Abstract
Given the presence of Src and PTP1B within rat brain mitochondria, we have investigated whether PTP1B regulates Src activity in mitochondria as in the cytosol. Results showed that Src was stimulated by in vitro addition of ATP to mitochondria, and this stimulation was reversed by a membrane-permeable allosteric inhibitor of PTP1B and by a potent selective Src inhibitor. They also indicated a direct action of PTP1B on phosphorylated tyrosine 527 residue of Src, thus implicating a role for PTP1B in the modulation of Src activity in mitochondria. Putative Src and PTP1B substrates were identified by liquid chromatography tandem mass spectrometry and two-dimensional blue native/SDS-PAGE. Both inhibitors inhibited ADP-stimulated respirations concurrently with Src activation and complex IV activation by ATP, while having no effect or increasing the activity of the other complexes. Our analysis emphasizes the regulatory function of Src and its modulation by PTP1B on oxidative phosphorylation in mitochondria.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0573-6) contains supplementary material, which is available to authorized users.
Keywords: Src kinase, PTP1B, Mitochondria, Oxidative phosphorylation, Regulation, Signaling, Respiration
Introduction
The main part of the energy used by the cell is produced by mitochondria through oxidative phosphorylation (OxPhos). During this process, reducing equivalents are transferred from the Krebs cycle to electron transport chain complexes. The course of electrons through the complexes I, II, III, and IV allows for the reduction of O2 to form H2O and the passage of protons (except for the second complex) to the mitochondrial intermembrane space to form a proton gradient that is used by the ATP synthase (complex V) to produce ATP [1].
Since the mitochondrial energetic production depends on various functions and demands of the cell, the OxPhos process needs tight regulation by different mechanisms [2]. One major known pathway of physiological regulation is phosphorylation of proteins. Numerous kinases are specifically targeted to mitochondria where they modulate fusion, fission, apoptosis, and metabolism and where they phosphorylate numerous proteins in all mitochondrial compartments (for reviews see [3, 4]). After being underestimated for years, phosphorylation/dephosphorylation on tyrosine residues is now coming to be seen as an important regulation pathway of OxPhos enzymes [5], with the discovery that many components of the respiratory chain complexes were tyrosine-phosphorylated. For example, the tyrosine kinase Src was shown to phosphorylate an unknown tyrosine (Y) residue of complex IV subunit II in osteoclasts, resulting in activation of the enzyme [6, 7]. Treatment of hepatocyte with tissue necrosis factor α stimulates an unknown tyrosine kinase to phosphorylate Y304 residue in complex IV subunit I, resulting in inactivation of the complex [8]. The flavoprotein of complex II was shown to be phosphorylated by Fgr, but the resulting effect was not known [9]. The γ chain of the ATP synthase monomers was shown to be phosphorylated on Y52, and this phosphorylation was postulated to regulate the central stalk rotation of the enzyme [10]. Complex IV activity is inhibited when its substrate cytochrome c is phosphorylated on Y48 [11]. Recently, Src and Lck-mediated phosphorylation of Y190 and Y194 in adenine nucleotide translocase was shown to be involved in its carrier function and in cell respiration [12]. Therefore, the data are in favor of tyrosine phosphorylation being a major factor in the regulation of mitochondrial bioenergetics.
Although further studies are required to identify other tyrosine-phosphorylated proteins and the kinases involved and to decipher their role in mitochondrial bioenergetics, an additional aspect worth studying is the interregulation of signaling proteins in mitochondria. Several members of the Src kinase family, including Src, Lyn, Fyn, Fgr, the regulatory C terminal Src kinase (Csk) [13, 14], and the tyrosine phosphatases PTP1B [15] and SHP2 [16] are present in rat brain mitochondria. It is well known that activation and inhibition of cytosolic Src kinases depend on the phosphorylation status of two key Y residues: Y416 and Y527 [17–19]. C-terminal Y527 is phosphorylated by Csk, enabling phosphorylated Y527 (pY527) to bind to the SH2 domain of Src kinases, thus resulting in the stabilization of a restrained and inactive form of these enzymes [17]. Dephosphorylation of pY527 by tyrosine phosphatases such as SHP2 or PTP1B induces a disruption of the SH2 domain–pY527 interaction [17, 18]. This opens the catalytic domain and facilitates autophosphorylation of Y416 (pY416) in the catalytic site leading to activation of Src kinases [19].
In this study, we used rat brain mitochondria as a model to study the occurrence of the regulation of mitochondrial Src by PTP1B, taking advantage of the fact that both enzymes are found in mitochondria from this tissue [20]. To address this issue, we used a specific membrane-permeable allosteric PTP1B inhibitor, 3-(3,5-dibromo-4-hydroxy-benzoyl)-2-ethyl-benzofuran-6-sulfonic acid-(4-(thiazol-2-ylsulfamyl)-phenyl)-amide (PTP1Bi) [21], and studied its effect on Src activation, using phosphospecific Src antibodies. The results were compared to the effect of 4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine (PP2), a well known potent and selective Src kinase inhibitor [22]. We provided evidence that PTP1B positively regulates Src activity by dephosphorylating the pY527 residue in mitochondrial Src. This was shown in in vitro experiments and also under more physiologic conditions as represented by studies of ADP-stimulated respiration linked to catalytic activities of the respiratory complexes. Finally, several mitochondrial proteins were shown to be putative Src and PTP1B substrates.
Materials and methods
Chemicals
The mouse monoclonal antibody to phosphotyrosine (PY20), PY20 directly coupled to agarose, the mouse monoclonal antibody to c-Src, and protein A/G PLUS agarose conjugate were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to phosphorylated and nonphosphorylated Src species (pY416-Src, pY527-Src, non-pY416-Src, and non-pY527-Src) were bought from Cell Signaling. The antibody to v-Src directly coupled to agarose, the Src kinase inhibitor PP2, and the PTP1B inhibitor (PTP1Bi) were from Calbiochem. All antibodies against OxPhos complexes were bought from Mitosciences. Protease and phosphatase cocktail inhibitors were bought from Roche. All other chemicals were from Sigma.
Isolation of mitochondria
Male Wistar rats were sacrificed by cervical shock and decapitation. Brain mitochondria were isolated from whole brain, as described by Clark and Nicklas [23] and further purified on a discontinuous Ficoll gradient. Protein content was measured by Biuret method. After isolation, 1 mg mitochondrial fractions were immediately stored at −80°C for further experiments. All oxygraphic measurements were carried out within 3 h with freshly isolated mitochondria.
In vitro Src regulation
Rat brain mitochondria were thawed and samples preincubated in parallel at 30°C for 10 min in kinase buffer (10 mM Tris-HCl, pH 7.5, 8 mM MgCl2) containing 20 mM oligomycin and 30 ng/ml rotenone, together with either 30 μM PP2, 50 μM PTP1Bi, or 10 mM NaF plus 2 mM orthovanadate. Then, 1 mM ATP was added, except in the control sample, and incubation continued for another 10 min period. The samples were supplemented with protease and phosphatase cocktail inhibitors and centrifuged at 12,000 rpm (4°C), and proteins in the pellet were immediately solubilized in 1× denaturing Laemmli buffer. In some experiments, mitochondrial Src was immunoprecipitated from samples as described below.
Oxygraphic experiments
Mitochondrial oxygen consumption was monitored with fresh mitochondria at 30°C in a 1 ml thermostatically controlled chamber equipped with a Clarke oxygen electrode, in the following respiration buffer: 75 mM mannitol, 25 mM sucrose, 100 mM KCl, 10 mM Tris-phosphate, 10 mM Tris-HCl, 50 mM EDTA, pH 7.4. Inhibitors (or DMSO for the control, never exceeding 0.8%) were incubated with mitochondria (1 mg/ml) in the oxygraphic cuvette for 3 min before addition of 10 mM pyruvate, 10 mM malate, and 2 mM ADP. Control values for respiration rates were 32.6 ± 2.4 for state 2, 56.8 ± 7.0 for state 3 (or ADP-stimulated respiration), with RCR = 1.8 ± 0.2 (means ± SD, n = 8 different mitochondrial preparations). Respiratory control ratios (RCR) were determined from the ratio between state 2 (pyruvate and malate) and state 3 (pyruvate and malate plus ADP). Preliminary experiments showed that states 2 and 4 were equivalent.
Src immunoprecipitation
Mitochondria were solubilized with n-dodecyl-maltoside (2 mg/mg protein) in the presence of protease and phosphatase cocktail inhibitors as described in [15]. After solubilization, samples were centrifuged and immunoprecipitation was performed on supernatant with the antibody to c-Src for 1 h at 4°C. Protein A/G agarose beads (20 μL) were then added and incubation continued overnight at 4°C with constant rocking. The beads were washed three times with PBS (4 mM KH2PO4, 8 mM Na2HPO4, 140 mM NaCl, 2.7 mM KCl, pH 7.3) containing protease and phosphatase inhibitors. Proteins were eluted from the beads with 1× Laemmli buffer, boiled with 5% β-mercaptoethanol, and processed for Western blotting.
Because pY527-Src was not immunodetected when immunoprecipitation was performed with the antibody to c-Src (whose epitope is between amino acids 506 and 537 at the C-terminus of c-Src), we also used an antibody to v-Src (that does not contain the C-terminus tail, and whose epitope is within the SH3 domain of Src, and therefore is not sensitive to the phosphorylation status of Y416 or Y527).
Enzymatic activities of electron transport system complexes and of ATP synthase
Enzymatic activities were measured as in [20]. When required, mitochondria were pre-incubated for 10 min at 30°C with 30 μM PP2 or 50 μM PTP1Bi, before addition of 1 mM ATP. The Src phosphorylation status was controlled at the end of complex I and V enzymatic assays, as these were performed with 40 and 30 μg/ml of mitochondria, respectively. The samples, supplemented with protease and phosphatase inhibitors, were taken from the spectrophotometric cuvettes and centrifuged at 6,000g (4°C) for 5 min. The pellets were solubilized in 1× denaturing Laemmli buffer, and Western blotted with pY416-Src antibody. This control could not be performed for complexes III and IV due to the small amount of mitochondria used in the assays (3 and 5 μg, respectively).
Two-dimensional electrophoresis
A 400 μg sample of purified mitochondria was subjected to in vitro treatments (see above), pelleted at 13,000 rpm at 4°C, and processed for BN-PAGE according to slightly modified protocols from [24, 25]. In brief, mitochondria were solubilized with n-dodecyl-maltoside (2.5 g mg/mg protein) in 1 M 6-aminocaproic acid, 50 mM BisTris, pH 7.0 buffer, supplemented with glycerol and Coomassie Blue, and subjected to 5–13% gradient BN-PAGE, which was directly transferred to PVDF membrane for immunodetection of tyrosine-phosphorylated proteins in the native complexes. Alternatively, the treated samples were processed for BN-PAGE, the lanes were cut out and processed for the second dimension on 12.5% SDS-PAGE after denaturation and reduction in 1% sodium dodecyl sulfate (w/v) and 1% (v/v) β-mercaptoethanol. The second-dimension gels were immunoblotted for detection of tyrosine-phosphorylated proteins. A second-dimension gel was kept for Coomassie blue coloration. Tyrosine-phosphorylated spots appearing in the Coomassie blue gel were excised and proteins identified by nano liquid chromatography tandem mass spectrometry (nano LC-MS/MS).
Gel electrophoresis and Western blotting
Samples were subjected to SDS-PAGE (12.5%), and separated proteins were transferred onto nitrocellulose or PVDF membranes as already described [15]. Phosphotyrosines were revealed with PY20 antibody while OxPhos complexes were revealed with antibody against complex I 39 kDa subunit, complex II 70 kDa subunit, complex III core 2 protein subunit, complex IV subunit III, and complex V α subunit. Primary antibodies were revealed with horseradish peroxidase-conjugated F(ab′)2 fragment of anti-mouse or anti-rabbit IgG, F(ab′)2 fragment-specific (Jackson ImmunoResearch Laboratories, West Grove, PA). Blots were visualized using enhanced chemiluminescence (ECL) Plus from Amersham. Labelings were quantified by densitometric analysis using Image J (NIH) software. Control and treated groups were run in parallel in order to minimize differences in handling throughout the experiments.
Proteomic analyses
The spots of interest were digested by trypsin as indicated in Pocaly et al. [26]. Peptides were further analyzed by nano liquid chromatography coupled to a MS/MS LTQ-Orbitrap XL mass spectrometer (Thermo-Finnigan, San Jose, CA). Peptides were identified with SEQUEST through the Bioworks 3.3.1 interface (Thermo-Finnigan, Torrence, CA, USA) against a subset of the UniProt database restricted to Rattus novergicus entries (UniProtKB, Release 2010_09, August 2010, 32,482 entries). Peptides were validated using the following criteria: DeltaCN ≥ 0.1, Xcorr ≥ 1.5 (single charge), 2.0 (double charge), 2.5 (triple charge), 3.0 (quadruple charge), peptide probability ≤ 0.001. Proteins were validated as soon as two different peptides were validated.
Statistical analyses
STATISTICA (7.1) was used for all statistical analyses. One-way ANOVA (with treatment as factor) was used to compare means among treatments for all parameters measured. The normality assumption was verified with the Kolmogorov-Smirnov test, and variance equality was confirmed with the Levene test. Significance was assessed at the 0.05 (or lower) level for all tests.
Results
In vitro study of mitochondrial Src kinase regulation
Assuming that PTP1B can upregulate Src activity in mitochondria, we examined the effect of PTP1Bi on the global tyrosine phosphorylation protein profile in mitochondria incubated in the presence of ATP, in comparison to the specific Src inhibitor PP2.
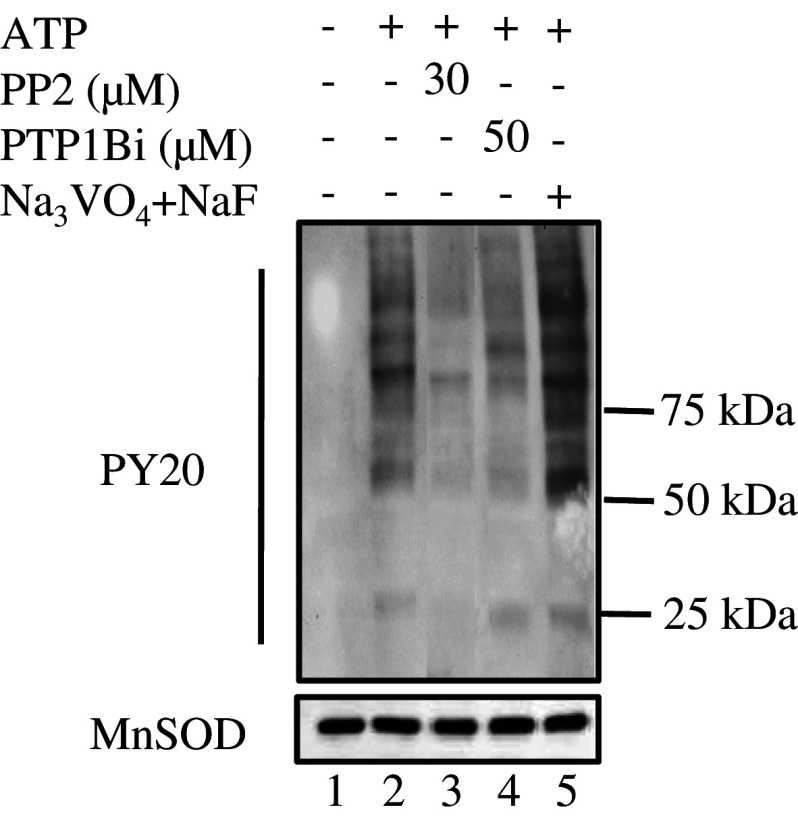
Incubation with exogenous ATP induced the appearance of tyrosine-phosphorylated mitochondrial proteins compared to control samples (Fig. 1; lanes 1 and 2). The maximum increase was observed in the presence of phosphatase inhibitors (lane 5). When compared to the ATP treatment, the global tyrosine phosphorylation level decreased with either 30 μM PP2 or 50 μM PTP1Bi (lanes 3–4). These inhibitor concentrations were the ones that gave a net and recurring decrease in the global tyrosine phosphorylation level.
Fig. 1.
Changes in protein tyrosine phosphorylation of rat brain mitochondria following exposure to ATP, PP2, PTP1Bi, or orthovanadate + NaF. Mitochondria (50 μg) were untreated (lane 1) or treated at 30°C for 10 min with 1 mM ATP alone (lane 2) or after pre-incubation at 30°C for 10 min with 30 μM PP2 (lane 3), 50 μM PTP1Bi (lane 4), or 2 mM orthovanadate plus 10 mM NaF (lane 5). The reaction was stopped by addition of 5× Laemmli buffer. Tyrosine-phosphorylated proteins on immunoblots were detected with an antibody to phosphotyrosine. The membrane was stripped and reprobed with an antibody to MnSOD as a loading control (below). Data are representative of at least three experiments
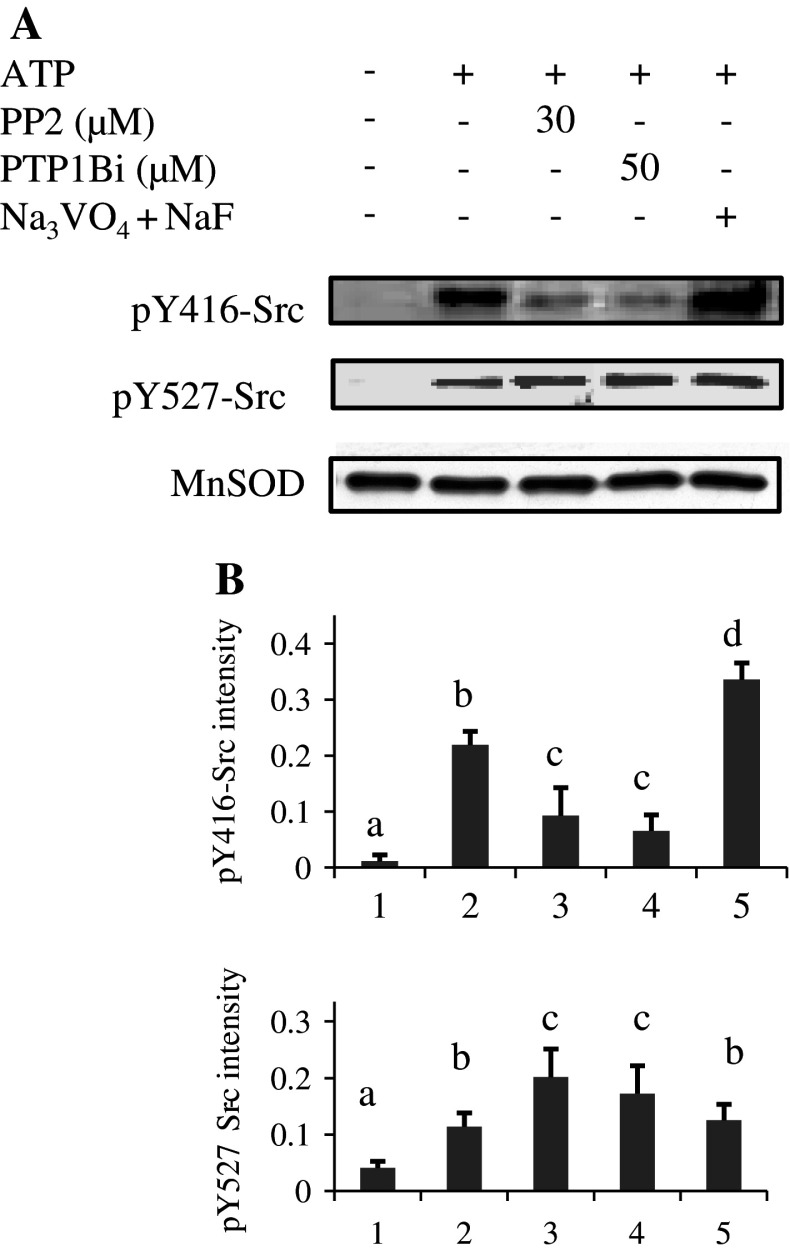
We next examined the phosphorylation status of Y416 and Y527 residues in Src with phosphospecific antibodies. The labeling intensities were quantified in relation to the MnSOD signal as a loading control. Results showed little labeling of pY416-Src in control samples (Fig. 2a, b; lane 1). Labeling increased in ATP-treated mitochondria and was greatest when mitochondria were incubated with phosphatase inhibitors prior to ATP (Fig. 2a, b; lanes 2 and 5). This latter result could reflect increased activity of Src family kinases in the presence of orthovanadate, as shown in several reports [27–29]. Indeed dephosphorylation of pY416 residue can be induced by unidentified tyrosine phosphatases and inhibited by orthovanadate [17].
Fig. 2a, b.
Phosphorylation status of mitochondrial Src in in vitro treatments. a Mitochondria, treated as described in the legend to Fig. 1, were Western blotted and immunodetected first with the anti-pY416-Src antibody, stripped and reprobed with the anti-pY527-Src antibody, stripped again and finally probed with an antibody to MnSOD as a loading control. b The intensity of pY416-Src and pY527-Src labeling was normalized over the MnSOD signal for each treatment. Data are representative of at least three experiments. Means that are not significantly different (P > 0.05) according to the Tukey test share the same letters
In the presence of PP2 or PTP1Bi, the labeling intensity of pY416-Src was clearly reduced (Fig. 2a, b; lanes 3 and 4). Concerning pY527-Src labeling, ATP treatment increased the signal compared to the control (Fig. 2a, b; lanes 1 and 2). A further significant increase was found when mitochondria were exposed to PP2 or PTP1Bi (Fig. 2a, b; lanes 3–4).
Evidence that the phosphospecific antibodies recognize mainly Src
As the phosphospecific antibodies can recognize other members of the Src kinase family, we analyzed the phosphorylation status in immunoprecipitated Src from samples collected after in vitro treatments.
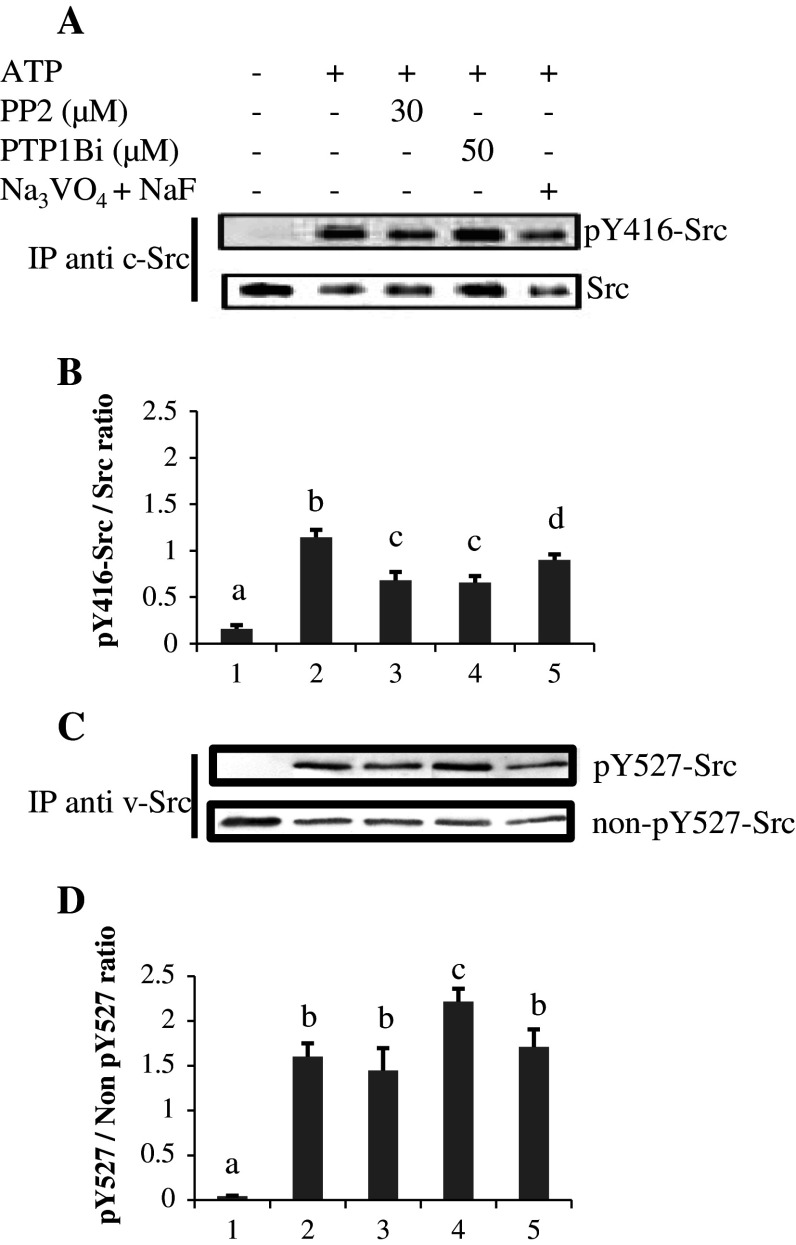
The results showed that pY416-Src labeling in immunoprecipitated Src differed in the same way as when immunodetection was performed in the whole lysate for mitochondria treated with ATP, PP2, and PTP1B (Figs. 2, 3). In the presence of phosphatase inhibitors (Fig. 3; lane 5), pY416-Src labeling was lower than in the mitochondrial lysates (Fig. 2; lane 5), reflecting here the level of active Src compared to mitochondrial lysates where several Src family kinases were probably labeled. The data also revealed that pY416 residue is not a target for PTP1B, since PTP1B inhibition would have led to increased pY416-Src labeling, as with orthovanadate.
Fig. 3a–d.
pY416-Src and pY527-Src labelings in immunoprecipitated Src from mitochondria in in vitro experiments. a Src was immunoprecipitated from mitochondria treated as described in the legend to Fig. 1, and immunoblotted first with the anti-pY416-Src antibody, stripped and reprobed with anti-Src antibody. b Normalizations of pY416-Src labeling versus Src signal. c For analysis of pY527-Src, Src was immunoprecipitated with an antibody to v-Src that recognizes an epitope in the SH3 region, and immunoblotted first with an antibody to pY527-Src, stripped and reprobed with an antibody to non-pY527-Src (see “Materials and methods”). d Normalizations of pY527-Src labeling versus non-pY527-Src signal. Data are representative of three experiments. Means that are not significantly different (P > 0.05) according to the Tukey test share the same letters
As pY527-Src was not detected when immunoprecipitation was performed with the antibody against c-Src, immunoprecipitation was performed with an antibody against v-Src (see “Materials and methods”), and the pY527-Src signals were quantified in relation to the non-phosphorylated Y527-Src signals (Fig. 3c, d). The data showed similar variations as in total lysates (Fig. 2). Normalization against v-Src recovery gave similar results (result not shown).
Taken together, the data confirmed that phosphorylated Y527 residue is a substrate of PTP1B, the inhibition of which resulted in an increased phosphorylation of Y527 residue, with a concomitant decrease in the pY416-Src labeling.
Determination of tyrosine-phosphorylated proteins
In an attempt to determine Src substrates in mitochondria, we compared the pattern of tyrosine phosphorylation according to the different treatments of native OxPhos complexes obtained by BN-PAGE followed by direct transfer and immunodetection of tyrosine phosphorylation (Fig. 4a). Results showed that the presence of ATP increased the tyrosine phosphorylation of complexes, identified by the labeling of one of their subunits (Fig. 4a) and by their catalytic staining by histochemical reactions (results not shown) as already performed [15]. PP2 decreased the tyrosine phosphorylation labeling to the level of the control sample, whereas PTP1Bi was less efficient.
Fig. 4a–c.
Identification of Src-dependent phosphorylation sites among mitochondrial proteins. Mitochondria were treated as described in the legend of Fig. 1. Proteins were then solubilized with n-dodecyl-maltoside (2.5 mg/mg protein), and 100 to 400 μg per lane was subjected to BN-PAGE (5–13%). a The BN-PAGE was directly transferred to PVDF membranes, and tyrosine-phosphorylated proteins were detected with an antibody to phosphotyrosine (PY20). Membranes were then stripped and reprobed with antibodies against one subunit of each OxPhos complex. b Individual lanes corresponding to each treatment (CTL, ATP, ATP + PP2, ATP + PTP1Bi) were cut, soaked with 1% sodium dodecyl sulfate (w/v) and 1% (v/v) β-mercaptoethanol, and subjected to a second-dimension SDS-PAGE (12.5%) for immunodetection of tyrosine phosphorylation. For each lane, one second-dimension gel was kept for Coomassie blue staining. c Spots (arrows) detected both in the Coomassie blue-stained gel and in the immunoblot corresponding to the ATP treatment were analyzed by LC-MS/MS. Data are representative of at least three experiments
We next separated subunits of the different complexes by performing a second-dimension SDS-PAGE after denaturation and reduction of the corresponding BN-PAGE, and studied their tyrosine phosphorylation state. Very low phosphotyrosine signals were detected in untreated mitochondria, while several spots appeared after incubation with ATP (Fig. 4b). The level of labeling of these spots was strongly attenuated in the presence of PP2 and less in the presence of PTP1Bi (Fig. 4b).
We analyzed a dozen spots by LC-MS/MS in the corresponding Coomassie blue-stained gel (Fig. 4c). The results show that except for spots 1, 2, 8, and 11, which corresponded at least at 88% to a single protein (clathrin, the 75 kDa subunit of complex I, the subunit 2 of complex III, and aconitase, respectively) (Table 1), the other spots were composed of several proteins (see supplemental data). In Table 1 for each spot, two or three proteins representing more than about 70% of the spot are shown. Interestingly, clathrin in spot 1 and the three subunits of sodium/potassium transporting ATPase in spot 12 are found in membrane fractions according to the UniProt database. This could suggest some contamination and/or mitochondrial-associated proteins. The other identified proteins are mitochondrial and most corresponded to OxPhos complex subunits and to matricial proteins. Therefore, this experiment enabled the detection of putative tyrosine-phosphorylated proteins in mitochondria that are sensitive to PP2 and also to PTP1Bi albeit to a lesser extent.
Table 1.
Proteins identified by LC-MS/MS in the different spots selected in Fig. 4c and amounting to at least 70% of the spot
| Spot | Accession number | Protein | MS/MS scans (n) | Peptides (n) | Protein sequence coverage of peptides (%) | Molecular weight | Relative abundancea |
|---|---|---|---|---|---|---|---|
| 1 | P11442 | Clathrin heavy chainb | 254 | 57 | 37.5 | 191,475 | 99 |
| 2 | Q66HF1 | NADH-ubiquinone oxidoreductase 75 kDa subunit | 224 | 28 | 44.7 | 79,362 | 89 |
| 3 | P15999 | ATP synthase subunit alpha | 515 | 25 | 50.4 | 59,716 | 78 |
| P10719 | ATP synthase subunit beta | 214 | 25 | 71 | 56,318 | 21 | |
| 4 | P10719 | ATP synthase subunit beta | 564 | 31 | 72.6 | 56,318 | 71 |
| P15999 | ATP synthase subunit alpha | 387 | 24 | 46.6 | 59,716 | 28 | |
| 5 | P10719 | ATP synthase subunit beta | 294 | 29 | 71.6 | 56,318 | 72 |
| P32551 | Cytochrome b-c1 complex subunit 2 | 97 | 17 | 42 | 48,366 | 27 | |
| 6 | P10719 | ATP synthase subunit beta | 99 | 24 | 68 | 56,318 | 40 |
| P35435 | ATP synthase subunit gamma | 44 | 9 | 23.4 | 30,171 | 40 | |
| 7 | Q68FY0 | Cytochrome b-c1 complex subunit 1 | 87 | 10 | 28.1 | 52,815 | 46 |
| P10719 | ATP synthase subunit beta | 87 | 24 | 63.5 | 56,318 | 17 | |
| P50554 | 4-Aminobutyrate aminotransferase | 54 | 14 | 38.4 | 56,419 | 23 | |
| 8 | P3255 | Cytochrome b-c1 complex subunit 2 | 119 | 19 | 40.5 | 48,366 | 89.5 |
| 9 | P17764 | Acetyl-CoA acetyltransferase | 52 | 10 | 28 | 44,666 | 19 |
| Q8VHF5 | Citrate synthase | 44 | 10 | 22.1 | 51,833 | 51 | |
| 10 | Q05962 | ATP/ADP translocase 1 | 49 | 9 | 23.5 | 32,968 | 32 |
| P00406 | Cytochrome c oxidase subunit 2 | 27 | 4 | 24.6 | 25,925 | 40 | |
| Q09073 | ATP/ADP translocase 2 | 6 | 2 | 8 | 32,880 | 10 | |
| 11 | Q9ER34 | Aconitase hydratase | 283 | 31 | 41.3 | 85,380 | 88 |
| 12 | P06685 | Na/K-transporting ATPase subunit alpha 1b | 132 | 26 | 31 | 112,982 | 31 |
| P06687 | Na/K-transporting ATPase subunit alpha 3b | 112 | 15 | 18 | 111,620 | 27 | |
| P06686 | Na/K-transporting ATPase subunit alpha 2b | 101 | 24 | 23.5 | 112,145 | 15 |
aCalculated from the MS signal. Corresponds to the ratio of the peak area under the reconstructed ion chromatogram (RIC) of each m/z identifying the protein to the peak area of the RIC of every identified m/z
bPlasma and other membranes
Functional study of mitochondrial Src kinase regulation
ATP induced an increase in the global tyrosine-phosphorylated protein profile concurrently with the labeling of active mitochondrial pY416-Src. PP2 and PTP1Bi inhibited these effects similarly, but PTP1Bi less efficiently inhibited tyrosine phosphorylation labeling in Western blots derived from 2D BN-PAGE/SDS/PAGE than from 1D SDS-PAGE. Since ATP is produced during ADP-stimulated respiration (state 3), we examined the effect of the inhibitors during this step of mitochondrial respiration. Results showed that both inhibitors inhibited ADP-stimulated respiration, with PTP1Bi being the most efficient (Table 2).
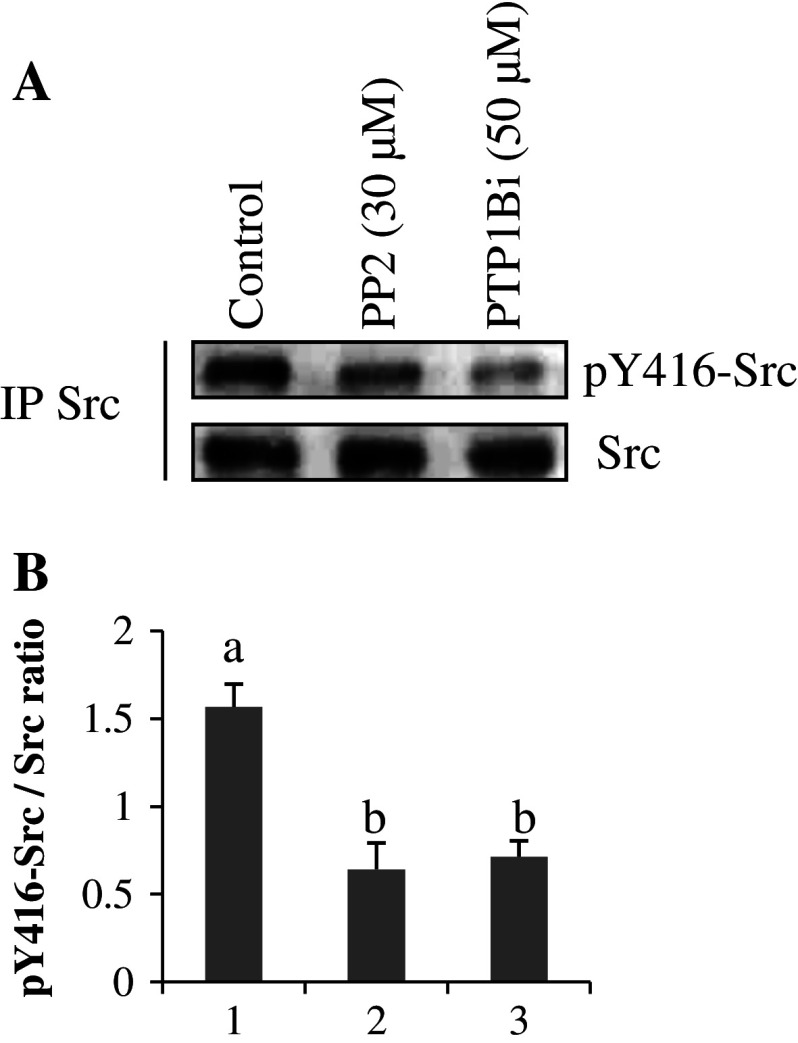
We then controlled the phosphorylation status of Y416 residue in mitochondria taken from the oxygraphic chamber. No pY416-Src labeling was observed when immunodetection was performed with mitochondrial lysates. Therefore, analysis was performed on Src immunoprecipitated from mitochondria pelleted from the oxygraphic samples. Results showed that PP2 and PTP1Bi decreased pY416-Src labeling during ADP-stimulated respiration to a similar degree (Fig. 5a, b; lanes 1 and 2).
Enzymatic activities of OxPhos complexes
The preceding results suggest a tight link between oxygen consumption and Src activity. As respiration depends on OxPhos activities, this could occur by means of Src-induced phosphorylation of OxPhos complexes. To address this question, we examined the effect of the inhibitors on the enzymatic activities of the OxPhos complexes in an attempt to identify potential targets. Enzymatic activities in control samples (Table 3) were in the range of previous results from our laboratory [20]. ATP addition significantly affected enzymatic activities of all complexes, decreasing the activities of complex I, III, I + III, and V while increasing the activity of complex IV (Table 3). PP2 and PTP1Bi reversed the effect of ATP on the enzymatic activities for all the complexes, except for complex III with PTP1Bi (Table 3). The inhibitors had no effect on enzymatic activities of the complexes measured in the absence of ATP (data not shown). Controls (results not shown) showed that the labeling of pY416-Src in the assays differed in a similar way to the in vitro experiments (Fig. 2).
Table 3.
Effects of ATP, PP2, and PTP1Bi on the enzymatic activities of the OxPhos complexes (μmol min−1 mg protein−1)
| Control | ATP | ATP + PP2 (30 μM) | ATP + PTP1Bi (50 μM) | |
|---|---|---|---|---|
| Complex I | 0.107 (0.003) a | 0.056 (0.001) b | 0.097 (0.001) c | 0.077 (0.003) d |
| Complex III | 2.209 (0.070) a | 1.745 (0.048) b | 2.149 (0.304) a | 1.708 (0.170) b |
| Complex I +III | 0.136 (0.003) a | 0.119 (0.001) b | 0.125 (0.001) c | 0.130 (0.003) c |
| Complex IV | 1 1.220 (0.081) a | 1.739 (0.200) b | 1.195 (0.204) a | 1.246 (0.133) a |
| Complex V | 0.439 (0.017) a | 0.343 (0.031) b | 0.366 (0.002) c | 0.419 (0.047) a |
Thawed mitochondria were incubated at 30°C for 10 min in kinase buffer with 1 μM cAMP-dependent protein kinase inhibitor, 30 ng/ml rotenone (except for complex I), 20 μM oligomycin (except for ATPase activity), and the different inhibitors as indicated. Then 1 mM ATP was added and incubation continued for 10 min. Control experiments were performed under the same conditions without ATP or PP2 and PTP1Bi. Results are the means (±SD) of five independent measurements
Means that are not significantly different (P > 0.05) according to the Tukey test are identified by the same letters
Discussion
Since the discovery of tyrosine kinases of the Src family in rat brain mitochondria [13, 14], Src-mediated tyrosine phosphorylation is emerging as an important pathway for the regulation of mitochondrial bioenergetics. In addition, the presence of the tyrosine phosphatase PTP1B in mitochondria represents a possible mechanism of Src regulation. In this study, we used a PTP1B inhibitor [21] to study PTP1B-mediated regulation of Src activity in brain mitochondria and the influence of this regulation on mitochondrial bioenergetics.
In vitro studies showed that ATP induced phosphorylation at Y416 and Y527 residues in Src, in parallel with the increase in the tyrosine phosphorylation profile of mitochondrial proteins. Although fully active Src is characterized by phosphorylated Y416 and unphosphorylated Y527 residues (Fig. 6b), numerous studies have shown that Src phosphorylated on Y416 is active, regardless of the phosphorylation state of the Y527 residue [30, 31]. Therefore, the phosphorylation of Y416 residue is a reliable indicator of active Src in mitochondria, as confirmed by the concurrent enhanced protein tyrosine phosphorylation pattern (Fig. 1; lane 2).
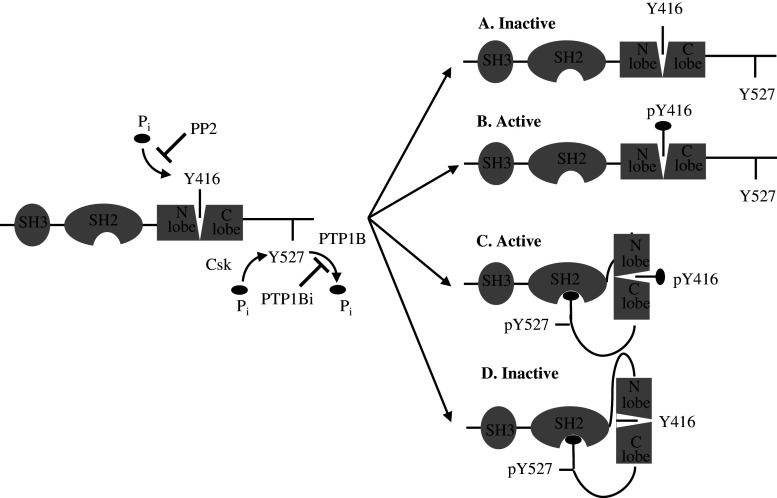
Fig. 6a–d.
Phosphorylation sites and organization of Src kinase family. Src kinases possess an SH3 domain directing binding to specific adaptors or substrates and an SH2 domain containing a phosphotyrosyl-binding pocket and linked to the N lobe. The autocatalytic Y416 residue in C-lobe lies in a pocket between the two lobes. The two lobes can move and open or close the catalytic site. Src can be non-phosphorylated (a), phosphorylated on Y416 residue (b), phosphorylated on both Y416 and Y527 residues (c), and phosphorylated on Y527 residue. Our data showed that a was found in thawed, untreated mitochondria, b and c in ATP-treated mitochondria, and d in PP2- and PTP1Bi-treated mitochondria. ATP induced both autophosphorylation of Y416 residue and Csk-mediated phosphorylation of Y527 residue. PP2 blocks the ATP binding site in the catalytic domain of Src thus leading to inhibition of autophosphorylation without preventing Csk-mediated phosphorylation of Y527. PTP1Bi inhibits PTP1B-mediated dephosphorylation of Y527 residue, leading to interaction of pY527 with the SH2 binding pocket, thus sequestering Y416 residue, which cannot be phosphorylated
In the presence of PP2, the labeling of tyrosine-phosphorylated mitochondrial proteins and of pY416-Src decreased while the signal of pY527-Src increased (Figs. 1–3; lane 3). PP2 blocks the ATP binding site of Src [14, 20, 22], preventing autophosphorylation of Y416 residue and activation of Src (Fig. 6d). The increase in pY527-Src labeling (Fig. 2a, b; lane 3) suggested an increased activity of mitochondrial Csk on Y527 in inactivated Src. This is supported by a study showing that a mutant of Lck, another member of the Src kinase family containing a K273R mutation, is defective in binding ATP and cannot autophosphorylate [32] but is nonetheless an efficient substrate for Csk [33].
In mitochondria pre-incubated with PTP1Bi, the data exhibited a similar variation as with PP2, i.e., a decrease in the tyrosine phosphorylation of mitochondrial proteins, a decrease in the labeling of pY416-Src, and an increase in pY527-Src signal (Figs. 1–3; lane 4). Previous studies reported that PTP1B dephosphorylates the pY527 residue and stimulates Src activity in several human cancer cell lines [34, 35]. Therefore, our experimental data indicated that PTP1Bi inhibited PTP1B-induced dephosphorylation of Y527 residue, thus enhancing pY527-Src labeling (Fig. 3d). The phosphorylation of this residue induces intramolecular interactions between the SH2 and SH3 domains and the pY527 residue, preventing the autophosphorylation of the Y416 residue [36], explaining the lower labeling of pY416-Src with PTP1Bi. Thus our results showed for the first time that the pY527 residue in mitochondrial Src is a PTP1B substrate, as in the cytosol, and its dephosphorylation results in the activation of this kinase.
In spite of the fact that PP2 and PTP1Bi induced similar inhibitory effects on Src activity, examination of the effect of both inhibitors on the tyrosine phosphorylation of OxPhos complexes showed that PP2 was more efficient than PTP1Bi in preventing tyrosine phosphorylation induced by the presence of ATP. This is particularly evidenced in the second-dimension SDS-PAGE following the BN-PAGE (Fig. 4b). Mass spectroscopy enabled the identification of some proteins that are tyrosine-phosphorylated in the presence of ATP (Table 1). However, it is difficult to assign a defined protein to a tyrosine-phosphorylated spot due to the fact that different proteins have sometimes been identified in a single spot. In addition, some tyrosine-phosphorylated spots have no counterparts in the corresponding Coomassie blue-stained second-dimension gel. Nonetheless, the higher level of tyrosine phosphorylation labeling in the presence of PTP1Bi compared to PP2 allows us to propose that PTP1Bi inhibits PTP1B-induced direct dephosphorylation of mitochondrial proteins, in addition to inhibiting Src, as shown in Figs. 2 and 3.
ADP-stimulated respiration and pY416-Src labeling were decreased in the presence of PP2 or PTP1Bi (Table 2, Fig. 5). These results indicate that Src kinase acivity is at least partly involved in the regulation of mitochondrial ADP-stimulated respiration. As ADP is progressively transformed to ATP during state 3 respiration, available ATP is therefore lower than in in vitro experiments, resulting in a lower amount of pY416-Src, which must be immunoprecipitated to be detected in oxygraphic experiments (Fig. 5).
Table 2.
Effect of PP2 and PTP1Bi on mitochondrial ADP-stimulated respiration rates
| Control | PP2 (30 μM) | PTP1Bi (50 μM) | |
|---|---|---|---|
| State 3 (%) | 100 (0.0) | 73.4 (3.5)* | 13.8 (0.90)* |
The inhibitors were incubated for 3 min at 30°C with mitochondria in respiration buffer before addition of pyruvate plus malate and ADP. ADP-stimulated respiration rates are presented as percentage [mean (SD), n = 3] of a control sample treated with DMSO
* P < 0.05 different from control, according to the Tukey test
Fig. 5a, b.
pY416-Src labeling in immunoprecipitated Src from oxygraphic assays. a Mitochondrial proteins were sampled from the oxygraphic chamber and immunoprecipitation of Src was immediately performed, as described in “Materials and methods.” The phosphorylation status of Src was detected with the anti-pY416-Src antibody. The membrane was stripped and reprobed with anti-Src antibody. b The intensity of pY416-Src labeling was normalized over the Src signal for each treatment. Data are representative of three experiments. Means that are not significantly different (P > 0.05) according to the Tukey test share the same letters
To find a potential link between respiration rates and OxPhos activities required that the analysis be performed on complex activities in the presence of ATP, since pY416-Src was labeled in ADP-stimulated respiration (Fig. 5; lane 2) and in enzymatic measurements performed in the presence of ATP (as in Fig. 2). The data are in favor of an effect of Src on the activities of OxPhos complexes, a result reinforced by the observation that several subunits of the complexes were detected in tyrosine-phosphorylated spots when mitochondria were incubated in the presence of ATP (Table 1). The data also showed that the inhibitors decreased complex IV activity (Table 3), while decreasing Src activity as demonstrated by the loss of labeling of pY416-Src. This result is in line with the finding that Src kinase is known to upregulate cytochrome c oxydase activity in osteoclasts [6, 7]. The inhibitors had no effect or increased enzymatic activities on the other complexes. Therefore, the inhibition of mitochondrial respiration observed in the presence of the inhibitors could be due (at least in part) to decreased Src-mediated phosphorylation of complex IV components. Interestingly, the subunit 2 of complex IV was found in spot 10 to probably be tyrosine-phosphorylated by Src. As a matter of fact, this subunit has been found to be directly phosphorylated by purified Src [6]. Notably, it is known that cytochrome c oxydase tightly controls OxPhos [37–39], emphasizing the importance of Src-mediated phosphorylation of complex IV components in the regulation of respiration.
The different effects of PP2 and PTP1Bi on state 3 respirations or tyrosine-phosphorylation pattern in 2D BN-PAGE/SDS-PAGE analysis suggest that, besides Src, PTP1B could have other mitochondrial substrates whose dephosphorylation is involved in state 3 respirations. These could be Oxphos complexes or other tyrosine-phosphorylated proteins involved in the mitochondrial metabolism such as aconitase found in spot 11 (Table 1), a citric acid component [5, 40], due to the localization of PTP1B in the mitochondrial matrix [15].
In conclusion, we provide the first experimental evidence of a positive regulation of Src by PTP1B in rat brain mitochondria, and of its involvement in ADP-stimulated respiration and in enzymatic activity of the OxPhos complexes. The data also underline that PTP1B could have other mitochondrial substrates. These experiments represent a significant step towards understanding the role of Src and PTP1B in the regulation of mitochondrial function.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Nadia Barrier for her technical support. Etienne Hébert Chatelain was financially supported by FQRNT (Fonds Québécois de la Recherche sur la Nature et les Technologies) and NSERC (National Sciences and Engineering Resaerch Council of Canada) scholarships. This work was supported in part by grants from INSERM, the University Victor Segalen-Bordeaux 2, and the Fondation de France (N°Engt 2008002720).
Abbreviations
- OxPhos
Oxidative phosphorylation
- Y
Tyrosine
- Csk
C terminal Src kinase
- PP2
4-Amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine
- PTP1Bi
3-(3,5-Dibromo-4-hydroxy-benzoyl)-2-ethyl-benzofuran-6-sulfonic acid-(4-(thiazol-2-ylsulfamyl)-phenyl)-amide inhibitor
- 2D BN/SDS-PAGE
Two-dimensional blue native/sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Nano LC-MS/MS
Nano liquid chromatography tandem mass spectrometry
References
- 1.Navarro A, Boveris A. The mitochondrial energy transduction system and the aging process. Am J Physiol Cell Physiol. 2007;292:C670–C686. doi: 10.1152/ajpcell.00213.2006. [DOI] [PubMed] [Google Scholar]
- 2.Huttemann M, Lee I, Samavati L, Yu H, Doan JW. Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochim Biophys Acta. 2007;1773:1701–1720. doi: 10.1016/j.bbamcr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Horbinski C, Chu CT. Kinase signaling cascades in the mitochondrion: a matter of life or death. Free Radic Biol Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 4.Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Salvi M, Brunati AM, Toninello A. Tyrosine phosphorylation in mitochondria: a new frontier in mitochondrial signaling. Free Radic Biol Med. 2005;38:1267–1277. doi: 10.1016/j.freeradbiomed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Miyazaki T, Neff L, Tanaka S, Horne WC, Baron R. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J Cell Biol. 2003;160:709–718. doi: 10.1083/jcb.200209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazaki T, Tanaka S, Sanjay A, Baron R. The role of c-Src kinase in the regulation of osteoclast function. Mod Rheumatol. 2006;16:68–74. doi: 10.1007/s10165-006-0460-z. [DOI] [PubMed] [Google Scholar]
- 8.Samavati L, Lee I, Mathes I, Lottspeich F, Huttemann M. Tumor necrosis factor alpha inhibits oxidative phosphorylation through tyrosine phosphorylation at subunit I of cytochrome c oxidase. J Biol Chem. 2008;283:21134–21144. doi: 10.1074/jbc.M801954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvi M, Morrice NA, Brunati AM, Toninello A. Identification of the flavoprotein of succinate dehydrogenase and aconitase as in vitro mitochondrial substrates of Fgr tyrosine kinase. FEBS Lett. 2007;581:5579–5585. doi: 10.1016/j.febslet.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Di Pancrazio F, Bisetto E, Alverdi V, Mavelli I, Esposito G, Lippe G. Differential steady-state tyrosine phosphorylation of two oligomeric forms of mitochondrial F0F1ATPsynthase: a structural proteomic analysis. Proteomics. 2006;6:921–926. doi: 10.1002/pmic.200500077. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, Lee I, Salomon AR, Yu K, Huttemann M. Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochim Biophys Acta. 2008;1777:1066–1071. doi: 10.1016/j.bbabio.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J, Lucchinetti E, Enkavi G, Wang Y, Gehrig P, Roschitzki B, Schaub MC, Tajkhorshid E, Zaugg K, Zaugg M. Tyrosine phosphorylation by Src within the cavity of the adenine nucleotide translocase 1 regulates ADP/ATP exchange in mitochondria. Am J Physiol Cell Physiol. 2010;298:C740–C748. doi: 10.1152/ajpcell.00310.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvi M, Brunati AM, Bordin L, La Rocca N, Clari G, Toninello A. Characterization and location of Src-dependent tyrosine phosphorylation in rat brain mitochondria. Biochim Biophys Acta. 2002;1589:181–195. doi: 10.1016/S0167-4889(02)00174-X. [DOI] [PubMed] [Google Scholar]
- 14.Tibaldi E, Brunati AM, Massimino ML, Stringaro A, Colone M, Agostinelli E, Arancia G, Toninello A. Src-Tyrosine kinases are major agents in mitochondrial tyrosine phosphorylation. J Cell Biochem. 2008;104:840–849. doi: 10.1002/jcb.21670. [DOI] [PubMed] [Google Scholar]
- 15.Augereau O, Claverol S, Boudes N, Basurko MJ, Bonneu M, Rossignol R, Mazat JP, Letellier T, Dachary-Prigent J. Identification of tyrosine-phosphorylated proteins of the mitochondrial oxidative phosphorylation machinery. Cell Mol Life Sci. 2005;62:1478–1488. doi: 10.1007/s00018-005-5005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salvi M, Stringaro A, Brunati AM, Agostinelli E, Arancia G, Clari G, Toninello A. Tyrosine phosphatase activity in mitochondria: presence of Shp-2 phosphatase in mitochondria. Cell Mol Life Sci. 2004;61:2393–2404. doi: 10.1007/s00018-004-4211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roskoski R., Jr Src kinase regulation by phosphorylation and dephosphorylation. Biochem Biophys Res Commun. 2005;331:1–14. doi: 10.1016/j.bbrc.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Roskoski R., Jr Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun. 2004;324:1155–1164. doi: 10.1016/j.bbrc.2004.09.171. [DOI] [PubMed] [Google Scholar]
- 19.Brown MT, Cooper JA. Regulation, substrates and functions of Src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 20.Arachiche A, Augereau O, Decossas M, Pertuiset C, Gontier E, Letellier T, Dachary-Prigent J. Localization of PTP-1B, SHP-2, and Src exclusively in rat brain mitochondria and functional consequences. J Biol Chem. 2008;283:24406–24411. doi: 10.1074/jbc.M709217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, Fahr BJ, Zhong M, Taylor L, Randal M, McDowell RS, Hansen SK. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat Struct Mol Biol. 2004;11:730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 22.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark JB, Nicklas WJ. The metabolism of rat brain mitochondria. Preparation and characterization. J Biol Chem. 1970;245:4724–4731. [PubMed] [Google Scholar]
- 24.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 25.Williams SL, Valnot I, Rustin P, Taanman JW. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J Biol Chem. 2004;279:7462–7469. doi: 10.1074/jbc.M309232200. [DOI] [PubMed] [Google Scholar]
- 26.Pocaly M, Lagarde V, Etienne G, Dupouy M, Lapaillerie D, Claverol S, Vilain S, Bonneu M, Turcq B, Mahon FX, Pasquet JM. Proteomic analysis of an imatinib-resistant K562 cell line highlights opposing roles of heat shock cognate 70 and heat shock 70 proteins in resistance. Proteomics. 2008;8:2394–2406. doi: 10.1002/pmic.200701035. [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa Y, Ohi N, Yagisawa A, Doi Y, Yamamoto Y, Yoshida M, Tokairin T, Yoshioka T, Omori Y, Enomoto K. Suppressive effect of orthovanadate on hepatic stellate cell activation and liver fibrosis in rats. Am J Pathol. 2009;174:881–890. doi: 10.2353/ajpath.2009.080261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shani V, Bromberg Y, Sperling O, Zoref-Shani E. Involvement of Src tyrosine kinases (SFKs) and of focal adhesion kinase (FAK) in the injurious mechanism in rat primary neuronal cultures exposed to chemical ischemia. J Mol Neurosci. 2009;37:50–59. doi: 10.1007/s12031-008-9113-3. [DOI] [PubMed] [Google Scholar]
- 29.Mohamed AS, Rivas-Plata KA, Kraas JR, Saleh SM, Swope SL. Src-class kinases act within the agrin/MuSK pathway to regulate acetylcholine receptor phosphorylation, cytoskeletal anchoring, and clustering. J Neurosci. 2001;21:3806–3818. doi: 10.1523/JNEUROSCI.21-11-03806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman DK. The Y’s that bind: negative regulators of Src family kinase activity in platelets. J Thromb Haemost. 2009;7(Suppl 1):195–199. doi: 10.1111/j.1538-7836.2009.03369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stover DR, Furet P, Lydon NB. Modulation of the SH2 binding specificity and kinase activity of Src by tyrosine phosphorylation within its SH2 domain. J Biol Chem. 1996;271:12481–12487. doi: 10.1074/jbc.271.21.12481. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi H, Hendrickson WA. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–489. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- 33.Sondhi D, Xu W, Songyang Z, Eck MJ, Cole PA. Peptide and protein phosphorylation by protein tyrosine kinase Csk: insights into specificity and mechanism. Biochemistry. 1998;37:165–172. doi: 10.1021/bi9722960. [DOI] [PubMed] [Google Scholar]
- 34.Bjorge JD, Pang A, Fujita DJ. Identification of protein-tyrosine phosphatase 1B as the major tyrosine phosphatase activity capable of dephosphorylating and activating c-Src in several human breast cancer cell lines. J Biol Chem. 2000;275:41439–41446. doi: 10.1074/jbc.M004852200. [DOI] [PubMed] [Google Scholar]
- 35.Zhu S, Bjorge JD, Fujita DJ. PTP1B contributes to the oncogenic properties of colon cancer cells through Src activation. Cancer Res. 2007;67:10129–10137. doi: 10.1158/0008-5472.CAN-06-4338. [DOI] [PubMed] [Google Scholar]
- 36.Harrison SC. Variation on an Src-like theme. Cell. 2003;112:737–740. doi: 10.1016/S0092-8674(03)00196-X. [DOI] [PubMed] [Google Scholar]
- 37.Villani G, Attardi G. In vivo control of respiration by cytochrome c oxidase in wild-type and mitochondrial DNA mutation-carrying human cells. Proc Natl Acad Sci USA. 1997;94:1166–1171. doi: 10.1073/pnas.94.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villani G, Greco M, Papa S, Attardi G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J Biol Chem. 1998;273:31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]
- 39.D’Aurelio M, Pallotti F, Barrientos A, Gajewski CD, Kwong JQ, Bruno C, Beal MF, Manfredi G. In vivo regulation of oxidative phosphorylation in cells harboring a stop-codon mutation in mitochondrial DNA-encoded cytochrome c oxidase subunit I. J Biol Chem. 2001;276:46925–46932. doi: 10.1074/jbc.M106429200. [DOI] [PubMed] [Google Scholar]
- 40.Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry. 2006;45:2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.