Abstract
CD4+CD25+Foxp3+ regulatory T cells (Treg cells) are critical for the maintenance of peripheral tolerance, and the suppression of autoimmune diseases and even tumors. Although Treg cells are well characterized in humans, little is known regarding their existence or occurrence in ancient vertebrates. In the present study, we report on the molecular and functional characterization of a Treg-like subset with the phenotype CD4-2+CD25-like+Foxp3-like+ from a pufferfish (Tetraodon nigroviridis) model. Functional studies showed that depletion of this subset produced an enhanced mixed lymphocyte reaction (MLR) and nonspecific cytotoxic cell (NCC) activity in vitro, as well as inflammation of the intestine in vivo. The data presented here will not only enrich the knowledge of fish immunology but will also be beneficial for a better cross-species understanding of the evolutionary history of the Treg family and Treg-mediated regulatory networks in cellular immunity.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0574-5) contains supplementary material, which is available to authorized users.
Keywords: Foxp3, CD4, CD25, Treg cells, Fish, Comparative immunology
Introduction
Regulatory T cells (Treg cells) and Treg-mediated regulation networks have recently become hot topics for study, because of their crucial roles in the induction and maintenance of peripheral tolerance, as well as in suppression of autoimmune diseases and tumors [1–3]. The naturally occurring CD4+CD25+ Treg cells are characterized as anergic to IL-2 stimulation or signaling through TCR alpha alone, which markedly suppress the polyclonal proliferation of T cells by an antigen nonspecific manner in vitro [4, 5]. Treg cells are also known to inhibit proliferation of B cells, cytotoxic function of NK cells, maturation of antigen presenting cells including DCs and macrophages, and cytokine production in both CD4+ and CD8+ T cells. Therefore, Treg cells can target several stages of the adaptive immune response, spanning the events of lymphocyte proliferation and activation, through to effector function [6]. In vivo, elimination of Treg cells correlates with an increase in inflammatory diseases, such as insulitis, arthritis, gastritis, and inflammatory bowel disease (IBD) [7, 8]. More than one mechanism of CD4+CD25+ Treg-mediated suppression exists, including involvement of immuno-suppressive cytokines (IL-10 and TGF-β1), contribution of cell-to-cell cognate interactions, and modification of APCs [1]. CD25 was first reported in 1995 as a phenotypic marker for CD4+ regulatory T cells [7], and studies since then have identified the transcription factor forkhead box P3 (Foxp3) as a key intracellular marker and also a crucial developmental and functional factor for CD4+CD25+ regulatory T cells [9, 10].
Treg cells have been well characterized in humans and several other mammals; however, little is known about their existence or occurrence in ancient vertebrates. It is generally accepted that a precise immune system might have become established in teleosts about 450 million years ago [11], based on the origin of hallmark molecules and cells for adaptive immunity, such as immunoglobulins, major histocompatibility complex (MHC), and T and B lymphocytes [12–15]. Nonetheless, the exact molecular and cellular mechanisms underlying teleost immunity remain elusive [16, 17]. Recently, a CD8 alpha positive leucocyte population from fugu (Takifugu rubripes) was found to proliferate in the same fashion as mammalian CD8+ T cells in response to PHA [18]. However, very little is known about the sub-populations of fish lymphocytes, such as Th and Treg cells, or whether they may function in regulation of cell-mediated immunity, since most of the hallmark molecules of these subsets in fish species have still not been well characterized.
In the present study, we report on the molecular and cellular identification and functional characterization of a Treg-like cell population from a pufferfish (Tetraodon nigroviridis). The molecular hallmarks of Treg cells, including CD4, CD25 and Foxp3, were identified from this fish model. Phenotypic identification showed that a CD4-2+CD25-like+Foxp3-like+ Treg-like subset existed in the Tetraodon leucocyte population. The elimination of this subset produced an enhanced mixed lymphocyte reaction (MLR) and nonspecific cytotoxic cell (NCC) activity in vitro, while in vivo, it evoked inflammation of intestine. Our results provide the first evidence for the existence of a functional Treg-like regulatory mechanism underlying cellular immunity in a fish species. This strongly suggests that precise cell-to-cell interactions and signal exchange mechanisms underlie the immunity of ancient vertebrates, and gives insights into the origin of a Treg-like subset during early vertebrate evolution. Given the central roles played by Treg cells in the regulation of cellular immunity, autoimmune diseases, and transplant tolerance, we hope that the identification of this subset in fish will not only enrich the knowledge of fish immunology but will also contribute to a better cross-species understanding of the evolutionary history of the Treg family and Treg-mediated cellular regulatory networks.
Materials and methods
Experimental fish
Spotted green pufferfish, T. nigroviridis, weighing approximately 4–6 g, were kept in running water at 25°C and fed with commercial pelleted food twice a day. The fish were held in laboratory for at least 2 weeks prior to experimental use, to allow acclimatization and evaluation of overall fish health. Only healthy fish, as determined by general appearance and level of activity, were used for these studies.
Cloning and sequence analysis of foxp3, CD4 and CD25
The sequences of foxp3, CD4 and CD25 were initially screened in silico from the genome databases at UCSC as previously described [19]. Total RNAs were extracted from spleen and the head kidney tissues using RNAiso Plus (TaKaRa). RT-PCR and RACE were used to obtain cDNA sequences using primers shown in Supplemental Table 1. The foxp3 distribution was detected by both RT-PCR and real time RT-PCR in several tissues, including spleen, kidney, gut, gill, and leucocytes, using primers shown in Supplemental Table 1. Genomic structures and organizations were determined by comparing these cloned sequences to the Tetraodon genome draft using the BLAST program at UCSC. The potential motifs were searched in the PROSITE database. Protein sequence alignment was generated using Clustal X (1.8), and phylogenetic analyses were conducted using MEGA version 4 [20].
Production of recombinant proteins
Sequences encoding Tetraodon mature IL-16 (C-terminal of pro-IL-16, GenBank accession no. AY944133), MHC-II β chain non-polymorphic region (GenBank accession no. FJ812037), IL-2 (GenBank accession no. EF513163), mature peptide of IL-15 (GenBank accession no. DQ059388), extracellular domains of CD4s (CD4-2, CD4-4) and CD25-like, were amplified with primers shown in Supplemental Table 1. Purified PCR products were ligated into digested pET28a, pET32a and pET41a vectors (Novagen) with sequences encoding 6×His, Trx and GST tags, respectively. The recombinant proteins were induced in E.coli Rosetta (Novagen) as soluble forms by isopropyl-b-d-thiogalactopyranoside (IPTG, 1 mmol/L), and were affinity-purified using Ni–NTA resin (QIAGEN) and GSTrap FF (GE Healthcare) according to the manufacturer’s instructions.
Binding assays
Quantitative ELISAs were utilized to analyze the interactions among CD4s/CD25 and their ligands. Briefly, different concentrations (0–100 pmol) of recombinant Trx-CD4-2, Trx-CD4-4 and Trx (negative control) were coated onto a 96-well plate and incubated overnight at 4°C. After blocking with 2% BSA at 37°C for 1 h, 25 pmol GST-IL-16, GST-MHC-II β chain or GST (negative control) were added to each well, and incubated at 37°C for 4 h. The wells were washed three times with PBST, then incubated with 1:5,000 diluted anti-GST antibody (GenScript) at 37°C for 1 h. Different concentrations (0–10 pmol) of recombinant GST-CD25-like or GST (negative control) were also coated, blocked, incubated with 3 pmol Trx-IL-2, Trx-IL-15 or Trx (negative control), and 1:5,000 diluted anti-Trx antibody (GenScript), as described above. After washing, the wells were incubated with secondary HRP-conjugated antibody (Santa Cruz Biotechnology) at 37°C for 1 h. Positive signals were developed with the TMB color reaction and analyzed with a plate reader at 630 nm.
Preparation of polyclonal antibodies against CD4s and CD25-like
Recombinant proteins containing only 6×-His tags were purified for polyclonal antibody (Abs) production. Briefly, male 6-week-old New Zealand White Rabbits (~3 kg) were immunized with 3 mg CD25-like, and Balb/c mice (~30 g) were immunized with 50 μg CD4-2 or CD4-4. The anti-sera were collected after six immunizations at biweekly intervals. The anti-CD4-2, anti-CD4-4 and anti-CD25-like Abs were subsequently purified into IgG isotypes by affinity chromatography with purified antigens and HiTrap Protein A HP column (Amersham Biosciences). Western blots and antigen-specific ELISAs were performed to characterize the specificity of the Abs to CD4-2, CD4-4 or CD25-like.
Preparation of leucocytes
Leucocytes were prepared from Tetraodon peripheral blood, spleen, and head kidney, as described previously [21]. Briefly, spleen and head kidney were removed after sacrifice of the fish. Single cell suspensions were obtained by teasing the tissue (in serum-free RPMI-1640 culture medium) through a nylon sieve. The cells were washed twice in ice-cold culture medium without serum, collected, and layered on 1.5 vol Histopaque (Sigma, density adjusted to 1.0808 g/mL). Following 25 min centrifugation at 900g, leucocytes were collected from the interface layer and washed three times with PBS. These cells were then cultured in RPMI-1640 medium (Gibco) supplemented with 5% fetal calf serum (Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin at 28°C. The cell number was determined with a hemacytometer.
Chemotaxis assay
The chemotactic responses of CD4-2+ or CD4-4+ cells to IL-16 were assessed by measuring the migration of leucocytes, isolated from head kidney as above, through a 3-μm-pore polycarbonate filter in Transwell chambers (Corning Costar). Briefly, 1.5 × 105 freshly isolated leucocytes suspended in 1640 medium were added to the top chamber, and recombinant GST-IL-16 and GST (negative control) in 1640 medium were added to the bottom chamber. After 4 h of incubation at 28°C, cells in the bottom chamber were harvested and counted. Migration index (MI) was defined as the ratio of cell migration in the presence of GST-IL-16 or GST to 1640 medium control. Limulus amoebocyte lysate (LAL) assay was performed using the Endotoxin Assay Kits (GenScript) to make sure that the recombinant proteins contain low concentration of LPS. Each chemotaxis assay was performed at a series of concentrations and repeated five times, independently. For blockage experiments, freshly isolated leucocytes were incubated for 30 min at 28°C with anti-CD4-2, anti-CD4-4, PBS (mock control) and mouse IgG (negative control) at saturating doses. After washing twice with PBS, blocked leucocytes were used for chemotaxis assays as described above.
Flow cytometry
Surface markers of fish leucocytes were analyzed with a fluorescence-activated cell sorter (FACSCALIBUR; BD Biosciences). Briefly, cells were blocked with 5% normal goat serum, then incubated with primary Abs against CD4-2 and/or CD25-like and secondary R-PE and/or FITC-conjugated Abs (Santa Cruz Biotechnology). At least 10,000 events were collected. The analysis was performed using Cell Quest Software (Becton–Dickinson).
Immunofluorescence staining
Cells for immunofluorescence staining were first fixed with cold methanol for 5 min, blocked with 5% normal goat serum at 37°C for 45 min, and incubated with primary Abs against CD4-2 and CD25-like at 37°C for 45 min. After washing, cells were then incubated with secondary TRITC- and FITC-conjugated anti-mouse or anti-rabbit Abs (Santa Cruz Biotechnology), and additional stainings with DAPI were performed before photomicrography. Samples were photomicrographed under a confocal laser-scanning microscope (LSM 510; Carl Zeiss).
Magnetic cell sorting
Approximately 1 × 109 leucocytes were incubated with mouse anti-CD4-2 Abs for 15 min at 10°C, and then incubated for 15 min at 10°C with anti-mouse IgG magnetic beads (Miltenyi Biotec) after 2 washes with MACS buffer. The washing procedure was repeated and cells were resuspended in MACS buffer. The cell suspension was applied to an LS separation column according to the manufacturer’s instructions. The CD4-2 positive (CD4-2+) cells were collected and then incubated in RPMI-1640 (5% FBS) at 28°C overnight to detach the magnetic beads. Approximately 1 × 107 CD4-2+ cell suspension was then subjected to secondary sorting for CD25-like+ cells using the same procedure. The sorted cell types were confirmed by FACs analysis. The fractionated CD4-2+CD25-like+ (~5 × 104) and CD4-2+CD25-like− (~1 × 105) cells were used for identification of cell types, and the expression of TCR-α, IgL, CD8, and foxp3 was detected by RT-PCR analysis, using the primers shown in Supplemental Table 1.
In situ hybridization
Head kidney paraffin sections and isolated leucocytes were processed for ISH using a probe specific to foxp3 mRNA, which was prepared according to the kit instructions (Boster). Briefly, foxp3 cDNA that spanned two exons was amplified using the primers shown in Supplemental Table 1, and the purified DNAs were then labeled with digoxigenin (DIG). Tissue sections and leucocytes prepared by standard methods (Boster) were hybridized with foxp3 probes in hybridization solution at 47°C for 15 h, while the hybridization solution was added as a negative control. The positive signals were revealed with DAB color reaction. Positive foxp3 cells showed a brown color under the light microscope (Eclipse 80i; Nikon).
Depletion of CD25-like+ cells in vitro and in vivo
In vitro depletion of Treg-like cells from the total leucocytes population was performed according to the method described previously, by treating the cells with anti-CD25-like antibody and complement [7]. Briefly, cell suspensions (5 × 106) were incubated with 20 μL of 1:10-diluted of anti-CD25-like (2 μg) for 45 min on ice, washed once with PBS, and incubated with 1 mL of nontoxic mouse serum diluted 1:5 with RPMI-1640 for 30 min at 37°C. After washing twice with PBS, cells were suspended in RPMI-1640 (5% FBS). A negative control used normal rabbit IgG (2 μg) for the incubation. For in vivo depletion, either 0.5, 2, or 5 μg anti-CD25-like Abs were injected i.p. into healthy Tetraodon weekly, according to previous reports, with slight modification [22]. Normal rabbit IgG (2 μg/fish) was also injected as a negative control. The effect of in vitro and in vivo depletion was confirmed by FACs analysis.
Functional assessment by NCC activity
Cytotoxic activity for NCC (as effector cells) to K562 target cells was assayed by measuring lactate dehydrogenase (LDH) release according to the manufacturer’s protocols (CytoTox 96 Non-radioactive Cytotoxicity Assay; Promega). Briefly, cell populations of total leucocytes (1 × 105) without any treatment, or CD25-like− (Treg-like depleted) leucocytes (1 × 105), were co-cultured with 1 × 104 K562 target cells for 4 h at 28°C. The percentage of cytotoxicity for NCC activity was computed as the ratio of absorbance of the experimental co-cultures minus the spontaneous release of both the effector and target cells to that of the target maximum release minus spontaneous target values. In parallel, a recruitment experiment was performed by adding magnetically sorted Treg-like (CD4-2+CD25-like+) cells back into a Treg-like depleted (CD25-like−) cell population at different proportions of 5:0, 4:1, 1:1, or 1:4, in a total cell population of 1 × 105, and the NCC activity was examined as described above. Each assay was carried five times in individual experiments.
Functional assessment by MLR
A two-way MLR was performed to characterize the Treg-derived suppressive functions [23]. Briefly, fish leucocytes were prepared from peripheral blood, spleen and head kidney of individuals as reported before. Cultures of leucocytes (5 × 105 cells/well) derived from one fish were mixed with equal numbers of total or CD25-like− leucocytes from another fish and incubated for 72 h at 28°C. The cellular proliferation was detected with a CCK-8 kit (Beyotime) according to the manufacturer’s instructions. The stimulus index (SI) was defined as the ratio of the increased number of mixed lymphocyte cultures to the sum of cells cultured alone. In addition, a recruitment experiment was performed by adding magnetic sorting Treg-like (CD4-2+CD25-like+) cells back into Treg-like depleted (CD25-like−) mixed cultures at four different proportions, 5:0, 4:1, 1:1, or 1:4, in a total cell of 5 × 105, and cellular proliferation was examined as described above. Each assay was carried five times in individual experiments.
Functional assessment in an IBD model by CD25-like+ cell depletion
Functional characterization of Treg-like cells was also determined using an IBD model. For this purpose, Treg-like cells were depleted by in vivo inoculation with anti-CD25-like as in the method described above. The guts were fixed in 10% formalin and embedded in paraffin, and 8-μm-thick sections were then stained with hematoxylin and eosin for histological assessment. In parallel, total RNAs of intestines were extracted, and RT-PCRs were performed to detect the expression of representative inflammation factors for IBD, including TNF-α and IFN-γ. In addition, IL-10 was also detected, which secreted by Treg cells as a regulator of IBD. The primers used in RT-PCR are shown in Supplemental Table 1.
Statistical analysis
Statistical analysis was performed using SPSS version 16.0 software, and data were expressed as mean ± SD. Differences between the values were determined by two-tailed paired t test. A value of p < 0.05 was considered to be significant. The sample number of each group was more than five fish of equal mean body weight. Data points were from at least three independent experiments.
Results
Identification of Tetraodon foxp3-like molecule
The molecular characterization of Tetraodon foxp3-like is shown in Supplemental Figure 1. It shared an overall structural consistency with the foxp3 molecules from mammals, including a similar gene organization pattern on chromosomes, a conserved forkhead domain at the C-terminal, and a single C2H2 zinc finger domain 108 amino acids upstream from the forkhead domain [24]. However, somewhat unlike the gene from human, the coding exons of Tetraodon foxp3-like were more similar to those of zebrafish [25, 26], as the last coding exon was divided into two segments corresponding to the last single exon in human foxp3. The putative Tetraodon Foxp3-like shared a similarity of 40% with its mammalian homologs, 35% with Xenopus Foxp3, and 49% with zebrafish Foxp3. Phylogenetic analysis showed that Tetraodon foxp3-like clustered most closely together with zebrafish foxp3, and then with foxp3s from other species. The remaining foxp family members (foxp1, 2, and 4) were more closely grouped, excluding the foxp3s (see Supplemental Figure 2).
Identification of Tetraodon CD4 molecules
The Tetraodon CD4 gene was identified by comparison with several reported fish CD4 sequences [27–31]. In this study, two CD4 genes were cloned from Tetraodon. Because they possessed structural hallmarks of two and four extracellular Ig domains, they were designated as CD4-2 and CD4-4, respectively. Molecular characterization analysis showed that these two molecules shared similarities in genomic organization and syntenic relationship with the human forms, and were conserved in the cytoplasmic tail (CYT) with a protein tyrosine kinase p56lck binding motif (C–X–C) similar to that from mammalian CD4s (see Supplemental Figure 3). Together with published CD4 amino acid sequences, a phylogenetic tree was built using lamprey (Petromyzon marinus) as an outgroup. The result showed that fish CD4-2 molecules formed a cluster in the CD4 family, indicating that these two-Ig-domain molecules were additional CD4 homologues in fish (see Supplemental Figure 4).
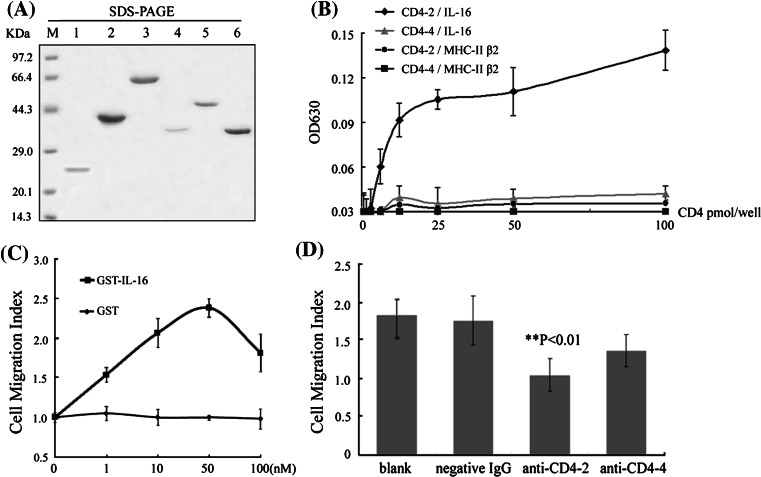
The functional diversity between the two fish CD4 molecules was characterized by their ligands (IL-16 and MHC-II β chain β2 domain) binding experiments and IL-16 chemotactic assays, according to the reports in mammals [32, 33]. The results showed that CD4-2 has a significantly higher binding capacity to IL-16 than did CD4-4 (Fig. 1a, b). CD4-2 also demonstrated its ability to bind with MHC-II β chain non-polymorphic region, while CD4-4 showed negligible binding. In contrast, the binding capacity of CD4-2 to IL-16 was stronger than to MHC-II β chain under the same receptor/ligand molar ratio conditions. The interactions between the molecules exhibited dose-dependent. When the molar ratio of the receptor/ligand exceeded 1, a plateau appeared due to an excess amount of receptor. The background binding of GST controls was much weaker and negligible. Similar results were also seen in chemotactic assays. As shown in Fig. 1c, IL-16 stimulated optimal leucocyte migration at a concentration of 5 × 10−8 mol/L. Before the blocking analyses, anti-CD4-2 and anti-CD4-4 Abs were pretested for their abilities of binding native CD4-2 and CD4-4 proteins on the cell surface by cellular staining experiments (data not shown). Blockade cells with anti-CD4-2 Abs significantly (p < 0.01) inhibited the migration of leucocytes in response to IL-16, as determined by the reduction of average MI from 1.76 to 1.05 (Fig. 1d). Little reduction in MI to IL-16 was seen in response to anti-CD4-4 Abs treatment. There was a little reduction of MI with the negative IgG, which might be due to some non-specific interference comparing to the blank control. Thus, IL-16 appeared to function as a chemoattractant factor that was preferential for CD4-2 positive leucocytes, suggesting that CD4-2 might play further essential roles in leucocyte functions.
Fig. 1.
Functional diversity between Tetraodon CD4-2 and CD4-4. a Purified fusion CD4s and their ligands were detected by SDS-PAGE. M protein marker; lanes 1–3 Trx (~21 kDa), Trx-CD4-2 (~42 kDa), and Trx-CD4-4 (~63 kDa) fusion proteins, respectively; lanes 4–6 GST (~34 kDa), GST-IL-16 (~52 kDa), and GST-MHC-II β (~38 kDa) fusion proteins, respectively. b Direct binding ELISAs were conducted for the detection of the interaction between solid-phase CD4s with GST fusion ligands. The fusion Trx-CD4s binding values minus the Trx background values are shown, and the background values of CD4s and GST tag are relatively low. The means of three experiments are presented for each set of experiments. c Optimal leucocyte migration was determined by incubation with different concentrations of IL-16 (squares) or GST (triangles). d Polyclonal antibodies were tested for their ability to block rIL-16 stimulated Tetraodon leucocyte migration. All data are expressed as migration indexes of five averaged experiments, with significance indicated as **p < 0.01
Identification of Tetraodon CD25-like molecule
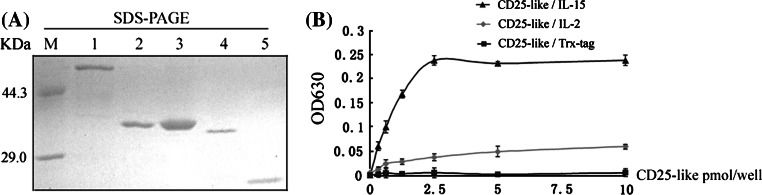
The Tetraodon CD25-like shared an overall consistency with the CD25 molecules from mammals and other species, including conserved chromosome synteny, genomic organization, and functional motifs such as the sushi domain and Pro/Thr rich domain (see Supplemental Figure 5). However, Tetraodon CD25-like contained only one sushi domain, instead of the two domains common to mammalian CD25 molecules. We suggested that fish CD25-like might serve as a common receptor (IL-2Rα/IL-15Rα) for IL-2 and IL-15, since only one gene was found at the corresponding locus of mammalian CD25 (IL-2Rα) and IL-15Rα by comparative genomic analysis (see Supplemental Figure 6). To verify this hypothesis, we performed a binding assay between CD25-like and IL-2/IL-15, and found that Tetraodon CD25-like could bind to both IL-2 and IL-15 in dose-dependent manners, with a ~fivefold preference for binding IL-15 (Fig. 2). In parallel, the background binding of Trx-tag controls was negligible.
Fig. 2.
Direct binding assays between Tetraodon CD25-like and IL-2/IL-15. a Purified fusion CD25-like, IL-2 and IL-15 were revealed by SDS-PAGE. M protein marker; lanes 1–2 GST-CD25-like and GST proteins, respectively; lanes 3–5 Trx-IL-2, Trx-IL-15 and Trx proteins, respectively. b Direct binding ELISAs were conducted for the detection of the interaction between solid-phase CD25-like with Trx fusion ligands. The fusion GST-CD25-like binding values minus the GST background values were exhibited here, and the background value of CD25-like and Trx tag is relatively low. The means of three experiments are presented for each set of experiments
To determine the phylogenic relationship of Tetraodon CD25-like molecule with other CD25 molecules, we aligned the first sushi domain of CD25 (IL-2Rα) with the only sushi domain of IL-15Rα, as these contribute to the majority of cytokine binding surfaces. As shown (see Supplemental Figure 7), the higher vertebrate IL-2Rα, IL-15Rα and fish IL-2Rα/15Rα formed distinct clades. The evolutionary distance appeared closer between higher vertebrate IL-2Rα and IL-15Rα than with teleost IL-2Rα/15Rα. Thus, the fish CD25-like gene might be an ancestral molecule of both mammalian IL-2Rα and IL-15Rα.
Identification of CD4-2+CD25-like+ cells
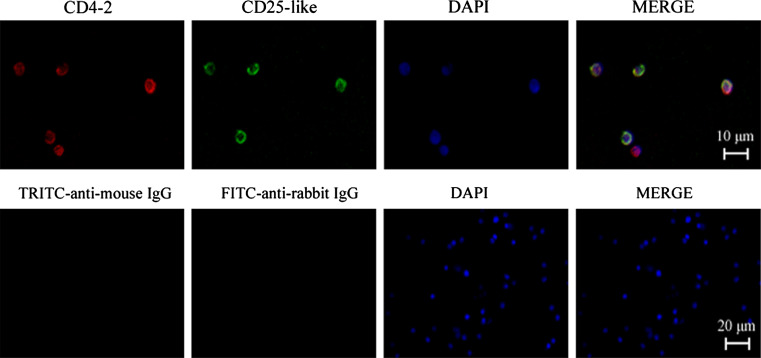
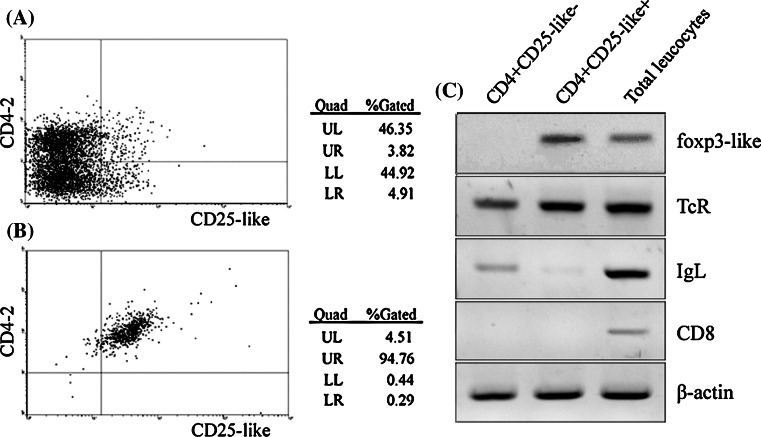
For phenotypic identification of Treg-like cells in Tetraodon, recombinant CD25-like and CD4-2 proteins and their polyclonal antibodies were developed (see Supplemental Figure 8). By dual immunofluorescence staining with anti-CD4-2 and anti-CD25-like, followed by TRITC- and FITC-conjugated secondary antibodies, it was showed that CD4-2 and CD25-like were co-located on the surface of considerable isolated leucocytes (Fig. 3). FACs analysis showed that about 3.82% of the peripheral leucocytes were CD4-2+CD25-like+ (Fig. 4a). This proportion was consistent overall with that seen for mammalian Treg cells. To dissect this subset, CD4-2+CD25-like+ cells were sorted from the total leucocytes by magnetic beads. About 95% of magnetically sorted cells were CD4-2+CD25-like+ according to the FACs analysis (Fig. 4b). These isolated CD4-2+CD25-like+ cells, together with CD4-2+CD25-like− cells and total leucocytes, were used for RT-PCR analysis for further phenotypic identification using several lymphocyte markers. As shown in Fig. 4c, TCR-α molecules were expressed in all three groups, while IgL transcripts were rich in total leucocytes, detectable in CD4-2+CD25-like− cells, but only faint in CD4-2+CD25-like+ cells. This confirmed that most of the CD4+ lymphocytes might be T cells. The CD8 molecules detected in total leucocytes were not expressed in CD4-2+ cells, which suggested that fish possessed a single positive style in common with mammalian T cells. Similar to human Treg cells, a strong signal of transcription factor foxp3-like was detected in CD4-2+CD25-like+ cells, but not in CD4-2+CD25-like− cells.
Fig. 3.
Immunofluorescence stains of CD4-2+CD25-like+ leucocytes. CD4-2 molecules were detected with TRITC-conjugated anti-mouse IgG that revealed red fluorescence, while CD25-like molecules were detected with FITC-conjugated anti-rabbit IgG that exhibited green fluorescence. DAPI stain shows the location of the nuclei. The negative controls are shown in the second row, and consisted of negative mouse and rabbit IgGs in place of primary anti-CD4-2 and anti-CD25-like Abs
Fig. 4.
CD4-2+CD25-like+ leucocytes sorted by magnetic beads. a FACs analysis of CD4-2+CD25-like+ cells before magnetically sorting. b FACs analysis of magnetically sorted CD4-2+CD25-like+ cells. c The isolated CD4-2+CD25-like−, CD4-2+CD25-like+ and total leucocytes were subjected to RT-PCR for detecting foxp3-like, TCR, IgL, and CD8 mRNA expression
The Tetraodon Treg-like cells were further examined with a foxp3-like probe (see Supplemental Figure 9). RT-PCR and real-time RT-PCR showed that foxp3-like mRNAs were detectable in all the tissues examined, including peripheral leucocytes, spleen, kidney, gut, and gill (a thymus containing tissue of fish), with the highest expression detected in gills, followed by kidney. Peripheral leucocytes and kidney tissues were selectively processed for ISH analysis. The result showed that foxp3-like signals were strongly focused on glomeruli of kidney, which were known to be full of blood cells. Foxp3-like signals were also detected in isolated leucocytes.
Inhibitory effects of CD25-like+ cells on NCC and MLR
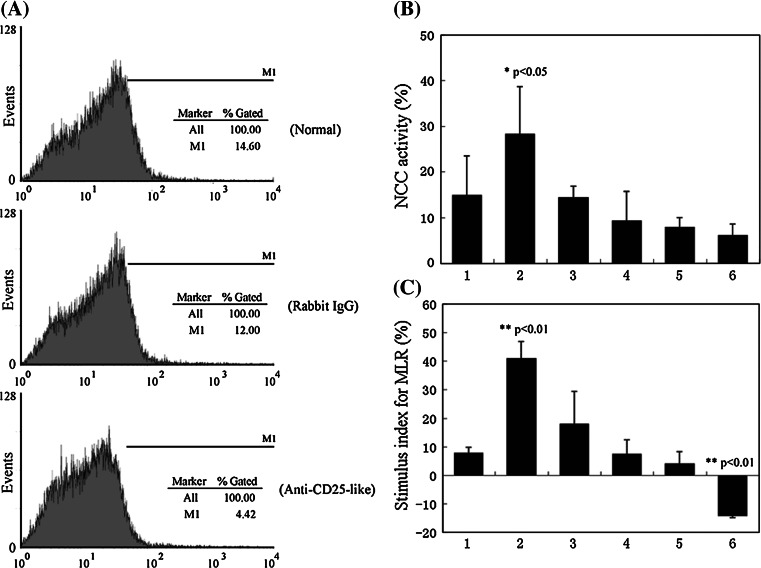
The Treg-like cells were also identified by their repressive functions. For this purpose, in vitro NCC and MLR activity assays were performed. The Treg-like cells were depleted by treatment of spleen and kidney leucocytes with anti-CD25-like plus complement. As shown in Fig. 5a, after this treatment, the proportion of CD25-like+ cells declined by 63.1%, from 10.9 ± 2.6 to 4.02 ± 0.4%, while this proportion was only slightly reduced in the negative control group treated with rabbit IgG.
Fig. 5.
Repressive function of Tetraodon CD25-like+ cells in vitro. a In vitro depletion of Tetraodon CD25-like+ leucocytes. The Tetraodon leucocytes were incubated with rabbit anti-CD25-like Abs and complement, and FACs was conducted to analyze the proportion of CD25-like positive cells. A negative control using normal rabbit IgG was devised for the incubation. b, c Functional identifications of Tetraodon Treg-like cells by non-specific cytotoxic cell (NCC) (b) and mixed lymphocyte reaction (MLR) (c) activities in vitro. Figures on the abscissa indicate different effector cells: 1 total leucocytes, 2 CD25-like+ depleted leucocytes, 3–5 CD25-like+ depleted leucocytes plus CD4-2+CD25-like+ cells with a proportion of 4:1, 1:1, 1:4, respectively, 6 CD4-2+CD25-like+ cells. NCC or MLR activities of treated cells had been compared with total leucocytes, with significance indicated by *p < 0.05 and most significant differences indicated by **p < 0.01
The NCC activity in this treated population (28.3 ± 10.4%) was significantly elevated (p < 0.05) compared with that of the total leucocyte population (15.0 ± 8.5%) that had no treatment (Fig. 5b). This suggested that NCC activity had been enhanced by the loss of the CD25-like+ population. As we expected, when CD4-2+CD25-like+ cells were added proportionally to a cell population from which the CD25-like+ cells had been depleted, the NCC activities gradually decreased from 14.4 ± 2.5 to 6.1 ± 2.5%, indicating recruitment of Treg-like suppressive activity in the depleted population.
Similar results were also observed in MLR assays. As shown in Fig. 5c, a significant (p < 0.01) increase in MLR activity was observed in CD25-like+ cell depleted groups compared to that of untreated control groups. In this case, the stimulus indexes were dramatically elevated from 7.9 ± 1.8 to 40.9 ± 5.8%. Meanwhile, in recruitment experiments, the stimulus indexes sharply dropped from 40.9 ± 5.8 to 18.1 ± 11.3%, accompanied by the supplement of different proportions of CD4-2+CD25-like+ cells to the population with CD25-like+ cells depleted. A negative stimulus index was detected in experiments in which the stimulator untreated leucocytes were co-cultured with extra CD4-2+CD25-like+ cells from another fish. In this case, the stimulus index was significantly (p < 0.01) decreased in the experimental group in comparison with that in normal untreated group. This provided further support for the repressive function of this Treg-like subset. Control tests in which leucocytes were incubated with negative rabbit IgG plus complement exhibited similar results to the normal untreated group in both NCC and MLR assays (data not shown). Thus, CD4-2+CD25-like+ cells could clearly dampen cellular immune activities in vitro.
Functional characterization by an in vivo autoimmune disease model
To further investigate the existence of Treg-like cells in Tetraodon, an in vivo elicitation of an inflammatory bowel disease model was developed. For this purpose, various dosages (0.5, 2, or 5 μg/fish) of anti-CD25-like antibody were injected into healthy Tetraodon to deplete their Treg-like cells. Normal rabbit IgG was injected into control fish for a negative control. The depletion of CD25-like+ cells was determined by flow cytometry.
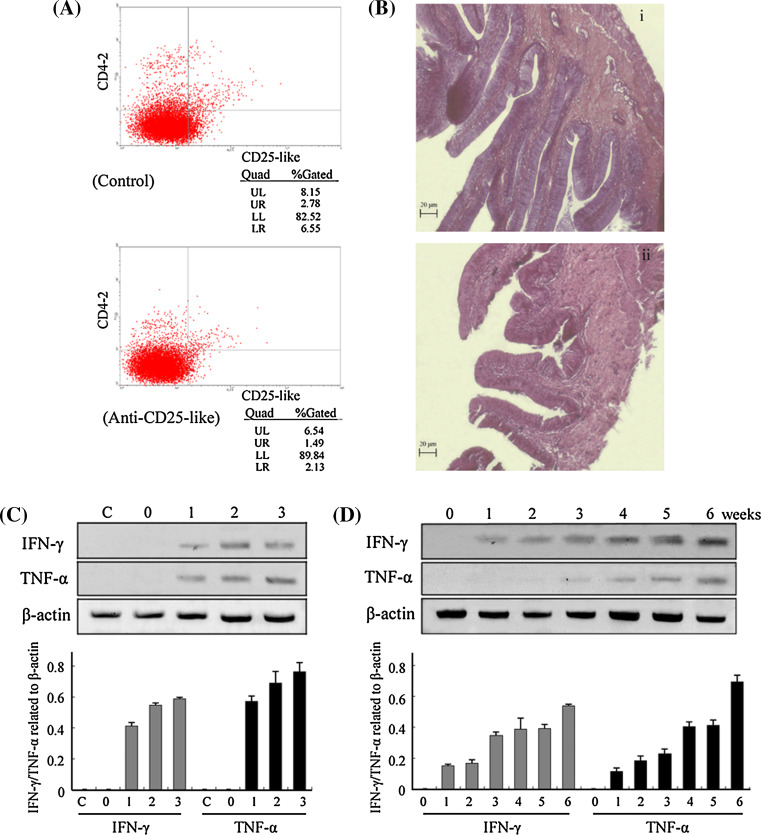
As shown in Fig. 6a, the CD4-2+CD25-like+ Treg-like cells were reduced by 46.4% (from 2.78 to 1.49%) after a 6-week inoculation of anti-CD25-like antibody at the dosage of 2 μg/fish. The guts from both experimental and control fish were sectioned for pathological analysis. As shown in Fig. 6b, inoculation with 2 μg anti-CD25-like antibodies for 6 weeks induced a severe intestinal wall damage with epithelial exfoliation, while no such pathological changes were observed in the control inoculation intestinal segment. The intestinal villi were blunted and broadened, with interstitial edema, and the infiltration of lymphocytes could be also observed in the submucosa. These are all typical features seen in inflammatory bowel disease. The guts from the three groups that received 0.5, 2, or 5 μg/fish anti-CD25-like antibodies were verified to have developed inflammatory lesions, by detecting the levels of IFN-γ and TNF-α, two typical inflammatory factors involved in inflammatory bowel disease. As shown in Fig. 6c, both IFN-γ and TNF-α were detectably expressed in experimental groups, but were negligibly expressed in normal fish or in fish injected with rabbit IgG. The expression levels of these two cytokines were increased with increasing levels of anti-CD25-like inoculum.
Fig. 6.
Assessment of Tetraodon inflammatory bowel disease after depletion of CD25-like+ cells. a Flow cytometry analysis for CD4-2+CD25-like+ cells of a fish that was injected with 2 μg anti-CD25-like Ab for 6 weeks comparing with a control fish injected with 2 μg rabbit IgG. b The guts of these two fish were fixed in 10% formalin and processed for paraffin sectioning and, HE staining: 1 for control fish injected with rabbit IgG, 2 for fish injected with anti-CD25-like antibodies. c Detection of intestine IFN-γ and TNF-α 6 weeks after injection with anti-CD25-like antibodies by RT-PCR analyses: C for normal fish, 0 injection with 2 μg rabbit IgG, 1 injection with 0.5 μg anti-CD25-like antibodies, 2 injection with 2 μg anti-CD25-like antibodies, 3 injection with 5 μg anti-CD25-like antibodies, and the corresponding semi-quantitative analyses of these two genes relative to β-actin were shown in lower panel. d Weekly detection of intestine IFN-γ and TNF-α for injection with 2 μg anti-CD25-like antibodies by RT-PCR, and the corresponding semi-quantitative analyses are shown in the lower panel
To further characterize the inflammatory response, fish that were injected with 2 μg anti-CD25-like antibody were sacrificed every week, and intestinal RNAs were extracted for RT-PCR. As shown in Fig. 6d, IFN-γ emerged in the first week, increased in the third week, and was substantially enhanced in the sixth week. TNF-α, however, was first observed in the first week, increased in the fourth week, and then showed a sharp increase in the sixth week. Taken together, these results indicated that depletion of CD4-2+CD25-like+ Treg-like cells in vivo contributed to intestinal inflammation in previously healthy Tetraodon.
Discussion
Regulatory T cells are critical for an effective immune system as they prevent aggressive and harmful immune responses. Increasingly, more evidence is now revealing that specific T cell populations with suppressive/regulatory properties tightly control autoaggressive immune responses. This suppression was first described in the 1970s, when a group of T cells was shown to inhibit various T cell-mediated reactions, notably the mixed lymphocyte reaction [34]. In 1995, CD25 was first reported as a phenotypic marker for CD4+ regulatory T cells [7]. More recent studies have shown that the transcription factor forkhead box P3 (foxp3) is not only a key intracellular marker but is also a crucial developmental and functional factor for CD4+CD25+ regulatory T cells [9, 10].
However, although numerous, the studies on Treg cells have tended to be confined to a few mammals, such as human and mouse. To explore the origin and evolution of the acquired immune system, especially for the specific regulatory T cell populations, it is essential that ancient vertebrates also be integral participants. Therefore, the results from this study on the Tetraodon, which has a clear genome background, can now begin to fill this void in our knowledge.
The identification of CD4, CD25-like and foxp3-like molecules from Tetraodon gives a preliminary observation of the existence of a Treg-like subset in this species. As an essential transmembrane glycoprotein expressed on the lymphocyte surface, CD4 molecules with various extracellular domains have been previously identified in several teleosts [27–31]. In Tetraodon, different CD4-like genes also exist that have a different number of extracellular Ig domains, indicating a diversity of fish CD4 molecules. Regarding the origin of this molecule, Laing et al. [28] suggested that the two-Ig-domain CD4REL was duplicated to generate the four-domain molecule of CD4, with CD4REL likely representing the primordial form in fish. However, why fish possess various CD4 molecules and how these molecules function are still unknown. In the present study, we found that CD4-2, rather than CD4-4, showed binding capacity with both MHC-II and IL-16 and was largely responsible for lymphocyte migration in response to the chemoattractant factor IL-16. These observations suggested that the ancestral CD4-2 plays an essential role in fish cellular immunity, and it might represent a Th/Treg marker in fish, corresponding to their mammalian CD4 orthologs. Nevertheless, further study is needed to decipher the biological significance of fish CD4-4.
In human and mammals, the full receptor complexes of both IL-2 and IL-15 are heteromeric, sharing co-utilized beta and gamma chains, and a specific unit for IL-2 and IL-15, respectively. CD25 (IL-2Rα) is the specific chain for IL-2, while IL-15Rα is the corresponding one for IL-15 [35, 36]. In general, IL-2Rα and IL-15Rα are believed to arise from a common ancestral gene, as there is much common genetic ground between them [36], but direct evidence to prove this hypothesis is lacking. In the present study, we have found that, unlike the close linkage of IL-2Rα and IL-15Rα in mammalian genomes, only one corresponding IL-2Rα/IL-15Rα-like gene is located at the same chromosomal site in teleost fish. Phylogenetic analysis showed that fish CD25-like (IL-2Rα/IL-15Rα-like) formed a distinct clade apart from the IL-2Rα and IL-15Rα of higher vertebrates, indicating that fish CD25-like might be the ancestral gene of the IL-2Rα/15Rα gene family. The fish CD25-like also indicated a potential tandem duplication between IL-2Rα and IL-15Rα, which probably had occurred locally in the four-limbed species after separation from teleost fish. On the other hand, it was retained as one single copy in teleost species, as shown in Supplemental Figure 10. The fish CD25-like gene shared somewhat more genetic similarities with the mammalian IL-15Rα gene, and the CD25-like molecule had affinity to both IL-15 and IL-2 (high to IL-15 and moderate to IL-2). We suggest that the CD25-like molecule in fish may be evolutionarily closer to IL-15Rα, and we envisage it as serving as a co-receptor component for both IL-2 and IL-15.
Based on the molecular identification of the hallmarks of Treg cells, we were successful in isolating a Treg-like subset from Tetraodon with a phenotype of CD4-2+CD25-like+foxp3-like+. This subset may represent ancestors of regulatory T cells from early vertebrate evolution. Similar to mammalian Treg cells, this CD4-2+CD25-like+ population expressed TCR, CD4-2, CD25-like, and foxp3-like, but not IgL or CD8. Another isolated population with phenotype of CD4-2+CD25-like− was found to express TCR and IgL, but not foxp3 or CD8, revealing a similar single positive style to that seen in mammalian T cells [37, 38]. The fish version of foxp3-like seemed to be specifically expressed in CD4-2+CD25-like+ lymphocytes, indicating its essential role throughout Treg cell evolution, with conserved structure and function. Therefore, fish may prove to be perfect models for investigations of both foxp3 and Treg cells.
The Treg-like cells were also functionally identified by their suppressive characterizations, both in vitro and in vivo. Healthy fish contained a low proportion of Treg-like cells, but they exerted strong suppressive effects on both MLR and NCC activity. In MLR assays, the cellular proliferation was dramatically increased by depleting Treg-like cells from the responding cells, while it could be significantly reduced by supplementation with sorted Treg-like cells. In NCC assays, we have also characterized stronger aggression of these Treg-like-depleted populations against tumor K562 cells.
In mammals, it has been well documented that Treg cells can prevent the development of pathogenic responses toward commensal bacteria [8]. The intestinal immune system must orchestrate a complex balance between Th and Treg cells, and disruptions in this balance can result in IBD [39, 40]. Therefore, the IBD model was used for functional evaluation of Treg-like cells in our study. The pathological changes of the intestine and the expression of IFN-γ and TNF-α were used to evaluate the levels of inflammatory bowel disease, and IL-10 was used to evaluate Treg-like activity. After 6 weeks following in vivo inoculation of anti-CD25-like antibody, a significantly reduced number of Treg-like cells were observed. This was accompanied with dramatic intestinal lesions, including histological lesions, upregulation of IFN-γ and TNF-α, and downregulation of IL-10. All these responses could have been elicited by dysregulation of Treg-like cells in the intestinal immune system. These results were similar to those seen in mammalian IBD models, in which the suppressive cytokine, IL-10, is important for the maintenance of T cell tolerance in the gut [41, 42]. The exact mechanism underlying the fish Treg-like immunity remains to be clarified.
In conclusion, our studies provide the first evidence that a regulatory mechanism mediated by T cells operates in ancient vertebrates. A precise cellular regulatory network might therefore have emerged during early vertebrate evolution. We hope that it will be beneficial for the better understanding of the evolutionary history of adaptive immunity. We also anticipate that fish will become a model for understanding T cell immunity and evolutionary relationships.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Tetraodon foxp3-like sequences, gene organization and chromosomal synteny analyses. (A) The cDNA sequence (1411 bp) of Tetraodon foxp3-like gene (GenBank accession no. GU592499), with decoded 413 amino acid (shown beneath), showing the full length ORF (1242 bp) and 3’ UTR with a polyA tail. The zinc finger domain is underlined, the forkhead domain is in bold, and the asterisk reveals a stop codon. (B) Comparison of foxp3 gene organizations between fish and human. The Tetraodon foxp3-like gene consisted of twelve exons and eleven introns, in which the fifth and sixth exons encoded the zinc finger domain, the seventh and eighth exons encoded the leu zipper, and the last four exons encoded the forkhead domain. The rectangles represent exons and lines between them indicate introns. The numbers above rectangles or below lines represent the corresponding lengths. (C) Chromosomal syntenic analysis of Foxp3 genes among different species, showing partial syntenic relationship between fish Foxp3 and Xenopus and human Foxps genes, although it slightly divers from species, possibly suggesting the different recombination events occurred among them, in which the Foxp3 in Xenopus seems to be at a transition stage (TIFF 210 kb)
Multiple alignment analysis (A) and phylogenetic analysis (B) of Foxp3 orthologs among different species. In the alignment, residues shaded in black are completely conserved across all species aligned, and the residues shaded in gray are similar with respect to side chains. The dashes in the amino acid sequences indicate gaps introduced to maximize alignment. The result showed that foxp3 proteins were well conserved at both zinc finger domains in the middle and forkhead domains at C-terminals, from fish to mammals. The phylogenetic tree was constructed by the neighbor-joining method, and the numbers at branch nodes indicate percent bootstrap confidence values derived from 2000 replications. GenBank accession numbers for these amino acid sequences are as follows: human foxp3 (ABQ15210), cat foxp3 (ABN79272), mouse foxp3 (CAM25950), Xenopus foxp3 (BAG12188) and zebrafish foxp3 (FJ906821); human foxp1 (Q9H334), mouse foxp1 (P58462), Xenopus foxp1 (Q5W1J5), zebrafish foxp1b (Q2LE08), Tetraodon foxp1b (FJ358692); human foxp2 (O15409), mouse foxp2 (P58463), Xenopus foxp2 (Q4VYS1), medaka foxp2 (B1NY82), zebrafish foxp2 (Q4JNX5); human foxp4 (Q8IVH2), mouse foxp4 (Q9DBY0), and Xenopus foxp4 (Q4VYR7). Phylogenetic analysis showed the orthology of fish foxp3 genes with others from different species (TIFF 554 kb)
Tetraodon CD4-2 (GenBank accession no. EF601918) and CD4-4 (GenBank accession no. EF601919) sequences, gene organization and chromosomal synteny analyses. (A) Nucleotide and predicted amino acid sequences of CD4-2. It spanned a 930 bp ORF encoding a polypeptide of 309 amino acids with a 24 amino acid signal peptide. The mature CD4-2 protein with a predicted molecular mass of 31.4 kDa and an expected isoelectric point of 9.38 consisted of a 210 amino acid extracellular region including two Ig domains, a 23 amino acid hydrophobic trans-membrane (TM) region, and a 52 amino acid cytoplasmic tail (CYT), comprising one N-glycosylation site at the position 175-177. The N-glycosylation site is underlined, the conserved p56lck site is in bold, and the asterisk reveals a stop codon. (B) Nucleotide and predicted amino acid sequences of CD4-4. It spanned a 1401 bp ORF encoding a polypeptide of 466 amino acids with a 22 amino acid signal peptide. The mature CD4-4 protein with a predicted molecular mass of 48.9 kDa and an expected isoelectric point of 9.20 consisted of a 390 amino acid extracellular region including four Ig domains, a 23 amino acid hydrophobic trans-membrane region, and a 31 amino acid cytoplasmic tail, comprising three N-glycosylation sites at positions 288-290, 342-344 and 395-397. The N-glycosylation sites are underlined, the conserved p56lck site is in bold, and the asterisk reveals a stop codon. (C) The comparison of CD4 gene organization in Tetraodon and humans. The Tetraodon CD4s shared similarities in genomic organization. The CD4-2 gene consisted of 8 exons and 7 introns, and the CD4-4 gene consisted of nine exons and eight introns. Each domain was encoded by a single exon, except that there were two D1 exons, D1A and D1B. The rectangles represent exons and lines between them indicate introns. The numbers above rectangles or below lines represent the corresponding lengths. Essential domains of Tetraodon CD4s are shown in simplified form in Sig, signal peptide; D, IG domain; H, hinge region; TM, transmembrane region; Int, intracellular region. (D) Chromosomal syntenic analysis of CD4 genes among different species, showing partial syntenic relationship among Tetraodon, chicken, human and mouse CD4 genes. Tetraodon CD4-2s are located on chromosome 8 with two copies while the adjacent CD4-4 has only one locus (TIFF 1519 kb)
Phylogenetic tree of CD4 molecules. This tree was constructed by the neighbor-joining method, and the numbers at branch nodes indicate percent bootstrap confidence values derived from 2000 replications. GenBank accession numbers for these amino acid sequences are as follows: catfish CD4-4 (DQ435302), zebrafish CD4-4 (EF601917), trout CD4-4 (AAY42070), fugu CD4-4 (BAD37153), Tetraodon CD4-4 (EF601919), fugu CD4-2 (predicted), Tetraodon CD4-2 (EF601918), trout CD4-2 (AY772711), catfish CD4-2 (DQ435301), zebrafish CD4-2 (EF601915), Xenopus CD4 (predicted), chicken CD4 (ABA55042), mouse CD4 (P06334), pig CD4 (AAT52342), and human CD4 (CAA60883) (TIFF 242 kb)
Tetraodon CD25-like sequences (GenBank accession no. EF143577), gene organization and structural characterization. The CD25-like gene spanned 4251 bp, and consisted of eight exons and seven introns. The CD25-like cDNA was composed of 2061 bp, including a 21 bp 5’ UTR, a 717 bp ORF and a 1323 bp 3’ UTR. The deciphered CD25-like amino acid sequence was a trans-membrane protein with 238 amino acids, a molecular weight of ~25.6 kDa, and a theoretical isoelectric point of 8.28. It contained a N-terminal extra-cellular region (position 1-180) with a 24 amino acid signal peptide, one sushi domain (position 34-99), a Pro/Thr rich domain (position 106-146), a trans-membrane domain (position 181-203), and a C-terminal intra-cellular domain (position 204-238). (A) Nucleotide and predicted amino acid sequences of CD25-like ORF. The sushi domain is in bold, and the asterisk reveals a stop codon. (B) The gene organizations of Tetraodon CD25-like. White and black rectangles represent UTRs (untranslated regions) and CDSs (coding sequences), respectively, while lines between them indicate introns. The numbers above rectangles or below lines represent the corresponding lengths. (C) Essential domains of CD25-like shown in simplified (TIFF 453 kb)
Chromosome syntenies of CD25 (IL-2Rα)/15Rα subfamily among vertebrates, including human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), the Western clawed frog (Xenopus tropicalis) and some teleost species (Tetraodon, Tetraodon nigroviridis; fugu, Takifugu rubripes; stickleback, Gasterosteus aculeatus; medaka, Oryzias latipes and zebrafish, Danio rerio). Genes are indicated by the rectangles with annotations, genomic positions and transcription orientations (TIFF 75 kb)
Phylogenetic tree of CD25 (IL-2Rα)/15Rα chains. These trees were constructed by the neighbor-joining method, based on the multiple alignment of the extracted sushi domains with main responsibility for cytokine binding (the first sushi of IL-2Rα or the only sushi of IL-15Rα). The numbers at branch nodes indicate percent bootstrap confidence values derived from 2000 replications. GenBank accession numbers for these amino acid sequences or genome data are as follows: IL-2Rα (CD25): human (NM000417), mouse (NM008367), chick (AF143806), Xenopus (EL724547); IL-15Rα: human (NM002189), mouse (NM008358), chick (AI980376), Xenopus (scaffold_11:3548294-3548476); teleost CD25-like (IL-2Rα/15Rα): fugu (CA846124); stickleback (EF513159), medaka (DK176609), trout (DQ381970), salmon (EG807345), zebrafish (EF143578) (TIFF 58 kb)
Recombinant proteins and antibody reactivity. The fusion CD4-2, CD4-4 and CD25-like recombinant proteins were purified using Ni-NTA resin and detected on SDS-PAGE as the expected 31, 45 and 28 kDa bands, respectively. (A) Purified fusion CD4-2 and CD25-like consisting of extracellular regions with 6×His, and protein marker were exhibited by SDS-PAGE (lane 1, 2 and M). Western blot analysis of anti-CD4-2 antibody to recombinant CD4-2 and CD25-like (lane 3 and 4), and anti-CD25-like antibody to CD4-2 and CD25-like (lane 5 and 6), which were then incubated with secondary HRP-conjugated Abs, and detected by ECL plus (Amersham Biosciences); B. Purified fusion CD4-2 and CD4-4 consisting of extracellular regions with 6×His, and protein marker were exhibited by SDS-PAGE (lane 1, 2 and M). Western blot analysis of anti-CD4-2 antibody to recombinant CD4-2 and CD4-4 (lane 3 and 4), and anti-CD4-4 antibody to CD4-2 and CD4-4 (lane 5 and 6), which were then incubated with secondary Abs, and detected by ECL plus as described above (TIFF 775 kb)
Distribution of Tetraodon transcriptional factor foxp3-like. Foxp3-like mRNAs were detected in several immune related tissues of healthy Tetraodon, including peripheral leucocytes (pl), spleen (sp), kidney (ki), gut (gu), and gill (gi) by RT-PCR (A). Quantitative analyses of foxp3-like relative to β-actin in corresponding tissues by real-time RT-PCR (B). ISH detection for foxp3-like probes (C): i, paraffin section of kidney with foxp3-like probes; ii, paraffin section of kidney with hybridization solution; iii, leucocytes with foxp3-like probes; iv, leucocytes with hybridization solution. Arrows indicate foxp3-like positive signals (TIFF 1335 kb)
A diagram of the hypothetical evolution of IL-15Rα and IL-2Rα gene family among vertebrate species. Gene tandem duplication had been involved in the emergence and divergence of IL-15Rα and IL-2Rα in tetrapods only, but not in teleost species (TIFF 53 kb)
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (973) (2006CB101805), Hi-Tech Research and Development Program of China (863) (2008AA09Z409), the National Natural Science Foundation of China (30871936, 31072234), and the Science and Technology Foundation of Zhejiang Province (2006C12038, 2006C23045, 2006C12005, 2007C12011).
Abbreviations
- MLR
Mixed lymphocyte reaction
- NCC
Nonspecific cytotoxic cell
- IBD
Inflammatory bowel disease
- ISH
In situ hybridization
- DIG
Digoxigenin
- LDH
Lactate dehydrogenase
- TM
Trans-membrane
- CYT
Cytoplasmic tail
References
- 1.Sakaguchi S. Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Von Herrath MG, Harrison LC. Antigen-induced regulatory T cells in autoimmunity. Nat Rev Immunol. 2003;3:223–232. doi: 10.1038/nri1029. [DOI] [PubMed] [Google Scholar]
- 3.Khazaie Boehmer K, Von Boehmer H. The impact of CD4+ CD25+ Treg on tumor specific CD8+ T cell cytotoxicity and cancer. Semin Cancer Biol. 2006;16:124–136. doi: 10.1016/j.semcancer.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Thornton AM, Shevach EM. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thornton AM, Shevach EM. Suppressor effector function of CD4+CD25+ immunoregulatory T cells is antigen nonspecific. J Immunol. 2000;164:183–190. doi: 10.4049/jimmunol.164.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Guerin LR, Prins JR, Robertson SA. Regulatory T-cells and immune tolerance in pregnancy: a new target for infertility treatment? Hum Reprod Update. 2009;15:517–535. doi: 10.1093/humupd/dmp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 8.Asseman C, Fowler S, Powrie F. Control of experimental inflammatory bowel disease by regulatory T cells. Am J Respir Crit Care Med. 2000;162:S185–S189. doi: 10.1164/ajrccm.162.supplement_3.15tac9. [DOI] [PubMed] [Google Scholar]
- 9.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8:457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 11.Benton MJ. Phylogeny of the major tetrapod groups: morphological data and divergence dates. J Mol Evol. 1990;30:409–424. doi: 10.1007/BF02101113. [DOI] [PubMed] [Google Scholar]
- 12.Zwollo P, Cole S, Bromage E, Kaattari S. B cell heterogeneity in the teleost kidney: evidence for a maturation gradient from anterior to posterior kidney. J Immunol. 2005;174:6608–6616. doi: 10.4049/jimmunol.174.11.6608. [DOI] [PubMed] [Google Scholar]
- 13.Levraud JP, Boudinot P. The immune system of teleost fish. Med Sci (Paris) 2009;25:405–411. doi: 10.1051/medsci/2009254405. [DOI] [PubMed] [Google Scholar]
- 14.Matsuo MY, Asakawa S, Shimizu N, Kimura H, Nonaka M. Nucleotide sequence of the MHC class I genomic region of a teleost, the medaka (Oryzias latipes) Immunogenetics. 2002;53:930–940. doi: 10.1007/s00251-001-0427-3. [DOI] [PubMed] [Google Scholar]
- 15.Solem ST, Stenvik J. Antibody repertoire development in teleosts—a review with emphasis on salmonids and Gadus morhua L. Dev Comp Immunol. 2006;30:57–76. doi: 10.1016/j.dci.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Laird DJ, De Tomaso AW, Cooper MD, Weissman IL. 50 million years of chordate evolution: seeking the origins of adaptive immunity. Proc Natl Acad Sci USA. 2000;97:6924–6926. doi: 10.1073/pnas.97.13.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langenau DM, Zon LI. The zebrafish: a new model of T cell and thymic development. Nat Rev Immunol. 2005;5:307–317. doi: 10.1038/nri1590. [DOI] [PubMed] [Google Scholar]
- 18.Araki K, Akatsu K, Suetake H, Kikuchi K, Suzuki Y. Characterization of CD8+ leukocytes in fugu (Takifugu rubripes) with antiserum against fugu CD8alpha. Dev Comp Immunol. 2008;32:850–858. doi: 10.1016/j.dci.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Wen Y, Shao JZ, Xiang LX, Fang W. Cloning, characterization and expression analysis of two Tetraodon nigroviridis interleukin-16 isoform genes. Comp Biochem Physiol B Biochem Mol Biol. 2006;144:159–166. doi: 10.1016/j.cbpb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 21.Xiang LX, Peng B, Dong WR, Yang ZF, Shao JZ. Lipopolysaccharide induces apoptosis in Carassius auratus lymphocytes, a possible role in pathogenesis of bacterial infection in fish. Dev Comp Immunol. 2008;32:992–1001. doi: 10.1016/j.dci.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Arck PC, Merali F, Chaouat G, Clark DA. Inhibition of immunoprotective CD8+ T cells as a basis for stress-triggered substance P-mediated abortion in mice. Cell Immunol. 1996;171:226–230. doi: 10.1006/cimm.1996.0197. [DOI] [PubMed] [Google Scholar]
- 23.Godfrey WR, Ge YG, Spoden DJ, Levine BL, June CH, Blazar BR, Porter SB. In vitro-expanded human CD4(+)CD25(+) T regulatory cells can markedly inhibit allogeneic dendritic cell-stimulated MLR cultures. Blood. 2004;104:453–461. doi: 10.1182/blood-2004-01-0151. [DOI] [PubMed] [Google Scholar]
- 24.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 25.Quintana FJ, Iglesias AH, Farez MF, Caccamo M, Burns EJ, Kassam N, Oukka M, Weiner HL. Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish. PLoS One. 2010;5:e9478. doi: 10.1371/journal.pone.0009478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra S, Alnabulsi A, Secombes CJ, Bird S. Identification and characterization of the transcription factors involved in T cell development, t-bet, stat6 and foxp3, within the zebrafish, Danio rerio . FEBS J. 2010;277:128–147. doi: 10.1111/j.1742-4658.2009.07460.x. [DOI] [PubMed] [Google Scholar]
- 27.Edholm ES, Stafford JL, Quiniou SM, Waldbieser G, Miller NW, Bengten E, Wilson M. Channel catfish, Ictalurus punctatus, CD4-like molecules. Dev Comp Immunol. 2007;31:172–187. doi: 10.1016/j.dci.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Laing KJ, Zou JJ, Purcell MK, Phillips R, Secombes CJ, Hansen JD. Evolution of the CD4 family: teleost fish possess two divergent forms of CD4 in addition to lymphocyte activation gene-3. J Immunol. 2006;177:3939–3951. doi: 10.4049/jimmunol.177.6.3939. [DOI] [PubMed] [Google Scholar]
- 29.Dijkstra JM, Somamoto T, Moore L, Hordvik I, Ototake M, Fischer U. Identification and characterization of a second CD4-like gene in teleost fish. Mol Immunol. 2006;43:410–419. doi: 10.1016/j.molimm.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Suetake H, Araki K, Suzuki Y. Cloning, expression, and characterization of fugu CD4, the first ectothermic animal CD4. Immunogenetics. 2004;56:368–374. doi: 10.1007/s00251-004-0694-x. [DOI] [PubMed] [Google Scholar]
- 31.Buonocore F, Randelli E, Casani D, Guerra L, Picchietti S, Costantini S, Facchiano AM, Zou J, Secombes CJ, Scapigliati G. A CD4 homologue in sea bass (Dicentrarchus labrax): molecular characterization and structural analysis. Mol Immunol. 2008;45:3168–3177. doi: 10.1016/j.molimm.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 32.Doyle C, Strominger JL. Interaction between CD4 and class II MHC molecules mediates cell adhesion. Nature. 1987;330:256–259. doi: 10.1038/330256a0. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Cruikshank WW, O′loughlin T, O′reilly P, Center DM, Kornfeld H. Identification of a CD4 domain required for interleukin-16 binding and lymphocyte activation. J Biol Chem. 1999;274:23387–23395. doi: 10.1074/jbc.274.33.23387. [DOI] [PubMed] [Google Scholar]
- 34.Innes JB, Kuntz MM, Kim YT, Weksler ME. Induction of suppressor activity in the autologous mixed lymphocyte reaction and in cultures with concanavalin A. J Clin Invest. 1979;64:1608–1613. doi: 10.1172/JCI109622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giri JG, Kumaki S, Ahdieh M, Friend DJ, Loomis A, Shanebeck K, Dubose R, Cosman D, Park LS, Anderson DM. Identification and cloning of a novel IL-15 binding protein that is structurally related to the alpha chain of the IL-2 receptor. EMBO J. 1995;14:3654–3663. doi: 10.1002/j.1460-2075.1995.tb00035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson DM, Kumaki S, Ahdieh M, Bertles J, Tometsko M, Loomis A, Giri J, Copeland NG, Gilbert DJ, Jenkins NA, Valentine V, Shapiro DN, Morris SW, Park LS, Cosman D. Functional characterization of the human interleukin-15 receptor alpha chain and close linkage of IL15RA and IL2RA genes. J Biol Chem. 1995;270:29862–29869. doi: 10.1074/jbc.270.50.29862. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, Mcgrady G, Wahl SM. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe N, Wang YH, Lee HK, Ito T, Wang YH, Cao W, Liu YJ. Hassall’s corpuscles instruct dendritic cells to induce CD4+CD25+ regulatory T cells in human thymus. Nature. 2005;436:1181–1185. doi: 10.1038/nature03886. [DOI] [PubMed] [Google Scholar]
- 39.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 40.Boden EK, Snapper SB. Regulatory T cells in inflammatory bowel disease. Curr Opin Gastroenterol. 2008;24:733–741. doi: 10.1097/MOG.0b013e328311f26e. [DOI] [PubMed] [Google Scholar]
- 41.Groux H, O′garra A, Bigler M, Rouleau M, Antonenko S, De Vries JE, Roncarolo MG. A CD4+ T cell subset inhibits antigen-specific T cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-P. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tetraodon foxp3-like sequences, gene organization and chromosomal synteny analyses. (A) The cDNA sequence (1411 bp) of Tetraodon foxp3-like gene (GenBank accession no. GU592499), with decoded 413 amino acid (shown beneath), showing the full length ORF (1242 bp) and 3’ UTR with a polyA tail. The zinc finger domain is underlined, the forkhead domain is in bold, and the asterisk reveals a stop codon. (B) Comparison of foxp3 gene organizations between fish and human. The Tetraodon foxp3-like gene consisted of twelve exons and eleven introns, in which the fifth and sixth exons encoded the zinc finger domain, the seventh and eighth exons encoded the leu zipper, and the last four exons encoded the forkhead domain. The rectangles represent exons and lines between them indicate introns. The numbers above rectangles or below lines represent the corresponding lengths. (C) Chromosomal syntenic analysis of Foxp3 genes among different species, showing partial syntenic relationship between fish Foxp3 and Xenopus and human Foxps genes, although it slightly divers from species, possibly suggesting the different recombination events occurred among them, in which the Foxp3 in Xenopus seems to be at a transition stage (TIFF 210 kb)
Multiple alignment analysis (A) and phylogenetic analysis (B) of Foxp3 orthologs among different species. In the alignment, residues shaded in black are completely conserved across all species aligned, and the residues shaded in gray are similar with respect to side chains. The dashes in the amino acid sequences indicate gaps introduced to maximize alignment. The result showed that foxp3 proteins were well conserved at both zinc finger domains in the middle and forkhead domains at C-terminals, from fish to mammals. The phylogenetic tree was constructed by the neighbor-joining method, and the numbers at branch nodes indicate percent bootstrap confidence values derived from 2000 replications. GenBank accession numbers for these amino acid sequences are as follows: human foxp3 (ABQ15210), cat foxp3 (ABN79272), mouse foxp3 (CAM25950), Xenopus foxp3 (BAG12188) and zebrafish foxp3 (FJ906821); human foxp1 (Q9H334), mouse foxp1 (P58462), Xenopus foxp1 (Q5W1J5), zebrafish foxp1b (Q2LE08), Tetraodon foxp1b (FJ358692); human foxp2 (O15409), mouse foxp2 (P58463), Xenopus foxp2 (Q4VYS1), medaka foxp2 (B1NY82), zebrafish foxp2 (Q4JNX5); human foxp4 (Q8IVH2), mouse foxp4 (Q9DBY0), and Xenopus foxp4 (Q4VYR7). Phylogenetic analysis showed the orthology of fish foxp3 genes with others from different species (TIFF 554 kb)
Tetraodon CD4-2 (GenBank accession no. EF601918) and CD4-4 (GenBank accession no. EF601919) sequences, gene organization and chromosomal synteny analyses. (A) Nucleotide and predicted amino acid sequences of CD4-2. It spanned a 930 bp ORF encoding a polypeptide of 309 amino acids with a 24 amino acid signal peptide. The mature CD4-2 protein with a predicted molecular mass of 31.4 kDa and an expected isoelectric point of 9.38 consisted of a 210 amino acid extracellular region including two Ig domains, a 23 amino acid hydrophobic trans-membrane (TM) region, and a 52 amino acid cytoplasmic tail (CYT), comprising one N-glycosylation site at the position 175-177. The N-glycosylation site is underlined, the conserved p56lck site is in bold, and the asterisk reveals a stop codon. (B) Nucleotide and predicted amino acid sequences of CD4-4. It spanned a 1401 bp ORF encoding a polypeptide of 466 amino acids with a 22 amino acid signal peptide. The mature CD4-4 protein with a predicted molecular mass of 48.9 kDa and an expected isoelectric point of 9.20 consisted of a 390 amino acid extracellular region including four Ig domains, a 23 amino acid hydrophobic trans-membrane region, and a 31 amino acid cytoplasmic tail, comprising three N-glycosylation sites at positions 288-290, 342-344 and 395-397. The N-glycosylation sites are underlined, the conserved p56lck site is in bold, and the asterisk reveals a stop codon. (C) The comparison of CD4 gene organization in Tetraodon and humans. The Tetraodon CD4s shared similarities in genomic organization. The CD4-2 gene consisted of 8 exons and 7 introns, and the CD4-4 gene consisted of nine exons and eight introns. Each domain was encoded by a single exon, except that there were two D1 exons, D1A and D1B. The rectangles represent exons and lines between them indicate introns. The numbers above rectangles or below lines represent the corresponding lengths. Essential domains of Tetraodon CD4s are shown in simplified form in Sig, signal peptide; D, IG domain; H, hinge region; TM, transmembrane region; Int, intracellular region. (D) Chromosomal syntenic analysis of CD4 genes among different species, showing partial syntenic relationship among Tetraodon, chicken, human and mouse CD4 genes. Tetraodon CD4-2s are located on chromosome 8 with two copies while the adjacent CD4-4 has only one locus (TIFF 1519 kb)
Phylogenetic tree of CD4 molecules. This tree was constructed by the neighbor-joining method, and the numbers at branch nodes indicate percent bootstrap confidence values derived from 2000 replications. GenBank accession numbers for these amino acid sequences are as follows: catfish CD4-4 (DQ435302), zebrafish CD4-4 (EF601917), trout CD4-4 (AAY42070), fugu CD4-4 (BAD37153), Tetraodon CD4-4 (EF601919), fugu CD4-2 (predicted), Tetraodon CD4-2 (EF601918), trout CD4-2 (AY772711), catfish CD4-2 (DQ435301), zebrafish CD4-2 (EF601915), Xenopus CD4 (predicted), chicken CD4 (ABA55042), mouse CD4 (P06334), pig CD4 (AAT52342), and human CD4 (CAA60883) (TIFF 242 kb)
Tetraodon CD25-like sequences (GenBank accession no. EF143577), gene organization and structural characterization. The CD25-like gene spanned 4251 bp, and consisted of eight exons and seven introns. The CD25-like cDNA was composed of 2061 bp, including a 21 bp 5’ UTR, a 717 bp ORF and a 1323 bp 3’ UTR. The deciphered CD25-like amino acid sequence was a trans-membrane protein with 238 amino acids, a molecular weight of ~25.6 kDa, and a theoretical isoelectric point of 8.28. It contained a N-terminal extra-cellular region (position 1-180) with a 24 amino acid signal peptide, one sushi domain (position 34-99), a Pro/Thr rich domain (position 106-146), a trans-membrane domain (position 181-203), and a C-terminal intra-cellular domain (position 204-238). (A) Nucleotide and predicted amino acid sequences of CD25-like ORF. The sushi domain is in bold, and the asterisk reveals a stop codon. (B) The gene organizations of Tetraodon CD25-like. White and black rectangles represent UTRs (untranslated regions) and CDSs (coding sequences), respectively, while lines between them indicate introns. The numbers above rectangles or below lines represent the corresponding lengths. (C) Essential domains of CD25-like shown in simplified (TIFF 453 kb)
Chromosome syntenies of CD25 (IL-2Rα)/15Rα subfamily among vertebrates, including human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), the Western clawed frog (Xenopus tropicalis) and some teleost species (Tetraodon, Tetraodon nigroviridis; fugu, Takifugu rubripes; stickleback, Gasterosteus aculeatus; medaka, Oryzias latipes and zebrafish, Danio rerio). Genes are indicated by the rectangles with annotations, genomic positions and transcription orientations (TIFF 75 kb)
Phylogenetic tree of CD25 (IL-2Rα)/15Rα chains. These trees were constructed by the neighbor-joining method, based on the multiple alignment of the extracted sushi domains with main responsibility for cytokine binding (the first sushi of IL-2Rα or the only sushi of IL-15Rα). The numbers at branch nodes indicate percent bootstrap confidence values derived from 2000 replications. GenBank accession numbers for these amino acid sequences or genome data are as follows: IL-2Rα (CD25): human (NM000417), mouse (NM008367), chick (AF143806), Xenopus (EL724547); IL-15Rα: human (NM002189), mouse (NM008358), chick (AI980376), Xenopus (scaffold_11:3548294-3548476); teleost CD25-like (IL-2Rα/15Rα): fugu (CA846124); stickleback (EF513159), medaka (DK176609), trout (DQ381970), salmon (EG807345), zebrafish (EF143578) (TIFF 58 kb)
Recombinant proteins and antibody reactivity. The fusion CD4-2, CD4-4 and CD25-like recombinant proteins were purified using Ni-NTA resin and detected on SDS-PAGE as the expected 31, 45 and 28 kDa bands, respectively. (A) Purified fusion CD4-2 and CD25-like consisting of extracellular regions with 6×His, and protein marker were exhibited by SDS-PAGE (lane 1, 2 and M). Western blot analysis of anti-CD4-2 antibody to recombinant CD4-2 and CD25-like (lane 3 and 4), and anti-CD25-like antibody to CD4-2 and CD25-like (lane 5 and 6), which were then incubated with secondary HRP-conjugated Abs, and detected by ECL plus (Amersham Biosciences); B. Purified fusion CD4-2 and CD4-4 consisting of extracellular regions with 6×His, and protein marker were exhibited by SDS-PAGE (lane 1, 2 and M). Western blot analysis of anti-CD4-2 antibody to recombinant CD4-2 and CD4-4 (lane 3 and 4), and anti-CD4-4 antibody to CD4-2 and CD4-4 (lane 5 and 6), which were then incubated with secondary Abs, and detected by ECL plus as described above (TIFF 775 kb)
Distribution of Tetraodon transcriptional factor foxp3-like. Foxp3-like mRNAs were detected in several immune related tissues of healthy Tetraodon, including peripheral leucocytes (pl), spleen (sp), kidney (ki), gut (gu), and gill (gi) by RT-PCR (A). Quantitative analyses of foxp3-like relative to β-actin in corresponding tissues by real-time RT-PCR (B). ISH detection for foxp3-like probes (C): i, paraffin section of kidney with foxp3-like probes; ii, paraffin section of kidney with hybridization solution; iii, leucocytes with foxp3-like probes; iv, leucocytes with hybridization solution. Arrows indicate foxp3-like positive signals (TIFF 1335 kb)
A diagram of the hypothetical evolution of IL-15Rα and IL-2Rα gene family among vertebrate species. Gene tandem duplication had been involved in the emergence and divergence of IL-15Rα and IL-2Rα in tetrapods only, but not in teleost species (TIFF 53 kb)