Abstract
Sarcomas are a heterogeneous group of tumors with mesenchymal origins. Sarcomas are broadly classified into bone and soft tissue sarcomas with over 50 subtypes. Despite recent advances in sarcoma classification and treatment strategies, the prognosis of some aggressive sarcoma types remains poor due to treatment infectiveness and development of drug resistance. A better understanding of sarcoma pathobiology will significantly increase the potential for the development of therapeutics and treatment strategies. Recently, expressions of microRNAs (miRNA), a class of small non-coding RNAs, have been found to be deregulated in many sarcomas and are implicated in sarcoma pathobiology. Comprehensive understanding of gene regulatory networks mediated by miRNAs in each sarcoma type and the conservation of some shared/conserved miRNA-gene networks could be potentially investigated in the prevention, diagnosis, prognosis and as multi-modal treatment options in these cancers. In this review, we will discuss the current knowledge of miRNA–gene regulatory networks in various sarcoma types and give a perspective of the complex multilayer miRNA-mediated gene regulation in sarcomas.
Keywords: Sarcoma, MicroRNA, Soft tissue sarcoma, Bone sarcoma, Markers, Expression, Gene networks, Osteosarcoma, Rhabdomyosarcoma, MPNSTs, GIST, Synovial sarcoma, Liposarcoma
Introduction
Sarcomas are mesenchymal tumors that accounts for about 1 % of all malignant tumor types in humans [1]. Approximately 15 % of all malignant tumors in children are pediatric sarcomas. Sarcomas can occur anywhere in the body, but the majority occur in the extremities and can be broadly classified into bone and soft tissue sarcoma with over 50 subtypes [2, 3]. Sarcoma nomenclature is generally based on the cell and/or tissue type, as in osteosarcoma (OS), angiosarcoma, rhabdomyosarcoma (RMS) and liposarcoma. Even within a specific sarcoma type, tumors are highly heterogeneous and the histopathological and clinical features are not always distinct, causing a diagnostic challenge. Several sarcoma types, however, are characterized by chromosomal translocations resulting in tumor-specific fusion transcripts [4, 5]. The presence or absence of such fusion transcripts are widely used as diagnostic markers for certain subtypes [6]. In addition, DNA copy number changes are used in sarcoma diagnosis [7]. Despite these markers, a significant number of sarcomas are characterized by complex karyotypes and remain unclassified [6]. The risk of metastasis and the prognosis also depends on the sarcoma type and grade [2]. Generally, the standard-of-care for high-grade tumors includes chemotherapy and/or radiation. Despite advances in treatment strategies, the 5-year survival rate remains static for certain sarcoma types [8]. This bleak prognosis could be attributed to the relative ineffectiveness of the treatment as well as the complex tumor biology leading to drug resistance [9]. A better understanding of sarcoma biology will significantly increase the potential for identification and development of novel therapeutic targets and treatment strategies.
Significant efforts have been made to generate gene expression profiles of various sarcoma types [10–14]. These profiles have enhanced our understanding of the biology and classification of various sarcoma types [15]. In recent years, microRNA (miRNA) expression patterns of several sarcoma types have been determined [16, 17]. MicroRNA signature-based classification and subgrouping of sarcomas were largely based on lines of differentiation [16]. It is well established that miRNAs play a significant role in posttranscriptional gene regulation [18]. Substantial expression data have been generated using sarcoma tumor tissues and cell lines, allowing us to integrate gene and miRNA expression patterns and perform complex correlative and functional studies [19] to better decipher the sarcoma biology. In this review, we will discuss our current understanding of miRNA-mediated gene regulatory networks in various sarcoma types and give our perspective on the complex layers of gene regulation mediated by miRNAs in these cancers.
MicroRNA biogenesis and function
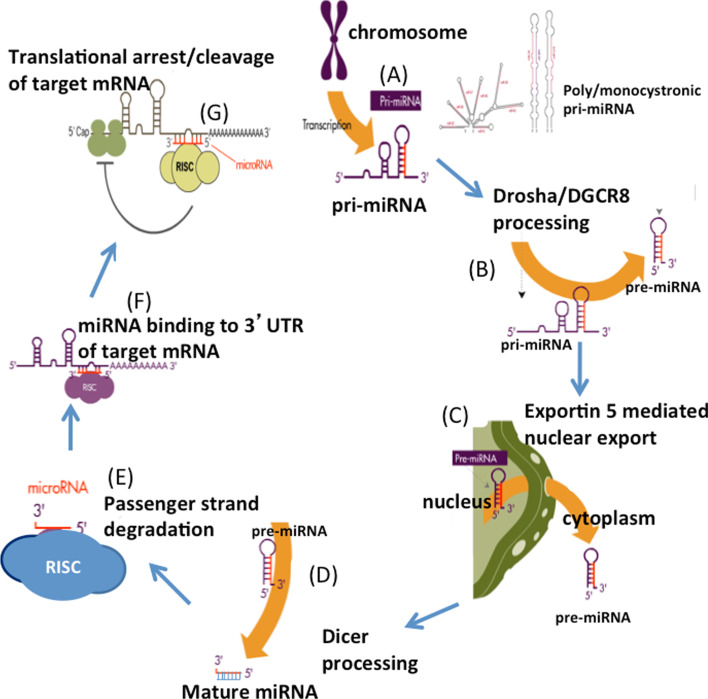
MicroRNAs play a fundamental but significant role in most aspects of cellular biology [20–25]. miRNA biogenesis pathways and the mechanisms of regulatory functions are described elsewhere in detail [25–28]. Here, we briefly outline the biogenesis of miRNA. miRNAs are evolutionarily conserved small non-coding RNAs that post-transcriptionally regulate gene expression. In humans, miRNAs are present in exons, introns or intergenic regions. miRNAs are initially transcribed as primary miRNAs (pri-miRNAs), either as monocistronic (single) or polycistronic (multiple miRNAs) transcripts [29]. These pri-miRNAs are processed by an RNAse III enzyme, Drosha, to become precursor miRNA (pre-miRNA), a stem loop structure of about 70 nucleotides. Pre-miRNAs are exported to cytoplasm by exportin 5 and subsequently processed by Dicer to form mature miRNAs, 18–24 nucleotides in length. The mature miRNA duplex is then bound to a miRNA-induced silencing complex (miRISC), where the passenger or star strand is selectively degraded, retaining the active strand. The ‘seed’ sequence in the active strand directs the miRISC to complementary sites in the mRNA transcripts (typically in the 3′UTRs) and negatively regulates gene expression [30, 31]. Protein translation regulation or mRNA degradation of the target is determined based on the miRNA complementarity with the target sequences [31, 32]. The biogenesis of miRNAs is graphically illustrated in Fig. 1.
Fig. 1.
Schematic representation of microRNA biogenesis. The figure is partly adapted from Subramanian and Steer [36]
miRNAs have emerged as major gene regulators and are implicated in many disease processes including cancers [25]. More than 60 % of human transcripts are directly or indirectly under the control of miRNAs. miRNAs associated with cancer are collectively known as oncomiRs [33]. A miRNA can function either as an oncogene or a tumor suppressor based on the cell types and the target genes it regulates [34, 35]. The role of miRNA can be attributed to the established hallmarks of cancer. For instance, over 40 miRNAs are implicated in cellular apoptosis [36] and some may have overlapping roles in other cancer mechanisms such as cell angiogenesis, invasion and metastasis [37–39].
The functional role of miRNAs has been extensively studied in many cancer types, especially carcinomas and hematopoietic malignancies [35]. In addition, these non-coding RNAs are also implicated in regulating human embryonic stem cells and cancer stem cells [40, 41]. Due to the rarity and modest availability of quality tumor tissues, there are only limited studies of miRNAs in sarcomas. Further, lack of sufficient sarcoma models has also dampened the miRNA studies in sarcomas. Despite these obstacles, a number of studies have been reported on various sarcoma types and on the mechanisms through which these miRNAs might contribute to sarcoma biology. For instance, work by Riggi et al. [42] provides insights into the mechanisms whereby a single oncogene can reprogram primary cells to display a cancer stem cell phenotype in Ewing’s sarcoma. They reported that fusion protein EWS/Fli1 and miR-145 form a mutually regulating feedback loop and identified SOX2 as their common target.
miRNA expression patterns in human sarcomas
Expression profiling of large numbers of miRNAs is a powerful technique widely used to understand gene regulatory networks in cancer. It has been reported that, compared to mRNA profiling, miRNA profiling can accurately classify human cancers [43]. Thus, miRNA expression patterns may be more closely linked to tumor differentiation. Our group conducted the first study to report global miRNA profiling in sarcomas [16]. This study included 27 sarcomas, representing 7 sarcoma types. Profiling was performed using microarray technology and/or small RNA cloning and sequencing. The different tumor types showed distinct miRNA expression profiles, as demonstrated by an unsupervised hierarchical clustering. Unique miRNA expression signatures were identified in each tumor class. Remarkably, the miRNA expression patterns were able to successfully distinguish two of the sarcomas that had been misdiagnosed. We confirmed this by reevaluating the tumors using histopathologic and molecular analyses. Further, using a cloning approach, we identified 14 novel miRNAs in the sarcomas examined. Our data showed that different histological types of sarcomas have distinct miRNA expression patterns, reflecting the apparent lineage and differentiation status of the tumors. The identification of unique miRNA signatures in each tumor type may indicate their role in tumorigenesis and may aid in diagnosis and/or prognosis of soft tissue sarcomas. Recently, Greither et al. [44] reported the expression of hypoxia-regulated miRNA, miR-210, to be significantly associated with the prognosis and the age of tumor onset in a gender-specific manner in soft-tissue sarcoma patients. This evidence on the correlation of expression of a single miRNA with prognosis in sarcoma sheds light on the importance of these small RNA molecules in sarcoma biology.
Recently, our group developed a web-accessible Sarcoma miRNA Expression Database (S-MED), which is a repository that describes the patterns of miRNA expression in 22 human sarcoma subtypes [17]. S-MED provides both basic and advanced data search options for exploration of the data in heat map and text/numerical formats. The database also provides statistical details such as fold changes and p values for differentially expressed miRNAs in each sarcoma type and corresponding normal tissue. This comprehensive database is available through the URL link http://www.oncomir.umn.edu/. Together, these high-throughput analyses reveal unique miRNA expression signatures for different sarcoma subtypes, indicating that miRNAs can regulate specific tissue lineages during tumor differentiation. As the cell of origin for many sarcomas remains uncertain, a key difficulty is in determining the normal tissue to which sarcomas expression can be compared. To circumvent this issue, some researchers concentrate mainly on differences in miRNA expression between different sarcoma types. Our database will aid investigators in comparing their sarcoma or miRNA of interest against other tumor subtypes or miRNAs. Further functional characterization of candidate miRNAs has identified the networks of several miRNAs and their target genes that can serve as potential diagnostic/prognostic biomarkers and as therapeutic targets. For the purpose of clarity, the studies are presented for each subtype, although for some extremely rare forms of sarcomas, no reports are yet available.
Bone sarcomas
Bone sarcomas are relatively rare and account for only 0.2 % of all neoplasms. Several subtypes of bone sarcomas such as OS, Ewing’s sarcoma and chondrosarcoma are identified based on the unique histology, cell of origin, clinical features and site distribution. The following are the major histological subtypes of bone sarcoma and their miRNA expression patterns and functions.
Osteosarcoma
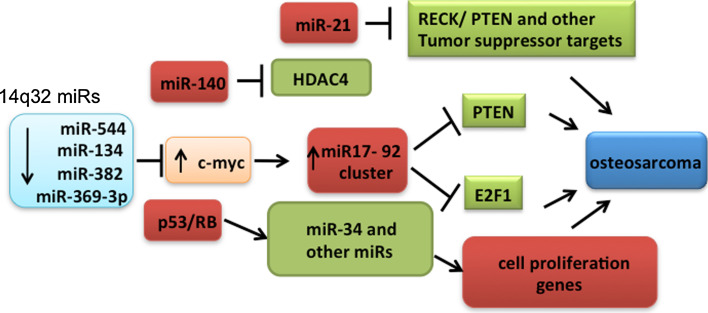
Osteosarcoma is the most common sarcoma and the primary malignant bone tumor with an incidence of 4–5 cases per million. It arises from the metaphysis of the long bones of adolescents and young adults. Two studies have shown significant downregulation of miRNAs at the chr.14q32 locus in OS compared to normal bone tissues [45, 46]. Interestingly, neither group observed DNA copy number changes in this locus, suggesting that additional epigenetic mechanisms have contributed to this downregulation. In addition, using bioinformatic predictions, we identified a subset of 14q32 miRNAs (miR-382, miR-369-3p, miR-544 and miR-134) that could potentially target cMYC transcript [45]. We also functionally characterized this regulatory network. Restoring the expression of these four 14q32 miRNAs decreased cMYC levels and synergistically induced apoptosis in Saos2 cells. Further exogenous expression of 14q32 miRNAs in Saos2 cells significantly downregulated miR-17-92, a transcriptional target of cMYC. The pro-apoptotic effect of 14q32 miRNAs in Saos2 cells was rescued either by overexpression of cMYC cDNA without the 3′UTR or with miR-17-92 cluster. Together, our data support a model where the deregulation of a network involving 14q32 miRNAs, cMYC and miR-17-92 miRNAs could contribute to OS pathogenesis [45]. Maire et al. [46] also identified miRNAs that are predicted to target genes involved in diverse intracellular signaling pathways, including Notch, RAS/p21, MAPK, Wnt, and the Jun/FOS pathways. Further, they developed a comprehensive molecular genetic map by integrating the miRNA profiles with previously published array comparative genomic hybridization DNA imbalance and mRNA gene expression profiles from a set of partially overlapping OS tumor samples. Among the different miRNAs examined, an miR-382 and cMYC regulatory circuit was observed to be deregulated in multiple studies and has been functionally validated [45–47]. miRNA-gene network deregulation in OS is illustrated in Fig. 2.
Fig. 2.
Deregulation of miRNA-mediated gene networks in osteosarcoma
Several other groups have compared miRNA expression in OS tissues, cell lines, and normal osteoblast cell lines. They have identified dysregulated miRNAs and their potential mRNA targets in OS [47–52]. For instance, Braun et al. [53] reported that p53-responsive miRNAs, miR-192 and miR-215 are capable of inducing cell cycle arrest in U2OS cells carrying wild-type p53. Recently, Yan et al. [54] demonstrated that over-expression of p53 transcriptional target, miR-34a, could inhibit tumor growth and metastasis of OS probably through downregulating cMet. Functional studies have enabled identification of miRNAs which have the potential to prevent disease progression. Creighton et al. [55] uncovered a p53-associated role for miR-31 in inhibiting proliferation of OS cell lines. miR-31 was demonstrated as being able to inhibit multiple steps in the metastatic development of breast cancer by downregulating mRNA targets such as intergrin A5, radixin and RhoA [55]. These results suggest that in vivo delivery of miR-31 may have the potential to prevent pulmonary metastasis in OS patients [56]. miRNAs can also be potential biomarkers and therapeutic targets in OS. Duan et al. [47] have reported that restoring miR-199a-3p function may provide therapeutic benefits in OS by decreasing mTOR and signal transducer and activator of transcription 3 (STAT3) expression. miR-125b is yet another miRNA that is frequently deregulated in OS samples and human OS cell lines. It suppresses proliferation and migration of OS cells through regulation of STAT3 [57]. Similarly, suppression of miR-21 which is overexpressed in OS, decreased invasion and migration in MG-63 OS cell lines [58]. miR-21 targets RECK which suppresses invasion of OS cells by decreasing the activity of MMPs [59]. Also, miR-183 plays a role in suppressing migration and invasion in OS cells by targeting Ezrin [60]. Further, the expression levels of miR-183 significantly correlate with lung metastasis as well as with local recurrence of OS. A recent study by Jones et al. [49] confirmed the tumor suppressive role of miR-16 and the pro-metastatic role of miR-27a by performing in vitro and in vivo functional validation in OS cell lines.
miRNAs can also act as biomarkers of chemotherapeutic responses in sarcomas. miR-140 is the first reported miRNA candidate associated with drug sensitivity in OS tumor xenografts treated with the chemotherapeutic agents Doxorubicin, cisplatin and ifosfamide (IFO)[61]. In this study, Song et al. [62] revealed consistent high expression of miR-140, which caused chemoresistance to methotrexate (MTX) and 5-Fluorouracil (5-FU) and suppressed cell proliferation in both U2OS and MG-63 OS cells. Further, miR-140 negatively regulates histone deacetylase 4 (HDAC4) which interacts with p21, resulting in 5-FU resistance. The same group also identified miR-215 as playing a significant role in inducing chemoresistance to MTX in U2OS cells. In another study, Gougelet et al. [63] examined miRNA expression profiles in 27 OS paraffin-embedded samples to determine the relevance of miRNA expression on chemoresistance to IFO. Supervised hierarchical clustering identified five candidate miRNAs (miR-92a, miR-99b, miR-132, miR-193a-5p, and miR-422a) that showed a statistically significant ability to discriminate between good and poor responders to IFO. The targets of these miRNAs detected by the in silico approach were involved in cell cycle regulation, invasion, and bone resorption through MAP kinase, TGF-β and Wnt pathways. Recently, Jones et al. [49] reported higher expression of miR-451 and miR-15b in pre-treatment biopsy OS samples that correlated with subsequent positive responses to chemotherapy. This indicates the ability of miRNAs to stratify patients, suggesting that personalized chemotherapy regimens may be viable in the near future. A recent review extensively covers miRNA studies reported in OS [56].
Ewing’s sarcoma
Ewing’s sarcoma is an aggressive pediatric malignancy that usually begins growing in a bone. It occurs primarily in children and young adults, often appearing during the teen years. It is primarily driven by a fusion of EWS/Ets oncoproteins, which are gain-of-function transcriptional regulators. Two studies analyzed global miRNA profile changes after stable silencing of EWS/Fli1 fusion protein [64, 65]. Mckinsey et al. [65] reported strong repression of a group of miRNAs by EWS/Fli1. Interestingly, these miRNAs have predicted targets in the insulin-like growth factor (IGF) signaling pathway, a pivotal driver of Ewing sarcoma oncogenesis. In the second study, Ban et al. [64] identified miR-145 as the top EWS/Fli1-repressed miRNA. miR-145 was expressed at low levels in primary Ewing’s sarcoma tumor samples as compared to mesenchymal progenitor cells. Surprisingly, miR-145 was found to target the EWS/Fli1 transcript directly, functioning in a mutually repressive feedback loop that regulated expression of this fusion protein and miRNA [42, 65]. This regulatory network represents an important component of the EWS-FLI1-mediated Ewing’s sarcomagenesis that may open a new avenue to future miRNA-mediated therapy of this devastating malignant disease. Yet another target of this fusion oncoprotein is let-7a [66]. In this study, miRNA arrays were used to compare the global miRNA expression profile of human mesenchymal stem cells and Ewing’s sarcoma cell lines, and showed that Ewing’s sarcoma displays a distinct miRNA signature that includes induction of the oncogenic miR-17-92 cluster and repression of the tumor suppressor let-7 family. In particular, the authors identified deregulation of let-7a and its target HMGA2 expression to be key events in the development of Ewing’s sarcoma.
miRNAs have also been investigated as potential biomarkers of survival in Ewing’s sarcoma [67]. In this study, miR-34a appeared to be associated with either event-free or overall survival and emerged as a significant predictor of outcomes. Patients with the highest expression of miR-34a did not experience adverse events in 5 years. The high miR-34a expression could be detected in paraffin-embedded tissues by in situ hybridization, thus contributing to an easy routine evaluation of this miRNA. Further, restoration of miR-34a activity may be useful to decrease malignancy and increase tumor sensitivity to current drugs, doxorubicin and vincristine, thus suggesting its role as a response to therapy marker. In a recent study, an integrated analysis of miRNA and gene copy numbers was performed in xenografts of Ewing’s sarcoma [68]. Twenty differentially expressed miRNAs were pinpointed in regions carrying altered copy numbers, attributing changes in miRNA expression to copy number changes.
Chondrosarcoma
Chondrosarcoma is a cancer composed of cells derived from transformed cells that produce cartilage. Peripheral chondrosarcoma is a malignant transformation of multiple osteochondroma (MO), which is a rare skeletal disease characterized by the formation of multiple benign cartilage-capped bone tumors. The most serious complication of this pathology is malignant transformation into chondrosarcoma, which is estimated to occur in 1–5 % of patients. Specific miRNAs have been reported to be involved in chondrogenesis and inflammatory cartilage diseases [69, 70]. Recently, Salvatore et al. [71] reported a miRNA profiling study in osteochondroma using 19 patient samples compared to normal cartilage growth plate controls. They observed an expression signature comprising of eight miRNAs that were able to further distinguish healthy growth plate controls from MO patients, which can regulate genes involved in MAPK and insulin signaling pathway, Wnt signaling, heparin sulfate and glycan structure and biosynthesis. This is in line with gene expression analyses previously performed in osteochondroma and chondrosarcoma samples showing modulation of signal transduction pathways like TGF-β/Wnt and IHH/PTHLH during osteochondroma formation and malignant progression. Other gene members of IGF family (e.g., IGF2 and IGFBP5) were also identified as differentially expressed between osteochondromas and human growth plates. The overlap between miRNA target predictions by Salvatore et al. [72] and signaling cascades previously related to MO pathogenesis support the potential involvement of the detected miRNAs in MO pathogenesis and malignant transformation.
Chordoma
Chordoma is a rare, slow-growing neoplasm that typically arises from bone in the skull base and anywhere along the spine. A subtype called chondroid chordoma shows histological features of chordoma and chondrosarcoma. Recently, miRNA expression was analyzed in chordoma-derived cell lines and chordoma tissue using miRNA microarray technology with unsupervised hierarchical clustering analysis [73]. In this study, the authors identified miR-1 and miR-206 as being expressed at a significantly lower level or absent in chordoma cells compared to normal cells. Reintroduction of miR-1 inhibited the growth of chordoma cells, with suppression of MET and HDAC4. MET is part of a receptor tyrosine kinase family of oncogenes overexpressed in many human cancers including sarcomas, particularly in chordoma (94.4 %), chondrosarcoma (54.2 %), and OS (23.3 %) [74]. Importantly, a recent study suggested that the MET oncoprotein plays a major role in the metastatic process in chordoma [75].
Soft tissue sarcomas
Cancers that primarily affect connective tissues such as muscle (smooth and skeletal), fat and blood vessels are generally grouped as soft tissue sarcomas. The majority of sarcoma subtypes are classified as soft tissue sarcomas. These malignant mesenchymal tumors account for about 1 % of all human malignant cancers. Soft tissue sarcomas such as gastrointestinal stromal tumor, synovial sarcoma (SS), RMS, leiomyosarcoma (LMS), liposarcoma, fibrosarcoma, and angiosarcoma are discussed in this section.
Gastrointestinal stromal tumor
Gastrointestinal stromal tumor (GIST) is one of the most common mesenchymal tumors of the gastrointestinal tract (1–3 % of all gastrointestinal malignancies). It is typically defined as a tumor with activating mutations in the c-kit gene or PDGFRA gene. We have profiled miRNA expression of GIST samples and observed that miR-221 and miR-222, which have been shown to target the 3′UTR region of KIT in experimental systems [76], were expressed at a lower level. Since 80 % of GIST harbors an activating mutation in KIT [77], lower expression of miR-221 and miR-222 may allow for increased translation of KIT and further enhance its oncogenic influence on the cell. This is consistent with a recent finding using 54 paraffin-embedded GIST tissues, where the authors observed significant repression of miR-221 and miR-222 in KIT-positive GISTs, compared to normal tissue, which was completely reversed in KIT-negative GISTs [78]. The authors suggest that miR-221/222 can be considered as a tool for future therapeutic strategies for GISTs, especially for tumors with secondary resistance to tyrosine kinase inhibitors. Another therapeutic target that has been investigated recently is miR-494 [79]. Using functional studies, Kim et al. [79] have established miR-494 to be a negative regulator of KIT in GISTs and have shown that overexpression of miR-494 in GISTs may be a promising approach to GIST treatment. Previously, we have shown that miRNA expression patterns can distinguish and identify a misdiagnosed GIST case [16].
Another interesting observation with regard to miRNA regulation in GISTs is the significant downregulation of 44 miRNAs clustered in a genetically imprinted region at 14q32.31 [80]. In this study involving 12 GISTs, the authors observed significantly lower expression of two candidate 14q32 miRNAs in GISTs with 14q loss, and also in GISTs with tumor progression, indicating a correlation between miRNA expression and tumor progression. This was further validated in another study using 20 frozen GISTs. In this study, Choi et al. [81] observed 6 GISTs without 14q loss form a separate cluster distinct from remaining 14 samples. In the 14 GISTs with 14q loss, 5 small bowel GISTs formed a separate cluster, whereas the 9 remaining GISTs divided into two groups according to frequent chromosomal losses and tumor risk. Moreover, overexpression of miR-196a was associated with high-risk grade GISTs showing metastasis and poor survival. The expression of miR-196a was also correlated with expression of HOXC and the lncRNA, HOTAIR [82]. Thus, these studies demonstrate the efficiency of miRNAs to classify GISTs to various subtypes and tumor grade.
Rhabdomyosarcoma
Rhabdomyosarcoma, a malignant, skeletal muscle-derived tumor is one of the most common soft-tissue sarcomas of childhood. It accounts for 6–8 % of all pediatric tumors and includes two main histological subtypes, embryonal (ERMS) and alveolar (ARMS) RMS [3, 83]. While ERMS has better prognosis (5-year overall survival >70 %), ARMS is more aggressive with poor outcomes [84]. ARMS is often associated with chromosomal translocations, the resulting oncogenic fusion protein (for, e.g., PAX3-FKHR) have greater prognostic values as they characterize a distinctly aggressive subgroup that is frequently unresponsive to conventional therapies and has a high rate of recurrence. Since RMS is widely thought to originate from myogenic precursors, the bulk of studies addressing the role of miRNAs in RMS have focused on myomiRs, i.e., the miR-1/miR-133/miR-206 family. These are miRNAs that specifically control cell fate determination of myogenic precursors and muscle tissue homeostasis.
In our initial study, we showed the distinction between ERMS and ARMS using miRNA profiling [16]. Similarly, Gougelet et al. [85] also demonstrated miRNA expression profiling as a useful strategy to discriminate RMS subtypes. Several studies have examined the potential of myomiRs as therapeutic targets in RMS [86]. The myomiRs are largely downregulated in RMS and their functional contribution has been confirmed by gain-of-function studies in which overexpression of myomiRs inhibited RMS cell proliferation [87–90]. Overexpression of miR-1 and miR-133a in an ERMS cell line revealed a tumor suppressor-like role for these myogenic miRNAs [87]. Similarly, re-expression of miR-206 in RMS cells promoted myogenic differentiation and blocked tumor growth in xenografted mice [89]. Further, it has been shown that miR-1 and miR-206 directly regulate MET proto-oncogene, the Met tyrosine-kinase receptor, which is overexpressed in RMS [88, 89].
We have also observed relative downregulation of myomiRs, miR-1 and miR-133 in RMS compared to normal skeletal muscle [16]. Further, in ARMS, miR-335 was found to be overexpressed. This is interesting, since miR-335 resides in the intron 2 of MEST (also known as PEG1). MEST has been indicated to play a role in muscle differentiation [91], and its mRNA expression is high in ARMS (Baer et al. 2004 [92]). MEST is a downstream target of PAX3, the gene involved in the PAX3–FKHR fusion [93]. It thus appears that the PAX3–FKHR fusion may influence the transcription of miR-335 that has several predicted myogenic targets including CHFR and HAND1.
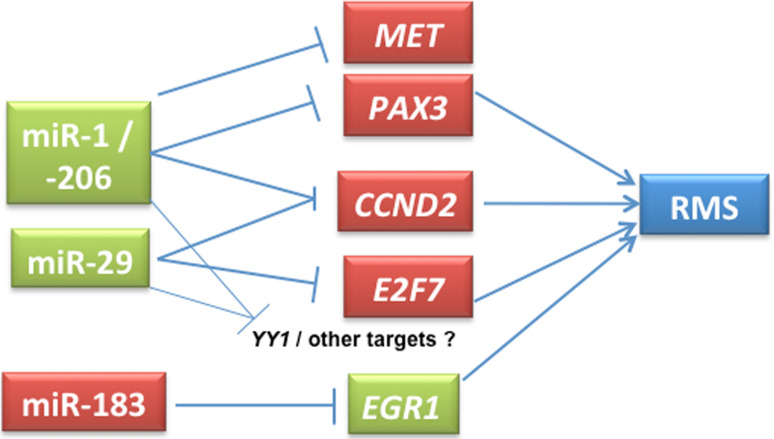
In addition to myomiRs, non-muscle-specific miRNAs have also been implicated in RMS. miR-29 has been shown to be downregulated in RMS compared to normal muscle tissues [16, 94]. miR-29 is a tumor suppressor in RMS, the overexpression of which leads to cell cycle arrest and differentiation of RMS cell lines. Wang et al. [94] characterized the miRNA regulatory circuit NFkB-YY1-miR-29, and indicated that the disruption of this circuit can lead to RMS tumorigenesis. NF-kB activation can lead to overexpression of YY1, which in turn interacts with EZH2, causing sustained downregulation of miR-29b/miR-29c leading to repression of myogenesis. Consistent with this, tumor tissues from RMS patients showed upregulation of YY1 and EZH2. miR-26a is yet another negative regulator of EZH2, the expression of which is downregulated in RMS [95]. We have observed high levels of miR-183 in RMS cell lines and primary tumors and have shown an oncogenic role for this miRNA using knockdown studies [34]. This study also highlights the conserved miRNA gene regulatory networks in several sarcoma types, including RMS and SS. Elevated levels of miR-183 in these tumors significantly reduce the transcripts and protein levels of EGR1, a tumor suppressor and a transcription factor. Since EGR1 is downregulated, its transcriptional target of PTEN is also significantly affected. Blocking the activity of miR-183 using antimiRs elevated the levels of both EGR1 and PTEN. Thus, a single miRNA miR-183 directly and indirectly regulates two tumor suppressors, EGR1 and PTEN [34].
Recently, we examined the regulatory networks underlying miR-1, miR-206 and miR-29 downregulation in RMS [90]. Our data indicate that deregulation of these miRNAs stabilizes the expression of PAX3 and cyclin D2 in both (ERMS and ARMS) RMS types. Ectopic expression of miR-1 and miR-206 shows significant downregulation of PAX3 protein expression only in ERMS, and not in ARMS cell lines. In ARMS, PAX3 forms a fusion transcript with FKHR, and the resultant loss of PAX3 3′UTR in the fusion transcript indicates an oncogenic mechanism to evade miRNA-mediated regulation of PAX3. In addition, miR-29 also targets E2F7, another cell cycle regulator. Overexpression of miR-29 downregulates the expression of these cell cycle genes and induces partial G1 arrest leading to decreased cell proliferation. Taken together, our data suggest that the RMS state is stabilized by the deregulation of multiple miRNAs and their target genes, supporting a tumor suppressor role for these miRNAs [90]. Similarly, miR-29 has been implicated in epigenetic regulation in other cancers. miR-29b can directly target HDAC4 during osteoblast differentiation [96], and the miR-29 family can potentially target DNA methyl transferases in lung cancer, thereby causing global downregulation of DNA methylation [97]. These are potential areas that can be investigated in RMS biology. Some of the experimentally validated miRNA target gene interactions in RMS are given in Fig. 3.
Fig. 3.
Representative functionally validated miRNA-target genes in RMS. Partly adapted from Li et al. [90]
The clinical relevance of these miRNAs has been highlighted in several studies. Amplification of 13q31-32 chromosomal region, which includes C13orf25 gene harboring miR-17-92 cluster, was observed in a fraction of ARMS patients [98]. Further, in a recent study, it was shown that five of six miRNAs in miR-17-92 cluster were overexpressed in PAX7-FKHR positive ARMS samples with 13q31 amplification [99]. The high levels of these five miRNAs correlated with worse outcome in 13q31-amplified cases indicating a prognostic value for these miRNAs. In a study involving a large cohort of RMS samples, Missiaglia et al. [100] reported lower levels of miR-206 in RMS compared to normal skeletal muscle, the lower miR-206 levels correlating with increased metastasis and poor survival at diagnosis in advanced-stage disease. Moreover, this lower expression was associated with activation of NFκB, ERK and JNK signaling. In an effort to identify non-invasive biomarkers for RMS, Miyachi et al. [101] investigated the levels of muscle-specific miRNAs in human serum samples. miR-206 levels were found to be higher in sera of RMS patients compared to healthy donors or pediatric patients with other tumors. Although miR-206 is a tumor suppressor and downregulated in RMS, it is unclear why miR-206 levels were elevated in serum. These studies indicate the potential of miR-206 expression as a diagnostic and prognostic marker. miRNAs can also regulate drug responsiveness. In RMS cell lines, the downregulation of miR-485-3p can cause NF-YB-dependent decrease in Topoisomerase II (Top2) [102]. NF-YB can bind Top2 promoter and inhibit its transcription, thereby reducing the effect of Top2 inhibitors. Overexpression of miR-485-3p reduces NF-YB and restores Top2 expression, thereby reversing drug resistance. Thus, miRNAs can be potential biomarkers for response to therapy.
Synovial sarcoma
Synovial sarcoma, representing about 8 % of all soft tissue sarcoma, occur near the joints of the arm, neck or leg, and is most common in adolescents and young adults. We performed the first study to examine miRNA expression in this sarcoma [16]. We observed miR-143 to be expressed at very low levels in SS relative to GIST and LMS, as demonstrated by microarray, cloning and northern analyses. miR-143 has been proven to target ERK5 (also known as MAPK7), which is known to promote cell growth and proliferation in response to tyrosine kinase signaling [103]. SSX1, a common 3′-fusion partner gene resulting from a t(X; 18) in SS, is predicted to be another target for miR-143. Recently, our group has identified miR-183 to have a potential oncogenic role in SS [34]. miR-183 targets the tumor suppressor, EGR1. In SS, the SS18-SSX fusion protein represses EGR1 expression through direct association with the EGR1 promoter. Further, miR-183 knockdown in SS cell lines revealed deregulation of a miRNA network composed of the miR-183–EGR1–PTEN pathway. Global miRNA expression profiles in this translocation-associated sarcoma have been examined by Hisaoka et al. [104]. In this study, 35 miRNAs were observed to be differentially expressed in SS in comparison to other tissue types. There were 21 significantly upregulated miRNAs, including some miRNAs, such as let-7e, miR-99b, and miR-125a-3p, clustered within the same chromosomal loci. Further, using antimiR inhibitors against let-7e and miR-99b, the authors demonstrated suppression in cell proliferation and modulation in expression of their targets, HMGA2 and SMARCA5, suggesting potential oncogenic role for these proteins. The unique miRNA expression patterns observed in this study warrant further investigation in order to develop a better understanding of the oncogenic mechanisms and future therapeutic strategies for SS.
Leiomyosarcoma
Leiomyosarcoma is a malignant cancer of uterine smooth muscle. The benign form of this tumor is leiomyoma. In our initial characterization of sarcoma, we included six LMS samples [16]. Significance analysis of microarray comparison of LMS and normal smooth muscle samples identified significant overexpression of miR-1, miR-133a and miR-133b in LMS. This is in contrast to what is observed in RMS. These miRNAs play a major role in myogenesis and myoblast proliferation [105]. Our analysis also indicated LMS to be molecularly heterogeneous [16]. Recently, miRNAs were shown to play a pivotal role in the control of smooth muscle differentiation. Danielson et al. [106] demonstrated the remarkable ability of miRNA patterns to subclassify different tumors of the smooth muscle lineage. They showed differential expression of >70 miRNAs, including members of the oncogenic miR-17-92 cluster, in LMS compared to normal myometrium. However, the role of these miRNAs was not functionally characterized in this study. Recently, Shi et al. [107] examined the functional correlation between let-7 and its target HMGA2. Overexpression of HMGA2 is common in uterine leiomyomas. This group observed that overexpression of HMGA2 and let-7-mediated HMGA2 repressions are relevant molecular alterations in LMS. Further, using in vitro assays, they demonstrated that disruption of the control of HMGA2 and let-7 pairs promotes LMS cell growth.
miRNAs have also been investigated as ideal biomarkers for diagnosing malignant phenotypes in LMS. For instance, in a recent study, Nuovo and Schmittgen [108] utilized upregulation of miR-221 expression as an accurate way to differentiate LMS from benign metastasizing leiomyoma. They observed that metastasizing leiomyomas are most likely benign lesions and not the malignant LMS. Moreover, miRNAs have also been investigated as a marker of response for hormonal therapy. Pan et al. [108] examined the expression, regulation and function of miR-21 in leiomyomas. They observed miR-21 to be aberrantly expressed and hormonally regulated in these benign tumors. Further, the authors propose a feedback regulatory interaction between miR-21, TGF-β and ovarian steroids, necessary to balance their different functions in a cell- and tissue-dependent context, influencing events such as inflammatory response, cell growth regulation and tissue turnover, leading either to cellular transformation and tumorigenesis or tissue fibrosis. These studies indicate the ability of miRNAs to modulate differentiation and act as biomarkers, thus rendering them attractive therapeutic agents against poorly differentiated sarcomas.
Liposarcoma
Liposarcoma is a malignant tumor that arises in fat cells in deep soft tissue. It is the most common mesenchymal cancer, with a mortality rate of 60 % among patients with this disease.
Urgas et al. [109] investigated the alterations in miRNA expression associated with liposarcomagenesis. They compared miRNA expression between normal adipose tissue, well-differentiated liposarcoma, and dedifferentiated liposarcoma using microarrays and deep sequencing studies. This analysis identified over 40 miRNAs that were dysregulated in dedifferentiated liposarcomas including upregulation of miR-21 and miR-26a and downregulation of miRNAs that are highly abundant in adipose tissue (miR-143 and miR-145). Further functional characterization of miR-143 demonstrated its tumor suppressor role in liposarcoma, potentially by decreasing expression of BCL2, topoisomerase 2A, protein regulator of cytokinesis 1 (PRC1), and polo-like kinase 1 (PLK1). In another study, Zhang et al. [110] identified important functions for miR-155 and β-catenin signaling in progression of liposarcoma. miR-155 was the most overexpressed miRNA in this analysis, and functional investigations assigned its role in the growth of dedifferentiated liposarcoma cell lines. Casein kinase 1α (CK1α) was identified as a direct target of miR-155 control which enhanced β-catenin signaling and cyclin D1 expression, promoting tumor cell growth. In a recent study, Taylor et al. [111] examined genetic alterations contributing to liposarcomagenesis by sequencing the genome, exome, transcriptome, and cytosine methylome of a primary and recurrent dedifferentiated liposarcoma from treatment-naive patients. These studies established a role for small RNAs in liposarcomagenesis, typified by methylation-induced silencing of miRNA-193b in dedifferentiated liposarcoma, but not in their well-differentiated counterpart. Patterns of miRNA expression in both dedifferentiated and well-differentiated liposarcomas are given in S-MED [17].
Malignant peripheral nerve sheath tumor
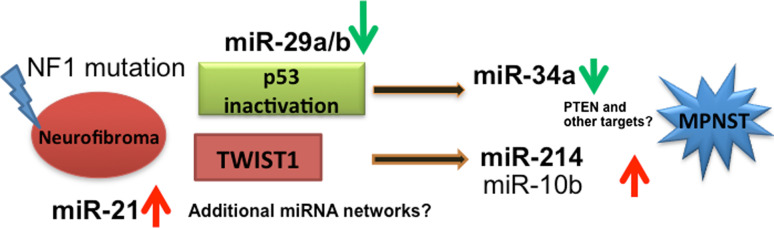
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive soft tissue tumors that occur, either sporadically or in patients with neurofibromatosis type 1. These tumors have a 5-year survival rate of about 40 % [3]. Gene expression analysis performed by our group has identified an expression signature indicating p53 inactivation in the majority of MPNSTs [112]. Subsequently, we performed miRNA profiling in benign and malignant PNSTs. This analysis indicated a relative downregulation of miR-34a in most MPNSTs compared to neurofibromas. Using functional studies, we demonstrated that exogenous expression of p53 or miR-34a promote apoptotic cell death in MPNSTs. In addition, miR-214 was highly upregulated in MPNSTs compared to neurofibromas. miR-214 is a direct transcriptional target of TWIST1 [113], a regulator of metastasis [114]. It is to be noted that TWIST1 is highly expressed in MPNSTs. miR-214 targets PTEN, hence the TWIST1-miR-214-PTEN pathway could be potentially explored to decipher MPNST pathogenesis. Collectively, our findings suggest that deregulation of miRNAs has a potential role in the malignant transformation process in PNSTs [112]. This was further confirmed by a series of studies examining the role of several candidate miRNAs by other groups. For instance, Chai et al. [115] reported upregulation of miR-10b in primary Schwann cells isolated from neurofibromas and in MPNST tumors and cell lines. Using functional studies in multiple cell lines, the authors demonstrated that the inhibition of miR-10b reduced cell proliferation, migration and invasion by affecting NF1 expression and RAS signaling. In another study, miR-21 was shown to play an important role in MPNST tumorigenesis and progression through its target, PDCD4 [116]. Subsets of MPNSTs are characterized by the presence of a 1.4-Mb microdeletion. Pasmant et al. [117] identified the presence of two miRNAs, miR-193A and miR-365-2, in this microdeletion. However, expression analysis of these miRNAs did not show any significant difference between human dermal and plexiform neurofibromas and MPNSTs.
Schwannomas are benign tumors characterized by mutations in the NF2 gene. Saydam et al. [118] reported miRNA profiling of schwannomas and characterized the tumor suppressor function of miR-7, which was one of the most downregulated miRNAs in these tumors. In addition to the known oncogene targets, epidermal growth factor receptor (EGFR) and p21-activated kinase 1 (Pak1) for miR-7, this group identified associated cdc42 kinase 1(Ack1) oncogene as a direct target. These findings suggest miR-7 as a potential therapeutic molecule for schwannoma treatment, and prompt clinical evaluation of drugs that can inhibit the EGFR, Pak1, and Ack1 signaling pathways to treat this tumor type. miR-21 was also observed to be consistently overexpressed in vestibular schwannomas. This was correlated with decreased levels of tumor suppressor, PTEN, a known molecular target of miR-21 [119]. Both gene and miRNA profiles of schwannomas were also compared with those of neurofibroma and MPNSTs. It can be noted that neurofibroma and schwannoma clustered separately compared to MPNSTs [112]. A schematic representation of miRNA deregulations in the malignant transformation of neurofibroma to MPNST is given is Fig. 4.
Fig. 4.
Potential miRNA gene networks in the malignant transformation of neurofibroma to MPNSTs
Fibrosarcoma
Fibrosarcoma (fibroblastic sarcoma) is a malignant tumor derived from fibrous connective tissue and is characterized by the presence of immature proliferating fibroblasts or undifferentiated anaplastic spindle cells. Not many studies have examined miRNA expression in this rare sarcoma. Recently, two studies have characterized miRNA expression and function in human fibrosarcoma cell line HT1080. Liu and Wilson [120] have demonstrated that miR-520c and miR-373 increases the expression of MMP9 by directly targeting the 3′UTRs of mRNAs of mTOR and SIRT1 and suppressing their translation. This results in activation of the Ras/Raf/MEK/Erk signaling pathway and NF-κB finally increasing MMP9 levels and activity, thereby enhancing cell migration and cell growth in these cells. In the second study, Weng et al. [121] showed that miR-409-3p inhibits tumor growth, vascularization and metastasis through downregulating angiogenin expression in HT1080 cells. Angiogenin is an angiogenic and tumorigenic factor that is elevated in various types of cancers. This was further confirmed by the inverse correlation of the expression of miR-409-3p with angiogenin in mouse xenograft tissues and in human fibrosarcoma samples.
Angiosarcoma and other sarcoma types
Our group has experimentally validated differentially expressed miRNAs in angiosarcoma. Using S-MED, we identified miR-515-3p, 515-5p, 517a, 517c, 518b, 519a, and 522 as upregulated specifically in this tumor type when compared to the other sarcomas, and this could be exploited to aid angiosarcoma diagnosis [17]. miRNA expression patterns of many sarcoma types have been characterized; however, the role of miRNAs in several sarcoma types is not yet understood. For instance, the role of miRNAs in sarcomas such as hemangiosarcoma and pleomorphic undifferentiated sarcoma have not yet been reported.
Conclusion
Although diagnosis and treatment of sarcoma pose many challenges, there are several opportunities for investigating sarcoma biology. For instance, by small RNA sequencing of various sarcomas and corresponding normal types, we have identified 14 novel human miRNAs [16]. Thus, the potential for discovering novel RNA transcripts, including small and long non-coding (lncRNAs), is high in sarcomas, since sarcoma genomics is not extensively studied. Further, these novel RNA transcripts may also play a role in other tissues and cancer types. This is relevant, especially in the characterization of conserved gene regulatory networks in sarcomas and other epithelial and hematopoietic malignancies. Common oncogenes and tumor suppressors are deregulated in most cancer types. Both p53 and cMYC can induce the expression of multiple miRNAs and can be regulated by sets of miRNAs that are conserved and found to be deregulated in multiple cancers [24, 122]. For instance, activation of miR-17-92 mediated by cMYC is a characteristic feature in OS and several other carcinomas [24, 45]. Similarly, deregulation of miR-29 is noticed in both sarcomas (RMS and MPNSTs) [90, 112] and other malignancies [123, 124]. miR-29 family members activate p53 [123], and their loss of expression can limit p53 levels when required in the cell. Subsequently, loss or inactivation of p53 expression leads to downregulation of miR-34, a transcriptional target of p53 [122]. Thus, understanding the factors that modulate cMYC and/or p53 expression in sarcomas can also provide valuable insights in the pathobiology of other malignancies.
miRNA-gene deregulations are conserved in certain types of sarcomas and other malignancies; however, investigating miRNA biology in sarcomas can be challenging. First, a single miRNA can regulate many genes, and, depending on the cell type and target availability, a miRNA can function either as an oncogene or tumor suppressor. Thus, roles of miRNAs cannot be generalized across multiple cancers or cell types. Second, modulation of miRNAs can also have off-target effects. Third, deregulation in miRNA expression leads to ‘noise’ in the cellular gene expression, and these noise levels vary in different cancers. Thus, based on miRNA deregulations, the resultant noise in tumor cells and in surrounding stromal cells, outcomes in terms of tumor initiation, progression, invasion and metastases can vary extensively and contribute to the heterogeneity within a specific sarcoma subtype.
miRNAs are known to have hundreds of targets, and many of them may function as passengers in the tumorigenic process [19]. Driver genes are key players in tumorigenesis, and, depending on their function, they can be both oncogenic and tumor suppressors. Investigating the interactions between miRNAs and driver genes (driver of driver genes) will significantly enhance our understanding of the sarcoma pathobiology. These miRNA–gene networks can also work cooperatively with the competing endogenous RNAs (ceRNAs) [125] and regulate/be regulated by a plethora of gene networks that are maintained in an intricate balance for normal cellular functioning. Understanding these multilayered gene regulatory mechanisms may pose a major challenge in understanding cancer biology such as drug resistance and tumor recurrence.
Focusing on key ‘driver of driver genes’ may reduce the miRNA-gene network complexities to a certain extent. The future research directions for the roles of miRNAs in sarcoma may include (1) development of body fluid (serum/plasma) based miRNA biomarkers, (2) miRNA-based targeting of genes in a cancer signaling pathway using miRNA mimics and or antimiRs/sponges instead of targeting single gene/protein product, (3) investigation of potential ceRNAs as alternative drug targets for cancer genes that are currently undruggable by regular therapeutic approaches, and (4) modulating miRNA(s) expression levels may be explored as differentiation, intercellular communication and stem cell-based cancer therapies.
Acknowledgments
Due to space restrictions, we could not cite many significant contributions made by numerous other investigators in this important and rapidly progressing field. We thank Ms. Jennie W. Knoot for assisting with manuscript editing. This work is supported by grants from Minnesota Medical Foundation, Academic Health Center, University of Minnesota and The Karen Wykoff Sarcoma Foundation and the Department of Defense (W81XwH10-1-0556).
References
- 1.Taylor BS, Barretina J, Maki RG, Antonescu CR, Singer S, Ladanyi M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011;11(8):541–557. doi: 10.1038/nrc3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher CDM, Unni KK, Mertens F. World Health Organization Classification of Tumors, Pathology and genetics of tumours of soft tissue and bone. Lyon: IARC Press; 2002. [Google Scholar]
- 3.Weiss S, Goldblum J. Soft tissue tumors. Mosby: Elsevier; 2008. [Google Scholar]
- 4.Dela Cruz F, Matushansky I. MicroRNAs in chromosomal translocation-associated solid tumors: learning from sarcomas. Discov Med. 2011;12(65):307–317. [PubMed] [Google Scholar]
- 5.Barr FG. Molecular genetics and pathogenesis of rhabdomyosarcoma. J Pediatr Hematol Oncol. 1997;19(6):483–491. doi: 10.1097/00043426-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Riggi N, Cironi L, Suva ML, Stamenkovic I. Sarcomas: genetics, signalling, and cellular origins part 1: the fellowship of TET. J Pathol. 2007;213(1):4–20. doi: 10.1002/path.2209. [DOI] [PubMed] [Google Scholar]
- 7.Brisset S, Schleiermacher G, Peter M, Mairal A, Oberlin O, Delattre O, Aurias A. CGH analysis of secondary genetic changes in Ewing tumors: correlation with metastatic disease in a series of 43 cases. Cancer Genet Cytogenet. 2001;130(1):57–61. doi: 10.1016/s0165-4608(01)00454-x. [DOI] [PubMed] [Google Scholar]
- 8.Mankin HJ, Hornicek FJ, Rosenberg AE, Harmon DC, Gebhardt MC. Survival data for 648 patients with osteosarcoma treated at one institution. Clin Orthop Relat Res. 2004;429:286–291. doi: 10.1097/01.blo.0000145991.65770.e6. [DOI] [PubMed] [Google Scholar]
- 9.Scurr M. Histology-driven chemotherapy in soft tissue sarcomas. Curr Treat Options Oncol. 2011;12(1):32–45. doi: 10.1007/s11864-011-0140-x. [DOI] [PubMed] [Google Scholar]
- 10.Subramanian S, West RB, Corless CL, Ou W, Rubin BP, Chu KM, Leung SY, Yuen ST, Zhu S, Hernandez-Boussard T, Montgomery K, Nielsen TO, Patel RM, Goldblum JR, Heinrich MC, Fletcher JA, van de Rijn M. Gastrointestinal stromal tumors (GISTs) with KIT and PDGFRA mutations have distinct gene expression profiles. Oncogene. 2004;23(47):7780–7790. doi: 10.1038/sj.onc.1208056. [DOI] [PubMed] [Google Scholar]
- 11.Subramanian S, West RB, Marinelli RJ, Nielsen TO, Rubin BP, Goldblum JR, Patel RM, Zhu S, Montgomery K, Ng TL, Corless CL, Heinrich MC, van de Rijn M. The gene expression profile of extraskeletal myxoid chondrosarcoma. J Pathol. 2005;206(4):433–444. doi: 10.1002/path.1792. [DOI] [PubMed] [Google Scholar]
- 12.West RB, Rubin BP, Miller MA, Subramanian S, Kaygusuz G, Montgomery K, Zhu S, Marinelli RJ, De Luca A, Downs-Kelly E, Goldblum JR, Corless CL, Brown PO, Gilks CB, Nielsen TO, Huntsman D, van de Rijn M. A landscape effect in tenosynovial giant-cell tumor from activation of CSF1 expression by a translocation in a minority of tumor cells. Proc Natl Acad Sci USA. 2006;103(3):690–695. doi: 10.1073/pnas.0507321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O’Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359(9314):1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- 14.Antonescu CR. Molecular profiling in the diagnosis and treatment of high grade sarcomas. Ultrastruct Pathol. 2008;32(2):37–42. doi: 10.1080/01913120801897174. [DOI] [PubMed] [Google Scholar]
- 15.van de Rijn M, Fletcher JA. Genetics of soft tissue tumors. Annu Rev Pathol. 2006;1:435–466. doi: 10.1146/annurev.pathol.1.110304.100052. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, Corless CL, Fire AZ, van de Rijn M. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27(14):2015–2026. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 17.Sarver AL, Phalak R, Thayanithy V, Subramanian S. S-MED: sarcoma microRNA expression database. Lab Invest J Techn Methods Pathol. 2010;90(5):753–761. doi: 10.1038/labinvest.2010.53. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5(5):396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 21.Hobert O. miRNAs play a tune. Cell. 2007;131(1):22–24. doi: 10.1016/j.cell.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 22.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim VN. Small RNAs: classification, biogenesis, and function. Mol Cell. 2005;19(1):1–15. [PubMed] [Google Scholar]
- 27.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9(11):831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 29.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 31.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 32.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 33.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 34.Sarver A, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Can Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 35.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian S, Steer CJ. MicroRNAs as gatekeepers of apoptosis. J Cell Physiol. 2010;223(2):289–298. doi: 10.1002/jcp.22066. [DOI] [PubMed] [Google Scholar]
- 37.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Pure E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 39.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res. 2011;71(18):5950–5954. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 42.Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suva D, Tercier S, Joseph JM, Guillou L, Stamenkovic I. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24(9):916–932. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 44.Greither T, Wurl P, Grochola L, Bond G, Bache M, Kappler M, Lautenschlager C, Holzhausen HJ, Wach S, Eckert AW, Taubert H. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int J Cancer. 2012;130(5):1230–1235. doi: 10.1002/ijc.26109. [DOI] [PubMed] [Google Scholar]
- 45.Thayanithy V, Sarver AL, Kartha RV, Li L, Angstadt AY, Breen M, Steer CJ, Modiano JF, Subramanian S. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50:171–181. doi: 10.1016/j.bone.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maire G, Martin JW, Yoshimoto M, Chilton-MacNeill S, Zielenska M, Squire JA. Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma. Cancer Genet. 2011;204(3):138–146. doi: 10.1016/j.cancergen.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 47.Duan Z, Choy E, Harmon D, Liu X, Susa M, Mankin H, Hornicek FJ. MicroRNA-199a-3p is down regulated in human osteosarcoma and regulates cell proliferation and migration. Mol Cancer Ther. 2011;10(8):1337–1345. doi: 10.1158/1535-7163.MCT-11-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lulla RR, Costa FF, Bischof JM, Chou PM, de FBM, Vanin EF, Soares MB. Identification of differentially expressed microRNAs in osteosarcoma. Sarcoma. 2011;2011:732690. doi: 10.1155/2011/732690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, Leblanc K, Palatini J, Randall RL, Volinia S, Stein GS, Croce CM, Lian JB, Aqeilan RI. MicroRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osaki M, Takeshita F, Sugimoto Y, Kosaka N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T, Ito H, Oshimura M, Ochiya T MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol Ther 19 (6):1123–1130. doi:10.1038/mt.2011.53 [DOI] [PMC free article] [PubMed]
- 51.Zhang H, Cai X, Wang Y, Tang H, Tong D, Ji F. microRNA-143, down-regulated in osteosarcoma, promotes apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol Rep. 2010;24(5):1363–1369. doi: 10.3892/or_00000994. [DOI] [PubMed] [Google Scholar]
- 52.He C, Xiong J, Xu X, Lu W, Liu L, Xiao D, Wang D. Functional elucidation of MiR-34 in osteosarcoma cells and primary tumor samples. Biochem Biophys Res Commun. 2009;388(1):35–40. doi: 10.1016/j.bbrc.2009.07.101. [DOI] [PubMed] [Google Scholar]
- 53.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68(24):10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q, Ma B. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS ONE. 2012;7(3):e33778. doi: 10.1371/journal.pone.0033778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Creighton CJ, Fountain MD, Yu Z, Nagaraja AK, Zhu H, Khan M, Olokpa E, Zariff A, Gunaratne PH, Matzuk MM, Anderson ML. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70(5):1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi E, Hornicek FJ, Duan Z. MicroRNA involvement in osteosarcoma. Sarcoma. 2012;2012:359739. doi: 10.1155/2012/359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu LH, Li H, Li JP, Zhong H, Zhang HC, Chen J, Xiao T. miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem Biophys Res Commun. 2011;416(1–2):31–38. doi: 10.1016/j.bbrc.2011.10.117. [DOI] [PubMed] [Google Scholar]
- 58.Ziyan W, Shuhua Y, Xiufang W, Xiaoyun L MicroRNA-21 is involved in osteosarcoma cell invasion and migration. Med Oncol doi:10.1007/s12032-010-9563-7 [DOI] [PubMed]
- 59.Kang HG, Kim HS, Kim KJ, Oh JH, Lee MR, Seol SM, Han I. RECK expression in osteosarcoma: correlation with matrix metalloproteinases activation and tumor invasiveness. J Orthop Res. 2007;25(5):696–702. doi: 10.1002/jor.20323. [DOI] [PubMed] [Google Scholar]
- 60.Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang X, Wang L. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting ezrin. Am J Pathol. 2012 doi: 10.1016/j.ajpath.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 61.Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, Fodstad O, Ju J. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene. 2009;28(46):4065–4074. doi: 10.1038/onc.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song B, Wang Y, Titmus MA, Botchkina G, Formentini A, Kornmann M, Ju J. Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol Cancer. 2010;9:96. doi: 10.1186/1476-4598-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gougelet A, Pissaloux D, Besse A, Perez J, Duc A, Dutour A, Blay JY, Alberti L. Micro-RNA profiles in osteosarcoma as a predictive tool for ifosfamide response. Int J Cancer. 2011;129(3):680–690. doi: 10.1002/ijc.25715. [DOI] [PubMed] [Google Scholar]
- 64.Ban J, Jug G, Mestdagh P, Schwentner R, Kauer M, Aryee DN, Schaefer KL, Nakatani F, Scotlandi K, Reiter M, Strunk D, Speleman F, Vandesompele J, Kovar H. Hsa-mir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene. 2011;30(18):2173–2180. doi: 10.1038/onc.2010.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKinsey EL, Parrish JK, Irwin AE, Niemeyer BF, Kern HB, Birks DK, Jedlicka P. A novel oncogenic mechanism in Ewing sarcoma involving IGF pathway targeting by EWS/Fli1-regulated microRNAs. Oncogene. 2011;30(49):4910–4920. doi: 10.1038/onc.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Vito C, Riggi N, Suva ML, Janiszewska M, Horlbeck J, Baumer K, Provero P, Stamenkovic I. Let-7a is a direct EWS-FLI-1 target implicated in Ewing’s sarcoma development. PLoS ONE. 2011;6(8):e23592. doi: 10.1371/journal.pone.0023592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S, Alberghini M, Grilli A, Knuutila S, Schaefer KL, Mattia G, Negrini M, Picci P, Serra M, Scotlandi K. miR-34a predicts survival of Ewing’s sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol. 2012;226(5):796–805. doi: 10.1002/path.3007. [DOI] [PubMed] [Google Scholar]
- 68.Mosakhani N, Guled M, Leen G, Calabuig-Farinas S, Niini T, Machado I, Savola S, Scotlandi K, Lopez-Guerrero JA, Llombart-Bosch A, Knuutila S. An integrated analysis of miRNA and gene copy numbers in xenografts of Ewing’s sarcoma. J Exp Clin Cancer Res CR. 2012;31(1):24. doi: 10.1186/1756-9966-31-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin EA, Kong L, Bai XH, Luan Y, Liu CJ. miR-199a, a bone morphogenic protein 2-responsive microRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284(17):11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580(17):4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 71.Zuntini M, Salvatore M, Pedrini E, Parra A, Sgariglia F, Magrelli A, Taruscio D, Sangiorgi L. MicroRNA profiling of multiple osteochondromas: identification of disease-specific and normal cartilage signatures. Clin Genet. 2010;78(6):507–516. doi: 10.1111/j.1399-0004.2010.01490.x. [DOI] [PubMed] [Google Scholar]
- 72.Hameetman L, Rozeman LB, Lombaerts M, Oosting J, Taminiau AH, Cleton-Jansen AM, Bovee JV, Hogendoorn PC. Peripheral chondrosarcoma progression is accompanied by decreased Indian hedgehog signalling. J Pathol. 2006;209(4):501–511. doi: 10.1002/path.2008. [DOI] [PubMed] [Google Scholar]
- 73.Duan Z, Choy E, Nielsen GP, Rosenberg A, Iafrate J, Yang C, Schwab J, Mankin H, Xavier R, Hornicek FJ. Differential expression of microRNA (miRNA) in chordoma reveals a role for miRNA-1 in Met expression. J Orthop Res. 2010;28(6):746–752. doi: 10.1002/jor.21055. [DOI] [PubMed] [Google Scholar]
- 74.Naka T, Iwamoto Y, Shinohara N, Ushijima M, Chuman H, Tsuneyoshi M. Expression of c-met proto-oncogene product (c-MET) in benign and malignant bone tumors. Mod Pathol. 1997;10(8):832–838. [PubMed] [Google Scholar]
- 75.Ostroumov E, Hunter CJ. Identifying mechanisms for therapeutic intervention in chordoma: c-Met oncoprotein. Spine. 2008;33(25):2774–2780. doi: 10.1097/BRS.0b013e31817e2d1e. [DOI] [PubMed] [Google Scholar]
- 76.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102(50):18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fletcher JA, Rubin BP. KIT mutations in GIST. Curr Opin Genet Dev. 2007;17(1):3–7. doi: 10.1016/j.gde.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 78.Official Publication of the European Dialysis and Transplant Association-European Renal Association (1990) Abstracts of the Nephrology, dialysis, transplantation. Combined meeting of the Dutch Society of Nephrology and the Renal Association, Amsterdam, 27–28 October 1989 [PubMed]
- 79.Kim WK, Park M, Kim YK, Tae YK, Yang HK, Lee JM, Kim H. MicroRNA-494 downregulates KIT and inhibits gastrointestinal stromal tumor cell proliferation. Clin Cancer Res. 2011;17(24):7584–7594. doi: 10.1158/1078-0432.CCR-11-0166. [DOI] [PubMed] [Google Scholar]
- 80.Haller F, von Heydebreck A, Zhang JD, Gunawan B, Langer C, Ramadori G, Wiemann S, Sahin O. Localization- and mutation-dependent microRNA (miRNA) expression signatures in gastrointestinal stromal tumours (GISTs), with a cluster of co-expressed miRNAs located at 14q32.31. J Pathol. 2009;220(1):71–86. doi: 10.1002/path.2610. [DOI] [PubMed] [Google Scholar]
- 81.Choi HJ, Lee H, Kim H, Kwon JE, Kang HJ, You KT, Rhee H, Noh SH, Paik YK, Hyung WJ, Kim H. MicroRNA expression profile of gastrointestinal stromal tumors is distinguished by 14q loss and anatomic site. Int J Cancer. 2010;126(7):1640–1650. doi: 10.1002/ijc.24897. [DOI] [PubMed] [Google Scholar]
- 82.Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, Nishida T, Bamba T, Kanda T, Ajioka Y, Taguchi T, Okahara S, Takahashi H, Nishida Y, Hosokawa M, Hasegawa T, Tokino T, Hirata K, Imai K, Toyota M, Shinomura Y. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72(5):1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 83.Gurney JG, Davis S, Severson RK, Fang JY, Ross JA, Robison LL. Trends in cancer incidence among children in the US. Cancer. 1996;78(3):532–541. doi: 10.1002/(SICI)1097-0142(19960801)78:3<532::AID-CNCR22>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 84.Qualman SJ, Coffin CM, Newton WA, Hojo H, Triche TJ, Parham DM, Crist WM. Intergroup rhabdomyosarcoma study: update for pathologists. Pediatr Dev Pathol. 1998;1(6):550–561. doi: 10.1007/s100249900076. [DOI] [PubMed] [Google Scholar]
- 85.Gougelet A, Perez J, Pissaloux D, Besse A, Duc A, Decouvelaere AV, Ranchere-Vince D, Blay JY, Alberti L. miRNA Profiling: how to bypass the current difficulties in the diagnosis and treatment of sarcomas. Sarcoma. 2011;2011:460650. doi: 10.1155/2011/460650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rota R, Ciarapica R, Giordano A, Miele L, Locatelli F. MicroRNAs in rhabdomyosarcoma: pathogenetic implications and translational potentiality. Mol Cancer. 2011;10:120. doi: 10.1186/1476-4598-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rao PK, Missiaglia E, Shields L, Hyde G, Yuan B, Shepherd CJ, Shipley J, Lodish HF Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J 24 (9):3427–3437. doi:10.1096/fj.09-150698 [DOI] [PMC free article] [PubMed]
- 88.Yan D, Dong XD, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009 doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119(8):2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li L, Sarver AL, Alamgir S, Subramanian S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab Invest. 2012;92(4):571–583. doi: 10.1038/labinvest.2012.10. [DOI] [PubMed] [Google Scholar]
- 91.Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem. 2003;278(10):8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 92.Baer C, Nees M, Breit S, Selle B, Kulozik AE, Schaefer KL, Braun Y, Wai D, Poremba C. Profiling and functional annotation of mRNA gene expression in pediatric rhabdomyosarcoma and Ewing’s sarcoma. Int J Cancer. 2004;110(5):687–694. doi: 10.1002/ijc.20171. [DOI] [PubMed] [Google Scholar]
- 93.Mayanil CS, George D, Freilich L, Miljan EJ, Mania-Farnell B, McLone DG, Bremer EG. Microarray analysis detects novel Pax3 downstream target genes. J Biol Chem. 2001;276(52):49299–49309. doi: 10.1074/jbc.M107933200. [DOI] [PubMed] [Google Scholar]
- 94.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14(5):369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, Giordano A. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8(1):172–175. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
- 96.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284(23):15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gordon AT, Brinkschmidt C, Anderson J, Coleman N, Dockhorn-Dworniczak B, Pritchard-Jones K, Shipley J. A novel and consistent amplicon at 13q31 associated with alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2000;28(2):220–226. [PubMed] [Google Scholar]
- 99.Reichek JL, Duan F, Smith LM, Gustafson DM, O’Connor RS, Zhang C, Dunlevy MJ, Gastier-Foster JM, Barr FG. Genomic and clinical analysis of amplification of the 13q31 chromosomal region in alveolar rhabdomyosarcoma: a report from the children’s oncology group. Clin Cancer Res. 2011;17(6):1463–1473. doi: 10.1158/1078-0432.CCR-10-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Missiaglia E, Shepherd CJ, Patel S, Thway K, Pierron G, Pritchard-Jones K, Renard M, Sciot R, Rao P, Oberlin O, Delattre O, Shipley J MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer 102 (12):1769–1777. doi:10.1038/sj.bjc.6605684 [DOI] [PMC free article] [PubMed]
- 101.Miyachi M, Tsuchiya K, Yoshida H, Yagyu S, Kikuchi K, Misawa A, Iehara T, Hosoi H. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem Biophys Res Commun. 2010;400(1):89–93. doi: 10.1016/j.bbrc.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 102.Chen CF, He X, Arslan AD, Mo YY, Reinhold WC, Pommier Y, Beck WT. Novel regulation of nuclear factor-YB by miR-485-3p affects the expression of DNA topoisomerase IIalpha and drug responsiveness. Mol Pharmacol. 2011;79(4):735–741. doi: 10.1124/mol.110.069633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, Dean NM, Freier SM, Bennett CF, Lollo B, Griffey R. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279(50):52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 104.Hisaoka M, Matsuyama A, Nagao Y, Luan L, Kuroda T, Akiyama H, Kondo S, Hashimoto H. Identification of altered microRNA expression patterns in synovial sarcoma. Genes Chromosomes Cancer. 2011;50(3):137–145. doi: 10.1002/gcc.20837. [DOI] [PubMed] [Google Scholar]
- 105.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Danielson LS, Menendez S, Attolini CS, Guijarro MV, Bisogna M, Wei J, Socci ND, Levine DA, Michor F, Hernando E. A differentiation-based microRNA signature identifies leiomyosarcoma as a mesenchymal stem cell-related malignancy. Am J Pathol. 2010;177(2):908–917. doi: 10.2353/ajpath.2010.091150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shi G, Perle MA, Mittal K, Chen H, Zou X, Narita M, Hernando E, Lee P, Wei JJ. Let-7 repression leads to HMGA2 overexpression in uterine leiomyosarcoma. J Cell Mol Med. 2009;13:3898–3905. doi: 10.1111/j.1582-4934.2008.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nuovo GJ, Schmittgen TD. Benign metastasizing leiomyoma of the lung: clinicopathologic, immunohistochemical, and micro-RNA analyses. Am J Surg Pathol B. 2008;17(3):145–150. doi: 10.1097/PDM.0b013e31815aca19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ugras S, Brill E, Jacobsen A, Hafner M, Socci ND, Decarolis PL, Khanin R, O’Connor R, Mihailovic A, Taylor BS, Sheridan R, Gimble JM, Viale A, Crago A, Antonescu CR, Sander C, Tuschl T, Singer S. Small RNA sequencing and functional characterization reveals microRNA-143 tumor suppressor activity in liposarcoma. Cancer Res. 2011;71(17):5659–5669. doi: 10.1158/0008-5472.CAN-11-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang P, Bill K, Liu J, Young E, Peng T, Bolshakov S, Hoffman A, Song Y, Demicco EG, Terrada DL, Creighton CJ, Anderson ML, Lazar AJ, Calin GG, Pollock RE, Lev D. MiR-155 is a liposarcoma oncogene that targets casein kinase-1alpha and enhances beta-catenin signaling. Cancer Res. 2012;72(7):1751–1762. doi: 10.1158/0008-5472.CAN-11-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Taylor BS, Decarolis PL, Angeles CV, Brenet F, Schultz N, Antonescu CR, Scandura JM, Sander C, Viale AJ, Socci ND, Singer S. Frequent alterations and epigenetic silencing of differentiation pathway genes in structurally rearranged liposarcomas. Cancer Discov. 2011;1(7):587–597. doi: 10.1158/2159-8290.CD-11-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Subramanian S, Thayanithy V, West RB, Lee CH, Beck AH, Zhu S, Downs-Kelly E, Montgomery K, Goldblum JR, Hogendoorn PC, Corless CL, Oliveira AM, Dry SM, Nielsen TO, Rubin BP, Fletcher JA, Fletcher CD, van de Rijn M. Genome-wide transcriptome analyses reveal p53 inactivation mediated loss of miR-34a expression in malignant peripheral nerve sheath tumours. J Pathol. 2010;220(1):58–70. doi: 10.1002/path.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37(1):123–128. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 115.Chai G, Liu N, Ma J, Li H, Oblinger JL, Prahalad AK, Gong M, Chang LS, Wallace M, Muir D, Guha A, Phipps RJ, Hock JM, Yu X. MicroRNA-10b regulates tumorigenesis in neurofibromatosis type 1. Cancer Sci. 2010;101(9):1997–2004. doi: 10.1111/j.1349-7006.2010.01616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Itani S, Kunisada T, Morimoto Y, Yoshida A, Sasaki T, Ito S, Ouchida M, Sugihara S, Shimizu K, Ozaki T. MicroRNA-21 correlates with tumorigenesis in malignant peripheral nerve sheath tumor (MPNST) via programmed cell death protein 4 (PDCD4) J Cancer Res Clin Oncol. 2012 doi: 10.1007/s00432-012-1223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pasmant E, Masliah-Planchon J, Levy P, Laurendeau I, Ortonne N, Parfait B, Valeyrie-Allanore L, Leroy K, Wolkenstein P, Vidaud M, Vidaud D, Bieche I. Identification of genes potentially involved in the increased risk of malignancy in NF1-microdeleted patients. Mol Med. 2011;17(1–2):79–87. doi: 10.2119/molmed.2010.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saydam O, Senol O, Wurdinger T, Mizrak A, Ozdener GB, Stemmer-Rachamimov AO, Yi M, Stephens RM, Krichevsky AM, Saydam N, Brenner GJ, Breakefield XO. miRNA-7 attenuation in schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 2011;71(3):852–861. doi: 10.1158/0008-5472.CAN-10-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cioffi JA, Yue WY, Mendolia-Loffredo S, Hansen KR, Wackym PA, Hansen MR. MicroRNA-21 overexpression contributes to vestibular schwannoma cell proliferation and survival. Otol Neurotol. 2010;31(9):1455–1462. doi: 10.1097/MAO.0b013e3181f20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu P, Wilson MJ. miR-520c and miR-373 upregulate MMP9 expression by targeting mTOR and SIRT1, and activate the Ras/Raf/MEK/Erk signaling pathway and NF-kappaB factor in human fibrosarcoma cells. J Cell Physiol. 2012;227(2):867–876. doi: 10.1002/jcp.22993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weng C, Dong H, Chen G, Zhai Y, Bai R, Hu H, Lu L, Xu Z. miR-409-3p inhibits HT1080 cell proliferation, vascularization and metastasis by targeting angiogenin. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 122.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16(1):23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 124.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113(25):6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, Denicola G, Webster KA, Weiss D, Perez-Mancera PA, Krauthammer M, Halaban R, Provero P, Adams DJ, Tuveson DA, Pandolfi PP. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]