Abstract
Coordination of cell division with growth and development is essential for the survival of organisms. Mistakes made during replication of genetic material can result in cell death, growth defects, or cancer. Because of the essential role of the molecular machinery that controls DNA replication and mitosis during development, its high degree of conservation among organisms is not surprising. Mammalian cell cycle genes have orthologues in plants, and vice versa. However, besides the many known and characterized proliferation genes, still undiscovered regulatory genes are expected to exist with conserved functions in plants and humans. Starting from genome-wide Arabidopsis thaliana microarray data, an integrative strategy based on coexpression, functional enrichment analysis, and cis-regulatory element annotation was combined with a comparative genomics approach between plants and humans to detect conserved cell cycle genes involved in DNA replication and/or DNA repair. With this systemic strategy, a set of 339 genes was identified as potentially conserved proliferation genes. Experimental analysis confirmed that 20 out of 40 selected genes had an impact on plant cell proliferation; likewise, an evolutionarily conserved role in cell division was corroborated for two human orthologues. Moreover, association analysis integrating Homo sapiens gene expression data with clinical information revealed that, for 45 genes, altered transcript levels and relapse risk clearly correlated. Our results illustrate how a systematic exploration of the A. thaliana genome can contribute to the experimental identification of new cell cycle regulators that might represent novel oncogenes or/and tumor suppressors.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0909-x) contains supplementary material, which is available to authorized users.
Keywords: Arabidopsis thaliana, MCF7, Cell cycle, Cancer genomics, Comparative genomics
Introduction
The cell cycle represents a precisely programmed series of events that enables a cell to duplicate its content and to generate two daughter cells. In all eukaryotes studied to date, the cell division process is controlled by cyclin-dependent kinases (CDKs) [1, 2]. The numerous components controlling the activity of these kinases form a complex molecular network that has not been fully dissected even 30 years after their initial discovery. All physiological signals and signaling pathways affecting cell proliferation are in some way connected to the cell cycle regulators. Therefore, it is not surprising that mutations in key steps within these signaling pathways provoke dramatic changes in DNA replication, DNA repair efficiency, and cell proliferation rate. In mammals, a deregulated cell cycle is directly linked with malignant transformation processes that lead to tumorigenesis and cancer.
A wide spectrum of strategies has been used to identify new oncogenes or cell malignancy modulators, from proteomics studies [3] and cytogenetics [4] to cancer epigenetics [5]. With the technological progress in gene expression techniques, methods such as digital differential display [6, 7] and serial analysis of gene expression (SAGE) [8] have been used as tools to discover new oncogenes and tumor suppressors. Microarrays have also been employed as a highly preferred technology to characterize cancer-specific expression patterns (cancer fingerprints) and cancer-deregulated pathways [9–13]. Additionally, recent technological advances have provided platforms that allow hundreds of thousands of single nucleotide polymorphisms (SNPs) to be analyzed in genome-wide association studies (GWAS), providing a basis for the identification of moderate-risk alleles that contribute to cancer progression [14–16]. Nevertheless, in spite of the invaluable information obtained with these tools, cancer persists as one of the major killing diseases in the world [17]. Therefore, it is desirable to develop additional approaches that allow us to get better and more systemic, insight into the origin, progression, and outcome of cancer.
Comparative genomics represents a complementary tool for cancer research [18–20]. Although 1.6 billion years ago the mammalian and plant clades had diverged, commonly shared pathways and signaling cascades inherited from their last common ancestor still persist. Correspondingly, Arabidopsis thaliana not only has had a great impact on the understanding of the plant kingdom itself but has also contributed extensively to the dissection of specific mechanisms that have been evolutionarily conserved. Innate immunity [21], circadian clock [22], DNA methylation [23], RNAi processing mechanisms [24], and G protein signaling [25] are some of the traits firstly studied in Arabidopsis. Similarly, the Arabidopsis and Homo sapiens genomes contain a highly comparable repertoire of “disease genes”. Almost 70% of the genes implicated in cancer have Arabidopsis homologues, which is comparable to the percentage found in Drosophila melanogaster (67%), Caenorhabditis elegans (72%), and Saccharomyces cerevisiae (41%) [26].
Regarding cancer, nowadays an old paradigm has been reinforced, namely that, underlying the variability among different tumors, only a relatively small number of critical events lie at the origin of their development. In most instances, deregulated cell proliferation provides the fundamental platform for neoplastic transformation [27, 28]. Through microarray expression analysis of different types of cancers, it has been possible to detect the cancer core mechanisms, represented by an early deregulation of the mitotic cell cycle, DNA replication, DNA repair, and chromatin assembly. Interestingly, all these processes are largely controlled by the RB-E2F pathway [29], in agreement with the common alteration of this pathway in cancer [30, 31].
The RB-E2F pathway is one of the most conserved pathways between plants and mammals, as illustrated by the large amount of E2F target genes that are shared by both organisms [32, 33]. Therefore, given that at its early stages abnormal cell proliferation is a cancer hallmark, new cell cycle regulators with a specific role in carcinogenesis might be identified by a systematic study of the cell replication machinery in Arabidopsis. Here, we applied a combination of functional prediction and comparative genomics strategies to identify evolutionarily conserved cell cycle genes. A subset of the computational identified genes was tested experimentally, both in plant and human cell cultures, to validate their role in cell cycle progression. A Cox survival analysis revealed a strong enrichment for genes that upon misexpression might result in cancer relapse, demonstrating that the designed integrative strategy had been successful in detecting novel cell division genes that were conserved between humans and plants.
Materials and methods
Arabidopsis microarray expression data analysis and clustering
Microarray data were retrieved from the NASC transcriptomics service [34]. Based on the Affymetrix ATH1 array, 20,777 A. thaliana genes were analyzed using 213 microarray CEL files covering different tissues and under different experimental conditions (Supplementary Table 1). To detect coexpressed genes, all 20,777 Arabidopsis genes were used as seed to detect coexpression neighborhoods using the complete expression compendium. The Pearson correlation coefficient (PCC) was calculated for each pair of genes within the dataset, generating a 20,777 × 20,777 data matrix. For all the pair-wise comparisons, a significance value of coexpression between the compared genes was established [35].
Gene ontology associations
Gene ontology (GO) associations for Arabidopsis proteins were retrieved from TAIR [36] and for human proteins from AmiGO [37]. The assignments of genes to the original GO categories were extended to include parental terms (i.e., a gene assigned to a given category was automatically also assigned to all the parent categories). Enrichment values for the GO terms DNA repair (GO:0006281) and DNA replication (GO:0006260) for both Arabidopsis and H. sapiens were calculated as the ratio of the relative occurrence in a set of genes (coexpression neighborhood) to the relative occurrence in the genome. The statistical significance of the functional enrichment within sets of genes was evaluated with the hypergeometric distribution adjusted by the Bonferroni correction for multiple hypothesis testing. Corrected P values smaller than 0.05 were considered as significant. GO enrichment analysis for validating the different filtering steps was performed using ATCOECIS (http://bioinformatics.psb.ugent.be/ATCOECIS/).
Cis-regulatory elements detection
One-kb promoter regions of the set of genes significantly enriched for the terms DNA repair and/or DNA replication were scanned for the presence of an E2F binding-site by means of a positional weight matrix (PWM), with TTTssCGC as consensus sequence (based on a set of E2F-upregulated genes; [38]). E2F motif instances were identified with MotifLocator and using a threshold of 0.95 [39].
Detection of orthologous genes
Orthologous genes between Arabidopsis and H. sapiens were identified with OrthoMCLDB [40], a comparative genomics resource hosting orthologous families based on protein clustering. Starting from the selected Arabidopsis genes, the corresponding orthologous gene families were retrieved and evaluated by phylogenetic inference. For each family, protein sequences were aligned using MUSCLE [41] and a neighbor-joining phylogenetic tree was constructed using TREECON [42], with the Poisson correction for evolutionary distance calculation. Highly supported nodes (bootstrap support >90%), indicating the speciation between plants and mammals, were used to identify orthologous genes and copy numbers.
Human microarray data analysis
The human microarray data analysis comprised CEL files of studies performed on Affymetrix array platforms compatible with the mRNA expression data (HG133A or HG133plus2), involving at least 50 breast tumor samples (Supplementary Table 2) published before September 2009 in the GEO or Array Express databases. Data were extracted, background-subtracted, normalized, and summarized (median polish option) using frozen (f)RMA, the new summarization Bioconductor package [43]. Data from the nine selected studies were merged in a pooled dataset. To avoid over-fitting, data corresponding to the same patient analyzed in different studies were included only once in the pooled dataset containing 1,400 patients. Statistical processing and Cox survival analysis were performed as given in the Supplemental Methods file.
Plant growth conditions and phenotypic analysis
Arabidopsis thaliana (L.) Heyhn. accession Columbia-0 and the mutant plants were grown under long-day conditions (16 h/8 h light/darkness) at 22°C on half-strength Murashige and Skoog (MS) agar plates. All the insertion T-DNA lines were obtained from the European Arabidopsis Stock Centre (NASC). To screen for homozygous insertion alleles, primers were designed following the instructions of the Salk Institute genomic analysis laboratory (http://signal.salk.edu/tdnaprimers.2.html). The complete list of the used primers for the selection of homozygous lines is detailed in Supplementary Table 3. For characterization of embryo-lethal mutants, independent seedpods (>10) from different plants were harvested and dissected. Pictures were taken with a Leica MZ16 stereoscope using a ×5 magnification factor. The number of aborted seeds was correlated with the proportion of expected homozygous seeds; the significance of this correlation was tested with the χ 2 statistical test. For DNA ploidy analysis, the first developed leaf (harvested 3 weeks after sowing) was chopped with a razor blade in 200 μl of nucleus extraction solution, supplemented with 800 μl of staining solution (http://www.partec.com). The homogenate was filtered through a 30-μm mesh. The nuclei were analyzed using a CyFlow cytometer and FloMax software (http://www.partec.com). The EI was calculated as the fraction of nuclei of each represented ploidy level multiplied by the number of endoreduplication cycles necessary to reach the corresponding ploidy level. Leaf cell number and cell size measurements and root growth analysis were performed as given in the Supplemental Methods file.
MCF7 cell culture and transfection
MCF7 cell cultures were grown in complete medium (Dulbecco’s modified MEM Eagle medium with 5% fetal calf serum, suplemented with L-Gln, NaPy, NEAA, and 6 ng/ml bovine insulin) at 37°C and 5% CO2. The following small interfering (si)RNA sequences (DharmaFECT; Thermo Fisher Scientific, Waltham, MA, USA), were used for the specific transfections: human HEATR6 (SMARTpool; J-015921-09, J-015921-10, J-015921-11, J-015921-12), human STATIP1 (SMARTpool; J-021064-05, J-021064-06, J-021064-07, J-021064-08), human C14ORF21 (SMARTpool; J-017798-09, J-017798-10, J-017798-11, J-017798-12) and control (SMARTpool non-targeting pool). Growth and ploidy content were measured as given in the Supplemental Methods file.
QPCR analysis of Homo sapiens siRNAs
MCF7 cells (250,000 cells approximately) were seeded in 5 ml of MCF7 medium without antibiotics in a 6-well plate and grown under the previously described conditions. STATIP1, C14ORF21, HEATR6, and control siRNAs were transfected into the cells according to the manufacturer’s instructions (DharmaFECT; Thermo Fisher Scientific). The final concentration of each siRNA was 30 nM. Cells were collected 48 h after transfection with a rubber policeman. RNA was extracted with an RNeasy animal Mini Kit (Qiagen) and cDNA was prepared with the cDNA synthesis system according to the manufacturer’s instructions (Roche Diagnostics, Indianapolis, USA). For quantitative PCR, a Light-Cycler 480 SYBR Green I Master (Roche Diagnostics) was used with 100 nM primers and 0.1 mg of reverse transcription reaction product. Reactions were run and analyzed on the LightCycler 480 Real Time PCR System according to the manufacturer’s instructions (Roche Diagnostics). All quantifications were normalized to the TATA binding protein (TBP) and Ubiquitin C (UBC) expression levels. Quantitative reactions were done in triplicate and averaged. Primers used for QPCR analysis are given in Supplementary Table 4.
Results
Selection of target genes using data integration and comparative genomics
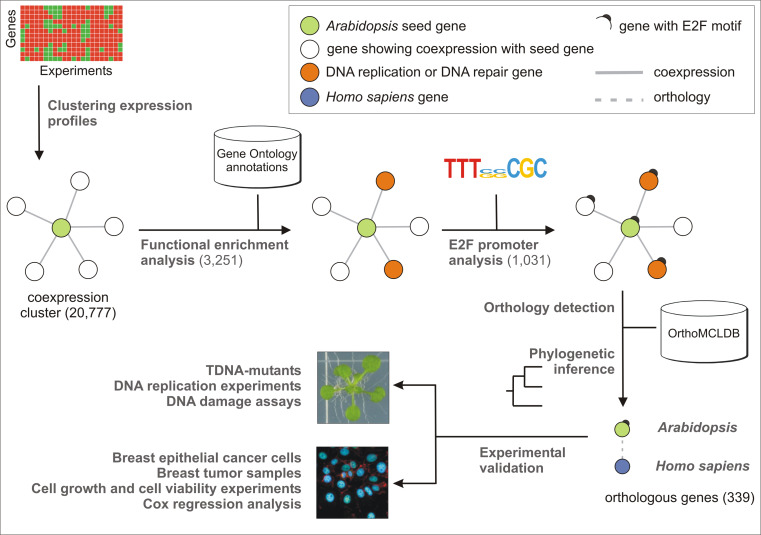
To identify new genes playing a putative role in the regulation of the cell cycle in plants and humans, we applied an integrative genomics strategy (Fig. 1). Starting from >200 microarray experiments (Supplementary Table 1), the expression levels for 20,777 Arabidopsis genes were used to identify gene coexpression neighborhoods based on the Pearson correlation coefficient (PCC) (see “Materials and methods”). Depending on the seed gene, neighborhood clusters of coexpressed genes contained between 10 and 450 genes. Subsequently, each gene cluster was tested for functional enrichment with GO. The terms “DNA replication” (GO:0006260) and “DNA repair” (GO:0006281) were scanned within the annotations of the coexpressed neighbors of all seed genes. In total, 3,251 genes were significantly enriched (P <0.05) for one or both terms (Supplementary Table 5). To identify within this list the genes with a putative role in DNA replication or DNA repair, the 1-kb promoter regions of the 3,251 genes were scanned for the presence of E2F cis-regulatory elements by means of a PWM with a consensus sequence TTTssCGC (see “Materials and methods”). A total of 1,031 Arabidopsis genes were found, harboring one or more predicted E2F-binding sites within their promoter region (Supplementary Table 6). Subsequently, to select only those genes with a putatively conserved role across species, plant genes with a mammalian orthologue were identified with the OrthoMCL database (http://www.orthomcl.org/cgi-bin/OrthoMclWeb.cgi). The sets of orthologues were verified by means of phylogenetic inference (see “Materials and methods”), and for 515 genes at least one human orthologue was identified. As functional redundancy might obscure downstream functional analysis upon gene knock-out, only those genes that were part of a low copy number family in both Arabidopsis and human were retained. A total of 339 genes fitted this criterion (Supplementary Table 7).
Fig. 1.
Schematic representation of the applied methodology for the selection of the target genes using data integration and comparative genomics. Starting from genome-wide Arabidopsis thaliana microarray data, an integrative strategy based on coexpression, functional enrichment analysis, and cis-regulatory element annotation was combined with a comparative genomics approach between plants and humans to detect conserved cell cycle genes involved in DNA replication and DNA repair processes. Numbers in parentheses report the number of genes that were retained after each step
A GO enrichment analysis was performed to validate the effectiveness of the used filters (see “Materials and methods”). This analysis demonstrated a progressive enrichment for both GO terms after each filter applied (Supplementary Table 8), illustrating that the application of the E2F and the Arabidopsis–H. sapiens orthology filters effectively resulted in an enrichment of the candidates genes with a putative role in DNA replication and/or DNA repair.
Validation of the putative cell cycle regulators using the plant model
To experimentally validate a subset of the above-identified genes as novel plant cell cycle genes, we screened for potential Arabidopsis knock-out lines in the available T-DNA insertion collections (http://signal.salk.edu/cgi-bin/tdnaexpress). Forty genes were randomly selected that harbored a T-DNA insertion inbetween the translational start and stop codons, either in an intron or in an exon (Table 1). No homozygous T-DNA insertion lines could be identified for three genes (AT1G06590, AT4G07410, and AT5G22370), indicating that their deficiency was embryonically lethal. Indeed, when the seedpods of the hemizygous lines were analyzed in detail, 25% of the embryos were aborted, indicative of an embryo lethal phenotype (P < 0.01 according to the statistical χ 2 test), and suggesting that the proteins encoded by these three genes are essential for embryogenesis (Supplementary Fig. 1).
Table 1.
Genes selected for downstream experimental validation
| Arabidopsis line | TAIR Annotation | T-DNA accession | HUGO |
|---|---|---|---|
| AT1G01940 | F22M8.7 | 061120.53.75.X-Intronic | PPIL3 |
| AT1G03110 | TRM82 | 025857.27.50.X-Exonic | WDR4 |
| AT1G03530 | ATNAF1 | 013589.53.50.X-Exonic | NAF1 |
| AT1G04020 | ATBARD1 | 031862.53.75.X-Exonic | BARD1 |
| AT1G06590 | F12K11.7 | 024997.29.40.X-Intronic | ANAPC5 |
| AT1G08410 | T27G7.9 | 119395.38.15.X-Exonic | LSG1 |
| AT1G10490 | T10O24.10 | 070262.56.00.X-Intronic | NAT10 |
| AT1G13330 | AHP2 | 136002.41.85.X-Exonic | PSMC3IP |
| AT1G49540 | ATELP2 | 106485.50.75.X-Intronic | ELP2-STATIP1 |
| AT1G72320 | APUM23 | 052992.53.50.X-Intronic | C14ORF21 |
| AT1G74150 | F9E11.8 | 088010.26.55.X-Exonic | KHLDC3 |
| AT1G76260 | DWA2 | 143341.50.65.X-Exonic | TSSC1 |
| AT2G15790 | CYCLOPHILIN 40 | 033511.51.20.X-Intronic | PPID |
| AT2G19430 | ATTHO6 | 051022.41.15.X-Exonic | THOC6 |
| AT2G28450 | T1B3.3 | 039998.52.40.X-Exonic | TRMT2A |
| AT2G40550 | ETG1 | 145460.18.05.X-Exonic | MCMBP |
| AT2G34260 | F13P17.10 | 063054.55.75.X-Intronic | WDR55 |
| AT3G02220 | F14P3.13 | 028532.34.35.X-Exonic | C9ORF85 |
| AT3G07050 | F17A9.21 | 099852.47.75.X-Exonic | GNL3 |
| AT3G26410 | ATTRM11 | 122158.32.05.X-Exonic | TRMT11 |
| AT3G42660 | T12K4.110 | 052512.12.95.X-Exonic | WDHD1 |
| AT3G49990 | F3A4.70 | 090801.18.60.X-Exonic | LTV1 |
| AT3G55160 | T26I12.40 | 006621.56.00.X-Exonic | THADA |
| AT3G56990 | EDA7 | 098429.45.45.X-Exonic | NOL10 |
| AT3G60660 | T4C21.70 | 041743.49.40.X-Exonic | C18ORF24-SKA1 |
| AT4G00850 | GIF3 | 052744.30.10.X-Exonic | SS18 |
| AT4G01270 | F2N1.19 | 056467.55.00.X-Exonic | TRAIP |
| AT4G07410 | F28D6.14 | 022607.45.25.X-Exonic | CIRH1A |
| AT4G15890 | DL3985 W | 094776.23.50.X-Intronic | NCAPD3 |
| AT4G20350 | F9F13.6 | 138864.18.85.X-Exonic | ALKBH6 |
| AT4G22970 | AESP | 037016.52.60.X-Intronic | ESPL1 |
| AT4G35910 | T19K4.40 | 030197.20.30.X-Intronic | CTU2 |
| AT4G38120 | F20D10.240 | 066582.56.00.X-Exonic | HEATR6 |
| AT5G05660 | ATNFXL2 | 017558.18.75.X-Exonic | NFXL1 |
| AT5G11240 | F2I11.130 | 052897.39.70.X-Exonic | WDR43 |
| AT5G14600 | T15N1.90 | 024680.34.10.X-Exonic | TRMT61B |
| AT5G22370 | EMB1705 | 059852.56.00.X-Intronic | GPN2 |
| AT5G40530 | MNF13.4 | 102154.30.95.X-Intronic | RRP8 |
| AT5G49110 | K20J1.8 | 055483.52.00.X-Exonic | FANCI |
| AT5G61770 | PAN-LIKE | 088929.56.00.X-Exonic | PPAN |
HUGO Gene nomenclature, Homo sapiens official symbol
For the available homozygous insertion lines, effects on overall cell division and DNA replication activity were determined for the first developed leaf pair harvested at maturity (3 weeks after sowing). As demonstrated previously, the first leaf of Arabidopsis is an excellent model system to study cell division and DNA replication parameters [44–47]. As the leaf grows, its cells progressively shift from a dividing mode to a phase during which they exit their cell cycle program and start to expand. Mutations that affect cell division affect the total number of cells formed at leaf maturity. Furthermore, the cell expansion phase is correlated with the onset of endoreduplication, an alternative cell cycle during which cells continue to replicate their DNA without cell division. Mutations that affect the endoreduplication index (EI; the mean number of endoreduplication cycles) of the leaf are indicative of a change in the cell differentiation timing, with a decreased or increased EI reflecting a delayed or premature cell cycle exit, respectively.
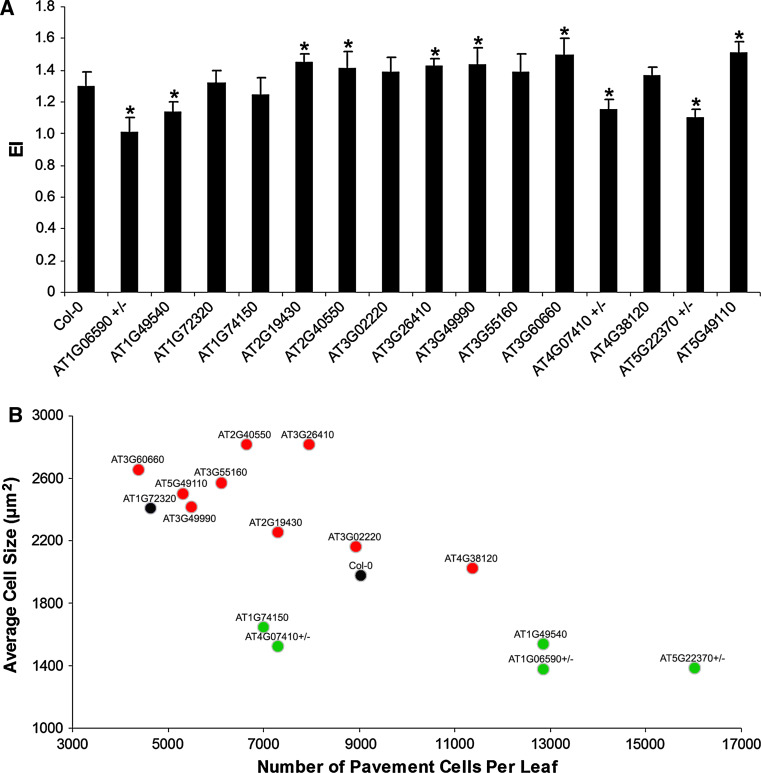
EI measurements revealed a shift in DNA ploidy distribution for 15 of the 40 knockout lines (37 homozygous knockouts and the 3 hemizygous mutants) (Fig. 2a; Supplementary Fig. 2). In contrast, among 11 randomly selected insertion lines, only 1 (AT5G46160) displayed a replication phenotype (Supplementary Fig. 3), illustrating a strong enrichment for replication mutants in the selected set of mutants. In five mutant lines, the EI was lower than that in wild-type plants, whereas for ten knockout lines it was higher (Table 2). Although the mutant line for AT1G72320 (APUM23) had an EI almost identical to that of the control plants, it displayed a totally different DNA ploidy distribution (Supplementary Fig. 2), which implies that proliferation in this line was both stimulated and inhibited, probably in a tissue-specific manner.
Fig. 2.
Experimental association of Arabidopsis thaliana candidate genes with cell replication. The Arabidopsis first leaf was used to measure cell division and DNA replication parameters. a The mean number of endoreduplication cycles denoted as Endoreduplication Index (EI) of the T-DNA insertion lines [*statistically different from the control (Col-0) plants, according to the t test P < 0.05 (n = 10); ± represents hemizygous mutants]. b Scatter plot of the analyzed mutants. Mutants were plotted according to their respective number of cells and cell size. Mutant lines are color-coded according to their DNA ploidy content phenotype. Green and red dots represent mutants with a reduced and increased EI, respectively
Table 2.
Analysis of endoreduplication index (EI), pavement cell size, and cell number in the first developed leaf pair of the studied T-DNA insertion mutants
| Line | EI | Average leaf area (mm2) | Average cell size (μm2) | Pavement cells per mm2 (Cell density) | Pavement cells per leaf |
|---|---|---|---|---|---|
| Col-0 | 1.29 | 28.2 | 1,976 | 320 | 9,040 |
| AT1G06590+/− | 1.01 | 23.0 | 1,380 | 559 | 12,864 |
| AT1G49540 | 1.13 | 38.7 | 1,537 | 332 | 12,850 |
| AT1G72320 | 1.32 | 17.9 | 2,409 | 258 | 4,626 |
| AT1G74150 | 1.24 | 20.1 | 1,643 | 347 | 6,987 |
| AT2G19430 | 1.45 | 27.0 | 2,252 | 270 | 7,289 |
| AT2G40550 | 1.42 | 21.5 | 2,819 | 309 | 6,651 |
| AT3G02220 | 1.39 | 40.0 | 2,165 | 223 | 8,937 |
| AT3G26410 | 1.43 | 33.0 | 2,816 | 241 | 7,939 |
| AT3G49990 | 1.44 | 26.4 | 2,419 | 208 | 5,499 |
| AT3G55160 | 1.39 | 29.9 | 2,569 | 204 | 6,110 |
| AT3G60660 | 1.50 | 26.6 | 2,651 | 165 | 4,376 |
| AT4G07410+/− | 1.16 | 18.3 | 1,524 | 398 | 7,286 |
| AT4G38120 | 1.37 | 30.1 | 2,020 | 377 | 11,359 |
| AT5G22370+/− | 1.10 | 29.1 | 1,387 | 550 | 16,016 |
| AT5G49110 | 1.51 | 26.2 | 2,498 | 203 | 5,320 |
+/− Hemizygous lines
Changes in the DNA content due to an altered cell differentiation timing should affect the total leaf cell number and cell size distribution, in which a delayed or premature onset of cell differentiation often correlates with smaller or bigger cells, respectively [48]. Therefore, cell number and cell size distribution analyses of the leaf epidermal cells were performed. When the average cell numbers and cell sizes were plotted, two main subgroups of mutants could be recognized: one characterized by more but smaller cells, and one with few but larger cells, than those of the wild-type plants (Fig. 2b). According to the flow cytometric measurements, a subgroup of mutant lines in the first group had a reduced EI (green dots), showing that the differences at the DNA ploidy level originated from enhanced cell proliferation or delayed cell differentiation. Conversely, the other subgroup of mutants comprised plants displaying an increased DNA ploidy content (red dots), indicative of premature cell cycle exit. The data were substantiated by cell size distribution analysis, with those mutants showing a decreased EI exhibiting an increased subpopulation of small cells, in comparison with control plants. Conversely, the mutants that displayed an increased EI were enriched in enlarged cells (Supplementary Fig. 4). In the mutant line for AT1G72320, the population of both small and large cells had increased, hinting again at a dual effect of this gene on cell proliferation.
DNA damage assays
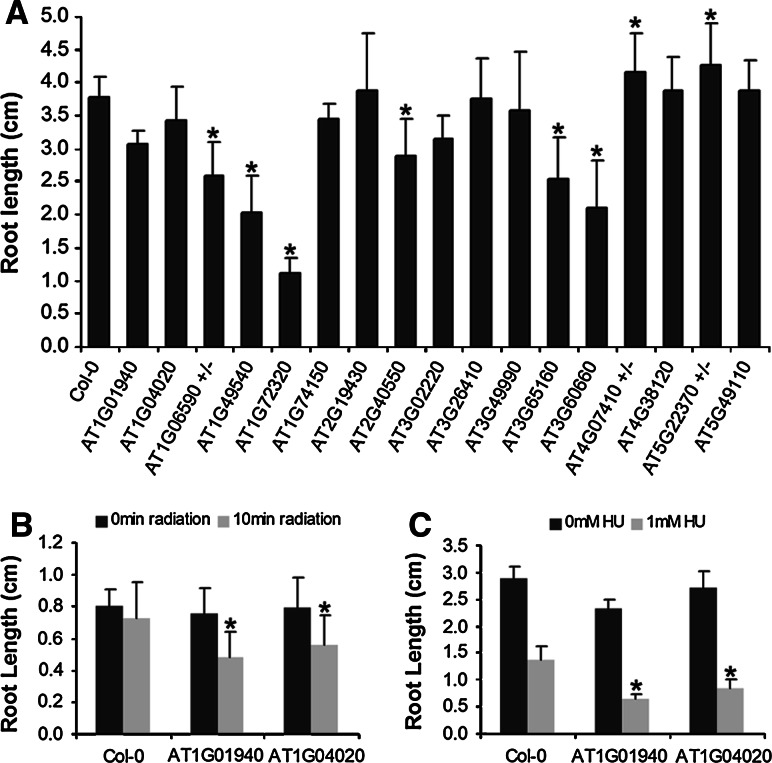
As the screening method involved a selection of genes displaying a significant enrichment of genes involved in DNA repair among coexpressed neighbors, the knock-out lines were tested for hypersensitivity toward DNA replication inhibiting stress treatments, including UV-B (UV) radiation and hydroxyurea (HU) treatment. UV-B radiation dimerizes adjacent pyrimidine bases, and inhibits replication and transcription, eventually causing a growth delay. Similarly, HU treatment causes a collapse of the replication fork, with inhibition of growth as a consequence. DNA damage was measured by comparing root growth under control and DNA-damaging growth conditions (see “Materials and methods”). Without any DNA stress treatment, the mutants for ATG06590 (hemizygous mutant), AT1G49540 (ATELP2), AT1G72320 (APUM23), AT2G40550 (ETG1), AT3G55160, and AT3G60660, showed a significant root growth reduction (P < 0.01 according to Student’s t test), when compared to wild-type Col-0 plants, displaying at 7 days after germination 35, 46, 67, 23, 33, and 44% of growth reduction, respectively. Conversely, the hemizygous mutants for AT4G07410 and AT5G22370 showed a significant increase in root growth (Fig. 3a). Wild-type plants were not hypersensitive towards UV-B (1.9 W/m2). In contrast, the lines mutant for AT1G01940 and AT1G04020 showed a clear growth inhibition 72 h after the treatment (Fig. 3b). Similarly, these two mutants displayed a root growth inhibition stronger than that observed for the wild-type plants when treated with 1 mM HU for 6 days (Fig. 3c).
Fig. 3.
Hypersensitivity of selected T-DNA insertion lines towards DNA replication-inhibiting treatments. a Root length under standard growth conditions for the analyzed T-DNA insertion lines. Roots were measured after 7 days of growth on vertical MS plates. b, c Mutants displaying a differential root growth response upon UV-B irradiation or in the presence of 1 mM HU, respectively [*Statistically different from the control (Col-0) plants according to the t test P < 0.05 (n = 30); ± represents hemizygous mutants]
Validation of putative cell cycle regulators in MCF7 cells
To test whether the obtained gene list had a predictive power for detecting cell cycle-related genes in the mammalian model, three human genes that, to our knowledge, had not been implicated in cancer origin or progression, were silenced in breast epithelial cancer cell cultures (MCF7 cells), including the orthologues of AT1G49540 (STATIP1), AT4G38120 (HEATR6), and AT1G72320 (C14ORF21). In Arabidopsis, knock-out of the AT1G49540 gene resulted into an enhanced cell division phenotype, and the knock-out of the AT4G38120 gene caused an early induction of the differentiation processes, whereas the knockout of the AT1G72320 gene was responsible for a dual phenotype. Similarly to its plant counterpart, the coexpression neighborhood of HEATR6 was enriched for the GO term “DNA repair” (P < 0.01 according to the hypergeometric distribution) (Table 3). This was not the case for STATIP1 and C14ORF21.
Table 3.
Gene ontology (GO) enrichment conservation
| Arabidopsis fold enrichments | Homo sapiens fold enrichments | ||||
|---|---|---|---|---|---|
| AGI code | DNA replication | DNA repair | HUGO | DNA replication | DNA repair |
| AT1G01940 | 3.86* | 2.92* | PPIL3 | 4.26* | 2.59** |
| AT1G04020 | 6.31* | 3.07** | BARD1 | 22.23* | 12.59* |
| AT1G06590 | 5.07* | 3.89* | APC5 | 1.89- | 1.85- |
| AT1G49540 | 3.73* | 2.59* | STATIP1 | 0.00- | 1.11- |
| AT1G72320 | 5.04* | 2.98* | C14ORF21 | 0.95- | 1.85- |
| AT1G74150 | 4.98* | 3.68* | KLHDC3 | 0.95- | 1.11- |
| AT2G19430 | 4.38* | 3.02* | THOC6 | 7.09* | 4.07* |
| AT2G40550 | 5.18* | 3.03* | MCMBP | 3.31* | 5.93* |
| AT3G02220 | 3.95* | 0.00- | C9ORF85 | 0.00- | 0.00- |
| AT3G26410 | 4.31* | 2.91* | TRMT11 | 3.31* | 3.33* |
| AT3G49990 | 3.95* | 0.00- | LTV1 | 3.31* | 1.85- |
| AT3G55160 | 3.58* | 2.72* | THADA | 0.47- | 0.00- |
| AT3G60660 | 3.88* | 2.69* | C18ORF24 | 23.65* | 13.7* |
| AT4G07410 | 4.06* | 2.57* | CIRH1A | 6.15* | 2.22** |
| AT4G38120 | 5.46* | 2.76* | HEATR6 | 1.42- | 3.33* |
| AT5G22370 | 4.04* | 2.69* | GPN2 | 0.95- | 1.85- |
| AT5G49110 | 3.94* | 2.81* | FANCI | 23.18* | 14.07* |
GO enrichment for the terms DNA repair and DNA replication was calculated for Arabidopsis and Homo sapiens and is given for each GO class
Statistical significance according to the hypergeometric distribution: *P < 0.01, **P < 0.05, - no significant enrichment
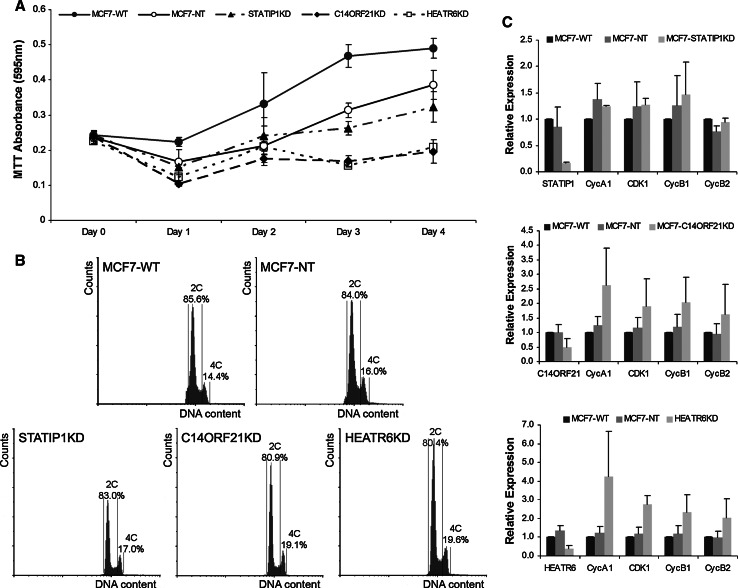
After transient knock-down of STATIP1, C14ORF21, and HEATR6 through specific siRNA pools, cell culture growth was monitored by the colorimetric MTT assay [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] [49]. In comparison with controls [untransfected cells (WT) and cells transfected with si-control (NT)], knock-down of C14ORF21 and HEATR6 clearly affected growth (Fig. 4a). The reduced number of cells might be caused by a cell cycle arrest. To corroborate this possibility, flow cytometric experiments revealed a larger number of G2/M cells in the knock-down cultures of the C14ORF21 and HEATR6 genes than that in the controls (Fig. 4b), indicative of a transient G2 arrest. In agreement with these results, the transcripts of the G2/M marker genes CDK1, CyclinB1, and CyclinB2 were up-regulated upon knock-down of C14ORF21 and HEATR6 (Fig. 4c).
Fig. 4.
Experimental association of human candidate genes with cell division in MCF7 cell cultures. Genes were silenced in breast epithelial cancer cell cultures (MCF7 cells) using small interfering (si) RNA sequences. a Growth curves of the siRNA knocked-down MCF7 cultures, illustrating growth inhibition by knock-down of C14ORF21 and HEATR6. b Ploidy distributions of the STATIP1, C14ORF21, and HEATR6 knocked-down cultures in comparison with controls assessed by flow cytometry, illustrating a significant increased number of G2/M cells in C14ORF21 and HEATR6 knock-down cultures (P < 0.05 (n = 9) according to a t test). c Expression levels of cell cycle phase makers measured by Q-PCR, illustrating transcriptional upregulation of the G2/M marker genes CDK1, CyclinB1, and CyclinB2 in C14ORF21 and HEATR6 knock-down cell cultures, indicative for a transient G2 arrest
Associations with cancer relapse probability
To assess the potential correlation between the phenotypes of the plant genes selected by means of the designed integrative approach with those of their corresponding human orthologues, we created a database of transcriptional profiles of 1,400 non-redundant breast cancer samples linked to well-annotated clinical information; including relapse events and relapse time (see “Materials and methods”). The significance of a particular association between gene expression and a relapse event was assessed by Cox regression analysis. To ensure that the increased statistical power of the analysis due to the great number of patients in the database did not lead to irrelevant association with relapse risk, we iteratively and randomly subdivided the initial patient set into two complementary subsets of 100 training sets of 75% of the samples (n = 1,050) and 100 validation sets of the corresponding remaining samples (n = 350). The Cox survival analysis was performed independently in parallel with both the training and validation sets. We considered for further analysis only the probe sets with significant association (at the 0.01 level) with increased or decreased risk in at least 95% of the corresponding training sets and validation sets. After stability evaluation, 182 out of the 9,976 available reliable probe sets (see Supplemental Methods file) were associated with decreased risk of relapse, while 995 probe sets were associated with an increased relapse risk. Among these, genes known to be associated with good disease outcome, such as the ESR1 estrogen and PGR progesterone receptors, were associated with a decreased risk of relapse. Conversely, genes known to be associated with poor disease outcome such as ERBB2 or TOP2A were correlated with a significantly increased relapse risk, proving the validity of our database (Supplemental Fig. 5).
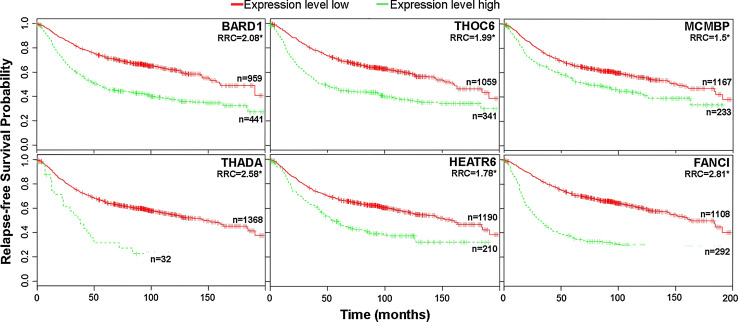
For the list of candidate genes resulting from the comparative analysis between plant and human, 211 reliable probe sets (corresponding to 169 human orthologues; Supplementary Table 9) were available, for which 162 were not significantly associated with relapse risk or their association with it was not stable upon cross-validation. Only one was stably associated with decreased risk of relapse. In contrast, 48 probe sets (corresponding to 45 genes) were stably associated with an increased relapse risk (Supplementary Table 9). Thus, compared to the 9,976 probe sets present in the whole database, the 221 probes were significantly enriched in probe sets associated with an increased risk of relapse (P = 1.14 × 10−7 according to the hypergeometric distribution). Interestingly, among the 15 analyzed Arabidopsis mutant lines that displayed a leaf growth phenotype upon mutation, 6 were associated with an increased relapse risk. For these genes, comprising four uncharacterized genes and the well-characterized replication genes BARD1 and FANCI, Cox survival curves showed a clear association between altered expression levels and a diminished probability of survival, indicating that they can be considered as good markers to predict disease outcome in human breast cancer (Fig. 5).
Fig. 5.
Association of human orthologues of Arabidopsis genes involved in cell replication with specific cancer outcomes (relapse risk). Cox survival plots for the human orthologues of Arabidopsis genes with a direct influence on cell proliferation were constructed. A clear association between increased gene expression levels and a diminished probability of relapse-free survival is shown. RRC relative risk coefficient, *statistically significant differences in the survival probability, P < 0.01
Discussion
The field of comparative genomics has been growing and evolving rapidly thanks to the massive amount of genomic data generated over the last decade. Here, we have integrated coexpression analysis with comparative genomics to identify putative new cell cycle genes. Previously, we had demonstrated that in Arabidopsis coexpression alone performs poorly to infer known biological gene functions [35]. To improve the predictive power of coexpression networks, we have combined different functional prediction elements (GO enrichment analysis and cis-regulatory element scoring) to create a reliable platform for the detection of novel conserved cell cycle regulators. Interestingly, recently available ChIP-Seq data [50] revealed that there is a highly significant overlap (P = 2.75 × 10−14 according to the hypergeometric distribution), between the E2F target genes detected by our strategy and the genes that are predicted to be direct E2F targets on basis of the ChIP analysis.
The success rate of the integrative approach was illustrated by the observation that among 11 randomly selected T-DNA insertion lines only 1 displayed a DNA ploidy distribution profile different from wild-type plants, generating an identification rate of mutants possibly involved in replication events of 9%. In contrast, out of 40 plant candidate genes selected for downstream functional analysis, 15 were experimentally proven to affect cell proliferation, representing a success rate fivefold higher than that of the random approach. Moreover, two Arabidopsis mutant lines could be related with DNA stress responses and two human selected orthologues clearly affected cell proliferation when knocked-down in breast epithelial cancer cells, emphasizing the highly significant predictive value of our integrative approach.
The importance of including Arabidopsis data in our search for novel cancer genes is illustrated by the observation that our final list of 339 genes retains 79 human genes that, according to the gene ontology classification (based on AMIGO), do not have a defined category (genes with unknown function). Similarly, there are 82 human genes that according to the GO classification are involved in functions totally unrelated to DNA replication and repair (Supplementary Table 7). This total of 161 genes represents half of the final list, illustrating the importance of the Arabidopsis–H. sapiens orthology relationship in order to give or detect new gene functions even in highly distant organisms. A good example of the importance of the Arabidopsis filtering process is that the genes THADA, HEATR6, and MCMBP, which, according to the analyses presented, might represent important predictors of breast carcinomas, were exclusively retained in the final list of candidate genes due to the fact that their respective Arabidopsis orthologues were strongly associated with DNA replication processes.
Known proliferation genes populate the list of 339 candidate genes, that encode cell division control proteins (CDC6, CDC7, and CDC27), the retinoblastoma protein RB, replication proteins (MCM1, MCM2, MCM3, MCM4, MCM, MCM8, ORC1L, ORC2L, ORC3L, ORC5L, ORC6L, and PCNA), repair proteins (WEE1, PARP1, RAD50, RAD51, DDB1, and MRE11A), and previously characterized oncogenes (BARD1, BRIP1, API5, and ESPL1). These genes can be considered as positive controls. It suggests that the new genes found with this approach might be new cell cycle regulators. Indeed, we showed that 48 of the candidate genes have a significant prognostic value, at least for breast cancer, being associated with specific clinical outcomes when deregulated. In other words, 30%, of the retained genes are putative cancer predictors and represent highly significant cancer associations (P < 0.01 according to the hypergeometric distribution). Interestingly, comparing the data of a cancer gene census study [51], the candidate list of 339 genes showed a largely similar set of GO categories, although at a slightly different relative abundance (Supplementary Fig. 6).
Different facts argue in favor of the list of new cell cycle regulators to hold important elements in the mammalian cell cycle. First, two of the orthologous genes that are embryo lethal in Arabidopsis have an important role in the origin and progression of different diseases, including cancer. APC5, the human orthologue of the mutant line AT1G06590, is part of the gene set that is commonly misregulated during the onset and progression of breast and colorectal cancers [52]. CIRH1A, the human orthologue of the Arabidopsis embryo-lethal line AT4G07410, is the cause of the North American Indian Childhood Cirrhosis (NAIC/CIRH1A), a severe autosomal recessive intrahepatic cholestasis. All NAIC patients have a homozygous mutation in the CIRH1A protein, of which the function is still unknown [53]. Nevertheless, CIRH1A can upregulate a canonical NF-κB element and might participate in the regulation of other genes containing NF-κB responsive elements [54]. Because the activities of genes regulated through NF-κB responsive elements are especially important during development, this interaction might explain not only the appearance of NAIC but it also suggests that CIRH1A misregulation is a new important element in the NF-κB pathway, alterations of which have been extensively proved to lie at the basis of cancer origin and progression [55, 56].
Secondly, three of the genes in the final list have been linked recently with cell proliferation or DNA repair in plants and/or mammals. SKA1, orthologue of the Arabidopsis AT3G60660 gene, plays a critical role in coupling chromosome movement to microtubule dynamics at the outer kinetochore [57]. The plant orthologue of the well-studied mammalian breast cancer associated RING domain protein 1 gene (BARD1), involved in DNA repair, also controls DNA repair in plants [58]. Whereas this gene had been established to be essential for responding to the DNA cross-linking agent mitomycin, our results reveal that BARD1 knocked-down plants are sensitive toward UV irradiation and HU. Another example is the E2F TARGET GENE 1 (ETG1) protein that had been identified recently as a novel evolutionarily conserved replisome factor. ETG1 is associated with the minichromosome maintenance complex, being crucial for efficient DNA replication [59]. Additionally, depletion of ETG1 or its human orthologue MCM-BP, results in a stringent late G2 cell cycle arrest that correlates with a partial loss of sister chromatids cohesion [60, 61], hinting at an equally important developmental role for this molecule in plants and mammals.
Here, we found that the knock-down of the genes C14ORF21 and HEATR6, which are orthologues of the Arabidopsis AT1G72320 and AT4G38120 genes, respectively, have an inhibitory effect on cell proliferation. We showed that depletion of C14ORF21 and HEATR6 resulted in an increase in the population of cells with a 4C DNA content, which is supported by an upregulation of G2/M cell cycle marker genes. Interestingly, HEATR6 is present on one of the most commonly amplified fragments in breast cancer [62] and, accordingly, its transcript is significantly overexpressed in gastric, brain, and breast carcinomas. Similarly, the C14ORF21 transcript is upregulated in colorectal, gastric, and prostate cancers (Supplementary Fig. 7).
Some of the genes found in the present study might at first sight not fit the classical picture of tumor suppressors or oncogenes, like those related to ribosomes and ribogenesis (such as AT2G28450, AT3G02220, and AT3G49990 genes, orthologues of the human genes TRMT2A, C9ORF85, and LTV1, respectively). Ribosomal proteins are ubiquitous, abundantly present, and mostly regarded as constants in the cells. Approximately 80 proteins have been reported to be part of the ribosomes, and many more are involved in their biogenesis and assembly. However, recent data showed that some of these proteins appear to have extra-ribosomal functions [63], and some are even linked to cancer [64, 65]. The imbalance of ribosomal subunits leads to p53 activation and apoptosis [66]. Additionally, in recent years, drugs that disrupt ribosome production, such as rapamycin, have been applied successfully to cancer treatments. As cell division requires the synthesis of a large amount of proteins, deregulation of ribosome biogenesis emerges as a novel strategy to control abnormal cell proliferation, given that without a protein synthesis machinery that can cope with an altered DNA replication process, no division can occur. The best example of this is that inactivation of SSF1 (orthologue of AT5G61770), involved in ribosome synthesis, leads to loss of contact inhibition [67].
The data presented argue in favor of an applied integrative approach as a powerful strategy to discover new conserved cell cycle regulators. Nevertheless, this strategy suffers from restrictions, especially because it is based on gene coexpression, and thus cannot provide a full perspective of molecular interactions, such as protein–protein interactions, as exemplified by the Arabidopsis gene AT1G49540 and its human orthologue STATIP1. Although the knockdown of the Arabidopsis gene triggered cell proliferation, the knock-down of STATIP1 did not. In contrast to the plant gene, the coexpression neighborhood of STATIP1 is not enriched for DNA replication or DNA repair (Table 3), indicating that despite their orthology relationship both molecules may have diverged functionally during evolution. The contrasting phenotypic effects between these two orthologous genes illustrate that not only the components belonging to a specific network are important but also their wiring.
Conclusions
To understand the origin and progression of the carcinogenic process, and to shed light onto the complex mechanisms that lead to tumorigenesis and cancer, different model organisms have been used. Some of them, like Mus musculus, are relatively closely related with humans, and several mouse models are currently used in cancer research [68–70], whereas some others, like Drosphila melanogaster or Saccharomyces cerevisiae, are distantly related. Nevertheless, they have also contributed extensively to the understanding of the disease [71–74]. With the data presented in this study, we demonstrated that through the use of comparative genomics the plant model species A. thaliana, but likely any model organism for which large expression datasets and genome data are available, can aid in the discovery of putative cancer genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank all members of the cell cycle and oncology groups for fruitful discussions and suggestions, the Arabidopsis Biological Research Center for providing the T-DNA insertion lines, and Martine De Cock and Lorena López for help in preparing the manuscript. This work was supported by grants from the Interuniversity Poles of Attraction Programne (IUAP VI/33), initiated by the Belgian State, Science Policy Office, the Research Foundation-Flanders (grant no. G008306), Ghent University (“Geconcerteerde Onderzoeksacties” no.01G013B7), the Stichting tegen Kanker (no. 189-2008), the Association for International Cancer Research (Scotland), the EU-FP6 framework program BRECOSM LSHC-CT-2004-503224, and the EU-FP7 framework program TuMIC 2008-201662. M.Q. is indebted with the VIB international PhD program. K.V. acknowledges the support by Ghent University (Multidisciplinary Research Partnership “Bioinformatics: from nucleotides to networks”) and the Interuniversity Attraction Poles Programme (IUAP P6/25), initiated by the Belgian State, Science Policy Office (BioMaGNet). S.D. is indebted to the Agency for Innovation through Science and Technology for a predoctoral fellowship.
Abbreviations
- CDK
Cyclin-dependent kinase
- EI
Endoreduplication index
- fRMA
Frozen Robust Multiarray Analysis
- GO
Gene ontology
- HU
Hydroxyurea
- PCC
Pearson correlation coefficient
- PWM
Positional Weight Matrix
- QPCR
Quantitative polymerase chain reaction
- siRNA
Small interfering RNA
References
- 1.Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Inze D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 3.Srinivas PR, Verma M, Zhao Y, Srivastava S. Proteomics for cancer biomarker discovery. Clin Chem. 2002;48:1160–1169. [PubMed] [Google Scholar]
- 4.Pekarsky Y, Zanesi N, Palamarchuk A, Huebner K, Croce CM. FHIT: from gene discovery to cancer treatment and prevention. Lancet Oncol. 2002;3:748–754. doi: 10.1016/S1470-2045(02)00931-2. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 6.Marone M, Scambia G, Giannitelli C, Ferrandina G, Masciullo V, Bellacosa A, Benedetti-Panici P, Mancuso S. Analysis of cyclin E and cdk2 in ovarian cancer: gene amplification and RNA overexpression. Int J Cancer. 1998;75:34–39. doi: 10.1002/(SICI)1097-0215(19980105)75:1<34::AID-IJC6>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Scheurle D, DeYoung MP, Binninger DM, Page H, Jahanzeb M, Narayanan R. Cancer gene discovery using digital differential display. Cancer Res. 2000;60:4037–4043. [PubMed] [Google Scholar]
- 8.Argani P, Rosty C, Reiter RE, Wilentz RE, Murugesan SR, Leach SD, Ryu B, Skinner HG, Goggins M, Jaffee EM, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Discovery of new markers of cancer through serial analysis of gene expression: prostate stem cell antigen is overexpressed in pancreatic adenocarcinoma. Cancer Res. 2001;61:4320–4324. [PubMed] [Google Scholar]
- 9.Alizadeh AA, Ross DT, Perou CM, van de Rijn M. Towards a novel classification of human malignancies based on gene expression patterns. J Pathol. 2001;195:41–52. doi: 10.1002/path.889. [DOI] [PubMed] [Google Scholar]
- 10.Korkola JE, DeVries S, Fridlyand J, Hwang ES, Estep ALH, Chen Y-Y, Chew KL, Dairkee SH, Jensen RM, Waldman FM. Differentiation of lobular versus ductal breast carcinomas by expression microarray analysis. Cancer Res. 2003;63:7167–7175. [PubMed] [Google Scholar]
- 11.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller LD, Liu ET. Expression genomics in breast cancer research: microarrays at the crossroads of biology and medicine. Breast Cancer Res. 2007;9:206. doi: 10.1186/bcr1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe J-P, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo W-L, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 15.Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, SEARCH Collaborators. Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen C-Y, Wu P-E, Wang H-C, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo K-Y, Noh D-Y, Ahn S-H, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low Y-L, Bogdanova N, Schürmann P, Dörk T, Tollenaar RAEM, Jacobi CE, Devilee P, Klijn JGM, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MWR, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den Ouweland A, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko Y-D, Spurdle AB, Beesley J, Chen X, kConFab. AOCS Management Group. Mannermaa A, Kosma V-M, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BAJ. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai Y–Y, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 18.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 19.Ala U, Piro RM, Grassi E, Damasco C, Silengo L, Oti M, Provero P, Di Cunto F. Prediction of human disease genes by human–mouse conserved coexpression analysis. PLoS Comput Biol. 2008;4:e1000043. doi: 10.1371/journal.pcbi.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, Marcotte EM. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci USA. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 22.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 23.Chan SW-L, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 24.Matzke MA, Matzke AJM, Pruss GJ, Vance VB. RNA-based silencing strategies in plants. Curr Opin Genet Dev. 2001;11:221–227. doi: 10.1016/S0959-437X(00)00183-0. [DOI] [PubMed] [Google Scholar]
- 25.Ma H. GTP-binding proteins in plants: new members of an old family. Plant Mol Biol. 1994;26:1611–1636. doi: 10.1007/BF00016493. [DOI] [PubMed] [Google Scholar]
- 26.Jones AM, Chory J, Dangl JL, Estelle M, Jacobsen SE, Meyerowitz EM, Nordborg M, Weigel D. The impact of Arabidopsis on human health: diversifying our portfolio. Cell. 2008;133:939–943. doi: 10.1016/j.cell.2008.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. Mol Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/S1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 31.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 32.Chen H-Z, Tsai S-Y, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen LJ, Jensen TS, de Lichtenberg U, Brunak S, Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- 34.Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S. NASCArrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2009;150:535–546. doi: 10.1104/pp.109.136028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole RL. The TAIR database. Methods Mol Biol. 2007;406:179–212. doi: 10.1007/978-1-59745-535-0_8. [DOI] [PubMed] [Google Scholar]
- 37.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, Hub AmiGO, Group Web Presence Working. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GTS, Gruissem W, Van de Peer Y, Inzé D, De Veylder L. Genome-wide identification of potential plant E2F target genes. Plant Physiol. 2005;139:316–328. doi: 10.1104/pp.105.066290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouzé P, Moreau Y. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Comput Biol. 2002;9:447–464. doi: 10.1089/10665270252935566. [DOI] [PubMed] [Google Scholar]
- 40.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL–DB: querying a comprehensive multi–species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: a multiple sequence alignment with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 43.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Veylder L, Beeckman T, Beemster GTS, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inzé D. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 2002;21:1360–1368. doi: 10.1093/emboj/21.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inzé D, De Veylder L. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell. 2004;16:2683–2692. doi: 10.1105/tpc.104.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L. The DP–E2F–like DEL1 gene controls the endocycle in Arabidopsis thaliana . Curr Biol. 2005;15:59–63. doi: 10.1016/j.cub.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 47.Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis . Plant Physiol. 2005;138:734–743. doi: 10.1104/pp.104.053884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsukaya H, Beemster GTS. Genetics, cell cycle and cell expansion in organogenesis in plants. J Plant Res. 2006;119:1–4. doi: 10.1007/s10265-005-0254-y. [DOI] [PubMed] [Google Scholar]
- 49.Cory AH, Owen TC, Barltrop JA, Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3:207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- 50.Cao AR, Rabinovich R, Xu M, Xu X, Jin VX, Farnham PJ. Genome-wide analysis of transcription factor E2F1 mutant proteins reveals that N- and C-terminal protein interaction domains do not participate in targeting E2F1 to the human genome. J Biol Chem. 2011;286:11985–11996. doi: 10.1074/jbc.M110.217158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puente XS, Velasco G, Gutierrez-Fernandez A, Bertranpetit J, King MC, Lopez-Otin C. Comparative analysis of cancer genes in the human and chimpanzee genomes. BMC Genomics. 2006;7:15. doi: 10.1186/1471-2164-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JKV, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 53.Chagnon P, Michaud J, Mitchell G, Mercier J, Marion J-F, Drouin E, Rasquin-Weber A, Hudson TJ, Richter A. A missense mutation (R565 W) in Cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am J Hum Genet. 2002;71:1443–1449. doi: 10.1086/344580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu B, Mitchell GA, Richter A. Cirhin up-regulates a canonical NF-κB element through strong interaction with Cirip/HIVEP1. Exp Cell Res. 2009;315:3086–3098. doi: 10.1016/j.yexcr.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 55.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. NF-KappaB functions as a tumour promoter in inflammation–associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 56.Huber MA, Azoitei N, Baumann B, Grünert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-κB is essential for epithelial–mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, III, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reidt W, Wurz R, Wanieck K, Chu HH, Puchta H. A homologue of the breast cancer–associated gene BARD1 is involved in DNA repair in plants. EMBO J. 2006;25:4326–4337. doi: 10.1038/sj.emboj.7601313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takahashi N, Lammens T, Boudolf V, Maes S, Yoshizumi T, De Jaeger G, Witters E, Inzé D, De Veylder L. The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 2008;27:1840–1851. doi: 10.1038/emboj.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi N, Quimbaya M, Schubert V, Lammens T, Vandepoele K, Schubert I, Matsui M, Inzé D, Berx G, De Veylder L. The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet. 2010;6:e1000817. doi: 10.1371/journal.pgen.1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishiyama A, Frappier L, Méchali M. MCM–BP regulates unloading of the MCM2–7 helicase in late S phase. Genes Dev. 2011;25:165–175. doi: 10.1101/gad.614411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu G-j, Sinclair C, Hinson S, Ingle JN, Roche PC, Couch FJ. Structural analysis of the 17q22–23 amplicon identifies several independent targets of amplification in breast cancer cell lines and tumors. Cancer Res. 2001;61:4951–4955. [PubMed] [Google Scholar]
- 63.Lai M-D, Xu J. Ribosomal proteins and colorectal cancer. Curr Genomics. 2007;8:43–49. doi: 10.2174/138920207780076938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindström MS, Zhang Y. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leontieva OV, Ionov Y. RNA-binding motif protein 35A is a novel tumor suppressor for colorectal cancer. Cell Cycle. 2009;8:490–497. doi: 10.4161/cc.8.3.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Welch PM, Gabal M, Betts DM, Whelan NC, Studer ME. In vitro analysis of antiangiogenic activity of fungi isolated from clinical cases of equine keratomycosis. Vet Ophthalmol. 2000;3:145–151. doi: 10.1046/j.1463-5224.2000.00132.x. [DOI] [PubMed] [Google Scholar]
- 68.White DE, Kurpios NA, Zuo D, Hassell JA, Blaess S, Mueller U, Muller WJ. Targeted disruption of β1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer Cell. 2004;6:159–170. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 69.Pearson T, Greiner DL, Shultz LD. Humanized SCID mouse models for biomedical research. Curr Top Microbiol Immunol. 2008;324:25–51. doi: 10.1007/978-3-540-75647-7_2. [DOI] [PubMed] [Google Scholar]
- 70.Vucur M, Roderburg C, Bettermann K, Tacke F, Heikenwalder M, Trautwein C, Luedde T. Mouse models of hepatocarcinogenesis: What can we learn for the prevention of human hepatocellular carcinoma? Oncotarget. 2010;1:373–378. doi: 10.18632/oncotarget.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hartwell LH. Role of yeast in cancer research. Cancer. 1992;69:2615–2621. doi: 10.1002/1097-0142(19920515)69:10<2615::AID-CNCR2820691036>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 72.Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, Margulies IMK, King CR, Liotta LA, Steeg PS. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–180. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- 73.Moberg KH, Bell DW, Wahrer DCR, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- 74.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster . Nat Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.