Abstract
The molecular features that dominate the binding mode of agonists by a broadly tuned olfactory receptor are analyzed through a joint approach combining cell biology, calcium imaging, and molecular modeling. The odorant/receptor affinities, estimated through statistics accrued during molecular dynamics simulations, are in accordance with the experimental ranking. Although in many systems receptors recognize their target through a network of oriented interactions, such as H-bonding, the binding by broadly tuned olfactory receptors is dominated by non-polar terms. We show how such a feature allows chemicals belonging to different chemical families to similarly activate the receptors through compensations of interactions within the binding site.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1116-0) contains supplementary material, which is available to authorized users.
Keywords: Molecular dynamics, Free energy, Odorant, GPCR, Structure
Introduction
Humans are able to perceive odorants through an extraordinary complex sense. The perception of smell is primarily triggered by the recognition of one or a mixture of odorants by olfactory neurons, which house a single type of olfactory receptor (OR) [1]. The recognition mode of a neuron is then rooted in the way its associated ORs are activated by a chemical. In ORs, however, the chemoreception process is far removed from the classical lock-and-key paradigm, where an odorant, acting as a key, selectively activates a given type of receptor, functioning as a lock. Although some ORs are very narrowly tuned for odorants [2], numerous ORs can be activated by very different odorants, revealing broad recognition abilities and large plasticity of their binding cavity [3]. This apparent fuzziness is in fact mandatory, since it allows a combinatorial mode of odorant recognition, endowing our olfactory system (based on less than 400 types of olfactory neurons, or receptors) the ability to recognize an almost infinite number of odorants with a spectacular discriminatory power.
Ligand-based structure–odor relationships consider that an odorant structure encompasses all the information needed to predict its odor. The sense of smell is, however, extraordinarily complex, and many different biological protagonists are involved during the perception of an odor, suggesting that it is virtually impossible to predict an odor based solely on the odorant’s structure [4, 5]. In addition, ORs are rather tuned to physico-chemical properties of odorants and not directly to odors [6], which arises from the neuro-processing of elicited signals. However, the perception of smell starts by no more than a series of chemical interactions involving odorants and ORs located at the neuron–olfactory mucus interface. These interactions are eventually modulated by peri-receptor events, such as those played by odorant-binding proteins or odorant-degrading enzymes. The main step among the series being the odorant–OR interaction.
Up to now, OR’s broad recognition potencies have been assessed by means of various experimental approaches, but atomic-level clues about the way an OR is activated by odorants with different shapes or chemical groups remain scarce. Human OR1G1 can be regarded as prototypical of broadly tuned human-ORs. It has already been shown to have a large recognition spectrum [7], and a structure–odor relationship emphasized that OR1G1 binds odorants that do not correspond to the same olfactophores [8]. It remains that the physico-chemical properties at the binding site, together with the way a receptor can recognize an odorant of different chemical classes, still remains poorly understood at the atomic level. To streamline the binding mode of ORs, we use a combination of experimental and theoretical work. Various studies have been devoted to build models of several ORs in interactions with odorants. They identified the residues that control odorants recognition by ORs and notably assessed the efficiency of molecular modeling to predict the structure of ORs, on the basis of either homology modeling or an ab initio approach [9–19].
To have an odor, a chemical must fulfill some criteria, of which hydrophobicity stands as a priority. As a consequence, our sense of smell had to adapt to such a feature by building a system dedicated to recognize odorants through interactions and binding modes that remain to be uncovered. In order to answer the question of how a broadly tuned receptor can equally recognize agonists belonging to different chemical classes, we used a joint approach combining cellular biology, calcium imaging, and state-of-the-art molecular modeling.
We have selected four odorants that were measured to similarly act as agonists for OR1G1 and compared their behavior to a known non-activating odorant. We built a full atomic-detail structure of the receptor and performed molecular dynamics simulations to analyze the binding mode on an energetic basis.
Results and discussion
Calcium imaging
The functional expression and the binding tests on hOR1G1 were assessed through fura-2 fluorescence Ca2+ imaging [20]. Stimulation of an endogenous GPCR by octopamine has permitted to control the responsiveness of cells expressing hOR1G1. Surface-receptor expression kinetics was measured to control that a sufficient number of receptors are present at the cell surface. This allows thorough statistics (Table 1). Fura-2 fluorescence Ca2+ imaging was then used to measure [Ca2+]intracellular in 72 H post-infected cells (see Supporting Information).
Table 1.
hOR1G1 expression kinetics
| Time post-infection (h) | Amount of cell surface receptors |
|---|---|
| 12 | 36 (3) |
| 24 | 60 (4) |
| 72 | 99.8 (11) |
The number of cells is expressed in pmoles/106 cells and the standard deviation is given in parenthesis
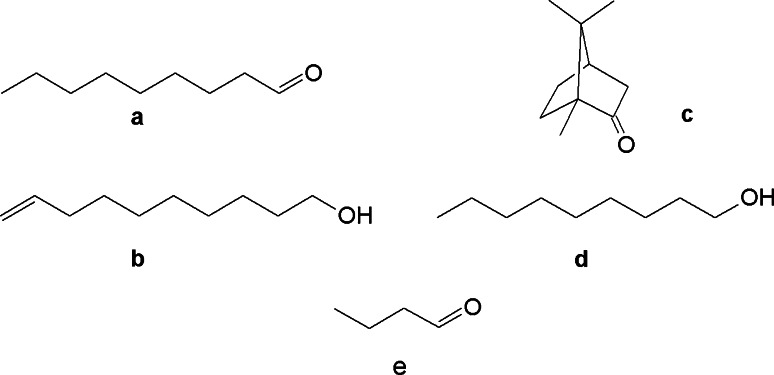
The calcium imaging experiments are performed for four odorants (Scheme 1), viz. nonanal (fatty-roselike) (a), 9-decen-1ol (fresh, dewy, rose) (b), camphor (aldehydic, green, camphor, pine) (c), and 1-nonanol (fresh, clean, fatty, floral, rose, orange, dusty, wet) (d) [21, 22]. Their performances are then compared to that of n-butanal, a non-binder of hOR1G1.
Scheme 1.
Chemical structures of nonanal (a), decenol (b), camphor (c), nonanol (d) and butanal (e)
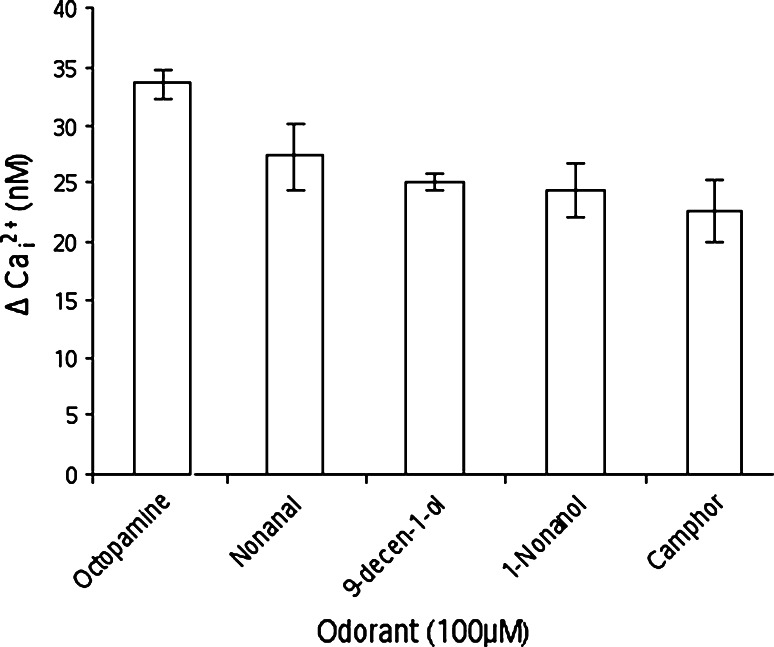
Figure 1 reports the binding of these four odorants with respect to octopamine, which constitutes a reference for full activation. All ligands strongly activate the Ca2+ discharge, showing that they all act as strong agonists.
Fig. 1.
Calcium imaging results for the four odorants compared to octopamine (the standard error is shown as error bars)
The Ca2+ concentrations are similar and fall within the same value if we consider the standard deviation. The intracellular calcium variation is measured to 27.3, 25.1, 24.4, and 22.6 nM for nonanal, 9-decen-1-ol, 1-nonanol and camphor, respectively. Counter-intuitively, even if all odorants bear a ~9 carbon atom skeleton, their chemical nature, size, or shapes do not strongly affect their binding strength.
Notice that other closely related odorants have been shown to elicit a much weaker hOR1G1 response. Expression of OR1G1 in HEK293-derivatives cells allowed performing a screening of many odorants belonging to different families [7, 8]. This approach is consistent with ours, regarding the agonist character of our odorants. In this study, 1-decanol or 1-octanol led to responses twice weaker than that of 1-nonanol. The same is true for decanal with respect to nonanal. In the next part of the article, we compare the behavior of these agonists with that of n-butanal, which was experimentally shown to elicit no response of hOR1G1 [8]. All these five systems were submitted to a multiple molecular modeling protocol. Two different initial docking poses are subjected to 20-ns molecular dynamics (MD) to analyze their dynamic behavior and to obtain a rescored affinity on a statistical basis.
hOR1G1 structure and binding pocket
To assess the accuracy of the docking protocol, we have conducted a test on the human beta-2-adrenergic receptor (pdb id: 2RH1), bound to a good and a weak binder, i.e., carazolol and metoprolol, respectively [23]. The docking protocol recovers the position of carazolol within the cavity and proposes a pose where the aromatic cycle of metoprolol is located in a position equivalent to that of carazolol. Carazolol is predicted to have a much better affinity for the receptor (docking score of −9 kcal/mol and −4.8 kcal/mol for carazolol and metoprolol, respectively). This justifies the use of a docking protocol to obtain a starting binding mode. However, in this test set, the receptor is already pre-organized to bind carazolol, which is probably not the case for our hOR1G1 model.
Indeed, according to best docking poses, the following ranking of affinity with hOR11G was obtained: decen-1-ol > nonanol > nonanal > camphor. All poses had the hydrophilic part of the odorant directed towards T202. To refine the scoring function, docking solutions were then clustered into poses where the hydrophobic part of the odorant was either directed towards residue T202 or towards residue T279. The most representative pose in each cluster was submitted to a 20-ns MD simulation in an explicit membrane model and the affinity was obtained by means of the MM-GBSA protocol (see Supp. Info.).
Our approach involves docking on an OR structure. To get insight into the accuracy of molecular modeling of hOR1G1, we built the structures by means of two totally different protocols. In a homology-based approach, hOR1G1 sequence was aligned following Man et al. protocol [24]. We particularly focused on the alignment of hOR1G1 with various GPCRs for which structures are either experimentally known or built on the basis of site-directed mutagenesis (see Supp. Info.). The alignment predicts TM helices in accordance with other structures (either experimental or modeled). Fifty structures were generated by Modeller software on the basis of the rhodopsin template, since, similarly to ORs, the latter also exhibits a hydrophobic binding pocket. The final model was chosen on the basis of two criteria, (i) having a maximum number of residues in favorable areas of the Ramachandran plot and (ii) showing a binding site with a maximum of residues found to be important on the basis of site-directed mutagenesis experiments on other ORs and GPCRs.
An ab initio model of the seven-helix bundle was built with the MembEnsemb program [25, 26]. This protocol has already proven successful in the identification of many GPRCs structures, and more particularly ORs [9, 15, 17, 27].
The seven TM domains of hOR1G1 were predicted by PredicTM, which combines hydrophobicity analysis and multiple sequence alignment. All characteristic GPCRs features are observed. The ab initio structure of the bundle has the classical interhelical H-bonding networks in TMs 1-2-7 (N 421.50, D 702.50, and N 2867.49), and TMs 2-4 (N 652.45 and W 1494.50) which are found in Rhodopsin family A GPCRs. The conserved salt-bridge between D 1213.49 and K 2346.30 in the D(E)RY region is recovered. In addition, in ORs, E 1113.39 and H 2446.40 are highly conserved (more than 95 %). Our most stable ab initio model exhibits a hydrogen-bond between these residues, suggesting that they are involved in the structure stability.
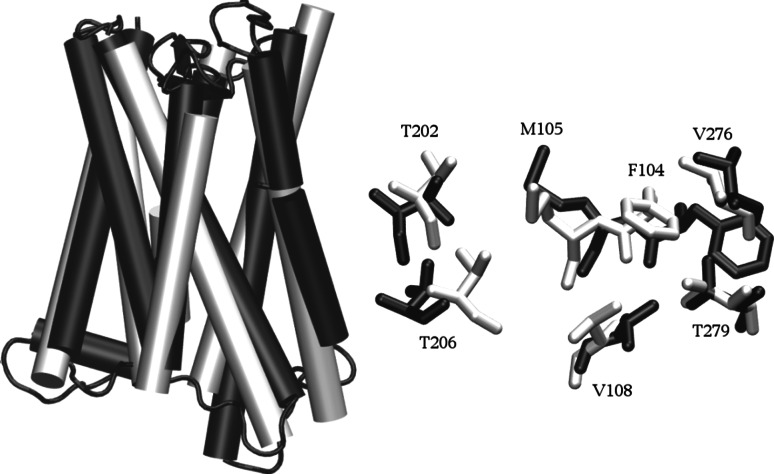
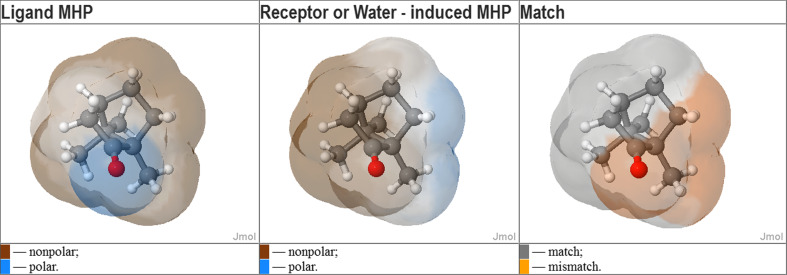
The resulting ab initio and homology models look very similar. hOR1G1 bundle is made up of seven transmembrane helices (TM 1–7) that go back and forth the membrane. A single binding cavity is made up of residues belonging to helices 3, 5, 6, and 7. Figure 2 compares the two models, emphasizing the convergence of both approaches concerning hOR1G1 cavity. Comparing the bundles, the root mean square deviation is computed to 6.8 Å. This deviation is mainly due to the top and bottom parts of the helices. Focusing on the binding cavity, we observe the same residues and the RMSd considering only the binding cavity is 1.6 Å. The convergence of these two totally different approaches strengthens the accuracy of our model. This cavity is mainly hydrophobic and matches well with the ligand hydrophobic property. Figure 3 shows the molecular hydrophobic potential (MHP) created on camphor surface by either camphor itself, or by the binding cavity of hOR1G1when camphor is in the best docking pose (see [28] for a review). This was done with the PLATINUM program [29]. One can observe a good match between the hydrophobic property of camphor and the hydrophobicity of the receptor, at the exception of a small part of the molecule, notably comprising the oxygen atom. This also suggests that the hydrophilic part of the ligand is not driving the position of the latter in the binding pocket.
Fig. 2.
Left: superposition of two models of hOR1G1 bundle. The homology model is shown in black and the ab initio model is in white. Right: position of residues lining the binding site within the two models (H atoms are omitted)
Fig. 3.
Complementarity of hydrophobic properties of camphor and hOR1G1-binding site. Left: molecular hydrophobicity potential (MHP) created by the ligands atoms on its own surface. Middle: MHP created by the receptor atoms on the ligand surface (which is the same). Right: match/mismatch between left and middle
The residues constituting the cavity wall are as follows: F104, M105, V108, T202, T206, F256, F260, and T279. Interestingly, in other OR or GPCR sequences, identical positions have been shown to strongly contribute to ligand recognition. This is notably the case for residues 104–105, which are found to be involved in several odorants’ recognition by hOR2AG1 and mOR283-2 [16], rOR5 [10], hOR1D2 [18], hOR1A2 [12], and mOR-EG [14, 19] for example. Residues in positions close to 206, 256, 260, and more particularly 279, have also been shown to be involved in binding in these human or rat ORs (hOR2AG1, rOR5). Residue T279 was shown to belong to the binding cavity of hOR2AG1 [16]. It is conserved in hOR1G1 and is located at the same place in the 3D structures (see Supp Info.).
The odorants were docked into the binding cavity of the homology model. For each odorant, a starting structure corresponding to the best docking pose is compared to an alternative pose where the hydrophilic part of the odorant is directed towards T279, since this residue was previously identified as crucial for hOR2AG1 activation [16].
The systems were embedded into a model membrane prior to two 20-ns molecular dynamics simulation that allowed relaxing the structures. A large statistical sampling of the odorant/OR interactions could thus be obtained.
Non-polar terms dominate binding
For the five odorant/receptor systems, the OR structure remains stable throughout the simulation (see Supp. Info.). For each odorant, two simulations have been performed. The only difference being the initial position of the ligand within the cavity. This rescoring protocol has allowed us to obtain two estimations of the binding energy, associated with two distinct odorant poses. We nonetheless observe large reorganization of the odorants within the cavity. Each simulation samples ligand orientations that are common in both models, emphasizing the large mobility of ligands when they are stabilized by hydrophobic contacts rather than by directional hydrogen bonds. Such behavior has already been shown on odorant-binding protein complexes, where odorants sample a large conformational space within the hydrophobic binding cavity [30, 31].
To assess the model quality, docking scores are evaluated by an estimation of a component of the binding free energy, computed through a molecular mechanics-generalized born solvent accessibility (MM-GBSA) protocol [32]. For each simulation, the pose corresponding to the largest affinity is used for further analysis. The binding free energy estimations (ΔG MM-GBSA Total) gathered in Table 2 are very similar numbers concerning the agonists. More importantly, they follow the trend observed experimentally, except for camphor, which is predicted to bind a bit more tightly than nonanol. This emphasizes that taking into account for dynamical effects and using a quite-elaborated free-energy estimation is able to propose an accurate model of OR/odorants affinity. For the agonists, the four values spread within a 3-kcal/mol range. This corresponds to their typical standard deviation. Butanal binds less tightly to the receptor, with an affinity more than twice as weak as those of the agonists. As for the Ca2+ imaging experiments on agonists, the affinities fall within the same range, suggesting that the model has accurately captured the physics of the system at the receptor–odorant-binding site and that these agonists are indeed strongly bound to the receptor, contrarily to butanal.
Table 2.
MM-GBSA free energy of binding and decomposition into polar and non-polar terms
| Odorant | ΔG MM-GBSA polar | ΔG MM-GBSA non-polar | ΔG MM-GBSA Total |
|---|---|---|---|
| 1-Nonanal | 4.6 | −33.3 | −28.7 (2.0) |
| 9-Decen-1-ol | 7.8 | −33.3 | −25.5 (2.0) |
| 1-Nonanol | 7.6 | −34.0 | −26.4 (2.0) |
| Camphor | 3.6 | −30.7 | −27.1 (2.0) |
| Butanal | 4.4 | −15.1 | −10.7 (2.5) |
Values are in kcal/mol. Standard deviations are shown in parenthesis
The binding free energy can be decomposed into polar and non-polar terms. In these systems, the interaction energy is driven by the hydrophobic term. The polar term represents a third of the total binding energy for decenol (32 %) and nonanol (29 %), respectively. For camphor and nonanal, this contribution decreases to 13 and 16 %, respectively, which corresponds to a weight of ~85 % for the non-polar term. For butanal, the energetics of binding is more balanced between hydrophilic and hydrophobic contributions. The polar term accounts for 41 % of the free energy of binding. In the agonists, most of the interactions indeed occur through hydrophobic contacts with non-polar residues of the binding cavity (vide infra). These hydrophobic contacts are not directional (contrarily to H-bonds) and allow a reorganization of the ligand within the receptor’s cavity with respect to its initial position.
The binding mode differs from one odorant to another
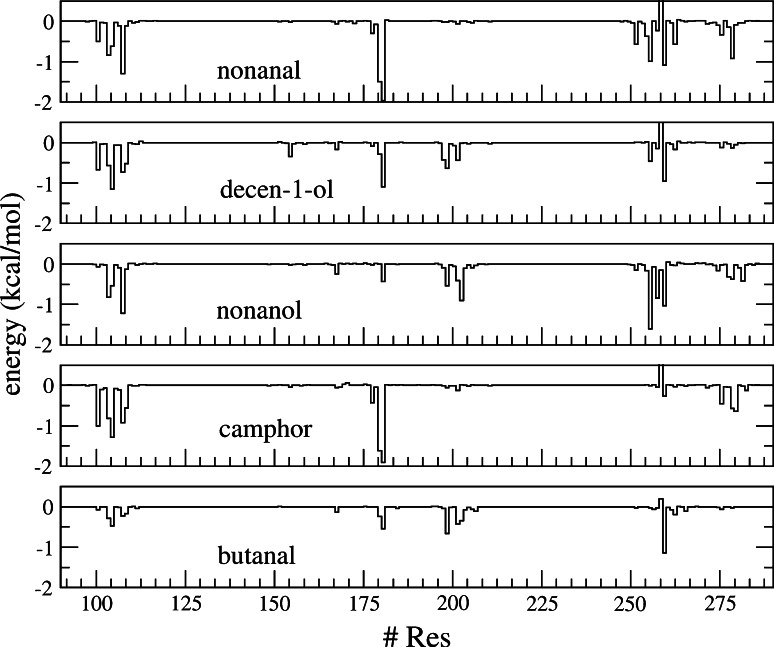
The identification of the binding mode, i.e., the interaction between the cavity and the odorants, is analyzed through the MM-GBSA protocol that allows decomposing the binding free energy on a per-residue basis (Fig. 4).
Fig. 4.
Decomposition of the free energy of binding on a per-residue basis
It provides a general view of the binding that takes into account for the interactions sampled during the whole MD simulations. It is more instructive than a classical geometrical analysis.
Most of the binding contributions lie between 0 and −2 kcal/mol. Although many zones in the sequence are equally involved in the binding, many other residues exhibit a strong contribution for a given ligand but a weaker one for another ligand, illustrating the difference of binding mode.
From a general point of view, the analysis recovers the typical parts of GPCRs known to form the binding cavity. Some residues are involved in binding, whatever the ligand, and constitute the main building blocks of the OR1G1 odorant-binding cavity.
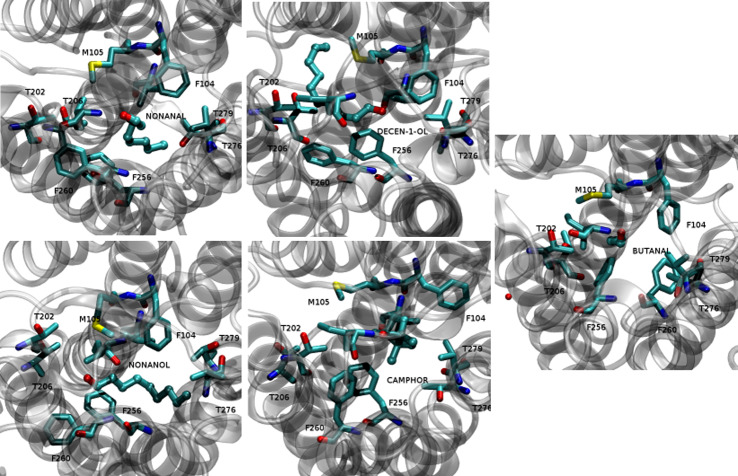
These residues are all hydrophobic (F104, M105, F168, I181, and F260), except T202 or T279, which can be engaged in a hydrogen bond with the odorants, as shown in Fig. 5.
Fig. 5.
Close-up view of the odorants’ positions within hOR1G1-binding cavity. The atomic representation is as follows: ligands in balls and stick, binding cavity residues in licorice. C cyan, O red, N blue, S yellow
Then, depending on the odorant’s chemical nature and shape, the binding is strengthened by other residues. A multimodal way of binding is observed.
Nonanal interacts quite strongly with F260 at the end of helix 6, I181 in the extra-cellular Loop 2 (ECL2) and F104 and V108 (helix 3). All of these interactions involve hydrophobic contacts, suggesting that the binding is mainly enslaved to non-polar terms.
The double bond of 9-decen-1-ol strongly interacts with F260. The π-staking interaction with the aromatic cycle of F260 dominates the binding mode, leading to a slight shift of decen-1-ol far from residues belonging to helix 3, notably V108. T202 and I199 are engaged in a hydrogen bond with the odorant’s alcohol moiety. The contribution of T279 is deemed negligible, and that of I181 is twice weaker than that of nonanal and camphor.
The binding mode of nonanol is regularly spread over many parts of the binding pocket. It nonetheless mostly involves hydrophobic contacts of the odorant aliphatic side chain with V108, F256, and F260. Compared to decenol, the lack of a double bond in the aliphatic part spreads the decomposition over a larger number of residues, more particularly those belonging to helix 7, such as T279.
For camphor, a large part of the binding comes from the contribution of I181 and from residues of helix 3 (F104, M105, and V108). T279 is involved in binding via hydrophilic interactions but the odorant is mostly recognized through non-polar contacts with residues belonging to helix 3. Contributions of helices 5, 6, and 7 are weaker than those found for the other agonists. Butanal binding is very poor. The pattern is similar to nonanol, but with much weaker binding. The largest part of the interaction energy comes from F260, but the other ones are negligible with respect to those found in the other systems.
An adaptation of the cavity to the odorant is observed. The classical paradigm would rather consider a model where odorants interact with a hydrogen-bond anchoring point (T202) with the remainder of the binding pocket serving as a hydrophobic space where the ligand must fit. Here, this model is somehow called into question. Both the odorant and the cavity undergo conformational fitting.
At the cavity, we observe conformational changes of residues side chains, which affect the binding cavity volume. Counter-intuitively, the cavity volume evolution within the series is not correlated to the odorant molecular volume. In the docked conformation, the ligands volumes are as follows: decenol > nonanol > nonanal > camphor > butanal, as shown in Table 3. The binding cavity is very plastic and breathes around different volumes depending on the bound odorant. Evolutions of the cavity volume all along the MD simulations are provided in the Supp. Info.
Table 3.
Volume of the odorants and of hOR1G1-binding cavity with the root mean square within parenthesis (in Å3)
| Ligand | Nonanal | Decen-1-ol | Nonanol | Camphor | Butanal |
|---|---|---|---|---|---|
| Ligand volume | 297 | 331 | 317 | 272 | 72 |
| Cavity volume | 325 (43) | 458 (90) | 579 (72) | 424 (52) | 189 (91) |
In the cases of camphor and nonanal, one observes much smaller variations of the volume, revealing a larger hydrophobic collapse of the binding pocket. In a general manner, for all the odorants, the cavity adapts to the odorant’s shape. In the butanal complex, residues of the binding pocket fill the cavity through a large hydrophobic collapse. The filling of the cavity is notably due to F256 and F260 side chains, which enter deep inside the cavity, as shown in Fig. 5. The case of butanal is a clear example of the plasticity and adaptability of the binding pocket.
A model for recognition of broadly tuned olfactory receptors
The sense of smell allows us to perceive volatile chemicals present in our environment. The almost infinite number of odorant molecules has to be accurately qualified and quantified. To this end, our sense of smell has to adapt to such a difficult task with a recognition paradigm that differs from the classical ones found in pharmacologically relevant systems. We perceive odors through a combinatorial code involving less than 400 receptors. Then, many of our receptors are broadly tuned and do not only respond to well-defined chemical classes. These so-called broadly tuned receptors use a recognition mode that differs from those encountered in more specific systems. hOR1G1 was considered as a model for such ORs.
The binding feature of this broadly tuned OR is typical of an opportunistic mode of binding. Hydrogen bonds’ role is deemed rather minor (although it is probably mandatory for receptor’s activation [16]) and the binding is dominated by hydrophobic contacts. The lack of an H-bonded interaction hampers any directionality in the binding and the cavity adapts to the odorant’s shape. Such a mode of interaction is well known through another protein involved in the perception of smell: the odorant-binding protein. In OBPs, the odorant is stabilized through a network of hydrophobic contacts within a solvent-occluded cavity [30, 31, 33]. Also, in some X-ray structures, two identical odorants can adopt alternative poses within the same binding cavity [33]. Here again, its shows how the olfactory systems have to adapt to the almost infinite number of odorant chemicals belonging to all families. Notice also that the concept of multiple binding mode leading to a GPCR activation has already been put forward [34]. Broadly tuned ORs combine these two features, with a domination of hydrophobic contacts that favor multiple binding modes through opportunistic interactions.
This domination of the binding by non-polar contacts endows the OR a spectacular adaptability, likely to bind several type of odorants with equivalent affinities. It is nonetheless also able to discriminate between very closely related odorants, as shown on ORI7 for example [13, 35].
Plasticity, together with the lack of a “same structure → same function” paradigm in broadly tuned OR allows us to perceive tens of thousands odors with less than 400 functional receptors. The sense of smell is indeed known to be the most difficult sense to decipher. This is mostly because of the combinatorial code that governs an odorant’s chemoreception. We show here that this complexity projects into the binding mode of ORs at the atomic level. A multimodal way of binding appears to be the main characteristic that makes some ORs so broadly tuned, as shown in Fig. 5, where one can observe a very different orientation of the residues belonging to the cavity. The plasticity of the binding cavity allows different ligands to interact optimally with various residues. ORs are indeed likely to adapt to an odorant’s structure and chemical family. The control of the binding mode by non-polar interactions leads to an equivalent OR activation by an aldehyde (nonanal) and its associated alcohol function (nonanol), since the hydrophilic functional group poorly governs binding. The addition of a methyl group that would decrease the affinity in the alcohol family (nonanol → decanol, see [7]) is compensated by a double bond (9-decen-1-ol) that interacts with a phenylalanine residue. These aliphatic skeletons can equivalently be replaced by the presence of a bulky group (camphor) bound through shape complementarities with the cavity, still via non-polar contacts. On the contrary, non-activating ligands do not fulfill such hydrophobic interactions, due to a smaller carbon skeleton, as observed with butanal. In this case, the ligand weakly binds to the receptor and no particular residue can be considered as highly involved in binding.
State-of-the-art molecular modeling approaches appear likely to predict the affinity of an odorant towards ORs. The agonist or antagonist character remains, however, a hard task to predict. Studies are on the way to get insight into GPCRs’ activation, although they require very massive computer power [36]. They are nevertheless far from being used daily as standard approaches, justifying the use of joint studies, where the agonist potency is experimentally addressed and the binding mode is analyzed by means of molecular modeling.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Note added in proof
A recent article reports a model of hOR1G1 [37]. Their model and our both put forward equivalent residues for the binding cavity. For example, their bound structure of 1-nonanol corresponds to one of our initial structures.
Acknowledgments
The CINES provided computer time. JG acknowledges the University of Nice Sophia Antipolis for funding the project Olfactome. Dr. Steffen Wolf and Pr. Klaus Gerwert are acknowledged for sending the structure of hOR2AG1. Dr. Ravinder Abrol helped in GPCR ab initio modeling.
References
- 1.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- 2.Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature. 2007;449:468–472. doi: 10.1038/nature06162. [DOI] [PubMed] [Google Scholar]
- 3.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/S0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 4.Sell C. Odor cannot be predicted by molecular shape. Chem Senses. 2009;34:181. doi: 10.1093/chemse/bjn069. [DOI] [PubMed] [Google Scholar]
- 5.Sell CS. On the unpredictability of odor. Angew Chem Int Ed Engl. 2006;45:6254–6261. doi: 10.1002/anie.200600782. [DOI] [PubMed] [Google Scholar]
- 6.Triller A, Boulden EA, Churchill A, Hatt H, Englund J, Spehr M, Sell CS. Odorant-receptor interactions and odor percept: a chemical perspective. Chem Biodivers. 2008;5:862–886. doi: 10.1002/cbdv.200890101. [DOI] [PubMed] [Google Scholar]
- 7.Sanz G, Schlegel C, Pernollet JC, Briand L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem Senses. 2005;30:69–80. doi: 10.1093/chemse/bji002. [DOI] [PubMed] [Google Scholar]
- 8.Sanz G, Thomas-Danguin T, Hamdani el H, Le Poupon C, Briand L, Pernollet JC, Guichard E, Tromelin A. Relationships between molecular structure and perceived odor quality of ligands for a human olfactory receptor. Chem Senses. 2008;33:639–653. doi: 10.1093/chemse/bjn032. [DOI] [PubMed] [Google Scholar]
- 9.Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Zamanakos G, Goddard WA., 3rd Prediction of structure and function of G protein-coupled receptors. Proc Natl Acad Sci USA. 2002;99:12622–12627. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singer MS, Shepherd GM. Molecular modeling of ligand-receptor interactions in the OR5 olfactory receptor. NeuroReport. 1994;5:1297–1300. doi: 10.1097/00001756-199406020-00036. [DOI] [PubMed] [Google Scholar]
- 11.Singer MS. Analysis of the molecular basis for octanal interactions in the expressed rat 17 olfactory receptor. Chem Senses. 2000;25:155–165. doi: 10.1093/chemse/25.2.155. [DOI] [PubMed] [Google Scholar]
- 12.Schmiedeberg K, Shirokova E, Weber HP, Schilling B, Meyerhof W, Krautwurst D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J Struct Biol. 2007;159:400–412. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Lai PC, Singer MS, Crasto CJ. Structural activation pathways from dynamic olfactory receptor-odorant interactions. Chem Senses. 2005;30:781–792. doi: 10.1093/chemse/bji070. [DOI] [PubMed] [Google Scholar]
- 14.Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall SE, Floriano WB, Vaidehi N, Goddard WA., 3rd Predicted 3-D structures for mouse I7 and rat I7 olfactory receptors and comparison of predicted odor recognition profiles with experiment. Chem Senses. 2004;29:595–616. doi: 10.1093/chemse/bjh063. [DOI] [PubMed] [Google Scholar]
- 16.Gelis L, Wolf S, Hatt H, Neuhaus EM, Gerwert K. Prediction of a ligand-binding niche within a human olfactory receptor by combining site-directed mutagenesis with dynamic homology modeling. Angew Chem Int Ed Engl. 2012;51:1274–1278. doi: 10.1002/anie.201103980. [DOI] [PubMed] [Google Scholar]
- 17.Floriano WB, Vaidehi N, Goddard WA, 3rd, Singer MS, Shepherd GM. Molecular mechanisms underlying differential odor responses of a mouse olfactory receptor. Proc Natl Acad Sci USA. 2000;97:10712–10716. doi: 10.1073/pnas.97.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doszczak L, Kraft P, Weber HP, Bertermann R, Triller A, Hatt H, Tacke R. Prediction of perception: probing the hOR17-4 olfactory receptor model with silicon analogues of bourgeonal and lilial. Angew Chem Int Ed Engl. 2007;46:3367–3371. doi: 10.1002/anie.200605002. [DOI] [PubMed] [Google Scholar]
- 19.Baud O, Etter S, Spreafico M, Bordoli L, Schwede T, Vogel H, Pick H. The mouse eugenol odorant receptor: structural and functional plasticity of a broadly tuned odorant-binding pocket. Biochemistry. 2011;50:843–853. doi: 10.1021/bi1017396. [DOI] [PubMed] [Google Scholar]
- 20.Matarazzo V, Clot-Faybesse O, Marcet B, Guiraudie-Capraz G, Atanasova B, Devauchelle G, Cerutti M, Etievant P, Ronin C. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem Senses. 2005;30:195–207. doi: 10.1093/chemse/bji015. [DOI] [PubMed] [Google Scholar]
- 21.Surburg H, Panten J (2006) Common fragrance and flavor materials: preparation, properties and uses. Wiley-VCH, Weinheim
- 22.The Good Scent Company, Nonanol (2011). http://www.thegoodscentscompany.com
- 23.Danyi S, Degand G, Duez C, Granier B, Maghuin-Rogister G, Scippo ML. Solubilisation and binding characteristics of a recombinant beta2-adrenergic receptor expressed in the membrane of Escherichia coli for the multianalyte detection of beta-agonists and antagonists residues in food-producing animals. Anal Chim Acta. 2007;589:159–165. doi: 10.1016/j.aca.2007.02.057. [DOI] [PubMed] [Google Scholar]
- 24.Man O, Gilad Y, Lancet D. Prediction of the odorant-binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 2004;13:240–254. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abrol R, Bray JK, Goddard WA. Bihelix: towards de novo structure prediction of an ensemble of G-protein coupled receptor conformations. Proteins. 2011;80:505–518. doi: 10.1002/prot.23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abrol R, Kim SK, Bray JK, Griffith AR, Goddard WA., 3rd Characterizing and predicting the functional and conformational diversity of seven-transmembrane proteins. Methods. 2011;55:405–414. doi: 10.1016/j.ymeth.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hummel P, Vaidehi N, Floriano WB, Hall SE, Goddard WA., 3rd Test of the Binding Threshold Hypothesis for olfactory receptors: explanation of the differential binding of ketones to the mouse and human orthologs of olfactory receptor 912–93. Protein Sci. 2005;14:703–710. doi: 10.1110/ps.041119705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Efremov RG, Chugunov AO, Pyrkov TV, Priestle JP, Arseniev AS, Jacoby E. Molecular lipophilicity in protein modeling and drug design. Curr Med Chem. 2007;14:393–415. doi: 10.2174/092986707779941050. [DOI] [PubMed] [Google Scholar]
- 29.Pyrkov TV, Chugunov AO, Krylov NA, Nolde DE, Efremov RG. PLATINUM: a Web tool for analysis of hydrophobic/hydrophilic organization of biomolecular complexes. Bioinformatics. 2009;25:1201–1202. doi: 10.1093/bioinformatics/btp111. [DOI] [PubMed] [Google Scholar]
- 30.Charlier L, Nespoulous C, Fiorucci S, Antonczak S, Golebiowski J. Binding free energy prediction in strongly hydrophobic biomolecular systems. Phys Chem Chem Phys. 2007;9:5761–5771. doi: 10.1039/b710186d. [DOI] [PubMed] [Google Scholar]
- 31.Golebiowski J, Antonczak S, Fiorucci S, Cabrol-Bass D. Mechanistic events underlying odorant-binding protein chemoreception. Proteins. 2007;67:448–458. doi: 10.1002/prot.21307. [DOI] [PubMed] [Google Scholar]
- 32.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE., 3rd Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 33.Vincent F, Spinelli S, Ramoni R, Grolli S, Pelosi P, Cambillau C, Tegoni M. Complexes of porcine odorant-binding protein with odorant molecules belonging to different chemical classes. J Mol Biol. 2000;300:127–139. doi: 10.1006/jmbi.2000.3820. [DOI] [PubMed] [Google Scholar]
- 34.Kahsai AW, Xiao K, Rajagopal S, Ahn S, Shukla AK, Sun J, Oas TG, Lefkowitz RJ. Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nat Chem Biol. 2011;7:692–700. doi: 10.1038/nchembio.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araneda RC, Kini AD, Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci. 2000;3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- 36.Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, Borhani DW, Shaw DE. Activation mechanism of the beta2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Launay G, Téletchéa S, Wade F, Pajot-Augy E, Gibrat JF, Sanz G. Automatic modeling of mammalian olfactory receptors and docking of odorants. Protein Eng Des Sel. 2012;25:377–386. doi: 10.1093/protein/gzs037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.