Abstract
Hypoxia-inducible factors (HIF) are transcription factors responding to reduced oxygen levels and are of utmost importance for regulation of a widespread of cellular processes, e.g., angiogenesis. In contrast to HIF-1α/HIF-2α, the relevance of HIF-3α for the regulation of the HIF pathway in human vascular cells is largely unknown. HIF-3α mRNA increases under hypoxia in endothelial and vascular smooth muscle cells. Analysis of HIF-3α isoforms revealed a cell type-specific pattern, but only one isoform, HIF-3α2, is hypoxia-inducible. Reporter gene assays of the appropriate promoter localized a 31-bp fragment, mediating this hypoxic regulation. The contribution of HIF-1/2 and NFκB to the HIF-3α induction was verified. Functional studies focused on overexpression of HIF-3α isoforms, which decrease the hypoxia-mediated expression of VEGFA and Enolase2. These data support the notion of a hypoxia-induced inhibitory function of HIF-3α and demonstrate for the first time the existence of this negative regulation of HIF-signaling in vascular cells.
Keywords: Hypoxia inducible factor, HIF-3α isoform, Hypoxia, Endothelial cells, Vascular smooth muscle cells
Introduction
Hypoxia is regarded as an important stimulus of physiological and pathophysiological angiogenesis. Beside the well-established impact of hypoxia-driven angiogenesis in tumor development, the presence of hypoxia and angiogenesis is shown in advanced atherosclerotic lesions [1–3]. The cellular effects of hypoxia are mostly mediated by the hypoxia-inducible factors (HIF), consisting of one of the tightly controlled α-subunits (HIF-1α, HIF-2α and HIF-3α) and the constitutively expressed HIF-1β. A possible role for HIF in atherosclerosis is supported by the presence of intraplaque angiogenesis and the expression of several known HIF-responsive genes in atherosclerosis, e.g., VEGFA. In human atherosclerosis HIF-1α and HIF-2α co-localize in hypoxic areas of the plaque with macrophages and are associated with expression of angiogenic and inflammatory factors [4–7]. However, the contribution of vascular cells such as endothelial cells (EC) and vascular smooth muscle cells (VSMC) to hypoxic response in atherosclerosis is undefined. In healthy tissues ECs are in a quiescent state and rarely divide and migrate. In contrast, under hypoxic conditions they are able to divide rapidly, to migrate and promote the formation of new vessels [8]. Beside the established function of HIF-1α in endothelial proliferation, migration and sprouting [9, 10], HIF-2α seems to participate more in remodeling and maturation of the microvasculature and maintaining the vessel integrity [11, 12]. In contrast, the distribution and especially the function of HIF-3α in the human vascular system is still unidentified.
HIF-α subunits and HIF-1β are structurally related proteins of basic helix-loop-helix (bHLH) and PER-ARNT-SIM (PAS) domain transcription factors [13]. Beside these domains, which are necessary for DNA binding and dimerization, α-subunits own an oxygen dependent degradation domain (ODDD) and two transactivation domains (NTAD and CTAD). The expression, stability and activity of the HIF-α subunits are closely regulated [14]. Posttranslational hydroxylation of α-subunits under normoxia by oxygen-dependent monooxygenases (prolylhydroxylases, PHD) permits subsequent degradation by the ubiquitin/proteasome pathway [15–17]. Upon inhibition of HIF-α degradation under hypoxia, the α-subunits translocate to the nucleus and dimerize with HIF-1β. This complex recognizes and binds to the hypoxia response element (HRE) [18], and thus induces a broad range of genes that are involved in multiple cellular processes like angiogenesis, glycolysis and cell survival, but also apoptosis [19, 20].
HIF-3α shows a high similarity to HIF-1α and HIF-2α in the bHLH and PAS domains, but lacks the C-terminal transactivation domain (CTAD) [21]. A splicing variant of HIF-3α, identified in mice and termed inhibitory PAS (IPAS) was shown to act as a dominant-negative regulator of the HIF pathway [22]. This transcript was observed in heart and lung of hypoxia-exposed mice, and a feedback inhibition of adaptive responses to hypoxia in these tissues was postulated [23]. A first characterization of human HIF-3α was presented in 2001 [24]. The authors showed that HIF-3α is expressed in human kidney and suppresses HIF-mediated reporter gene expression. HIF-3α, as detected in a human alveolar epithelial cell line (A549), shows a strong hypoxic induction at mRNA and protein level because of transcriptional activation and enhanced protein stability [25]. To date several human HIF-3α isoforms, generated by alternative splicing, have been described [26–28]. Among them, especially short isoforms similar to mouse IPAS are specified to operate as dominant-negative regulators of HIF signaling. Recently published data described HIF-3α isoforms in a renal carcinoma cell line; one of them is upregulated by hypoxia. A target gene- and cell-specific impact of HIF-3α for HIF target gene expression was postulated [29].
The present study shows that in vascular cells hypoxia upregulates HIF-3α in a newly defined isoform-specific fashion. This induction is dependent on HIF-signaling itself. Adenoviral overexpression of HIF-3α in vascular cells exerts an antagonistic effect on hypoxia-dependent induction of HIF-1/HIF-2 target genes. These data support the notion of counteractive, i.e., stimulatory and inhibitory elements of the HIF-pathway in vascular cells potentially contributing to the genesis of atherosclerosis.
Materials and methods
Cell culture
All culture media and supplements were purchased from PAA Laboratories (Germany) unless otherwise specified. FCS was provided by Sigma-Aldrich (Germany). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. For hypoxic conditions cells were cultivated in a humidified chamber at 0.5% O2 balanced with 95% N2 and 5% CO2 at 37°C. HEK293 and HELA cells were purchased from DSMZ (Germany) and maintained in DMEM (high glucose) and RPMI1640, respectively, with 10% FCS and antibiotic/antimycotic solution (PAA Laboratories, Germany). Human umbilical venous endothelial cells (HUVEC) were isolated according to a modified method of Jaffe et al. [30] using enzymatic digestion with collagenase type 2 from umbilical cords. Cells were grown in endothelial cell growth medium MV (ECGM; PromoCell, Germany) with antibiotic/antimycotic solution and used from passages 1–4. Primary cultures of vascular smooth muscle cells were obtained by explant technique from human aortic sections derived from coronary artery bypass surgery. VSMCs were cultivated in DMEM (low glucose) with 10% FCS and antibiotic/antimycotic solution.
RNA isolation and real-time RT-PCR
Total RNA was isolated using the Invisorb Spin Cell RNA Mini Kit (Invitek, Germany). cDNA was synthesized with the Revert Aid™ H Minus First Strand Synthesis Kit (MBI Fermentas, Germany) from 1–5 μg total RNA using oligo-dT. Real-time PCR was performed using the iCycler IQ Real Time PCR System (BioRad, USA) and SYBR® Premix ExTaq™ (Takara Bio Inc./Lonza, Germany). PCR conditions for all primer sets (Table 1) were as follows: 95°C for 2 min followed by 40 amplification cycles, each consisting of 95°C for 20 s, 58°C for 30 s and 72°C for 20 s with a final extension step at 72°C for 2 min. Identity of PCR products was proved by melting point analysis, electrophoresis and sequencing of resulting PCR products (sequencing facility MPI CBG Dresden, Germany). Relative quantification of gene expression was calculated with the ΔΔCT method using Gene Expression Macro™ Version 1.1 (BioRad Laboratories, USA) with HPRT1 as housekeeping gene.
Table 1.
Primers used in real-time RT-PCR
| Gen | Sequence | GenBank acc. no. |
|---|---|---|
| ENO2 |
5′-CTG AAG CCA TCC AAG CGT-3′ 5′-CAG ACG TTC AGA ACG GCA C-3′ |
NM_001975 |
| HIF-1α |
5′-GTT CAC CTG AGC CTA ATA GTC C-3′ 5′-GGA ACG TAA CTG GAG GTC ATC-3′ |
NM_001530 |
| HIF-2α |
5′-AGT CAC CAG AAC TTG TGC ACC-3′ 5′-CTT CTC AAT CTC ACT CAG GAC G-3′ |
NM_001430 |
| HIF-3α |
5′-TGT ACT CTT CAT GCG CAA GG-3′ 5′-TGT GAC CAA GAG GAG CTT CAG-3′ |
|
| HPRT1 |
5′-TTGCGACCTTGACCATCTTTG-3′ 5′-CTTTGCTGACCTGCTGGATTAC-3′ |
NM_000194 |
| VEGFA |
5′-GCC TTG CCT TGC TGC TCT AC-3′ 5′-GAA GAT GTC CAC CAG GGT CTC-3′ |
NM_001025370 |
Detection of HIF-3a isoforms
Touchdown-PCR was performed with GoTaq Polymerase (Promega, Germany) and isoform-specific primers (95°C for 5 min, 12 cycles with 95°C for 30 s, decreasing annealing temperature from 60 to 54°C for 30 s, 72°C for 30 s; 30 cycles with 95°C for 30 s, 54°C for 30 s and 72°C for 30 s). PCR fragments were analyzed by electrophoresis, cloned with pGEM®-T Easy Vector System (Promega, Germany) and sequenced. For semiquantitative evaluation of HIF-3α isoform expression, aliquots of PCR reactions were removed after defined cycles and analyzed on agarose gels. Signal intensities of DNA bands were estimated with “Quantity One” software from BioRad (USA) and normalized by parallel detection of housekeeping gene HPRT1. The following PCR primers were used: HIF-3α1 sense 5′-CAT GGA CTG GCA AGA CCA C-3′; HIF-3α2 sense 5′-CTG CAG CGC GCA AGG TC-3′; HIF-3α3 sense 5′-ACC CAC TCG TAA CTC GCA CC-3′; universal antisense 5′-TCC CAC CTG GTT CCA CTCC-3′.
Transient transfection of siRNA into HUVEC
Transfections of HUVEC with siRNA (siMAX siRNA, MWG Biotech, Germany) were done using Turbofect™ siRNA transfection reagent (Fermentas GmbH, Germany) according to the manufacturer's instructions. Briefly, HUVECs were seeded with 1 × 105 cells/cm2 in endothelial cell growth medium MV (ECGM, Promocell, Heidelberg, Germany). Before transfection, the medium was replaced by Opti-MEM I (Invitrogen GmbH, Darmstadt, Germany) with 2% FCS. For transfection of 1 well in a 24-well plate, 0.5 μg (37.5 pmol) of siRNA was diluted in 100 μl Opti-MEM I; 1.5 μl Turbofect™ siRNA transfection reagent was added, mixed and incubated for 20 min at room temperature. Subsequently complexes were added drop-wise to each well. After 5 h the medium was replaced by ECGM. Gene silencing was assayed 24–72 h later. siRNA sequences were as follows: HIF-1α (sense 5′-AGUUCACCUGAGCCUAAUATT-3′, antisense 5′-UAUUAGGCUCAGGUGAACUTT-3′); HIF-2α (sense 5′-GGAGCUAACAGGACAUAGUTT-3′, antisense 5′-ACUAUGUCCUGUUAGCUCCTT-3′); non-specific control (sense 5′-AGGUAGUGUAAUCGCCUUGTT-3′, antisense 5′-CAAGGCGAUUACACUACCUTT-3′).
Construction of adenoviruses and adenoviral infection
Recombinant adenoviruses were produced with the Adeno-X™ Expression System (Clontech, USA) according to the manufacturer's protocol. Primers for plasmid construction are summarized in Table 2. HIFαdn was amplified from HUVEC cDNA, and HIF-3α2 and HIF-3α3 were amplified from plasmid harboring cDNA coding for HIF-3α3 (German Resource Centre for Genome Research RZPD, Germany). Wild-type IκBα was synthesized from HUVEC cDNA. The construction of dominant active IκBα (IκBαm) was done by site-directed mutagenesis of both serine phosphorylation sites at position 32 and 36 to alanine. The resulting PCR fragments were cloned into pShuttle with appropriate restriction enzymes. To construct an adenovirus expressing small hairpin RNA (shRNA) under control of H1 promoter, a synthetic double-stranded DNA fragment (HIF-3αsh1083 or shscr as control) with sticky ends for BamHI/HindIII was cloned into pshuttle-H1. The expression cassette was then transferred into pAdeno-X™ and transformed into E. coli Stbl2. Recombinant pAdeno-X™ plasmid was digested with PacI and transfected in HEK293 cells using a calcium phosphate transfection kit (Sigma-Aldrich, Germany). After evidence of a cytopathic effect, cells were lysed by three freeze-thaw cycles, and primary viral stock was stored at −80°C. Recombinant virus was amplified in HEK293 cells, and purification was performed by cesium chloride density gradient centrifugation. Briefly, clarified lysate was overlaid onto a three-step gradient (1 ml CsCl with 1.245 g/cm3, 2.5 ml CsCl with 1.32 g/cm3 and 1.5 ml CsCl with 1.44 g/cm3 in 25 mM Tris-Cl, 137 mM NaCl, 5 mM KCl, 0.5 mM Na2HPO4, pH 7.4) and centrifuged at 220,000g at 10°C for 2 h. The virus-containing band was separated, dialyzed against 10 mM Tris-Cl, pH 8.0, 135 mM NaCl, 3 mM KCl, 1 mM MgCl2, 1% (v/v) glycerol and stored after addition of glycerol to 10% at −80°C. Viral titer was determined with the AdenoX™ Rapid Titer Kit (Clontech). HUVECs (VSMC) were grown to 60–80% confluence for about 24 h. Then cells were transfected with adenovirus at a multiplicity of infection (M.O.I.) of 200 (100). The virus-containing medium was removed after 24 h, and cells were cultivated with fresh medium for a further 24 h before hypoxic treatment.
Table 2.
Primers used for plasmid constructions and competition experiments
| Primer set | Sequence |
|---|---|
| HIF-3α2 |
5′-GCTCTAGAATGCGCCTCACCATCAGC-3′ (XbaI) 5′-AAGGTACCGGAGGAGAAGGCAGATG-3′ (KpnI) |
| HIF-3α3 |
5′-AATCTAGACCGAGCCATGGCGCTG-3′ (XbaI) 5′-AAGGTACCGGAGGAGAAGGCAGATG-3′ (KpnI) |
| HIF-3αpromI |
5′-ATAAGCTTAATTATTCATC-3′ (HindIII) 5′-AGCGCCATGGCTCGCCAGTC-3′ (NcoI) |
| HIF-3αpromII |
5′-TAGCTAGCAGAAGGCGCGTTCAG-3′ (NheI) 5′-AACTCGAGGCACGGCGGTGAGATG-3′ (XhoI) |
| HIF-3αpromIII |
5′-TTGCTAGCCTCCTGCCTCAGCCTTTC-3′ (NheI) 5′-TGGTCTTGCCAGTCCATGGTG-3′ (NcoI) |
| HIFαdn |
5′-GGGCTAGCTCTGAACGTCTCAAAGG-3′ (NheI) 5′-GGTACCTTACAGGGCTATTGGGCGT-3′ (KpnI) |
| IκBα |
5′-AAGCTAGCATGTTCCAGGCGGCCGAG-3′ (NheI) 5′-CACGGTACCTCATAACGTCAGACGCTG-3′ (KpnI) |
| IκBαm |
5′-CGCCACGACGCCGGCCTGGACGCCATGAAAGAC-3′ 5′-GCGTCCAGGCCGGCGTCGTGGCGG-3′ |
| pGL-34 |
5′-CTAGGCCTCCCCACCCAAGGCCGGCCCTTTCCTGTGGAG-3′ 5′-TCGAGCTCCACAGGAAAGGGCCGGCCTTGGGTGGGGAGGC-3′ |
| pGL-41 |
5′-CTAGGCTGAGCTCGGGTGCCCCCCCTCCCCACCCAAGGCCGGCC-3′ 5′-TCGAGGCCGGCCTTGGGTGGGGAGGGGGGGCACCCGAGCTCAGC-3′ |
| pGL-30 |
5′-CTAGGCTTTCCTGTGGAGTCATCTCACCGCCGTGCC-3′ 5′-TCGAGGCACGGCGGTGAGATGACTCCACAGGAAAGC-3′ |
| pGL-41A |
5′-CTAGGCCTCCCCACCCAAGGCCGGCCCTTTCCTGTGGAGTCATC-3′ 5′-TCGAGATGACTCCACAGGAAAGGGCCGGCCTTGGGTGGGGAGGC-3′ |
| pGL-51 |
5′-CTAGGCCTCCCCACCCAAGGCCGGCCCTTTCCTGTGGAGTCATCTCA CCGCCGTGCC-3′ 5′-TCGAGGCACGGCGGTGAGATGACTCCACAGGAAAGGGCCGGCCTT GGGTGGGGAGGC-3′ |
| pGL-31 |
5′-CTAGGCAAGGCCGGCCCTTTCCTGTGGAGTCATC-3′ 5′-TCGAGATGACTCCACAGGAAAGGGCCGGCCTTGC-3′ |
| pGL-31m1 |
5′-CTAGGCAAGGCCGGCCCTTTGCTGTGGAGTCATC-3′ 5′-TCGAGATGACTCCACAGCAAAGGGCCGGCCTTGC-3′ |
| pGL-31m2 |
5′-CTAGGCAAGGCCGGCCCTTTCCTGTGGTTTCATC-3′ 5′-TCGAGATGAAACCACAGGAAAGGGCCGGCCTTGC-3′ |
| HIF-3αsh1083 |
5′-GATCCCGAGTATCGTCTGTGTCCATTTCTCGAGAAATGGACACAGAC GATACTCTTTTTTCCAAA-3′ 5′-AGCTTTTGGAAAAAAGAGTATCGTCTGTGTCCATTTCTCGAGAAATG GACACAGACGATACTCGG-3′ |
| shscr |
5′-GATCGCGTGATGAGGCGGTTAACGTTCCTTGATATCCGGGAACGTTA ACCGCCTCATCATTTTTTCCAAA-3′ 5′-AGCTTTTGGAAAAAATGATGAGGCGGTTAACGTTCCCGGATATCAAG GAACGTTAACCGCCTCATCACGC-3′ |
Recognition sites for restriction enzymes are underlined and identification noted behind sequences
Construction of HIF3α promoter-driven luciferase reporter plasmids
DNA fragments from the 5′-flanking regions of HIF-3α exon 1a, 1b and 1c were amplified from BAC plasmid RZPDB737A022172 (German Resource Centre for Genome Research RZPD, Germany). The PCR products were cloned in pGL3-basic vector (Promega, Germany) using appropriate restriction enzymes. Otherwise chemically synthesized double-stranded oligonucleotides corresponding to short promoter sequences were ligated directly into pGL3-basic vector (Table 2).
Transient transfection of HUVEC and luciferase assay
HUVECs were transfected using jetPEI™HUVEC (Polyplus transfection, PEQLAB Germany) according to the manufacturer's instructions. Briefly, HUVECs were seeded in 48-well cell culture plates at a density of 20,000 cells/cm2 the day before transfection in ECGM. Medium was changed before transfection to Opti-MEM I (Invitrogen, Germany) supplemented with 2% FCS; 1 μg Plasmid-DNA was used with 2 μl jetPEI™HUVEC per well (10 ng/well). pRL-CMV vector (10 ng/well, Promega, Germany) expressing Renilla reniformis luciferase was co-introduced as internal control for normalization of transfection efficiency. Cells were incubated for 3 h and thereafter washed and placed in ECGM for a further 3 h. Hypoxic or normoxic conditions were applied for 24 h, and cellular extracts were prepared for Dual Luciferase® Reporter Assay (Promega, Germany) following the manufacturer's instructions. Luciferase was quantified with a Lumat BL9507 luminometer (EG&G Berthold, Germany). Promoter activities were calculated as quotient of firefly luciferase and renilla luciferase luminescence levels relative to pGL3-basic.
Western blot analysis
Western blot analysis was performed as described previously [31] using the following antibodies and dilutions: mouse monoclonal anti-HIF-1α (BD Biosciences, Germany) 1:500; rabbit polyclonal anti-HIF-2α (Novus Biologicals, USA) 1:1,000; rabbit polyclonal HIF-3α (ab10134, Abcam, UK) 1:1,000; mouse anti-β-actin (Santa Cruz, USA) 1:1,000; sheep anti-mouse-HRP (GE Healthcare, USA) 1:10,000; goat anti-rabbit-HRP (Santa Cruz, USA) 1:2,500. Intensities of protein bands were estimated with “Quantity One” software from BioRad (USA). Equal loading of proteins was determined by detection of β-actin.
TransAM™ ELISA
Nuclear protein extraction was performed with the Nuclear Extract Kit (Active Motif Europe, Belgium). ELISA-based kits for detection of HIF-1 (TransAM™ HIF-1) and AP-1 (TransAM™ AP-1 family including c-Fos, FosB, Fra-1, c-Jun, JunB and JunD) were used according to the manufacturer's instructions (Active Motif Europe, Belgium). TransAM kits contain a 96-well plate on which an oligo containing a binding site for HIF-1 (5′-TACGTGCT-3′) or AP-1 (5′-TGAGTCA-3′), respectively, has been immobilized. Transcription factors contained in nuclear extract specifically bind to this oligonucleotide. Primary antibodies recognize accessible epitopes on HIF-1 or c-Fos, FosB, Fra-1, c-Jun, JunB and JunD upon DNA binding. Addition of secondary HRP-conjugated antibody provides a colorimetric read-out that is quantified by spectrophotometry.
Database searches and statistical analysis
Searches of GenBank™ were performed using BLAST service at the National Center for Biotechnology Information (NCBI) home page (http://www.ncbi.nlm.nih.gov). Searches for transcription factor binding sites were performed with Genomatix software (http://www.genomatix.de), PROMO 3.0.2 (http://alggen.lsi.upc.es) and TESS (http://www.cbil.upenn.edu/tess) [32] .
For statistical analysis all values were normalized as indicated and expressed as mean ± standard deviation. Data were analyzed using unpaired Student’s t-test between two groups. P-values below 0.05 and 0.01 were considered to be statistically significant (*) and very significant (**), respectively. If applicable the additional consideration of Bonferroni correction of k = 2 is indicated; hence α/k = 0.025 (*) and 0.005 (**).
Results
Expression profile of HIF-3α
The computational analysis of expressed sequence tag (EST) counts allows the generation of an approximate expression pattern for HIF-3α (http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.420830). It shows, in comparison to the widely expressed HIF-1α and HIF-2α, a more restricted HIF-3α expression to certain organs. HIF-3α ESTs were detected for example in spleen, nerve, heart, placenta and umbilical cord, but not in blood, bone marrow or lymph. To get more insight into the involved cell types different primary cells were analyzed for HIF-3α expression. Human endothelial cells (venous and arterial cells from umbilical cord, aortic endothelial cells) and vascular smooth muscle cells from human aorta express HIF-3α mRNA, which is also inducible by hypoxic culture conditions. CD34+ hematopoietic progenitor cells, monocytes from peripheral blood and monocyte-derived macrophages did not show detectable amounts of HIF-3α, in agreement with the mentioned EST data and underlining the limited tissue/cell-specific expression of HIF-3α.
Expression of HIF-3α subunits during hypoxia in vascular cells
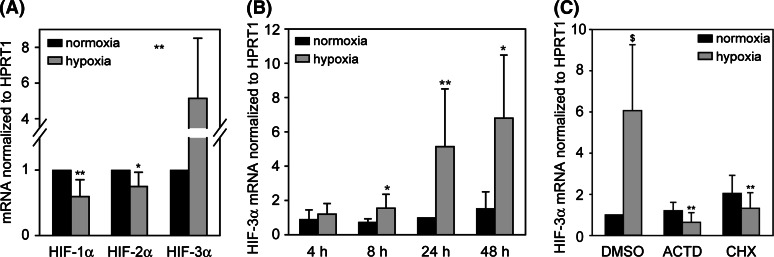
HIF-α subunit expression was determined under normoxic and hypoxic treatment of HUVEC (Fig. 1). As is already known, HIF-1α and HIF-2α mRNA are expressed in high amounts in endothelial cells. Whereas HIF-1α and HIF-2α mRNA are decreased by hypoxia, a significant increase of HIF-3α mRNA is detectable from 8 h up to 48 h of hypoxic treatment (Fig. 1a, b). Comparable results were achieved with VSMC (data not shown). To investigate the role of active transcriptional and translational processes for hypoxic induction of HIF-3α mRNA, HUVECs were pretreated with actinomycin D (ACTD) or cycloheximide (CHX) prior to hypoxia. Both inhibitors prevent the upregulation of HIF-3α mRNA completely, indicating that the hypoxic increase is caused rather by mRNA synthesis than stabilization and depends on active protein synthesis (Fig. 1c).
Fig. 1.
a Differential regulation of α-subunit mRNA in HUVEC after 24 h hypoxia. *P < 0.05, **P < 0.01 hypoxia versus normoxia. b Hypoxia increases HIF-3α expression in HUVEC significantly after 8 h up to 48 h. *P < 0.025, **P < 0.005 hypoxia versus normoxia. Bonferroni correction considered. c Hypoxic upregulation of HIF-3α in HUVEC is inhibited after treatment with inhibitors of transcription and translation. Cells were preincubated for 2.5 h with solvent (DMSO), 0.5 μg/ml actinomycin D (ACTD) or 1 μg/ml cycloheximide (CHX) and then subjected to hypoxia for 22 h. Expression of mRNA was analyzed by real-time RT-PCR and calculated as relative values to normoxia (24 h). $ P < 0.01 versus DMSO/normoxia. **P < 0.005 versus DMSO/hypoxia, Bonferroni correction considered. Means and standard deviations are shown
Hypoxic HIF-3α induction is dependent on HIF and NFκB signaling
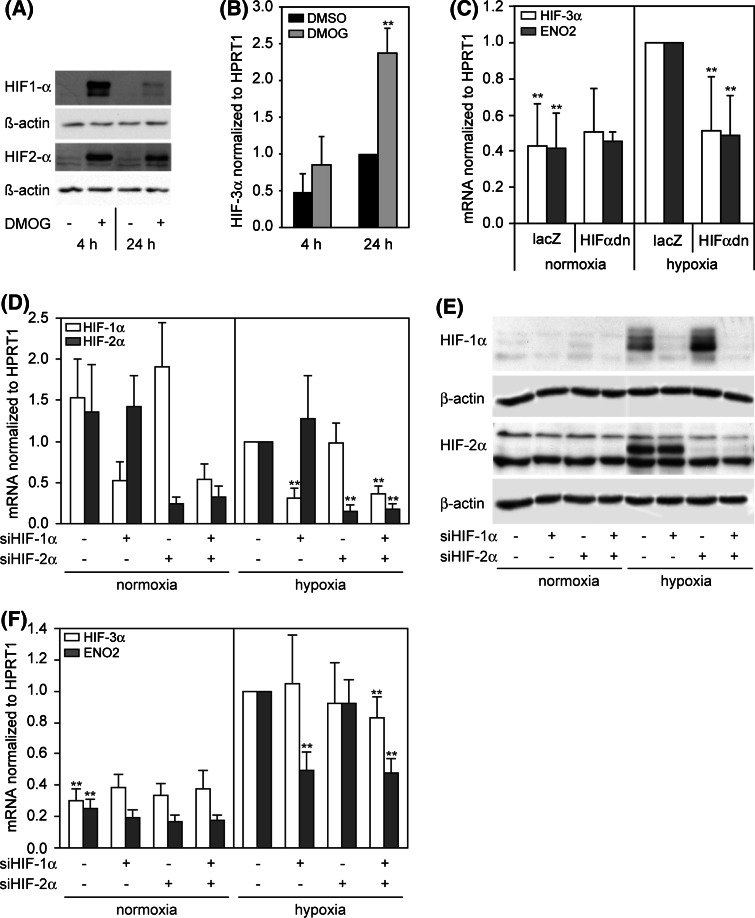
Dimethyloxalylglycine (DMOG), inhibiting the PHDs and stabilizing Hif-α subunits, is a well-established hypoxia mimetic. In fact, the treatment of HUVEC with DMOG under normoxic conditions leads to stabilization of both proteins, HIF-1α and HIF-2α. Subsequently, HIF-3α mRNA is upregulated significantly after 24 h of DMOG treatment and shows a similar behavior as under hypoxia, suggesting an involvement of HIF signaling on HIF-3α regulation (Fig. 2a, b).
Fig. 2.
a, b Dimethyloxalylglycine (DMOG) caused stabilization of HIF-1α and HIF-2α under normoxia and HIF-3α mRNA induction. HUVECs were incubated with solvent (DMSO) or 2.5 mM DMOG for 4 and 24 h. a DMOG causes a non-hypoxic stabilization of HIF-1α and HIF-2α protein as detected by Western blot. b HIF-3α mRNA is induced by DMOG comparable to hypoxia (n = 5). Expression of HIF-3α mRNA was analyzed by real-time RT-PCR and calculated as relative values to DMSO (24 h). **P < 0.01 DMOG versus DMSO. c Adenoviral overexpression of dominant-negative HIF-2α (HIFαdn) in HUVEC under normoxia/hypoxia. Cells were infected with virus expressing lacZ (control) or HIFαdn for 24 h, further incubated with fresh medium for 24 h and exposed to normoxia/hypoxia for 24 h. Data were calculated as relative values to hypoxia/lacZ. Hypoxic induction of HIF-3α mRNA is inhibited under overexpressed HIFαdn (n = 14). *P < 0.05, **P < 0.01 versus lacZ/hypoxia. d–e Transfection of HUVEC with siRNA targeting HIF-1α and HIF-2α. A siRNA with scrambled non-specific sequence was used in control samples. d Silencing of HIF-1α and HIF-2α mRNA 72 h posttransfection of respective siRNA relative to hypoxia. e Western blot detecting HIF-1α and HIF-2α 72 h posttransfection of respective siRNA shows clearly reduced protein levels under hypoxia. f Influence of HIF-1α and HIF-2α inhibition on expression of HIF-3α and ENO2 under normoxia and hypoxia (n = 4). *P < 0.05, **P < 0.01 versus hypoxia without specific siRNA. Means and standard deviations are shown
To determine whether the regulation of HIF-3α depends on the key factors of hypoxic gene regulation, HIF-1 and HIF-2, a dominant-negative mutant of HIF-2α (HIFαdn) was adenovirally overexpressed in HUVEC. HIFαdn has a C-terminal deleted HIF-2α, which lacks the transactivation domain, but retains the ability to form heterodimers with HIF-1β and therefore acts as a competitive inhibitor for both HIF-1α and HIF-2α subunits [33]. Cells expressing the inhibitor HIFαdn show a complete downregulation of hypoxia-induced HIF-3α, whereas the normoxic expression is not affected, suggesting that HIF-1 and/or HIF-2 is involved in the hypoxic regulation (Fig. 2c). As a proof of principle the expression of the well-known HIF target gene ENO2 was measured, and similar results were obtained.
Further experiments were done with siRNA against HIF-1α and HIF-2α to verify the HIF-dependent HIF-3α upregulation under hypoxia (Fig. 2d–e). Therefore, siRNA targeting HIF-1α, HIF-2α and the combination of both was transiently transfected into HUVEC. Transfection efficiency was estimated by in parallel transfected FITC-labeled siRNA and was generally about 90%. Control samples were transfected with non-specific scrambled siRNA. Silencing of HIF-1α and HIF-2α was proven on the mRNA and protein level. The resulting expression of HIF-3α and the known HIF target gene ENO2 was measured under normoxia and hypoxia (Fig. 2e). It is shown that hypoxic ENO2 upregulation depends on HIF-1, but not HIF-2. In contrast, HIF-3α cannot be influenced by siHIF-1α or siHIF-2α alone. Only the combination of both siRNA causes a slightly, but significantly decreased HIF-3α expression under hypoxia. Therefore, both HIF-α subunits are able to drive the hypoxic induction of HIF-3α in HUVEC.
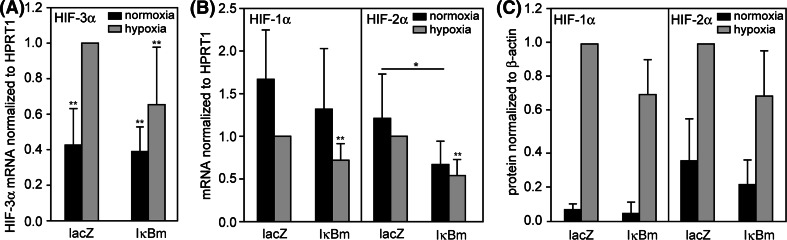
Another transcription factor often involved in hypoxia-driven signaling is NFκB. To determine the engagement of this factor in HIF-3α regulation, constitutive active IκBα (IκBαm) was adenovirally overexpressed (Fig. 3a–c). IκBαm acts as irreversible inhibitor of NFκB [34, 35]. Overexpression of IκBαm causes a significant inhibition of HIF-3α upregulation under hypoxia, whereas the expression of HIF-3α under normoxia is not altered, suggesting an at least partial regulation by NFκB. Interestingly, the mRNA expression and protein amount of HIF-1α and HIF-2α show a tendency to decrease under IκBαm overexpression; therefore, the inhibition of HIF-3α under hypoxia could be a secondary effect of NFκB inhibition.
Fig. 3.
Adenoviral overexpression of constitutive active IκBα (IκBαm) in HUVEC under normoxia/hypoxia. Cells were infected with virus expressing lacZ (control) or IκBαm for 24 h, further incubated with fresh medium for 24 h and exposed to normoxia/hypoxia for 24 h. Data were calculated as relative values to hypoxia/lacZ. a Hypoxic expression of HIF-3α mRNA is decreased under overexpressed IκBαm. b Relative abundance of HIF-1α and HIF-2α mRNA is decreased under overexpressed IκBαm (n = 5). c Influence of overexpressed IκBαm on HIF-1α and HIF-2α protein levels (n = 4). *P < 0.05, **P < 0.01 versus lacZ/hypoxia. Means and standard deviations are shown
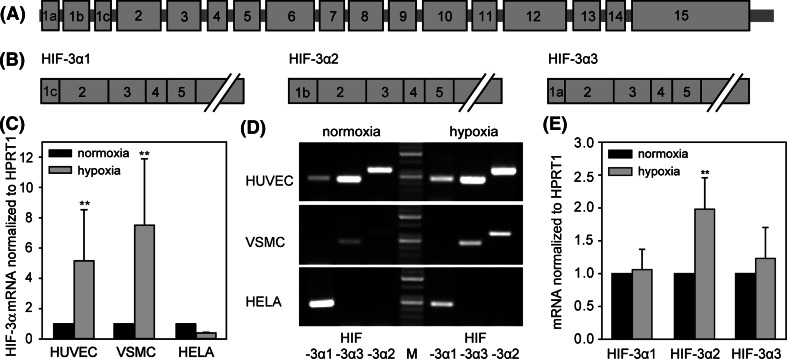
Detection of the hypoxia-inducible HIF-3α isoform
As formerly mentioned the occurrence of HIF-3α is cell-specific, and only few data exist for the identification of distinct HIF-3α isoforms [15, 26–29], thereby the expression in vascular cells is completely unknown. At least five human HIF-3α isoforms have been described. The isoforms HIF-3α1, HIF-3α2 and HIF-3α3 differ only in the first exon, resulting in proteins owning individual N-termini (Fig. 4b). As shown in Fig. 4c, HUVEC and VSMC exhibit a hypoxia-dependent expression of HIF-3α mRNA. In contrast, HIF-3α is detectable, but not inducible by hypoxia in HELA cells. To determine distinct HIF-3α isoforms and their inducibility by hypoxia in different cell types, isoform-specific RT-PCR was performed. As shown in Fig. 4d, HUVECs express all three long isoforms compared to VSMC. In VSMC only HIF-3α2 and HIF-3α3 are detectable. In contrast, HELA cells express HIF-3α1 exclusively. Semiquantitative RT-PCR was performed with cDNA from HUVEC (Fig. 4e) and VSMC (data not shown). Under hypoxia a significantly elevated expression was observed only for isoform HIF-3α2. These data support the notion of a hypoxia-non-inducible HIF-3α expression in HELA cells. HELA cells express only HIF-3α1. Differences in induction values of total HIF-3α (Fig. 4c) and HIF-3α2 (Fig. 4e) could be explained by using different measurement methods. The detection of different isoforms required a special touch-down PCR protocol. This allowed only the detection of PCR fragments on agarose gels with subsequent estimation of band intensities, but not a real-time detection of PCR fragment amplification. In contrast to real-time RT-PCR used for data of Fig. 4c, this method is more insensitive, but the results showed the same kind of regulation.
Fig. 4.
Cell-specific expression of HIF-3α. a Genomic organization of HIF-3α gene locus on chromosome 19. b Organization of isoform-specific mRNA for HIF-3α1, HIF-3α2 and HIF-3α3. c Variable regulation of HIF-3α in different cell types after 24 h hypoxia. Expression of HIF-3α mRNA was calculated as relative values to HUVEC/normoxia. d Analysis of qualitative isoform-specific RT-PCR products shows cell-specific expression of HIF-3α isoforms. (M) DNA ladder. e Estimation of hypoxia-modulated HIF-3α isoform. HUVECs were incubated under normoxic or hypoxic conditions for 24 h; HIF-3α isoforms were detected by isoform-specific RT-PCR and evaluated by agarose gel electrophoresis. Calculation of relative HIF-3α signal intensities was carried out with the in parallel measured housekeeping gene HPRT1 (n = 6). Expression of mRNA was calculated as relative values to normoxia. **P < 0.01 hypoxia versus normoxia. Means and standard deviations are shown
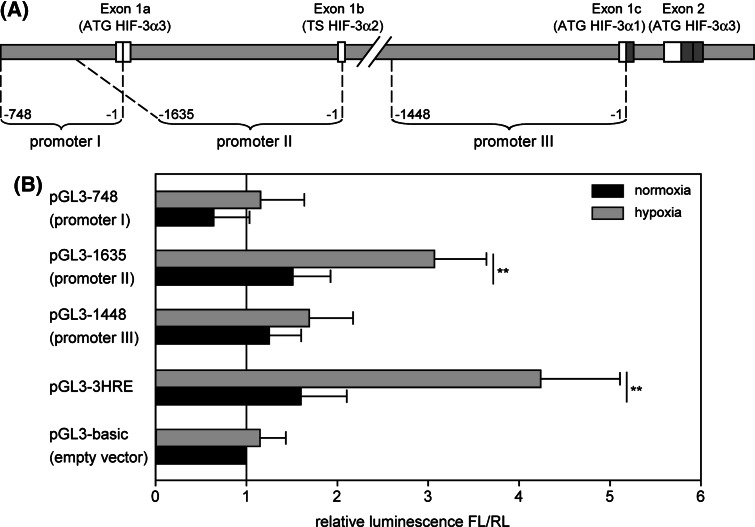
Determination of functional promoter elements responsible for the hypoxia-mediated HIF-3α induction
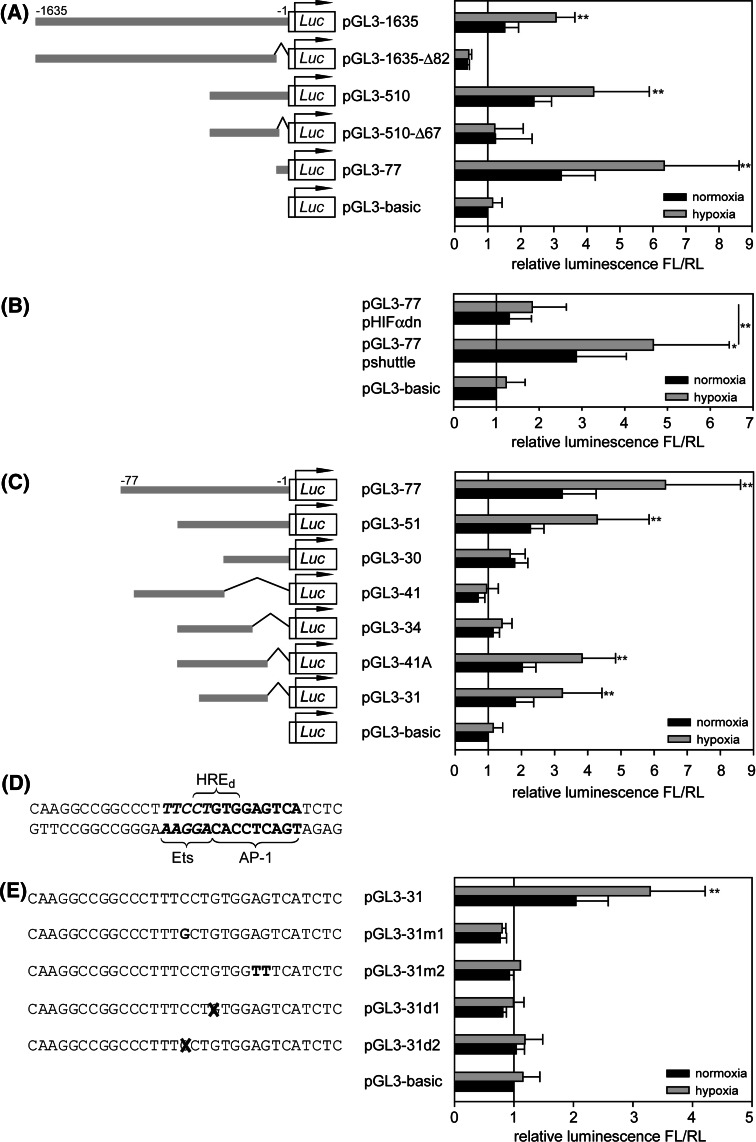
Computational analysis of the human HIF-3α gene locus with the Genomatix software package (Genomatix Software, Germany) identified three potential promoter regions (Fig. 5a). These regions were cloned into the firefly luciferase reporter gene vector pGL3-basic and transiently transfected into HUVEC. pGL3-HRE, a luciferase vector with three consecutive conserved hypoxia response elements (HRE) from phosphoglycerate kinase gene, served as positive control. The luciferase expression was determined under normoxic and hypoxic conditions. Only one promoter sequence was identified to be able to drive hypoxic induction of luciferase (Fig. 5b). This sequence, labeled promoter II, encloses 1,635 bp upstream of the transcription start point of exon 1b and could be identified to control the expression of the isoform HIF-3α2. To precisely map the cis-acting element of HIF-3α promoter II, successive 5′- and 3′-deletions of promoter sequence were generated. HUVECs, transfected with constructs pGL3-1635, pGL3-510 and pGL3-77, which are 5′-truncated promoter sequences from 1,635 to 77 bp length, always show an intact hypoxia-inducible luciferase expression (Fig. 6a). On the other hand, removal of the 77-bp region in construct pGL3-1635-Δ82 and pGL3-510-Δ67 results in a complete loss of hypoxic luciferase induction, documenting the importance of this sequence as a promoter in hypoxia. Cotransfection of pGL3-77 with pHIFαdn, overexpressing dominant-negative HIFαdn, leads to significant loss of promoter activity, underlining the necessity of HIF signaling for luciferase expression (Fig. 6b). To further characterize the 77-bp region, additional deletion constructs were generated. Measurement of luciferase expression shows that hypoxic response is still present in a 31-bp subfragment (Fig. 6c). Searches for transcription factor binding sites with various algorithms (Genomatix, TESS and PROMO) were performed. Surprisingly, the fragment does not contain a conserved, but only a degenerated HRE with two sequence mismatches. Furthermore, potential binding sites for Ets and AP-1 transcription factors were detected (Fig. 6d). Point mutations in either the Ets (pGL3-31m1) or the AP-1 (pGL3-31m2) core sequence and single base deletions (pGL3-31d1, pGL3-31d2) were generated. The analysis of luciferase expression shows abolished promoter activities for all of these constructs, leading to the conclusion that both transcription factors could be involved in HIF-3α regulation (Fig. 6e).
Fig. 5.
Detection of hypoxia-inducible HIF-3α promoter. a Putative promoter sequences upstream of exon 1a, exon 1b and exon 1c were cloned into pGL3-basic encoding firefly luciferase. b Luciferase activity of transfected HUVEC under normoxic and hypoxic conditions. pGL3-3HRE expressing firefly luciferase under control of three consecutive hypoxia response elements is used as positive control. Promoter activities are calculated as quotient of firefly luciferase and renilla luciferase luminescence levels relative to pGL3-basic (n ≥ 4). *P < 0.025, **P < 0.005 hypoxia versus normoxia, Bonferroni correction (k = 2) considered. TS transcription start. Means and standard deviations are shown
Fig. 6.
Identification of functional transcription factor binding sites of HIF-3α promoter II. Sequences corresponding to specific promoter fragments were cloned into pGL3-basic and transfected into HUVEC. Promoter activities are calculated as quotient of firefly luciferase and renilla luciferase luminescence levels relative to pGL3-basic. a Successive deletions of HIF-3α promoter II (n ≥ 3). b Contribution of HIF pathway to promoter activity of the 77-bp element. pHIFαdn or pshuttle (control) were co-transfected with pGL3-77 into HUVEC (n = 7). c Deletions of 77-bp promoter fragment (n ≥ 3). d Sequence of 31-bp promoter element with labeled potential transcription factor binding sites. HREd, degenerated HRE; e mutations and deletions of the 31-bp sequence in the potential Ets and AP-1 recognition sites revealed inhibition of luciferase expression to basic level (n ≥ 3). *P < 0.025, **P < 0.005 hypoxia versus normoxia, Bonferroni correction (k = 2) considered. Means and standard deviations are shown
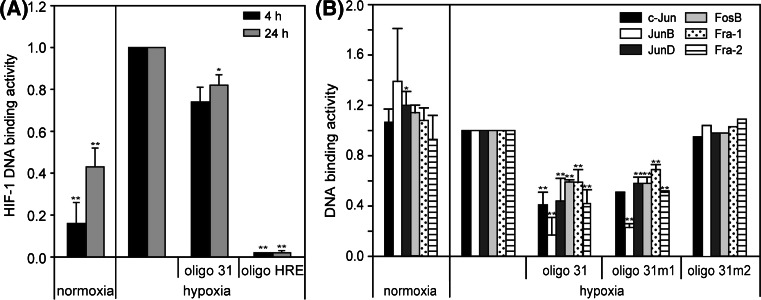
To verify whether the degenerated HRE has the ability to bind HIF-1, an ELISA-based HIF-1 DNA binding assay (TransAM™ HIF-1, Active Motif, Belgium) was performed (Fig. 7a). Using nuclear extracts from HUVEC treated with normoxia/hypoxia, a significant induction of HIF-1 activity under hypoxia is shown. Applying the 31-bp promoter element as competitor, HIF-1 DNA binding activity is only slightly decreased. In contrast, competitor with a conserved HRE from phosphoglycerate kinase promoter causes a nearly complete inhibition of HIF-1 DNA binding. Concluding from this experiment, HIF-1 does not effectively bind the 31-bp fragment, and the degenerated HRE is in all likelihood not responsible for direct promoter activation.
Fig. 7.
TransAM™ DNA binding assay. Binding activities were calculated as relative values to hypoxia. a HUVECs were cultivated under normoxia and hypoxia for 4 and 24 h, nuclear protein extracts were isolated, and HIF-1 DNA binding activity was measured. Data were shown as values relative to hypoxia. For competitive binding experiments oligo 31 and oligo HRE were applied. b HUVECs were cultivated under normoxia and hypoxia for 24 h, nuclear protein extracts were isolated, and AP-1 family DNA binding activity was measured. For competitive binding experiments oligo 31, oligo 31m1 and oligo 31m2 were applied. *P < 0.05, **P < 0.01 versus hypoxia. Means and standard deviations are shown
For the estimation of the involvement of AP-1 family members in hypoxic regulation, nuclear protein extracts of normoxic and hypoxic HUVEC were analyzed for activity of c-Jun, JunB, JunD, FosB, c-Fos, Fra-1 and Fra-2 with the TransAM™ AP-1 family transcription factor assay kit (Active Motif, Belgium). As shown in Fig. 7b, all transcription factors except for c-Fos are detectable, but neither of them shows a hypoxia-dependent regulation after 24 h hypoxia. Using the 31-bp fragment as competitor results in a partial, but significant decrease of DNA-binding activity. Competition with oligo 31m1, which is mutated in the Ets, but not in the AP-1 site, shows the same result. In contrast, applying oligo 31m2 with a mutated AP-1 recognition sequence fails in competition with the assay-delivered oligo. The results suggest that members of the AP-1 family could bind to the 31-bp promoter key element, but in consequence of the missing hypoxia responsiveness of measured AP-1 proteins, the specific mechanism of HIF-3α induction remains unclear.
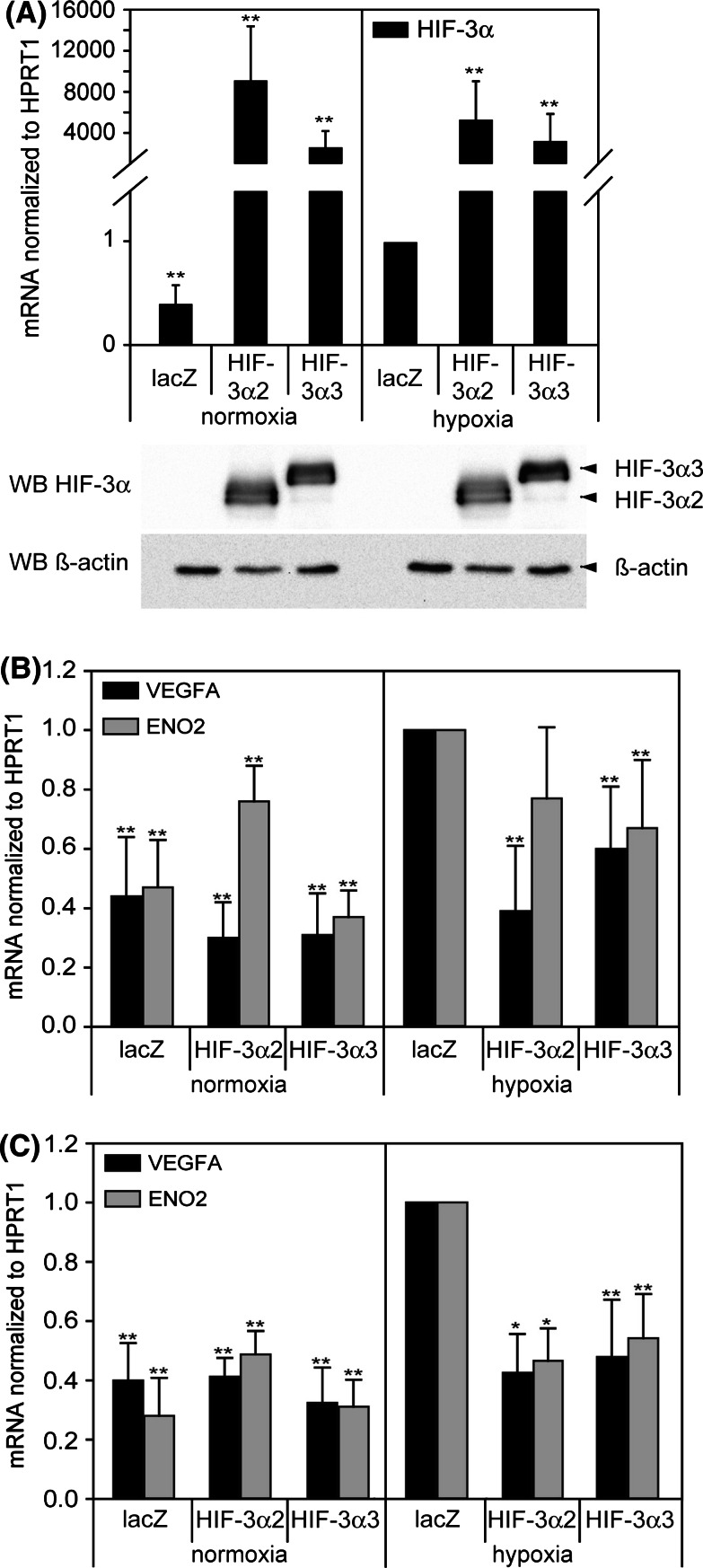
Overexpression of HIF-3α isoforms
Overexpression of HIF-3α isoforms was performed to verify the functionality as negative modulators of the HIF-signaling pathway in HUVEC and VSMC (Fig. 8). Adenoviral overexpression of isoforms HIF-3α2 and HIF-3α3, found to be expressed in HUVEC and VSMC (Fig. 8a), was performed, and the expression of known HIF-regulated genes (VEGFA, ENO2) was measured under normoxic and hypoxic conditions. As shown in Fig. 8b and c, normoxic expression of HIF-target genes is not altered by HIF-3α overexpression. In contrast, overexpression of both HIF-3α isoforms under hypoxia causes a significant reduction of hypoxia-induced VEGFA and ENO2 expression. This finding indicates that both Hif-3α isoforms are able to operate as antagonists of the HIF-pathway in human vascular cells. Only one exception to this principle could be detected: overexpression of HIF-3α2 in HUVECs revealed increased ENO2 expression under normoxia und only slightly decreased levels of ENO2 under hypoxia.
Fig. 8.
Confirmation of inhibitory behavior of overexpressed HIF-3α2 and HIF-3α3 on expression of HIF target genes VEGFA and ENO2 in HUVEC (b) and VSMC (c). Cells were infected with adenovirus overexpressing lacZ (control), HIF-3α2 or HIF-3α3, and subsequently exposed to normoxia/hypoxia for 24 h. a Overexpression of HIF-3α isoforms in HUVEC was measured by real-time RT-PCR and Western blot. b, c Expression of VEGFA and ENO2 mRNA was analyzed by real-time RT-PCR and calculated as relative values to hypoxia/lacZ. *P < 0.05, **P < 0.01 versus hypoxia/lacZ. Means and standard deviations are shown
Inhibition of HIF-3α in endothelial cells by shRNA
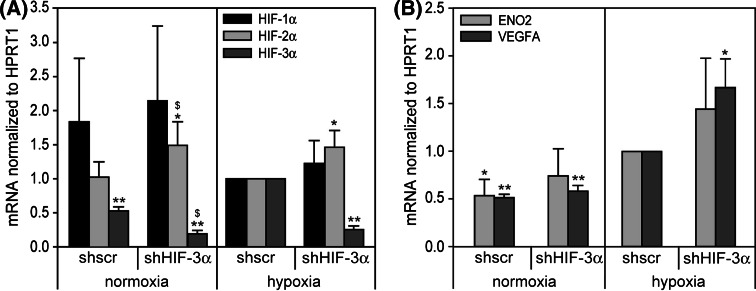
To support the finding of an inhibitory function of HIF-3α in endothelial cells, the endogenously expressed HIF-3α was silenced by adenovirally expressed small hairpin RNA (shRNA). Various shRNAs were tested for their ability to knock down HIF-3α in endothelial cells. One of the shRNAs (target sequence from 1,083 to 1,103 of NM_022462), targeting all long HIF-3α isoforms, showed a knock-down of HIF-3α mRNA to about 25% (Fig. 9a) and was used for further experiments. Subsequently the expression of HIF-target genes VEGFA and ENO2 was measured under normoxic and hypoxic conditions (Fig. 9b). The graph shows elevated expressions of the HIF target VEGFA, supporting the inhibitory function of HIF-3α. Interestingly, ENO2 is slightly but not significantly induced.
Fig. 9.
Adenoviral expression of shRNA targeting HIF-3α in HUVEC. Cells were infected with adenovirus expressing shscr (scrambled control shRNA) or shHIF-3α for 48 h and subsequently exposed to normoxia/hypoxia for 24 h. a Expression of HIF-α subunits was measured by real-time RT-PCR calculated as relative values to hypoxia/lacZ. b Expression of ENO2 and VEGFA mRNA was analyzed by real-time RT-PCR and calculated as relative values to hypoxia/lacZ. *P < 0.05, **P < 0.01 versus hypoxia/shscr. $ P < 0.05 vs.normoxia/shscr. Means and standard deviations are shown
Discussion
The expression and regulation of human HIF-3α in vascular cells is characterized for the first time in this article. The salient finding of this study is the characterization of an autoregulatory function of HIF-3α2 via a hypoxia-dependent process.
HIF-3α was detected in various, especially strongly vascularized tissues such as heart, lung and kidney [21, 23, 24, 36], but the contribution of distinct cell types to the measured overall HIF-3α expression is poorly characterized. A more comprehensive analysis exists only for mice HIF-3α. Yamashita et al. [36] addressed the effects of mouse NEPAS/HIF-3α disruption to endothelial cells of vessels and capillaries in alveoli of lung and alveolar epithelium. In contrast, in human tissues the characterization of HIF-3α shows only a few details [25]. A helpful tool for the association of HIF-3α expression to different human tissues is the analysis of EST counts, showing HIF-3α expression in tissues such as heart, placenta or lung. Nevertheless, the in silico analysis of tissue distribution gives no hint of specific cell types engaged in HIF-3α expression. The present study revealed for the first time that HIF-3α is expressed in human endothelial cells (EC) as well as vascular smooth muscle cells (VSMC), indicating the important role of vascular cells for a response to changed oxygen levels.
EC and VSMC exhibit a significant, hypoxia-mediated upregulation of HIF-3α mRNA (Fig. 1). Using HUVEC as a model, the regulation of this induction was examined in more detail. In agreement with other authors [29, 37], the present study found a HIF-dependent upregulation of HIF-3α using a dominant-negative HIF-2α mutant (HIFαdn) to block the HIF-pathway (Fig. 2c). In addition, activation of the HIF-pathway by normoxic stabilization of α-subunits with the PHD inhibitor DMOG causes a normoxic stabilization of HIF-1α and HIF-2α and subsequently increased HIF-3α mRNA levels in the same time frame compared to hypoxia (Fig. 2b). Additionally, the silencing of both α-subunits, HIF-1α and HIF-2α, results in decreased HIF-3α levels (Fig. 2f). The discrepancy between the complete inhibition of HIF-3α to normoxic levels by HIFαdn and the only partial inhibition by siRNA might be explainable by a different mechanism of action. The siRNA reduces absolute mRNA and protein levels, but it is not definitely a complete inhibition. In contrast, the overexpression of HIFαdn with an adenovirus has two effects: the transfection efficiency is nearly 100%, and HIFαdn blocks the HIF signaling on functional level.
Nevertheless, the hypoxic induction of HIF-3α could not depend on HIF-pathway alone. As mentioned before, the expression of HIF-3α is restricted to certain cell types: macrophages do not express HIF-3α, and HELA cells show only low level HIF-3α mRNA without hypoxic upregulation, although both cell types express HIF-1α and HIF-2α in high amounts. It seems that additional cell-specific factors play a role in HIF-3α regulation. As shown in Fig. 3a-c, the inhibition of another transcription factor, NFκB, leads to a partial downregulation of hypoxia-dependent HIF-3α induction. Significantly decreased mRNA levels of HIF-1α and -2α and slightly reduced HIF-1α and HIF-2α protein amounts during hypoxia were observed under NFκB repression. As recently published, NFκB is able to regulate basal HIF-1α expression, and complete inhibition of NFκB impairs hypoxia-induced HIF-1α levels [38–40]. Additionally, the inhibition of PHD-1 by hypoxia, DMOG or siRNA leads to an activation of NFκB. This activation is independent of the known stabilization of HIF-α subunits [41]. Therefore, it is conceivable that more than one factor may contribute to the HIF-3α induction in HUVEC. Hypoxia-induced PHD inhibition represents the first step on this path. On the one hand HIF-1α and HIF-2α are stabilized leading to increased HIF-3α expression; on the other hand NFκB is activated and in turn able to induce HIF-1 activity. NFκB, however, also could activate transcription factors other than HIF-1, at last responding with upregulation of HIF-3α under hypoxia.
The expression and regulation of distinct human HIF-3α isoforms in vascular cells were completely unknown until now. The characterization of the long isoforms HIF-3α1, HIF-3α2 and HIF-3α3 was performed in HUVEC, VSMC and compared to HELA cells (Fig. 4). It shows, that different isoforms contribute to the basic HIF-3α mRNA expression in HUVEC and VSMC, but only HIF-3α2 is inducible by hypoxia. In the following, these results were verified by analysis of the hypoxia-responsible promoter elements of human HIF-3α (Fig. 5). Makino et al. [37] showed that only one promoter of mouse HIF-3α gene locus is susceptible to hypoxia: IPAS, but not HIF-3α is upregulated under hypoxia. Furthermore, the recently discovered HIF-3α isoform NEPAS contains the same first exon as IPAS and undergoes a similar hypoxic upregulation [36]. Analogous to our results, Tanaka et al. recently identified one human isoform, HIF-3α2, which is hypoxia-inducible. Analysis of the responsible HIF-3α promoter in Hep3B cells revealed a functional hypoxia response element, which is bound by HIF-1 [29]. This HRE is also present in our promoter sequence of HIF-3α2 (pGL3-1635) at position −1,133 to −1,137. Interestingly, this potential HRE is not functional for hypoxic upregulation of HIF-3α2 in our cell model. This is documented by the fact that deletion of this element in construct pGL3-510 does not abolish hypoxic luciferase expression (Fig. 6a). Nevertheless, subcloning of fragments from HIF-3α2 promoter shows another sequence of 77 bp, which mediates hypoxic induction in HUVEC. In agreement with data for overexpression of HIFαdn (Fig. 2c), the hypoxic induction of reporter gene expression under control of this 77-bp sequence depends on HIF-signaling (Fig. 6b). Further deletions of this fragment reveal a 31-bp stretch, which is still responsive to hypoxia. Sequence analysis with different databases detecting potential transcription factor binding sites identified conserved motifs for Ets and AP-1 factors, but not for HIF-1/HIF-2. In comparison, the mouse IPAS promoter permits a direct interaction of HIF-1 with an HRE-like motif (5′-GGGTG-3′)[37]. This motif does not represent a perfect match to the HRE consensus sequence; therefore the 31-bp fragment was analyzed for a degenerated HRE, which is indeed located within this region. This sequence (5′-CTGTG-3′) differs in two bases from conventional core sequence 5′-RCGTG-3′. A deletion of the G at the third position abrogates luciferase expression (pGL3-31d1, Fig. 6e), strongly suggesting a pivotal role of this element in the hypoxic response of the HIF-3α promoter. In contrast, the investigation of published HIF-1/HIF-2 binding sequences of human, mouse and rat promoters (BIOBASE Knowledge Library and Transfac database) shows that beside the strictly conserved bases on position 3–5, also the first and second position are highly conserved [42]. Only 2 out of 70 hypoxia response elements vary at the second position; one of them is the previously described element of mouse IPAS promoter [37]. Additionally, none of the HRE shows two exceptions from the consensus, which leads to the conclusion that the degenerated HRE of the 31-bp fragment may not be functional. Nevertheless, to test this hypothesis the binding activity of HIF-1 to a conserved and to the degenerated HRE was determined (Fig. 7). It was shown that HIF-1 does not effectively bind to the 31-bp sequence. This result opened the question of whether other transcription factors may be involved in the hypoxic regulation of HIF-3α.
As mentioned above, two other potential transcription factor binding sites were identified in the HIF-3α promoter element, i.e., AP-1 and Ets (Fig. 6d). The AP-1 family is a prominent representative of transcription factors contributing to hypoxic signaling. Activation of AP-1 has been reported for a wide variety of cell lines and primary cell cultures [43–49]. Furthermore, the Ets family has been described as participating in hypoxic signaling and plays an important role in the association with HIF-2 target gene selection [50–55]. The mutation of core nucleotides of either the AP-1 or Ets recognition sequence completely abrogated promoter activation (Fig. 6e), suggesting that both transcription factors may be involved in HIF-3α regulation. As shown in Fig. 7, the 31-bp promoter fragment is able to compete partially with a consensus AP-1 motif for AP-1 transcription factors; a 31-bp sequence with a mutated AP-1 site is not able to mediate this effect. Nevertheless, none of the AP-1 proteins analyzed seems to be activated by hypoxia under conditions used in the present model. These data clearly suggest that AP-1 alone does not suffer for HIF-3α modulation.
Taken together, the regulation of HIF-3α was shown to be cell-specific, and the data indicate that transcription factors in addition to HIF-1/-2 are engaged in the hypoxia-mediated regulation of HIF-3α in HUVEC: (1) Highly expressed and in hypoxia stabilized HIF-1α and HIF-2α, as detected in HUVEC and in VSMC, but also in macrophages and in HELA cells (data not shown), are not sufficient for HIF-3α regulation. These conclusions are based on the finding that no relevant amounts of HIF-3α mRNA in macrophages were detected. This holds true for normoxic and hypoxic conditions. Furthermore, we could not find a hypoxic upregulation of HIF-3α in HELA cells. (2) Promoter analysis in HUVEC failed to detect of a functional HIF binding site, but revealed other motifs, potentially involved in HIF-3α regulation. Summarizing these data of the promoter analysis, it is shown that increased HIF-3α2 expression under hypoxia depends on HIF-signaling, but in contrast to mouse IPAS/NEPAS, HIF-1 could not bind directly to the HIF-3α2 promoter in endothelial cells. Therefore HIF-1/HIF-2 is hypothesized to be involved in other ways, which might be the transcriptional induction of other transcription factors or the interaction of HIF-1/HIF-2 with other factors. Whether these are cell-specific and independent from hypoxia remains to be elucidated.
For the first time, this study estimates the function of HIF-3α in primary EC and VSMC by overexpression of the endogenously found long isoforms HIF-3α2 and HIF-3α3. Compared to HIF-3α3, the inclusion of exon 1b in HIF-3α2 causes a shift of the translation start point to exon 2 and leads to a deletion of the bHLH DNA binding region. This allows the elucidation of whether the truncation of the bHLH domain causes a modification of HIF-3α function. As a functional read-out the expression of the typical HIF target genes VEGFA and ENO2 was used (Fig. 8b, c). These experiments revealed that both HIF-3α isoforms act as negative regulators of HIF signaling. It is noticeable that binding to the HRE mediated by the bHLH domain is not crucial for the negative effect of HIF-3α in vascular smooth muscle cells. The specific effect of HIF-3α2 in HUVEC with increasing ENO2 levels under normoxia and only slightly decreased levels under hypoxia needs further elucidation. HIF-3α isoforms are described as dimerizing with HIF-1β, but also with HIF-1α or HIF-2α [22, 26, 27]. Until now the binding partners of specific HIF-3α isoforms in specific cells have been largely unknown. ENO2 is predominantly regulated by HIF-1 and not HIF-2 (Fig. 2e). Possibly HIF-3α2 has a higher affinity to HIF-2α than to HIF-1α or HIF-1β in HUVEC. Interestingly, after inhibition of HIF-3α by shRNA expression the ENO2 expression under hypoxia in HUVEC shows in turn no significant increase (Fig. 9b). Because only HIF-3α2 expression depends on hypoxia in HUVEC and this isoform does not effectively decrease the expression of ENO2 as shown by overexpression, we would speculate that ENO2 regulation in HUVEC is largely non-influenceable by HIF-3α2. Nevertheless, targeting HIF-3α with shRNA in HUVEC causes an increase of VEGFA expression, supporting the inhibitory behavior of HIF-3α on HIF signaling (Fig. 9b).
In summary, these data show that HIF-3α2 is the only long isoform that is induced by hypoxia in vascular cells. In comparison to recently published data [29], the novel finding is an alternative regulation of HIF-3α2. This regulation seems specific for endothelial cells and is mediated by more than one transcription factor involving HIF and NFκB. Also members of the AP-1 family may be involved. Finally, this study proves that HIF-3α2 and HIF-3α3 are negative regulators able to abolish an overactivation of the HIF pathway. This finding underlines the mechanisms of a multiple-level controlled HIF-signaling in vascular cells under low oxygen tensions. The pathophysiological impact of this newly characterized mechanism in processes such as atherosclerosis or angiogenesis remains to be elucidated.
Acknowledgments
We thank the Cardiosurgical Department at the Heart Center Dresden for providing tissue of human aortas at the time of coronary bypass grafting and Anita Männel for excellent technical assistance.
Conflict of interest
None declared.
Abbreviations
- HIF
Hypoxia-inducible factor
- HRE
Hypoxia response element
- HUVEC
Human umbilical venous endothelial cell
- VSMC
Vascular smooth muscle cell
- DMSO
Dimethylsulfoxid
- DMOG
Dimethyloxalylglycine
- PHD
Prolylhydroxylases
- Ets
e26 transformation-specific transcription factor
- AP-1
Activating protein-1 transcription factor
References
- 1.Bjornheden T, Levin M, Evaldsson M, Wiklund O. Evidence of hypoxic areas within the arterial wall in vivo. Arterioscler Thromb Vasc Biol. 1999;19:870–876. doi: 10.1161/01.ATV.19.4.870. [DOI] [PubMed] [Google Scholar]
- 2.Celletti FL, Waugh JM, Amabile PG, Brendolan A, Hilfiker PR, Dake MD. Vascular endothelial growth factor enhances atherosclerotic plaque progression. Nat Med. 2001;7:425–429. doi: 10.1038/86490. [DOI] [PubMed] [Google Scholar]
- 3.Chen F, Eriksson P, Kimura T, Herzfeld I, Valen G. Apoptosis and angiogenesis are induced in the unstable coronary atherosclerotic plaque. Coron Artery Dis. 2005;16:191–197. doi: 10.1097/00019501-200505000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Sluimer JC, Gasc JM, van Wanroij JL, Kisters N, Groeneweg M, Sollewijn G, Cleutjens JP, van den Akker LH, Corvol P, Wouters BG, Daemen MJ, Bijnens AP. Hypoxia, hypoxia-inducible transcription factor, and macrophages in human atherosclerotic plaques are correlated with intraplaque angiogenesis. J Am Coll Cardiol. 2008;51:1258–1265. doi: 10.1016/j.jacc.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 5.Luque A, Turu M, Juan-Babot O, Cardona P, Font A, Carvajal A, Slevin M, Iborra E, Rubio F, Badimon L, Krupinski J. Overexpression of hypoxia/inflammatory markers in atherosclerotic carotid plaques. Front Biosci. 2008;13:6483–6490. doi: 10.2741/3168. [DOI] [PubMed] [Google Scholar]
- 6.Vink A, Schoneveld AH, Lamers D, Houben AJ, van der GP, van Diest PJ, Pasterkamp G. HIF-1 alpha expression is associated with an atheromatous inflammatory plaque phenotype and upregulated in activated macrophages. Atherosclerosis. 2007;195:e69–e75. doi: 10.1016/j.atherosclerosis.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Higashida T, Kanno H, Nakano M, Funakoshi K, Yamamoto I. Expression of hypoxia-inducible angiogenic proteins (hypoxia-inducible factor-1alpha, vascular endothelial growth factor, and E26 transformation-specific-1) and plaque hemorrhage in human carotid atherosclerosis. J Neurosurg. 2008;109:83–91. doi: 10.3171/JNS/2008/109/7/0083. [DOI] [PubMed] [Google Scholar]
- 8.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 9.Jiang M, Wang B, Wang C, He B, Fan H, Guo TB, Shao Q, Gao L, Liu Y. Inhibition of hypoxia-inducible factor-1 alpha and endothelial progenitor cell differentiation by adenoviral transfer of small interfering RNA in vitro. J Vasc Res. 2006;43:511–521. doi: 10.1159/000095964. [DOI] [PubMed] [Google Scholar]
- 10.Yamakawa M, Liu LX, Date T, Belanger AJ, Vincent KA, Akita GY, Kuriyama T, Cheng SH, Gregory RJ, Jiang C. Hypoxia-inducible factor-1 mediates activation of cultured vascular endothelial cells by inducing multiple angiogenic factors. Circ Res. 2003;93:664–673. doi: 10.1161/01.RES.0000093984.48643.D7. [DOI] [PubMed] [Google Scholar]
- 11.Skuli N, Liu L, Runge A, Wang T, Yuan L, Patel S, Iruela-Arispe L, Simon MC, Keith B. Endothelial deletion of hypoxia-inducible factor-2{alpha} (HIF-2{alpha}) alters vascular function and tumor angiogenesis. Blood. 2009;114:469–477. doi: 10.1182/blood-2008-12-193581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng J, Zhang L, Drysdale L, Fong GH. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc Natl Acad Sci USA. 2000;97:8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp Mol Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 15.Maynard MA, Qi H, Chung J, Lee EH, Kondo Y, Hara S, Conaway RC, Conaway JW, Ohh M. Multiple splice variants of the human HIF-3 alpha locus are targets of the von Hippel–Lindau E3 ubiquitin ligase complex. J Biol Chem. 2003;278:11032–11040. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- 16.Semenza GL. HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell. 2001;107:1–3. doi: 10.1016/S0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 17.Siddiq A, Aminova LR, Ratan RR. Hypoxia inducible factor prolyl 4-hydroxylase enzymes: center stage in the battle against hypoxia, metabolic compromise and oxidative stress. Neurochem Res. 2007;32:931–946. doi: 10.1007/s11064-006-9268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J Biol Chem. 2006;281:15215–15226. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- 20.Scheurer SB, Rybak JN, Rosli C, Neri D, Elia G. Modulation of gene expression by hypoxia in human umbilical cord vein endothelial cells: a transcriptomic and proteomic study. Proteomics. 2004;4:1737–1760. doi: 10.1002/pmic.200300689. [DOI] [PubMed] [Google Scholar]
- 21.Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr. 1998;7:205–213. [PMC free article] [PubMed] [Google Scholar]
- 22.Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 23.Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3alpha locus. J Biol Chem. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 24.Hara S, Hamada J, Kobayashi C, Kondo Y, Imura N. Expression and characterization of hypoxia-inducible factor (HIF)-3alpha in human kidney: suppression of HIF-mediated gene expression by HIF-3alpha. Biochem Biophys Res Commun. 2001;287:808–813. doi: 10.1006/bbrc.2001.5659. [DOI] [PubMed] [Google Scholar]
- 25.Li QF, Wang XR, Yang YW, Lin H. Hypoxia upregulates hypoxia inducible factor (HIF)-3alpha expression in lung epithelial cells: characterization and comparison with HIF-1alpha. Cell Res. 2006;16:548–558. doi: 10.1038/sj.cr.7310072. [DOI] [PubMed] [Google Scholar]
- 26.Jang MS, Park JE, Lee JA, Park SG, Myung PK, Lee dH, Park BC, Cho S. Binding and regulation of hypoxia-inducible factor-1 by the inhibitory PAS proteins. Biochem Biophys Res Commun. 2005;337:209–215. doi: 10.1016/j.bbrc.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 27.Maynard MA, Evans AJ, Shi W, Kim WY, Liu FF, Ohh M. Dominant-negative HIF-3 alpha 4 suppresses VHL-null renal cell carcinoma progression. Cell Cycle. 2007;6:2810–2816. doi: 10.4161/cc.6.22.4947. [DOI] [PubMed] [Google Scholar]
- 28.Maynard MA, Evans AJ, Hosomi T, Hara S, Jewett MA, Ohh M. Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J. 2005;19:1396–1406. doi: 10.1096/fj.05-3788com. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka T, Wiesener M, Bernhardt W, Eckardt KU, Warnecke C. The human HIF (hypoxia-inducible factor)-3alpha gene is a HIF-1 target gene and may modulate hypoxic gene induction. Biochem J. 2009;424:143–151. doi: 10.1042/BJ20090120. [DOI] [PubMed] [Google Scholar]
- 30.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest. 1973;52:2745–2756. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willert M, Augstein A, Poitz DM, Schmeisser A, Strasser RH, Braun-Dullaeus RC. Transcriptional regulation of Pim-1 kinase in vascular smooth muscle cells and its role for proliferation. Basic Res Cardiol. 2010;105:267–277. doi: 10.1007/s00395-009-0055-x. [DOI] [PubMed] [Google Scholar]
- 32.Schug J (2003) Using TESS to predict transcription factor binding sites in DNA sequence. In: Baxevanis AD (ed) Current protocols in bioinformatics. Wiley, Hoboken, NJ [DOI] [PubMed]
- 33.Maemura K, Hsieh CM, Jain MK, Fukumoto S, Layne MD, Liu Y, Kourembanas S, Yet SF, Perrella MA, Lee ME. Generation of a dominant-negative mutant of endothelial PAS domain protein 1 by deletion of a potent C-terminal transactivation domain. J Biol Chem. 1999;274:31565–31570. doi: 10.1074/jbc.274.44.31565. [DOI] [PubMed] [Google Scholar]
- 34.Traenckner EB, Pahl HL, Henkel T, Schmidt KN, Wilk S, Baeuerle PA. Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita T, Ohneda O, Nagano M, Iemitsu M, Makino Y, Tanaka H, Miyauchi T, Goto K, Ohneda K, Fujii-Kuriyama Y, Poellinger L, Yamamoto M. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol Cell Biol. 2008;28:1285–1297. doi: 10.1128/MCB.01332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makino Y, Uenishi R, Okamoto K, Isoe T, Hosono O, Tanaka H, Kanopka A, Poellinger L, Haneda M, Morimoto C. Transcriptional up-regulation of inhibitory PAS domain protein gene expression by hypoxia-inducible factor 1 (HIF-1): a negative feedback regulatory circuit in HIF-1-mediated signaling in hypoxic cells. J Biol Chem. 2007;282:14073–14082. doi: 10.1074/jbc.M700732200. [DOI] [PubMed] [Google Scholar]
- 38.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Uden P, Kenneth NS, Rocha S. Regulation of hypoxia-inducible factor-1alpha by NF-kappaB. Biochem J. 2008;412:477–484. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonello S, Zahringer C, BelAiba RS, Djordjevic T, Hess J, Michiels C, Kietzmann T, Gorlach A. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler Thromb Vasc Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 41.Cummins EP, Berra E, Comerford KM, Ginouves A, Fitzgerald KT, Seeballuck F, Godson C, Nielsen JE, Moynagh P, Pouyssegur J, Taylor CT. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci USA. 2006;103:18154–18159. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamashita K, Discher DJ, Hu J, Bishopric NH, Webster KA. Molecular regulation of the endothelin-1 gene by hypoxia. Contributions of hypoxia-inducible factor-1, activator protein-1, GATA-2, AND p300/CBP. J Biol Chem. 2001;276:12645–12653. doi: 10.1074/jbc.M011344200. [DOI] [PubMed] [Google Scholar]
- 44.Minet E, Michel G, Mottet D, Piret JP, Barbieux A, Raes M, Michiels C. c-JUN gene induction and AP-1 activity is regulated by a JNK-dependent pathway in hypoxic HepG2 cells. Exp Cell Res. 2001;265:114–124. doi: 10.1006/excr.2001.5180. [DOI] [PubMed] [Google Scholar]
- 45.Michiels C, Minet E, Michel G, Mottet D, Piret JP, Raes M. HIF-1 and AP-1 cooperate to increase gene expression in hypoxia: role of MAP kinases. IUBMB Life. 2001;52:49–53. doi: 10.1080/15216540252774766. [DOI] [PubMed] [Google Scholar]
- 46.Fradette C, Du SP. Hypoxia-inducible factor-1 and activator protein-1 modulate the upregulation of CYP3A6 induced by hypoxia. Br J Pharmacol. 2003;140:1146–1154. doi: 10.1038/sj.bjp.0705543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fantozzi I, Zhang S, Platoshyn O, Remillard CV, Cowling RT, Yuan JX. Hypoxia increases AP-1 binding activity by enhancing capacitative Ca2+ entry in human pulmonary artery endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1233–L1245. doi: 10.1152/ajplung.00445.2002. [DOI] [PubMed] [Google Scholar]
- 48.Bandyopadhyay RS, Phelan M, Faller DV. Hypoxia induces AP-1-regulated genes and AP-1 transcription factor binding in human endothelial and other cell types. Biochim Biophys Acta. 1995;1264:72–78. doi: 10.1016/0167-4781(95)00116-x. [DOI] [PubMed] [Google Scholar]
- 49.Ausserer WA, Bourrat-Floeck B, Green CJ, Laderoute KR, Sutherland RM. Regulation of c-jun expression during hypoxic and low-glucose stress. Mol Cell Biol. 1994;14:5032–5042. doi: 10.1128/mcb.14.8.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aprelikova O, Wood M, Tackett S, Chandramouli GV, Barrett JC. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 2006;66:5641–5647. doi: 10.1158/0008-5472.CAN-05-3345. [DOI] [PubMed] [Google Scholar]
- 51.Dutta D, Ray S, Vivian JL, Paul S. Activation of the VEGFR1 chromatin domain: an angiogenic signal-ETS1/HIF-2alpha regulatory axis. J Biol Chem. 2008;283:25404–25413. doi: 10.1074/jbc.M804349200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elvert G, Kappel A, Heidenreich R, Englmeier U, Lanz S, Acker T, Rauter M, Plate K, Sieweke M, Breier G, Flamme I. Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1) J Biol Chem. 2003;278:7520–7530. doi: 10.1074/jbc.M211298200. [DOI] [PubMed] [Google Scholar]
- 53.Li B, Lager J, Wang D, Li D. Ets-1 participates in and facilitates developmental expression of hypoxia-induced mitogenic factor in mouse lung. Front Biosci. 2007;12:2269–2278. doi: 10.2741/2229. [DOI] [PubMed] [Google Scholar]
- 54.Oikawa M, Abe M, Kurosawa H, Hida W, Shirato K, Sato Y. Hypoxia induces transcription factor ETS-1 via the activity of hypoxia-inducible factor-1. Biochem Biophys Res Commun. 2001;289:39–43. doi: 10.1006/bbrc.2001.5927. [DOI] [PubMed] [Google Scholar]
- 55.Salnikow K, Aprelikova O, Ivanov S, Tackett S, Kaczmarek M, Karaczyn A, Yee H, Kasprzak KS, Niederhuber J. Regulation of hypoxia-inducible genes by ETS1 transcription factor. Carcinogenesis. 2008;29:1493–1499. doi: 10.1093/carcin/bgn088. [DOI] [PMC free article] [PubMed] [Google Scholar]