Abstract
Structural symmetry is observed in the majority of fundamental protein folds and gene duplication and fusion evolutionary processes are postulated to be responsible. However, convergent evolution leading to structural symmetry has also been proposed; additionally, there is debate regarding the extent to which exact primary structure symmetry is compatible with efficient protein folding. Issues of symmetry in protein evolution directly impact strategies for de novo protein design as symmetry can substantially simplify the design process. Additionally, when considering gene duplication and fusion in protein evolution, there are two competing models: “emergent architecture” and “conserved architecture”. Recent experimental work has shed light on both the evolutionary process leading to symmetric protein folds as well as the ability of symmetric primary structure to efficiently fold. Such studies largely support a “conserved architecture” evolutionary model, suggesting that complex protein architecture was an early evolutionary achievement involving oligomerization of smaller polypeptides.
Keywords: Protein evolution, Protein symmetry, Protein design, Proteogenesis, Peptide motif, Tandem duplication
Symmetry in protein structure
Proteins, the essential workhorse molecules of living organisms, comprise typical proteome sizes of 1–4 × 104 unique proteins [1, 2]. However, examination of protein tertiary structure reveals the diverse application of a small number of common structural folds. Notably, the majority of such fundamental protein folds exhibit internal (i.e., rotational, Cn) structural symmetry. Among ten fundamental protein superfolds proposed by Thornton [3], six exhibit structural symmetry. The evolutionary origin of structural symmetry (in carp muscle calcium binding protein) was proposed by McLachlan [4] as having emerged from an ancient homo-oligomeric assembly into a single polypeptide chain via gene duplication and fusion events. Subsequent analyses of other proteins with evidence of primary or tertiary structure symmetry led to an increasing number of similar proposals (most notably by McLachlan) [5–13]. The discovery, for example, of four-helix up-down bundles in both homo-tetramer and homo-dimer forms provided compelling evidence for its evolution by a gene duplication and fusion event [13]. The rotational symmetry, Cn, observed in symmetric protein folds typically varies from n = 2–8 with examples of both even and odd values. A single gene duplication and fusion event can yield structural twofold (C2) symmetry, the simplest level of symmetry (e.g., type I dockerin [14]). Higher symmetry (e.g., n = 3–8), requires subsequent duplication and fusion events. Odd-number symmetry (e.g., n = 3, 5, 7) requires either incomplete (i.e., sub-domain) duplication and fusion, or duplication and fusion followed by a truncation event (which can also lead to structural circular permutation) [15, 16]. Domain repeats are thought to arise via tandem duplication within a gene, where a segment is duplicated and the copy is inserted next to its origin. Although the exact mechanism of tandem duplication is not fully understood, nonhomologous recombination appears most likely, and tandem duplication has been identified as a major evolutionary force in the emergence of extant protein architecture [17, 18]. Analyses show that proteins with domain repeats are abundant, with ~5 % of the proteome of Archaea and bacteria containing proteins with domain repeats (typically 2–9 repeats) and ~7–17 % of the proteome of eukaryotes containing proteins with domain repeats (typically 2–20+ repeats) [17]. Duplicated but non-fused genes are even more abundant, with 15–30 % of Archaea and bacteria genes, and 30–65 % of eukaryotic genes, showing evidence of duplication [19].
Symmetry in protein structure is of central interest not only to understanding protein evolution but also in the application of symmetric design principles to de novo protein design. The application of symmetry in de novo protein design substantially reduces the complexity of the design process; however, there remains uncertainty as to the limits of primary structure symmetry that will yield a foldable polypeptide [20–22]. Uncertainty regarding the foldability of symmetric primary structure calls into question the viability of tandem gene duplication in evolutionary processes, since immediately subsequent to gene duplication and fusion exact primary structure symmetry would exist. However, recent experimental work from several groups provides compelling support for gene duplication and fusion in the evolution of symmetric protein architecture, with support for an evolutionary model that is capable of yielding complex protein architecture with simple peptide motifs. Recent experimental work also shows that exact primary structure symmetry is not de facto incompatible with efficient protein folding, and is a critical result for both de novo protein design and models of protein evolution. Such experimental studies have been postulated to probe aspects of protein architecture and evolution prior to the last universal common ancestor (LUCA) of living systems.
Emergent versus conserved architecture evolutionary models
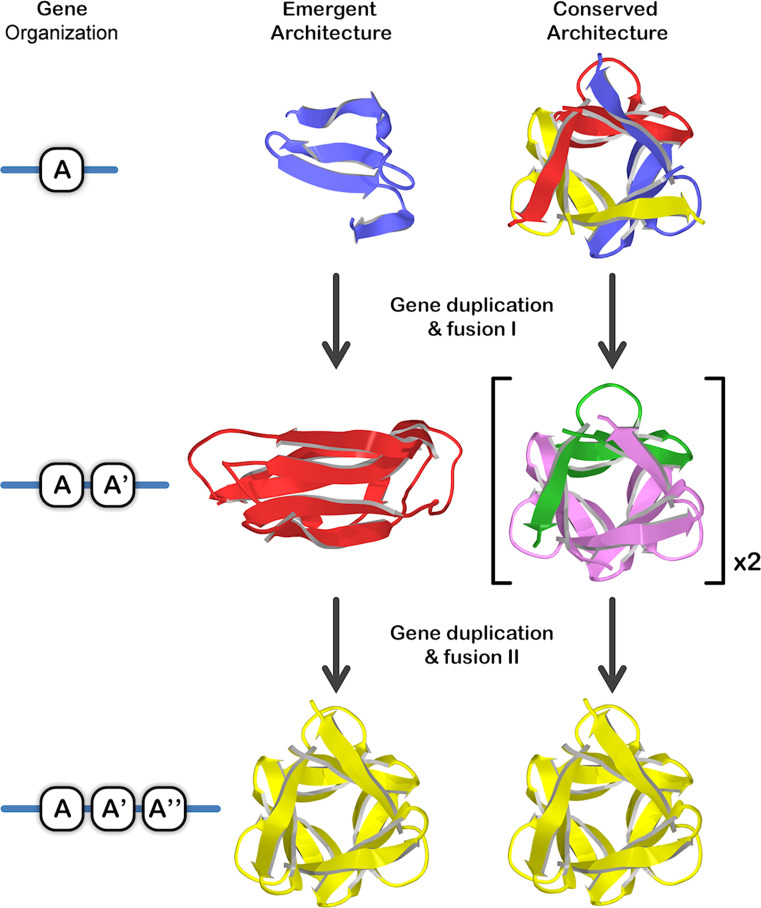
Two distinctly different types of models for the evolution of symmetric protein architecture from a primordial peptide motif, via gene duplication and fusion processes, have been proposed [23–29] and describe either an “emergent architecture” or a “conserved architecture” evolutionary pathway. The most detailed descriptions for the emergent and conserved architecture evolutionary models were proposed simultaneously by Mukhopadhyay [23] and Ponting and Russell [24], respectively, and each utilized the β-trefoil as the example symmetric protein fold. In the emergent architecture model for the β-trefoil fold a simple β4 peptide motif (i.e., comprising two ββ-hairpins [13]) autonomously folded to yield a correspondingly simple architecture, similar to the epidermal growth factor (EGF) fold (Fig. 1). A gene duplication and fusion event yielded a twofold symmetric (β4)2 architecture as an independently folding polypeptide, similar to the ecotin fold. A subsequent gene duplication and fusion event involving a β4 motif yielded the extant (β4)3 threefold symmetric β-trefoil fold. In contrast, in the conserved architecture model, the β4 peptide motif was hypothesized not to fold as an independent simple architecture, rather, it oligomerized as a (β4)3 homo-trimer to produce a complete β-trefoil fold (Fig. 1). A gene duplication and fusion event yielded a tandem repeat of this motif, which also folded as a homo-trimer to generate two intact, yet inter-connected (via some form of domain-swapping) β-trefoil folds. A subsequent gene duplication and fusion event involving a β4 motif (either by partial duplication or duplication followed by truncation) yielded the extant (β4)3 threefold symmetric β-trefoil fold as a single polypeptide. Corresponding structural details would hold for these alternative evolutionary models applied to other symmetric protein architectures such as the β-propeller and (βα)8-barrel symmetric protein folds. Whereas the emergent architecture model implies “simple gene, simple polypeptide motif, simple architecture”, the conserved architecture model implies “simple gene, simple polypeptide motif, complex architecture” (achieved via oligomerization); thus, one primary distinction of the conserved architecture model is that the extant symmetric architecture is a comparatively early evolutionary achievement enabling simple genes to nonetheless code for structurally complex protein folds. Experimental testing of such evolutionary models of symmetric protein architecture is based upon characterization of the folding and structural properties of isolated peptide motifs derived from extant symmetric proteins.
Fig. 1.
Comparison of emergent and conserved architecture evolutionary models (using the threefold symmetric β-trefoil fold as example) [23, 24]. A fundamental difference between these models is the ability of simple peptide motifs to achieve complex (symmetric) architecture (via oligomerization) in the conserved architecture model. Coloring is used to indicate individual polypeptide chains. The relative order of the repeated domains A, A’, and A” is potentially variable and dependent upon the particular mechanism of duplication [18]
Experimental support for evolutionary models of symmetric protein folds
The β-propeller fold
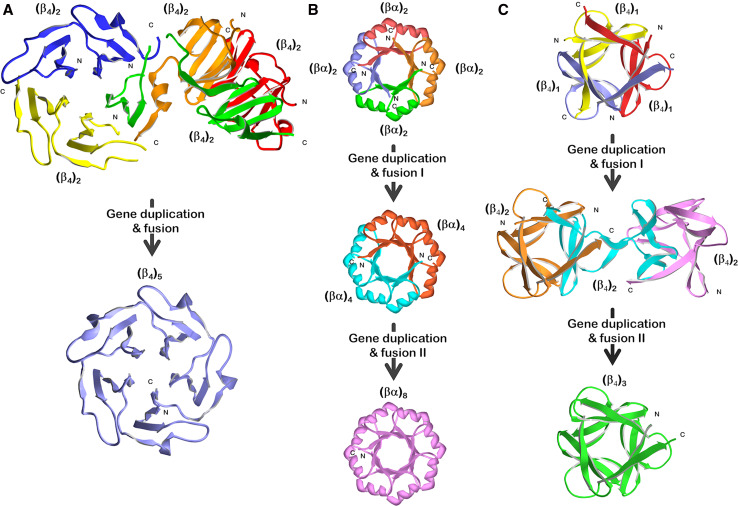
The β-propeller fold has rotational symmetry typically involving 4–8 repeats (i.e., n = 4–8) of a (β4) structural motif of ~50 amino acids known as a “blade” (Fig. 2, bottom of panel a). In addition to the variability in number of repeating blade motifs, the sequence identity between β-propeller proteins is often quite low, suggesting an ancient protein fold. Fragmentation studies of a five-bladed (C5 rotational symmetry) β-propeller (Tachylectin 2) by Tawfik and coworkers yielded a tandem repeat of the fundamental (β4) motif that oligomerized as a pentamer to yield two integral copies of the (β4)5 β-propeller (Fig. 2, panel a) and with two distinct domain-swapped arrangements [26, 30]. Further studies utilizing consensus sequence analysis and random mutagenesis produced a near-symmetric pentameric repeat of a 47 amino acid blade subdomain that was soluble and exhibited lectin affinity (indicating correct folding into a β-propeller) [21].
Fig. 2.
Experimental studies of the folding and structural properties of simple peptide motifs derived from three common symmetric protein folds and the relationship to plausible gene duplication and fusion events in their evolution. a Tandem repeats of a ~50-amino-acid (β4) motif from a five-bladed β-propeller fold [21, 26, 30]; b tandem and quadruplicate repeats of a ~35-amino-acid (βα) motif from a (βα)8-barrel fold [28, 35, 38, 59–61]; c single, tandem, and triple repeats of a ~40-amino-acid (β4) motif from a β-trefoil fold [29, 47]. Coloring is used to indicate individual polypeptide chains
The (βα)8-barrel (TIM barrel) fold
The (βα)8-barrel (“TIM barrel”) fold has a symmetric architecture with eightfold (C8) rotational symmetry of a repeating (βα) structural motif of ~35 amino acids comprising a central β-strand, reverse-turn, outer α-helix, and reverse turn [31] (Fig. 2, bottom of panel b). Because of a general lack of sequence homology, the ancestry of this molecule is difficult to trace by sequence analyses alone and is presumed to be a very ancient fold. The evolutionary pathway of the (βα)8-barrel has been the subject of debate, and arguments have been made in favor of both convergent [32] and divergent evolutionary processes [33]. Kirschner and colleagues demonstrated the ability of phosphoribosylanthranilate isomerase (PRAI; a (βα)8-barrel protein) to efficiently fold with circularly permuted termini [34] including the transposition of the last two (βα) motifs (i.e., (βα)7–8) onto the N terminus. Subsequently, wild-type PRAI was fragmented to yield separate amino terminus (βα)1–6 and carboxyl-terminus (βα)7–8 domains, which were capable of stoichiometric assembly to yield an intact functional enzyme [35]. These results raised the prospect of subdomain oligomerization in the evolutionary pathway of the (βα)8 barrel.
Sterner and coworkers in studying the molecular structure of the HisA and HisF (βα)8-barrel proteins of the histidine biosynthetic pathway of Thermotoga maritima noted a pronounced twofold structural relationship between reverse turn regions of the N and C terminus half-barrels [11]. This structural similarity led to the proposal of a specific evolutionary pathway for the emergence of the (βα)8-barrel architecture via gene duplication and fusion of an ancient (βα)4-half-barrel. In later work published by this group, it was hypothesized that this putative ancient (βα)4-half-barrel was unlikely to have existed as an independently folding motif (due to exposure of hydrophobic groups); thus, the (βα)4-half-barrel likely existed as a homo-dimer forming an intact (βα)8-barrel [36]. A soluble, stable, and cooperatively folding (βα)8-barrel protein with an almost exact primary structure symmetry was constructed from duplicated C-termini (βα)4-half-barrels of T. maritima HisF [37]. Working with Escherichia coli N-(5′-phosphoribosyl)anthranilate isomerase (TrpF), a (βα)8-barrel protein of the tryptophan biosynthetic pathway, and evaluating circular permutations of (βα)4-half-barrels definitions, Akanuma and Yamagishi showed that a (βα)3–6-half-barrel fragment could both autonomously-fold as a (βα)4-half-barrel monomer and also oligomerize as a homo-dimer [38]. Furthermore, a single polypeptide containing a tandem duplication of the TrpF (βα)3–6 sequence yielded a stable monomeric (βα)8-barrel structure. These results provide additional clues to the potential evolutionary process, indicating that (βα)4-half-barrels dimerized to produce an intact (βα)8-barrel (although single (βα)4-half-barrels could have existed as independent simpler structural motifs), and that circular permutation may have occurred as part of the duplication and fusion process in the emergence of the (βα)8-barrel.
Sequence analysis of the T. maritima HisF protein suggests that its (βα)2-quarter barrel subdomains may have a common evolutionary origin, and that the first (βα)2-quarter barrel (HisF-N1) within this protein is the slowest evolving and most conserved subdomain [28]. Expression and refolding of the HisF-N2 subdomain yielded evidence for formation of a stable disulfide-linked (βα)8-barrel tetramer. Based upon these results, Sterner and coworkers have proposed an evolutionary pathway for the (βα)8-barrel architecture that involves two sequential gene duplication and fusion events starting from a (βα)2-quarter barrel subdomain, and describing a conserved architecture evolutionary model (Fig. 2, panel b).
The β-trefoil fold
The β-trefoil fold has threefold (C3) rotational symmetry of a repeating (β4) structural motif of ~40 amino acids that forms an overall six-stranded β-barrel capped at one end by three β-hairpins [8, 39, 40] (Fig. 2, bottom of panel c). As with the β-propeller and (βα)8-barrel architectures, the general lack of sequence homology between repeating domains, as well as between different members of this family, suggest it is an ancient fold [41, 42]. The β-trefoil fold is widely held to have evolved via gene duplication and fusion processes; however, within this framework both an emergent architecture [23] and a conserved architecture evolutionary pathway have been proposed [24] (Fig. 1).
Starting with fibroblast growth factor-1 (FGF-1), a β-trefoil protein, Blaber and coworkers introduced cumulative symmetric constraint mutations, ultimately producing a purely threefold symmetric primary structure that efficiently folded into a thermostable β-trefoil architecture [29, 43–47]. Notably, all the intermediary mutations (14 total) in this symmetric design also efficiently folded, demonstrating a plausible pathway through foldable sequence space from the extant (and highly asymmetric) FGF-1 protein to a purely symmetric β-trefoil protein (representative of a putative ancient form). Subsequent fragmentation of this threefold symmetric β-trefoil protein into single (42-amino-acid) and tandem (84-amino-acid) peptide motifs successfully yielded, in both cases, spontaneously folding and thermostable homo-trimer oligomeric β-trefoil folds (Fig. 2, panel c). Meiering and coworkers [48] pursued a symmetric β-trefoil protein design starting with ricin and utilizing a consensus sequence computational approach. This effort yielded a soluble and thermostable β-trefoil protein with exact primary structure symmetry (although, subsequent fragmentation did not yield foldable polypeptides). The central hydrophobic core packing group in the β-trefoil fold is formed from a set of 12–18 residues [48–50] and is postulated to be a key determinant of the structural symmetry [42]. Notably, the symmetric solutions for the β-trefoil folds determined independently starting from FGF-1 or ricin have an essentially identical core-packing arrangement, suggesting the possibility of a common ancestor for these β-trefoil proteins.
Informatics studies suggest repeated emergence of β-propeller and β-trefoil protein folds
An informatics study of the evolution of the β-propeller fold [51] concluded that this fold has risen repeatedly throughout evolution by amplification of single blades (and not dimeric or trimeric repeats). Underlying symmetry (e.g., twofold or threefold within six-bladed propellers) is not observed, suggesting that the evolutionary pathway may be distinctly different from that of the (βα)8-barrel (where evidence of an underlying twofold or fourfold symmetry typically exists). This analysis also suggested that the single ancestral blade may likely have had the capacity to fold as an oligomer; although extant examples of oligomeric assemblies yielding a β-propeller fold are currently limited to a single example of a trimer of double-bladed units [52]. This analysis therefore implies a likely evolutionary jump from a multi-bladed oligomeric structure to multiple (4–8) repeat arrangements within a single polypeptide (involving multiple gene duplication and fusion events within an evolutionarily brief period). Assuming the hypothesis that a single ancestral blade had the capacity to oligomerize is correct, the evolutionary pathway of the β-propeller is consistent with a conserved architecture model. Similarly, an analysis of the β-trefoil fold by Meiering and coworkers [48] concluded that the β-trefoil fold has arisen repeatedly throughout evolution (and that this is an ongoing evolutionary process). The mechanism of this emergence appears similar to the β-propeller, in that single (or in some cases double) trefoil-fold subdomains within existing β-trefoil proteins have undergone duplication events spawning new β-trefoil proteins (as concluded from identity analyses between trefoil-folds within a single β-trefoil protein, and between the trefoil-folds of different β-trefoil proteins). This proposed “modular” evolutionary process is also consistent with a conserved architecture model.
Conclusions
Although it cannot be absolutely ruled out that symmetric protein architectures emerged through a convergent evolutionary process (and not by gene duplication and fusion events), the preponderance of experimental data, involving several different symmetric protein folds, are consistent with gene duplication and fusion evolutionary processes. The folding of the tandem (β4)2 motif of the five-bladed β-propeller protein Tachylectin 2 to form two intact five-bladed β-propeller folds supports a conserved architecture model of evolution. Similarly, the folding properties of (βα)2-quarter barrel and (βα)4-half barrel domains of HisF and TrpF also support a conserved architecture model for evolution (although some evidence exists for soluble, independently folded (βα)4-half barrel folds). The folding properties of the single and tandem repeating (β4) β-trefoil sub-domains are remarkably consistent with the hypothesized conserved architecture model of Ponting and Russell. Overall, therefore, the above experimental studies, involving three different symmetric protein folds, provide broad support for a conserved architecture evolutionary model. An important hypothesis emerging from these results is that complex protein architecture is possible in primitive organisms having a simple genome and capable of encoding only comparatively small (i.e., 35–50mer) peptides. However, no such examples of oligomeric assemblies of simple peptides yielding β-propeller, (βα)8-barrel or β-trefoil folds (as shown in Fig. 2) have been identified in any extant proteome. Such architecture is therefore postulated to represent an extremely primitive type of protein structure, possibly preceding the LUCA of bacteria and Archaea [28, 29].
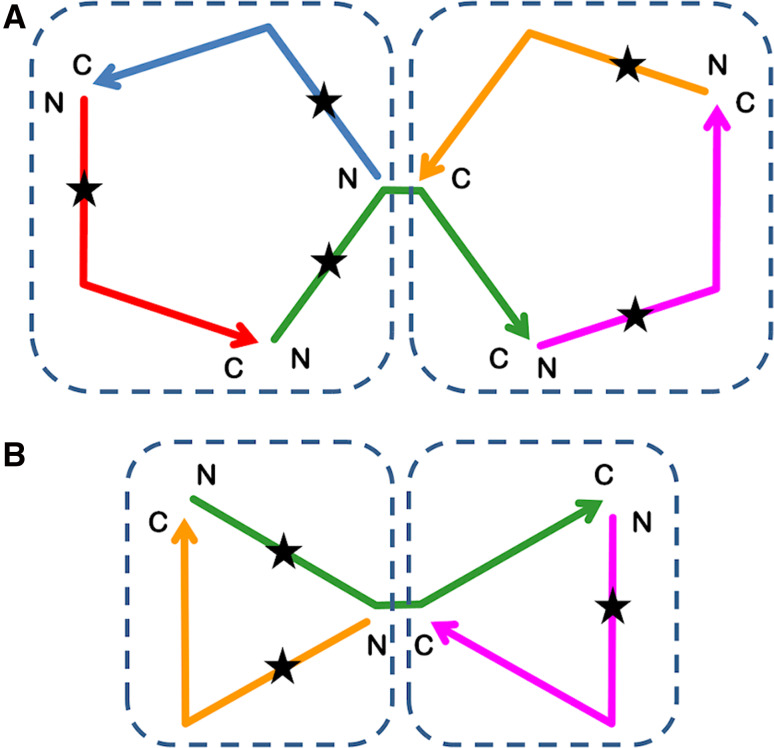
If β-propeller, (βα)8-barrel and β-trefoil symmetric protein architecture were present at an evolutionarily early time point, then in one sense subsequent tandem duplication played no role in the evolution of these protein folds per se—tandem duplication simply enabled the structure to be contained within a single polypeptide chain. Tandem duplication would, however, have enabled an increase in the information content (i.e., Shannon’s entropy [53]) of the architecture; in this sense, the conserved architecture evolutionary model is not associated with conserved entropy. Tandem duplication would subsequently also enable markedly diverse functional evolution since it would permit the emergence of asymmetry (and functional adaptation) not possible in homo-oligomeric (i.e., purely symmetric) assemblies. Furthermore, an intriguing aspect of the conserved architecture evolutionary model for protein folds with odd number symmetry (i.e., Cn, n = 1, 3, 5, 7) is the potential for intermediate forms with multiple integral folds (e.g., the dimer architectures observed for the β-trefoil and five-bladed β-propeller folds in Fig. 2). These would not be strictly identical domains as point mutations would be unequally distributed throughout the twofolds (Fig. 3). In such cases, the resulting physical proximity of two related yet distinctly different proteins provides a structural foundation for the emergence of efficient biochemical pathways (i.e., there would be a high local concentration of both folds and a “metabolite” produced by onefold could be efficiently bound yet differentially acted upon by the second-fold) [11]. Thus, intrinsic to the conserved architecture model for odd symmetry protein architecture is an associated mechanism that potentially supports the emergence of efficient biochemical pathways.
Fig. 3.
Intermediates in the conserved architecture evolutionary model [24, 29] of the (five-bladed) β-propeller (top) and β-trefoil folds (bottom) (see Fig. 2 a, c). The star indicates the asymmetric effect of a point mutation upon the two integral folds formed by homo-trimer assembly. Coloring is used to indicate individual polypeptide chains
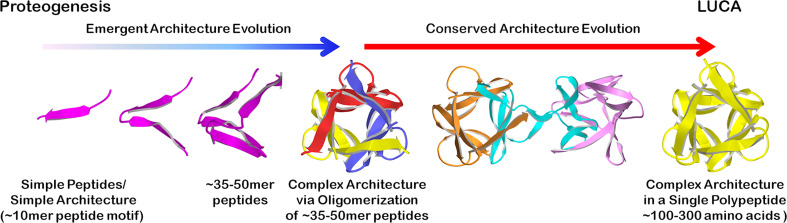
The symmetric protein folds discussed above contain, as fundamental repeating motifs, peptides that comprise 35–50 amino acids and encode (β4) or (βα) structural elements. Such structural motifs can be conceptually divided further (e.g., the (β4) motif can be separated into two (β2) β-hairpins). However, the oligomerization of such smaller domains to construct the extant complex symmetric folds is associated with a substantially increased entropic penalty. Thus, an emergent architecture evolutionary pathway may be more likely when generating the 35–50 amino acid motifs from yet simpler polypeptides and represents a more fundamental and major evolutionary step in the development of complex protein structure (Fig. 4).
Fig. 4.
One possible evolutionary pathway leading from proteogenesis and simple peptide motifs, via gene duplication and fusion, to the emergence of complex architecture present at the time of the LUCA. Coloring is used to indicate individual polypeptide chains
Tracing the evolutionary pathway of symmetric protein architecture back in time can be considered akin to a “top-down” approach to protein design [54] with the goal of identifying a simple peptide motif having the capacity to spontaneously self-assemble to achieve the desired target architecture. Recent experimental work along these lines has identified a 47-amino-acid peptide motif that can oligomerize (with limited asymmetry) to yield an intact 5-bladed β-propeller [55]; a 74-amino-acid peptide that can oligomerize to yield an intact (αβ)8-barrel [28]; and a 42-amino acid peptide that can oligomerize to yield an intact β-trefoil [29, 47]. Typically, such solutions are shown to be thermophile proteins [28, 48, 56]; thus having the capacity to tolerate mutational change in order to introduce novel function (at the expense of stability [57, 58]). Thus, elucidating protein evolutionary pathways can also present new opportunities in de novo protein design.
References
- 1.Harrison PM, Kumar A, Lang N, Snyder M, Gerstein M. A question of size: the eukaryotic proteome and problems in defining it. Nucleic Acids Res. 2002;30:1083–1090. doi: 10.1093/nar/30.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schad E, Tompa P, Hegyi H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011;12:R120. doi: 10.1186/gb-2011-12-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thornton JM, Orengo CA, Todd AE, Pearl FM. Protein folds, functions and evolution. J Mol Biol. 1999;293(2):333–342. doi: 10.1006/jmbi.1999.3054. [DOI] [PubMed] [Google Scholar]
- 4.McLachlan AD. Gene duplication in carp muscle calcium binding protein. Nat New Biol. 1972;240(98):83–85. doi: 10.1038/newbio240083a0. [DOI] [PubMed] [Google Scholar]
- 5.McLachlan AD. Evidence for gene duplication in collagen. J Mol Biol. 1976;107(2):159–174. doi: 10.1016/S0022-2836(76)80024-1. [DOI] [PubMed] [Google Scholar]
- 6.McLachlan AD, Walker JE. Evolution of serum albumin. J Mol Biol. 1977;112(4):543–558. doi: 10.1016/S0022-2836(77)80163-0. [DOI] [PubMed] [Google Scholar]
- 7.Tang J, James MN, Hsu IN, Jenkins JA, Blundell TL. Structural evidence for gene duplication in the evolution of the acid proteases. Nature. 1978;271:618–621. doi: 10.1038/271618a0. [DOI] [PubMed] [Google Scholar]
- 8.McLachlan AD. Three-fold structural pattern in the soybean trypsin inhibitor (Kunitz) J Mol Biol. 1979;133:557–563. doi: 10.1016/0022-2836(79)90408-X. [DOI] [PubMed] [Google Scholar]
- 9.McLachlan AD. Gene duplications in the structural evolution of chymotrypsin. J Mol Biol. 1979;128:49–79. doi: 10.1016/0022-2836(79)90308-5. [DOI] [PubMed] [Google Scholar]
- 10.McLachlan AD. Repeated structure and possible gene duplications in high potential iron protein and rubredoxin. J Mol Evol. 1980;15(4):309–315. doi: 10.1007/BF01733137. [DOI] [PubMed] [Google Scholar]
- 11.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion. Science. 2000;289(5484):1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 12.Hocker B, Schmidt S, Sterner R. A common evolutionary origin of two elementary enzyme folds. FEBS Lett. 2002;510(3):133–135. doi: 10.1016/S0014-5793(01)03232-X. [DOI] [PubMed] [Google Scholar]
- 13.Soding J, Lupas AN. More than the sum of their parts: on the evolution of proteins from peptides. Bioessays. 2003;25:837–846. doi: 10.1002/bies.10321. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho AL, Dias FM, Prates JA, Nagy T, Gilbert HJ, Davies GJ, Ferreira LM, Romão MJ, Fontes CM. Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc Nat Acad Sci USA. 2003;100:13809–13814. doi: 10.1073/pnas.1936124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeltsch A. Circular permutations the molecular evolution of DNA methyltransferases. J Mol Evol. 1999;49:161–164. doi: 10.1007/PL00006529. [DOI] [PubMed] [Google Scholar]
- 16.Peisajovich SG, Rockah L, Tawfik DS. Evolution of new protein topologies through multistep gene rearrangements. Nat Genet. 2006;38:168–174. doi: 10.1038/ng1717. [DOI] [PubMed] [Google Scholar]
- 17.Bjorklund AK, Ekman D, Elofsson A. Expansion of protein domain repeats. PLoS One. 2006;2:959–969. doi: 10.1371/journal.pcbi.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nacher JC, Hayashida M, Akutsu T. The role of internal duplication in the evolution of multi-domain proteins. Biosystems. 2010;101:127–135. doi: 10.1016/j.biosystems.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 20.Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 21.Yadid I, Tawfik DS. Functional β-propeller lectins by tandem duplications of repetitive units. Prot Eng Des Sel. 2011;24(1–2):185–195. doi: 10.1093/protein/gzq053. [DOI] [PubMed] [Google Scholar]
- 22.Borgia MB, Borgia A, Best RB, Steward A, Nettels D, Wunderlich BS, Schuler B, Clarke J. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature. 2011;474:662–665. doi: 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay D. The molecular evolutionary history of a winged bean α-chymotrypsin inhibitor and modeling of its mutations through structural analysis. J Mol Evol. 2000;50:214–223. doi: 10.1007/s002399910024. [DOI] [PubMed] [Google Scholar]
- 24.Ponting CP, Russell RB. Identification of distant homologues of fibroblast growth factors suggests a common ancestor for all beta-trefoil proteins. J Mol Biol. 2000;302:1041–1047. doi: 10.1006/jmbi.2000.4087. [DOI] [PubMed] [Google Scholar]
- 25.Liu L, Iwata K, Yohda M, Miki K. Structural insight into gene duplication, gene fusion and domain swapping in the evolution of PLP-independent amino acid racemases. FEBS Lett. 2002;528:114–118. doi: 10.1016/S0014-5793(02)03264-7. [DOI] [PubMed] [Google Scholar]
- 26.Yadid I, Tawfik DS. Reconstruction of functional β-propeller lectins via homo-oligomeric assembly of shorter fragments. J Mol Biol. 2007;365:10–17. doi: 10.1016/j.jmb.2006.09.055. [DOI] [PubMed] [Google Scholar]
- 27.Akanuma S, Matsuba T, Ueno E, Umeda N, Yamagishi A. Mimicking the evolution of a thermally stable monomeric four-helix bundle by fusion of four identical single-helix peptides. J Biochem. 2010;147:371–379. doi: 10.1093/jb/mvp179. [DOI] [PubMed] [Google Scholar]
- 28.Richter M, Bosnali M, Carstensen L, Seitz T, Durchschlag H, Blanquart S, Merkl R, Sterner R. Computational and experimental evidence for the evolution of a (βα)8-barrel protein from an ancestral quarter-barrel stabilized by disulfide bonds. J Mol Biol. 2010;398:763–773. doi: 10.1016/j.jmb.2010.03.057. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Blaber M. Experimental support for the evolution of symmetric protein architecture from a simple peptide motif. Proc Nat Acad Sci USA. 2011;108:126–130. doi: 10.1073/pnas.1015032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS. Metamorphic proteins mediate evolutionary transitions of structure. Proc Nat Acad Sci USA. 2010;107:7287–7292. doi: 10.1073/pnas.0912616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banner DW, Bloomer A, Petsko GA, Phillips DC, Wilson IA. Atomic coordinates for triose phosphate isomerase from chicken. Biochem Biophys Res Commun. 1976;72(1):146–155. doi: 10.1016/0006-291X(76)90972-4. [DOI] [PubMed] [Google Scholar]
- 32.Lesk AM, Branden CL, Chothia C. Structural principles of alpha/beta barrel proteins: the packing of the interior of the sheet. Proteins. 1989;5(2):139–148. doi: 10.1002/prot.340050208. [DOI] [PubMed] [Google Scholar]
- 33.Reardon D, Farber GK. The structure and evolution of α/β barrel proteins. FASEB J. 1995;9:497–503. doi: 10.1096/fasebj.9.7.7737457. [DOI] [PubMed] [Google Scholar]
- 34.Luger K, Hommel U, Herold M, Hofsteenge J, Kirschner K. Correct folding of circularly permuted variants of a βα barrel enzyme in vivo. Science. 1989;243:206–210. doi: 10.1126/science.2643160. [DOI] [PubMed] [Google Scholar]
- 35.Eder J, Kirschner K. Stable substructures of eightfold βα-barrel proteins: fragment complementation of phosphoribosylanthranilate isomerase. Biochemistry. 1992;31:3617–3625. doi: 10.1021/bi00129a010. [DOI] [PubMed] [Google Scholar]
- 36.Henn-Sax M, Hocker B, Wilmanns M, Sterner R. Divergent evolution of (βα)8-barrel enzymes. Biol Chem. 2001;382:1315–1320. doi: 10.1515/BC.2001.163. [DOI] [PubMed] [Google Scholar]
- 37.Seitz T, Bocola M, Claren J, Sterner R. Stabilization of a (beta-alpha)8-barrel protein designed from identical half barrels. J Mol Biol. 2007;372:114–129. doi: 10.1016/j.jmb.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 38.Akanuma S, Yamagishi A. Experimental evidence for the existence of a stable half-barrel subdomain in the (β/α)8-barrel fold. J Molec Biol. 2008;382:458–466. doi: 10.1016/j.jmb.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 39.Blow DM, Janin J, Sweet RM. Mode of action of soybean trypsin inhibitor (Kunitz) as a model for specific protein–protein interactions. Nature. 1974;249:54–57. doi: 10.1038/249054a0. [DOI] [PubMed] [Google Scholar]
- 40.Sweet RM, Wright HT, Janin J, Chothia CH, Blow DM. Crystal structure of the complex of porcine trypsin with soybean trypsin inhibitor (Kunitz) at 2.6 angstrom resolution. Biochemistry. 1974;13:4212–4228. doi: 10.1021/bi00717a024. [DOI] [PubMed] [Google Scholar]
- 41.Rutenber E, Ready M, Robertus JD. Structure and evolution of ricin B chain. Nature. 1987;326:624–626. doi: 10.1038/326624a0. [DOI] [PubMed] [Google Scholar]
- 42.Feng J, Li M, Huang Y, Xiao Y. Symmetric key structural residues in symmetric proteins with beta-trefoil fold. PLoS One. 2010;5:e14138. doi: 10.1371/journal.pone.0014138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brych SR, Blaber SI, Logan TM, Blaber M. Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core region of a β-trefoil. Protein Sci. 2001;10:2587–2599. doi: 10.1110/ps.ps.34701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brych SR, Kim J, Logan TM, Blaber M. Accommodation of a highly symmetric core within a symmetric protein superfold. Protein Sci. 2003;12:2704–2718. doi: 10.1110/ps.03374903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brych SR, Dubey VK, Bienkiewicz E, Lee J, Logan TM, Blaber M. Symmetric primary and tertiary structure mutations within a symmetric superfold: a solution, not a constraint, to achieve a foldable polypeptide. J Mol Biol. 2004;344(3):769–780. doi: 10.1016/j.jmb.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 46.Dubey VK, Lee J, Blaber M. Redesigning symmetry-related “mini-core” regions of FGF-1 to increase primary structure symmetry: thermodynamic and functional consequences of structural symmetry. Protein Sci Publ Protein Soc. 2005;14(9):2315–2323. doi: 10.1110/ps.051494405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J, Blaber SI, Dubey VK, Blaber M. A polypeptide “building block” for the β-trefoil fold identified by “top-down symmetric deconstruction”. J Mol Biol. 2011;407:744–763. doi: 10.1016/j.jmb.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Broom A, Doxey AC, Lobsanov YD, Berthin LG, Rose DR, Howell PL, McConkey BJ, Meiering EM. Modular evolution and the origins of symmetry: reconstruction of a three-fold symmetric globular protein. Structure. 2012;20:1–11. doi: 10.1016/j.str.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 49.Murzin AG, Lesk AM, Chothia C. β-Trefoil fold. Patterns of structure and sequence in the Kunitz inhibitors interleukins-1β and 1β and fibroblast growth factors. J Mol Biol. 1992;223:531–543. doi: 10.1016/0022-2836(92)90668-A. [DOI] [PubMed] [Google Scholar]
- 50.Blaber M, DiSalvo J, Thomas KA. X-ray crystal structure of human acidic fibroblast growth factor. Biochemistry. 1996;35:2086–2094. doi: 10.1021/bi9521755. [DOI] [PubMed] [Google Scholar]
- 51.Chaudhuri I, Soding J, Lupas AN. Evolution of the β-propeller fold. Proteins. 2008;71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 52.Kostlánová N, Mitchell EP, Lortat-Jacob H, Oscarson S, Lahmann M, Gilboa-Garber N, Chambat G, Wimmerová M, Imberty A. The fucose-binding lectin from Ralstonia solanacearum. A new type of beta-propeller architecture formed by oligomerization and interacting with fucoside, fucosyllactose, and plant xyloglucan. J Biol Chem. 2005;280:27839–27849. doi: 10.1074/jbc.M505184200. [DOI] [PubMed] [Google Scholar]
- 53.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27(3):379–423. [Google Scholar]
- 54.Blaber M, Lee J. Designing proteins from simple motifs: opportunities in top-down symmetric deconstruction. Curr Opin Structural Biol. 2012 doi: 10.1016/j.sbi.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 55.Nikkhah M, Jawad-Alami Z, Demydchuk M, Ribbons D, Paoli M. Engineering of β-propeller protein scaffolds by multiple gene duplication and fusion of an idealized WD repeat. Biomol Eng. 2006;23:185–194. doi: 10.1016/j.bioeng.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Alsenaidy MA, Wang T, Kim JH, Joshi SB, Lee J, Blaber M, Volkin DB, Middaugh CR. An empirical phase diagram approach to investigate conformational stability of “second-generation” functional mutants of acidic fibroblast growth factor (FGF-1) Protein Sci Publ Protein Soc. 2012;21(3):418–432. doi: 10.1002/pro.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beadle BM, Shoichet BK. Structural basis of stability: function tradeoffs in enzymes. J Mol Biol. 2002;321:285–296. doi: 10.1016/S0022-2836(02)00599-5. [DOI] [PubMed] [Google Scholar]
- 58.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLoS Comput Biol. 2008;4(2):e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hocker B, Beismann-Driemeyer S, Hettwer S, Lustig A, Sterner R. Dissection of a (βα)8-barrel enzyme into two folded halves. Nat Struct Biol. 2001;8:32–36. doi: 10.1038/83021. [DOI] [PubMed] [Google Scholar]
- 60.Hocker B, Claren J, Sterner R. Mimicking enzyme evolution by generating new (beta-alpha)8-barrels from (beta-alpha)4-half-barrels. Proc Nat Acad Sci USA. 2004;101:16448–16453. doi: 10.1073/pnas.0405832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hocker B, Lochner A, Seitz T, Claren J, Sterner R. High-resolution crystal structure of an artificial (betaalpha)(8)-barrel protein designed from identical half-barrels. Biochemistry. 2009;48(6):1145–1147. doi: 10.1021/bi802125b. [DOI] [PubMed] [Google Scholar]