.1073/pnas.0510283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zegzouti H, Li W, Lorenz TC, Xie M, Payne CT, Smith K, Glenny S, Payne GS, Christensen SK. Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J Biol Chem. 2006;281(46):35520–35530. doi: 10.1074/jbc.M605167200. [DOI] [PubMed] [Google Scholar]
- 28.Camehl I, Drzewiecki C, Vadassery J, Shahollari B, Sherameti I, Forzani C, Munnik T, Hirt H, Oelmuller R. The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis . PLoS Pathog. 2011;7(5):e1002051. doi: 10.1371/journal.ppat.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deak M, Casamayor A, Currie RA, Downes CP, Alessi DR. Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett. 1999;451(3):220–226. doi: 10.1016/S0014-5793(99)00556-6. [DOI] [PubMed] [Google Scholar]
- 30.Mahfouz MM, Kim S, Delauney AJ, Verma DP. Arabidopsis target of rapamycin interacts with raptor, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18(2):477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otterhag L, Gustavsson N, Alsterfjord M, Pical C, Lehrach H, Gobom J, Sommarin M. Arabidopsis PDK1: identification of sites important for activity and downstream phosphorylation of S6 kinase. Biochimie. 2006;88(1):11–21. doi: 10.1016/j.biochi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Robaglia C, Thomas M, Meyer C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol. 2012;15(3):301–307. doi: 10.1016/j.pbi.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Anderson GH, Veit B, Hanson MR. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 2005;3:12. doi: 10.1186/1741-7007-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deprost D, Truong HN, Robaglia C, Meyer C. An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem Biophys Res Commun. 2005;326(4):844–850. doi: 10.1016/j.bbrc.2004.11.117. [DOI] [PubMed] [Google Scholar]
- 36.Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA. 2002;99(9):6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreau M, Azzopardi M, Clement G, Dobrenel T, Marchive C, Renne C, Martin-Magniette ML, Taconnat L, Renou JP, Robaglia C, Meyer C. Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell. 2012;24(2):463–481. doi: 10.1105/tpc.111.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meyer C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8(9):864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong Y, Pan X, Welti R, Wang X. The effect of phospholipase Dalpha3 on Arabidopsis response to hyperosmotic stress and glucose. Plant Signal Behav. 2008;3(12):1099–1100. doi: 10.4161/psb.3.12.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, Ren JM, Hornbeck P, Cantley LC, Gygi SP, Rush J, Comb MJ. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal. 2010;3(136):ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang F, Zago MK, Abas L, van Marion A, Galvan-Ampudia CS, Offringa R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell. 2010;22(4):1129–1142. doi: 10.1105/tpc.109.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anthony RG, Khan S, Costa J, Pais MS, Bogre L. The Arabidopsis protein kinase PTI1-2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J Biol Chem. 2006;281(49):37536–37546. doi: 10.1074/jbc.M607341200. [DOI] [PubMed] [Google Scholar]
- 43.Forzani C, Carreri A, de la Fuente van Bentem S, Lecourieux D, Lecourieux F, Hirt H. The Arabidopsis protein kinase Pto-interacting 1–4 is a common target of the oxidative signal-inducible 1 and mitogen-activated protein kinases. FEBS J. 2011;278(7):1126–1136. doi: 10.1111/j.1742-4658.2011.08033.x. [DOI] [PubMed] [Google Scholar]
- 44.Zhou J, Loh YT, Bressan RA, Martin GB. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell. 1995;83(6):925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 45.Sessa G, D’Ascenzo M, Martin GB. The major site of the pti1 kinase phosphorylated by the pto kinase is located in the activation domain and is required for pto-pti1 physical interaction. Eur J Biochem. 2000;267(1):171–178. doi: 10.1046/j.1432-1327.2000.00979.x. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi A, Agrawal GK, Yamazaki M, Onosato K, Miyao A, Kawasaki T, Shimamoto K, Hirochika H. Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell. 2007;19(9):2940–2951. doi: 10.1105/tpc.106.047142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howden AJ, Salek M, Miguet L, Pullen M, Thomas B, Knight MR, Sweetlove LJ. The phosphoproteome of Arabidopsis plants lacking the oxidative signal-inducible1 (OXI1) protein kinase. New Phytol. 2010;190(1):49–56. doi: 10.1111/j.1469-8137.2010.03582.x. [DOI] [PubMed] [Google Scholar]
- 48.Dittrich AC, Devarenne TP. An ATP analog-sensitive version of the tomato cell death suppressor protein kinase Adi3 for use in substrate identification. Biochim Biophys Acta. 2012;1824(2):269–273. doi: 10.1016/j.bbapap.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 49.Pitzschke A, Schikora A, Hirt H. MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol. 2009;12(4):421–426. doi: 10.1016/j.pbi.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 50.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415(6875):977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 51.Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis . J Biol Chem. 2006;281(48):36969–36976. doi: 10.1074/jbc.M605319200. [DOI] [PubMed] [Google Scholar]
- 52.Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18(12):1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- 53.del Pozo O, Pedley KF, Martin GB. MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 2004;23(15):3072–3082. doi: 10.1038/sj.emboj.7600283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262(5138):1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 55.Kim YJ, Lin NC, Martin GB. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109(5):589–598. doi: 10.1016/S0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 56.Ek-Ramos MJ, Avila J, Cheng C, Martin GB, Devarenne TP. The T-loop extension of the tomato protein kinase AvrPto-dependent Pto-interacting protein 3 (Adi3) directs nuclear localization for suppression of plant cell death. J Biol Chem. 2010;285(23):17584–17594. doi: 10.1074/jbc.M110.117416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh CS, Martin GB. Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 2011;16(3):132–140. doi: 10.1016/j.tplants.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 58.Vossen JH, Abd-El-Haliem A, Fradin EF, van den Berg GC, Ekengren SK, Meijer HJ, Seifi A, Bai Y, ten Have A, Munnik T, Thomma BP, Joosten MH. Identification of tomato phosphatidylinositol-specific phospholipase-C (PI-PLC) family members and the role of PLC4 and PLC6 in HR and disease resistance. Plant J. 2010;62(2):224–239. doi: 10.1111/j.1365-313X.2010.04136.x. [DOI] [PubMed] [Google Scholar]
- 59.Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, Lipka V, Kogel KH, Schafer P. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica . Plant Physiol. 2011;156(2):726–740. doi: 10.1104/pp.111.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torres MA, Dangl JL, Jones JD. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA. 2002;99(1):517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, Chen S, Tang X, Zhou JM. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe. 2007;1(3):175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 62.Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, Shibuya N. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64(2):204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammond RW, Zhao Y. Characterization of a tomato protein kinase gene induced by infection by Potato spindle tuber viroid. Mol Plant Microbe Interact. 2000;13(9):903–910. doi: 10.1094/MPMI.2000.13.9.903. [DOI] [PubMed] [Google Scholar]
- 64.Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4(7):851–865. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 65.Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, Kadowaki T, Hay N. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15(17):2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jorgensen LB, Jones JD, Mundy J, Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis . Cell. 2009;137(4):773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 67.Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M, Bassham DC, Vierstra RD, Parker JE, Bautor J, Molina A, Escudero V, Shindo T, van der Hoorn RA, Gust AA, Nurnberger T. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011;66(5):818–830. doi: 10.1111/j.1365-313X.2011.04546.x. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5(7):954–963. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- 69.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X. Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G. Autophagy. 2008;4(2):151–175. [Google Scholar]
- 70.Liu Y, Bassham DC. TOR is a negative regulator of autophagy in Arabidopsis thaliana . PLoS One. 2010;5(7):e11883. doi: 10.1371/journal.pone.0011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez-Perez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii . Plant Physiol. 2010;152(4):1874–1888. doi: 10.1104/pp.109.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Devarenne TP. The plant cell death suppressor Adi3 interacts with the autophagic protein Atg8h. Biochem Biophys Res Commun. 2011;412(4):699–703. doi: 10.1016/j.bbrc.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 73.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121(4):567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 74.Qiang X, Weiss M, Kogel KH, Schafer P. Piriformospora indica-a mutualistic basidiomycete with an exceptionally large plant host range. Mol Plant Pathol. 2011;13(5):508–518. doi: 10.1111/j.1364-3703.2011.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vadassery J, Ranf S, Drzewiecki C, Mithofer A, Mazars C, Scheel D, Lee J, Oelmuller R. A cell wall extract from the endophytic fungus Piriformospora indica promotes growth of Arabidopsis seedlings and induces intracellular calcium elevation in roots. Plant J. 2009;59(2):193–206. doi: 10.1111/j.1365-313X.2009.03867.x. [DOI] [PubMed] [Google Scholar]
- 76.Shahollari B, Vadassery J, Varma A, Oelmuller R. A leucine-rich repeat protein is required for growth promotion and enhanced seed production mediated by the endophytic fungus Piriformospora indica in Arabidopsis thaliana . Plant J. 2007;50(1):1–13. doi: 10.1111/j.1365-313X.2007.03028.x. [DOI] [PubMed] [Google Scholar]
- 77.Stein E, Molitor A, Kogel KH, Waller F. Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol. 2008;49(11):1747–1751. doi: 10.1093/pcp/pcn147. [DOI] [PubMed] [Google Scholar]
- 78.Schafer P, Pfiffi S, Voll LM, Zajic D, Chandler PM, Waller F, Scholz U, Pons-Kuhnemann J, Sonnewald S, Sonnewald U, Kogel KH. Phytohormones in plant root-Piriformospora indica mutualism. Plant Signal Behav. 2009;4(7):669–671. doi: 10.4161/psb.4.7.9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vadassery J, Ritter C, Venus Y, Camehl I, Varma A, Shahollari B, Novak O, Strnad M, Ludwig-Muller J, Oelmuller R. The role of auxins and cytokinins in the mutualistic interaction between Arabidopsis and Piriformospora indica . Mol Plant Microbe Interact. 2008;21(10):1371–1383. doi: 10.1094/MPMI-21-10-1371. [DOI] [PubMed] [Google Scholar]
- 80.Camehl I, Sherameti I, Venus Y, Bethke G, Varma A, Lee J, Oelmuller R. Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana . New Phytol. 2010;185(4):1062–1073. doi: 10.1111/j.1469-8137.2009.03149.x. [DOI] [PubMed] [Google Scholar]
- 81.Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bogre L. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J. 2010;29(17):2979–2993. doi: 10.1038/emboj.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dhonukshe P, Huang F, Galvan-Ampudia CS, Mahonen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, Offringa R. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS (N/S) motifs to direct apical PIN recycling. Development. 2010;137(19):3245–3255. doi: 10.1242/dev.052456. [DOI] [PubMed] [Google Scholar]
- 83.Zourelidou M, Muller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C. The polarly localized D6 protein kinase is required for efficient auxin transport in Arabidopsis thaliana . Development. 2009;136(4):627–636. doi: 10.1242/dev.028365. [DOI] [PubMed] [Google Scholar]
- 84.Pflieger D, Bigeard J, Hirt H. Isolation and characterization of plant protein complexes by mass spectrometry. Proteomics. 2011;11(9):1824–1833. doi: 10.1002/pmic.201000635. [DOI] [PubMed] [Google Scholar]