Abstract
Poly(ADP-ribose) polymerase (PARP)-2 is a nuclear enzyme that belongs to the PARP family and PARP-2 is responsible for 5–15 % of total cellular PARP activity. PARP-2 was originally described in connection to DNA repair and in physiological and pathophysiological processes associated with genome maintenance (e.g., centromere and telomere protection, spermiogenesis, thymopoiesis, azoospermia, and tumorigenesis). Recent reports have identified important rearrangements in gene expression upon the knockout of PARP-2. Such rearrangements heavily impact inflammation and metabolism. Metabolic effects are mediated through modifying PPARγ and SIRT1 function. Altered gene expression gives rise to a complex phenotype characterized primarily by enhanced mitochondrial activity that results both in beneficial (loss of fat, enhanced insulin sensitivity) and in disadvantageous (pancreatic beta cell hypofunction upon high fat feeding) consequences. Enhanced mitochondrial biogenesis provides protection in oxidative stress-related diseases. Hereby, we review the recent developments in PARP-2 research with special attention to the involvement of PARP-2 in transcriptional and metabolic regulation.
Keywords: PARP-2, ARTD2, SIRT1, DNA repair, Differentiation, Metabolism, Mitochondria
The PARP superfamily
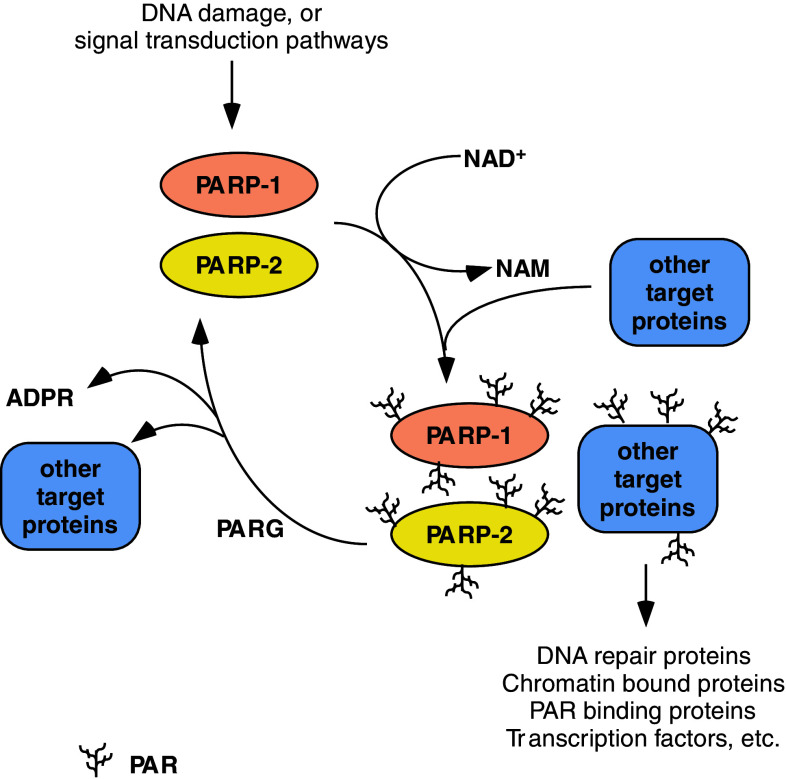
Poly(ADP-ribosyl)ation is a transient post-translational modification of proteins mediated by poly(ADP-ribose) polymerase (PARP) enzymes. This is a dynamic process during which the enzymes catalyze the formation of ADP-ribose polymers onto different acceptor proteins using NAD+ as a substrate. The half-life of the polymer is very short since it is quickly degraded by poly(ADP-ribose) glycohydrolase (PARG). PARPs constitute a family of 17 members, encoded by 17 different genes sharing a conserved sequence coding for the catalytic domain that contains the PARP signature motif, a highly conserved sequence that forms the active site [1]. Based on sequence and structural homologies and the similarity of the reactions catalyzed, Hottiger and colleagues recently proposed to unite all ADP-ribose transferases (PARPs and mono-ADP-ribosyl transferases) in one protein family [2]. The same study proposed a new nomenclature for these enzymes, where PARP-2 was renamed ARTD2 (ADP-ribosyltransferase diphtheria toxin-like 2).
The prototypical enzyme of the PARP family is PARP-1 (ARTD1). PARP-1 cleaves NAD+ to ADP-ribose and nicotinamide followed by the attachment of the first ADP-ribose moiety to a glutamate or aspartate residue of target proteins. During the elongation of the polymer further ADP-ribose moieties are attached to these protein-bound monomers. In the absence of DNA damage the constitutive polymer levels are usually very low and appear as mono- or oligo(ADP-ribose). However, in response to DNA strand breaks, the levels of poly(ADP-ribose) (PAR) polymers increase 10–500 fold and large [3, 4] branched PAR polymers occur on different acceptor proteins and PARP-1 itself (auto-PARylation) (Fig. 1). Upon extensive PARP-1 activation, cellular NAD+ levels are markedly reduced [5]. Poly(ADP-ribosyl)ation, at any level, is likely to have important effects on the acceptor’s properties, hence PARylation and PARPs are involved in the regulation of various cellular processes [6]. In cells, polymers can be detected as early as 2 to 3 min after the one-off induction of DNA damage, and then PAR polymers are quickly degraded by PARG. In tissues, PAR levels can be detected on a longer timeline as reflecting a steady state between synthesis and degradation. Some PARP enzymes carry mutations in the catalytic domain and hence are either inactive or perform only mono-, or oligo(ADP-ribosyl)ation [1, 2, 7].
Fig. 1.
The poly(ADP-ribosyl)ation cycle. NAM nicotinamide, ADPR ADP-ribose, PAR poly(ADP-ribose), all other abbreviations are in the text
The structure of the PARP-2 gene and PARP-2 protein
PARP-2 was discovered when residual DNA-dependent PARP activity was detected in PARP-1 −/− murine embryonic fibroblasts (MEFs) [8]. So far PARP-1, PARP-2, and PARP-3 are the only PARP enzymes whose catalytic activity is stimulated by DNA strand breaks suggesting that they function as crucial members in the cellular pathways responding to DNA damage [8–11].
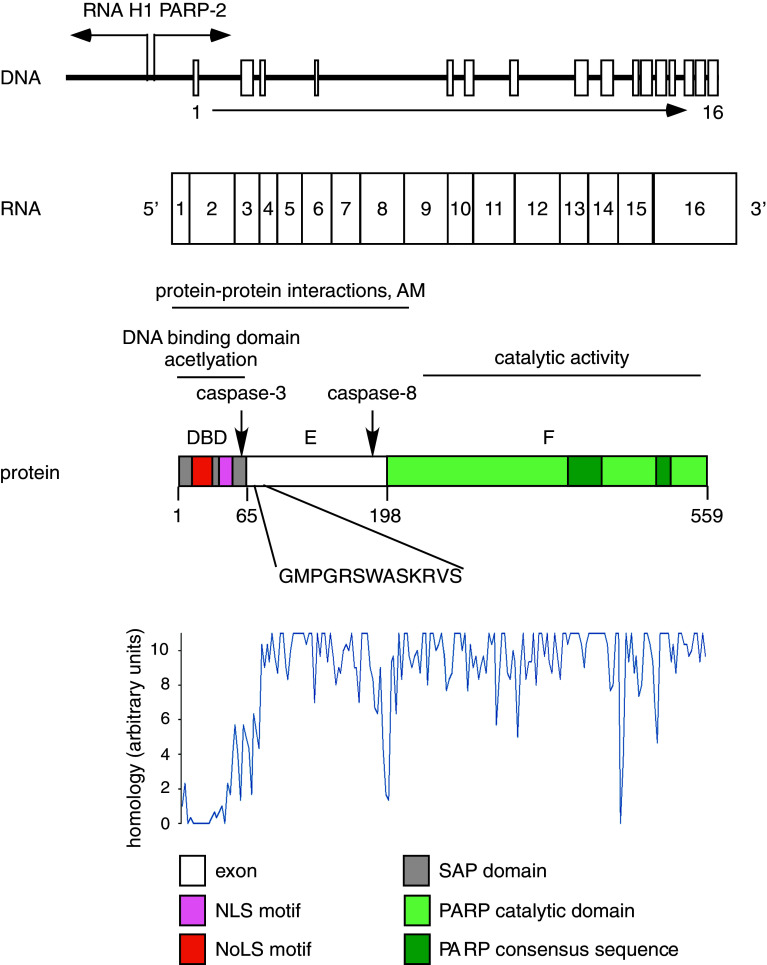
The PARP-2 gene is located on chromosome 14 in humans. The gene is driven by a bidirectional promoter that PARP-2 shares with RNase P [12]. Such a combination of RNA polymerase II and RNA polymerase III genes is relatively rare. A functional TATA box and DSE/Oct-1 expression control elements were identified in the promoter regulating PARP-2 expression [12]. Due to alternative splicing, two isoforms of PARP-2 exist with the longer isoform containing an extra set of 13 amino acids on the border between the DNA binding domain and domain E. The longer isoform has been identified, or predicted in humans [13], common chimpanzee (Pan troglodytes) [14], northern white-cheeked gibbon (Nomascus leucogenys) [15] and sumatran orangutan (Pongo abelii) [16] according to the NCBI database. The sequences of different mammalian PARP-2 genes are highly homologous (Fig. 2). Although PARP-2 is absent in birds, sequences similar to PARP-2 can be found in lower vertebrates (Danio rerio, Xenopus), lower animals (e.g., sponges) and in Arabidopsis thaliana [17].
Fig. 2.
The structure of the PARP-2 gene and PARP-2 protein. The PARP-2 gene is driven by a bidirectional promoter and consists of 16 exons. The protein product of the gene can be divided into three domains: DBD, domain E, domain F. Numbers below the protein product indicate amino acids on the border between domains. The arrows point to caspases-3 and caspases-8 cleavage sites. The highlighted amino acid sequence is the conserved 13 amino acid sequence of the longer PARP-2 isoform. Eighteen mammalian PARP-2 sequences of the shorter isoform (Ailuropoda melanoleuca [118], Bos taurus [119], Callithrix jacchus [120], Canis lupus familiaris [121], Cavia porcellus [122], Cricetulus griseus [123], Equus caballus [124], Homo sapiens [13], Loxodonta africana [125], Macaca mulatta [126], Monodelphis domestica [127], Mus musculus [128], Nomascus leucogenys [15], Oryctolagus cuniculus [129], Pan troglodytes [14], Pongo abelii [16], Rattus norvegicus [130], and Sus scrofa [131]) were compared with Clustal W2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and relative conservation of the amino acids were plotted. Higher values indicate higher levels of conservation. DBD DNA binding domain, AM automodification, NLS nuclear localization signal, NoLS nucleolar localization signal, SAP SAP domain
The tissue-specific expression of PARP-2 was primarily characterized by in situ hybridization. Liver expression of PARP-2 was high at fetal age 12.5 days, decreased at 18.5 days fetal age, and was even lower in newborn mice [8, 18]. In adult mice, the expression of PARP-2 is low in the liver (the lowest among the metabolic tissues; Bai P, unpublished data). It is tempting to speculate that the gradual decrease in PARP-2 expression by age during fetal and postnatal development points toward the possible involvement of PARP-2 in early stage hemopoiesis that takes place in the liver.
In the central nervous system, PARP-2 content was high in the spinal ganglia and in certain parts of the brain. In the neocortical areas, PARP-2 expression is elevated as compared to lower brain regions. High PARP-2 expression was detected in stratum granulosum of the dentate gyrus and the stratum pyramidale of the hippocampus and was even higher in the cortex and the olfactory bulb [18]. Apart from the previously mentioned tissues, PARP-2 is highly expressed in the cortical region of the kidneys, the spleen, adrenal glands, stomach, thymus, and intestinal epithelium [18]. The testis was also positive for PARP-2 expression.
In humans, a slightly different expression pattern was detected. PARP-2 was very abundant in the skeletal muscle, brain, heart, testis, it was high in pancreas, kidney, placenta, ovary, spleen, and low PARP-2 expression was detected in the lungs, leukocytes, gastrointestinal tract (both colon and small intestine), thymus, and liver [19].
Translation of the PARP-2 mRNA yields a protein product of 62-kDa apparent molecular weight. The PARP-2 protein can be divided into similar functional regions as PARP-1: the N-terminus of mouse PARP-2 contains the DNA binding domain (DBD), followed by domain E and the catalytic domain (domain F) [8]. The DBD is formed by a SAP domain that is responsible for DNA binding [20], and contains a functional nuclear localization signal (NLS) [21] and a nucleolar localization signal (NoLS) [22]. A caspase-3 cleavage site defines the border between the DBD and domain E, which is homologous to the caspase-3 site in the E domain of PARP-1 [23]. Domain E serves as a homodimerization interface, an automodification domain and a protein–protein interaction domain as well [24]. Auto-poly(ADP-ribosyl)ation of PARP-2 takes place on domain E [25] and on lysine 36 and 37 that are targets of simultaneous acetylation [26, 27]. The PARP-2 interactome was mapped by Isabelle and coworkers [28], who identified a large number of proteins. These proteins covered a wide array of functions such as cell cycle, cell death, DNA repair, DNA replication, transcription, metabolism, energy homeostasis, and RNA metabolism.
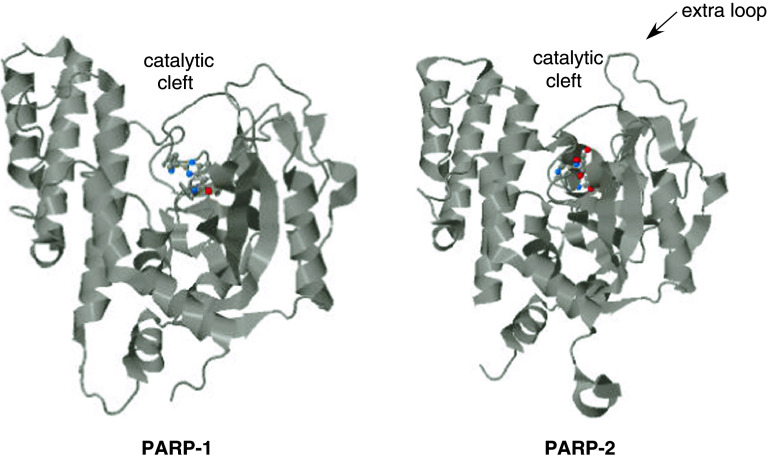
Domain F on the C-terminus of PARP-2 harbors the PARP signature motif carrying the essential amino acid residues for catalysis [8]. Domain F is separated from domain E by a caspase-8 cleavage site [29]. PARP-2 and PARP-1 share a catalytic domain of 69 % similarity, with the exception that PARP-2 contains an additional three-amino-acid insertion in the loop connecting the β-strands k and l in PARP-1 [8, 30, 31] (Fig. 3). The three-dimensional structure of the catalytic domain also shows high similarity; however, the catalytic domain of PARP-2 has a narrower catalytic cleft that likely explains the lower substrate affinity and turnover rate of PARP-2 as compared to PARP-1 (K M for NAD+ 50/130 μM; k cat/K M 6,000 s−1 M−1/323 s−1 M−1 for PARP-1/-2, respectively) [8, 30]. PARP-2 accounts for 5–15 % of total PARP activity in cells depending on the model used [8, 32, 33]. PARP-2 performs auto [18] and hetero-PARylation of proteins. Troiani and coworkers have identified possible targets of PARP-2 activity that covered proteins involved in transcription, translation and mitochondrial organization [34].
Fig. 3.
Three-dimensional structure of the catalytic domain of PARP-1 and PARP-2. Crystal structure of PARP-1 (3GN7) and PARP-2 (3KJD) were retrieved from the protein data bank (PDB, http://www.rcsb.org). Both structures contain an inhibitor (in color), the PARP-1 catalytic domain is in complex with A861696, while the PARP-2 catalytic domain is in complex with ABT-888 [31]. The catalytic cleft and the PARP-2-specific loop is indicated
PARP-2 in DNA repair and genomic integrity
PARP-1 is a well-established DNA-repair protein [6], therefore the functional similarity with PARP-2 suggested a role for PARP-2 in the maintenance of DNA integrity. Upon the induction of DNA damage (ionizing irradiation or laser irradiation), PARP-2 accumulates at the damage foci [35] with a slower kinetics than PARP-1, and PARP-2 persisted longer at DNA damage sites [36]. In murine embryonic fibroblasts (MEFs), the loss of PARP-2 leads to hypersensitivity to ionizing irradiation and cell-cycle arrest in G1 [23], although PARP-2−/− cells are less sensitive to ionizing radiation than PARP-1−/− cells [23, 24]. In line with these observations, female lethality due to X chromosome instability was observed in PARP-1 +/− PARP-2 −/− mice [23].
PARP-2 preferentially binds to one-nucleotide gaps [25] and it is involved in single-strand repair processes. As shown in murine models, upon the loss of PARP-2, base excision repair (BER) slows down [18]. Moreover, PARP-2 interacts with numerous members of the BER machinery such as XRCC1, PARP-1, DNA polβ, and DNA ligase III [18] that further signifies its importance in BER. It is tempting to hypothesize that the early embryonic lethality of the PARP-1/PARP-2 double knockout mice [23] might be due to the strong impairment of DNA repair processes.
PARP-1 has been described to participate in double-strand break repair [37]. Nicolás and coworkers [38] have identified the accumulation of double-strand breaks in PARP-2 −/− murine thymocytes. This observation is in line with a previous report by Yelamos and colleagues [24] who suggested that PARP-2 interacts with the Ku proteins, mediators of double-strand break repair. Moreover, Robert and colleagues [39] have identified PARP-2 as a suppressor of recombination during immunoglobulin class switch events in murine and human B cells, while Bryant et al. have suggested that both PARP-1 and -2 are essential in resolving blocked replication forks by homologous recombination in CHO and murine embryonic fibroblasts (MEFs) [37, 40]. The fact that the ATM/PARP-2 double-knockout genotype is embryonic lethal [20] further supports the involvement of PARP-2 in double-strand break repair during replication. Moreover, the fact that mitomycin C treatment that leads to DNA double-strand breakage provoked the induction of PARP-2 expression and other double-strand break repair proteins in human cervical carcinoma cells also underlines the involvement of PARP-2 in double-strand break repair [41].
Appropriate telomere and centromere maintenance requires PARP-2. PARP-2 binds to and negatively regulates the DNA binding of telomere-binding protein, TRF-2 in different rodent and human cell models. The loss of PARP-2 expression increased the frequency of spontaneous chromosome and chromatid breaks and the number of DNA ends lacking detectable telomere repeats [42].
PARP-2 localizes to centromeres in human and murine cells in a cell-cycle-dependent manner and interacts with the kinetochore proteins centromere protein A (CENPA), centromere protein B (CENPB), and mitotic spindle checkpoint protein BUB3 in prometaphase and metaphase [43]. Interestingly, this centromeric accumulation of PARP-2 is increased when microtubule dynamics are disrupted, suggesting a dominant role of PARP-2 in accurate chromosome segregation [44].
Incomplete or insufficient DNA repair may ultimately lead to either cell death or cellular transformation and tumorigenesis. PARP-1 has been associated with both cell death [45] and tumorigenesis [46]. PARP-2 seems to be involved in cell death regulation similarly to PARP-1 [44, 47]. However, Cohausz and colleagues have found differences in the expression of cell death genes upon N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) treatment in PARP-1 and -2 knockdown cells. Furthermore, PARP-2 is also engaged in tumorigenesis. In mice, the double deletion of PARP-2 and p53-induced spontaneous lymphomas and certain sarcomas [38] and decreased expression of PARP-2 correlated with increased susceptibility to alkylator-induced acute myeloid leukemia (AML) [48]. Results from these experimental models suggest that PARP-2 has a dominant role in suppressing leukemias [49].
Role of PARP-2 in chromatin remodeling and genome maintenance during spermiogenesis
PARP-2 is expressed in the testis of mice [18] and rats [50], and is highly expressed in human testis [19]. Moreover, PARP-2 is found in the ejaculated spermatozoa in both mice and in humans [44, 51]. However, PARP-2 seems to be responsible for a smaller portion of PARP activity than PARP-1 in rat testis [52]. These observations prompted the study of the possible testicular functions of PARP-2 in mice. Dantzer and coworkers have revealed that upon crossing of PARP-2−/− males and females litter size was lower than in colonies bred by crossing wild-type mice. A smaller testis size and high number of abnormal spermatids in the distal epidymis have also been reported [44].
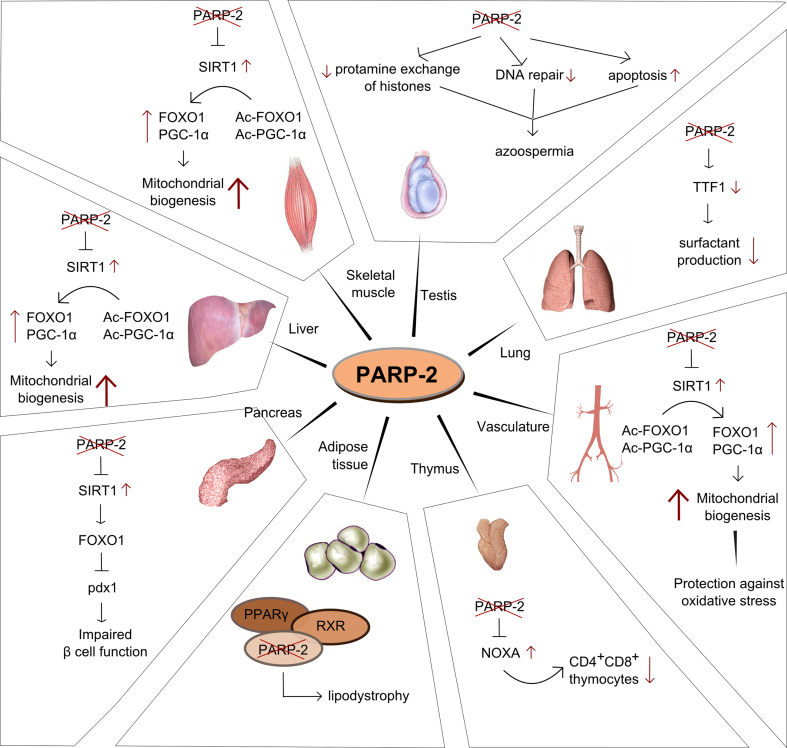
Decreased spermatogenesis is likely to have multiple roots that all trace back to insufficient maintenance of genomic integrity during spermatocyte differentiation (Fig. 4). The differentiation of spermatozoa was found to be hampered and large numbers of apoptotic cells were detected in murine testis [44]. Cell death is probably linked to hampered meiotic sex chromosome inactivation, and the block of cell division in meiosis I, whereby chromosome missegregation was detected [44]. Jha and colleagues [53], based on studies on 18 healthy and 12 infertile humans, also proposed a role for PARP-2 in the preservation of genomic integrity by protecting DNA against oxidative stress. Spermiogenesis involves the compaction of DNA and the exchange of histones to different protamines [54]. In this process PARP-2 (and PARP-1) regulate the activity of topoisomerase IIβ that is essential for appropriate DNA organization (e.g., removal of histone 1) [55], transition protein 2 (TP2) and the transition chaperone HSPA2 [56] as shown in mice.
Fig. 4.
Tissue-specific functions of PARP-2
Different SNPs impacting PARP-2 functionality may also hamper human spermiogenesis. Sakugawa and colleagues [51] have shown on a cohort of 18 Japanese men that such PARP-2 SNPs coincide with azoospermia in humans. Sakugawa and colleagues identified five SNPs. Three of them fall into the coding region, while two into the 3′ UTR. The SNPs in the coding region are all in the catalytic domain. One is synonymous (1159C/T), while two others (1359G/A and 1469A/C) lead to an amino acid change: Arg/Gln and Asn/His, respectively. It is tempting to speculate that changes in the catalytic domain may affect the catalytic activity of PARP-2. SNPs in the 3′ UTR (1789A/C and 1790T/C) may interfere with mRNA stability leading to the reduction of PARP-2 mRNA copy number and consequently decreasing PARP-2 protein levels. Lower PARP-2 levels, or lower PARP-2 activity may interfere with spermiogenesis as described above.
The role of PARP-2 in thymopoiesis and inflammatory regulation
The earliest reports on PARP-2 described high PARP-2 expression in the subcapsular zone of the thymus where lymphocyte proliferation is the most intense. PARP-2 expression gradually decreases towards the center of the thymus as lymphocytes differentiate and mature [18, 47]. PARP-2 transcripts were detected in the white pulp of the spleen and Peyer patches in mice, which also points toward the involvement of PARP-2 in the proliferation of lymphocytes [18].
In line with these observations, the deletion of PARP-2 in mice led to decreases in the weight of thymus and in the total cell numbers and number of CD4+, CD8+ thymocytes in thymus [47] (Fig. 4). The loss of the double-positive thymocytes was due to enhanced p53-mediated apoptosis [47]. Increased expression of a pro-apoptotic, bcl-2 homolog NOXA showed correlation with the enhanced apoptosis [47]. Apoptosis can be reversed by the removal of p53 [38], suggesting that cell death is induced by unresolved DNA damage. In line with this observation, when PARP-2 −/− mice were bred on a p53 −/− background, spontaneous lymphomas and to a smaller extent other sarcomas developed in the double-knockout mice [38], indicating a functional interplay between these two proteins in protecting genome integrity.
It remains to be seen whether atrophy of thymus and the higher rate of thymocyte apoptosis in PARP-2 −/− mice results in a restricted T cell repertoire and altered T cell responses. In fact, PARP inhibition or PARP-1 depletion has provided marked protection in most animal models of inflammation with many of them being dependent on T cell functions [45, 57]. The defect of PARP-2 seems to be associated with a narrower spectrum of diseases in murine models. The lack of PARP-2 impairs astrocyte activation [58] and provides protection against colitis [59], while it has no effect in models of contact hypersensitivity [60], irritative dermatitis [60], or pancreatitis [61]. Interestingly, a common set of genes (iNOS, Il-1β, TNFα) has been shown to be regulated by both PARP-1 and PARP-2, suggesting similar or overlapping mechanisms in inflammatory regulation by the two PARP isoforms. However, the exact mechanism of protection by genetic PARP-2 deletion is unknown [58, 59].
PARP-2 in the regulation of gene expression
Recent reports revealed that the depletion of PARP-2 modifies the activity of multiple transcription factors [62–64]. In HepG2 cells depleted of PARP-2 by shRNA, we have found the dysregulation of more than 600 genes in microarray experiments (Szántó and Bai, unpublished data), indicating an important role for PARP-2 in the regulation of gene expression.
PARP-2 acts at multiple levels on gene transcription. PARP-2 might be capable of modifying chromatin through regulating transcriptional intermediary factor (TIF)-1β and heterochromatin protein (HP)-1α [65]: depletion of PARP-2 modified the expression of two genes (Mest and HNF4) that are dependent on the TIF1β-HP1α complex [65]. Poly(ADP-ribosyl)ation and PARP-1 have eminent roles in epigenetic control [66–69]. Based on the similarities of PARP-1 and PARP-2-catalyzed reactions, and partially overlapping interactome and acceptor protein profile, it is tempting to assume that similar epigenetic roles may also be assigned to PARP-2.
PARP-2 can influence gene expression through more direct interactions. PARP-2 interacts with topoisomerase I and topoisomerase IIβ [40, 55] and may thus regulate the rearrangement of DNA structure in conjunction with RNA transcription. Moreover, PARP-2 has been shown to interact with nucleophosmin/B23 [22] that is involved in rRNA transcription [70]. RNA polymerase I inhibition removes PARP-2 from the nucleolus; however, the deletion of PARP-2 does not change rRNA expression. Thus, the exact mechanism whereby PARP-2 regulates rRNA expression requires further investigation. On the course of mRNA expression, also known as RNA polymerase II-mediated transcription, PARP-2 can act as either a positive co-factor, or a repressor of gene expression. Transcription factors regulated by PARP-2 are summarized in Table 1.
Table 1.
Transcription factors directly regulated by PARP-2
| Name | Mode of action | Effects | Model system | Known tissue specificity | References |
|---|---|---|---|---|---|
| ERα | Unknown | Depletion of PARP-2 suppress ERα activation | Luciferase reporter system in PARP-2-specific shRNA-treated HEK293T cells | Unknown | – |
| RXR/PPARα | Unknown | Depletion of PARP-2 enhance PPARα activation | Luciferase reporter system in PARP-2-specific shRNA-treated HEK293T cells | Unknown | [59] |
| RXR/PPARδ | Unknown | Depletion of PARP-2 enhance PPARδ activation | Luciferase reporter HEK293T in PARP-2-specific shRNA-treated HEK293T cells | Unknown | [59] |
| RXR/PPARγ | Cofactor of receptor | Modulates transcription of PPARγ target genes, Depletion of PARP-2 leads to WAT hypofunction | Luciferase reporter system in PARP-2-specific shRNA-treated HEK293T cells; PARP-2 knockout mice; embryonic fibroblasts from PARP-2 knockout mice | White adipose tissue | [59] |
| SIRT1 | Transcriptional repressor of the SIRT1 promoter. | PARP-2 depletion induces SIRT1 and consequently enhance mitochondrial biogenesis in skeletal muscle and liver | PARP-2 knockout mice; Luciferase reporter system in PARP-2-specific shRNA-treated HEK293T cells; PARP-2 knockdown C2C12 cells | Skeletal muscle, liver | [58, 83] |
| TTF1 | Transcriptional cofactor | Regulates the expression of surfactant protein B | Luciferase reporter system in PARP-2-specific shRNA-treated HeLa/MLE15 cells; interaction mapping in mice and in cells | Lungs | [60] |
Nuclear receptor signaling
PARP-2 has been shown to interact with several members of the nuclear receptor superfamily such as the peroxisome proliferator-activated receptors (PPARs) and estrogen receptor (ER)α.
The group of PPARs has three members, PPARα, PPARδ, and PPARγ [71] that heterodimerize with the retinoid X receptor (RXR) and thus binds to DNA [72, 73]. PPARs bind different lipophilic ligands [74] that regulate their transcriptional activity. PPARs control the expression of a large set of genes involved in the regulation of energy, lipid, and glucose homeostasis [75]. The binding of ligands to the receptors leads to receptor activation and the release of corepressor proteins and the subsequent binding of activators [76]. PARP-1 has been suggested to be involved in nuclear receptor function. Ju and colleagues [77] have shown that upon estrogen receptor activation, topoisomerase IIβ creates DNA strand breaks that are resolved through the action of PARP-1. Moreover, inhibition of topoisomerase IIβ or PARP-1 hampered efficient gene expression [77].
PARP-2 serves as a cofactor for the members of the PPAR transcription factor family. The absence of PARP-2 impairs PPARγ activation but enhances PPARα and PPARδ activation [63]. PARP-2 binds to PPARγ-driven promoters and its absence decreases the expression of genes such as adipocytes protein 2 (aP2), CD36, lipoprotein lipase (LPL), and fatty acid synthase (FAS) [63]. Since PARP-2 is a DNA repair protein and can interact with topoisomerase IIβ [55], it is possible that PARP-2 may also play a role in resealing transcription-related DNA breaks. The effects of PARP-2 depletion on PPARα and PPARδ activation were demonstrated only in reporter assays [63], therefore further molecular and in vivo verification of these interactions is necessary.
Estrogen receptor (ER)α activation is repressed by the depletion of PARP-2 in luciferase reporter assays (P. Bai, unpublished data). The effect of PARP-2 on PPARγ and ERα may share similar molecular characteristics. Further investigation is required to reveal possible physiological consequences of the reduced ERα activity upon PARP-2 ablation. It is important to note that PARP-2 does not interfere with the activation of ERβ, therefore ERβ and its target genes (e.g., keratin 19) are ideal negative controls in studies addressing the role of PARP-2 in nuclear receptor-mediated gene expression [63].
Interaction with SIRT1
SIRT1 belongs to the family of sirtuins. Sirtuins have seven homologs in humans and mice (SIRT1-7) [78, 79]. SIRT1 is considered to be a nuclear enzyme [80], although it may also appear in the cytosol [81]. SIRT1 is an NAD+-dependent protein deacetylase [82] that enables SIRT1 to sense the energetic status of cells (e.g., changes in NAD+/NADH ratio) [83]. SIRT1 is activated by increases in NAD+ levels, or indirectly by different small molecule activators such as resveratrol [84], SIRT1720 [85], AMPK activators [86], or PARP inhibitors [87]. SIRT1 activation leads to the deacetylation and activation of numerous metabolic transcription factors such as PPAR gamma coactivator (PGC)-1α [88], FOXOs [89], and p53 [90]. Their activation leads to increased mitochondrial biogenesis and oxidative metabolism through enhancing the expression of key mitochondrial enzymes involved in terminal oxidation, fatty acid degradation, and mitochondrial uncoupling in several target tissues [88, 91].
It has been shown that PARP-2 can directly regulate the expression of SIRT1 [62]. PARP-2 serves as a negative regulator of SIRT1 expression, as the absence of PARP-2 induces SIRT1 expression and results in higher SIRT1 activity [33, 62]. PARP-2 binds to the murine SIRT1 promoter in a region between −1 and −91, which is a highly conserved region among mammals, showing homology even in Xenopus [62]. It must be noted that the ablation, or pharmacological inhibition of PARP-1 also induces SIRT1 activity. However, SIRT1 activation in the absence of PARP-1 depends on enhanced NAD+ availability and the ablation of PARP-1 does not alter the activity of the SIRT1 promoter [87].
SIRT1 induction upon PARP-2 ablation causes the deacetylation of PGC-1α and FOXO1, which in turn boost mitochondrial biogenesis by enhancing the expression of PGC-1α, uncoupling protein (UCP)-2, muscle isoform of carnitine O-palmitoyltransferase 1 (mCPT1), acyl coenzyme A oxidase I (ACOX1), medium-chain specific acyl-CoA dehydrogenase (MCAD), malonyl-CoA decarboxylase (MCD), Ndufa2, cytochrome c (cyt c) and COX IV [62]. The action of PARP-2 has been shown in multiple organs and tissues such as skeletal muscle, liver, smooth muscle [33, 62], and an unexpected disadvantageous effect has been shown in the pancreas [62]. The depletion of PARP-2 was found not to interfere with SIRT2 or SIRT3 activation [62].
Similar to PARP-2 gene inactivation, SIRT1 activation has been shown to inhibit the production of inflammatory mediators and suppress certain forms of inflammation [92–94]. It is therefore plausible that the induction of SIRT1 may be responsible for the antiinflammatory effect of PARP-2 depletion in colitis [59] and in astrocyte activation [58].
Thyroid transcription factor-1
Nkx-2 transcription factors constitute a family of homeodomain-containing transcription factors. Thyroid transcription factor (TTF)-1 belongs to the Nkx-2 family and TTF-1 plays a dominant role in lung morphogenesis, respiratory epithelial cell morphogenesis, and differentiation [95, 96]. In cultured lung epithelial cells, PARP-2 interacts with TTF1 [64]. By affecting TTF1 activity PARP-2 may regulate the expression of surfactant protein-B (Fig. 4).
The role of PARP-2 in metabolic regulation
Alterations in gene expression accompany various biological phenomena ranging from inflammatory responses (NOXA, TNFα, IL-1β, etc.) to metabolic regulation. The transcriptional regulatory role of PARP-2 has been linked to cellular metabolism. PARP-2 −/− mice are smaller and leaner as they have less body fat than their wild-type littermates [62, 63]. At the same time, PARP-2 −/− mice showed higher oxygen consumption rates and lower respiratory quotients during the active (dark) phase, which points toward higher fatty acid oxidation [62].
When examining the skeletal muscle of PARP-2 −/− mice, increased mitochondrial content was observed in line with higher expression of genes related to oxidative metabolism and fatty acid oxidation, which is in line with the above-described phenotype [62]. The increase in oxidative metabolism can be explained by higher SIRT1 expression due the loss of the transcriptional repressor activity of PARP-2. It is the increase in SIRT1 expression that induces mitochondrial biogenesis through PGC-1α and FOXO1 deacetylation [62] (Fig. 4).
The liver of the PARP-2 −/− mice displayed characteristics similar to the ones in skeletal muscle: SIRT1 induction and consequently enhanced mitochondrial biogenesis and oxidative metabolism [62] (Fig. 4). Interestingly, the brown adipose tissue was not involved in the development of the energy expenditure phenotype in contrast to PARP-1 −/− mice [62, 87].
The increased energy expenditure fueled by enhanced mitochondrial biogenesis in skeletal muscle and liver had beneficial effects on the metabolism of PARP-2 −/− mice. PARP-2 −/− mice are protected against diet-induced obesity, and insulin resistance of the animals was retained even after high-fat feeding [62]. Interestingly, PARP-2 −/− mice proved to be glucose intolerant after high-fat feeding [62]. The pancreas in PARP-2 −/− mice failed to appropriately respond to diet-induced insulin resistance as it showed no signs of hyperproliferation or reduction in pancreas weight, islet size, and pancreatic insulin content [62]. Reduced expression of pancreatic and duodenal homeobox 1 (pdx-1) is likely to be responsible for the pancreatic hypofunction in PARP-2 −/− mice [62] (Fig. 4).
Functions of the white adipose tissue (WAT) are orchestrated by the RXR/PPARγ receptor [72]. We have shown that PARP-2 acts a cofactor of the RXR/PPARγ dimer [63, 97]. The loss of PARP-2 hampers RXR/PPARγ receptor activation and decreases the expression of certain PPARγ-driven genes (e.g., LPL, CD36, etc.). Due to that alterations in gene expression, the WAT of PARP-2 −/− mice turned hypomorphic and hypofunctional [63] (Fig. 4). Moreover, in cellular models of adipocyte differentiation, the lack of PARP-2 resulted in decreased adipocytic differentiation [63]. It is of note that SIRT1 induction may inhibit PPARγ [98] that may provide an auxiliary mechanism underlying WAT hypofunction in the absence of PARP-2.
PARP-2 in oxidative stress-related diseases
PARP-1 depletion, or pharmacological PARP inhibition is protective against numerous oxidative stress-related diseases [45]. Depletion of PARP-2 also resulted in a protective phenotype against diseases associated with increased oxidative stress. Genetic deletion or silencing of PARP-2 has provided protection in models of focal and global cerebral ischemia [99, 100], colitis [59], and doxorubicin-induced vascular smooth muscle damage [33]. Since PARP-2 accounts for a small fraction of total cellular PARP activity [8, 32, 33], it is unlikely that the ablation of PARP-2 could protect against the loss of cellular NAD+ and ATP, suggesting different mechanisms of cell death as compared to the case of PARP-1 ablation.
Cerebral ischemia and doxorubicin-induced vascular impairment involve mitochondrial damage [101, 102] and preventing mitochondrial damage proved to be a successful novel treatment in these pathologies [103–108]. Since SIRT1 has been demonstrated to enhance or restore mitochondrial activity in various tissues [62, 87, 91, 109–111], it is logical to assume that the interference between PARP-2 and SIRT1 expression [62] could be key for the protective phenotype. Indeed, in the case of doxorubicin-induced vascular damage, enhanced SIRT1 expression and consequent stabilization of the mitochondrial membrane potential was proposed to be responsible for the protection provided by the PARP-2 −/− phenotype [33] which might be a prototypical mechanism by which PARP-2 mediates oxidative stress-related pathologies (Fig. 4). Moreover, ablation of PARP-2 led to the mitochondrial retention of apoptosis inducing factor (AIF) [100]. AIF is a mitochondrial protein that shuttles to the nucleus upon oxidative stress-evoked cell death in a PARP-1-dependent manner [112]. In a model of focal cerebral ischemia, ablation of PARP-2 only slightly reduced PAR formation but markedly inhibited the nuclear translocation of AIF [100]. This interesting finding may also be linked to the stabilization of mitochondrial membrane upon SIRT1 induction.
The involvement of PARP-2 in oxidative stress-related pathologies points towards the applicability and hence the development of PARP-2-specific inhibitors. However, all known PARP inhibitors are capable of inhibiting both PARP-1 and -2, which is not surprising since the catalytic domain of the enzymes are very similar and most PARP inhibitors bind there [30, 31]. In the quest for synthesizing PARP-2-specific inhibitors, the laboratory of Gilbert de Murcia suggested the targeting of a loop that is unique in PARP-2 [25, 30]. Efforts to develop highly PARP-2-selective compounds have given rise to inhibitors that have 10–60 fold higher affinity for PARP-2 as compared to PARP-1 [113–117]. One of these inhibitors, UPF-1069, which has 60-fold higher affinity towards PARP-2 than PARP-1, was shown to provide protection against cerebral ischemia [117]. Although at the moment such selectivity is the highest achievable, it is possible that in cellular models or in in vivo settings these inhibitors may partially inhibit PARP-1, too. Nevertheless, the development of highly PARP-2-specific inhibitors is of current interest. Since PARP-2 is a minor PARP isoform, its inhibition is an attractive way to counteract certain drawbacks of pan-PARP inhibition or PARP-1-specific inhibitors. Since PARP-2 accounts for only 5–15 % of PARP activity [8, 18, 32, 33], it is therefore tempting to speculate that its loss probably would not drastically hamper PARylation-dependent DNA repair. Thus, highly PARP-2-specific inhibitors may provide a preferable alternative for the treatment of metabolic diseases, whereas pan-PARP inhibitors may be superior in severe oxidative injury. However, DNA damage assessment in such cases is an absolute necessity.
Conclusions and perspectives
PARP-2 has been shown to participate in multiple cellular processes such as DNA repair, maintenance of genomic integrity, spermiogenesis, and thymopoiesis. On the other hand, PARP-2 is involved in transcriptional regulation of metabolism and oxidative stress response. In fact, this plethora of functions partly overlaps with the functions of PARP-1 [24]. A better understanding of the similarities and differences between the actions of PARP-1 and -2 is of outmost importance. On the one hand, PARP-1 and -2 can act synergistically, while on the other hand isoform-specific functions of the PARP enzymes also exist. Specific targeting of PARP-2 may help overcome unwanted side-effects of pan-PARP inhibition.
Understanding the role of PARP-2 in DNA repair may hold importance in tumor biology. The better understanding of the role of PARP-2 in DNA repair may provide new knowledge on tumorigenesis, and can be capitalized in inducing synthetic lethality by joint inhibition of parallel DNA repair pathways [49].
The metabolic effects of PARP-2 can be exploited in multiple manners. Obviously, the depletion of PARP-2 can be utilized in combating metabolic diseases and mitochondrial stabilization may overcome oxidative stress-evoked damage. Better understanding the properties of PARP-2 may in turn facilitate the development of PARP-2-specific inhibitors that may have advantages over pan-PARP inhibitors.
Acknowledgments
This work was supported by Bolyai fellowship to PB, grants from the National Innovation Office (TéT_09-2010-0023, Baross program Seahorse grant), OTKA CNK80709, K82009, K75864, PD83473, TÁMOP-4.2.2/B-10/1-2010-0024 and TÁMOP-4.2.2. A-11/1/KONV-2012-0025 projects and Medical and Health Science Center (Mecenatura Mec-8/2011). We acknowledge the helpful corrections of Dr. György Haskó.
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 2.Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Miwa M, Sugimura T. Quantification of in vivo levels of poly(ADP-ribose): tritium labeling method and radioimmunoassay. Methods Enzymol. 1984;106:495–500. doi: 10.1016/0076-6879(84)06053-5. [DOI] [PubMed] [Google Scholar]
- 4.Miwa M, Sugimura T. Structure of poly(ADP-ribose) Methods Enzymol. 1984;106:441–450. doi: 10.1016/0076-6879(84)06048-1. [DOI] [PubMed] [Google Scholar]
- 5.Berger NA. Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res. 1985;101:4–15. doi: 10.2307/3576299. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 7.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 9.Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol Cell. 2011;41:33–45. doi: 10.1016/j.molcel.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci USA. 2011;108:2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menissier-de Murcia J, Molinete M, Gradwohl G, Simonin F, de Murcia G. Zinc-binding domain of poly(ADP-ribose)polymerase participates in the recognition of single strand breaks on DNA. J Mol Biol. 1989;210:229–233. doi: 10.1016/0022-2836(89)90302-1. [DOI] [PubMed] [Google Scholar]
- 12.Ame JC, Schreiber V, Fraulob V, Dolle P, de Murcia G, Niedergang CP. A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. Structure and expression of the mouse PARP-2 gene. J Biol Chem. 2001;276:11092–11099. doi: 10.1074/jbc.M007870200. [DOI] [PubMed] [Google Scholar]
- 13.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Homo sapiens] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/10038. Accessed 17 April 2012
- 14.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Pan troglodytes] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/465197. Accessed 17 April 2012
- 15.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Nomascus leucogenys] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100591855. Accessed 17 April 2012
- 16.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Pongo abelii] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100452137. Accessed 17 April 2012
- 17.Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol Genet Genomics. 2001;265:954–963. doi: 10.1007/s004380100506. [DOI] [PubMed] [Google Scholar]
- 18.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 19.Johansson M. A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. G. Genomics. 1999;57:442–445. doi: 10.1006/geno.1999.5799. [DOI] [PubMed] [Google Scholar]
- 20.Huber A, Bai P, Menissier-de Murcia J, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst) 2004;3:1103–1108. doi: 10.1016/j.dnarep.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Haenni SS, Altmeyer M, Hassa PO, Valovka T, Fey M, Hottiger MO. Importin alpha binding and nuclear localization of PARP-2 is dependent on lysine 36, which is located within a predicted classical NLS. BMC Cell Biol. 2008;9:39. doi: 10.1186/1471-2121-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 23.Menissier-de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Schreiber V, Ricoul M, Amé JC, Dantzer F, Meder VS, Spenlehauer C, Stiegler P, Niedergang C, Sabatier L, Favaudon V, Ménissier-de Murcia J, de Murcia G (2004) PARP-2: structure–function relationship. In: Burkle A (ed) Poly(ADP-ribosyl)ation, Springer, New York, pp 13–31
- 26.Haenni SS, Hassa PO, Altmeyer M, Fey M, Imhof R, Hottiger MO. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int J Biochem Cell Biol. 2008;40:2274–2283. doi: 10.1016/j.biocel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isabelle M, Moreel X, Gagne JP, Rouleau M, Ethier C, Gagne P, Hendzel MJ, Poirier GG. Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci. 2010;8:22. doi: 10.1186/1477-5956-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benchoua A, Couriaud C, Guegan C, Tartier L, Couvert P, Friocourt G, Chelly J, Menissier-de MJ, Onteniente B. Active caspase-8 translocates into the nucleus of apoptotic cells to inactivate poly(ADP-ribose) polymerase-2. J Biol Chem. 2002;277:34217–34222. doi: 10.1074/jbc.M203941200. [DOI] [PubMed] [Google Scholar]
- 30.Oliver AW, Ame JC, Roe SM, Good V, de Murcia G, Pearl LH. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlberg T, Hammarstrom M, Schutz P, Svensson L, Schuler H. Crystal structure of the catalytic domain of human PARP2 in complex with PARP inhibitor ABT-888. Biochemistry. 2010;49:1056–1058. doi: 10.1021/bi902079y. [DOI] [PubMed] [Google Scholar]
- 32.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 33.Szanto M, Rutkai I, Hegedus C, Czikora A, Rozsahegyi M, Kiss B, Virag L, Gergely P, Toth A, Bai P. Poly(ADP-ribose) polymerase-2 depletion reduces doxorubicin-induced damage through SIRT1 induction. Cardiovasc Res. 2011;92:430–438. doi: 10.1093/cvr/cvr246. [DOI] [PubMed] [Google Scholar]
- 34.Troiani S, Lupi R, Perego R, Re Depaolini S, Thieffine S, Bosotti R, Rusconi L. Identification of candidate substrates for poly(ADP-ribose) polymerase-2 (PARP2) in the absence of DNA damage using high-density protein microarrays. FEBS J. 2011;278:3676–3687. doi: 10.1111/j.1742-4658.2011.08286.x. [DOI] [PubMed] [Google Scholar]
- 35.Chalmers A, Johnston P, Woodcock M, Joiner M, Marples B. PARP-1, PARP-2, and the cellular response to low doses of ionizing radiation. Int J Radiat Oncol Biol Phys. 2004;58:410–419. doi: 10.1016/j.ijrobp.2003.09.053. [DOI] [PubMed] [Google Scholar]
- 36.Mortusewicz O, Ame JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicolas L, Martinez C, Baro C, Rodriguez M, Baroja-Mazo A, Sole F, Flores JM, Ampurdanes C, Dantzer F, Martin-Caballero J, Aparicio P, Yelamos J. Loss of poly(ADP-ribose) polymerase-2 leads to rapid development of spontaneous T-cell lymphomas in p53-deficient mice. Oncogene. 2010;29:2877–2883. doi: 10.1038/onc.2010.11. [DOI] [PubMed] [Google Scholar]
- 39.Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J Exp Med. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malanga M, Althaus FR. Poly(ADP-ribose) reactivates stalled DNA topoisomerase I and Induces DNA strand break resealing. J Biol Chem. 2004;279:5244–5248. doi: 10.1074/jbc.C300437200. [DOI] [PubMed] [Google Scholar]
- 41.Kang YH, Lee KA, Kim JH, Park SG, Yoon DY. Mitomycin C modulates DNA-double strand break repair genes in cervical carcinoma cells. Amino Acids. 2010;39:1291–1298. doi: 10.1007/s00726-010-0568-5. [DOI] [PubMed] [Google Scholar]
- 42.Dantzer F, Giraud-Panis MJ, Jaco I, Ame JC, Schultz I, Blasco M, Koering CE, Gilson E, Menissier-de Murcia J, de Murcia G, Schreiber V. Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2: PARP activity negatively regulates TRF2. Mol Cell Biol. 2004;24:1595–1607. doi: 10.1128/MCB.24.4.1595-1607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saxena A, Wong LH, Kalitsis P, Earle E, Shaffer LG, Choo KH. Poly(ADP-ribose) polymerase 2 localizes to mammalian active centromeres and interacts with PARP-1, Cenpa, Cenpb and Bub3, but not Cenpc. Hum Mol Genet. 2002;11:2319–2329. doi: 10.1093/hmg/11.19.2319. [DOI] [PubMed] [Google Scholar]
- 44.Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, Murcia G, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc Natl Acad Sci USA. 2006;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 46.Miwa M, Masutani M. PolyADP-ribosylation and cancer. Cancer Sci. 2007;98:1528–1535. doi: 10.1111/j.1349-7006.2007.00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yelamos J, Monreal Y, Saenz L, Aguado E, Schreiber V, Mota R, Fuente T, Minguela A, Parrilla P, de Murcia G, Almarza E, Aparicio P, Menissier-de Murcia J. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J. 2006;25:4350–4360. doi: 10.1038/sj.emboj.7601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cahan P, Graubert TA. Integrated genomics of susceptibility to alkylator-induced leukemia in mice. BMC Genomics. 2010;11:638. doi: 10.1186/1471-2164-11-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: new players in tumour development. Am J Cancer Res. 2011;1:328–346. [PMC free article] [PubMed] [Google Scholar]
- 50.Tramontano F, Di MS, Quesada P. Co-localization of poly(ADPR)polymerase 1 (PARP-1) poly(ADPR)polymerase 2 (PARP-2) and related proteins in rat testis nuclear matrix defined by chemical cross-linking. J Cell Biochem. 2005;94:58–66. doi: 10.1002/jcb.20295. [DOI] [PubMed] [Google Scholar]
- 51.Sakugawa N, Miyamoto T, Tsujimura A, Koh E, Miyagawa Y, Sato H, Namiki M, Okuyama A, Sengoku K. LMTK2 and PARP-2 gene polymorphism and azoospermia secondary to meiotic arrest. J Assist Reprod Genet. 2009;26:545–552. doi: 10.1007/s10815-009-9347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tramontano F, Malanga M, Quesada P. Differential contribution of poly(ADP-ribose)polymerase-1 and -2 (PARP-1 and -2) to the poly(ADP-ribosyl)ation reaction in rat primary spermatocytes. M. Mol Hum Reprod. 2007;13:821–828. doi: 10.1093/molehr/gam062. [DOI] [PubMed] [Google Scholar]
- 53.Jha R, Agarwal A, Mahfouz R, Paasch U, Grunewald S, Sabanegh E, Yadav SP, Sharma R. Determination of poly(ADP-ribose) polymerase (PARP) homologues in human ejaculated sperm and its correlation with sperm maturation. Fertil Steril. 2009;91:782–790. doi: 10.1016/j.fertnstert.2007.12.079. [DOI] [PubMed] [Google Scholar]
- 54.Fuentes-Mascorro G, Serrano H, Rosado A. Sperm chromatin. Arch Androl. 2000;45:215–225. doi: 10.1080/01485010050193995. [DOI] [PubMed] [Google Scholar]
- 55.Meyer-Ficca ML, Lonchar JD, Ihara M, Meistrich ML, Austin CA, Meyer RG. Poly(ADP-ribose) polymerases PARP1 and PARP2 modulate topoisomerase II beta (TOP2B) function during chromatin condensation in mouse spermiogenesis. Biol Reprod. 2011;84:900–909. doi: 10.1095/biolreprod.110.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quenet D, Mark M, Govin J, van Dorsselear A, Schreiber V, Khochbin S, Dantzer F. Parp2 is required for the differentiation of post-meiotic germ cells: identification of a spermatid-specific complex containing Parp1, Parp2, TP2 and HSPA2. Exp Cell Res. 2009;315:2824–2834. doi: 10.1016/j.yexcr.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 57.Peralta-Leal A, Rodriguez-Vargas JM, Aguilar-Quesada R, Rodriguez MI, Linares JL, de Almodovar MR, Oliver FJ. PARP inhibitors: new partners in the therapy of cancer and inflammatory diseases. Free Radic Biol Med. 2009;47:13–26. doi: 10.1016/j.freeradbiomed.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 58.Phulwani NK, Kielian T. Poly(ADP-ribose) polymerases (PARPs) 1-3 regulate astrocyte activation. J Neurochem. 2008;106:578–590. doi: 10.1111/j.1471-4159.2008.05403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Popoff I, Jijon H, Monia B, Tavernini M, Ma M, McKay R, Madsen K. Antisense oligonucleotides to poly(ADP-ribose) polymerase-2 ameliorate colitis in interleukin-10-deficient mice. J Pharmacol Exp Ther. 2002;303:1145–1154. doi: 10.1124/jpet.102.039768. [DOI] [PubMed] [Google Scholar]
- 60.Brunyanszki A, Hegedus C, Szanto M, Erdelyi K, Kovacs K, Schreiber V, Gergely S, Kiss B, Szabo E, Virag L, Bai P. Genetic ablation of PARP-1 protects against oxazolone-induced contact hypersensitivity by modulating oxidative stress. J Invest Dermatol. 2010;130:2629–2637. doi: 10.1038/jid.2010.190. [DOI] [PubMed] [Google Scholar]
- 61.Mota RA, Sanchez-Bueno F, Saenz L, Hernandez-Espinosa D, Jimeno J, Tornel PL, Martinez-Torrano A, Ramirez P, Parrilla P, Yelamos J. Inhibition of poly(ADP-ribose) polymerase attenuates the severity of acute pancreatitis and associated lung injury. Lab Invest. 2005;85:1250–1262. doi: 10.1038/labinvest.3700326. [DOI] [PubMed] [Google Scholar]
- 62.Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, Gergely P, Menissier-de Murcia J, Schreiber V, Sauve AA, Auwerx J. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011;13:450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma heterodimer. J Biol Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- 64.Maeda Y, Hunter TC, Loudy DE, Dave V, Schreiber V, Whitsett JA. PARP-2 interacts with TTF-1 and regulates expression of surfactant protein-B. J Biol Chem. 2006;281:9600–9606. doi: 10.1074/jbc.M510435200. [DOI] [PubMed] [Google Scholar]
- 65.Quenet D, Gasser V, Fouillen L, Cammas F, Sanglier-Cianferani S, Losson R, Dantzer F. The histone subcode: poly(ADP-ribose) polymerase-1 (Parp-1) and Parp-2 control cell differentiation by regulating the transcriptional intermediary factor TIF1beta and the heterochromatin protein HP1alpha. Faseb J. 2008;22:3853–3865. doi: 10.1096/fj.08-113464. [DOI] [PubMed] [Google Scholar]
- 66.Quenet D, El Ramy R, Schreiber V, Dantzer F. The role of poly(ADP-ribosyl)ation in epigenetic events. Int J Biochem Cell Biol. 2009;41:60–65. doi: 10.1016/j.biocel.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 67.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. doi: 10.1042/0264-6021:3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caiafa P, Guastafierro T, Zampieri M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. Faseb J. 2009;23:672–678. doi: 10.1096/fj.08-123265. [DOI] [PubMed] [Google Scholar]
- 69.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 70.Derenzini M. The AgNORs. Micron. 2000;31:117–120. doi: 10.1016/S0968-4328(99)00067-0. [DOI] [PubMed] [Google Scholar]
- 71.Forman BM, Chen J, Evans RM. The peroxisome proliferator-activated receptors: ligands and activators. Ann NY Acad Sci. 1996;27:266–275. doi: 10.1111/j.1749-6632.1996.tb18621.x. [DOI] [PubMed] [Google Scholar]
- 72.Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, Vidal-Puig A, Flier J, Briggs MR, Staels B, Vidal H, Auwerx J. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 73.Bardot O, Aldridge TC, Latruffe N, Green S. PPAR-RXR heterodimer activates a peroxisome proliferator response element upstream of the bifunctional enzyme gene. Biochem Biophys Res Commun. 1993;192:37–45. doi: 10.1006/bbrc.1993.1378. [DOI] [PubMed] [Google Scholar]
- 74.Dreyer C, Keller H, Mahfoudi A, Laudet V, Krey G, Wahli W. Positive regulation of the peroxisomal beta-oxidation pathway by fatty acids through activation of peroxisome proliferator-activated receptors (PPAR) Biol Cell. 1993;77:67–76. doi: 10.1016/S0248-4900(05)80176-5. [DOI] [PubMed] [Google Scholar]
- 75.Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 76.McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/S0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 77.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 78.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 79.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 82.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 83.Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD+? Pharmacol Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 85.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 89.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 90.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 91.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Z, Lowry SF, Guarente L, Haimovich B. Roles of SIRT1 in the acute and restorative phases following induction of inflammation. J Biol Chem. 2010;285:41391–41401. doi: 10.1074/jbc.M110.174482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh DY, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2009;298:E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nayagam VM, Wang X, Tan YC, Poulsen A, Goh KC, Ng T, Wang H, Song HY, Ni B, Entzeroth M, Stunkel W. SIRT1 modulating compounds from high-throughput screening as anti-inflammatory and insulin-sensitizing agents. J Biomol Screen. 2006;11:959–967. doi: 10.1177/1087057106294710. [DOI] [PubMed] [Google Scholar]
- 95.Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14:5671–5681. doi: 10.1128/MCB.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 97.Bai P, Houten SM, Auwerx J, Menisser-de Murcia J, de Murcia GM. Impaired fat storage in PARP-2 knockout mice. Med Sci Monit. 2005;11(1):15. [Google Scholar]
- 98.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, hado De OR, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kofler J, Otsuka T, Zhang Z, Noppens R, Grafe MR, Koh DW, Dawson VL, Menisser-de Murcia J, Hurn PD, Traystman RJ. Differential effect of PARP-2 deletion on brain injury after focal and global cerebral ischemia. J Cereb Blood Flow Metab. 2006;26:135–141. doi: 10.1038/sj.jcbfm.9600173. [DOI] [PubMed] [Google Scholar]
- 100.Li X, Klaus JA, Zhang J, Xu Z, Kibler KK, Andrabi SA, Rao K, Yang ZJ, Dawson TM, Dawson VL, Koehler RC. Contributions of poly(ADP-ribose) polymerase-1 and -2 to nuclear translocation of apoptosis-inducing factor and injury from focal cerebral ischemia. J Neurochem. 2010;113:1012–1022. doi: 10.1111/j.1471-4159.2010.06667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yen HC, Oberley TD, Vichitbandha S, Ho YS, St Clair DK. The protective role of manganese superoxide dismutase against adriamycin-induced acute cardiac toxicity in transgenic mice. J Clin Invest. 1996;98:1253–1260. doi: 10.1172/JCI118909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sims NR, Muyderman H. Mitochondria, oxidative metabolism and cell death in stroke. Biochim Biophys Acta. 2010;1802:80–91. doi: 10.1016/j.bbadis.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 103.Wen Y, Li W, Poteet EC, Xie L, Tan C, Yan LJ, Ju X, Liu R, Qian H, Marvin MA, Goldberg MS, She H, Mao Z, Simpkins JW, Yang SH. Alternative mitochondrial electron transfer as a novel strategy for neuroprotection. J Biol Chem. 2011;286:16504–16515. doi: 10.1074/jbc.M110.208447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ye R, Zhang X, Kong X, Han J, Yang Q, Zhang Y, Chen Y, Li P, Liu J, Shi M, Xiong L, and Zhao G Ginsenoside Rd attenuates mitochondrial dysfunction and sequential apoptosis after transient focal ischemia. Neuroscience 178:169–180 [DOI] [PubMed]
- 105.Hasinoff BB, Schnabl KL, Marusak RA, Patel D, Huebner E. Dexrazoxane (ICRF-187) protects cardiac myocytes against doxorubicin by preventing damage to mitochondria. Cardiovasc Toxicol. 2003;3:89–99. doi: 10.1385/CT:3:2:89. [DOI] [PubMed] [Google Scholar]
- 106.Tao R, Karliner JS, Simonis U, Zheng J, Zhang J, Honbo N, Alano CC. Pyrroloquinoline quinone preserves mitochondrial function and prevents oxidative injury in adult rat cardiac myocytes. Biochem Biophys Res Commun. 2007;363:257–262. doi: 10.1016/j.bbrc.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu M, Ashraf M. Melatonin protection against lethal myocyte injury induced by doxorubicin as reflected by effects on mitochondrial membrane potential. J Mol Cell Cardiol. 2002;34:75–79. doi: 10.1006/jmcc.2001.1485. [DOI] [PubMed] [Google Scholar]
- 108.Panickar KS, Anderson RA. Effect of polyphenols on oxidative stress and mitochondrial dysfunction in neuronal death and brain edema in cerebral ischemia. Int J Mol Sci. 2011;12:8181–8207. doi: 10.3390/ijms12118181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Danz ED, Skramsted J, Henry N, Bennett JA, Keller RS. Resveratrol prevents doxorubicin cardiotoxicity through mitochondrial stabilization and the Sirt1 pathway. Free Radic Biol Med. 2009;46:1589–1597. doi: 10.1016/j.freeradbiomed.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 110.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 111.Morris KC, Lin HW, Thompson JW, Perez-Pinzon MA. Pathways for ischemic cytoprotection: role of sirtuins in caloric restriction, resveratrol, and ischemic preconditioning. J Cereb Blood Flow Metab. 2011;31:1003–1019. doi: 10.1038/jcbfm.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 113.Ishida J, Yamamoto H, Kido Y, Kamijo K, Murano K, Miyake H, Ohkubo M, Kinoshita T, Warizaya M, Iwashita A, Mihara K, Matsuoka N, Hattori K. Discovery of potent and selective PARP-1 and PARP-2 inhibitors: SBDD analysis via a combination of X-ray structural study and homology modeling. Bioorg Med Chem. 2006;14:1378–1390. doi: 10.1016/j.bmc.2005.09.061. [DOI] [PubMed] [Google Scholar]
- 114.Iwashita A, Hattori K, Yamamoto H, Ishida J, Kido Y, Kamijo K, Murano K, Miyake H, Kinoshita T, Warizaya M, Ohkubo M, Matsuoka N, Mutoh S. Discovery of quinazolinone and quinoxaline derivatives as potent and selective poly(ADP-ribose) polymerase-1/2 inhibitors. FEBS Lett. 2005;579:1389–1393. doi: 10.1016/j.febslet.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 115.Pellicciari R, Camaioni E, Costantino G, Formentini L, Sabbatini P, Venturoni F, Eren G, Bellocchi D, Chiarugi A, Moroni F. On the way to selective PARP-2 inhibitors. Design, synthesis, and preliminary evaluation of a series of isoquinolinone derivatives. ChemMedChem. 2008;3:914–923. doi: 10.1002/cmdc.200800010. [DOI] [PubMed] [Google Scholar]
- 116.Sunderland PT, Woon EC, Dhami A, Bergin AB, Mahon MF, Wood PJ, Jones LA, Tully SR, Lloyd MD, Thompson AS, Javaid H, Martin NM, Threadgill MD. 5-Benzamidoisoquinolin-1-ones and 5-(omega-carboxyalkyl)isoquinolin-1-ones as isoform-selective inhibitors of poly(ADP-ribose) polymerase 2 (PARP-2) J Med Chem. 2011;54:2049–2059. doi: 10.1021/jm1010918. [DOI] [PubMed] [Google Scholar]
- 117.Moroni F, Formentini L, Gerace E, Camaioni E, Pellegrini-Giampietro DE, Chiarugi A, Pellicciari R. Selective PARP-2 inhibitors increase apoptosis in hippocampal slices but protect cortical cells in models of post-ischaemic brain damage. Br J Pharmacol. 2009;157:854–862. doi: 10.1111/j.1476-5381.2009.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Ailuropoda melanoleuca] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100483143. Accessed 17 April 2012
- 119.(2012) LOC505828 poly[ADP-ribose] polymerase 2-like [Bos taurus] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/505828. Accessed 17 April 2012
- 120.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Callithrix jacchus] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100388484. Accessed 17 April 2012
- 121.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Canis lupus familiaris] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/475392. Accessed 17 April 2012
- 122.(2012) LOC100720452 poly[ADP-ribose] polymerase 2-like [Cavia porcellus]. NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene?term=PARP2%20Cavia%20porcellus. Accessed 17 April 2012
- 123.(2012) Parp2 poly(ADP-ribose) polymerase family, member 2 [Cricetulus griseus] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100753655. Accessed 17 April 2012
- 124.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Equus caballus] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100072572. Accessed 17 April 2012
- 125.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Loxodonta africana] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100661865. Accessed 17 April 2012
- 126.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Macaca mulatta] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/701955. Accessed 17 April 2012
- 127.(2012) LOC100029480 poly[ADP-ribose] polymerase 2-like [Monodelphis domestica] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100029480. Accessed 17 April 2012
- 128.(2012) Parp2 poly(ADP-ribose) polymerase family, member 2 [Mus musculus] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/11546. Accessed 17 April 2012
- 129.(2012) LOC100351185 poly(ADP-ribose) polymerase family, member 2-like [Oryctolagus cuniculus] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100351185. Accessed 17 April 2012
- 130.(2012) Parp2 poly(ADP-ribose) polymerase 2 [Rattus norvegicus] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/290027. Accessed 17 April 2012
- 131.(2012) PARP2 poly(ADP-ribose) polymerase 2 [Sus scrofa] NCBI Pubmed. http://www.ncbi.nlm.nih.gov/gene/100518917. Accessed 17 April 2012