Abstract
Directed evolution of proteins for improved or modified functionality is an important branch of modern biotechnology. It has traditionally been performed using various in vitro methods, but more recently, methods of in vivo artificial evolution come into play. In this review, we discuss and compare prokaryotic and eukaryotic-based systems of directed protein evolution in vivo, highlighting their benefits and current limitations and focusing on the biotechnological potential of vertebrate immune cells for the generation of protein diversity by means of the immunoglobulin diversification machinery.
Keywords: Directed evolution, Protein engineering, Mutator strain, Cell culture-based systems, Somatic hypermutation, Gene conversion
Introduction
Directed protein evolution is a powerful approach to optimize functions and characteristics of diverse proteins or even develop proteins with totally new features [1–4]. The most widespread directed evolution techniques are based on various in vitro methods such as random and site-directed mutagenesis or DNA shuffling, as well as mRNA, ribosome, and phage displays [5–11]. In recent years, however, new in vivo methods of directed evolution have been brought to life, with their popularity increasing and having a potential for further development.
The in vitro systems of directed protein evolution have proven to be a powerful tool for optimizing and modifying proteins [12]. The strategies of in vitro protein evolution can be, in general, divided into two major parts: pure in vitro systems, where both the generation of diversity and the selection of the improved variants are performed in vitro, and combined approaches, where the mutagenesis performed in vitro is followed by cell transformation/transfection with the subsequent screening in vivo. Typical examples of the combined approach are phage display [11] (in which case both generation of diversity and selection are typically performed in vitro, with the step of bacterial transformation in between) and generation of mutagenized gene libraries through DNA shuffling or error-prone PCR followed by bacterial transformation and selection [13, 14]. Such in vitro-based diversification methods with the in vivo sequel provide impressive results. In a recent example, a combination of error-prone PCR, bacterial transformation, and FACS-based screening for fluorescently labeled agonist binding has evolved the amino acid sequence of a G protein-coupled receptor (GPCR) towards enhanced protein production and stability upon bacterial expression—an achievement which may significantly ease GPCR crystallization and structure analysis [15]. However, the combined in vitro/in vivo approaches have certain limitations, intrinsic to several steps of the existing artificial evolution protocols. For example, the analyzable size of a gene library is restricted by the host cell transformation efficiency. Further, the iterative application of the in vitro diversification methods is laborious if the screening is performed in transfected eukaryotic cells.
The pure in vitro systems of directed protein evolution are exemplified by mRNA [16] and ribosome [17] displays. These techniques are based on the in vitro translation of a diversified nucleic acid library with subsequent isolation of the synthesized proteins with the highest affinity for a desired ligand. These techniques rely on the simultaneous isolation of the corresponding mRNA molecules encoding the selected protein, which is achieved by a covalent mRNA-protein linkage with puromycin in the case of mRNA display, and through isolation of the non-covalent protein–mRNA–ribosome complexes in the ribosome display [16, 17]. In both setups, the isolated mRNAs are applied to more rounds of mutagenesis, translation, and selection. These assays do not suffer from the handicaps of low transformation efficiency, therefore the screenable library size is expanded by several orders of magnitude in comparison to the phage or bacterial displays [10]. Another advantage of pure in vitro systems is the unique possibility to work with protein sequences that are unstable or toxic in cells [10, 18]. Pure in vitro protein evolution systems are efficient when a simple affinity-based selection of modified proteins is desired, but are difficult to apply when a more complex optimization of proteins (e.g., involved in metabolic pathways) is required.
The limitations mentioned above can be overcome by application of the in vivo-only methods of directed evolution. Artificial evolution in vivo combines the benefits of generating high genetic diversity and conducting selection of improved protein variants, both performed in a living cell culture [2]. Performing all manipulations within the real-life environment of a cell is important, as many proteins are affected by the intracellular parameters such as ion concentrations and pH, as well as peculiarities of protein folding, post-translational modifications, and degradation. The natural cellular environment is also of high significance for directed evolution of target proteins which participate in complex multi-protein interactions, since the latter can be difficult to reproduce in more artificial systems. The overall potential for the generation of diversity is fairly high in living cell-based systems, as a cell culture can be expanded to an amount of hundreds of millions of single cells (or even billions in the case of bacteria) carrying individual mutations that accumulate in parallel with cell divisions.
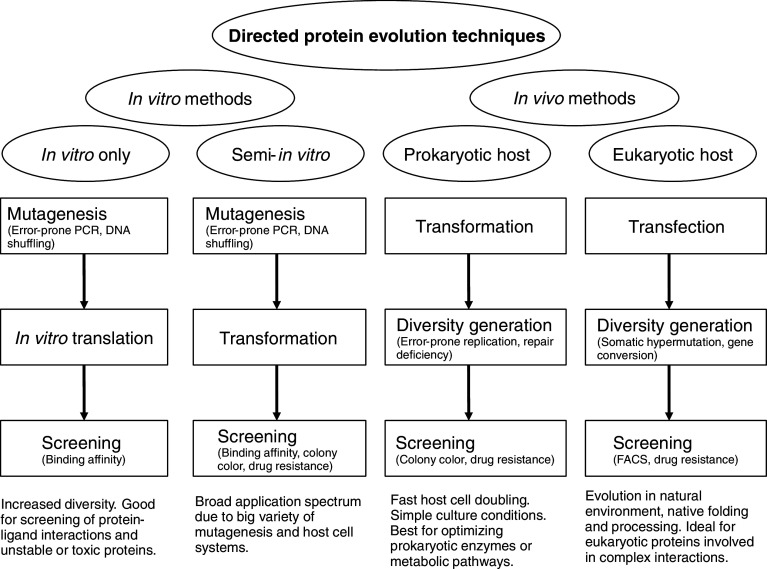
Of course, living cells also posses certain limitations as a tool for directed protein evolution. One of them is the difficulty to efficiently mutagenize the sequence of interest without doing much damage to the cell itself; another one is the problem of selecting the cells with acquired beneficial mutations—for not all mutant phenotypes are measurable in a cell culture. Although the in vivo technologies of directed evolution are not yet as widespread as the methods described above based on the generation of diversity in vitro, the in vivo technologies are expected to soon develop into the broadest biotechnological applications. In this review, we discuss diverse methods of artificial protein evolution in vivo compared to each other and to the in vitro systems, with their benefits, handicaps, and examples of successful solutions to the problems mentioned above (Fig. 1).
Fig. 1.
Schematic diagram depicting the main directed protein evolution strategies, highlighting their benefits and possible applications (see text for details)
Prokaryotic in vivo evolution systems
As prokaryotic organisms have been a “workhorse” for molecular biologists since long ago, usage of Escherichia coli and other bacteria for in vivo protein engineering seems to be the most straightforward approach. However, the natural mutation rate in bacteria, despite their short doubling time, is too low for this purpose, and special mutator strains have been generated to facilitate the mutagenesis process. Most often, DNA repair-deficient bacterial strains are used, such as the commercially available E. coli XL1-Red, deficient for mutD, mutS, and mutT genes [19]. Upon transformation of XL1-Red with a plasmid encoding the target DNA, mutations are accumulated at each round of DNA replication. The mutation frequency of this method is reported to be 1 base change per 2,000 nucleotides [19], which is optimal when a relatively low amount of mutations randomly distributed in the target sequence is expected to bring about the desired changes in performance of the protein of interest [20]. As an example, the XL1-Red strain has been used to change the substrate specificity of an esterase from Pseudomonas fluorescens, obtaining variants capable of hydrolyzing a sterically hindered 3-hydroxy ester, which serves as a component in the synthesis of epothilones [21, 22]. Screening has been performed by plating XL1-Red E.coli expressing the P. fluorescens esterase on minimal media agar plates containing indicator substances (Neutral Red and Crystal Violet). The esterase-catalyzed hydrolysis of the 3-hydroxy ester (ethyl or glycerol ester) has been detected by formation of red-colored colonies due to a pH decrease caused by the released acid.
The XL1-Red mutator strain has also been successfully applied to shift the activity conditions of ADH beta-glucuronidase from Lactobacillus gasseri [23]. The original enzyme exhibited maximal activity at the acidic pH 5.0, but was poorly active at the neutral pH 6–7. Usage of the XL1-Red system produced variants of ADH beta-glucuronidase with increased activity at the neutral pH. The modified enzyme could then be utilized as a reporter in non-acidophilic expression hosts, in addition to its previous usage in lactic acid bacteria and other microorganisms growing in acidifying environments. The XL1-Red mutator system has also been applied to the entire viral genome of the Citrus tatter leaf virus to select for mutants with altered pathogenicity in inoculated plants [24].
There are reports of additional bacterial mutator strains with different DNA repair or replication deficiencies. For example, an E. coli strain allowing IPTG-induced expression of antisense RNAs simultaneously silencing three genes involved in DNA repair and synthesis—mutS, mutD (dnaQ), and ndk—increased the spontaneous mutation frequency over 2,000-fold [25]. A mutator phenotype has also been shown in a strain of Bacillus anthracis deficient for the dnaN1 gene that encodes the beta subunit of a replicative DNA polymerase [26]. However, these strains have not so far been used in directed evolution applications.
An elegant approach to preferentially mutagenize the target DNA has been developed based on an E. coli strain with mutations in structural domains of DNA polymerase I responsible for Pol I fidelity [27]. As Pol I is predominantly active in replication of the endogenous ColE1 plasmid, this error-prone polymerase mostly affects target sequences provided by the plasmid. The resulting mutation rate reaches 8.1 × 10−4 mutations per base pair, giving an 80,000-fold increase over the natural value. The mutation range extends for about 3 kb from the replication origin of the plasmid, with the maximal frequency in the first ca. 700-bp region. This mutational profile results from the role of Pol I in initiation of replication of the ColE1 plasmid; subsequent replication steps are largely dependent on the more accurate Pol III [28]. In the region of maximal mutation frequency, mutations are distributed rather evenly and show a variety of base pair substitutions, predominantly transitions. Mutagenesis induced by the error-prone Pol I is most active in bacterial cultures maintained at the stationary phase. This system has been tested for enzyme modification by targeting TEM-1 β-lactamase for mutagenesis and selecting for resistance to a third-generation lactam antibiotic, aztreonam, which is very low for the wild-type enzyme. As a result, aztreonam resistance has increased 150-fold [27].
The applications described above deal with improvements of single target genes. A novel method called multiplex automated genome engineering (MAGE) has been developed to target and simultaneously optimize a series of genes responsible for a complex biosynthetic pathway [29]. The method utilizes the E.coli strain EcNR2 expressing bacteriophage l-Red ssDNA-binding protein b. This strain allows incorporation of homologous oligonucleotides into the lagging DNA strand during DNA replication; a pool of oligonucleotides homologous to the targeted genes but carrying nucleotide substitutions is repeatedly introduced into the cultured cells [29, 30]. Under optimal conditions, ca. 30% of the cell population accumulate new genetic modifications introduced in this manner. The overall capacity of mutation accumulation could reach as many as 15 billion genetic variants (4.3 × 108 bp variations per cycle for 35 MAGE cycles) in a propagating bacterial culture [29]. The correct selection of homologous oligonucleotides allows targeting chosen genes responsible for a desired biochemical pathway. This technology has been applied to the E. coli 1-deoxy-d-xylulose-5-phosphate (DXP) biosynthesis pathway in order to increase production of the industrially important isoprenoid lycopene [29]. Within 3 days of bacterial culture, overproduction strains were isolated with a more than fivefold increase in the lycopene yield; 24 genes involved in the DXP pathway were found to be modified in these strains [29].
In general, artificial evolution of proteins in prokaryotic systems is attractive due to simple culturing conditions and the fast doubling time of host cells; for example, the XL1-Red system takes an overnight culture to create a broad spectrum of mutations. The most suitable screening systems for bacteria are those that allow distinguishing between bacterial colonies by means of drug resistance or some color manifestations. However, the biotechnological potential of prokaryotes is limited in the sense of the range of possible directed evolution targets. It is clear that bacterial enzymes or even metabolic networks can be directly targeted in prokaryotes. In contrast, eukaryotic proteins, which should undergo complicated folding and post-translational modifications, unavailable in bacteria, are often unsuitable for directed evolution in prokaryotic hosts.
Eukaryotic in vivo evolution systems
Eukaryotic in vivo artificial evolution systems provide many advantages compared to the prokaryotic ones (Fig. 1). In these systems, the eukaryotic protein evolves in a natural environment, which ensures proper folding, post-translational modifications, and intracellular localization (or secretion). Thus establishment of eukaryotic systems able to successfully mutagenize target proteins is of high importance for modern biotechnology.
Talking about eukaryotic in vivo evolution systems, it is logical to start with yeast as a simple eukaryote highly popular in numerous biotechnological applications. In a recent usage of the yeast homologous recombination for directed protein evolution, simultaneous modification of several loops in a target enzyme by means of recombination with a synthetic diversified oligonucleotide library was achieved [31]. A mixture of diversified oligonucleotides containing flanking regions complementary to the target sequence was used together with a linearized plasmid carrying the gene of interest for yeast transformation. The system was tested for the stop-codon reversion in a classic yeast selection marker TRP1, which allowed selecting positive clones based on their ability to grow in the absence of tryptophan. The average efficiency of oligonucleotide recombination at a single position was ca. 5%, although it decreased from 20 to 25-fold when another stop codon was modified simultaneously (giving an average efficiency of 0.2% for two and 0.01% for three positions) [31]. In general, this oligonucleotide-based strategy resembles the bacterial approach MAGE, described above [29] and seems to be useful, though the decrease in efficiency when modifying multiple sites is a handicap.
A simple approach to directed eukaryotic in vivo evolution appears to be the usage of mismatch repair-deficient cells, similarly to the way it is often done with the prokaryotic systems (see above). In this case, however, the same disadvantage as that relevant for the prokaryotic systems remains—such methods normally randomize the entire genome wastefully and often deleteriously rather than focusing on the gene of interest [19]. So, eukaryotic systems capable of autonomously diversifying the target transgenes are much desired for directed evolution in vivo as an accurate instrument for protein engineering and functional studies. An excellent solution for the mutation target specificity problem is offered by the natural mechanisms of antibody affinity improvement—somatic hypermutation and gene conversion.
Somatic hypermutation is a unique mechanism that takes place during development of the immune response when activated B cells encounter antigens [32–34]. This process is highly specific for B cells and results in a drastic increase in spontaneous point mutations in variable regions of the immunoglobulin light and heavy chain genes [35–38]. The mutation rate in somatic hypermutation is about 1 × 10−3 mutations per base pair per generation, 106 times higher than that in the rest of the genome [39]. The process requires activation-induced cytidine deaminase (AID) [40]. It is currently believed that AID deaminates deoxycytidine residues in actively transcribed genes [41–43]. As a consequence of AID activity, point mutations are introduced as the DNA repair mechanisms attempt to fix the AID-mediated DNA damage in an error-prone manner [38, 44–46]. The possibility to hypermutate a non-immunoglobulin transgene in AID-producing cell lines has been shown using neomycin phosphotransferase [47] or EGFP [48, 49].
The first attempt to use somatic hypermutation as a tool for directed protein evolution was performed on the monomeric red fluorescent protein (mRFP) [50] expressed in Burkitt lymphoma Ramos cells, a human B cell line that hypermutates its IgV genes continuously during culturing [51]. It was possible to deliver the target gene by random integration of a retroviral vector into the vicinity of a hypermutable immunoglobulin heavy chain locus, which resulted in hypermutation of the highly expressed transgene [52]. Lower levels of hypermutation were also detected with the transgene in several other integration sites [52]. Transcription of the transgenic mRFP was controlled by a doxycycline-inducible promoter, which permitted turning transcription on or off at will to start or stop hypermutation. The hypermutating cell culture was sorted by FACS for cells with the most far-red emission spectrum. Twenty-three rounds of repetitive cell sorting resulted in generation of mRFP variants with increased photostability and far-red emissions (649 nm), leaving behind the best results of the structure-based design [52].
Another mechanism used by B cells for immunoglobulin gene diversification is the pseudogene-templated gene conversion, which naturally exists in birds and some mammals like swine or rabbit and takes place simultaneously with somatic hypermutation, being controlled by the same key enzyme—the AID deaminase [53]. Deletion of the pseudo-V-gene conversion donor sequences in the chicken B cell line DT40 results in complete substitution of gene conversion by somatic hypermutation [54]. The DT40 cell line thus appears to be an excellent platform for in vivo protein engineering with the possibility of utilization of both mutagenizing mechanisms. Another advantage of DT40 over murine and human hypermutating B cell lines is the very high targeted-to-random transgene integration ratio achieved by means of homologous recombination [55] which allows controlling the target gene integration site and delivering the target gene directly to the immunoglobulin loci. Further enhancement of gene conversion efficiency in the DT40 cell line by addition of a histone deacetylase inhibitor trichostatin A (TSA) has been successfully used for upregulation of the native immunoglobulin variable gene segment diversification activity [56]. This effect has been applied to select DT40 cells producing specific antibodies with antigen-conjugated magnetic beads. This autonomously diversifying library (ADLib) selection system enabled the quick establishment (approximately 1 week from selection of bulk cells producing antibodies against an antigen of interest to screening by ELISA or FACS after subcloning by limited dilution and clonal expansion) of various clones producing monoclonal IgMs with specificity and affinity sufficient for immunological assays [57]. Antibody libraries obtained by means of this technology are applicable for subsequent rational antibody design. This method also permits creation of antibodies with specificity to autoantigens, thereby overcoming the problem of immune tolerance [58].
Another successful application of the DT40 line has been performed with artificial non-immunoglobulin gene conversion donor sequences and a non-immunoglobulin transgene [59]. The green fluorescent protein (GFP) gene served as a donor and the blue fluorescent protein (BFP) gene as an acceptor for gene conversion. A small percent of the initially BFP-expressing cells shifted their fluorescence from blue to green after being cultured for 2–3 weeks when the substrate constructs were integrated in the Ig light chain locus (but not in the ovalbumin locus, indicating the strong specificity of AID-mediated gene diversification in the DT40 line to the immunoglobulin loci). This fluorescence shift resulted from AID-dependent gene conversion where the GFP gene served as a template for the BFP gene diversification, thereby leading to introduction of variously sized GFP-derived gene segments into the BFP gene [59]. Thus, the gene conversion machinery in DT40 cells can serve as a useful tool to generate altered or improved non-Ig proteins by a type of in vivo DNA shuffling.
A variant of the DT40 cell line lacking the pseudo-V-genes (and thus unable to perform gene conversion) has been used for somatic hypermutation of the traditional example—the GFP gene, which was delivered to the immunoglobulin light chain locus using a specially designed homologous recombination-based targeting vector [60]. Three rounds of FACS sorting during 2 months culturing were sufficient to obtain variants with increased green fluorescence intensity. Analysis of the acquired mutations and their combination with rational design allowed generating GFP variants with more than threefold increased brightness [60].
In comparison to optimization of proteins in the hypermutating mammalian B cell lines such as Ramos or 18-81, DT40 offers advantages that transgenes can be inserted in a targeted manner as single copies into the Ig light chain locus and AID expression can be shut down by tamoxifen induction via the Cre–loxP system [54, 61]. In the future, transgene diversification in DT40 may be accomplished by addition of gene conversion on top of hypermutation if homologous non-immunoglobulin conversion donor sequences are inserted upstream of the transgene, thus resembling the natural process of antibody diversity generation in avian species. Studies on AID-dependent gene diversification have revealed that natural restriction of hypermutation and gene conversion activities to the immunoglobulin loci is controlled by cis-regulatory sequences usually overlapping with immunoglobulin enhancers [62–65]. These findings pave the way to creation of non B cell hypermutating cell lines as platforms for directed protein evolution by means of modifying a cell line of interest. Expression of AID in such a line and co-transfection with a target gene “armed” with the cis-regulatory sequence is expected to elicit efficient protein evolution in such non B cell lines.
Conclusions
Prokaryotic in vivo systems of artificial protein evolution were established more than a decade ago, and found a series of useful biotechnological applications such as improvement of activity or a shift in substrate specificity of bacterial enzymes as well as optimization of production of certain metabolites. Prokaryotic systems continue to improve, with the recent developments exemplified by the automated MAGE system described above [29]. Nevertheless, they possess a series of natural handicaps, such as a moderate mutation rate of hypermutable strains (just 3–4 orders of magnitude above the wild-type) and low compatibility with eukaryotic genes due to folding and modification problems.
Eukaryotic systems of directed in vivo evolution, on the other hand, are a good alternative to prokaryotes, as well as to the in vitro evolution systems, when it comes to optimizing eukaryotic proteins involved in complex intracellular interactions (Fig. 1). Due to high mutation rates (up to 106 times higher than the wild-type rates), convenient and fast evolution process, and the natural protein environment they offer, these systems have a strong potential for future development. Among the eukaryotic in vivo protein evolution platforms, the chicken cell line DT40 should be highlighted and is becoming more and more popular [66]. Increased frequency of targeted gene integration in DT40 gives a possibility to standardize the directed evolution process, escaping from the uncontrollable influence of the nearby genomic environment unavoidable upon non-targeted integration.
Being a relatively new approach compared to other protein evolution systems, somatic hypermutation and gene conversion have not so far found a broad application, focusing mostly on artificial improvement of immunoglobulins and the easily selectable non-immunoglobulin targets like fluorescent proteins. The real routine of protein improvement in vivo has to follow after these pioneering efforts. We should expect that future developments will endow researchers with the full potential of directed protein evolution in the natural vertebrate cell environment.
Acknowledgments
The work was supported by grant No. 02.740.11.5016 from the Russian Ministry of Science and by Deutsche Forschungsgemeinschaft (TR SFB-11) to V.L.K. A.B. is a recipient of the Russian state scholarship No. 8084 p/12612 “Participant of the Youth Contest for Science and Innovation”.
Abbreviations
- AID
Activation-induced deaminase
- BFP
Blue fluorescent protein
- EGFP
Enhanced GFP
- FACS
Fluorescence-activated cell sorting
- GFP
Green fluorescent protein
- GPCR
G protein-coupled receptor
- IPTG
Isopropyl β-d-1-thiogalactopyranoside
- mRFP
Monomeric red fluorescent protein
- PCR
Polymerase chain reaction
- TSA
Trichostatin A
References
- 1.Kaur J, Sharma R. Directed evolution: an approach to engineer enzymes. Crit Rev Biotechnol. 2006;26(3):165–199. doi: 10.1080/07388550600851423. [DOI] [PubMed] [Google Scholar]
- 2.Jackel C, Kast P, Hilvert D. Protein design by directed evolution. Annu Rev Biophys. 2008;37:153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- 3.Bottcher D, Bornscheuer UT Protein engineering of microbial enzymes. Curr Opin Microbiol 13 (3):274–282. S1369-5274(10)00015-9 [pii] [DOI] [PubMed]
- 4.Dougherty MJ, Arnold FH. Directed evolution: new parts and optimized function. Curr Opin Biotechnol. 2009;20(4):486–491. doi: 10.1016/j.copbio.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370(6488):389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 6.Ness JE, Welch M, Giver L, Bueno M, Cherry JR, Borchert TV, Stemmer WP, Minshull J. DNA shuffling of subgenomic sequences of subtilisin. Nat Biotechnol. 1999;17(9):893–896. doi: 10.1038/12884. [DOI] [PubMed] [Google Scholar]
- 7.Harayama S. Artificial evolution by DNA shuffling. Trends Biotechnol. 1998;16(2):76–82. doi: 10.1016/S0167-7799(97)01158-X. [DOI] [PubMed] [Google Scholar]
- 8.Shen B. PCR approaches to DNA mutagenesis and recombination. An overview. Methods Mol Biol. 2002;192:167–174. doi: 10.1385/1-59259-177-9:167. [DOI] [PubMed] [Google Scholar]
- 9.Matsuura T, Yomo T. In vitro evolution of proteins. J Biosci Bioeng. 2006;101(6):449–456. doi: 10.1263/jbb.101.449. [DOI] [PubMed] [Google Scholar]
- 10.Lipovsek D, Pluckthun A. In vitro protein evolution by ribosome display and mRNA display. J Immunol Methods. 2004;290(1–2):51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Smith GP, Petrenko VA. Phage display. Chem Rev. 1997;97(2):391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 12.Labrou NE. Random mutagenesis methods for in vitro directed enzyme evolution. Curr Protein Pept Sci. 2010;11(1):91–100. doi: 10.2174/138920310790274617. [DOI] [PubMed] [Google Scholar]
- 13.Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91(22):10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirino PC, Mayer KM, Umeno D. Generating mutant libraries using error-prone PCR. Methods Mol Biol. 2003;231:3–9. doi: 10.1385/1-59259-395-X:3. [DOI] [PubMed] [Google Scholar]
- 15.Sarkar CA, Dodevski I, Kenig M, Dudli S, Mohr A, Hermans E, Pluckthun A. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc Natl Acad Sci USA. 2008;105(39):14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Barrick JE, Szostak JW, Roberts RW. Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods Enzymol. 2000;318:268–293. doi: 10.1016/S0076-6879(00)18058-9. [DOI] [PubMed] [Google Scholar]
- 17.Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94(10):4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spirin AS. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotechnol. 2004;22(10):538–545. doi: 10.1016/j.tibtech.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Greener A, Callahan M, Jerpseth B. An efficient random mutagenesis technique using an E. coli mutator strain. Methods Mol Biol. 1996;57:375–385. doi: 10.1385/0-89603-332-5:375. [DOI] [PubMed] [Google Scholar]
- 20.Chusacultanachai S, Yuthavong Y. Random mutagenesis strategies for construction of large and diverse clone libraries of mutated DNA fragments. Methods Mol Biol. 2004;270:319–334. doi: 10.1385/1-59259-793-9:319. [DOI] [PubMed] [Google Scholar]
- 21.Bornscheuer UT, Altenbuchner J, Meyer HH. Directed evolution of an esterase for the stereoselective resolution of a key intermediate in the synthesis of epothilones. Biotechnol Bioeng. 1998;58(5):554–559. doi: 10.1002/(SICI)1097-0290(19980605)58:5<554::AID-BIT12>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 22.Bornscheuer UT, Altenbuchner J, Meyer HH. Directed evolution of an esterase: screening of enzyme libraries based on pH indicators and a growth assay. Bioorg Med Chem. 1999;7(10):2169–2173. doi: 10.1016/S0968-0896(99)00147-9. [DOI] [PubMed] [Google Scholar]
- 23.Callanan MJ, Russell WM, Klaenhammer TR. Modification of Lactobacillus beta-glucuronidase activity by random mutagenesis. Gene. 2007;389(2):122–127. doi: 10.1016/j.gene.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Lu X, Hirata H, Yamaji Y, Ugaki M, Namba S. Random mutagenesis in a plant viral genome using a DNA repair-deficient mutator Escherichia coli strain. J Virol Methods. 2001;94(1–2):37–43. doi: 10.1016/S0166-0934(01)00270-1. [DOI] [PubMed] [Google Scholar]
- 25.Nakashima N, Tamura T. Conditional gene silencing of multiple genes with antisense RNAs and generation of a mutator strain of Escherichia coli . Nucleic Acids Res. 2009;37(15):e103. doi: 10.1093/nar/gkp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Miller JH. Deletion of dnaN1 generates a mutator phenotype in Bacillus anthracis . DNA Repair (Amst) 2008;7(3):507–514. doi: 10.1016/j.dnarep.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci USA. 2003;100(17):9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh T, Tomizawa J. Initiation of replication of plasmid ColE1 DNA by RNA polymerase, ribonuclease H, and DNA polymerase I. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):409–417. doi: 10.1101/sqb.1979.043.01.047. [DOI] [PubMed] [Google Scholar]
- 29.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc Natl Acad Sci USA. 2001;98(12):6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirakitikulr N, Ostrov N, Peralta-Yahya P, Cornish VW PCR less library mutagenesis via oligonucleotide recombination in yeast. Protein Sci 19(12):2336–2346. doi:10.1002/pro.513 [DOI] [PMC free article] [PubMed]
- 32.Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228(5276):1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Davis M, Sinn E, Patten P, Hood L. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981;27(3 Pt 2):573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- 34.McKean D, Huppi K, Bell M, Staudt L, Gerhard W, Weigert M. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc Natl Acad Sci USA. 1984;81(10):3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milstein C, Rada C. The maturation of the antibody response. In: Honjo T, Alt F, editors. Immunoglobulin genes. London: Academic Press Limited; 1995. pp. 57–83. [Google Scholar]
- 36.Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes: merging mechanisms for genetic diversity. Cell. 2002;109(Suppl):S35–S44. doi: 10.1016/S0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 37.Martin A, Scharff MD. AID and mismatch repair in antibody diversification. Nat Rev Immunol. 2002;2(8):605–614. doi: 10.1038/nri858. [DOI] [PubMed] [Google Scholar]
- 38.Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK. Immunity through DNA deamination. Trends Biochem Sci. 2003;28(6):305–312. doi: 10.1016/S0968-0004(03)00111-7. [DOI] [PubMed] [Google Scholar]
- 39.Rajewsky K, Forster I, Cumano A. Evolutionary and somatic selection of the antibody repertoire in the mouse. Science. 1987;238(4830):1088–1094. doi: 10.1126/science.3317826. [DOI] [PubMed] [Google Scholar]
- 40.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/S0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 41.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100(7):4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424(6944):103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 43.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4(1):57–65. doi: 10.1016/S1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 44.Cascalho M, Wong J, Steinberg C, Wabl M. Mismatch repair co-opted by hypermutation. Science. 1998;279(5354):1207–1210. doi: 10.1126/science.279.5354.1207. [DOI] [PubMed] [Google Scholar]
- 45.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419(6902):43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 46.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418(6893):99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 47.Yelamos J, Klix N, Goyenechea B, Lozano F, Chui YL, Gonzalez Fernandez A, Pannell R, Neuberger MS, Milstein C. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 1995;376(6537):225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- 48.Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J Immunol. 2001;166(8):5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- 49.Yoshikawa K, Okazaki IM, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296(5575):2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 50.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99(12):7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sale JE, Neuberger MS. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9(6):859–869. doi: 10.1016/S1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci USA. 2004;101(48):16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295(5558):1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 54.Arakawa H, Saribasak H, Buerstedde JM. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2004;2(7):E179. doi: 10.1371/journal.pbio.0020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buerstedde JM, Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67(1):179–188. doi: 10.1016/0092-8674(91)90581-I. [DOI] [PubMed] [Google Scholar]
- 56.Seo H, Masuoka M, Murofushi H, Takeda S, Shibata T, Ohta K. Rapid generation of specific antibodies by enhanced homologous recombination. Nat Biotechnol. 2005;23(6):731–735. doi: 10.1038/nbt1092. [DOI] [PubMed] [Google Scholar]
- 57.Seo H, Hashimoto S, Tsuchiya K, Lin W, Shibata T, Ohta K. An ex vivo method for rapid generation of monoclonal antibodies (ADLib system) Nat Protoc. 2006;1(3):1502–1506. doi: 10.1038/nprot.2006.248. [DOI] [PubMed] [Google Scholar]
- 58.Seo H, Yamada T, Hashimoto S, Lin W, Ohta K. Modulation of immunoglobulin gene conversion in chicken DT40 by enhancing histone acetylation, and its application to antibody engineering. Biotechnol Genet Eng Rev. 2007;24:179–193. doi: 10.1080/02648725.2007.10648099. [DOI] [PubMed] [Google Scholar]
- 59.Kanayama N, Todo K, Takahashi S, Magari M, Ohmori H. Genetic manipulation of an exogenous non-immunoglobulin protein by gene conversion machinery in a chicken B cell line. Nucleic Acids Res. 2006;34(2):e10. doi: 10.1093/nar/gnj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arakawa H, Kudo H, Batrak V, Caldwell RB, Rieger MA, Ellwart JW, Buerstedde JM. Protein evolution by hypermutation and selection in the B cell line DT40. Nucleic Acids Res. 2008;36(1):e1. doi: 10.1093/nar/gkm616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arakawa H, Lodygin D, Buerstedde JM. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77(2):239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 63.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6(8):573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 64.Kothapalli N, Norton DD, Fugmann SD. Cutting edge: a cis-acting DNA element targets AID-mediated sequence diversification to the chicken Ig light chain gene locus. J Immunol. 2008;180(4):2019–2023. doi: 10.4049/jimmunol.180.4.2019. [DOI] [PubMed] [Google Scholar]
- 65.Blagodatski A, Batrak V, Schmidl S, Schoetz U, Caldwell RB, Arakawa H, Buerstedde JM. A cis-acting diversification activator both necessary and sufficient for AID-mediated hypermutation. PLoS Genet. 2009;5(1):e1000332. doi: 10.1371/journal.pgen.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arakawa H, Buerstedde JM. Activation-induced cytidine deaminase-mediated hypermutation in the DT40 cell line. Philos Trans R Soc Lond B Biol Sci. 2009;364(1517):639–644. doi: 10.1098/rstb.2008.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]