Abstract
Resurrection plants are a small but diverse group of land plants characterized by their tolerance to extreme drought or desiccation. They have the unique ability to survive months to years without water, lose most of the free water in their vegetative tissues, fall into anabiosis, and, upon rewatering, quickly regain normal activity. Thus, they are fundamentally different from other drought-surviving plants such as succulents or ephemerals, which cope with drought by maintaining higher steady state water potential or via a short life cycle, respectively. This review describes the unique physiological and molecular adaptations of resurrection plants enabling them to withstand long periods of desiccation. The recent transcriptome analysis of Craterostigma plantagineum and Haberlea rhodopensis under drought, desiccation, and subsequent rehydration revealed common genetic pathways with other desiccation-tolerant species as well as unique genes that might contribute to the outstanding desiccation tolerance of the two resurrection species. While some of the molecular responses appear to be common for both drought stress and desiccation, resurrection plants also possess genes that are highly induced or repressed during desiccation with no apparent sequence homologies to genes of other species. Thus, resurrection plants are potential sources for gene discovery. Further proteome and metabolome analyses of the resurrection plants contributed to a better understanding of molecular mechanisms that are involved in surviving severe water loss. Understanding the cellular mechanisms of desiccation tolerance in this unique group of plants may enable future molecular improvement of drought tolerance in crop plants.
Keywords: Drought stress, Craterostigma plantagineum, Haberlea rhodopensis, Next generation sequencing, Proteome, Metabolome analysis
Introduction
Water deficiency is the most common abiotic stress factor for land plants. Prolonged drought can lead to cell and tissue damage, impaired development, and in the case of crop plants, reduced productivity and crop yield [1]. Ultimately, severe drought leads to plant death.
Vegetative tissues of land plants have different levels of tolerance to water deficiency. While some plants are extremely sensitive to water deprivation and already show signs of damage after 10 % water loss, others are more tolerant to drought, and this tolerance may vary even within a species between different cultivars [1–3]. Extreme loss of water or desiccation (10 % RWC and below) is tolerated only by seeds, some pollen grains, and by a small group of so-called resurrection plants.
Plants adapt to drought by a number of physiological and morphological mechanisms. Immediate responses and slower adaptation responses can be distinguished. One of the first responses is stomatal closure, governed mainly by the plant hormone abscisic acid (ABA) [4]. Stomatal closure is modulated by a number of factors, including ion channels, protein kinases and phosphatases, lipid messengers, reactive oxygen species (ROS), and positive and negative transcriptional regulators [5–9]. This is followed by downregulation of photosynthesis, which also serves to minimize ROS production [2]. Osmoprotectants such as late embryogenesis abundant (LEA) proteins, polyols, proline, sucrose, and other sugars rapidly accumulate in many tissues [10–12]. Aquaporins play an important role for water redistribution among different tissues and cellular compartments [13, 14]. Functional antioxidant systems are also essential for protection against excessive ROS production under drought [15]. Much slower responses include biochemical alterations in the cell wall and changes in root architecture [16–18]. However, proliferation of the root system in response to water deficit is often coupled with reduced above-ground plant growth [4].
Signaling events that lead to these responses involve activation of ion channels, Na+/H+ antiporters, Ca2+-binding proteins such as calmodulin and calcium-dependent protein kinases, receptor-like kinases, and mitogen-activated protein kinases [1, 4]. These early signaling events eventually regulate transcriptional factors and coregulators that govern the global transcriptional re-programming necessary for the execution of the above-mentioned physiological and morphological changes, resulting in adjustment to drought stress [3].
Accumulating data in recent years indicate that the perception of drought in resurrection plants is probably similar to the non-resurrecting plants. However, the final output is different: tolerance to desiccation of vegetative tissues. This suggests that resurrection plants possess unique protective mechanisms. Furthermore, there is increasing evidence that some of the drought-protective mechanisms inducible in most plants by mild drought are constantly active in resurrection plants (T. Gechev and co-workers, unpublished results); [19, 20]. Here, we summarize the current knowledge of the molecular mechanisms that contribute to desiccation tolerance in resurrection plants.
Resurrection plants: overview
Characteristics and geographic diversity of flowering resurrection plants
Resurrection plants are unique in that they are able to lose more than 95 % of the water in vegetative tissues, fall into anabiosis for long periods, and regain full functions after rehydration. Vegetative desiccation tolerance is more common in lower plants such as bryophytes, rare in pteridophytes and angiosperms, and absent in gymnosperms [2, 21]. It has been estimated that the total number of desiccation-tolerant plants is at least c. 1,300 (1,000 pteridophytes and 300 angiosperm plants) [21]. While the mechanisms of desiccation tolerance in bryophytes are mainly related to cellular repair, the more complex tissues in angiosperms require mechanisms that prevent desiccation-induced cell- and tissue damage in the first place [22].
The small group of angiosperm resurrection plants displays remarkable habitat and geographic diversity throughout both the northern and the southern hemispheres. Resurrection plants can be found among both monocots and dicots. Most occur in dry and desert areas or in more temperate areas with sufficient rain precipitation but periods of drought or/and cold winters (like the European resurrection plants Haberlea rhodopensis and Ramonda serbica) [2, 21, 23]. A resurrection plant, Lindernia brevidens, was even discovered in the tropical rainforests of Africa, where humidity is always high [24]. Most of the resurrection species are herbaceous plants [21, 25]. A list with the most well-studied resurrection plants and their origin was recently published by Dinakar et al. [23].
The resurrection plants are interesting not only because of the desiccation tolerance and as a source for gene discovery but also because they have unique metabolites, some of which have potential uses in biotechnology and medicine. For example, the South African woody resurrection species Myrothamnus flabellifolia has long been known for its medicinal properties [26]. Its extracts, rich in polyphenols and essential oils, are used to treat various disorders, including influenza, kidney diseases, and gingivitis [26, 27]. The predominant polyphenol 3,4,5-tri-O-galloylquinic acid has been shown to inhibit M-MLV and HIV-1 reverse transcriptases [28]. Myconoside, a glycoside abundantly present in extracts of H. rhodopensis, can strongly stimulate antioxidant skin defenses and extracellular matrix protein synthesis [29]. Extracts from H. rhodopensis, which are also rich in polyphenols, can stimulate the synthesis of elastin in a dose-dependent manner and also possess radioprotective, anticlastogenic, and antioxidant effects on rabbit blood samples exposed to gamma radiation in vitro [29, 30]. These results suggest that the strong medicinal properties of some resurrection plants should be the aim of further extensive experiments which could provide strategies and solutions in combating various human diseases.
Desiccation-imposed types of stress and protection mechanisms in resurrection species
Physiological and metabolic processes associated with desiccation tolerance resemble those observed during drought stress and some processes associated with seed maturation [12]. Severe drought and desiccation impose metabolic, mechanical, and oxidative stresses, which are addressed by complex protective mechanisms.
Initial physiological responses driven by mild water deficiency include rapid increases in ABA concentrations, which lead to ABA-directed transcription of stress-associated genes encoding protective proteins such as aldehyde dehydrogenases, heat shock factors, and LEA proteins [31–33]. ABA-mediated gene expression is highly complex and involves both positive and negative transcriptional regulators [32, 34]. Eventually, as the water deficiency progresses, photosynthesis is completely inhibited [21, 35]. Shutting down photosynthesis is not just a consequence of severe drought stress but also serves a protective role, minimizing excessive ROS production always observed during water deficiency [12]. Some resurrection plants lose their chlorophyll and degrade their thylakoid membranes during dehydration (poikilochlorophyllous plants), which prevents photosynthetic-related ROS production [36]. Other resurrection plants such as Craterostigma plantagineum and H. rhodopensis retain their chlorophyll and thylakoid structure (homoiochlorophyllous plants) [25, 37]. The photosynthetic systems in homoiochlorophyllous plants are therefore just reversibly inactivated, not destroyed, which enables them to recover fast after rehydration [36, 37]. The downside of this strategy, a possible ROS production, is minimized by additional morphological and biochemical mechanisms such as leaf folding to reduce absorbed radiation or/and accumulation of anthocyanins and other phenolic compounds to protect against solar radiation [12]. Homoiochlorophyllous plants therefore accumulate higher concentrations of anthocyanins than poikilochlorophyllous plants [2]. Further protection against photo-oxidative damage may be provided by accumulation of ELIPs (early light-inducible proteins), which inactivates chlorophyll by binding to it, as is proposed for the 22-kDa ELIP dsp22 in C. plantagineum [38, 39].
The progressive decrease in water content imposes considerable mechanical stress on plant cell architecture and exposes the macromolecules to a risk of dehydration and inactivation. The mechanical stress is counteracted by reversible changes in cell wall architecture [2]. Changes in the plant cell wall polysaccharides and proteins take place, making the cell wall more flexible [40–42]. These changes also enable rolling and reversible folding of the leaves (Fig. 1; [42, 43]). Such changes could be species-specific and could involve incorporation of specific proteins in the cell wall as observed for the glycine-rich protein BhGRP1 in Boea hygrometrica [18], substitution of glucose residues with galactose residues in the xyloglucan molecules in Craterostigma wilmsii [41], activation of expansins resulting in a more extendable cell wall in C. plantagineum [40], or arabinose-rich pectin polymers in Myrothamnus flabellifolia [42]. In addition to the leaf folding, mechanical stress is also alleviated by increased vacuolation, a process of water replacement in vacuoles by non-aqueous substances. Depending on the species, some resurrection plants utilize either of the two strategies or both, while neither mechanism has been described for desiccation-sensitive species [2].
Fig. 1.
Unstressed fully hydrated (a, d), desiccated (b, e), and rehydrated (c, f) Haberlea rhodopensis (a, b, c) and Craterostigma plantagineum (d, e, f) plants
As water content further decreases, both the mechanical and the metabolic stresses become more prominent, with subcellular perturbations and cellular content becoming more concentrated, which in turn increases the chances of unwanted molecular interactions, altered biochemical activities, and macromolecular denaturation. Sugars, LEA proteins, and heat shock proteins are responsible for water replacement and stabilization of proteins and other macromolecular structures [12]. Sucrose is the principal sugar that accumulates in most resurrection plants during desiccation [11, 44–47]. Sucrose and trehalose can serve as osmoprotectants of biological membranes and can stabilize micromolecular structures [11, 48]. In addition to their protective roles, these sugars may have signaling functions, regulating important metabolic pathways. Trehalose and trehalose-6-phosphate in particular are central metabolite regulators, influencing carbohydrate status, growth, and energy metabolism [49]. Trehalose-6-phosphate stimulates ADP-glucose pyrophosphorylase, promoting starch synthesis, whereas trehalose has the opposite effect, stimulating starch breakdown [49, 50]. Raffinose family oligosaccharides, such as raffinose and stachyose, also accumulate during desiccation in many angiosperm resurrection plants and may have prominent roles protecting the cells by water replacement and vitrification [2, 45]. Raffinose is synthesized by raffinose synthase from galactinol and sucrose [51]. It has recently been shown that both galactinol and raffinose can protect from paraquat-induced oxidative damage [51].
Resurrection plants retain photosynthetic activity during mild drought but photosynthesis is compromised during severe desiccation. Sucrose, the principal osmoprotectant in many species, is then synthesized from carbon originating from reserve sugars such as starch in most resurrection plants or from octulose in C. plantagineum [44]. Stachyose may also provide a source of carbon for sucrose synthesis [45].
Hydrophilins and LEA proteins in particular are ubiquitous proteins rapidly synthesized during desiccation in vegetative tissues and seeds of both desiccation-sensitive and resurrection plants [35, 52, 53]. One of their possible roles is to provide a water hydration “shell” to certain proteins and macromolecules, thus preventing damage due to dehydration [10]. In contrast to desiccation-sensitive plants, expression of certain desiccation-responsive LEA genes is constitutive in H. rhodopensis under normal (water-sufficient) growth conditions (T. Gechev and co-workers, unpublished results). Likewise, small heat shock proteins (sHSPs) are expressed in unstressed tissues of desiccation-tolerant C. plantagineum and further induced upon drought stress [20]. The sHSPs have similar properties to hydrophilins but some of them also act as molecular chaperones for other proteins, thus preventing proteins from both aggregation and denaturation [2]. Other heat shock genes are rapidly induced upon dehydration in various resurrection plants ([54]; T. Gechev and co-workers, unpublished results). The importance of dehydrins for desiccation tolerance has been demonstrated in the moss Physcomitrella patens, where a knockout of the dehydrin gene PpDHNA by homologous recombination impairs growth after salt and osmotic stress [55]. In addition, novel types of hydrophilic proteins are expressed during desiccation in resurrection plants, such as CpEdi-9 from the resurrection plant C. plantagineum [56]. Cellular compartmentalization of LEA proteins is also an important determinant of stress tolerance [53]. Taken together, these cases substantiate the role of LEA proteins and hydrophilins in desiccation tolerance of resurrection plants.
Progressive water loss leads to excessive production of ROS, alcohols, and carbonyls. This is alleviated by induction of ROS scavenging enzymes like 1-cys peroxiredoxin or aldehyde dehydrogenases [31, 57]. Other antioxidant enzymes, maintaining a tight balance of reactive oxygen species, are equally important for acquisition of drought stress tolerance. Indeed, the ability of Myrothamnus flabellifolia to survive desiccation has been correlated with its antioxidant status [15].
Signaling mechanisms, transcription factors and downstream reactions
Many of the changes in response to water deficit result from massive transcriptional reprogramming. Signaling molecules could be ABA, lipid messengers, or/and the alterations in the redox components [32, 58]. ABA-dependent gene expression is modulated by both positive and negative factors [32, 34]. However, ABA-independent mechanisms of gene regulation also exist [3] (Fig. 2).
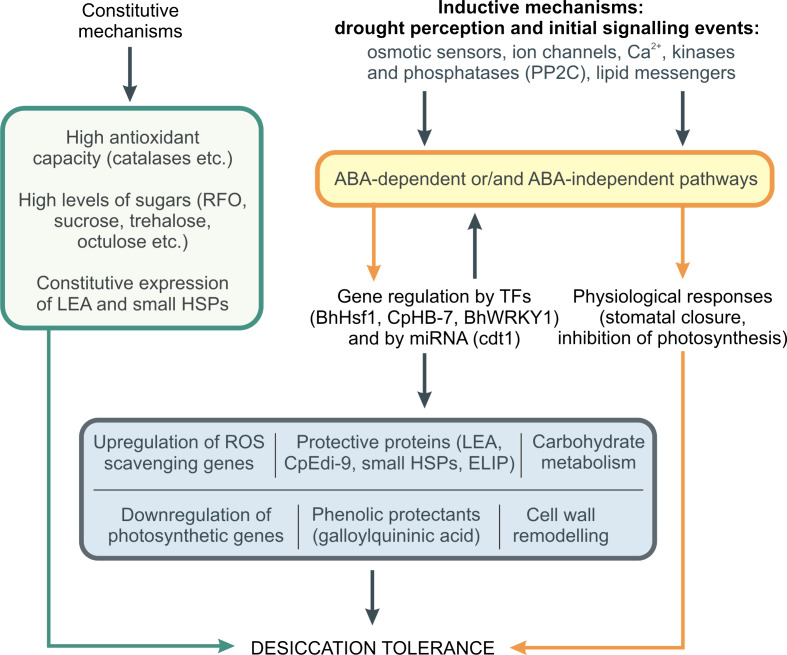
Fig. 2.
Diagrammatic representation of signaling events leading to acquisition of desiccation tolerance in resurrection plants. Both constitutive and inducible mechanisms are employed by the resurrection plants to maintain cellular homeostasis and protect from desiccation-induced damage. The constitutive mechanisms include high activity of antioxidant enzymes (such as catalases, superoxide dismutases, etc.), high levels of particular non-reducing sugars (sucrose, raffinose family oligosaccharides), and constitutive expression of late embryogenesis abundant and heat shock proteins. The inducible mechanisms, relayed by osmotic sensors, Ca2+ fluxes, kinases, phosphatases, and lipid messengers, activate rapid physiological responses (stomatal closure, inhibition of photosynthesis) and regulate gene expression via abscisic acid (ABA)-dependent and -independent pathways, resulting in upregulation of antioxidant genes, further accumulation of protective sugars and proteins, cell remodeling, synthesis of secondary metabolites related to defence, and eventually conferring desiccation tolerance
Transcription factors from different families have been implicated in governing the desiccation-induced gene re-programming leading to induction of protective mechanisms, inhibition of photosynthesis, and, in many cases, growth retardation [32, 34, 58–61]. For example, a WRKY transcription factor binds to a W-box promoter element of the galactinol synthase gene to induce synthesis of raffinose and raffinose family oligosaccharides (RFOs) in Boea hygrometrica, possibly in an ABA-dependent manner [60]. Another transcription factor from the Myb family, CpMYB10, binds to and regulates its own promoter as well as the promoter of the LEA gene Cp11-24 in C. plantagineum [62]. Interestingly, it may function as a repressor in non-stressed tissues and as an activator in tissues experiencing water deficit. Transgenic A. thaliana overexpressing CpMYB10 are less sensitive to drought, which correlates with elevated levels of the alcohol dehydrogenase ADH1 and two hydrophilic proteins, RD29A and COR15A [62]. The drought-responsive transcriptional regulator CpHB-7, a leucine zipper protein, is a negative modulator of ABA and targets the desiccation-inducible dehydrin CDeT6-19 [32].
In addition to transcription factors, RNA silencing adds to the complexity of expression control. The unique CDT1 gene confers desiccation tolerance in C. plantagineum [63]. It by-passes any ABA requirement for desiccation tolerance, as its overexpression in C. plantagineum callus cultures is sufficient to induce desiccation tolerance without ABA treatment [63, 64]. CDT1 is a member of a retrotransposon-like gene family encoding a small regulatory RNA that is responsible for the transcriptional reprogramming during drought stress in C. plantagineum [34]. However, the molecular mechanism is still not known.
Some of the downstream targets in the signaling cascade are genes that bring new insights into the mechanisms of desiccation tolerance, such as the member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily upregulated in leaves and roots during desiccation in Xerophyta humilis [65] or the plastid-targeted protein with DNA-binding activity in C. plantagineum [58]. One such novel gene isolated from Xerophyta viscosa encodes a hydrophobic protein named XvSAP1 [66]. XvSAP1 is induced by a variety of stresses, including drought, salinity, and extreme temperatures [67]. When overexpressed in A. thaliana, it conferred tolerance against drought, salt, and high temperature stress [66] (Table 1).
Table 1.
Genes from resurrection species that alter stress tolerance in transgenic plants
| Gene, gene product, and resurrection species source | Transgenic plant and observed phenotype | Reference |
|---|---|---|
|
BhGolS1 Galactinol synthase from B. hygrometrica |
Transgenic tobacco is more tolerant to drought stress; no phenotype under normal conditions | Wang et al. [60] |
|
BcBCP1 Phytocyanin-related early nodulin-like gene from Boea crassifolia |
Tobacco expressing BcBCP1 is more tolerant to osmotic stress | Wu et al. [33] |
|
BhHsf1 Heat shock transcription factor from B. hygrometrica |
Over-expression of BhHsf1 in Arabidopsis and tobacco results in increased thermo-tolerance but decreased cell proliferation and reduced size of aerial organs | Zhu et al. [59] |
|
XVSAP1 Hydrophobic membrane protein from X. viscosa |
Expression of XVSAP1 in E. coli results in osmotic stress tolerance; expression in Arabidopsis improves performance against osmotic, heat, and salt stress | Garwe et al. [66, 67] |
|
CDT-1 Regulatory RNA from C. plantagineum |
Constitutive expression in C. plantagineum results in desiccation tolerance of callus tissues without ABA treatment | Smith-Espinoza et al. [64] |
|
CpHB-7 Homeodomain leucine zipper transcription factor from C. plantagineum |
Transgenic tobacco germinate early and grow faster under normal conditions. Reduced sensitivity to ABA during seed germination and stomatal closure. Plants are not more tolerant to dehydration and chilling but seeds are more tolerant to salinity | Deng et al. [32] |
|
CpMYB10 Transcription factor from C. plantagineum |
Transgenic Arabidopsis is more tolerant to drought and salt stress. Plants are insensitive to glucose and hypersensitive to ABA | Villalobos et al. [62] |
|
BhLEA1 and BhLEA2 Late embryogenesis abundant proteins from B. hygrometrica |
Transgenic tobacco have higher relative water content, increased PSII activity, decreased electrolyte leakage, increased peroxidase and superoxide dismutase activities during drought stress | Liu et al. [88] |
|
PpDHNA Dehydrin gene from Physcomitrella patens |
A knockout of the dehydrin gene PpDHNA in Physcomitrella patens results in impaired growth after salt and osmotic stress treatments. | Saavedra et al. [55] |
New insights on the molecular mechanisms of desiccation tolerance from recent transcriptome profiling of resurrection plants
Recent advances in next generation sequencing has enabled a comprehensive analysis of the genes and biochemical pathways regulated during dehydration and subsequent recovery of two dicot resurrection plants. C. plantagineum and H. rhodopensis grown with sufficient water to maintain a fully hydrated state were subjected to mild drought stress, severe desiccation (less than 5 % relative water content), and then rehydration (T. Gechev and co-workers, unpublished results; [54]). Transcripts were sequenced from the different physiological stages. This allows full genome coverage of transcripts in species with unsequenced genomes, which is particularly useful, not only for following dynamics in gene expression but also for discovering unknown genes, which have not been reported before.
Blast analysis of transcripts obtained from Haberlea and C. plantagineum revealed that most of the transcript sequences are similar to genes from grapevine (Vitis vinifera), castor bean (Ricinus communis) and poplar (Populus trichocarpa). Gene ontology analysis identified categories prominently represented in particular conditions. Genes highly abundant in unstressed samples of both species were related to photosynthesis, growth, and cell wall organization. Many of these genes are also abundantly represented in the rehydrated samples, confirming the functional recovery of the plants. Genes encoding a sucrose synthase, chromosome scaffold proteins, and many stress-related proteins, including several LEA proteins, a GABA transaminase, lipocalin, and pathogenesis-related proteins, were induced at the early stages of dehydration in C. plantagineum [54]. Signaling-related genes encoding a calcium channel protein and proteins involved in ABA signaling were also induced, confirming the role of ABA in early events required for acquisition of desiccation tolerance in C. plantagineum [25]. Genes related to thiamine metabolism and stress, such as LEA proteins, cysteine proteases and desaturases, were abundantly present in desiccated tissues [54]. Genes involved in metabolism of vitamin K-related compounds and reactive oxygen species were overrepresented in the rehydrated plants.
In H. rhodopensis, strong constitutive expression of LEA and catalase genes in unstressed plants supports the notion that the transcriptome of H. rhodopensis is primed for drought stress events (T. Gechev and co-workers, unpublished results). Two catalase genes maintain substantial levels of expression during all conditions, and several other catalase genes are upregulated by drought stress. One LEA gene is further upregulated during drought and desiccation. Genes exclusively expressed in water-deficient samples include a stachyose synthase, two genes encoding early light-inducible proteins (ELIP), heat shock factors, more LEA genes, signaling components such as protein phosphatases, and a number of transcription factors and transcriptional regulators (T. Gechev and co-workers, unpublished results). These events coincided with induction of beta-amylase, presumably activated to utilize starch reserves. Induction of ELIPs has been demonstrated in Tortula ruralis and C. plantagineum [38, 39, 68]. The ELIP proteins are believed to protect chloroplasts from photooxidative damage [69]. The most abundantly induced gene in H. rhodopensis was a protein phosphatase with high similarity to protein phosphatase type 2C (PP2C) from Ricinus communis (T. Gechev and co-workers, unpublished results). This gene was virtually not expressed in unstressed plants, but transcript levels were high under mild drought and even more in desiccated samples, and then disappeared in rehydrated samples. Protein phosphorylation and de-phosphorylation is believed to be an essential part of signaling events leading to acquisition of drought/desiccation tolerance, and changes in the phosphorylated status of a number of proteins have been shown for C. plantagineum [70]. Photosynthesis-related genes are downregulated in both species, underlining a common response to stress. This, together with induction of ELIPs and catalases, may serve to control ROS levels during drought and desiccation. The large number of genes with no sequence similarities to other species are potential targets for drought/desiccation tolerance studies.
In addition to the transcriptome analysis by next generation sequencing, a recent AFLP analysis of H. rhodopensis transcriptome identified cDNAs of genes encoding early- and late-desiccation regulated proteins, including a protein kinase and a bZIP transcription factor [71].
Proteome and metabolome analysis in desiccation-tolerant species
Studies of the proteome in resurrection plants
Comprehensive proteome studies are more difficult to conduct relative to the transcriptome studies as only a limited number of proteins (primarily the most abundant) can be identified. Nevertheless, several groups have quantified proteome changes during desiccation in resurrection plants [72–74]. Recent analysis of Sporobolus stapfianus proteins revealed a decrease in Rubisco large subunit and associated proteins, indicating a decrease in photosynthesis during dehydration [74]. At the same time, protein kinases, enzymes of glycolysis, and enzymes of the Calvin cycle increased. This study also identified upregulation of proteins involved in maintaining the chromatin structure, indicating that chromatin structures may undergo substantial changes as a result of desiccation [74].
Dehydration-responsive proteins were also identified in Xerophyta viscosa [73]. Analysis of the whole proteome at 35 % RWC (when induction of late protective mechanisms are initiated) revealed dehydration-induced antioxidants, glycolytic enzymes, a chloroplast FtsH protease, and an RNA-binding protein, while photosynthetic proteins decreased in accordance with the targeted catabolism of the photosynthetic machinery in this poikilochlorophyllous species [73].
In B. hygrometrica leaves, dehydration and subsequent rehydration induced alterations in a number of proteins related to photosynthesis (Rubisco large subunit, oxygen-evolving complex of PSII), transporters (vacuolar H + ATPase A subunit, ABC transporter), and antioxidant metabolism (glutathione peroxidase, glutathione-S-transferase) [72]. However, in this case, some photosynthetic-related proteins increased in contrast to the other observations.
A proteome study of the resurrection fern ally Selaginella tamariscina revealed regulation of proteins related to photosynthesis, energy and sugar metabolism, protein synthesis and folding, and stress response proteins [75]. As in most other cases, photosynthesis-related proteins, including the Rubisco large subunit, chl a/b binding protein, and oxygen-evolving complex, were downregulated. Seven members of the ATP synthase family were highly expressed during desiccation, possibly reflecting the requirement of increased energy supply during dehydration. Not surprisingly, the abundance of a number of proteins related to sugar metabolism, including transketolases, sucrose synthase, ADP-glucose pyrophosphorylase, and GDP-mannose 3,5-epimerase, were altered during the different stages of desiccation and rehydration. Most of the proteins involved in protein synthesis, folding, and degradation were downregulated at the early stages of desiccation but upregulated at later stages [75]. Fourteen of the 103 differentially expressed proteins had two or more spots on 2D gels, which suggests that posttranslational modifications may take place during desiccation and subsequent rehydration. This notion was further supported by the reversible phosphorylation of a number of proteins during dehydration and subsequent rehydration of C. plantagineum [70].
In comparison to the transcriptomic analyses only a small number of proteins have been studied in proteomic approaches in resurrection plants. However, there are several examples of posttranslational modifications of dehydration-induced proteins. In particular, there is evidence for phosphorylation of LEA proteins, and it remains a challenge for future studies to understand the role of phosphorylation in the context of desiccation. Other posttranslational modifications such as sumoylation or nitrosylation have not yet been addressed in resurrection plants, although these modifications are important regulatory mechanisms in cellular metabolism.
Metabolite changes during desiccation in resurrection species
Most of the data on metabolite changes observed under different water regimes are related to sugar metabolism. Accumulation of many types of sugars, as noted above, is observed in resurrection species [2, 76–78]. The most abundantly accumulating sugars are sucrose and RFOs. Both are believed to act as osmoprotectants, but they could also protect against oxidative damage [51]. In addition, some RFOs are storage carbohydrates and are mobilized during drought as sources of energy and for sucrose synthesis [45]. Octulose, a rare C8 sugar [79], accumulates as a storage carbohydrate in C. plantagineum and is believed to serve as carbon source for sucrose formation during desiccation, as octulose concentrations decrease with progression of drought in parallel with increases in sucrose concentrations [25].
A few studies using comprehensive metabolomic approaches were recently conducted in resurrection plants. The desiccation-tolerant Sporobolus stapfianus has higher concentrations of osmolytes and nitrogen metabolites but lower concentrations of metabolites associated with energy metabolism than the desiccation-sensitive Sporobolus pyramidalis under normal, well-watered conditions [19]. Desiccation of the two species resulted in production of protective compounds in S. stapfianus related to ROS and ammonia detoxification, nitrogen remobilization, and soluble sugar production, but such responses were not observed in the sensitive S. pyramidalis. This suggests that the metabolome of S. stapfianus is already primed for drought stress and responds adequately to desiccation.
Metabolome analysis of H. rhodopensis revealed dramatic changes in specific metabolites during dehydration, desiccation, and subsequent rehydration (T. Gechev and co-workers, unpublished results). Principal component analysis of the four conditions forms two separate clusters: one with the drought-stressed and desiccated plants and another with control and rehydrated plants, confirming that metabolic activities return to normal after rehydration. This notion is supported by the physiological data and transcriptome analysis of H. rhodopensis (T. Gechev and co-workers, unpublished results). Increases in maltose, verbascose, and massive accumulation of sucrose were observed during drought and desiccation. The levels of these sugars in rehydrated samples returned to levels similar to unstressed controls. In contrast, glucose, fructose, myo-inositol, and galactinol decreased in water-deficient samples. This could partly be explained by their use for synthesis of sucrose and complex RFOs such as verbascose. Interestingly, high concentrations of galactinol, myo-inositol, and pyruvate were accumulated after rehydration, probably due to activation of sugar catabolic pathways to enhance energy production by respiration supporting the recovery from desiccation. Furthermore, drought and particularly desiccation resulted in increased accumulation of two stress-related metabolites, spermidine and GABA. Spermidine, a polyamine implicated in osmotic stress defense, also increases in C. plantagineum during desiccation [80], while GABA is a well-known signaling molecule with a role in controlling stress responses and growth [81, 82]. Proline is another compound implicated in osmotic stress tolerance. Accumulation of amino acids including proline during dehydration is observed in many resurrection plants [83, 84]. Proline, however, does not seem to be involved in the mechanism of desiccation tolerance in H. rhodopensis, as it does not accumulate under stress and only low concentrations are found in unstressed plants (T. Gechev and co-workers, unpublished results).
Much higher concentrations of sucrose, trehalose, and GABA were found in the desiccation-tolerant H. rhodopensis compared to the desiccation-sensitive A. thaliana (T. Gechev and co-workers, unpublished results). These findings further support a role of these metabolites in desiccation tolerance and corroborate the notion that both the transcriptome and the metabolome of H. rhodopensis are ready to cope with desiccation.
General conclusions
Why are resurrection plants different from other plants? What makes them tolerant to desiccation in contrast to plants that are sensitive to drought or drought tolerant but desiccation sensitive? At least three reasons emerge from the cumulative physiological, transcriptional, proteomic, and metabolomic data on desiccation-tolerant resurrection plants.
First, some resurrection plants are already primed for desiccation as deduced from molecular mechanisms. Their transcriptomes, proteomes, and metabolomes in normal, non-stress conditions are characterized by much higher transcript levels of particular antioxidant genes, genes encoding cell remodeling proteins, hydrophilins, heat shock proteins and chaperones, dehydrogenases, and other protectants, compared with desiccation-sensitive plants (T. Gechev and co-workers, unpublished results; [20, 74]). This makes them better prepared to meet the adverse consequences of drought, extreme dehydration, oxidative stress, mechanical stress, protein denaturation, and formation of toxic products. Particular cyclophilins, dehydrins, and other LEA proteins are already present during normal growth conditions in resurrection plants. In contrast, such proteins are expressed only in desiccating seeds and during drought in other, desiccation-sensitive species. Likewise, many resurrection plants contain high amounts of anthocyanins and other polyphenols in optimal growth conditions whereas other plants accumulate these compounds only upon stress exposure. Resurrection plants maintain their antioxidant system, including antioxidant metabolites and enzymes at high capacity throughout all kinds of environmental conditions, protecting themselves against adverse fluctuations in water content. This is a unique feature of the resurrection plant antioxidant machinery, as desiccation-sensitive species do not appear to maintain functional antioxidant systems during extreme drought.
Secondly, most of the resurrection plants possess similar mechanisms and utilize similar strategies to counteract water deficiency as do other plants, but many of these protective mechanisms are more substantially activated in the resurrection plants upon sensing water deficiency. Upon water stress, cyclophilins, dehydrins, and other LEA proteins can reach as much as 40 % of the dry weight in some resurrection plants, and the powerful phenolic antioxidant 3,4,5-tri-O-galloylquinic acid can accumulate to more than 70 % of dry weight [85]. Furthermore, concentrations of stress signals such as GABA and protective molecules such as particular heat shock proteins, polyphenols, and sugars such as sucrose, stachyose, raffinose, and trehalose can reach levels several times higher in magnitude than levels observed in sensitive plants (T. Gechev and co-workers, unpublished results; [86]).
Thirdly, resurrection plants contain so far unknown genes, proteins, and metabolites with putative protective properties. Examples of such novel proteins and metabolites are the hydrophilic protein CpEdi-9 from C. plantagineum and 3,4,5-tri-O-galloylquinic acid, isolated from the resurrection plant M. flabellifolia, respectively [56, 85]. The large number of unidentified genes in C. plantagineum and H. rhodopensis suggest that some of these could be unique, such as the CDT-1 gene important in acquisition of desiccation tolerance in callus of C. plantagineum [87]. Supporting this notion, 41 % of all H. rhodopensis transcripts have no sequence homology to genes from other species (T. Gechev and co-workers, unpublished results).
Due to these three distinctive features, physiological adaptations characteristic for the resurrection species evolved to protect against severe drought. For example, poikilochlorophyllous resurrection plants disassemble their photosynthetic machinery and degrade their chlorophyll during desiccation, downregulating photosynthesis and eliminating photosynthesis-related production of ROS [2]. Even more surprisingly, homoiochlorophyllous resurrection plants switch off photosynthesis without breaking down their photosynthetic machinery [37]. They are capable of resuming photosynthesis rapidly upon re-watering, thanks to their preserved photosynthetic apparatus. High concentrations of anthocyanins and other polyphenol antioxidants together with ELIPs and with mechanical adaptations such as rolling of leaves protect from excessive radiation [2].
Many resurrection plants grow slowly. Perhaps this is the price they have to pay in exchange of their extreme stress tolerance; some of the energy resources are directed towards synthesis of stress-protective compounds at the expense of energy needed for faster growth [4]. Consistent with this, Arabidopsis and tobacco plants overexpressing the dehydration-inducible transcription factor BhHsf1 from the resurrection plant B. hygrometrica displayed enhanced stress tolerance but retarded growth [59].
Genetic adaptations such as the novel genes mentioned above or the RNAi control of gene expression via expansion of retroelement-amplified regulatory RNA (CDT-1 gene in C. plantagineum) further illustrates the complex control of desiccation tolerance.
Drought stress is nowadays the most common threat to agriculture. The potential for molecular improvement has already been shown by expressing genes from resurrection species origin in model and crop species [32, 62, 66]. However, a complex approach is probably needed in order to engineer desiccation tolerance in other species. Future research on resurrection plants such as detailed kinetics studies on the molecular events during desiccation and rehydration, comparative analysis with non-resurrection plants, and functional analysis of genes with presumed roles in dehydration/rehydration responses can help us to understanding the cellular mechanisms of desiccation tolerance in the unique group of resurrection plants, and serve as a basis for future molecular improvement of drought tolerance in crop species.
Acknowledgments
D.B. is a member of the COST action FA090 “Putting halophytes to work”. M.B., V.T., and T.G. acknowledge EC FP7 (project BioSupport, 245588).
Abbreviations
- ABA
Abscisic acid
- ELIP
Early light-inducible proteins
- GABA
γ-Aminobutyric acid
- LEA
Late embryogenesis abundant genes/proteins
- RFOs
Raffinose family oligosaccharides
- ROS
Reactive oxygen species
- RWC
Relative water content
References
- 1.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrant J, Brandt W, Lidsey GG. An overview of mechanisms of desiccation tolerance in selected angiosperm resurrection plants. Plant Stress. 2007;1:72–84. [Google Scholar]
- 3.Bartels D, Souer E. Molecular responses of higher plants to dehydration. In: Hirt H, Shinozaki K, editors. Plant responses to abiotic stress. Berlin: Springer; 2003. pp. 9–38. [Google Scholar]
- 4.Harb A, Krishnan A, Ambavaram MM, Pereira A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010;154:1254–1271. doi: 10.1104/pp.110.161752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al Rasheid KA, Grill E, Romeis T, Hedrich R. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters C, Li M, Narasimhan R, Roth M, Welti R, Wang X. Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis . Plant Cell. 2010;22:2642–2659. doi: 10.1105/tpc.109.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Li Y, Li H, Wu G. Overexpression of AtNHX5 improves tolerance to both salt and drought stress in Broussonetia papyrifera (L.) Vent. Tree Physiol. 2011;31:349–357. doi: 10.1093/treephys/tpr003. [DOI] [PubMed] [Google Scholar]
- 8.Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009;23:1805–1817. doi: 10.1101/gad.1812409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, Romeis T, Hedrich R. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA. 2009;106:21425–21430. doi: 10.1073/pnas.0912021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olvera-Carrillo Y, Campos F, Reyes JL, Garciarrubio A, Covarrubias AA. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis . Plant Physiol. 2010;154:373–390. doi: 10.1104/pp.110.158964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinelli T. In situ localization of glucose and sucrose in dehydrating leaves of Sporobolus stapfianus . J Plant Physiol. 2008;165:580–587. doi: 10.1016/j.jplph.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Farrant JM, Moore JP. Programming desiccation-tolerance: from plants to seeds to resurrection plants. Curr Opin Plant Biol. 2011;14:340–345. doi: 10.1016/j.pbi.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Almeida-Rodriguez AM, Cooke JE, Yeh F, Zwiazek JJ. Functional characterization of drought-responsive aquaporins in Populus balsamifera and Populus simonii X balsamifera clones with different drought resistance strategies. Physiol Plant. 2010;140:321–333. doi: 10.1111/j.1399-3054.2010.01405.x. [DOI] [PubMed] [Google Scholar]
- 14.Rae L, Lao NT, Kavanagh TA. Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta. 2011;234:429–444. doi: 10.1007/s00425-011-1414-z. [DOI] [PubMed] [Google Scholar]
- 15.Kranner I, Beckett RP, Wornik S, Zorn M, Pfeifhofer HW. Revival of a resurrection plant correlates with its antioxidant status. Plant J. 2002;31:13–24. doi: 10.1046/j.1365-313X.2002.01329.x. [DOI] [PubMed] [Google Scholar]
- 16.Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Hofer R, Schreiber L, Chory J, Aharoni A. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol. 2007;145:1345–1360. doi: 10.1104/pp.107.105676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de CB, De C, I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, Genty B, Stubbs KA, Inzé D, Van Breusegem F (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22:2660-2679 [DOI] [PMC free article] [PubMed]
- 18.Wang L, Shang H, Liu Y, Zheng M, Wu R, Phillips J, Bartels D, Deng X. A role for a cell wall localized glycine-rich protein in dehydration and rehydration of the resurrection plant Boea hygrometrica . Plant Biol. 2009;11:837–848. doi: 10.1111/j.1438-8677.2008.00187.x. [DOI] [PubMed] [Google Scholar]
- 19.Oliver MJ, Guo L, Alexander DC, Ryals JA, Wone BW, Cushman JC. A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus . Plant Cell. 2011;23:1231–1248. doi: 10.1105/tpc.110.082800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alamillo J, Almoguera C, Bartels D, Jordano J. Constitutive expression of small heat shock proteins in vegetative tissues of the resurrection plant Craterostigma plantagineum . Plant Mol Biol. 1995;29:1093–1099. doi: 10.1007/BF00014981. [DOI] [PubMed] [Google Scholar]
- 21.Porembski S. Evolution, diversity and habitats of poikilohydrous plants. In: Luttge U, Beck E, Bartels D, editors. Plant desiccation tolerance. Berlin: Springer; 2011. pp. 139–156. [Google Scholar]
- 22.Martinelli T, Whittaker A, Masclaux-Daubresse C, Farrant JM, Brilli F, Loreto F, Vazzana C. Evidence for the presence of photorespiration in desiccation-sensitive leaves of the C4 ‘resurrection’ plant Sporobolus stapfianus during dehydration stress. J Exp Bot. 2007;58:3929–3939. doi: 10.1093/jxb/erm247. [DOI] [PubMed] [Google Scholar]
- 23.Dinakar C, Djilianov D, Bartels D. Photosynthesis in desiccation-tolerant plants: energy metabolism and antioxidative stress defense. Plant Sci. 2012;182:29–41. doi: 10.1016/j.plantsci.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Phillips, Fischer E, Baron M, van den DN, Facchinelli F, Kutzer M, Rahmanzadeh R, Remus D, Bartels D. Lindernia brevidens: a novel desiccation-tolerant vascular plant, endemic to ancient tropical rainforests. Plant J. 2008;54:938–948. doi: 10.1111/j.1365-313X.2008.03478.x. [DOI] [PubMed] [Google Scholar]
- 25.Bartels D. Desiccation tolerance studied in the resurrection plant Craterostigma plantagineum . Integr Compar Biol. 2005;45:696–701. doi: 10.1093/icb/45.5.696. [DOI] [PubMed] [Google Scholar]
- 26.Moore JP, Lindsey GG, Farrant JM, Brandt WF. An overview of the biology of the desiccation-tolerant resurrection plant Myrothamnus flabellifolia . Ann Bot. 2007;99:211–217. doi: 10.1093/aob/mcl269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohr G, Beikler T, Podbielski A, Standar K, Redanz S, Hensel A. Polyphenols from Myrothamnus flabellifolia Welw. inhibit in vitro adhesion of Porphyromonas gingivalis and exert anti-inflammatory cytoprotective effects in KB cells. J Clin Periodontol. 2011;38:457–469. doi: 10.1111/j.1600-051X.2010.01654.x. [DOI] [PubMed] [Google Scholar]
- 28.Kamng’ona A, Moore JP, Lindsey G, Brandt W. Inhibition of HIV-1 and M-MLV reverse transcriptases by a major polyphenol (3,4,5-tri-O-galloylquinic acid) present in the leaves of the South African resurrection plant, Myrothamnus flabellifolia . J Enzyme Inhib Med Chem. 2011;26:843–853. doi: 10.3109/14756366.2011.566220. [DOI] [PubMed] [Google Scholar]
- 29.Dell’Acqua G, Schweikert K. Skin benefits of a myconoside-rich extract from resurrection plant Haberlea rhodopensis . Int J Cosm Sci. 2011;34:132–139. doi: 10.1111/j.1468-2494.2011.00692.x. [DOI] [PubMed] [Google Scholar]
- 30.Popov B, Georgieva S, Gadjeva V, Petrov V. Radioprotective, anticlastogenic and antioxidant effects of total extract of Haberlea rhodopensis on rabbit blood samples exposed to gamma radiation in vitro. Revue Med Vet. 2011;1:34–39. [Google Scholar]
- 31.Kirch HH, Nair A, Bartels D. Novel ABA- and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantagineum and Arabidopsis thaliana . Plant J. 2001;28:555–567. doi: 10.1046/j.1365-313X.2001.01176.x. [DOI] [PubMed] [Google Scholar]
- 32.Deng X, Phillips J, Brautigam A, Engstrom P, Johannesson H, Ouwerkerk PB, Ruberti I, Salinas J, Vera P, Iannacone R, Meijer AH, Bartels D. A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Mol Biol. 2006;61:469–489. doi: 10.1007/s11103-006-0023-x. [DOI] [PubMed] [Google Scholar]
- 33.Wu H, Shen Y, Hu Y, Tan S, Lin Z. A phytocyanin-related early nodulin-like gene, BcBCP1, cloned from Boea crassifolia enhances osmotic tolerance in transgenic tobacco. J Plant Physiol. 2011;168:935–943. doi: 10.1016/j.jplph.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 34.Hilbricht T, Salamini F, Bartels D. CpR18, a novel SAP-domain plant transcription factor, binds to a promoter region necessary for ABA mediated expression of the CDeT27-45 gene from the resurrection plant Craterostigma plantagineum Hochst . Plant J. 2002;31:293–303. doi: 10.1046/j.1365-313X.2002.01357.x. [DOI] [PubMed] [Google Scholar]
- 35.Bartels D, Hussain SS. Resurrection plants: physiology and molecular biology. In: Luttge U, Beck E, Bartels D, editors. Plant desiccation tolerance. Berlin: Springer; 2011. pp. 339–364. [Google Scholar]
- 36.Ingle RA, Collett H, Cooper K, Takahashi Y, Farrant JM, Illing N. Chloroplast biogenesis during rehydration of the resurrection plant Xerophyta humilis: parallels to the etioplast-chloroplast transition. Plant Cell Environ. 2008;31:1813–1824. doi: 10.1111/j.1365-3040.2008.01887.x. [DOI] [PubMed] [Google Scholar]
- 37.Strasser RJ, Tsimilli-Michael M, Qiang S, Goltsev V. Simultaneous in vivo recording of prompt and delayed fluorescence and 820-nm reflection changes during drying and after rehydration of the resurrection plant Haberlea rhodopensis . Biochim Biophys Acta. 2010;1797:1313–1326. doi: 10.1016/j.bbabio.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Alamillo JM, Bartels D. Effects of desiccation on photosynthesis pigments and the ELIP-like dsp 22 protein complexes in the resurrection plant Craterostigma plantagineum . Plant Sci. 2001;160:1161–1170. doi: 10.1016/S0168-9452(01)00356-9. [DOI] [PubMed] [Google Scholar]
- 39.Bartels D, Hanke C, Schneider K, Michel D, Salamini F. A desiccation-related Elip- like gene from the resurrection plant Craterostigma plantagineum is regulated by light and ABA. EMBO J. 1992;11:2771–2778. doi: 10.1002/j.1460-2075.1992.tb05344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones L, McQueen-Mason S. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum . FEBS Lett. 2004;559:61–65. doi: 10.1016/S0014-5793(04)00023-7. [DOI] [PubMed] [Google Scholar]
- 41.Vicre M, Lerouxel O, Farrant J, Lerouge P, Driouich A. Composition and desiccation-induced alterations of the cell wall in the resurrection plant Craterostigma wilmsii . Physiol Plant. 2004;120:229–239. doi: 10.1111/j.0031-9317.2004.0234.x. [DOI] [PubMed] [Google Scholar]
- 42.Moore JP, Farrant JM, Driouich A. A role for pectin-associated arabinans in maintaining the flexibility of the plant cell wall during water deficit stress. Plant Signal Behav. 2008;3:102–104. doi: 10.4161/psb.3.2.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore JP, Nguema-Ona E, Chevalier L, Lindsey GG, Brandt W, Lerouge P, Farrant J, Driouich A. Response of the leaf cell wall to desiccation in the resurrection plant Myrothamnus flabellifolius . Plant Physiol. 2006;141:651–662. doi: 10.1104/pp.106.077701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norwood M, Truesdale MR, Richter A, Scott P. Photosynthetic carbohydrate metabolism in the resurrection plant Craterostigma plantagineum . J Exp Bot. 2000;51:159–165. doi: 10.1093/jexbot/51.343.159. [DOI] [PubMed] [Google Scholar]
- 45.Norwood M, Toldi O, Richter A, Scott P. Investigation into the ability of roots of the poikilohydric plant Craterostigma plantagineum to survive dehydration stress. J Exp Bot. 2003;54:2313–2321. doi: 10.1093/jxb/erg255. [DOI] [PubMed] [Google Scholar]
- 46.Drennan PM, Smith MT, Goldsworthy D, van Staden J. The occurrence of trehalose in the leaves of the desiccation-tolerant angiosperm Myrothamnus flabellifolius Welw. J Plant Physiol. 1993;142:493–496. doi: 10.1016/S0176-1617(11)81257-5. [DOI] [Google Scholar]
- 47.Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 48.Crowe JH, Hoekstra FA, Crowe LM. Anhydrobiosis. Annu Rev Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- 49.Smeekens S, Ma J, Hanson J, Rolland F. Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol. 2010;13:274–279. doi: 10.1016/j.pbi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana . Proc Natl Acad Sci USA. 2003;100:6849–6854. doi: 10.1073/pnas.1132018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishizawa A, Yabuta Y, Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez MC, Edsgard D, Hussain SS, Alquezar D, Rasmussen M, Gilbert T, Nielsen BH, Bartels D, Mundy J. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum . Plant J. 2010;63:212–228. doi: 10.1111/j.1365-313X.2010.04243.x. [DOI] [PubMed] [Google Scholar]
- 55.Saavedra L, Svensson J, Carballo V, Izmendi D, Welin B, Vidal S. A dehydrin gene in Physcomitrella patens is required for salt and osmotic stress tolerance. Plant J. 2006;45:237–249. doi: 10.1111/j.1365-313X.2005.02603.x. [DOI] [PubMed] [Google Scholar]
- 56.Rodrigo MJ, Bockel C, Blervacq AS, Bartels D. The novel gene CpEdi-9 from the resurrection plant C. plantagineum encodes a hydrophilic protein and is expressed in mature seeds as well as in response to dehydration in leaf phloem tissues. Planta. 2004;219:579–589. doi: 10.1007/s00425-004-1264-z. [DOI] [PubMed] [Google Scholar]
- 57.Mowla SB, Thomson JA, Farrant JM, Mundree SG. A novel stress-inducible antioxidant enzyme identified from the resurrection plant Xerophyta viscosa Baker. Planta. 2002;215:716–726. doi: 10.1007/s00425-002-0819-0. [DOI] [PubMed] [Google Scholar]
- 58.Phillips JR, Hilbricht T, Salamini F, Bartels D. A novel abscisic acid- and dehydration-responsive gene family from the resurrection plant Craterostigma plantagineum encodes a plastid-targeted protein with DNA-binding activity. Planta. 2002;215:258–266. doi: 10.1007/s00425-002-0755-z. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Y, Wang Z, Jing Y, Wang L, Liu X, Liu Y, Deng X. Ectopic over-expression of BhHsf1, a heat shock factor from the resurrection plant Boea hygrometrica, leads to increased thermotolerance and retarded growth in transgenic Arabidopsis and tobacco. Plant Mol Biol. 2009;71:451–467. doi: 10.1007/s11103-009-9538-2. [DOI] [PubMed] [Google Scholar]
- 60.Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta. 2009;230:1155–1166. doi: 10.1007/s00425-009-1014-3. [DOI] [PubMed] [Google Scholar]
- 61.Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D. Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum . Plant Cell. 2000;12:111–124. doi: 10.1105/tpc.12.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Villalobos MA, Bartels D, Iturriaga G. Stress tolerance and glucose insensitive phenotypes in Arabidopsis overexpressing the CpMYB10 transcription factor gene. Plant Physiol. 2004;135:309–324. doi: 10.1104/pp.103.034199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furini A, Koncz C, Salamini F, Bartels D. High level transcription of a member of a repeated gene family confers dehydration tolerance to callus tissue of Craterostigma plantagineum . EMBO J. 1997;16:3599–3608. doi: 10.1093/emboj/16.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith-Espinoza CJ, Phillips JR, Salamini F, Bartels D. Identification of further Craterostigma plantagineum cdt mutants affected in abscisic acid mediated desiccation tolerance. Mol Genet Genomics. 2005;274:364–372. doi: 10.1007/s00438-005-0027-2. [DOI] [PubMed] [Google Scholar]
- 65.Mulako I, Farrant JM, Collett H, Illing N. Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is upregulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz. J Exp Bot. 2008;59:3885–3901. doi: 10.1093/jxb/ern226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garwe D, Thomson JA, Mundree SG. XVSAP1 from Xerophyta viscosa improves osmotic-, salinity- and high-temperature-stress tolerance in Arabidopsis . Biotechnol J. 2006;1:1137–1146. doi: 10.1002/biot.200600136. [DOI] [PubMed] [Google Scholar]
- 67.Garwe D, Thomson JA, Mundree SG. Molecular characterization of XVSAP1, a stress-responsive gene from the resurrection plant Xerophyta viscosa Baker. J Exp Bot. 2003;54:191–201. doi: 10.1093/jxb/erg013. [DOI] [PubMed] [Google Scholar]
- 68.Zeng Q, Chen X, Wood AJ. Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J Exp Bot. 2002;53:1197–1205. doi: 10.1093/jexbot/53.371.1197. [DOI] [PubMed] [Google Scholar]
- 69.Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc Natl Acad Sci USA. 2003;100:4921–4926. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Röhrig H, Colby T, Schmidt J, Harzen A, Facchinelli F, Bartels D. Analysis of desiccation-induced candidate phosphoproteins from Craterostigma plantagineum isolated with a modified metal oxide affinity chromatography procedure. Proteomics. 2008;8:3548–3560. doi: 10.1002/pmic.200700548. [DOI] [PubMed] [Google Scholar]
- 71.Georgieva T, Christov N, Djilianov D. Identification of desiccationregulated genes by cDNA-AFLP in Haberlea rhodopensis: a resurrection plant. Acta Physiol Plant. 2012;34:1055–1066. doi: 10.1007/s11738-011-0902-x. [DOI] [Google Scholar]
- 72.Jiang G, Wang Z, Shang H, Yang W, Hu Z, Phillips J, Deng X. Proteome analysis of leaves from the resurrection plant Boea hygrometrica in response to dehydration and rehydration. Planta. 2007;225:1405–1420. doi: 10.1007/s00425-006-0449-z. [DOI] [PubMed] [Google Scholar]
- 73.Ingle RA, Schmidt UG, Farrant JM, Thomson JA, Mundree SG. Proteomic analysis of leaf proteins during dehydration of the resurrection plant Xerophyta viscosa . Plant Cell Environ. 2007;30:435–446. doi: 10.1111/j.1365-3040.2006.01631.x. [DOI] [PubMed] [Google Scholar]
- 74.Oliver MJ, Jain R, Balbuena TS, Agrawal G, Gasulla F, Thelen JJ. Proteome analysis of leaves of the desiccation-tolerant grass, Sporobolus stapfianus, in response to dehydration. Phytochemistry. 2011;72:1273–1284. doi: 10.1016/j.phytochem.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 75.Wang X, Chen S, Zhang H, Shi L, Cao F, Guo L, Xie Y, Wang T, Yan X, Dai S. Desiccation tolerance mechanism in resurrection fern-ally Selaginella tamariscina revealed by physiological and proteomic analysis. J Proteome Res. 2010;9:6561–6577. doi: 10.1021/pr100767k. [DOI] [PubMed] [Google Scholar]
- 76.Peters S, Mundree SG, Thomson JA, Farrant JM, Keller F. Protection mechanisms in the resurrection plant Xerophyta viscosa (Baker): both sucrose and raffinose family oligosaccharides (RFOs) accumulate in leaves in response to water deficit. J Exp Bot. 2007;58:1947–1956. doi: 10.1093/jxb/erm056. [DOI] [PubMed] [Google Scholar]
- 77.Bianchi G, Gamba A, Murelli C, Salamini F, Bartels D. Low molecular weight solutes in desiccated and ABA-treated calli and leaves of Craterostigma plantagineum . Phytochemistry. 1992;31:1917–1922. doi: 10.1016/0031-9422(92)80334-B. [DOI] [Google Scholar]
- 78.Bianchi G, Gamba A, Limiroli R, Pozzi N, Elster R, Salamini F, Bartels D. The unusual sugar composition in leaves of the resurrection plant Myrothamnus flabellifolia . Physiol Plant. 1993;87:223–226. doi: 10.1111/j.1399-3054.1993.tb00146.x. [DOI] [Google Scholar]
- 79.Bianchi G, Gamba A, Murelli C, Salamini F, Bartels D. Novel carbohydrate metabolism in the resurrection plant Craterostigma plantagineum . Plant J. 1991;1:355–359. doi: 10.1046/j.1365-313X.1991.t01-11-00999.x. [DOI] [PubMed] [Google Scholar]
- 80.Alcazar R, Bitrian M, Bartels D, Koncz C, Altabella T, Tiburcio AF. Polyamine metabolic canalization in response to drought stress in Arabidopsis and the resurrection plant Craterostigma plantagineum . Plant Signal Behav. 2011;6:243–250. doi: 10.4161/psb.6.2.14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Renault H, Roussel V, El AA, Arzel M, Renault D, Bouchereau A, Deleu C. The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 2010;10:20. doi: 10.1186/1471-2229-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Renault H, El AA, Palanivelu R, Updegraff EP, Yu A, Renou JP, Preuss D, Bouchereau A, Deleu C. GABA accumulation causes cell elongation defects and a decrease in expression of genes encoding secreted and cell wall-related proteins in Arabidopsis thaliana . Plant Cell Physiol. 2011;52:894–908. doi: 10.1093/pcp/pcr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaff DF, McGregor GR. The effect of dehydration and rehydration in the nitrogen content of various fractions from resurrection plants. Biol Plant. 1979;21:92–99. doi: 10.1007/BF02909453. [DOI] [Google Scholar]
- 84.Tymms MJ, Gaff DF. Proline accumulation during water stress in resurrection plants. J Exp Bot. 1978;30:165–168. doi: 10.1093/jxb/30.1.165. [DOI] [Google Scholar]
- 85.Moore JP, Westall KL, Ravenscroft N, Farrant J, Lindsey GG, Brandt W. The predominant polyphenol in the leaves of the resurrection plant Myrothamnus flabellifolius, 3,4,5-tri-O-galloylquinic acid, protects membranes against desiccation and free radical-induced oxidation. Biochem J. 2005;385:301–308. doi: 10.1042/BJ20040499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Djilianov D, Ivanov S, Moyankova D, Miteva L, Kirova E, Alexieva V, Joudi M, Peshev D, Van den Ende W. Sugar ratios, glutathione redox status and phenols in the resurrection species Haberlea rhodopensis and the closely related non-resurrection species Chirita eberhardtii . Plant Biol. 2011;13:767–776. doi: 10.1111/j.1438-8677.2010.00436.x. [DOI] [PubMed] [Google Scholar]
- 87.Hilbricht T, Varotto S, Sgaramella V, Bartels D, Salamini F, Furini A. Retrotransposons and siRNA have a role in the evolution of desiccation tolerance leading to resurrection of the plant Craterostigma plantagineum . New Phytol. 2008;179:877–887. doi: 10.1111/j.1469-8137.2008.02480.x. [DOI] [PubMed] [Google Scholar]
- 88.Liu X, Wang Z, Wang L, Wu R, Phillips J, Deng X. LEA 4 group genes from the resurrection plant Boea hygrometrica confer dehydration tolerance in transgenic tobacco. Plant Sci. 2009;176:90–98. doi: 10.1016/j.plantsci.2008.09.012. [DOI] [Google Scholar]