Abstract
Metabolic engineering is the enabling science of development of efficient cell factories for the production of fuels, chemicals, pharmaceuticals, and food ingredients through microbial fermentations. The yeast Saccharomyces cerevisiae is a key cell factory already used for the production of a wide range of industrial products, and here we review ongoing work, particularly in industry, on using this organism for the production of butanol, which can be used as biofuel, and isoprenoids, which can find a wide range of applications including as pharmaceuticals and as biodiesel. We also look into how engineering of yeast can lead to improved uptake of sugars that are present in biomass hydrolyzates, and hereby allow for utilization of biomass as feedstock in the production of fuels and chemicals employing S. cerevisiae. Finally, we discuss the perspectives of how technologies from systems biology and synthetic biology can be used to advance metabolic engineering of yeast.
Keywords: Metabolic engineering, Yeast, Substrate range, Biobutanol, Isoprenoids, Industrial biotechnology
Introduction
Microbial fermentations have been used for the production of fermented food and beverages since ancient times. Already around 1920 microbial fermentation was introduced for the production of citric acid, and this was the first large-scale industrial production process of a chemical compound based on microbial fermentation. With the development of genetic engineering in the 1970s, it became possible to produce compounds that are not native to microbes, such as pharmaceutical proteins like human insulin and human growth hormone using fermentation technology. Genetic engineering also allowed the transformation of microbes into “cell factories” for the production of chemicals through so-called metabolic engineering [1–3], a field dedicated to design of microbial metabolism to efficiently convert cheap raw materials like glucose, sucrose, and biomass-derived sugars into fuels and chemicals. With the further development of genomics and omics analysis and advanced modeling tools in the field of systems biology [4–8], it has become possible to perform very detailed phenotypic characterization of microorganisms that can serve as efficient cell factories for the production of fuels and chemicals. Thus, the last 10 years have witnessed a substantial technology push in terms of cell factory design, and with the recent desire to develop more sustainable processes for the production of fuels, chemicals, and materials, the chemical industry is trying to exploit these technological developments. The net result is the development of what is generally referred to as industrial biotechnology. There are already several examples of how the fuel and chemical industry is trying to develop novel bioprocesses that can change the primary feedstock from oil to agricultural-based products:
Dupont, one of the largest chemical companies in the world, has launched a process for production of 1,3-propanediol using a recombinant Escherichia coli. 1,3-propanediol is used as one of the key chemicals in the production of the polymer Sorona®, which is used for the manufacturing of fabrics, carpets, and a wide range of plastic-based materials. The process was developed through close collaboration of Dupont, Genencor, and Tate & Lyle.
DSM has launched a completely biotech route for production of the antibiotic cephalexin, which was earlier produced based on chemical conversion of penicillin.
BASF has launched a completely biotech route for production of the vitamin B2, riboflavin. The previous process relied on several chemical synthesis steps and the biotech route resulted in both a reduction in raw materials and energy usage [9].
Dupont has entered into a joint venture with British Petroleum (BP), Butamax, on developing a bio-based production of butanol as a sustainable biofuel.
Dupont has also entered into another joint venture with Danisco for the development of a second-generation bioethanol production plant that will rely on the use of lignocellulosics as raw materials for ethanol production. BP recently acquired Verenium also with the objective of developing second-generation bioethanol production, but using a different technology.
ExxonMobil has entered into a joint venture with Synthetic Genomics to develop a novel microalgae-based process for the production of biodiesel.
Novozymes has entered a joint venture with Cargill with the objective of developing a bio-based process for the production of 3-hydroxypropionic acid, which is to be used for the production of acrylates for the production of a range of personal care products, e.g., dipers and other hygienic products.
Gevo has launched a process for production of isobutanol that can find application as a biofuel and a commodity chemical.
Amyris has developed a yeast-based fermentation process for production of farnese that can be used as biodiesel as well as be converted into squalene, which is used in cosmetics.
These and many other examples clearly demonstrate two key points: (1) the large chemical and fuel companies are turning to biotech as the solution to develop sustainable processes for the production of fuels and chemicals, and (2) most novel processes are developed through close collaboration/joint ventures involving two or more companies, and often also involve academia or small technology-based companies as a provider of novel technologies. The reason for the latter is that the development of a novel bioprocess requires a wide range of competences. Traditional chemical companies hold the necessary engineering competence required for scale-up and plant construction, but they may lack competence on the biotech part.
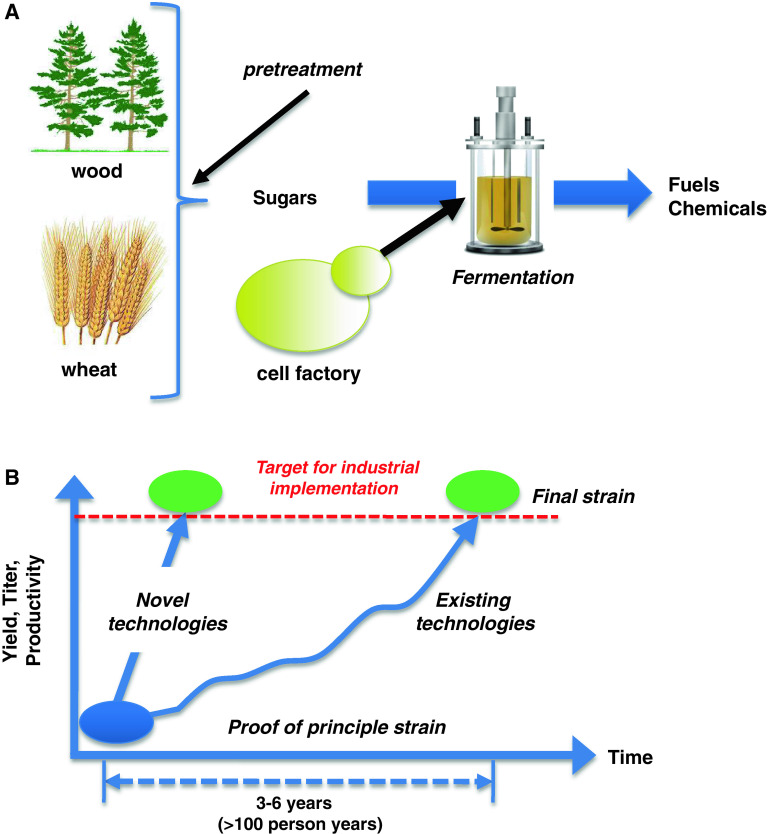
The overall idea in the above-mentioned and many other ventures is to develop bio-based processes that use wheat, corn, sugarcane, or biomass as a raw material for production of fuels and chemicals using so-called biorefineries (Fig. 1a). In a biorefinery, there is pre-treatment of the plant-based raw materials, and this has been well implemented for sugar and starch-based raw materials, i.e., sugarcane, sugar beet, wheat, and corn, and today there is very large scale production of bioethanol and other chemicals using these raw materials. Very efficient enzymes have been developed for the degradation of starch and today many processes are based on simultaneous saccharification and fermentation, where the enzymes are added directly to the fermentation process. Despite the success of sugar- and starch-based bioprocesses, these raw materials will not be able to meet the increasing demand for bio-based products, and there is therefore a need to move to biomass-based raw materials. Here, the pre-treatment is far more complicated and it depends on the raw material to be used, but recently there has been progress on several different fronts, including the development of efficient enzymes that can hydrolyze celluloses and hemicelluloses [10–12]. A key part of developing novel bioprocesses for production of fuels and chemicals is the construction of the cell factory. This cell factory has to meet commercial requirements for yield, productivity, and titer. Often it is possible to quite rapidly develop a proof-of-principle strain that produces the product of interest, whereas it is generally far more time consuming to develop a strain that meets the commercial targets for yield, productivity, and titer (Fig. 1b). There is therefore much interest to develop novel technologies that may speed up this development process, and here it is expected that tools from systems biology may assist. Advancement in the field of synthetic biology may also allow faster development of efficient cell factories, as synthetic biology may provide novel tools for controlled expression of genes, assembly of complete pathways on scaffold proteins, and completely novel enzymatic functions [13]. Nielsen and Keasling [2] recently discussed the synergies between synthetic biology and metabolic engineering.
Fig. 1.
Illustration of the biorefinery concept and the development time of novel bioprocesses. a In a biorefinery, plant-based feed-stocks such as sugarcane, corn, wheat, or biomass are converted into sugars that are subsequently used for microbial fermentations. In the fermentation process, cell factories convert the sugars into fuels and chemicals. b The development of cell factories is the central research and development process in connection with the development of a novel bioprocess. Construction of an efficient cell factory requires large investment, in particular in connection with bringing the cell factory from proof-of-principle stage where it is producing small amounts of the desired product to a final strain that produces the product at yields, titers, and productivities that make the process financially competitive with fossil fuel-based processes
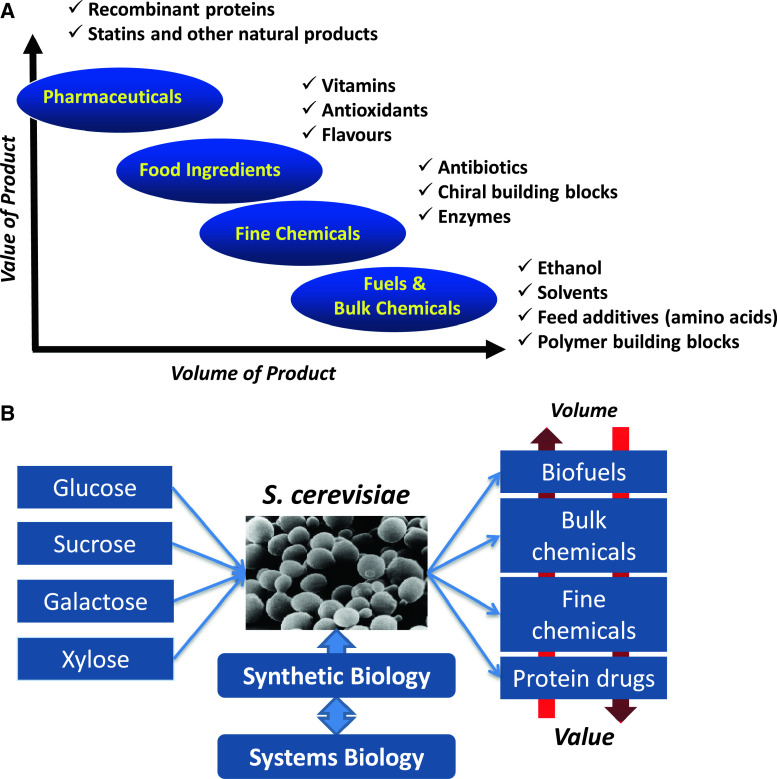
Microbial fermentation is already today used for the production of a whole range of products (Fig. 2a), but not all of these will fit naturally into the biorefinery concept, which is primarily geared towards fuels and bulk chemicals where there is a requirement for cheap raw materials that are available in large quantities. However, also for products that do not fit into the biorefinery concept, there is a need for decreasing the development time such that novel products to be used as food and pharmaceutical ingredients can be brought to the market faster. In this context, the type of cell factory plays a very important role, and in recent years there has been some consolidation towards the use of a few industrial platform cell factories that include (not an exhaustive list) the yeast Saccharomyces cerevisiae, the bacteria Escherichia coli, Corynebacterium glutamicum and Bacillus subtilis, and the filamentous fungi Aspergillus niger and Aspergillus oryzae. S. cerevisiae is a very attractive cell factory, as it has been demonstrated to be very well suited for industrial production of a range of products due to its robustness and tolerance towards industrial conditions. Thus, it is used for the production of bioethanol, the largest-volume fermentation product by far, and it is also used for the production of several pharmaceuticals, e.g., human insulin, hepatitis vaccines, and human papillomavirus vaccines, and the production of nutraceuticals, e.g., resveratrol, has been announced to be produced by this cell factory (Fig. 2b). Furthermore, a number of academic studies have illustrated the suitability of this cell factory for the production of a range of chemicals [5, 14], e.g., lactic acid, glycerol, and malic acid. Several recent reviews provide an overview of the many different metabolic engineering examples using yeast as a cell factory [15, 16], and Table 1 provides a summary of some of these key developments. In addition, the wide use of this organism is illustrated by the very large number of patents filed on the use of yeast and/or S. cerevisiae for production of fuels and chemicals (Table 2). Yeast also serves as an important model eukaryote, and many fundamental studies have therefore been performed on this organism [54]. It was also the first eukaryotic organism to have its genome sequenced and a number of high-throughput studies have been pioneered using this organism as a model [55–57]. Thus, there is an extensive technology platform in terms of systems biology and synthetic biology available for this organism (Fig. 2b), and this makes it a promising host for rapid development as a cell factory for production of novel fuels and chemicals. S. cerevisiae, however, has one major limitation in its use, and that is its lack of ability to efficiently grow and metabolize pentoses that are present in hemicelluloses, and therefore result from biomass hydrolysis. However, there has been much interest in metabolic engineering of S. cerevisiae for improving its ability to use pentoses, in particular xylose.
Fig. 2.
Range of products and illustration of the key research problems associated with cell factory design and development. a Biotech products range from high-value-added to low-value-added products, with the latter being produced in large quantities and the former in small quantities. Examples of the different types of products are indicated. The yeast S. cerevisiae is used for the production of products in the whole spectrum. b In connection with the development of yeast for the production of different types of products using different sugars as feedstock, there is a need for an extensive platform of technologies from synthetic and systems biology
Table 1.
Example of products and strains of S. cerevisiae
| Categories | Products | Specific applications | Strains | References |
|---|---|---|---|---|
| Biofuels | Ethanol | Redox balance problem by inhibiting glycerol formation in anaerobic culture was solved by combining gene deletion (GPD1 and GPD2) and integration (mhpF from E. coli) with acetic acid supplementation, which was presented at substantial quantities in lignocellulosic hydrolysates of agricultural residues | CEN.PK102-3A (MATa ura3 leu2) | [17] |
| Biobutanol | Overexpression of genes in valine metabolism, ILV2, ILV3, ILV5, and BAT2 showed an increased production of isobutanol in S. cerevisiae, which strain was decided as a host because of relative tolerance to alcohols, and robustness in industrial fermentation | CEN.PK 2-1C (MATα leu2-3, 112 his3-Δ1 ura3-52 trp1-289 MAL2-8(Con) MAL3 SUC3) | [18] | |
| Biodiesels | Glycerol utilization for production of fatty acid ethyl esters (FAEEs) was done by amplification of ethanol production pathway, which is used for the transesterification in FAEEs synthesis, with overexpression of an unspecific acyltransferase from Acinetobacter baylyi | YPH499 (MATa ura3-52 lys2-801_amber ade2-101_ochre trp1-D63 his3-D200 leu2-D1) | [19] | |
| Bisabolene (D2 diesel fuel, bisabolane) | Bisabolene, the immediate precursor to bisabolane, was produced by (1) using the strategy for increasing pool of farnesyl diphosphate (FPP) in artemisinic acid production [20] and (2) screening and codon-optimizing bisabolene synthases (sesquiterpene synthases). The final titers were over 900 mg/l in shake flasks | BY4742 (MATα his3D1 leu2D0 lys2D0 ura3D0) | [21] | |
| Bulk chemicals | 1,2-propanediol | The combination effects of different copy number (from 0 to 3) of two E. coli genes (mgs and gldA) ware studied. Although the three copy numbers of two genes showed the highest level of 1,2-propanediol, specific activity of Mgs and inhibitory relationship by GldA was considered more importantly for the production of 1,2-propanediol |
NOY386αA (MATα ura3-52 lys2-801 trp1-Δ63 his3-Δ200 leu2-Δ1) BWG1-7a (MATa ade1-100 his4-519 leu2-3,112 ura3-52 GAL +) |
[22] |
| d-ribose and ribitol | The flux from glucose to pentose phosphate pathway was amplified by inactivation of both phosphoglucose isomerase and transketolase with overexpression of sugar phosphate phosphatase (DOG1). Fructose was supplied and redox balance was controlled by overexpression of NAD+-specific glutamate dehydrogenase (GDH2) of S. cerevisiae or NADPH-utilizing glyceraldehyde-3-phosphate dehydrogenase (gapB) of Bacillus subtilis | CEN.PK2-1D (VW-1B; MATα, leu2-3/112 ura3-52 trp1-289 his3Δ1 MAL2-8c SUC2) | [23] | |
| l-lactic acid | Improved production of l-lactic acid was achieved by overexpression of LDH gene coding l-lactic acid dehydrogenase from bovine and knocked out a PDC1 gene coding pyruvate decarboxylase to redirect the fluxes to l-lactic acid; and overexpression of an NADH oxidase (nox) from Streptococcus pneumoniae into the cytoplasm to reduce the ratio of NADH/NAD+ | CEN. PK2-1C (MATa ura3-52 trp1-289 leu2-3,112 his3Ä1 MAL2-8C SUC2) | [24] | |
| Polyhydroxy-alkanoates | The synthesis of diverse size of PHA polymer (C4 to C14) was investigated by cytosolic expression of mcl-PHA synthase from Pseudomonas oleovorans or peroxisomal expression of scl-PHA synthase from Ralstonia eutropha | BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0) | [25] | |
| Pyruvic acid | Pyruvate decarboxylase-negative [Pdc(−)] strains were evolved in glucose-limited chemostat cultivation by progressively lowering the acetate content in the feed to obtain an acetate-independent Pdc (−) mutant. Maximum yield was 0.54 g of pyruvate/g glucose | CEN.PK113-7D (MATa MAL2-8C, SUC2) | [26] | |
| Succinic acid | The deletion of the genes SDH1, SDH2, IDH1, and IDP1 made higher flux to succinic acid production. Maximum yield was 0.11 mol of succinic acid/mol of glucose | AH22ura3 (MATa ura3Δ leu2-3 leu2-112 his4-519 can1) | [27] | |
| Fine chemicals | β-amyrin | The differences of phenotype and genotype in two yeast strains, CEN.PK113-7D and S288C, were compared. CEN.PK113-7D had more contents of ergosterol and fatty acids with non-silent SNPs in relative metabolism, ERG8, ERG9, and HFA1. Amplification of those genes exhibited a fivefold increase of β-amyrin | CEN.PK113-7D (MATa MAL2-8C SUC2) | [28] |
| β-carotene | Genomic integration and overexpression of carotenogenic genes from X. dendrorhous (crtYB, crtE, and crtI) and S. cerevisiae (BTS1 and truncated HMG1) with change of copy number achieved high levels of β-carotene, up to 5.9 mg/g dry cell weight | CEN.PK113-7D (MATa MAL2-8C SUC2) | [29] | |
| Amorpha-4, 11- diene | Amplification of mevalonate pathway in CEN.PK2 was engineered and compared to previously constructed strain S288C [20]. Artemisinic acid production was doubled, while amorpha-4, 11-diene was tenfold higher, over 40 g/l |
CEN.PK2-1C (MATa ura3-52 trp1-289 leu2-3,112 his3Ä1 MAL2-8C SUC2) CEN.PK2-1D (MATα ura3-52 trp1-289 leu2-3,112 his3Ä1 MAL2-8C SUC2) |
[30] | |
| Valencene and amorphadiene | Co-expression of heterologous enzymes, farnesyl diphosphate synthases (FDPSs), and sesquiterpene synthase (ex. Citrus sinensis valencene synthase CsTPS1, Artemisia annua terpene synthase, amorpha-4,11-diene synthase ADS) in mitochondria and cytosol improved the production of valencene and amorphadiene | W303-1A (MATa, ade2-1 trp1-1 leu2-3, 112 his3-11, 15 ura3-1) mBDXe (a uracilauxotroph derivative of strain BDX, Lallemand, Rexdale, Ontario, Canada) | [31] | |
| Casbene (an anti-fungal diterpene) | Genes of putative Casbene synthases from different Euphorbiaceae species were isolated and applied for production of diterpenes. Maximum concentration of Casbene was 31 mg/l | BY4742 (MATα his3D1 leu2D0 lys2D0 ura3D0) | [32] | |
| Cinnamoyl anthranilates | Twenty-six different cinnamoyl anthranilates molecules were produced by co-expressing a 4-coumarate/CoA ligase (4CL, EC 6.2.1.12) from Arabidopsis thaliana and a hydroxycinnamoyl/benzoyl-CoA/anthranilate N-hydroxycinnamoyl/benzoyltransferase (HCBT, EC 2.3.1.144) from Dianthus caryophyllus | BY4742 (MATα his3D1 leu2D0 lys2D0 ura3D0) | [33] | |
| Cubebol | Overexpression of GFTpsC (a sesquiterpene synthase isolated from Citrus paradisi and encoding for a cubebol synthase) with integration of tHMG1 into genome and reduction of ERG9 gene expression produced cubebol up to 10 mg/l | CEN.PK113-5D (MATa MAL2-8c SUC2 ura3-52) | [34] | |
| Eicosapentaenoic acid (EPA) | Five heterologous fatty acid desaturases and an elongase were identified by a BLAST search and assayed their substrate preferences activity. Without supplement of fatty acids, EPA/ARA were produced | CEN.PK113-5D (MATa MAL2-8c SUC2 ura3-52) | [35] | |
| Farnese and geranyl geraniol | ERG9 deletion and overexpression of two isozymes of HMGCoA reductases (HMG1 and HMG2) was implemented in a host strain with overexpression of diverse FPP synthases and GGPP synthases | FL100 (MATa, ATCC: 28383) | [36] | |
| l-ascorbic acid | About 100 mg of l-ascorbic acid per liter was produced by overexpression of d-arabionono-1,4-lactose oxidase from S. cerevisiae and l-galactose dehydrogenase from Arabidopsis thaliana |
GRF18U (MATα his3 leu2 ura3; NRRL Y-30320) W303 1B (MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100) |
[37] | |
| Linalool | Overexpression of Clarkia breweri linalool synthase gene (LIS) in wine strain T73 showed higher levels of linalool than conventional laboratory strains. Combining with deregulation of HMG-CoA reductase improved linalool yield | BQS252 (MATa ura3-52 (derivative of FY1679)) | [38] | |
| Methylmalonyl-coenzyme A | Polyketide precursor (Methylmalonyl-CoA) pathway was constructed by introducing propionyl-CoA carboxylase and malonyl/methylmalonyl-CoA ligase from Streptomyces coelicolor |
InvSC1 (MATa, his3delta1, leu2, trp1-289, ura3-52 (Invitrogen, Carlsbad, CA, USA)) BJ5464 (MATα, ura3-52, trp1, leu2-delta1, his3-delta200, pep4::HIS3, prb1-delta1.6R, can1, GAL). |
[39] | |
| Patchoulol | A physical fusion between native (farnesyl diphosphate synthase) and heterologous enzymes (patchoulol synthase of plant origin, Pogostemon cablin was successfully applied to produce patchoulol, 25 mg/l | CEN.LA100 (MATa/MATα ERG20/erg20::hph MAL2-8c/MAL2-8c SUC2/SUC2 ura3-52/ura3-52) | [40] | |
| Resveratrol | Co-expression of the coenzyme-A ligase-encoding gene (4CL216) from a hybrid poplar and the grapevine resveratrol synthase gene (vst1) from Vitis vinifera with supplement of p-coumaric acid produced resveratrol, 1.45 mg/L | FY23 (MATa ura3-52 trplA63 leu2A1) | [41] | |
| Vanillin | Knock-out targets, PDC1 and GDH1, suggested by in silico metabolic model was applied and production of vanillin was improved up to fivefold | X2180-1A (MATa his3D1 leu2D0 met15D0 ura3D0 adh6::LEU2 bgl1::KanMX4 PTPI1::3DSD [AurC]::HsOMT [NatMX]::ACAR [HphMX]) | [42] | |
| Se-methylselenocysteine | Combination of metabolic (codon optimization of heterologous selenocysteine methyltransferase) and bioprocess (tuning carbon-and sulfate-limited fed-batch) engineering achieved 24-fold increase in Se-methylselenocysteine production | CEN.PK113-7D (MATa MAL2-8C SUC2) | [43] | |
| Non-ribosomal peptides | Separated non-ribosomal peptide synthetase modules with compatible communication-mediating domains showed functional interaction, which meant that new module combinations could produce novel non-ribosomal peptides | CEN.PK113-11C (MAT a MAL2-8c SUC2 ura3-52 his3-D1) | [44] | |
| Protein drugs | Insulin-like growth factor 1 (fhlGF-1) | Inactivation of GAS1 increased the yield of human insulin-like growth factor1, from 8 to 55 mg/l | GcP3 (MAT a pep4-3 prb1-1122 ura3-52 leu2 gal2 cir°) | [45] |
| Glucagon | Disruption of YPS1 encoded aspartic protease increased glucagon, 17.5 mg/l | SY107 (MATα YPS1 Δtpi::LEU2 pep4-3 leu2 Δura3 cir +) | [46] | |
| Single-chain antibodies (scFv) | Production of an anti-transferrin receptor single-chain antibody (OX26 scFv) was optimized by adjusting expression temperature and gene dosage and final yield was 0.5 mg/l | BJ5464 (MATa ura3-52 trp1 leu2D1 his3D200 pep40HIS3 prb1D1.6R can1 GAL) | [47] | |
| Hepatitis surface antigen (HBsAg) | Glyceraldehyde-3-phosphate dehydrogenase (GAP) promoter of Pichia pastoris was used for HBsAg production and final yield was 19.4 mg/l | INVSc1 (MATa his3D1 leu2 trp1-289 ura3-52) | [48] | |
| Parvovirus B19 VP2 | The major-capsid protein VP2 of Parvovirus B19 produced in S. cerevisiae showed similar properties to native virus or produced by baculovirus system in size, molecular weight, and antigenicity. The yield was 400 mg/l | HT393 (MATa leu2-3 leu2-112 ura3Δ5 prb1-1 prc1-1 pra1-1 pre1-1) | [49] | |
| Epidermal growth factor (EGF) | O-glycosylation pathway was constructed by introduction of GFR (GDP-fucose transporter), POFUT1 (O-fucosyltransferase 1), manic fringe gene (β1,3-N-acetylglucosaminyltransferase) from human and MUR1 (GDP-mannose-4,6-dehydratase), AtFX/GER1(GDP-4-keto-6-deoxy-mannose-3,5-epimerase/4-reductase) from Arabidopsis thaliana producing O-glycosylated EGF protein |
W303-1A (MATa leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100) W303-1B (MATα leu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-100) |
[50] | |
| Immunoglobulin G | Leader peptides for the enhanced secretion of proteins constructed by directed evolution allowed for a 180-fold increase in secretion of full-length, functional, glycosylated human IgG | BJ5464a (MATα ura3-52 leu2~1 his3~200 pep4::HIS3 prb1~1.6Rcan1 GAL) | [51] | |
| Hepatitis B virus surface antigen (HBsAg) | The yield of S domain of hepatitis B virus surface antigen (sHBsAg) was increased by co-expression of disulfide isomerase (PDI1) with adjusting fermentation mode | S. cerevisiae 2805 (MATα pep4::HIS3 prb-Δ1.6 his3 ura3-52 gal2 can1) | [52] | |
| L1 protein of human papillomavirus (HPV) type16 | Optimization of the secondary structure of HPV16 L1 mRNA increased the expression level of that protein up to fourfold than of wild-type | S. cerevisiae 2805 (MATα pep4::HIS3 prb-Δ1.6 his3 ura3-52 gal2 can1) | [53] |
Table 2.
Overall analysis of patents
| Search words or phrases in title and abstract | S. cerevisiae | Yeast | E. coli | Fermentation |
|---|---|---|---|---|
| Strains or fermentation | 4,630 | 13,769 | 14,914 | 9,065 |
| Productiona | 3,080 | 7,812 | 9,306 | 5,211 |
| Pharmaceuticalsa | 985 | 2,686 | 3,343 | 956 |
| Food ingredientsa | 24 | 41 | 21 | 60 |
| Chemicala | 1,702 | 4,830 | 5,683 | 2,651 |
| Production and chemicala | 1,342 | 3,367 | 4,003 | 1,899 |
| Production and fine chemicala | 58 | 64 | 80 | 19 |
| Fuela | 66 | 145 | 110 | 369 |
| Production and fuela | 59 | 126 | 94 | 321 |
US Granted 5,266,192 patents searched (July 31, 2011)
aSearch term: Strains or fermentation and a keyword
Here we will review recent advances in the use of S. cerevisiae for production of novel fuels and chemicals with a focus on bio-butanol and isoprenoids. Considering the importance of using pentoses in future biorefineries we will, however, also review work on expanding the substrate range of yeast. Much of the work we will be reviewing has been carried out within companies, and there is therefore limited information in the published literature, and we will therefore to a large extent rely on patents and patent applications. We chose to focus on butanol as an example of an advanced biofuel and isoprenoids as examples of a class of valuable biochemical, as there have been much development within several companies in the production of these compounds recently.
Extended substrate range
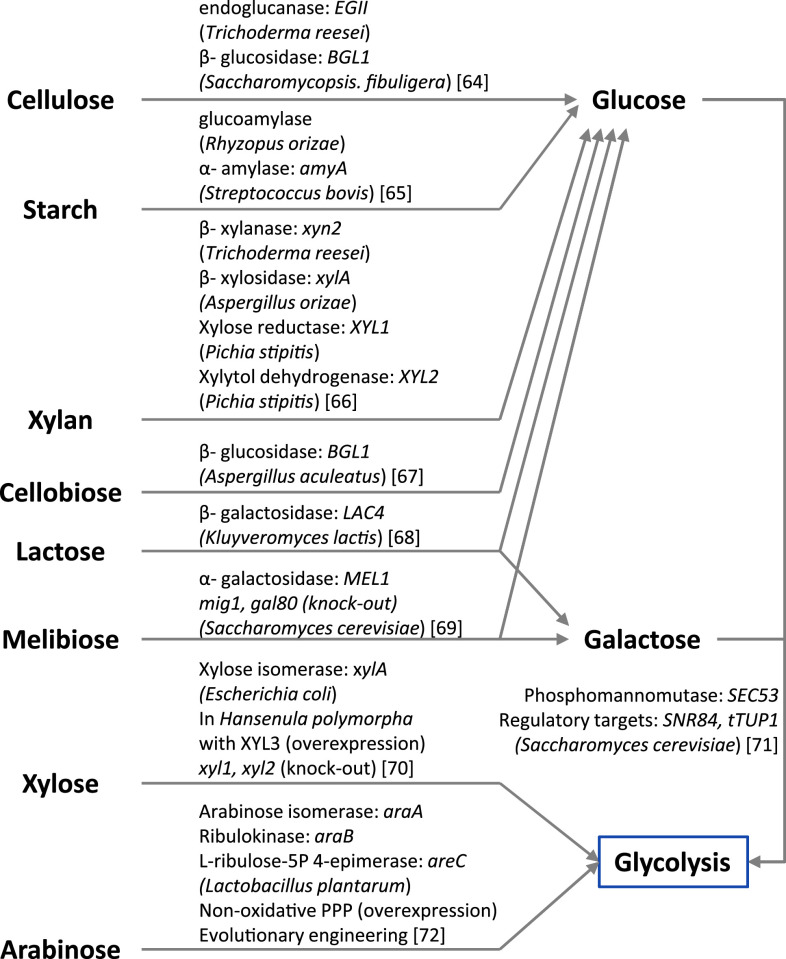
Ethanol is a first-generation biofuel that is being used either in pure form or blended with gasoline, and it is primarily being produced using S. cerevisiae. Most of the currently produced fuel ethanol is produced in Brazil and the US. In 2009, about 55% of the world’s ethanol production was in Brazil, where the main carbon source was sugarcane, while about 35% is produced in the USA through corn fermentation [58]. To avoid the resource competition between food and fuel, there are demands for the use of the abundant and sustainable non-food resource such as switchgrass and waste cheese whey as well as agricultural by-products like corn-cob and bagasse. Currently available non-food resources are not sufficient to fully replace the fuel produced from oil; however, several efforts to increase non-food biomass have been implemented [59, 60]. These biomass resources comprise diverse types of carbon structures such as polymers (cellulose, starch, xylan), dimers (cellobiose, melibiose, lactose) and monomers (glucose, fructose, galactose, arabinose, xylose). Except for the hexoses (glucose, fructose, galactose) and a few dimmers (sucrose and maltose), most of these compounds are not naturally metabolized by S. cerevisiae. Even among the hexoses there are wide differences in terms of uptake, e.g., the uptake rate of galactose is much lower than for the other hexoses. Therefore, the extension of substrate range in S. cerevisiae has been a major priority in connection with the use of yeast for biofuel and biochemical production, and this has recently been covered by several reviews [14, 61–63]. An overview of alternative substrates is provided in Fig. 3. Recently, since tools from systems biology have been available, they have been applied to identify new target genes and understand metabolism at the whole-cell level. Comparative genomics among xylose-fermenting fungi were used to identify new pathways or genes for increasing xylose utilization [73]. The capacity of xylose utilization by those genes was demonstrated by engineering S. cerevisiae, and genome-scale modeling was implemented to assess that global flux analysis could predict the effect of co-factor balancing for pentose utilization [74]. In the following section, we review recent work improving galactose utilization and co-fermentation of cellobiose/xylose.
Fig. 3.
Illustration of relevant substrates that have been considered for yeast fermentation. Heterologous enzymes that are currently applied are summarized for non-utilizable carbon sources in S. cerevisiae such as polymers (cellulose, starch, xylan), disaccharide (cellobiose, lactose, melibiose), pentose sugar (xylose, arabinose). In case of galactose, which is utilized slowly compared to glucose, over-expression targets of innate enzymes for improving galactose availability were screened
The modification targets to improve galactose utilization have been well elucidated in S. cerevisiae, which include engineering of the regulatory network (inactivation of repressors and up-regulation of activator) and over-expression of the final enzyme, phosphoglucomutase (PGM2) in the Leloir pathway responsible for galactose catabolism [75–77]. All these targets were directly associated with the galactose metabolic pathway, but using a cDNA library, another target that is not part of galactose metabolism was also found [71]. In this study, three beneficial over-expression targets, SEC3, tTUP1, and SNR84 were identified. Although two of them were confirmative with previous works due to the function of those genes, SEC3 (phosphomannomutase having activity as phosphoglucomutase) and truncated TUP1 (repressor); the last target was unpredicted. SNR84 codes for H/ACA box small nucleolar RNA, and higher activity of phosphoglucomutase in the transformant over-expressing SNR84 proposed a relationship between this gene and galactose metabolism. Recently, evolutionary engineering was also implemented to find unforeseen targets using systems biology [78]. The combination of different systems biology techniques enabled linking phenotype and genotype. Also, to identify true-positive targets, three different evolved clones on galactose were compared, and all evolved mutants showed higher transcripts and metabolite level in storage carbohydrate metabolism together with up-regulation of phophoglucomutase, whereas there were no mutations in any of the GAL-genes and PGM2 including 1kb up- and downstream. However, based on analysis of all three mutants, the Ras2/PKA signaling pathway was strongly suggested to induce the observed phenotypic changes because this signaling pathway has mutations commonly in all three evolved mutants. In another study, co-fermentation of cellobiose and xylose was implemented by co-expression of cellodextrin transporter (cdt-1), β-glucosidase (gh1-1), and xylose enzymes (XYL1, mXYL1, XYL2, and XKS1) [79]. mXYL1 is a mutant enzyme that exhibits much higher preference for NADH as co-factor unlike the wild-type XYL1 that prefers the use of NADPH as co-factor. Introduction of these genes minimized glucose repression of xylose fermentation, since glucose was generated after transporting of cellobiose by the cellodextrin transporter and degraded by β-glucosidase inside the cell. These modifications allowed S. cerevisiae to co-ferment cellobiose and xylose with improved ethanol yield.
There are also indications of industrial application of pentose metabolism by yeast, in particular the utilization of the pentose sugars xylose and arabinose, as several patents cover the metabolism of these sugars [80–87]. There are two Dutch companies that have patents on the use of these two carbon sources, namely DSM and Royal Nedalco. Both focus on isomerase-based pathways to avoid the co-factor balancing problem in xylose and arabinose utilization. DSM over-expressed xylose isomerase from Thermotoga maritime MSB8 and arabinose genes (araA, araB, and araC) from Lactobacillus plantarum with codon optimization and constitutive expression of the genes in the non-oxidative pentose phosphate pathway [82, 83, 86]. Royal Nedalco employed xylose isomerase of Piromyces sp. E2 (ATCC 76762) and arabinose genes of Arthrobacter aurescens, Clavibacter michiganensis, or Gramella forsetii [80, 81, 85]. The main focus of both approaches was application of higher activity of isomerases from heterologous sources (Fig. 4).
Fig. 4.
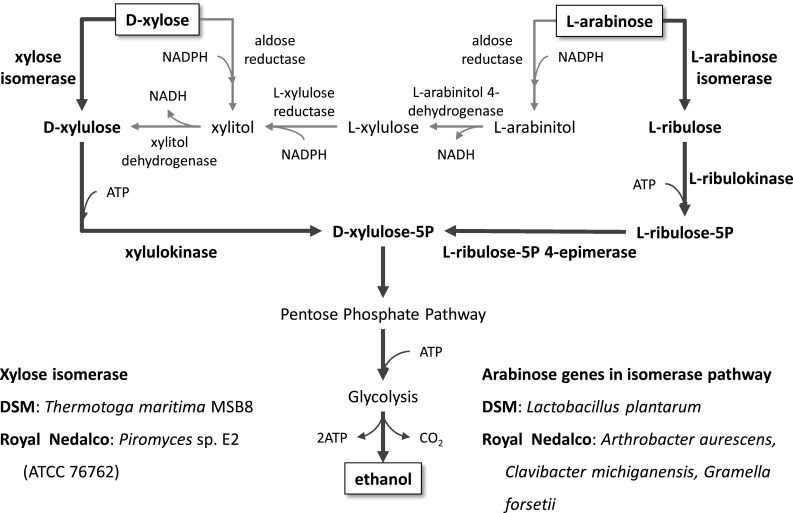
Overview of pathways for pentose utilization covered by patent applications of DSM and Royal Nedalco. d-xylose and l-arabinose can be utilized by two pathways: (1) aldose reductase NADPH-dependent and (2) isomerase cofactors-independent. In case of the latter, requirement of cofactor balance is eliminated and enhancing activity of isomerase remains main issue. Two Dutch companies, DSM and Royal Nedalco, claim over-expression of heterologous xylose isomerases from Thermotoga maritime MSB8 and Piromyces sp. E2 (ATCC 76762), respectively. Xylulokinase and enzymes in pentose phosphate pathway were also amplified simultaneously. Arabinose genes in isomerase pathway such as l-arabinose isomerase, l-ribulokinase, and l-ribulose-5P 4-epimerase originated from Lactobacillus plantarum in DSM and Arthrobacter aurescens, Clavibacter michiganensis, Gramella forsetii in Royal Nedalco were amplified with enzymes in pentose phosphate pathways
Bio-butanol production
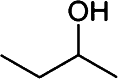
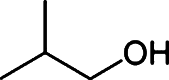
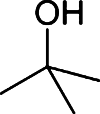
Butanol has gained much attention as a potential biofuel to replace ethanol, currently by far the dominating biofuel. Butanol has a number of advantages as a biofuel compared to ethanol. It has a higher energy density than ethanol and only around 4% less than that of gasoline [88–92]. Furthermore, butanol blends better with gasoline than with ethanol and it is non-corrosive, which allows it to be used in the existing petrochemical infrastructure. There are four different types of butanol (Table 3): n-butanol, sec-butanol, isobutanol, and tert-butanol. n-butanol and sec-butanol have a stretched carbon chain and a hydroxyl group at position 1 or 2. Isobutanol and tert-butanol have two or three branched carbon chains, respectively. The branched structure results in a higher octane number [93] (Table 3), which means a higher anti-knock property. Although tert-butanol has a higher octane number than isobutanol, its much higher melting temperature (25°C) than that of isobutanol (−101.9°C) prohibits its use as a pure fuel source. Isobutanol is therefore preferred over the other butanols.
Table 3.
Comparison of butanol isomers [49]
|
|
|
|
|
|
|---|---|---|---|---|
| n-butanol | sec-butanol | Isobutanol | tert-butanol | |
| Research octane number (RON) | 96 | 101 | 113 | 105 |
| Motor octane number (MON) | 78 | 32 | 94 | 89 |
| Melting temperature (°C) | −89.5 | −114.7 | −108 | 25.7 |
| Boiling temperature (°C) | 117.7 | 99.5 | 108 | 82.4 |
| Enthalpy of vaporization at Tboil (kJ/kg) | 582 | 551 | 566 | 527 |
Butanol is naturally produced by different species of Clostridia, but most of these processes are limited by relatively low yields and titers, and therefore much interest has been expressed in the development of novel cell factories that can be used for bio-based production of butanol [89, 94, 95]. Mainly, two strategies for producing bio-butanol have been used, (1) use of a host that has an innate butanol pathway and improving its yield and productivity, and (2) re-construction of an efficient butanol pathway in strains that are already widely used for industrial production of other fuels and chemicals. Yeast is one of the hosts that produce butanol naturally through the so-called Ehrlich pathway for fusel alcohol production [96, 97]. Also, yeast is widely used for industrial ethanol production due to its high ethanol tolerance and its robustness towards harsh industrial conditions, e.g., high osmotic stress and low pH [14, 98]. In academic papers, however, most results presented have been based on work using Clostridia or E. coli [99, 100]. So far, only two papers have been published on the use of yeast for bio-butanol production, one describing n-butanol production based on reconstruction of a pathway from Clostridia and the other describing iso-butanol production based on engineering of yeast’s natural pathway, and both presented very low yields and productivities [18, 101]. There are several recent reviews that summarized these academic studies [88, 94], and from comparisons of results on different organisms yeast do not seem to be an attractive host for bio-butanol production because of its low yield and productivities, especially compared to metabolically engineered E. coli. However, in contrast to academic research, most of the companies announcing work towards commercial production of bio-butanol are using yeast as a production organism [90, 91, 102–104]. We will therefore here review the main strategies employed in industry based on analysis of information provided in patents and patent applications. Based on patent applications and issued patents, the three dominating companies for producing bio-butanol by yeast are presently Gevo, Butamax Advanced Biofuels, and Butalco.
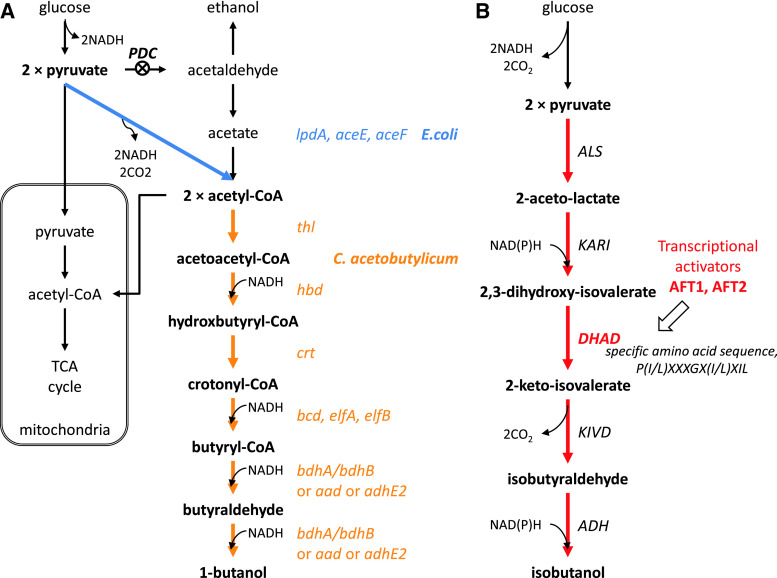
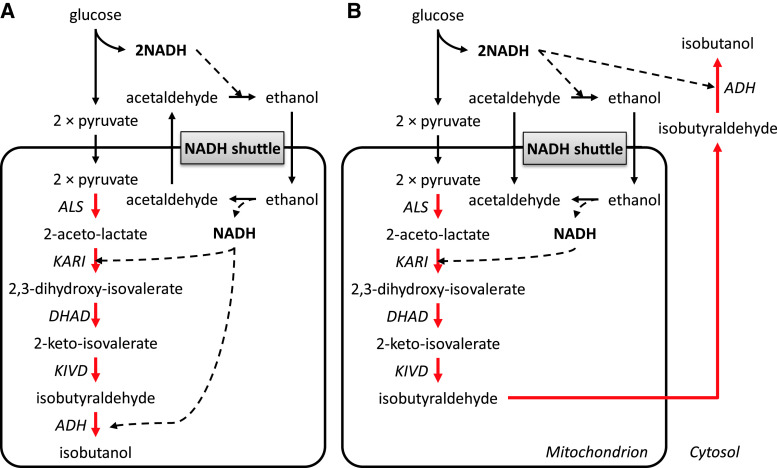
Gevo is one of the pioneers in bio-butanol production, and they received a combined grant from the US Department of Agriculture and the Department of Energy for developing a yeast fermentation system to produce iso-butanol from cellulosic-derived sugars. Gevo’s patents describe different strategies to produce n-butanol and isobutanol. First, to produce n-butanol, they propose to increase cytosolic acetyl-CoA pool and incorporate the butanol synthetic pathway from Clostridia species into yeast [91]. To increase the cytosolic acetyl-CoA pool, the pyruvate dehydrogenase multienzyme complex (lpdA, aceE, aceF) from E. coli was expressed to establish a direct pathway from pyruvate to acetyl-CoA not passing via acetaldehyde that can be converted to ethanol. In addition, the activity of pyruvate decarboxylase (PDC), which converts pyruvate to acetaldehyde, is reduced (Fig. 5a). Second, isobutanol production has been tried intensively using different strategies [105, 106]. As mentioned previously, yeast can naturally produce isobutanol, and this pathway shares valine synthesis from pyruvate to 2-keto-isovalerate in the mitochondria. 2-keto-isovalerate can be exported from the mitochondria to the cytosol where decarboxylation by pyruvate decarboxylase (PDC) and dehydrogenation by alcohol dehydrogenases (ADH) can lead to production of isobutanol [18]. Gevo worked on reconstructing the isobutanol pathway in the mitochondria and the cytosol separately using innate yeast or heterologous genes. Using a mitochondrial targeting sequence, all enzymes involved in isobutanol synthesis were localized in the mitochondria and isobutanol was produced there (Fig. 6a). In this strategy, cofactor balancing was considered important, since NADH is produced in the glycolysis and NADPH is needed for isobutanol production. First, NADPH-dependent enzymes were engineered to an NADH-dependent form, and then NADH was supplied by using a NADH shuttle concept [107]. Acetaldehyde and ethanol produced by fermentation can freely transport across membranes, and alcohol dehydrogenase in the mitochondria (encoded by ADH3) can provide one NADH by conversion of ethanol to acetaldehyde. Moreover, isobutyraldehyde, an intermediate of the isobutanol pathway, can be transported to the cytosol where it can be converted to isobutanol under consumption of NADH generated by glycolysis in the cytosol (Fig. 6b). To avoid cofactor balancing in the mitochondria, the whole isobutanol pathway was in another strategy expressed in the cytosol [90, 105, 108]. In this case, modification or amplification of dihydroxyacid dehydratase (DHAD) was emphasized, and obtained from Lactococcus lactis and Neurospora crassa. Additionally, the transcriptional activators AFT1/AFT2 are over-expressed to increase DHAD activity (Fig. 5b). Gevo is currently developing 18 million gallons per year (MGPY) plant in the USA and have plans to develop 350 MPGY of new capacity by 2015.
Fig. 5.
Illustration of Gevo’s strategies for n-butanol and isobutanol production in the cytosol [90, 91, 105]. a n-butanol production was attempted by amplification of heterologous genes such as the pyruvate dehydrogenase multienzyme complex (lpdA, aceE, aceF) from E. coli for increasing the cytosolic acetyl-CoA pool, and the genes in butanol synthetic pathway from Clostridia species. Moreover, the activity of pyruvate decarboxylase (PDC) was reduced. b Isobutanol was produced in the cytosol to avoid cofactor balancing in the mitochondria; all the genes in isobutanol pathway were over-expressed in cytosol. Especially, dihydroxyacid dehydratases (DHAD) from Lactococcus lactis and Neurospora crassa were used, which had specific amino sequence, P(I/L)XXXGX(I/L)XIL. Also, the transcriptional activators AFT1/AFT2 were over-expressed to increase DHAD activity
Fig. 6.
Gevo’s isobutanol production strategy in the mitochondria [106]. a All enzymes involved in isobutanol synthesis were localized in the mitochondria; KIVD and ADH being in cytosol were expressed in mitochondria with signal sequence. NADPH-dependent enzymes were engineered to an NADH-dependent form, and then NADH was supplied by using an NADH shuttle concept. Acetaldehyde and ethanol produced by fermentation were transported across membranes, and alcohol dehydrogenase in the mitochondria (encoded by ADH3) provided NADH by conversion of ethanol to acetaldehyde. b Isobutyraldehyde was transferred to the cytosol from mitochondria where it is converted to isobutanol under consumption of NADH generated by glycolysis in the cytosol to make more precise cofactor balance
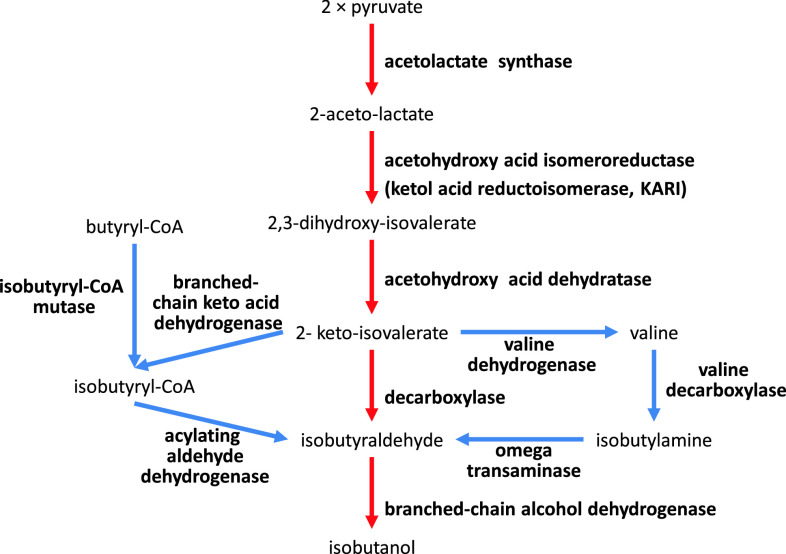
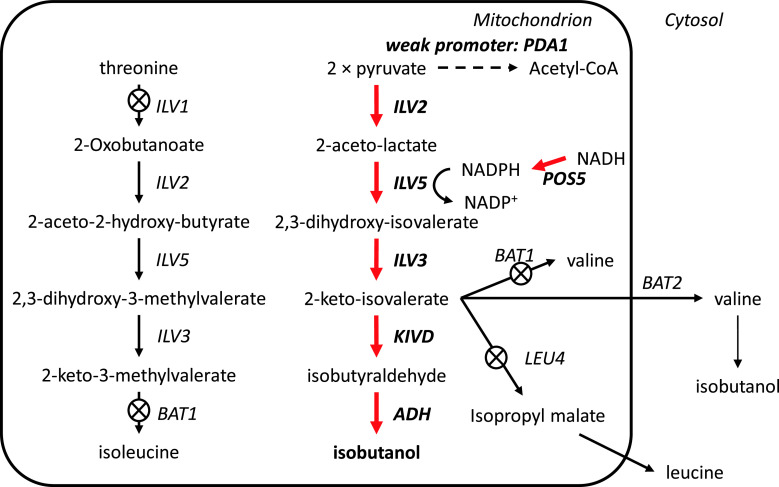
Butamax Advanced Biofuels was formed in 2009 as a joint venture between Dupont and BP, two large companies in the chemical and energy industry, respectively. Before establishing Butamax, Dupont investigated many different approaches in strain development for n-butanol, sec-butanol, and isobutanol production [109–111]. Consequently, one of their foundational patents for isobutanol production was officially granted in August 2011 by the US Patent and Trademark Office [102]. Dupont’s (and now Butamax’s) strategy is to find and introduce many different heterologous genes related to butanol biosynthesis and introduce these into yeast. For isobutanol production they considered four different pathways, which include conversion of valine to isobutylamine and butyryl-CoA to isobutyryl-CoA (Fig. 7). A total of 11 enzyme reactions were considered and each reaction could be catalyzed by at least three to four heterologous enzymes. For example, acetolactate synthase (ALS), which catalyzes the first step, could be Klebsiella pneumoniae budB, Bacillus subtilis alsS, or Lactococcus lactis als. Key findings claimed were the use of Pseudomonas ilvC (ketol acid reductioisomerase: KARI) and a method to amplify ilvD (DHAD), which contains a Fe–S cluster. Isobutanol production in mitochondria was also considered [109, 112, 113]. Here, the genes involved in substrate competing reactions, BAT1, ILV1, and LEU4 were deleted and the activity of the E1 alpha subunit of the pyruvate dehydrogenase (PDH) complex (PDA1), which converts pyruvate to acetyl-CoA, was reduced by promoter exchange. Furthermore, NADH kinase (POS5) was over-expressed to ensure sufficient supply of NADPH required by the KARI enzyme (Fig. 8). Butamax also investigated the butanol tolerance by modification of the regulatory network [114–118]. Yeast, especially S. cerevisiae, is known to have higher butanol tolerance than other microorganisms and it can grow in butanol concentrations higher than 20 g/l [119], but still several targets were identified for improving butanol tolerance and these are summarized in Table 4. The first operational plant for commercial production of isobutanol by Butamax is scheduled to be operational by 2013.
Fig. 7.
Butamax’s strategies for isobutanol production in cytosol [102]. Four different pathways for isobutanol production were suggested: (1) pyruvate to isobutanol directly (red arrows), (2) pyruvate through valine bypass (blue arrows), (3) pyruvate through isobutyryl-CoA bypass (blue arrows), and (4) butyryl-CoA to isobutanol (blue arrows). A total of 11 enzyme reactions were considered and at least three to four heterologous enzymes in each step were claimed in patents of Butamax
Fig. 8.
Butamax’s isobutanol production strategies in the mitochondria [103]. To block substrate-competing reactions BAT1, ILV1, and LEU4 were deleted and the activity of the E1 alpha subunit of the pyruvate dehydrogenase (PDH) complex (PDA1) was reduced by promoter exchange to a weak one. NADH kinase (POS5) was over-expressed to ensure sufficient supply of NADPH required by the KARI enzyme. Red arrows mean over-expression of genes
Table 4.
Targets for increasing butanol tolerance in yeast (Butamax)
| Targeting | Modified genes | Butanol tolerance [growth yield improvement in butanol % (w/v)] | References |
|---|---|---|---|
| Multidrug resistance ATP-binding cassette transporter | Pdr5p, CDR1, BFR1 | ~1.8-fold in 0.75% | [114] |
| Cell wall integrity pathway | SLT2p | ~25% in 1% | [115] |
| Osmolality/glycerol response pathway | PBS2p | ~40% in 1% | [116] |
| Filamentous growth response pathway | MSS11p | ~2-fold in 1.5% | [117] |
| Amino acid starvation |
Gcn1p, Gcn2p, Gcn3p, Gcn4p, Gcn5p, Gcn20p |
~1.8-fold in 2.0% | [118] |
The third company working actively on bio-butanol production by yeast is Butalco, which is a biofuel company that also develops ethanol producing yeast that can use xylose and arabinose as carbon sources. Isobutanol production strategies by this company are not to use heterologous genes; but rely solely on endogenous yeast genes [104]. As mentioned above, yeast has all enzymes necessary for isobutanol production; and Butalco is employing its core technology for genetic optimization of yeast. Four points are mainly considered in their strategy: (1) different intracellular location of the isobutanol synthetic enzyme, (2) the weak activity of the enzymes, (3) cofactor imbalance, and (4) the formation of secondary products. All isobutanol synthetic genes were suggested to be express in the cytosol using strong promoters and the activity of pyruvate decarboxylase (PDC), which catalyzes pyruvate to ethanol reactions, were removed or reduced. To ensure cofactor balance, the acetohydroxy acid reducto-isomerase (KARI) and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were proposed to ensure balancing of the co-factors. Either NADH-dependent yeast KARI enzyme (ILV5 NADH) could use NADH from the glycolysis or NADP+-dependent yeast GAPDH (GLDs) could produce NADPH required by NADPH-dependent KARI enzyme (ILV5).
Isoprenoids production
Isoprenoids are a chemically diverse group of natural compounds that have many different biological functions, and they have found applications as medicines, perfumes, food additives, and fine chemical intermediates [120]. Recently, the possibility that they can be applied as fuels has been discussed [121]. Although the standard definition of isoprenoids has not been not clearly set, the compounds that have isopentenyl diphosphate (IPP) or dimethylallyl diphosphate (DMAPP) as a unit molecule are typically categorized into isoprenoids. Those unit molecules have five carbons and the combination of them can generate bigger unit molecules such as geranyl diphosphate (GPP, ten carbons), farnesyl diphosphate (FPP, 15 carbons), and geranylgeranyl diphosphate (GGPP, 20 carbons), which are precursors for monoterpenes, sesquiterpenes and diterpenes, or carotenoids, respectively [122].
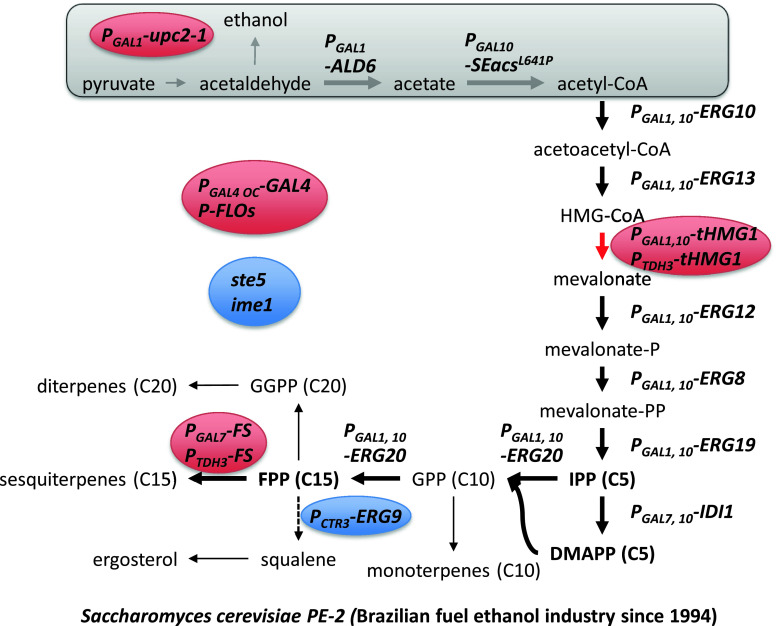
The production of isoprenoids has been limited by insignificant quantity in their natural sources, mostly plants and low yield of extraction. Moreover, the diversity of isoprenoids is enormous, which means that new bioactive compounds can be produced by finding new enzymes and they can even be used in combination to make chimeric pathways resulting in novel compounds [123]. For these reasons, there has been much interest in developing microbial-based production of isoprenoids by re-creating a plant-like pathway in yeast with genetic modifications of leader sequence and codon-optimization for re-locating and overexpression of relative genes. Two biosynthetic pathways for IPP and DMAPP are known—the mevalonate-dependent (MVA) pathway that converts acetyl-CoA to IPP and the deoxyxylulose-5-phosphate (DXP) pathway that converts glyceraldehyde-3-phosphate and pyruvate to IPP/DMAPP. The MVA pathway is present in the cytosol and mitochondria of plants and yeast, while the DXP pathway is found in bacteria. In higher plants, both pathways are present; with the DXP pathway being present in chloroplast, probably of bacterial origin [120]. Extensive endeavors have focused on (1) optimization of these pathways for increasing the metabolic flux of isoprenoids and (2) discovery of heterologous enzymes for production of different isoprenoid-based products. As a result, there have been remarkable achievements; especially, elucidation of key targets for metabolic engineering of the MVA pathway in yeast such as over-expression of truncated HMG1 (tHMG1), upc2-1, and ERG20 combined with repression of ERG9 [20] (Fig. 9). In addition to optimization of the MVA pathway, a strategy for the increase of the acetyl-CoA pool has also been implemented by over-expression of ALD6 of S. cerevisiae and a constantly active acetyl-CoA synthase mutant from Salmonella enterica [124]. In order to overcome regulation of the MVA pathway, attempts to reconstruct the DXP pathway in yeast have also been evaluated [125, 126]. Much of the work on metabolic engineering of yeast for isoprenoid production has been reviewed recently [122, 123, 127, 128], and we will therefore here focus on industrial applications using analysis of the patent literature.
Fig. 9.
Overview of Amyris metabolic engineering strategies. Industrial strain Saccharomyces cerevisiae PE-2 was used as a production host because of its higher tolerance to the industrial environment [129]. All promoters of mevalonate genes were exchanged to strong one in chromosome. Gray box means the strategies that were used in a scientific article [124] but not in the patent. Red color circles mean even higher expression than other overexpressed genes. Blue color circles mean knock-out of genes or reduction of expression level. Thick arrows mean amplified steps based on plasmids in a scientific article [20]. The dotted arrow indicates reduction of flux
Isoprenoid and yeast were used as keywords in the Delphion™ Web site that analyzes and clusters relative patents. A list of companies and their patents were generated. Most of patents that are related to the flux increase of isoprenoid precursors (FPP) are assigned to University of California and Amyris, and is mainly derived from work of Jay Keasling’s research group. Many of the strategies described in the patents are very similar to published scientific articles, but there are a few additional important strategies described for improving isoprenoid production at an industrial scale [129, 130] (Fig. 9). Firstly, the production host was selected among industrial strains, resulting in selection of Saccharomyces cerevisiae PE-2, which has been used in the Brazilian fuel ethanol industry since 1994. This strain was selected because of its higher tolerance to industrial fermentation conditions and its ability to tolerate yeast recycling typically used in Brazilian ethanol production plants. The range of tolerance includes high ethanol concentration, high cell density, high temperature, osmotic stress, low pH and sulfite, and bacterial contamination [131]. Secondly, all genetic modifications were done at the chromosomal DNA level and not using plasmids as is often done in academic research groups [20, 132]. In order to ensure strong expression of the genes, the promoters of all the genes in the MVA pathway were changed. In the case of tHMG1, which should be higher expressed than HMG-CoA synthase to avoid accumulation of HMG-CoA [133], one more copy with another strong promoter (PTDH3) was integrated into the chromosome. These changes are comparable to previously published papers, which used over-expression of upc2-1 to increase the expression level of genes in the MVA pathway. Over-expression of GAL4 and knock-out of GAL80 were constructed to induce expression of the genes under GAL1, 7, and 10 promoters. Especially, the modified GAL4 promoter (PGAL4 OC) that has no MIG1 binding site was used for induction of GAL4 expression in fermentation media containing glucose [134]. Repression of ERG9 (squalene synthase) was done by replacing the native promoter to the PCTR3, which is controlled by copper. Formerly, this repression was performed by using the methionine repressible promoter PMET3, a strategy that has been used by different research groups in academia [20, 34, 135]. Ergosterol is the end-product from IPP/DMAPP condensation in yeast (Fig. 9). Reducing the activity of Erg5 theoretically enables an increase in flux to other terpenes while cellular growth can be maintained. Thirdly, additional modifications were performed in other pathways besides the MVA pathway. To facilitate the purification process, flocculation proteins (FLOs) were over-expressed. Furthermore, sporulation (IME1) and endogenous mating were impaired by disruption of responsible genes. The pheromone response genes (STEs) were also functionally inactivated in order to prevent mating. These three strategies were simultaneously implemented in one patent application [129]. Other strategies are also described in the patent literature, e.g., to increase C1 metabolism serine, which is a precursor of this metabolism, were supplied to the media [136] and GTR reductase and ALA synthase were over-expressed to increase the heme pool [137]. Besides genetic modification, Amyris also developed a monitoring method that can concurrently quantify cofactors, energy molecules, and intermediates in the MVA pathway by using LC/MS/MS [138]. This technique allows observation of the metabolite concentration change in the overall pathway during the fermentation process, and this was found very useful in finding steps that needed further modification.
Other companies that have patents on increasing isoprenoid production are Arkion Life Science and Allylix, and these patents mostly focus on the development of highly active HMGR (HMG-CoA reductase, HMG1, HMG2) and reduction of ERG9, which have already been recognized as critical targets [139, 140]. HMGR is the main enzyme controlling the flux through the MVA pathway. ERG9 encodes squalene synthase that converts FPP to squalene, and by attenuating the ERG9 expression, the FPP pool can be increased and flux can be directed towards sesquiterpenes. However, yeast cannot grow without ERG9 at aerobic condition, even if ergosterol is fed to the medium, and generation of erg9 mutants that can grow aerobically with ergosterol in the medium were developed by these companies. Dupont also holds many patents related to isoprenoids production, but most of these are related to the identification of novel enzymes for conversion of FPP to different isoprenoids [141–144].
Conclusions
Based on our review of ongoing metabolic engineering projects in industry and academy, it is clear that there are several high-profile projects ongoing on using S. cerevisiae for the production of novel fuels and chemicals. The vast knowledge on this organism combined with the robustness of this organism to harsh industrial conditions makes it a preferred organism for production of many fermentation-derived products. Compared to E. coli, which is widely used in academia, yeast cannot be contaminated by phages. It is very osmo-tolerant and can hence tolerate very high sugar concentrations, and it tolerates a lower pH than most bacteria. These advantages typically outweigh the fact that compared to E. coli it is more difficult to engineer yeast and that the rates of conversion typically are lower in yeast. Despite the extensive synthetic and systems biological toolbox available for yeast, it is necessary to further advance technologies that can be used in metabolic engineering. Still strain development is time-consuming and often serial in nature, meaning that development time cannot be reduced through increased resources for a short time. Among the novel technologies that would allow for faster development are tools for deletion of several genes in one round of transformation, a larger promoter library, in particular better characterization of promoters and their activity at different industrially relevant fermentation conditions, inducible promoters, and tools for rapid identification of how flux through different metabolic pathways are controlled [4, 13]. With the development of such tools combined with experience from development of yeast-based fermentation processes for novel biofuels like isobutanol and farnesene we are, however, confident that the way will be paved for faster development of other processes for sustainable production of fuels and chemicals to the benefit of society. This may lead to the establishment of yeast platform strains that can use a range of relevant sugars efficiently, i.e., sucrose, glucose, galactose, xylose, and arabinose, and efficiently convert each of these sugars to relevant building blocks that serve as precursors for different types of industrially relevant products. Such yeast platforms will have great prospects for application in biorefineries, where the same yeast platform can then be combined with different product formation pathways, and hereby the biorefinery can use a plug-and-play solution for production of a range of different products, and hence allow for easy adjustment to the market developments. Key factors for major advancement towards this scenario will be the ability to construct new strains faster, the improved ability to establish stable strains that can be used in industrial conditions, and focus on using yeast strains that have industrial relevance. Concerning the latter, there has fortunately been development towards a wider use of the CEN.PK yeast strain background, which, besides serving as a good laboratory strain, has excellent performance for industrial use [145]. Even though in the last 10 years there have been increased interactions between academia and industry, it will be necessary to strengthen these ties, as this will ensure faster implementation of novel developments as well as gaining fundamental understanding of cellular processes that are relevant for industrial processes.
References
- 1.Nielsen J. Metabolic engineering. Appl Microbiol Biotechnol. 2001;55(3):263–283. doi: 10.1007/s002530000511. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen J, Keasling JD. Synergies between synthetic biology and metabolic engineering. Nat Biotechnol. 2011;29(8):693–695. doi: 10.1038/nbt.1937. [DOI] [PubMed] [Google Scholar]
- 3.Tyo KE, Alper HS, Stephanopoulos GN. Expanding the metabolic engineering toolbox: more options to engineer cells. Trends Biotechnol. 2007;25(3):132–137. doi: 10.1016/j.tibtech.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Tyo KE, Kocharin K, Nielsen J. Toward design-based engineering of industrial microbes. Curr Opin Microbiol. 2010;13(3):255–262. doi: 10.1016/j.mib.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen J, Jewett MC. Impact of systems biology on metabolic engineering of Saccharomyces cerevisiae . FEMS Yeast Res. 2008;8(1):122–131. doi: 10.1111/j.1567-1364.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- 6.Conrad TM, Lewis NE, Palsson BO. Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol. 2011;7:509. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petranovic D, Vemuri GN. Impact of yeast systems biology on industrial biotechnology. J Biotechnol. 2009;144(3):204–211. doi: 10.1016/j.jbiotec.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, Kim TY, Jang YS, Choi S, Lee SY. Systems metabolic engineering for chemicals and materials. Trends Biotechnol. 2011;29(8):370–378. doi: 10.1016/j.tibtech.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Saling P. Eco-efficiency analysis of biotechnological processes. Appl Microbiol Biotechnol. 2005;68(1):1–8. doi: 10.1007/s00253-005-1951-0. [DOI] [PubMed] [Google Scholar]
- 10.Chandel AK, Singh OV. Weedy lignocellulosic feedstock and microbial metabolic engineering: advancing the generation of ‘Biofuel’. Appl Microbiol Biotechnol. 2011;89(5):1289–1303. doi: 10.1007/s00253-010-3057-6. [DOI] [PubMed] [Google Scholar]
- 11.Elkins JG, Raman B, Keller M. Engineered microbial systems for enhanced conversion of lignocellulosic biomass. Curr Opin Biotechnol. 2010;21(5):657–662. doi: 10.1016/j.copbio.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Wilson DB. Microbial diversity of cellulose hydrolysis. Curr Opin Microbiol. 2011;14(3):259–263. doi: 10.1016/j.mib.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Krivoruchko A, Siewers V, Nielsen J. Opportunities for yeast metabolic engineering: lessons from synthetic biology. Biotechnol J. 2011;6(3):262–276. doi: 10.1002/biot.201000308. [DOI] [PubMed] [Google Scholar]
- 14.Nevoigt E. Progress in metabolic engineering of Saccharomyces cerevisiae . Microbiol Mol Biol Rev. 2008;72(3):379–412. doi: 10.1128/MMBR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Jong B, Siewers V, Nielsen J (2011) Systems biology of yeast: enabling technology for development of cell factories for production of advanced biofuels. Curr Opin Biotechnol (in press) [DOI] [PubMed]
- 16.Kim IK, Roldao A, Siewers V, Nielsen J. A systems-level approach for metabolic engineering of yeast cell factories. FEMS Yeast Res. 2012;12(2):228–248. doi: 10.1111/j.1567-1364.2011.00779.x. [DOI] [PubMed] [Google Scholar]
- 17.Guadalupe Medina V, Almering MJ, van Maris AJ, Pronk JT. Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microbiol. 2010;76(1):190–195. doi: 10.1128/AEM.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K. Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol Biofuels. 2011;4:21. doi: 10.1186/1754-6834-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu KO, Jung J, Kim SW, Park CH, Han SO. Synthesis of FAEEs from glycerol in engineered Saccharomyces cerevisiae using endogenously produced ethanol by heterologous expression of an unspecific bacterial acyltransferase. Biotechnol Bioeng. 2012;109(1):110–115. doi: 10.1002/bit.23311. [DOI] [PubMed] [Google Scholar]
- 20.Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 21.Peralta-Yahya PP, Ouellet M, Chan R, Mukhopadhyay A, Keasling JD, Lee TS. Identification and microbial production of a terpene-based advanced biofuel. Nat Commun. 2011;2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee W, Dasilva NA. Application of sequential integration for metabolic engineering of 1,2-propanediol production in yeast. Metab Eng. 2006;8(1):58–65. doi: 10.1016/j.ymben.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Toivari M, Maaheimo H, Penttilä M, Ruohonen L. Enhancing the flux of d-glucose to the pentose phosphate pathway in Saccharomyces cerevisiae for the production of d-ribose and ribitol. Appl Microbiol Biot. 2010;85(3):731–739. doi: 10.1007/s00253-009-2184-4. [DOI] [PubMed] [Google Scholar]
- 24.Zhao L, Wang J, Zhou J, Liu L, Du G, Chen J. Modification of carbon flux in Saccharomyces cerevisiae to improve l-lactic acid production. Wei Sheng Wu Xue Bao. 2011;51(1):50–58. [PubMed] [Google Scholar]
- 25.Zhang B, Carlson R, Srienc F. Engineering the monomer composition of polyhydroxyalkanoates synthesized in Saccharomyces cerevisiae . Appl Environ Microbiol. 2006;72(1):536–543. doi: 10.1128/AEM.72.1.536-543.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Maris AJ, Geertman JM, Vermeulen A, Groothuizen MK, Winkler AA, Piper MD, van Dijken JP, Pronk JT. Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl Environ Microbiol. 2004;70(1):159–166. doi: 10.1128/AEM.70.1.159-166.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raab AM, Gebhardt G, Bolotina N, Weuster-Botz D, Lang C. Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab Eng. 2010;12(6):518–525. doi: 10.1016/j.ymben.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Madsen KM, Udatha GD, Semba S, Otero JM, Koetter P, Nielsen J, Ebizuka Y, Kushiro T, Panagiotou G. Linking genotype and phenotype of Saccharomyces cerevisiae strains reveals metabolic engineering targets and leads to triterpene hyper-producers. PLoS One. 2011;6(3):e14763. doi: 10.1371/journal.pone.0014763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verwaal R, Wang J, Meijnen JP, Visser H, Sandmann G, van den Berg JA, van Ooyen AJ. High-level production of beta-carotene in Saccharomyces cerevisiae by successive transformation with carotenogenic genes from Xanthophyllomyces dendrorhous . Appl Environ Microbiol. 2007;73(13):4342–4350. doi: 10.1128/AEM.02759-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A, Fickes S, Diola D, Benjamin KR, Keasling JD, Leavell MD, McPhee DJ, Renninger NS, Newman JD, Paddon CJ. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci USA. 2012;109(3):E111–118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farhi M, Marhevka E, Masci T, Marcos E, Eyal Y, Ovadis M, Abeliovich H, Vainstein A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab Eng. 2011;13(5):474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Kirby J, Nishimoto M, Park JG, Withers ST, Nowroozi F, Behrendt D, Rutledge EJG, Fortman JL, Johnson HE, Anderson JV, Keasling JD. Cloning of casbene and neocembrene synthases from Euphorbiaceae plants and expression in Saccharomyces cerevisiae . Phytochemistry. 2010;71(13):1466–1473. doi: 10.1016/j.phytochem.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Eudes A, Baidoo E, Yang F, Burd H, Hadi M, Collins F, Keasling J, Loqué D. Production of tranilast [N-(3′,4′-dimethoxycinnamoyl)-anthranilic acid] and its analogs in yeast Saccharomyces cerevisiae . Appl Microbiol Biotechnol. 2011;89(4):989–1000. doi: 10.1007/s00253-010-2939-y. [DOI] [PubMed] [Google Scholar]
- 34.Asadollahi MA, Maury J, Schalk M, Clark A, Nielsen J. Enhancement of farnesyl diphosphate pool as direct precursor of sesquiterpenes through metabolic engineering of the mevalonate pathway in Saccharomyces cerevisiae . Biotechnol Bioeng. 2010;106(1):86–96. doi: 10.1002/bit.22668. [DOI] [PubMed] [Google Scholar]
- 35.Tavares S, Grotkjaer T, Obsen T, Haslam RP, Napier JA, Gunnarsson N. Metabolic engineering of Saccharomyces cerevisiae for production of eicosapentaenoic acid, using a novel {delta}5-desaturase from Paramecium tetraurelia . Appl Environ Microbiol. 2011;77(5):1854–1861. doi: 10.1128/AEM.01935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Millis JRKWI, Maurina-Brunker JAWI, McMullin TWMWI (2003) Production of farnesol and geranylgeraniol. US Patent US 2003/0092144 A1
- 37.Sauer M, Branduardi P, Valli M, Porro D. Production of l-ascorbic acid by metabolically engineered Saccharomyces cerevisiae and Zygosaccharomyces bailii . Appl Environ Microbiol. 2004;70(10):6086–6091. doi: 10.1128/AEM.70.10.6086-6091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rico J, Pardo E, Orejas M. Enhanced production of a plant monoterpene by overexpression of the 3-hydroxy-3-methylglutaryl coenzyme A reductase catalytic domain in Saccharomyces cerevisiae . Appl Environ Microbiol. 2010;76(19):6449–6454. doi: 10.1128/AEM.02987-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mutka SC, Bondi SM, Carney JR, Da Silva NA, Kealey JT. Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae . FEMS Yeast Res. 2006;6(1):40–47. doi: 10.1111/j.1567-1356.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- 40.Albertsen L, Chen Y, Bach LS, Rattleff S, Maury J, Brix S, Nielsen J, Mortensen UH. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl Environ Microbiol. 2011;77(3):1033–1040. doi: 10.1128/AEM.01361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker JV, Armstrong GO, van der Merwe MJ, Lambrechts MG, Vivier MA, Pretorius IS. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res. 2003;4(1):79–85. doi: 10.1016/S1567-1356(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 42.Brochado AR, Matos C, Moller BL, Hansen J, Mortensen UH, Patil KR. Improved vanillin production in baker’s yeast through in silico design. Microb Cell Fact. 2010;9:84. doi: 10.1186/1475-2859-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mapelli V, Hillestrøm PR, Kápolna E, Larsen EH, Olsson L. Metabolic and bioprocess engineering for production of selenized yeast with increased content of seleno-methylselenocysteine. Metab Eng. 2011;13(3):282–293. doi: 10.1016/j.ymben.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Siewers V, San-Bento R, Nielsen J. Implementation of communication-mediating domains for non-ribosomal peptide production in Saccharomyces cerevisiae . Biotechnol Bioeng. 2010;106(5):841–844. doi: 10.1002/bit.22739. [DOI] [PubMed] [Google Scholar]
- 45.Vai M, Brambilla L, Orlandi I, Rota N, Ranzi BM, Alberghina L, Porro D. Improved secretion of native human insulin-like growth factor 1 from gas1 mutant Saccharomyces cerevisiae cells. Appl Environ Microbiol. 2000;66(12):5477–5479. doi: 10.1128/AEM.66.12.5477-5479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egel-Mitani M, Andersen AS, Diers II, Hach M, Thim L, Hastrup S, Vad K. Yield improvement of heterologous peptides expressed in yps1-disrupted Saccharomyces cerevisiae strains. Enzyme Microb Technol. 2000;26(9–10):671–677. doi: 10.1016/S0141-0229(00)00158-7. [DOI] [PubMed] [Google Scholar]
- 47.Hackel BJ, Huang D, Bubolz JC, Wang XX, Shusta EV. Production of soluble and active transferrin receptor-targeting single-chain antibody using Saccharomyces cerevisiae . Pharm Res. 2006;23(4):790–797. doi: 10.1007/s11095-006-9778-7. [DOI] [PubMed] [Google Scholar]
- 48.Vellanki RN, Komaravelli N, Tatineni R, Mangamoori LN. Expression of hepatitis B surface antigen in Saccharomyces cerevisiae utilizing glyceraldehyde-3-phosphate dehydrogenase promoter of Pichia pastoris . Biotechnol Lett. 2007;29(2):313–318. doi: 10.1007/s10529-006-9242-0. [DOI] [PubMed] [Google Scholar]
- 49.Lowin T, Raab U, Schroeder J, Franssila R, Modrow S. Parvovirus B19 VP2-proteins produced in Saccharomyces cerevisiae: comparison with VP2-particles produced by baculovirus-derived vectors. J Vet Med B Infect Dis Vet Public Health. 2005;52(7–8):348–352. doi: 10.1111/j.1439-0450.2005.00871.x. [DOI] [PubMed] [Google Scholar]
- 50.Chigira Y, Oka T, Okajima T, Jigami Y. Engineering of a mammalian O-glycosylation pathway in the yeast Saccharomyces cerevisiae: production of O-fucosylated epidermal growth factor domains. Glycobiology. 2008;18(4):303–314. doi: 10.1093/glycob/cwn008. [DOI] [PubMed] [Google Scholar]
- 51.Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae . Biotechnol Bioeng. 2009;103(6):1192–1201. doi: 10.1002/bit.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim E-J, Park Y-K, Lim H-K, Park Y-C, Seo J-H. Expression of hepatitis B surface antigen S domain in recombinant Saccharomyces cerevisiae using GAL1 promoter. J Biotech. 2009;141(3–4):155–159. doi: 10.1016/j.jbiotec.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 53.Kim HJ, Lee SJ, Kim H-J. Optimizing the secondary structure of human papillomavirus type 16 L1 mRNA enhances L1 protein expression in Saccharomyces cerevisiae . J Biotechnol. 2010;150(1):31–36. doi: 10.1016/j.jbiotec.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 54.Petranovic D, Tyo K, Vemuri GN, Nielsen J. Prospects of yeast systems biology for human health: integrating lipid, protein and energy metabolism. FEMS Yeast Res. 2010;10(8):1046–1059. doi: 10.1111/j.1567-1364.2010.00689.x. [DOI] [PubMed] [Google Scholar]
- 55.Snyder M, Gallagher JE. Systems biology from a yeast omics perspective. FEBS Lett. 2009;583(24):3895–3899. doi: 10.1016/j.febslet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Canelas AB, Harrison N, Fazio A, Zhang J, Pitkanen JP, van den Brink J, Bakker BM, Bogner L, Bouwman J, Castrillo JI, Cankorur A, Chumnanpuen P, Daran-Lapujade P, Dikicioglu D, van Eunen K, Ewald JC, Heijnen JJ, Kirdar B, Mattila I, Mensonides FI, Niebel A, Penttila M, Pronk JT, Reuss M, Salusjarvi L, Sauer U, Sherman D, Siemann-Herzberg M, Westerhoff H, de Winde J, Petranovic D, Oliver SG, Workman CT, Zamboni N, Nielsen J. Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat Commun. 2010;1:145. doi: 10.1038/ncomms1150. [DOI] [PubMed] [Google Scholar]
- 57.Alberghina L, Cirulli C. Proteomics and systems biology to tackle biological complexity: yeast as a case study. Proteomics. 2010;10(24):4337–4341. doi: 10.1002/pmic.201000114. [DOI] [PubMed] [Google Scholar]
- 58.Josling T, Blandford D, Earley J (2010) Biofuel and biomass subsidies in the US, EU and Brazil: towards a transparent system of notification. IPC position paper
- 59.Vennestrom PN, Osmundsen CM, Christensen CH, Taarning E. Beyond petrochemicals: the renewable chemicals industry. Angew Chem Int Ed Engl. 2011;50(45):10502–10509. doi: 10.1002/anie.201102117. [DOI] [PubMed] [Google Scholar]
- 60.Perlack RD, Wright LL, Turhollow AF, Graham RL, Stokes BJ, Erbach DC (2005) Biomass as feedstock for a bioenergy and bioproducts industry: the technical feasibility of a billion-ton annual supply. (Tech Rep ORNL/TM-2006/66, Oak Ridge National Laboratory, Oak Ridge, TN). Also available at http://feedstockreviewornlgov/pdf/billion_ton_visionpdf
- 61.Hahn-Hagerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74(5):937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- 62.Pronk JT, van Maris AJA, Abbott DA, Bellissimi E, van den Brink J, Kuyper M, Luttik MAH, Wisselink HW, Scheffers WA, van Dijken JP. Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Anton Leeuw Int J G. 2006;90(4):391–418. doi: 10.1007/s10482-006-9085-7. [DOI] [PubMed] [Google Scholar]
- 63.Bettiga M, Gorwa-Grauslund MF, Hahn-Hägerdal B (2009) Metabolic engineering in yeast. In: The metabolic pathway engineering handbook. Metabolic pathway engineering handbook. CRC Press, Boca Raton pp 22-21–22-48
- 64.Den Haan R, Rose SH, Lynd LR, van Zyl WH. Hydrolysis and fermentation of amorphous cellulose by recombinant Saccharomyces cerevisiae . Metab Eng. 2007;9(1):87–94. doi: 10.1016/j.ymben.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Shigechi H, Koh J, Fujita Y, Matsumoto T, Bito Y, Ueda M, Satoh E, Fukuda H, Kondo A. Direct production of ethanol from raw corn starch via fermentation by use of a novel surface-engineered yeast strain codisplaying glucoamylase and alpha-amylase. Appl Environ Microbiol. 2004;70(8):5037–5040. doi: 10.1128/AEM.70.8.5037-5040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katahira S, Fujita Y, Mizuike A, Fukuda H, Kondo A. Construction of a xylan-fermenting yeast strain through codisplay of xylanolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. Appl Environ Microbiol. 2004;70(9):5407–5414. doi: 10.1128/AEM.70.9.5407-5414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujita Y, Ito J, Ueda M, Fukuda H, Kondo A. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl Environ Microbiol. 2004;70(2):1207–1212. doi: 10.1128/AEM.70.2.1207-1212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siso MIG, Beccerra M, Prado SD, Rodriguez-Belmonte E, Cerdan ME. Metabolic engineering for direct lactose utilization by Saccharomyces cerevisiae . Biotechnol Lett. 2002;24(17):1391–1396. doi: 10.1023/A:1019886112076. [DOI] [Google Scholar]
- 69.Ostergaard S, Roca C, Ronnow B, Nielsen J, Olsson L. Physiological studies in aerobic batch cultivations of Saccharomyces cerevisiae strains harboring the MEL1 gene. Biotechnol Bioeng. 2000;68(3):252–259. doi: 10.1002/(SICI)1097-0290(20000505)68:3<252::AID-BIT3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 70.Dmytruk OV, Voronovsky AY, Abbas CA, Dmytruk KV, Ishchuk OP, Sibirny AA. Overexpression of bacterial xylose isomerase and yeast host xylulokinase improves xylose alcoholic fermentation in the thermotolerant yeast Hansenula polymorpha . FEMS Yeast Res. 2008;8(1):165–173. doi: 10.1111/j.1567-1364.2007.00289.x. [DOI] [PubMed] [Google Scholar]
- 71.Lee KS, Hong ME, Jung SC, Ha SJ, Yu BJ, Koo HM, Park SM, Seo JH, Kweon DH, Park JC, Jin YS. Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol Bioeng. 2011;108(3):621–631. doi: 10.1002/bit.22988. [DOI] [PubMed] [Google Scholar]
- 72.Wisselink HW, Toirkens MJ, Del Rosario Franco Berriel M, Winkler AA, van Dijken JP, Pronk JT, van Maris AJ. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl Environ Microbiol. 2007;73(15):4881–4891. doi: 10.1128/AEM.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wohlbach DJ, Kuo A, Sato TK, Potts KM, Salamov AA, Labutti KM, Sun H, Clum A, Pangilinan JL, Lindquist EA, Lucas S, Lapidus A, Jin M, Gunawan C, Balan V, Dale BE, Jeffries TW, Zinkel R, Barry KW, Grigoriev IV, Gasch AP. Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc Natl Acad Sci USA. 2011;108(32):13212–13217. doi: 10.1073/pnas.1103039108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghosh A, Zhao H, Price ND. Genome-scale consequences of cofactor balancing in engineered pentose utilization pathways in Saccharomyces cerevisiae . PLoS One. 2011;6(11):e27316. doi: 10.1371/journal.pone.0027316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garcia Sanchez R, Hahn-Hagerdal B, Gorwa-Grauslund MF. PGM2 overexpression improves anaerobic galactose fermentation in Saccharomyces cerevisiae . Microb Cell Fact. 2010;9:40. doi: 10.1186/1475-2859-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bro C, Knudsen S, Regenberg B, Olsson L, Nielsen J. Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol. 2005;71(11):6465–6472. doi: 10.1128/AEM.71.11.6465-6472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ostergaard S, Olsson L, Johnston M, Nielsen J. Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat Biotechnol. 2000;18(12):1283–1286. doi: 10.1038/82400. [DOI] [PubMed] [Google Scholar]
- 78.Hong KK, Vongsangnak W, Vemuri GN, Nielsen J. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc Natl Acad Sci USA. 2011;108(29):12179–12184. doi: 10.1073/pnas.1103219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ha SJ, Galazka JM, Kim SR, Choi JH, Yang X, Seo JH, Glass NL, Cate JH, Jin YS. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc Natl Acad Sci USA. 2011;108(2):504–509. doi: 10.1073/pnas.1010456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Teunissen AWRH, De Bont JAM (2010) Xylose isomerase genes and their use in fermentation of pentose sugars. WO Patent WO 2010/074577 A1
- 81.Op Den Camp HJMO, Harhangi HR, Van Der Drift C, Pronk JT (2009) Transformed eukaryotic cells that directly convert xylose to xylulose. US Patent US 7622284
- 82.Klaassen P, van der Laan JM, Gielesen BEM, van Suylekom GP (2009) A pentose sugar fermenting cell. WO Patent WO 2009/109630 A1
- 83.Klaassen P, van der Laan JM, Gielesen BEM, van Suylekom GP (2009) A pentose sugar fermenting cell. WO Patent WO 2009/109631 A1
- 84.Ho NWY, Chen Z-D (2009) Stable recombinant yeasts for fermenting xylose to ethanol. US Patent US 7527927
- 85.De Bont JAM (2009) Novel arabinose-fermenting eukaryotic cells. WO Patent WO 2009/011591 A2
- 86.Van Maris AJA, Pronk JT, Wisselink W, Van Dijken JP, Winkler AA, De Winde H (2008) Metabolic engineering of arabinose- fermenting yeast cells. WO Patent WO 2008/041840 A1
- 87.Hughes SR, Butt TR (2010) Saccharomyces cerevisiae engineered for xylose utilization. WO Patent WO 2010/039692 A2
- 88.Mainguet SE, Liao JC. Bioengineering of microorganisms for C3 to C5 alcohols production. Biotechnol J. 2010;5(12):1297–1308. doi: 10.1002/biot.201000276. [DOI] [PubMed] [Google Scholar]
- 89.Yan Y, Liao JC. Engineering metabolic systems for production of advanced fuels. J Ind Microbiol Biotechnol. 2009;36(4):471–479. doi: 10.1007/s10295-009-0532-0. [DOI] [PubMed] [Google Scholar]
- 90.Feldman RMR, Gunawardena U, Urano J, Meinhold P, Aristidou A, Dundon CA, Smith C (2011) Yeast organism producing isobutanol at a high yield. US Patent US 2011/0020889 A1
- 91.Gunawardena U, Meinhold P, Peters MW, Urano J, Feldman RMR (2010) Butanol production by metabolically engineered yeast. US Patent US 2010/0062505 A1
- 92.Ramey DE (2008) Butanol: the other alternative fuel. agricultural biofuels: technology, sustainability and profitability, 137–147. (http://nabc.cals.cornell.edu/pubs/nabc_19/NABC19_5Plenary2_Ramey.pdf)
- 93.Wallner T, Miers SA, McConnell S. A comparison of ethanol and butanol as oxygenates using a direct-injection, spark-ignition engine. J Eng Gas Turb Power. 2009;131(3):032802. [Google Scholar]
- 94.Weber C, Farwick A, Benisch F, Brat D, Dietz H, Subtil T, Boles E. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl Microbiol Biotechnol. 2010;87(4):1303–1315. doi: 10.1007/s00253-010-2707-z. [DOI] [PubMed] [Google Scholar]
- 95.Nielsen DR, Leonard E, Yoon SH, Tseng HC, Yuan C, Prather KL. Engineering alternative butanol production platforms in heterologous bacteria. Metab Eng. 2009;11(4–5):262–273. doi: 10.1016/j.ymben.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 96.Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol. 2008;74(8):2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dickinson JR, Harrison SJ, Hewlins MJ. An investigation of the metabolism of valine to isobutyl alcohol in Saccharomyces cerevisiae . J Biol Chem. 1998;273(40):25751–25756. doi: 10.1074/jbc.273.40.25751. [DOI] [PubMed] [Google Scholar]
- 98.Porro D, Gasser B, Fossati T, Maurer M, Branduardi P, Sauer M, Mattanovich D. Production of recombinant proteins and metabolites in yeasts: when are these systems better than bacterial production systems? Appl Microbiol Biotechnol. 2011;89(4):939–948. doi: 10.1007/s00253-010-3019-z. [DOI] [PubMed] [Google Scholar]
- 99.Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. Fermentative butanol production by Clostridia. Biotechnol Bioeng. 2008;101(2):209–228. doi: 10.1002/bit.22003. [DOI] [PubMed] [Google Scholar]
- 100.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 101.Steen EJ, Chan R, Prasad N, Myers S, Petzold CJ, Redding A, Ouellet M, Keasling JD. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Fact. 2008;7:36. doi: 10.1186/1475-2859-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Donaldson GK, Eliot AC, Flint D, Maggio-Hall LA, Nagarajan V (2011) Fermentive production of four carbon alcohols. US Patent US 7993889 B1
- 103.Anthony LC, Huang LL, Ye RW (2010) Production of isobutanol in yeast mitochondria. US Patent US 2010/0129886 A1
- 104.Festel G, Boles E, Weber C, Brat D (2011) Fermentative production of isobutanol with yeast. US Patent US 2011/0053235 A1
- 105.Urano J, Dundon CA, Meinhold P, Feldman RMR, Aristidou A, Hawkins A, Buelter T, Peters M, Lies D, Porter-Scheinman S, Berry R, Kalra I (2011) Cytosolic isobutanol pathway localization for the production of isobutanol. US Patent US 2011/0076733 A1
- 106.Buelter T, Meinhold P, Smith C, Aristidou A, Dundon CA, Urano J (2010) Engineered microorganisms for the production of one or more target compounds. WO Patent WO 2010/075504 A2
- 107.Bakker BM, Overkamp KM, van Maris AJ, Kotter P, Luttik MA, van Dijken JP, Pronk JT. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae . FEMS Microbiol Rev. 2001;25(1):15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 108.Dundon CA, Aristidou A, Hawkins A, Lies D, Albert LH (2011) Methods of increasing dihydroxy acid dehydratase activity to improve production of fuels, chemicals, and amino acids. US Patent US 2011/0183393 A1
- 109.Liao D-I, Nelson MJ, Bramucci MG (2008) Fermentive production of isobutanol using highly active ketol-acid reductoisomerase enzymes. US Patent US 2008/0261230 A1
- 110.Donaldson GK, Huang LL, Maggio-Hall LA, Nagarajan V, Nakamura CE, Suh W (2008) Fermentive production of four carbon alcohols. US Patent US 2008/0182308 A1
- 111.Donaldson GK, Eliot AC, Nagarajan V, Nakamura CE, Huang LL (2007) Fermentive production of four carbon alcohols. US Patent US 2007/0259410 A1
- 112.Flint D, Rothman SC, Suh W, Tomb J-F, Ye RW (2010) Identification and use of bacterial [2Fe-2S] dihydroxy-acid dehydratases. US Patent US 2010/0081154 A1
- 113.Anthony LC, Maggio-Hall LA, Rothman SC, Tomb J-F (2010) Increased heterologous Fe-S enzyme activity in yeast. US Patent US 2010/0081179 A1
- 114.Van Dyk TK (2010) Yeast with increased butanol tolerance involving a multidrug efflux pump gene. US Patent US 2010/0221801 A1
- 115.Bramucci MG, Larossa RA, Smulski DR (2010) Yeast with increased butanol tolerance involving cell wall integrity pathway. US Patent US 2010/0167364 A1
- 116.Bramucci MG, Larossa RA, Smulski DR (2010) Yeast with increased butanol tolerance involving high osmolarity/glycerol response pathway. US Patent US 2010/0167365 A1
- 117.Bramucci MG, Larossa RA, Singh M (2010) Yeast with increased butanol tolerance involving filamentous growth response. US Patent US 2010/0167363 A1
- 118.Larossa RA (2009) Yeast strain for production of four carbon alcohols. US Patent US 2009/0280546 A1
- 119.Knoshaug EP, Zhang M. Butanol tolerance in a selection of microorganisms. Appl Biochem Biotechnol. 2009;153(1–3):13–20. doi: 10.1007/s12010-008-8460-4. [DOI] [PubMed] [Google Scholar]
- 120.Maury J, Asadollahi MA, Moller K, Clark A, Nielsen J. Microbial isoprenoid production: an example of green chemistry through metabolic engineering. Adv Biochem Eng Biotechnol. 2005;100:19–51. doi: 10.1007/b136410. [DOI] [PubMed] [Google Scholar]
- 121.Zhang F, Rodriguez S, Keasling JD. Metabolic engineering of microbial pathways for advanced biofuels production. Curr Opin Biotechnol. 2011;22(6):775–783. doi: 10.1016/j.copbio.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 122.Withers ST, Keasling JD. Biosynthesis and engineering of isoprenoid small molecules. Appl Microbiol Biotechnol. 2007;73(5):980–990. doi: 10.1007/s00253-006-0593-1. [DOI] [PubMed] [Google Scholar]
- 123.Kirby J, Keasling JD. Metabolic engineering of microorganisms for isoprenoid production. Nat Prod Rep. 2008;25(4):656–661. doi: 10.1039/b802939c. [DOI] [PubMed] [Google Scholar]
- 124.Shiba Y, Paradise EM, Kirby J, Ro DK, Keasling JD. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab Eng. 2007;9(2):160–168. doi: 10.1016/j.ymben.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 125.Clark A, Maury J, Asadollahi MA, MØLler K, Nielsen J, Schalk M (2009) Method for producing terpenes and MEP-transformed microorganisms therefore. US Patent US 2009/0155874 A1
- 126.Maury J, Asadollahi MA, Moller K, Schalk M, Clark A, Formenti LR, Nielsen J. Reconstruction of a bacterial isoprenoid biosynthetic pathway in Saccharomyces cerevisiae . FEBS Lett. 2008;582(29):4032–4038. doi: 10.1016/j.febslet.2008.10.045. [DOI] [PubMed] [Google Scholar]
- 127.Muntendam R, Melillo E, Ryden A, Kayser O. Perspectives and limits of engineering the isoprenoid metabolism in heterologous hosts. Appl Microbiol Biotechnol. 2009;84(6):1003–1019. doi: 10.1007/s00253-009-2150-1. [DOI] [PubMed] [Google Scholar]
- 128.Chang MC, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol. 2006;2(12):674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- 129.Ubersax JA, Platt DM (2010) Genetically modified microbes producing isoprenoids. WO Patent WO 2010/141452 A1
- 130.Tsuruta H, Lenihan JR, Regentin R (2009) Production of isoprenoids. WO Patent WO 2009/042070 A2
- 131.Basso LC, de Amorim HV, de Oliveira AJ, Lopes ML. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res. 2008;8(7):1155–1163. doi: 10.1111/j.1567-1364.2008.00428.x. [DOI] [PubMed] [Google Scholar]
- 132.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 133.Keasling JD, Newman JD, Pitera DJ (2010) Method for enhancing production of isoprenoid compounds. US Patent US 7670825
- 134.Johnston M, Flick JS, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae . Mol Cell Biol. 1994;14(6):3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Asadollahi MA, Maury J, Moller K, Nielsen KF, Schalk M, Clark A, Nielsen J. Production of plant sesquiterpenes in Saccharomyces cerevisiae: effect of ERG9 repression on sesquiterpene biosynthesis. Biotechnol Bioeng. 2008;99(3):666–677. doi: 10.1002/bit.21581. [DOI] [PubMed] [Google Scholar]
- 136.Kizer J (2009) Methods of increasing isoprenoid or isoprenoid precursor production. WO Patent WO 2009/005704 A1
- 137.Chang M, Krupa RA, Ro D-K, Yoshikuni Y, Keasling JD (2009) Nucleic acids encoding modified cytochrome P450 enzymes and methods of use thereof. US Patent US 2009/0098626 A1
- 138.Bajad S, Leavell M (2009) Methods of monitoring metabolic pathways. WO Patent WO 2009/097339 A1
- 139.Millis JR, Maurina-Brunker J, McMullin TW (2011) Production of isoprenoids. US Patent US 7927862
- 140.Julien B, Burlingame R (2010) Method for production of isoprenoids. US Patent US 2010/0151519 A1
- 141.Kinney AJ, Ni H, Rouviere PE, Suh W (2007) Method to increase hydrophobic compound titer in a recombinant microorganism. US Patent US 7256014
- 142.Brzostowicz PC, Rouviere PE, Pollak DM (2006) Biological production of tetradehydrolycopene. US Patent US 7087403
- 143.Bramucci MG, Brzostowicz PC, Cheng Q, Kostichka KN, Rouviere PE, Nagarajan V, Tao L, Thomas SM (2006) Genes involved in isoprenoid compound production. US Patent US 7034140
- 144.Cheng Q, Koffas M, Norton KC, Odom JM, Picataggio SK, Rouviere PE, Schenzle A, Tomb J-F (2002) Genes involved in isoprenoid compound production. WO Patent WO 2002/020733 A2
- 145.van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin ML, Heijnen JJ, Hoare M, Lange HC, Madden EA, Niederberger P, Nielsen J, Parrou JL, Petit T, Porro D, Reuss M, van Riel N, Rizzi M, Steensma HY, Verrips CT, Vindelov J, Pronk JT. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol. 2000;26(9–10):706–714. doi: 10.1016/S0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]