Abstract
A member of the cellulose synthase-like (subfamily D) gene family of Arabidopsis, AtCSLD3, has been identified by T-DNA tagging. The analysis of the corresponding mutant, csld3-1, showed that the AtCSLD3 gene plays a role in root hair growth in plants. Root hairs grow in phases: First a bulge is formed and then the root hair elongates by polarized growth, the so-called “tip growth.” In the mutant, root hairs were initiated at the correct position and grew into a bulge, but their elongation was severely reduced. The tips of the csld3-1 root hairs easily leaked cytoplasm, indicating that the tensile strength of the cell wall had changed at the site of the tip. Based on the mutant phenotype and the functional conservation between CSLD3 and the genuine cellulose synthase proteins, we hypothesized that the CSLD3 protein is essential for the synthesis of polymers for the fast-growing primary cell wall at the root hair tip. The distinct mutant phenotype and the ubiquitous expression pattern indicate that the CSLD3 gene product is only limiting at the zone of the root hair tip, suggesting particular physical properties of the cell wall at this specific site of the root hair cell.
Cellulose, the most abundant biopolymer, is part of the primary and secondary cell wall of plant cells. The presence of cellulose in secondary walls of cotton (Gossypium hirsutum) fibers and xylem vessels is economically exploited to produce fabric and paper. Cellulose consists of Glc-building blocks β-1,4-linked into polymers and crystallized into microfibrils in the primary cell wall. These microfibrils are organized into macrofibrils in the secondary cell walls. Cellulose is synthesized in complexes that are thought to consist of catalytic subunits of cellulose synthase (CelA, renamed CeSA; Delmer, 1999) of which the glycosyltransferase activity uses UDP-Glc that is delivered from associated membrane-bound Suc synthases as the substrate for polymerization into a growing β-1,4-d-glucan chain. Accessory proteins may facilitate the secretion of the growing cellulose chain through the plasma membrane. Putative regulatory components and crystallization units may be part of the complex (Delmer, 1999). Cellulose synthase complexes are visible as rosette-like structures in freeze-fractured plasma membranes.
CeSA genes initially had been cloned from bacteria, i.e. Acetobacter xylinum (Aloni et al., 1982; Saxena et al., 1990) and Agrobacterium tumefaciens (Matthysse et al., 1995). In plants, CeSA genes have been cloned from cotton, based on bacterial DNA sequence information. These clones were isolated as fiber-specific cDNAs for which glycosyltransferase activity has been shown (Pear et al., 1996). RADIAL SWOLLEN1 (RSW1) was the first CeSA gene isolated in Arabidopsis by mutational analysis (AtCeSA1; Arioli et al., 1998). To date, multiple CeSA genes have been identified in the Arabidopsis genome that are closely related to the cotton CeSA cDNAs (Cutler and Somerville, 1997; Holland et al., 2000; Richmond and Somerville, 2000; http://cellwall.stanford.edu/ cellwall). Based on sequence comparison, CeSA proteins contain highly conserved domains around the D, D, D, and QXXRW motifs, which span the catalytic domain, interspersed by less conserved regions and divergent amino and carboxyl termini. This variability suggests multiple ways of regulation, activity, or interaction with other proteins. The presence of a large gene family also suggests transcriptional regulation of gene expression in relation to cell type, developmental timing, and even environmental or stress conditions. A few mutants with cell-specific defects in secondary wall cellulose synthesis have been described in Arabidopsis. The trichome birefringence mutation affects cellulose synthesis only in trichomes (Potikha and Delmer, 1995). The irregular xylem (irx) mutant has an altered cellulose content specifically in the xylem (Turner and Somerville, 1997). The IRX3 gene is one of the CeSA genes (AtCeSA7; Taylor et al., 1999). These mutants confirm that different members of the CeSA gene family may be used during morphogenesis to diversify cell function.
A group of cellulose synthase-like (CSL) genes was identified in Arabidopsis by in silico analysis of the genome (Cutler and Somerville, 1997). By cluster analysis the AtCSL genes have been classified into six subfamilies, AtCSLA through AtCSLG (Richmond and Somerville, 2000; http://cellwall.stanford.edu/cellwall/Arabidopsis). All the AtCSL genes share a high protein similarity with the CeSA genes. The function of the CSL genes is still unclear: Aside from the amino acid conservation and the preservation of the UDP-glycosyltransferase catalytic domain, no information exists on their biological function. The Arabidopsis AtCSLD subfamily currently consists of six genes and is mostly related to the AtCeSA gene family. We isolated a member of the AtCSLD family, AtCSLD3, by using a molecular-genetic approach and we show that the biological function of AtCSLD3 is related to root hair elongation. The line homozygous for the defective AtCSLD3 gene is affected in “tip growth” of root hairs. Therefore, we postulated that the CSLD3 gene product is important for primary cell wall synthesis specifically in the tip-growing zone of root hairs.
RESULTS
Identification of the AtCSLD3 Gene by T-DNA Tagging
The CT-17 line was obtained from in planta transformation with the vector pGKB5 (Bouchez et al., 1993). The plant DNA that flanks the right T-DNA border in CT-17 was amplified by inverse PCR using an ApoI digest. An 800-bp amplified fragment contained 287 bp of plant DNA. Database search showed that 152 bp of these were 100% identical with the expressed sequence tag (EST) clone Z35212 comprised within the 5′ end of the cDNA clone FAI228 (Fig. 1). The complete insert of 700 bp in the FAI228 clone was sequenced accordingly. The FAI228 clone was highly homologous to the cotton CeSA cDNAs, U58284 and U58283, as well as to the Arabidopsis RSW1 (AtCeSA1) gene (Arioli et al., 1998). The FAI228 clone has been mapped at 4.2 centiMorgan (cM) from the top of chromosome 3 by using recombinant inbred lines and is flanked by the SGCSNP297 and the CA1 markers (http://nasc.nott.ac.uk/ RI_data/html). The sequence of this genomic region has been released in October 1999 as part of a bacterial artificial chromosome (BAC) clone T17B22 (accession no. AC012328). We concluded that the T-DNA had inserted into the transcribed region of a CeSA homolog for which no function had been described yet.
Figure 1.
AtCSLD3 gene structure, T-DNA insertion site, and restriction sites used for sub-cloning. Boxes are exons and lines are introns. The sizes (bp) are indicated. The position of the T-DNA in the last exon is indicated by an arrowhead. L, Left border; R, right border. Direction of transcription is indicated as a horizontal arrow below the AtCSLD3 gene structure. The clones used for complementation were an XbaI (X) clone of 12 kb and a SalI (S) clone of 5.38 kb. The inversed PCR fragment obtained after an ApoI (A) digest is shown in a full line; this fragment was 100% homologous with the FAI228 cDNA clone and EST Z35212.
A 12-kb XbaI genomic clone containing the entire CeSA homolog was isolated from the BAC clone T16F11 and sequenced. To determine the gene structure the genomic sequence was compared with a cDNA clone amplified from a cDNA library (CLONTECH) of 3-week-old vegetative tissue by using the primers no. 49 and no. 39 (Fig. 2). The gene consisted of three exons and two introns as predicted also by the program GeneMark.hmm (Figs. 1 and 2). The first (AG/GC) and second (AG/GT) intron/exon splice site sequences conform to the consensus sequence for plant introns. The ATG translation initiation codon is the first codon of the first exon and the open reading frame is 1,145 amino acids. The CeSA homolog was identified as AtCSLD3, a member of the cellulose synthase-like subfamily D, the AtCSL subfamily most closely related to the AtCeSA genes (Richmond and Somerville, 2000; http://cellwall.stanford.edu/ cellwall/Arabidopsis). Between plant CSL and CeSA genes three variable regions and three conserved regions are spaced around the D, D, D, and QXXRW motifs of the catalytic glycosyltransferase domain according to Delmer (1999) and Richmond and Somerville (2000). The alignment of AtCSLD3 with the other members of the AtCSLD family and with the two AtCeSA genes, RSW1 and IRX3, is shown in Figure 3. Within the AtCSLD subfamily, only two variable regions are found, VR1 at the NH2 terminus and VR3 between the second and the third D residue. The VR1 region in AtCSLD3 spans almost the entire exon 1 (Figs. 2 and 3). The VR3 region is longer between the AtCSLD subfamily and the AtCeSA genes than within the D subfamily (Fig. 3). Because there is no obvious VR2 variable region between the first and second D residue within the D subfamily, two blocks of conservation instead of three are present (Fig. 3). The T-DNA integrated into the third highly conserved region among plant CeSA genes that is part of the catalytic glycosyltransferase domain, at the end of the third membrane-spanning domain (Figs. 2 and 3); as a consequence, the catalytic domain is probably intact, but the six predicted membrane-spanning domains at the carboxyl terminus will be missing in the mutated CSLD3 gene product.
Figure 2.
Nucleotide and deduced amino acid sequence of the AtCSLD3 gene. The SalI genomic fragment was sequenced. Numbers on the left correspond to the nucleotide (top) and amino acid (bottom) position. The asterisk indicates the stop codon. The primers for reverse transcription (RT)-PCR are nos. 38, 60, and 42, and for cDNA amplification are nos. 49 and 39. The positions of the three Asp residues (D) and QXXRW motifs are underlined. The T-DNA position is indicated with an arrowhead.
Figure 3.
Alignment of the deduced amino acid sequences of the members of the AtCSLD gene family and two AtCeSA genes, RSW1 and IRX3. The identical amino acids are in red, the conservative amino acid changes are in blue, and the divergent ones in black. The positions of the three Asp residues (D) and the QXXRW motifs are indicated by asterisks. The three variable regions in the proteins are indicated by VR1, VR2, and VR3. The eight membrane-spanning domains are indicated with lines above the protein sequences.
T-DNA Induces the csld3-1 Mutation
A root hair-defective mutation, csld3-1 segregated 128 wild-type and 39 mutant seedlings in an F2 population derived from a cross of the CT-17 line with wild type. These data showed that the mutation was nuclear recessive (χ2 (3:1) = 0.29, P > 0.5). The root hair phenotype was obvious when mutant seedlings were grown for 3 to 4 d in vitro in a vertical orientation. csld3-1 mutants initiated root hair bulges from the primary root but their elongation was severely reduced (Fig. 4, A and B). At the root-hypocotyl junction (collet), fewer and shorter root hairs were formed in the mutant (Fig. 4, C and D).
Figure 4.
Phenotype of the csld3-1 mutant visualized by stereomicroscope (A, B, M, and N), differential interference optics (E and F), confocal microscope after propidium iodide (PI) staining (C, D, and G–L), and by dark-field microscope (O and P). Wild-type (A) and mutant 4-d-old seedlings (B); root-hypocotyl junction of wild type (C) and of mutant 4- to 7-d-old seedlings germinated in vitro (D); E, wild-type root hair initiation (E1) and elongation at consecutive positions along the apical-basal axis (E2–E4); F, mutant root hair initiation (F1) and elongation at consecutive positions along the apical-basal axis (F2–F6). Tip growth is illustrated in F1 and F3, leaking root hair tips in F2 and F5, and highly vacuolated root hairs in F4 and F6. G, Wild-type elongating root hair; H–L, mutant root hairs: root hair initiation zone (K), root hair elongation zone (G, H, and L), root hair differentiation zone (I and J), reduced root hair growth (H–J), bulges are indicated by arrowheads (I and L), cytoplasm leakage at the root hair tip is indicated by arrows (F2, F5, H, and L), wavy root hair (H and I), and branched root hairs (J). Additional growing points are indicated with asterisks (J); cells in root hair files of the mutant are stained with PI (indicated by white arrowhead) at the beginning of the differentiation zone (K); root hairs from callus of wild type (M) and mutant (N); pollen tube growth in stigma of wild type (O) and mutant (P). c, Callose plugs; h, hypocotyl; r, primary root. The bar in the upper left corner represents 1.6 mm (A, B), 500 μm (M, N), 150 μm (O, P), 120 μm (C, D), 50 μm (K), 40 μm (H–J, L), 36 μm (G), and 19 μm (E, F).
Southern analysis showed the presence of a single T-DNA copy in the csld3-1 genome (data not shown). Two mutant lines derived from a mutant crossed to wild type were analyzed in the F3 population: 2,000 individuals all displayed the root hair-defective phenotype and were resistant to kanamycin (Km), indicating that the mutation and the T-DNA were linked. F2 individuals (142) derived from a mutant crossed to wild type segregated 66 wild-type KmR, 32 wild-type KmS, 44 csld3-1 KmR, and no KmS mutants, implying that no recombination between T-DNA and mutation had occurred. The maximum genetic distance between csld3-1 and KmR was calculated as 6.7 ± 0.4 cM according to Koornneef and Stam (1987).
The genomic region of AtCSLD3 was comprised within a 12-kb XbaI and in a 5.38-kb SalI genomic fragment (Figs. 1 and 2). These fragments were cloned into the plant transformation vector PGSC1704 and introduced into the csld3-1 mutant. Ten transgenic lines were obtained with either construct. T2 seeds were germinated on germination medium (see “Materials and Methods”) and sensitivity and resistance to hygromycin (the selectable marker of the PGSC1704 plant transformation vector) as well as the root hair phenotype were scored. Each one of the 1,884 hygromycin-resistant T2 individuals obtained formed normal root hairs. The 5.38-kb genomic AtCSLD3 region containing 1,200 bp of the 5′ upstream region was sufficient to restore the mutant root hair phenotype to wild type.
The csld3-1 Mutation Specifically Affects Root Hair Growth
The root hair phenotype was analyzed in detail using differential interference contrast optics. Phytagel blocks containing the seedlings were removed from the tissue culture plates and mounted onto a slide. In this way the tissue remained in situ and the only manipulation was the addition of a coverslip. In wild type, root hairs originate at the basal part of the hair cell and grow into a bulge. The hairs subsequently elongate by polarized growth, the so-called tip growth, apparent by the vesicle-rich cytoplasmic region at the root hair tip, the reverse fountain-type of cytoplasmic streaming, and a central vacuole at the basal part of the root hair. The initiation and growth of a root hair in wild type in four consecutive cells of a cell file is shown in Figure 4 (E1–E4). The mutant formed bulges at the right position, but growth at consecutive positions along the apical-basal axis was severely reduced (Fig. 4, F1–F6; number of primary roots analyzed per line is n = 20). Mutant hairs contained a vesicle-rich tip at initiation (Fig. 4, F1 and F3) and rapidly became highly vacuolated (Fig. 4, F4 and F6). A high number of mutant hairs leaked cytoplasm from their tips, namely 175 out of 200 (78%) in the root hair initiation and elongation zone (n = 10), indicating that the tensile strength of the cell wall was reduced (Fig. 4, F2, F5, H, and L). No leaking tips were observed in wild type. Leaking tips were never observed in mutant root hairs at the differentiation zone, implying that leakage was correlated with root hair elongation. The mutant root hair phenotype was similar when grown on higher concentrations of phytagel (5 or 7 g L−1).
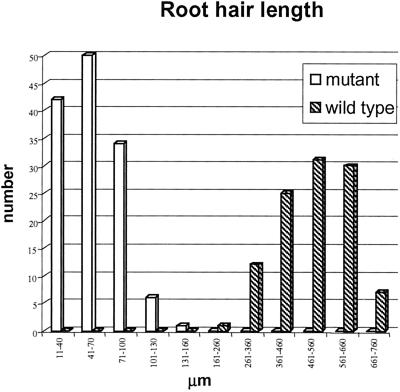
Confocal microscopy was done on primary roots stained with propidium iodide (PI) to visualize the cell's periphery. The meristematic and elongation zone of the mutant primary root was indistinguishable from wild type. In the differentiation zone where root hairs initiate, elongate, and maturate along an apical-basal gradient, the epidermal pattern of root hair cell files alternating with non-hair cell files was correct in the mutant. We measured root hair length starting 10 cells upward from the root hair initiation zone, in which normally rapid tip growth occurs, until the region of fully expanded root hairs. Figure 5 summarizes the distribution of the root hair lengths measured in mutant and wild type. Length intervals of 30 and 100 μm were taken for the mutant and for wild type, respectively. Wild-type root hairs had a length of approximately 200 μm at the start of the region analyzed and were growing fast to a maximum length of 700 μm when fully elongated. Root hair length increased along the apical-basal axis of the root. Mutant root hairs remained much shorter and did not increase in length along the apical-basal axis. Full-grown mutant root hairs in the differentiation zone, where root hair elongation had stopped, never reached the size of wild-type hairs (Fig. 4, I and J). Instead, their length varied between 10 and 134 μm at random positions (Fig. 5). In wild type, root hair bulges were only present at the beginning of the root hair initiation zone, below the region analyzed. In the mutant, bulges (10–40 μm) were detected throughout the differentiation zone, indicating that no tip growth had occurred (Fig. 4, I and L). Root hair bulges ectopically present in the differentiation zone, the leaking root hair tips, and the severely reduced root hair length in the mutant demonstrated that specifically root hair elongation had been affected by the mutation. The ectopic presence of root hair bulges in the differentiation zone suggested that threshold values of mutant proteins had not been reached to sustain tip growth in a subset of cells.
Figure 5.
Distribution of root hair lengths in mutant and wild type. Measurements were done in the root hair elongation zone at 10 cells distal from the root hair initiation up to the differentiation zone where tip growth had ceased. Length intervals of 30 and 100 μm were defined for the mutant and the wild type, respectively.
At the region of root hair initiation, mutant root hair cells were slightly stained by PI, probably through plasma membrane damage (Fig. 4, K). A few cells were observed with collapsed cytoplasm, an indication of cell death. In the upper part of the differentiation zone where tip growth had ceased, the PI staining was restricted to the cell periphery, implying that the plasma membrane defect had been restored.
The morphology of mutant root hairs was altered. Mutants formed slightly wavy instead of straight root hairs (Fig. 4, H and I); occasionally, branched root hairs were observed (Fig. 4 J). Root hair growth, which was induced from callus by a tissue culture procedure, was also severely reduced in the mutant (Fig. 4, M and N).
The tip growth of mutant pollen tubes was analyzed both in vivo and in vitro. Upon self-fertilization, pollen grains germinated on the stigma, and pollen tubes grew down through the transmitting tract of the style toward the ovary. Callose plugs that were formed at regular distances along the growing pollen tube were taken as a measure for pollen tube growth. The same growth rate was observed between mutant and wild-type pollen tubes (Fig. 4, O and P). In an in vitro pollen germination assay, the germination and growth of csld3-1 pollen could not be distinguished from that of wild-type pollen. These data show that the tip growth of the pollen tubes is not affected by the csld3-1 mutation.
The csld3-1 Mutation Has No Effect on Salt Tolerance
The function of root hairs is to aid nutrient and water uptake and to anchor the plant in the soil. Root hairs also might be important to create a microclimate to attract microorganisms, whose symbiosis is beneficial for plant growth in general. Nevertheless, some plants do not form root hairs, which creates a dilemma with respect to their function and their utility in plant growth. csld3-1 mutant seedlings grew into normal fertile plants with normal seed set under greenhouse conditions. Normal vegetative growth has been reported for other root hair-defective mutants (Schiefelbein and Somerville, 1990) and confirms that root hairs are not essential for growth. The root hair phenotype of the csld3-1 mutant is similar to that of the root hair phenotype obtained by treatment of seedlings with abscisic acid, which plays a role in the drought response (Schnall and Quatrano, 1992). This might indicate that AtCSLD3 has an effect on the drought or salt tolerance of plants. We investigated whether plant growth in the mutant would suffer more than wild type under salt stress conditions.
In a root-bending assay (Wu et al., 1996) a slight and strong inhibition of bending was noticed at 50 and 100 mm NaCl, respectively, in both the csld3-1 mutant and the wild type. A second experiment monitored the inhibition of expansion of the vegetative tissue (cotyledons and leaves) according to Zhu et al. (1998). Seeds were germinated onto the germination medium supplemented with 0, 50, 75, 100, 125, and 150 mm NaCl in horizontal position. After 2 weeks, germination of the seed and expansion of the leaves were scored. Expansion of the leaves in both the mutant and the wild type was slightly inhibited at 50 mm, more at 75 mm, and strongly at 100 mm NaCl. In a third experiment the growth rate of the primary roots of 4-d-old seedlings was monitored during 2 weeks of culture on petri dishes grown in vertical position on medium containing increasing concentrations of NaCl: 0, 50, 100, 125, and 150 mm. The growth rate was severely reduced at 100, 125, and 150 mm NaCl in the mutant and the wild type. This reduction in growth rate coincided with swelling of the elongation zone of the primary root and root hairs in that zone in differential interference contrast optics. Both phenotypes were slightly more pronounced in the mutant (n = 10). In conclusion, the growth of the primary root of the csld3-1 mutant was not more sensitive to NaCl than the wild type and the defective cell wall in the root hair tip did not hamper the water balance in the mutant.
Expression Analysis of the AtCSLD3 Gene in Arabidopsis
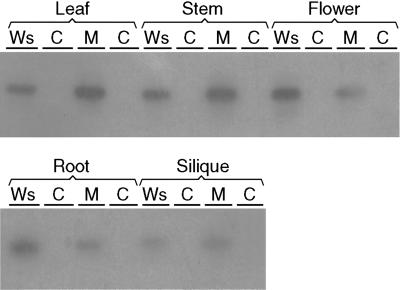
RT-PCR was performed on poly(A+)RNA of 4-d-old intact seedling as well as on the poly(A+)RNA of 3-week-old plant organs, such as roots, leaves, flower buds and flowers, stems, and siliques. cDNA was synthesized using the gene-specific primer number 42 (Fig. 2). Alignment of the six members of the AtCSLD family showed that the 5′ parts of the coding sequences were divergent and this part was used to design the AtCSLD3-specific primer no. 38 (Fig. 2). At the 3′ end of primer number 38 (5′-GGCGTCTAATAATCATTTCATGA-3′), four out of five bases differed from the corresponding sequence in AtCSLD2 (5′-GGCATCTAATAAGCATTTTGATA-3′), which is the most conserved copy with AtCSLD3 in that region. This divergence is sufficient for gene specificity. The AtCSLD3 transcripts were specifically amplified from the cDNA with the gene-specific primer nos. 38 and 60 located in a conserved part of the gene (Fig. 2). Figure 6 shows the gel blots of the RT-PCR products hybridized with the AtCSLD3 gene-specific probe amplified with primer numbers 38 and 42 (Fig. 2). PCR reactions on DNAse-treated mRNA samples were negative, indicating that no DNA contamination had occurred and that the positive signals were derived from transcripts. The AtCSLD3 gene expression was not restricted to wild-type roots but was present in all organs tested: leaves, stems, flowers, roots, and siliques (Fig. 6). The number of EST clones corresponding to AtCSLD3 to date originated from cDNAs isolated from different organs: Seven ESTs were derived of roots, two of aboveground parts, and two of developing siliques (http://cellwall.stanford.edu/cellwall). We conclude that in every plant organ there is functional redundancy with the CSLD3 gene product because no phenotype was detected except for the tip growth of the root hairs where CSLD3 is limiting. In the mutant transcripts were detectable in every organ tested. The phenotype in the mutant is probably not due to a lack of CSLD3 gene product, but to an altered or less efficient gene product.
Figure 6.
Gel blots showing transcripts of AtCSLD3 in wild type (Ws) and mutant (M) plant organs. Controls consisted of PCR amplification on DNAse-treated mRNAs of each tissue. RT-PCR analysis on poly(A+) mRNA of wild-type tissue was performed with the AtCSLD3 gene-specific primer no. 38 and the primer no. 60 (see Fig. 2). Bands are visualized by autoradiography after Southern analysis with the PCR product amplified with primer nos. 38 and 42 as a probe (indicated in Fig. 2).
DISCUSSION
We have isolated the AtCSLD3 gene by T-DNA tagging. The glycosyltransferase enzymatic domain of cellulose synthases is highly conserved in the CSLD3 protein. The CSLD3 gene product most probably plays a role in cell wall synthesis. Whether it catalyzes the synthesis of cellulose or another non-cellulosic cell wall polysaccharide remains to be determined. In the corresponding mutant line, csld3-1, the T-DNA is inserted at the end of the third highly conserved region, which is common to bacterial and plant CeSA genes. This region is predicted to function in UDP-Glc binding and catalysis. The mutated CSLD3 protein is probably present because the transcript is detected in the csld3-1 mutant. The catalytic function may not be affected by the mutation, but the six carboxyl-terminal membrane-spanning domains are absent. The membrane-spanning domains presumably form a pore in the plasma membrane through which the growing glucan polymers are excreted (Delmer, 1999). Because membrane anchoring is probably affected in the mutated CSLD3 protein, excretion of the polymers may be hampered and their number reduced in the cell wall. The mutant phenotype suggests that the CSLD3 protein is an essential catalytic enzyme for the synthesis of polymers for the primary cell wall at the tip of the root hair. The root hair tips consist only of the primary cell wall and is thinner than the lateral wall of the root hair at which the secondary cell wall is deposited 25 μm below the tip (Newcomb and Bonnett, 1965). In the csld3-1 mutant the tensile strength of the cell wall of the root hair tip zone is reduced when compared with wild type. This characteristic indicates that either the type or the number of polymers represented in microfibrils of the cell wall confer distinct mechanical properties to the hair tip zone. Earlier observations showed that a reduced number of cellulose polymers affect the mechanical properties of plant cell walls (Kokubo et al., 1991; Shedletzky et al., 1992).
AtCSLD3 expression is expected to be present only in the roots because the mutant phenotype is restricted to the root hair tip. However, RT-PCR data showed that the AtCSLD3 gene is expressed in every plant organ tested. In addition, a cDNA has been isolated from green vegetative tissue and several ESTs originated from cDNA libraries of different organs. This observation suggests gene redundancy in every organ except for the root hair tip, the only site where CSLD3 appears to be limiting. The recessivity of the csld3 mutation supports the hypothesis that there is no redundancy in gene function at the root hair tip. The presence of over 40 members in the Arabidopsis CeSA superfamily (Richmond and Somerville, 2000) implies redundancy in gene function but also refers to different levels of gene regulation and genetic interactions (Delmer, 1999). Because the RT-PCR detection method does not distinguish between tissue- or cell-specific gene expression, it is still possible that the AtCSLD3 gene is transcriptionally regulated and is only expressed in certain cell types. RT-PCR expression analysis of the CeSA genes of maize and Arabidopsis showed a ubiquitous expression pattern as well, but some of the genes appeared to be cell type-specific when refined methods were used (Holland et al., 2000).
The root epidermis specification pattern is normal in the csld3-1 mutant. The root hair-forming cell files are alternated with one or two non-hair cell files. Therefore, the AtCSLD3 gene is probably not a component of the root hair cell specification pathway that is controlled by the regulatory genes TTG, GL2, and CPC (Galway et al., 1994; Di Cristina et al., 1996; Wada et al., 1997). In the mutant, root hairs initiate at the proper position in the root hair cell and the first phase of bulge formation is normal, but the subsequent vesicle-driven tip growth is reduced. Several other mutants defective in root hair formation have been identified (Schiefelbein and Somerville, 1990). These mutants have been classified with respect to their site of initiation in the root hair cell (RHD6), their localized outgrowth (RHD1), their initial growth phase (RHD2), and their polarized and straight extension (TIP1, RHD3, and RHD4; Hülskamp et al., 1998). The RHD loci probably identify components of the tip-focused growth machinery in which exocytosis, Ca2+ gradients, the cytoskeleton, and the cytoskeleton-plasma membrane-cell wall continuum play a major role (Miller et al., 1997). The reduced growth and the wavy and branched structure of a subset of root hairs in the csld3-1 mutant resemble the root hair phenotype of certain rhd mutants, such as tip1, rhd3, and rhd4. rhd3 is not allelic to csld3-1 because the RHD3 gene encodes a protein with GTP-binding motifs (Wang et al., 1997). Based on the map position, tip1 (Ryan et al., 1998) can be excluded as an allele of csld3-1. RHD4 and FAI228 (CSLD3) both map on chromosome 3 and have very similar root hair phenotypes; whether they are allelic or identify independent loci in the root hair growth pathway remains to be tested. An indication that CSLD3 might be a novel locus comes from a particular feature of the csld3-1 root hair tips: They easily leak cytoplasm, a feature not reported for the rhd mutants (Schiefelbein and Somerville, 1990).
By genetic and pharmacological analyses the ethylene and auxin signaling pathways have been demonstrated to regulate root hair growth (Masucci and Schiefelbein, 1996; Pitts et al., 1998). Based on the mutant phenotype, the AtCSLD3 gene might be a downstream target of components in these pathways. It is a late gene in the process of root hair formation because the phenotype of the corresponding mutation clearly shows a defect in the last part of the process, namely root hair elongation.
Pollen tubes and root hairs are the only two types of plant cells that enlarge by tip growth. Certain gene products, such as TIP1, are required for the directional growth of both cell types (Ryan et al., 1998). However, both cell types have distinct growth features. Root hairs grow in distinct phases (Dolan et al., 1994; Miller et al., 1997; Gilroy and Jones, 2000), in contrast to pollen tubes, which form callose plugs behind the growing point that prevent flow back of cytoplasm and enforce directional growth. These differences in growth suggest that a number of loci have to be expected that are specific for tip growth either in pollen tubes or in root hairs. Normal pollen tube germination and growth have been observed in in vivo and in vitro assays in the mutant. In the csld3-1 mutant, the root hair growth process is thus specifically affected. Some root hair cells in the mutant developed more than a bulge; others grew into a small root hair. This plasticity in the mutant phenotype indicates that the machinery of polarized growth (i.e. the tip-focused Ca2+ gradient and the actin cytoskeleton) in itself is not affected in the mutant root hairs. The position of the T-DNA in the AtCSLD3 gene interferes with the membrane anchoring of the protein so that the main effect of the mutation is probably a reduction in delivery of building polymers at the primary cell wall in the growing root hair tip.
In Arabidopsis, root hairs grow perpendicularly to the surface of the root, maintaining a single growing point. It has been demonstrated that the cytoskeleton is important for the straight growth of the root hairs. Stabilization or depolymerization of the microtubuli results in root hairs with a wavy morphology and multiple growing points (Bibikova et al., 1999). The mutant root hairs were not straight, but wavy, and some of them were branched. The altered morphology of the csld3-1 root hairs indicates that the cytoskeleton is somehow affected by the defective cell wall at the growing tip. Analogy might exist with polarization processes in other single cells, such as the fertilized egg in Fucus distichus. Local differences in cell wall composition seem to play a role in the axis fixation part of the polarization process via the formation of transmembrane bridges between cell wall fibrils and cytoskeleton filaments (Kropf et al., 1988; Quatrano and Shaw, 1997). From the phenotypic analysis of the csld3-1 mutant we postulate that the composition, number, or arrangement of cell wall polymers at the root hair tip might feed back to the cytoskeleton through the cytoskeleton-plasma membrane-cell wall continuum to sustain the directionality of the root hair tip growth.
MATERIALS AND METHODS
Plant Material, Media, and Culture Conditions
Arabidopsis ecotype Wassilewskija (Ws) was used in all experiments and was provided by Kenneth Feldmann (University of Arizona, Tucson). The line CT-17 was provided by Jan Traas (Institut National de la Recherche Agronomique, Versailles, France). CT-17 was backcrossed to wild-type Ws. One of the F2 progenies, CT–17-7, which is heterozygous for the root hair-defective mutation csld3-1, was used to obtain homozygous lines. A line homozygous for the csld3-1 mutation has been registered as CS899 and N899 at the Arabidopsis Biological Resource Center (Columbus, OH) and Nottingham Seed Stock Centre (Nottingham, UK), respectively.
Root hair formation was scored 4 to 7 d after germination of seeds on 10-cm2 petri plates, grown in a vertical orientation. A week later, young plants were transferred to soil in a growth chamber to obtain seeds and to check pollen tube growth. The plant growth conditions were as described by Van Lijsebettens et al. (1991).
DNA Isolation and Southern Analysis
Genomic DNA was isolated from the plant tissue by using standard procedures and purified on CsCl gradients (Sambrook et al., 1989). One microgram of DNA was digested with different enzymes, separated on 0.8% (w/v) agarose gel, and blotted onto Hybond N+ (Amersham Pharmacia Biotech, Little Chalfont, UK). Blots were hybridized to DNA probes labeled by random priming (Amersham Pharmacia Biotech) according to Sambrook et al. (1989). The blots were exposed overnight to x-ray film (Kodak) using an intensifying screen.
Isolation of T-DNA Flanking Sequences and Genomic Clones of AtCSLD3
The sequence that flanks the right T-DNA border was isolated by inverse PCR. The mutant DNA was digested by ApoI and self-ligated. A PCR (94°C for 45 s, 58°C for 45 s, and 72°C for 60 s) was performed with oligonucleotide primer no. 4 (GTATTGCCAACGAACCGGATACCCG) and primer no. 3 (GACTGAATGCCCACAGGCCGTCGAG). The PCR cycle parameters, dependent on the length of the amplified product, melting temperature of the primers, and primer template stability were determined by the computer program OLIGO 4 primer analysis software (Rychlik et al., 1990). The 287-bp product generated was cloned into pGEM-T vector (Promega, Madison, WI), in which 152 bp had 100% homology with an EST clone, FAI228. The FAI228 clone was ordered from the Arabidopsis Biological Resource Center and the sequence of the entire insert was determined.
A genomic restriction map was constructed of the AtCSLD3 genomic region with the BamHI, DraI, EcoRI, EcoRV, SacII, SpeI, SspI, and XbaI restriction enzymes on wild-type genomic DNA and FAI228 as a probe. To sub-clone the wild-type AtCSLD3 gene region, three-dimensional DNA pools of an Arabidopsis genomic BAC library (Texas A&M University, College Station) were screened by PCR. A first PCR was performed with a primer designed to the T-DNA-flanking sequence (primer no. 27, CAGGCCAGTTCATAGTCCAAA) combined with one complementary to the FAI228 sequence (primer no. 33, CCCAACCAAATAAAAAAACAA). A second PCR was done on positive clones with nested primers (primer no. 7, GGTGGCGAAACGAGCAGTTCTG and primer no. 32, GGCCAATCTCTGTCTCCATCT). One positive BAC clone T16F11 was recovered from this library. The genomic region corresponding to the AtCSLD3 gene was sub-cloned from this BAC clone as a 12-kb XbaI fragment into pDRSF01 (kindly provided by Peter Breyne, Ghent University, Belgium) and sequenced. A 5.38-kb genomic fragment containing the entire AtCSLD3 gene has been deposited with accession no. AJ297948. The same gene was cloned independently (Favery et al., 2001).
DNA Sequencing
The DNA sequencing analysis was performed by the dideoxy chain termination method of Sanger et al. (1977). Genomic subclones were sequenced in a cycle-sequencing protocol on both DNA strands with ABI373A and ABI377 automatic DNA sequencers (Applied Biosystems, Foster City, CA) through primer walking by fluorescent dye terminators and AmpliTaq FS (Perkin Elmer, Norwalk, CT). Primers were designed by using the program OLIGO 4 (Rychlik et al., 1990) and were synthesized on an ABI392 DNA/RNA synthesizer. The sequence readings and the corresponding electropherograms were assembled into contigs with a homemade computer program called Sequence Assembly Facility Environment. For the analysis of the T-DNA flanking sequence, the inverse PCR product amplified from the mutant DNA was sequenced by using the T7 and SP6 primers of pGEM-T.
Plant Transformation
For complementation analysis, Agrobacterium tumefaciens- mediated transformation was performed on leaf explants by hygromycin selection (Van Lijsebettens et al., 1991). The AtCSLD3 gene region was sub-cloned as a 12-kb XbaI fragment or a 5.3-kb SalI fragment into the binary plant transformation vector pGSC1704 in the Escherichia coli strain JM109 (Promega). These clones were combined with the pMP90 plasmid in the A. tumefaciens strain C58C1RifR. Bacterial cultures were grown in Luria Broth, with shaking at 200 rpm at 37°C for E. coli and 28°C for A. tumefaciens PGSC1704 is a binary plant transformation vector (provided by Johan Botterman, Plant Genetic Systems N.V., Gent, Belgium) containing a pVS1 origin of replication and a hygromycin phosphotranferase-coding region under the control of the nopaline synthase promoter of A. tumefaciens cloned between the left and right T-DNA border. A multicloning site is between the Pnos-hpt gene and the right border of the T-DNA.
Mapping
Gel-blot analyses of genomic DNA from three Arabidopsis ecotypes (Col, Ler, and Ws), which were digested with different restriction enzymes, were performed with FAI228 as a probe. Because this clone corresponds to the 3′-divergent region of the AtCSLD3 gene, single bands were obtained. An RFLP was found between Landsberg erecta and Columbia ecotypes by using a MunI restriction enzyme digest and FAI228 (an EST corresponding to the 3′-divergent end of the AtCSLD3 gene) as a probe. The AtCSLD3 gene has been mapped by using a set of 100 recombinant inbred lines (Lister and Dean, 1993).
Microscopy
Seeds were germinated on mineral salts (De Greef and Jacobs, 1979), 1% (w/v) Suc, 0.5 g L−1 2-(N-morpholino)ethanesulfonic acid (pH 5.8), and 3.5 g L−1 phytagel (Sigma-Aldrich, St. Louis) and grown vertically. Phytagel blocks containing the seedlings were removed from the tissue culture plates and mounted onto a slide. A drop of liquid medium and a coverslip were added for differential interference contrast optics to analyze tip growth (Leica DMLB, Heerbrugg, Switzerland). Primary roots of 4- to 7-d-old seedlings mounted in the same way were stained for 5 to 60 min by adding a drop of PI solution (10 μg mL−1 dissolved in water) and visualized with a confocal microscope (LSM510, Zeiss, Jena, Germany) with 543-nm excitation and 505- to 530-nm emission lines to measure root hair length. The images were processed by using the laser scanning microscope images assembled using Adobe Photoshop 5.
Callus Induction Assay
A callus induction assay was used to test resistance versus sensitivity of segregating seedlings for the genetic linkage analysis. Root explants of 3-week-old seedlings were excised and transferred to callus-inducing medium (Valvekens et al., 1988) with 50 mg L−1 kanamycin. After 2 weeks of in vitro culture, resistant roots produced callus with root hairs, whereas sensitive roots did not. This assay was also used to test root hair formation from callus in the csld3-1 mutant.
RT-PCR Analysis
Poly(A+) RNA was prepared with the help of QuickPrep Micro mRNA Purification Kit (Amersham Pharmacia Biotech). After the mRNA was isolated, it was treated with DNAse at 37°C for 2 h and at 65°C for 30 min. For RT-PCR, first-strand cDNA was synthesized with approximately 60 to 90 ng of mRNA sample in a reaction using the Superscript Preamplification System for first Strand cDNA synthesis kit (Gibco/BRL Life Technologies, Gaithersburg, MD). cDNA synthesis was performed with the gene-specific primer no. 42 (5′-CCGGGATTGAATCGATAAGGAAA-3′). To detect the AtCSLD3 gene transcript, the gene-specific primers no. 38 (5′-GGCGTCTAATAATCATTTCATGA-3′; just downstream of the AtCslD3 translation start site) and no. 60 (5′-CCCTACAGATTTTGAAATCACAC-3′) were used (Fig. 2). As negative control PCR was done on DNAse-treated mRNA for each tissue sample. PCR conditions were: 95°C for 5 min followed by 40 cycles at 94°C for 60 s, 54°C for 60 s, and 72°C for 3 min, and 72°C for 2 min. RT-PCR products were blotted to Hybond N+ (Amersham Pharmacia Biotech) and hybridized with a PCR product of primers nos. 38 and 42 (Fig. 2).
Pollen Assays
Anthers were removed from wild-type (Ws) or mutant (csld3-1) plants and used to transfer pollen to the stigma of wild-type flowers. The stigma surface was completely saturated with pollen. After 5 h of pollination, the stigmas were detached and immersed overnight in 70% (v/v) ethanol to stop pollen tube growth. Stigmas were then placed in a drop of fresh 0.05% (w/v) water-soluble aniline blue in 50 mm sodium phosphate buffer (pH 8.5). Samples were squashed prior to mounting on glass slides and observed with the UV fluorescent microscope. For the in vitro pollen tube growth assays, pollen grains were incubated in vitro at 25°C for 20 h on a microscopical slide with a drop of pollen growth medium: 10% (w/v) Suc, 0.01% (w/v) boric acid, and 3 mm calcium nitrate. The microscope slides were stored in humid chambers. Pollen germination was analyzed with a stereomicroscope.
Salt Tolerance
Bending assay of the roots were done according to the method described by Wu et al. (1996). Seeds were germinated on Murashige and Skoog salts, 0.5 g L−1 2-(N-morpholino) ethanesulfonic acid, 1% (w/v) Suc (pH 5.7), 0.9% (w/v) agar (Lab M Plant tissue culture) without NaCl in horizontal position for 7 d. The seedlings were transferred to plates containing the same medium but supplemented with 0, 50, 100, or 150 mm NaCl and grown upside down in vertical position; 24 h later the capacity of the roots to bend and grow downward was scored.
ACKNOWLEDGMENTS
The authors thank Tom Gerats and Dominique Van Der Straeten for helpful suggestions and critical reading of the manuscript and Martine De Cock for help in preparing it.
Footnotes
This work was supported by a grant from the Fund for Scientific Research, Flanders (grant no. G.0075.97).
LITERATURE CITED
- Aloni Y, Delmer DP, Benziman M. Achievement of high rates of in vitro synthesis of 1,4-β-d-glucan: activation by cooperative interaction of the Acetobacter xylinum enzyme system with GTP, polyethylene glycol, and a protein factor. Proc Natl Acad Sci USA. 1982;79:6448–6452. doi: 10.1073/pnas.79.21.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 1999;17:657–665. doi: 10.1046/j.1365-313x.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C, Caboche M. A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C R Acad Sci Paris Life Sci. 1993;316:1188–1193. [Google Scholar]
- Cutler S, Somerville C. Cellulose synthesis: cloning in silico. Curr Biol. 1997;7:R108–R111. doi: 10.1016/s0960-9822(06)00050-9. [DOI] [PubMed] [Google Scholar]
- De Greef W, Jacobs M. In vitro culture of the sugar beet: description of a cell-line with high regeneration capacity. Plant Sci Lett. 1979;17:55–61. [Google Scholar]
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. Clonal relationship and cell patterning in the root epidermis of Arabidopsis. Development. 1994;120:2465–2474. [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonk K, Steer M, Shaw P, Dolan L. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001;15:79–89. doi: 10.1101/gad.188801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- Gilroy S, Jones DL. Through form to function: root hair development and nutrient uptake. Trends Plant Sci. 2000;5:56–60. doi: 10.1016/s1360-1385(99)01551-4. [DOI] [PubMed] [Google Scholar]
- Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP. A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol. 2000;123:1313–1323. doi: 10.1104/pp.123.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M, Folkers U, Grini PE. Cell morphogenesis in Arabidopsis. BioEssays. 1998;20:20–29. doi: 10.1002/(SICI)1521-1878(199801)20:1<20::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kokubo A, Sakurai N, Kuraishi S, Takeda K. Culm brittleness of barley (Hordeum vulgare L.) mutants is caused by smaller number of cellulose molecules in cell wall. Plant Physiol. 1991;97:509–514. doi: 10.1104/pp.97.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Stam P. Procedure for mapping by using F2 and F3 populations. Arabidopsis Inf Serv. 1987;25:35–40. [Google Scholar]
- Kropf DL, Kloareg B, Quatrano RS. Cell wall is required for fixation of the embryonic axis in Fucus zygotes. Science. 1988;239:187–189. doi: 10.1126/science.3336780. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DD, de Ruijter NCA, Emons AMC. From signal to form: aspects of the cytoskeleton-plasma membrane-cell wall continuum in root hair tips. J Exp Bot. 1997;48:1881–1896. [Google Scholar]
- Newcomb EH, Bonnett HT., Jr Cytoplasmic microtubule and wall microfibril orientation in root hairs of radish. J Cell Biol. 1965;27:575–589. doi: 10.1083/jcb.27.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1998;16:553–560. doi: 10.1046/j.1365-313x.1998.00321.x. [DOI] [PubMed] [Google Scholar]
- Potikha T, Delmer DP. A mutant of Arabidopsis thaliana displaying altered patterns of cellulose deposition. Plant J. 1995;7:453–460. [Google Scholar]
- Quatrano RS, Shaw SL. Role of the cell wall in the determination of cell polarity and the plane of cell division in Fucus embryos. Trends Plant Sci. 1997;2:15–21. [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan E, Grierson CS, Cavell A, Steer M, Dolan L. TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol. 1998;138:49–58. [Google Scholar]
- Rychlik W, Spencer WJ, Rhoads RE. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990;18:6409–6412. doi: 10.1093/nar/18.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Lin FC, Brown RM., Jr Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol. 1990;15:673–683. doi: 10.1007/BF00016118. [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C. Genetic control of root hair development in Arabidopsis thaliana. Plant Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnall JA, Quatrano RS. Abscisic acid elicits the water-stress response in root hairs of Arabidopsis thaliana. Plant Physiol. 1992;100:216–218. doi: 10.1104/pp.100.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedletzky E, Shmuel M, Trainin T, Kalman S, Delmer D. Cell wall structure in cells adapted to growth on the cellulose-synthesis inhibitor 2,6-dichlorobenzonitrile: a comparison between two dicotyledonous plants and a graminaceous monocot. Plant Physiol. 1992;100:120–130. doi: 10.1104/pp.100.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible W-R, Cutler S, Somerville CR, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens M, Vanderhaeghen R, Van Montagu M. Insertional mutagenesis in Arabidopsis thaliana: isolation of a T-DNA-linked mutation that alters leaf morphology. Theor Appl Genet. 1991;81:277–284. doi: 10.1007/BF00215734. [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 1997;11:799–811. doi: 10.1101/gad.11.6.799. [DOI] [PubMed] [Google Scholar]
- Wu S-J, Ding L, Zhu J-K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]