Abstract
The nematode Caenorhabditis elegans has been used to study genetics and development since the mid-1970s. Over the years, the arsenal of techniques employed in this field has grown steadily in parallel with the number of researchers using this model. Since the introduction of C. elegans transgenesis, nearly 20 years ago, this system has been extensively used in areas such as rescue experiments, gene expression studies, and protein localization. The completion of the C. elegans genome sequence paved the way for genome-wide studies requiring higher throughput and improved scalability than provided by traditional genetic markers. The development of antibiotic selection systems for nematode transgenesis addresses these requirements and opens the possibility to apply transgenesis to investigate biological functions in other nematode species for which no genetic markers had been developed to date.
Keywords: Caenorhabditis elegans, Nematodes, Antibiotics, Transgenesis, Genetics, Model organisms
Introduction
Since Sydney Brenner deliberately chose Caenorhabditis elegans as a simple multicellular eukaryotic model organism [1], the number of researchers using this model has steadily grown. Over the past four decades, the worm has become a powerful system with which to study a large spectrum of biological processes in many fields, such as genetics, development, aging, neurobiology, immunology, and evolution [2]. The success of C. elegans is due to favorable characteristics with respect to laboratory study (including small size, 3 day life cycle, self-fertilization, several hundred progeny, ease of cultivation, and cryopreservation), a completely sequenced genome, and last but not least, the ease of DNA transformation. Since its development in the 1980s [3], DNA transformation of C. elegans has proven to be an extremely powerful tool to study gene function in the nematode, and it rapidly became an invaluable technique of C. elegans research.
C. elegans transgenesis: what for?
Transgenesis of C. elegans has been widely used for various purposes: to validate gene function by rescuing a mutant phenotype using a wild-type copy of the DNA [4–6], to study gene expression patterns without the need to obtain specific antibodies [7–11], to interfere with biological processes by overexpression or ectopic expression of wild-type or mutant genes [9], to analyze the site of gene action by mosaic analysis [12, 13], and also to screen for regulatory promoter sequences using a promoter-trap approach [14]. With the introduction of the green fluorescent protein (GFP) in 1994 [10], C. elegans transgenesis gained in popularity and found new applications [15]. The animal’s transparency and thin diameter (70 μm) allow the direct visualization of GFP and GFP variants in live animals. As a result, some otherwise tedious studies became easier to achieve, for example, the study of gene expression patterns in living animals [16, 17], the localization of proteins to specific cells and subcellular compartments [18–20], and the study of spatial relationships of individual cells [21], an achievement only previously possible through electron-micrograph reconstructions [22]. After the completion of the C. elegans genome sequence [23], large-scale projects also benefited from reporter genes and the ease of C. elegans transgenesis. The generation of hundreds of reporter gene fusions also allowed the analysis of DNA sequences and gene expression patterns at a genomic scale [24–27]. In short, transgenesis is an indispensable tool for almost every study of C. elegans biology.
C. elegans transgenesis: how?
Stable transformation of C. elegans was first achieved by microinjection of the DNA of interest into the syncytial gonad of the hermaphrodite [3, 28]. Exogenous DNA (plasmid or PCR product) is directly introduced into the cytoplasm of the germline. Then, as individual oocytes separate from the syncytium, it is incorporated into fertilized eggs. The exogenous DNA undergoes intermolecular ligation leading to the formation of tandem repeats of about 80–300 plasmid copies, also referred to as multicopy extrachromosomal arrays [3, 28]. These arrays are semi-stable, thus displaying non-Mendelian segregation and leading to mosaic animals. An array that is transmitted to the F2 generation is generally inherited by the following generations with a characteristic transmission rate ranging from 10 to 90% [28] (Figs. 1, 2). It is also possible to inject the DNA of interest with high concentrations of single-stranded oligonucleotides containing 50 bases [28]. Oligonucleotides seem to promote integration of the plasmid into the genome in a random fashion, leading to stable integrated lines. However, integrants are obtained at low frequency ranging from 1 in 20 to 1 in 100 integrants per injected parent, making the technique relatively inefficient. Another variant consists of injecting the exogenous DNA directly into oocyte nuclei [29]. This technique combined with the use of poisonous DNA sequences that are lethal at high copy number leads to the formation of low-copy extrachromosomal arrays or the random integration of a low-copy number of the transgene into the genome. Although strains obtained by this method are stable, injection into the nuclei of oocytes is technically difficult and integrants are obtained at a comparatively low frequency.
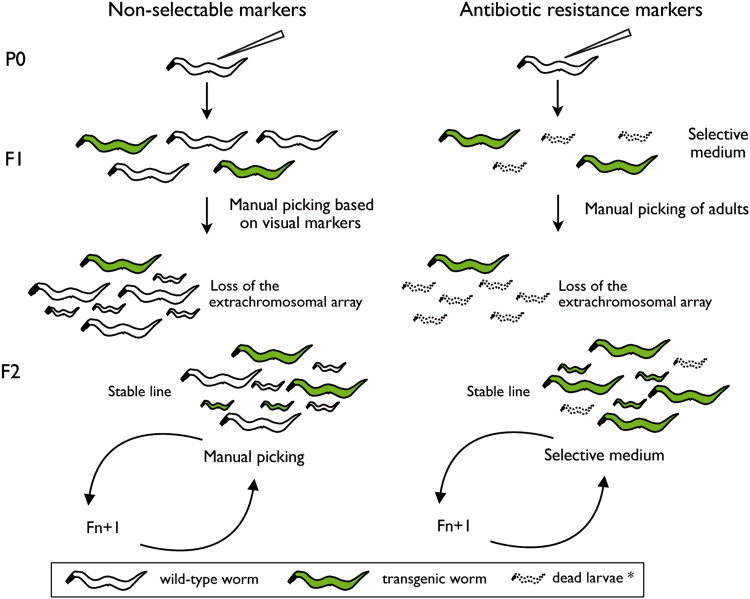
Fig. 1.
Comparison between traditional non-selectable genetic markers (left) and antibiotic resistance markers (right) in the context of microinjection. When visual phenotypic markers are used for nematode transgenesis by microinjection, transgenic F1 worms need to be screened and isolated by hand by an experimentalist (left), while antibiotic selection offers the possibility of hands-off selection and maintenance of stable transgenic lines (right). In the case of antibiotic resistance markers, F1 isolation by hand ensures clonal homogeneity of isolated strains, but further maintenance can be done by chunking without further visual screening as long as the animals are kept on antibiotic-containing medium. *In the presence of antibiotic, wild-type larvae are unable to develop and die after a few days
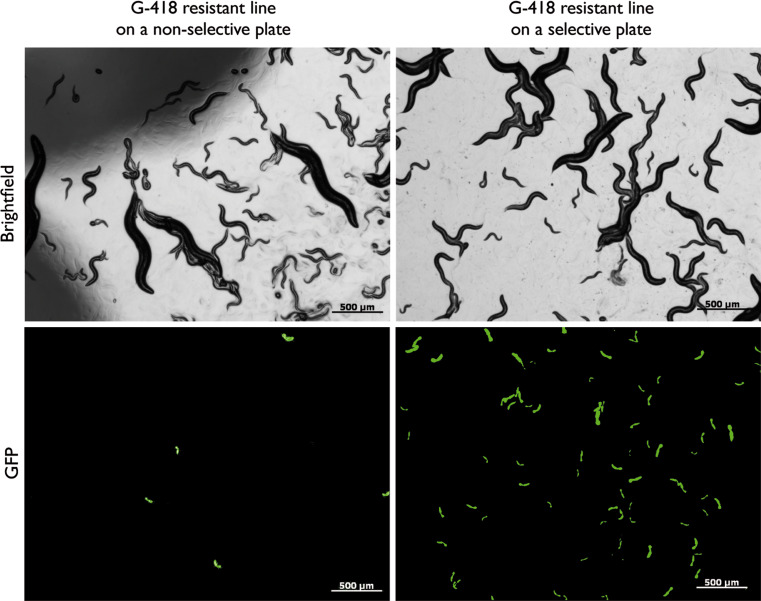
Fig. 2.
Hands-off maintenance of an extrachromosomal C. elegans line on antibiotic-selective medium. An extrachromosomal C. elegans line carrying the NeoR nematode cassette and a p myo-2::gfp reporter (worms carrying p myo-2::gfp express GFP in the pharynx) amplified on a plate without G-418 (left) and on a plate supplemented with G-418 at the critical concentration (selective plate) (right). After several generations and in the absence of a selective agent, the extrachromosomal array is lost and transgenic animals are rare (left) while on selective plates wild-type worms are eliminated at the L1 stage and the transgenic population is enriched without manual maintenance
A second technique for C. elegans transgenesis, microparticle bombardment [30, 31], was popularized about 10 years ago and is a relatively easy method for the generation of integrated lines. With the use of commercially available particle delivery systems, it is possible to bombard more than 100,000 worms at a time with DNA-coated beads. Transgenic animals are identified through unc-119(ed3) rescue, a gene marker that allows for the partial selection of transgenic animals in starvation conditions where mutants are dying due to their inability to enter the Dauer stage (worms in this state of diapause and dormancy are able to survive in starvation conditions) [32]. Transgenic worms obtained this way also carry extrachromosomal arrays, but random integration events are observed in around 1/4 to 1/8 of the transgenic lines thus obtained [26, 30]. These lines carry a lower copy number of the transgene than lines carrying extrachromosomal arrays.
C. elegans genetic markers
Three main issues arise during the establishment and analysis of transgenic C. elegans: (1) the identification of transformed animals in the F1 generation, (2) the maintenance of nonintegrated transgenic lines, and (3) obtaining large numbers of transgenic animals to carry out further experiments. The identification of transformed animals is achieved by the use of transformation markers that are co-injected with the DNA of interest. Co-injected DNAs contribute to the extrachromosomal array in proportions relative to their concentration in the injection mix [28]. In the case of microparticle bombardment, the transgene and transformation marker are generally carried by the same plasmid [30]. Transformation markers belong to three different categories. The first group includes the rescue of nonlethal mutations by wild-type copies of the corresponding gene, such as rescue of dpy-20(e1282ts), dpy-5(e907) (both Dumpy) [33, 34], pha-1(e2123ts) (embryonic lethal at 25°C) [35], lin-15(n765ts) (multivulva at 22.5°C or higher temperatures) [36], and unc-119(ed3) (uncoordinated and unable to enter Dauer stage upon starvation) [32]. Secondly, there are dominant phenotype markers such as unc-22 antisense DNA, which leads to lower mobility, twitching, and egg-laying defect [37], and rol-6(su1006), a gain of function mutation that results in a right-hand roller phenotype [28]. The final category consists of fluorescent reporter constructs, such as sur-5::gfp (expresses GFP in all cell nuclei) [13] or myo-2::gfp (expresses GFP in the pharynx) [38]. All these genetic markers have their own relative advantages and limitations. Rescue of nonlethal mutations requires the use of specific mutant strains that are often more difficult to inject than wild-type animals. Dominant phenotype markers confer phenotypes that may interfere with subsequent experiments and may not be compatible with every mutant background. Finally, most transformation markers do not provide a selective advantage over the wild-type population, thus requiring hands-on maintenance of nonintegrated transgenic lines. A few exceptions to the latter are pha-1 and unc-119, which allow a partial positive selection of transgenic animals. However, the selective capacity of pha-1 is limited to specific temperatures [35, 36], and unc-119 can only provide a selective advantage in starving conditions [30] (Table 1).
Table 1.
Genetic markers for C. elegans transformation
| Marker | Recipient strain | Selection | Picking | Final phenotype |
|---|---|---|---|---|
| Phenotypic rescue | ||||
| unc-119 a | unc-119 -/- | Upon starvation* | Yes | Rescued* |
| pha-1 | pha-1 -/- | Selective temperature* | No* | Rescued* |
| dpy-5 | dpy-5 -/- | No | Yes | Rescued* |
| dpy-20 | dpy-20 -/- | No | Yes | Rescued* |
| lin-15 | lin-15-/- | Selective temperature* | Yes | Rescued* |
| Dominant phenotype | ||||
| unc-22 | unc-22 wt | No | Yes | Mutant |
| rol-6 a,b | rol-6 wt | No | Yes | Mutant |
| Fluorescent markers | ||||
| GFP a,b | GFP−* | No | Yes | GFP + |
| Antibiotic resistance | ||||
| NeoR a | G-418 sensitive | +G-418* | No* | Wt or mutantc* |
| PuroR a | Puromycin sensitive | +Puromycin* | No* | Wt or mutantc* |
Asterisks indicate advantageous features
aAlso amenable for Caenorhabditis briggsae transformation
bEquivalent markers exist for Pristionchus pacificus
cAntibiotic resistance markers do not rely on endogenous nematode genes, they can therefore be used on wild-type animals as well as any mutants of interest without changing the initial phenotype
The limits of these selectable markers make most nonintegrated lines unsuitable for experiments where a large number of transgenic worms is needed, for example, for protein purification [39]. Therefore, past researchers wanting to perform biochemistry on transgenic animals had to generate integrated transgenic lines.
Genomic integration of transgenes
The most popular technique to obtain integrated lines, apart from microparticle bombardment as mentioned above, is via mutagenesis-induced integration [40]. After exposure of nonintegrated lines to X-rays or gamma-rays, extrachromosomal arrays can be incorporated into the genome in a random fashion. However, as with all random integration events, there may be accompanying modifications of the expression of the array by genomic enhancers, a mutant phenotype may be caused by integration at a particular loci, and because other mutations can be introduced by the mutagenesis procedure, all integrants have to be backcrossed several times after integration, making the technique tedious and time-consuming.
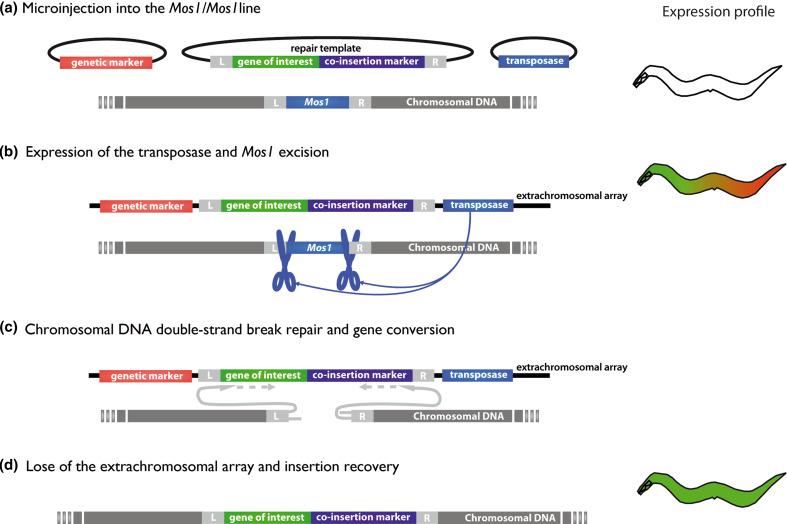
A new method for transgene integration was recently developed that allows the insertion of a single copy of a transgene into a defined site [41]. This method, called Mos1-mediated single-copy insertion (MosSCI), is based on the use of the fruit fly transposon Mos1 and its mobilization by a transposase (Fig. 3). Briefly, a C. elegans strain carrying a single insertion of Mos1 is injected with a transposase source, a repair template vector carrying the transgene, and transformation markers for the identification of worms carrying the extrachromosomal array thus formed. The repair template is composed of the gene of interest and a co-insertion marker, both flanked by 1.5 kb of genomic DNA corresponding to the right and left sequences adjacent to the Mos1 insertion locus. After expression of the transposase, Mos1 excision creates a double-strand break in the genomic DNA that is repaired by gene conversion using the repair template in the extrachromosomal array, leading to the insertion of the gene of interest and the co-insertion marker into the genome. Insertion events can then be distinguished from nonintegrated transgenes by identifying animals expressing the co-insertion marker while lacking the transformation markers contained solely in the extrachromosomal array. Because the technique has been developed using unc-119(ed3) rescue as a co-insertion marker, the strains carrying the desired Mos1 insertion have to be previously crossed with unc-119(ed3) [41]. This method, therefore, is not suitable for cases where the unc-119 (ed3) mutant background may interfere with the study or if the desired Mos1 insertion site is too close to the unc-119 locus [42].
Fig. 3a–d.
Transgene integration by Mos1 single-copy insertion (MosSCI) mechanism. The main steps of MosSCI are represented. On the right is a schematic view of the expression profile of the worm at each step. a Extrachromosomal array formation: a C. elegans strain carrying a single characterized insertion of Mos1 is injected with a transposase source, a repair template vector, and genetic markers for the identification of worms carrying the extrachromosomal array. The repair template is composed of the gene of interest and a co-insertion marker, both flanked by 1.5 kb of genomic DNA corresponding to the right (R) and left (L) sequences flanking the Mos1 insertion locus. b Expression of the transposase: Mos1 excision by the transposase creates a double-strand break in the genomic DNA. c Gene conversion: the double-strand break is repaired by gene conversion using the repair template in the extrachromosomal array, leading to the insertion of the gene of interest and the co-insertion marker into the genome. d Loss of the extrachromosomal array: insertion events are discriminated from nonintegrated transgenes by identifying animals expressing the co-insertion marker but lacking the transformation markers contained solely in the extrachromosomal array. Adapted from [41]
The creation of integrated lines, no matter the technique of choice, represents a laborious process. Most of the difficulties inherent in the generation of integrated transgenes in C. elegans can potentially be avoided if a transformation marker could provide a direct selective advantage when present in extrachromosomal arrays without requiring any specific genetic background.
Antibiotic markers
Antibiotics, combined with antibiotic resistance genes, have been widely used as genetic selection systems in transformation processes. Antibiotic selection was first developed for bacteria in the 1970s and rapidly proved its usefulness for molecular biologists [43]. Thanks to the availability of broad-spectrum antibiotics, this stringent selection system was expanded to simple eukaryotic model organisms, and although most antibiotic resistance genes are of bacterial origin, they were easily adapted to eukaryotes by using 3′ and 5′ untranslated regulatory sequences [44]. Indeed, antibiotic selection systems have been developed for yeast [45–48], mammalian and insect cells [49–54], plant cells [55–57], fungus [58–61], amoeba [62], and protozoans [63–66]. In 1985, Steller and Pirrotta [67] used the broad-spectrum antibiotic G-418 and its corresponding resistance gene to select transformed Drosophila larvae and thus demonstrated that it was also possible to expand antibiotic resistance to metazoans. The resistance gene was expressed under a drosophilid promoter, and the progeny of injected individuals were selected at the larval stage in the presence of food containing the antibiotic. At a concentration lethal for wild-type larvae, transformed larvae were able to grow as fast as untransformed individuals in the absence of the drug. This system was shown to be a fairly efficient method to avoid mass screening for transgenic individuals in Drosophila and inspired its use for nondrosophilid insect transformation. Attempts were made to use this system for three different species of mosquitos [68–70], a Mediterranean fruit fly, and a migratory locust [71] for which genetic markers were not available at the time. In all of these experiments, antibiotic selection highly reduced the size of the population to screen, and transgenic individuals could be found in the resistant pool. However, the presence of numerous false positives (nontransgenic resistant individuals) warranted verification of transformation events by molecular biology methods. Since conditions for a strict selection of transgenic insects could not be determined, the method was not further developed. Yet, this work demonstrated that antibiotic resistance markers provide a versatile transformation system compatible with metazoan transgenesis and allowing the elimination of most nontransformed individuals.
Antibiotic markers for C. elegans
As with other metazoans, nematodes are sensitive to broad-spectrum antibiotics, such as G-418, puromycin, and phleomycin [72, 73]. G-418 and puromycin interfere with the function of ribosomes and block protein synthesis in eukaryotes and prokaryotes [51, 74], while phleomycin inhibits DNA synthesis, again both in eukaryotes and prokaryotes [75]. G-418 has been shown to be toxic to C. elegans in liquid and solid culture at concentrations higher than 0.1 mg/ml [72, 73]. As had already been shown in Drosophila [67], hatchlings are more sensitive to the drug than young adults, probably due to a higher dependency on their protein synthesis machinery for development. Accordingly, Giordano-Santini and co-workers defined a critical concentration of G-418 for solid culture (0.4 mg/ml), at which L1 larvae are unable to develop and die after a few days while young adults are unaffected and are able to lay eggs. Puromycin is also toxic to C. elegans both in liquid and solid culture, at concentrations similar to that of G-418 [73]. Finally, the sensitivity of C. elegans to phleomycin has so far only been shown in liquid culture and is relatively low, presumably because cells in intact worms are relatively inaccessible to the drug [73]. This finding is supported by the observation that the addition of mild detergents in liquid culture enhances nematode sensitivity to all three tested antibiotics [73].
Once sensitivity had been established, antibiotic resistance genes were adapted for their expression in C. elegans [72, 73]. Antibiotic resistance cassettes for nematodes are composed of three elements: (1) a strong endogenous promoter from a gene with an ubiquitous expression pattern, to allow for continuous gene expression in all tissues (this ensures the detoxification of every cell during development, regardless of the drug intake pathway), (2) the corresponding antibiotic resistance gene, and (3) an endogenous 3′ untranslated region (UTR) to ensure efficient processing of transcripts and mRNA stability. Following this scheme, the puromycin N-acetyl-transferase (PuroR) and the aminoglycoside phosphotransferase 3′(II) (NeoR) genes, which detoxify puromycin and G-418 respectively, were put under the control of promoters for C. elegans ribosomal protein genes, rpl-28 and rps-27, and included well characterized endogenous 3′ UTR. C. elegans transformed with p rpl-28::PuroR::3′ UTR let-858 and p rps-27::NeoR::3′ UTR unc-54 are resistant to puromycin and G-418, respectively [72, 73]. Indeed, C. elegans carrying extrachromosomal arrays containing these cassettes as well as single copies of the genes are able to survive and proliferate in the presence of lethal concentrations of antibiotics. This procedure enables the use of antibiotic resistance cassettes as selection markers for nematode transgenesis [72, 73].
A stringent and convenient selection
When transforming C. elegans by microinjection, only a part of the F1 progeny carries the exogenous DNA. The percentage of transgenic F1 can vary depending on the skills of the researcher, but rarely exceeds 20 transgenic F1 per injected P0 parent. Thus, when using nonselective transformation markers, large populations have to be screened under the dissecting microscope to look for transgenic individuals. Since extrachromosomal arrays are semi-stable, transgenic worms have to be manually selected under the microscope in order to maintain nonintegrated lines (Fig. 1).
By contrast, selection after microinjection of transgenic F1 worms carrying antibiotic resistance cassettes can be done on NGM plates supplemented with antibiotic at the critical concentration (selective plates) (Fig. 1) [72]. In this condition, injected P0 parents are able to lay eggs, and while wild-type L1 do not develop and die after a few days, transgenic L1 are able to reach adulthood. After isolation of adult F1 onto new selective plates, it is possible to identify at a glance plates in which transmission of the array occurred, as only transgenic worms can give rise to mixed-stage populations. Similarly, nonintegrated lines can be easily maintained on selective plates without the need to manually pick transgenic individuals [72, 73]. They can be maintained endlessly by chunking from plate to plate when food is depleted. In short, antibiotic markers allow a clear-cut selection of transgenic F1 and hands-off maintenance of nonintegrated lines (Table 1).
Antibiotic selection can thus be used to rapidly obtain large pure populations of transgenic worms as required for biological assays [72, 73]. Nonintegrated transgenic populations can be enriched in the presence of antibiotics in solid and liquid culture. On selective plates, it is possible to accumulate several generations until food depletion and obtain almost pure populations with all developmental stages from L2 to adults. Only arrested and dead wild-type L1 accumulate on these plates. It has been shown that enrichment of transgenic populations from L2 to adults can reach >93% on average, independent of the transmission rate [72]. Another method of enrichment is with selection in liquid medium from a mixed population of synchronized transgenic and wild-type L1. The selection process consists of a 4 day incubation in liquid NGM or M9 buffer supplemented with the corresponding antibiotic, in the presence or absence of food [73]. Because worms can be incubated at high densities (up to 50 worms per μl), large transgenic populations can be obtained in cultures of a few milliliters in less than a week, without the need for sophisticated equipment or tedious manipulation. Enriched synchronized populations thus obtained can be transferred to nonselective plates for further experiments in cases where the presence of the antibiotic might interfere with experimental procedures. Selection of nonintegrated transgenic worms using antibiotic resistance has been shown to be as efficient as using integrated lines in the context of quantitative RT-PCR experiments measuring the expression levels of a heat-shock-inducible transgene [73].
There is no indication that antibiotic resistance markers have any detrimental effects on nematodes. No phenotypic abnormality has been observed after G-418 or puromycin selection over several generations, aside from a slight delay in the development of resistant worms from L1 to adulthood on selective medium, as occurs with other mutant and transgenic lines [72, 73]. G-418 and puromycin do not seem to cause harm to worms that are properly protected by the corresponding resistance gene. The only adverse phenotypes observed so far are germline morphological defects and lower fertility in some animals carrying a high copy number of NeoR resistance cassette [72]. This phenotype seems to be avoided by the use of small doses of the resistance cassette in the injection mixture that is sufficient to insure proper G-418 protection [72].
MosSCI-biotics: antibiotic resistance cassettes as co-insertion markers for single-copy integration
While the use of antibiotic markers makes extrachromosomal array strains convenient and easy to manipulate, there may be cases where integration of a low copy number of the transgene may be desirable. Integration indeed prevents mosaicism [3], overexpression due to high copy number of the transgene [4], silencing in the germline [76], and changes in the expression of the transgene over several generations [30, 76, 77]. Each of these limitations can be avoided using different methods, but single-copy insertion of the gene of interest by MosSCI has been shown to sidestep all of these limitations at once (Fig. 3) [41].
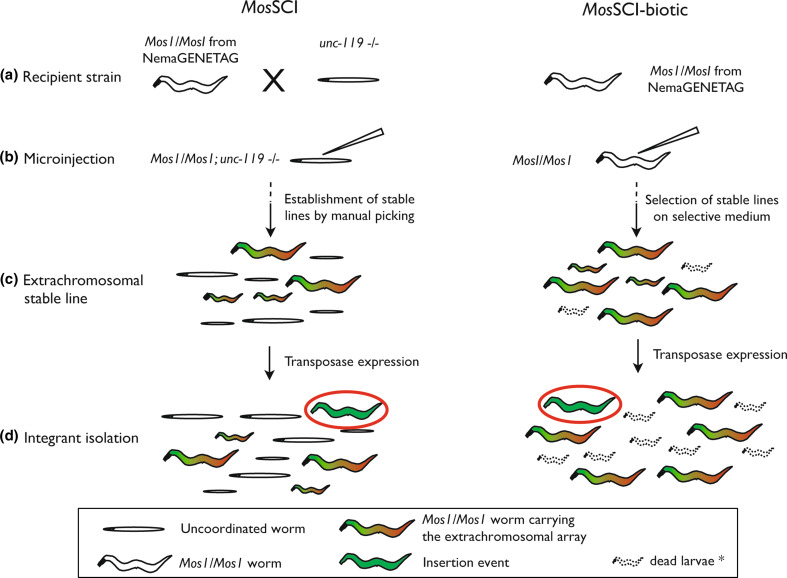
MosSCI has been developed using unc-119 as co-insertion marker (see above), and therefore recipient strains carrying Mos1 insertion also need to carry the unc-119(ed3) allele. Two intergenic Mos1 insertion sites have been used to date, for which repair template vectors carrying unc-119 were created [41]. In theory, every Mos1 intergenic insertion from NemaGENETAG, a library of Mos1 single copy insertion lines [42] could be used for gene conversion and single-copy integration. However, the use of unc-119 as a co-insertion marker necessitates the introduction of unc-119(ed3) in the genetic background beforehand.
Almost every step of the MosSCI protocol can be facilitated by the use of antibiotic resistance systems (Fig. 4) [72]. First, every Mos1 recipient strain from NemaGENETAG can be used without the need for crossing into an unc-119 mutant background, making host strains easier to amplify and to inject. Second, when using the transposase source under the control of a heat-shock promoter (heat shock protocol [41]), large populations of nonintegrated adults can be easily obtained for heat-shock by hands-off enrichment on selective medium. Third, screening for insertion events is done in populations that are not crowded by nontransgenic animals, thus reducing screen populations (when using unc-119, insertion events can be isolated after selection under starvation conditions—unc-119(ed3) individuals can not enter Dauer stage—but the procedure implies longer culture times). Finally, once an insertion event is isolated, worms obtained by MosSCI-biotic do not carry the unc-119(ed3) allele. Some further experiments may be simplified by this feature, for example, genetic crosses. Worms carrying unc-119 as a co-insertion marker can only be crossed with other unc-119 mutant strains in order to allow phenotypic selection. On the other hand, antibiotic-resistant lines are convenient to cross with most strains, and selection is easily accomplished using selective plates. The availability of different antibiotic markers makes possible the use of multiple-resistance selection (i.e., when crossing a puromycin resistant insertion strain with a G-418 resistant insertion strain, double-resistant progeny should be easily selected on G-418/puromycin plates). The combination of MosSCI and antibiotic selection systems (MosSCI-biotic) represents a convenient and effective single-copy insertion system. Indeed, the benefits of antibiotic selection also extend to single-copy transgene applications and should contribute to the development of more flexible and efficient techniques for nematode transgenesis.
Fig. 4a–d.
Comparison between MosSCI using unc-119 as co-insertion marker (left) and MosSCI-biotic (right). The use of antibiotic resistance genes as co-insertion markers facilitates almost every step of MosSCI. a, b Every Mos1 recipient strain from NemaGENETAG can be used without the need to cross into an unc-119 mutant background, making host strains easier to amplify and to inject. c When using the transposase source under the control of a heat-shock promoter, large populations of nonintegrated adults can be easily obtained for heat-shock by hands-off enrichment on selective medium. d When using an antibiotic resistance gene as co-insertion marker, screening for insertion events is done in smaller populations that are not crowded by nontransgenic worms
More recently, another method has been developed that takes advantage of the Mos1 excision-repair process to delete up to 25 kb of genomic DNA flanking the Mos1 insertion site [78]. The method is called Mos1-mediated deletion (MosDEL) and has been designed to create gene-specific knockouts and simultaneous insertion of a positive selection marker. Methods to obtain knockout worms by MosDEL are very similar to those of MosSCI. Again, unc-119 has been used as a marker for insertion and for positive selection of knockout animals. Therefore, as with MosSCI, a C. elegans strain carrying the Mos1 insertion at the desired site has to be first crossed in the unc-119(ed3) mutant background. But, while with MosSCI the integration site was chosen to be intergenic, in MosDEL the transposon is by definition located near a gene locus and either its insertion or the desired deletion may have phenotypic effects susceptible to interaction with the unc-119(ed3) background. Replacing unc-119 with an antibiotic resistance cassette as a marker should therefore dramatically enable MosDEL applications.
Application to other nematodes
The tremendous power of C. elegans as a genetic model comes from the accumulated knowledge and tools assembled by the research community over several decades. If similar tools could be developed for a number of the great variety of known nematode species it would provide an extraordinary opportunity to study the evolution of biological processes. While the process of gene discovery in free-living and parasitic nematodes has been greatly accelerated by an increasing number of nematode genome sequences, transgenesis techniques allowing functional genetic studies have only been developed for a few nematode species. One of the major obstacles for the development of transgenesis techniques has been the lack of suitable transformation markers in species other than C. elegans and Caenorhabditis briggsae.
Developing genetic markers based on fluorescent reporters [79, 80], mutant strains, or dominant alleles [80] for new species usually implies extensive genetic analyses. On the other hand, antibiotic resistance genes can be used as transformation markers without the prerequisite for any specific genetic background. It has been shown that several nematode species including helminth parasites are sensitive to broad-spectrum antibiotics [72, 81]. Antibiotic resistance markers might therefore prove useful for researchers wanting to develop projects on nematodes lacking the C. elegans transgenesis toolbox. Indeed, G-418 and puromycin resistance cassettes have been shown to work also in C. briggsae [72, 73]. The use of NeoR and PuroR driven by C. elegans regulatory sequences in C. briggsae likely indicates that these cassettes might be used in any species from the Caenorhabditis familiy. Moreover, C. elegans regulatory sequences of let-858, hsp-16, and unc-54 genes have been shown to work in more distant species such as Parastrongyloides trichosuri [82] and Heterorhabditis bacteriophora [83], suggesting that antibiotic resistance cassettes within C. elegans 5′ and 3′ regulatory sequences might also be expressed in other nematodes. For more distantly related species where the activity of C. elegans regulatory sequences is not preserved, the use of endogenous regulatory sequences should enable the use of these selection systems [79, 80]. With an increasing interest in using nematodes as a model for evolutionary biology and the need for reliable transgenesis techniques for parasitic nematodes, the availability of such versatile markers will be highly advantageous. The demonstration that it is possible to use antibiotic systems as genetic markers for metazoan transgenesis might also open opportunities to develop new avenues for genetic studies in previously inaccessible small model organisms, maybe even outside of nematodes. Indeed there is a growing interest in developing transgenesis techniques for a variety of metazoan species such as Ciona intestinalis, Plasmodium falciparum, and silkworms [84–88].
Conclusion
Antibiotic resistance markers provide a new approach to nematode transgenesis forming a new category of transformation markers that circumvent most of the limitations previously displayed. They provide a strongly selectable advantage in normal growth conditions at permissive temperatures, they do not require specific mutant backgrounds, and they do not have any detectable detrimental effects on transgenic nematodes that are properly protected by the resistance cassette. They allow clear-cut selection of transgenic individuals from the F1 generation, hands-off maintenance of nonintegrated transgenic lines, and the creation of large populations of transgenic individuals without tedious manipulation or sophisticated equipment. In a nutshell, extrachromosomal lines carrying antibiotic selection markers present the advantages of integrated lines but can be obtained more rapidly. Moreover, resistance cassettes also work in the context of single-copy insertions, making these markers compatible with transposon excision-repair methods.
To make full use of this new category of transformation markers, antibiotic selection should be adapted to microparticle bombardment. This could greatly facilitate the development of biolistic transformation in nematodes lacking nonlethal mutation rescue markers such as unc-119(ed3). So far, early attempts to use resistance cassettes as a selection system for microparticle bombardment have failed because clear-cut selection cannot be obtained in excessive crowding conditions [72]. Still, it is possible that the development of adapted procedures could bring together antibiotic selection and microparticle bombardment in the future.
So far, only G-418 and puromycin and their corresponding resistance genes have been used as selective markers for nematode transgenesis. The sensitivity of wild-type nematodes to other antibiotics, potentially in combination with mild detergents, should allow the development of new antibiotic selection markers for nematodes. For instance, a new C. elegans cassette is available, p rpl-28::ZeoR::3′UTR let-858, that could be used to confer resistance to phleomycin in wild-type worms and should provide all the advantages of antibiotic selection already described (Semple and Lehner, personal communication). Following the methodology defined for the development of G-418 and puromycin selection systems, the resistance markers toolkit should be easily expanded to other drugs and other organisms [89].
Acknowledgments
We thank J. I. Semple, B. Lehner, C. Mackereth, and K. Rebora for comments on the manuscript.
References
- 1.Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riddle DL, Blumenthal T, Meyer BJ, Preiss JR. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 3.Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans . Mol Cell Biol. 1985;5(12):3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fire A, Waterston RH. Proper expression of myosin genes in transgenic nematodes. EMBO J. 1989;8(11):3419–3428. doi: 10.1002/j.1460-2075.1989.tb08506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goetinck S, Waterston RH. The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J Cell Biol. 1994;127(1):79–93. doi: 10.1083/jcb.127.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronoff R, Baran R, Hodgkin J. Molecular identification of smg-4. required for mRNA surveillance in C. elegans . Gene. 2001;268(1–2):153–164. doi: 10.1016/S0378-1119(01)00414-0. [DOI] [PubMed] [Google Scholar]
- 7.Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54(1):5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- 8.Stone S, Shaw JE. A Caenorhabditis elegans act-4::lacZ fusion: use as a transformation marker and analysis of tissue-specific expression. Gene. 1993;131(2):167–173. doi: 10.1016/0378-1119(93)90290-J. [DOI] [PubMed] [Google Scholar]
- 9.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4(5):815–826. doi: 10.1016/S1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 10.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 11.Fire A, Kondo K, Waterston R. Vectors for low copy transformation of C. elegans . Nucleic Acids Res. 1990;18(14):4269–4270. doi: 10.1093/nar/18.14.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baum PD, Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19(1):51–62. doi: 10.1016/S0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- 13.Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149(3):1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hope IA. ‘Promoter trapping’ in Caenorhabditis elegans . Development. 1991;113(2):399–408. doi: 10.1242/dev.113.2.399. [DOI] [PubMed] [Google Scholar]
- 15.Hobert O, Loria P (2005) Uses of GFP in C. elegans. In: Chalfie M, Kain S (eds) Green fluorescent protein: properties, applications and protocols. Wiley, New York
- 16.Bacaj T, Shaham S. Temporal control of cell-specific transgene expression in Caenorhabditis elegans . Genetics. 2007;176(4):2651–2655. doi: 10.1534/genetics.107.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulin T, Etchberger JF, Hobert O (2006) Reporter gene fusions. WormBook. doi:10.1895/wormbook.1.106.1 [DOI] [PMC free article] [PubMed]
- 18.Vogel BE, Hedgecock EM. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development. 2001;128(6):883–894. doi: 10.1242/dev.128.6.883. [DOI] [PubMed] [Google Scholar]
- 19.Loria PM, Duke A, Rand JB, Hobert O. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr Biol. 2003;13(15):1317–1323. doi: 10.1016/S0960-9822(03)00532-3. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Serricchio AS, Sternberg PW. Visualization of C. elegans transgenic arrays by GFP. BMC Genet. 2006;7:36. doi: 10.1186/1471-2156-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutter H. New ways to look at axons in Caenorhabditis elegans . Microsc Res Tech. 2000;48(1):47–54. doi: 10.1002/(SICI)1097-0029(20000101)48:1<47::AID-JEMT6>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 22.White JG, Albertson DG, Anness MA. Connectivity changes in a class of motoneurone during the development of a nematode. Nature. 1978;271(5647):764–766. doi: 10.1038/271764a0. [DOI] [PubMed] [Google Scholar]
- 23.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282(5396):2012–2018 [DOI] [PubMed]
- 24.Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, Carvunis AR, Pulak R, Shingles J, Reece-Hoyes J, Hunt-Newbury R, Viveiros R, Mohler WA, Tasan M, Roth FP, Le Peuch C, Hope IA, Johnsen R, Moerman DG, Barabasi AL, Baillie D, Vidal M. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans . Nat Biotechnol. 2007;25(6):663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 25.Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans . PLoS Biol. 2007;5(9):e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJ, Vidal M, Hope IA. Insight into transcription factor gene duplication from Caenorhabditis elegans promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH. Unlocking the secrets of the genome. Nature. 2009;459(7249):927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10(12):3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fire A. Integrative transformation of Caenorhabditis elegans . EMBO J. 1986;5(10):2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans . Genetics. 2001;157(3):1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilm T, Demel P, Koop HU, Schnabel H, Schnabel R. Ballistic transformation of Caenorhabditis elegans . Gene. 1999;229(1–2):31–35. doi: 10.1016/S0378-1119(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 32.Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141(3):977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark DV, Suleman DS, Beckenbach KA, Gilchrist EJ, Baillie DL. Molecular cloning and characterization of the dpy-20 gene of Caenorhabditis elegans . Mol Gen Genet. 1995;247(3):367–378. doi: 10.1007/BF00293205. [DOI] [PubMed] [Google Scholar]
- 34.Thacker C, Sheps JA, Rose AM. Caenorhabditis elegans dpy-5 is a cuticle procollagen processed by a proprotein convertase. Cell Mol Life Sci. 2006;63(10):1193–1204. doi: 10.1007/s00018-006-6012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22(9):1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark SG, Lu X, Horvitz HR. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 1994;137(4):987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fire A, Albertson D, Harrison SW, Moerman DG. Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Development. 1991;113(2):503–514. doi: 10.1242/dev.113.2.503. [DOI] [PubMed] [Google Scholar]
- 38.Thatcher JD, Haun C, Okkema PG. The DAF-3 Smad binds DNA and represses gene expression in the Caenorhabditis elegans pharynx. Development. 1999;126(1):97–107. doi: 10.1242/dev.126.1.97. [DOI] [PubMed] [Google Scholar]
- 39.Polanowska J, Martin JS, Fisher R, Scopa T, Rae I, Boulton SJ. Tandem immunoaffinity purification of protein complexes from Caenorhabditis elegans . Biotechniques. 2004;36(5):778–780, 782. doi: 10.2144/04365BM05. [DOI] [PubMed] [Google Scholar]
- 40.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. doi: 10.1016/S0091-679X(08)61399-0. [DOI] [PubMed] [Google Scholar]
- 41.Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans . Nat Genet. 2008;40(11):1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bazopoulou D, Tavernarakis N. The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans . Genetica. 2009;137(1):39–46. doi: 10.1007/s10709-009-9361-3. [DOI] [PubMed] [Google Scholar]
- 43.Cohen SN, Chang AC, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro . Proc Natl Acad Sci USA. 1973;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies J, Jimenez A. A new selective agent for eukaryotic cloning vectors. Am J Trop Med Hyg. 1980;29(5 Suppl):1089–1092. doi: 10.4269/ajtmh.1980.29.1089. [DOI] [PubMed] [Google Scholar]
- 45.Jimenez A, Davies J. Expression of a transposable antibiotic resistance element in Saccharomyces . Nature. 1980;287(5785):869–871. doi: 10.1038/287869a0. [DOI] [PubMed] [Google Scholar]
- 46.Gatignol A, Baron M, Tiraby G. Phleomycin resistance encoded by the ble gene from transposon Tn 5 as a dominant selectable marker in Saccharomyces cerevisiae . Mol Gen Genet. 1987;207(2–3):342–348. doi: 10.1007/BF00331599. [DOI] [PubMed] [Google Scholar]
- 47.Kimura M, Kamakura T, Tao QZ, Kaneko I, Yamaguchi I. Cloning of the blasticidin S deaminase gene (BSD) from Aspergillus terreus and its use as a selectable marker for Schizosaccharomyces pombe and Pyricularia oryzae . Mol Gen Genet. 1994;242(2):121–129. doi: 10.1007/BF00391004. [DOI] [PubMed] [Google Scholar]
- 48.Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae . Gene. 1983;25(2–3):179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 49.Colbere-Garapin F, Horodniceanu F, Kourilsky P, Garapin AC. A new dominant hybrid selective marker for higher eukaryotic cells. J Mol Biol. 1981;150(1):1–14. doi: 10.1016/0022-2836(81)90321-1. [DOI] [PubMed] [Google Scholar]
- 50.Blochlinger K, Diggelmann H. Hygromycin B phosphotransferase as a selectable marker for DNA transfer experiments with higher eucaryotic cells. Mol Cell Biol. 1984;4(12):2929–2931. doi: 10.1128/mcb.4.12.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de la Luna S, Soria I, Pulido D, Ortin J, Jimenez A. Efficient transformation of mammalian cells with constructs containing a puromycin-resistance marker. Gene. 1988;62(1):121–126. doi: 10.1016/0378-1119(88)90585-9. [DOI] [PubMed] [Google Scholar]
- 52.Pfeifer TA, Hegedus DD, Grigliatti TA, Theilmann DA. Baculovirus immediate-early promoter-mediated expression of the Zeocin resistance gene for use as a dominant selectable marker in dipteran and lepidopteran insect cell lines. Gene. 1997;188(2):183–190. doi: 10.1016/S0378-1119(96)00756-1. [DOI] [PubMed] [Google Scholar]
- 53.Okano K, Miyajima N, Takada N, Kobayashi M, Maekawa H. Basic conditions for the drug selection and transient gene expression in the cultured cell line of Bombyx mori . In Vitro Cell Dev Biol. 1992;28A(11–12):779–781. doi: 10.1007/BF02631067. [DOI] [PubMed] [Google Scholar]
- 54.Lopez MG, Alfonso V, Carrillo E, Taboga O. Trans-complementation of polyhedrin by a stably transformed Sf9 insect cell line allows occ- baculovirus occlusion and larval per os infectivity. J Biotechnol. 2010;145(2):199–205. doi: 10.1016/j.jbiotec.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 55.Herrera-Estrella L, Block MD, Messens E, Hernalsteens JP, Montagu MV, Schell J. Chimeric genes as dominant selectable markers in plant cells. EMBO J. 1983;2(6):987–995. doi: 10.1002/j.1460-2075.1983.tb01532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez P, Tiraby G, Kallerhoff J, Perret J. Phleomycin resistance as a dominant selectable marker for plant cell transformation. Plant Mol Biol. 1989;13(4):365–373. doi: 10.1007/BF00015548. [DOI] [PubMed] [Google Scholar]
- 57.Waldron C, Murphy EB, Roberts JL, Gustafson GD, Armour SL, Malcolm SK. Resistance to hygromycin B: a new marker for plant transformation studies. Plant Mol Biol. 1985;5:103–108. doi: 10.1007/BF00020092. [DOI] [PubMed] [Google Scholar]
- 58.Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli . Gene. 1987;56(1):117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 59.Gastmann S, Dunkler A, Walther A, Klein K, Wendland J. A molecular toolbox for manipulating Eremothecium coryli . Microbiol Res. 2007;162(4):299–307. doi: 10.1016/j.micres.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 60.Cullen D, Leong SA, Wilson LJ, Henner DJ. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene. 1987;57(1):21–26. doi: 10.1016/0378-1119(87)90172-7. [DOI] [PubMed] [Google Scholar]
- 61.Jain S, Durand H, Tiraby G. Development of a transformation system for the thermophilic fungus Talaromyces sp. CL240 based on the use of phleomycin resistance as a dominant selectable marker. Mol Gen Genet. 1992;234(3):489–493. doi: 10.1007/BF00538710. [DOI] [PubMed] [Google Scholar]
- 62.Dubin M, Nellen W. A versatile set of tagged expression vectors to monitor protein localisation and function in Dictyostelium . Gene. 2010;465(1–2):1–8. doi: 10.1016/j.gene.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 63.Brunk CF, Navas P. Transformation of Tetrahymena thermophila by electroporation and parameters effecting cell survival. Exp Cell Res. 1988;174(2):525–532. doi: 10.1016/0014-4827(88)90322-9. [DOI] [PubMed] [Google Scholar]
- 64.Freedman DJ, Beverley SM. Two more independent selectable markers for stable transfection of Leishmania . Mol Biochem Parasitol. 1993;62(1):37–44. doi: 10.1016/0166-6851(93)90175-W. [DOI] [PubMed] [Google Scholar]
- 65.Goyard S, Beverley SM. Blasticidin resistance: a new independent marker for stable transfection of Leishmania . Mol Biochem Parasitol. 2000;108(2):249–252. doi: 10.1016/S0166-6851(00)00210-3. [DOI] [PubMed] [Google Scholar]
- 66.Suarez CE, McElwain TF. Stable expression of a GFP-BSD fusion protein in Babesia bovis merozoites. Int J Parasitol. 2009;39(3):289–297. doi: 10.1016/j.ijpara.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 67.Steller H, Pirrotta V. A transposable P vector that confers selectable G418 resistance to Drosophila larvae. EMBO J. 1985;4(1):167–171. doi: 10.1002/j.1460-2075.1985.tb02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller LH, Sakai RK, Romans P, Gwadz RW, Kantoff P, Coon HG. Stable integration and expression of a bacterial gene in the mosquito Anopheles gambiae . Science. 1987;237(4816):779–781. doi: 10.1126/science.3039658. [DOI] [PubMed] [Google Scholar]
- 69.Morris AC, Eggleston P, Crampton JM. Genetic transformation of the mosquito Aedes aegypti by micro-injection of DNA. Med Vet Entomol. 1989;3(1):1–7. doi: 10.1111/j.1365-2915.1989.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 70.McGrane V, Carlson JO, Miller BR, Beaty BJ. Microinjection of DNA into Aedes triseriatus ova and detection of integration. Am J Trop Med Hyg. 1988;39(5):502–510. doi: 10.4269/ajtmh.1988.39.502. [DOI] [PubMed] [Google Scholar]
- 71.Handler AM, O’Brochta DA. Prospects for gene transformation in insects. Annu Rev Entomol. 1991;36:159–183. doi: 10.1146/annurev.en.36.010191.001111. [DOI] [PubMed] [Google Scholar]
- 72.Giordano-Santini R, Milstein S, Svrzikapa N, Tu D, Johnsen R, Baillie D, Vidal M, Dupuy D. An antibiotic selection marker for nematode transgenesis. Nat Methods. 2010;7(9):721–723. doi: 10.1038/nmeth.1494. [DOI] [PubMed] [Google Scholar]
- 73.Semple JI, Garcia-Verdugo R, Lehner B. Rapid selection of transgenic C. elegans using antibiotic resistance. Nat Methods. 2010;7(9):725–727. doi: 10.1038/nmeth.1495. [DOI] [PubMed] [Google Scholar]
- 74.Bar-Nun S, Shneyour Y, Beckmann JS. G-418, an elongation inhibitor of 80s ribosomes. Biochim Biophys Acta. 1983;741(1):123–127. doi: 10.1016/0167-4781(83)90018-0. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka N, Yamaguchi H, Umezawa H. Mechanism of action of phleomycin, a tumor-inhibitory antibiotic. Biochem Biophys Res Commun. 1963;10:171–174. doi: 10.1016/0006-291X(63)90045-7. [DOI] [PubMed] [Google Scholar]
- 76.Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146(1):227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hammarlund M, Palfreyman MT, Watanabe S, Olsen S, Jorgensen EM. Open syntaxin docks synaptic vesicles. PLoS Biol. 2007;5(8):e198. doi: 10.1371/journal.pbio.0050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frokjaer-Jensen C, Davis MW, Hollopeter G, Taylor J, Harris TW, Nix P, Lofgren R, Prestgard-Duke M, Bastiani M, Moerman DG, Jorgensen EM. Targeted gene deletions in C. elegans using transposon excision. Nat Methods. 2010;7(6):451–453. doi: 10.1038/nmeth.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, Massey HC, Jr, Nolan TJ, Schad GA, Kraus K, Sundaram M, Lok JB. Successful transgenesis of the parasitic nematode Strongyloides stercoralis requires endogenous non-coding control elements. Int J Parasitol. 2006;36(6):671–679. doi: 10.1016/j.ijpara.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 80.Schlager B, Wang X, Braach G, Sommer RJ. Molecular cloning of a dominant roller mutant and establishment of DNA-mediated transformation in the nematode Pristionchus pacificus . Genesis. 2009;47(5):300–304. doi: 10.1002/dvg.20499. [DOI] [PubMed] [Google Scholar]
- 81.Shiomi K, Omura MJA. Antiparasitic agents produced by microorganisms. Proc Jpn Acad. 2004;80:245–258. doi: 10.2183/pjab.80.245. [DOI] [Google Scholar]
- 82.Grant WN, Skinner SJ, Newton-Howes J, Grant K, Shuttleworth G, Heath DD, Shoemaker CB. Heritable transgenesis of Parastrongyloides trichosuri: a nematode parasite of mammals. Int J Parasitol. 2006;36(4):475–483. doi: 10.1016/j.ijpara.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 83.Hashmi S, Hashmi G, Gaugler R. Genetic transformation of an entomopathogenic nematode by microinjection. J Invertebr Pathol. 1995;66(3):293–296. doi: 10.1006/jipa.1995.1103. [DOI] [PubMed] [Google Scholar]
- 84.Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian Ciona intestinalis . Development. 1997;124(3):589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 85.Wilson DW, Crabb BS, Beeson JG. Development of fluorescent Plasmodium falciparum for in vitro growth inhibition assays. Malar J. 2010;9:152. doi: 10.1186/1475-2875-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsuoka T, Awazu S, Shoguchi E, Satoh N, Sasakura Y. Germline transgenesis of the ascidian Ciona intestinalis by electroporation. Genesis. 2005;41(2):67–72. doi: 10.1002/gene.20096. [DOI] [PubMed] [Google Scholar]
- 87.Guo X-Y, Dong L, Wang S-P, Guo T-Q, Wang J-Y, Lu C-D. Introduction of foreign genes into silkworm eggs by electroporation and its application in transgenic vector test. Acta Biochim Biophys Sin. 2004;36(5):323–330. doi: 10.1093/abbs/36.5.323. [DOI] [PubMed] [Google Scholar]
- 88.Maher B. Evolution: biology’s next top model? Nature. 2009;458(7239):695–698. doi: 10.1038/458695a. [DOI] [PubMed] [Google Scholar]
- 89.Chamberlin HM. C. elegans select. Nat Methods. 2010;7(9):693–695. doi: 10.1038/nmeth0910-693. [DOI] [PubMed] [Google Scholar]