Abstract
Microbes have a fascinating repertoire of bioenergetic enzymes and a huge variety of electron transport chains to cope with very different environmental conditions, such as different oxygen concentrations, different electron acceptors, pH and salinity. However, all these electron transport chains cover the redox span from NADH + H+ as the most negative donor to oxygen/H2O as the most positive acceptor or increments thereof. The redox range more negative than −320 mV has been largely ignored. Here, we have summarized the recent data that unraveled a novel ion-motive electron transport chain, the Rnf complex, that energetically couples the cellular ferredoxin to the pyridine nucleotide pool. The energetics of the complex and its biochemistry, as well as its evolution and cellular function in different microbes, is discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0555-8) contains supplementary material, which is available to authorized users.
Keywords: Acetobacterium woodii, Ferredoxin, Rnf, Energy conservation, Electron transport
The acetogenic bacterium Acetobacterium woodii, a paradigm for an organism that has based its bioenergetics on a sodium ion current
Acetogenic bacteria such as A. woodii are strictly anaerobic and defined by their ability to use CO2 as an electron acceptor that is reduced to acetate. Acetogens are characterized by a special pathway for CO2 reduction, the acetyl-CoA pathway, also named the Wood–Ljungdahl pathway [1–3]. Electrons for CO2 reduction are derived from various donors such as sugars (fructose, glucose; oxidation via the Embden–Meyerhof pathway), alcohols such as propanediol or methanol, methoxy groups from lignin-derived aromatic compounds such as ferulate, or from molecular hydrogen [4–7]. The pathway of carbon dioxide reduction is coupled to energy conservation via a chemiosmotic mechanism, but the enzymes involved are obscure [8]. However, some acetogens have cytochromes, and a proton-motive electron transport chain is assumed in those whereas others do not have cytochromes. For the latter group, A. woodii is a bioenergetic paradigm. It couples carbon dioxide reduction to acetate production with the generation of an electrochemical sodium ion gradient across the membrane that is then used to drive ATP synthesis [9–13] and flagella rotation [9, 14], but again, the enzyme(s) involved is/are unknown [15].
Recently, some light was shed on the mystery of bioenergetics in acetogens. It turned out that instead of CO2 acetogens can also use alternative electron acceptors such as nitrate (Moorella thermoacetica) [16, 17] or arylacrylates such as caffeate (A. woodii) [18]. The enzymology and bioenergetics of caffeate respiration in A. woodii was unraveled very recently and led to the discovery of a novel Na+-translocating, electron-transferring ferredoxin:NAD+ oxidoreductase with similarity to the Rnf complex present in a wide variety of prokaryotes. The enzymology of caffeate respiration is briefly summarized below; for detailed reviews, the reader is referred to [8, 19].
Enzymology of caffeate respiration in A. woodii
Phenylacrylates such as caffeate are derived from lignin degradation and are a major constituent of the global carbon storage in soils [20]. The acetogen A. woodii cannot use caffeate as a carbon source but is known to use it as an electron acceptor by reducing the carbon–carbon double bond of caffeate according to:
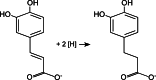 (eq. 1)
(eq. 1)
The electrons for caffeate reduction can be derived from various donors such as fructose, methanol or hydrogen [9, 18]. Caffeate reduction with hydrogen as electron donor is coupled to energy conservation in whole cells [18]. Clear evidence for ATP synthesis coupled to caffeate reduction was obtained with resting cells of A. woodii in which hydrogen-dependent caffeate reduction was accompanied by the synthesis of ATP [21], and later it was shown that ATP synthesis occurred by a chemiosmotic mechanism [9]. Most interestingly, like CO2 reduction, hydrogen-dependent caffeate reduction was strictly Na+-dependent and ATP synthesis relied on a transmembrane Na+ gradient. These studies led to the hypothesis that caffeate reduction is coupled to the generation of a transmembrane electrochemical Na+ gradient that is then used for ATP synthesis by the well-established Na+ F1FO ATP synthase [9–13]. However, the nature of the Na+-translocating enzyme remained enigmatic until very recently.
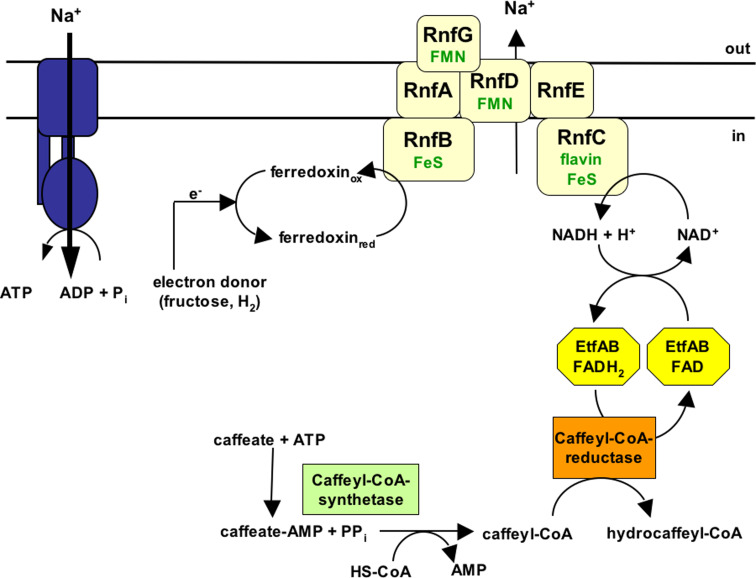
To identify the Na+-translocating enzyme, we have unraveled the entire pathway of electrons derived from the donor, molecular hydrogen, to the acceptor, caffeate (Fig. 1). Based on enzymatic activities, proteomics and also genetic evidence, we have suggested that caffeate is activated to caffeyl-CoA and then reduced to hydrocaffeyl-CoA by NADH via soluble enzymes involving an electron-transferring flavoprotein (Etf) and a caffeyl-CoA reductase/hydrocaffeyl-CoA dehydrogenase. This entire reaction sequence from NADH to caffeyl-CoA was found in the soluble fraction excluding any of these reactions in Na+ transport [22]. Furthermore, hydrogenase activity in A. woodii is cytosolic and the purified enzyme uses ferredoxin as an electron acceptor [23]. This left the electron transfer from reduced ferredoxin (Fdred) to NAD+ as the only membrane-bound and potentially Na+-translocating reaction [19, 22]. Indeed, a membrane-bound ferredoxin:NAD+ oxidoreductase (Fno) activity was found in washed membranes of A. woodii [22].
Fig. 1.
Model of caffeate respiration in A. woodii, showing the flow of electrons from electron donors (fructose or hydrogen) to the acceptor caffeate. Fno/Rnf couples the flow of electrons from reduced ferredoxin (Fdred) to NAD+ thereby generating a sodium ion gradient across the cytoplasmic membrane. For explanations, see text. ox oxidized, red reduced
A membrane-bound, Na+-translocating ferredoxin:NAD+ oxidoreductase, the Rnf complex
To test whether the Fno activity is coupled to Na+ translocation, studies with inverted membrane vesicles were performed. These vesicles catalyzed ferredoxin-dependent NAD+ reduction and electron flow was accompanied by 22Na+ transport into the lumen of the vesicles [24]. 22Na+ transport was dependent on NAD+ as electron acceptor and ferredoxin and titanium (III) citrate as electron donor. 22Na+ transport was electrogenic and accumulation prevented by the Na+ ionophore ETH2120 but not by protonophores, indicating a primary event. Addition of ETH2120 during steady state of transport led to a rapid efflux of 22Na+ from the vesicles. The data revealed that the ferredoxin:NAD+ oxidoreductase of A. woodii catalyzed a primary Na+ transport energized by electron flow from ferredoxin to NAD+ [24].
What is the enzyme catalyzing the Fno activity? We have partially purified an enzyme from A. woodii membranes that has Fno activity as well as ferricyanide-dependent NADH oxidation activity. The preparation contained a number of polypeptides [25]. Using N-terminal sequencing, RnfC was identified as one of the major polypeptides. With an antibody directed against RnfG and RnfB, these proteins were also shown to be part of the preparation. Rnf is suggested to be a membrane-bound electron transport complex with some similarity to the Na+-translocating NADH:ubiquinone oxidoreductase (Nqr) (see below, including Table 2) that is known to couple electron flow from NADH to quinone with the electrogenic translocation of Na+ outside the cell. Therefore, Rnf was assumed to be a Na+ pump since its first discovery [26] and was speculated to be the Na+-translocating coupling site in caffeate respiration of A. woodii [22]. With the partially purified complex in hand we searched for inhibitors. Fno activity catalyzed by the partially purified Rnf complex was inhibited by AgNO3, CuSO4, 1,10-phenanthroline, diphenyliodonium chloride, and diphenyleniodonium chloride. At the same time, these inhibitors also abolished Na+ transport coupled to Fno activity as catalyzed by inverted vesicles [24]. This is consistent with the hypothesis that the Rnf complex catalyzes the observed Na+-motive ferredoxin:NAD+ oxidoreductase activity. Although the final proof that Rnf is a sodium pump has to await its functional reconstitution into liposomes, we will, for the sake of clarity, refer in the following to Fno as Rnf.
Table 2.
Sequence identity of A. woodii Rnf subunits to V. alginolyticus Nqr subunits
| RnfD (%) | RnfE (%) | RnfA (%) | RnfG (%) | |
|---|---|---|---|---|
| NqrB | 27 | |||
| NqrD | 37 | 20 | ||
| NqrE | 24 | 45 | ||
| NqrC | 18 |
ClustalW was used for alignments and score determination
Genomic organization of the rnf genes
The rnf genes were first discovered in Rhodobacter capsulatus and were shown to be involved in nitrogen fixation (Rhodobacter nitrogen fixation). When the rnf genes were published for the first time only five genes were described to be in one cluster, rnfABCDE, and an additional gene, rnfF, was detected outside this cluster [26]. It was also noted that the rnfABCDE cluster is transcribed in the opposite direction to other genes of the nif region A such as rnfF. Later corrections were made to the published DNA sequence of R. capsulatus leading now to seven genes in the Rnf cluster, rnfABCDGEH [27]. The role of rnfH is not clear since it is not present in most rnf clusters. Since the rnf genes were first described, homologues have also been found in many prokaryotic and so far in two archaeal genomes. However, the organization of the genes differs and three major clusters can be distinguished that are commonly found in bacteria. Clusters similar to the one found in R. capsulatus rnfABCDGE are present in Escherichia coli [28], Vibrio fischeri MJ11 (VFMJ11_0974-0969) Klebsiella pneumoniae (KP1_3036-3041), Haemophilus influenzae (NTHI1989-1990), Haemophilus ducreyi (HD0396-0403), Shigella dysenteriae (SD1012_2262-2267), Pseudomonas stutzeri [29], Photobacterium profundum (CAG20939-20944), Azotobacter vinelandii [30], and Vibrio cholerae (VC0395_A0538-A0532) (Fig. S1A). Interestingly, in some species (Alteromonas, Azotobacter, Escherichia, Citrobacter, Erwinia, Marinobacter, Shewanella, Serratia, Photobacterium, Shigella), downstream of rnfE is an endonuclease III (oxidative base excision repair protein) encoded in the same orientation. It has been shown that the gene nth encoding for endonuclease III in E. coli is co-transcribed with the rnf genes [31, and see “Physiological roles of Rnf in different bacteria”]. As can be seen in Fig. S1A, the size of RnfC varies. All RnfC subunits contain the conserved motifs (FeS center, flavin- and NADH-binding site), but some have a longer C-terminus.
The next type of cluster rnfCDGEAB that was first described in Clostridium tetani [32] is also found in A. woodii [25], Clostridium kluyveri [33], Clostridium difficile (CD1137-1142), Clostridium botulinum (CBO0368-0373), Clostridium phytofermentans (Cphy_0211-0216), Clostridium thermocellum (Cther_0911-0906), Clostridium novyi (NT01CX_1579-1584), Clostridium beijerinckii (Cbei_2449-2454), Alkaliphilus oremlandii (Clos_1775-1770), Alkaliphilus metalliredigens QYMF (Amet2280-2285), Thermanaerobacter pseudethanolicus (Teth39_2124-2119), Ruminococcus torques (RUMTOR_10017-10032) and Ruminococcus obeum (RUMOBE_01198-01206) (Fig. S1B). Interestingly, this second variation of the Rnf cluster is similar to the order of the genes of the Nqr cluster of Vibrio alginolyticus (nqrABCDEF).
In addition, the variation rnfBCDGEA is found in some organisms like Bacteroides vulgatus (BVU_3890-3885), Chlorobium limicola (Clim_0989-0984), Chlorobium luteolum (Plut_1154-1159), Parabacteroides sp. (HMPREF0619_02359-02355), Prosthecochloris aestuarii (Paes_1309-1314) and Porphyromonas uenonis (PORUE0001_0056-0061) (Fig. S1C).
To address the transcriptional organization of the rnf genes in A. woodii, mRNA was extracted from fructose-grown cells and molecular analyses revealed that all six rnf genes are indeed found on a single transcript. The same can be expected for the rnf cluster found in other organisms. The genomic organization of the rnf genes in one operon suggests that the gene products are in a functional context [25].
Most organisms only have one Rnf cluster. However, in A. vinelandii, two rnf clusters were found in the genome; rnf1 expression is part of the nitrogen regulon and rnf2 expression is independent of the nitrogen source of the growth medium [30]. Also, Desulfobacterium autotrophicum HRM2 contains two rnf clusters in the genome. Furthermore single rnf genes like RnfC homologues that are not part of a complete cluster are found in some genomes.
Another interesting observation is that in some species (Serratia proteamaculans, Yersinia enterocolitica, Yersinia pseudotuberculosis), the genes rnfABC are followed by a gene for a transcriptional regulator araC, followed by rnfDGE. Furthermore, in two Thermosipho species (T. africanus and T. melanesiensis), rnfB is replaced by a gene coding for a putative arsenate or citrate transporter, respectively.
In the archaea Methanosarcina acetivorans and Methanococcoides burtonii, the cluster is organized as in C. tetani. However, upstream of rnfC is a gene that encodes a predicted multi-heme c-type cytochrome. The predicted protein is membrane-associated and could, therefore, contribute to the Rnf complex of both organisms (Fig. S1D). RT-PCR data showed that the corresponding gene in M. acetivorans is indeed co-transcribed with the rnf genes [34]. Downstream of rnfB is another gene that is co-transcribed with the rnf genes. The deduced amino acid sequence shows no identity to known proteins but is predicted to be membrane-integral [34].
Properties of the Rnf complex
Rnf subunits of R. capsulatus physically interact with each other [35]. An antibody directed against RnfC co-immunoprecipitated with the target antigen and at least five other proteins, one of which was identified as RnfB. RnfC and two unkown polypeptides co-immunoprecipitated using the antibody against RnfB. It was further shown that depletion of one subunit causes destabilization of the complex [36]. A rnfA mutant of R. capsulatus had almost no RnfB and only a little RnfC, while a rnfB mutant had reduced amounts of RnfC, and a rnfC mutant had almost no detectable amounts of RnfB. This was evidence that the presence of RnfA was required for stable existence of both RnfB and RnfC, indicating that these proteins actually form a complex [35, 36].
To date, integrated data on the biochemistry of the Rnf complex are still lacking. The first native purifications of Rnf complexes were mentioned for Clostridium tetanomorphum and Fusobacterium nucleatum in review papers [37, 38]. The complex was purified from membranes of C. tetanomorphum, enriched 30-fold and is composed of six subunits. N-terminal sequencing showed that RnfC, RnfD, RnfG and RnfE were part of the preparation [37]. Proteins with a molecular mass corresponding to those of RnfG and RnfD were found to be fluorescent under UV-illumination indicating the presence of FMN in these subunits. A phosphodiesterase treatment of the denatured protein increased fluoresence intensity but removed the fluorescent band of RnfG, indicating the release of the flavin that might be connected to the protein via phosphodiester linkage as has been shown for NqrC [37, 39]. The purified complex also had a NADH-dependent ferricyanide reduction activity [37].
So far, the subunit stoichiometry in the complex remains unknown, although preliminary experiments performed in our laboratory indicate a molecular mass of the complex of 186 kDa that would argue for each subunit being present as a single copy in the complex.
In summary, the biochemical analysis of the complex is still in its infancy and, therefore, most of the properties of the complex are deduced from bioinformatics analyses of the single subunits. The properties of the subunits are summarized in Table 1.
Table 1.
Properties of Rnf subunits of A. woodii
| Subunit | ||||||
|---|---|---|---|---|---|---|
| RnfC | RnfD | RnfG | RnfE | RnfA | RnfB | |
| Size (bp) | 1,332 | 960 | 624 | 591 | 588 | 1,002 |
| Mass (kDa) | 48.7 | 35 | 22.8 | 21.6 | 21.4 | 36.6 |
| Predicted localization | Soluble | Membrane integral | Membrane associated | Membrane integral | Membrane integral | Membrane associated |
| TMHa | 0 | 6–9 | 1 | 6 | 6 | 1–2 |
| Experimental localization | Membrane | Membrane | Membrane | Membrane | Membrane | Membrane |
| Cofactors predicted | FeS | FMN | FMN | – | – | FeS |
| Flavin | ||||||
| Cofactors experimentally found | FeSb | FMNc | FMNc | – | – | FeSb |
Properties of the subunits
RnfC
RnfC of A. woodii is predicted to have a molecular mass of 48.7 kDa. Hydropathy analyses predict that RnfC is soluble although Western blotting analysis with antibodies directed against RnfC showed that it is membrane-bound in A. woodii [25], suggesting a tight association with the membrane-integral Rnf subunits. It was shown by immunological studies that RnfC of R. capsulatus is only membrane-associated, since a fraction was released from the membrane by alkaline treatment [36]. Cysteine motifs (C-XX-C-XX-C-XXX-C-P) typical for coordination of 4Fe4S clusters were described in the amino acid sequence of RnfC [26]. The heterologously produced RnfC of R. capsulatus indeed harbored at least one FeS cluster, as determined by EPR spectroscopy and an absorbance spectrum with a maximum at 419 nm and a shoulder at 330 nm [27].
The presence of flavins in Rnf subunits was first suggested by Kumagai et al., and conserved amino acids similar to proposed FMN-binding sites were found in RnfC [36, 40, 41]. So far, the presence of flavin in RnfC has not been shown experimentally. Conserved residues with strong similarity to proposed NADH binding sites in mitochondrial complex I [42, 43], bacterial NAD+-reducing hydrogenases [44–46], and bacterial complex I [40, 41] are present in RnfC, indicating that it may bind NADH. This is also supported by a βαβ-fold in the conserved region of RnfC [36].
RnfD
A. woodii RnfD is predicted to have a molecular mass of 35 kDa and 6–9 transmembrane helices supporting that RnfD is a membrane-integral protein (Fig. S2). The purified Rnf complex of C. tetanomorphum contains a fluorescent subunit with the same retention time in an SDS PAGE as RnfD [37], and Backiel et al. presented biochemical evidence for the presence of FMN in RnfD from V. cholerae. They produced and purified RnfD of V. cholerae and the protein showed fluorescence under UV-illumination and a typical flavin spectrum. Mass spectrometry analysis showed that FMN is covalently attached to threonine-187 in RnfD [47]. This conserved threonine was also detected in the amino acid sequences of RnfD of A. woodii at position T157 as well as in others such as Vibrio harveyi, V. fischeri and V. alginolyticus [25], indicating that FMN is conserved across disparate phylogenetic groups. Topological analyses (reporter fusions and computer predictions) indicated that FMN in V. cholerae RnfD is located in the periplasm [47]. This is not only in contrast to Nqr where all the cofactors are located on the cytoplasmic side of the membrane [48] but also very unusual. However, additional biochemical data are required to confirm the cellular localization of the flavin binding site in RnfD and to hypothesize on its biological significance.
RnfG
The deduced protein has a molecular mass of 22.8 kDa and is mainly hydrophilic but has a stretch of 30 hydrophobic amino acids at the N-terminus, which led to the conclusion that this subunit is anchored in the membrane (Fig. S3), which was verified experimentally [25]. The purified Rnf complex of C. tetanomorphum contains a fluorescent subunit with the identical SDS gel migration as RnfG [37]. Treatment of the complex with phosphodiesterase removed this band, indicating that the flavin is attached to the protein via phosphodiester linkage [37]. Backiel et al. [47] presented biochemical evidence for the presence of FMN in RnfG of V. cholerae. The protein showed fluorescence under UV-illumination and a spectrum typical of flavins. FMN is covalently attached to threonine-175 in RnfG of V. cholerae [47]. This residue is conserved at position T185 in A. woodii RnfG [25]. Topological analysis (reporter fusions and computer predictions) indicated that the FMN in RnfG is also situated in the periplasmic space, again as opposed to Nqr where its RnfG homologue is localized on the cytoplasmic side of the membrane [48].
The structure of RnfG has been solved from Thermotoga maritima in the course of a structural genome project [Protein Data Bank (PDB) ID: 3DCZ, Joint Center for Structural Genomics (JCSG)]. Interestingly, the T. maritima RnfG homologue did not contain the flavin cofactor when heterologously produced in E. coli.
RnfE
RnfE of A. woodii is a membrane-integral protein with a molecular mass of 21.6 kDa. Hydropathy analysis suggests six transmembrane helices for RnfE of A. woodii (Fig. S4) with no conserved cofactor binding sites. For membrane topology, see next section on RnfA.
RnfA
A. woodii RnfA is likely to be membrane-integral (six transmembrane helices) (Fig. S5) with no recognized cofactor binding sites. Topological analyses with PhoA fusions of R. capsulatus RnfA confirmed its membrane localization, and it was shown that it spans the chromatophore membrane with its odd-numbered hydrophilic regions exposed to the periplasm [36]. The membrane topology of RnfE and RnfA of E. coli has been addressed experimentally by PhoA fusions to the homologous E. coli proteins YdgQ and Orf193 [49]. These studies revealed that both proteins have six transmembrane helices each but have evolved opposite orientations in the membrane. RnfA is proposed to have N- and C-termini reaching into the periplasm, whereas in RnfE both termini are in the cytoplasm. It is proposed by the authors that this leads to an unusual symmetric complex in the membrane which might have interesting implications for the function in electron transfer or ion channel formation [49] within the Rnf complex.
RnfB
RnfB is predicted to be 36.6 kDa and mainly hydrophilic but has a stretch of 30 hydrophobic amino acids at the N-terminus to anchor to the membrane through one or two transmembrane helices ([27, 36]; Fig. S6). RnfB contains six cysteine motifs typical for six 4Fe4S clusters. Starting at positions C172, C246 and C279, the motif C-XX-C-XX-C-XXX-C-P is present (Fig. S6). Starting at positions C138, C217 and C310, conserved cysteines are found but they do not form the classical motif, although deviations of the motif have been described [50]. Also, two cysteine clusters contain arginine instead of proline, but substitutions of proline by other amino acids do not seem to alter electron transfer rates [51]. R. capsulatus RnfB purified from E. coli had a red-brown color, and EPR analysis, absorption spectroscopy and iron content determination suggest the presence of one 2Fe2S cluster [27]. Recombinant RnfB might aggregate in E. coli and fold improperly. A reconstitution of the holoprotein was not successful and it is speculated that RnfB alone might not be stable but only as part of the complex [27]. It was also shown that, when cells of R. capsulatus were grown under iron limitation, the amount of RnfB was reduced significantly (4-fold less) indicating an iron-dependent regulation of the rnf genes in R. capsulatus [27]. Since RnfB contains FeS clusters, it is also termed polyferredoxin.
Pathways of electron and ion flow in Rnf
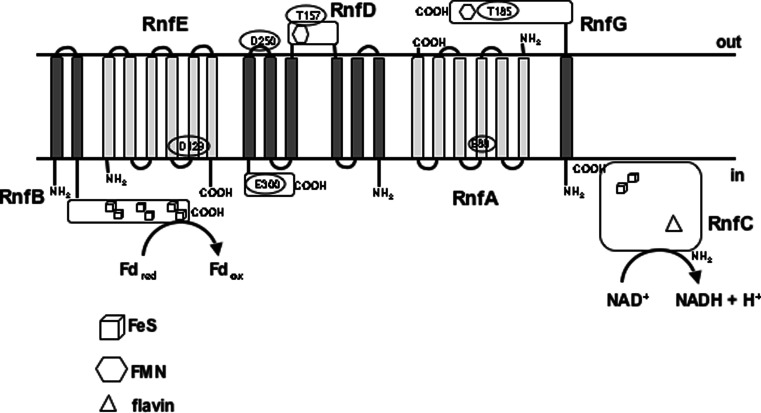
The Rnf complex of A. woodii catalyzes oxidation of Fdred with concomitant reduction of NAD+ [22, 24]. RnfB is soluble and has a “polyferredoxin”-type feature and, therefore, is presumably the entry point in the cytoplasm for electrons derived from ferredoxin oxidation. The membrane-attached RnfC has an NADH binding site and is, most likely, the exit point for electrons and therefore should be located on the cytoplasmic side. RnfG is anchored to the membrane and the cofactor FMN is predicted to be on the periplasmic side [47]. Nothing is known about the relative topology of the three membrane integral subunits A, D and E. Since RnfE and RnfA do not contain binding sites for cofactors, their role in electron transport is unknown and therefore they might just stabilize the complex or be part of the ion translocation domain.
In summary, we suggest that electrons derived from Fdred enter the complex at RnfB and travel via the other subunits to RnfC. There, the electron pair is generated at the flavin site to reduce NAD+ to NADH + H+ (Fig. 2). If the predicted topology of subunits is correct, then it is obvious that additional electron carriers have to be present to allow passage from RnfB to the membrane-bound subunits (RnfA, RnfD, and RnfE), to the periplasm (RnfG) and back to the cytoplasm (RnfC).
Fig. 2.
Topology model of Rnf subunits. Rnf subunits are indicated. N- and C-termini are as indicated. Fd ferredoxin, red reduced, ox oxidized. For explanations, see text
Electron flow from RnfB to RnfC in A. woodii is accompanied by the electrogenic transport of sodium ions out of the cell into the medium and thus establishing an electrochemical sodium ion potential that is used to drive endergonic reactions such as ATP synthesis or flagella rotation [24]. How this is achieved is unknown, but a conformational pump mechanism is most likely since Na+ cannot be transported by a classical redox loop-type mechanism. In line with this argument, the membrane-integral subunits RnfA, RnfD and RnfE may be involved in ion pumping and their pumping action energized by electron flow. The principle may be the same as used by complex I of the respiratory chain of mitochondria or bacteria [52]. Such a mechanism has the additional charm that it would also allow for proton transport and, as we shall discuss later, proton transport may indeed be true for Rnf complexes from other organisms.
So far, only speculations can be made on subunits or amino acids that are involved in Na+ binding and/or transport. The membrane-embedded subunits RnfA, RnfD and RnfE might be major players in binding sodium ions and forming a channel to transport the ion across the membrane. Recently, acidic residues in the corresponding Nqr subunits of V. cholerae have been found to be important for sodium binding (NqrB-D397, NqrD-D133 and NqrE-E95) [53]. Substitutions of these amino acids for alanine had a large effect on Nqr activity and on the K m value for Na+ (increase of 9-fold). All three mutations abolished the ability to form a membrane potential (Δψ). Other mutants (NqrB-E28, NqrB-E144, NqrB-D346 and NqrD-D88) showed altered turnover but no change in K m for Na+, indicating that these subunits are not directly involved in Na+ binding but in forming the channel [53]. It has been shown that, in V. cholerae, some of the acidic residues in Nqr are also conserved in the corresponding Rnf subunits of this organism [53]. Alignments of homologous subunits of Nqr revealed that NqrB-D397 is changed to glutamate in RnfD of A. woodii (RnfD-E300). NqrB-D346 is also conserved in RnfD at position D250 (Fig. S7). In the topology model, these amino acids are not part of a transmembrane helix but are situated on the opposite sides of the membrane (Fig. 2). NqrD-D133 is conserved in RnfE of A. woodii at position D129 (Fig. S8). The topology model shows that RnfE-D129 lies in transmembrane helix 5 (Fig. 2). NqrE-E95 is also conserved in RnfA of A. woodii at position E88 (Fig. S9). The topology model predicts that RnfA-E88 lies within helix 3 (Fig. 2). In conclusion, all the predicted Na+ binding sites of Nqr are conserved in Rnf, whereas only one of the amino acids that may be part of the sodium channel in Nqr is conserved in Rnf. Additional residues certainly play a role in binding and transport of Na+.
From the data available to date, it is not possible to identify the Na+ or H+ binding site in Rnf. They are known, however, in ATP synthase. In the c ring rotor of the FO motor, the Na+ binding site is sandwiched in between two c subunits by five amino acids [54]. At least three residues (Q…ES(T)) are highly conserved in Na+-ATP synthases [54–58]. This motif is not found in Rnf or in Na+-motive decarboxylases or Na+-motive methyltransferases. However, it is important to note that one residue of the Na+-binding motif is sufficient and essential for H+ transport [59–61].
Energetics and reversibility of the Rnf complex
As outlined above, so far only three Rnf complexes have been (partially) purified. All are from anaerobic bacteria and have ferredoxin:NAD+ oxidoreductase activity. The difference in redox potential between ferredoxin (E 0′ = −500 to −420 mV) and NADH (E 0′ = −320 mV) [62] is equivalent to approximately −20 to −35 kJ/mol. Assuming an electrochemical ion potential of around −200 mV across the cytoplasmic membrane, the free energy change of this reaction would allow for the translocation of one to two ions across the membrane. A lower electrochemical ion potential would increase the number of ions that can be pumped. At any rate, if one assumes the energetically most unfavorable value of one ion exported per mol of ferredoxin oxidized, 3 mol of Fdred must be oxidized to yield the three ions required by the ATP synthase to make 1 mol of ATP.
The Rnf complex described in A. woodii uses the energy released by the exergonic electron transfer from Fdred to NAD+ for outward Na+ transport and thus the generation of an electrochemical Na+ gradient across the cytoplasmic membrane. However, bacterial electron transport chains are largely reversible and often the physiology of the bacterial cell requires reverse electron flow [63]. This is, for example, encountered in chemolithoautotrophs that grow by oxidation of an electron donor such as nitrite that is too electropositive to allow direct reduction of NAD+ required for biosynthetic reactions. In this case, reverse electron flow drives energetic uphill transport of electrons to the redox level of NAD+ [64–68]. A reversal of the ferredoxin-dependent NAD+ reduction is essential in many metabolic scenarios (see examples below). Ferredoxin is used as a low potential reductant not only in anaerobes, but also in aerobic bacteria that may use reverse electron flow driven by ΔμNa+ or ΔμH+ to overcome the energy barrier in the endergonic reduction of ferredoxin with NADH as reductant. Ferredoxin is then used as electron donor for, for example, nitrogenase or regulatory proteins (see below). Anaerobes that grow autotrophically have to produce Fdred with very low redox potential from H2 as a reductant for anabolic reactions, i.e. acetyl-CoA carboxylation to pyruvate or formylmethanofuran reduction (in methanogenic archaea). Thus, mechanistically, ferredoxin:NAD+ oxidoreductase activity is freely reversible and may only be controlled by the magnitude of the thermodynamic parameters. In addition, it may be possible that the direction of reaction is also controlled by intrinsic properties of the proteins involved.
Evolution of the Rnf complex
Ferredoxin as “high energy” intermediate in electron transfer reactions
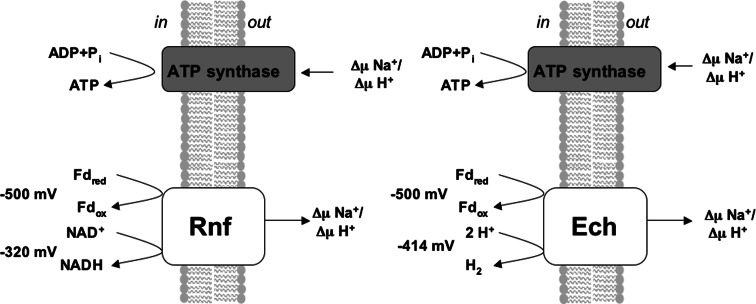
Ferredoxin plays a very central role in the energy metabolism of many anaerobic bacteria and archaea and has evolved as an electron carrier very early in life history [69]. However, its role in the bioenergetics of anaerobes was underestimated for a long time, and only recently did the hypothesis arise that anaerobes do even use the potential difference between ferredoxin at −500 to −420 mV and pyridine nucleotides at −320 mV to establish an ion gradient, which drives endergonic reactions such as ATP synthesis. This is equivalent to one-third to one-half of an ATP. Enzymes such as the pyruvate:ferredoxin oxidoreductase, pyruvate:formate lyase [70, 71], hydrogenase [23], CO dehydrogenase [72] and some formate dehydrogenases [73, 74] use ferredoxin as an electron acceptor. Therefore, enzymes must be present that connect the cellular ferredoxin pool to that of reduced pyridine nucleotides. Apart from Rnf, another membrane-bound ferredoxin-dependent oxidoreductase has been described, the Ech hydrogenase (Fig. 3).
Fig. 3.
Model of Ech and Rnf complexes. The potential difference between electron donor and acceptor is used to build up a transmembrane ion potential that is then used by ATP synthase to synthesize ATP. Fd ferredoxin, ox oxidized, red reduced
Ech and complex I of the respiratory chain, two evolutionary related, energy conserving complexes
The energy converting hydrogenase (Ech) is part of the family of the NiFe-hydrogenases and is found in bacteria as well as in archaea [75]. The biochemically most thoroughly studied enzyme is from the archaeon Methanosarcina barkeri, and its biochemical function and physiological role has been described in a review [76]. Briefly, the enzyme from M. barkeri has six subunits encoded by the echABCDEF operon. Two of the subunits (A and B) are apparently membrane-integral and have no cofactors bound whereas the others are located on the cytoplasmic side of the membrane, have FeS centers, and catalyze the oxidation of Fdred coupled to the reduction of protons to hydrogen. As outlined above, this reaction is exergonic and assumed to be the driving force for vectorial ion transport across the membrane. Back in 1986, Bott et al. [77] demonstrated that CO oxidation (which is coupled to ferredoxin reduction) is accompanied by the generation of a membrane potential, and later, evidence was presented that this reaction drives outward proton translocation [78]. Recently, direct experimental evidence for ion (proton) transport coupled to proton reduction with ferredoxin as reductant was obtained [79].
As discussed above for Rnf, Ech is a reversible enzyme. The methanogenic archaeon M. barkeri grows chemolithoautotrophically on H2 + CO2 that are converted to methane [80, 81]. The first step in methanogenesis from H2 + CO2, the formation of formylmethanofuran, requires ferredoxin as electron carrier and is highly endergonic with hydrogen as reductant. The energy barrier is overcome by reverse electron flow from hydrogen to ferredoxin catalyzed by Ech hydrogenase and driven by the electrochemical ion gradient across the membrane. Other endergonic reactions that require Fdred are CO2 reduction to CO, mediated by acetyl-CoA synthase/CO dehydrogenase, and the carboxylation of acetyl-CoA to pyruvate, mediated by the pyruvate:ferredoxin oxidoreductase. Both reactions are essential for CO2 fixation into biomass. The reverse reaction, oxidation of Fdred with concomitant release of hydrogen coupled to the generation of a proton or sodium motive force is encountered during methanogenesis from acetate or CO [82].
Direct evidence for proton transport coupled to the Ech reaction is available [79]. However, protons as well as sodium ions, depending on the organism, may be used as coupling ion. Some methanogens with their documented sodium bioenergetics may use Na+. The same may be true for Pyrococcus furiosus that has hydrogenases of the Eha- and Ehb-type [80, 83] and a Na+-driven A1AO ATP synthase [84]. Others like Rhodospirillum rubrum or Carboxydothermus hydrogenoformans that have no documented sodium bioenergetics may use the proton motive force [76].
With the first sequences available for hydrogenases, it was immediately evident that hydrogenases (including Ech) and complex I are evolutionary related and evolved from a common ancestor. The evolution of hydrogenases and complex I was discussed recently in excellent reviews [76, 85–88]. The membrane-bound Ech subunits EchA and EchB are highly homologous to Nuo subunits. EchA is homologous to NuoL, NuoM and NuoN, and EchB is homologous to NuoH [76]. These subunits form the catalytic core of complex I. However, it is interesting to note that neither complex I nor Ech show any homology to subunits of Rnf, indicating two different lines of evolution that resulted in different solutions to the same problem.
Rnf and Nqr, two evolutionary related, energy conserving complexes
When the rnf genes of R. capsulatus were described for the first time, database searches revealed that some subunits of the at that time already established Na+-translocating Nqr were the closest homologues. Nqr was first described in the facultative anaerobic marine bacterium V. alginolyticus by Tokuda and Unemoto [89]. This bacterium is dependent on Na+ for solute transport and flagella rotation and, therefore, a primary sodium ion pump that generates a sodium ion potential was assumed. Elegant experiments using whole cells, inverted vesicles and purified enzymes identified this novel type of energy conserving NADH:quinone oxidoreductase as not being related to complex I in this organism [90]. Nqr feeds electrons into the electron transport chain and it was shown unequivocally by different methods that this electron transfer reaction is coupled to vectorial, electrogenic Na+ transport out of the cell [91, 92]. That was the first example of a Na+-pumping NADH:quinone oxidoreductase, an enzyme later also found in several other bacteria [93, 94].
Nqr is an enzyme complex with an approximate molecular mass of 200 kDa consisting of six subunits (NqrA-F) encoded by the nqr operon. The complex channels electrons from NADH to quinone and thereby translocates sodium ions out of the cell [91, 92]. Three subunits (NqrB, NqrD, NqrE) are membrane-bound, NqrA is hydrophilic, and NqrC and NqrF are connected to the membrane via transmembrane helices. NqrF contains the NADH-binding site, the binding site for FAD and iron-sulfur clusters and is therefore characterized as the electron entry site [52, 93]. The two subunits NqrB and NqrC contain covalently bound FMN that is attached by an ester bond through a threonine residue of the protein [39, 95]. NqrB is suggested to contain part of the quinone binding site. Also, the presence of a riboflavin was experimentally shown for the Nqr complex [96]. In addition to flavins and iron-sulfur clusters, the Nqr complex contains a tightly bound ubiquinone-8 [52, 91, 93, 97, 98].
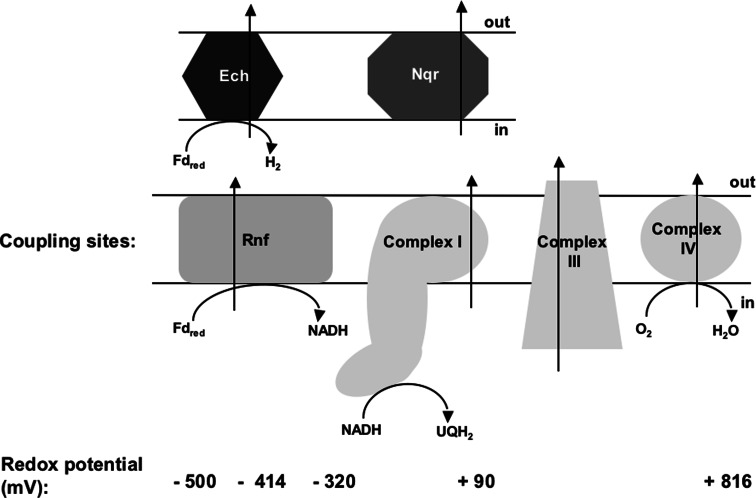
It is important to note that the mode of action, their physiological roles and also the electron input and output modules are quite different in Rnf and Nqr. Rnf mediates electron transfer from ferredoxin to NAD+, but Nqr from NADH to quinone. Energetically, Rnf is located “upstream” of Nqr and covers redox spans between −500 and −320 mV, whereas Nqr covers the −320 to +90 mV [99] region (Fig. 4). RnfC is the electron output module and does not share significant sequence similarity with any of the Nqr subunits, but like NqrF, the electron input module of Nqr, it contains an NADH binding site, flavin and FeS clusters [100]. Therefore, it is considered the functional homologue of NqrF. RnfB is a polyferredoxin-type protein and the electron input module of Rnf. Apparently, there is no functional homologue in Nqr. The only subunits with significant sequence similarity are the membrane-bound subunits and a FMN-containing subunit (Table 2). RnfD is similar to NqrB, RnfG to NqrC, RnfE to NqrD, and RnfA to NqrE. In addition, there is modest sequence identity between RnfE and NqrE and RnfA and NqrD.
Fig. 4.
The redox span used for energy conserving reactions in biology and the enzymes used. Rnf and Ech cover the most reduced range from −500 to −320 mV, complex I and Nqr the range from −320 to +90 mV, complex III from +90 to +240 mV, complex IV from +240 to +816 mV. For the sake of clarity, the region from −320 to +816 mV is shown by only four respiratory enzyme complexes. Please note that there are a couple of enzymes that work in this region. Fd ferredoxin, red reduced, UQ ubiquinone
Phylogenetic analyses of Rnf genes
Phylogenetic analyses of RnfD revealed that A. woodii RnfD groups with other clostridia (Clostridium, Thermoanaerobacter, Eubacterium) and Thermanaerovibrio. The RnfDs from proteobacteria are a monophyletic group, except for members of the sulfate-reducing bacteria, while the Thermotogaceae also form a robust cluster. The phylogenetic affinities for other phyla (Archaea, Fusobacteria, Dictyoglomi, Spirochaetes, Bacteroidetes/Chlorobi) are less straightforward. The genome of the clostridium Natranaerobius thermophilus stands out since it has four copies of the rnfD gene with amino acid percent identities among each other in the range from 30 to 62% suggesting it might have different functions or regulations to adapt to its unique environment (Fig. 5). In the case of RnfC, the proteobacteria also group together, except for the sulfate reducers. A. woodii RnfC is closest to Thermoanaerobacter as well as other clostridia (Fig. 6). Both RnfD and RnfC were not only found in multiple copies in the genomes of clostridia, especially in the case of Natranaerobius, but also had greater sequence diversity when compared to other phylogenetic groups.
Fig. 5.
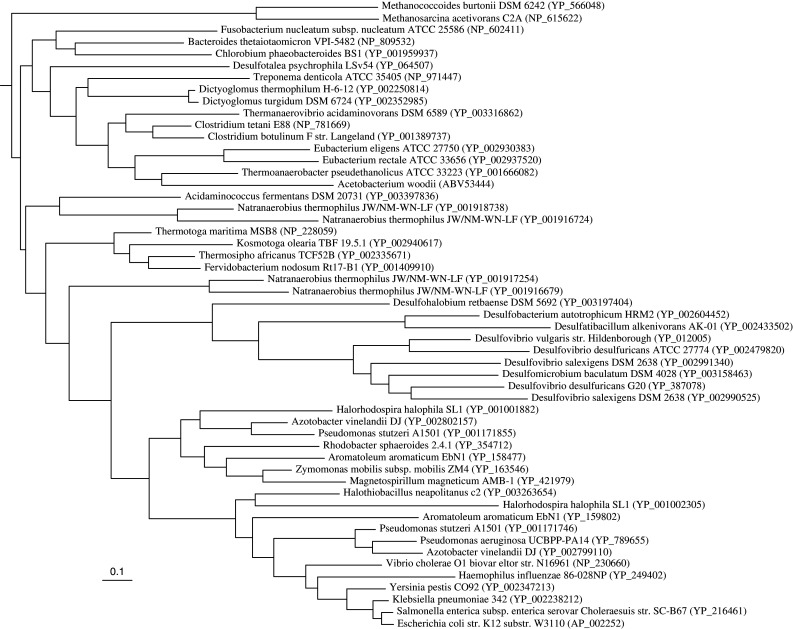
Maximum likelihood tree of the RnfD homologs. The tree contains representative sequences from the phyla where RnfD is found (Proteobacteria, Firmicutes, Methanosarcinaceae, Fusobacteria, Dictyoglomi, Spirochaetes, Bacteroidetes/Chlorobi, Synergistales, Thermotogales). The sequence of the NqrB homolog of Pseudomonas aeruginosa (accession number NP_251688) served as the outgroup. Number between parentheses is the Genbank accession number. Scale bar represents the number of substitutions per site
Fig. 6.
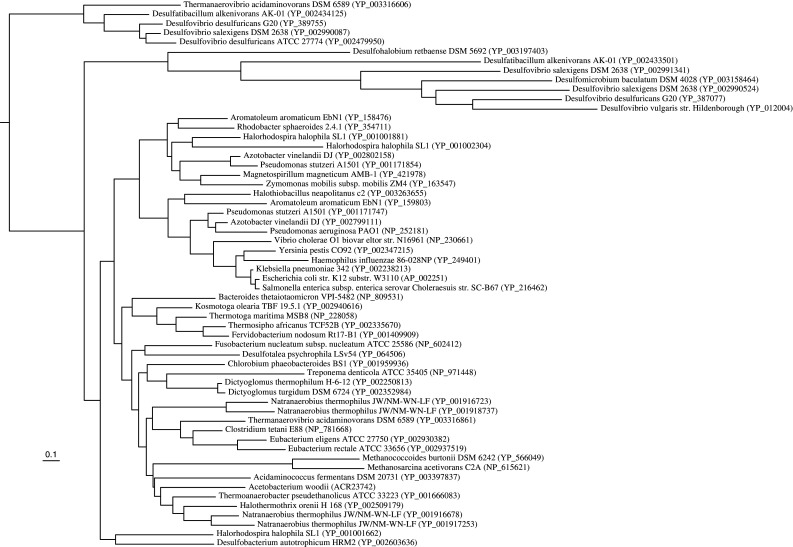
Maximum likelihood tree of the RnfC homologs from representative sequences. The sequence of Desulfatibacillum alkenivorans AK-01 (accession number YP_002433501) served as the outgroup. Number between parentheses is the Genbank accession number. Scale bar represents the number of substitutions per site
Distribution of rnf genes
A search of gene sequences deposited in databases revealed that 154 organisms contain rnf but no nqr operons (1,026 genomes; January 2010). It is noteworthy to discriminate between the presence of single, rnf-like genes and the presence of the entire rnf operon in the genome, since, for example, rnfC-like genes can be found in addition to the one in the rnf operon in another genomic context. In contrast, only 31 genomes contain the nqr operon alone, mostly strains in the families Chlamydiaceae and Neisseriaceae. In addition, 109 genomes have both Nqr and Rnf, most in the family enterobacteria, Shewanella spp., Vibrio spp. and Pasteurellaceae. The majority of enterobacteria, however, only contain the rnf operon.
As shown above, the rnf genes are widely distributed and are present in different phylogenetic and trophic groups, for example in chemolithoautotrophs, photolithoautotrophs and chemoorganoheterotrophs. There is also no correlation to salinity, pH or temperature of the environment since the genes are also present in a number of extremophiles. They are found in aerobes, facultative aerobes and anaerobes, but there is a very strong bias towards anaerobic life style. Only a handful of strictly aerobic species have the rnf genes, most being obligate intracellular symbionts like Buchnera spp, in contrast to more than 150 facultative anaerobic/anaerobic rnf-containing species (see Tables 3 and 4). This certainly reflects the essential role of rnf genes in energy conservation in strict anaerobes and may suggest a role of rnf in anaerobic metabolism in the facultative anaerobes. However, at least for enterobacteria, ferredoxin is not known to play a major role in fermentation or anaerobic respiration which questions the role of Rnf in energy metabolism. Instead, a role of rnf in keeping a certain redox potential in the cell and oxygen stress response (see below) is suggested [28].
Table 3.
Aerobes and facultative anaerobes containing rnf
| Acholeplasma laidlawii | Magnetococcus sp. MC1 | Shewanella denitrificans |
| Actinobacillus pleuropneumoniae | Magnetospirillum magneticum | Shewanella frigidimarina |
| Actinobacillus succinogenes | Magnetospirillum magnetotacticum | Shewanella halifaxensis |
| Aeromonas hydrophila | Mannheimia succiniciproducens | Shewanella loihica |
| Aeromonas salmonicida | Marinobacter algicola | Shewanella sp. MR4 |
| Aliivibrio salmonicida | Marinobacter aquaeolei | Shewanella MR7 |
| Alkalilimnicola ehrlichei | Marinobacter sp. | Shewanella oneidensis |
| Alcanivorax borkumensis | Marinomonas sp. MWYL1 | Shewanella pealeana |
| Alteromonas macleodii | Methylococcus capsulatus | Shewanella piezotolerance |
| Aromatoleum aromaticum | Nitrosococcus oceani | Shewanella putrefaciens |
| Azotobacter vinelandii | Pasteurella dagmatis | Shewanella sediminis |
| Buchnera aphidicola | Pasteurella multocida | Shewanella W3181 |
| Buchnera sp. | Photobacterium damselae | Shewanella woodyi |
| Cellvibrio japonicus | Photobacterium profundum | Shigella boydii |
| Citrobacter koseri | Photobacterium sp. | Shigella dynsenteriae |
| Citrobacter rodentium | Photorhabdus luminescens | Shigella flexneri |
| Colwellia psychrerythraea | Proteus mirabilis | Shigella sonnei |
| Dechloromonas aromaticum | Pseudoalteromonas atlantica | Sodalis glossinidius |
| Enterobacter 638 | Pseudoalteromonas haloplanktis | Symbiobacterium thermophilum |
| Enterobacter sakazakii | Pseudomonas aeruginosa | Thioalkalivibrio HL_EbGR7 |
| Erwinia carotovora | Pseudomonas mendocina | Thiobacillus denitrificans |
| Erwinia pyrifoliae | Pseudomonas stutzeri | Thiomicrospira crunogena |
| Erwinia tasmaniensis | Psychromonas ingrahamii | Vibrio cholerae |
| Escherichia coli | Rhodobacter capsulatus | Vibrio fischeri |
| Escherichia fergusonii | Rhodobacter sphaeroides | Vibrio harveyi |
| Haemophilus ducreyi | Saccharophagus degradans | Vibrio parahaemolyticus |
| Haemophilus influenzae | Salmonella enterica | Vibrio splendidus |
| Haemophilus parasuis | Salmonella typhi | Vibrio vulnificus |
| Haemophilus somnus | Salmonella typhimurium | Yersina enterocolitica |
| Hahella chejuensis | Serratia proteamaculans | Yersina pestis |
| Halorhodospira halophila | Shewanella amazoniensis | Yersina pseudotuberculosis |
| Idiomarina loihiensis | Shewanella ANA3 | Zymomonas mobilis |
| Idiomarina baltica | Shewanella baltica | |
| Klebsiella pneumoniae | Shewanella benthica |
Table 4.
Anaerobes containing Rnf
| Alkaliphilus oremlandii | Clostridium thermocellum | Petrotoga mobilis |
| Alkaliphilus metalliredigens | Desulfatibacillum alkenivorans | Porphyromonas endodontalis |
| Acetobacterium woodii | Desulfobacterium autotrophicum | Porphyromonas gingivalis |
| Azoarcus BH72 | Desulfotalea psychrophila | Porphyromonas uenonis |
| Bacteroides fragilis | Dethiobacter alkaliphilus | Prosthecochloris aestuarii |
| Bacteroides thetaiotaomicron | Dichelobacter nodosus | Prosthecochloris vibrioformis |
| Bacteroides vulgatus | Dictyoglomus thermophilum | Ruminococcus obeum |
| Brachyspira hyodysenteriae | Dictyoglomus furgidum | Ruminococcus torques |
| Chlorobium limicola | Fervidobacterium nodosum | Syntrophobacter fumaroxidans |
| Chlorobium luteolum | Finegoldia magna | Syntrophus aciditrophicus |
| Chlorobium phaeobacteroides | Fusobacterium nucleatum | Thermoanaerobacter pseudoethanolicus |
| Chloroherpeton thalassium | Halothermothrix orenii | Thermoanaerobacter X514 |
| Clostridium beijerinckii | Methanococcoides burtonii a | Thermosipho africanus |
| Clostridium botulinum | Methanosarcina acetivorans a | Thermosipho melanesiensis |
| Clostridium difficile | Natranaerobius thermophilus | Thermotoga lattingae |
| Clostridium kluyveri | Parabacteroides distasonis | Thermotoga maritima |
| Clostridium novyi | Parabacteroides johnsonii | Thermotoga neapolitana |
| Clostridium perfringens | Parabacteroides merdae | Thermotoga petrophila |
| Clostridium phytofermentans | Parabacteroides sp. | Thermotoga RQ2 |
| Clostridium tetani | Pelobacter carbinolicus | Treponema denticola |
| Clostridium tetanomorphum | Pelodictyon luteolum |
a Archaea
Another interesting observation is the presence of rnf genes in a number of pathogenic bacteria (Table 5). Again, this may be for different reasons. The clostridia are fermenting organisms that produce butyrate. As described below, Rnf is a key enzyme in electron flow and energy conservation in the fermentation pathway. Whether or not the Rnf complex is involved in pathogenicity as a virulence factor itself or part of a sensory/regulatory circuit is a highly interesting question that needs to be addressed in the future. This question strikes another, more general one: is the sodium ion circuit across the membrane involved in induction of virulence factors [101]? Indeed, studies performed with V. cholerae suggest an intimate linkage of the sodium ion potential to the expression of virulence factors. As mentioned above, V. cholerae contains both, Nqr and Rnf. Nqr is the site of entry for electrons into the electron transport chain, whereas the role of Rnf is unknown. Nqr mutants have an elevated expression of ToxT, a regulator for the expression of virulence genes [101, 102].
Table 5.
Pathogens containing Rnf
| Actinobacillus pleuropneumoniae | Fusobacterium nucleatum | Salmonella typhimurium |
| Aeromonas hydrophila | Haemophilus ducreyi | Serratia proteamaculans |
| Aeromonas salmonicida | Haemophilus influenzae | Shewanella loihica |
| Bacteroides fragilis | Haemophilus parasuis | Shewanella putrefaciens |
| Bacteroides thetaiotaomicron | Haemophilus somnus | Shigella boydii |
| Bacteroides vulgatus | Klebsiella pneumoniae | Shigella dysenteriae |
| Brachyspira hyodysenteriae | Mannheimia succiniciproducens | Shigella flexneri |
| Citrobacter koseri | Pasteurella multocida | Shigella sonnei |
| Clostridium botulinum | Photorhabdus luminescens | Vibrio cholerae |
| Clostridium difficile | Porphyromonas gingivalis | Vibrio parahaemolyticus |
| Clostridium perfringens | Proteus mirabilis | Vibrio splendidus |
| Clostridium tetani | Pseudoalteromonas atlantica | Vibrio vulnificus |
| Dichelobacter nodosus | Pseudoalteromonas haloplanktis | Yersina enterocolitica |
| Enterobacter sakazii | Pseudomonas aeruginosa | Yersina pestis |
| Erwinia carotovora | Pseudomonas mendocina | Yersina pseudotuberculosis |
| Escherichia coli | Pseudomonas stutzeri | |
| Escherichia fergusonii | Salmonella enterica | |
| Finegoldia magna | Salmonella typhi |
Evolution of the coupling ion used by Rnf
So far, the Rnf complex of A. woodii is the first Rnf complex for which ion transport has been demonstrated experimentally. There, the coupling ion is Na+, and this was actually expected since this anaerobe uses exclusively Na+ in its bioenergetics, i.e. as coupling ion for ATP synthesis, flagella rotation and solute symport [8, 14, 15, 19, 103]. The same may be true for others, for example C. tetani or V. cholerae that have established Na+ bioenergetics. However, Rnf complexes are widespread in prokaryotes and found in organisms that are neither known nor suggested to have Na+-based bioenergetics such as, for example, R. capsulatus or E. coli. In these organisms, protons are likely to be the coupling ions in Rnf. Moreover, acetogenic bacteria were classified bioenergetically into the H+-dependent subgroup that has cytochromes and the cytochrome-free, Na+-dependent subgroup [8, 104]. However, it may turn out that some Na+-independent acetogens without cytochromes have a proton-motive Rnf complex. Indeed, based on genome sequence data and a non-observed Na+-dependence of growth, it is speculated that the acetogen Clostridium ljungdahlii has a H+-coupled Rnf complex [105]. Therefore, the old bioenergetic differentiation of acetogens may no longer be valid. Instead, we would have two groups based on the type of electron transfer reactions: one which has cytochromes and the other which has Rnf. Both groups may turn out to contain H+ as well as Na+ pumping specimens. Actually, the Rnf complex is the first electron transport complex in bioenergetics that may use both, sodium ions or protons, depending on the organism. This has been suggested for complex I [106–108] and IV [109] of the bacterial electron transport chain but not yet shown unequivocally. Furthermore, there is no H+ translocating Nqr known.
Physiological roles of Rnf in different bacteria
Rhodobacter capsulatus
The rnf genes were first discovered in the non-sulfur, photosynthetic purple bacterium R. capsulatus [26]. Insertion and deletion mutants of R. capsulatus were created focusing on genes in the nifA region to unravel novel genes involved in nitrogen fixation. Indeed, six novel genes rnfABCDEF involved in nitrogen fixation were discovered by analyzing diazotrophic growth. The in vivo and in vitro nitrogenase activities were reduced in a ΔrnfE mutant (1.4% in vivo, 26.3% in vitro compared to wild-type). The mutants ΔrnfE, ΔrnfC, ΔrnfA and ΔrnfF contained significantly reduced amounts of nitrogenase (only 30% compared to wild-type) as shown by Western blotting analysis [26]. The same mutants had higher growth yields in the presence of metronidazole than the wild-type suggesting that these mutants contained lower amounts of Fdred [26]. Metronidazole leads to toxic derivatives in presence of high amounts of ferredoxin [110, 111]. Later, Saeki and Kumagai [112] demonstrated that Rnf is essential for nitrogen fixation in the light but also has an essential role in nitrogen fixation during anaerobic growth in the dark with dimethylsulfoxide as alternative electron acceptor. Furthermore, overproduction of the rnf operon in R. capsulatus resulted in higher levels of nitrogenase activity (50–100%) supporting once again the role of Rnf in nitrogen fixation in this organism [35]. In summary, it is concluded that the Rnf system in R. capsulatus could use the electrochemical ion potential generated by the photosynthetic apparatus to drive a reverse electron flow to reduce ferredoxin which serves as the ultimate donor for the dinitrogenase reductase. Therefore, the Rnf complex of R. capsulatus was suggested as a new electron transfer system that links known electron transport systems in the membrane to the nitrogenase system in the cytoplasm [26]. This function, production of “low redox” electrons, may be a general theme for the function of Rnf in aerobic organisms.
Pseudomonas stutzeri
Upstream of the fdx and nifENX genes that are all involved in nitrogen fixation in P. stutzeri, a rnf gene cluster (rnfABCDGEH) was detected. rnf insertion mutants were created in this organism and were found to display strongly reduced nitrogenase activities [113], as was shown for R. capsulatus rnf mutants. Therefore, also in P. stutzeri, Rnf could drive reverse electron flow from NADH generated by substrate oxidation to the reduction of ferredoxin that then delivers electrons to dinitrogenase reductase.
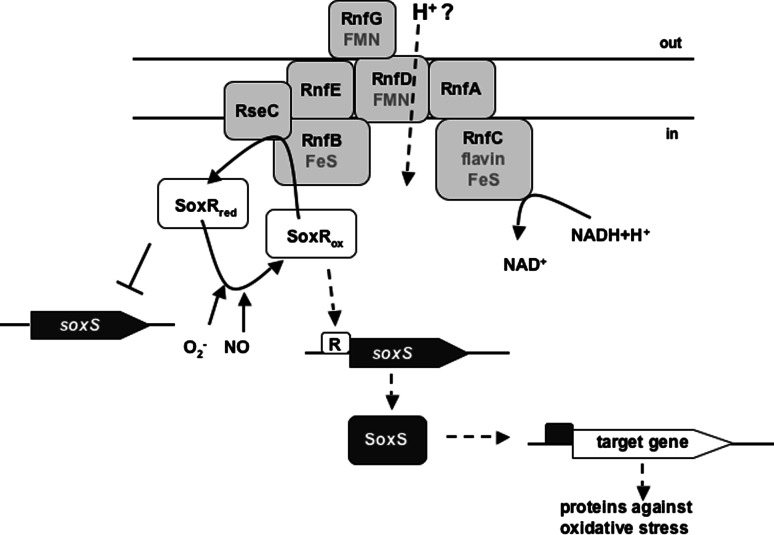
Escherichia coli
In E. coli, a system homologous to Rnf has been found and called Rsx (reducer of SoxR). The rsx locus contains the genes rsxABCDGE [28]. SoxR is a 17-kDa transcription factor that senses oxidant levels (mainly superoxide and nitric oxide) by its FeS center and activates oxygen defense systems. Oxidized SoxR increases synthesis of SoxS which activates target gene expression and SoxR is reduced upon removal of oxidative stress. Rnf/Rsx might maintain this reduced state during aerobic growth. A random insertional mutation screen for constitutive SoxS expression phenotype in the absence of oxidants revealed that the rnf/rsx genes (rnf/rsxA–E) were affected. The redox level of SoxR was significantly decreased in these rnf/rsx mutants [28] indicating that Rnf keeps SoxR reduced. Therefore, in contrast to the Rhodobacter/Pseudomonas system where Rnf transfers electrons to nitrogenase, in E. coli it is involved in oxidative stress response and transfers electrons to SoxR. As depicted in Fig. 7, it was believed that SoxR has a much more negative redox potential than NADH and the energy barrier is overcome by the proton-motive force as driving force. However, experimental evidence points to a more electropositive potential of −285 mV for SoxR [114]. How this fits into the picture remains to be established.
Fig. 7.
Model of proposed electron flow in E. coli. Electrons deriving from NADH are channeled through the complex to keep SoxR in its reduced state. When oxidized SoxR activates target genes against oxidative stress. For explanation, see text. red reduced, ox oxidized
Azotobacter vinelandii
A. vinelandii is a nitrogen-fixing bacterium and contains two rnf clusters that are differentially regulated [30]. rnf2 is constitutively expressed regardless of the nitrogen source. rnf1 and nif expression was found to be activated 10-fold after 6 h of ammonium deprivation. Deletion mutants in either rnf1 or rnf2 and a double mutant Δrnf1/rnf2 were constructed. It was shown that the nitrogen fixation activity was reduced in all mutants. Seven hours after nitrogenase derepression (ammonium-free growth conditions), the activity was 15 (Δrnf1), 35 (Δrnf2) and 5% (Δrnf1/rnf2) of the level of the wild-type. Furthermore, the nifHDK (nitrogenase) expression was analysed by reporter assays using β-galactosidase. Both single mutants showed lower β-galactosidase activity than the wild-type and lower protein levels after 3 h of nitrogenase derepression. After 7 h, the protein levels reached wild-type levels but the activity was still impaired. It was further shown that rnf mutants accumulate a Fe-deficient and inactive form of dinitrogenase reductase (NifH) which could explain the reduced activities. Therefore, Curatti et al. [30] suggest that the Rnf complex (Rnf1 and Rnf2) in A. vinelandii is required for early expression of nifHDK (nitrogenase) during nitrogenase derepression and stable accumulation of the 4Fe4S cluster in NifH. In contrast to R. capsulatus, the data do not support a role of Rnf in electron transport to nitrogenase in A. vinelandii.
Vibrio cholerae
V. cholerae is a facultative anaerobic chemo-organoheterotrophic bacterium pathogenic to human but it is also able to persist in the absence of the human host [115]. Like other Vibrio strains, it does not have complex I of the respiratory chain (NADH dehydrogenase) but the Na+-motive Nqr. In addition, it has a Rnf complex that has been analyzed biochemically [47]. Its role may be as in aerobes to produce “low redox” electrons to reduce cellular components involved in signaling, stress response, or other physiological activities. However, the function of Rnf is unknown but it would be interesting to see expression profiles of Rnf under different growth conditions and in planktonic versus host-associated cells.
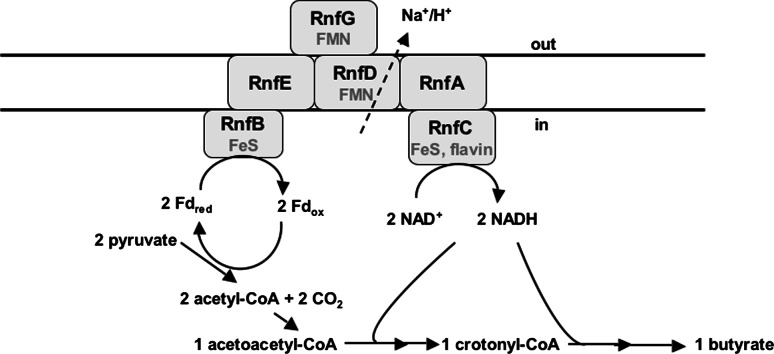
Clostridia
The Rnf complex was discovered in a number of clostridial genomes: C. tetani, C. tetanomorphum, C. kluyveri, C. botulinum, C. beijerinckii, C. difficile, C. novyi, C. perfringens, C. phytofermentans and C. thermocellum. The Rnf complex in C. tetani is involved in electron flow during butyrate fermentation (Fig. 8). Fermentation of sugars occurs via glycolysis to pyruvate, which is then oxidized by pyruvate:ferredoxin oxidoreductase to carbon dioxide, acetyl-CoA and Fdred. Two mol of acetyl-CoA are converted to acetoacetyl-CoA which is then converted via hydroxybutyryl-CoA and crotonyl-CoA to butyryl-CoA and subsequently to butyrate. The Fdred generated by the pyruvate:ferredoxin oxidoreductase is thought to be reoxidized by the Rnf complex that drives export of ions (Na+/H+) from the cytoplasm, thus establishing an ion gradient across the membrane that can be used for ion (Na+/H+)-dependent symporters to take up sugars and amino acids into the cell. Whether Rnf of C. tetani generates a Na+ or H+ potential remains to be established. In contrast to Clostridium acetobutylicum and C. perfringens, C. tetani does not harbor the genes for an F1Fo ATP synthase. C. tetani has the genes for a V-type ATPase and another ATPase that is annotated as archaeal “Na+-V-type” [32, 116]. However, both do not have the conserved Na+-binding motif of ATPases and both have a c subunit with one or no ion binding in four transmembrane helices suggesting a role in ATP hydrolysis and making a function in ATP synthesis unlikely. Therefore, the ion gradient established by the Rnf complex is not used as driving force for ATP synthesis but, most likely, substrate transport. Whether Rnf uses H+ or Na+ remains to be addressed experimentally.
Fig. 8.
Model of proposed electron flow in C. tetani. For explanation, see text. Fd ferredoxin, red reduced, ox oxidized. C. tetani is a peptolytic Clostridium that preferentialy ferments amino acids and the degradation of many amino acids results in the formation of pyruvate
Rnf is also found in the glutamate-fermenting C. tetanomorphum. Glutamate is oxidized to pyruvate and acetate via 3-methylaspartate. Pyruvate is then converted to butyrate as described above. The function of Rnf in this organism might be equivalent to that in C. tetani. Rnf is an Na+-pumping enzyme accepting electrons from Fdred (derived from pyruvate oxidation or crotonyl-CoA reduction) and generating NADH for butyrate synthesis [37].
Clostridium kluyveri ferments ethanol and acetate according to:
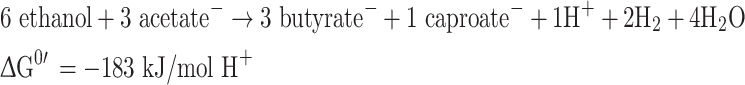 |
The mechanism of H2 formation from NADH via ferredoxin could not be explained for a long time since this reaction is endergonic. The reductant for crotonyl-CoA and acetoacetyl-CoA reduction is NAD(P)H that derives, for example, from glycolysis. In contrast to acetoacetyl-CoA reduction (E 0′ = −240 mV), reduction of crotonyl-CoA (E0′ = −10 mV) with NADH + H+ is highly exergonic. Recently, it was demonstrated for C. kluyveri that the energy released during crotonyl-CoA reduction with NADH + H+ as reductant is used to drive the endergonic reduction of ferredoxin via a process termed “electron bifurcation” which offers a mechanism for H2 production in C. kluyveri [117, 118]. A butyryl-CoA dehydrogenase/Etf complex has been described that is able to bifurcate electrons via FAD (bound to Etf) from the oxidant NADH. Of the two electrons from NADH, one proceeds to the more positive electron acceptor crotonyl-CoA, which is converted to butyryl-CoA, while the other is transported in the opposite direction to the more negative acceptor ferredoxin that is used for H2 production. Another NADH delivers the second electron to complete the reduction of each acceptor. This way, the endergonic reduction of ferredoxin is driven by the exergonic reduction of crotonyl-CoA. The bifurcation is accomplished by the prosthetic group FAD, which is reduced to its hydroquinone form by NADH. One electron is then transferred to the next FAD and further to ferredoxin, whereas the other is used by the dehydrogenase [118]. The same principle is known for the bc 1 complex of the respiratory chain. The complex oxidizes ubihydroquinone and transfers electrons to cytochrome c. The electrons from hydroquinone are bifurcated; one electron is transferred via a Rieske FeS cluster to cytochrome c, while the second electron is transferred via hemes to quinone, which is reduced to hydroquinone. This reaction is strictly coupled and is completed by the bifurcation of the second pair of electrons from another hydroquinone. In total, half the electrons are recycled from another hydroquinone, back to the quinone, thus the Q-cycle leads to a doubling of the proton-motive efficiency [119]. This sort of direct coupling in which the downhill transport of one electron drives the uphill transport of another is probably seen more often in anaerobes. Examples are the bifurcating hydrogenase of Thermotoga maritima [120] and the heterodisulfide reductase in Methanothermobacter [80].
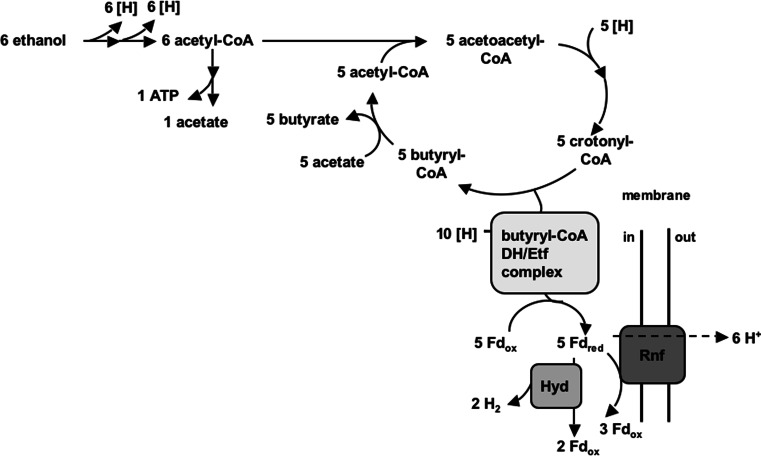
Fdred is used by Rnf that is encoded in the genome of C. kluyveri to pump ions across the cytoplasmic membrane while generating NADH [33]. In Fig. 9, a simplified version of the metabolism of C. kluyveri is shown. Ethanol is oxidized to acetyl-CoA. Through a cycle, the electrons are transferred to the butyryl-CoA dehydrogenase/Etf complex and butyrate is formed. At the end, the Fdred is split and used by hydrogenase to produce hydrogen and by Rnf to generate an ion gradient. The coupling ion of Rnf is unknown at present, but since the ATP synthase of C. kluyveri has no conserved Na+ binding site, one would expect a H+-based Rnf complex.
Fig. 9.
Metabolism of C. kluyveri. For the sake of simplicity, only the formation of butyrate and H2 is shown. Reactions involved in caproate formation are analogous (butyryl-CoA condenses with acetyl-CoA). Ten mol NADH are consumed by the butyryl-CoA-DH complex. They are bifurcated to produce 5 mol butyryl-CoA and 5 mol Fdred. Fdred is either used by hydrogenase to produce H2 or by Rnf to produce NADH and thereby generating an ion gradient. For further explanation, see text. [H] reducing equivalents, Fd ferredoxin, red reduced, ox oxidized
Syntrophus aciditrophicus and other synthrophic bacteria
Syntrophus aciditrophicus is a syntrophic bacterium that grows on fatty acids, benzoate, cyclohexane, carboxylate, cyclohex-1-ene carboxylate and crotonate in coculture with methanogens or sulfate reducers [121]. Degradation of substrates with H2 production is
thermodynamically unfavorable (propionate ΔG0′ = +76.1 kJ/mol, butyrate ΔG0′ = +48.6 kJ/mol, benzoate ΔG0′ = +70.1 kJ/mol), therefore the syntrophic partner (methanogen) is needed that uses the produced H2 as a substrate and thus lowers the hydrogen partial pressure in the ecosystem. Mechanistically, the syntrophic bacterium must have a way to overcome the energy barrier between NADH + H+ (generated during the oxidation reaction) and Fdred (the ultimate precursor of hydrogen). The genome sequence of S. aciditrophicus revealed a couple of membrane-bound FeS proteins and oxidoreductases as well as a rnf gene cluster. The latter is assumed to be the key enzyme in the bioenergetics of the syntrophic metabolism since it provides the cell with the precursor of hydrogen. The driving force for NADH-dependent ferredoxin reduction by Rnf is assumed to be an Na+ gradient established by a glutaconyl-CoA decarboxylase.
Desulfobacterium autotrophicum and other sulphate reducers
Desulfobacterium autotrophicum is a chemolithoautotrophic sulfate-reducing bacterium that can conserve energy by sulfate reduction to sulfide with H2 as reductant [122, 123]. Uptake of sulfate is an energy-dependent process and, in addition, sulfate is activated at the expense of 2 mol ATP prior to reduction. Sulfate reduction to sulfide requires eight electrons, provided by periplasmic hydrogenases. This results in eight scalar protons that would allow for the synthesis of 2.6 mol ATP (considering 3 H+/ATP). Clearly, additional mechanisms for the generation of a proton (ion) motive force must exist. This is where Rnf comes into play. Electron flow to sulfate could be carried out by a soluble, ferredoxin-reducing hydrogenase and the Rnf complex which would lead to NADH production, the reductant for sulfate. This would allow for additional ATP synthesis by an Rnf-mediated ion motive force [124].
Methanosarcina acetivorans
Like anaerobic bacteria, the anaerobic methanogenic archaea have developed mechanisms to conserve energy from the redox difference of ferredoxin and the central electron carrier, F420. However, it should be noted that the genomes of only two methanogens contain rnf genes (M. acetivorans and M. burtonii). M. acetivorans produces methane from acetate according to:
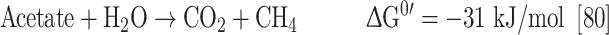 |
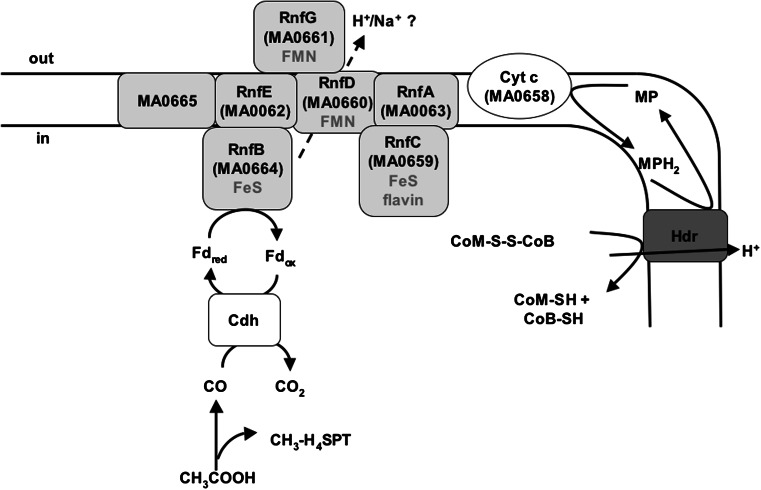
Methanogenesis from acetate has the lowest ΔG0′ of any methanogenic pathway and the free energy change may not even allow the synthesis of 1 mol ATP. However, methanogens have developed highly efficient mechanisms to conserve energy [82]. Acetate is oxidized by CO dehydrogenase/acetyl-CoA synthase to an enzyme-bound carbonyl group and an enzyme-bound methyl group [125, 126]. The methyl group is transferred to tetrahydromethanopterin. The next step is a methyl group transfer from methyltetrahydromethanopterin to coenzyme M. This reaction is catalyzed by a membrane-bound methyltransferase composed of a hydrophobic membrane translocator and a hydrophilic, corrinoid-containing subdomain that catalyzes the methyl transfer. The energy liberated by the methyl transfer reaction is used to pump Na+ out of the cell [127, 128]. The next step, methane formation, is catalyzed by a soluble enzyme. Interestingly, this enzyme uses coenzyme B (CoB) as a reductant for methane formation thereby generating a disulfide of coenzyme M (CoM) and CoB, the so-called heterodisulfide. This is used as an electron acceptor for anaerobic respiration. Electrons for the reduction derive from oxidation of enzyme-bound CO which yields Fdred. The electron transport chain involves methanophenazine as electron carrier and an ion-motive heterodisulfide reductase [80, 129]. The question is whether there is an additional coupling site between ferredoxin and methanophenazine. In a proteomic study, Rnf was detected at a higher concentration (2.5- to 3.5-fold) in acetate-grown cells than in methanol-grown cells [130]. Therefore, it was speculated that Rnf presents a coupling site in acetate metabolism of M. acetivorans (Fig. 10). Due to the recognized role of Na+ bioenergetics in methanogens, it is speculated that Rnf is a sodium ion pump in M. acetivorans but again, one should consider that the ion specificity is not known, and also not for the heterodisulfide reductase of M. acetivorans. Whether Rnf is indeed an ion (Na+) pump in M. acetivorans needs to be addressed experimentally. Moreover, if F420 is the electron acceptor of the Rnf complex, there may be an additional coupling site in between F420 and the methanophenazine. Most interestingly, the Rnf complexes of M. acetivorans and M. burtonii are the only Rnf complexes so far known that contain a cytochrome c. Therefore, one may hypothesize that the cytochrome c is involved in F420 reduction. Moreover, it may also be possible that F420 is not the electron acceptor of the Rnf complex, but that the electrons are channeled into the membrane via cytochrome c. Moreover, it may also be possible that the cytochrome c has a completely different function such as channeling electrons into the system from yet unknown electron donors.
Fig. 10.
Model of involvement of Rnf in metabolism of M. acetivorans. A possible way of electron transport is shown. Note that other ways of electron transport are possible (see “Physiological roles of Rnf in different bacteria”). For explanation, see text. MP methanophenazine, Cdh CO-dehydrogenase/acetyl-CoA synthase, Hdr heterodisulfide reductase, Cyt c cytochrome c, CoM coenzyme M, CoB coenzyme B, H 4 SPT tetrahydrosarcinapterin. Fd ferredoxin, ox oxidized, red reduced
During growth on trimethylamine or other methylated C1-compounds, one-quarter of the methyl group is oxidized to CO2. One step of the oxidation pathway, the oxidation of formyl-methanofuran to CO2, is exergonic and coupled to the reduction of ferredoxin. Fdred may be oxidized through Rnf, either with concomitant reduction of F420, the central electron carrier of methanogens, or directly with reduction of the heterodisulfide [80]. In any case, Rnf might be used to generate an electrochemical ion potential. Like M. acetivorans, M. burtonii is a member of the cytochrome-containing Methanosarcinales [80]. It does not grow on acetate but on methanol [131], and rnf and cytochrome c genes are present in its genome. The function of Rnf may have to do with the electron flow during methyl group oxidation as in M. acetivorans.
Conclusion
The Rnf complex is a novel ion-motive electron transport chain widely distributed in prokaryotes that deserves further attention. Its physiological role has been demonstrated in A. woodii as the key bioenergetic enzyme in caffeate respiration. Indeed, in this type of respiration, it is the only coupling site. The same may be true for acetogenesis. In some other anaerobes, it is likely to have the same function, and in aerobes, it may be involved driving reverse, endergonic electron flow. However, future work has to concentrate on defining the physiological role of the Rnf complex in the different bacteria and archaea. Moreover, the biochemical analyses is in its infancy and nothing is known about how electron flow is coupled to ion transport. These are the challenging tasks for future work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Genomic organization of rnf genes. nth encodes for endonuclease III. Membrane-bound subunits are indicated by an asterisk. A rnf cluster ABCDGE, B rnf cluster CDGEAB, C rnf cluster BCDGEA, D rnf organization in archaea. x encodes for a hypothetical protein. R. capsulatus: Rhodobacter capsulatus, A. vinelandii: Azotobacter vinelandii, E. coli: Escherichia coli, V. cholerae: Vibrio cholerae, K. pneumoniae: Klebsiella pneumoniae, A. woodii: Acetobacterium woodii, C. tetani: Clostridium tetani, A. metalliredigens: Alkaliphilus metalliredigens, T. pseudethanolicus: Thermoanaerobacter pseudethanolicus, C. kluyveri: Clostridium kluyveri, R. torques: Ruminococcus torques, B. vulgatus: Bacteroides vulgatus, C. limicola: Chlorobium limicola, P. sp.: Parabacteroides sp., P. aestuarii: Prosthecochloris aestuarii, P. uenonsis: Porphyromonas uenonsis, M. acetivorans: Methanosarcina acetivorans, M. burtonii: Methanococcoides burtonii (TIFF 79 kb)
Topology model of RnfD (based on SOSUI prediction, [132]). The N- and C-termini are indicated. Threonine 157 is highlighted in grey as potential FMN-binding site. Aspartate 250 and glutamate 300 are highlighted (black) as potential amino acids involved in Na+ binding (TIFF 124 kb)
Topology model of RnfG (based on SOSUI prediction, [132]). The N- and C-termini are indicated. Threonine 185 is highlighted in grey as potential FMN-binding site (TIFF 97 kb)
Topology model of RnfE (based on Sääf et al. 1999). The N- and C-termini are indicated. Aspartate 129 is highlighted (black) as potential Na+ binding site (TIFF 106 kb)
Topology model of RnfA (based on Sääf et al. 1999). The N- and C-termini are indicated. Glutamate 88 is highlighted (black) as potential Na+ binding site (TIFF 105 kb)
Topology model of RnfB (based on SOSUI prediction, [132]). The N- and C-termini are indicated. Conserved cysteines are highlighted in grey, that might form the FeS cluster binding sites (TIFF 132 kb)
Alignment of RnfD and NqrB. Alignment was done using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html [133]). An asterisk indicates complete amino acid conservation. The arrow shows the potential Na+-binding site. VC-Vibrio cholerae, AW-Acetobacterium woodii (TIFF 91 kb)
Alignment of RnfE and NqrD. Alignment was done using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html [133]). An asterisk indicates complete amino acid conservation. The arrow shows the potential Na+-binding site. VC-Vibrio cholerae, AW-Acetobacterium woodii (TIFF 62 kb)
Alignment of RnfA and NqrE. Alignment was done using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html [133]). An asterisk indicates complete amino acid conservation. The arrow shows the potential Na+-binding site. VC-Vibrio cholerae, AW-Acetobacterium woodii (TIFF 59 kb)
Acknowledgments
Work from the authors’ laboratory was supported by the Deutsche Forschungsgemeinschaft. We are grateful to Profs. U. Deppenmeier (Bonn), B. Ludwig (Frankfurt), J. R. Andreesen (Halle) and M. Rother (Frankfurt) for critical reading of the manuscript.
References
- 1.Drake HL, Daniel S, Küsel K, Matthies C, Kuhner C, Braus-Strohmeyer S. Acetogenic bacteria: what are the in situ consequences of their diverse metabolic diversities? Biofactors. 1997;1:13–24. doi: 10.1002/biof.5520060103. [DOI] [PubMed] [Google Scholar]
- 2.Müller V, Imkamp F, Rauwolf A, Küsel K, Drake HL. Molecular and cellular biology of acetogenic bacteria. In: Nakano MM, Zuber P, editors. Strict and facultative anaerobes. Medical and environmental aspects. Norfolk: Horizon Biosciences; 2004. pp. 251–281. [Google Scholar]
- 3.Ragsdale SW. Enzymology of the Wood–Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci. 2008;1125:129–136. doi: 10.1196/annals.1419.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diekert G, Wohlfarth G. Metabolism of homoacetogens. Antonie van Leeuwenhoek Int J Gen M. 1994;66:209–221. doi: 10.1007/BF00871640. [DOI] [PubMed] [Google Scholar]
- 5.Ragsdale SW, Pierce E. Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta. 2008;1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichler B, Schink B. Oxidation of primary aliphatic alcolhols by Acetobacterium carbinolicum sp. nov., a homoacetogenic anaerobe. Arch Microbiol. 1984;140:147–152. [Google Scholar]
- 7.Bache R, Pfennig N. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol. 1981;130:255–261. [Google Scholar]
- 8.Müller V. Energy conservation in acetogenic bacteria. Appl Environ Microbiol. 2003;69:6345–6353. doi: 10.1128/AEM.69.11.6345-6353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imkamp F, Müller V. Chemiosmotic energy conservation with Na+ as the coupling ion during hydrogen-dependent caffeate reduction by Acetobacterium woodii . J Bacteriol. 2002;184:1947–1951. doi: 10.1128/JB.184.7.1947-1951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heise R, Müller V, Gottschalk G. Presence of a sodium-translocating ATPase in membrane vesicles of the homoacetogenic bacterium Acetobacterium woodii . Eur J Biochem. 1992;206:553–557. doi: 10.1111/j.1432-1033.1992.tb16959.x. [DOI] [PubMed] [Google Scholar]
- 11.Reidlinger J, Müller V. Purification of ATP synthase from Acetobacterium woodii and identification as a Na+-translocating F1FO-type enzyme. Eur J Biochem. 1994;223:275–283. doi: 10.1111/j.1432-1033.1994.tb18992.x. [DOI] [PubMed] [Google Scholar]
- 12.Fritz M, Klyszejko AL, Morgner N, Vonck J, Brutschy B, Müller DJ, Meier T, Müller V. An intermediate step in the evolution of ATPases: a hybrid F1FO rotor in a bacterial Na+ F1FO ATP synthase. FEBS J. 2008;275:1999–2007. doi: 10.1111/j.1742-4658.2008.06354.x. [DOI] [PubMed] [Google Scholar]
- 13.Fritz M, Müller V. An intermediate step in the evolution of ATPases: the F1FO-ATPase from Acetobacterium woodii contains F-type and V-type rotor subunits and is capable of ATP synthesis. FEBS J. 2007;274:3421–3428. doi: 10.1111/j.1742-4658.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 14.Müller V, Bowien S. Differential effects of sodium ions on motility in the homoacetogenic bacteria Acetobacterium woodii and Sporomusa sphaeroides . Arch Microbiol. 1995;164:363–369. [Google Scholar]
- 15.Schmidt S, Biegel E, Müller V. The ins and outs of Na+ bioenergetics in Acetobacterium woodii . Biochim Biophys Acta. 2009;1787:691–696. doi: 10.1016/j.bbabio.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 16.Seifritz C, Daniel SL, Gössner A, Drake HL. Nitrate as a preferred electron sink for the acetogen Clostridium thermoaceticum . J Bacteriol. 1993;175:8008–8013. doi: 10.1128/jb.175.24.8008-8013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fröstl JM, Seifritz C, Drake HL. Effect of nitrate on the autotrophic metabolism of the acetogens Clostridium thermoautotrophicum and Clostridium thermoaceticum . J Bacteriol. 1996;178:4597–4603. doi: 10.1128/jb.178.15.4597-4603.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tschech A, Pfennig N. Growth yield increase linked to caffeate reduction in Acetobacterium woodii . Arch Microbiol. 1984;137:163–167. [Google Scholar]
- 19.Müller V, Imkamp F, Biegel E, Schmidt S, Dilling S. Discovery of a ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium woodii: a novel potential coupling site in acetogens. Ann N Y Acad Sci. 2008;1125:137–146. doi: 10.1196/annals.1419.011. [DOI] [PubMed] [Google Scholar]
- 20.Blum U, Wentworth TR, Klein K, Worsham AD, King LD, Gerig TM, Lyu S-W. Phenolic acid content of soils from wheat-no till, wheat-conventional till, and fallow-conventional till soybean cropping systems. J Chem Ecol. 1991;17:1045–1068. doi: 10.1007/BF01402933. [DOI] [PubMed] [Google Scholar]
- 21.Hansen B, Bokranz M, Schönheit P, Kröger A. ATP formation coupled to caffeate reduction by H2 in Acetobacterium woodii NZva16. Arch Microbiol. 1988;150:447–451. [Google Scholar]
- 22.Imkamp F, Biegel E, Jayamani E, Buckel W, Müller V. Dissection of the caffeate respiratory chain in the acetogen Acetobacterium woodii: indications for a Rnf-type NADH dehydrogenase as coupling site. J Bacteriol. 2007;189:8145–8153. doi: 10.1128/JB.01017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragsdale SW, Ljungdahl LG. Hydrogenase from Acetobacterium woodii . Arch Microbiol. 1984;139:361–365. doi: 10.1007/BF00408380. [DOI] [PubMed] [Google Scholar]
- 24.Biegel E, Müller V. A bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc Natl Acad Sci USA. 2010;107:18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biegel E, Schmidt S, Müller V. Genetic, immunological and biochemical evidence of a Rnf complex in the acetogen Acetobacterium woodii . Environ Microbiol. 2009;11:1438–1443. doi: 10.1111/j.1462-2920.2009.01871.x. [DOI] [PubMed] [Google Scholar]
- 26.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 27.Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus: characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem. 1998;251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 28.Koo MS, Lee JH, Rah SY, Yeo WS, Lee JW, Lee KL, Koh YS, Kang SO, Roe JH. A reducing system of the superoxide sensor SoxR in Escherichia coli . EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Y, Yang J, Dou Y, Chen M, Ping S, Peng J, Lu W, Zhang W, Yao Z, Li H, Liu W, He S, Geng L, Zhang X, Yang F, Yu H, Zhan Y, Li D, Lin Z, Wang Y, Elmerich C, Lin M, Jin Q. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci USA. 2008;105:7564–7569. doi: 10.1073/pnas.0801093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curatti L, Brown CS, Ludden PW, Rubio LM. Genes required for rapid expression of nitrogenase activity in Azotobacter vinelandii . Proc Natl Acad Sci USA. 2005;102:6291–6296. doi: 10.1073/pnas.0501216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gifford CM, Wallace SS. The genes encoding endonuclease VIII and endonuclease III in Escherichia coli are transcribed as the terminal genes in operons. Nucleic Acids Res. 2000;28:762–769. doi: 10.1093/nar/28.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brüggemann H, Bäumer S, Fricke WF, Wiezer A, Liesegang H, Decker I, Herzberg C, Martinez-Arias R, Merkl R, Henne A, Gottschalk G. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci USA. 2003;100:1316–1321. doi: 10.1073/pnas.0335853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seedorf H, Fricke WF, Veith B, Brüggemann H, Liesegang H, Strittmatter A, Miethke M, Buckel W, Hinderberger J, Li F, Hagemeier C, Thauer RK, Gottschalk G. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci USA. 2008;105:2128–2133. doi: 10.1073/pnas.0711093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Li L, Rejtar T, Lessner DJ, Karger BL, Ferry JG. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans . J Bacteriol. 2006;188:702–710. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong HS, Jouanneau Y. Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes. J Bacteriol. 2000;182:1208–1214. doi: 10.1128/jb.182.5.1208-1214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumagai H, Fujiwara T, Matsubara H, Saeki K. Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry. 1997;36:5509–5521. doi: 10.1021/bi970014q. [DOI] [PubMed] [Google Scholar]
- 37.Boiangiu CD, Jayamani E, Brügel D, Herrmann G, Kim J, Forzi L, Hedderich R, Vgenopoulou I, Pierik AJ, Steuber J, Buckel W. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J Mol Microbiol Biotechnol. 2005;10:105–119. doi: 10.1159/000091558. [DOI] [PubMed] [Google Scholar]
- 38.Kim J, Hetzel M, Boiangiu CD, Buckel W. Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of alpha-amino acids by anaerobic bacteria. FEMS Microbiol Rev. 2004;28:455–468. doi: 10.1016/j.femsre.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama Y, Yasui M, Sugahara K, Hayashi M, Unemoto T. Covalently bound flavin in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus . FEBS Lett. 2000;474:165–168. doi: 10.1016/s0014-5793(00)01595-7. [DOI] [PubMed] [Google Scholar]
- 40.Yagi T. The bacterial energy-transducing NADH-quinone oxidoreductases. Biochim Biophys Acta. 1993;1141:1–17. doi: 10.1016/0005-2728(93)90182-f. [DOI] [PubMed] [Google Scholar]
- 41.Yagi T, Yano T, Matsuno-Yagi A. Characteristics of the energy-transducing NADH-quinone oxidoreductase of Paracoccus denitrificans as revealed by biochemical, biophysical, and molecular biological approaches. J Bioenerg Biomembr. 1993;25:339–345. doi: 10.1007/BF00762459. [DOI] [PubMed] [Google Scholar]
- 42.Walker JE. The NADH: ubiquinone oxidoreductase (Complex-I) of respiratory chains. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 43.Fearnley IM, Walker JE. Conservation of sequences of subunits of mitochondrial complex I and their relationships with other proteins. Biochim Biophys Acta. 1992;1140:105–134. doi: 10.1016/0005-2728(92)90001-i. [DOI] [PubMed] [Google Scholar]
- 44.Tran-Betcke A, Warnecke U, Böcker C, Zaborosch C, Friedrich B. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16. J Bacteriol. 1990;172:2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz O, Boison G, Hilscher R, Hundeshagen B, Zimmer W, Lottspeich F, Bothe H. Molecular biological analysis of a bidirectional hydrogenase from cyanobacteria. Eur J Biochem. 1995;233:266–276. doi: 10.1111/j.1432-1033.1995.266_1.x. [DOI] [PubMed] [Google Scholar]
- 46.Malki S, Saimmaime I, De Luca G, Rousset M, Dermoun Z, Belaich JP. Characterization of an operon encoding an NADP-reducing hydrogenase in Desulfovibrio fructosovorans . J Bacteriol. 1995;177:2628–2636. doi: 10.1128/jb.177.10.2628-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Backiel J, Juarez O, Zagorevski DV, Wang Z, Nilges MJ, Barquera B. Covalent binding of flavins to RnfG and RnfD in the Rnf complex from Vibrio cholerae . Biochemistry. 2008;47:11273–11284. doi: 10.1021/bi800920j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duffy EB, Barquera B. Membrane topology mapping of the Na+-pumping NADH: quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis. J Bacteriol. 2006;188:8343–8351. doi: 10.1128/JB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sääf A, Johansson M, Wallin E, von Heijne G. Divergent evolution of membrane protein topology: the Escherichia coli RnfA and RnfE homologues. Proc Natl Acad Sci USA. 1999;96:8540–8544. doi: 10.1073/pnas.96.15.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otaka E, Ooi T. Examination of protein sequence homologies: IV. Twenty-seven bacterial ferredoxins. J Mol Evol. 1987;26:257–267. doi: 10.1007/BF02099857. [DOI] [PubMed] [Google Scholar]
- 51.Quinkal I, Davasse V, Gaillard J, Moulis JM. On the role of conserved proline residues in the structure and function of Clostridium pasteurianum 2[4Fe-4S] ferredoxin. Protein Eng. 1994;7:681–687. doi: 10.1093/protein/7.5.681. [DOI] [PubMed] [Google Scholar]
- 52.Kerscher S, Dröse S, Zickermann V, Brandt U. The three families of respiratory NADH dehydrogenases. Results Probl Cell Differ. 2008;45:185–222. doi: 10.1007/400_2007_028. [DOI] [PubMed] [Google Scholar]
- 53.Juarez O, Athearn K, Gillespie P, Barquera B. Acid residues in the transmembrane helices of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae involved in sodium translocation. Biochemistry. 2009;48:9516–9524. doi: 10.1021/bi900845y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meier T, Krah A, Bond PJ, Pogoryelov D, Diederichs K, Faraldo-Gomez JD. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J Mol Biol. 2009;391:498–507. doi: 10.1016/j.jmb.2009.05.082. [DOI] [PubMed] [Google Scholar]
- 55.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus . Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 56.Rahlfs S, Aufurth S, Müller V. The Na+-F1FO-ATPase operon from Acetobacterium woodii. Operon structure and presence of multiple copies of atpE which encode proteolipids of 8- and 18-kDa. J Biol Chem. 1999;274:33999–34004. doi: 10.1074/jbc.274.48.33999. [DOI] [PubMed] [Google Scholar]
- 57.Rahlfs S, Müller V. Sequence of subunit c of the Na+-translocating F1FO ATPase of Acetobacterium woodii: proposal for determinants of Na+ specificity as revealed by sequence comparisons. FEBS Lett. 1997;404:269–271. doi: 10.1016/s0014-5793(97)00088-4. [DOI] [PubMed] [Google Scholar]
- 58.Rahlfs S, Müller V. Sequence of subunit a of the Na+-translocating F1FO-ATPase of Acetobacterium woodii: proposal for residues involved in Na+ binding. FEBS Lett. 1999;453:35–40. doi: 10.1016/s0014-5793(99)00576-1. [DOI] [PubMed] [Google Scholar]
- 59.Altendorf K, Siebers A, Epstein W (1992) The Kdp ATPase of Escherichia coli. In: Scarpa A, Carafoli E, Papa S (eds) Ion: motive ATPases: structure, function, and regulation, vol 671. Annals of the New York Academy of Sciences, New York, NY, USA, pp 228–243 [DOI] [PubMed]
- 60.Deckers-Hebestreit G, Altendorf K. The F0F1-type ATP synthases of bacteria: structure and function of the FO complex. Annu Rev Microbiol. 1996;50:791–824. doi: 10.1146/annurev.micro.50.1.791. [DOI] [PubMed] [Google Scholar]
- 61.Fillingame RH. Coupling H+ transport and ATP synthesis ln F1F0-ATP synthases: glimpses of interacting parts in a dynamic molecular machine. J Exp Biol. 1997;200:217–224. doi: 10.1242/jeb.200.2.217. [DOI] [PubMed] [Google Scholar]
- 62.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bact Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anthony C. Quinoproteins and energy transduction. In: Anthony C, editor. Bacterial energy transduction. New York: Academic; 1988. pp. 293–316. [Google Scholar]
- 64.Hooper AB, Vannelli T, Bergmann DJ, Arciero DM. Enzymology of the oxidation of ammonia to nitrite by bacteria. Antonie Van Leeuwenhoek. 1997;71:59–67. doi: 10.1023/a:1000133919203. [DOI] [PubMed] [Google Scholar]
- 65.Poughon L, Dussap CG, Gros JB. Energy model and metabolic flux analysis for autotrophic nitrifiers. Biotechnol Bioeng. 2001;72:416–433. [PubMed] [Google Scholar]
- 66.Aleem MI. Generation of reducing power in chemosynthesis. II. Energy-linked reduction of pyridine nucleotides in the chemoautotroph, Nitrosomonas europaea . Biochim Biophys Acta. 1966;113:216–224. doi: 10.1016/s0926-6593(66)80062-0. [DOI] [PubMed] [Google Scholar]
- 67.Arp DJ, Stein LY. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit Rev Biochem Mol Biol. 2003;38:471–495. doi: 10.1080/10409230390267446. [DOI] [PubMed] [Google Scholar]
- 68.Wood PM. Nitrification as a bacterial energy source. In: Prosser JI, editor. Nitrification. Oxford: IRL; 1986. pp. 39–62. [Google Scholar]
- 69.Eck RV, Dayhoff MO. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science. 1966;152:363–366. doi: 10.1126/science.152.3720.363. [DOI] [PubMed] [Google Scholar]
- 70.Blaschkowski HP, Neuer G, Ludwig-Festl M, Knappe J. Routes of flavodoxin and ferredoxin reduction in Escherichia coli. CoA-acylating pyruvate: flavodoxin and NADPH: flavodoxin oxidoreductases participating in the activation of pyruvate formate-lyase. Eur J Biochem. 1982;123:563–569. [PubMed] [Google Scholar]
- 71.Furdui C, Ragsdale SW. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood–Ljungdahl pathway. J Biol Chem. 2000;275:28494–28499. doi: 10.1074/jbc.M003291200. [DOI] [PubMed] [Google Scholar]
- 72.Shanmugasundaram T, Wood HG. Interaction of ferredoxin with carbon monoxide dehydrogenase from Clostridium thermoaceticum . J Biol Chem. 1992;267:897–900. [PubMed] [Google Scholar]
- 73.Jungermann K, Kirchniawy H, Thauer RK. Ferredoxin dependent CO2 reduction to formate in Clostridium pasteurianum . Biochem Biophys Res Commun. 1970;41:682–689. doi: 10.1016/0006-291x(70)90067-7. [DOI] [PubMed] [Google Scholar]
- 74.Thauer RK, Rupprecht E, Jungermann K. The synthesis of one-carbon units from CO2 via a new ferredoxin dependent monocarboxylic acid cycle. FEBS Lett. 1970;8:304–307. doi: 10.1016/0014-5793(70)80293-9. [DOI] [PubMed] [Google Scholar]
- 75.Thauer RK, Kaster AK, Goenrich M, Schick M, Hiromoto T, Shima S. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem. 2010;79:507–536. doi: 10.1146/annurev.biochem.030508.152103. [DOI] [PubMed] [Google Scholar]
- 76.Hedderich R. Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J Bioenerg Biomembr. 2004;36:65–75. doi: 10.1023/b:jobb.0000019599.43969.33. [DOI] [PubMed] [Google Scholar]
- 77.Bott M, Eikmanns B, Thauer RK. Coupling of carbon monoxide oxidation to CO2 and H2 with the phosphorylation of ADP in acetate-grown Methanosarcina barkeri . Eur J Biochem. 1986;159:393–398. doi: 10.1111/j.1432-1033.1986.tb09881.x. [DOI] [PubMed] [Google Scholar]
- 78.Bott M, Thauer RK. Proton translocation coupled to the oxidation of carbon monoxide to CO2 and H2 in Methanosarcina barkeri . Eur J Biochem. 1989;179:469–472. doi: 10.1111/j.1432-1033.1989.tb14576.x. [DOI] [PubMed] [Google Scholar]
- 79.Welte C, Krätzer C, Deppenmeier U. Involvement of Ech hydrogenase in energy conservation of Methanosarcina mazei. FEBS J. 2010;277:3396–3403. doi: 10.1111/j.1742-4658.2010.07744.x. [DOI] [PubMed] [Google Scholar]
- 80.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 81.Deppenmeier U. The unique biochemistry of methanogenesis. Prog Nucleic Acid Res Mol Biol. 2002;71:223–283. doi: 10.1016/s0079-6603(02)71045-3. [DOI] [PubMed] [Google Scholar]
- 82.Deppenmeier U, Müller V. Life close to the thermodynamic limit: how methanogenic archaea conserve energy. Results Probl Cell Differ. 2008;45:123–152. doi: 10.1007/400_2006_026. [DOI] [PubMed] [Google Scholar]
- 83.Sapra R, Bagramyan K, Adams MWW. A simple energy-conserving system: proton reduction coupled to proton translocation. Proc Natl Acad Sci USA. 2003;100:7545–7550. doi: 10.1073/pnas.1331436100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pisa KY, Huber H, Thomm M, Müller V. A sodium ion-dependent A1AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus . FEBS J. 2007;274:3928–3938. doi: 10.1111/j.1742-4658.2007.05925.x. [DOI] [PubMed] [Google Scholar]
- 85.Friedrich T, Scheide D. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 2000;479:1–5. doi: 10.1016/s0014-5793(00)01867-6. [DOI] [PubMed] [Google Scholar]
- 86.Friedrich T, Weiss H. Modular evolution of the respiratory NADH:ubiquinone oxidoreductase and the origin of its modules. J Theor Biol. 1997;187:529–540. doi: 10.1006/jtbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 87.Vignais PM, Colbeau A. Molecular biology of microbial hydrogenases. Curr Issues Mol Biol. 2004;6:159–188. [PubMed] [Google Scholar]
- 88.Brandt U, Kerscher S, Dröse S, Zwicker K, Zickermann V. Proton pumping by NADH:ubiquinone oxidoreductase. A redox driven conformational change mechanism? FEBS Lett. 2003;545:9–17. doi: 10.1016/s0014-5793(03)00387-9. [DOI] [PubMed] [Google Scholar]
- 89.Tokuda H, Unemoto T. Na+ is translocated at NADH:quinone oxidoreductase segment in the respiratory chain of Vibrio alginolyticus . J Biol Chem. 1984;259:7785–7790. [PubMed] [Google Scholar]
- 90.Tokuda H, Unemoto T. The Na+-motive respiratory chain of marine bacteria. Microbiol Sci. 1985;2:65–71. [PubMed] [Google Scholar]
- 91.Barquera B, Hellwig P, Zhou W, Morgan JE, Hase CC, Gosink KK, Nilges M, Bruesehoff PJ, Roth A, Lancaster CR, Gennis RB. Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae . Biochemistry. 2002;41:3781–3789. doi: 10.1021/bi011873o. [DOI] [PubMed] [Google Scholar]
- 92.Tokuda H, Udagawa T, Unemoto T. Generation of the electrochemical potential of Na+ by the Na+-motive NADH oxidase in inverted membrane vesicles of Vibrio alginolyticus . FEBS Lett. 1985;183:95–98. doi: 10.1016/0014-5793(85)80961-3. [DOI] [PubMed] [Google Scholar]
- 93.Steuber J. Na+-translocation by bacterial NADH:quinone oxidoreductases: an extension to the complex-I family of primary redox pumps. Biochim Biophys Acta. 2001;1505:45–56. doi: 10.1016/s0005-2728(00)00276-0. [DOI] [PubMed] [Google Scholar]
- 94.Hayashi M, Nakayama Y, Unemoto T. Recent progress in the Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus . Biochim Biophys Acta. 2001;1505:37–44. doi: 10.1016/s0005-2728(00)00275-9. [DOI] [PubMed] [Google Scholar]
- 95.Hayashi M, Nakayama Y, Yasui M, Maeda M, Furuishi K, Unemoto T. FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus . FEBS Lett. 2001;488:5–8. doi: 10.1016/s0014-5793(00)02404-2. [DOI] [PubMed] [Google Scholar]
- 96.Barquera B, Zhou W, Morgan JE, Gennis RB. Riboflavin is a component of the Na+-pumping NADH-quinone oxidoreductase from Vibrio cholerae . Proc Natl Acad Sci USA. 2002;99:10322–10324. doi: 10.1073/pnas.162361299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pfenninger-Li XD, Albracht SPJ, Vanbelzen R, Dimroth P. NADH:Ubiquinone oxidoreductase of Vibrio alginolyticus: Purification, properties, and reconstitution of the Na+ pump. Biochemistry. 1996;35:6233–6242. doi: 10.1021/bi953032l. [DOI] [PubMed] [Google Scholar]
- 98.Zhou W, Bertsova YV, Feng B, Tsatsos P, Verkhovskaya ML, Gennis RB, Bogachev AV, Barquera B. Sequencing and preliminary characterization of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio harveyi . Biochemistry. 1999;38:16246–16252. doi: 10.1021/bi991664s. [DOI] [PubMed] [Google Scholar]
- 99.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 100.Türk K, Puhar A, Neese F, Bill E, Fritz G, Steuber J. NADH oxidation by the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae: functional role of the NqrF subunit. J Biol Chem. 2004;279:21349–21355. doi: 10.1074/jbc.M311692200. [DOI] [PubMed] [Google Scholar]
- 101.Häse CC, Barquera B. Role of sodium bioenergetics in Vibrio cholerae . Biochim Biophys Acta. 2001;1505:169–178. doi: 10.1016/s0005-2728(00)00286-3. [DOI] [PubMed] [Google Scholar]
- 102.Häse CC, Mekalanos JJ. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae . Proc Natl Acad Sci USA. 1999;96:3183–3187. doi: 10.1073/pnas.96.6.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Müller V, Aufurth S, Rahlfs S. The Na+ cycle in Acetobacterium woodii: identification and characterization of a Na+-translocating F1FO-ATPase with a mixed oligomer of 8 and 16 kDa proteolipids. Biochim Biophys Acta. 2001;1505:108–120. doi: 10.1016/s0005-2728(00)00281-4. [DOI] [PubMed] [Google Scholar]
- 104.Müller V, Gottschalk G. The sodium ion cycle in acetogenic and methanogenic bacteria: generation and utilization of a primary electrochemical sodium ion gradient. In: Drake HL, editor. Acetogenesis. New York: Chapman & Hall; 1994. pp. 127–156. [Google Scholar]
- 105.Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, Wollherr A, Ehrenreich A, Liebl W, Gottschalk G, Dürre P. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc Natl Acad Sci USA. 2010;107:13087–13092. doi: 10.1073/pnas.1004716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stolpe S, Friedrich T. The Escherichia coli NADH:ubiquinone oxidoreductase (complex I) is a primary proton pump but may be capable of secondary sodium antiport. J Biol Chem. 2004;279:18377–18383. doi: 10.1074/jbc.M311242200. [DOI] [PubMed] [Google Scholar]
- 107.Krebs W, Steuber J, Gemperli AC, Dimroth P. Na+-translocation by the NADH:ubiquinone oxidoreductase (complex I) from Klebsiella pneumoniae . Mol Microbiol. 1999;33:590–598. doi: 10.1046/j.1365-2958.1999.01506.x. [DOI] [PubMed] [Google Scholar]
- 108.Gemperli AC, Dimroth P, Steuber J. Sodium ion cycling mediates energy coupling between complex I and ATP synthase. Proc Natl Acad Sci USA. 2003;100:839–844. doi: 10.1073/pnas.0237328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Efiok BJ, Webster DA. A cytochrome that can pump sodium ion. Biochem Biophys Res Commun. 1990;173:370–375. doi: 10.1016/s0006-291x(05)81067-8. [DOI] [PubMed] [Google Scholar]
- 110.Hallenbeck PC, Vignais PM. The effect of electron transport inhibitors on nitrogenase activity in the photosynthetic bacterium, Rhodopseudomonas capsulata . FEMS Microbiol Lett. 1981;12:15–18. [Google Scholar]
- 111.Schmidt GW, Matlin KS, Chua NH. A rapid procedure for selective enrichment of photosynthetic electron transport mutants. Proc Natl Acad Sci USA. 1977;74:610–614. doi: 10.1073/pnas.74.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saeki K, Kumagai H. The rnf gene products in Rhodobacter capsulatus play an essential role in nitrogen fixation during anaerobic DMSO-dependent growth in the dark. Arch Microbiol. 1998;169:464–467. doi: 10.1007/s002030050598. [DOI] [PubMed] [Google Scholar]
- 113.Desnoues N, Lin M, Guo X, Ma L, Carreño-Lopez R, Elmerich C. Nitrogen fixation genetics and regulation in a Pseudomonas stutzeri strain associated with rice. Microbiology. 2003;149:2251–2262. doi: 10.1099/mic.0.26270-0. [DOI] [PubMed] [Google Scholar]
- 114.Cunningham RP, Asahara H, Bank JF, Scholes CP, Salerno JC, Surerus K, Munck E, McCracken J, Peisach J, Emptage MH. Endonuclease III is an iron-sulfur protein. Biochemistry. 1989;28:4450–4455. doi: 10.1021/bi00436a049. [DOI] [PubMed] [Google Scholar]
- 115.Faruque SM, Nair GB. Molecular ecology of toxigenic Vibrio cholerae . Microbiol Immunol. 2002;46:59–66. doi: 10.1111/j.1348-0421.2002.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 116.Brüggemann H, Gottschalk G. Insights in metabolism and toxin production from the complete genome sequence of Clostridium tetani . Anaerobe. 2004;10:53–68. doi: 10.1016/j.anaerobe.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 117.Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri . J Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Herrmann G, Jayamani E, Mai G, Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol. 2008;190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brandt U. Bifurcated ubihydroquinone oxidation in the cytochrome bc1 complex by proton-gated charge transfer. FEBS Lett. 1996;387:1–6. doi: 10.1016/0014-5793(96)00436-x. [DOI] [PubMed] [Google Scholar]
- 120.Schut GJ, Adams MW. The iron hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol. 2009;191:4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McInerney MJ, Rohlin L, Mouttaki H, Kim U, Krupp RS, Rios-Hernandez L, Sieber J, Struchtemeyer CG, Bhattacharyya A, Campbell JW, Gunsalus RP. The genome of Syntrophus aciditrophicus: life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci USA. 2007;104:7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Badziong W, Thauer RK, Zeikus JG. Isolation and characterization of Desulfovibrio growing on hydrogen plus sulfate as the sole energy source. Arch Microbiol. 1978;116:41–49. doi: 10.1007/BF00408732. [DOI] [PubMed] [Google Scholar]
- 123.Badziong W, Thauer RK. Growth yields and growth rates of Desulfovibrio vulgaris (Marburg) growing on hydrogen plus sulfate and hydrogen plus thiosulfate as the sole energy sources. Arch Microbiol. 1978;117:209–214. doi: 10.1007/BF00402310. [DOI] [PubMed] [Google Scholar]
- 124.Strittmatter AW, Liesegang H, Rabus R, Decker I, Amann J, Andres S, Henne A, Fricke WF, Martinez-Arias R, Bartels D, Goesmann A, Krause L, Pühler A, Klenk HP, Richter M, Schüler M, Glöckner FO, Meyerdierks A, Gottschalk G, Amann R. Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ Microbiol. 2009;11:1038–1055. doi: 10.1111/j.1462-2920.2008.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ferry JG. Biochemistry of methanogenesis. Crit Rev Biochem Mol Biol. 1992;27:473–503. doi: 10.3109/10409239209082570. [DOI] [PubMed] [Google Scholar]
- 126.Rother M. Methanogenesis. In: Timmis KN, editor. Handbook of hydrocarbon and lipid microbiology. Berlin: Springer; 2010. pp. 483–499. [Google Scholar]
- 127.Müller V, Winner C, Gottschalk G. Electron transport-driven sodium extrusion during methanogenesis from formaldehyde + H2 by Methanosarcina barkeri . Eur J Biochem. 1988;178:519–525. doi: 10.1111/j.1432-1033.1988.tb14478.x. [DOI] [PubMed] [Google Scholar]
- 128.Gottschalk G, Thauer RK. The Na+-translocating methyltransferase complex from methanogenic archaea. Biochim Biophys Acta. 2001;1505:28–36. doi: 10.1016/s0005-2728(00)00274-7. [DOI] [PubMed] [Google Scholar]
- 129.Deppenmeier U. Redox-driven proton translocation in methanogenic archaea. Cell Mol Life Sci. 2002;59:1–21. doi: 10.1007/s00018-002-8526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rohlin L, Gunsalus RP. Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol. 2010;10:62. doi: 10.1186/1471-2180-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Franzmann PD, Springer N, Ludwig W, Conway de Macario E, Rohde M. A methanogenic archaeon from Ace lake, antarctica: Methanococcoides burtonii sp. nov. Syst Appl Microbiol. 1992;15:573–581. [Google Scholar]
- 132.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 133.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic organization of rnf genes. nth encodes for endonuclease III. Membrane-bound subunits are indicated by an asterisk. A rnf cluster ABCDGE, B rnf cluster CDGEAB, C rnf cluster BCDGEA, D rnf organization in archaea. x encodes for a hypothetical protein. R. capsulatus: Rhodobacter capsulatus, A. vinelandii: Azotobacter vinelandii, E. coli: Escherichia coli, V. cholerae: Vibrio cholerae, K. pneumoniae: Klebsiella pneumoniae, A. woodii: Acetobacterium woodii, C. tetani: Clostridium tetani, A. metalliredigens: Alkaliphilus metalliredigens, T. pseudethanolicus: Thermoanaerobacter pseudethanolicus, C. kluyveri: Clostridium kluyveri, R. torques: Ruminococcus torques, B. vulgatus: Bacteroides vulgatus, C. limicola: Chlorobium limicola, P. sp.: Parabacteroides sp., P. aestuarii: Prosthecochloris aestuarii, P. uenonsis: Porphyromonas uenonsis, M. acetivorans: Methanosarcina acetivorans, M. burtonii: Methanococcoides burtonii (TIFF 79 kb)
Topology model of RnfD (based on SOSUI prediction, [132]). The N- and C-termini are indicated. Threonine 157 is highlighted in grey as potential FMN-binding site. Aspartate 250 and glutamate 300 are highlighted (black) as potential amino acids involved in Na+ binding (TIFF 124 kb)
Topology model of RnfG (based on SOSUI prediction, [132]). The N- and C-termini are indicated. Threonine 185 is highlighted in grey as potential FMN-binding site (TIFF 97 kb)
Topology model of RnfE (based on Sääf et al. 1999). The N- and C-termini are indicated. Aspartate 129 is highlighted (black) as potential Na+ binding site (TIFF 106 kb)
Topology model of RnfA (based on Sääf et al. 1999). The N- and C-termini are indicated. Glutamate 88 is highlighted (black) as potential Na+ binding site (TIFF 105 kb)
Topology model of RnfB (based on SOSUI prediction, [132]). The N- and C-termini are indicated. Conserved cysteines are highlighted in grey, that might form the FeS cluster binding sites (TIFF 132 kb)
Alignment of RnfD and NqrB. Alignment was done using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html [133]). An asterisk indicates complete amino acid conservation. The arrow shows the potential Na+-binding site. VC-Vibrio cholerae, AW-Acetobacterium woodii (TIFF 91 kb)
Alignment of RnfE and NqrD. Alignment was done using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html [133]). An asterisk indicates complete amino acid conservation. The arrow shows the potential Na+-binding site. VC-Vibrio cholerae, AW-Acetobacterium woodii (TIFF 62 kb)
Alignment of RnfA and NqrE. Alignment was done using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html [133]). An asterisk indicates complete amino acid conservation. The arrow shows the potential Na+-binding site. VC-Vibrio cholerae, AW-Acetobacterium woodii (TIFF 59 kb)