Abstract
MicroRNAs provide an additional layer in the regulation of gene expression acting as repressors with several targets at the posttranscriptional level. This study describes microRNA expression patterns during differentiation and activation of mast cells. The expression levels of 567 different mouse miRNAs were compared by microarray between c-Kit+ committed progenitors, mucosal mast cells, resting and IgE-crosslinked BMMCs in vitro. The strongest upregulation of miR-132 upon IgE-mediated activation was validated in human cord blood-derived mast cells as well. HB-EGF growth factor also upregulated upon activation and was ranked high by more prediction algorithms. Co-transfection of miR-132 mimicking precursor and the 3′UTR of human Hbegf-containing luciferase vector proves that the predicted binding site is functional. In line with this, neutralization of miR-132 by anti-miR inhibitor leads to sustained production of HB-EGF protein in activated mast cells. Our data provide a novel example for negative regulation of a growth factor by an upregulated miRNA.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0786-3) contains supplementary material, which is available to authorized users.
Keywords: Mast cells, MicroRNAs, HB-EGF, Activation, IgE, Differentiation
Introduction
MicroRNAs (miRNAs) represent a new layer of gene expression regulation by interacting with mRNAs and subsequently repressing translation in animals. The mature form of miRNAs consists of approximately 21–25 nucleotides, which can initiate either the suppression of translation or cleavage of their targets by binding to the 3′-untranslated region of the mRNA. This recognition is based on complex and not well understood sequence complementary rules. miRNAs influence a wide variety of developmental events, such as the appropriate establishment of tissue- or cell type-specific expression patterns during the multicellular differentiation processes. As they are able to adjust the concentration of many proteins in a rapid and synchronized manner, miRNAs are thought to play an important role in cell responses and adaptation to the environment [1, 2].
The regulation of mammalian hematopoiesis and immune functions also seems to be coordinated by miRNAs, which may represent a gene regulatory network beneath the well-characterized signaling pathways. Some miRNAs are preferentially expressed in distinct subsets of immune cells, such as miR-142, miR-150, miR-155, miR-181, miR-221/222 and miR-223, indicating their possible role during differentiation. Among these miRNAs, miR-181a and miR-150 are regulators of adaptive immunity, while the more pleiotropic function of miR-155 was underlined also in T and B cells and monocyte/macrophage lineages [3]. In addition, the role of microRNAs is not restricted to the cell fate decisions, but they are active participants in the regulation of mature cell responses.
During the past few years many computational algorithms have been developed to predict miRNA targets with more or less reliability. The target prediction criteria vary widely among the different computational methods, but in general they include (1) Watson-Crick basepairing of the 5′ 2–8 nucleotides of the miRNA to a complementary site in the 3′UTR of the mRNA, (2) calculation of minimum free energy (MFE) of the interacting RNA–RNA duplexes and (3) evolutional conservation of the miRNA binding site [4]. Although the prediction of interactions between RNA molecules by using pure sequence data seems to be more feasible compared to transcription factor-DNA predictions, more as yet unidentified factors should be taken into consideration, since the overall performance is typically just acceptable with an approximately two-thirds false-positive rate [5].
Although mast cells are primarily known as effectors of allergic inflammatory reactions, they play a critical role in host defense mechanisms by orchestrating the innate and adaptive immune responses, too [6, 7]. In marked contrast to other hematopoietic lineages, mast cells leave the bone marrow as immature precursors and complete their maturation in the local mucosal or connective tissues. Depending on the tissue context, they may undergo differentiation to give rise to morphologically and functionally identifiable mature mast cell subtypes. There are at least two distinct subpopulations of mast cells in rodents: the connective tissue-type (CTMCs) and the mucosal mast cells (MMCs). While the former cell population can be characterized with a high level of Mcpt1 and Mcpt2, the latter mast cell group expresses predominantly Mcpt4, Mcpt5 (chymase1, cma1), Mcpt6 (tryptase beta2, Tpsb2), Mcpt7 (tryptase alpha/beta1, Tpsab1) and carboxypeptidase 1 [8, 9]. These subtypes differ in location, staining characteristics, mediator content and dependency of T cell-derived cytokines. In humans, mast cell subpopulations are usually classified based on their protease content. In contrast to the MCT type, which produces only tryptase, MCTC cells contain both tryptase and chymase, members of two protease families that contribute critically to various pathological conditions either as pro-inflammatory or protective players (e.g., in parasitic and bacterial infections, allergic inflammation and arthritis) [10]. According to their tissue localization, the MCT type corresponds to MMCs and the MCTC cells to CTMCs in rodents [11].
The activation of mast cells results in releasing a wide range of mediators in allergic reactions, which can be evoked by the contribution of IgE and its high-affinity receptor (FcεRI) on the surface of mast cells. This process is regulated by crosslinking the FcεRI receptors, which is initiated by an interaction between the antigen and the receptor-bound IgE. Subsequently kinases, such as FYN, LYN or SYK, are rapidly phosphorylated at tyrosine residues, and their activation induces PLC-γ, leading to the production of inositol triphosphate and to the increase of intracellular calcium concentration. Mast cells are able to secrete preformed mediators upon stimulation, such as biogenic amines (e.g., histamine), proteoglycans and proteases. At the same time, their activation also initiates the de novo synthesis of lipid mediators, cytokines, chemokines and specific growth factors (TGF-β, NGF, VEGF, FGF2, etc.) [12]. Importantly, some of the growth factors, such as activin A, amphiregulin and heparin-binding epidermal-like growth factor (HB-EGF), known for its ability to induce the growth of fibroblasts and smooth muscle cells, were found to be upregulated by IgE crosslinking in mast cells of various origin [13, 14].
HB-EGF, a member of the EGF family, is implicated in various biological processes, like wound healing [15–17], blastocyst implantation [18], heart function [19] and diseases, such as cancer [20] and arteriosclerosis [21]. HB-EGF is initially synthesized as a transmembrane protein (pro-HB-EGF), which has been identified as the receptor for diphteria toxin. Pro-HB-EGF is then cleaved by matrix metalloproteinases, resulting in the release of the mature, soluble form. HB-EGF binds to its receptors HER2 and HER4 in the presence of heparan sulfate as cofactor. Many cell types, including epithelial cells, skeletal muscle cells, macrophages, keratinocytes and T cells, express HB-EGF, and it shows potent mitogenic and chemotactic activity for fibroblasts and epithelial cells [22, 23].
In the present study, we performed miRNA gene expression profiling using microarray technology to gain insight into the differentiation process of mature MMCs and the IgE-mediated activation of BMMCs, a widely accepted in vitro model system. From among genes expressed differentially upon IgE-crosslinking, miR-132 microRNA showed the highest upregulation; thus, we selected it for further analysis. To investigate the role of IgE-induced miR-132 expression, we carried out a bioinformatic search in different target prediction databases to select functionally relevant targets. According to three prediction algorithms, HB-EGF turned out to be a highly ranked candidate for miR-132 targets, which was experimentally confirmed. As HB-EGF is also upregulated upon mast cell activation, miR-132 may play an important negative regulatory role in adjusting the appropriate level of this mitogenic mediator, thus influencing the growth of resident tissue cells. This feedback loop becomes especially interesting in physiological responses and diseases at the intersection of tissue remodeling and inflammatory activity of mast cells, such as wound healing or asthma, nasal polyposis and psoriasis.
Materials and methods
Cell lines and transfection
CHO epithelial cell line (ATCC, Manassas, VA) was propagated in Ham’s F12 medium with 10% fetal bovine serum (FBS). The mouse macrophage cell line J774.2 was cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS. MC/9 mouse mast cell line (ATCC no. CRL-8306) was maintained in high glucose (4.5 g/l) DMEM supplemented with 6 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM non-essential amino acid solution (Sigma-Aldrich, St. Louis, MO), 0.05 mM 2-mercaptoethanol, 10% rat T-STIM (Becton Dickenson, Mountain View, CA) or conditioned medium of concanavalin A-stimulated (Sigma-Aldrich) mouse splenocytes and 10% FBS.
Then 4 × 106 MC/9 cells or BMMCs were transfected using a Nucleofector II device (Lonza, Cologne, Germany, program X-005) in 100 μl Solution V according to the manufacturer’s protocol. To determine the transfection efficiency (varying between 55 and 85% in MC/9 determined by flow cytometry), control cells were transfected with pmaxGFP vector. For transfections, miR-132 miRNA precursor (Pre-miR-132) and Pre-miR-132 negative control, Anti-miR-132, as well as Anti-miRNA negative control oligonucleotides, were purchased from Applied Biosystem.
Generation of bone marrow-derived mast cells
BMMCs were generated from bone marrow of 6- to 8-week-old male BALB/c mice. Briefly, mice were killed, and femora with intact medullary cavities were removed. Sterile, endotoxin-free medium was flushed repeatedly through the bone shaft using a needle and syringe. The suspension of bone marrow cells was centrifuged at 320×g for 10 min and cultured at a concentration of 0.5 × 106 cells/ml in RPMI 1640 with 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and a combination of 5 ng/ml interleukin-3 (IL-3) and 20 ng/ml stem cell factor (SCF) (Sigma-Aldrich) for 3–4 weeks at 37°C in a humidified atmosphere with 5% CO2. Nonadherent cells were transferred to fresh medium at least twice a week. After 3–4 weeks when a mast cell purity of >90% was achieved (as assessed by flow cytometric analysis), the cells were used for experiments. To compare microRNA gene expression changes during mast cell differentiation, c-Kit positive mast cell progenitors were magnetically isolated on the 6th day. To establish an in vitro model of MMC differentiation, the culture medium of 4-week-old BMMCs was supplemented with 10 ng/ml IL-9 (Immunotools, Friesoythe, Germany) and 2 ng/ml TGF-β (Sigma-Aldrich) for 5 days. The maturation of BMMCs to MMCs was verified by measuring the marked upregulation of mast cell protease-1 (mMcpt1). To investigate the activation, BMMCs were sensitized with 3 μg/ml anti-dinitrophenyl (DNP) IgE (clone SPE7, Sigma-Aldrich) for 2–4 h, washed and then challenged with DNP-HSA (Sigma-Aldrich) for 2 h unless otherwise indicated. The antigen concentration for the maximal mast cell activation was determined by IL-13 qRT-PCR (for more details, see the Supplementary methods).
Human cord blood-derived mast cells
Human mast cells were differentiatied from umbilical cord blood with the permission of the local ethics committee of the Semmelweis University. Whole blood was diluted with RPMI 1640, and the mononuclear cells were separated with Histopaque-1077 (Sigma-Aldrich). Residual erythrocytes were removed by hypotonic lysis, and the mononuclear cells were resuspended in 0.5% BSA and 2 mM EDTA. After labeling with anti-CD34 microbeads, the cells were magnetically isolated twice (Miltenyi Biotech, Bergisch Gladbach, Germany). The CD34+ cells were then resuspended in RPMI 1640 with 10% FBS, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 100 U/ml penicillin and 100 μg/ml streptomycin. The cells were seeded at 106 cells/ml and cultured in the presence of 100 ng/ml SCF, 50 ng/ml IL-6 and 3 μM lysophosphatidic acid (LPA, Sigma-Aldrich). Cells were cultured for up to 8 weeks, and the culture medium was replaced weekly. To examine the IgE-mediated activation of human mast cells, cells were sensitized with human myeloma IgE (Serotec, Düsseldorf, Germany) at 37°C overnight. After washing, they were re-suspended in 0.5% BSA in DMEM, and challenged with 1.5 μg/ml of rabbit anti-human IgE (Dako Corp., Glostrup, Denmark) at 37°C for 2 h.
Flow cytometry
Cells were washed twice with 0.5% BSA in PBS and then labeled at room temperature for 30 min. The following antibodies were used: anti-mouse CD117 APC, anti-mouse B220 PE, anti-mouse CD11b PE, anti-mouse TER119 PE, anti-mouse CD5 PE (all from Pharmingen, San Diego, CA) and anti-mouse FcεRIα biotin (eBioscience, San Diego, CA). Then 10,000 cells were measured by FACSCalibur (BD Biosciences, San Jose, CA), data were collected with CellQuest (Becton-Dickinson Immunocytometry Systems, San Jose, CA) and further analyzed by the FlowJo software (TreeStar, Inc., Palo Alto, CA).
RNA isolation, quality determination, reverse transcription and real-time PCR
RNA from cell samples was extracted by miRNeasy Mini Kit (Qiagen, Valencia, CA), the quantity and quality of total RNA were assessed by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA), respectively. Only those samples were used for microarray experiments that gave >8.0 for RNA integrity number and showed a clear gel image, and for which no DNA contamination was observed on the histogram.
To determine mRNA expression, 1 µg RNA was converted to cDNA by 1.25U MuLV reverse transcriptase (Applied Biosystems, Foster City, CA) with random primers in the presence of 20 U RNasin ribonuclease inhibitor (Promega, Madison, WI) at 42°C for 55 min. MuLV was then inactivated at 95°C for 5 min. The real-time PCR reactions were carried out in an ABIPrism 7000 instrument according to the manufacturer’s instructions with 2 μl RT-product in each well and in 25 μl final volume.
To measure microRNA expression, primer sets for specific microRNA assays and sno135 endogenous control and the MicroRNA Reverse Transcription Kit were utilized following the manufacturer’s protocol. Briefly, 10 ng of each total RNA sample was transcribed by Multiscribe Reverse Transcriptase. qRT-PCR was carried out using Applied Biosystems 7000 Real-Time PCR system. The relative expression of each miRNA and mRNA was calculated from the equation 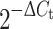 , where ΔC
t = mean C
t (miRNA) – mean C
t (internal control) (where C
t is the threshold cycle for a sample). The applied TaqMan gene expression and microRNA assays are listed in Supplementary Table 1. All reagents and instruments for RT-PCR were purchased from Applied Biosystems, Inc., except when otherwise indicated.
, where ΔC
t = mean C
t (miRNA) – mean C
t (internal control) (where C
t is the threshold cycle for a sample). The applied TaqMan gene expression and microRNA assays are listed in Supplementary Table 1. All reagents and instruments for RT-PCR were purchased from Applied Biosystems, Inc., except when otherwise indicated.
Microarray
For microRNA profiling, the Agilent Mouse miRNA Microarray Kit (G4472A, 8 × 15 k) was applied according to the manufacturer’s instruction (version 1.0) with 100–100 ng quality-checked total RNA. The labeled samples were hybridized for 20 h at 55°C. The arrays were scanned with an Agilent DNA Microarray Scanner BA, the signal quantification was carried out by Feature Extraction 10.7 Image Analysis Software, and data were further analyzed by Genespring GX10.0. Raw data were normalized to the 75th percentile signal intensity, and entities showing present call in all samples of a condition were filtered out. Differentially expressed genes were selected when passing the signal intensity filter (entities where at least 100% of samples in any one out of four conditions have values within cutoff) and showing at least twofold statistically significant change (ANOVA and Tukey HSD post-hoc test, with Benjamini-Hochberg Multiple Testing Correction p value <0.05) between BMMC and any other groups. The microarray data are deposited in NCBI Gene Expression Omnibus with accession no. GSE24321.
Luciferase assay
The effects of miR-132 mimics on endogenous human HB-EGF 3′UTRs were measured using the luciferase reporter construct (SwitchGear Genomics, Menlo Park, CA). A vector containing a random genomic sequence was used to control for overall signal variation caused by the cellular non-specific response (non-target 3′UTR). Transient transfection assays were conducted in CHO cells in 96-well plates. Transfection efficiency was adjusted with FAM-conjugated negative control pre-miR oligonucleotides and typically yielded ~80% (Fig. S1) with >95% viability as determined by propidium-iodide staining at 24 h. Briefly, 6,000 cells per well were seeded in culture medium 24 h before transfection. Next, 0.15 μl Dharmafect Duo (Thermo Fisher Scientific, Waltham, MA) was used in each well to transfect 100 ng of plasmid DNA and either a miRNA mimic or a non-targeting control according to the manufacturer’s protocol. Pre-miR-132 and non-targeting controls (Pre-miR negative control, Ambion, Austin, TX) were added at a final concentration of 3, 30 and 90 nM. After 24 h, 100 μl of the Steady-Glo Luciferase Assay System reagent (Promega, Madison, WI) was added into each well, incubated at room temperature for 30 min, and then signals were determined in a standard plate luminometer (Labsystems Luminoskan RS Microplate Reader, Helsinki, Finnland). Each transfection was conducted and assayed in triplicate.
Western blotting
For Western blotting, protein extracts were prepared by resuspending cell pellets in 1% NP-40 (Igepal CA-630, Sigma-Aldrich) lysis buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EGTA, 5 mM NaF, 2 mM phenyl-methyl-sulfonyl-fluoride, 1 mM Na-orthovanadate, 10 μg/ml leupeptin and 10 μg/ml aprotinin. Debris was removed by centrifugation, and the protein yield was assessed by spectrophotometry (Total Protein Kit, Micro Lowry, Peterson’s Modification, Sigma-Aldrich). Then 30-μg aliquots of heat-denatured, β-mercaptoethanol-treated protein samples were loaded on precast Ready Gels (Bio-Rad, Hercules, CA). Gels were blotted onto polyvinylidene difluoride membranes (Bio-Rad), the membranes were blocked with 5% milk powder in TBS and 0.1% Tween-20 for 1 h, and then incubated with 1.5 μg/ml goat anti-human HB-EGF (R&D Systems, Minneapolis, MN) overnight at 4°C. Blots were washed, the secondary antibody (1:3,000 HRP-conjugated anti-goat IgG) was applied for 45 min and the immunoreactive bands were visualized with the ECL-Plus Western blotting Detection System (GE Healthcare-Amersham Biosciences, Piscataway, NJ). Specific band size was determined with the Fermentas Plus Prestained Protein Ladder (Fermentas, Glen Burnie, MD). As a housekeeping control the beta-actin level was determined by using mouse anti-actin antibody (Sigma-Aldrich, 1:1,000 dilution) and HRP-conjugated anti-mouse IgG (Dako Corp., Glostrup, Denmark, 1:10,000 dilution). Image analysis was carried out with the ImageJ software (http://rsb.info.nih.gov/ij/).
Target predictions
To improve the robustness of target prediction for miR-132, data sets of the three most commonly used target prediction databases were used simultaneously. Using the outputs of the target prediction algorithms from PicTar (http://pictar.mdc-berlin.de/), MirBase (http://www.ebi.ac.uk/enright-srv/microcosm/) and TargetScan (version 5.1, http://www.targetscan.org/), lists of putative target genes were generated. As different IDs are applied by the three target prediction algoritms, the identifiers of BioMART (http://www.ensembl.org/Multi/martview) were used to generate a comprehensive list. If more transcript IDs correspond to one HUGO identifier, the ones included in the other two data sets were chosen. In order to collate the annotated data sets, the corresponding scores (proportional with the likelihood of interaction between mRNA and microRNA) were ranked in a range from 0 to 100. These procedures were carried out both with the mouse and human data sets.
Results
Marked change in the microRNA pattern during mast cell differentiation
Although BMMCs are a widely accepted in vitro model system for studying mast cell functions, they represent an immature cell population that needs other factors to complete their differentiation. Previous studies have provided a detailed description for generating MMCs in vitro, a close homolog of the mast cell population existing in the intestinal mucosa [24, 25], and this protocol has been successfully implemented in our laboratory [26, 27].
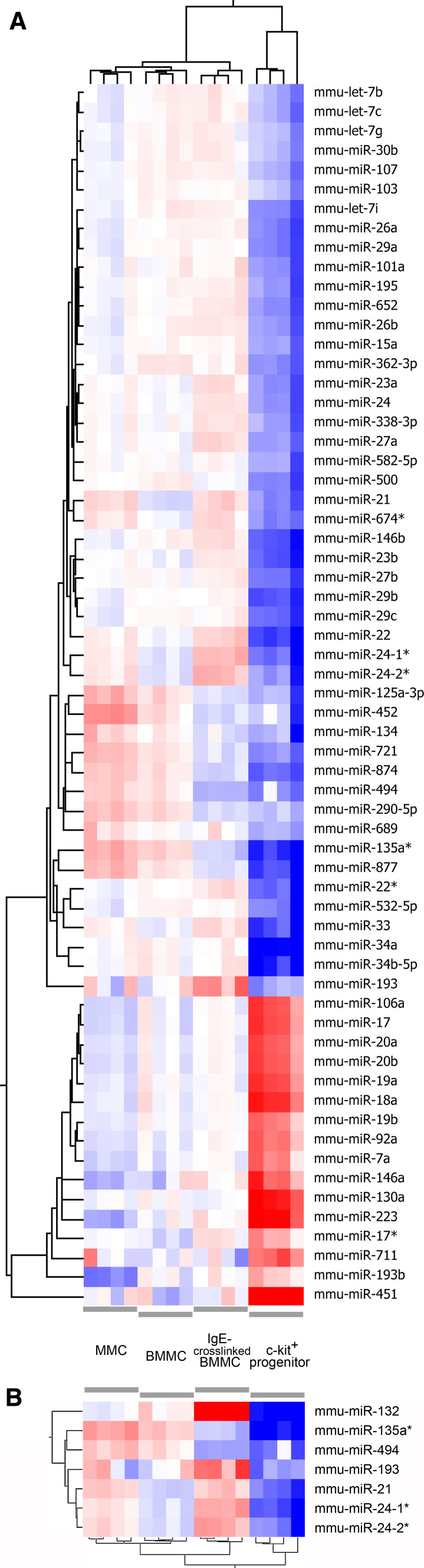
To identify microRNA genes with a characteristic expression profile during the differentiation and activation of murine mast cells, putative committed mast cell progenitors (c-Kit+ cells isolated on the 6th day of differentiation), immature mast cells (BMMC), mucosal-type mast cells (MMC) and IgE-activated BMMCs (Fcε receptor-bound IgE were cross-linked by DNP-HSA for 2 h) were produced and applied to single channel microarray experiments. Next, genes showing a >2.0-fold up- or downregulation at p < 0.05 significance level were selected. Since miRNAs are well known to play an outstanding role in the differentiation in various cell types, we expected the most significant changes to take place between the c-Kit+ progenitors and the other three groups containing immature or differentiated mast cells. Unexpectedly, mucosal differentiation did not result in major changes of microRNA levels, although it can be characterized with a marked phenotypic and gene expression alteration [26] (Fig. 1a).
Fig. 1.
The heat map of differentially expressed miRNA genes among murine mast cell samples representing mucosal mast cell differentiation and activation. a Data passing the signal intensity filter and showing at least two-fold significant changes between BMMCs and c-Kit+ progenitors were hierarchically clustered (entities and conditions, Euclidean metric centroid linkage). b Differentially expressed miRNAs upon IgE-crosslinking were clustered in a similar way. Blue color indicates the relative downregulation, red shows upregulation in the respective group
Our microarray data analysis resulted in 64 miRNAs with significant expression difference among the experimental groups. miRNAs with a higher signal intensity than the average background and with an at least twofold change between the BMMC and any other group were further processed for statistical analyses. During differentiation, when BMMCs were compared to the c-Kit+ progenitors and MMCs, 63 miRNAs showed significant changes (among them, only three expressed differently between BMMCs and MMCs). Two hours after IgE-crosslinking of BMMCs, 7 miRNAs up- or downregulated significantly (Supplementary Table S2–4). Several miRNAs that showed alteration between c-Kit+ progenitor cells (6th day of culture) and BMMCs are known to be involved in the differentiation or function of other hematopoietic cells (Table 1). Interestingly, miRNAs of the miR-17–92 and miR-106a–363 polycistronic clusters, which are well known for their central role in the lymphoid differentiation, showed an intensive and coordinated downregulation [28, 29]. In our model the expression of the myeloid lineage-specific miR-223 was also downregulated during mast cell differentiation. Similarly, miR-451, which is characteristic for the erythroid cells, showed a strikingly lower (~66-fold) expression in BMMCs compared to the progenitors. Taken together, these results show that mast cells coordinately downregulate many miRNAs during their differentiation that are specific for other cell lineages.
Table 1.
Differentially expressed miRNAs involved in the hemotopoietic differentiation and functions
| Differently expressed miRNAs between c-Kit+ progenitor cells (6th day of culture) and BMMCs | Involvement in differentiation or function of hematopoietic cells | ||||||
|---|---|---|---|---|---|---|---|
| miRNAs (systematic name) | BMMC vs c-Kit + progenitor cells | Cells (direction of alteration) | Relevant function | Target | Reference | ||
| Fold change | Regulation | ||||||
| mmu-miR-34a | 14.8 | Up | Dendritic cell (up) | Upregulated during monocyte-derived dendritic cell differentiation | JAG1, WNT1 | [30] | |
| Megakaryocyte (up) | Promotes phorbol esther-induced megakaryocyte differentiation | MYB | [31] | ||||
| B cells | Constitutive expression perturbs B cell development by causing an increase in cells at the pro-B to pre-B cell transition | FOXP1 | [32] | ||||
| mmu-miR-21 | 2.26 | Up | Monocyte/macrophage (up) | Induced in three mouse models of asthma largely through an IL-13Rα1-independent pathway, expressed in cells of monocyte/macrophage lineage | IL12-p35 | [33] | |
| Dendritic cell (up) | Upregulated during monocyte-derived dendritic cell differentiation | JAG1, WNT1 | [30] | ||||
| mmu-miR-451 | 66 | Down | Erythroid cells (up) | Promotes erythroid differentiation | gata2 (zebrafish) | [34, 35] | |
| mmu-miR-223 | 8.91 | Down | Neutrophil granulocytes (up) | Myeloid cell-specific expression | MEF2 c | [36–38] | |
| Enhances granulocytic differentiation, negative regulation of maturation | NFI-A | ||||||
| Enhances T cell differentiation | |||||||
| mmu-miR-146a | 4.65 | Down | Monocytes (up) | Response to LPS as part of TLR-NFκB signaling | IRAK1,IRAK2 and TRAF6 | [39] | |
| mmu-miR-146b | 5.83 | Up | |||||
| miR-17–92 cluster | mmu-miR-17 | 4.77 | Down | B cells | Mice deficient for miR-17–92 die shortly after birth (lung hypoplasia and ventricular septal defect) | BIM | [28] |
| Essential for fetal and adult B-cell development (progression from the pro-B to pre-B cell stage), overexpressed cluster results in the expansion of CD4+ T cells | |||||||
| mmu-miR-18a | 5.67 | Down | |||||
| mmu-miR-19a | 4.47 | Down | PTEN | [29] | |||
| mmu-miR-19b | 2.8 | Down | |||||
| mmu-miR-20a | 3.96 | Down | T cells monocytes (down) | miR-17-5p-20a-106a downregulation unblocks AML1 translation (causes M-CSFR upregulation) in an in vitro monocytic differentiation model | AML1 (RUNX1) | [40] | |
| mmu-miR-92a | 3.43 | Down | |||||
| miR-106a–363 cluster | mmu-miR-106a | 4.58 | Down | Functionally cooperates with miR-17~92 cluster in regulating mouse embryonic development | [28] | ||
| mmu-miR-20b | 4.19 | Down | |||||
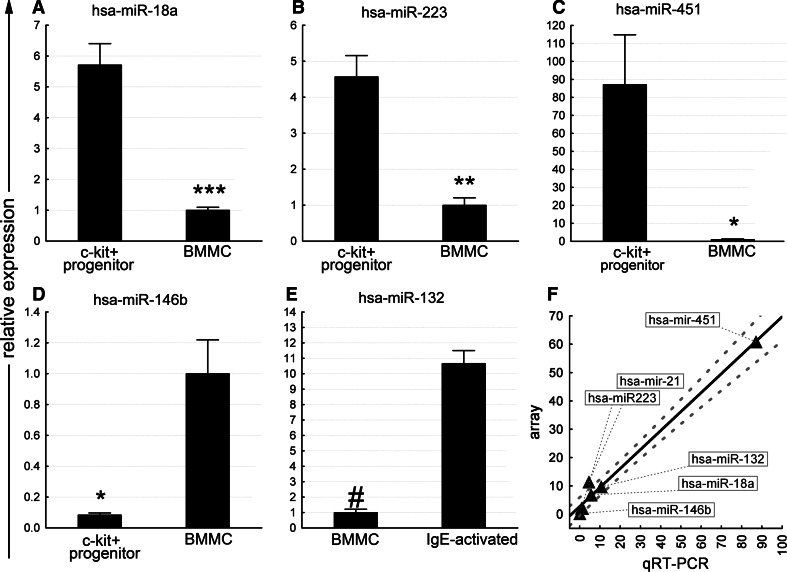
Importantly, when validating the results of the microarray platform by real time RT-PCR, we found a strong correlation between the two types of data (Fig 2).
Fig. 2.
Validation of microarray data by qRT-PCR. a–e qRT-PCR validation of a selected set of miRNA probes normalized to snoRNA135. *, **, ****, # indicate p-values <0.05, <0.01, <0.001, <10−5, respectively. Data are represented as mean ± SEM; n = 4–6 samples per group. f Correlation of miRNA microarray and qRT-PCR data. Dashed line represents 95% confidence interval (Spearman rank order correlation R = 0.857, p < 0.05)
miR-132 is upregulated upon IgE-crosslink in mouse and human mast cells
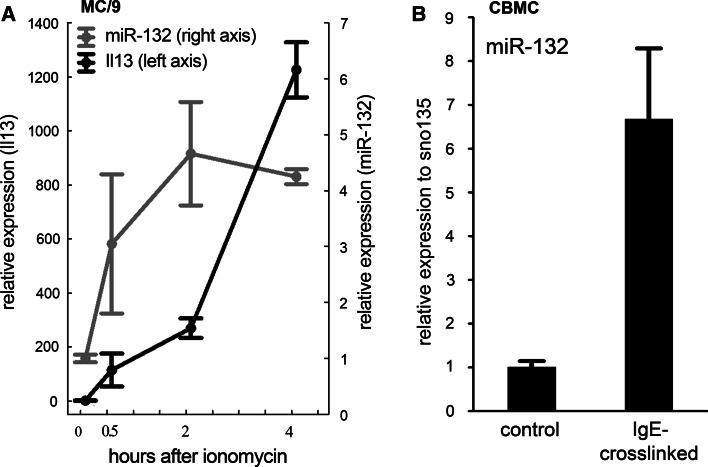
Since miR-132 showed the highest upregulation upon activation (Fig. 1b), and it has been implicated in the regulation of the innate immune response, we selected this miRNA for futher investigation. Mir-132 was previously recognized as an endotoxin-responsive gene in human THP-1 monocytes [39], and BMMCs express TLR4, the major receptor for LPS [41]. In order to exclude the possibility that miR-132 is upregulated because of an LPS contamination of our reagents, first we carried out a control experiment. According to our results IgE and DNP-HSA, but not LPS, specifically induce the transcription of mir-132 in BMMCs (Fig. S2), which not only proves the lack of LPS in our system, but also rules out the possibility that miR-132 upregulation is caused by a minor contamination of the monocyte/macrophage lineage. At the same time, the treatment with LPS caused a marked induction of proinflammatory cytokines (IL-1β, IL-6 and TNF-α) in BMMCs, and it had (but IgE + DNP not at all) a similar effect on these cytokines in the J774.2 macrophage cell line in a parallel experiment (Fig. S3). Importantly, the induction of miR-132 in response to IgE-crosslinking suggested the involvement of an increase in the cytoplasmic Ca2+ concentration, a crucial event of the activation process. Indeed, when using the Ca-ionophor ionomycin in MC/9 mouse mast cells, miR-132 expression started to increase at 30 min and reached a plateau phase already at 2 h after treatment, much earlier than the well-known transcriptional activation marker IL-13 (Fig. 3). In a recent publication, upregulation of miR-221 and miR-222 was found upon ionomycin-induced activation in BMMCs [52]. In our system, miR-221 was induced with a similar kinetics upon IgE-crosslinking and ionomycin; however, miR-132 increased to a greater extent compared to miR-221 (Fig. S4-5).
Fig. 3.
miR-132 is upregulated upon IgE-crosslink in mouse and human mast cells. a Kinetics of Il13 and miR-132 upregulation after ionomycin treatment in MC/9 mouse mast cell line. The induction of miR-132 reaches its plateau phase 2 h after ionomycin treatment (N = 3). b miR-132 is upregulated in human cord blood-derived mast cells 2 h after IgE-crosslinking. (n = 4, p < 0.05, unpaired T test; mean ± SEM)
To confirm the activation-induced miR-132 expression in a human system, mast cells were differentiated in vitro from the CD34+ fraction of cord blood, and the purity of the cultures was checked after 6 weeks. Importantly, when human mast cells were activated by IgE and an agonist anti-IgE antibody, we observed a marked increase in the miR-132 level, thus proving that our findings are not restricted to mouse mast cells.
Target prediction of miR-132
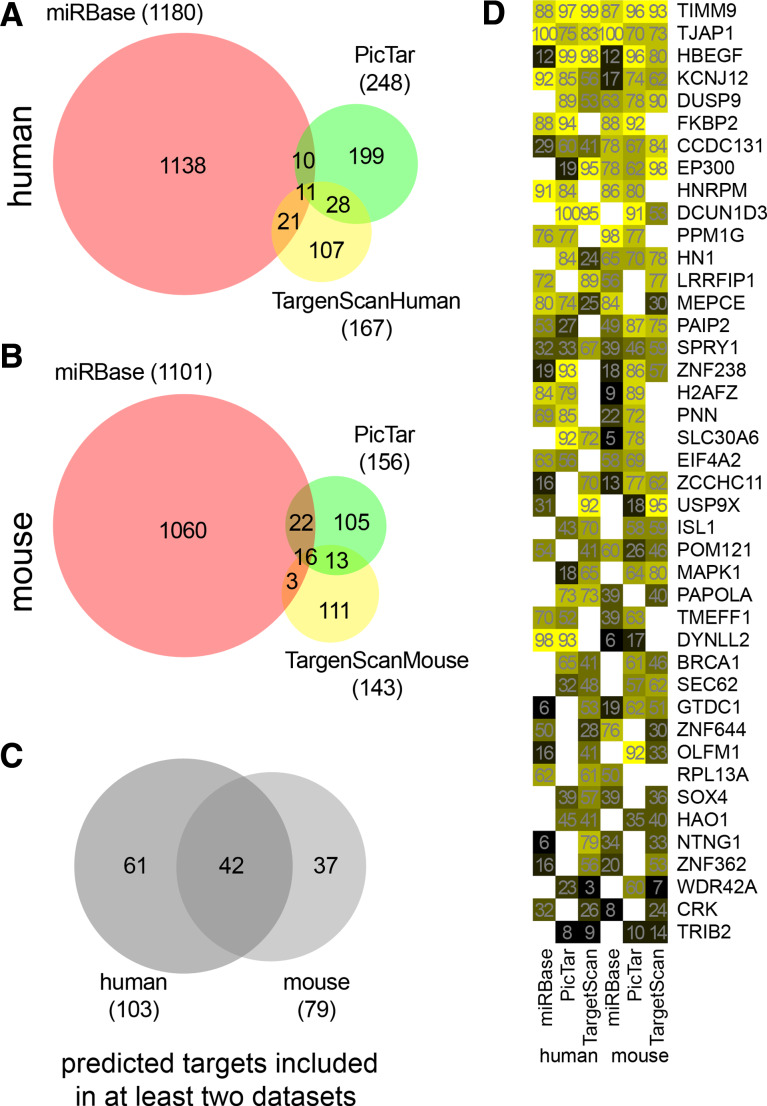
The significant upregulation of miR-132 upon mast cell activation suggested that this microRNA may participate in the modulation of genes with a significant role in mast cell activation. Using bioinformatic approaches we searched for potential mRNA targets of miR-132 by analyzing the mouse and human predicted data sets. To improve the robustness of target prediction for miR-132, three different algorithms were applied simultaneously. The target prediction algorithms assign scores that reflect the probability of interaction between the given microRNA and its target. In order to collate the annotated data sets, the corresponding scores were ranked in a range from 0 to 100 (rank 1 was assigned to the less likely, and the further values were fractionated as percentages).
As was expected, the diverse inputs and analysis stringency of the prediction algorithms resulted in significant differences both in the size and in the composition of the outputs. The number of targets predicted by at least two algorithms is very low compared to the size of the outputs generated by one single method, which is extremely obvious in case of the MiRBase database (Fig. 4a, b). However, when using only those targets that occurred in the output of at least two algorithms, the mouse and human data showed a significant overlap (Fig. 4c, 41 and 53% of human and mouse targets, respectively), reflecting the high degree of conservation in binding sites for miR-132. Next we graded all the records that were present in the output of at least two algorithms, thus identifying several genes as potential targets of miR-132 (Fig. 4d), among them HB-EGF. Interestingly, the functional annotation of the human data set led to a list of 96 targets, with an overrepresentation of the TGF-beta signaling pathway (7 elements, Benjamini-corrected p = 1, 8E–4), ErbB signaling pathway (4 genes, p = 7, 4E−2), adherens junction (4 targets, p = 6, 2E–2) and chemokine signaling pathway (5 genes, p = 6, 8E−2) of the KEGG pathways determined by DAVID v6.7 (http://david.abcc.ncifcrf.gov).
Fig. 4.
Target prediction of miR-132 using mouse and human data sets of three different target prediction databases. The size of outputs from the predictions and their overlap for the human (a) and mouse (b) database show surprisingly minimal overlap. c Targets included in at least two outputs. d Genes from c were graded according to the sum of ranked scores. The color intensity indicates the relative probability of the interaction between miR-132 and the respective targets. Values in each box show the fractionated ranks in the given data set with HUGO identifiers on the right side
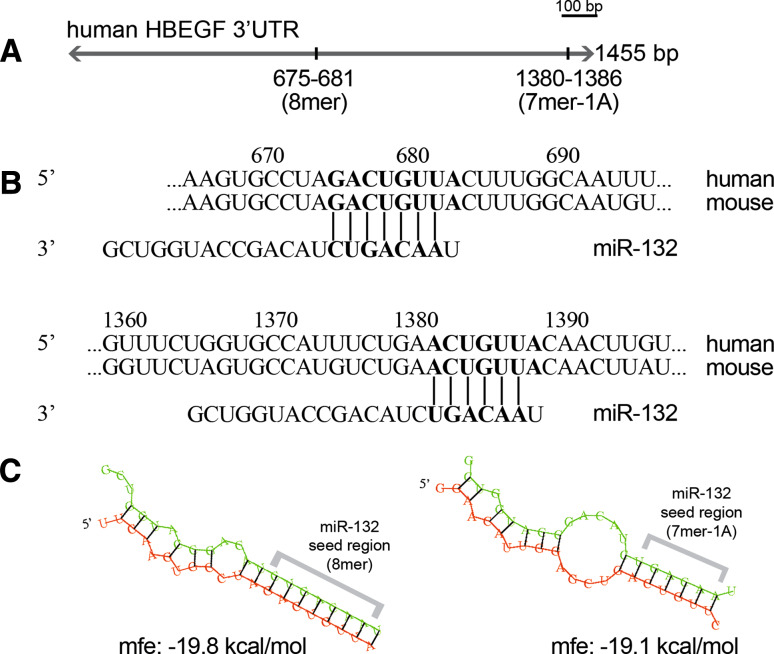
High-throughput approaches used to study the regulatory effects of microRNAs on the protein level have substantiated the hierarchy of types of binding sites and proved the significance of multiple sites in the same 3′UTRs. Importantly, the 8-mer and 7-mer matches seemed to have the strongest impact on expression, and an additive effect was found if a given miRNA had more than one binding site in the target 3′UTR [5, 42]. Interestingly, the localization of binding sites was found to have importance, too; thus a site is more likely to be funtional if localized near the two ends of long UTRs [43]. As HB-EGF has been anticipated to play a critical role in the tissue environment where mast cells reside, furthermore, the two ends of 3′UTR of HB-EGF possess two predicted binding sites for miR-132, which are conserved in mouse and human (Fig. 5); we chose this gene for further evaluation.
Fig. 5.
MiR-132 has two predicted binding sites in the 3′UTR of mouse and human HB-EGF. a The target prediction database TargetScan (version 5.1) shows an 8mer and a 7mer-1A predicted pairing between human HB-EGF 3′UTR and miR-132. b The predicted target sites are identical in the mouse and human HB-EGF 3′UTRs. c Minimum free energy values and structures of the hybridized miRNA/target duplexes calculated by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/)
HB-EGF is upregulated in both mouse and human IgE/antigen-activated mast cells
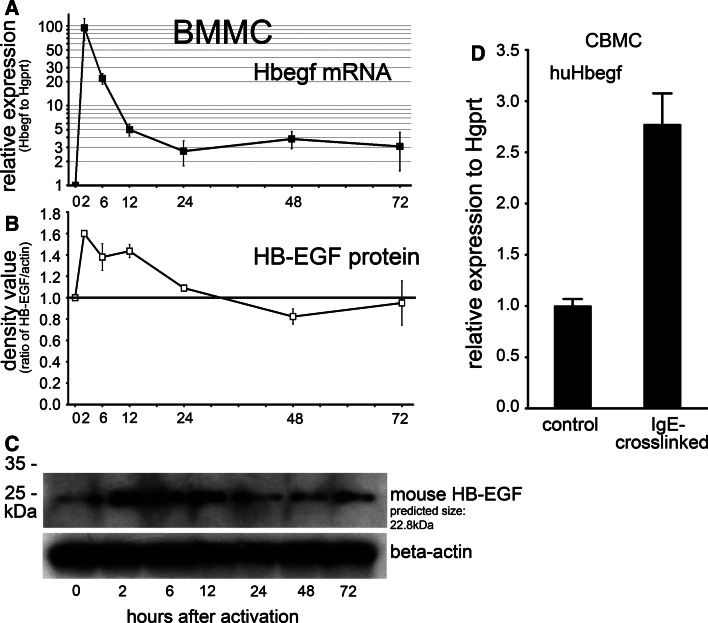
Similar to other growth factors, HB-EGF is upregulated in IgE/antigen-induced mast cells [29]. To follow up the kinetics of HB-EGF expression during mast cell activation, we performed both real-time RT-PCR and Western blot experiments. Already 2 h after activation via IgE/antigen, we could detect a significant increase in the mRNA level of HB-EGF both in BMMCs (Fig. 6a) and human mast cells (Fig. 6d). Interestingly, MC/9 cells showed a more pronounced upregulation of this growth factor compared to the primary cells (Fig. 7b). In line with this, HB-EGF protein showed an increment after 2 h and dropped back to the initial value at 24–48 h after activation (Fig. 6b–c).
Fig. 6.
Kinetic analysis of HB-EGF production in activated mouse mast cells. IgE-presensitized BMMCs were stimulated by 10 ng/ml DNP-HSA, and samples were collected both for RNA and protein analysis from the same cultures at the indicated time points. Note that the marked upregulation of Hbegf mRNA (a, relative expression on a logarithmic scale) is followed by subsequent protein production (b), which returned to baseline after 24 h. The densitometry results of three biological parallels are shown (mean ± SEM). A representative Western blot is depicted on c. d Hbegf is upregulated in human cord blood-derived mast cells 2 h after IgE-crosslinking, as well (n = 5, p < 0.001, unpaired T test, mean ± SEM)
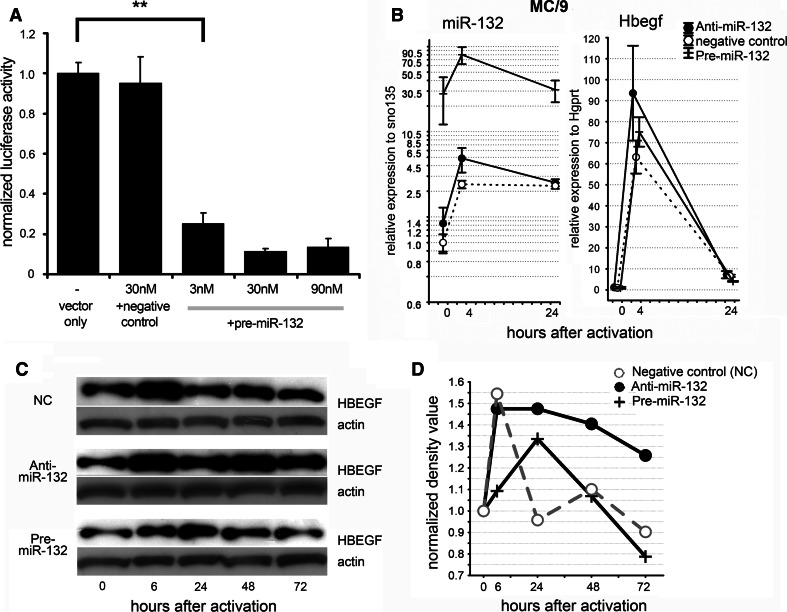
Fig. 7.
miR-132 regulates HB-EGF expression at protein level. a Pre-miR-132 represses the activity of HB-EGF 3′UTR luciferase vector. CHO cells were co-transfected with a luciferase vector containing HB-EGF 3′UTR or control sequence and negative control precursor oligos or Pre-miR-132. The measured luciferase activity signals were normalized to values of the only with vector transfected cells, and then these values were plotted as ratios of correspondingly treated groups, dividing normalized signals derived from HB-EGF 3′UTR and control sequences containing luciferase (n = 3, means ± SEM are shown, **p < 0.01 by ANOVA and Tukey-HSD post-hoc). b Kinetics of miR-132 and Hbegf expression in MC/9 mast cells transfected with miR-132 mimicking (Pre-miR) and neutralizing (Anti-miR) oligonucleotides. MC/9 cells were activated by ionomycin for the indicated time intervals (n = 3, mean ± SEM). Note that miR-132 did not influence the expression of Hbegf at the RNA level in MC/9 mouse cell line. c The effect of modulated expression of miR-132 on HB-EGF protein level in BMMCs. Three independently differentiated BMMCs were pooled and transfected with 100 nM Anti-miR negative control (NC), Anti-miR-132 and Pre-miR-132 oligonucleotides with Amaxa Nucleofector. After 24 h, cells were presensitized with IgE, and then 10 ng/ml DNP-HSA was added. The cells were lysed after the indicated incubation time, and then equal amounts of total proteins were subjected to Western blotting. The blots were incubated with the same buffer with corresponding antibodies (anti-HB-EGF or anti-actin) and exposed to the same X-ray film in order to compare the inter-transfection baseline alterations. d Quantification of the bands on Fig. 7c. These bands were quantified with ImageJ, and the HBEGF/actin density values were plotted after normalization to the corresponding controls. The x axis shows the time point after IgE+ antigen activation. Note that the HB-EGF protein expression was still sustained in the Anti-miR-132-transfected samples even after 72-h-long stimulation
miR-132 regulates HB-EGF expression at the translational level
To validate the interaction between miR-132 and HB-EGF, the miR-132 level was regulated by transiently transfecting Pre-miR and Anti-miR nucleotides into MC/9 mouse mast cells and BMMCs 24 h before activation. We then monitored the effect of miR-132 level on Hbegf in activated cells by using RT-qPCR. The delivery of miR-132 precursor oligonucleotides did not consistently influence the expression of Hbegf at the RNA level upon activation (Fig. 7b and Fig. S6).
First, we checked the functionality of miR-132 binding sites in the 3′-UTR of human Hbegf. For this experiment the luciferase reporter gene was fused with the 3′UTR sequences of human Hbegf, and this vector was co-transfected with Pre-miR-132 or negative control oligos into CHO cells. As the expression of the Hbegf 3′UTR fused luciferase was 4–5-fold less in samples co-transfected with Pre-miR than in the controls (Fig. 7a), this proves that the binding sites in the human 3′UTR of Hbegf are really functional.
Next, we examined whether HBEGF is regulated by miR-132 at the protein level in mouse mast cells. To accomplish this aim, BMMCs were transfected with miRNA-mimicking and antagonizing nucleotides (Pre-miR and Anti-miR, respectively), and were stimulated by cross-linking the high-affinity IgE receptor. In order to analyze the effect of modulated miR-132 levels on kinetics of HB-EGF protein expression, samples were subjected to immunoblotting at different time points after activation (Fig. 7c). The negative control-transfected cells showed a similar HBEGF expression profile as it was observed in untransfected cells (Fig. 6b). In addition, HBEGF protein levels mirrored miR-132 expression if blots were detected and exposed under the same circumstances. In the presence of Pre-miR-132 precursor oligos, a decreased baseline expression of HBEGF was found compared to the negative control (Fig. 7d). Interestingly, the peak of HBEGF production developed tardily and also was completed at a lower level. More importantly, neutralization of endogenously produced miR-132 by Anti-miR-132 results in sustained HBEGF production, and its level remains elevated even after when downregulated in transfection controls. Thus, the exogenous delivery and knockdown of miR-132 clearly prove that miR-132 controls HBEGF protein levels in activated mast cells.
Discussion
Our study determined a microRNA gene expression pattern by microarray technology to gain insight into the differentiation process of mature mucosal-type mast cells and IgE-mediated activation of a commonly used in vitro mouse mast cell model, the BMMCs. The role of microRNAs in the differentiation and functions of cells, such as most hematopoietic lineages, is underlined by an increasing number of studies. Importantly, to our knowledge this is the first comprehensive study that uncovered the specific microRNA pattern characteristic for differentiated and activated mast cells. Surprisingly, our data indicated that most differences could be detected between the progenitor stage and the immature form of mast cells, while only a few miRNAs changed when mucosal mast cells were differentiated from BMMCs in vitro. The relatively low number of miRNAs associated with the MMC maturation process is unexpected as previous studies showed the marked switch of gene expression pattern and phenotypic differences between these two cell populations.
To follow up the miRNA expression changes during mast cell differentiation, data had to be compared to a progenitor population; however, the early steps of this process are still not well understood [44–48]. To obtain these cells for gene expression studies, we separated the c-Kit+ cells at an early time point from differentiating culture committed to the mast cell lineage. These cells, characterized by the CD34+, lineage−, FcεRIα+ phenotype, represent a very early phase of mast cell maturation after 6-day exposition to a defined cytokine environment.
Our microarray analysis showed that during mast cell differentiation many miRNAs, which are involved in haemopoietic differentiation, such as miR-223 or miR-451, are downregulated. Interestingly, miR-223 was found to be specific for the myeloid lineage and negatively regulates the proliferation of the progenitors and granulocyte differentiation and activation [38]. Expression of miR-223 decreases as granulocyte-monocyte progenitors differentiate to monocytes, indicating the role of miR-223 in fate decisions of the myeloid lineage. In addition, miR-451 is known to play an essential positive regulatory role in the differentiation of erythroid cells, whereas it changes in the opposite direction when activated by GATA-1 directly [49]. In vivo studies identified that miR-451 regulates the expression of gata2 in zebrafish [34]. Interestingly, GATA-1 and GATA-2 are shared in erythropoiesis/megakaryopoiesis and mast cell differentiation, and a strict regulation of these transcription factors is important for cell fate decisions of the hematopoietic lineages [50].
In the frame of the first miRNA profiling of hematopoietic cells, the mast cell differentiation was also examined. In this study hematopoietic progenitors, derived from Pu.1−/− mice, were compared to in vitro differentiated BMMCs and to cells representing the different developmental stages of lymphoid lineage. The authors concluded that the expression patterns of differentiated effector cells (Th1, Th2 and BMMC) were more similar to each other than to their respective precursor cells (DN T and Pu.1−/− precursor cells) [51]. The upregulation of miR-26a, miR-24 and miR-27a, and the downregulation of miR-223 have been considered as specific changes during mast cell differentiation. In our system we got similar results about these miRNA changes as there were significant differences in their expression (increased 3.72-fold, 2.87-fold, 2.57-fold and decreased 8.9-fold, respectively).
Our miRNA profiling data may raise many, not yet investigated questions, which can make the role of miRNAs in the differentiation of this hematopoietic lineage understandable. In order to confirm these findings, other studies are needed. In fact, gain of function (ectopic expression of a microRNA gene) and loss of function (knockout of a microRNA gene) experiments help to reveal the functional relevancy of the spatiotemporal expression of these microRNAs [36].
Next we focused on miRNAs differentially regulated upon mast cell activation, and we selected miR-132, which showed the highest upregulation in IgE/antigen-stimulated mast cells. In a previous publication, the upregulation of miR-221 and miR-222 was demonstrated upon mast cell activation in BMMCs in vitro [52]. It was found that these were the only miRNAs that were identified as being differentially expressed in activated compared to resting mast cells. As these upregulated miRNAs were shown to affect cell cycle regulation, they are suggested to play a role in proliferation of mast cells after their activation. In our microarray experiment, during the differentiation and activation we missed detecting the alteration of these miRNAs. This may be due to the different ways to model the activation process, e.g., ionomycin and PMA were applied because of their stronger observed effect on mast cells. Nevertheless, in our system, the crosslinking of Fcε receptor-bound IgE represents a milder but more specific way to induce the activation of mast cells. In fact, in ionomycin-stimulated BMMCs we also observed a significant upregulation of miR-221, but the miR-132 level change was more pronounced upon activation, especially if their alterations in IgE-activated cells are compared to unstimulated ones. The appropriate time for measurements probably has an important role as well, but in response to different activating stimuli similar kinetics were observed in the expression of miR-132 and miR-221 between 6 and 72 h post activation.
The majority of miRNAs are expressed in a wide variety of tissues, and only a relatively small subset of miRNAs accounts for most of the differences in miRNA profiles between cell lineages and tissues. Nevertheless, in many cases miRNAs, originally thought to be tissue-specific, later turned out to be expressed more broadly. Interestingly, miR-132 belongs to this group: first, it was described as a brain-related microRNA. Its function was initially discovered in the brain [53–56], but later its crucial role was proved in other tissues, such as in adipocytes [57], ovarian granulosa cells [58] or normal mammary gland development [59]. Furthermore, recently miR-132 has been discovered to be critical in the maturation and function of cell types involved in the immune system, like monocytes [39] and primary lymphatic endothelial cells [60]. Moreover, this miRNA can serve as a possible link between the neuronal and immune systems. Recently, it has been reported that inflammatory stimuli induced the expression of the miR-132, which mediates an anti-inflammatory effect via targeting acetylcholinesterase in leukocytes. Since the vagal secretion of acetylcholine suppresses inflammation in the periphery, miR-132 was hypothetized to attenuate the inflammation. Furthermore, these studies also showed the involvement of microRNAs in the regulation of the brain-immune axis [61]. Importantly, the emerging role of miR-132 in the immune system is indicated in models of bacterial [39] and viral infection [60], and malignant hematological [62] and chronic inflammatory diseases [63].
Interestingly, our data clearly showed that the upregulation of miR-132 can be induced by a Ca-influx, which is a downstream step of FcεR-signaling. MiR-132 is known to be upregulated in the neuronal context in a CREB-mediated way [55]. Other transcription factors have been indicated to be involved in mast cell activation, like GATA1 and GATA2 (required also in development) [64]. Interestingly, GATA proteins interact with the MeCP2 [54]) and the CREB-binding proteins CBP/p300 [60], both of which are experimentally validated targets of miR-132, and the latter one is also a high-ranked gene in our predicted list (Ep300). These interactions may lead to the accessibility of chromatin for expression from the Th2 locus [65]. With computational methods several cis-regulatory motives (24!) have been identified that may regulate miR-132 expression. The large number of predicted sites for transcription factor binding may indicate the functional importance of miR-132. Furthermore, the combinatorial interaction of these factors is suggested to result in the wide dynamic range of miRNA expression [66]. One of these predicted transcription factors, the cAMP-response element binding protein (CREB), has been validated in a study. Interestingly, miR-132 was found to contribute to neural morphogenesis through the translational repression of a GTPase-activating protein, p250GAP [55].
To find relevant targets of this microRNA, we performed a bioinformatic search by using both mouse and human data sets of three different algorithms. Among the probable targets, the growth factor HB-EGF was further analyzed, as it is upregulated upon IgE crosslinking and is a high-ranked candidate predicted by all three algorithms both in the mouse and human. In luciferase assay we could demonstrate the functionality of miR-132 binding sites in the 3′UTR of human Hbegf. By using miR-132 mimicking and neutralizing oligonucleotides, we confirmed the regulatory effect of miR-132 on HB-EGF expression at the protein level in mast cells. Interestingly, HB-EGF would not be the only target of the “gatekeeper” miR-132, because it was observed to play a role in induction of neovascularization via targeting p120RasGAP in tumor endothelium [67].
HB-EGF is expressed in many types of cells, and it functions as a mitogenic and chemotactic factor for cells expressing the ErbB4 receptor, such as vascular smooth muscle cells, fibroblasts, keratinocytes and hepatocytes. As HB-EGF is also upregulated upon mast cell activation, we suggest that miR-132 is a negative regulator of this mast cell-derived growth factor to avoid inadequate remodeling in chronic allergen exposure. Especially, it may have great importance in physiological responses and diseases, such as wound healing, asthma, nasal polyposis and psoriasis, as all of them can be characterized by an extensive tissue remodeling and inflammatory activity of mast cells. HB-EGF has also been implicated in the pathogenesis of bronchial asthma. Asthma is a chronic respiratory disease that is generally characterized by inflamed airways and reversible bronchoconstriction. A relatively small proportion of patients suffers from a severe type of asthma with permanent airway constricton and the unresponsiveness to corticosteroid therapy. Interestingly, in severe asthma neither the presence of inflammatory cells nor the extent of epithelial damage, but rather the elevated number of fibroblasts and larger mucous glands and airway smooth muscle areas correlate with the severity of the disease [68]. The complex remodeling of the airways and lung tissue has been suggested to be mediated by EGFR-signaling [69] in a rat model of allergic asthma. Very recently, the involvement of EGFR signaling was suggested in the IL-6-dependent recruitment of T cells to the airways in an OVA-induced rat asthma model [70]. HB-EGF is also implicated in heart tissue remodeling after myocardial infarct [71]. Thus, all these data point to the critical importance of proper regulation of EGFR signaling and HB-EGF.
In summary, here we attempted to gain a comprehensive view of the miRNA pattern and its alterations during mast cell differentiation and IgE-mediated activation. These data were obtained by a standard microarray platform, and therefore they can be easily integrated into miRNA gene expression data of other hematopoietic lineages. The interplay between miR-132 upregulation and the regulation of HB-EGF expression suggests a negative feedback mechanism that may take part in silencing the activated mast cells and thus contributes to avoiding the prolonged stimulation of the tissue environment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work is supported by the National Research Foundation of the Hungarian Academy of Sciences (grant OTKA 67955).
Abbreviations
- 3′-UTR
3′-Untranslated region
- BMMC
Bone marrow-derived mast cell
- CREB
Cyclic AMP-response-element-binding protein
- CTMC
Connective tissue-type mast cells
- DNP-HSA
Dinitrophenyl-human serum albumin
- FAM
Fluorescein amidite
- FBS
Fetal bovine serum
- FGF2
Fibroblast growth factor 2
- FYN
Oncogene related to SRC, FGR, YES
- HB-EGF
Heparin-binding EGF-like growth factor
- HER2
Human epidermal growth factor receptor 2
- IL-3
Interleukin-3
- LYN
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog
- MeCP2
Methyl CpG-binding domain protein-2
- MMC
Mucosal mast cells
- mMcpt1
Mouse Mast Cell Protease-1
- NGF
Nerve growth factor
- PLC-γ
Phospholipase C-γ
- SCF
Stem cell factor
- SYK
Spleen tyrosine kinase
- TGF-β
Transforming growth factor beta
- VEGF
Vascular endothelial growth factor
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 4.Rajewsky N. MicroRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 5.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraham SN, St John AL. Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010;10:440–452. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli SJ, Grimbaldeston M, Tsai M. Immunomodulatory mast cells: negative, as well as positive, regulators of immunity. Nat Rev Immunol. 2008;8:478–486. doi: 10.1038/nri2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurish MF, Boyce JA. Mast cells: ontogeny, homing, and recruitment of a unique innate effector cell. J Allergy Clin Immunol. 2006;117:1285–1291. doi: 10.1016/j.jaci.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Kashiwakura J, Xiao W, Kitaura J, Kawakami Y, Maeda-Yamamoto M, Pfeiffer JR, Wilson BS, Blank U, Kawakami T. Pivotal advance: IgE accelerates in vitro development of mast cells and modifies their phenotype. J Leukoc Biol. 2008;84:357–367. doi: 10.1189/jlb.1207841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pejler G, Ronnberg E, Waern I, Wernersson S. Mast cell proteases: multifaceted regulators of inflammatory disease. Blood. 2010;115:4981–4990. doi: 10.1182/blood-2010-01-257287. [DOI] [PubMed] [Google Scholar]
- 11.Metcalfe DD, Baram D, Mekori YA. Mast cells. Physiol Rev. 1997;77:1033–1079. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 12.Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008;454:445–454. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho SH, Yao Z, Wang SW, Alban RF, Barbers RG, French SW, Oh CK. Regulation of activin A expression in mast cells and asthma: its effect on the proliferation of human airway smooth muscle cells. J Immunol. 2003;170:4045–4052. doi: 10.4049/jimmunol.170.8.4045. [DOI] [PubMed] [Google Scholar]
- 14.Wang SW, Oh CK, Cho SH, Hu G, Martin R, Demissie-Sanders S, Li K, Moyle M, Yao Z. Amphiregulin expression in human mast cells and its effect on the primary human lung fibroblasts. J Allergy Clin Immunol. 2005;115:287–294. doi: 10.1016/j.jaci.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 15.Marikovsky M, Breuing K, Liu PY, Eriksson E, Higashiyama S, Farber P, Abraham J, Klagsbrun M. Appearance of heparin-binding EGF-like growth factor in wound fluid as a response to injury. Proc Natl Acad Sci U S A. 1993;90:3889–3893. doi: 10.1073/pnas.90.9.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoll S, Garner W, Elder J. Heparin-binding ligands mediate autocrine epidermal growth factor receptor activation In skin organ culture. J Clin Invest. 1997;100:1271–1281. doi: 10.1172/JCI119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokumaru S, Higashiyama S, Endo T, Nakagawa T, Miyagawa JI, Yamamori K, Hanakawa Y, Ohmoto H, Yoshino K, Shirakata Y, Matsuzawa Y, Hashimoto K, Taniguchi N. Ectodomain shedding of epidermal growth factor receptor ligands is required for keratinocyte migration in cutaneous wound healing. J Cell Biol. 2000;151:209–220. doi: 10.1083/jcb.151.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie H, Wang H, Tranguch S, Iwamoto R, Mekada E, Demayo FJ, Lydon JP, Das SK, Dey SK. Maternal heparin-binding-EGF deficiency limits pregnancy success in mice. Proc Natl Acad Sci USA. 2007;104:18315–18320. doi: 10.1073/pnas.0707909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto R, Yamazaki S, Asakura M, Takashima S, Hasuwa H, Miyado K, Adachi S, Kitakaze M, Hashimoto K, Raab G, Nanba D, Higashiyama S, Hori M, Klagsbrun M, Mekada E. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc Natl Acad Sci USA. 2003;100:3221–3226. doi: 10.1073/pnas.0537588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ongusaha PP, Kwak JC, Zwible AJ, Macip S, Higashiyama S, Taniguchi N, Fang L, Lee SW. HB-EGF is a potent inducer of tumor growth and angiogenesis. Cancer Res. 2004;64:5283–5290. doi: 10.1158/0008-5472.CAN-04-0925. [DOI] [PubMed] [Google Scholar]
- 21.Nakata A, Miyagawa J, Yamashita S, Nishida M, Tamura R, Yamamori K, Nakamura T, Nozaki S, Kameda-Takemura K, Kawata S, Taniguchi N, Higashiyama S, Matsuzawa Y. Localization of heparin-binding epidermal growth factor-like growth factor in human coronary arteries. Possible roles of HB-EGF in the formation of coronary atherosclerosis. Circulation. 1996;94:2778–2786. doi: 10.1161/01.cir.94.11.2778. [DOI] [PubMed] [Google Scholar]
- 22.Higashiyama S, Abraham JA, Klagsbrun M. Heparin-binding EGF-like growth factor stimulation of smooth muscle cell migration: dependence on interactions with cell surface heparan sulfate. J Cell Biol. 1993;122:933–940. doi: 10.1083/jcb.122.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raab G, Klagsbrun M. Heparin-binding EGF-like growth factor. Biochim Biophys Acta. 1997;1333:F179–F199. doi: 10.1016/s0304-419x(97)00024-3. [DOI] [PubMed] [Google Scholar]
- 24.Knight PA, Brown JK, Wright SH, Thornton EM, Pate JA, Miller HR. Aberrant mucosal mast cell protease expression in the enteric epithelium of nematode-infected mice lacking the integrin alphavbeta6, a transforming growth factor-beta1 activator. Am J Pathol. 2007;171:1237–1248. doi: 10.2353/ajpath.2007.061245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller HR, Wright SH, Knight PA, Thornton EM. A novel function for transforming growth factor-beta1: upregulation of the expression and the IgE-independent extracellular release of a mucosal mast cell granule-specific beta-chymase, mouse mast cell protease-1. Blood. 1999;93:3473–3486. [PubMed] [Google Scholar]
- 26.Gilicze A, Kohalmi B, Pocza P, Keszei M, Jaeger J, Gorbe E, Papp Z, Toth S, Falus A, Wiener Z. HtrA1 is a novel mast cell serine protease of mice and men. Mol Immunol. 2007;44:2961–2968. doi: 10.1016/j.molimm.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Wiener Z, Pocza P, Racz M, Nagy G, Tolgyesi G, Molnar V, Jaeger J, Buzas E, Gorbe E, Papp Z, Rigo J, Falus A. IL-18 induces a marked gene expression profile change and increased Ccl1 (I-309) production in mouse mucosal mast cell homologs. Int Immunol. 2008;20:1565–1573. doi: 10.1093/intimm/dxn115. [DOI] [PubMed] [Google Scholar]
- 28.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood. 2009;114:404–414. doi: 10.1182/blood-2008-09-179150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarro F, Gutman D, Meire E, Caceres M, Rigoutsos I, Bentwich Z, Lieberman J. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood. 2009;114:2181–2192. doi: 10.1182/blood-2009-02-205062. [DOI] [PubMed] [Google Scholar]
- 32.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 33.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhan M, Miller CP, Papayannopoulou T, Stamatoyannopoulos G, Song CZ. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp Hematol. 2007;35:1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 37.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 38.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 39.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 41.McCurdy JD, Lin TJ, Marshall JS. Toll-like receptor 4-mediated activation of murine mast cells. J Leukoc Biol. 2001;70:977–984. [PubMed] [Google Scholar]
- 42.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 43.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, Tenen DG, Austen KF, Akashi K. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci USA. 2005;102:18105–18110. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CC, Grimbaldeston MA, Tsai M, Weissman IL, Galli SJ. Identification of mast cell progenitors in adult mice. Proc Natl Acad Sci USA. 2005;102:11408–11413. doi: 10.1073/pnas.0504197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamur MC, Grodzki AC, Berenstein EH, Hamawy MM, Siraganian RP, Oliver C. Identification and characterization of undifferentiated mast cells in mouse bone marrow. Blood. 2005;105:4282–4289. doi: 10.1182/blood-2004-02-0756. [DOI] [PubMed] [Google Scholar]
- 47.Kitamura Y, Shimada M, Hatanaka K, Miyano Y. Development of mast cells from grafted bone marrow cells in irradiated mice. Nature. 1977;268:442–443. doi: 10.1038/268442a0. [DOI] [PubMed] [Google Scholar]
- 48.Rodewald HR, Dessing M, Dvorak AM, Galli SJ. Identification of a committed precursor for the mast cell lineage. Science. 1996;271:818–822. doi: 10.1126/science.271.5250.818. [DOI] [PubMed] [Google Scholar]
- 49.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, Hardison RC, Paw BH, Weiss MJ. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Migliaccio AR, Rana RA, Sanchez M, Lorenzini R, Centurione L, Bianchi L, Vannucchi AM, Migliaccio G, Orkin SH. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J Exp Med. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, Rao A, Kosik KS. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayoral RJ, Pipkin ME, Pachkov M, van Nimwegen E, Rao A, Monticelli S. MicroRNA-221–222 regulate the cell cycle in mast cells. J Immunol. 2009;182:433–445. doi: 10.4049/jimmunol.182.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. MicroRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 55.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23:1876–1884. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiedler SD, Carletti MZ, Hong X, Christenson LK. Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod. 2008;79:1030–1037. doi: 10.1095/biolreprod.108.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ucar A, Vafaizadeh V, Jarry H, Fiedler J, Klemmt PA, Thum T, Groner B, Chowdhury K. miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nat Genet. 2010;42:1101–1108. doi: 10.1038/ng.709. [DOI] [PubMed] [Google Scholar]
- 60.Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 61.Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, Soreq H. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 62.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell’Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Masuda A, Hashimoto K, Yokoi T, Doi T, Kodama T, Kume H, Ohno K, Matsuguchi T. Essential role of GATA transcriptional factors in the activation of mast cells. J Immunol. 2007;178:360–368. doi: 10.4049/jimmunol.178.1.360. [DOI] [PubMed] [Google Scholar]
- 65.Gregory GD, Raju SS, Winandy S, Brown MA. Mast cell IL-4 expression is regulated by Ikaros and influences encephalitogenic Th1 responses in EAE. J Clin Invest. 2006;116:1327–1336. doi: 10.1172/JCI27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee J, Li Z, Brower-Sinning R, John B. Regulatory circuit of human microRNA biogenesis. PLoS Comput Biol. 2007;3:e67. doi: 10.1371/journal.pcbi.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, King PD, Weis SM, Cheresh DA. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benayoun L, Druilhe A, Dombret MC, Aubier M, Pretolani M. Airway structural alterations selectively associated with severe asthma. Am J Respir Crit Care Med. 2003;167:1360–1368. doi: 10.1164/rccm.200209-1030OC. [DOI] [PubMed] [Google Scholar]
- 69.Tamaoka M, Hassan M, McGovern T, Ramos-Barbon D, Jo T, Yoshizawa Y, Tolloczko B, Hamid Q, Martin JG. The epidermal growth factor receptor mediates allergic airway remodelling in the rat. Eur Respir J. 2008;32:1213–1223. doi: 10.1183/09031936.00166907. [DOI] [PubMed] [Google Scholar]
- 70.Tsuchiya K, Jo T, Takeda N, Al Heialy S, Siddiqui S, Shalaby KH, Risse PA, Maghni K, Martin JG. EGF receptor activation during allergic sensitization affects IL-6-induced T-cell influx to airways in a rat model of asthma. Eur J Immunol. 2010;40:1590–1602. doi: 10.1002/eji.200939907. [DOI] [PubMed] [Google Scholar]
- 71.Ushikoshi H, Takahashi T, Chen X, Khai NC, Esaki M, Goto K, Takemura G, Maruyama R, Minatoguchi S, Fujiwara T, Nagano S, Yuge K, Kawai T, Murofushi Y, Fujiwara H, Kosai K. Local overexpression of HB-EGF exacerbates remodeling following myocardial infarction by activating noncardiomyocytes. Lab Invest. 2005;85:862–873. doi: 10.1038/labinvest.3700282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.