Abstract
Tumor-initiating cells (TICs) have emerged as the driving force of carcinomas, which appear as hierarchically structured. TICs as opposed to the tumor bulk display tumor forming potential, which is linked to a certain degree of self-renewal and differentiation, both major features of stem cells. Markers such as CD44, CD133, CD24, EpCAM, CD166, Lgr5, CD47, and ALDH have been described, which allow for the prospective enrichment of TICs. It is conspicuous that the same markers allow for an enrichment of TICs in various entities and, on the other hand, that different combinations of these markers were independently reported for the same tumor entity. Potential functions of these markers in the regulation of TIC phenotypes remained somewhat neglected although they might give insights in common molecular themes of TICs. The present review discusses major TIC markers with respect to their function and potential contributions to the tumorigenic phenotype of TICs.
Keywords: Tumor-initiating cells, CD133, CD44, CD47, CD166, EpCAM
Introduction
Two models for the development of solid tumors have been proposed and are intensively discussed nowadays. The stochastic model suggests an accumulation of successive mutations and the clonal selection of tumorigenic cells. The second and more recently elaborated model can be referred to as the hierarchical model, which implements tumor-initiating (stem) cells as the origin of cancer. The concept of tumor-initiating cells (TICs) emanates from the fact that, in some cancer types, seemingly not every single cancer cell is empowered with oncogenic potential in vivo in animal models. Rather, a subpopulation of tumor cells with characteristics reminiscent of stem cells, including self-renewal and the potential to aberrantly differentiate into non-TICs, initiates tumors [1]. Accordingly, tumors would be organized in a hierarchical manner very much comparable to normal tissue, however, with a loss of control of homeostasis in the tissue stem cell compartment. State-of-the-art characterization of TICs relies on serial xenotransplantations of human tumor cells into immunocompromised mice. To this end, cellular antigens such as CD44, CD133, EpCAM, CD24, CD166, CD47, Lgr5, and ALDH1 serve as markers for the enrichment of cells bearing tumorigenic potential [1]. Marker-positive but not marker-negative cells, can be serially transplanted and give rise to new tumors with a cellular composition reminiscent of the primary malignancy. Obviously, this seeming paradigma shift in the general view on carcinogenesis was accompanied by strong arguments, debate, and even disbelief in the mere existence of TICs. Beyond legitimate issues concerning the lack of accuracy of mouse models of xenotransplantation to depict frequencies of TICs in primary carcinomas [2, 3], numerous questions relate to the amplitude of the differentiation potential of TICs and to their actual cellular origin, both of which have not been satisfyingly answered until now. A prudent forecast is that neither of the two dogmatic models will exclusively be able to explain tumorigenesis in every given cancer entity and, even less so, in every patient. It can nonetheless be seen as a consensus that, at a given time point, some cells but not others are in an epigenetic and signaling state, which renders the cells permissive to tumor growth. In other words, these cells are TICs at that particular time point and in that given microenvironment. The eventuality of a substantial plasticity in these cells and an associated inter-conversion between TICs and non-TICs further adds depth to our view of tumorigenesis [4]. Plasticity can be the result of a trans-differentiation between two cell types without any hierarchical traits, or of the differentiation of stem-like cells into more differentiated progeny, and vice-versa. Knowledge of determinants of TIC phenotypes and plasticity might eventually be decoupled from a gridlocked definition of models of tumor development, and hence be more informative. However it is triggered, TIC phenotype and associated plasticity will ultimately be provided by changes in the transcription or repression of key regulators of TIC phenotypes as well as by the regulation of key switches at the functional level. Three general ways to regulate the expression and function of genes exist in cells: a genetic compound, which refers to sequence differences or mutations in the genome of selected cells, an epigenetic compound in which semi-stable imprinting of DNA generates diversity, and lastly a signaling compound, which allows for rapidly inducible changes in gene expression and function. With lessons learned from the field of embryonic stem cell research and, more recently, from the molecular prerequisites to generate induced pluripotent stem cells (iPS), regulators of stemness became more evident. A manageable set of transcription factors including c-myc, nanog, klf4, oct4, and sox2 has been demonstrated to be sufficient in order to reprogram terminally differentiated cells into stem-like cells, although with very low efficiency [5]. Together with signaling pathways known to be instrumental during embryonic development, in stem cell and cancer development, such as the Wnt and TGFβ pathways, these regulators of iPS might eventually play essential roles in TIC identity and plasticity [6].
Prerequisites to become a tumor-initiating cell
The knowledge of prerequisites, which determine the phenotype of TICs, should substantially enhance our insights into molecular requirements and key regulators of TICs. In principle, two major levels of prerequisites can be defined: (1) regulators of pluripotency, differentiation, and self-renewal, and (2) regulators of migration, invasion, engraftment, and communication. The first regulators are mandatory for cells to become TICs and will hence be termed primary traits in the following (Fig. 1), while the second regulators are elective and rather serve the purpose of fine-tuning capacities of TICs (Fig. 2). Priming of TICs from differentiated cells, progenitors, or stem cells will rely on the induction of primary traits and result in TICs with various capacities including the formation of large tumors and the generation of circulating/disseminated tumor cells (Fig. 2). Secondary traits can be observed in more differentiated, non-TIC tumor cells, too, and might even be the final outcome of TIC differentiation or of dissemination from primary tumors. Accordingly, TICs and their progeny must be seen as smooth transitions of various states of tumor cells, which all ultimately rely on regulation of gene expression and function.
Fig. 1.
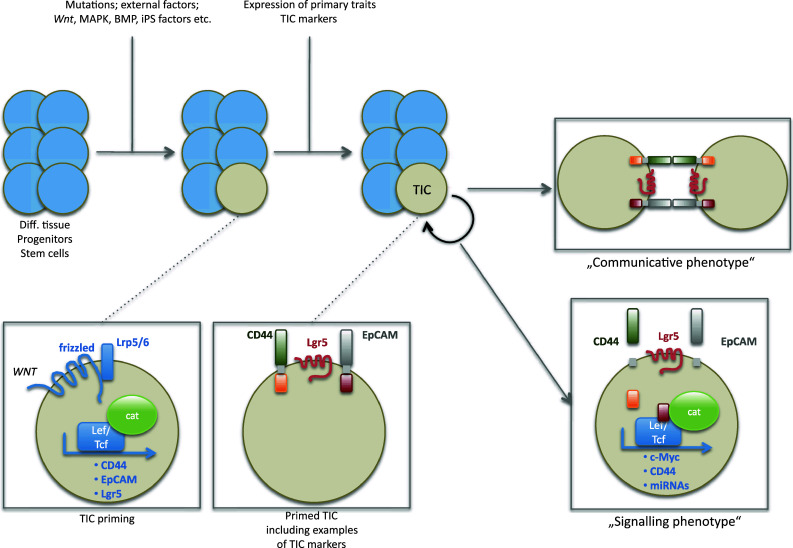
Priming and regulation of TICs. Differentiated cells, progenitors, and/or stem cells require mutations and/or the activity of external factors to prime them to become TICs. Amongst these stimuli are Wnt, MAPK, and BMP signaling, and expression of iPS factors. Upon induction of these signals, cells de novo or over-express primary traits TIC markers such as CD133, CD44, and EpCAM, and thus become primed. In this primed state, TICs can opt between various sub-phenotypes such as a communicative phenotype, in which markers like CD44 and EpCAM mediate cell adhesion, and a signaling phenotype based on regulated intramembrane proteolysis and nuclear translocation of intracellular domains. Target genes of these TIC markers include members of the iPS genes, anti-apoptotic genes, and genes involved in direct cell cycle and proliferation regulation
Fig. 2.
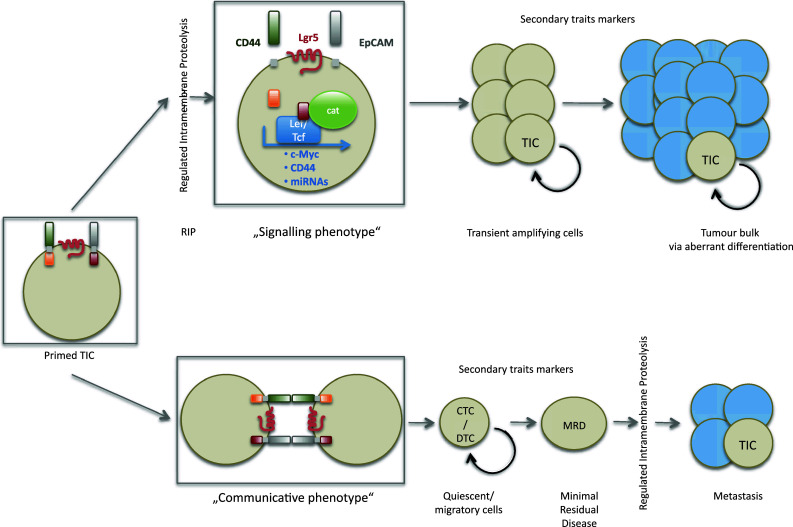
Different fates of primed TICs. Primed TICs expression various markers of primary traits, which can be regulated at the functional level upon regulated intramembrane proteolysis. Upon RIP, cells become activated to proliferate and differentiate to a tumor bulk in which TIC will represent a minor fraction of cells. Under conditions of low RIP activity and/or lack of inducing ligands, cells can adopt a communicative and rather quiescent phenotype. Such cells can acquire additional (secondary) trait markers and advance to circulating (CTC) or disseminated tumor cells (DTC) and be cells of the minimal residual disease (MRD). After engraftment, reactivation of proliferation, for example upon RIP and induction of primary trait markers, will lead to the formation of secondary tumors at distant sites or metastases
The following features will be referred to as primary traits and must be unleashed from the tight control during homeostasis in healthy counterparts in order to generate TICs, which will act as tumor founders:
Self-renewal
Pluripotency and differentiation capacity (plasticity)
Switch between quiescence and proliferation.
Secondary traits represent:
Low apoptosis and high chemoresistance
Communication with microenvironment
Autonomy from tissue integrity (disseminated/circulating TICs)
Migratory capacity and engraftment (metastasizing TICs).
Obviously, a cell will not receive signals incorporating all these aspects from a single molecule or after a singular mutation of the genome, and will not necessarily display all secondary traits either. Rather will orchestrated functions of various proteins associated with primary traits result in the phenotype of TICs and fine-tuning of their capacity (e.g., as a circulating cell or with respect to metastasis formation) occur at the level of secondary traits and as a consequence of genuine tumor formation. In the following, reported TIC markers will be reviewed according to their molecular functions and potential role in the regulation of TIC phenotypes. Each marker will be classified with respect to its potential level of involvement in the regulation of TICs. Unless otherwise mentioned, markers discussed herein have been defined to be bona fide TIC-associated proteins upon serial transplantations of human marker-positive tumor cells into immunodeficient mouse strains. Additional in vitro experiments including serial formation of sphere-like structures have been performed for some (e.g., CD44 and CD133), but not all markers.
CD44
Proteins encoded by the CD44 gene constitute a large family of at least 20 variants, based on differential splicing and post-translational glycosylation. CD44 is a single transmembrane protein with a comparably short intracellular domain (72 amino acids), whose expression is regulated by the Wnt signaling pathway via β-catenin [7], as is the case for other TIC markers such as EpCAM and Lgr5 (Fig. 1). CD44 has been described as part of the signature of TICs from colon carcinomas [8], head and neck carcinomas [9], non-small cell lung cancer [10], hepatocellular carcinoma [11], and breast cancer [12]. The actual expression pattern of CD44 in TICs of head and neck squamous cell carcinomas (HNSCC) is a matter of debate. Initial publications described a subpopulation of HNSCC, which represented less than 10% of the tumor bulk in average and retained tumor-inducing capacities [9]. These findings collide with reports on the almost ubiquitous expression of the standard form of CD44 (CD44s) and alternative splice variants (e.g., CD44v6) in HNSCC and in most normal epithelia of the head and neck area [13, 14], and, until now, remain an unresolved issue. Whatever the expression levels and patterns of CD44 in TICs actually are, it is striking that CD44 is one of the most frequently described markers of TICs in numerous different malignancies [15], raising the question if this potentially abundant protein fulfils essential tasks in TICs (for review see [16]). CD44 comprises dual functions as it promotes adhesion to and communication with extracellular matrix of the microenvironment as the major hyaluronan receptor. Further, CD44 transmits signals from the plasma membrane to the nucleus upon regulated intramembrane proteolysis [17]. Activation of the signaling pathway of CD44 relies on regulated intramembrane proteolysis (RIP) to release the intracellular domain of CD44 (CD44-ICD) into the nucleus (Figs. 1, 2). There, CD44-ICD regulates transcription via binding to TPA-responsive elements within promoters of target genes, including CD44 itself [18] and is involved in cell transformation of rat fibroblasts [19]. The following interesting model for CD44-mediated migration was proposed: At the leading edge, activation of ADAM17 by PKC and small GTPase Rac results in cleavage and shedding of the ectodomain of CD44, allowing cells to detach and creep on extracellular matrix. Ensuing stretching of cells provokes an influx of calcium, inducing the activity of ADAM10 and the cleavage of CD44 at the rear pole of the cells, and additional detachment of cells from their support. Ectodomain shedding is in turns a prerequisite for cleavage and release of CD44-ICD into the nucleus, and the associated up-regulation of CD44 gene transcripts. Newly generated CD44 protein will sustain re-attachment of cells and thus cycles of migration will be completed [18]. Interestingly, leukemia-initiating cells appeared to depend more strongly on CD44 for homing to the bone marrow than normal hematopoietic stem cells [20]. Hence, it is conceivable that CD44 is involved in the migratory phenotype of TICs to their respective niche, for example along with CXCR-4, which would allow for the sensing of chemoattractant gradients [21]. Additionally, CD44 plays a role in the balance of survival, apoptosis, and chemoresistance in colorectal cancers as shown in the Apcmin/+ mouse model, while it did not impact proliferation [22]. Activation of CD44 upon interaction with hyaluronan (HA) results in induction of the acetyltransferase p300, acetylation of β-catenin and NF-κB, and eventually in an up-regulation of genes involved in multidrug resistance and protection from apoptosis [23]. Furthermore, HA-mediated activation of CD44 induces PKCε and Src kinase, resulting in the activating phosphorylation of Nanog and Twist, respectively. These transcription factors are involved in the transcription of two microRNAs (mir21 and mir10b), themselves regulators of tumor suppressors and GTPases, which are active in cytoskeleton reorganization during metastasis formation, and of inhibitor of apoptosis and multidrug-resistant protein 1 [24]. Resistance of cells towards pro-apoptotic signals, which increase upon release from a united cell structure due to a lack of anti-apoptotic signals from neighboring cells, will play an essential role for disseminated and circulating tumor cells (Fig. 2). Additionally, Twist is involved in the process of EMT, leading to an up-regulation of ALDH1 and CD44, and in the activation of the Akt pathway and β-catenin [25]. Hence, CD44 has the potential to regulate the number and function of vital TICs, and consequently the pool of TICs in vivo, and can be ranked as a primary trait regulator.
| Relevance to TICs: ••••• |
|---|
| Primary trait regulator |
| • Activation of Nanog (stemness) and Twist (EMT) |
| • Related to Wnt, PKC, Src kinase, and NFκB |
| • Low apoptosis and high chemoresistance |
| • Communication with microenvironment |
| • Migratory capacity and engraftment (metastasizing TICs) |
| • Autonomy from tissue integrity (disseminated/circulating TICs) |
CD133
CD133 (prominin-1) is a ~120-kDa glycoprotein with an N-terminal extracellular domain, two large extracellular loops, which are strongly N-glycosylated, and an intracellular C-terminus [26]. The AC133 antigen, which represents a hyper-glycosylated version of CD133, is primarily expressed in stem and progenitor cells [27] such as embryonic epithelium [28], brain stem cells [29], hematopoietic stem cells [30], and in cancers such as leukemias [31] and retinoblastomas [32]. AC133 served to isolate TICs from colon [33, 34], pancreas [35], gallbladder [36], ovarian [37], lung [38], and brain cancers [39], childhood malignant melanoma [40] and Ewing′s sarcoma [41]. This led to the notion that the CD133 protein is exclusively expressed in the TIC subpopulation in cancers. This notion was vigorously challenged when Shmelkov et al. demonstrated that both, CD133+ and CD133− cells have tumor seeding capacity in metastatic colon cancers, questioning the validity of CD133 as a marker [42]. In a similar contradictory manner, a CD133-negative cancer cell line has been generated from colon carcinoma, which further lacked CD44 expression but retained tumor-initiating and aberrant differentiation potential [43]. In glioblastomas, although initial reports focused on CD133-positive TICs, more recent data support the existence of CD133-negative cells with TIC properties, too [44].
Due to hypo-glycosylation, the AC133-specific antibody does not react with the CD133 protein any longer and hence mimics a loss of CD133 antigen in more differentiated tumor cells. Actually, a superior expression in stem cells and TICs only holds true for the AC133 antigen, while a hypo-glycosylated variant of CD133 still remains expressed in more differentiated tumor cells [45]. Although used as a marker for TICs from numerous cancer entities, CD133 is not strictly related to traits of stemness. Controversy is underscored by various publications including data on the over-expression of CD133 under stress conditions, e.g., as a result of hypoxia in gliomas [46]. Generally, it is of utmost importance to reckon that a substantial number of TIC markers also become over-expressed in the case of cellular stress or restrictive culture conditions, and do by no means represents purely specific TIC markers.
Functional analysis of CD133 is hindered by the existence of several splice variants [47] and differentially glycosylated forms [45], the respective impact of which on the function of CD133 is so far unexplored. No relation to any signaling pathways has been described so far. In hematopoietic stem cells (HSCs), CD133 expression correlates with a more pronounced multipotent phenotype compared to CD34+/CD133− cells [48]. During asymmetric division of HSCs, CD133 is primarily distributed to daughter cells with a stem cell phenotype [30], as is the case for neuroepithelial cells. Furthermore, budding of CD133-containing membranous particles, termed prominosomes, has been reported during differentiation of neuroepithelia to radial glial cells and neuron-generating progenitors [28]. Release of CD133-positive particles from the apical membrane of neuroepithelia might be a means of intercellular communication and/or disposal of proteins that regulate stemness. Hence, CD133 appears as a determinant of the stem cell-like phenotype, which is often distributed in a polar fashion in cells and might allow for a differential communication with the surrounding niche, both in normal and malignant situations [30]. This is especially apparent in migrating HSCs, where CD133 localization is confined to a structure at the rear pole termed uropod, which protrudes, and does not contact the stem cell niche [30]. Directed locomotion relies on the expression of CXCR4 at the leading edge and a gradient of its ligand, SDF1. Noteworthy, CXCR4 efficiently marks invasive and metastatic CD133+ TICs in pancreas carcinomas [49]. In contrast, after contact of the uropod with mesenchymal stromal cells, HSCs adopt a more sessile, “communicative” phenotype (Fig. 1).
In colon carcinoma cells, in which CD133 is a published TIC marker with prognostic significance, the repression of CD133 expression had no effect whatsoever on proliferation, colony formation, migration, and invasion [50]. Thus, even though it is a valuable prognostic marker, CD133 seems to lack a functional role in colorectal cancer development [50]. In the murine model, CD133 marks intestinal cells, which, upon hyper-activation of the Wnt signaling pathway, are prone to transformation and will populate the intestine with neoplastic cells [51]. In summary, it is at present not possible to draw solid conclusions on the molecular function of CD133 during malignancy, although implications in the regulation of stemness qualify it as a very interesting target molecule and as a primary trait regulator.
| Relevance to TICs: ••• |
|---|
| Primary trait regulator |
| • Pluripotency and differentiation capacity (asymmetric division, plasticity) |
| • Communication with microenvironment |
| • Migratory capacity and engraftment (metastasizing TICs) |
EpCAM
Epithelial cell adhesion molecule (EpCAM) is an integral transmembrane protein composed of a large extracellular domain, one transmembrane region, and a small intracellular domain of 26 amino acids. High over-expression of EpCAM in combination with CD44+ or CD44+/CD24− was described as the signature of TICs from carcinoma entities such as breast [12], colorectal [8], and pancreatic carcinomas [52]. Two features of EpCAM most probably led to its poor consideration as a possible “driver” of TICs: (1) EpCAM was perceived as a cell adhesion molecule without transforming potential, and (2) EpCAM is per se expressed in most carcinomas, as is CD44, and does, as a single marker, not allow for the discrimination of TICs versus non-TICs. Things changed with the growing body of evidence, which characterized EpCAM as an oncogenic signaling receptor. De novo expression of EpCAM results in enhanced proliferation along with the induction of the proto-oncogene c-Myc [53], while siRNA-mediated reduction of EpCAM expression in breast carcinoma cell lines induced a loss of proliferation and invasion capacity [54]. Furthermore, EpCAM transforms cells such that they become anchorage-independent and form colonies in soft agar [53]. Constitutive knock-out of murine EpCAM is lethal at embryonic stages, which underscores essential roles in regular development and morphoregulation in vivo [55]. Elucidation of EpCAM′s signaling pathway, which incorporates regulated intramembrane proteolysis and interactions with β-catenin and Lef-1 to deliver the intracellular part of EpCAM (EpICD) to the nucleus [56], has essentially contributed to the understanding of EpCAM′s role. Regulated intramembrane proteolysis (RIP), as described for CD44, can be seen as a common means to switch between adhesive and signaling properties of the molecule (Fig. 2). In addition to the expression levels of TIC markers themselves, regulation of their function can occur at a second level. Expression and activity of essential components of RIP will impact on selected TIC marker functions and distinguish between a “signaling phenotype” and a “communicative phenotype” after cells have been primed to TICs (Figs. 1, 2). For the case of EpCAM, proteases required for proteolytic activation (i.e., TACE and presenilin) and interaction partner FHL2 are substantially up-regulated in various malignant tissue [57–62], and might hence explain the strong differences in EpCAM activation observed in colon carcinoma as compared to normal colonic mucosa [56].
Components of the Wnt pathway crucially participate in the maintenance of the stem cell phenotype and are involved in various malignant situations [63, 64], The epcam gene itself is under the control of Tcf4 [65], suggesting feedbacks between EpCAM expression and EpCAM signaling. c-Myc is an early transcriptional target gene of both, Wnt [66] and EpCAM signaling [53], and represents one of the essential switches of the transcriptional programs from adult to embryonic stem (ES) cells [67]. Expression of c-Myc was sufficient to induce an ES cell-like transcriptional signature in normal and cancer cells. c-Myc expression also enhanced the proportion of TICs in human keratinocytes transformed by Ras and IκBα 150-fold [67]. Together with the fact that EpCAM is highly expressed and an essential factor in the maintenance of stemness in murine and human ES cells [68, 69] through its ability to regulate the promoters of reprogramming genes such as Oct4, c-Myc, Sox2, Nanog, and KLF4 [70], these data strongly suggest a functional role in TICs. It is tempting to speculate that TICs necessitate high expression of EpCAM with sustained and hyper-activated signaling for the maintenance of their phenotype via the induction of stemness genes known from the field of iPS generation [71]. Since homotypic interaction of EpCAM molecules on opposing cells is regarded as one route of activation of signaling [72], de novo expression of EpCAM in stromal cells surrounding carcinoma cells, but not healthy epithelium, as has been demonstrated for prostate cancer, is of great interest with respect to EpCAM activation in tumors [73]. In summary, EpCAMhigh emerged as an important primary trait regulator of proliferation and pluripotency via its capacity to signal in combination with members of the Wnt pathway. In line with this notion, EpCAM expression marked fully reprogrammed induced pluripotent stem cells and allowed for their enrichment following the expression of Oct4, Sox2, Klf4, and n- or c-Myc in murine fibroblasts [74]. In this respect, EpCAM appears very much comparable to CD44, as it is frequently over-expressed in TICs, alone does not allow for the differentiation of TICs and non-TICs, but emerges as a central regulator of various aspects of the TIC phenotype.
| Relevance to TICs: ••••• |
|---|
| Primary trait regulator |
| • Self-renewal |
| • Pluripotency and differentiation capacity (asymmetric division, plasticity) |
| • Switch between quiescence and proliferation |
| • Related to Wnt and PI3 kinase signaling |
| • Communication with microenvironment |
CD24
CD24, initially termed heat-stable antigen (HSA), is a comparably small and strongly glycosylated adhesion molecule that was first described in normal B and T cells [75]. It is a mucin-type protein that is anchored into the plasma membrane via glycosylphosphatidylinositol (GPI anchor) and interacts with P-selectin. Heavy glycosylation of CD24 generates proteins ranging from 20 to 70 kDa, depending on the tissue and cell type of origin. Heterogeneity is also reflected in the functions that have been attributed to the molecule. CD24 is implicated in T cell co-stimulation, regulation of homeostatic proliferation of dendritic and T cells, growth and metastasis of cancer cells, and apoptosis (for review see [76]). Ligand specificity varies strongly with the organ in which CD24 is expressed. Natural ligands are P-selectin, CD24 itself, fibronectin, and the L1 receptor [76]. The picture of an association of CD24 with the signature of TICs is not uniform. For the case of breast cancer-derived TICs, the lack or low expression of CD24 along with high expression of CD44/EpCAM is characteristic [12]. In contrast, pancreatic TICs have been defined as CD44+/CD24+/EpCAM+ cells [52].
Morel et al. reported on the generation of CD44+/CD24−/low cells with TIC capacities from non-tumorigenic mammary CD44low/CD24+ cells [77]. Trans-differentiation was achieved after expression of oncogenic Ras in human mammary epithelial cells additionally over-expressing human TERT, SV40 large and small T antigen, and similarly in MCF10A breast carcinoma cells. This is suggestive of a central role for the MAP-kinase pathway in the generation of CD24− TICs. Using single-cell cloning, the authors could demonstrate that CD24+ cells trans-differentiate into heterogeneous CD24+/CD24− and homogeneous CD24− populations. The resulting CD24− cells displayed features of TICs in vitro (formation of mammospheres) and in vivo (tumor generation) [77]. Interestingly, these cells also underwent changes attributed to the process of epithelial-to-mesenchymal transition (EMT) and TGF-β, a known inducer of EMT, sped up the appearance of CD24− cells from CD24+ precursors. Absence of CD24 in breast cancer TICs correlated with an invasive phenotype, although the actual contribution of the lack of CD24 expression remains unexplored. CD24 expression is a prognostic marker for ovarian, breast, prostate, and non-small cell lung carcinomas. Accordingly, conditional expression of CD24 in mammary carcinoma cell lines resulted in an enhancement of tumorigenic and metastatic potentials of the cells [78]. CD24 expression was associated with increased proliferation, adherence to fibronectin, spreading, and migration. These controversial results are hardly reconcilable. However, if CD24+ cells represent a pool of precursors of CD44+/CD24−/low TICs [77], ectopic expression of CD24 might trigger differentiation/signaling pathways, which yield potent TICs even in the presence of CD24 and therefore increase tumor formation. Trans-differentiation of non-TICs to TICs, as monitored upon their expression of CD24, further underscores the notion that TICs display some degree of plasticity. Owing to these strongly controversial findings, classification of CD24 into primary or secondary trait regulator is barely practicable.
| Relevance to TICs: ••• |
|---|
| Primary/secondary trait regulator |
| • Switch between quiescence and proliferation |
| • Pluripotency and differentiation capacity (plasticity) |
| • Migratory capacity and engraftment (metastasizing TICs) |
CD166
CD166 is a member of the immunoglobulin superfamily, which is also known as activated leukocyte cell adhesion molecule (ALCAM), KG-CAM, BEN/DM-GRASP, hematopoietic cell antigen (HCA), and neurolin [79]. Expression of CD166 was described on normal hematopoietic cells, mesenchymal stem cells, neuronal and stromal cells [80]. CD166 has the capacity to undergo homotypic and heterotypic interactions with CD6. CD166 is over-expressed in head and neck tumors, invasive melanomas, pancreas and prostate carcinomas, and is an independent prognostic marker for some entities [81]. CD166 is part of the signature of colon TICs [8]. Most information available on the function of CD166 in tumor cells relates to adhesive properties based on homotypic interactions. In vitro and in vivo homotypic interaction is counter-regulated upon shedding of the extracellular domain by ADAM17/TACE [82]. Blocking of adhesion upon treatment of cells with an antagonizing antibody resulted in enhanced migration of ovarian carcinoma cells, while the inhibition of TACE displayed the opposite effect along with reduced wound-healing capacities in vitro. It is tempting to speculate that TICs first express CD166 in order to allow for the adherence of small numbers of TICs at a first site in vivo to generate an initial tumor “seed” and avoid dispersion of cells before they have acquired migratory and invasive potential, and/or a critical minimal tumor size. When tumor cells acquire traits of invasiveness, adhesive functions of CD166 can be obviated upon ectodomain shedding (exemplified in Fig. 2), which can be monitored in vivo as the presence of soluble CD166 (sCD166) in sera and ascites of tumor patients to a higher degree than in normal donors [82]. Although the release of sCD166 is a rather permanently ongoing phenomenon, treatment of cells with growth factors such as EGF strongly enhanced shedding. As for the case of CD44 and EpCAM, it is conceivable that shedding of the ectodomain of CD166 further induces regulated intramembrane cleavage of the intracellular domain via proteases such as the gamma-secretase complex. Regulated intramembrane proteolysis would generate an intracellular domain (ICD) of at least 34 amino acids, which is in a comparable size range as CD44 and EpCAM. Additionally, CD166 displayed protective effects in breast carcinoma cells in vitro, correlated with the expression of the anti-apoptotic molecule Bcl2, and counteracted apoptosis in these cells [83]. Whether this phenotype relies on adhesive or potential signaling capacities of CD166 remains entirely unknown.
| Relevance to TICs: ••• |
|---|
| Secondary trait regulator |
| • Autonomy from tissue integrity |
| • Low apoptosis and high chemoresistance |
| • Migratory capacity and engraftment (metastasizing TICs) |
Lgr5
Leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5) is an orphan seven-span transmembrane protein and a target of the Wnt signaling pathway [84]. In line with the long-known implication of Wnt signaling in various stem cells, including ES cells, hematopoietic stem cells, colonic crypt stem cells, hair follicle progenitors, and other adult stem/progenitor cells, Lgr5 efficiently marks most of these stem cell types [85]. Lgr5 is expressed in adult intestinal stem cells at the crypt bottom, most probably in crypt base columnar cells, which are cycling cells believed to represent intestinal stem cells. Deletion of the adenomatous poliposis coli (APC) gene, a central regulator of β-catenin stability, in Lgr5-positive cells resulted in fast and progressive transformation [86]. In vitro, Lgr5-positive stem cells form crypt-like structures independently of mesenchymal stromal cells, and Lgr5-positive cells remain at the apex of the stem cell hierarchy in these newly generated crypts [87]. Being a seven-span membrane receptor, it seems conceivable that Lgr5-mediated signaling itself contributes to morphoregulation and to initiation of transformation. In this respect, the exploration of an association of Lgr5 with G-proteins and a potential involvement in cAMP and or phosphatidylinositol signaling appears of great interest. Based on the available knowledge, it is not possible to strictly classify Lgr5 into primary or secondary trait regulator. However, if, as expected, Wnt signaling is of paramount importance for the priming of cells to become TICs, then over-expression of Lgr5 will automatically occur and potentially influence the phenotype of TICs (Fig. 1).
| Relevance to TICs: ••• |
|---|
| Primary/secondary trait regulator |
| • Communication with microenvironment |
| • Pluripotency and differentiation capacity (plasticity) |
| • Self-renewal |
CD47
CD47 was initially named IAP for integrin-associated protein based on its interaction with various integrins via the β3 subunit. CD47 represents a ligand for thrombospondin-1 (TSP-1) on opposing cells and a G-protein coupled receptor (GPCR) [88, 89]. CD47 displays an unconventional structure with a single immunoglobulin-like extracellular domain and five, instead of the classical seven, transmembrane domains. An interesting view of the integrins/CD47 interaction covers the possibility that the five transmembrane domains of CD47 together with the two transmembrane domains of integrins might form a more classical seven-span GPCR. Via its ability to activate the signal inhibitory receptor protein-α (SIRPα) on monocytic cells, CD47 is involved in the regulation of the innate immunity, allowing for the recognition of self and the prevention of engulfment of cells of the body. Ligation of CD47 with agonistic antibodies, TSP-1, or activating peptides of TSP-1 results in chemotaxis, spreading, migration, and proliferation of various cell types including platelets and smooth muscle cell [90]. For the case of bladder cancer, a subpopulation of CD44+/CK5+/CK20− cells was identified as the TIC subpopulation. When comparing the gene signature in non-muscle invasive versus muscle invasive tumors, CD47 was determined as an important marker of aggressive cells with prognostic potential [91]. This is in line with a role as secondary trait marker, which is involved in the regulation of the metastatic potential (Fig. 2). In vitro, these CD44+/CD47high tumor cells were significantly less sensitive towards engulfment by macrophages, a feature that possibly helps carcinoma cells to escape from immune surveillance.
Additional integrin-independent features associated with cell proliferation might further explain the strong over-expression of CD47 in TICs. Astrocytoma but not normal astrocytes displayed increased proliferation upon engagement of CD47. This phenotype was associated with an induction of downstream signaling via G-proteins, leading to the activation of the PI3K/Akt pathway [92], which is anti-apoptotic and fosters proliferation. Based on these findings, CD47 emerges as a novel putative therapeutic target of interest [93] and, consequently, monoclonal antibodies against CD47 are effective in combination with Rituximab for the eradication of non-Hodgkin lymphoma cells [94].
| Relevance to TICs: •• |
|---|
| Secondary trait regulator |
| • Communication with microenvironment |
| • Switch between quiescence and proliferation |
Aldehyde dehydrogenase ALDH
Human aldehyde dehydrogenases represent a family of at least 19 genes subdivided in 11 families and four subfamilies [95]. The cognate proteins are NAD(P)+-dependent enzymes, which catalyze the oxidation of aldehydes to carboxylic acids and are crucial for cellular detoxification owing to the detrimental effects of aldehydes, including cytotoxic and carcinogenic features. The “gold standard” for the assessment of ALDH1 enzymatic activity relies on the fluorescent substrate BODIPY aminoacetaldehyde, better known as the Aldefluor® Assay. Together with the expression of the catalytic subunit of telomerase and the ATP-binding cassette membrane transporter ABCG2, ALDH1high is characteristic of normal stem cells of most tissues investigated. ALDH1high is also a marker for TICs of cancers of the breast, lung, head and neck, pancreas, cervix, prostate, liver, colon, and bladder (reviewed in [95]). Numerous cellular processes such as differentiation, proliferation, morphoregulation, and development, are directly and/or indirectly regulated by the enzymatic activity of ALDH1 family members, which might account for the frequent over-expression of ALDH1 in TICs (exemplified for other TIC markers in Fig. 1). ALDH1 members catalyze the final and irreversible step in the conversion of retinol to retinoic acid (RA), regulating the production of RA as a central molecule involved in gene transcription and in stem cell differentiation [96]. ALDH1 enzymes confer cytoprotective effects and resistance to alkylating chemotherapeutic drugs such as cyclophosphamide via direct recycling of aldehydes [95] and indirect pathways involving the production of cytokines. Following priming of cells to become TICs and support self-renewal and pluripotency, their refractory capacity towards therapies will be essential and allow them to prevail (Fig. 2). As for the case of CD133 and very much in accordance with its role in detoxification, ALDH1 is also up-regulated as a result of cellular stress [97]. Nonetheless, the relative proportion of ALDH1high but not of CD44+/CD24− cells increased in patients suffering from breast carcinomas after neoadjuvant chemotherapeutic treatment with paclitaxel and epirubicin [98]. A subset of cells from colitic patients with an ALDH+/EpCAM+ phenotype have been described to be the cells of origin of the transition from a chronic ulcerative colitis to an overt colorectal cancer [99]. Hence, ALDH+/EpCAM+ potentially mark pre-malignant TICs in a situation associated with higher predispositions to develop certain cancer types. ALDH can be classified as a primary and secondary trait regulator.
| Relevance to TICs: •• |
|---|
| Primary/secondary trait regulator |
| • Switch between quiescence and proliferation |
| • Low apoptosis and high chemoresistance |
Conclusions and outlook
The discovery of TICs as a driving force of malignancies of tumors is of paramount importance for the understanding and treatment of the disease. With the emergence of a variety of cell-surface markers of TICs, it was possible to prospectively isolate the cells of origin of tumors and to demonstrate their remarkable oncogenic potential in vivo, as opposed to the bulk of tumor cells. These studies suggested that single or, rarely, combinations of 2-3 TIC markers differentiate between oncogenic and non-oncogenic cells, between TICs and non-TICs. Actually, the belief in single markers discriminating these two tumor cell populations appears as the first misleading notion: rather is the concomitant expression of several TIC markers to be expected in order to cover the needs of TICs on the one hand, and thorough distinction of cell subpopulations on the other hand. The notion of single markers originates from the way studies of TICs have been conducted, where each group focuses on single molecules without investigating more than 1-3 different markers. The actual rationale to study a given marker is rarely delineated in publications and mostly does not relate to its possible functions in TICs. More in-depth analysis of TICs from solid tumors will necessitate the implementation of numerous potential markers, as is the standard for the study of hematopoietic stem cells. Different combinations of TIC markers could eventually provide tumor cells with primary traits of the TIC phenotype to support sufficient self-renewal and differentiation (Fig. 1 and Table 1), and the assimilation of substantial numbers of TIC markers in descriptive and functional studies are highly desirable. Assuming that a subset of cells within a tumor bear higher oncogenic capacity and further agreeing on the fact that genetic and epigenetic modifications are the basis for these intercellular differences, a more general concept can be postulated. That is, TICs require a minimal subset of functions, which can be provided to them by various combinations of reported TIC markers (Table 1 and exemplified in Fig. 1) and yet unknown proteins. Expression of TIC markers relies on dysregulation of gene function and/or major signaling pathways, which are commonly affected in cancer, as is the case for the Wnt pathway (Fig. 1). Amongst these regulators of primary traits, common and frequent TIC markers emerge: Recurrence of TIC markers such as CD44, CD133, EpCAM, and ALDH, whose functions across entities cover the basic requirements for self-renewal and pluripotency, supports this notion and speaks in favor of a general concept of TIC formation based on primary traits (Fig. 1). Interestingly, a substantial number of these markers are regulated upon regulated intramembrane proteolysis at the functional level. Hence, depending on the availability and activation status of essential components of RIP, different phenotypes of TICs can be modulated and smooth transitions thereof generated (Fig. 2). With the knowledge of essential transcription factors required for iPS reprogramming, the common over-expression of TIC markers such as EpCAM, which have the capacity to induce a subset of these central molecular switches, appears consequential.
Table 1.
Regulators of primary and secondary traits of TICs
| Primary traits | |
| Self-renewal | EpCAM [68, 69]; CD133 [48]; Lgr5 [86] |
| Pluripotency | CD44 [23, 24, 100]; EpCAM [68–70, 74]; CD133 [30, 45, 48]; CD24 [77]; Lgr5 [85–87]; |
| Switch quiescence/proliferation | CD44 [19]a; EpCAM [56]* [53, 101]; CD47 [92]; ALDH [96] |
| Secondary traits | |
| Low apoptosis | CD44 [22], CD47 [92]; ALDH [95] |
| Chemoresistance | CD44 [22, 23]; ALDH [95] |
| Communication with microenvironment | CD44 [16]; CD133 [30]; EpCAM [102]; CD166 [82]*; CD47 [88, 89] |
| Autonomy from tissue integrity | CD44 [24]; EpCAM [53], Lgr5 [87] |
| Migration and engraftment | CD44 [18]* [20, 23, 103]; EpCAM [54]; CD133 [49]; CD24 [78]; CD166 [82]* |
* These functions have been demonstrated to depend upon regulated intramembrane proteolysis
In summary, a substantial number of markers for TICs and normal stem cells share common features:
They are Wnt signaling target genes (CD44, Lgr5, EpCAM).
They display dual functions as cell adhesion molecules and signaling receptors (CD44, EpCAM, CD47, and possibly CD166).
They are modulated in their function upon ectodomain shedding and regulated intramembrane proteolysis (i.e., CD44, CD166, EpCAM).
Hence, co-regulation of expression and of function of TIC markers by Wnt signaling and RIP, respectively, emerges as a potential common theme for a decent number of TIC markers (Figs. 1, 2). Possibly, Wnt signaling represents one of the key initial prerequisites for the priming of pre-malignant cells to become TICs (Fig. 1). As a result, TIC markers such as Lgr5, CD44, and EpCAM are expressed and endow cells with a panoply of adhesive and signaling properties, which may result in either communicative but rather quiescent cells (“adhesive phenotype”) or dividing cells (“proliferative phenotype”) (Fig. 1). The ability to switch between both functions of CD44, EpCAM, and potentially also CD166, is accomplished by sheddases and γ-secretases upon ectodomain shedding and regulated intramembrane proteolysis, resulting in the generation of soluble ectodomains and signaling intracellular domains. Expression levels and activation of these membrane-associated proteases and other essential components of signaling include another level to the regulation of TIC capacities and especially to their plasticity. Not only do we need to study the expression levels of TIC markers, but the additionally activation status and the expression of components of their respective signaling pathways. This knowledge is especially important with respect to the generation of specific inhibitors of central TIC markers. Strikingly, CD44 and EpCAM signal via components of the Wnt pathway, too. As demonstrated for the intracellular domain of EpCAM, EpICD is essential for the assembly of a protein-DNA complex at Lef1 consensus sites [56] and might hence modulate the specificity and promoter choice of β-catenin-Lef nuclear complexes. It is attractive to think of a specific inhibition of the expression/function of TIC markers, which are “drivers” in the process of tumor seeding and recurrence. Combinations of inhibitors of Wnt signaling and potent inhibitors of ADAM proteases and gamma-secretase emerge as novel treatment options along with therapeutic antibodies. Additionally, interruption of central signaling cascades of TIC markers such as CD44, CD133, and EpCAM with specific peptides, which inhibit essential interactions, is a highly promising future approach.
Conflict of interest
OG is a consultant for Micromet Inc.
References
- 1.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 2.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317(5836):337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- 4.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15(9):1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 5.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132(4):567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, Sprick MR, Kemper K, Richel DJ, Stassi G, Medema JP. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12(5):468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 7.Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, Clevers H, Pals ST. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154(2):515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, Shelton AA, Parmiani G, Castelli C, Clarke MF. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung EL, Fiscus RR, Tung JW, Tin VP, Cheng LC, Sihoe AD, Fink LM, Ma Y, Wong MP. Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS One. 2010;5(11):e14062. doi: 10.1371/journal.pone.0014062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J, Li J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer. 2010;126(9):2067–2078. doi: 10.1002/ijc.24868. [DOI] [PubMed] [Google Scholar]
- 12.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mack B, Gires O. CD44s and CD44v6 expression in head and neck epithelia. PLoS ONE. 2008;3(10):e3360. doi: 10.1371/journal.pone.0003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawano T, Nakamura Y, Yanoma S, Kubota A, Furukawa M, Miyagi Y, Tsukuda M. Expression of E-cadherin, and CD44s and CD44v6 and its association with prognosis in head and neck cancer. Auris Nasus Larynx. 2004;31(1):35–41. doi: 10.1016/j.anl.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 16.Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 17.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 18.Nagano O, Saya H. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 2004;95(12):930–935. doi: 10.1111/j.1349-7006.2004.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelletier L, Guillaumot P, Freche B, Luquain C, Christiansen D, Brugiere S, Garin J, Manie SN. Gamma-secretase-dependent proteolysis of CD44 promotes neoplastic transformation of rat fibroblastic cells. Cancer Res. 2006;66(7):3681–3687. doi: 10.1158/0008-5472.CAN-05-3870. [DOI] [PubMed] [Google Scholar]
- 20.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 21.Avigdor A, Goichberg P, Shivtiel S, Dar A, Peled A, Samira S, Kollet O, Hershkoviz R, Alon R, Hardan I, Ben-Hur H, Naor D, Nagler A, Lapidot T. CD44 and hyaluronic acid cooperate with SDF-1 in the trafficking of human CD34+ stem/progenitor cells to bone marrow. Blood. 2004;103(8):2981–2989. doi: 10.1182/blood-2003-10-3611. [DOI] [PubMed] [Google Scholar]
- 22.Zeilstra J, Joosten SP, Dokter M, Verwiel E, Spaargaren M, Pals ST. Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 2008;68(10):3655–3661. doi: 10.1158/0008-5472.CAN-07-2940. [DOI] [PubMed] [Google Scholar]
- 23.Bourguignon LY, Xia W, Wong G. Hyaluronan-mediated CD44 interaction with p300 and SIRT1 regulates beta-catenin signaling and NFkappaB-specific transcription activity leading to MDR1 and Bcl-xL gene expression and chemoresistance in breast tumor cells. J Biol Chem. 2009;284(5):2657–2671. doi: 10.1074/jbc.M806708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourguignon LY, Wong G, Earle C, Krueger K, Spevak CC. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, MicroRNA-10b expression, and RhoA/RhoC up-regulation, leading to rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J Biol Chem. 2010;285(47):36721–36735. doi: 10.1074/jbc.M110.162305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Zhou BP. Activation of beta-catenin and Akt pathways by Twist are critical for the maintenance of EMT associated cancer stem cell-like characters. BMC Cancer. 2011;11:49. doi: 10.1186/1471-2407-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corbeil D, Roper K, Fargeas CA, Joester A, Huttner WB. Prominin: a story of cholesterol, plasma membrane protrusions and human pathology. Traffic. 2001;2(2):82–91. doi: 10.1034/j.1600-0854.2001.020202.x. [DOI] [PubMed] [Google Scholar]
- 27.Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90(12):5002–5012. [PubMed] [Google Scholar]
- 28.Marzesco AM, Janich P, Wilsch-Brauninger M, Dubreuil V, Langenfeld K, Corbeil D, Huttner WB. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci. 2005;118(Pt 13):2849–2858. doi: 10.1242/jcs.02439. [DOI] [PubMed] [Google Scholar]
- 29.Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV, Tsukamoto AS, Gage FH, Weissman IL. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97(26):14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freund D, Bauer N, Boxberger S, Feldmann S, Streller U, Ehninger G, Werner C, Bornhauser M, Oswald J, Corbeil D. Polarization of human hematopoietic progenitors during contact with multipotent mesenchymal stromal cells: effects on proliferation and clonogenicity. Stem Cells Dev. 2006;15(6):815–829. doi: 10.1089/scd.2006.15.815. [DOI] [PubMed] [Google Scholar]
- 31.Horn PA, Tesch H, Staib P, Kube D, Diehl V, Voliotis D. Expression of AC133, a novel hematopoietic precursor antigen, on acute myeloid leukemia cells. Blood. 1999;93(4):1435–1437. [PubMed] [Google Scholar]
- 32.Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, Bridges RJ, Kumaramanickavel G, John S, Nancarrow D, Roper K, Weigmann A, Huttner WB, Denton MJ. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet. 2000;9(1):27–34. doi: 10.1093/hmg/9.1.27. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 34.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 35.Hermanek P, Hutter RV, Sobin LH, Wittekind C. International Union Against Cancer. Classification of isolated tumor cells and micrometastasis. Cancer. 1999;86(12):2668–2673. doi: 10.1002/(SICI)1097-0142(19991215)86:12<2668::AID-CNCR11>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 36.Shi C, Tian R, Wang M, Wang X, Jiang J, Zhang Z, Li X, He Z, Gong W, Qin R CD44(+)CD133(+) population exhibits cancer stem cell-like characteristics in human gallbladder carcinoma. Cancer Biol Ther 10(11):1182–1190 [DOI] [PubMed]
- 37.Curley MD, Therrien VA, Cummings CL, Sergent PA, Koulouris CR, Friel AM, Roberts DJ, Seiden MV, Scadden DT, Rueda BR, Foster R. CD133 expression defines a tumor-initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27(12):2875–2883. doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 38.Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S, Roz E, Caserini R, Lo Vullo S, Camerini T, Mariani L, Delia D, Calabro E, Pastorino U, Sozzi G. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci USA. 2009;106(38):16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour-initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 40.Al Dhaybi R, Sartelet H, Powell J, Kokta V. Expression of CD133+ cancer stem cells in childhood malignant melanoma and its correlation with metastasis. Mod Pathol. 2010;23(3):376–380. doi: 10.1038/modpathol.2009.163. [DOI] [PubMed] [Google Scholar]
- 41.Suva ML, Riggi N, Stehle JC, Baumer K, Tercier S, Joseph JM, Suva D, Clement V, Provero P, Cironi L, Osterheld MC, Guillou L, Stamenkovic I. Identification of cancer stem cells in Ewing’s sarcoma. Cancer Res. 2009;69(5):1776–1781. doi: 10.1158/0008-5472.CAN-08-2242. [DOI] [PubMed] [Google Scholar]
- 42.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, Chadburn A, Murphy AJ, Valenzuela DM, Gale NW, Thurston G, Yancopoulos GD, D’Angelica M, Kemeny N, Lyden D, Rafii S. CD133 expression is not restricted to stem cells, and both CD133 and CD133 metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118(6):2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navarro-Alvarez N, Kondo E, Kawamoto H, Hassan W, Yuasa T, Kubota Y, Seita M, Nakahara H, Hayashi T, Nishikawa Y, Hassan RA, Javed SM, Noguchi H, Matsumoto S, Nakaji S, Tanaka N, Kobayashi N, Soto-Gutierrez A. Isolation and propagation of a human CD133(-) colon tumor-derived cell line with tumorigenic and angiogenic properties. Cell Transpl. 2010;19(6):865–877. doi: 10.3727/096368910X508997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beier CP, Beier D. CD133-negative cancer stem cells in glioblastoma. Front Biosci (Elite Ed) 2011;3:701–710. doi: 10.2741/e280. [DOI] [PubMed] [Google Scholar]
- 45.Kemper K, Sprick MR, de Bree M, Scopelliti A, Vermeulen L, Hoek M, Zeilstra J, Pals ST, Mehmet H, Stassi G, Medema JP. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010;70(2):719–729. doi: 10.1158/0008-5472.CAN-09-1820. [DOI] [PubMed] [Google Scholar]
- 46.Griguer CE, Oliva CR, Gobin E, Marcorelles P, Benos DJ, Lancaster JR, Jr, Gillespie GY. CD133 is a marker of bioenergetic stress in human glioma. PLoS One. 2008;3(11):e3655. doi: 10.1371/journal.pone.0003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fargeas CA, Huttner WB, Corbeil D. Nomenclature of prominin-1 (CD133) splice variants—an update. Tissue Antigens. 2007;69(6):602–606. doi: 10.1111/j.1399-0039.2007.00825.x. [DOI] [PubMed] [Google Scholar]
- 48.Bauer N, Fonseca AV, Florek M, Freund D, Jaszai J, Bornhauser M, Fargeas CA, Corbeil D. New insights into the cell biology of hematopoietic progenitors by studying prominin-1 (CD133) Cells Tissues Organs. 2008;188(1–2):127–138. doi: 10.1159/000112847. [DOI] [PubMed] [Google Scholar]
- 49.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 50.Horst D, Scheel SK, Liebmann S, Neumann J, Maatz S, Kirchner T, Jung A. The cancer stem cell marker CD133 has high prognostic impact but unknown functional relevance for the metastasis of human colon cancer. J Pathol. 2009;219(4):427–434. doi: 10.1002/path.2597. [DOI] [PubMed] [Google Scholar]
- 51.Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457(7229):603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 53.Munz M, Kieu C, Mack B, Schmitt B, Zeidler R, Gires O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene. 2004;23(34):5748–5758. doi: 10.1038/sj.onc.1207610. [DOI] [PubMed] [Google Scholar]
- 54.Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, Cole DJ, Gillanders WE. EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 2004;64(16):5818–5824. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]
- 55.Nagao K, Zhu J, Heneghan MB, Hanson JC, Morasso MI, Tessarollo L, Mackem S, Udey MC. Abnormal placental development and early embryonic lethality in EpCAM-null mice. PLoS One. 2009;4(12):e8543. doi: 10.1371/journal.pone.0008543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, Kieu C, Papior P, Baeuerle PA, Munz M, Gires O. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11(2):162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 57.Johannessen M, Moller S, Hansen T, Moens U, Mc-rp-p VanGhelue. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell Mol Life Sci. 2006;63(3):268–284. doi: 10.1007/s00018-005-5438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li M, Wang J, Ng SS, Chan CY, Chen AC, Xia HP, Yew DT, Wong BC, Chen Z, Kung HF, Lin MC. The four-and-a-half-LIM protein 2 (FHL2) is overexpressed in gliomas and associated with oncogenic activities. Glia. 2008;56(12):1328–1338. doi: 10.1002/glia.20701. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Yang Y, Xia HH, Gu Q, Lin MC, Jiang B, Peng Y, Li G, An X, Zhang Y, Zhuang Z, Zhang Z, Kung HF, Wong BC. Suppression of FHL2 expression induces cell differentiation and inhibits gastric and colon carcinogenesis. Gastroenterology. 2007;132(3):1066–1076. doi: 10.1053/j.gastro.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 60.Kenny PA. TACE: a new target in epidermal growth factor receptor dependent tumors. Differentiation. 2007;75(9):800–808. doi: 10.1111/j.1432-0436.2007.00198.x. [DOI] [PubMed] [Google Scholar]
- 61.Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131(2):215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 62.Merchant NB, Voskresensky I, Rogers CM, Lafleur B, Dempsey PJ, Graves-Deal R, Revetta F, Foutch AC, Rothenberg ML, Washington MK, Coffey RJ. TACE/ADAM-17: a component of the epidermal growth factor receptor axis and a promising therapeutic target in colorectal cancer. Clin Cancer Res. 2008;14(4):1182–1191. doi: 10.1158/1078-0432.CCR-07-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 64.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423(6938):409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 65.Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67(22):10831–10839. doi: 10.1158/0008-5472.CAN-07-0908. [DOI] [PubMed] [Google Scholar]
- 66.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 67.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2(4):333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez B, Denzel S, Mack B, Conrad M, Gires O. EpCAM is involved in maintenance of the murine embryonic stem cell phenotype. Stem Cells. 2009;27(8):1782–1791. doi: 10.1002/stem.97. [DOI] [PubMed] [Google Scholar]
- 69.Ng VY, Ang SN, Chan JX, Choo AB (2009) Characterization of epithelial cell adhesion molecule as a surface marker on undifferentiated human embryonic stem cells. Stem Cells. doi:10.1002/stem.221 [DOI] [PubMed]
- 70.Lu TY, Lu RM, Liao MY, Yu J, Chung CH, Kao CF, Wu HC. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J Biol Chem. 2010;285(12):8719–8732. doi: 10.1074/jbc.M109.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69(14):5627–5629. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 72.Denzel S, Maetzel D, Mack B, Eggert C, Barr G, Gires O. Initial activation of EpCAM cleavage via cell-to-cell contact. BMC Cancer. 2009;9:402. doi: 10.1186/1471-2407-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mukherjee S, Richardson AM, Rodriguez-Canales J, Ylaya K, Erickson HS, Player A, Kawasaki ES, Pinto PA, Choyke PL, Merino MJ, Albert PS, Chuaqui RF, Emmert-Buck MR. Identification of EpCAM as a molecular target of prostate cancer stroma. Am J Pathol. 2009;175(6):2277–2287. doi: 10.2353/ajpath.2009.090013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen HF, Chuang CY, Lee WC, Huang HP, Wu HC, Ho HN, Chen YJ, Kuo HC (2011) Surface marker epithelial cell adhesion molecule and E-cadherin facilitate the identification and selection of induced pluripotent stem cells. Stem Cell Rev. doi:10.1007/s12015-011-9233-y [DOI] [PubMed]
- 75.Kay R, Rosten PM, Humphries RK. CD24, a signal transducer modulating B cell activation responses, is a very short peptide with a glycosyl phosphatidylinositol membrane anchor. J Immunol. 1991;147(4):1412–1416. [PubMed] [Google Scholar]
- 76.Liu Y, Zheng P. CD24: a genetic checkpoint in T cell homeostasis and autoimmune diseases. Trends Immunol. 2007;28(7):315–320. doi: 10.1016/j.it.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 77.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3(8):e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, Yagita H, Sleeman JP. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 2005;65(23):10783–10793. doi: 10.1158/0008-5472.CAN-05-0619. [DOI] [PubMed] [Google Scholar]
- 79.Ohneda O, Ohneda K, Arai F, Lee J, Miyamoto T, Fukushima Y, Dowbenko D, Lasky LA, Suda T. ALCAM (CD166): its role in hematopoietic and endothelial development. Blood. 2001;98(7):2134–2142. doi: 10.1182/blood.V98.7.2134. [DOI] [PubMed] [Google Scholar]
- 80.Swart GW. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. Eur J Cell Biol. 2002;81(6):313–321. doi: 10.1078/0171-9335-00256. [DOI] [PubMed] [Google Scholar]
- 81.Weidle UH, Eggle D, Klostermann S, Swart GW. ALCAM/CD166: cancer-related issues. Cancer Genomics Proteomics. 2010;7(5):231–243. [PubMed] [Google Scholar]
- 82.Rosso O, Piazza T, Bongarzone I, Rossello A, Mezzanzanica D, Canevari S, Orengo AM, Puppo A, Ferrini S, Fabbi M. The ALCAM shedding by the metalloprotease ADAM17/TACE is involved in motility of ovarian carcinoma cells. Mol Cancer Res. 2007;5(12):1246–1253. doi: 10.1158/1541-7786.MCR-07-0060. [DOI] [PubMed] [Google Scholar]
- 83.Jezierska A, Matysiak W, Motyl T. ALCAM/CD166 protects breast cancer cells against apoptosis and autophagy. Med Sci Monit. 2006;12(8):BR263–BR273. [PubMed] [Google Scholar]
- 84.Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M, Van Gijn ME, Suijkerbuijk S, Van de Wetering M, Marra G, Clevers H. The intestinal Wnt/TCF signature. Gastroenterology. 2007;132(2):628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 85.Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174(3):715–721. doi: 10.2353/ajpath.2009.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 87.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 88.Lindberg FP, Gresham HD, Schwarz E, Brown EJ. Molecular cloning of integrin-associated protein: an immunoglobulin family member with multiple membrane-spanning domains implicated in alpha v beta 3-dependent ligand binding. J Cell Biol. 1993;123(2):485–496. doi: 10.1083/jcb.123.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gao AG, Frazier WA. Identification of a receptor candidate for the carboxyl-terminal cell binding domain of thrombospondins. J Biol Chem. 1994;269(47):29650–29657. [PubMed] [Google Scholar]
- 90.Wang XQ, Frazier WA. The thrombospondin receptor CD47 (IAP) modulates and associates with alpha2 beta1 integrin in vascular smooth muscle cells. Mol Biol Cell. 1998;9(4):865–874. doi: 10.1091/mbc.9.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, Gill H, Presti J, Jr, Chang HY, van de Rijn M, Shortliffe L, Weissman IL. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci USA. 2009;106(33):14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sick E, Boukhari A, Deramaudt T, Ronde P, Bucher B, Andre P, Gies JP, Takeda K. Activation of CD47 receptors causes proliferation of human astrocytoma but not normal astrocytes via an Akt-dependent pathway. Glia. 2011;59(2):308–319. doi: 10.1002/glia.21102. [DOI] [PubMed] [Google Scholar]
- 93.Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, Jr, van Rooijen N, Weissman IL. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chao MP, Alizadeh AA, Tang C, Myklebust JH, Varghese B, Gill S, Jan M, Cha AC, Chan CK, Tan BT, Park CY, Zhao F, Kohrt HE, Malumbres R, Briones J, Gascoyne RD, Lossos IS, Levy R, Weissman IL, Majeti R. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010;142(5):699–713. doi: 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma I, Allan AL (2010) The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. doi:10.1007/s12015-010-9208-4 [DOI] [PubMed]
- 96.Gudas LJ, Wagner JA. Retinoids regulate stem cell differentiation. J Cell Physiol. 2011;226(2):322–330. doi: 10.1002/jcp.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang M, Shoeb M, Goswamy J, Liu P, Xiao TL, Hogan D, Campbell GA, Ansari NH. Overexpression of aldehyde dehydrogenase 1A1 reduces oxidation-induced toxicity in SH-SY5Y neuroblastoma cells. J Neurosci Res. 2010;88(3):686–694. doi: 10.1002/jnr.22230. [DOI] [PubMed] [Google Scholar]
- 98.Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, Tamaki Y, Noguchi S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15(12):4234–4241. doi: 10.1158/1078-0432.CCR-08-1479. [DOI] [PubMed] [Google Scholar]
- 99.Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69(20):8208–8215. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bourguignon LY, Spevak CC, Wong G, Xia W, Gilad E. Hyaluronan-CD44 interaction with protein kinase C(epsilon) promotes oncogenic signaling by the stem cell marker Nanog and the Production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance in breast tumor cells. J Biol Chem. 2009;284(39):26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maaser K, Borlak J. A genome-wide expression analysis identifies a network of EpCAM-induced cell cycle regulators. Br J Cancer. 2008;99(10):1635–1643. doi: 10.1038/sj.bjc.6604725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Litvinov SV, Balzar M, Winter MJ, Bakker HA, Briaire-de Bruijn IH, Prins F, Fleuren GJ, Warnaar SO. Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J Cell Biol. 1997;139(5):1337–1348. doi: 10.1083/jcb.139.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gunthert U, Hofmann M, Rudy W, Reber S, Zoller M, Haussmann I, Matzku S, Wenzel A, Ponta H, Herrlich P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65(1):13–24. doi: 10.1016/0092-8674(91)90403-L. [DOI] [PubMed] [Google Scholar]


