Abstract
The majority of human cancers are initiated when a single cell in an epithelial sheet becomes transformed. Cell transformation arises from the activation of oncoproteins and/or inactivation of tumor suppressor proteins. Recent studies have independently revealed that interaction and communication between transformed cells and their normal neighbors have a significant impact on the fate of the transformed cell. Several reports have shown that various phenomena occur at the interface between normal and transformed epithelial cells following the initial transformation event. In epithelia of Drosophila melanogaster, transformed and normal cells compete for survival in a process termed cell competition. This review will summarize current research and discuss the impact of these studies on our understanding of how primary tumors emerge and develop within a normal epithelium.
Keywords: Epithelial cells, Ras, Apical extrusion, Cell competition, Drosophila melanogaster, Field cancerization
Introduction
In the majority of human cancers, transformation occurs in single cells within an epithelial cell sheet [1]. Normal epithelial cells become transformed by acquiring genetic mutations that lead to the activation of oncogenes and inactivation of tumor suppressor genes. The multi-step process by which transformed cells develop into a primary tumor is well established, and many autonomous signaling pathways have been described [2]. The role of the extracellular microenvironment on tumor cell growth, proliferation, and metastasis is also well established. A plethora of research has shown that the stromal microenvironment significantly contributes to the development of invasive tumors (reviewed in [3]). Many in vitro and in vivo studies have shown that epithelial tumor cell growth, survival, and metastasis is significantly enhanced by the presence of stromal and cancer-associated fibroblasts (reviewed in [4]). Moreover, senescent fibroblasts promote the development of tumors from transformed or preneoplastic epithelial cells [5, 6]. Remodeling and stiffening of the extracellular matrix both in vitro and in vivo also enhances cancer cell growth and survival, and co-operates with oncogenesis to promote malignancy [7–9]. What remains poorly understood are the cellular and molecular events that arise between transformed cells and their normal neighbors within an epithelium during early tumor development. In this review, I will summarize recent work from several independent groups, describing how interaction with normal neighboring epithelial cells can influence the fate of a transformed cell.
Fibroblast-epithelial cell growth studies
The interaction between normal mesenchymal and transformed epithelial cells has been studied for almost 50 years [10]. Early research examining the effect of transformation of fibroblasts in culture by viral oncogenes showed that growth of the transformed cells was inhibited when in contact with confluent monolayers of normal cells [10] (and recently reviewed in [11]). Early studies described a role of gap junctional intercellular communication (GJIC) as a mechanism of cell–cell communication between transformed and normal cells. By inhibiting gap junction function and activity, normal cells were no longer able to inhibit transformed cell growth [12, 13]. Gap junctions are defined as cell–cell junctions between two apposing plasma membranes that contain channels formed by a family of proteins termed connexins. GJIC describes the process by which gap junction channels exchange ions (K+, Ca2+), second messengers (cAMP, IP3), metabolites such as glucose and electrical signals [14]. Based on these studies, connexins were described as having a tumor suppressive role. Since then it has been shown in vitro and in vivo that inhibition of GJIC activity and/or loss of connexin expression leads to an increase in tumor cell growth. However, the identities of the molecules that are exchanged between cells within normal healthy tissue are not clearly defined. Therefore, whether this exchange is altered or how this exchange is regulated between tumor and normal cells is unknown. Moreover, the molecular mechanisms that regulate tumor cell growth downstream of connexins and gap junction function in intercellular communication remains complex and is not yet fully understood. There is also evidence to suggest that inhibitory effects of normal cells on transformed cell growth occurs via mechanisms independent of gap junctional communication [15, 16], suggesting that additional, unidentified molecules are also required for this process. Most of the work describing a role of normal cells on restricting growth of cancer cells has focused on homotypic interactions between mesenchymal cells, or heterotypic interactions between mesenchymal and epithelial cells. The effect of normal epithelial cell interaction on transformed epithelial cell growth has not yet been examined.
Role of the neighboring epithelial cells
Two major studies have recently shown that novel cellular processes occur at the interface between transformed and normal mammalian epithelial cells [17, 18].
Non-transformed Madin-Darby canine kidney (MDCK) epithelial cells form polarized cell sheets in culture and offer a useful tool to study the interaction between epithelial cells within a confluent monolayer [19]. In these studies, inducible MDCK cell lines expressing constitutively active mutants of the oncogenes ras (RasV12) or src (v-src) were first established. Transformed cells retain parental epithelial characteristics in the absence of the inducible signal. This allows the mixing of transformed cells with non-transformed cells to generate mosaic epithelial cell sheets. Following the establishment of a monolayer and proper cell–cell adhesion between neighboring cells, oncogene expression is induced, and the fate of the transformed cell is monitored over time. This cell culture model system allows direct examination of the cellular processes that arise at the interface between transformed and normal epithelial cells.
Ras
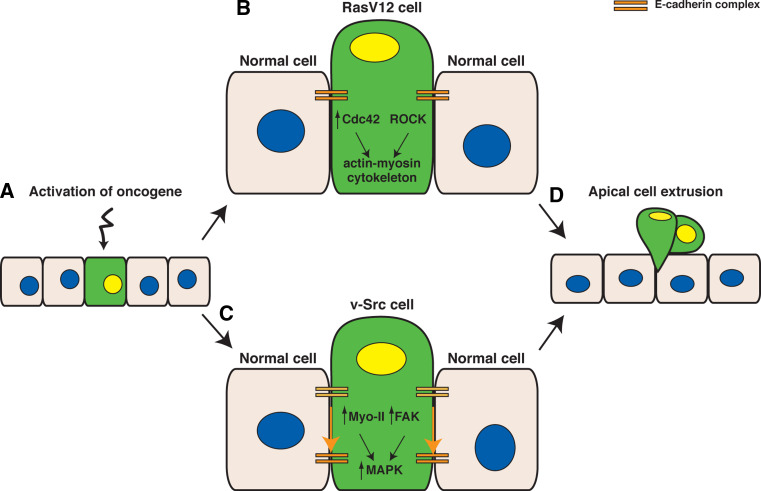
Ras GTPase is the founding member of the Ras superfamily of small GTPases and plays a role in many cellular processes including cell growth and proliferation, cell survival, actin cytoskeletal reorganization, and cell polarity [20]. Ras is mutated to a constitutive active form in approximately 33% of all human cancers, which occurs as one of the earliest transformation events [20, 21]. To examine the fate of a single RasV12 cell in a normal epithelial cells sheet, MDCK epithelial cells expressing GFP-tagged constitutively active oncogenic Ras (RasV12) in a tetracycline-inducible manner were generated (MDCK-pTR GFP-RasV12 cells, hereafter referred as RasV12 cells) [17]. RasV12 cells were mixed with non-transformed MDCK cells at a ratio of 1:100 in the absence of tetracycline. Cells were seeded on collagen gels, and once cell–cell adhesion was restored and cells formed an epithelial monolayer, RasV12 expression was induced by the addition of tetracycline. When RasV12 cells were surrounded by normal cells, the majority of RasV12 cells were apically extruded from a normal epithelial cell sheet. Apical extrusion did not occur when RasV12 cells surround RasV12 cells. This suggests that the process of apical extrusion is not a direct consequence of activation of cell autonomous signaling downstream of oncogenic RasV12, but requires interaction with the surrounding normal cells.
Prior to being extruded from normal monolayers, RasV12 cells significantly increased cell height, accumulated F-actin at cell–cell contacts, and had higher levels of phosphorylated myosin light chain (pMLC). This suggests that interaction with the surrounding normal cells induces a change in cell shape and a remodeling of the actin–myosin cytoskeleton in RasV12 cells. The frequency of apical extrusion of RasV12 cells was significantly reduced using inhibitors of actin–myosin contractility, suggesting that actin polymerization and myosin II activity are required for this process. Furthermore, Cdc42 (a member of the Rho GTPase subfamily) is activated to a greater extent in RasV12 cells when surrounded by normal cells and this activity is required to promote apical extrusion of RasV12 cells.
However, not all RasV12 cells are extruded from normal monolayers, so what is the fate of RasV12 cells that are not extruded? Non-extruded RasV12 cells form large, dynamic basal protrusions beneath the surrounding normal cells and eventually delaminate basally and invade a collagen matrix. Importantly, the formation of large basal protrusions, and basal delamination of RasV12 cells are not observed in monolayers of RasV12 cells, suggesting that this process also requires the interaction with normal cells. Protrusions were observed at the interface between RasV12 cells and normal cells and not between adjacent RasV12 cells. Interestingly, adherens junctions are specifically disrupted at cell–cell contacts between the RasV12 cell that has formed a protrusion and its normal neighbor. Furthermore, basal protrusion formation and basal delamination of RasV12 cells occur to a significantly higher level when RasV12 cells are surrounded by E-cadherin-deficient cells, suggesting that loss of E-cadherin in the surrounding cells promotes basal delamination of RasV12 cells. However, loss of E-cadherin alone is not sufficient to promote basal protrusion formation, nor basal delamination, suggesting that an additional RasV12-dependent signal is required for these phenomena to occur.
How is the fate of RasV12 cells decided; eliminated apically or basally? What partly determines the outcome is the activity of Cdc42 and a downstream effector of Rho, ROCK (also known as Rho kinase). Coexpression of constitutively inactive Cdc42 (Cdc42 N17) or dominant negative ROCK (ROCK DN) in RasV12 cells reduces the frequency of apical extrusion and promotes basal protrusion formation, suggesting that both Cdc42 and ROCK are crucial regulators of these phenomena. Pharmacological inhibition of MAPK signaling reduces the frequency of both apical extrusion and basal protrusion formation. Co-expression of a dominant negative Raf suppresses apical extrusion of RasV12 cells, but expression of a constitutively active Raf (RafCAAX) has no effect, suggesting that MAPK signaling (downstream of Raf) in RasV12 cells is required but not sufficient for apical extrusion to occur. Interestingly, apical extrusion is significantly reduced and basal delamination is significantly enhanced when E-cadherin-deficient cells surround RasV12 cells. This implies that the integrity of an epithelial monolayer can influence the fate of a RasV12-transformed cell; under pathological conditions where E-cadherin-based cell–cell adhesions are disrupted (e.g., chronic inflammation), basal delamination of RasV12 cells would occur more frequently. On the other hand, an intact epithelial cell sheet would contribute to apical extrusion of RasV12 cells, which could be tumor suppressive.
Src
In a second study, Kajita and colleagues used MDCK cells that express a temperature-sensitive mutant of v-Src (ts-v-Src) to examine the interaction between Src-transformed and normal epithelial cells [18]. c-Src encodes a non-receptor tyrosine kinase, which regulates cell survival and proliferation, as well as cell migration and adhesion. Oncogenic Src (v-Src) is the oldest oncogene and deregulated Src activity is implicated in many human cancers [22].
In MDCK ts-Src cells (hereafter referred to as Src cells), Src activation is controlled by a temperature shift from 40.5°C (inactive) to 35°C (active). In order to examine the fate of single Src-transformed cells, Kajita and colleagues pre-stained Src cells with a fluorescent tracker dye and mixed with normal cells at a ratio of 1:100. Following the establishment of cell–cell contacts, cells were shifted to 35°C to induce Src activity. The authors showed that Src-transformed MDCK cells are apically extruded from a monolayer of normal cells but not when surrounded by Src cells, suggesting that apical extrusion of Src cells also occurs following interaction with normal cells. Indeed similar to that observed for RasV12 cells, Src cells also increase cell height and accumulate higher levels of pMLC when surrounded by normal cells but not when surrounded by Src cells. These data suggest that Src-transformed cells also recognize that they are surrounded by normal cells and subsequently alter cell shape and morphology. Moreover, Src-transformed epithelial cells are also apically extruded in vivo. Using zebrafish embryos, oncogenic v-Src was expressed in a mosaic manner within the enveloping layer (EVL) of epithelial cells that surround zebrafish embryos during gastrulation [18]. When surrounded by non-transformed cells, Src-expressing cells remained viable, increased in cell height, and were apically extruded from the surface of the epithelial monolayer. This is the first in vivo demonstration in vertebrates that Src-transformed cells are extruded from normal epithelial monolayers.
Both of these studies reveal that the process of apical extrusion of transformed epithelial cells from normal monolayers requires specific common features (Fig. 1); (i) the interaction with normal cells, (ii) myosin II-dependent increase in cell height and cell shape of the transformed cells, and (iii) activation of MAPK signaling pathways. Unlike previous studies showing extrusion as a process to eliminate apoptotic cells [23], extrusion of transformed cells from normal MDCK monolayers occurs independent of apoptosis. Following extrusion RasV12 and Src cells remain viable, and extruded RasV12 cells proliferate to form multi-cellular aggregates [17, 18]. Taken together, these data suggest that common signaling pathways may be activated at the interface between transformed and normal cells that promote apical extrusion. However, while non-extruded RasV12-expressing cells form basal protrusions beneath the normal neighbors, Src-transformed cells do not. Furthermore, interaction between Src-transformed and normal cells, but not RasV12-expressing cells, leads to increased activation of focal adhesion kinase (FAK) in Src cells as well as basal relocalization of the E-cadherin complex at cell–cell contacts [18] (Fig. 1c). Thus, while some common molecular mechanisms are required, distinct signaling pathways are also induced following interaction between normal and transformed cells.
Fig. 1.
Model of molecular mechanisms of apical extrusion of transformed cells from normal epithelial monolayers. a When expression of an oncogene (RasV12 or v-Src) is induced in a single epithelial cell (green) within a monolayer of normal cells, transformed cells recognize they are surrounded by normal cells and modulate their cell height and cell signaling. b The fate of a RasV12 cell is influenced by the activity of Cdc42 and ROCK in RasV12 cells and/or by the presence of intact E-cadherin-based cell–cell adhesions in the surrounding normal cells. Cdc42 activity is increased in RasV12 cells surrounded by normal cells. Both Cdc42 and ROCK signaling is required for the increase in cell height and remodeling of the actin–myosin cytoskeleton. c The activity of both myosin-II (myo-II) and FAK are increased in Src cells surrounded by normal cells, which leads to the activation of downstream MAPK signaling pathways. Activation of myosin-II, FAK, and MAPK pathways are required for apical extrusion. Apical extrusion of Src cells also requires basal relocalization of the E-cadherin complex in a myosin-II- and FAK-dependent manner. d Oncogene-transformed cells are apically extruded from normal epithelial monolayers
These two studies have taken a unique approach to understanding how oncogene expression in cells affects cell behavior and cell fate. Through the interaction with the normal neighbors, transformed cells modulate their cell shape, cell signaling, and cell behavior, suggesting that transformed and normal cells recognize differences between them. What remain unclear are the molecular mechanisms of cell–cell recognition. This may involve differences in physical properties of cells, or differences in the composition of the plasma membranes (e.g., lipids and proteins). Indeed, cells expressing either RasV12 or v-Src have altered physical properties compared to normal cells (i.e., higher membrane elasticity and cell viscosity) [17, 18]. F-actin accumulates to a greater extent at cell–cell contacts between RasV12 cells in a monolayer of normal cells, which in turn increases cell–cell adhesion between cells. This, together with an increase in phosphorylated myosin-II, would contribute towards an increase in cell surface tension and intercellular adhesion. Interestingly, both parameters have been shown to play a role in cell sorting [24], a process by which populations of cells physically separate from each other to form distinct tissues or compartments [25]. In a similar process, cells with higher surface tension and/or intercellular adhesions are encompassed by cells with lower surface tension and/or intercellular adhesions. Thus, segregation of transformed cells, in particular RasV12 cells, from normal cells could be a thermodynamically favorable process. However, whether differences in physical properties are involved in the cell–cell recognition machinery between transformed and normal cells remains to be determined. Future studies will reveal whether additional, unidentified molecules are also required for cell–cell recognition and apical extrusion.
Interaction between transformed and normal cells in Drosophila melanogaster
Epithelial monolayers of wing imaginal discs of Drosophila melanogaster provide ideal in vivo models to examine the interaction between transformed and normal cells. Using genetic tools and an inducible site-specific recombination event (FLP-FRT system), researchers can generate mosaic tissues of patches of mutant cells surrounded by wild-type cells. Using this system in combination with a Gal4-UAS expression system [26], specific genes or RNAi constructs are targeted in cells within these patches. Using these genetic tools, researchers have independently shown that transformed cells adopt distinct cell responses when surrounded by wild-type cells compared to when an entire tissue is composed of transformed cells.
Oncogene-induced transformation in vivo
Clones of cells expressing constitutively active, oncogenic Ras (RasV12) are apically and basally eliminated from wild-type wing imaginal discs of Drosophila [17, 27, 28], suggesting that extrusion of RasV12 cells from normal epithelia is an evolutionary conserved phenomenon between flies and mammalian cells.
The major inhibitor of Src-family kinases is the C-terminal Src kinase (Csk). Global loss of Drosophila Csk (dCsk) expression by RNAi in the developing eye or wing imaginal discs leads to overproliferation, inhibition of apoptosis, and decreased cell adhesion. However, when dCsk-deficient cells are surrounded by wild-type cells, the dCsk mutant cells are basally excluded from the normal epithelium [29]. Moreover, eliminated dCsk cells migrate from the anterior to posterior compartment of the wing disc, and eventually die by apoptosis. Apoptosis of dCsk mutant cells requires activation of JNK and Rho1 signaling pathways. Basal exclusion, invasive migration, and apoptosis of dCsk mutant cells also require dE-cadherin and dP120-catenin [29]. Interestingly, in Drosophila wing imaginal discs, expression of components of E-cadherin-based cell–cell adhesions including dE-cadherin and β-catenin (Armadillo) are enhanced within clones of RasV12 cells surrounded by wild-type cells [30]. Taken together, these studies suggest that E-cadherin complexes may provide a cell-sensing mechanism at the interface between transformed and wild-type cells. On the other hand, a cell–cell recognition mechanism may function between oncogene-transformed and wild-type cells, which directly alters the level of E-cadherin-based cell–cell adhesions. Future studies are required to fully elucidate the mechanism of cell–cell recognition and interaction between RasV12- or Src-transformed and normal cells.
Cell competition in Drosophila melanogaster
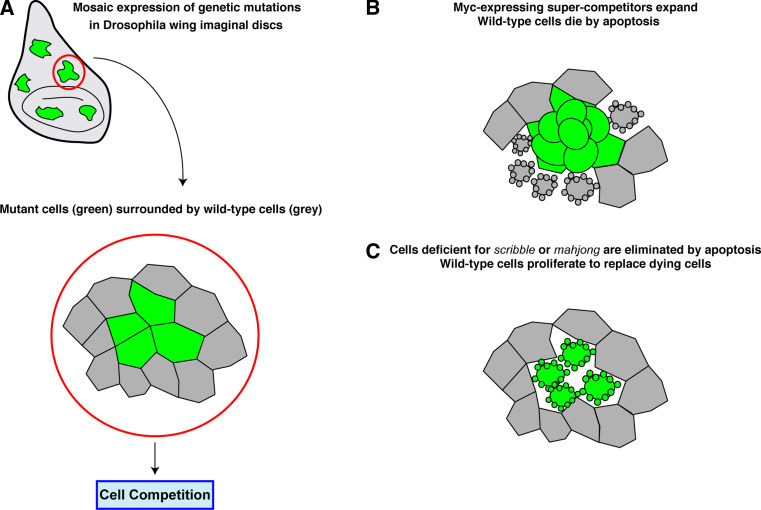
Cell competition describes the process by which one cell type in a tissue survives (‘winner’) over another (‘loser’) cell and is also used to describe cell fitness. Cell competition requires short-range contact between cells to occur and always leads to the death of the ‘loser’ cells, even though out-competed cells are autonomously viable.
In 1975, Morata and Ripoll showed in Drosophila wing epithelia that cells heterozygous for a set of mutations in ribosomal protein genes, known collectively as Minute, proliferate more slowly than wild-type cells [31]. Minute homozygous cells do not survive because of defective ribosomal function. Wild-type cells in a Minute heterozygous (M/+) background expand to cover larger areas of the adult wing than Minute cells, while Minute cells are eliminated by apoptosis from a wild-type tissue [32]. This suggested that cell–cell interaction between wild-type and Minute cells induced competition between cells based on proliferation rates; slower proliferating cells are actively removed from the developing tissue. This study opened a new field in Drosophila research known as cell competition.
Many genes that regulate cell growth and proliferation have been shown to play a role in cell competition. For example, cells with reduced expression of the proto-oncogene dMyc are out-competed by wild-type cells, whereas wild-type cells are eliminated when in contact with cells overexpressing dMyc [33, 34]. Moreover, cells expressing elevated levels of dMyc become ‘super-competitors’, suggesting that the competition between cells is not due to an intrinsic defect of the ‘loser’ cells but is directly linked to the level of dMyc between cells; cells with lower levels are eliminated [35]. Mutations in the conserved Salvador/Warts/Hippo (SWH) pathway that regulates cell growth and organ size [36], rescue the M/+ cell competition phenotype and prevent loss of M/+ cells, suggesting that these mutant cells can act as super-competitors [37]. However, not all genes that regulate cell growth are involved in cell competition. For example, mosaic overexpression of phosphoinositide 3-kinase (PI3 K) or cyclin d/Cdk4, both of which promote cell growth, do not cause cell competition. Under these conditions, patches of cells expressing PI3 K or Cdk4 mutants grow at a higher rate to the surrounding wild-type cells, which remain unaffected, suggesting that a difference in cell growth speed alone is not sufficient to induce cell competition [33]. It is generally accepted that an important feature of cell competition is that proliferation and expansion of the ‘winner’ cells requires the activation of apoptosis-dependent pathways and the elimination of ‘loser’ cells. Thus, the overall size of the developing organ is unchanged. However, a recent study describes how regulation of tissue size control does not require cell competition; expansion of faster growing cells does not require the elimination of a weaker cell population [38]. In this study, slow-dividing M/+ cells are eliminated by apoptosis at borders between M/+ clones and wild-type tissue, although this is relatively infrequent as the number of apoptotic cells is overall low. Moreover, if apoptosis is inhibited, M/+ clone size and tissue size remain unaffected, suggesting that the rate of proliferation of M/+ cells is not dependent on interaction with its neighbors [38]. The authors argue that M/+ clones expand because they have a higher proliferation rate and that the mechanism that controls overall size control of a tissue arrests cell growth once the final tissue size has been reached [38]. However, this study does not rule out a role of cell competition in removing non-viable or developmentally abnormal cells from tissues.
Polarity genes
The polarity genes, scribble, lethal-giant larvae (lgl), and discs large (dlg) are required for establishment and maintenance of apicobasal polarity in epithelial tissue of both flies and mammals. Drosophila scribble, lgl, and dlg have been shown to negatively regulate cell proliferation and are therefore classed as tumor suppressor genes (reviewed in [39]). A similar role has been described for scribble and lgl in mammals; loss of scribble induces disruption of three-dimensional architecture of mammary epithelial cells and inhibits apoptosis [40]. In mice, knockout of Lgl1, a mammalian homologue of Lgl, leads to hyperproliferation and loss of cell polarity in neuroepithelial cells [41].
In Drosophila imaginal discs, loss of both copies of any of the three polarity genes induces epithelia to acquire tumor characteristics including loss of apicobasal polarity, uncontrolled proliferation and loss of apoptosis [39]. However, when scribble or dlg are depleted in patches of cells within wild-type eye imaginal discs, scribble −/− or dlg −/− mutant cells die by apoptosis and are eliminated by cell competition that is dependent on interaction with the surrounding wild-type cells [42]. Recent studies have shown that the fate of Lgl mutant cells is more complicated and depends on context as well as cell competition. In the developing eye epithelium, clones of lgl −/− cells survive and proliferate by deregulating the SWH pathway, although mutant cells directly in contact with wild-type tissue at clonal borders are eliminated by JNK-mediated apoptosis [43]. Depletion of Lgl results in mislocalization of Hippo and RASSF (Ras-associated domain family protein) and a decrease in phosphorylated (inactive) Yorkie (Yki), which ultimately leads to activation of SWH target genes such as cyclin E and E2F1 [43, 44]. Depletion of Lgl in the developing wing epithelia has different effects depending on the timing of induction; clones induced in early development are eliminated, whereas clones induced later survive [43]. These differences can be attributed to differences in endogenous dMyc levels between mutant and wild-type cells [43, 45]. Indeed, when dMyc levels are downregulated in lgl −/− clones compared to the surrounding wild type tissue, lgl mutant cells are eliminated by JNK-dependent cell competition, whereas when lgl −/− cells have higher dMyc levels, lgl −/− cells survive and proliferate to form invasive tumors [45]. The precise mechanism for how loss of Lgl induces mislocalization of SWH components, and alters dmyc expression is still not clear, although dMyc has been shown to be a transcriptional target of the SWH pathway [45, 46]. Moreover, when depletion of Lgl results in complete disruption of apico-basal polarity, the SWH pathway is deregulated and dmyc is upregulated [45]. Taken together, these studies suggest that the fate of lgl −/− cells is dependent on the nature of the surrounding wild-type tissue and the context of the cell–cell interactions between mutant cells and their neighbors.
Interestingly, groups of cells deficient for either Scribble (scrib −/−) or Lgl (lgl −/−) survive and expand to form tumors if they also express oncogenic RasV12 [47–49]. Indeed, cooperation between expression of oncogenic RasV12 and inactivation of any of the genes that regulate cell polarity (e.g., discs large (dlg), bazooka (baz), and cdc42) induce metastasis of the mutant cells into neighboring tissues [48]. While expression of genes that promote cell proliferation and/or survival induce expansion of scrib-mutant tissue, they cannot induce metastasis, suggesting that undefined mechanisms downstream of RasV12, other than promoting cell proliferation and survival are required for metastatic behavior of scrib-mutant cells [48].
Cooperation between oncogenic RasV12 and loss of scribble also promote tumor growth even when expressed in adjacent epithelial cells [47]. Interclonal cooperation between RasV12 and lgl −/− also produced tumors, indicating that other polarity genes also cooperate with RasV12 expression between adjacent cells. Mechanistically, scrib −/− cells induce neoplastic overgrowth of RasV12 cells through activation of stress-induced JNK pathway, which in turn leads to expression of JAK/STAT-dependent cytokine gene expression in RasV12 cells [47]. Importantly, this study highlights the importance of cell–cell interactions in oncogenic cooperation in tumor development.
Conversely, a recent study has described how single cells deficient for lgl but expressing oncogenic RasV12 cells are eliminated by cell competition [49]. Within small groups of lgl −/− rasV12 cells, individual cells at boundaries between mutant and wild-type cells are eliminated by apoptosis, while cells within the middle of the clone are protected. This also occurs for small clones of scrib −/− rasV12 cells surrounded by normal tissue, suggesting that individual lgl −/− rasV12 or scrib −/− rasV12 cells are eliminated by cell competition, even though the mutant cells were shown to proliferate at a higher rate than the wild-type cells. This study also reveals that small clones of lgl −/− rasV12 cells merge to form larger clones. Together with a higher proliferation rate (as shown for lgl −/− rasV12 cells through downregulation of the Hippo pathway), the merging of clones generates a microenvironment that prevents tumor cells from being eliminated and contributes towards tumor formation.
Recently, the Lgl-binding protein, Mahjong, has also been shown to play a role in cell competition in both Drosophila and in mammalian epithelial cells [50]. In fact, this study is the first to describe a cell competition phenotype using a mammalian cell culture system. In Drosophila, loss of expression of Mahjong (mahj −/−) in a mosaic manner in the wing disc epithelium results in cell competition, and mahj −/− are eliminated by apoptosis when surrounded by wild-type cells. Similarly, when Mahjong-knockdown MDCK epithelial cells undergo apoptosis and are eliminated by apical extrusion only when surrounded by non-transformed cells, suggesting that the process of cell competition is conserved between flies and mammals.
Mechanisms of cell competition
Early studies of Minute and dMyc mutants in Drosophila focused on cellular fitness and differences in growth rates as a measure of competitiveness between cells. The level of ribosomal protein (Rp) is directly regulated by Minute [51], while Myc is involved in regulating the expression of various ribosomal genes that all contribute to ribosomal biogenesis [52]. Thus, cells with mutations in either Minute or Myc have slower proliferation rates due to less efficient ribosomal function. Interaction between mutant and wild-type cells would result in cells with different ribosomal activities in direct contact with each other, and hence ribosomal vigor could be a mechanism for cell competition. Indeed, dMyc-expressing super-competitors that are heterozygously mutant for a ribosomal protein (RpL19) can no longer out-compete their neighbors, suggesting that functional ribosomes and protein synthesis machinery provide a competitive advantage to dMyc-expressing cells [35].
A second mechanism proposed to regulate cell competition was the ‘ligand-capture’ hypothesis, whereby cells compete for available survival factors. In Drosophila imaginal discs, the morphogen Decapentaplegic (Dpp), a member of the transforming growth factor (TGFβ) superfamily, is required for correct cell patterning, cell survival, and growth [53]. Using the Minute (M/+) and the dMyc models of cell competition, Moreno and colleagues showed that inefficient Dpp signaling in cells correlates with expression of the transcriptional repressor brinker (Brk) and activation of c-jun N-terminal kinase (JNK) and apoptosis [32, 35]. They proposed that cells compete for Dpp through efficient capture of the ligand via receptor-mediated endocytosis, and therefore cells with weaker ribosomal activity may be less efficient at Dpp uptake and/or Dpp-dependent signal transduction and are triggered to die by apoptosis. However, the role of Dpp is not required for all cell competition [33], suggesting that additional survival factors may play a role in this process.
How do cells sense differences in growth rates that lead to one cell being targeted as ‘loser’ and the other as ‘winner’ cell? The majority of studies have shown that cell competition primarily occurs at boundaries between mutant and wild-type tissue, suggesting that direct cell–cell interaction is required for this process. Yet, cell competition between Myc-expressing winner cells and wild-type loser cells can occur at distance of up to ten cell diameters away [33], suggesting a role for short-range diffusible factors. A recent study has suggested that soluble factors may also play a role [54]. Using an in vitro S2 cell culture system, Senoo-Matsuda and colleagues showed that cell competition occurs in vitro between cells expressing different levels of Myc and this process does not require direct cell–cell contact. Moreover, conditioned medium from mixed populations of cells induces cell competition when supplemented to naive cocultures, and winners and losers are determined depending on the level of Myc-expression; high Myc-expressers increase proliferation to outcompete low Myc-expressing cells, which are committed to die via apoptosis. Interestingly, neither of the populations of cells cultured alone (i.e., winners or losers) secrete these factors—both populations in a mixed culture are required. However, the identity of these factors and how they signal in cells to activate cell death or cell survival and proliferation still remain unclear. Moreover, whether diffusible factors play a role in cell competition in vivo remains to be established.
In the majority of examples, the outcome of cell competition is that one population of cells with higher cell fitness and/or proliferation rate triggers a death response in another weaker population of cells. This leads to a reciprocal response in both cells; the loser cell is induced to die by caspase-dependent apoptosis, while the winner cell is triggered to increase proliferation and expand, so that the overall cell number in a tissue remains unchanged (Fig. 2). This reciprocal relationship between winners and losers is demonstrated by inhibiting apoptosis by expressing a caspase inhibitor p35, which blocks cell death of Minute loser cells but also reduces cell growth of wild-type winner cells [55]. Genetic analysis in flies has shown that activation of the JNK pathway is often required for apoptosis and elimination of loser cells expressing various mutations including mahjong and scribble [42, 50].
Fig. 2.
Cell competition in Drosophila melanogaster. a Mosaic expression of different genetic mutations in patches of cells within wing imaginal discs of Drosophila creates epithelial monolayers where mutant cells (green) are surrounded by wild-type cells (grey). In many cases, this results in cell competition. b If mutant cells are expressing high levels of dMyc, these cells become super-competitors and expand by proliferation at the expense of the neighboring wild-type cells, which die by apoptosis. c Loss of expression of scribble or mahjong in mutant cells induces apoptosis-dependent elimination of mutant cells, while surrounding wild-type cells expand and proliferate to replace dying cells
How are ‘winner’ cells triggered to proliferate and expand following cell competition? Several recent studies describe a link between activation of JNK signaling and the induction of compensatory proliferation in tissue regeneration that requires repression of the Salvador/Warts/Hippo (SWH) pathway and concomitant activation of the transcriptional co-activator Yorkie (Yki) [56–59]. Following tissue damage, Yki is activated in cells neighboring apoptotic cells [58]. Repression of the SWH pathway and activation of Yki is also observed in cells juxtaposed to cells harboring mutations in genes required for cell viability, including scribble [56]. Activation of JNK signaling is both necessary and sufficient to induce Yki activation in response to cell damage in the developing wing [58], suggesting that active JNK signaling in dying cells may act as an early modulator of the SWH pathway in response to tissue damage to promote compensatory proliferation and tissue regeneration [58]. Hippo inactivation or overexpression of Yki in the Drosophila midgut also induces regenerative growth and proliferation of intestinal stem cells (ISC) [57]. In this context, Yki activation is triggered by a stress response following bacterial infection, suggesting that Yki acts as a stress sensor. In addition, increasing JNK signaling also drives Yki activation [57]. Yki activity leads to the transcription of unpaired (Upd) cytokines and known targets involved in proliferation including cyclin E [59]. Signals additional to JNK activity may also play a role in Yki activation, including Fat-Dachsous (Ft-Ds) signaling [59], and regulators of apico-basal polarity such as scribble and lgl [56, 58].
However, the role of JNK-dependent apoptosis is not always observed in cell competition [33]. Another regulator of apoptosis in Drosophila is the prodeath protein Hid, which plays a role in cell competition between cells expressing different levels of Myc [33]. However, what upstream signals induce Hid expression remain unknown.
Unlike other developmental processes that require the clearing of apoptotic cells by phagocytosis, in Drosophila, apoptotic loser cells are not extruded from the epithelium and are not cleared by phagocytic hemocytes, but are engulfed by the winner cells. Corpse engulfment by winner cells is an active process and is required for cell competition to occur [55]. A set of engulfment genes including draper (drp), Wiskott–Aldrich syndrome protein (WASp), the phosphatidylserine receptor (psr) and rac1 are required in winner cells to outcompete and engulf loser cells. Loss of gene expression of drp, WASp or psr in wild-type cells significantly decreases the ability of wild-type cells to compete with M/+ cells [55]. Moreover, apoptosis of M/+ cells is significantly reduced between M/+ and drp loss-of-function mutant cells compared to M/+ cells and wild-type, suggesting that engulfment genes are required for the apoptosis of loser cells at boundaries between loser and winner cells.
How are winner cells distinguished from loser cells? Two recent studies shed some light on this question [60, 61]. Using Myc-expressing super-competitors surrounding wild-type cells as a model for cell competition, Rhiner et al. [60] performed microarray analysis to identify changes in gene expression between winner and loser cells during cell competition [60]. Using in situ hybridization experiments, they identified genes that were specifically upregulated in loser cells at an early time-point prior to activation of apoptosis. One of the genes identified encodes a membrane protein termed Flower (Fwe), which has been shown to act as a calcium channel protein and is involved in regulating endocytosis and exocytosis in vertebrates. In Drosophila, alternative splicing of Flower produces three isoforms, which differ in the extracellular C-terminal region; Fweubi, FweLoseA and FweLoseB. Fweubi is ubiquitously expressed in all imaginal disc cells. The other two isoforms are not expressed in the absence of cell competition. However, when wild-type cells are confronted with clones of cells mutant for Minute (M/+), dMyc, or scribble −/−, FweLose isoforms were specifically expressed in the loser cells. Overexpression of FweLose isoforms in a clone of cells surrounded by wild-type cells induces apoptosis of FweLose-expressing cells, and loss of expression of FweLose in out-competed cells prevents cell elimination. Moreover, if all three isoforms are deleted in patches within an imaginal disc, cells depleted for Fwe are eliminated by cell competition by neighboring wild-type cells, suggesting that Fwe isoforms are required and sufficient to induce cell competition. On the other hand, global expression of any of the three isoforms throughout an epithelium has no effect on cells. This indicates that cells compare the relative level of isoforms of Fwe protein (level of Fweubi to FweLose) expressed on the cell surface, and those that express higher levels of FweLose are committed to die by apoptosis. Thus, expression of FweLose at the cell surface labels cells as losers. However, the molecular mechanism that triggers activation of apoptosis-dependent machinery downstream of FweLose is not yet clear. Furthermore, how FweLose expression is regulated in out-competed cells remains unknown.
A second marker for cell competition also identified by microarray analysis is the Drosophila homologue of mammalian SPARC proteins, dSPARC [61]. SPARC (secreted protein acidic and rich in cysteine) proteins are highly conserved matricellular glycoproteins that have diverse biological roles including remodeling of the extracellular matrix [62]. SPARC protein expression is also associated with tumor progression in many human cancers [63]. In Drosophila, expression of dSPARC is specifically upregulated in cells that are undergoing cell competition, and functions to protect cells from apoptosis [61]. Importantly, dSPARC was shown to be a marker of cell competition and is not expressed as a general response to inducers of apoptosis. dSPARC and Fwe do not function in the same pathway but appear to have opposing roles in cell competition; Fwe labels loser cells whereas expression of dSPARC in loser cells provides a temporal protection from apoptosis by inhibiting caspase activation downstream of survival factor withdrawal [61]. This suggests that in the early stages of cell competition, expression of dSPARC provides a mechanism by which cells can recover from transient stress in the microenvironment and prevent cells from elimination by their neighbors.
Field cancerization and cell competition
A large body of work has described cell competition between cells expressing a range of genetic mutations and wild-type cells (Fig. 2), and is now suggested to be a model for understanding the early stages of cancer in humans (reviewed in [52, 64, 65]). Accumulating evidence suggests that cell competition also occurs in mammals in vivo (recently reviewed in [11]). Several recent reports have described how transformed cells are removed from normal epithelial cell sheets in a cell death-independent manner [17, 18]. How is this relevant to early cancer development? The majority of human cancers arise from epithelial tissues and during the initial stages of tumorigenesis, transformed cells proliferate and expand as clones surrounded by normal cells. There is increasing evidence that for many human cancers, early pre-neoplastic lesions exist undetected within normal tissue, and at the margins of a primary tumor [66]. Indeed, the presence of these abnormal cells may be responsible for the recurrence of cancers following surgical removal of primary tumors. Field cancerization broadly describes the clonal expansion of a patch of mutant cells during the initial stages of many epithelial cancers. This can occur as a result of multiple cells throughout a tissue independently acquiring mutations following exposure to a carcinogen, or due to the clonal expansion of a single mutated cell [67]. It has recently been proposed that mutant precursor cells clonally expand within a tissue by processes similar to cell competition. Therefore, with the identification of markers of cell competition, we may also uncover novel biomarkers of early pre-neoplastic cancer lesions, leading to new diagnostic tools and prevent the onset of an invasive and malignant disease.
Concluding remarks
Recent studies in Drosophila and in mammalian cells demonstrate that the interaction between transformed epithelial cells with their normal neighboring cells has a significant impact on the fate of the transformed cell. This is a newly emerging field in cancer biology. To fully understand the significance of this interaction, a comprehensive examination of the cellular responses that arise between transformed and normal cells is required, as well as an understanding of the molecular mechanisms underlying these processes. An improved understanding of how transformed and normal cells interact and communicate at the initial stages of epithelial cancer, at the single-cell level, will significantly contribute to our knowledge of how primary tumors emerge and develop within a normal epithelium. This improved knowledge may lead to the development of novel therapeutic and diagnostic tools and prevent progression of the disease towards malignancy.
Acknowledgments
I am grateful to Y. Fujita and M. Norman for the critical reading of the manuscript. This work is supported by MRC funding to the Cell Biology Unit.
Abbreviations
- GJIC
Gap junctional intercellular communication
- cAMP
Cyclic adenosine monophosphate
- IP3
Inositol triphosphate
- MDCK
Madin-Darby canine kidney
- pMLC
Phosphorylated myosin light chain
- ROCK
Rho kinase
- MAPK
Mitogen-activated protein kinase
- ts-Src
Temperature-sensitive mutant of v-Src
- v-Src
Sarcoma virus Src
- EVL
Enveloping layer
- FAK
Focal adhesion kinase
- Csk
C-terminal Src kinase
- Rp
Ribosomal protein
- JNK
c-Jun N-terminal kinase
- Lgl
Lethal giant larvae
- Dlg
Discs large
- DPP
Decapentaplegic
- TGFβ
Transforming growth factor β
- SWH
Salvador/Warts/Hippo
- Yki
Yorkie
- Drp
Draper
- WASp
Wiskott–Aldrich syndrome protein
- PSR
Phosphatidylserine receptor
- Fwe
Flower
- SPARC
Secreted protein acidic and rich in cysteine
References
- 1.Nowell PC. Tumor progression: a brief historical perspective. Semin Cancer Biol. 2002;12:261–266. doi: 10.1016/S1044-579X(02)00012-3. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 3.Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp Cell Res. 2001;264:169–184. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 4.Xing F, Saidou J, Watabe K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front Biosci. 2010;15:166–179. doi: 10.2741/3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrenson K, Grun B, Benjamin E, Jacobs IJ, Dafou D, et al. Senescent fibroblasts promote neoplastic transformation of partially transformed ovarian epithelial cells in a three-dimensional model of early stage ovarian cancer. Neoplasia. 2010;12:317–325. doi: 10.1593/neo.91948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Stoker MG, Shearer M, O’Neill C. Growth inhibition of polyoma-transformed cells by contact with static normal fibroblasts. J Cell Sci. 1966;1:297–310. doi: 10.1242/jcs.1.3.297. [DOI] [PubMed] [Google Scholar]
- 11.Hogan C, Kajita M, Lawrenson K, Fujita Y. Interactions between normal and transformed epithelial cells: their contributions to tumourigenesis. Int J Biochem Cell Biol. 2011;43:496–503. doi: 10.1016/j.biocel.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Mehta PP, Bertram JS, Loewenstein WR. Growth inhibition of transformed cells correlates with their junctional communication with normal cells. Cell. 1986;44:187–196. doi: 10.1016/0092-8674(86)90497-6. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg GS, Martyn KD, Lau AF. A connexin 43 antisense vector reduces the ability of normal cells to inhibit the foci formation of transformed cells. Mol Carcinog. 1994;11:106–114. doi: 10.1002/mc.2940110208. [DOI] [PubMed] [Google Scholar]
- 14.Naus CC, Laird DW. Implications and challenges of connexin connections to cancer. Nat Rev Cancer. 2010;10:435–441. doi: 10.1038/nrc2841. [DOI] [PubMed] [Google Scholar]
- 15.Alexander DB, Ichikawa H, Bechberger JF, Valiunas V, Ohki M, et al. Normal cells control the growth of neighboring transformed cells independent of gap junctional communication and SRC activity. Cancer Res. 2004;64:1347–1358. doi: 10.1158/0008-5472.CAN-03-2558. [DOI] [PubMed] [Google Scholar]
- 16.Martin W, Zempel G, Hulser D, Willecke K. Growth inhibition of oncogene-transformed rat fibroblasts by cocultured normal cells: relevance of metabolic cooperation mediated by gap junctions. Cancer Res. 1991;51:5348–5351. [PubMed] [Google Scholar]
- 17.Hogan C, Dupre-Crochet S, Norman M, Kajita M, Zimmermann C, et al. Characterization of the interface between normal and transformed epithelial cells. Nat Cell Biol. 2009;11:460–467. doi: 10.1038/ncb1853. [DOI] [PubMed] [Google Scholar]
- 18.Kajita M, Hogan C, Harris AR, Dupre-Crochet S, Itasaki N, et al. Interaction with surrounding normal epithelial cells influences signalling pathways and behaviour of Src-transformed cells. J Cell Sci. 2010;123:171–180. doi: 10.1242/jcs.057976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behrens J, Vakaet L, Friis R, Winterhager E, Van Roy F, et al. Loss of epithelial differentiation and gain of invasiveness correlates with tyrosine phosphorylation of the E-cadherin/beta-catenin complex in cells transformed with a temperature-sensitive v-SRC gene. J Cell Biol. 1993;120:757–766. doi: 10.1083/jcb.120.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aleshin A, Finn RS. SRC: a century of science brought to the clinic. Neoplasia. 2010;12:599–607. doi: 10.1593/neo.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenblatt J, Raff MC, Cramer LP. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr Biol. 2001;11:1847–1857. doi: 10.1016/S0960-9822(01)00587-5. [DOI] [PubMed] [Google Scholar]
- 24.Krieg M, Arboleda-Estudillo Y, Puech PH, Kafer J, Graner F, et al. Tensile forces govern germ-layer organization in zebrafish. Nat Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 25.Tepass U, Godt D, Winklbauer R. Cell sorting in animal development: signalling and adhesive mechanisms in the formation of tissue boundaries. Curr Opin Genet Dev. 2002;12:572–582. doi: 10.1016/S0959-437X(02)00342-8. [DOI] [PubMed] [Google Scholar]
- 26.Heidmann D, Lehner CF. Reduction of Cre recombinase toxicity in proliferating Drosophila cells by estrogen-dependent activity regulation. Dev Genes Evol. 2001;211:458–465. doi: 10.1007/s004270100167. [DOI] [PubMed] [Google Scholar]
- 27.Prober DA, Edgar BA. Ras1 promotes cellular growth in the Drosophila wing. Cell. 2000;100:435–446. doi: 10.1016/S0092-8674(00)80679-0. [DOI] [PubMed] [Google Scholar]
- 28.Prober DA, Edgar BA. Interactions between Ras1, dMyc, and dPI3 K signaling in the developing Drosophila wing. Genes Dev. 2002;16:2286–2299. doi: 10.1101/gad.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal M, Larson DE, Cagan RL. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell. 2006;10:33–44. doi: 10.1016/j.devcel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 30.O’Keefe DD, Prober DA, Moyle PS, Rickoll WL, Edgar BA. Egfr/Ras signaling regulates DE-cadherin/Shotgun localization to control vein morphogenesis in the Drosophila wing. Dev Biol. 2007;311:25–39. doi: 10.1016/j.ydbio.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morata G, Ripoll P. Minutes: mutants of Drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 32.Moreno E, Basler K, Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416:755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- 33.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/S0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 34.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/S0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/S0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 36.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 37.Tyler DM, Li W, Zhuo N, Pellock B, Baker NE. Genes affecting cell competition in Drosophila . Genetics. 2007;175:643–657. doi: 10.1534/genetics.106.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin FA, Herrera SC, Morata G. Cell competition, growth and size control in the Drosophila wing imaginal disc. Development. 2009;136:3747–3756. doi: 10.1242/dev.038406. [DOI] [PubMed] [Google Scholar]
- 39.Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, et al. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- 40.Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila . EMBO J. 2003;22:5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grzeschik NA, Parsons LM, Richardson HE. Lgl, the SWH pathway and tumorigenesis: It’s a matter of context & competition! Cell Cycle. 2010;9:3202–3212. doi: 10.4161/cc.9.16.12633. [DOI] [PubMed] [Google Scholar]
- 44.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 45.Froldi F, Ziosi M, Garoia F, Pession A, Grzeschik NA, et al. The lethal giant larvae tumour suppressor mutation requires dMyc oncoprotein to promote clonal malignancy. BMC Biol. 2010;8:33. doi: 10.1186/1741-7007-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziosi M, Baena-Lopez LA, Grifoni D, Froldi F, Pession A, et al. (2010) dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet 6 [DOI] [PMC free article] [PubMed]
- 47.Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;302:1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- 49.Menendez J, Perez-Garijo A, Calleja M, Morata G. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proc Natl Acad Sci USA. 2010;107:14651–14656. doi: 10.1073/pnas.1009376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamori Y, Bialucha CU, Tian AG, Kajita M, Huang YC, et al. Involvement of Lgl and Mahjong/VprBP in cell competition. PLoS Biol. 2010;8:e1000422. doi: 10.1371/journal.pbio.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv Genet. 1998;38:69–134. doi: 10.1016/S0065-2660(08)60142-X. [DOI] [PubMed] [Google Scholar]
- 52.Johnston LA. Competitive interactions between cells: death, growth, and geography. Science. 2009;324:1679–1682. doi: 10.1126/science.1163862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burke R, Basler K. Dpp receptors are autonomously required for cell proliferation in the entire developing Drosophila wing. Development. 1996;122:2261–2269. doi: 10.1242/dev.122.7.2261. [DOI] [PubMed] [Google Scholar]
- 54.Senoo-Matsuda N, Johnston LA. Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc Natl Acad Sci USA. 2007;104:18543–18548. doi: 10.1073/pnas.0709021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Baker NE. Engulfment is required for cell competition. Cell. 2007;129:1215–1225. doi: 10.1016/j.cell.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 56.Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster . Dev Biol. 2011;350:255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 57.Shaw RL, Kohlmaier A, Polesello C, Veelken C, Edgar BA, et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol. 2011;350:139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rhiner C, Lopez-Gay JM, Soldini D, Casas-Tinto S, Martin FA, et al. Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila . Dev Cell. 2010;18:985–998. doi: 10.1016/j.devcel.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 61.Portela M, Casas-Tinto S, Rhiner C, Lopez-Gay JM, Dominguez O, et al. Drosophila SPARC is a self-protective signal expressed by loser cells during cell competition. Dev Cell. 2010;19:562–573. doi: 10.1016/j.devcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Bradshaw AD. The role of SPARC in extracellular matrix assembly. J Cell Commun Signal. 2009;3:239–246. doi: 10.1007/s12079-009-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chlenski A, Cohn SL. Modulation of matrix remodeling by SPARC in neoplastic progression. Semin Cell Dev Biol. 2010;21:55–65. doi: 10.1016/j.semcdb.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 64.Baker NE. Cell competition. Curr Biol. 2011;21:R11–R15. doi: 10.1016/j.cub.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 65.Rhiner C, Moreno E. Super competition as a possible mechanism to pioneer precancerous fields. Carcinogenesis. 2009;30:723–728. doi: 10.1093/carcin/bgp003. [DOI] [PubMed] [Google Scholar]
- 66.Braakhuis BJ, Brakenhoff RH, Leemans CR. Second field tumors: a new opportunity for cancer prevention? Oncologist. 2005;10:493–500. doi: 10.1634/theoncologist.10-7-493. [DOI] [PubMed] [Google Scholar]
- 67.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]