Abstract
Plasticity is a well-known property of macrophages that is controlled by different changes in environmental signals. Macrophage polarization is regarded as a spectrum of activation phenotypes adjusted from one activation extreme, the classic (M1), to the other, the alternative (M2) activation. Here we show, in vitro and in vivo, that both M1 and M2 macrophage phenotypes are tightly coupled to specific patterns of gene expression. Novel M2-associated markers were characterized and identified as genes controlling the extracellular metabolism of ATP to generate pyrophosphates (PPi). Stimulation of M1 macrophages with PPi dampens both NLR and TLR signaling and thus mediates cytokine production. In this context extracellular PPi enhanced the resolution phase of a murine peritonitis model via a decrease in pro-inflammatory cytokine production. Therefore, our study reveals an additional level of plasticity modulating the resolution of inflammation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0609-y) contains supplementary material, which is available to authorized users.
Keywords: Alternatively activated macrophages, Resolution of inflammation, Toll-like receptors, Nucleotide-binding domain and leucin-rich repeat receptors, Extracellular pyrophosphates, Ectonucleotidases, Cytokines, Peritonitis
Introduction
Macrophage-derived cytokines are crucial for the coordination of appropriate inflammatory responses [1]. Macrophage priming may be rapidly affected by signals from the surrounding microenvironment. These signals polarize the macrophage phenotype to produce a plethora of pro- or anti-inflammatory mediators [1]. Macrophages display a spectrum of phenotypes which allows them to be equally critical in both the initiation and the resolution of inflammation [1, 2]. The two major macrophage phenotypes reported are the classic pro-inflammatory M1 and the alternative anti-inflammatory M2 phenotypes [3, 4]. Macrophages activated with Th1 cytokines (IFNγ) and bacterial endotoxins polarize to M1. In contrast, macrophages activated with Th2 cytokines (IL-4, IL-13), IL-10, or glucocorticoid hormones polarize to M2 [3, 4]. Activation of macrophages to the M1 phenotype leads to (1) upregulation of several pro-inflammatory cytokines and chemokines (such as TNFα, IL-12, IL-6, CCL2, IL-1β) and (2) increase in the production of reactive oxygen species (ROS) and nitrogen intermediates [3, 4]. Activation of macrophages to the M2 phenotype leads to upregulation of scavenger, mannose and galactose receptors, arginase-1, Fizz1, and Ym1 [3, 4]. It is becoming accepted that macrophages are able to reversibly and dynamically switch from one activation state to the other [5–8]. This plasticity allows macrophages to adapt to different environmental signals and to present dual activity at the inflammatory loci.
Inflammation is characterized by an initial influx of polymorphonuclear leukocytes followed by monocyte-derived macrophages, finishing with the resolution phase after normal tissue homeostasis has been restored [9–11]. Despite a vast literature describing the mechanisms that drive inflammation, we do not yet fully understand the mechanisms that mediate the resolution of inflammation and the restoration of tissue homeostasis.
One potential mechanism to increase the resolution of inflammation would be to downregulate important pro-inflammatory signaling pathways. The nucleotide-binding domain and leucine-rich repeat receptors (NLRs), a type of pattern recognition receptor (PRR), are emerging as key regulators of innate immunity, since their activation is key to inflammasome formation and control of caspase-1 activation and the release of cytokines of the IL-1 family [12]. NLRs are activated by pathogen-derived and endogenous danger molecules [12]. One of the best-studied signaling pathways leading to NLR activation and inflammasome formation is the activation of P2X7 receptors by extracellular ATP [13, 14]. We have previously reported that, in macrophages undergoing alternative activation, extracellular metabolism of ATP to pyrophosphates (PPi) potently inhibits inflammasome formation and IL-1 release [8].
In this paper, we examined in detail the gene expression profile of murine macrophages during a dynamic macrophage activation protocol in vitro and in vivo. We discovered that not only NLR genes were downregulated towards M2 macrophage phenotypes, but also that the expression of genes involved in extracellular PPi production was increased. In addition, we report that extracellular PPis were able to further block toll-like receptor (TLR) signaling in M1 macrophages, ultimately leading to a similar phenotype to the M2-activated macrophages. In vivo, PPis were effective in decreasing inflammatory mediators produced during a zymosan-induced peritonitis model. These data suggest that PPis are a family of compounds capable of blocking the two main types of PRR signaling, TLR and NLR, and we suggest that this discovery opens an exciting avenue for novel anti-inflammatory drug design.
Materials and methods
Mice, cells, and reagents
Key reagents and their sources were as follows: E. coli LPS O55:B5, IFNγ and PPi (Sigma); clodronate (Calbiochem); murine recombinant IL-4 (BD Biosciences). Antibodies (Abs) for ELISAs were from R&D, for IκBα and β-actin from Santa Cruz Biotechnology, and for IL-1β mAb from the Biological Resources Branch, National Cancer Institute. All HRP-conjugated secondary Abs were from DAKO Cytomation.
Male C57BL/6 mice (6–8 weeks old, 24–27 g body weight) were obtained from Harlan and maintained in SPF conditions at a room temperature of 20 ± 2°C, and a 12 h:12 h light/dark cycle. The mice were fed a sterile commercial pellet diet and sterile tap water ad libitum. All experiments were approved by the local ethics committee. Peritoneal macrophages were obtained as previously described [15]. Briefly, the peritoneal cavity was gently lavaged with 5 ml phosphate-buffered saline (PBS, Invitrogen). The recovered buffer from two to three mice was pooled, and cells were collected by centrifugation (250×g, 5 min) and plated in 12-well plates at a density of 106 cells/well in RPMI 1640 medium (Lonza) supplemented with 10% fetal calf serum (Invitrogen), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). The macrophages were allowed to adhere overnight (37°C, 5% CO2) and washed with fresh medium to remove unattached cells before use.
Cell culture and stimulation
Macrophages were primed with fresh medium supplemented with different stimuli to obtain a gradient of polarity phenotypes as already reported [8]. The different polarity phenotypes from M1 to M2 are represented in the figures below with a gradient ranging from black (M1) to light gray (M2). LPS (1 μg/ml) and IFNγ (20 ng/ml) were used for 4 h to differentiate to M1 phenotype (black, state 1). IL-4 (20 ng/ml) was used for 4 h to differentiate to M2 phenotype (light gray, state 5). A combination of LPS/IFNγ/IL-4 for 4 h was used to differentiate to an M1/M2 intermediate macrophage polarization phenotype (state 3). To study the polarity changes and to achieve intermediate polarity states, cells were stimulated first for 4 h with IL-4, washed, and then stimulated for a further 4 h with LPS/IFNγ (M2 → M1, state 4); or stimulated first for 4 h with LPS/IFNγ, washed, and then stimulated for a further 4 h with IL-4 (M1 → M2, state 2). Alternatively, to examine the blocking of TLR signaling, macrophages were incubated with 5 mM of PPi or 1 mM of clodronate together with 1 μg/ml of LPS for 4 h.
Western blot and ELISA
Detailed methods used for Western blot analysis have been described previously [15], and ELISAs were performed following manufacturer’s instructions (R&D). Blots from three independent experiments were analyzed by densitometry measurements using NIH ImageJ software (http://rsb.info.nih.gov/ij/) normalizing data to β-actin.
Quantitative reverse transcriptase-PCR analysis and gene expression clustering
Detailed methods used for qRT-PCR have been described previously [8]. Specific primers were purchased from Qiagen (QuantiTech Primer Assays). For each primer set the efficiency was >95%, and a single product was seen on melt curve analysis. The expression of four housekeeping genes was evaluated (GAPDH, YWHAZ, HPRT1, and SDHA) across in vitro M1/M2 macrophage polarization protocol [see Fig. 1 in the Electronic Supplementary Material (ESM)]. No significant differences were observed in the normalized data to each of the four different housekeeping genes. Therefore the presented relative gene expression levels were calculated using the 2−ΔΔCt method normalizing to GAPDH expression levels for each treatment and the fold increase in expression was relative to the smallest expression level or to control basal levels [16]. To calculate the hierarchical (k-means) clustering algorithm, Cluster 3.0 software was used [17], and the output was visualized using Java TreeView software [18]. Quantitative RT-PCR data from the different macrophage polarity states calculated as fold increase or fold decrease relative to untreated macrophages were used as input for the gene clustering. Results were colored to indicate their relative expression: increasingly positive log ratios were colored with red of increasing intensity and increasingly negative log ratios were colored with greens of increasing intensity [19].
Zymosan A-induced peritonitis
Peritonitis was induced as described previously [20, 21]. Briefly 40 mg/kg of type A zymosan (Sigma) freshly prepared in 0.5 ml sterile 0.9% w/v saline was injected i.p. Some animals were co-injected with 5 mM PPi in the 18 h peritonitis group or were injected with 5 mM PPi in 0.2 ml sterile saline 18 h before killing in the 72 h peritonitis group. Control animals were treated with 0.5 ml sterile saline. Inflammatory cells were retrieved at selected time points by injecting 3 ml of PBS into the peritoneal cavity. Cells were centrifuged, and IL-1β in the supernatants was measured by ELISA following manufacturer’s instructions (R&D Systems). Cells were counted by hemocytometer, and CD11b+ cells were isolated using a MACS isolation kit according to the manufacturer’s instructions (Miltenyi Biotec) and subsequently used for RNA extraction. Data were obtained from four animals per group.
In vivo macrophage polarization
LPS-induced peritonitis was evoked by i.p. injection of 0.2 ml of PBS containing 1 μg of LPS and 100 ng of IFNγ for 2 h. In some animals 100 ng of IL-4 was used together with LPS/IFNγ to achieve the intermediate macrophage polarization phenotype (M1/M2). Injection of 100 ng of IL-4 alone was used as an extreme in vivo M2 polarization model to compare with the classical M1 LPS/IFNγ-induced peritonitis group. After 2 h, the animals were killed and the peritoneal cavity was lavaged with 5 ml of medium containing RPMI with 10% FBS. This peritoneal exudate containing cells was retrieved and seeded in a 6-well plate, incubated at 37°C for 30 min, washed twice with PBS, and the adherent macrophages were used for RNA extraction.
Statistical analysis
Results are expressed as the mean ± SEM from the number of assays indicated (from at least three separate experiments). Data were analyzed using an unpaired two-tailed Student’s t test to determine differences between groups using Prism software (GraphPad).
Results
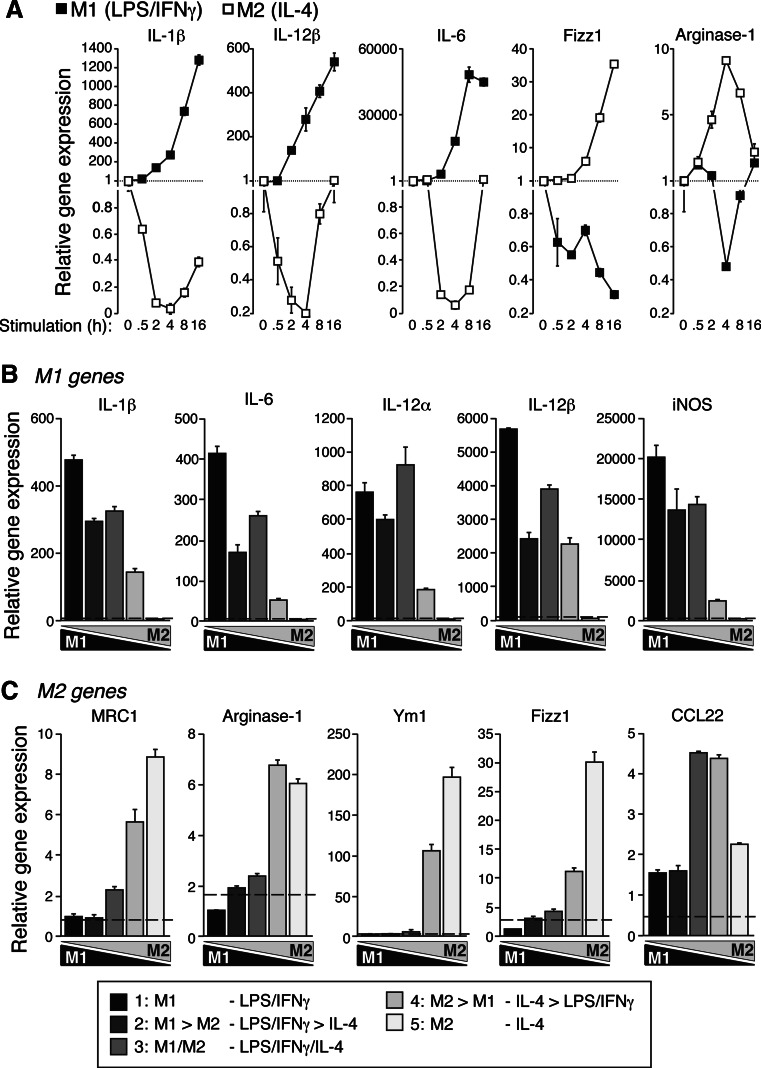
We previously developed a novel polarization gradient for macrophages to switch from classical M1 to alternative M2 extremes [8]. In vitro treatment of peritoneal macrophages with LPS and IFNγ or IL-4 induces a gene expression pattern associated with classical M1 macrophage whilst IL-4 treatment induces an alternative macrophage polarization, which is dynamic and changes during the stimulation time (Fig. 1a). As expected, pro-inflammatory gene expression (IL-1β, TNFα, IL-12α, IL-12β, IL-6, and iNOS) dramatically increased with time during M1 polarization, whereas the same set of genes decreased during M2 polarization (Figs. 1a, 2a). The inverse gene expression pattern was found for M2-related genes, since alternative activation increased the expression of arginase-1, Fizz1, Ym1, and MRC1, whereas classical M1 activation induced downregulation of these genes (Figs. 1a, 2a).
Fig. 1a–c.
Expression of representative M1 and M2 genes during macrophage polarization gradient. a Quantitative real-time (qRT)-PCR for representative M1 (IL-1β, IL-12β, and IL-6) and M2 (Fizz1 and arginase-1) genes during a kinetic of LPS/IFNγ (M1, black squares) or IL-4 (M2, white squares) macrophage polarization. Data are relative to basal expression levels and normalized to GAPDH. b, c qRT-PCR for classical M1 (b) and M2 (c) genes during the in vitro polarization gradient protocol detailed in the “Materials and methods” section. Extreme M1 is represented by a black bar and extreme M2 by a light gray bar; the gradient of intermediate phenotypes is represented by different gray tones. Data are normalized to GAPDH and relative to M2 (b) or to M1 (c) expression levels. Basal control expression level is represented with a dashed line. Data are average of triplicate reactions and representative of three independent experiments
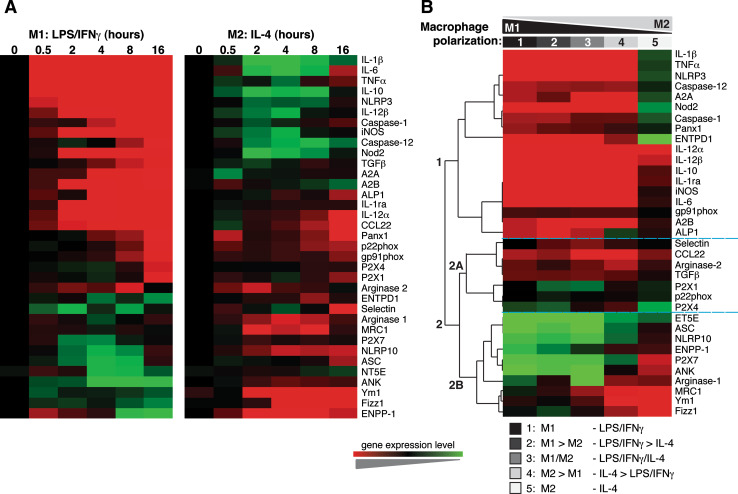
Fig. 2a, b.
Gene expression clusters during macrophage polarization. a Hierarchical clustering of the median-centered quantitative RT-PCR gene expression values from LPS/IFNγ (M1) or IL-4 (M2) polarized macrophages over time. b Hierarchical clustering of the differential transcription induced among a polarity gradient from LPS/IFNγ (extreme M1) to IL-4 (extreme M2). Three major clusters were observed, with a similar pattern of regulation. Cluster 1 principally contained pro-inflammatory genes, cluster 2A was a mix of genes, and cluster 2B contained alternative M2 marker genes. Expression is indicated by a color scale from low (green) to high (red)
It is now being appreciated that macrophages are able to reversibly and dynamically switch from one activation state to the other [7, 8], and we have recently reported an in vitro polarization gradient of stimulation to rapidly generate a five-stage model of macrophage polarization from extreme M1 to extreme M2 [8]. We used this gradient after 4 h of stimulation, when M1 or M2 polarization presented the maximum differences in gene expression (Figs. 1a, 2a) and confirmed that, in fact, the gradient protocol established a real molecular polarization gradient, where IL-4 was able to modulate classic activation of macrophages by altering pro-inflammatory gene expression (Fig. 2b). M1-related genes gradually decreased to near zero in extreme M2 macrophages (Figs. 1b, 2b), whereas typical M2 gene expression patterns gradually increased over the gradient towards M2 phenotypes (Figs. 1c, 2b). No significant changes were observed among four different housekeeping genes during the polarization gradient (see Fig. 1a in the ESM). Interestingly, intermediate polarized macrophages presented a unique gene expression pattern (Figs. 1b, c; 2b) with a trend to upregulate diverse M1 and M2 genes at the same time. Hierarchical clustering of log2-transformed gene expression values of the 35 key genes analyzed in this study revealed three major clusters with a similar pattern of regulation (Fig. 2b). Cluster 1 principally contained pro-inflammatory genes, whereas cluster 2A contained a mix of genes, including pro-inflammatory and M2 macrophage genes. Cluster 2B mainly contained alternative M2 genes (Fig. 2b). Notably, these clusters grouped genes differentially regulated in M1 or M2 polarities (see Figs. 2, 3 in the ESM).
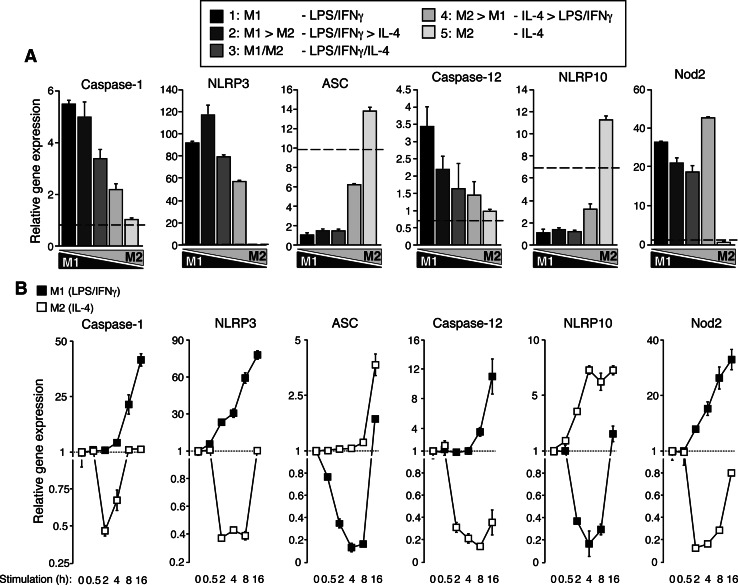
An important family of genes regulated during macrophage polarization is the NLR family of receptors (Fig. 3). We previously found that in intermediate activated macrophages, inflammasome activation was altered and activating molecules, such as extracellular ATP, became inflammasome inhibitors [8]. We studied the expression of inflammasome components and other NLRs during the macrophage polarization gradient and found that caspase-1, NLRP3, Nod2, and Caspase-12 followed a classic pro-inflammatory M1 profile, with a high expression in M1 polarity that gradually decreased towards M2 (Fig. 3a). Also, this expression gradually increased during the M1 stimulus over time and was accompanied by a decrease during the M2 stimulus (Fig. 3b). Surprisingly, the expression of the inflammasome key adaptor ASC (apoptosis-associated speck-like protein containing a C-terminal CARD) was significantly reduced after M1 stimulation and its expression increased during the dynamic stimulation gradient towards M2 states. The unexpected “M2 behavior” of ASC expression was also patent during M1 or M2 stimulation kinetics, and while M1 stimulation decreased its expression, M2 stimulation upregulated ASC expression over time (Fig. 3b). This pattern was similar to the one expected for the inflammasome repressor molecule NLRP10 (Fig. 3a, b), which clearly presents an M2 expression profile.
Fig. 3a, b.
Expression of NLR and inflammasome-related genes during macrophage polarization gradient. a Quantitative real-time (qRT)-PCR for the indicated genes during the polarization gradient protocol. A black bar represents extreme M1 and a light gray bar extreme M2; the gradient of intermediate phenotypes is represented by different gray tones. Data are normalized to GAPDH and relative to the lowest expression level. Basal control expression level is represented with a dashed line. b qRT-PCR for NLR and inflammasome-related genes during a kinetic of LPS/IFNγ (M1, black squares) or IL-4 (M2, white squares) macrophage polarization. Data are relative to basal expression levels and normalized to GAPDH. Data are average of triplicate reactions and representative of three independent experiments
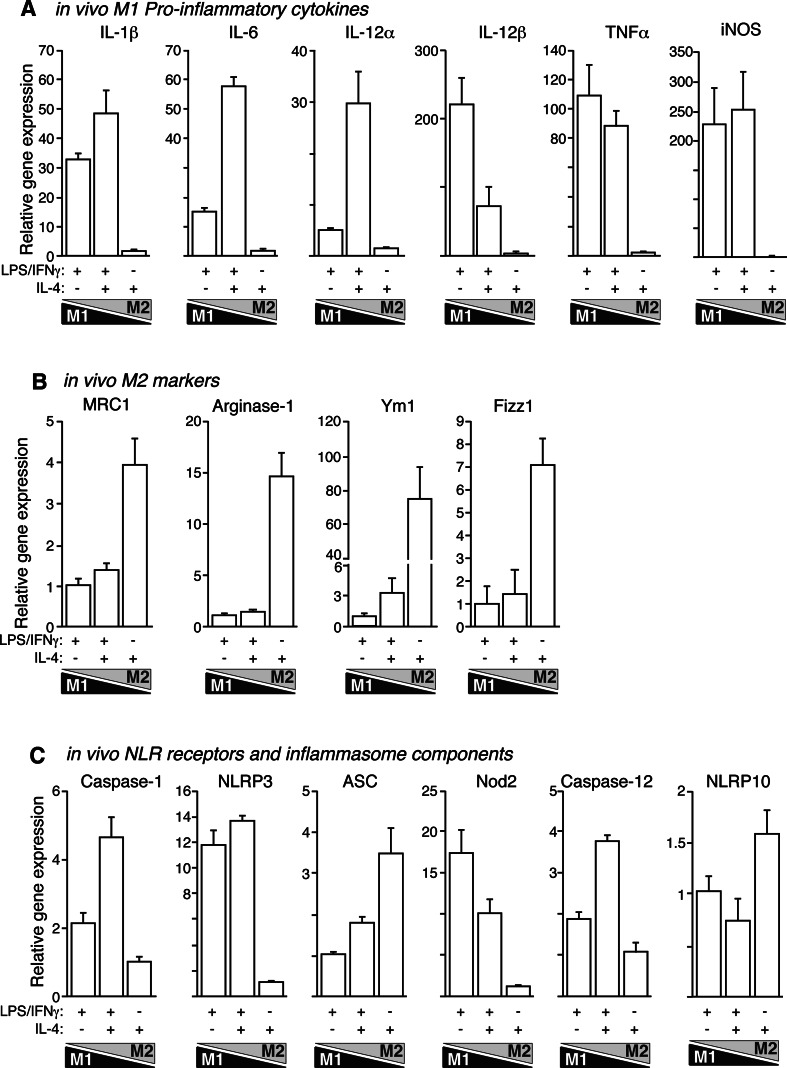
The identification of M2 macrophages in vivo is a controversial and unresolved issue. In order to clarify if M2 phenotype switching could be obtained in vivo, we designed a model of LPS-induced peritonitis where IL-4 was used to alter macrophage polarity in a similar way to our in vitro polarization model. IL-4 dramatically altered the LPS-induced gene expression pattern in vivo and surprisingly upregulated pro-inflammatory gene expression, especially for IL-1β, IL-6, IL-12α, NLRP3, caspase-1, and caspase-12 (Fig. 4a, c). This pattern matched the observed in vitro trend of intermediate M1/M2 polarized macrophages to increase IL-1β, IL-6, and IL-12α gene expression compared with the previous M1 → M2 polarization state (Fig. 1b). This result could not be explained by a macrophage-protective effect of IL-4, since the extracellular presence of lactate dehydrogenase (LDH) was similar among treatments (6.3 ± 0.4% in M1, 7.1 ± 0.9% in intermediate M1/M2, 7.1 ± 0.3% in M2 macrophages, and 7.9 ± 0.1% in resting macrophages, n = 3). IL-4 alone was also able to induce an increase in the gene expression of M2 markers (Fig. 4b), but in combination with LPS, this increase was almost abolished.
Fig. 4a–c.
Expression of M1 and M2 genes during LPS-induced peritonitis is altered by IL-4. Real-time quantitative RT-PCR for M1 (a), M2 (b), and NLR-related genes (c) as indicated during in vivo macrophage polarization gradient. Data are normalized to GAPDH and relative to the lowest expression level to clearly represent relative gene expression increase towards M1 or M2 phenotypes. Data are average of four mice per group and representative of two independent experiments
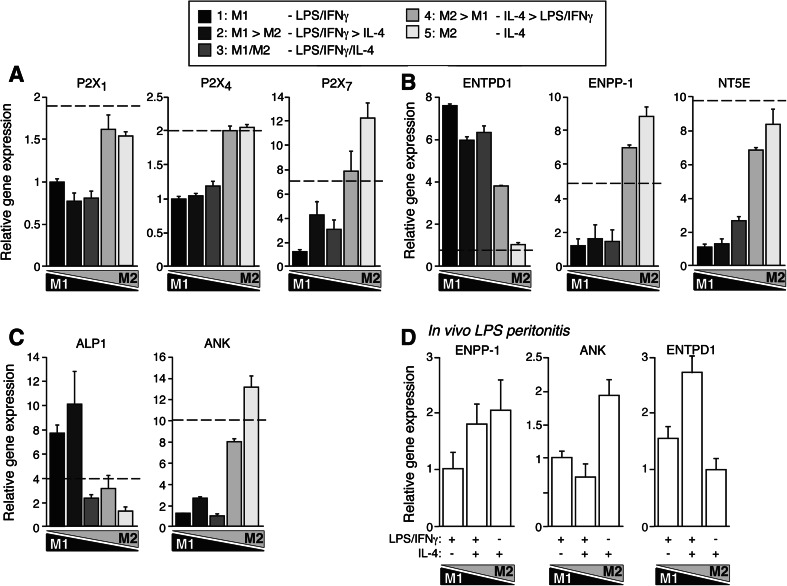
Activation of an alternative M2 macrophage phenotype by IL-4 had previously revealed an inflammasome-blocking action of PPi with the consequent inhibition of mature IL-1β release [8]. Thus, we decided to explore the expression of macrophage ATP-sensing P2X receptors, among them the P2X7 receptors that are involved in the activation of the inflammasome [13, 14], and ectonucleotidases involved in the generation of PPi [22]. P2X1, P2X4, and P2X7, the three P2X receptors expressed by macrophages [23], exhibited an M2-like expression profile, being markedly downregulated by LPS, but having an enhanced expression profile through M2 phenotypes (Fig. 5a). This expression profile is similar to ASC and other alternative M2 genes, which probably limits the action of these molecules in pro-inflammatory conditions to attenuate their amplification function. Interestingly, our in vitro polarization gradient revealed an increase towards M2 phenotypes in the gene expression of ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) and progressive ankylosis disease susceptibility gene product (ANK) molecules that are related to the production and accumulation of extracellular PPi [22, 24] (Fig. 5b, c). However, NTPDase-1 (ENTPD1, ecto-diphosphohydrolase or CD39) and alkaline phosphatase (ALP1), enzymes that degrade extracellular PPi and nucleotides by removing one inorganic phosphate group (Pi) in each reaction step [22], presented an increase in M1 phenotypes and decreased towards M2 polarization (Fig. 5b, c), suggesting that the degradation of extracellular PPi would be induced in pro-inflammatory M1 macrophages whilst its accumulation would be promoted in alternatively activated macrophages. NT5E (ecto-5’-nucleotidase or CD73) expression was strongly downregulated in M1 macrophages with the expression levels recovering through M2 polarities (Fig. 5b), suggesting a possible enhancement of extracellular adenosine accumulation in M2 polarities. This expression profile was also found in vivo when macrophages were obtained from our model of LPS-induced peritonitis co-stimulated with IL-4 (Fig. 5d). However, we were not able to detect any ENPP activity in macrophages (data not shown) and therefore confirm that macrophages were the source of PPi as our data suggested.
Fig. 5a–d.
Expression of purine P2X receptors and ectonucleotidases during in vitro and in vivo macrophage polarization gradient. a–c Quantitative real-time (qRT)-PCR for purine P2X1, P2X4, and P2X7 receptor (a), ectonucleotidases (b), and alkaline phosphatase (ALP1) and ANK (c) gene expression during the polarization gradient protocol. A black bar represents extreme M1, and extreme M2 is represented by light gray bar; the gradient of intermediate phenotypes is represented by different gray tones. Data are normalized to GAPDH and relative to the lowest expression level; basal control expression level is represented with a dashed line. Data are average of triplicate reactions and representative of three independent experiments. d qRT-PCR for ENPP1, ANK, and NTPDase1 (ENTPD1) gene expression during in vivo macrophage polarization gradient. Data are normalized to GAPDH and relative to the lowest expression level. Data are average of four mice per group and representative of two independent experiments
Due to the gene expression data obtained and because it was recently shown that extracellular PPi could be anti-inflammatory in macrophages [8], we decided to further investigate the effect of extracellular PPi in the macrophage polarization gradient.
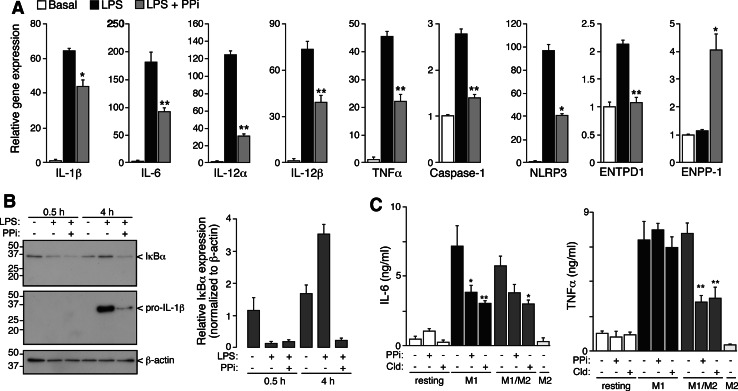
We investigated whether extracellular PPi could alter the M1 gene expression profile in a similar way to the polarization action of IL-4. Interestingly, PPi was able to dampen LPS-induced pro-inflammatory gene expression (Fig. 6a), with a decrease ranging from 75 to 30% depending on the analyzed gene. Surprisingly, extracellular PPi was able to upregulate ENPP1 expression in M1 macrophages, suggesting a positive feedback loop for extracellular PPi accumulation and the initiation of pro-resolving phenotypes. However, extracellular PPi alone was not able to modulate the expression of any analyzed gene in macrophages and was not able to polarize macrophages to M2 states or to alter M2 gene expression profile (not shown). Interestingly, extracellular PPi did not affect LPS-induced IκBα degradation (Fig. 6b) or NF-κB p65 nuclear translocation (data not shown), suggesting that PPi was not affecting cytosolic TLR signaling pathway. PPi inhibitory actions were also observed at the protein level, being able to significantly inhibit LPS-induced production of proIL-1β, IL-6, and TNF-α (Fig. 6b, c) and preventing the recovery of IκBα protein levels, which is also controlled by NF-κB (Fig. 6b). The inhibitory effects of PPi on TNF-α protein levels were only apparent in intermediate M1/M2 polarized macrophages, being ineffective in extreme M1 phenotypes (Fig. 6c). Similar inhibitory effects were obtained using clodronate (Fig. 6c), a synthetic nonhydrolyzable bisphosphonate analogue of PPi.
Fig. 6a–c.
Extracellular PPi dampens NF-κB response after TLR signaling. a Real-time quantitative RT-PCR for the indicated genes after 4 h of incubation with LPS (1 μg/ml) or LPS with PPi (5 mM). Data are normalized to GAPDH and relative to basal unprimed cells. Data are average of triplicate reactions and representative of three independent experiments. b IκBα degradation in response to LPS or LPS/PPi after 30 min or 4 h stimulation. Immunoblots for intracellular IκBα (top panel), pro-IL-1β (middle panel), or β-actin (bottom panel). Relative IκBα expression normalized to β-actin of n = 3 independent experiments (right panel). c ELISA for released IL-6 (left panel) or TNF-α (right panel) in resting, M1, M2, or intermediate M1/M2 polarized macrophages in the presence or absence of PPi (5 mM) or clodronate (Cld, 1 mM); n = 3 independent experiments. *p < 0.05 and **p < 0.001
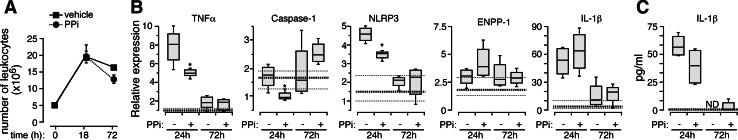
Next, we studied the effects of extracellular PPi in vivo during a pro-inflammatory model of resolving peritonitis induced by zymosan [20, 21]. As expected, zymosan increased the number of total leukocytes in the peritoneal cavity after 18 h with a subsequent decrease after 72 h, indicating the beginning of the resolution phase (Fig. 7a). Extracellular PPi supplementation together with zymosan did not significantly alter leukocyte infiltration during this resolving peritonitis model (Fig. 7a), however, PPi did significantly decrease the expression of TNF-α, caspase-1, and NLRP3 genes induced by zymosan (Fig. 7b). Also, in agreement with our in vitro results, extracellular PPi was able to increase the expression of ENPP1 (Fig. 7b). Interestingly, while IL-1β gene expression was not decreased by extracellular PPi, the total amount of released IL-1β detected by ELISA was lower in the animals treated with PPi (Fig. 7c). This result matches with the fact that PPis are potent blockers of inflammasome formation [8] and are able to downregulate caspase-1 and NLRP3 expression in our in vivo model (Figs. 6a and 7b). All gene expression returned to almost basal levels 72 h after zymosan injection (Fig. 7b, c).
Fig. 7a–c.
Extracellular PPi protects during in vivo zymosan-induced peritonitis. a Number of total infiltrated leukocytes into the peritoneum after 18 or 72 h of a peritonitis induced by zymosan A in the presence or absence of PPi. b Real-time quantitative RT-PCR for indicated gene expression in peritoneal CD11b positive cells after 18 or 72 h of a peritonitis induced by zymosan A in the presence or absence of PPi. Data are normalized to GAPDH expression. c ELISA for the detection of IL-1β in peritoneal fluid after 18 or 72 h of a peritonitis induced by zymosan A in the presence or absence of PPi. No IL-1β was detected after 72 h (ND). The mean (heavy dotted line) and 25th and 75th percentile (light dotted lines) values for animals injected with the basal vehicle are shown. Data are the average of four animals and representative of two independent experiments. *p < 0.05 and **p < 0.005
Discussion
Macrophages are dynamic cells that are involved in both the initiation and the resolution of inflammation, and this plasticity is important to tailor immune responses to different situations [1, 25]. However, little is known about the gene expression or signaling molecules that govern this plasticity, especially those controlling the different macrophage activation states between classic M1 and alternative M2 activated phenotypes. Here, we have identified a specific gene expression pattern that controls both M1 and M2 phenotypes with opposite regulation through a macrophage polarity gradient. In contrast to classic studies of genes upregulated through M2 phenotypes [26, 27], our study reveals that the specific gene cluster upregulated in M1 is in fact downregulated in M2 macrophages, showing that alternative activation triggers an anti-inflammatory or pro-resolving response, where the inflammatory mediators are not only not induced but are downregulated. The inverse gene expression pattern was found in M1 macrophages, where all M2 related genes were downregulated. This finding is in line with recent publications where a close phenotype was found among alternative and pro-resolving macrophages [21], suggesting similar gene expression patterns between these two types of macrophages and presenting M2 macrophages as a good in vitro model to study pro-resolving macrophages. Our novel in vitro polarization gradient that rapidly switches from one activation state to the other [8] revealed that IL-4 was able to modulate classic activation of macrophages by altering pro-inflammatory gene expression. In this model, intermediate polarized macrophages presented a distinctive gene expression pattern grouped in a defined cluster, with a trend to upregulate diverse M1 and M2 genes. This specific expression pattern explains the mixed functional phenotype of this population when responding to diverse pathogen and danger molecular patterns [8, 21].
Notably, typical pro-inflammatory genes, such as purinergic P2X7 receptor and ASC [28, 29], were found to be downregulated towards M1 polarities. This expression pattern could be associated with a tight control of excessive inflammation and macrophage cell death through pyroptosis/apoptosis [30, 31]. This might be species dependent since, in the human monocytic cell line THP-1, a pro-inflammatory stimulus such as LPS increases P2X7 receptor expression [28, 32], while in murine bone marrow–derived macrophages LPS does not seem to alter the response of this receptor to ATP [8, 33]. These data suggest that the high pro-inflammatory potency of these molecules needs to be tightly controlled in M1 macrophages.
Recently, different groups have been trying to characterize alternatively activated macrophages in vivo by administering exogenous IL-4 [34–36]. Similar to a recent publication [36], our data demonstrate that, in vivo, the intermediate macrophage phenotype between M1 and M2 appears unbalanced towards M1. This could be due to LPS attenuating M2 gene expression and IL-4 synergizing with LPS to promote pro-inflammatory gene expression. These data suggest that, in vivo, alternative pathways other than IL-4/IL-13 stimulation could be present to turn macrophages into M2 phenotypes, such as the actions of glucocorticoid hormones or phagocytosis of apoptotic cells [3, 25].
Our work has also revealed that genes involved in the extracellular generation of PPi behave as M2-associated markers. M2 polarities upregulate the transcript accumulation of ecto-nucleotidase ENPP1, which degrades ATP to produce AMP and PPi [22] as well as ANK, a plasma membrane protein that directly or indirectly mediates efflux of cytosolic PPi [24]. These data, together with a previous study showing that PPi dampens inflammasome activity [8], point to a crucial role of extracellular PPi in the physiology of M2 polarization. We could not, however, detect any ENPP activity or extracellular PPi accumulation in macrophages, and although experimental problems cannot be excluded, this could suggest that macrophages are not the main cell type producing PPi during the course of an inflammatory response. It has been previously reported that endothelia or vascular smooth muscle cells show a high ability to produce extracellular PPi [37], and so it is tempting to speculate that extracellular PPi produced by other cell types during resolution of the inflammation and tissue repair phase generates a microenvironment that dampens macrophage pro-inflammatory pathways to enhance resolution.
We found that extracellular PPi were not able to polarize macrophages to M2 states but were able to alter LPS-induced M1 gene expression profile in a similar way to IL-4 action. Interestingly, PPi were also able to upregulate ENPP1 expression after LPS stimulation, suggesting that during the time course of an infection, production of extracellular PPi could promote the initiation of the resolving state. Surprisingly, PPi and its analogue clodronate did not directly interfere with the cytosolic TLR signaling to activate NF-κB-dependent transcription. However, both molecules significantly inhibited LPS-induced production of pro-inflammatory cytokines, IL-1β, IL-6, and TNF-α, and the recovery of IκBα protein levels, all of which are controlled by NF-κB [38]. Clodronate and other bisphosphonates have been in clinical use for decades, primarily for the treatment of osteoarthritis and other inflammatory or metastatic bone diseases [39, 40]. However, several studies have provided evidence that they also exert an anti-inflammatory action in bone and immune cells [39, 41]. The PPi actions to inhibit the inflammasome [8] and NF-κB gene transcription identified in the present study may well underlie these anti-inflammatory actions of bisphosphonates. Both PPi and clodronate have been shown to directly or indirectly act as oxygen radical scavengers [42–44], and they also inhibit ROS production in macrophages [8]. Redox balance has been extensively implicated in NF-κB activation [45], as ROSs contribute to the cytoplasmic signaling pathways leading to NF-κB nuclear translocation. It can also control several key steps in the nuclear phase of the NF-κB program, including chromatin remodeling, recruitment of co-activators, and DNA binding [45, 46]. Our results suggest that PPi chelating actions of ROS do not affect NF-κB cytoplasmic signaling, as normal IκBα degradation was found to, but could specifically affect nuclear actions of translocated NF-κB to dampen pro-inflammatory gene expression and enhance ENPP gene expression.
The role of extracellular PPi interfering with both TLR and NLR signaling was also apparent in vivo during a pro-inflammatory model of resolving peritonitis induced by zymosan [20, 21]. In this context PPi was able to enhance the peritonitis resolution phase through the decrease in pro-inflammatory cytokine production without affecting leukocyte infiltration. Interestingly, the expression of IL-1β transcript was not decreased by PPi treatment. However, the total amount of released cytokine was decreased in the peritoneal cavity of these animals, suggesting that PPi was able to block the inflammasome formation in vivo.
The present study has direct and immediate clinical and physiological relevance since it shows, for the first time, the actions of alternative macrophages in vivo and the successful use of PPi dampening inflammatory mediators in a peritonitis model. PPi has already been proven to block the inflammasome and the release of IL-1β [8], and here we show that it can also dampen TLR-induced signaling similarly to M2 macrophage polarization. Altogether, our results suggest that extracellular PPi and its metabolism are key triggers in the switch from a pro-inflammatory macrophage towards its alternative functions in the resolution of inflammation and open an exciting research horizon for the development of PPi analogue drugs to use as novel anti-inflammatory compounds.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1. Expression of different housekeeping genes during macrophage polarization gradient. A Real-time quantitative (qRT)-PCR for the indicated house-keeping genes (GAPDH, YWHAZ, HPRT1, and SDHA) during the polarization gradient protocol. B qRT-PCR for TNF-α gene expression in M1, M1/M2, or M2 polarized macrophages normalized to the four different housekeeping genes. No significant differences were found in the fold increase/decrease trend among the different macrophage phenotypes. (TIFF 1259 kb)
Supplemental Fig. 2. Expression of the 39 analyzed genes in extreme M1 or M2 macrophages. Real-time quantitative RT-PCR for the indicated genes in extreme M1 (A) or M2 (B) polarization. M2 genes are represented in green, M1 in red, and housekeeping genes in blue. Relative expression normalized to YWHAZ was log2 transformed. (TIFF 1398 kb)
Supplemental Fig. 3. Gene expression in extreme M1 or M2 macrophages. Dot plot representing log2 transformed expression data from real-time quantitative RT-PCR normalized values for all genes included in this study (Suppl. Fig. 2). Data for each individual gene are represented in accordance with its expression in extreme M1 or M2 genes. R 2 = 0.341. (TIFF 2157 kb)
Acknowledgments
We thank Dr. Olga Fernández and Ms. Ana I. Gomez for in vivo models, technical support, and advice. We appreciate Prof. Annmarie Surprenant and Prof. Alex Verkhratsky for their support during this work. We are indebted to Dr. David Brough for carefully revising the manuscript. This work was supported by grants from Instituto Salud Carlos III FEDER (EMER07/049 and PFIS09/00120) and Fundación Séneca (11922/PI/09), managed by Fundación Formación Investigación Sanitaria Región de Murcia (FFIS).
Conflict of interest
The authors declare no competing financial interests.
Abbreviations
- ANK
Progressive ankylosis disease susceptibility gene product
- ASC
Apoptotic speck-like protein with a caspase-activating recruiting domain
- ENPP
Ectonucleotide pyrophosphatase/phosphodiesterase
- LDH
Lactate dehydrogenase
- M1
Classically activated macrophages
- M2
Alternatively activated macrophages
- NLR
Nucleotide-binding domain and leucin-rich repeat receptors
- NTPDase-1
Ecto-diphosphohydrolase
- PPi
Pyrophosphates
- ROS
Reactive oxygen species
- TLR
Toll-like receptors
Footnotes
G. Lopez-Castejón and A. Baroja-Mazo contributed equally to this work.
References
- 1.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Sica A, Locati M. New vistas on macrophage differentiation and activation. Eur J Immunol. 2007;37:14–16. doi: 10.1002/eji.200636910. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 4.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 5.Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J Leukoc Biol. 2004;76:509–513. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porcheray F, Viaud S, Rimaniol AC, Léone C, Samah B, Dereuddre-Bosquet N, Dormont D, Gras G. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol. 2005;142:481–489. doi: 10.1111/j.1365-2249.2005.02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gratchev A, Kzhyshkowska J, Köthe K, Muller-Molinet I, Kannookadan S, Utikal J, Goerdt S. Mphi1 and Mphi2 can be re-polarized by Th2 or Th1 cytokines, respectively, and respond to exogenous danger signals. Immunobiology. 2006;211:473–486. doi: 10.1016/j.imbio.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Pelegrin P, Surprenant A. Dynamics of macrophage polarization reveal new mechanism to inhibit IL-1beta release through pyrophosphates. EMBO J. 2009;28:2114–2127. doi: 10.1038/emboj.2009.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilroy D, Lawrence T, Perretti M, Rossi A. Inflammatory resolution: new opportunities for drug discovery. Nat Rev Drug Discov. 2004;3:401–416. doi: 10.1038/nrd1383. [DOI] [PubMed] [Google Scholar]
- 10.Serhan CN, Brain SD, Buckley CD, Gilroy D, Haslett C, O’Neill LA, Perretti M, Rossi A, Wallace JL. Resolution of inflammation: state of the art, definitions and terms. FASEB J. 2007;21:325–332. doi: 10.1096/fj.06-7227rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serhan CN, Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol. 2005;6:1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- 12.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, Panther E, Di Virgilio F. The P2X7 receptor: a key player in IL-1 processing and release. J Immunol. 2006;176:3877–3883. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 14.Pelegrin P, Surprenant A. The P2X(7) receptor-pannexin connection to dye uptake and IL-1beta release. Purinergic Signal. 2009;5:129–137. doi: 10.1007/s11302-009-9141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelegrin P, Barroso-Gutierrez C, Surprenant A. P2X7 receptor differentially couples to distinct release pathways for IL-1beta in mouse macrophage. J Immunol. 2008;180:7147–7157. doi: 10.4049/jimmunol.180.11.7147. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 18.Saldanha AJ. Java treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolaczkowska E, Koziol A, Plytycz B, Arnold B. Inflammatory macrophages, and not only neutrophils, die by apoptosis during acute peritonitis. Immunobiology. 2010;215:492–504. doi: 10.1016/j.imbio.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Bystrom J, Evans I, Newson J, Stables M, Toor I, Van Rooijen N, Crawford M, Colville-Nash P, Farrow S, Gilroy DW. Resolution-phase macrophages possess a unique inflammatory phenotype that is controlled by cAMP. Blood. 2008;112:4117–4127. doi: 10.1182/blood-2007-12-129767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann H. Extracellular metabolism of ATP and other nucleotides. Naunyn Schmiedebergs Arch Pharmacol. 2000;362:299–309. doi: 10.1007/s002100000309. [DOI] [PubMed] [Google Scholar]
- 23.Sim J, Park C, Oh S, Evans RJ, North RA. P2X1 and P2X4 receptor currents in mouse macrophages. Br J Pharmacol. 2007;152:1283–1290. doi: 10.1038/sj.bjp.0707504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho AM, Johnson MD, Kingsley DM. Role of the mouse ank gene in control of tissue calcification and arthritis. Science. 2000;289:265–270. doi: 10.1126/science.289.5477.265. [DOI] [PubMed] [Google Scholar]
- 25.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 26.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 27.Scotton CJ, Martinez FO, Smelt MJ, Sironi M, Locati M, Mantovani A, Sozzani S. Transcriptional profiling reveals complex regulation of the monocyte IL-1 beta system by IL-13. J Immunol. 2005;174:834–845. doi: 10.4049/jimmunol.174.2.834. [DOI] [PubMed] [Google Scholar]
- 28.Humphreys BD, Dubyak GR. Modulation of P2X7 nucleotide receptor expression by pro- and anti-inflammatory stimuli in THP-1 monocytes. J Leukoc Biol. 1998;64:265–273. doi: 10.1002/jlb.64.2.265. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi S, Sagara J. Regulatory molecules involved in inflammasome formation with special reference to a key mediator protein, ASC. Semin Immunopathol. 2007;29:231–238. doi: 10.1007/s00281-007-0082-3. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes-Alnemri T, Wu J, Yu J, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri E. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKenzie AB, Young MT, Adinolfi E, Surprenant A. Pseudoapoptosis induced by brief activation of ATP-gated P2X7 receptors. J Biol Chem. 2005;280:33968–33976. doi: 10.1074/jbc.M502705200. [DOI] [PubMed] [Google Scholar]
- 32.Humphreys BD, Dubyak GR. Induction of the P2z/P2X7 nucleotide receptor and associated phospholipase D activity by lipopolysaccharide and IFN-gamma in the human THP-1 monocytic cell line. J Immunol. 1996;157:5627–5637. [PubMed] [Google Scholar]
- 33.Lopez-Castejon G, Theaker J, Pelegrin P, Clifton AD, Braddock M, Surprenant A. P2X7 receptor-mediated release of cathepsins from macrophages is a cytokine-independent mechanism potentially involved in joint diseases. J Immunol. 2010;185:2611–2619. doi: 10.4049/jimmunol.1000436. [DOI] [PubMed] [Google Scholar]
- 34.Milner JD, Orekov T, Ward JM, Cheng L, Torres-Velez F, Junttila I, Sun G, Buller M, Morris SC, Finkelman FD, Paul WE. Sustained IL-4 exposure leads to a novel pathway for hemophagocytosis, inflammation and tissue macrophage accumulation. Blood. 2010;116:2476–2483. doi: 10.1182/blood-2009-11-255174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varin A, Mukhopadhyay S, Herbein G, Gordon S. Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood. 2010;115:353–362. doi: 10.1182/blood-2009-08-236711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gea-Sorli S, Closa D. In vitro, but not in vivo, reversibility of peritoneal macrophages activation during experimental acute pancreatitis. BMC Immunol. 2009;10:42. doi: 10.1186/1471-2172-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prosdocimo DA, Douglas DC, Romani A, O’Neill WC, Dubyak GR. Autocrine ATP release coupled to extracellular pyrophosphate accumulation in vascular smooth muscle cells. Am J Physiol Cell Ph. 2009;296:C828–C839. doi: 10.1152/ajpcell.00619.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun SC, Ley SC. New insights into NF-kappaB regulation and function. Trends Immunol. 2008;29:469–478. doi: 10.1016/j.it.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maksymowych W. Bisphosphonates–anti-inflammatory properties. Curr Med Chem. 2002;1:15–28. [Google Scholar]
- 40.Russell RG, Watts NB, Ebetino FH, Rogers MJ. Mechanisms of action of bisphosphonates: similarities and differences and their potential influence on clinical efficacy. Osteoporos Int. 2008;19:733–759. doi: 10.1007/s00198-007-0540-8. [DOI] [PubMed] [Google Scholar]
- 41.Pennanen N, Lapinjoki S, Urtti A, Mönkkönen J. Effect of liposomal and free bisphosphonates on the IL-1 beta, IL-6 and TNF alpha secretion from RAW 264 cells in vitro. Pharm Res. 1995;12:916–922. doi: 10.1023/A:1016281608773. [DOI] [PubMed] [Google Scholar]
- 42.Serretti R, Core P, Muti S, Salaffi F. Influence of dichloromethylene diphosphonate on reactive oxygen species production by human neutrophils. Rheumatol Int. 1993;13:135–138. doi: 10.1007/BF00301259. [DOI] [PubMed] [Google Scholar]
- 43.Kachur AV, Manevich Y, Biaglow JE. Effect of purine nucleoside phosphates on OH-radical generation by reaction of Fe2 + with oxygen. Free Radic Res. 1997;26:399–408. doi: 10.3109/10715769709084476. [DOI] [PubMed] [Google Scholar]
- 44.Dombrecht EJ, De Tollenaere CB, Aerts K, Cos P, Schuerwegh AJ, Bridts CH, Van Offel JF, Ebo DG, Stevens WJ, De Clerck LS. Antioxidant effect of bisphosphonates and simvastatin on chondrocyte lipid peroxidation. Biochem Biophys Res Commun. 2006;348:459–464. doi: 10.1016/j.bbrc.2006.07.075. [DOI] [PubMed] [Google Scholar]
- 45.Gloire G, Piette J. Redox regulation of nuclear post-translational modifications during NF-kappaB activation. Antioxid Redox Signal. 2009;11:2209–2222. doi: 10.1089/ars.2009.2463. [DOI] [PubMed] [Google Scholar]
- 46.Kabe Y, Ando K, Hirao S, Yoshida M, Handa H. Redox regulation of NF-kappaB activation: distinct redox regulation between the cytoplasm and the nucleus. Antioxid Redox Signal. 2005;7:395–403. doi: 10.1089/ars.2005.7.395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Expression of different housekeeping genes during macrophage polarization gradient. A Real-time quantitative (qRT)-PCR for the indicated house-keeping genes (GAPDH, YWHAZ, HPRT1, and SDHA) during the polarization gradient protocol. B qRT-PCR for TNF-α gene expression in M1, M1/M2, or M2 polarized macrophages normalized to the four different housekeeping genes. No significant differences were found in the fold increase/decrease trend among the different macrophage phenotypes. (TIFF 1259 kb)
Supplemental Fig. 2. Expression of the 39 analyzed genes in extreme M1 or M2 macrophages. Real-time quantitative RT-PCR for the indicated genes in extreme M1 (A) or M2 (B) polarization. M2 genes are represented in green, M1 in red, and housekeeping genes in blue. Relative expression normalized to YWHAZ was log2 transformed. (TIFF 1398 kb)
Supplemental Fig. 3. Gene expression in extreme M1 or M2 macrophages. Dot plot representing log2 transformed expression data from real-time quantitative RT-PCR normalized values for all genes included in this study (Suppl. Fig. 2). Data for each individual gene are represented in accordance with its expression in extreme M1 or M2 genes. R 2 = 0.341. (TIFF 2157 kb)