Abstract
Lysozymes are antibacterial effectors of the innate immune system in animals that hydrolyze peptidoglycan. Bacteria have evolved protective mechanisms that contribute to lysozyme tolerance such as the production of lysozyme inhibitors, but only inhibitors of chicken (c-) and invertebrate (i-) type lysozyme have been identified. We here report the discovery of a novel Escherichia coli inhibitor specific for goose (g-) type lysozymes, which we designate PliG (periplasmic lysozyme inhibitor of g-type lysozyme). Although it does not inhibit c- or i-type lysozymes, PliG shares a structural sequence motif with the previously described PliI and MliC/PliC lysozyme inhibitor families, suggesting a common ancestry and mode of action. Deletion of pliG increased the sensitivity of E. coli to g-type lysozyme. The existence of inhibitors against all major types of animal lysozyme and their contribution to lysozyme tolerance suggest that lysozyme inhibitors may play a role in bacterial interactions with animal hosts.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0507-3) contains supplementary material, which is available to authorized users.
Keywords: Goose-type lysozyme, Lysozyme inhibitor, Escherichia coli, Lysozyme tolerance, Peptidoglycan
Introduction
Lysozyme (EC 3.2.1.17) is a key player in the innate immune system of most, if not all, animals. It hydrolyzes the peptidoglycan wall of bacterial cells by cleaving the β-1,4-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine, resulting in cell lysis. Based on primary structure, three major classes of animal lysozymes have been identified. Although the overall sequence similarity between lysozymes of different classes is low, they share a similar overall three-dimensional structure [1, 2], and they have been proposed to have common ancestry [3–5]. Phylogenetic studies indicate that several major groups of animals produce lysozymes of two different classes, or at least have the open reading frames potentially encoding them [6]. Chicken (c-) and goose (g-) type lysozyme are found in all vertebrates, while i-type lysozyme is characteristic for invertebrates. The latter sometimes also have c-type (e.g., arthropods) or g-type (e.g., molluscs) in addition to i-type lysozyme [7, 8]. The expression of lysozyme genes typically shows a distinct spatial and temporal pattern and is species dependent. For example, the dominant lysozyme in birds’ egg white is c-type in chicken, but g-type in goose [9]. In the chicken intestine, on the other hand, c-type lysozyme is produced only in young birds up to 8 days after hatching, while g-type lysozyme is expressed at all ages [10]. The contribution of each lysozyme type to antibacterial defense is therefore also likely to be more pertinent in specific stages and tissues than in others.
Given the highly specific antibacterial mode of action and old evolutionary age of lysozymes, it is not surprising that bacteria have developed specific lysozyme-resistance mechanisms, for instance the production of chemical variants of peptidoglycan that are resistant to lysozyme [11] or the production of lysozyme inhibitors. The first bacterial lysozyme inhibitor was fortuitously discovered as a periplasmic protein of Escherichia coli, and was designated Ivy (inhibitor of vertebrate lysozyme) because of its specificity against vertebrate (c-type) lysozyme [12]. Ivy protects E. coli against lysozyme in the presence of an outer membrane permeabilizing compound like lactoferrin, and is essential for the ability of E. coli to grow in human saliva and contributes to its survival in the egg white of chicken eggs [13, 14]. Ivy is only found in about a dozen proteobacterial genera, but the use of a functional screening approach led to the discovery of a new family of c-type lysozyme inhibitors unrelated to Ivy that is more widely distributed among Gram-negative bacteria [15]. This novel family shares a common conserved PF09864 domain [16] and, depending on the location of the inhibitors in the cell, they were named MliC (membrane-bound lysozyme inhibitor of c-type lysozyme) or PliC (periplasmic lysozyme inhibitor of c-type lysozyme). Knock-out of pliC rendered Salmonella enteritidis more sensitive to lysozyme in the presence of lactoferrin and overexpression of mliC conferred enhanced lysozyme tolerance in E. coli. Moreover, before its function was known, mliC had already been picked up as one of the genes that are induced upon entry of S. typhi in macrophages and that are necessary for macrophage survival [17]. Using the same screening approach that led to the discovery of the MliC/PliC family, Van Herreweghe et al. [18] recently succeeded in isolating a bacterial i-type lysozyme inhibitor from Aeromonas hydrophila and designated it as PliI (periplasmic lysozyme inhibitor of i-type lysozyme). Despite the low overall relatedness of the PliI protein family to the PliC/MliC family (PliI from A. hydrophila is only 4.5 and 21.5% identical to PliC from Salmonella typhimurium LT2 and MliC from Pseudomonas aeruginosa PAO1, respectively), both families share a common motif that in MliC of P. aeruginosa was shown to form a loop that protrudes into the active site cavity of hen egg white lysozyme. Nevertheless, both PliI and MliC/PliC type inhibitors are highly specific and do not show any cross-inhibition towards other lysozyme families [18]. Ivy, on the other hand, weakly inhibited g-type lysozyme from goose egg white (GEWL) [19] but not from salmon (SalG). Using homology modeling, protein–protein docking and molecular dynamics simulations, Kyomuhendo et al. [20] were able to explain the weak interaction of Ivy with GEWL, and predicted that none of the known fish g-type lysozymes would be inhibited because of their different electrostatic surface properties and curvature. Further, a unique feature of all g-type lysozymes, both from terrestrial and from aquatic organisms, is the involvement of three active residues Glu73, Asp86 en Asp97 (numbering as in GEWL) [7, 20–22]. While Glu73 has obvious counterparts in c- and i-type lysozyme (respectively Glu35 and Glu11) [2, 21], the latter have only one aspartate residue (respectively Asp52 and Asp30) contributing to catalysis. Finally, g-type lysozyme also has a distinct substrate selectivity compared to both other lysozymes, with a relative preference for peptidoglycan chains with a peptide side chain [23–25], and a higher activity on peptidoglycan from Gram-negative than from Gram-positive bacteria [26].
The discovery of highly specific bacterial inhibitors of two of the three major types of lysozyme that exist in the animal kingdom, and the demonstration that these inhibitors confer bacterial lysozyme tolerance and thus may be involved in bacterial interactions with animal hosts, provided the basis of the current work, in which we conducted a search for a bacterial g-type lysozyme inhibitor. We succeeded in isolating such an inhibitor from E. coli, and we report on its characterization, relatedness to the known c- and i-type inhibitors, and contribution to bacterial g-type lysozyme tolerance.
Materials and methods
Bacterial strains and plasmids
The bacteria screened for g-type lysozyme inhibitory activity and their cultivation conditions are listed in Table 1. Specific E. coli mutant strains and plasmids used or constructed in this work are described in Table 2. Antibiotics (Sigma-Aldrich, Bornem, Belgium) were added when appropriate at the following final concentrations: ampicillin (Amp), 100 μg/ml; kanamycin (Km), 50 μg/ml; chloramphenicol (Cm), 20 μg/ml.
Table 1.
Inhibitory activity against GEWL in periplasmic extracts of Gram-negative bacteria
| Strains | Growth medium | Growth temperature (°C) | GEWL activity inhibition (%) |
|---|---|---|---|
| Aeromonas hydrophila ATCC7966 | LB | 30 | 99 |
| Bordetella avium 197 N | LB | 37 | 0 |
| Enterobacter aerogenes ATCC13048 | NB | 37 | 3 |
| Erwinia amylovora CFBP1430 | NB | 37 | 0 |
| E. coliivy::Cm mliC::Km | LB | 37 | 100 |
| Klebsiella pneumoniae ATCC13883 | NB | 30 | 7 |
| Proteus mirabilis LMM2010 | NB | 37 | 18 |
| Salmonella enteritidis ATCC13076 | LB | 37 | 17 |
| Salmonella typhimurium LT2 ∆pliC | NB | 37 | 26 |
| Vibrio harveyi ATCC BAA-1116 | LB + 1.5% NaCl | 37 | 19 |
| Yersinia enterocolitica ATCC9610 | BHI | 30 | 0 |
LB Luria Bertani (10 g/l Trypton, 5 g/l Yeast extract, 5 g/l NaCl); NB nutrient broth (Oxoid, Basingstoke, UK); BHI brain heart infusion (Oxoid); ATCC American type culture collection; CFBP Collection Française de Bactéries Phytopathogènes, Beaucouzé Cedex, France; LMM Collection Labo Levensmiddelenmicrobiologie, Katholieke Universiteit Leuven
Table 2.
Bacterial strains and plasmids
| Strain or plasmid | Description | References or source |
|---|---|---|
| E. coli BL21 GOLD (DE3) pQM64 | Expression host for pET series vectors, containing IPTG-inducible T7 RNA polymerase gene and carrying the pQM64 plasmid mith the salG gene of g-type salmon lysozyme under controle of a PT7 promotor | [27] |
| E. coli BW25113 pKD46 | ApR; repA101(ts) temperature-sensitive replicon; The lambdaRed recombinase genes are under control of the araB promoter | Coli Genetic Stock Center (CGSC) |
| E. coli MG1655 | Wild-type E. coli | |
| E. coli ∆pliG | E. coli MG1655; pliG gene deleted without leaving an antibiotic resistance marker | This study |
| E. coli GL113 ΔtolA::Km | E. coli GL113; tolA gene replaced by aph gene cassette from pKD4; Kmr | [45] |
| E. colitolA::Km | E. coli MG1655; tolA gene replaced by aph gene cassette from pKD4; Kmr | [15] |
| E. colitolA::Km ∆pliG | E. coli MG1655; tolA gene replaced by aph gene cassette from pKD4; pliG gene deleted without leaving an antibiotic resistance marker; Kmr | This study |
| E. coli tolA::Km ∆pliG PBAD-pliG | E. colitolA::Km ∆pliG; pliG gene was then exchanged with the araBAD genes to bring pliG under control of the chromosomal PBAD promotor | This study |
| E. coli BL21 (DE3) | Expression host for pET series vectors, containing IPTG-inducible T7 RNA polymerase gene | Novagen, Merck Biosciences, Darmstadt, Germany |
| pET28b(+) | Expression vector, Kmr | Novagen |
| pET28b(+) (PT7-pliG-Ec) | pliG from E. coli under control of PT7 promotor in pET28b; KmR | This study |
| pET28b(+) (PT7-pliG-Ah) | pliG from A. hydrophila under control of PT7 promotor in pET28b; KmR | This study |
| pET28b(+) (PT7-pliG-St) | pliG from S. typhimurium under control of PT7 promotor in pET28b; KmR | This study |
| pkD3 | Vector containing aph gene flanked by FRT sites, CmR | [29] |
| pKD4 | Vector containing aph gene flanked by FRT sites, KmR | [29] |
| pKD46 | Plasmid expressing γ, β and exo recombination genes of phage λ under control of PBAD; temperature-sensitive replicon; Ampr | [29] |
| pCP20 | Plasmid expressing the FLP (flippase) gene, directing recombination of FRT sites; temperature-sensitive replicon, Ampr; Cmr | [29] |
G-type lysozymes: isolation, enzymatic assay and inhibition assay
Goose egg white lysozyme (GEWL) was purified from the white of eggs of the Embden goose (Anser anser) as previously described [26]. The expression of recombinant g-type lysozyme from Atlantic salmon (SalG) in E. coli BL21 GOLD (DE3) pQM64 cells was performed according to Kyomuhendo et al. [27], except that no protease inhibitors were used and that the hexahistidine (His6) tag was not removed, but used to perform an additional purification step using Ni-Sepharose™ affinity chromatography with a HisTrap™ HP column (GE Healthcare Life sciences, Uppsala, Sweden). Protein concentrations were determined using the BCA protein assay kit (Novabiochem, Merck Biosciences, Darmstadt, Germany). G-type lysozyme activity, and inhibitory activities of periplasmic extracts or purified inhibitor fractions, were measured with a turbidity assay using Yersinia enterocolitica ATCC9610 cell wall material prepared by treatment of cells with chloroform-saturated 50 mM Tris pH 7.0 buffer as a substrate [26]. This material is a better substrate for g-type lysozymes than the Micrococcus luteus cells which are used in the standard lysozyme assay. Standardized inhibition assays [19] were performed by adding an inhibitor-containing sample to an amount of g-type lysozyme that produced a linear OD600nm decrease of 0.27 ± 0.04 over 2 h at 25°C in the absence of inhibitor, and measuring the inhibition percentage (I %) as follows: I (%) = [(L0−L)−(R0−R)]/[(L0−L)−(B0−B)] × 100 with L0−L, R0−R and B0−B representing the OD600nm decrease over a period of 2 h at 25°C, respectively, in the presence of lysozyme but with buffer instead of the inhibitor sample, in the presence of lysozyme and the inhibitor sample, and in the presence of the inhibitor sample but with buffer instead of lysozyme. One inhibitory unit (IU) was defined as the amount of inhibitor causing a 50% decrease of lysozyme activity in this assay.
Inhibitory activity against hen egg white lysozyme (HEWL; Fluka, 66,000 U/mg protein) or Tapes japonica lysozyme (TjL), purified as described by Van Herreweghe et al. [18], was measured as described above but using freeze-dried Micrococcus luteus ATCC4698 cells (Sigma-Aldrich) as a substrate [19].
Purification and identification of g-type lysozyme inhibitor
For purification, 150 ml of periplasmic protein extracts of stationary phase E. coli ivy::Cm mliC::Km cells, isolated as described by Deckers et al. [14], were passed over a weak cation exchange CM Sepharose Fast Flow resin equilibrated with 50 mM potassium phosphate buffer (PPB) pH 7.0 using an ÄKTA-FPLC (Amersham Pharmacia Biotech, Uppsala, Sweden) at room temperature. Elution with a linear gradient from 0 to 100% 2.0 M KCl in 50 mM PPB (pH 7.0) yielded fractions with lysozyme inhibitory activity, which were pooled, dialyzed against deionized water and lyophilized. The freeze-dried sample was dissolved in 200 μl of 50 mM PPB (pH 7.0) with 0.15 M KCl and further purified using size exclusion chromatography on a Superdex 75 HR 10/30 column (GE Healthcare) using the same buffer. Active fractions were pooled and lyophilized. A sample was then used for purity analysis by SDS-PAGE (4% concentrating and 15% separating gel) and silver staining [28]. Another sample was trypsin digested and subjected to capillary LC/MS/MS for identification, using an Ultimate 3000 system run in nano-lc set-up (Dionex, US) coupled to a Q-TOF hybrid quadrupole time-of-flight mass spectrometer (Micro-tofQ, Bruker, Germany). All MS and MS/MS spectra were automatically processed (background subtraction, smoothing and peak picking) using the FlexAnalysis software (Bruker Daltonics) to generate a peak list file in the MGF format. MGF files were submitted to a “Mascot” search (Matrix Science) against the NCBI-database with no taxonomy selected to identify the protein.
Recombinant expression of g-type lysozyme inhibitor (PliG) from E. coli and of related proteins/protein domains from other bacteria
The E. coli gene (ycgK) encoding the PliG protein (ordered locus name: b1178; NCBI RefSeq: NP_415696.1) including its signal peptide was amplified without stop codon using Phusion DNA-polymerase (Finnzymes, Espoo, Finland) with the primers ycgK_eco_pET_Fw (5′-CGGCCCATGGAAATGAAAATCAAGAGCATCAG-3′) and ycgK_eco_pET_Re (5′-CCGACTCGAGCTTAATTTGAATATCGACGTT-3′). After digestion with NcoI and XhoI (recognition sites in primers are underlined), the resulting fragment was ligated into pET28b(+) which provides a C-terminal His6 tag followed by a stop codon, and transformed to E. coli BL21 (DE3). The recombinant protein PliG-His6 was expressed by growing this strain in LB broth at 37°C to late exponential phase (OD600nm = 0.6), addition of 1 mM IPTG (Acros Organics, Geel, Belgium), and further growth during 2 h at 37°C. PliG-His6 was subsequently purified from a periplasmic extract of the bacteria using a HisTrap™ HP column (GE Healthcare) according to the manufacturer’s guidelines and using an ÄKTA-FPLC system. The pliG homolog of Salmonella typhimurium (STM2610; NP_461546.1) was cloned in a similar way. The homologs AHA_0125 (YP_854659.1) from Aeromonas hydrophila and BAV1242 (YP_785764.1) from Bordetella avium were amplified without their signal peptide, and then cloned into pET26b(+) to provide them with an endogenous E. coli N-terminal signal peptide, besides a C-terminal His6-tag. Further, bacterial pre-peptidase C-terminal domains (PPC-domains) of the zinc metalloprotease VIBHAR_05515 (NYP_001447646.1) from Vibrio harveyi and the elastase AHA_0851 (YP_855393.1) from Aeromonas hydrophila, to which PliG is related, were also cloned in pET26b(+) in a similar way. Each of these constructs was transformed to E. coli BL21(DE3), and recombinant proteins were expressed and purified as described for PliG from E. coli.
Construction of a PliG-affinity column
About 5.42 mg pure PliG-His6 was immobilized on 1 ml N-hydroxysuccinimide-activated HiTrap™ column (GE Healthcare) following the instructions of the manufacturer. The column was equilibrated with 10 column volumes 0.1 M Tris–HCl pH 7.0 and loaded with 5 ml of 0.2 mg/ml SalG. The column was washed with 10 column volumes 0.1 M Tris–HCl pH 7.0 and then eluted with a linear gradient from 0.1 M Tris–HCl pH 7.0 to 0.1 M Tris–NaOH pH 12.0 with 2.0 M KCl. Elution fractions of 1 ml were immediately neutralized with 100 μl 1.0 M Tris–HCl pH 7.0 and dialyzed against 10 mM PPB pH 7.0 for lysozyme activity testing.
Construction of E. coli pliG deletion and overexpression strains
A deletion mutant E. coli ∆pliG was constructed using the one-step inactivation method described by Datsenko et al. [29], followed by removal of the introduced antibiotic resistance cassette using the FRT/FLP recombination system. Briefly, 70-bp PCR primers were designed comprising a 50-bp 5′ part complementary to the region down- or upstream of the E. coli ycgK gene and a 20-bp 3′ part allowing amplification of the FRT-flanked Cm resistance cassette present in the plasmid pKD3. Purified PCR products were electrotransformed into E. coli MG1655 containing the pKD46 plasmid providing the lambda red recombinase, and Cm-resistant transformant colonies were analyzed for correct introduction of the pliG::Cm allele by PCR. The Cm resistance cassette was subsequently removed by expression of the flippase recombination enzyme (FLP) of the FRT/FLP recombination system from the temperature-sensitive pCP20 plasmid. Antibiotic resistance testing and PCR were used to finally select one ∆pliG strain from which the Cm resistance cassette had been properly excised. For overexpression of PliG, we replaced the genomic araBAD genes by pliG to bring the latter under control of the arabinose-inducible PBAD promotor. To this end, pliG was amplified and cloned next to the FRT-flanked Cm-resistance gene in pKD3. Next, pliG and the FRT-flanked Cm resistance gene were amplified with primers carrying 50-bp 5′ extensions complementary to the region downstream of the E. coli araB and araD genes, respectively. The purified PCR product was electrotransformed into E. coli ∆pliG pKD46 and Cm-resistant transformants were analyzed by PCR for correct gene replacement. Arabinose-dependent PliG production was confirmed and the newly constructed strain was designated E. coli ∆pliG PBAD-pliG. An additional tolA knock-out was introduced into E. coli MG1655, E. coli ∆pliG and E. coli ∆pliG PBAD-pliG by P1 transduction from E. coli GL113 tolA::Km.
Surface plasmon resonance analysis
Inhibition specificity and affinity constants for binding between lysozymes and inhibitors were assessed by surface plasmon resonance (SPR) analysis using a Biacore 3000 analytical system (Biacore, Uppsala, Sweden). Random amine coupling of HEWL, SalG and TjL was carried out by injecting the proteins (5 μg/ml) in 10 mM sodium acetate buffer, pH 4.5, following preactivation of the carboxymethylated dextran matrix (CM5 sensor chip; GE Healthcare) using N-hydroxysuccinimide (NHS) 1-Ethyl-3-[3-(dimethylaminopropyl)] carbodiimide hydrochloride (EDC). After coupling of the proteins, the residual NHS esters were deactivated by the injection of ethanolamine (1.0 M, pH 8.5). Each lysozyme was covalently coupled to approximately 1,200 resonance units (RU) with one RU corresponding to 1 pg of bound protein/mm2. PliG was diluted in HBS-EP buffer (Biacore) to concentrations between 5 and 200 nM and injected at a flow rate of 30 μl/min. Association and dissociation data were both collected for 6 min. After each cycle, the chip was regenerated with 10 μl of 0.1 M NaOH. Analysis of the association and dissociation phases was performed using Biaevaluation version 3.1 software (Langmuir binding, local fit; Biacore). Data obtained from a parallel flow cell to which a nonrelated monoclonal antibody against human Plasminogen Activator Inhibitor 1 (MA-31C9) was coupled, served as blank sensorgrams for substraction of changes in the bulk refractive index.
Effect of PliG on g-type lysozyme tolerance of E. coli
Lysozyme tolerance was assayed under two different conditions to distinguish cell killing (inactivation) and growth inhibition. For the inactivation assay, exponential phase (OD600nm = 0.3) wild-type and ∆pliG cells grown at 37°C in LB were harvested by centrifugation (4,000g, 5 min), washed and resuspended in an equivalent volume of 10 mM PPB pH 7.0 with either no additions or with 15 μg/ml SalG and/or 1 mM EDTA for outer membrane permeabilization. Alternatively, the assay was performed with E. coli tolA::Km, E. coli tolA::Km ∆pliG and E. coli tolA::Km ∆pliG PBAD-pliG, grown as described above but with addition of 0.1% arabinose for induction of the pliG gene under control of the araBAD promotor. Lysozyme challenge was done in 10 mM PPB pH 7.0 with 36 μg/ml SalG but without EDTA in this case. After 24 h of incubation at 22°C, survivors were counted on LB agar. For the growth inhibition assay, overnight cultures were diluted (1/100) in fresh LB and OD600 was followed at 30°C in a microplate reader (Multiskan Ascent; Thermo, Aalst, Belgium). As for inactivation, wild-type and ∆pliG strains were used in one variant of the assay, and challenged by addition of 1 mM EDTA and/or 150 μg/ml SalG to the growth medium. In the other variant, the three tolA strains were used and SalG was added at 35 μg/ml and 3.75 μg/ml without EDTA.
Results
Screening of bacterial periplasmic extracts for g-type lysozyme inhibition
Periplasmic extracts of Gram-negative bacteria were screened for inhibitory activity against GEWL. Most extracts exhibited low inhibitory activities (<30%), except those from Aeromonas hydrophila and E. coli ivy::Cm mliC::Km (≥99%) (Table 1). Since the latter strain is devoid of both its c-type lysozyme inhibitors, and its periplasmic extract shows no residual inhibition against c-type lysozyme (data not shown), the observed g-type lysozyme inhibition was considered to be indicative of a separate specific g-type inhibitor. Also, in the A. hydrophila extract, no c-type lysozyme inhibitory activity was detected, although this organism harbors ivy and mliC homologs (data not shown). The two periplasmic protein extracts inhibiting GEWL also showed strong inhibition of SalG (>90%), and because the latter was more convenient to produce in a pure form and in sufficient amounts, it was used in the remainder of this work as representative g-type lysozyme. The periplasmic protein extract of E. coli ivy::Cm mliC::Km was chosen to further isolate and identify a potential g-type lysozyme inhibitor.
Isolation and identification of E. coli inhibitor of g-type lysozyme
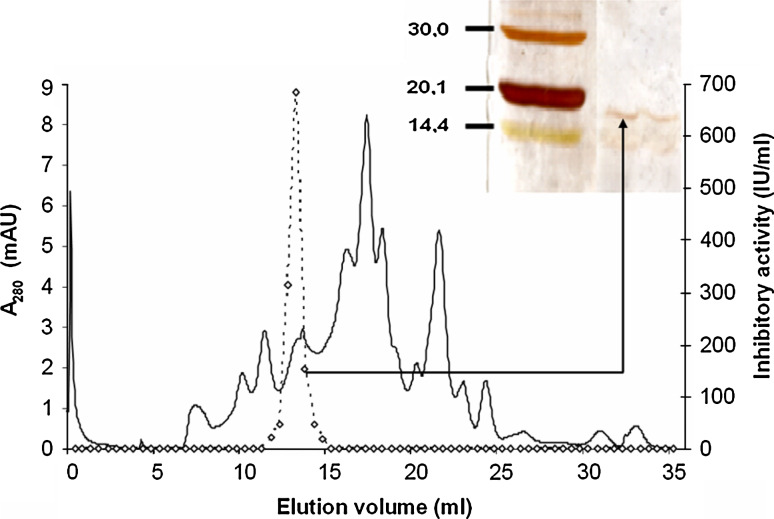
Approximately 150 ml of periplasmic extract of E. coli ivy::Cm mliC::Km was subjected to weak cation exchange. Lysozyme inhibitory activity was recovered at 1.6–2.0 M KCl during gradient elution. Active fractions were combined, dialyzed, lyophilized, redissolved in 200 μl of deinonized water and further purified by size exclusion chromatography (Fig. 1). As a result, 2 ml of putative inhibitor was obtained with an activity of 684 IU/ml. The yield of five independent isolations was then pooled and one part was analyzed on SDS-PAGE (Fig. 1). Although the presence of two bands indicated that the putative inhibitor was not yet pure, we submitted the rest of the sample to trypsin digestion and further identification. Tandem mass spectrometry allowed to identify with high confidence four peptides (YSPELDSHGQYSLPASGKYELR, GHSSAQYSGEIKGYDYDTYTFYAK, KYNVDIQIK and VHVSISNEGADTYLFGPGIDDSVDLSR), which correspond to the uncharacterized E. coli protein YcgK (ordered locus name: b1178; NCBI RefSeq: NP_415696.1). This protein has a predicted signal peptide of 22 residues upon analysis with Signal P 3.0 ([30]; http://www.cbs.dtu.dk/services/SignalP/), and the mature peptide has a predicted pI and molecular weight of, respectively, 9.01 and 12,518 Da as calculated with ProtParam (http://www.expasy.org/tools/protparam.html) (Fig. 2). This molecular weight is less than the estimate from gel migration (±17 kDa), but such deviations are not uncommon for small proteins, and a similar observation was reported for the PliC lysozyme inhibitor [15]. YcgK contains a conserved bacterial pre-peptidase C-terminal domain (PPC), corresponding to the Pfam domain PF04151 [16]. This domain is commonly found at the C-terminus of secreted bacterial peptidases, but also occurs as an independent ORF in several bacteria. The YcgK protein, extended with a C-terminal His6 tag, was recombinantly expressed in E. coli BL21(DE3) and purified by Ni-Sepharose™ Affinity Chromatography. The purified protein showed a single band after SDS-PAGE with an estimated molecular weight of 17.1 kDa. This corresponds to the value estimated for the major band in the purified periplasmic extract (Fig. 1), confirming this band to be YcgK. The recombinant protein displayed strong inhibitory activity against SalG, since addition of an equimolar amount of YcgK to 0.75 μg/ml SalG resulted in 96% inhibition. This result confirmed that YcgK is indeed a g-type lysozyme inhibitor, and the protein was named PliG (periplasmic lysozyme inhibitor of the g-type lysozyme), analogous to the previously characterized c- and i-type lysozyme inhibitors PliC and PliI. PliG showed low overall sequence similarity with PliC from Salmonella typhimurium LT2 (12.6%), MliC from Pseudomonas aeruginosa PAO1 (26.4%), PliI from Aeromonas hydrophila (28.2%) and Ivy from E. coli MG1655 (5.6%) (analysis performed with mature sequences without signal peptide using EMBOSS Pairwise Alignment Algorithms with default parameters [31]).
Fig. 1.
Gel filtration chromatogram showing the last step of purification of PliG from E. coli ivy::Cm mliC::Km periplasmic extract. Protein concentration in eluate was monitored by absorption at 280 nm (A 280) and expressed in milli absorption units (mAU) (solid line), inhibitory activity (IU/ml) of fractions against GEWL was monitored by the lysozyme inhibitor assay (dashed line). Photograph in inset shows a silver-stained SDS-PAGE gel with molecular weight markers (lane 1, and indices in kDa at the left) and a concentrated fraction containing PliG (lane 2)
Fig. 2.
Amino acid sequence alignment (Clustalw [46]; http://www.ebi.ac.uk/Tools/clustalw2/) of PliG of E. coli, S. typhimurium and A. hydrophila. Residues that are identical in all sequences in the alignment are marked with ‘*’ in the bottom row, conserved and semi-conserved substitutions with ‘:’ and ‘.’, respectively. The signal sequence is highlighted in bold, the PPC domain in shaded in gray, the region spanning the common conserved motif in active PliG, PliI, PliC and MliC homologs is underlined and the four peptides identified by mass spectrometry that led to the identification of the inhibitor are highlighted in italics
Specificity and binding affinity of PliG for g-type lysozyme
Using the standard bacteriolytic assays, PliG did not show any inhibition of HEWL (c-type) or TjL (i-type) lysozyme, even when added in tenfold molar excess (data not shown). A more quantitative study of PliG–lysozyme binding was performed by SPR analysis. The three lysozymes, together with an unrelated monoclonal antibody as a negative control were immobilized on the four channels of a sensor chip (CM5), and the interaction with different concentrations of PliG-His6 was analyzed. PliG was confirmed not to interact with HEWL and TjL, and showed a strong interaction with SalG, characterized by a high association rate constant [k a = 2.02 ± 0.47 × 106 (1/Ms)], a low dissociation rate constant [k d = 3.54 ± 0.69 × 10−3 (1/s)], and hence a high affinity constant [K A = k a/k d = 5.81 ± 1.29 × 108 (1/M)] (Fig. 5 in supplementary material). In comparison, with an affinity constant of 2.15 ± 0.61 × 1010 (1/M), the binding of PliI to TjL appears to be stronger [18], at least under the experimental conditions used. However, this comparison should be interpreted with caution, since protein–protein interactions are usually strongly influenced by pH and ionic strength. Furthermore, in the current work, the affinity of PliG was determined only against SalG, and it is possible that g-type lysozymes from mammals, birds, invertebrates or even some bacterial lytic transglycosylases with a g-type lysozyme domain [32] will show a higher affinity. In this context, it is noteworthy that we observed inhibition of the g-type lysozymes in extracts of the Urochordate Oikopleura dioica [33] (data not shown).
The specific interaction of PliG and SalG was also investigated by affinity chromatography using the inverse configuration as in SPR, i.e. by passing SalG in solution over an affinity matrix with immobilized PliG. No detectable lysozyme activity was retained in the flow through fractions, indicating that SalG was effectively bound on the column. Upon elution, a UV-absorption peak was detected near the end of the gradient (pH 12.0 and 2.0 M KCl), and the eluted material was confirmed to be SalG by SDS-PAGE, zymogram and lysozyme inhibition assay (data not shown).
Distribution and inhibitory activity of PliG homologs
PliG homologs were found in a limited number of bacterial genera by tBlastn search ([34]; http://blast.ncbi.nlm.nih.gov/Blast.cgi). They could be divided in two major groups, the first comprising PliG homologs existing as a separate ORF encoding a predicted periplasmic protein, and the second in which the PliG homolog is a PPC domain of a protease or peptidase. Homologs of the first group are restricted to about 15–20 genera within the α, β, γ, and δ Proteobacteria with few exceptions, whereas PPC-containing peptidases or proteases are more widely distributed, and are, for example, also found in several Gram-positive bacteria. The screening of periplasmic extracts had already suggested the presence of a g-type inhibitor in A. hydrophila, but not in S. typhimurium and B. avium, although all three species have a separately occurring PliG homolog. Therefore, these three ORF’s, which showed sequence identities with PliG from E. coli of, respectively, 36.0, 82.9 and 31.0% (analysis performed with mature sequences without signal peptide using EMBOSS Pairwise Alignment Algorithms with default parameters [31]) were cloned and expressed with a His6 tag in E. coli. After purification, the proteins showed a single band in SDS-PAGE, and the homologs of S. typhimurium and A. hydrophila exhibited inhibitory activity against SalG. The B. avium homolog, in contrast, lacked inhibitory activity although it was succesfully expressed. In addition, the PPC domains of the zinc metalloprotease and the elastase from, respectively, V. harveyi and A. hydrophila were cloned and expressed as separate proteins, but showed no SalG inhibitory activity. These results reveal that the PPC-domain of these proteins as such is not able to inhibit g-type lysozyme.
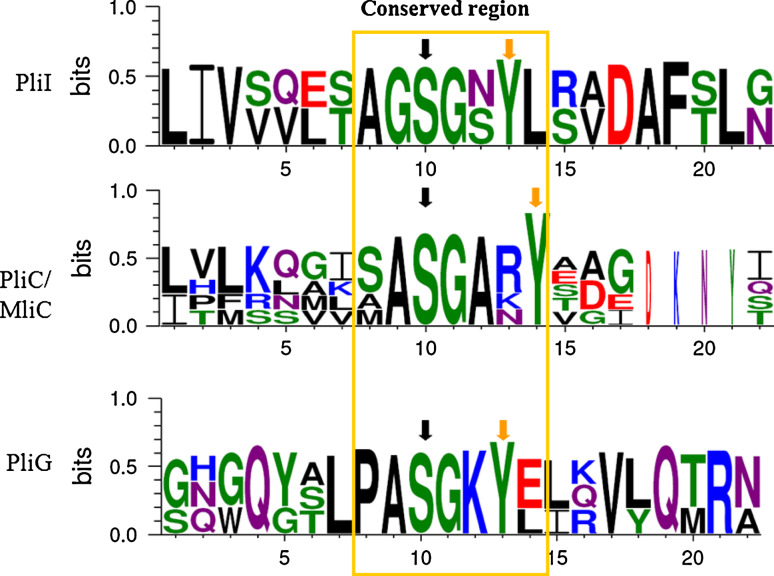
Two conserved key regions have been shown to be important in the interaction between MliC of P. aeruginosa and the active site of HEWL [35]. One of these regions was also found to be present in PliI and PliC [18]. Motif detection using GLAM2 (gapped local alignment of motifs [36]) now shows that this region also exists in the active PliG proteins (Fig. 3). In particular, the key residues Ser89 and Tyr92 within this motif of MliC that were demonstrated to be crucial for its inhibitory activity [35] are strictly conserved among the active PliG, PliC and PliI homologs.
Fig. 3.
Sequence logo of the conserved motif (using GLAM2) in three families of lysozyme inhibitors built with Weblogo 3.0 [47]. Input is a Clustal W multiple alignment of three active PliG inhibitors (E. coli, A. hydrophila and S. typhimurium), two active PliI inhibitors (A. hydrophila and Yersinia enterocolitica) and five active PliC/MliC inhibitors (PliC from S. typhimurium and Proteus mirabilis, MliC from S. typhimurium, Pseudomonas aeruginosa and E. coli). Amino acids are colored by chemistry (green, polar; blue, basic; red, acidic; and black, hydrophobic). The black and the orange arrows indicate, respectively, the conserved serine and tyrosine residues
PliG bestows g-type lysozyme tolerance on E. coli
To examine the function of PliG in E. coli, a deletion knock-out mutant (E. coli ∆pliG) was constructed. The residual SalG inhibitory activity in the periplasmic extract of the deletion strain (1 IU/ml) was strongly reduced in comparison to the wild-type strain (14 IU/ml). Since the antibiotic resistance cassette in the site of the deletion has been excised and the open reading frame downstream of pliG is oppositely orientated, this loss of inhibitory activity cannot be due to a polar effect of the deletion. This result confirms once more that the lysozyme inhibitory activity in the periplasmic extracts can be ascribed to the PliG protein.
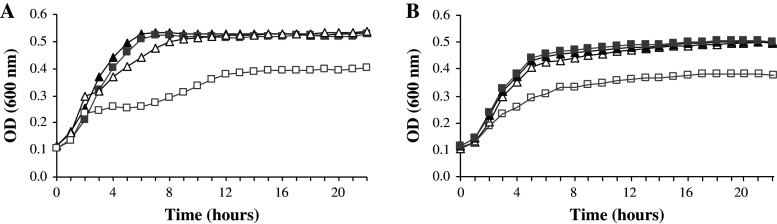
A difficulty in testing the lysozyme sensivity of Gram-negative bacteria in vitro is that they are surrounded by an outer membrane that is impermeable to lysozyme. In this work, E. coli was rendered sensitive to lysozyme with EDTA or by introducing a tolA knock-out mutation that increases outer membrane permeability. First, an inactivation assay was conducted by challenging cell suspensions for 24 h in buffer. Control treatments with only EDTA or SalG did not inactivate either strain, as expected. In fact, the buffer supported some growth when EDTA was not present, probably because of residual growth medium. However, the combination of EDTA and SalG caused inactivation, which was eightfold higher for E. coli ∆pliG (about 300-fold) than for the wild-type (about 35-fold) (Table 3). This increased sensitivity of the ∆pliG strain, specifically to the combined treatment, was also observed in the growth inhibition assay, which was performed with growing cultures in LB broth (Fig. 4a). Detailed analysis of the growth curves indicates that growth inhibition takes place immediately after the addition of SalG and EDTA. When destabilization of the outer membrane by tolA knock-out was used as an alternative to EDTA addition, growth inhibition experiments showed a similar pattern, with the tolA::Km ∆pliG exhibiting a growth retardation compared to the tolA::Km strain in the presence of SalG (Fig. 4b).
Table 3.
Sensitivity of E. coli strains to g-type lysozyme in the presence of EDTA
| Condition | Viability reduction factor (N0/N24)a | |
|---|---|---|
| E. coli | E. coli ∆pliG | |
| 10 mM PPB pH 7.0 (buffer) | 0.055 ± 0.051 | 0.219 ± 0.095 |
| Buffer + 1 mM EDTA | 2.00 ± 1.69 | 1.14 ± 0.13 |
| Buffer + 15 μg/ml SalG | 0.055 ± 0.024 | 0.140 ± 0.009 |
| Buffer + 1 mM EDTA and 15 μg/ml SalG | 37.3 ± 25.23 | 297.5 ± 10.6 |
aInactivation was expressed by the viability reduction factor, with N0 the initial colony count and N24 the colony count after 24 h of incubation at 22°C. Mean values were calculated from three replicate treatments
Fig. 4.
Influence of PliG inhibitors on E. coli growth inhibition by SalG. a Growth curves of E. coli and E. coli ΔpliG at 30°C in LB broth, with addition of 1 mM EDTA (respectively, filled triangle/gray square) or 1 mM EDTA and 150 μg/ml SalG (respectively, open triangle/open square) after 2 h. b Growth curves at 30°C in LB broth of E. coli tolA::Km with 35 μg/ml (open triangle) or 3.75 μg/ml (filled triangle) SalG, and E. coli tolA::Km ΔpliG with 35 μg/ml (open square) or 3.75 μg/ml (gray square) SalG
To further confirm that these differences between wild-type and ∆pliG are due the failure to produce PliG protein, a genomic inducible pliG overexpression strain with a tolA::Km and ∆pliG background was constructed (E. coli tolA::Km ∆pliG PBAD-pliG), and subjected to the inactivation assay (with cells grown in the presence of 0.1% arabinose). The tolA::Km ∆pliG strain underwent a 600-fold inactivation in the presence of SalG, while the tolA::Km ∆pliG PBAD-pliG strain (PliG overexpressor) showed complete survival, like the tolA::Km strain (wild-type with respect to PliG) (Table 4).
Table 4.
Sensitivity of E. coli strains with a tolA background to g-type lysozyme
| Strain | Viability reduction factor (N0/N24)a | |
|---|---|---|
| 10 mM PPB pH 7.0 (buffer) | Buffer + 36 μg/ml SalG | |
| E. coli tolA::Km | 5.05 ± 2.08 | 4.6 ± 1.66 |
| E. coli tolA::Km ∆pliG | 13.9 ± 6.87 | 751 ± 55.8 |
| E. coli tolA::Km ∆pliG PBAD-pliG | 12.4 ± 9.51 | 7.60 ± 6.09 |
Strains were grown in the presence of 0.1% arabinose
aInactivation was expressed by the viability reduction factor, with N0 the initial colony count and N24 the colony count after 24 h of incubation at 22°C. Mean values were calculated from three replicate treatments
Discussion
In this work, we report the first discovery of a specific bacterial lysozyme inhibitor against g-type lysozyme (PliG). This finding extends the recent discoveries of specific bacterial inhibitors of c-type lysozyme (Ivy, PliC/MliC) [12, 15] and i-type lysozyme (PliI) [18]. We isolated PliG initially from E. coli, but putative homologs can be found in about 15–20 other proteobacterial genera, and at least those from Salmonella typhimurium and Aeromonas hydrophila were shown to be active. The earlier described lysozyme inhibitors also occur predominantly in the Proteobacteria, but although many bacteria have more than one inhibitor homolog, the occurrence of different inhibitors does not appear to be correlated. For example, based on available genome sequences, all E. coli strains carry ivy, mliC and pliG but not pliI, while Salmonella has pliC, mliC and pliG, and A. hydrophila has homologs of all four inhibitors. In the hypothesis that lysozyme inhibitors are involved in bacterial interactions with animals, the production of multiple inhibitors against different types of lysozyme by most bacteria could be linked to the fact that most animals produce two types of lysozyme, and that bacteria often have more than one host, vertebrate or invertebrate. An overview of the specificity of the hitherto known lysozyme inhibitors based on published data, and on a few additional tests in case enzyme and inhibitor were available, is shown in Table 5. It can be seen that all inhibitors are highly specific, except for a weak cross-inhibition observed for both c-type lysozyme inhibitors (Ivy and MliC/PliC) against the g-type lysozyme of bird origin (GEWL). Interestingly, PliG inhibits not only the g-type lysozyme from goose and salmon but also from the larvacean Urochordate Oikopleura.
Table 5.
Overview of the inhibition spectrum of lysozyme inhibitors
| Inhibitor | Lysozyme | |||||||
|---|---|---|---|---|---|---|---|---|
| c-type | g-type | i-type | Other* | |||||
| HEWL | Human Lysozyme | SalG | GEWL | Oikopleuraa | TjL | Bacteriophage T7 lysozymea | Mutanolysin (Sigma-Aldrich) | |
| Ivy | + | + | −* | +/− | − | −* | − | − |
| PliC/MliC | + | + | −* | +/− | n.t. | −* | − | − |
| PliI | − | n.t. | − | n.t. | n.t. | + | − | − |
| PliG | − | n.t. | + | + | + | − | − | − |
Data were compiled from Callewaert et al. [15, 19], Monchois et al. [12], Nilsen et al. [33] and Van Herreweghe et al. [18] except those marked with ‘*’, which are from unpublished data from the authors
+ Strong inhibition, ± weak inhibition, − no inhibition, n.t. not tested
aPartially purified extract
A peculiarity of pliG is its similarity to the so-called bacterial prepeptidase C-terminal (PPC) domain. This domain occurs in some bacterial peptidases and is cleaved from the mature enzyme, but its function, if any, remains speculative [37, 38]. Two such PPC domains were cloned and expressed as separate proteins in this work, but failed to inhibit g-type lysozyme. It remains interesting, however, to speculate that some PPC domains may have PliG activity, in particular because peptidases with PPC domains also frequently occur in Gram-positive bacteria, in which no lysozyme inhibitors have been described thus far. Although PliG shows little overall similarity to the other lysozyme inhibitors, it shares a common motif that was earlier reported to exist in PliC/MliC and PliI and that was shown to be important for lysozyme inhibition by X-ray crystallography and mutagenesis [18, 35]. Taken together, the common motif and the limited distribution of PliC/MliC, PliI and PliG suggest that these inhibitors share a common origin and have arisen after branching of the Proteobacteria.
Like for the c-type and i-type inhibitors, we have been able to demonstrate that PliG confers increased lysozyme tolerance to the bacteria producing it. Although these experiments are somewhat artificial, involving an ex-vivo challenge of the bacteria with lysozyme in the presence of an outer membrane destabilizing agent (EDTA or lactoferrin), or the use of bacterial mutants with a destabilized outer membrane, they suggest a role for these inhibitors in bacteria that closely interact with a lysozyme-producing animal host. Most of the bacterial genera having a putative PliG homolog have indeed documented relationships with animals known or predicted to produce g-type lysozyme. For example, Escherichia, Salmonella, Shigella and Acinetobacter comprise notable human and other vertebrate pathogens; B. avium causes respiratory infections in birds; and Edwardsiella, Photobacterium, Vibrio angustum, and A. hydrophila are more typically found in aquatic habitats and include several fish pathogens. Conversely, it is true that several prominent pathogens appear to lack a PliG homolog. However, taking in consideration the existence of two different inhibitors of c-type lysozyme (Ivy and PliC/MliC), the possible existence of additional, unrelated, g-type lysozyme inhibitors should not be excluded.
While an answer to the question of whether and to what extent PliG contributes to host colonization and bacterial virulence in vivo will have to await challenge studies in animal models, it is worthwhile to consider the evidence for a role of g-type lysozyme in antibacterial defense of animals in the first place. G-type lysozyme is generally considered to be less important than c-type in mammals, although data on its occurrence and expression are scarce. The highest concentrations of lysozyme are found in the tears, milk and saliva of mammals, but this is c-type lysozyme. The expression of the g-type lysozyme gene is tissue-specific and differs between animals. For example, expression has been reported in the human adult kidney, which can be affected by uropathogenic E. coli (UPEC), in the mouse tongue and skin and in the chicken intestine, both of which are important barriers for bacterial invasion [39]. In fish, the innate immune system is considered to be more important than in mammals as the first line of defense against pathogens, and several studies have indicated a more important role of g-type lysozyme in fish compared to mammals. In several fish species, g-type lysozyme is expressed in tissues exposed to the external environment and/or that are involved in the immune response, such as the gills, the head kidney, the intestine, and the liver, and its expression is strongly enhanced upon exposure to bacterial pathogens [40]. Similarly, g-type lysozyme has been linked to innate immune defense in bivalves. In the Zhikong scallop Chlamys farreri, specific nucleotide polymorphisms in the promoter region of the g-type lysozyme gene were found to be associated with resistance against Listonella anguillarum [41], and transcripts were most abundant in the tissues of gills, hepatopancreas and gonad. Thus, while g-type lysozyme is generally less well known than c-type, its role in innate immunity in several animals is supported by strong circumstantial evidence. A specific difficulty that, until now, has hampered studies to establish the role of different lysozymes in immune defense, is that lysozyme activity assays do not distinguish between c-, g- or i-type lysozymes. With the discovery of PliG, a complete toolbox of three highly specific lysozyme inhibitors is now available, which makes it possible to assess the presence and/or activity of individual lysozymes in homogenous enzyme assays, affinity blotting, ELISA-type assays, etc.
Besides protecting bacteria against exogenous lysozymes, another possible function of lysozyme inhibitors that has been suggested is to control the activity of endogenous bacterial autolysins. Autolysins are vital peptidoglycan-degrading enzymes that create local cell wall lesions that are necessary for cell growth and division, flagellar assembly and some types of macromolecular transport. In support of this alternative function, MltB from Pseudomonas aeruginosa, a member of the lytic transglycosylase class of autolysins, was found to be inhibited by Ivy in vitro [42]. While MltB does not show any evident similarity to animal lysozymes, the so-called family 1 lytic transglycosylases are clearly related to g-type lysozymes [43], and it will be of interest to see whether representatives of this family, like E. coli Slt70, are inhibited by PliG. In the E. coli PliG knock-out created in this work, we did not observe any obvious phenotype related to growth, cell elongation, cell division or motility that could be associated with uncontrolled autolysin activity (data not shown). However, assuming that the inhibitors would function to control autolysins, it cannot be excluded that they are functionally redundant, just like several of the autolysins themselves [44].
In conclusion, with the current discovery of PliG, bacterial inhibitors of the three major types of lysozyme that occur in animals have now been described. The majority of Proteobacteria appear to produce at least one or more inhibitors, but the known inhibitors are absent in most other bacteria. Indications are emerging to support a role of lysozyme inhibitors in interactions with animal hosts and in controlling autolysis, but these need to be further substantiated.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
L.V. holds a doctoral fellowship from the Flemish Institute for the Promotion of Scientific Technological Research (IWT). J.M.V.H. was supported a doctoral and L.C. a postdoctoral fellowship of the Research Foundation-Flanders (F.W.O.-Vlaanderen). We thank Griet Compernolle for conducting SPR analysis. We also acknowledge P. Orndorff (Department Microbiology, Pathology and Parasitology, North Carolina State University, College of Veterinary Medicine, Raleigh, NC, USA) and M.A. Valvano (Departments of Microbiology and Immunology, University of Western Ontario, London, ON N6A5C1, Canada) for providing the bacterial strains Bordetella avium 197N and E. coli GL113 ΔtolA::Km, respectively. A construct for production of recombinant T7 lysozyme was kindly donated by Prof. Dr. Smita Patel (Departement of Biochemistry, Robert Wood Johnson Medical School University of Medicine and Dentisitry, NJ, USA).
References
- 1.Weaver L, Grütter M, Matthews B. The refined structures of goose lysozyme and its complex with a bound trisaccharide show that the “goose-type” lysozymes lack a catalytic aspartate residue. J Mol Biol. 1995;245(1):54–68. doi: 10.1016/S0022-2836(95)80038-7. [DOI] [PubMed] [Google Scholar]
- 2.Goto T, Abe Y, Kakuta Y, Takeshita K, Imoto T, Ueda T. Crystal structure of Tapes japonica lysozyme with substrate analogue: structural basis of the catalytic mechanism and manifestation of its chitinase activity accompanied by quaternary structural change. J Biol Chem. 2007;282(37):27459–27467. doi: 10.1074/jbc.M704555200. [DOI] [PubMed] [Google Scholar]
- 3.Savan R, Aman A, Sakai M. Molecular cloning of G type lysozyme cDNA in common carp (Cyprinus carpio L.) Fish Shellfish Immunol. 2003;15(3):263–268. doi: 10.1016/S1050-4648(02)00159-6. [DOI] [PubMed] [Google Scholar]
- 4.Grütter M, Weaver L, Matthews B. Goose lysozyme structure: an evolutionary link between hen and bacteriophage lysozymes? Nature. 1983;303(5920):828–831. doi: 10.1038/303828a0. [DOI] [PubMed] [Google Scholar]
- 5.Bachali S, Jager M, Hassanin A, Schoentgen F, Jollès P, Fiala-Medioni A, Deutsch J. Phylogenetic analysis of invertebrate lysozymes and the evolution of lysozyme function. J Mol Evol. 2002;54(5):652–664. doi: 10.1007/s00239-001-0061-6. [DOI] [PubMed] [Google Scholar]
- 6.Callewaert L, Michiels CW. Lysozymes in the animal kingdom. J Biosci. 2010;35(1):127–160. doi: 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J, Song L, Li C, Zou H, Ni D, Wang W, Xu W. Molecular cloning of an invertebrate goose-type lysozyme gene from Chlamys farreri, and lytic activity of the recombinant protein. Mol Immunol. 2007;44(6):1198–1208. doi: 10.1016/j.molimm.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Hikima S, Hikima J, Rojtinnakorn J, Hirono I, Aoki T. Characterization and function of kuruma shrimp lysozyme possessing lytic activity against Vibrio species. Gene. 2003;2003(316):187–195. doi: 10.1016/S0378-1119(03)00761-3. [DOI] [PubMed] [Google Scholar]
- 9.Canfield R, McMurry S. Purification and characterization of a lysozyme from goose egg white. Biochem Biophys Res Commun. 1967;26(1):38–42. doi: 10.1016/0006-291X(67)90249-5. [DOI] [PubMed] [Google Scholar]
- 10.Nile C, Townes C, Michailidis G, Hirst B, Hall J. Identification of chicken lysozyme g2 and its expression in the intestine. Cell Mol Life Sci. 2004;61(21):2760–2766. doi: 10.1007/s00018-004-4345-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32(2):287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 12.Monchois V, Abergel C, Sturgis J, Jeudy S, Claverie J. Escherichia coli ykfE ORFan gene encodes a potent inhibitor of C-type lysozyme. J Biol Chem. 2001;276(21):18437–18441. doi: 10.1074/jbc.M010297200. [DOI] [PubMed] [Google Scholar]
- 13.Deckers D, Vanlint D, Callewaert L, Aertsen A, Michiels CW. Role of the lysozyme inhibitor Ivy in growth or survival of Escherichia coli and Pseudomonas aeruginosa bacteria in hen egg white and in human saliva and breast milk. Appl Environ Microbiol. 2008;74(14):4434–4439. doi: 10.1128/AEM.00589-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deckers D, Masschalck B, Aertsen A, Callewaert L, Van Tiggelen CGM, Atanassova M, Michiels CW. Periplasmic lysozyme inhibitor contributes to lysozyme resistance in Escherichia coli . Cell Mol Life Sci. 2004;61(10):1229–1237. doi: 10.1007/s00018-004-4066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callewaert L, Aertsen A, Deckers D, Vanoirbeek KGA, Vanderkelen L, Van Herreweghe JM, Masschalck B, Nakimbugwe D, Robben J, Michiels CW. A new family of lysozyme inhibitors contributing to lysozyme tolerance in gram-negative bacteria. PLoS Pathogens. 2008;4(3):e1000019. doi: 10.1371/journal.ppat.1000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn R, Tate J, Mistry J, Coggill P, Sammut S, Hotz H, Ceric G, Forslund K, Eddy S, Sonnhammer E, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daigle F, Graham J, Rr Curtiss. Identification of Salmonella typhi genes expressed within macrophages by selective capture of transcribed sequences (SCOTS) Mol Microbiol. 2001;41(5):1211–1222. doi: 10.1046/j.1365-2958.2001.02593.x. [DOI] [PubMed] [Google Scholar]
- 18.Van Herreweghe J, Vanderkelen L, Callewaert L, Aertsen A, Compernolle G, Declerck P, Michiels C (2010) Lysozyme inhibitor conferring bacterial tolerance to invertebrate type lysozyme. Cell Mol Life Sci. doi:10.1007/s00018-009-0241-x [DOI] [PMC free article] [PubMed]
- 19.Callewaert L, Masschalck B, Deckers D, Nakimbugwe D, Atanassova M, Aertsen A, Michiels CW. Purification of Ivy, a lysozyme inhibitor from Escherichia coli, and characterisation of its specificity for various lysozymes. Enzyme Microb Technol. 2005;37(2):205–211. doi: 10.1016/j.enzmictec.2005.03.001. [DOI] [Google Scholar]
- 20.Kyomuhendo P, Nilsen I, Brandsdal B, Smalås A. Structural evidence for lack of inhibition of fish goose-type lysozymes by a bacterial inhibitor of lysozyme. J Mol Model. 2008;14(9):777–788. doi: 10.1007/s00894-008-0317-9. [DOI] [PubMed] [Google Scholar]
- 21.Kawamura S, Ohno K, Ohkuma M, Chijiiwa Y, Torikata T. Experimental verification of the crucial roles of Glu73 in the catalytic activity and structural stability of goose type lysozyme. J Biochem. 2006;140(1):75–85. doi: 10.1093/jb/mvj125. [DOI] [PubMed] [Google Scholar]
- 22.Helland R, Larsen R, Finstad S, Kyomuhendo P, Larsen A. Crystal structures of g-type lysozyme from Atlantic cod shed new light on substrate binding and the catalytic mechanism. Cell Mol Life Sci. 2009;66(15):2585–2598. doi: 10.1007/s00018-009-0063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hikima J, Minagawa S, Hirono I, Aoki T. Molecular cloning, expression and evolution of the Japanese flounder goose-type lysozyme gene, and the lytic activity of its recombinant protein. Biochim Biophys Acta. 2001;1520(1):35–44. doi: 10.1016/s0167-4781(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 24.Arnheim N, Inouye M, Law L, Laudin A. Chemical studies on the enzymatic specificity of goose egg white lysozyme. J Biol Chem. 1973;248(1):233–236. [PubMed] [Google Scholar]
- 25.Jollès P, Jollès J. What’s new in lysozyme research? Always a model system, today as yesterday. Mol Cell Biochem. 1984;63(2):165–189. doi: 10.1007/BF00285225. [DOI] [PubMed] [Google Scholar]
- 26.Nakimbugwe D, Masschalck B, Deckers D, Callewaert L, Aertsen A, Michiels CW. Cell wall substrate specificity of six different lysozymes and lysozyme inhibitory activity of bacterial extracts. FEMS Microbiol Lett. 2006;259(1):41–46. doi: 10.1111/j.1574-6968.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 27.Kyomuhendo P, Myrnes B, Nilsen I. A cold-active salmon goose-type lysozyme with high heat tolerance. Cell Mol Life Sci. 2007;64(21):2841–2847. doi: 10.1007/s00018-007-7372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988;9(1):28–32. doi: 10.1002/elps.1150090106. [DOI] [PubMed] [Google Scholar]
- 29.Datsenko K, Wanner B. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendtsen J, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 32.Thunnissen A, Isaacs N, Dijkstra B. The catalytic domain of a bacterial lytic transglycosylase defines a novel class of lysozymes. Proteins. 1995;22(3):245–258. doi: 10.1002/prot.340220305. [DOI] [PubMed] [Google Scholar]
- 33.Nilsen I, Myrnes B, Edvardsen R, Chourrout D. Urochordates carry multiple genes for goose-type lysozyme and no genes for chicken- or invertebrate-type lysozymes. Cell Mol Life Sci. 2003;60(10):2210–2218. doi: 10.1007/s00018-003-3252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Yum S, Kim M, Xu Y, Jin X, Yoo H, Park J, Gong J, Choe K, Lee B, Ha N. Structural basis for the recognition of lysozyme by MliC, a periplasmic lysozyme inhibitor in Gram-negative bacteria. Biochem Biophys Res Commun. 2009;378(2):244–248. doi: 10.1016/j.bbrc.2008.11.039. [DOI] [PubMed] [Google Scholar]
- 36.Frith M, Saunders N, Kobe B, Bailey T. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol. 2008;4(4):e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeats C, Bentley S, Bateman A. New knowledge from old: in silico discovery of novel protein domains in Streptomyces coelicolor . BMC Microbiol. 2003;3:3. doi: 10.1186/1471-2180-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi L, Sowdhamini R. Genome-wide survey of prokaryotic serine proteases: analysis of distribution and domain architectures of five serine protease families in prokaryotes. BMC Genomics. 2008;9:549. doi: 10.1186/1471-2164-9-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irwin D, Gong Z. Molecular evolution of vertebrate goose-type lysozyme genes. J Mol Evol. 2003;56(2):234–242. doi: 10.1007/s00239-002-2396-z. [DOI] [PubMed] [Google Scholar]
- 40.Caipang C, Brinchmann M, Kiron V. Profiling gene expression in the spleen of Atlantic cod, Gadus morhua upon vaccination with Vibrio anguillarum antigen. Comp Biochem Physiol B Biochem Mol Biol. 2009;153(3):261–267. doi: 10.1016/j.cbpb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Zhao J, Wang L, Qiu L, Zhang H, Dong C, Cong M, Song L. The polymorphism of lysozyme gene in Zhikong scallop (Chlamys farreri) and its association with susceptibility/resistance to Listonella anguillarum . Fish Shellfish Immunol. 2009;27(2):136–142. doi: 10.1016/j.fsi.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Clarke C, Scheurwater E, Clarke A. The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J Biol Chem. 2010;285(20):14843–14847. doi: 10.1074/jbc.C110.120931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheurwater E, Reid C, Clarke A. Lytic transglycosylases: bacterial space-making autolysins. Int J Biochem Cell Biol. 2008;40(4):586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 44.Heidrich C, Ursinus A, Berger J, Schwarz H, Höltje J. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli . J Bacteriol. 2002;184(22):6093–6099. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinés E, Marolda C, Balachandran A, Valvano M. Defective O-antigen polymerization in tolA and pal mutants of Escherichia coli in response to extracytoplasmic stress. J Bacteriol. 2005;187:3359–3368. doi: 10.1128/JB.187.10.3359-3368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crooks G, Hon G, Chandonia J, Brenner S. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.