Abstract
Islet 1 (ISL1), a marker of cardiac progenitors, plays a crucial role in cardiogenesis. However, the precise mechanism underlying the activation of its expression is not fully understood. Using the cardiac differentiation model of P19CL6 cells, we show that POU homeodomain protein, OCT1, modulates Isl1 expression in the process of cardiac differentiation. Oct1 knock-down resulted in reduction of Isl1 expression and downregulated mesodermal, cardiac-specific, and signal pathway gene expression. Additionally, the octamer motif located in the proximal region of Isl1 promoter is essential to Isl1 transcriptional activation. Mutation of this motif remarkably decreased Isl1 transcription. Although both OCT1 and OCT4 bound to this motif, it was OCT1 rather than OCT4 that modulated Isl1 expression. Furthermore, the correlation of OCT1 in regulation of Isl1 was revealed by in situ hybridization in early embryos. Collectively, our data highlight a novel role of OCT1 in the regulation of Isl1 expression.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0544-y) contains supplementary material, which is available to authorized users.
Keywords: Isl1, Cardiac progenitors, Differentiation, P19CL6 cells, Octamer motif, OCT1
Introduction
Islet 1 (ISL1), a LIM-homeodomain transcription factor, was originally identified as an insulin gene-enhancer binding protein [1]. ISL1 is important for the motor neuron specification and pancreatic development [2–4]. Recently, it has been reported that ISL1 also plays a crucial role in regulation of early-stage cardiogenesis [5–10]. Studies from Moretti and Bu et al. have documented that the multipotent ISL1+ cardiovascular progenitors derived from embryonic stem cells resemble developmental precursors in the embryonic heart and exhibit the pluripotent properties that can give rise to diverse cardiovascular cell types, such as cardiomyocytes, endothelial cells, and smooth muscle cells in vitro [7, 11]. Therefore, ISL1+ cells authentically represent a native population of cardiac progenitors and have great potential in regenerative cardiovascular medicine [12].
Fate mapping evidence demonstrated that ISL1 mainly marks the second heart field (SHF) progenitors, which contributed to the outflow tract, right ventricle, and much of the atria. Ablation of Isl1 results in lethality at embryonic day 10.5 (ED10.5) with defects in cardiac structures derived from the SHF [5, 9]. These defects are believed to be caused by severe decrease in proliferation or even apoptosis of ISL1+ progenitor cells [5]. RNA transcript analysis further reveals that Isl1 is expressed early in the cardiac crescent stage (ED7.5) and continues to be expressed until ED10.5. Its expression is restricted in the splanchnic and pharyngeal mesoderm, but not within myocardium. During early cardiac crescent stages, ISL1+ progenitors migrate into the forming heart from dorsal to medial regions, progressively differentiating as they converge with the cells of the first myocardial precursor lineage and finally forming the majority of cells of the right ventricle, outflow tract, and remainder of the atria. Once precursors differentiate, Isl1 expression is progressively downregulated and extinguished from ED12.5 to ED18.5 [5, 6]. Moreover, ISL1+ progenitors also contribute to specific regions of the left ventricle, including a junctional region between left and right ventricles, trabeculae, and the inner curvature [5]. Therefore, it is worth noting that during embryonic development, ISL1 function is required to maintain an undifferentiated state and/or may be incompatible with a differentiated state. Isl1 expression may delineate the undifferentiated progenitor state from differentiated cardiomyocytes.
As a transcription factor, ISL1 has been validated to lie on the top of a conservative transcriptional network governing SHF development. It interacts with other transcriptional factors such as GATA zinc-finger and forkhead transcripton factors, setting in motion a cascade of downstream transcriptional events involving MEF2c, NKX2.5, HAND2, and ultimately influencing SHF development [13]. Disruption of ISL1 resulted in more severe defects in the right ventricle and outflow tract formation than disruption of its downstream factors, suggesting that ISL1 served as a master regulator and played a leading role in embryonic heart development [13]. Moreover, Isl1 expression also requires inputs from extrinsic factors to facilitate the development of the SHF [14, 15]. For example, loss of FGF8 in the cardiac crescent mesoderm led to aberrant phenotype of Isl1 and its target Mef2c in the SHF [14]. Ablation of Wnt/β-catenin signaling resulted in significantly decreased expression of FGF ligands, which inhibited the expansion of ISL1+ progenitors [15]. A recent report showed that Notch pathway is negatively involved in the β-catenin-mediated Isl1 regulation for the maturation and differentiation of ISL1+ progenitors [16].
Although the ISL1-dependent transcriptional network for SHF development has been delineated and multiple signaling pathways have been integrated to elicit the effect on the expression of Isl1, the mechanisms, especially the upstream transcription factors, that regulate Isl1 transcriptional activity remain virtually unknown. In the current study, using the in vitro cardiac differentiation model of P19CL6 cells, we focused on determining the regulatory elements and trans-acting factors involved in Isl1 expression. We found that a POU homeodomain family member, OCT1, but not OCT4, favored Isl1 expression and transcriptional activation in the early stage of cardiogenesis, and that downregulation of OCT1 in the later stage was implicated in the silencing of Isl1 expression. Moreover, data from in situ hybridization also showed a good correlation in the distribution of Oct1 and Isl1 mRNAs in early developmental embryos. Thus, our data provide a novel insight into how Isl1 expression is controlled within the defined program of cardiac differentiation.
Materials and methods
Cell culture and induction of differentiation
P19CL6 cells were cultured and induced to cardiac differentiation as described by Habara-Ohkubo et al. [17]. Differentiation efficiency was calculated by counting α-actinin-positive cells as described below in the immunofluorescence assay.
RT-PCR and real-time RT-PCR
RNA extraction was performed using Trizol reagent (Invitrogen) in accordance with the manufacturer’s instructions. For RT-PCR, Taq DNA polymerase (Promega) and GeneAmp PCR System 9700 (Applied Biosystems) were utilized. The following thermal profile was used for all PCR experiments: 95°C for 5 min; and then appropriate cycles at 95°C for 30 s, annealing temperature (online supplemental material, Table S1) for 30 s, and 72°C for 30 s; and terminated by a final extension at 72°C for 8 min. For real-time RT-PCR, ABI Prism 7700 sequence detector and SYBR® Green real-time Master Mix (Toyobo) were utilized. The mRNA level was quantified as described by Livak et al. [18]. Relative PCR signals were normalized to the average expression levels of the undifferentiated samples, and normalized ratios were used to indicate up- and downregulation. Primers for RT-PCR and real-time RT-PCR are listed in the supplemental material, Table S1.
Immunofluorescence
P19CL6 cells on coverslips were induced for 8 or 12 days by DMSO and then fixed with 4% paraformaldehyde for 10 min at room temperature. After being rinsed with PBS, cells were permeabilized with 0.3% Triton X-100 (Sigma) for 15 min, and then blocked with normal goat serum for 30 min. In order to find out whether cells differentiated into cardiomyocytes, they were incubated overnight at 4°C with mouse monoclonal antibody against α-actinin (1:200 Sigma) and subsequently incubated with TRITC-conjugated goat anti-mouse IgG (1:200). Nuclei were counterstained with Hoechst 33342 (Sigma). Quantitative evaluation of differentiation efficiency was performed by counting α-actinin-positive cells. A minimum of five randomly imaged fields of each coverslip were counted from at least five coverslips. The percentage of α-actinin-positive cells out of the total number of cells counted represents the differentiation efficiency.
Primer extension analysis
Total RNA was isolated from induced day 4 P19CL6 cells using Trizol Reagent according to the manufacturer’s instructions. An antisense oligodeoxynucleotide, 5′-AAC TGG GGG AAA CAG CAG CC-3′, corresponding to the Isl1 sequence from −138 to −119 bp upstream of the translation start codon was labeled with [γ-32P]ATP (3000 mCi/mmol; FuRui, Beijing, China). Primer extension assay was performed following the manufacturer’s instructions (Promega). The transcriptional start site was determined by comparison with the Isl1 mRNA sequence from the GenBank database.
Cloning of the Isl1 5′ flanking region and construction of a series of luciferase reporter vectors
A 1,058 bp putative Isl1 promoter region was amplified by PCR using primers based on the genomic DNA sequence of Isl1 (accession number NM_021459). The amplified PCR fragment was cloned into pGEM-T Easy vector (Promega), digested with KpnI/HindIII, and transferred into the pGL3-basic reporter vector (Promega). This construct was designated as pGL(−909/+133), relative to the transcriptional start site (+1), and used for generating a series of deletion constructs. For 5′ nested truncation, the unique restriction sites BalI at position −563, BlpI at −406, and AatII at −122 in the Isl1 promoter region were chosen, and the pGL(−909/+133) was digested with KpnI/BalI, KpnI/BlpI, and KpnI/AatII, respectively. Finally, the large fragments containing the vector sequence were end-blunted with T4 DNA polymerase (Promega) and self-ligated with T4 DNA ligase (Promega), designated pGL(−563/+133), pGL(−406/+133), or pGL(−122/+133). The other deletion constructs were generated by PCR through specific primers and designated pGL(−246/+133) and pGL(−57/+133). All the constructs were confirmed by DNA sequencing. The primers are shown in the online supplemental material, Table S2.
Transfection and luciferase assays
Cells were seeded at a density of 10 × 104 per well in a 24-well plate and transfected using LipofectamineTM 2000 (Invitrogen) when they reached 80% confluence. pRL-CMV (Promega) was co-transfected in all experiments as an internal control to standardize transfection efficiency. For co-transfection experiments, the total amount of DNA was kept constant with empty plasmid DNA (maximum 600 ng/well). After 24–48 h of transfection, cell lysates were assayed for firefly and renilla luciferase activity on a Microplate Luminometer (FOLAR star reporter assay system, BMG) using the Dual-luciferase Reporter Assay System (Promega) and compared with that of the control vector. Each assay was performed in triplicate and repeated at least three times. The data were statistically analyzed using the unpaired, two-tailed Student’s t test, and differences were considered significant when P < 0.05.
Electrophoretic mobility shift assays
Nuclear extracts (NE) from uninduced or induced P19CL6 cells at indicated time points were prepared as described by Zhu et al. [19]. Oligonucleotides used in EMSA were labeled by [γ-32P]ATP. EMSA was carried out at room temperature for 30 min in 20 μl reaction mixtures containing 10 μg of NE, 20 μCi 32P-labeled oligonucleotides, 500 ng of poly (dI–dC) (Sigma), 20 mM HEPES, pH 7.9, 1 mM dithiothreitol, and 10% glycerol. For competition assay, an unlabeled competitor was added at 100-fold excess prior to the labeled probe. For supershift EMSA, 2 μg of anti-OCT1 (sc-25399; Santa Cruz) or anti-OCT3/4 (sc-9081, Santa Cruz) antibodies were incubated with NE prior to the labeled probe. Protein-DNA complexes were resolved by nondenaturing 6% polyacrylamide gel electrophoresis. The sequences of the sense strand of these oligonucleotides, mutants of Oligo-3 with two base substitutions, and competitors of Oligo-3 with mutated or deleted octamer motifs are depicted below.
DNase I footprinting
DNase I footprinting experiments were carried out with NE from induced day 4 P19Cl6 cells. The DNA fragment containing the −246 to −122 bp of Isl1 minimal promoter was amplified by PCR and then 3′-end-labeled (coding strand) with [α-32P]dATP. After incubation with the labeled probe and NE at room temperature for 30 min, DNase I was added for exactly 1 min. Footprinting was carried out using a core footprinting system kit (Promega). The size of the DNA bands was estimated by referring to a “G + A” Maxam-Gilbert sequencing ladder of the same labeled probe.
Site-directed mutagenesis
The luciferase reporter constructs harboring the double point substitutions of the octamer motif were generated using Site-Directed Mutagenesis Kit (Stratagene). Each mutant primer was synthesized and annealed to the template of pGL(−246/+133) vector. PfuTurbo® DNA Polymerase was used to synthesize the mutagenic promoter, followed by digestion of the parental plasmid by DpnI according to the manufacturer’s instructions. All resulting plasmid constructs were sequenced to confirm the mutations.
Chromatin immunoprecipitation (ChIP) assays
ChIP experiments were performed according to the method described by Shang et al. [20]. After crosslink reversal, precipitated DNA was analyzed by PCR for a fragment of the Isl1 promoter (−246 to +133). The primers are 5′-TAA TCC TCT CCT GCG GGC TCG C-3′ and 5′-AGA GGA TGC TGG TGC TGT GGC TAG G-3′.
Western blotting
Nuclear protein (20 μg) was prepared and subjected to 10% SDS polyacrylamide gel electrophoresis and subsequently transferred onto nitrocellulose membranes (Amersham), which were then blocked in 5% nonfat milk and incubated with anti-OCT1 or anti-OCT4 antibodies. Horseradish-peroxidase-conjugated anti-human or mouse antibodies (1:2,000) were used as secondary antibodies. Visualization was performed using ECL Western blotting detection reagents (Amersham).
RNA interference (RNAi)
P19CL6 cells were induced by DMSO for 2 days, then transfected with chemically synthesized specific Oct1 siRNA (sc-36120, Santa Cruz) in Opti-MEM® I Reduced Serum Medium (Invitrogen). After 6 h, the culture medium was replaced with differentiation medium (containing 1%DMSO). At induced day 4, P19CL6 cells were harvested and subjected to real-time PCR assay to evaluate mesodermal, cardiac-specific, and signal pathway gene expression under the condition of Oct1 knock-down. Oct1 irrelevant control RNA was used as a control. In order to evaluate the effect of Oct1 siRNA on the differentiation efficiency of P19CL6 cells, cells were plated on coverslips and subjected to immunofluorescence analysis as described above. To increase the Oct1 knock-down effect, Oct1 siRNA was transfected twice. At induced day 2, P19CL6 cells were transfected with Oct1 siRNA for the first time, and at day 4, the cells were transfected with Oct1 siRNA again. At day 8, the cells on coverslips were subjected to immunofluorescence analysis, and differentiation efficiency was evaluated by counting α-actinin-positive cells as described above.
In situ hybridization
In order to prepare the mRNA probes used for in situ hybridization, the specific fragments of Isl1 and Oct1 DNA sequences were cloned by PCR using the following primers: Isl1 primers are 5′-TGT GGT GGA GAG AGC CAG CCT-3′ and 5′-GCA TAC CAG GTC CGC AAG GT-3′; Oct1 primers are 5′-GCA CAG TGC TGC TGG GGC CAC-3′ and 5′-CCT GGG GGG TCT GTG AGA TGA-3′. The amplified products were then subcloned into the pGEM-T vectors and a 201- or 206-nucleotide complementary RNA for Isl1 or Oct1 antisense probes was prepared by in vitro transcription and labeled with digoxigenin (Roche). Subsequently, in situ hybridization was performed to investigate the distribution of Oct1 and Isl1 at the mRNA level in the early embryos. To visualize hybridization signals, consecutive paraffin-embedded tissue sections from embryos of ED8.5, ED9.5, ED10.5, ED11.5, and ED12.5 were incubated for 1 h with anti-digoxigenin-AP antibody (1:500; Roche) and the reaction products were colorized with nitro blue tetrazolium together with 5-bromo-4-choloro-3-indolyl phosphate (NBT/BCIP, Promega), resulting in a blue signal. Signal specificity was assessed by substituting the probe using the sense probe specific for Isl1 or Oct1 mRNAs. To further demonstrate the co-localization of Isl1 and Oct1 mRNA, a double staining in situ hybridization for sectioned embryos of ED8.5 was performed using digoxigenin-labeled Isl1 probe and biotin-labeled (Roche) Oct1 probe simultaneously. Staining reaction was performed for Isl1 mRNA with NBT/BCIP (blue) after incubating with alkaline phosphatase (AP)-conjugated anti-digoxigenin antibody, while Oct1 mRNA was detected with aminoethylcarbazole (AEC, Invitrogen) peroxidase substrate (red) after incubating with horseradish peroxidase (HRP)-conjugated streptavidin (Vector Lab). If Isl1 mRNA was co-localized with Oct1 mRNA, the overlapping regions would be dark purple. Signal specificity was assessed by substituting the probe using the sense probe specific for Isl1 or Oct1.
Statistical analysis
All data were expressed as the mean ± SD from at least three independent experiments. The statistical significances of difference between groups were calculated using Student’s two tailed t test or ANOVA with SPSS10.0 software. Differences were considered to be significant at P value < 0.05.
Results
The expression of Isl1 during cardiac differentiation of P19CL6 cells
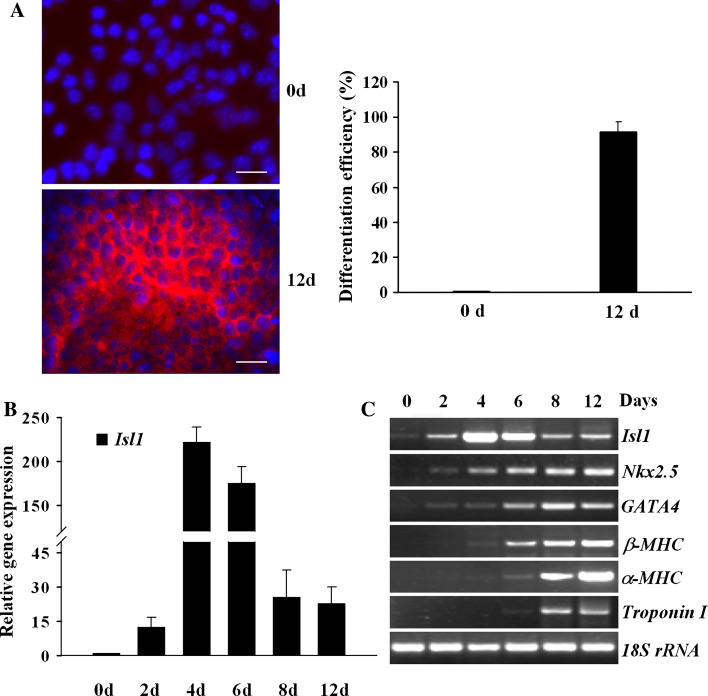
In our effort to understand the regulation of Isl1 expression, P19CL6 cells induced by DMSO in adherent culture were employed as a model of cardiogenesis [17]. After exposure to DMSO for 8 days, P19CL6 cells efficiently differentiated into mononucleated, rhythmically contracting cardiomyocytes. The dispersed clones of spontaneous contractions further expanded and fused together as multiple large-scale beating areas at day 12. About 90% of P19CL6 cells differentiated into cardiomyocytes as confirmed by immunostaining of sarcomeric α-actinin (Fig. 1a). Isl1 exhibited a dynamic expression profile during the differentiation process (Fig. 1b, c). It was elevated after DMSO induction and peaked at 4 days of differentiation, reaching over 200-fold that found in noninduced cells. Noticeably, the level of Isl1 mRNAs was attenuated 8 days after differentiation. In contrast, the expression of cardiac-specific transcription factors Nkx2.5 and Gata4 was upregulated after 2 days of differentiation. The expressions of cardiac contractile protein genes β-MHC, α-MHC, and Troponin I were detected at day 6 after induction and remained positive throughout the late stage of differentiation (Fig. 1c). These results reveal a stage-specific kinetic of Isl1 expression during cardiogenesis, suggesting that Isl1 characterizes a progenitor state/early cardiogenic state and becomes downregulated when the cells differentiate into cardiomyocytes.
Fig. 1.
The expression of Isl1 during cardiomyocytic differentiation of P19CL6 cells. a Uninduced and induced 12 day P19CL6 cells were analyzed by immunofluorescence staining with a monoclonal antibody against sarcomeric α-actinin (red in cytoplasm). Nuclei were counterstained with Hoechst33342 (blue). Scale bars 20 μm. Differentiation efficiency is shown on the right panel. b Real-time RT-PCR was performed to evaluate Isl1 expression during P19CL6 cell differentiation at indicated time points. Data are shown as relative Isl1 mRNA level compared with that of uninduced cells (0d) with mean ± SD from three independent experiments, each in triplicate. c RT-PCR was used to analyze the expression of Isl1 and cardiac-specific genes with RNA extracted from P19CL6 cells at indicated time points
Characterization of the 5′ flanking region of Isl1 gene
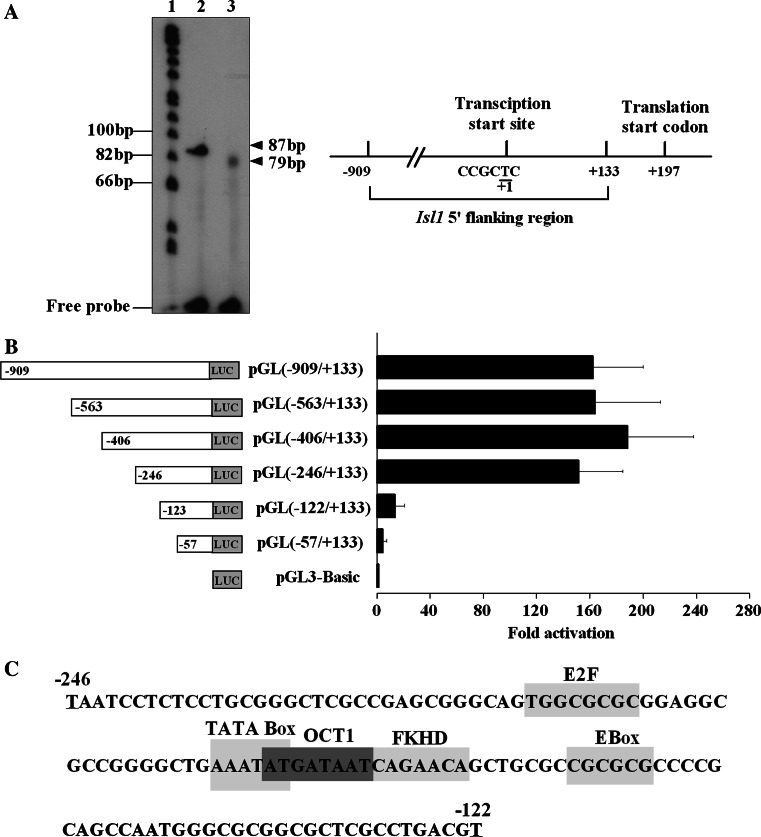
As the first step toward the study of the transcriptional control mechanism of Isl1, we mapped the transcriptional start site of Isl1 using primer extension analysis. As shown in Fig. 2a, a single extension product of 79 bp was detected, suggesting that only one Isl1 transcription initiation site exists. Subsequent comparison with the genomic sequence defined the T nucleotide in the sequence CCGCTC as the transcription start site located at 197 bp upstream of ATG in the Isl1 promoter.
Fig. 2.
Analysis of the 5′ flanking region of Isl1 gene. a Primer extension assay was used to identify the transcriptional start site of Isl1. Lane 1 ϕ×174 Hinf1 DNA marker, lane 2 control RNA, lane 3 induced day 4 P19CL6 RNA. The arrowheads indicate the products of the primer extension from control (87 bp) and experimental groups (79 bp). The map of Isl1 transcriptional start site is shown in the right panel. b Luciferase assay was performed to evaluate the activities of Isl1 promoter and its truncated constructs. Data are shown as fold activation over that of PGL3-basic promoter with means ± SD (n = 3). c Potential binding sites of transcription factors between the nucleotide −246 and −122 are indicated by boxes
To characterize the 5′ flanking region of Isl1, a series of luciferase reporter constructs containing successive 5′ deletions of Isl1 promoter were transiently transfected into induced day 4 P19CL6 cells. As shown in Fig. 2b, the luciferase activity caused by pGL(−909/+133) promoter was 160-fold greater than the basal level of pGL3-basic vector. Deletions of the segment from the 5′ end −909 to −246 had no significant effect on promoter activity. However, further deletion of the region from −246 to −122 caused a sharp drop of Isl1 promoter activity, indicating that the region between −246 and −122 harbored critical positive regulatory elements and defined the region as a minimal promoter of the Isl1 gene. The putative binding sites for transcriptional factors, such as E2F, TATA box, OCT, FKHD, and E Box, on Isl1 minimal promoter were revealed by professional MatInspector Database (Fig. 2c).
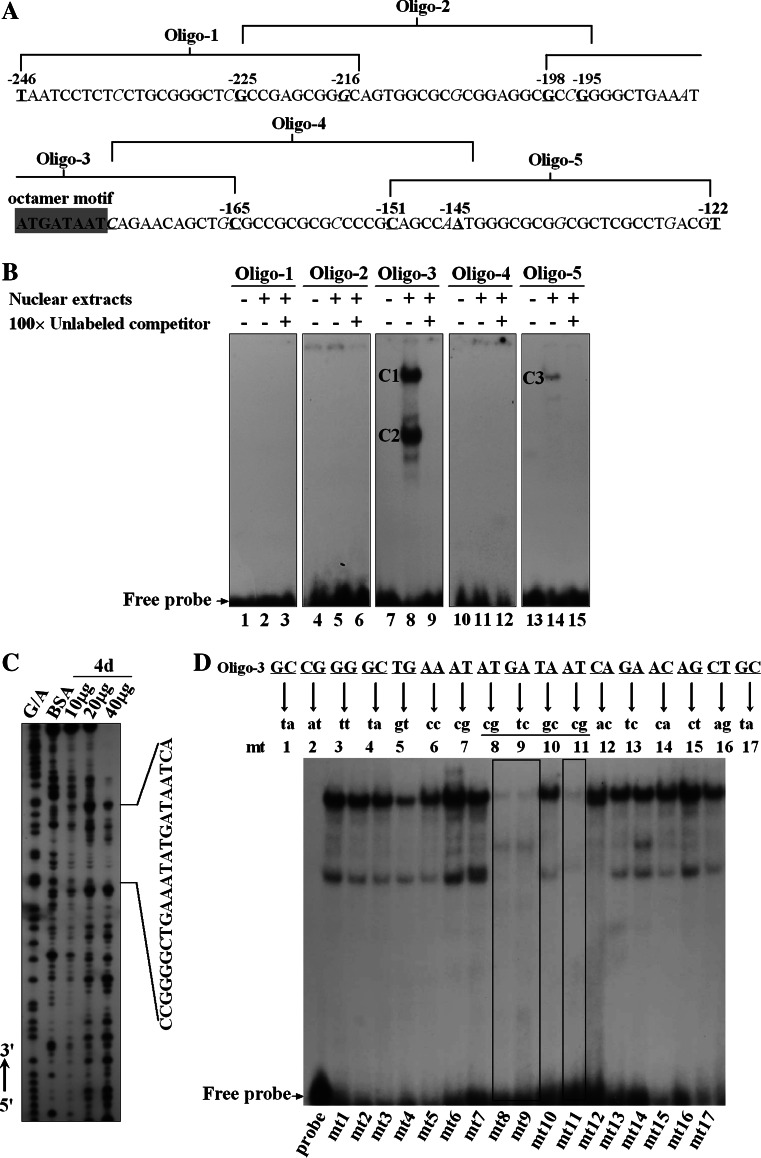
In order to identify the cis elements involved in the regulation of Isl1 expression, EMSA was performed. The results showed that two specific complexes, C1 and C2, were formed with Oligo-3 probe, one of five overlapping oligonucleotides spanning the region between −246 and −122 (Fig. 3a, b, lane 8). A weak binding complex, C3, was formed to Oligo-5 (Fig. 3b, lane 14), while no significant retarded band was detected using Oligo-1, -2, and -4 as a probe (Fig. 3b, lanes 2, 5, and 11, respectively). Specificity of the complexes was confirmed by incubation with a 100-fold excess of unlabeled oligonucleotide prior to the addition of the labeled probe (Fig. 3b, lanes 3, 6, 9, 12, and 15). Oligo-3 produced two stronger migrated bands than Oligo-5, indicating the corresponding transactivators were more abundant or had higher binding affinities. In addition, labeled Oligo-3 failed to induce the formation of any specific complexes when incubated with nuclear extracts from newborn rat cardiomyocytes (data not shown). It seemed that the corresponding transactivators of Oligo-3 were present in cardiac stem/progenitor cells rather than in mature cardiomyocytes. Thus, it was assumed that the Oligo-3 contained critical elements for regulating Isl1 expression during cardiogenesis. Furthermore, DNase I footprinting was performed to determine the exact binding sites of transcription factors. As shown in Fig. 3c, the nuclear extracts from induced day 4 P19CL6 cells gave rise to a region protected from DNase I digestion in the minimal promoter. Interestingly, the protected area was not observed within nuclear extracts derived from differentiated P19CL6 cells at day 12 (data not shown). Precise positioning of the protected area by comparison of the footprints with Maxam-Gilbert sequencing ladder revealed that this segment was CCGGGGCTGAAATATGATAATCA, which shared core nucleotides with Oligo-3.
Fig. 3.
Identification of cis elements involved in the regulation of Isl1 expression. a The 32P-ATP labeled oligonucleotides are indicated as Oligo-1, -2, -3, -4, and -5. Octamer motif is highlighted by dark gray box. b EMSA was performed using the labeled probes and nuclear extracts (NE) prepared from induced day 4 P19CL6 cells. The DNA–protein complexes formed are indicated as C1, C2, and C3. c DNase I footprinting identified the protected region by NE on the Isl1 minimal promoter. d The bases responsible for DNA binding activities on the Isl1 minimal promoter were determined by EMSA. The mutant probes are labeled as mt1 to mt17
To further clarify the bases within Oligo-3 responsible for protein binding, 17 sets of 2 bp substitutions were synthesized and subjected to EMSA. As shown in Fig. 3d, when the bases AT were mutated to cg, GA to tc, and AT to cg, all the migrated bands were completely eliminated, suggesting that the motif “ATGATAAT” was indispensable for DNA binding affinity. Bioinformatic analysis identified the motif “ATGATAAT” as an octamer-binding sequence recognized by POU-homeodomain family factors. Taken together, our data identified the octamer motif “ATGATAAT” in the minimal promoter of Isl1 gene as, at least, one of the major protein-binding sites.
The requirement of the octamer motif on the activation of Isl1 promoter
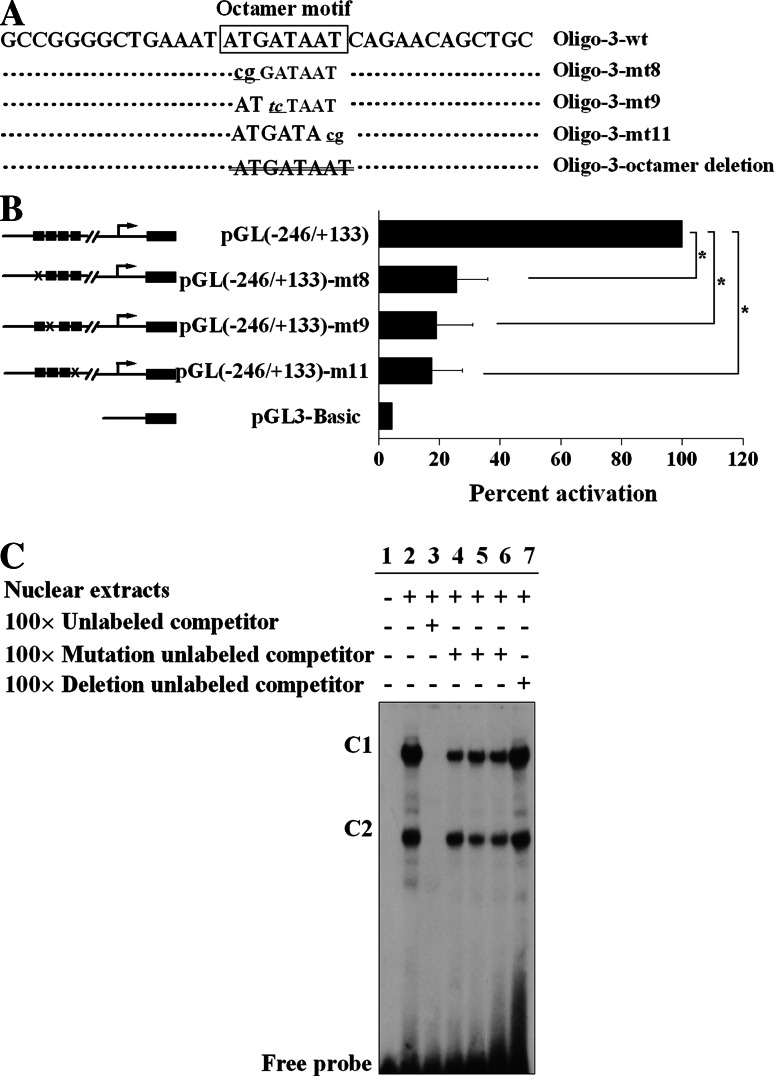
To functionally elucidate the contribution of the octamer motif acquired from DNase I footprinting and EMSA toward the Isl1 promoter activity, site-directed mutagenesis was carried out to introduce corresponding double-point mutations into the Isl1 minimal promoter, destroying the octamer motif (Fig. 4a). All the mutants transfected into induced day 4 P19CL6 cells led to a sharp drop in luciferase activity (Fig. 4b). The mutant constructs pGL(−246/+133) mt8, mt9, and mt11 exhibited levels of luciferase activity decreased to 25.6, 19.0, and 17.5%, respectively, of the wild-type promoter. Subsequently, DNA binding affinities further confirmed that all the mutant sequences partially blocked complex formations (Fig. 4c, lanes 4–6), while truncation of the octamer motif completely eliminated the DNA binding activity. These results suggest that an intact octamer motif is indispensable for the interaction with transactivators to activate the Isl1 promoter activity.
Fig. 4.
The requirement of octamer motif for activation of Isl1 minimal promoter. a A series of 2 bp mutants of octamer motif (lowercase letters) and octamer deletion (crossed-out lines) from Oligo-3 were used for EMSA in c. b Luciferase assay was performed to evaluate the transcriptional activity of Isl1 minimal promoter with mutants of octamer motif. × represents two mutant bases and black boxes represent the wild-type bases. Data are shown as percent activation over that of PGL(−246/+133) with mean ± SD (n = 3) [*P < 0.05, vs. PGL(246/+133)]. c EMSA was performed to evaluate the DNA binding affinity of mutant bases within the octamer motif. The binding complexes are indicated as C1 and C2
Regulation mechanisms of OCT1 on Isl1 expression
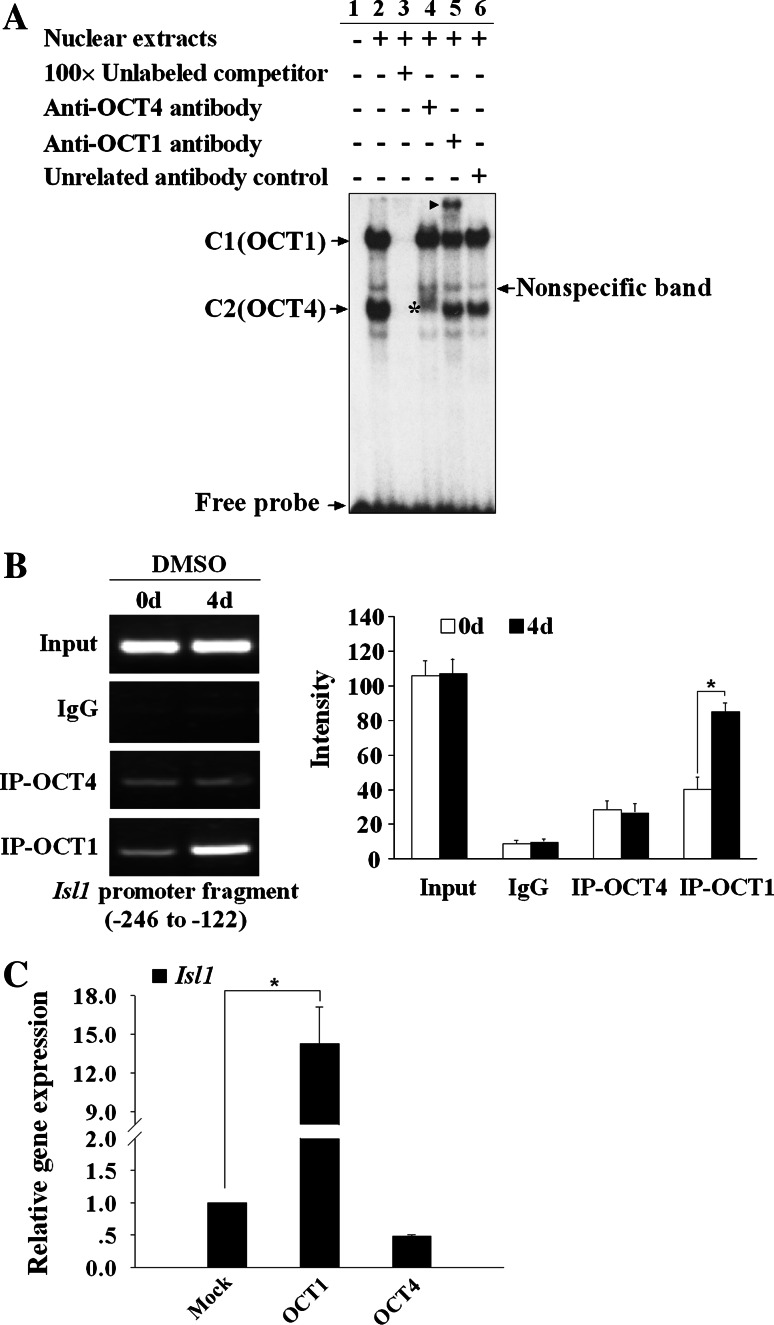
It is assumed that OCT transcription factors preferentially bound to the octamer motif through the POU domain [21]. EMSA was conducted to identify the binding factors using nuclear extract from induced day 4 P19CL6 cells. As shown in Fig. 5a, addition of anti-OCT1 antibody led to a super-shift of C1, indicating that C1 might represent OCT1 complex. In contrast, addition of anti-OCT4 antibody did not yield any super-shifted bands, but instead significantly attenuated the formation of C2 (lane 4). This suggests that the protein-DNA complex of C2 might contain OCT4 protein. Thus, our results determined that nuclear proteins that recognize the octamer motif are comprised of at least OCT1 and OCT4.
Fig. 5.
Regulation of OCT1 on the Isl1 expression. a EMSA was performed using the labeled Oligo-3 with NE from induced day 4 P19CL6 cells and antibodies against OCT1 or OCT4. Addition of anti-OCT1 antibody yields a supershift band as indicted by the arrowhead (lane 5), while addition of anti-OCT4 antibody produced a weakened band as indicated by the asterisk (lane 4). Nonspecific band and free probes are also indicated by arrows. b ChIP assay was performed with antibodies against OCT1 or OCT4 in day 0 or induced day 4 P19CL6 cells. The immunoprecipitated DNA fragments were amplified by PCR for the Isl1 promoter region from −246 to +133. Input represents 10% of the total input chromatin, and IgG served as a negative control. The quantitative presentation of the bands in the left panel, based on the ChIP data in c was analyzed by Multi Gauge V3.0 software and is shown with mean ± SD (*P < 0.05) (right panel). c Real-time RT-PCR analysis of effect by overexpression of OCT1 or OCT4 on the Isl1 expression. Data are shown as relative Isl1 gene expression over that of mock with means ± SD (n = 3) (*P < 0.05, vs. mock group)
Subsequently, we employed ChIP assay to evaluate the cellular binding property of OCT1 and OCT4 during cardiac differentiation of P19CL6 cells. As shown in Fig. 5b, OCT1 recruited onto Isl1 proximal promoter was dramatically augmented after DMSO induction for 4 days. A weak binding of OCT4 was detected at day 0, and DMSO-induced differentiation did not alter the binding amount of OCT4 on Isl1 proximal promoter. Obviously, the octamer motif was prominently occupied by OCT1 rather than by OCT4 during the differentiation of P19CL6 cells. Our data reveal the possible roles of OCT1 in Isl1 expression regulation during the process of DMSO-induced cardiac differentiation of P19CL6 cells.
Given the affinity of OCT1 and OCT4 with the octamer motif, we further tested whether OCT1 and/or OCT4 could activate Isl1 gene transcription in P19CL6 cells. As shown in Fig. 5c, forced expression of OCT1 evoked a 14-fold increase in the Isl1 mRNA level compared with the control. However, over-expression of OCT4 failed to activate Isl1 transcription, though it produced a weak repression. Taken together, our results strongly suggest that OCT1, not OCT4, is the predominant transactivator responsible for Isl1 expression.
The inhibition of Isl1 gene expression and cardiogenesis by Oct1 knock-down
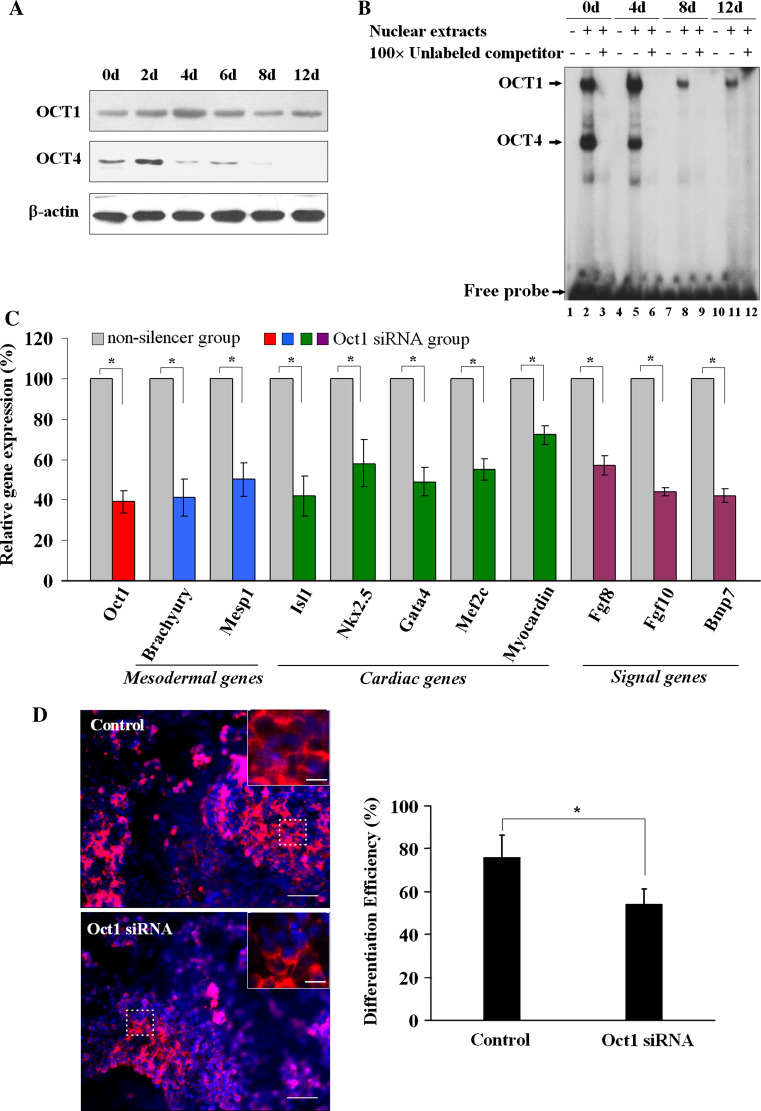
As demonstrated above, Isl1 displayed a stage-specific expression pattern during cardiac differentiation. In order to elucidate the correlation between OCT1 and the dynamic expression of Isl1, Western blot was performed to investigate the OCT1 variation. As expected, the protein level of OCT1 was increased at the early stage and reached a maximum at day 4 before declining to a lower level at the later stage of differentiation (Fig. 6a). The changes in OCT1 expression during cardiogenesis exhibited a high correlation with those of Isl1. Similar DNA binding activity alteration of OCT1 throughout the entire process of differentiation was observed (Fig. 6b), while Isl1 transcriptional activity did not correlate with OCT4 binding affinity. To further elucidate whether OCT1 is a crucial upstream factor of Isl1, the effects of Oct1 knock-down on the expression of Isl1; mesodermal genes including Brachyury and Mesp1; cardiac transcription factors such as Nkx2.5, Gata4, Mef2c, Myocardin; and some important signaling molecules including Fgf8, Fgf10, and Bmp7, were examined. As shown in Fig. 6c, siRNA-dependent reduction of Oct1 expression led to a marked decrease in the mRNA levels of the genes mentioned above. Furthermore, Oct1 knock-down in P19CL6 cells inhibited cardiogenesis to some extent. The differentiation efficiency was less in the Oct1 knock-down group (54.33 ± 6.73%) compared to the control group (75.93 ± 10.55%, P < 0.05, Fig. 6d). Therefore, our results established a close link between OCT1 and Isl1 expression regulation and suggest that OCT1 plays an important role in the differentiation of P19CL6 cells.
Fig. 6.
The influence of Oct1 knock-down on cardiogenesis during P19CL6 differentiation. a Western blot was performed to analyze OCT1 and OCT4 protein levels during P19CL6 cell differentiation at indicated time points. b EMSA was performed to assess the DNA binding activities with labeled Oligo-3 and NE from different time points of induced P19CL6 cells. c Real-time RT-PCR was used to evaluate a series of mesodermal (blue), cardiac-specific (green), and signal pathway genes (purple) expression in P19CL6 cells under the condition of Oct1 knock-down (as shown in red bar) at induced day 4. Data are normalized to 18S expression and presented as percent expression versus that of the nonsilencer group with mean ± SD (n = 3) (*P < 0.05). d Immunofluorescence staining with a monoclonal antibody against sarcomeric α-actinin (red) was analyzed after Oct1 knock-down. Nuclei were counterstained with Hoechst33342 (blue). The outlined areas are shown magnified in the upper right corners. Scale bars 10 μm. Differentiation efficiency is expressed as the percentage of α-actinin-positive cells out of the total number of cells counted (right panel) (*P < 0.05)
The correlation between Oct1 and Isl1 during early embryo development
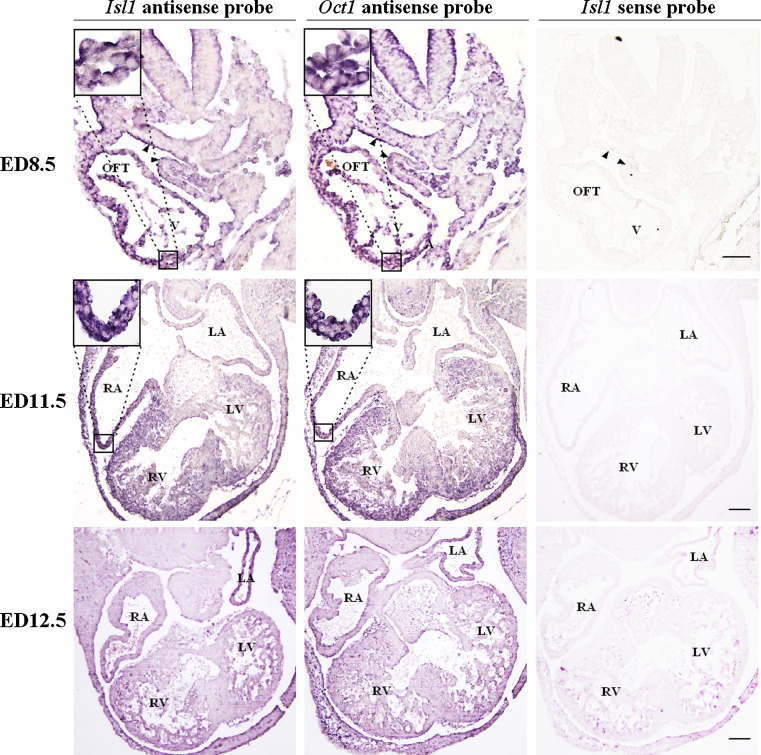
To further investigate the possible link between Oct1 and Isl1 in heart development, an in situ hybridization assay was performed on embryos from ED8.5 to ED12.5 to examine the spatial expression of Isl1 and Oct1 at the mRNA level during embryonic heart development. As shown in Fig. 7, at ED8.5, a heart tube stage, both Isl1 and Oct1 were expressed in the splanchnic mesoderm (SM), outflow tract (OFT), and ventricles (V). As development progressed to ED9.5, Isl1 and Oct1 expression were also overlapped in the OFT and V (data not shown). Furthermore, at ED11.5, Isl1 and Oct1 were both expressed in right atria, right ventricle, and a specific region of the left ventricle, and then their expression progressively decreased. However, at ED12.5, Isl1 and Oct1 mRNAs were hardly detected as the myocardium of the heart virtually came to maturity. Double staining in situ hybridization analysis of Isl1 and Oct1 on sectioned embryos of ED8.5 further confirmed the co-localization of Isl1 and Oct1 mRNA in SM, OFT, and V (Figure S1). Thus, Oct1 is directly correlated with Isl1, which strongly suggests that OCT1 as an activator participates in the regulation of Isl1 expression.
Fig. 7.
In situ hybridization analysis of Isl1 and Oct1 mRNAs on the embryos at ED8.5, ED11.5, and ED12.5. Serial sections of embryos were hybridized in situ with an antisense probe of Isl1 or Oct1. Strong positive signals were seen in SM, OFT, and V at ED8.5 and RA, RV, and a specific area of LV at ED11.5, but no signals were detected at ED12.5. Corresponding sense probes were used as negative controls. The results of Isl1 sense probe are shown. Black triangles indicate SM. The outlined areas are shown in the upper left corners. Scale bars 100 μm. SM Splanchnic mesoderm, OFT outflow tract, A atria, LA left atria, RA right atria, V ventricles, RV right ventricle, LV left ventricle
Discussion
Isl1 has been characterized as an important marker for cardiovascular progenitors, which give rise to cardiomyocytes and endothelial and smooth muscle lineages in vitro and in vivo [5–9]. However, the precise mechanism by which Isl1 expression is activated in cardiovascular progenitors remains unclear. In this report, we found a stage-specific expression profile of the Isl1 gene during in vitro cardiac differentiation of P19CL6 cells. Based on promoter analysis and EMSA, our data proved for the first time that OCT1 is a vital transcription factor that modulates the pattern of Isl1 expression during cardiac differentiation.
OCT1 belongs to the POU family of transcription factors, which recognize target sequences through bipartite POU DNA-binding domains. With the exception of OCT1, all of the known POU domain factors are expressed in restricted temporal and spatial patterns during development. Transgenic experiments demonstrate that POU protein families play a crucial role in the determination of cell fates, as well as cell proliferation, migration, and survival [22]. For example, OCT4 is required for maintaining self-renewal and pluripotency of stem cells [23]. However, the biological function of OCT1 is more complicated. Mice harboring two disrupted Oct1 alleles died in gestation, implying the essential role of OCT1 during the embryonic development [24]. The fibroblasts isolated from Oct1-deficient mice were sensitive to cytotoxic agents, and the expression of a number of stress/oxidative genes were influenced [25]. In embryos of Xenopus, a delineation of restricted spatiotemporal distribution of OCT1 implied that dynamical modulation of OCT1 is relevant to tissue morphogenesis [26]. OCT1 regulates target gene expression as either an activator or a repressor. In our study, the expression of Oct1 and Isl1 was associated with undifferentiated cell phenotypes. Overexpression or knock-down Oct1 led to upregulated or downregulated Isl1 expression, suggesting that OCT1 acts as an activator participating in the Isl1 expression. Kang et al. demonstrated that OCT1 and OCT4 were associated with cellular growth and stress responsiveness. Genotoxic and oxidative stress-induced OCT1 binding to physiological targets regulated native gene expression [27]. It has also been reported that OCT1 functions as a transcriptional repressor [28–31]. Recently, Shakya et al. reported that several genes involved in the Krebs-TCA cycle, including pdk4, pgc-1α, and ppargc1a, RNA, and protein increased in Oct1 −/− MEFs and thus resulted in a coordinated metabolic shift, which opposed tumorigenicity [25]. Therefore, it is assumed that OCT1 plays a pivotal role in modulating cell proliferation. However, to our knowledge, few studies have been investigated the role of OCT1 in mesoderm development. An early observation found that the expression of Brachyury, a marker of mesodermal gene expressed in early embryogenesis, was significantly downregulated in early gastrula stage embryos when injected with RNA of truncated OCT1 lacking N-terminal activation domain [32]. This result indicates that OCT1 may be related to mesoderm development.
Our data show that OCT1 is involved in cardiogenesis during P19CL6 cell differentiation. Oct1 knock-down, to some extent, inhibited differentiation efficiency and decreased the expressions of Isl1 and some mesodermal and cardiac-specific genes, suggesting a novel role of OCT1 in mesodermal and early cardiac differentiation. Notably, the content and DNA affinity of OCT1 is also dynamic. The variations of OCT1 at the protein level and DNA binding activity parallel the alteration of Isl1 mRNA, establishing a good correlation between Oct1 and Isl1 expression. The significant decrease of OCT1 at day 8 of differentiation could account for synchronous Isl1 downregulation. Intriguingly, our findings from in situ hybridization show that Oct1 and Isl1 mRNAs have similar distributions in the SM, OFT, and V at ED8.5, and in the right atria, right ventricle, and the specific area of the left ventricle at ED11.5, but simultaneously became undetectable at ED12.5 (Fig. 7), when the four-chamber heart gradually matures. Thus, the high level of OCT1 protein seems to be essential at the early stage of development but is dispensable for fulfilling the entire differentiation program.
OCT1 is associated with the switch of growth and maturity status of diverse cells. It has been documented that OCT1 modulates the expression of cell cycle-specific genes, such as histone H2B gene [33] and cyclinD1 [34]. OCT1 is present in virtually all proliferating cell lines of neuronal origin but is undetectable in mature nondividing neurons. Cell cycle arrest in G0/G1 and morphological differentiation of neuronal cells are accompanied by a decline in DNA affinity and the transcription level of Oct1 [35]. In addition, quiescent vascular smooth muscle cells (SMCs) in the mature vessel display an undetectable Oct1. Upon disruption of cell–extracellular matrix interactions, the proliferation potential of SMCs is evoked in parallel with a rapidly induced and constitutive expression of Oct1 [36]. It seems that the expression of Oct1 may exert its effect on maintaining the proliferation and undifferentiated status of cardiac progenitors during P19CL6 differentiation, indicating a function similar to Isl1. In combination with our observation, we hypothesize that further differentiation occurs with the cessation of proliferation and the expression of cardiac contractile genes, attenuated Oct1 activity, and the turning off of Isl1 expression.
Besides OCT1, an appropriate dose of OCT4 sustained in early heart development is also important to maintain the status of ES cells. OCT4 is considered a self-renewal marker. Once ES cells differentiate, the expression of Oct4 is markedly downregulated, which is consistent with our results. Therefore, it is not surprising that Isl1 does not seem to be under the control of OCT4, though OCT1 and OCT4 share identical DNA binding patterns.
Our results show that the octamer motif is requisite but not sufficient for triggering Isl1 expression in undifferentiated P19CL6 cells. Upon the addition of DMSO, Isl1 expression was elicited. It has been reported that WNT, BMP, and FGF8 are key upstream factors that drive the expression of Isl1 in cardiac progenitors [14, 15, 37]. Ablation of β-catenin caused aberrant production of ISL1+ progenitors and multiple malformations of the heart [15, 37, 38]. Promoter analysis also suggests that there are several putative Smads binding sites on Isl1 promoter mediating BMP signals [37]. Therefore, a coordinated program composed of OCT1 and signaling proteins activated by extracellular stimuli may exist. Additionally, the upregulation of Oct1 at the early stage of development could activate the downstream cascades resulting from extracellular stimuli. Thereby, augmented Oct1 is available to partially confer a signaling responsiveness to Isl1 promoter. However, the precise relationship between the initiation of Isl1 expression and the upregulation of Oct1 in the early stage needs further investigation.
On the other hand, Oct1 is frequently involved in transcriptional repression in an HDAC-dependent manner [28]. Previous documents had found that MyoD in partnership with HDAC1 was functionally linked to the silencing of muscle-specific genes prior to skeletal myogenesis [39]. We hypothesize that an analogous molecular mechanism including epigenetic modifications may act in cardiac myogenesis. Further investigations will focus on the effect of DNA methylation and histone modifications in Isl1 expression, and Oct1 in the epigenetic reconstruction of Isl1 promoter.
In the present study, we found that Isl1 and Oct1 positively stained in the outflow tract, ventricle, and atria of earlier embryos, such as ED8.5 and ED11.5, and Isl1 and Oct1 expression were more localized in right atria, right ventricle, and a specific region of the left ventricle. A number of papers showed Isl1 + precursors remained embedded in the embryonic mouse heart after its formation and their number decreases progressively from ED12.5 to ED18.5. After birth, relatively few Isl1 + cardioblasts were still detectable, averaging 500–600 in the myocardium of a 5-day-old rat [6]. In a recent report performed with rat embryo from ED11 (corresponding to ED9.5 in mice), the pool of Isl1 + cells was expanding after entering the heart with staining in right atrium. At ED15 (corresponding to ED13.5 in mice), more Isl1 + cells seemed to coexpress cardiac markers such as TnT, while few Isl1 + cells were actively dividing and remained undifferentiated [40].
During the preparation of this manuscript, two groups reported the identification of regulatory elements downstream of the translational stop codon of ISL1. Kang et al. demonstrated that ISL1 is a direct target of Forkhead transcription factors [41], while Kappen et al. revealed that Foxo1 as well as Gata4 contribute to the activity of the enhancer in the developing embryonic heart [42]. However, the core promoter region of Isl1 has not been described so far. The direct regulation of the proximal promoter of Isl1 has not been reported. The core promoter is required for proper initiation of a gene transcription. The identification of the core promoter of Isl1 will form a basis to study the transcription regulation of Isl1 and therefore provide important information for further investigation.
Taken together, OCT1 is a critical regulator for the expression of Isl1. Considering the temporal pattern of Oct1 expression, we presume that OCT1 is a major activator that dynamically regulates Isl1 expression and plays a pivotal role in early cardiomyocyte differentiation. Based on the extensive expression and multiplex function of ISL1, the role of OCT1 in regulating Isl1 expression may contribute to cardiogenesis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1 Single and double in situ hybridization analyses of Isl1 and Oct1 mRNA on the series sectioned embryos of ED8.5. The representative images of single (A) and double (B and C) staining using biotin-labeled Oct1 probe and digoxigenin-labeled Isl1 probe are shown. Isl1 mRNAs was detected with NBT and BCIP (blue) after incubating with AP-conjugated anti-digoxigenin antibody (Figure 7, top left). While, Oct1 mRNAs were detected with AEC peroxidase substrate (brick red) after incubating with HRP-conjugated streptavidin (A). The overlapping of Isl1 with Oct1 can be observed in some regions of SM, OFT and V (B, dark purple). Scale bar: 100 μm. Corresponding sense probes were used as negative controls (C). Black triangles indicate splanchic mesoderm. The magnified squared areas are shown on the upper left corners. Abbreviations: AP, alkaline phosphatase; HRP, horseradish peroxidase; SM: splanchic mesoderm; OFT, outflow tract; V, ventricles. Supplementary material 1 (TIFF 955 kb)
Table S1 Supplementary material 2 (DOC 58 kb)
Table S2 Supplementary material 3 (DOC 26 kb)
Acknowledgments
This work was supported by the National Natural Sciences Foundation of China (30871253, 90919022) and the 111 Project of Ministry of Education of China (B07001). We thank Prof. Yunzeng Zou, Fu Dan University, for providing us the P19CL6 cell line. We are grateful to Dr. Jason Wong, University of Cambridge, UK, for his kind help in the preparation of this manuscript.
Footnotes
Y. Liu and Y. Li equally contributed to this work.
References
- 1.Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990;344:879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- 2.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/S0092-8674(00)80985-X. [DOI] [PubMed] [Google Scholar]
- 3.Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 4.Du A, Hunter CS, Murray J, Noble D, Cai CL, Evans SM, Stein R, May CL. Islet 1 is required for the maturation, proliferation, and survival of the endocrine pancreas. Diabetes. 2009;58:2059–2069. doi: 10.2337/db08-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, Chien KR. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moretti A, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet 1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- 10.Brade T, Gessert S, Kuhl M, Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol. 2007;311:297–310. doi: 10.1016/j.ydbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 12.Lam JT, Moretti A, Laugwitz KL. Multipotent progenitor cells in regenerative cardiovascular medicine. Pediatr Cardiol. 2009;30:690–698. doi: 10.1007/s00246-009-9450-1. [DOI] [PubMed] [Google Scholar]
- 13.Black BL. Transcriptional pathways in second heart field development. Semin Cell Dev Biol. 2007;18:67–76. doi: 10.1016/j.semcdb.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11:951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habara-Ohkubo A. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct Funct. 1996;21:101–110. doi: 10.1247/csf.21.101. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Zhu WG, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, Yee L, Villalona-Calero MA, Plass C, Otterson GA. Methylation of adjacent CpG sites affects SP1/SP3 binding and activity in the P21(Cip1) promoter. Mol Cell Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/S0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 21.Verrijzer CP, Alkema MJ, Van WW, Van LH, Strating MJ, Van VP. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen B, Rosenfeld MG. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr Rev. 2001;22:2–35. doi: 10.1210/er.22.1.2. [DOI] [PubMed] [Google Scholar]
- 23.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 24.Wang VE, Schmidt T, Chen J, Sharp PA, Tantin D. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol Cell Biol. 2004;24:1022–1032. doi: 10.1128/MCB.24.3.1022-1032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shakya A, Cooksey R, Cox JE, Wang V, McClain DA, Tantin D. Oct1 loss of function induces a coordinate metabolic shift that opposes tumorigenicity. Nat Cell Biol. 2009;11:320–327. doi: 10.1038/ncb1840. [DOI] [PubMed] [Google Scholar]
- 26.Veenstra GJ, Beumer TL, Peterson-Maduro J, Stegeman BI, Karg HA, Van VP, Destree OH. Dynamic and differential Oct-1 expression during early Xenopus embryogenesis: persistence of Oct-1 protein following down-regulation of the RNA. Mech Dev. 1995;50:103–117. doi: 10.1016/0925-4773(94)00328-K. [DOI] [PubMed] [Google Scholar]
- 27.Kang J, Gemberling M, Nakamura M, Whitby FG, Handa H, Fairbrother WG, Tantin D. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev. 2009;23:208–222. doi: 10.1101/gad.1750709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hitomi T, Matsuzaki Y, Yasuda S, Kawanaka M, Yogosawa S, Koyama M, Tantin D, Sakai T. Oct-1 is involved in the transcriptional repression of the p15(INK4b) gene. FEBS Lett. 2007;581:1087–1092. doi: 10.1016/j.febslet.2007.01.092. [DOI] [PubMed] [Google Scholar]
- 29.Cheng CK, Yeung CM, Hoo RL, Chow BK, Leung PC. Oct-1 is involved in the transcriptional repression of the gonadotropin-releasing hormone receptor gene. Endocrinology. 2002;143:4693–4701. doi: 10.1210/en.2002-220576. [DOI] [PubMed] [Google Scholar]
- 30.Schwachtgen JL, Remacle JE, Janel N, Brys R, Huylebroeck D, Meyer D, Kerbiriou-Nabias D. Oct-1 is involved in the transcriptional repression of the von Willebrand factor gene promoter. Blood. 1998;92:1247–1258. [PubMed] [Google Scholar]
- 31.Wu GD, Lai EJ, Huang N, Wen X. Oct-1 and CCAAT/enhancer-binding protein (C/EBP) bind to overlapping elements within the interleukin-8 promoter. The role of Oct-1 as a transcriptional repressor. J Biol Chem. 1997;272:2396–2403. doi: 10.1074/jbc.272.4.2396. [DOI] [PubMed] [Google Scholar]
- 32.Veenstra GJ, Peterson-Maduro J, Mathu MT, Van VP, Destree OH. Non-cell autonomous induction of apoptosis and loss of posterior structures by activation domain-specific interactions of Oct-1 in the Xenopus embryo. Cell Death Differ. 1998;5:774–784. doi: 10.1038/sj.cdd.4400416. [DOI] [PubMed] [Google Scholar]
- 33.Fletcher C, Heintz N, Roeder RG. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987;51:773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- 34.Boulon S, Dantonel JC, Binet V, Vie A, Blanchard JM, Hipskind RA, Philips A. Oct-1 potentiates CREB-driven cyclin D1 promoter activation via a phospho-CREB- and CREB binding protein-independent mechanism. Mol Cell Biol. 2002;22:7769–7779. doi: 10.1128/MCB.22.22.7769-7779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakin ND, Palmer R, Lillycrop KA, Howard MK, Burke LC, Thomas NS, Latchman DS. Down regulation of the octamer binding protein Oct-1 during growth arrest and differentiation of a neuronal cell line. Brain Res Mol Brain Res. 1995;28:47–54. doi: 10.1016/0169-328X(94)00183-F. [DOI] [PubMed] [Google Scholar]
- 36.Weiser MC, Grieshaber NA, Schwartz PE, Majack RA. Perlecan regulates Oct-1 gene expression in vascular smooth muscle cells. Mol Biol Cell. 1997;8:999–1011. doi: 10.1091/mbc.8.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klaus A, Saga Y, Taketo MM, Tzahor E, Birchmeier W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc Natl Acad Sci USA. 2007;104:18531–18536. doi: 10.1073/pnas.0703113104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, Rosenfeld MG, Chen J, Evans SM. Beta-catenin directly regulates Islet 1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci USA. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mal A, Harter ML. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc Natl Acad Sci USA. 2003;100:1735–1739. doi: 10.1073/pnas.0437843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genead R, Danielsson C, Andersson AB, Corbascio M, Franco-Cereceda A, Sylven C, Grinnemo KH. Islet-1 cells are cardiac progenitors present during the entire lifespan: from the embryonic stage to adulthood. Stem Cells Dev. 2010;19:1601–1615. doi: 10.1089/scd.2009.0483. [DOI] [PubMed] [Google Scholar]
- 41.Kang J, Nathan E, Xu SM, Tzahor E, Black BL. Isl1 is a direct transcriptional target of Forkhead transcription factors in second-heart-field-derived mesoderm. Dev Biol. 2009;334:513–522. doi: 10.1016/j.ydbio.2009.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kappen C, Salbaum JM. Identification of regulatory elements in the Isl1 gene locus. Int J Dev Biol. 2009;53:935–946. doi: 10.1387/ijdb.082819ck. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Single and double in situ hybridization analyses of Isl1 and Oct1 mRNA on the series sectioned embryos of ED8.5. The representative images of single (A) and double (B and C) staining using biotin-labeled Oct1 probe and digoxigenin-labeled Isl1 probe are shown. Isl1 mRNAs was detected with NBT and BCIP (blue) after incubating with AP-conjugated anti-digoxigenin antibody (Figure 7, top left). While, Oct1 mRNAs were detected with AEC peroxidase substrate (brick red) after incubating with HRP-conjugated streptavidin (A). The overlapping of Isl1 with Oct1 can be observed in some regions of SM, OFT and V (B, dark purple). Scale bar: 100 μm. Corresponding sense probes were used as negative controls (C). Black triangles indicate splanchic mesoderm. The magnified squared areas are shown on the upper left corners. Abbreviations: AP, alkaline phosphatase; HRP, horseradish peroxidase; SM: splanchic mesoderm; OFT, outflow tract; V, ventricles. Supplementary material 1 (TIFF 955 kb)
Table S1 Supplementary material 2 (DOC 58 kb)
Table S2 Supplementary material 3 (DOC 26 kb)