Abstract
The long-wavelength-sensitive (LWS) opsins form one of four classes of vertebrate cone visual pigment and exhibit peak spectral sensitivities (λmax) that generally range from 525 to 560 nm for rhodopsin/vitamin-A1 photopigments. Unique amongst the opsin classes, many LWS pigments show anion sensitivity through the interaction of chloride ions with a histidine residue at site 197 (H197) to give a long-wavelength spectral shift in peak sensitivity. Although it has been shown that amino acid substitutions at five sites (180, 197, 277, 285 and 308) are useful in predicting the λmax values of the LWS pigment class, some species, such as the elephant shark and most marine mammals, express LWS opsins that possess λmax values that are not consistent with this ‘five-site’ rule, indicating that other interactions may be involved. This study has taken advantage of the natural mutation at the chloride-binding site in the mouse LWS pigment. Through the use of a number of mutant pigments generated by site-directed mutagenesis, a new model has been formulated that takes into account the role of charge and steric properties of the side chains of residues at sites 197 and 308 in the function of the chloride-binding site in determining the peak sensitivity of LWS photopigments.
Keywords: Visual pigments, Mouse, Colour vision, Spectral tuning, Anion sensitivity
Introduction
Vertebrate cone visual pigments are assigned to four classes on the basis of amino acid sequence and peak spectral sensitivity (λmax) that comprise a middle- or long-wavelength-sensitive (MWS/LWS) class, two short-wavelength-sensitive (SWS1 and SWS2) classes, and a middle-wavelength-sensitive rod opsin-like (RH2) class. The LWS class of pigments have λmax values generally between 525 and 565 nm for rhodopsin/vitamin-A1 photopigments with spectral shifts attributed to amino acid substitutions at five sites, 180, 197, 277, 285 and 308 [1]. The anion sensitivity that is unique to the LWS cone visual pigments [2–6] arises from the interaction of chloride ions with a histidine residue at site 197 (H197) in the second extracellular loop of the opsin protein [7]. The effect of this interaction is to long-wavelength shift the λmax of the pigment by around 30 nm [7].
The pigments of the mouse (Mus musculus), rat (Rattus norvegicus) and rabbit (Oryctolagus cuniculus) encoded by the LWS gene have a natural mutation at site 197 that encodes a H197Y substitution [8, 9]. The λmax of these pigments is short-wavelength shifted to around 510 nm, although only part of this shift is the result of the presence of Y197. As the mouse LWS photopigment exhibits a λmax at 508 nm [8, 9], which is in the middle of the visible light spectrum, it is more appropriately classified as MWS. The murine MWS pigment is also unusual in having serine rather than alanine at site 308 (S308), which additionally contributes to the short-wavelength shift. This has been demonstrated by a S308A substitution that results in a long-wavelength shift in the peak to 526 nm [9]. These two substitutions, therefore, combine to achieve the short-wavelength shifts of these pigments. The presence of S308 in combination with a functional chloride-binding site may lead to pigment instability, as found for the mouse pigment [9] but not in the MWS pigment of the bottlenose dolphin (Tursiops truncates) [10] where S308 is present alongside a functional chloride-binding site. With this pigment, a S308A substitution was found to long-wavelength shift the λmax from 524 to 552 nm [11].
Site 308 has also been implicated in the tuning of the MWS/LWS pigments of the elephant shark, Callorhinchus milii, a cartilaginous fish belonging to the subclass Holocephali. In this species, two LWS pigment genes were identified giving rise to pigments with λmax values at 499 nm (LWS1 and called MWS herein after) and 548 nm (LWS2 and called LWS herein after) [12]. The former pigment is the most short-wavelength shifted LWS photopigment so far reported. Both C. milii MWS and LWS pigments retain a chloride-binding site at H197 but the MWS photopigment possesses a serine residue at site 308, whilst the LWS pigment has A308. The predicted λmax for the MWS pigment based on its substitutions at sites 180, 197, 277 and 285 is 521 nm, which implies that S308 results in a 22 nm short-wavelength shift. However, the experimentally determined λmax of the C. milii MWS photopigment is 499 nm [12], which is considerably more short-wavelength shifted than expected. It is clear, therefore, that site 308 is important for MWS/LWS pigment tuning, but its functional interaction with chloride ion binding has yet to be fully clarified. The study reported here aims to investigate the mechanisms of vertebrate MWS/LWS spectral tuning by taking advantage of the natural mutation at the chloride-binding site in the mouse pigment.
Methods
Generation and cloning of mouse wild-type and mutant MWS opsin sequences
Ocular tissues were collected from adult mice that were sacrificed accordingly to local guidelines and frozen immediately at −80°C. Retinal messenger RNA (mRNA) was purified using the Illustra Quickprep Micro mRNA purification kit (GE Healthcare, UK), and 1–2 μg was converted to oligo-dT-primed complementary DNA (cDNA) following the manufacturer’s instructions contained within the 5′/3′ Rapid Amplification of cDNA Ends (RACE) kit (Roche Applied Science, UK). Once generated, mouse retinal cDNA was used to polymerase chain reaction (PCR) amplify the full-length mouse MWS opsin sequence, using forward (Mm_MWS_F: 5′-GCGCGAATTCCACCATGGCCCAAAGGCTTACAGG-3′) and reverse (Mm_MWS_R: 5′-GAAGGTCGACGCAGGTGACACTGAAGAGACAGA-3′) primers, Thermococcus kodakaraensis (KOD) DNA polymerase (Novogen, UK) and applying the following PCR conditions: an initial denaturation step at 95°C for 2 min and 40 cycles of 95°C for 30 s, 60°C for 15 s and 70°C for 1 min.
Both primers were designed to incorporate restriction sites; Eco RI in the forward primer and Sal I in the reverse primer to facilitate cloning of the full-length opsin sequence into the pMT4 expression vector [13]. A translation start codon (AUG) was presented within a Kozak consensus sequence to ensure efficient translation of the recombinant visual pigment (forward primer). In addition, a DNA tag was included that replaced the native opsin stop codon with a short sequence encoding the C-terminal bovine (Bos taurus) rod opsin 1D4 epitope (ETSQVAPA) (reverse primer), which is recognised by the anti-Rho1D4 monoclonal antibody [14].
Three mutant mouse opsin clones (Y192H, S308A and the double mutant Y192H/S308A) were obtained by site-directed mutagenesis of the wild-type clone using a QuikChange Site Directed Mutagenesis kit (Stratagene, UK) and one of the following mutagenesis primer pairs as appropriate (Mm_MWS_Y192H_F: 5′-AGCAGGTACTGGCCTCATGGCCTGAAGACATCC-3′; Mm_MWS_Y192H_R: 5′-GGATGTCTTCAGGCCATGAGGCCAGTACCTGCT-3′; Mm_MWS_S308A_F: 5′-GTGGCCTCCCTACCAGCCTACTTTGCCAAAAGTG-3′; Mm_MWS_S308A_R: 5′-CACTTTTGGCAAAGTAGGCTGGTAGGGAGGCCAC-3′). A double mutant (Y192H/S308A) was generated in a single step using both primer pairs simultaneously. All wild-type and mutant clones were sequenced to ensure that PCR amplification had not introduced any mutations.
Expression of wild-type and mutant mouse visual photopigments
All UV-visible spectrophotometric experiments were conducted in triplicate, where 210 μg of each pMT4-opsin construct, containing full-length wild-type or mutant opsin coding sequences, was used to transfect 12 140-mm plates of human embryonic kidney (HEK293T) cells using GeneJuice transfection reagent (Novagen, UK) at a DNA:GeneJuice ratio of 3:1, according to the manufacturer’s instructions. After 48 h post-transfection, the transfected cells were harvested and washed four times with 1× phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4) (pH 7.4) (Invitrogen, UK). Visual pigments were generated using excess 11-cis retinal in the dark, either in 1× PBS containing chloride ions or under chloride-free conditions (where 2.7 mM KCl and 137 mM NaCl were replaced with 2.7 mM KOH and 137 mM NaClO4, respectively, as previously described [5, 12]). Photopigments were isolated by affinity immunochromatography [15].
Absorbance spectra were recorded in triplicate in the dark using a Shimadzu UV-visible spectrophotometer (UV-2550) (Shimadzu, UK) and bleached by exposure to broad-spectrum white fluorescent light for 1 h before a second spectrum was recorded. The peak sensitivity, λmax, for each pigment was determined by subtracting the bleached spectrum from the dark spectrum of each pigment to produce a difference spectrum. This was then fitted to a standard Govardovskii rhodopsin/vitamin-A1 template [16] with a subtracted bleached retinal curve. Best-fit curves were obtained using the Solver add-in function in Microsoft Excel to determine the λmax by the method of least squares as previously described [17].
Modelling of spectral tuning in wild-type and mutant mouse visual photopigments
Homology modelling of the mouse MWS opsin was created by using Swiss-Model [18–21] and DeepView/Swiss-PdbViewer software v.3.7 (http://www.expasy.org/spdbv/), where the murine sequence was aligned to the crystal structure of bovine rhodopsin (RH1) [22, 23]. As a representation of the putative spatial properties of the triform binding pocket, the planar area of a scalene triangle created by residues at positions 197, 308 and 312 was calculated by measuring the distance of the three sides of the triangle (a, b and c) and applying Heron’s formula, where the area (A) = √s(s−a)(s−b)(s−c) and s = (a + b + c)/2.
Results
Peak spectral sensitivity and protein stability of expressed wild-type and mutant mouse pigments
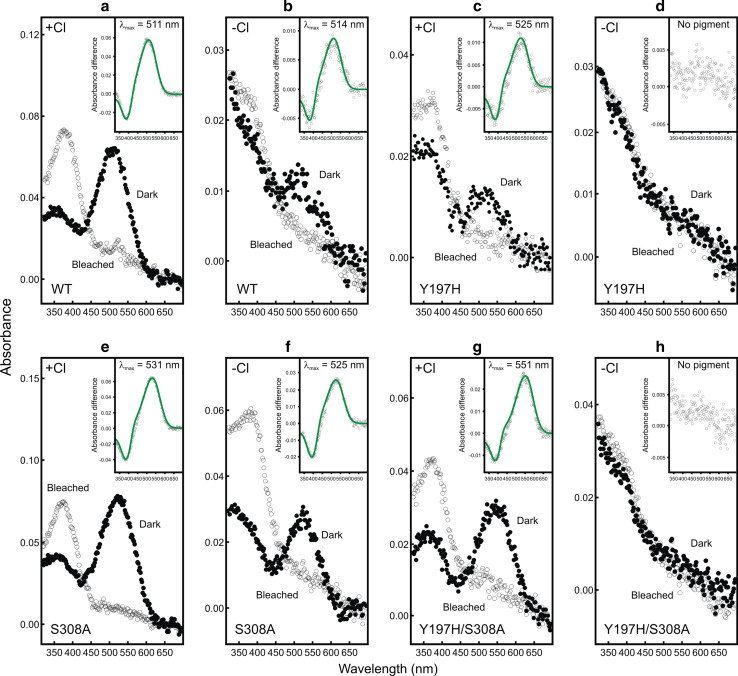
The wild-type mouse MWS pigment gave a λmax at 511 nm (Fig. 1a), which is similar to the previously reported value of 508 nm [9]. An estimate of the protein structural fidelity of the wild-type and mutant photopigments produced by our expression system was calculated by an approach that is routinely used in similar studies (e.g. [24]). Specifically, we determined the ratio of total protein (measured at A280) to opsin (measured at the λmax) (i.e. Aprotein/Aopsin). This allowed for a comparison of the amount of viable pigment generated for the mutant pigments with the wild-type pigment in the presence or absence of chloride ions, expressed as a percentage (i.e. % of the wild-type) (see Table 1). As shown in Fig. 1a, b, the wild-type pigment was spectrally refractory to the presence or absence of chloride ions (i.e. λmax = 511 nm vs. a value of 514 nm) but there was a significant reduction in viable photopigment generated (50.7% of wild-type) in the absence of chloride ions (Table 1). A previous investigation, using iodide ions instead of chloride ions, also reported a small, yet insignificant, change in the λmax value (~1 nm) of the wild-type mouse MWS pigment [9]. In the experiments presented here, a substitution of chloride ions with perchlorate ions resulted in a similarly small spectral shift of 3 nm, which is likely due to experimental error and not intrinsic to the type of ion used to substitute the chloride ions.
Fig. 1.
Absorption spectra of generated mouse (Mus musculus) wild-type (WT, a, b) and single (Y197H, c, d; S308A, e, f) and double (Y197H/S308A, g, h) mutant MWS photopigments, under chloride (+Cl−) (a, c, e, g) and chloride-free (−Cl−) (b, d, f, h) conditions. For all pigments, representative dark (closed circles) and light bleached (open circles) spectra are shown, with difference spectra (open grey circles) that have been fitted with a Govardovskii [16] rhodopsin/vitamin-A1 template (green line) in the inset to determine the λmax
Table 1.
A summary of the spectral peaks of absorbance and relative levels of opsin produced by wild-type (WT) and mutant mouse MWS photopigments, under chloride and chloride-free conditions, calculated by comparing the ratio of total protein absorbance (A280) to the normalised absorbance of opsin protein determined at the λmax (i.e. Aprotein/Aopsin)
| Opsin | +Cl− | −Cl− | ||||
|---|---|---|---|---|---|---|
| Peak (nm) | Ratio (Aprotein/Aopsin) | % of WT | Peak (nm) | Ratio (Aprotein/Aopsin) | % of WT | |
| WT (Y197/S308) | 511 | 7.2 | 100 | 514 | 14.2 | 50.7 |
| H197/S308 | 525 | 33.2 | 21.7 | No pigment | 86.2 | No pigment |
| Y197/A308 | 531 | 7.5 | 95.8 | 525 | 7.8 | 92.4 |
| H197/A308 | 551 | 14.9 | 48.1 | No pigment | 88.0 | No pigment |
Each resultant ratio was converted to a percentage of the wild-type ratio to produce a realtive measure of opsin-specific protein for comparison
The restoration of the chloride-binding site in this pigment by site-directed mutagenesis to give a Y197H substitution was previously reported to give an unstable pigment [9]. However, in our experimental system, an identical substitution, also generated by site-directed mutagenesis, resulted in a pigment with a λmax at 525 nm (Fig. 1c), although the amount of pigment was substantially reduced to 21.7% of wild-type levels (Table 1). This mutant opsin, however, failed to generate a pigment in the absence of chloride ions (Fig. 1d; Table 1).
Site-directed mutagenesis was used to produce a S308A substitution and the resulting pigment gave a 20 nm long-wavelength shift to a λmax of 531 nm (Fig. 1e), with only a small reduction in the total amount of pigment produced (95.8% of wild-type). This, therefore, represents the tuning effect of serine/alanine at site 308 on LWS pigments, which is consistent with a previously reported shift of 18 nm [9]. When chloride ions were omitted, the long-wavelength shift was reduced by 6 nm to give a λmax at 525 nm (Fig. 1f), with a small decrease in the amount of pigment produced to 92.4% of wild-type (Table 1). The introduction of both mutations by site-directed mutagenesis to give the double mutant of Y197H and S308A resulted in a reduction of pigment to 48.1% of the wild-type level and a long-wavelength shift to 551 nm (Fig. 1g). Similar to the Y197H mutant with S308 discussed above, the double mutant (Y197H/S308A) pigment, however, also failed to generate in the absence of chloride ions (Fig. 1h; Table 1).
As shown in Table 2, the combination of A180, Y277 and T285 found in the mouse, rat and rabbit MWS pigments is also present in the elephant (Loxodonta africana) pigment [25] and in the pigments of two marsupials, the honey possum (Tarsipes rostratus) and quenda (Isoodon obesulus) [26]. These pigments gave λmax values of 552, 557 and 551 nm, respectively. As the mouse MWS pigment exhibits a λmax of 511 nm, the combination of Y197 and S308 must be responsible for short-wavelength shifting of the peak by around 40 nm.
Table 2.
Residues present at five spectral tuning sites (180, 197, 277, 285 and 308) found in transmembrane helices IV, VI and VII and the second extracellular loop of the MWS/LWS pigments expressed in a subset of vertebrates, known to influence spectral sensitivity [1]
| Species | Residue at tuning site | Peak (nm) | ||||
|---|---|---|---|---|---|---|
| 180 | 197 | 277 | 285 | 308 | ||
| Human (Homo sapiens) (LWS) | Ser | His | Tyr | Thr | Ala | 565 [38] |
| Human (Homo sapiens) (MWS) | Ala | His | Phe | Ala | Ala | 530 [38] |
| Mouse (Mus musculus) | Ala | Tyr | Tyr | Thr | Ser | 508a [9]/511b |
| Rat (Rattus norvegicus) | Ala | Tyr | Tyr | Thr | Ser | 509 [8] |
| Rabbit (Oryctolagus cuniculus) | Ala | Tyr | Tyr | Thr | Ser | 509 [8] |
| Manatee (Trichechus manatus) | Ser | His | Tyr | Thr | Ala | 556 [39] |
| Elephant (Loxodonta africana) | Ala | His | Tyr | Thr | Ala | 552 [25] |
| Pilot whale (Globicephala melas) | Ala | His | Tyr | Thr | Ser | 531 [39] |
| Harbor porpoise (Phocoena phocoena) | Ala | His | Tyr | Thr | Ser | 522 [39] |
| Bottlenosed dolphin (Tursiops truncates) | Ala | His | Tyr | Thr | Ser | 524 [40] |
| Harbor seal (Phoca vitulina) | Ser | His | Phe | Thr | Ala | 548 [39] |
| Harp seal (Phoca groenlandica) | Ser | His | Phe | Thr | Ala | 548 [39] |
| Honey possum (Tarsipes rostratus) | Ala | His | Tyr | Thr | Ala | 557 [41] |
| Fat-tailed dunnart (Sminthopsis crassicaudata) | Ala | His | Phe | Ala | Ala | 530 [41] |
| Quenda (Isoodon obesulus) | Ala | His | Tyr | Thr | Ala | 551 [26] |
| Quokka (Setonix brachyurus) | Ala | His | Tyr | Ala | Ala | 538 [26] |
|
Tammar wallaby (Macropus eugenii) Elephant shark (Callorhinchus milii) (LWS) Elephant shark (Callorhinchus milii) (MWS) |
Ala Ser Ser |
His His His |
Phe Phe Phe |
Ala Thr Ala |
Ala Ala Ser |
530 [42] 548 [12] 499 [12] |
The peak spectral sensitivities of photopigments determined for each species is also indicated. Bottlenosed dolphin (Tursiops truncates) (GenBank accession no.: AAC12941); elephant (Loxodonta africana) (GenBank accession no.: AAT95415); elephant shark (Callorhinchus milii) MWS (GenBank accession no.: ABU84863) and LWS (GenBank accession no.: ABU84864); fat-tailed dunnart (Sminthopsis crassicaudata) (GenBank accession no.: ACA28596); harbor porpoise (Phocoena phocoena) (GenBank accession no.: AAP13019); harbor seal (Phoca vitulina) (GenBank accession no.: AAP13017); harp seal (Phoca groenlandica) (GenBank accession no.: AAP13018); honey possum (Tarsipes rostratus) (GenBank accession no.: AAX11257); human (Homo sapiens) MWS (GenBank accession no.: NP000504) and LWS (GenBank accession no.: NP064445); manatee (Trichechus manatus) (GenBank accession no.: AAP13016); mouse (Mus musculus) (GenBank accession no.: NP032132); pilot whale (Globicephala melas) (GenBank accession no.: AAP13015); quenda (Isoodon obesulus) (GenBank accession no.: AAW69833); quokka (Setonix brachyurus) (GenBank accession no.: AAW69832); rabbit (Oryctolagus cuniculus) (GenBank accession no.: NP001164614); rat (Rattus norvegicus) (GenBank accession no.: NP446000); tammar wallaby (Macropus eugenii) (GenBank accession no.: AAP37945)
bλmax calculated in this study using a 1% (w/v) dodecyl-maltoside/phenylmethanesulphonylfluoride/phosphate-buffered saline (DDM/PMSF/PBS) buffer containing 137 mM NaCl, 2.7 mM KCl
aλmax determined using a 1% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) buffer containing 140 mM NaCl [9]
Putative homology modelling of the chloride ion interaction with sites 197 and 308
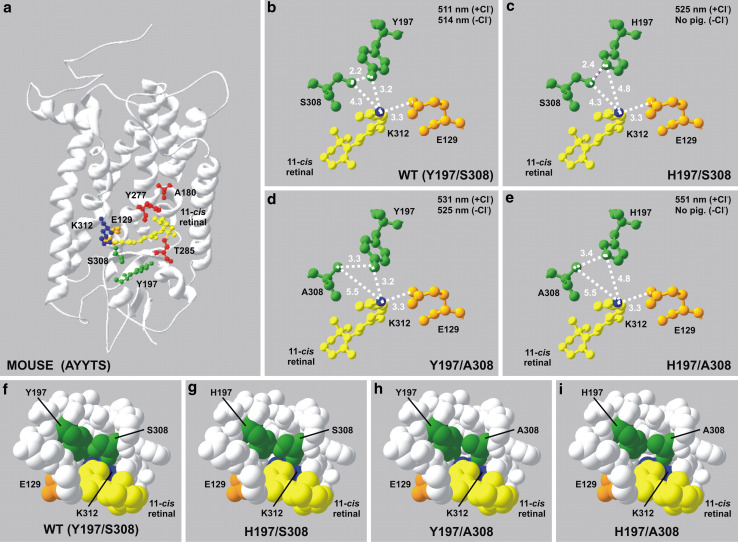
It is well established that cone and rod photopigments interact with different, yet related, isoforms of proteins of the phototransduction cascade [27], leading to differences in their activation and thermal properties [28–34]. Taken together with variation in spectral sensitivities [35, 36] and biochemical characteristics, it is possible that the overall tertiary structures of cone photopigments subtly differ from those expressed in rod photoreceptors. However, the rod opsin found in bovine retinae is the only vertebrate photopigment for which a crystal structure has been successfully determined [22, 23]. Despite not being able to utilise an experimentally determined structure of any cone opsin, the predicted positions of the known tuning sites for MWS/LWS photopigments may be investigated by homology modelling [18–21] their structures onto the three-dimensional crystal backbone structure of bovine (RH1) rhodopsin [22, 23] as shown in Fig. 2a. This places residues 197 and 308 in close proximity to each other and adjacent to both the Schiff base that links the retinylidene chromophore to the opsin protein via a covalent bond at K312 and the negative charge of the glutamate counterion at site 129 (E129). In contrast, the other major MWS/LWS tuning sites at positions 180, 277 and 285 are located distally toward the β-ionone ring of the chromophore (Fig. 2a). Thus, the residues that cause the largest shifts in the λmax (i.e. at sites 197 and 308) are located near the Schiff base, with those that result in smaller changes (i.e. 180, 277 and 285) positioned closer to the β-ionone ring.
Fig. 2.
a Putative structural model of the wild-type mouse MWS photopigment generated by homology modelling with the crystal structure of bovine rhodopsin [22, 23] modified to show the relative position of the five key tuning sites (A180, Y197, Y277, T285 and S308) in relation to the retinal-binding pocket containing the retinylidene chromophore (yellow), the K312 chromophore attachment site via a Schiff base (blue) and the E129 counterion (orange). Tuning sites are colour coded whether they are situated proximal (green) or distal (red) to the Schiff base. Enlarged views detailing the relative positions of sites 197 and 308, the retinal chromophore, the Schiff base and the glutamate counterion at 113 are shown for mouse wild-type (WT, Y197/S308, b, f) and single (H197/S308, c, g; Y197/A308, d, h) and double (H197/A308, e, i) mutant MWS photopigments. Intramolecular hydrogen bond formation (purple) is shown in panel c. Panels f - i indicate a horizontal view, parallel to the chromophore backbone, into the putative triform chloride-binding site consisting of the side chain of residues present at sites 197 and 308 and the Schiff base. All interatomic distances are shown in angstroms (Å). Spectral peak absorbance (λmax) values determined under chloride (+Cl−) and chloride-free (−Cl−) conditions are included. All homology models were created using Swiss Model [18–21] and DeepView/Swiss-PdbViewer software v.3.7 (http://www.expasy.org/spdbv/) and are based on comparisons with the crystal structure of bovine rhodopsin (RH1) [22, 23]
Closer inspection of the predicted structural relationships between the positively charged Schiff base and residues H197 (also positively charged) and A308 as found in all LWS photopigments with λmax values close to 560 nm (e.g. human LWS) suggests the presence of a putative compact triform binding pocket that allows entry of a chloride ion to act as a counterion to the positive charges of both H197 and the Schiff base and thereby stabilise a long-wavelength shift in the λmax (Fig. 2e, l). The H197Y substitution (as present in the mouse, rat and rabbit pigments) introduces a hydroxyl group into the chloride-binding site that results in a reduction of both the local positively charged environment and the binding area of the triform pocket (8.1 to 4.8 Å2) (Fig. 2d, h; Table 3). Together, the change in charge density (from one that is positive to a δ− of the hydroxyl side group) and a more limited spatial arrangement of the pocket are likely to weaken and prevent the binding of a chloride ion, resulting in a short-wavelength shift, presumably with the ionic shift having the greater effect. When H197 is present in combination with S308, the area of the scalene binding pocket is again reduced (5.2 Å2) (Fig. 2c, g; Table 3) to a similar size to that found in photopigments with a Y197/A308 combination (4.8 Å2) (Fig. 2d, h; Table 3). Moreover, the reduced relative spatial position of the amino acid residues present within this contracted triform binding pocket, in addition to the extra hydroxyl group introduced by S308, introduces the possibility of hydrogen bond formation between H197 and S308 (Fig. 2c). Together, these interactions may function to stabilise the three-dimensional space of the triform pocket and impede the binding of a chloride ion despite the presence of the positively charged H197, with the overall effect of short-wavelength shifting the λmax in a similar manner to a H197Y substitution. However, when the mutant mouse photopigments possessing H197 were generated in the absence of chloride ions, no pigment was formed (Fig. 1d, h). This suggests that a negatively charged chloride ion not only differentially affects spectral tuning, depending on the presence of either A308 or S308, but also plays a role in protein stabilisation by counteracting the positive charge of the H197 irrespective of the residue at site 308. By contrast, photopigments containing Y197 were successfully generated in the presence (Fig. 1a, b) or absence (Fig. 1e, f) of chloride ions, which is consistent with the redundancy of the chloride counterion and a minimal effect on protein stability. Under these circumstances, the triform binding pocket is minimal at 3.4 Å2 (Table 3) in the wild-type mouse MWS photopigment with S308 (Fig. 2b, f) and 4.8 Å2 (Table 3) in the mutant with A308 (Fig. 2d, h); thus, spectral tuning is solely dependent on the residue present at site 308. Interestingly, the elephant shark MWS photopigment (with a H197/S308 combination) forms a functional pigment in the presence and absence of chloride ions [12] so it would appear that the binding of chloride ions is not required for pigment production in this ancient fish. In this case, therefore, other residues must be responsible for counteracting the positive charge of H197 and stabilising the three-dimensional space of the binding pocket in the absence of chloride ions.
Table 3.
A summary of the calculated area of the predicted scalene triform binding pocket for wild-type and mutant mouse MWS photopigments
| Opsin | Calculated area of scalene triform binding pocket (Å2) | % of WT |
|---|---|---|
| WT (Y197/S308) | 3.4 | 100 |
| H197/S308 | 5.2 | 153 |
| Y197/A308 | 4.8 | 141 |
| H197/A308 | 8.1 | 238 |
Each resultant area was converted to a percentage of the wild-type value to produce a realtive measure of spatial change
Discussion
In this study, we have established that the residue present at site 308 in MWS/LWS photopigments has two effects on spectral tuning. Firstly, the presence of S308 removes anion sensitivity by inhibiting the binding of a chloride ion to H197. In the MWS photopigments of the mouse, rat and rabbit, the loss of chloride ion binding occurs directly due to the replacement of H197 with Y197. Thus, both mechanisms are functionally equivalent in preventing anion binding and result in a short-wavelength spectral shift. The second effect of S308 is to cause a 20 nm short-wavelength shift in the λmax compared to the much more common A308. Furthermore, this tuning effect of site 308 appears to be independent of the presence of a functional chloride-binding site.
The MWS photopigments of a subset of mammalian species, which include three deeper dwelling cetaceans, the bottlenose dolphin (Tursiops truncates), the pilot whale (Globicephala melas) and the harbor porpoise (Phocoena phocoena) [37], naturally possess S308; in all cases, this is associated with a shift in the λmax of the pigment to a shorter wavelength. Direct confirmation of the role of S308 in this shift has been demonstrated in the bottlenose dolphin pigment by a S308A replacement that resulted in a pigment that was long-wavelength shifted from the wild-type λmax of 524 to 552 nm, a change of 28 nm [10]. Thus, the presence of S308 in the LWS pigments of all of the deeper-dwelling cetaceans identified so far may act to inactivate the chloride-binding site. The elephant shark also possesses a MWS pigment with S308 and mutagenesis studies in this case have shown that the chloride-binding site is completely inactive [12]. This pigment also combines S308 with F277 and A285 to generate a λmax at 499 nm, making it the most short-wavelength shifted LWS pigment so far reported.
As a crystallographic structure has not been determined for any vertebrate cone opsin, three-dimensional modelling based on the bovine rhodopsin crystal [22, 23], though with its intrinsic limitations, may yield insights into the structure–function relationships of MWS/LWS photopigments. From our predicted homology modelling studies, it is uncertain whether the inactivation of the chloride-binding site by S308 is caused directly by steric hindrance by the side chain of the residue present at 308 or indirectly by changing the overall conformation of the visual pigment, and thus access of a chloride ion to the retinal binding pocket. Closer inspection of the opsin three-dimensional structure suggests that sites 197 and 308 form a tight triform binding pocket with the Schiff base that may house a chloride ion when a H197/A308 combination is present. The mutation of either (H197Y or A308S) or both (H197Y/A308S) sites introduces larger side chains that impinge on the binding pocket and may thereby sterically hinder local access by a chloride ion. Under circumstances where a histidine residue is present at site 197, local steric hindrance of a chloride ion by site 308 may be the predominant effect influencing spectral tuning. In the elephant shark MWS photopigment, S308 in conjunction with H197 appears to completely eliminate chloride sensitivity as the generated pigments are spectrally unaltered by the presence or absence of chloride ions. In the mouse MWS photopigment, however, the intermolecular interactions would appear to be somewhat different since although a 20 nm short-wavelength shift is caused by the loss of the chloride-binding site in H197Y mutants, a combination of H197 and S308 resulted in a short-wavelength shift of 26 nm (compared to a photopigment possessing H197/A308) that only occurred in the presence of chloride ions. In the absence of chloride ions, mouse mutant photopigments with H197 failed to generate, irrespective of the presence of either S308 or A308. Together, these data suggest that a negatively charged chloride ion remains essential for protein structural fidelity but paradoxically an A308S substitution appears to inhibit chloride ion binding altogether. Thus, we propose that chloride ions may exhibit a dual purpose in the mouse, rat and rabbit MWS photopigments. Firstly, the chloride ion may function directly as a counterion to H197 when access within the triform pocket is spatially possible (i.e. when combined with A308) and permit a large shift of the λmax to longer wavelengths. Secondly, it may act indirectly to stabilise the pigment when S308 sterically inhibits the binding of a chloride ion to H197, resulting in a large short-wavelength shift. These mechanistic differences between the mouse and elephant shark MWS photopigments presumably arise from interaction with other residues in close proximity to the chloride-binding site that differ between the mouse and elephant shark photopigments.
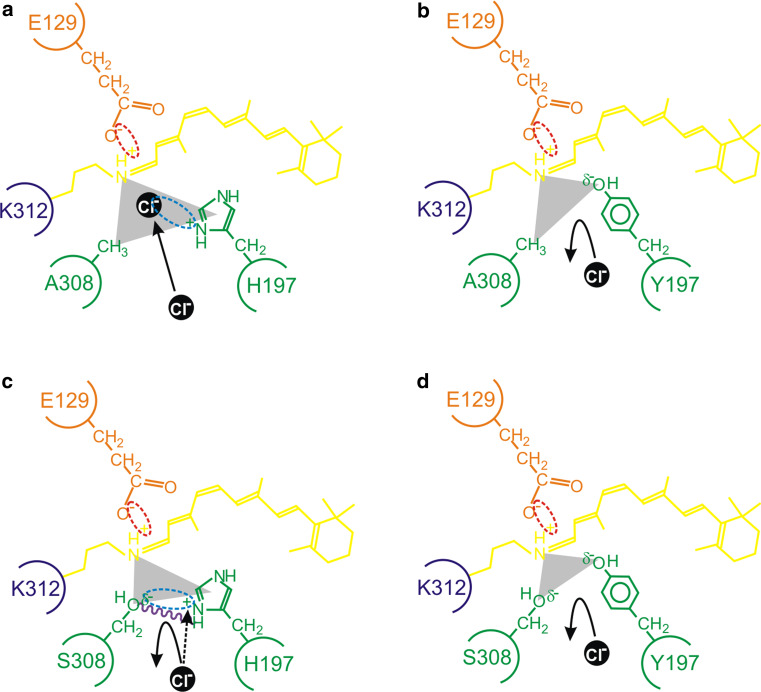
Collectively, our data suggest that both the charge and steric properties of the side chains of residues at sites 197 and 308 are important for determining the spectral tuning of vertebrate MWS/LWS photopigments by differentially interacting with chloride ions. The general model that we propose is shown in Fig. 3 and is based on a negatively charged chloride ion acting as a counterion to the positive charge of H197, but only when A308 is present. When H197 is replaced with Y197, the positive charge is lost and chloride ion binding is abolished, irrespective of the residue present at site 308. However, when both H197 and S308 are present, we propose that the spatial property of the hydroxyl group of S308 and the structural rigidity imposed by hydrogen bond formation between H197 and S308 may sterically hinder access of the chloride ion to the binding pocket. Concomitantly, the negative polarity of this hydroxyl side chain may repel the negatively charged chloride ion directly or itself function to partially counteract the effects of the positive charge of the histidine residue at site 197.
Fig. 3.
A schematic representation of the proposed model for the interaction between residues 197 and 308 (green) in close proximity to the Schiff base (dark blue) of the chromophore (yellow), showing a a chloride ion (black) acting as a counterion (interaction shown by a light blue dotted ellipse) to the positive charge present on H197; b the failure of binding and repulsion of a chloride ion due to a H197Y substitution that results in the loss of a positive charge (from H197) and the gain of a slight negative charge (δ−) (from Y197); c the δ− of the hydroxyl side chain of S308 repelling the negatively charged chloride ion and serving to reduce the strength of the positive charge of H197 (interaction shown by a light blue dotted ellipse); d the complete repulsion and loss of chloride ion binding due to the lack of a positive charge at site 197, the steric hindrance of S308, and the overall increase of a negative charge (δ−) from the side groups of Y197 and S308 (as observed in the wild-type mouse MWS photopigment). In panel c, putative intramolecular hydrogen bond formation (purple wavy line) further restricts the access of a chloride ion. As discussed in the main text, diverse vertebrate species (e.g. the mouse compared to the elephant shark) exhibit subtly different mechanistic roles for chloride ion binding. In some cases (e.g. the elephant shark), the presence of a serine residue at site 308 completely disrupts the chloride-binding site and photopigments generate in the presence or absence of chloride ions. By contrast, the absence of chloride ions in mouse mutant photopigments possessing H197 (whether site 197 is occupied by an alanine or serine residue) fail to generate successfully. This suggests that a chloride ion that should be repelled from binding directly within the binding pocket may exert some influence over protein structural fidelity and may partly act as a counterion (black dotted arrow) to the positive charge of H197 from a position that is external to the regular location of the chloride-binding site. In all cases, the E129 counterion (orange), the Schiff base linkage at K312 (dark blue), the retinylidene chromophore (yellow) and the triform spatial representation of the chloride-binding pocket (grey scalene triangle) are shown. Electrostatic attractions are highlighted between the side chain of E129 and the Schiff base (red dotted ellipse), the side chain of a histidine residue present at site 197 and a chloride ion (light blue dotted ellipse), and the amino acid side chains of H197 and S308 (light blue dotted ellipse)
Although our general model is consistent with the spectral tuning of MWS/LWS photopigments from disparate vertebrate species (e.g. mouse and elephant shark), other mechanistic differences may exist that impact either via steric hindrance or by changes in the charge density of residues lining the binding pocket on the access and binding of chloride ions. Whether these differences exist between mammals versus non-mammals or more broadly between terrestrial versus aquatic species remains to be determined. In this respect, high-resolution X-ray crystallography, in conjunction with site-directed mutagenesis, is needed to resolve these issues.
Acknowledgments
This work was supported by a grant from the BBSRC. We are grateful to Dr. Rosalie Crouch of the Storm Eye Institute, Medical University of South Carolina, USA, for the kind gift of 11-cis-retinal.
Footnotes
Wayne I. L. Davies and Susan E. Wilkie contributed equally to this research.
References
- 1.Yokoyama S, Radlwimmer FB. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crescitelli F. Ionochromic behavior of Grecko visual pigments. Science. 1977;195:187–188. doi: 10.1126/science.831267. [DOI] [PubMed] [Google Scholar]
- 3.Crescitelli F, Karvaly B. The gecko visual pigment: the anion hypsochromic effect. Vis Res. 1991;31:945–950. doi: 10.1016/0042-6989(91)90202-G. [DOI] [PubMed] [Google Scholar]
- 4.Fager RS, Goldman SL, Abrahamson EW. Ethanolamine attack of the bovine rhodopsin chromophore. Exp Eye Res. 1979;29:393–399. doi: 10.1016/0014-4835(79)90055-1. [DOI] [PubMed] [Google Scholar]
- 5.Kleinschmidt J, Harosi FI. Anion sensitivity and spectral tuning of cone visual pigments in situ. Proc Natl Acad Sci USA. 1992;89:9181–9185. doi: 10.1073/pnas.89.19.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shichida Y, Kato T, Sasayama S, Fukada Y, Yoshizawa T. Effects of chloride on chicken iodopsin and the chromophore transfer reactions from iodopsin to scotopsin and B-photopsin. Biochemistry. 1990;29:5843–5848. doi: 10.1021/bi00476a028. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Asenjo AB, Oprian DD. Identification of the Cl(−)-binding site in the human red and green color vision pigments. Biochemistry. 1993;32:2125–2130. doi: 10.1021/bi00060a001. [DOI] [PubMed] [Google Scholar]
- 8.Radlwimmer FB, Yokoyama S. Genetic analyses of the green visual pigments of rabbit (Oryctolagus cuniculus) and rat (Rattus norvegicus) Gene. 1998;218:103–109. doi: 10.1016/S0378-1119(98)00359-X. [DOI] [PubMed] [Google Scholar]
- 9.Sun H, Macke JP, Nathans J. Mechanisms of spectral tuning in the mouse green cone pigment. Proc Natl Acad Sci USA. 1997;94:8860–8865. doi: 10.1073/pnas.94.16.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasick JI, Cronin TW, Hunt DM, Robinson PR. The visual pigments of the bottlenose dolphin (Tursiops truncatus) Vis Neurosci. 1998;15:643–651. doi: 10.1017/S0952523898154056. [DOI] [PubMed] [Google Scholar]
- 11.Fasick JI, Robsinson PR. Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry. 1998;37:433–438. doi: 10.1021/bi972500j. [DOI] [PubMed] [Google Scholar]
- 12.Davies WL, Carvalho LS, Tay BH, Brenner S, Hunt DM, Venkatesh B. Into the blue: gene duplication and loss underlie color vision adaptations in a deep-sea chimaera, the elephant shark Callorhinchus milii. Genome Res. 2009;19:415–426. doi: 10.1101/gr.084509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke RR, Sakmar TP, Oprian DD, Khorana HG. A single amino acid substitution in rhodopsin (lysine 248 to leucine) prevents activation of transducin. J Biol Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- 14.Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 15.Davies WL, Collin SP, Hunt DM. Adaptive gene loss reflects differences in the visual ecology of basal vertebrates. Mol Biol Evol. 2009;26:1803–1809. doi: 10.1093/molbev/msp089. [DOI] [PubMed] [Google Scholar]
- 16.Govardovskii VI, Fyhrquist N, Reuter T, Kuzmin DG, Donner K. In search of the visual pigment template. Vis Neurosci. 2000;17:509–528. doi: 10.1017/S0952523800174036. [DOI] [PubMed] [Google Scholar]
- 17.Davies WL, Cowing JA, Carvalho LS, Potter IC, Trezise AE, Hunt DM, Collin SP. Functional characterization, tuning, and regulation of visual pigment gene expression in an anadromous lamprey. FASEB J. 2007;21:2713–2724. doi: 10.1096/fj.06-8057com. [DOI] [PubMed] [Google Scholar]
- 18.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 19.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 21.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 22.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 23.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 24.Kaushal S, Khorana HG. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama S, Takenaka N, Agnew DW, Shoshani J. Elephants and human color-blind deuteranopes have identical sets of visual pigments. Genetics. 2005;170:335–344. doi: 10.1534/genetics.104.039511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arrese CA, Beazley LD, Ferguson MC, Oddy A, Hunt DM. Spectral tuning of the long wavelength-sensitive cone pigment in four Australian marsupials. Gene. 2006;381:13–17. doi: 10.1016/j.gene.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Lamb TD, Pugh EN., Jr Phototransduction, dark adaptation, and rhodopsin regeneration the proctor lecture. Invest Ophthalmol Vis Sci. 2006;47:5137–5152. doi: 10.1167/iovs.06-0849. [DOI] [PubMed] [Google Scholar]
- 28.Baylor DA, Matthews G, Yau KW. Two components of electrical dark noise in toad retinal rod outer segments. J Physiol. 1980;309:591–621. doi: 10.1113/jphysiol.1980.sp013529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebrey T, Koutalos Y. Vertebrate photoreceptors. Prog Retin Eye Res. 2001;20:49–94. doi: 10.1016/S1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- 30.Kefalov VJ, Estevez ME, Kono M, Goletz PW, Crouch RK, Cornwall MC, Yau KW. Breaking the covalent bond—a pigment property that contributes to desensitization in cones. Neuron. 2005;46:879–890. doi: 10.1016/j.neuron.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JL, Picones A, Korenbrot JI. Differences in transduction between rod and cone photoreceptors: an exploration of the role of calcium homeostasis. Curr Opin Neurobiol. 1994;4:488–495. doi: 10.1016/0959-4388(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 32.Pugh EN, Jr, Nikonov S, Lamb TD. Molecular mechanisms of vertebrate photoreceptor light adaptation. Curr Opin Neurobiol. 1999;9:410–418. doi: 10.1016/S0959-4388(99)80062-2. [DOI] [PubMed] [Google Scholar]
- 33.Travis GH. DISCO! Dissociation of cone opsins: the fast and noisy life of cones explained. Neuron. 2005;46:840–842. doi: 10.1016/j.neuron.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Yau KW. Phototransduction mechanism in retinal rods and cones. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1994;35:9–32. [PubMed] [Google Scholar]
- 35.Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retin Eye Res. 2000;19:385–419. doi: 10.1016/S1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 36.Davies WL (2011) Adaptive gene loss in vertebrates: photosensitivity as a model case. Encyclopedia of life sciences
- 37.Newman LA, Robinson PR. Cone visual pigments of aquatic mammals. Vis Neurosci. 2005;22:873–879. doi: 10.1017/S0952523805226159. [DOI] [PubMed] [Google Scholar]
- 38.Merbs SL, Nathans J. Absorption spectra of human cone pigments [see comments] Nature. 1992;356:433–435. doi: 10.1038/356433a0. [DOI] [PubMed] [Google Scholar]
- 39.Newman LA, Robinson PR. The visual pigments of the West Indian manatee (Trichechus manatus) Vis Res. 2006;46:3326–3330. doi: 10.1016/j.visres.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Fasick JI, Cronin TW, Hunt DM, Robinson PR. The visual pigments of the bottlenose dolphin (Tursiops truncatus) Vis Neurosci. 1998;15:643–651. doi: 10.1017/S0952523898154056. [DOI] [PubMed] [Google Scholar]
- 41.Arrese CA, Hart NS, Thomas N, Beazley LD, Shand J. Trichromacy in Australian marsupials. Curr Biol. 2002;12:657–660. doi: 10.1016/S0960-9822(02)00772-8. [DOI] [PubMed] [Google Scholar]
- 42.Deeb SS, Wakefield MJ, Tada T, Marotte L, Yokoyama S, Marshall Graves JA. The cone visual pigments of an Australian marsupial, the tammar wallaby (Macropus eugenii): sequence, spectral tuning, and evolution. Mol Biol Evol. 2003;20:1642–1649. doi: 10.1093/molbev/msg181. [DOI] [PubMed] [Google Scholar]