Abstract
Matricellular proteins interact with the extracellular matrix (ECM) and modulate cellular processes by binding to cell surface receptors and initiating intracellular signal transduction. Their association with the ECM and the ability of some members of this protein family to regulate cell motility have opened up new avenues of research to investigate their functions in normal and diseased cells. In this review, we summarize the research on CyrA, an ECM calmodulin-binding protein in Dictyostelium. CyrA is proteolytically cleaved into smaller EGF-like (EGFL) repeat containing cleavage products during development. The first EGFL repeat of CyrA binds to the cell surface and activates a novel signalling pathway that modulates cell motility in this model organism. The similarity of CyrA to the most well-characterized matricellular proteins in mammals allows it to be designated as the first matricellular protein identified in Dictyostelium.
Keywords: Matricellular, EGF-like repeat, Extracellular matrix, Signal transduction, Cell motility, Dictyostelium, CyrA
Matricellular proteins
Matricellular proteins belong to a group of extracellular matrix (ECM) proteins that function as adaptors and modulators of cell–matrix interactions [1, 2]. The most well-studied examples of these proteins include tenascin cytotactin (tenascin C), thrombospondin (TSP) 1 and 2, and SPARC (secreted protein acidic and rich in cysteine) [3–5]. The evolutionary history of these proteins as well as the ECM was recently reviewed [2, 6]. A matricellular protein is defined by a number of characteristics. They associate with extracellular proteases and growth factors can function as both soluble and insoluble proteins are expressed at high levels during development, and unlike other components of the ECM do not appear to contribute directly to the organization of extracellular structures or their physical properties [1, 2, 7]. Matricellular proteins tend to be rapidly turned over and contain binding sites for some major structural elements in the ECM and specific cell surface receptors [3]. Another important attribute of matricellular proteins is their ability to modulate cellular processes by binding to cell surface receptors and initiating intracellular signal transduction [1, 3, 7–10].
The proteolytic cleavage of matricellular proteins to produce small signalling polypeptides and peptides has received much attention in recent in years and has been observed in many systems. Members of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) superfamily of matricellular proteins contain a metalloprotease domain that can cleave specific substrates in the ECM or on the cell surface [3, 11]. As evident by recent studies on tenascin C and TSP-1 (discussed below), the literature on protein processing to release functional cleavage products is currently on the rise.
EGF-like repeats as motility-enhancing domains
Many of the matricellular proteins that have been characterized contain epidermal growth factor-like (EGFL) repeats, which are cysteine-rich motifs that share sequence similarity with EGF [12]. EGF is a 6-kDa, 53 amino acid, cysteine-rich polypeptide that binds to an EGF receptor (EGFR) and activates signalling pathways involved in mediating a diversity of cellular processes (e.g., cell motility, chemotaxis, proliferation, growth, survival, differentiation, morphogenesis) [13, 14]. The EGFL repeat is a widespread, highly variable domain that contains amino acids in specific positions relative to conserved cysteine residues [12, 15]. EGFL repeats can appear as single entities or be arranged as multiple tandem repeats [12]. They are found extensively in human proteins and have also been detected in lower eukaryotes, including the model organisms Drosophila melanogaster and Dictyostelium discoideum [16–21].
The most well-studied examples of EGFL repeat-containing ECM proteins include tenascin C, TSP-1, and laminin-5, which have all been shown to modulate cell movement via one or more of their EGFL repeats. Tenascin C is a member of an extensive family of ECM glycoproteins. In its monomeric form, this 190–300 kDa protein contains 14 EGFL repeats (Ten 1–14) [22]. It is expressed during tissue development and regeneration, is involved in cell proliferation and migration, and has been linked to tumor progression [22–25]. Some EGFL repeats of tenascin C (e.g., Ten14) have been shown to increase cell motility by binding to the EGFR and activating EGFR-dependent signalling [26, 27]. Although the sequence and structure of Ten14 is similar to EGF, this 31 amino acid polypeptide has been shown to be a low affinity ligand for the EGFR, since micromolar concentrations are required to increase the rate of cell motility [26, 28]. Activation at a micromolar concentration contrasts with the nanomolar concentrations of classical growth factors such as EGF that are required to elicit similar responses [26]. Unlike EGF, binding of Ten14 to the EGFR is transient and the Ten14–EGFR complex is not internalized, thus restricting the activated receptor to the cell surface [27, 28]. Internalization of receptor–ligand complexes functions as an attenuation mechanism for the signal. In the case of transient binding of Ten14 to the EGFR, the lack of internalization allows for a continuous activation of the receptor thus maintaining the signal for cell motility [27].
TSP-1, an angiogenesis inhibitor, is a 420-kDa ECM glycoprotein composed of three identical 145-kDa monomers that are held together via disulphide linkages [29, 30]. Each 145-kDa monomer possesses three EGFL repeats. Several cell types such as smooth muscle and endothelial cells secrete TSP-1 into the ECM, where it participates in processes related to tissue regeneration and cell differentiation such as re-epithelialization after injury [29]. Its activity has also been linked to cancer progression [30]. The EGFL repeats of TSP-1 have recently been shown to increase epithelial cell migration by activating intracellular signalling; however, the binding of the repeats to the cell surface has not yet been shown [29]. Although the TSP-1 EGFL repeats activate the EGFR by inducing autophosphorylation of the receptor, they do not directly bind to the receptor suggesting that not all EGFL repeats function by binding to the EGFR [29].
Laminin-5 is an ECM glycoprotein that possesses EGFL repeats in its ectodomain [31]. Although its function as major structural component of basement membranes prevents it from being classified as a matricellular protein, the EGFL repeats of laminin-5 have also been shown to modulate cell movement. Intact laminin-5 has been shown to support cell adhesion, but when cleaved by matrix metalloproteinase 2 (MMP2), the EGFL repeat-containing cleavage products promote cell migration by binding to the EGFR and activating downstream signalling pathways [31, 32]. Several studies have reported the enhanced expression of laminin-5, especially of the cleaved EGFL repeat-containing products at sites of tumor cell penetration [33, 34]. Together, studies on tenascin C, TSP-1, and laminin-5 have indicated that a primary function of cysteine-rich, EGFL repeat-containing ECM proteins may be to modulate cell motility.
Dictyostelium as a model eukaryote for studying cell motility
Dictyostelium discoideum is a fascinating organism that is used as a model system for studying a number of cell and developmental processes [35]. During feeding, Dictyostelium amoebae find their food source by chemotactically responding to folic acid that is secreted by bacterial cells. Upon starvation, Dictyostelium cells begin to secrete cAMP which acts as a chemoattractant causing individual cells to aggregate into mounds. Aggregated cells then develop into a motile, multicellular structure known as a pseudoplasmodium or slug that can migrate on the substratum for an indefinite period of time in response to light and temperature. When conditions are suitable, the slug will culminate and develop into a fruiting body composed of a mass of spores that is supported by a stalk of dead stalk cells [36].
The mechanisms underlying cell motility and chemotaxis in Dictyostelium, as well as the molecular components and interactions involved are similar to those observed in mammalian cells making it a very useful system for studying this cellular process [37]. The movement of Dictyostelium amoebae has been extensively studied; however, all lines of evidence suggest that movement in this model eukaryote is a highly complex process likely involving a number of pathways that have yet to be identified [38]. It is thought that these unidentified pathways may also involve redundant components.
Phosphatidylinositol-3-kinase (PI3K) and phospholipase A2 (PLA2) are two proteins that have been shown to mediate cAMP chemotaxis in parallel compensatory pathways [39, 40]. PI3K controls F-actin polymerization, which has been linked to pseudopod extension [41, 42]; however, the activation mechanism and the targets of PLA2 are still unknown [39, 43]. In addition, studies have shown that phospholipase C (PLC) and Ca2+ are essential for PI3K- and PLA2-mediated signalling, respectively [40]. In mammalian cells, PLA2 induces Ca2+ signalling through the release of arachidonic acid; however, the ability of this fatty acid to mediate the PLA2-dependent chemotactic response in Dictyostelium has not yet been verified [39]. In addition to PI3K and PLA2 activity, Ca2+- and calmodulin (CaM)-mediated signalling via interaction with specific CaM-binding proteins (CaMBPs) have both been shown to be required for efficient cell motility and chemotaxis in Dictyostelium [44, 45]. A recent study described the ability of aggregation-competent Dictyostelium amoebae to directionally respond towards a gradient of Ca2+ [46], suggesting the existence of other motility regulatory components and mechanisms in Dictyostelium that have yet to be identified.
Although much less well studied compared to cAMP chemotaxis, Dictyostelium can also chemotax towards folic acid, which is secreted by bacteria and allows Dictyostelium amoebae to find their food source during growth. The signal transduction regulating folic acid chemotaxis is different from the signalling that mediates chemotaxis towards cAMP. For instance, tyrosine kinase activity has been shown to be required for chemotaxis towards folic acid, but not cAMP, and unique CaM-dependent phosphoproteins have been shown to be linked solely to folic acid chemotaxis [45, 47]. These findings, therefore, provide further support for the existence of multiple motility-regulating pathways in this model eukaryote.
The Dictyostelium slime sheath
The Dictyostelium slug is covered by a thin ECM (i.e., slime sheath) that is continuously synthesized from the tip of the slug forming a sheath around the migrating slug cells [48]. As the slug migrates along the substratum, it leaves behind a trail of slime sheath that provides clues into the cell–ECM interactions that occurred during slug movement. Vegetative amoebae have also been shown to secrete ECM material; however, little is known at the molecular level about the ECM during the growth phase. The Dictyostelium slime sheath is similar in structure and composition to both animal and plant ECMs. It is composed of cellulose fibers and polysaccharide embedded within a protein-containing matrix that contains both structural and non-structural proteins. The most well-studied structural protein of the slime sheath EcmA is distributed throughout the slug ECM and has been shown to be an integral structural protein of the sheath. EcmA knockout cells are still able to form slugs that migrate normally; however, the structure of the ECM that surrounds the slug cells is weakened [49]. A group of glycoproteins called the sheathins (i.e., EcmC, EcmD, EcmE) co-localize with cellulose and have been shown to be involved in regulating slug migration [50]. Research on the non-structural components of the slime sheath, however, is limited. Although previous studies have identified a group of soluble, mobile glycoproteins within the slug ECM, the identity of these proteins remains unknown indicating that much still remains to be discovered about the Dictyostelium slug ECM [51, 52].
Identification of EGFL repeat-containing proteins in Dictyostelium
Bioinformatic analyses suggest that the Dictyostelium genome encodes a higher percentage of EGFL domains than any other sequenced eukaryote, including humans [18]. Despite their abundance, the function of EGFL repeats and of EGFL repeat-containing proteins in Dictyostelium has been relatively unstudied. The most well-studied structural proteins of the Dictyostelium ECM, EcmA, and EcmB possess extensive cysteine-rich regions composed of tandem arrays of a cysteine-rich 24 amino acid repeat that shares some sequence similarity with EGF [48]. SadA, the novel adhesion receptor in Dictyostelium, contains three conserved EGFL repeats in the predicted extracellular domain of the protein that are similar to regions in cell adhesion proteins such as integrins and tenascins [19]. SadA associates with the actin cytoskeleton, possibly by interacting with cortexillin I, a known actin bundling protein [53]. sadA knockout cells possess a disrupted actin cytoskeleton and cytokinesis defect. They also move with increased speed during vegetative conditions, suggesting that SadA-mediated cell–substrate adhesion acts as a brake during cell movement [19]. Therefore, as in mammals, these data suggest that a major function of EGFL repeats in Dictyostelium may be to modulate cell movement.
CyrA is a matricellular protein in Dictyostelium
CyrA is the first extracellular CaMBP identified and characterized in Dictyostelium. This 63-kDa cysteine-rich protein possesses four tandem EGFL repeats in its C-terminus and possesses attributes that allows it to be classified as a matricellular protein (Table 1) [21]. The expression and secretion of CyrA increases at a constant rate during the early stages of development, but peaks during mid-development which coincides with the time that cells are developing into a motile, multicellular slug [21, 54]. Consistent with the fact that CyrA possesses an N-terminal signal sequence and is secreted, the protein has been shown to localize to the endoplasmic reticulum (ER), particularly to the region of the ER that surrounds the nucleus (i.e., perinuclear region) [54]. CyrA-GFP was also shown to co-localize with calnexin an integral transmembrane protein of the ER [54, 55].
Table 1.
Comparison of CyrA to the matricellular proteins tenascin C and thrombospondin-1
| Characteristics | Tenascin C | Thrombospondin-1 | CyrA |
|---|---|---|---|
| Secreted | Yes [1] | Yes [1] | Yes [21, 54] |
| Localizes to the ECM | Yes [1] | Yes [1] | Yes; slime sheath [21, 54] |
| Expressed at high levels during development | Yes [1] | Yes [1] | Yes [21, 54] |
| Contains EGFL repeats | Yes [28] | Yes [29] | Yes [21] |
| Releases cleavage products | Yes [23] | Unknown | Yes [21] |
| EGFL repeats modulate cell motility | Yes [27] | Yes [29] | Yes [20, 21, 60] |
| EGFL repeats bind to the cell surface | Yes [26, 28] | Unknown | Yes [54] |
| Receptor | Yes; EGFR [26, 28] | Unknown | Yes; uncharacterized [54] |
| Initiates intracellular signal transduction | Yes [27, 28] | Yes [29] | Yes [20, 21, 60] |
Like other matricellular proteins (e.g., tenascin C), CyrA is proteolytically cleaved during Dictyostelium development to release 45- and 40-kDa EGFL repeat-containing cleavage products (CyrA-C45 and CyrA-C40, respectively) (Table 1; Fig. 1) [21]. In Dictyostelium, this type of protein processing has previously been reported to occur in the acyl-CoA binding protein (AcbA), which is secreted and cleaved by the membrane-bound serine protease TagC to generate the bioactive peptide spore differentiation factor-2 (SDF-2) [56]. SDF-2 binds to a receptor on prespore cells in order to induce spore cell differentiation. During mid- to late Dictyostelium development, the multicellular mass of cells is covered by a thin ECM called a slime sheath, which is composed of protein, polysaccharide, and cellulose [48]. Full-length CyrA and its cleavage products localize to the slime sheath of the migrating slug (Fig. 1) [21, 54]. Interestingly, CaM has also been shown to be secreted and to localize to the slime sheath (Fig. 1) [21, 54, 57]. Full-length CyrA and its cleavage products bind CaM both intra- and extracellularly, and CaM antagonism has been shown to increase CyrA cleavage indicating that CaM regulates the release of EGFL repeats from CyrA and possibly other EGFL repeat-containing proteins during development (Fig. 1) [21, 54]. These findings are supported by studies that have shown that the activity and function of some EGFL repeats is dependent on Ca2+ binding, which is interesting considering that CaM is the primary sensor of Ca2+ within the cell [58]. Binding of Ca2+ to some EGFL repeats such as the EtMIC4 protein of Eimeria tenella has been shown to affect both the conformation of the repeat as well as its susceptibility to proteases [59]. Together, these data support an interaction between CyrA, CaM, and Ca2+ in Dictyostelium.
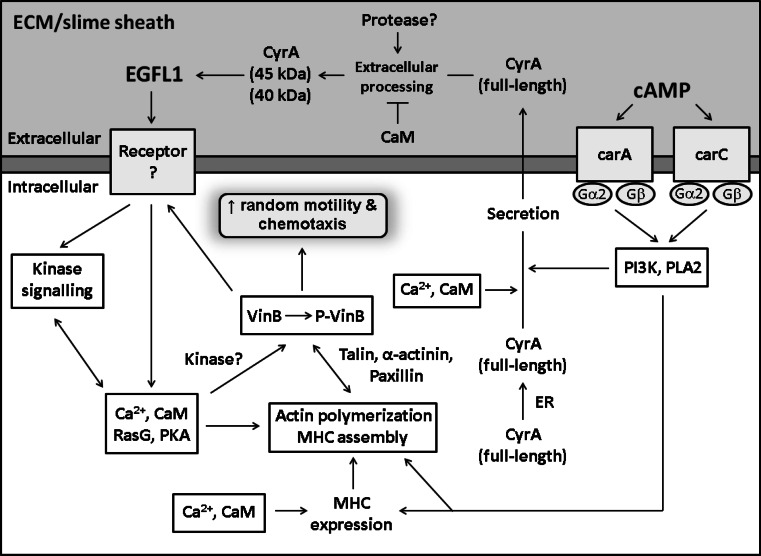
Fig. 1.
Model for CyrA EGFL repeat signal transduction in Dictyostelium discoideum. Refer to the text for a thorough explanation of the signal transduction mediating the response of cells to DdEGFL1
CyrA EGFL1 increases cell motility
A synthetic EGFL peptide (DdEGFL1), whose sequence is identical to the first 18 amino acids of the first EGFL (EGFL1) repeat of CyrA, functions extracellularly to increase both random cell motility and cAMP-mediated chemotaxis in a number of wild-type and parental strains of Dictyostelium (e.g., NC4, AX2, AX3, KAX3, DH1; Fig. 1) [20, 60]. The regulation of cell motility gives a matricellular function to CyrA (Table 1). DdEGFL1 shares sequence similarity with EGF, Ten14, and another C-terminal EGFL repeat in CyrA [20]. The DdEGFL1 sequence is highly conserved in Dictyostelium and shares sequence similarity with many regions of EcmA and EcmB, as well as many putative ECM proteins in Dictyostelium. A recent study showed that DdEGFL1 is not a chemoattractant for Dictyostelium cells, but instead functions in a supportive role to increase the rate of random cell movement and chemotaxis during development [61]. The response of cells to DdEGFL1 increases during starvation, supporting a function for DdEGFL1 during early developmental events (e.g., cAMP chemotaxis) [21]. CyrA over-expression has also been shown to increase the rate of cAMP-mediated chemotaxis, providing in vivo evidence linking CyrA function to Dictyostelium cell movement and confirming in vitro studies performed using DdEGFL1 [20, 21, 54, 60, 61]. In addition, since CyrA localizes to the Dictyostelium slime sheath during the later stages of multicellular development, this suggests that DdEGFL1 is involved in regulating the movement of cells within the slug during slug movement [21, 54]. DdEGFL1 also inhibits the cleavage of CyrA supporting the existence of a signalling pathway regulating the release of EGFL peptides (e.g., DdEGFL1) and/or CyrA cleavage products possessing functional EGFL domains (Fig. 1) [21].
Signalling pathways regulating the expression, secretion, and function of CyrA
Another criterion for establishing a protein as matricellular involves the ability of the protein and/or cleavage products to bind to the cell surface and initiate intracellular signal transduction (Table 1) [1]. DdEGFL1 increases the rate of Dictyostelium cell movement via a novel signalling pathway that does not require the heterotrimeric G-protein utilized in cAMP signalling or either of the two cAMP receptors that are active during early development cAMP receptor A or C (carA and carC, respectively; Fig. 1) [60]. The signalling pathway mediating cell movement in response to DdEGFL1 requires CaM activity and intracellular Ca2+ release, and DdEGFL1 stimulation increases the amount of polymeric actin and myosin II heavy chain (MHC) in the cytoskeleton (Fig. 1) [60]. In addition to actin and MHC, DdEGFL1 signalling has also been shown to require the cytoskeletal proteins talin B (TalB) and paxillin B (PaxB), which are homologues of mammalian talin and paxillin, respectively (Fig. 1) [62].
DdEGFL1-enhanced cell movement requires the activity of both PI3K and PLA2, two signalling proteins that have been shown to mediate the chemotaxis of Dictyostelium amoebae in parallel compensatory pathways (Fig. 1) [20, 39, 40]. Although the activity of both proteins is required, PLA2 appears to be the more dominant regulator of DdEGFL1-enhanced random cell motility, since inhibition of PLA2 alone significantly suppresses the increased movement, whereas PI3K inhibition alone has no significant effect [20, 61]. Interestingly, the secretion of CyrA has also been shown to be dependent on intracellular Ca2+ release and the activity of PI3K, PLA2, and CaM, indicating that a common mechanism regulates the function of CyrA (Fig. 1) [54]. The dependence of DdEGFL1 function on PI3K and PLA2 signalling is likely due to the regulation of CyrA secretion by both of these proteins.
Both inter- and intracellular Ca2+ signalling as well as Ras signalling have been shown to be required for EGF-induced cell movement in normal and cancerous cells [63, 64]. In Dictyostelium, two members of the Ras protein family, RasC and RasG, have been shown to regulate chemotaxis towards cAMP [65]. Our research has shown that DdEGFL1-increased movement partially requires the activity of RasG, but not RasC (Fig. 1) [60].
Protein kinase A (PKA, cAMP-dependent protein kinase), a serine/threonine kinase whose activity is dependent on cellular levels of cAMP, and which has been shown to be involved in regulating cell movement and chemotaxis in Dictyostelium, is also required for DdEGFL1 function, showing that PKA kinase activity is required for DdEGFL1 signal transduction (Fig. 1) [62, 66]. PKA kinase activity was also shown to be required for the DdEGFL1-induced phosphorylation of a 90-kDa phosphotyrosine during cell starvation (Fig. 1) [62]. Finally, Huber and O’Day [62] reported the detection of two phosphotyrosine proteins in DdEGFL1 pull-down assays. Together, these results show that the motility-enhancing pathway activated by DdEGFL1 involves protein kinases, which fits with observations of EGFR-mediating signalling in higher eukaryotes (Fig. 1) [67, 68].
Prior to studies describing DdEGFL1 function and signalling, it was reported that Dictyostelium cells could undergo random migration in the absence of functional heterotrimeric G proteins [69, 70]. A previous study showed that PI3K and Ras were activated in Gβ null cells during random cell migration, suggesting the existence of a Gβ-independent signalling pathway regulating random cell motility [70]. Activation occurred in the absence of external stimuli (i.e., folic acid) and resulted in F-actin polymerization. Interestingly, like the DdEGFL1-mediated pathway, that study also implicated RasG function [60, 70]. Based on their data, Sasaki et al. [70] suggested the existence of two pathways involving activated PI3K and Ras. One was a chemoattractant/Gβ-dependent pathway that regulated chemotaxis and the other was a Gβ-independent positive feedback circuit that regulated random cell migration. During chemotaxis, the authors suggested that the Gβ-dependent pathway overrides or disrupts the positive feedback loop involving activated PI3K and Ras, thereby allowing the cell to directionally respond to the chemoattractant (i.e., folic acid) [70]. Although Sasaki et al. [70] showed that the Gβ-independent pathway functioned in the absence of chemoattractants, the authors did not address whether the pathway was activated by other secreted components (e.g., EGFL repeat-containing proteins). Subsequent research on DdEGFL1 function and signalling suggests that activation of this pathway may occur via the binding of secreted EGFL repeat-containing proteins to a surface receptor that initiates intracellular signal transduction.
CyrA EGFL1 has differential effects on chemotaxis to folic acid versus cAMP
As discussed above, the pathway regulating folic acid chemotaxis is different from the one that mediates chemotaxis towards cAMP. DdEGFL1 does not significantly increase folic acid chemotaxis, possibly due to cells moving at or near their maximal rate [45, 47, 61, 71]. However, DdEGFL1 is able to completely restore the normal chemotactic response to folic acid when one of PI3K, PLA2, or tyrosine kinase activity is inhibited and partially restore the response when CaM is inhibited [61]. This restoration of chemotactic ability was also observed in mutants of cAMP signalling. Untreated carA-, carC-, and carA-/carC-cells possess lower rates of random motility when compared to parental cells [60]. However, DdEGFL1 was able to significantly increase the movement of all three strains to amounts greater than that observed in parental cells [60]. Together, these results provide further support for the existence of a novel motility-enhancing pathway regulated by EGFL repeats in this model organism. When components of either the folic acid or cAMP-mediated pathways are inactivated (e.g., in a knockout mutant or by pharmacological means), the pathway regulated by EGFL repeats/peptides is allowed to function as the dominant motility regulating pathway.
Vinculin B is a downstream target of CyrA EGFL1 signalling
DdEGFL1 sustains the threonine phosphorylation of vinculin B (VinB) during cell starvation (Fig. 1) [20, 62]. VinB localizes to the cytoplasm and cytoskeleton of Dictyostelium amoebae and has been shown to interact with DdEGFL1, CyrA-C45, and established vinculin-binding cytoskeletal proteins (e.g., MHC, α-actinin, talin, and actin; Fig. 1) [54, 62]. In higher organisms, vinculin over-expression has been shown to reduce cell migration, whereas down-regulation of the protein increases cell motility [72, 73]. VinB over-expression suppresses DdEGFL1-increased cell movement fitting with observations in mammalian cells [62]. Although PI3K and PKA have both been shown to be important for Dictyostelium cell motility, VinB threonine phosphorylation was shown to be independent of PI3K and PKA kinase activity and PI3K/PLA2 signalling, therefore indicating that another kinase is responsible for phosphorylating VinB (Fig. 1) [62]. In addition, these results suggest that the phosphorylation of VinB is regulated upstream of both PI3K and PLA2.
Future directions
DdEGFL1 has been shown to localize extracellularly, and was not observed inside cells even after prolonged incubation (Fig. 1) [20]. A recent study showed that DdEGFL1-FITC could be detected on the surface of cells capped with concanavalin A, suggesting that a receptor for EGFL repeats/peptides exists in Dictyostelium [54]. The identification of an EGFR-like (EGFRL) protein in Dictyostelium would be an important, evolutionarily significant finding, since disturbances in EGFR signalling have been associated with the development of malignant tumors and many forms of cancer [67, 74]. Therefore, understanding the evolution of this signalling may provide insight for future biomedical research. The existence of a membrane-bound EGFL repeat/peptide receptor possessing kinase activity is strengthened by the fact that DdEGFL1-activated signalling does not require the cAMP receptor or the heterotrimeric G-protein utilized in cAMP-mediated signalling [60]. A putative EGFRL protein was previously identified by inputting the amino acid sequence of EGFR from 14 different organisms into the BLASTp server of the online Dictyostelium resource dictyBase (http://www.dictybase.org/tools/blast). This candidate receptor possessed a putative kinase domain that shared sequence similarity with the intracellular tyrosine kinase domain of mammalian EGFR, suggesting that it may be an evolutionary precursor to mammalian EGFR (DDB0229955, http://www.dictybase.org) [20]. However, preliminary studies into the localization and binding partners of this protein suggest it is not the EGFL repeat/peptide receptor in Dictyostelium [62]. Although the search for the EGFL repeat/peptide receptor in Dictyostelium remains elusive, its identification would yield further insight into the attributes and evolution of receptors involved in mediating matricellular function.
To fully understand ECM dynamics in Dictyostelium, future research should be aimed at identifying the extracellular proteases that cleave CyrA and potentially other proteins in the ECM of Dictyostelium to generate EGFL repeat-containing cleavage products or peptides. The presence of a large number of extracellular proteases was reported in a recent study describing the secreted proteome profile of developing Dictyostelium cells [57]. Of the 349 proteins identified, 29 were linked to proteolysis including a putative dipeptidyl aminopeptidase (DDB0001159), a putative vitellogenic-like carboxypeptidase (DDB0167727), cysteine protease 4 (DDB0214999), cysteine proteinase 5 (DDB0185092), cysteine proteinase 6 (DDB0001517), and cysteine proteinase 7 (DDB0215005). These findings suggest that future studies should be performed to determine the function of these proteins during Dictyostelium development.
Conclusion
CyrA is the first extracellular CaMBP identified in Dictyostelium. Since it fulfills the criteria required to establish a protein as matricellular, it is designated as the first matricellular protein to be identified in Dictyostelium (Table 1) [1–3]. The first EGFL repeat (EGFL1) of CyrA has been well characterized. Our current knowledge of DdEGFL1-enhanced cell movement has allowed us to develop a model for the signal transduction mediating the response of cells to DdEGFL1 (Fig. 1). The dependence of CyrA secretion and DdEGFL1-enhanced cell movement on PI3K, PLA2, CaM, and intracellular Ca2+ release shows that these intracellular signalling components mediate the function of CyrA and form the basis of a novel motility-regulating pathway in Dictyostelium. In order to fully characterize the function of CyrA, it will be necessary to assess the ability of the other EGFL repeats of the protein to increase the rate of cell motility. The presence of repeated DdEGFL1-like sequences in EcmA, EcmB, and many putative ECM proteins in Dictyostelium suggest that these highly conserved sequences possess an important function for ECM proteins in this model organism. The abundance of EGFL domains in Dictyostelium and the ability of some EGFL repeats/peptides to increase cell movement suggests that this simple eukaryote could be used as a model system for studying the evolution of matricellular proteins, their receptors, and EGFL repeat signalling. The findings described here set the stage for further study into ECM proteins in Dictyostelium, specifically those that modulate cell motility.
Acknowledgments
This review was supported by a Discovery Grant (D.H.O’D.; A6807) and a Canada Graduate Scholarship (R.J.H.) from the Natural Sciences and Engineering Research Council of Canada.
Abbreviations
- AcbA
Acyl-CoA binding protein A
- ADAMTS
A disintegrin and mettaloproteinase with thrombospondin motifs
- carA
cAMP receptor A
- carC
cAMP receptor C
- CaM
Calmodulin
- CaMBP
CaM-binding protein
- CyrA
Cysteine-rich protein A
- CyrA-C40
40 kDa CyrA cleavage product
- CyrA-C45
45 kDa CyrA cleavage product
- ECM
Extracellular matrix
- EGFL
Epidermal growth factor-like
- EGFL1
CyrA EGFL repeat 1
- EGFR
EGF receptor
- ER
Endoplasmic reticulum
- MHC
Myosin II heavy chain
- MMP2
Matrix metalloproteinase 2
- PaxB
Paxillin B
- PI3K
Phosphatidylinositol-3-kinase
- PKA
Protein kinase A
- PLA2
Phospholipase A2
- PLC
Phospholipase C
- SDF-2
Spore differentiation factor 2
- SPARC
Secreted protein acidic and rich in cysteine
- TalB
Talin B
- Ten14
14th EGFL repeat of tenascin C
- Tenascin C
Tenascin cytotactin
- TSP
Thrombospondin
- VinB
Vinculin B
References
- 1.Bornstein P, Sage EH. Matricellular proteins: extracellular modulators of cell function. Curr Opin Cell Biol. 2002;14:608–616. doi: 10.1016/S0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 2.Mosher DF, Adams JC. Adhesion-modulating/matricellular ECM protein families: a structural, functional and evolutionary appraisal. Matrix Biol. 2012;31:155–161. doi: 10.1016/j.matbio.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Roberts DD. Emerging functions of matricellular proteins. Cell Mol Life Sci. 2011;68:3133–3136. doi: 10.1007/s00018-011-0779-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams JC, Lawler J. The thrombospondins. Cold Spring Harb Perspect Biol. 2011;3:a009712. doi: 10.1101/cshperspect.a009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol. 2012;44:480–488. doi: 10.1016/j.biocel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Özbek S, Balasubramanian PG, Chiquet-Ehrismann R, Tucker RP, Adams JC. The evolution of extracellular matrix. Mol Biol Cell. 2010;21:4300–4305. doi: 10.1091/mbc.E10-03-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy-Ullrich JE. The de-adhesive activity of matricellular proteins: is intermediate cell adhesion an adaptive state? J Clin Invest. 2001;107:785–790. doi: 10.1172/JCI12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brekken RA, Sage EH. SPARC, a matricellular protein: at the crossroads of cell-matrix communication. Matrix Biol. 2000;19:569–580. doi: 10.1016/S0945-053X(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 9.Yanagisawa H, Schluterman MK, Brekken RA. Fibulin-5, an integrin-binding matricellular protein: its function in development and disease. J Cell Commun Signal. 2009;3:337–347. doi: 10.1007/s12079-009-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weaver MS, Workman G, Cardo-Vila M, Arap W, Pasqualini R, Sage EH. Processing of the matricellular protein hevin in mouse brain is dependent on ADAMTS4. J Biol Chem. 2010;285:5868–5877. doi: 10.1074/jbc.M109.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apte SS. A disintegrin-like and metalloproteinase (reprolysin-type) with thrombospondin type I motif (ADAMTS) superfamily-functions and mechanisms. J Biol Chem. 2009;284:31493–31497. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Appella E, Weber I, Blasi F. Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett. 1988;231:1–4. doi: 10.1016/0014-5793(88)80690-2. [DOI] [PubMed] [Google Scholar]
- 13.Grotendorst GR, Soma Y, Takehara K, Charette M. EGF and TGF-alpha are potent chemoattractants for endothelial cells and EGF-like peptides are present at sites of tissue regeneration. J Cell Physiol. 1989;139:617–623. doi: 10.1002/jcp.1041390323. [DOI] [PubMed] [Google Scholar]
- 14.Linggi B, Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Rao Z, Handford P, Mayhew M, Knott V, Browniee GC, Stuart D. The structure of a Ca2+-binding epidermal growth factor-like domain: its role in protein–protein interactions. Cell. 1995;82:131–141. doi: 10.1016/0092-8674(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 16.Campbell I, Bork P. Epidermal growth factor-like modules. Curr Opin Struct Biol. 1993;3:385–392. doi: 10.1016/S0959-440X(05)80111-3. [DOI] [Google Scholar]
- 17.Kurucz E, Márkus R, Zsámboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, Udvardy A, Krausz I, Lukacsovich T, Gateff E, Zettervall CJ, Hultmark D, Andó I. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–654. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Glöckner G, Eichinger L, Szafranski K, Pachebat JA, Bankier AT, Dear PH, Lehmann R, Baumgart C, Parra G, Abril JF, Guigó R, Kumpf K, Tunggal B, Cox E, Quail MA, Platzer M, Rosenthal A, Noegel AA. Sequence and analysis of chromosome 2 of Dictyostelium discoideum . Nature. 2002;418:79–85. doi: 10.1038/nature00847. [DOI] [PubMed] [Google Scholar]
- 19.Fey P, Stephens S, Titus MA, Chisholm RL. SadA, a novel adhesion receptor in Dictyostelium . J Cell Biol. 2002;159:1109–1119. doi: 10.1083/jcb.200206067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber RJ, O’Day DH. An EGF-like peptide sequence from Dictyostelium enhances cell motility and chemotaxis. Biochem Biophys Res Commun. 2009;379:470–475. doi: 10.1016/j.bbrc.2008.12.081. [DOI] [PubMed] [Google Scholar]
- 21.Suarez A, Huber RJ, Myre MA, O’Day DH. An extracellular matrix, calmodulin-binding protein from Dictyostelium with EGF-like repeats that enhance cell motility. Cell Signal. 2011;23:1197–1206. doi: 10.1016/j.cellsig.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Jones FS, Jones PL. The tenascin family of ECM glycoproteins: structure, function, and regulation during embryonic development and tissue remodelling. Dev Dyn. 2000;218:235–259. doi: 10.1002/(SICI)1097-0177(200006)218:2<235::AID-DVDY2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 23.Wallner K, Li C, Shah PK, Wu KJ, Schwartz SM, Sharifi BG. EGF-Like domain of tenascin-C is proapoptotic for cultured smooth muscle cells. Arterioscler Thromb Vasc Biol. 2004;24:1416–1421. doi: 10.1161/01.ATV.0000134299.89599.53. [DOI] [PubMed] [Google Scholar]
- 24.Chen P, Gupta K, Wells A. Cell movement elicited by epidermal growth factor receptor requires kinase and autophosphorylation but is separable from mitogenesis. J Cell Biol. 1994;124:547–555. doi: 10.1083/jcb.124.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsunoda T, Inada H, Kalembeyi I, Imanaka-Yoshida K, Sakakibara M, Okada R, Katsuta K, Sakakura T, Majima Y, Yoshida T. Involvement of large tenascin-C splice variants in breast cancer progression. Am J Pathol. 2003;162:1857–1867. doi: 10.1016/S0002-9440(10)64320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer AK, Kien TT, Borysenko CW, Cascio M, Camacho CJ, Blair HC, Bahar I, Wells A. Tenascin cytotactin epidermal growth factor-like repeat binds epidermal growth factor receptor with low affinity. J Cell Physiol. 2007;211:748–758. doi: 10.1002/jcp.20986. [DOI] [PubMed] [Google Scholar]
- 27.Iyer AKV, Tran KT, Griffith L, Wells A. Cell surface restriction of EGFR by a tenascin cytotactin-encoded EGF-like repeat is preferential for motility-related signalling. J Cell Physiol. 2008;214:504–512. doi: 10.1002/jcp.21232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swindle CS, Tran KT, Johnson TD, Banerjee P, Mayes AM, Griffith L, Wells A. Epidermal growth factor (EGF)-like repeats of human tenascin-C as ligands for EGF receptor. J Cell Biol. 2001;154:459–468. doi: 10.1083/jcb.200103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu A, Garg P, Yang S, Gong P, Pallero MA, Annis DS, Liu Y, Passaniti A, Mann D, Mosher DF, Murphy-Ullrich JE, Goldblum SE. Epidermal growth factor-like repeats of thrombospondins activate phospholipase Cgamma and increase epithelial cell migration through indirect epidermal growth factor receptor activation. J Biol Chem. 2009;284:6389–6402. doi: 10.1074/jbc.M809198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitchev PP, Wcislak SM, Lee C, Bergh A, Brendler CB, Stellmach VM, Crawford SE, Mavroudis CD, Cornwell ML, Doll JA. Thrombospondin-1 regulates the normal prostate in vivo through angiogenesis and TGF-β activation. Lab Invest. 2010;90:1078–1090. doi: 10.1038/labinvest.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenk S, HIntermann E, Bilban M, Koshikawa N, Hojilla C, Khokha R, Quaranta V. Binding to EGF receptor of a laminin-5 EGF-like fragment liberated during MMPdependent mammary gland involution. J Cell Biol. 2003;161:197–209. doi: 10.1083/jcb.200208145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin 5. Science. 1997;277:225–228. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto H, Itoh F, Iku S, Hosokawa M, Imai K. Expression of the gamma (2) chain of laminin-5 at the invasive front is associated with recurrence and poor prognosis in human esophageal squamous cell carcinoma. Clin Cancer Res. 2001;7:896–900. [PubMed] [Google Scholar]
- 34.Niki T, Kohno T, Iba S, Moriya Y, Takahashi Y, Saito M, Maeshima A, Yamada T, Matsumo Y, Fukayama M, Yokota J, Hirohashi S. Frequent co-localization of cox-2 and laminin-5 gamma2 chain at the invasive front of early-stage lung adenocarcinomas. Am J Pathol. 2002;160:1129–1141. doi: 10.1016/S0002-9440(10)64933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams JG. Dictyostelium finds new roles to model. Genetics. 2010;185:717–726. doi: 10.1534/genetics.110.119297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaap P. Evolutionary crossroads in developmental biology: Dictyostelium discoideum . Development. 2011;138:387–396. doi: 10.1242/dev.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin T, Hereld D. Moving toward understanding eukaryotic chemotaxis. Eur J Cell Biol. 2006;85:905–913. doi: 10.1016/j.ejcb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Insall R, Andrew N. Chemotaxis in Dictyostelium: how to walk straight using parallel pathways. Curr Opin Microbiol. 2007;10:578–581. doi: 10.1016/j.mib.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Chen LF, Iijima M, Tang M, Landree MA, Huang YE, Xiong Y, Iglesias PA, Devreotes PN. PLA2 and PI3K/PTEN pathways act in parallel to mediate chemotaxis. Dev Cell. 2007;12:603–614. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Haastert PJM, Keizer-Gunnink I, Kortholt A. Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J Cell Biol. 2007;177:809–816. doi: 10.1083/jcb.200701134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Janetopoulos C, Huang YE, Iijima M, Borleis J, Devreotes PN. Two phases of actin polymerization display different dependencies on PI (3, 4, 5) P3 accumulation and have unique roles during chemotaxis. Mol Biol Cell. 2003;14:5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeda K, Sasaki AT, Ha H, Seung H-A, Firtel RA. Role of phosphatidylinositol 3-kinases in chemotaxis in Dictyostelium . J Biol Chem. 2007;282:11874–11884. doi: 10.1074/jbc.M610984200. [DOI] [PubMed] [Google Scholar]
- 43.Veltman DM, Keizer-Gunnik I, Van Haastert PJM. Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum . J Cell Biol. 2008;180:747–753. doi: 10.1083/jcb.200709180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lusche DF, Wessels D, Soll DR. The effects of extracellular calcium on motility, pseudopod and uropod formation, chemotaxis, and the cortical localization of myosin II in Dictyostelium discoideum . Cell Motil Cytoskeleton. 2009;66:567–587. doi: 10.1002/cm.20367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gauthier ML, O’Day DH. Detection of calmodulin-binding proteins and calmodulin-dependent phosphorylation linked to calmodulin-dependent chemotaxis to folic and cAMP in Dictyostelium . Cell Signal. 2001;13:575–584. doi: 10.1016/S0898-6568(01)00187-5. [DOI] [PubMed] [Google Scholar]
- 46.Scherer A, Kuhl S, Wessels D, Lusche DF, Raisley B, Soll DR. Ca2+ chemotaxis in Dictyostelium discoideum . J Cell Sci. 2010;123:3756–3767. doi: 10.1242/jcs.068619. [DOI] [PubMed] [Google Scholar]
- 47.Browning DD, The T, O`Day DH. Comparative analysis of chemotaxis in Dictyostelium using a radial bioassay method: protein tyrosine kinase activity is required for chemotaxis to folate but not to cAMP. Cell Signal. 1995;7:481–489. doi: 10.1016/0898-6568(95)00016-I. [DOI] [PubMed] [Google Scholar]
- 48.Wilkins MR, Williams KL. The extracellular matrix of the Dictyostelium discoideum slug. Experientia. 1995;51:1189–1196. doi: 10.1007/BF01944736. [DOI] [PubMed] [Google Scholar]
- 49.Morrison A, Blanton RL, Grimson M, Fuchs M, Williams KL, Williams J. Disruption of the gene encoding the EcmA, extracellular matrix protein of Dictyostelium, alters slug morphology. Dev Biol. 1994;163:457–466. doi: 10.1006/dbio.1994.1162. [DOI] [PubMed] [Google Scholar]
- 50.Breen EJ, Vardy PH, Williams KL. Movement of the multicellular slug stage of Dictyostelium discoideum: an analytical approach. Development. 1987;101:313–321. [Google Scholar]
- 51.Grant WN, Williams KL. Monoclonal antibody characterization of the slime sheath: the extracellular matrix of Dictyostelium discoideum . EMBO J. 1983;2:935–940. doi: 10.1002/j.1460-2075.1983.tb01524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breen EJ, Williams KL. Movement of the Dictyostelium discoideum slug: models, musings and images. Dev Genet. 1988;9:539–548. doi: 10.1002/dvg.1020090430. [DOI] [PubMed] [Google Scholar]
- 53.Kowal AS, Chisholm RL. Uncovering a role for the tail of the Dictyostelium discoideum SadA protein in cell-substrate adhesion. Eukaryot Cell. 2011;10:662–671. doi: 10.1128/EC.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber RJ, Suarez A, O’Day DH. CyrA, a matricellular protein that modulates cell motility in Dictyostelium discoideum . Matrix Biol. 2012;31:271–280. doi: 10.1016/j.matbio.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Müller-Taubenberger A, Lupas AN, Li H, Ecke M, Simmeth E, Gerisch G. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 2001;20:6772–6782. doi: 10.1093/emboj/20.23.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anjard C, Loomis WF. Peptide signaling during terminal differentiation of Dictyostelium . Proc Natl Acad Sci USA. 2005;102:7607–7611. doi: 10.1073/pnas.0501820102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bakthavatsalam D, Gomer RH. The secreted proteome profile of developing Dictyostelium discoideum cells. Proteomics. 2010;10:2556–2559. doi: 10.1002/pmic.200900516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Handford PA, Mayhew M, Baron M, Winship PR, Campbell ID, Brownlee GG. Key residues involved in calcium-binding motifs EGF-like domains. Nature. 1991;351:164–167. doi: 10.1038/351164a0. [DOI] [PubMed] [Google Scholar]
- 59.Periz J, Gill AC, Knott V, Handford PA, Tomley FM. Calcium binding activity of the epidermal growth factor-like domains of the apicomplexan microneme protein EtMIC4. Mol Biochem Parasitol. 2005;143:192–199. doi: 10.1016/j.molbiopara.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 60.Huber RJ, O’Day DH. EGF-like peptide-enhanced cell motility in Dictyostelium functions independently of the cAMP-mediated pathway and requires active Ca2+/calmodulin signaling. Cell Signal. 2011;23:731–738. doi: 10.1016/j.cellsig.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 61.Nikolaeva I, Huber RJ, O’Day DH. EGF-like peptide of Dictyostelium discoideum is not a chemoattractant but it does restore folate-mediated chemotaxis in the presence of signal transduction inhibitors. Peptides. 2012;34:145–149. doi: 10.1016/j.peptides.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 62.Huber RJ, O’Day DH. EGF-like peptide-enhanced cell movement in Dictyostelium is mediated by protein kinases and the activity of several cytoskeletal proteins. Cell Signal. 2012;24:1770–1780. doi: 10.1016/j.cellsig.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 63.Bryant JA, Finn RS, Slamon DJ, Cloughesy TF, Charles AC. EGF activates intracellular and intercellular calcium signaling by distinct pathways in tumor cells. Cancer Biol Ther. 2004;3:1243–1249. doi: 10.4161/cbt.3.12.1233. [DOI] [PubMed] [Google Scholar]
- 64.Kato K, Ueoka Y, Tamura T, Nishida J, Wake N. Oncogenic Ras modulates epidermal growth factor responsiveness in endometrial carcinomas. Eur J Cancer. 1998;34:737–744. doi: 10.1016/S0959-8049(97)10124-1. [DOI] [PubMed] [Google Scholar]
- 65.Bolourani P, Spiegelman GB, Weeks G. Delineation of the roles played by RasG and RasC in cAMP-dependent signal transduction during the early development of Dictyostelium discoideum . Mol Biol Cell. 2006;17:4543–4550. doi: 10.1091/mbc.E05-11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H, Heid PJ, Wessels D, Daniels KJ, Pham T, Loomis WF, Soll DR. Constitutively active protein kinase A disrupts motility and chemotaxis in Dictyostelium discoideum . Eukaryot Cell. 2003;2:62–75. doi: 10.1128/EC.2.1.62-75.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zandi R, Larsen AB, Andersen P, Stockhausen MT, Poulsen HS. Mechanisms for oncogenic activation of the epidermal growth factor receptor. Cell Signal. 2007;19:2013–2023. doi: 10.1016/j.cellsig.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 68.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 69.Wu L, Valkema R, Van Haastert PJ, Devreotes PN. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium . J Cell Biol. 1995;129:1667–1675. doi: 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, Firtel RA. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varnum A, Soll DR. Chemoresponsiveness to cAMP and folic acid during growth, development, and dedifferentiation in Dictyostelium discoideum . Differentiation. 1981;18:151–160. doi: 10.1111/j.1432-0436.1981.tb01116.x. [DOI] [PubMed] [Google Scholar]
- 72.Gallant ND, Michael KE, García AJ. Cell adhesion strengthening: contributions of adhesive area, integrin binding, and focal adhesion assembly. Mol Biol Cell. 2005;16:4329–4340. doi: 10.1091/mbc.E05-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 74.Diasio RB, Fourie J. Targeting the epidermal growth factor receptor in the treatment of colorectal cancer. Drugs. 2006;66:1441–1463. doi: 10.2165/00003495-200666110-00003. [DOI] [PubMed] [Google Scholar]