Abstract
Signal strength evoked by ligand stimulation is crucial for cellular responses such as fate decision, cell survival/death, secretion, and migration. For example, morphogens are secreted signaling molecules that form concentration gradients within tissues and induce distinct cell fates in a signal strength-dependent manner. In addition to extracellular ligand abundance, the sensitivity of signal-receiving cells to ligands also influences signal strength. Cell sensitivity to ligands is controlled at various levels: receptor presentation at the cell surface, positive/negative regulation of signal transduction, and target gene activation/repression. While the regulation of signal transduction and gene transcription is well studied, receptor presentation is still not fully understood. Recently, it was reported that cellular sensitivity to the Wingless (Wg)/Wnt morphogen is regulated by balanced ubiquitination and deubiquitination of its receptor Frizzled (Fz). In this review, we review how ubiquitination regulates receptor presentation at the cell surface for the detection of extracellular signal strength.
Keywords: Endocytosis, Trafficking, Wg/Wnt, Frizzled, Ubiquitin
Introduction
Signal strength plays essential roles in various biological processes including pattern formation [1–3] and cell migration in development, and chemotaxis of leukocyte and tumor cells [4, 5]. Signal strength is controlled at the levels of extracellular signaling ligands, cell surface receptors, intracellular signal transduction molecules, transcription factors, and chromatin regulators. The amount of extracellular signaling ligands is the primary determinant of signal strength. The levels of extracellular ligands depend on the expression levels, transport efficiencies to target cells, and degradation rates of the ligands. Once the ligands reach the target cells, they are detected by specific receptors presented at the cell surface. Therefore, regulation of receptor presentation is crucial for precise detection of extracellular signal strength and is controlled by the level of expression and by subcellular localization.
Some receptors are constitutively cycling between the cell surface and the endosomes. Localization of the majority of receptors is determined by a balance in internalization and recycling rates. For example, the epidermal growth factor receptor (EGFR) is mainly located at the cell surface because its recycling rate is about ten times higher than its internalization rate [6–8]. When EGF binds to EGFR, the internalization rate of EGF–EGFR complexes is increased [6], and these internalized complexes are sorted to different cellular destinations, such as the cell surface or multi vesicular bodies (MVBs), where they are further degraded in lysosomes. Given that sorting to the lysosome requires the ubiquitination of EGFR [9], its subcellular localization may be determined by its ubiquitination. However, ubiquitin-mediated control of receptor dynamics differs depending on receptor species and biological context. Here, we review ubiquitin-mediated control of receptor localization and cellular responsiveness to extracellular ligands.
Ubiquitination and deubiquitination
Ubiquitination, one of the major post-translational modifications, regulates cellular signaling and homeostasis [10, 11]. Ubiquitin is conjugated to target proteins in a series of reactions conducted by ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes [12–14]. There are two types of ubiquitin-protein conjugations: monoubiquitination and polyubiquitination. The former is the addition of a single ubiquitin to a target protein and, when it occurs at multiple sites on a single target protein, the term multi-monoubiquitination is employed. The latter is the attachment of ubiquitin chains to a target protein. Ubiquitin chains are generated by the formation of an isopeptide bond between a carboxyl group of ubiquitin Gly76 and an ε-amino group of another ubiquitin Lys. Among the seven Lys residues in ubiquitin, Lys48- and Lys11-linked ubiquitin chains are used as tags for degradation by the 26S proteasome [15–19]. Lys63-linked chains are involved in the activation of nuclear factor-κB (NF-κB), the orchestration of different steps during DNA repair, and the targeting of the modified protein to the lysosome [11, 20–26]. Monoubiquitination serves to regulate the internalization of cell-surface proteins, the sorting of internalized proteins, as well as other cellular processes [23, 27–29].
Removal of ubiquitin moieties from modified proteins is catalyzed by a number of deubiquitination enzymes (DUBs) [30–34]. DUBs comprise two classes of proteolytic enzymes: cysteine proteases and metalloproteases. The cysteine protease class includes four families: the ubiquitin-specific protease (USP), the ubiquitin C-terminal hydrolase (UCH), the ovarian tumor protease (OTU), and the Machado–Joseph disease protein domain protease (MJD) families. The metalloprotease class of DUBs comprises one family: the JAB1/MPN/Mov34 (JAMM) motif protease family.
There is a large number of E3 enzymes and DUBs, including 600–1,000 E3s and more than 100 DUBs in humans. Because different enzymes have different spectrums of substrates, a large number of target proteins are regulated by these enzymes in different contexts. This suggests that the balance of ubiquitination and deubiquitination activities may play essential roles in each protein’s function.
Role of ubiquitination in internalization, lysosomal trafficking, and degradation of receptors
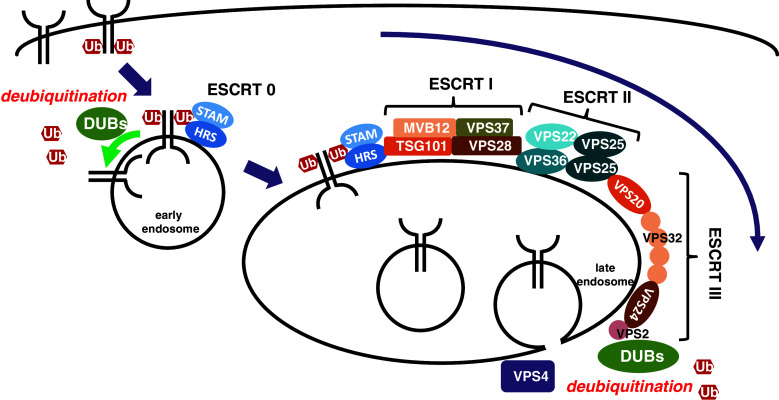
Monoubiquitinated or Lys63-linked polyubiquitinated membrane proteins are transported to lysosomes for degradation through the endocytic pathway. As there are excellent reviews of membrane protein degradation available [35–37], we will only briefly summarize the degradation process here (Fig. 1). Upon stimulation, receptors are ubiquitinated at the plasma membrane by specific E3 enzymes and internalized into endosomes via membrane vesicles. Ubiquitinated receptors at endosomes are recognized by ubiquitin binding-domains (UBDs) of hepatocyte growth factor-regulated substrate (HRS) and signal-transduction adaptor molecule 1/2 (STAM1/2), the main components of the endosomal sorting complex required for transport (ESCRT)-0 complex. ESCRT-0 complex recruits ESCRT-I, which is composed of Tsg101, Vps28, Vps37, and Mvb12. While Tsg101 interacts with HRS through its common ubiquitin- and ESCRT-0-binding UEV domain, Vps28 binds to GRAM-like ubiquitin-binding in Eap45 (GLUE) domain of Vps36 that forms ESCRT-II complex with Vps22 and Vps25. Therefore, ESCRT-I complex acts as a bridge between ESCRT-0 and -II complexes. Vps25 of ESCRT II, in turn, binds Vps20, a component of ESCRT-III, to recruit ESCRT-III complex. ESCRT-III complex clusters ubiquitinated receptors into the invaginated endosomal membrane, retrieves ubiquitins from those receptors by recruiting DUBs, and forms intraluminal vesicles filled with receptors by recruiting Vps4 ATPase. Endosomes with intraluminal vesicles, called multi-vesicular bodies (MVBs), are fused to lysosomes for degradation of their contents.
Fig. 1.
Endocytic transport of membrane receptors for degradation. Receptors ubiquitinated at the plasma membrane are internalized and recognized by ESCRT-0. Recognized receptors are sequentially transported by ESCRT-I, -II and -III complexes, and internalized into luminal side of late endosomes to form multi-vesicular bodies for degradation. Deubiquitination occurs at early and late endosomes to regulate receptor degradation and ubiquitin retrieval, respectively
The roles of ubiquitination and deubiquitination have been extensively studied in yeast and mammalian cells. DUBs located at early and late endosomes promote deubiquitination of receptors in the degradation pathway in at least two ways (Fig. 1). First, DUBs at the early endosome regulate degradation rates of internalized receptors before they are trapped by ESCRT-0. When ubiquitinated receptors are deubiquitinated, they are recycled back to the plasma membrane or retained at endosomes, resulting in delayed degradation. However, as described below, the effects of deubiquitination of EGFR are still under debate. Second, DUBs at the late endosome, as described above, retrieve ubiquitins from ubiquitinated receptors just before interluminal vesicle formation.
The role of ubiquitination in intracellular trafficking was first demonstrated in yeast, in which ubiquitin is required for the first step of receptor internalization, as well as for trafficking to lysosomes [38, 39]. In mammalian cells, however, the role of ubiquitination in receptor trafficking is more complex. The epidermal growth factor receptor (EGFR), G protein-coupled receptors (GPCRs), and Notch have been intensively studied; therefore, here, we use these proteins as examples to discuss the roles of ubiquitination in intracellular trafficking.
EGFR
Binding of EGF to EGFR induces EGFR internalization and localization to the lysosome. Initially, monoubiquitination of EGFR was shown to be sufficient for its internalization [29]. Furthermore, mass-spectrometric analysis identified Lys63-linked polyubiquitin chains, as well as monoubiquitin, in wild-type EGFR [40], suggesting that polyubiquitination may also contribute to the efficient internalization of EGFR. Conversely, it was also shown that ubiquitination of EGFR is not required for its internalization. Non-ubiquitinated forms of EGFR, which had been mutagenized either at the E3 enzyme binding site or at the acceptor Lys residues for ubiquitination, exhibited normal internalization [9, 41, 42]. This discrepancy may be due to the existence of alternative internalization pathways, including clathrin-dependent and -independent routes [42, 43]. Interestingly, the clathrin-dependent and -independent internalization pathways are used differentially depending on the level of EGFR ubiquitination, which is regulated by the local concentration of EGF. When the EGF concentration is low, EGFR is not ubiquitinated and is instead internalized through the clathrin-dependent pathway. By contrast, at high EGF concentrations, EGFR is ubiquitinated and internalized through both clathrin-dependent and clathrin-independent pathways. Interestingly, this suggests that different EGF signal strengths evoke different cellular responses depending on the level of ubiquitination and the internalization pathway of its receptor.
Although the precise roles of EGFR ubiquitination in its internalization remain obscure, it is widely accepted that ubiquitination is essential for lysosomal trafficking and degradation of EGFR. As described above, ubiquitinated EGFR, internalized into the early endosome, is recognized by HRS and STAM1/2, and then incorporated into the lumen of MVBs by ESCRT-I, -II, and -III complexes [37]. Therefore, deubiquitination of EGFR is thought to play a critical role in the regulation of EGFR degradation. However, the effects of deubiquitination are still under debate. While it is known that UBPY is involved in EGFR deubiquitination [44, 45], several reports show an opposing effect of UBPY on EGFR degradation. One particular study showed that overexpression or knockdown of UBPY results in delay or acceleration of EGF-induced EGFR degradation, respectively [44]. By contrast, another study showed that knockdown of UBPY blocks EGFR down-regulation [45, 46]. The involvement of the associated molecule with the SH3 domain of STAM (AMSH) in EGFR deubiquitination is also controversial. Whereas overexpression of AMSH has no effect on the level of EGFR ubiquitination in EGF-stimulated cells [44], a previous study showed that knockdown of AMSH increases the level of EGFR ubiquitination [47]. Further investigation is necessary to resolve these discrepancies.
GPCRs
GPCRs, also known as seven-transmembrane receptors, represent the largest family of membrane proteins in mammalian cells [48, 49]. In humans, there are about 700 GPCRs that control a wide range of physiological events. Ligand-dependent activation of GPCRs occurs at the cell surface and evokes intracellular signaling, mainly through heterotrimeric G proteins. Activated GPCRs are then internalized and transported to lysosomes where they are degraded. Therefore, signaling via GPCRs is at least partially regulated by internalization of the receptors. Interestingly, ligand-independent constitutive internalization has also been reported [50]. It is thought that cellular signaling responsiveness is attenuated by GPCR internalization and restored by its recycling to the cell surface. When internalized GPCRs are transported to lysosomes, responsiveness may be downregulated.
The role of ubiquitination in intracellular trafficking of GPCRs was first reported to be required for the efficient internalization of Ste2p in yeast [38]. The addition of α-mating factor, the ligand for Ste2p, increases the level of ubiquitination and internalization of Ste2p.
By contrast, it is generally agreed that direct ubiquitination is not essential for efficient internalization of mammalian GPCRs, but it does affect intracellular trafficking of GPCRs after internalization. Similar to the case of EGFR, ubiquitinated GPCRs are transported to lysosomes, mainly by interaction with ESCRT complexes. A requirement for direct ubiquitination in lysosome trafficking was shown for rhodopsin [51], β2-adrenergic receptor (β2AR) [52], chemokine receptor 4 (CXCR4) [53], vasopressin receptor 2 (V2R) [54], proteolytically activated receptor 2 (PAR2) [55], neurokinin receptor 1 (NK1) [56], and κ-opioid receptor (KOR) [57]. Lysosomal degradation of these receptors has been shown to be inhibited or delayed by mutations of lysine residues that are modified by ubiquitin. By contrast, ubiquitin-deficient PAR1, in which all lysine residues are replaced with arginines, resulted in its increased constitutive internalization. Moreover, direct fusion of ubiquitin to PAR1 with no lysine residues inhibited its constitutive internalization [58]. The ubiquitination of PAR1 may block the interaction between PAR1 and the AP2 adaptor complex, which is essential for the constitutive internalization of PAR1. Consistently, PAR1 is ubiquitinated under unstimulated conditions and deubiquitinated upon ligand stimulation. Similar to PAR1, the δ-opioid receptor (DOR) is effectively downregulated when it is not ubiquitinated [59]. This was recently found to be because ubiquitination of DOR is not involved in the early sorting of internalized DOR out of the recycling pathway [60]. Therefore, the effects of ubiquitination vary depending on GPCR species.
To date, among the many E3 enzymes and DUBs currently known, only a few have been found to regulate the ubiquitination level of GPCRs. Members of the E3 enzyme family involved in GPCR regulation are neuronal precursor cell-expressed developmentally downregulated 4-1 (Nedd4-1) for β2AR [61, 62], AIP4 for CXCR4 [63–65] and δ-opioid receptor (DOR) [66], and c-Cbl for platelet-activating factor (PAF) [67] and PAR2 [55]. The DUB family of proteins contain USP33 and USP20 for β2AR [68], USP14 for CXCR4 [69], UBPY and AMSH for DOR [66] and PAR2 [70], and cylindromatosis tumor suppressor gene (CYLD) for KOR [57]. Interestingly, knockdown of some DUBs exhibited different results. When expression of both USP20 and USP33 was reduced by RNAi, ubiquitination, lysosomal trafficking, and degradation of β2AR increased, while β2AR recycling was completely inhibited [68]. By contrast, knockdown of UBPY expression stabilized CXCR4 on the cell surface and attenuated receptor degradation [71]. In this context, UBPY promoted deubiquitination of HRS and AMSH1, but not of CXCR4. As described below, UBPY directly deubiquitinated Drosophila Fz2 (DFz2) and mammalian Fz4 to promote its own recycling to the plasma membrane. Regulation of receptor dynamics by ubiquitination and deubiquitination is therefore dependent on receptor species and biological context.
Notch
Notch is an evolutionarily conserved receptor for transmembrane protein ligands: Dl and Ser in Drosophila and Delta-like (Dll) and Jagged in mammals [72]. In contrast to other signaling systems that involve soluble ligands, ubiquitination of Notch ligands plays an essential role in conventional Notch signaling. As this issue has already been covered in excellent reviews [73, 74], here we will only address the ubiquitination roles of Notch.
Upon binding to the ligands, Notch is activated by a sequence of proteolytic processes [72]. Notch is cleaved by a disintegrin and metalloprotease (ADAM) proteases at site 2 (S2) in the juxtamembrane region of the extracellular domain (between Ala1710 and Val1711 of mouse Notch1) [75, 76] followed by further cleavage by γ-secretase within the transmembrane domain at site 3 (S3) (between Gly1743 and Val1744) [77] and site 4 (S4) (between Ala1731 and Ala1732) [78] to release the Notch intracellular domain (NICD). Soluble NICD translocates to the nucleus and regulates the expression of downstream target genes.
S2 cleavage takes place at the cell surface upon Notch interaction with its ligands, and whether the following S3 and S4 cleavages require Notch internalization has been extensively investigated. Recently, the requirement of endocytosis in this process was supported by the results of several studies. Ligand-dependent activation of Notch in the signal-receiving cell is impaired by loss of the endocytic protein dynamin [79], early endosomal protein rab5 [80], endocytic syntaxin avalanche [81], or vacuolar ATPase (V-ATPase), which is required for endosomal/lysosomal acidification [82, 83]. However, it remains unclear whether the ubiquitination of Notch is essential for its internalization and activation. The non-ubiquitinated form of Notch in which Lys at position 1749 is replaced to Arg (K1749R) compromises its internalization, cleavage, and activation [84]. However, the cleaved site of this mutant (K1749R) is different from the S3/S4 cleavage site [85], suggesting that impairment of Notch activation may be caused by side-effects of mutagenesis.
Ligand-independent internalization of Notch, followed by either recycling back to the cell surface or lysosomal trafficking, is important for preventing ectopic activation of Notch. In mutants of components of the ESCRT complex involved in the intracellular trafficking of ubiquitinated proteins [80, 86–89] or endosomal protein lethal giant discs (lgd) [90–92], Notch is improperly cleaved and activated in a ligand-independent manner. Ubiquitination of Notch is catalyzed by RING finger E3 enzymes, Deltex (Dx) [93, 94] and c-Cbl [95, 96], and two HECT domain-containing E3 enzymes, Suppressor of deltex [Su(dx)] [97–99] and Nedd4 [100, 101]. Su(dx)/Itch/AIP4, Nedd4, and c-Cbl are involved in lysosomal trafficking for the degradation of unactivated Notch, suggesting that Notch ubiquitination prevents ligand-independent ectopic activation of Notch. In contrast to E3 enzymes, DUBs that directly deubiquitinate Notch have not been identified.
Graded activity of Wg/Wnt as morphogen
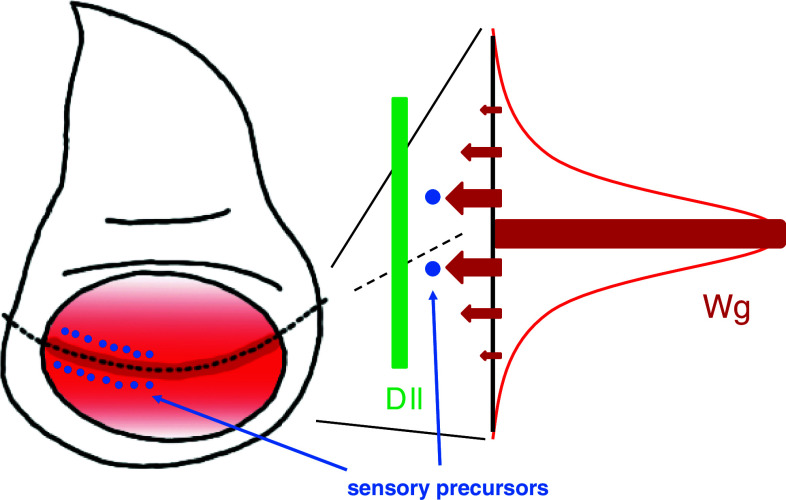
Pattern formation is mediated by morphogens, which are extracellular signaling molecules that are expressed in and secreted from a subset of cells in developing organs. Morphogens are distributed within tissues in a gradient manner and induce distinct cell fates in a dose-dependent manner to form genetically programmed patterns. Concentration plays an essential role in cell fate specification, cell proliferation, and cell polarity in the Wingless (Wg)/mammalian Wnt signaling pathway [102, 103]. Thus, defects in Wg/Wnt signaling cause developmental defects and metastatic tumors. In Drosophila wing discs, Wg is expressed in cells at the dorsoventral boundary and the cells show a concentration gradient of Wg distribution (Fig. 2). A high concentration of Wg induces sensory precursor cells near the dorsoventral boundary that develops the adult wing margin [104]. A lower threshold of Wg concentration for Distalless (Dll) expression ensures its broad expression in wing discs to shape the adult wings [105, 106]. Accordingly, mutants defective in Wg signaling exhibit deformation of the sensory cells in the wing margin as well as shrunken wings.
Fig. 2.
Wg morphogen in the Drosophila wing disc. (Left) Wg is expressed in cells (dark red) at the dorsoventral boundary (a dotted line). Secreted Wg is distributed in a concentration gradient manner (red). Sensory organ precursor cells (blue dots) are induced near the Wg-expressing region. (Right) Different levels of signal strength (arrows) are evoked depending on extracellular Wg concentration (red). A strong signal induces sensory precursor cells (blue dots) in the proximal region to the Wg-expressing cells. Dll expression (green) is induced over a broad area because it requires a lower threshold of Wg concentration
Wg/Wnt signals are mediated by their seven-transmembrane receptor family, Frizzled (Fz). Fz further activates three major pathways in a context-dependent manner: the canonical, planar cell polarity, and Ca2+ pathways [107]. As a morphogen, Wg/Wnt regulates tissue patterning depending on the strength of the canonical signaling pathway generated in target cells.
While the mechanisms of graded distribution of morphogens have been intensively investigated [108], the regulation of morphogen receptors is only partially understood. For example, forced expression of Fz in the Drosophila wing disc was shown to perturb the graded activity of Wg, indicating that the level of Fz must be tightly regulated to sense extracellular Wg signal strength [109]. The Fz level is thought to be regulated by its expression, localization, and degradation. While the regulation of Fz transcription has been shown, regulation of its localization and degradation remains to be fully elucidated.
Regulation of Fz by ubiquitination
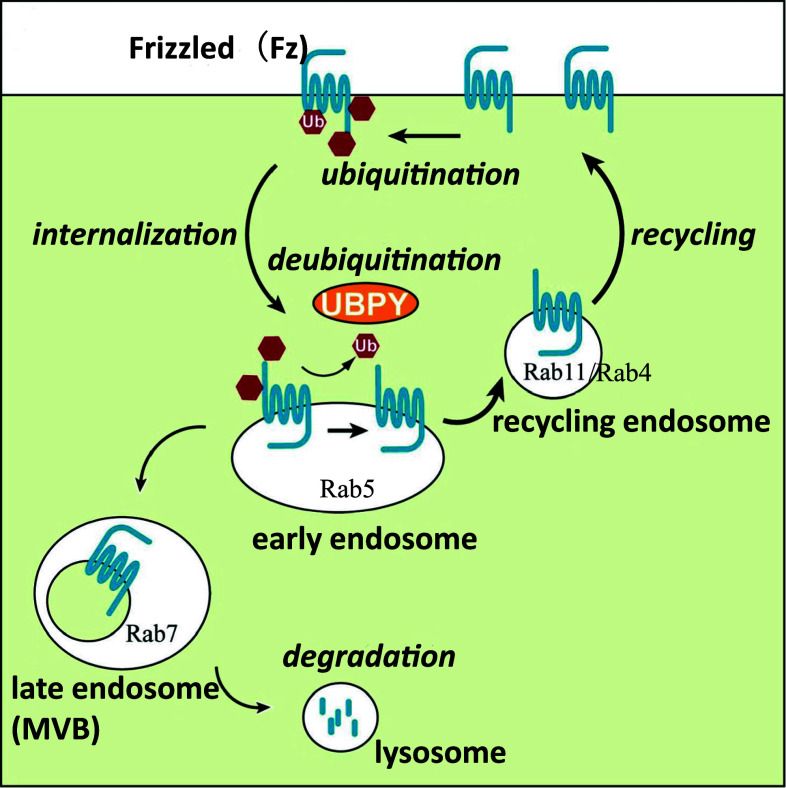
Fz receptors are internalized by machineries involving Dishevelled (Dvl), β-arrestin, protein kinase C, Arrow (Arr)/LRP 5 and 6, dynamin, and AP2 [110–112]. Following intracellular trafficking of Fz, receptors are regulated by small G proteins such as Rab5, Rab4, and Rab11 [113, 114]. While Rab5 is involved in the endocytic pathway, Rab4 and Rab11 participate in recycling routes, suggesting that Fz receptors are cycling between the cell surface and endosomes. Indeed, expression of a dominant negative form of Rab11 results in intracellular accumulation of Fz2 in Drosophila wing discs [114], and the aberrant activation of Wg/Wnt signaling by expression of activated Rab5 is restored by co-expression of activated forms of Rab4 or Rab11 [113]. This restoration may be due to the proper balance between endocytosis and recycling rates. It was recently revealed that the amount of Fz receptors (Drosophila Fz2 and mammalian Fz4 in this chapter) recycled back to the cell surface is determined by the balance between ubiquitination and deubiquitination (Fig. 3) [114]. Ubiquitination of Fz receptors is independent of ligand stimulation, suggesting that it is involved in constitutive cycling of Fz receptors. Although the E3 enzyme for ubiquitination of Fz receptors has not been identified, UBPY was found to directly deubiquitinate Fz receptors. When the level of ubiquitination is decreased by expression of the wild-type or activated form of UBPY, the cell surface level of Fz receptors is increased as a result of accelerated recycling of Fz receptors to the cell surface. Conversely, when the ubiquitination level is increased by expression of a dominant negative form or double-stranded RNA of UBPY, the cell surface level of Fz receptors is decreased by elevated trafficking to lysosomes. Importantly, the increase and decrease of cell surface levels of Fz receptors lead to the up- and down-regulation, respectively, of canonical Wg/Wnt signaling. Furthermore, in vivo studies have shown that inappropriate levels of Fz ubiquitination result in defects in wing patterning regulated by Wg morphogen. Collectively, it can be concluded that ubiquitination of Fz receptors regulates cellular responsiveness to Wg/Wnt signaling by the cell surface level of Fz receptors.
Fig. 3.
A model for the regulation of the cell surface level of Fz. Fz is constitutively ubiquitinated and internalized to the early endosome where UBPY is localized Fz. Deubiquitination of Fz is mediated by UBPY and deubiquitinated Fz is recycled back to the cell surface via the recycling endosome. Fz not deubiquitinated by UBPY is transported to the lysosome for degradation through the MVB/late endosome. The balance between ubiquitination at the cell surface and deubiquitination in the early endosome determines cellular responsiveness to Wg through the cell surface level of Fz
Regulation of cell surface levels of other receptors by ubiquitination
It was recently reported that cell surface levels of other receptors are also regulated by ubiquitination. Like the Fz family, receptor-like tyrosine kinase (RYK) that directly binds to Wnt molecules is involved in the Wnt canonical and other pathways. Mindbomb 1 (MIB1), an E3 ligase, promotes RYK ubiquitination and degradation by the proteasome and lysosome [115]. Interestingly, whereas upon Wnt-3a stimulation, RYK is cleaved by γ-secretase and its intracellular domain accumulates in the nucleus [116], RYK cleavage is not required for its ubiquitination and degradation by MIB1 [115]. This suggests that MIB1-mediated ubiquitination of RYK does not depend on the activation of RYK. The cell surface level of RYK is decreased by overexpression of MIB1, while it is increased by siRNA-mediated inhibition of MIB1. In addition, MIB1 activates the Wnt pathway in an RYK-dependent manner. Collectively, MIB1 is both necessary and sufficient for the cell surface level of RYK to sense extracellular Wnt signals.
The cell surface level of the glutamate receptor GLR-1 in C. elegans is also controlled by ubiquitination. A mutation of usp-46, which encodes a deubiquitination enzyme for GLR-1, causes decreased surface level of GLR-1 and defective glutamate-dependent behavior [117]. The AMPA-type glutamate receptor, GluA1, is also ubiquitinated by Nedd4, an E3 ligase [118]. When Nedd4 is overexpressed in neurons, the cell surface level of GluA1 is decreased and GluA1-mediated excitatory synaptic transmission is suppressed. As synaptic transmission efficiency plays an essential role in learning, memory, and behavior, ubiquitination-mediated regulation of the cell surface levels of neurotransmitter receptors are pivotal for normal brain function.
Perspective of Fz ubiquitination in human diseases
It is well established that Wg/Wnt signaling is involved in various cancers, including blood and colon cancers [102, 103]. For example, the adenomatous polyposis coli (APC) gene, encoding a negative regulator of Wg/Wnt signaling, is mutated in colorectal cancer, indicating that aberrant activation of Wg/Wnt signaling causes human cancer. The search for human diseases in which UBPY expression is altered has shown the up-regulation of UBPY mRNA in chronic lymphocytic leukemia (CLL) [114]. Moreover, mRNAs of STAM1 and AMSH-like protein (AMSH-LP) [119], a functionally related protein to UBPY [114], and Fz [120] are frequently elevated in CLL. Therefore, although UBPY is involved in multiple processes that are linked to tumorigenesis, deubiquitination of Fz by elevated activity of UBPY and its related protein may partially contribute to tumorigenesis in CLL. Appropriate Wg/Wnt signaling strength, mediated by the balance between ubiquitination and deubiquitination, plays a pivotal role in the development and homeostasis of animal cells.
Conclusions and prospects
Cellular responsiveness to extracellular signals is affected by signal strength, which is at least partly regulated by cell surface presentation of receptors through their ubiquitination and deubiquitination. However, not all enzymes responsible for ubiquitination and deubiquitination of cell surface receptors have been identified, and this remains a major obstacle. Another interesting question is whether the regulation of expression and/or activity of these enzymes is dependent on development, extracellular stimulation, and pathological context. UBPY expression increases in pathological conditions such as CLL, but whether the expression and/or activity of UBPY changes in physiological conditions remains unknown. It was recently reported that the level of UBPY is regulated by a signaling cascade comprised of diacylglycerol kinase δ (DGK δ), protein kinase C (PKC), and Akt/protein kinase B (PKB) to modulate EGFR degradation, but the physiological significance of this process remains unknown [121]. Studying the involvement of this kinase cascade in Fz regulation and its significance in physiological and pathological conditions is therefore of interest.
Ubiquitination of DFz2 and Fz4 is independent of ligand stimulation whereas ubiquitination of EGFR and other receptors is ligand-dependent. Ubiquitination of DFz2 and Fz4 regulate cell surface levels of receptors in the absence of ligands. This means that cellular responsiveness to ligands can be controlled by the level of receptors presented at the cell surface before the presence of ligands. By contrast, ligand-dependent ubiquitination of EGFR and other receptors decreases cell surface levels of receptors after their activation, suggesting that this process is involved in desensitization in response to excess stimulation. It is interesting to address whether these two ubiquitination processes occur for the same receptors in different contexts, whether these two processes conduct different types of ubiquitination at different lysine residues in receptor polypeptides, and how these ubiquitination processes are regulated.
Acknowledgments
This work was supported by Grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to SG.
Footnotes
A. Mukai and M. Yamamoto-Hino contributed equally to this work.
References
- 1.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/S0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg TB, Guha A. Understanding morphogen gradients: a problem of dispersion and containment. Curr Opin Genet Dev. 2007;17:264–271. doi: 10.1016/j.gde.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez-Camacho C, Bovolenta P. Emerging mechanisms in morphogen-mediated axon guidance. Bioessays. 2009;31:1013–1025. doi: 10.1002/bies.200900063. [DOI] [PubMed] [Google Scholar]
- 4.Jin T, Xu X, Hereld D. Chemotaxis, chemokine receptors and human disease. Cytokine. 2008;44:1–8. doi: 10.1016/j.cyto.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiley HS, Herbst JJ, Walsh BJ, Lauffenburger DA, Rosenfeld MG, Gill GN. The role of tyrosine kinase activity in endocytosis, compartmentation, and down-regulation of the epidermal growth factor receptor. J Biol Chem. 1991;266:11083–11094. [PubMed] [Google Scholar]
- 7.Chang CP, Lazar CS, Walsh BJ, Komuro M, Collawn JF, Kuhn LA, Tainer JA, Trowbridge IS, Farquhar MG, Rosenfeld MG, et al. Ligand-induced internalization of the epidermal growth factor receptor is mediated by multiple endocytic codes analogous to the tyrosine motif found in constitutively internalized receptors. J Biol Chem. 1993;268:19312–19320. [PubMed] [Google Scholar]
- 8.Resat H, Ewald JA, Dixon DA, Wiley HS. An integrated model of epidermal growth factor receptor trafficking and signal transduction. Biophys J. 2003;85:730–743. doi: 10.1016/S0006-3495(03)74516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, Reddi AL, Ghosh A, Fernandes N, Zhou P, Mullane-Robinson K, Rao N, Donoghue S, Rogers RA, Bowtell D, Naramura M, Gu H, Band V, Band H. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J Biol Chem. 2003;278:28950–28960. doi: 10.1074/jbc.M304474200. [DOI] [PubMed] [Google Scholar]
- 10.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 12.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 15.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 19.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci USA. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 21.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 24.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hicke L. Gettin’ down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/S0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 27.Polo S, Confalonieri S, Salcini AE, Di Fiore PP (2003) EH and UIM: endocytosis and more. Sci STKE 2003(213):re17 [DOI] [PubMed]
- 28.Brooks CL, Li M, Gu W. Monoubiquitination: the signal for p53 nuclear export? Cell Cycle. 2004;3:436–438. [PubMed] [Google Scholar]
- 29.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 30.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson KD, Tashayev VL, O’Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 34.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Komada M. Controlling receptor downregulation by ubiquitination and deubiquitination. Curr Drug Discov Technol. 2008;5:78–84. doi: 10.2174/157016308783769469. [DOI] [PubMed] [Google Scholar]
- 37.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 38.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/S0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 39.Kolling R, Hollenberg CP. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Huang F, Goh LK, Sorkin A. EGF receptor ubiquitination is not necessary for its internalization. Proc Natl Acad Sci USA. 2007;104:16904–16909. doi: 10.1073/pnas.0707416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci USA. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J Cell Biol. 2010;189:871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- 46.Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, Lehner PJ, Luzio JP. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J Biol Chem. 2006;281:5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- 47.Kyuuma M, Kikuchi K, Kojima K, Sugawara Y, Sato M, Mano N, Goto J, Takeshita T, Yamamoto A, Sugamura K, Tanaka N. AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct Funct. 2007;31:159–172. doi: 10.1247/csf.06023. [DOI] [PubMed] [Google Scholar]
- 48.Jacoby E, Bouhelal R, Gerspacher M, Seuwen K. The 7 TM G-protein-coupled receptor target family. ChemMedChem. 2006;1:761–782. doi: 10.1002/cmdc.200600134. [DOI] [PubMed] [Google Scholar]
- 49.Fredholm BB, Hokfelt T, Milligan G. G-protein-coupled receptors: an update. Acta Physiol (Oxf) 2007;190:3–7. doi: 10.1111/j.1365-201X.2007.01689.x. [DOI] [PubMed] [Google Scholar]
- 50.Chen L, Davis NG. Ubiquitin-independent entry into the yeast recycling pathway. Traffic. 2002;3:110–123. doi: 10.1034/j.1600-0854.2002.030204.x. [DOI] [PubMed] [Google Scholar]
- 51.Obin MS, Jahngen-Hodge J, Nowell T, Taylor A. Ubiquitinylation and ubiquitin-dependent proteolysis in vertebrate photoreceptors (rod outer segments). Evidence for ubiquitinylation of Gt and rhodopsin. J Biol Chem. 1996;271:14473–14484. doi: 10.1074/jbc.271.24.14473. [DOI] [PubMed] [Google Scholar]
- 52.Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 53.Marchese A, Benovic JL. Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J Biol Chem. 2001;276:45509–45512. doi: 10.1074/jbc.C100527200. [DOI] [PubMed] [Google Scholar]
- 54.Martin NP, Lefkowitz RJ, Shenoy SK. Regulation of V2 vasopressin receptor degradation by agonist-promoted ubiquitination. J Biol Chem. 2003;278:45954–45959. doi: 10.1074/jbc.M308285200. [DOI] [PubMed] [Google Scholar]
- 55.Jacob C, Cottrell GS, Gehringer D, Schmidlin F, Grady EF, Bunnett NW. c-Cbl mediates ubiquitination, degradation, and down-regulation of human protease-activated receptor 2. J Biol Chem. 2005;280:16076–16087. doi: 10.1074/jbc.M500109200. [DOI] [PubMed] [Google Scholar]
- 56.Cottrell GS, Padilla B, Pikios S, Roosterman D, Steinhoff M, Gehringer D, Grady EF, Bunnett NW. Ubiquitin-dependent down-regulation of the neurokinin-1 receptor. J Biol Chem. 2006;281:27773–27783. doi: 10.1074/jbc.M603369200. [DOI] [PubMed] [Google Scholar]
- 57.Li JG, Haines DS, Liu-Chen LY. Agonist-promoted Lys63-linked polyubiquitination of the human kappa-opioid receptor is involved in receptor down-regulation. Mol Pharmacol. 2008;73:1319–1330. doi: 10.1124/mol.107.042846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfe BL, Marchese A, Trejo J. Ubiquitination differentially regulates clathrin-dependent internalization of protease-activated receptor-1. J Cell Biol. 2007;177:905–916. doi: 10.1083/jcb.200610154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanowitz M, Von Zastrow M. Ubiquitination-independent trafficking of G protein-coupled receptors to lysosomes. J Biol Chem. 2002;277:50219–50222. doi: 10.1074/jbc.C200536200. [DOI] [PubMed] [Google Scholar]
- 60.Henry AG, White IJ, Marsh M, von Zastrow M, Hislop JN. The role of ubiquitination in lysosomal trafficking of delta-opioid receptors. Traffic. 2011;12:170–184. doi: 10.1111/j.1600-0854.2010.01145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shenoy SK, Xiao K, Venkataramanan V, Snyder PM, Freedman NJ, Weissman AM. Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. J Biol Chem. 2008;283:22166–22176. doi: 10.1074/jbc.M709668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 2010;11:605–611. doi: 10.1038/embor.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell. 2003;5:709–722. doi: 10.1016/S1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- 64.Bhandari D, Robia SL, Marchese A. The E3 ubiquitin ligase atrophin interacting protein 4 binds directly to the chemokine receptor CXCR4 via a novel WW domain-mediated interaction. Mol Biol Cell. 2009;20:1324–1339. doi: 10.1091/mbc.E08-03-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhandari D, Trejo J, Benovic JL, Marchese A. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J Biol Chem. 2007;282:36971–36979. doi: 10.1074/jbc.M705085200. [DOI] [PubMed] [Google Scholar]
- 66.Hislop JN, Henry AG, Marchese A, von Zastrow M. Ubiquitination regulates proteolytic processing of G protein-coupled receptors after their sorting to lysosomes. J Biol Chem. 2009;284:19361–19370. doi: 10.1074/jbc.M109.001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dupre DJ, Chen Z, Le Gouill C, Theriault C, Parent JL, Rola-Pleszczynski M, Stankova J. Trafficking, ubiquitination, and down-regulation of the human platelet-activating factor receptor. J Biol Chem. 2003;278:48228–48235. doi: 10.1074/jbc.M304082200. [DOI] [PubMed] [Google Scholar]
- 68.Berthouze M, Venkataramanan V, Li Y, Shenoy SK. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO J. 2009;28:1684–1696. doi: 10.1038/emboj.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mines MA, Goodwin JS, Limbird LE, Cui FF, Fan GH. Deubiquitination of CXCR4 by USP14 is critical for both CXCL12-induced CXCR4 degradation and chemotaxis but not ERK ativation. J Biol Chem. 2009;284:5742–5752. doi: 10.1074/jbc.M808507200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hasdemir B, Murphy JE, Cottrell GS, Bunnett NW. Endosomal deubiquitinating enzymes control ubiquitination and down-regulation of protease-activated receptor 2. J Biol Chem. 2009;284:28453–28466. doi: 10.1074/jbc.M109.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berlin I, Higginbotham KM, Dise RS, Sierra MI, Nash PD. The deubiquitinating enzyme USP8 promotes trafficking and degradation of the chemokine receptor 4 at the sorting endosome. J Biol Chem. 2010;285:37895–37908. doi: 10.1074/jbc.M110.129411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinmaster G, Fischer JA. Notch ligand ubiquitylation: what is it good for? Dev Cell. 2011;21:134–144. doi: 10.1016/j.devcel.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le Bras S, Loyer N, Le Borgne R. The multiple facets of ubiquitination in the regulation of notch signaling pathway. Traffic. 2011;12:149–161. doi: 10.1111/j.1600-0854.2010.01126.x. [DOI] [PubMed] [Google Scholar]
- 75.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/S1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 76.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/S1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 77.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 78.Okochi M, Steiner H, Fukumori A, Tanii H, Tomita T, Tanaka T, Iwatsubo T, Kudo T, Takeda M, Haass C. Presenilins mediate a dual intramembranous gamma-secretase cleavage of Notch-1. EMBO J. 2002;21:5408–5416. doi: 10.1093/emboj/cdf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seugnet L, Simpson P, Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev Biol. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- 80.Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster . J Cell Biol. 2008;180:755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila . Nat Cell Biol. 2005;7:1232–1239. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- 82.Vaccari T, Duchi S, Cortese K, Tacchetti C, Bilder D. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development. 2010;137:1825–1832. doi: 10.1242/dev.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan Y, Denef N, Schupbach T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila . Dev Cell. 2009;17:387–402. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gupta-Rossi N, Six E, LeBail O, Logeat F, Chastagner P, Olry A, Israel A, Brou C. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J Cell Biol. 2004;166:73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tagami S, Okochi M, Yanagida K, Ikuta A, Fukumori A, Matsumoto N, Ishizuka-Katsura Y, Nakayama T, Itoh N, Jiang J, Nishitomi K, Kamino K, Morihara T, Hashimoto R, Tanaka T, Kudo T, Chiba S, Takeda M. Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol Cell Biol. 2008;28:165–176. doi: 10.1128/MCB.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 87.Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila . Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 88.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 89.Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Childress JL, Acar M, Tao C, Halder G. Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis. Curr Biol. 2006;16:2228–2233. doi: 10.1016/j.cub.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gallagher CM, Knoblich JA. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila . Dev Cell. 2006;11:641–653. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 92.Jaekel R, Klein T. The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev Cell. 2006;11:655–669. doi: 10.1016/j.devcel.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 93.Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, Baron M, Matsuno K. Drosophila deltex mediates suppressor of hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- 94.Fuwa TJ, Hori K, Sasamura T, Higgs J, Baron M, Matsuno K. The first deltex null mutant indicates tissue-specific deltex-dependent Notch signaling in Drosophila . Mol Genet Genomics. 2006;275:251–263. doi: 10.1007/s00438-005-0087-3. [DOI] [PubMed] [Google Scholar]
- 95.Jehn BM, Dittert I, Beyer S, von der Mark K, Bielke W. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J Biol Chem. 2002;277:8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- 96.Wang Y, Chen Z, Bergmann A. Regulation of EGFR and Notch signaling by distinct isoforms of D-cbl during Drosophila development. Dev Biol. 2010;342:1–10. doi: 10.1016/j.ydbio.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fostier M, Evans DA, Artavanis-Tsakonas S, Baron M. Genetic characterization of the Drosophila melanogaster suppressor of deltex gene: a regulator of notch signaling. Genetics. 1998;150:1477–1485. doi: 10.1093/genetics/150.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chastagner P, Israel A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, Fang D, Hunter T, Liu YC. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 100.Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, Hayashi S. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14:2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 101.Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, Myat A, Evans DA, Cornell M, Baron M. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol. 2004;14:2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 102.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 103.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 104.Couso JP, Bishop SA, Martinez Arias A. The wingless signalling pathway and the patterning of the wing margin in Drosophila . Development. 1994;120:621–636. doi: 10.1242/dev.120.3.621. [DOI] [PubMed] [Google Scholar]
- 105.Zecca M, Basler K, Struhl G. Direct and long-range action of a wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/S0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- 106.Neumann CJ, Cohen SM. Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- 107.Schulte G, Bryja V. The Frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:518–525. doi: 10.1016/j.tips.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 108.Muller P, Schier AF. Extracellular movement of signaling molecules. Dev Cell. 2011;21:145–158. doi: 10.1016/j.devcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cadigan KM, Fish MP, Rulifson EJ, Nusse R. Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell. 1998;93:767–777. doi: 10.1016/S0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- 110.Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE, Caron MG, Barak LS, Nusse R, Lefkowitz RJ. Dishevelled 2 recruits beta-arrestin 2 to mediate Wnt5A-stimulated endocytosis of Frizzled 4. Science. 2003;301:1391–1394. doi: 10.1126/science.1082808. [DOI] [PubMed] [Google Scholar]
- 111.Yamamoto H, Komekado H, Kikuchi A. Caveolin is necessary for Wnt-3a-dependent internalization of LRP6 and accumulation of beta-catenin. Dev Cell. 2006;11:213–223. doi: 10.1016/j.devcel.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 112.Yu A, Rual JF, Tamai K, Harada Y, Vidal M, He X, Kirchhausen T. Association of Dishevelled with the clathrin AP-2 adaptor is required for Frizzled endocytosis and planar cell polarity signaling. Dev Cell. 2007;12:129–141. doi: 10.1016/j.devcel.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Purvanov V, Koval A, Katanaev VL. A direct and functional interaction between Go and Rab5 during G protein-coupled receptor signaling. Sci Signal. 2010;3:ra65. doi: 10.1126/scisignal.2000877. [DOI] [PubMed] [Google Scholar]
- 114.Mukai A, Yamamoto-Hino M, Awano W, Watanabe W, Komada M, Goto S. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 2010;29:2114–2125. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berndt JD, Aoyagi A, Yang P, Anastas JN, Tang L, Moon RT. Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/beta-catenin signaling. J Cell Biol. 2011;194:737–750. doi: 10.1083/jcb.201107021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lyu J, Yamamoto V, Lu W. Cleavage of the Wnt receptor Ryk regulates neuronal differentiation during cortical neurogenesis. Dev Cell. 2008;15:773–780. doi: 10.1016/j.devcel.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 117.Kowalski JR, Dahlberg CL, Juo P. The deubiquitinating enzyme USP-46 negatively regulates the degradation of glutamate receptors to control their abundance in the ventral nerve cord of Caenorhabditis elegans . J Neurosci. 2011;31:1341–1354. doi: 10.1523/JNEUROSCI.4765-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lin A, Hou Q, Jarzylo L, Amato S, Gilbert J, Shang F, Man HY. Nedd4-mediated AMPA receptor ubiquitination regulates receptor turnover and trafficking. J Neurochem. 2011;119:27–39. doi: 10.1111/j.1471-4159.2011.07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakamura M, Tanaka N, Kitamura N, Komada M. Clathrin anchors deubiquitinating enzymes, AMSH and AMSH-like protein, on early endosomes. Genes Cells. 2006;11:593–606. doi: 10.1111/j.1365-2443.2006.00963.x. [DOI] [PubMed] [Google Scholar]
- 120.Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, Leoni LM, Kipps TJ, Corr M, Carson DA. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cai J, Crotty TM, Reichert E, Carraway KL, 3rd, Stafforini DM, Topham MK. Diacylglycerol kinase delta and protein kinase C(alpha) modulate epidermal growth factor receptor abundance and degradation through ubiquitin-specific protease 8. J Biol Chem. 2010;285:6952–6959. doi: 10.1074/jbc.M109.055731. [DOI] [PMC free article] [PubMed] [Google Scholar]