Abstract
Sequence-specific protein-DNA interactions (PDIs) are critical for regulating many cellular processes, including transcription, DNA replication, repair, and rearrangement. We review recent experimental advances in high-throughput technologies designed to characterize PDIs and discuss recent studies that use these tools, including ChIP-chip/seq, SELEX-based approaches, yeast one-hybrid, bacterial one-hybrid, protein binding microarray, and protein microarray. The results of these studies have challenged some long-standing concepts of PDI and provide valuable insights into the complex transcriptional regulatory networks.
Keywords: Protein DNA interactions, Transcription factors, Unconventional DNA-binding proteins, Protein microarray, DNA-binding specificity
Introduction
Genetic information encoded in DNA has to be precisely transcribed to execute cellular functions that are required for cells to grow, differentiate, divide, and respond to environmental stimuli. Sequence-specific protein-DNA interactions (PDIs) play a central role in regulating gene transcription; mutations in DNA-binding proteins such as transcription factors (TFs) can often cause diseases and cancer in humans [1]. Therefore, identifying the underlying mechanistic rules that control sequence-specific PDIs is a necessary step towards understanding how gene expression is regulated.
Biochemical approaches have long been the primary mechanism by which PDIs have been characterized. Most studies have traditionally focused on characterizing interactions of one or a few proteins with a relatively limited number of candidate DNA sequences using methods such as electrophoretic mobility shift assay (EMSA) and nuclease footprinting [2–4]. Years of studies have accumulated much important PDI data, much of which is accessible in the TRANSFAC and JASPAR databases [5, 6]. However, such conventional approaches have several drawbacks. They are generally low throughput and laborious, and more importantly, they normally generate consensus DNA-binding sites of lower resolution because of the limited number of DNA sequences tested.
In the last decade, the development of high-throughput technologies has revolutionized the characterization of PDIs and generated unprecedented datasets of the DNA sequence preferences for a large number of DNA-binding proteins. These technologies can be categorized into two different but complementary approaches [7]. On the one hand, protein-centered biochemical approaches are used to determine DNA-binding specificities of individual proteins. Some widely used protein-centered approaches include SELEX-based approaches, chromatin immunoprecipitation (ChIP)-based approaches, and protein binding microarray (PBM). On the other hand, DNA-centered genetic approaches have also been developed that determine the binding proteins for DNA sequences, and these approaches include the bacterial/yeast one-hybrid (B1H/Y1H) and protein microarray-based approaches.
The most intensively studied DNA-binding proteins are TFs, which specifically activate and/or repress expression of their target genes. In the eukaryotic kingdom, 347 known DNA-binding domains (DBDs) have been annotated so far, and most TFs (with some exceptions) contain one or more of these DBDs [8]. However, the consensus binding sites for many TFs remain largely uncharacterized. Aside from TFs, the target DNA sequences of the larger universe of DNA-binding proteins have not been extensively explored.
In this review, we provide an overview of high-throughput technologies recently developed for characterizing PDIs. We specifically discuss the advantages and disadvantages associated with each approach, with a summary in Table 1. In addition, we also review large-scale PDI studies and some widely used PDI databases, which are summarized in Tables 2 and 3, respectively.
Table 1.
High-throughput technologies for detecting PDIs
| Advantages | Disadvantages | |
|---|---|---|
| ChIP-chip/ChIP-seq | In vivo | Availability of TF-specific antibody |
| PDI can be examined under different cellular conditions and at different time points | Difficult to retrieve DNA-binding motifs as genomic sites may associate directly or indirectly with the protein examined | |
| PBM | Unbiased survey of DNA-binding sites | In vitro |
| Require relatively high amounts of purified proteins | ||
| Semi-quantitative measurement | DNA position effects | |
| Currently limited to oligomers up to 10-mer | ||
| Detection of interactions with high affinity because of extensive washing | ||
| SELEX | Require low amounts of purified proteins | In vitro |
| Characterize full-length proteins or proteins requiring post-translational modification | Large pools of clones to be screened | |
| Positive clones have to be sequenced | ||
| Identify DNA-binding sequences more than 10-mer | ||
| Protein microarray | Characterize full-length proteins | In vitro |
| Not for proteins requiring post-translational modification | ||
| Depending on the completeness of protein microarray, possible proteome-scale | Monomer only | |
| Detection of interactions with high affinity because of extensive washing | ||
| Y1H | Characterize full-length proteins | Detects monomer only |
| Depending on the completeness of screened TF library, possible proteome-scale | Performed in yeast, outside of endogenous context | |
| B1H | Selection procedure is fast | In vitro |
| Detects monomer only | ||
| Single round of selection is required | Performed in bacteria, outside of endogenous context | |
| Some baits are toxic in bacteria |
TF Transcription factor
Table 2.
Summary of high-throughput PDI studies
| Study | Species | TF family | Protein length | No. of TFs | Technology |
|---|---|---|---|---|---|
| Noyes [36] | Drosophila | Homeobox | DBD | 84 | B1H |
| Hu [41] | Human | All the major human TFs and uDBPs | Full | 1013a | Protein microarray |
| Berger [24] | Mouse | Homeobox | DBD | 168 | PBM |
| Badis [27] | Mouse | Major mouse TFs | DBD | 104 | PBM |
| Deplancke [34] | Worm | Worm TFs | DBD | 112 | Y1H |
| Wei [67] | Human and mouse | ETS | DBD/full | 27 | PBM and TF DNA-binding specificity assay |
| Zhu [26] | Yeast | Major yeast TFs | DBD | 89 | PBM |
| Badis [25] | Yeast | Major yeast TFs | DBD | 112 | PBM |
| Harbison [15] | Yeast | Major yeast TFs | Full | 203b | ChIP-chip |
PDI Protein-DNA interaction,TF transcription factor, DBD DNA-binding domains, uDBP unconventional DNA-binding protein
aA total of 4,191 human TFs tested, 1,013 with DNA-binding activity and 437 with generated DNA-binding motifs
bA total of 203 yeast TFs tested, 147 DNA-binding motifs generated and 116 with high confidence
Table 3.
Summary of PDI database
| Database | Species | DNA-binding protein | No. of defined binding motifs (logos) | Cost | Approach |
|---|---|---|---|---|---|
| TRANSFAC [18] | Multiple | TFs | 1,300 | Commercial and free versions | Multiple |
| JASPAR [77]a | Multiple | TFs | 457a | Free | Multiple |
| UniPROBE [78] | Multiple, mainly mouse, worm, yeast | TFs | 393 | Free | PBM |
| hPDI [55] | Human | TFs and uDBPs | 437 | Free | Protein microarray |
TF Transcription factor, uDBP unconventional DNA-binding protein
aOverlapped with UniPROBE
High-throughput PDI techniques and studies
ChIP-chip/ChIP-seq
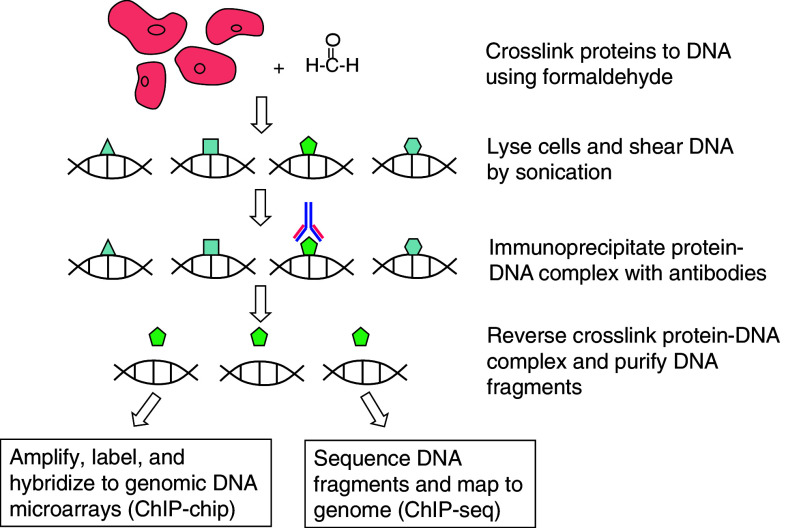
Chromatin immunoprecipitation (ChIP) is a well established method used to characterize PDIs in vivo. A comprehensive review on the recent progress of all the ChIP-based approaches can be found in [9]. In brief, ChIP-based approaches include the following steps (Fig. 1). First, chemicals such as formaldehyde are applied to cells to covalently crosslink proteins and DNA that are in direct contact. Chromosomal DNA is then fragmented, and specific antibodies are used to immunoprecipitate their target TFs along with any crosslinked DNA fragments. Finally, the bound DNA fragments are analyzed using polymerase chain reaction (ChIP-PCR), for which is it necessary to have some prior knowledge of candidate regions. Alternatively, the genomic regions bound in vivo can be characterized in a comprehensive and unbiased manner using chromatin immunoprecipitation coupled with either microarray readout (ChIP-chip) [10, 11] or high-throughput sequencing (ChIP-seq) [12–14]. Compared to ChIP-chip, ChIP-seq is a more unbiased method for precisely mapping transcription factor binding sites, as it is not restricted by predetermined probe sets. Furthermore, recent improvements in DNA sequencing technology now allow tens of millions of individual sequence reads to be generated in a single experiment, greatly increasing both the sensitivity and resolution of ChIP-seq.
Fig. 1.
Workflows of ChIP-chip and ChIP-seq
The unique strength of ChIP is that it captures PDIs in vivo in the context of cellular chromatin and is at present the only readily useable method that can reliably measure PDIs in vivo. However, it is technically challenging, and high-quality ChIP-grade antibodies are available for only a limited number of TFs, particularly for metazoan model organisms. Although the limited availability of ChIP-grade antibodies is one of the most seriously limiting factors for ChIP-base approaches, it may be circumvented by using epitope-tagged TFs, although these modified genes typically must be introduced into the endogenous genetic locus via homologous targeting in order to be expressed at normal physiological levels [15]. ChIP-based approaches may also be ineffective at detecting TFs that are expressed at low levels or in a small fraction of cells in a tissue sample. Finally, ChIP-based data does not discriminate between proteins that directly bind DNA and those that merely interact with proteins that do, thus adding a note of caution when interpreting any results obtained.
Applications
Large-scale ChIP-chip study was first applied to yeast where the genomic binding sites of 106 yeast TFs were surveyed [16]. Later, the same group surveyed DNA-binding sites of the epitope-tagged yeast TFs using ChIP-chip [15]. They determined the genomic occupancy of 203 yeast epitope-tagged TFs, and 11,000 unique interactions were identified between regulators and promoter regions. Computational analysis of the binding data from this study yielded sequence-specific binding motifs for 147 yeast TFs, 116 with high confidence. DNA-binding specificities of 102 TFs are in good agreement with previously reported in vitro data. For the TFs that did not display a consensus binding motif or do not agree with the in vitro data, this may be caused by low levels of TF expression. Furthermore, some predicted TFs may not selectively bind DNA at all, but rather interact with TFs that demonstrate highly selective DNA binding. DNA motifs identified by ChIP analysis are thus often different from the in vitro DNA-binding motifs of the TFs themselves [17].
SELEX-based high-throughput approaches
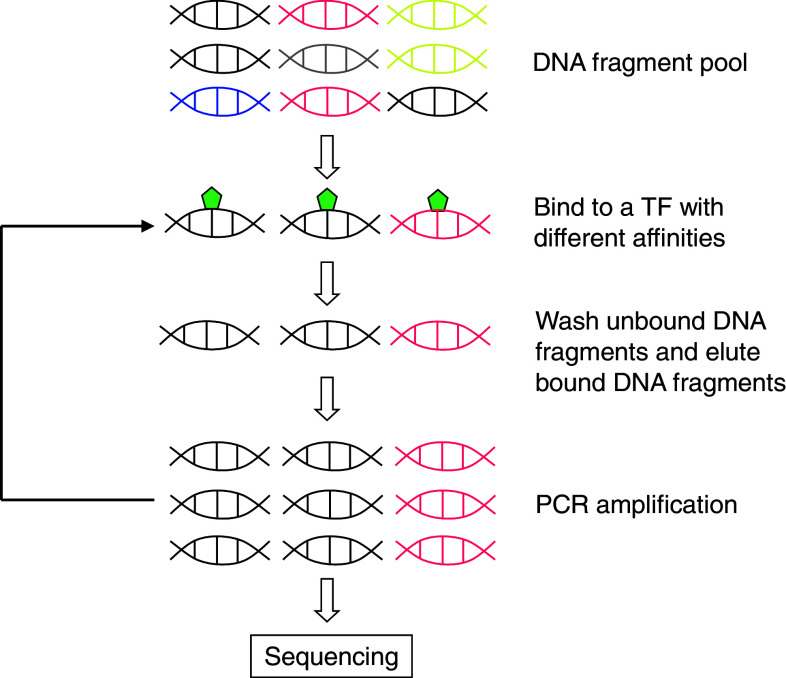
The systematic evolution of ligands by exponential enrichment (SELEX) is a well-established approach that enables in vitro selection of enriched sequences from libraries of random DNA sequences that are bound by a recombinant TF (Fig. 2). This has been used more extensively than any other experimental approach to obtain the preferred in vitro TF binding motifs for most entries in the TRANSFAC database [18]. However, it is difficult to use this approach for analysis of large numbers of DNA-binding proteins, as it requires multiple rounds of selection to complete. It is also difficult to apply this method for obtaining DNA-binding motifs with high resolution as SELEX contains typically 20–70 sites. To overcome the problems, a modified protocol called SELEX-SAGE was proposed, where SAGE stands for serial analysis of gene expression [19]. SELEX-SAGE includes two improvements that make this approach high-throughput. Firstly, a radiolabeled oligonucleotide probe is used to monitor binding conditions to prevent selections of only high-affinity binding sites. Secondly, SAGE is used to increase the sequencing throughput by concatenation of DNA sequences obtained through in vitro selection of TF binding sites. More recently, an improved SELEX-based protocol was reported that enables a higher-throughput detection of PDIs [20]. Instead of concatenation of bound DNA sequences, the method instead utilizes massively parallel sequencing technology of individual TF binding sites, which eliminates all costly and labor-intensive cloning steps. Furthermore, the modified protocol can generate a very large number of individual sequencing reads, resulting in dramatically increased sequence yield and throughput.
Fig. 2.
Workflow of systematic evolution of ligands by exponential enrichment (SELEX)
Applications
In the original proof-of-principle study for parallel sequencing-based SELEX, the approach was validated by determining binding specificities of 14 TFs from different gene families [20]. For all the TFs tested, the number of sequences was of the same order of magnitude as the ones using SELEX-SAGE, with lower cost and simpler procedure. The results are in good agreement with the previous knowledge of the DNA-binding motifs of these TFs. The DNA-binding preferences of two TFs were also successfully validated using ChIP-seq.
Protein binding microarray
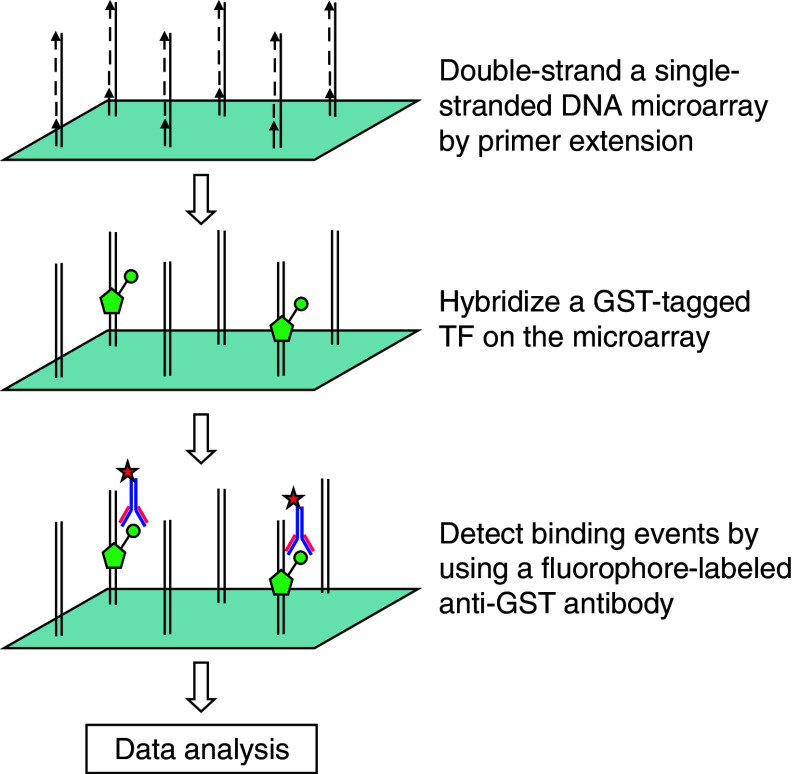
Protein binding microarray (PBM) analysis allows high-throughput characterization of PDIs by directly probing purified proteins to a double-stranded DNA (dsDNA) microarray (Fig. 3). The basic idea is that a given recombinant TF protein is labeled and then bound directly to the dsDNA microarray. The DNA-binding specificity of that protein can be directly determined by measuring the signal intensity of bound dsDNAs.
Fig. 3.
Workflow of protein binding microarray (PBM)
In an early proof-of-principle study, Bulyk et al. [21] synthesized and printed short dsDNAs onto a microarray and then probed the microarray with C2H2 zinc finger DNA-binding domain of Egr1. They found that spots with higher signal intensities contained higher affinity binding sites, which suggested that the dsDNA microarray can indeed be used to determine specific PDIs. Later, Warren et al. [22] created microarrays with short synthetic dsDNAs covering all possible sequence variants of 10 base pairs (bp). Exhaustive search of the entire 10-bp DNA space allows one to detect subtle differences in DNA-binding activity, and thus a highly accurate consensus DNA-binding site is generated. This DNA microarray design permits a rapid and unbiased approach to determine DNA-binding specificity given a protein without any prior knowledge about its possible DNA-binding profiles. However, as the length of DNA-binding sites to be examined increases, it becomes difficult to fit all possible binding sequences on a single microarray. To overcome this limitation, a compact universal DNA microarray design was proposed [23], where a number of DNA-binding sites are allowed to overlap within a given DNA spot. For example, 31 overlapping 10-bp DNA sequences can be packed in a 40-bp DNA molecule. Such design permits a comprehensive examination of all possible sequence variants of a given length, which saves great space on a chip and therefore lowers experiment costs.
Compared to the SELEX-based in vitro selection, PBM provides a larger number of sequence variants to be examined and therefore defines more precise DNA-binding motifs. However, as the PBM method needs relatively high amounts of recombinant protein, it is difficult to analyze proteins that do not express well, such as many full-length TFs, or proteins that require post-translational modifications. Because extensive washing is required to avoid non-specific binding, PBM can normally only detect interactions with high affinity. Moreover, PBM cannot currently effectively determine binding specificities for TFs that prefer longer than 10-bp DNA-binding sequences due to the limited number of sequences that can be placed on a chip.
Applications
The power of the PBM approach was dramatically demonstrated in a large-scale PDI study by Bulyk, Hughes, and their colleagues, in which the sequence preferences of the majority mouse homeodomain-containing TFs (168) were characterized via probing all possible 8-mer dsDNA sequences [24]. A total of 168 DNA-binding motifs were identified and clustered into 65 distinct groups, showing a rich and diverse sequence preference in homeodomain TFs. Using the identified binding profiles, the authors sought to predict the DNA profiles of a TF from the primary amino acid sequence of the protein. The authors compared previously published chromatin immunoprecipitation data for individual homeodomain TFs with the PBM data obtained in this study and found a considerable amount of overlap.
After these first successful efforts, the Bulyk and Hughes’ groups continued characterizing DNA-binding specificities for the majority of yeast TFs. The Hughes’ group identified binding specificities for 112 DNA-binding proteins representing 19 different gene families [25]. More than half of the identified binding motifs corresponded to previously identified ones. In addition, new consensus sites were identified for 36 TFs whose binding preference was not previously characterized. Using previously published ChIP-chip datasets, the authors further characterized two TFs in greater detail and found that their binding sequences tend to cluster at roughly −100 bp relative to the transcription start site (TSS), illustrating the biological relevance of the in vitro PDI. Using PBMs that cover all possible 8-mers, the Bulyk group also characterized the DNA-binding preferences for 89 yeast TFs [26]. The authors predicted the potential target genes, regulatory roles, and condition specificities of these TFs using their 8-mer binding profiles. Finally, the authors proposed that these PBM data may be used to interpret ChIP-chip data in order to distinguish direct versus indirect binding targets of immunoprecipitated TFs.
More recently, the Bulyk and Hughes’s groups together profiled the binding site preferences for 104 mouse TFs of different TF families [27]. When the DNA-binding preferences for 21 members of the Sox (SRY-related high-mobility group box)/TCF (T cell factor) family were compared, most of them (14/21) preferentially bound to an identical sequence, indicating conserved DNA-binding activity in this subfamily. For the homeodomain family, most of the members recognized the canonical TAAT core sequence, as demonstrated previously [24]. However, when examining the top 100 8-mer DNA sequences bound by each individual protein, one-third of them contained sequences that were substantially different from this canonical consensus homeodomain binding site. Even many well studied homeodomain TFs were found to be able to recognize two distinct binding sites. This observation is consistent with a previous study in which Nkx2-5 was shown to recognize two distinct consensus sites, though binding affinity for each site was significantly different [28].
Yeast one-hybrid
The SELEX and PBM approaches are well suited for identifying the preferred DNA-binding sequences for a given protein. However, when the goal is to identify proteins that can specifically recognize a given DNA sequence (e.g., a piece of DNA in the promoter of a gene of interest), the yeast or bacterial one-hybrid (Y1H or B1H) system would be a proper choice [29, 30]. More recently, a large number of metazoan TFs have been cloned into expression constructs suitable for Y1H analysis, increasing the extent to which it is possible to systematically interrogate TFs that bind a specific target sequence [31, 32].
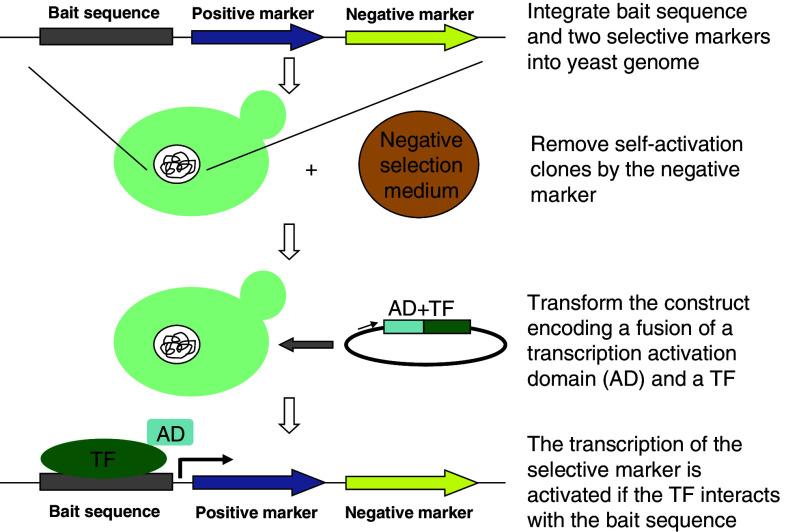
Originally, the Y1H system used DNA cloning methods based on restriction digestion for generating the plasmid constructs required, a substantial roadblock for scaling up to high-throughput analysis. This problem was recently solved by using the Invitrogen Gateway-compatible Y1H system, which allows high-throughput subcloning of multiple DNA baits (cis-regulatory DNA elements) into a Y1H destination vector and detects PDIs based on the selection of reporter gene expression in yeast (Fig. 4) [33]. This provides a high-throughput method for the identification of interactions between a DNA “bait” and a protein “prey.” In brief, DNA baits are subcloned to two reporter genes and the two bait::reporter constructs are integrated into the genome of the host yeast strain, where one reporter (HIS3) is used for positive selection of constructs that drive reporter expression, while the other (lacZ) can be measured for a more quantitative readout of transcriptional activation levels [33]. As a control, the baits are first tested for their ability to activate reporter gene expression in the absence of prey proteins, which consist of a fusion of a protein of interest and a transcription activation domain. In the presence of prey proteins, when the protein of interest can recognize and bind to the DNA bait, expression of the reporter gene is activated, and PDIs between the prey protein and its DNA targets can thus be identified.
Fig. 4.
Workflow of yeast one-hybrid (Y1H)
Applications
Using the Y1H approach, the Walhout group used the promoters of 72 C. elegans digestive tract gene as “baits” and 117 proteins as “preys” and identified 283 specific PDIs [34]. This study identified target sites for 10% of all worm TFs, many of which were previously uncharacterized, and the newly annotated TFs are enriched in the digestive tract. Interestingly, they found that 10 proteins not possessing any known DBDs also bound to specific DNA sequences. Using ChIP-PCR assays, eight of these ten proteins were confirmed to bind these sequences in yeast, suggesting that there might be additional uncharacterized DBDs in worms. Because large DNA fragments can be used in the one-hybrid approach (e.g., promoter sequences in this study), an obvious advantage is that the identified TFs are readily connected to their target genes, resulting in a more informative PDI network from which testable hypotheses about transcriptional regulatory circuitry can be more readily generated.
Bacteria one-hybrid
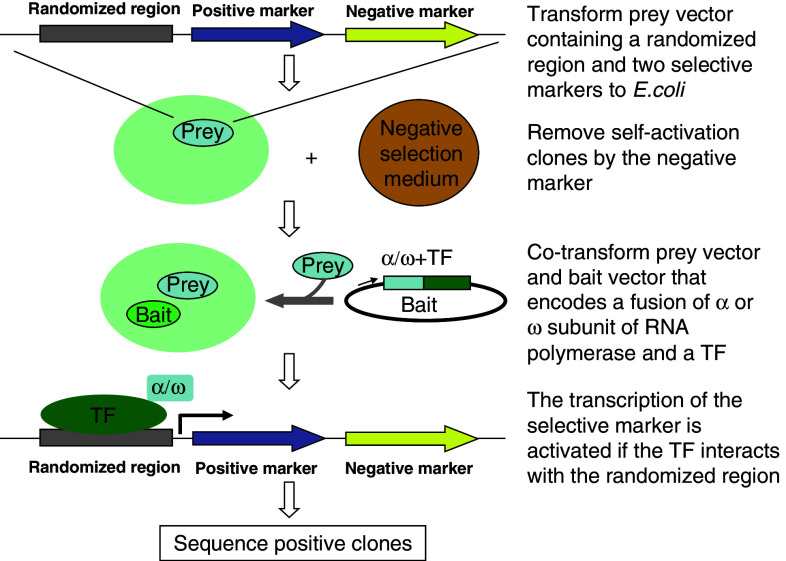
Similar to the Y1H method, which is derived from yeast two-hybrid technology, the B1H system arose as a variation of the bacterial two-hybrid platform aimed towards identification of specific PDIs (Fig. 5) [35]. The B1H system contains three components: a protein of interest is expressed as a fusion to a RNA polymerase subunit, a library of randomized oligonucleotides is cloned upstream of the promoter of two selectable markers, and the bacterial strain is used for selection of PDIs. When a DNA bait is recognized by the prey protein, RNA polymerase will be recruited to the promoter and will activate the reporter gene expression. To reduce false positives, self-activating baits are selected from the library of randomized oligonucleotides using URA3 reporter.
Fig. 5.
Workflow of bacteria one hybrid (B1H)
The selection procedure of B1H is rapid because only a single round of selection is required to identify a set of TF binding sites. However, the B1H system may not be suited for determining the specificity for all the DBDs or TFs, as some foreign TFs may be toxic to the bacteria, resulting in an insufficient number of clones. Furthermore, the use of a prokaryotic selection system implies that eukaryotic-specific protein cofactors, such as histones, and a whole host of post-translational modifications will be absent. This may result in PDIs that differ from those measured in the Y1H system.
Applications
In the manuscript that first described the B1H system, the Wolfe group analyzed 84 homeodomains from Drosophila [36]. The majority of those factors can be organized into 11 different DNA-binding specificity groups, with additional six homeodomain TFs that display unique specificities. They further tested DNA-binding specificities for 16 mutant homeodomain TFs containing point mutations in residues that contribute to DNA recognition and yielded reasonable agreement with the prediction. The authors also predicted with a high success rate the binding specificities of homeodomain TFs from other organisms based on the DNA-binding profiles of Drosophila TFs.
Protein microarray
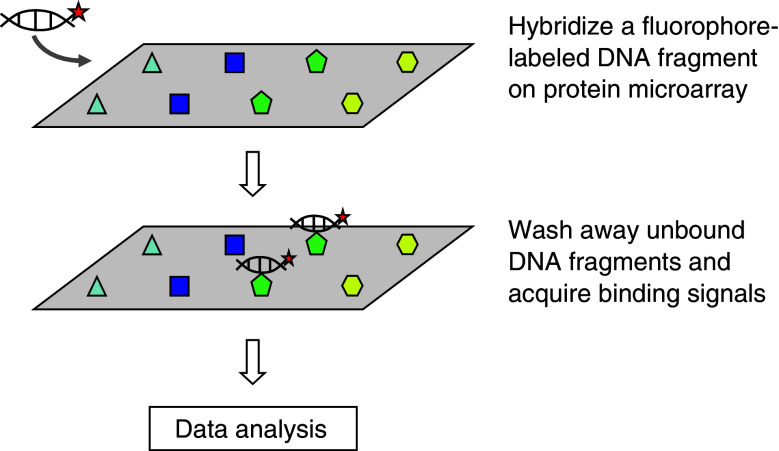
In contrast to the PBM approach, a labeled DNA motif is queried against thousands of proteins on a glass slide using a functional protein microarray approach (Fig. 6). This approach is complementary to the PBM in terms of the methodology and questions to ask. By definition, a functional protein microarray is formed by immobilizing thousands of individually purified proteins in a single glass slide [37, 38]. A functional protein microarray offers a flexible platform that has been applied to characterize many different types of protein activity, including protein-DNA, protein-RNA, protein-lipid, protein-drug, protein-protein, and protein-antibody interactions [39–42]. Moreover, a unique application of the functional protein microarrays is to identify substrates of various enzymes. Protein arrays have been used to identify proteins modified by phosphorylation, ubiquitylation, acetylation, and SUMOylation [41, 43–48].
Fig. 6.
Workflow of protein microarray
When a functional protein microarray is used for analysis of PDIs, it is not intended to comprehensively survey the entire 8-mer DNA space as in the PBM approach; rather, it is designed to simultaneously screen a large number of proteins for their ability to selectively interact with an individual DNA sequence. For example, when one seeks to identify TFs that can bind to the promoter region of a gene of interest, or to a predicted DNA motif, one can label the DNA fragment and probe it to a protein microarray composed of thousands of TF proteins. In addition, one need not limit one’s search simply to annotated TFs but can also interrogate proteins from a broad range of other functional classes to determine whether they might also show unexpected sequence-specific DNA-binding activity.
Though it is still technically challenging to fabricate a functional protein microarray of high content and high density, once it is made, it becomes rather easy to screen hundreds of labeled DNA probes, generated either by PCR reactions or DNA oligo synthesis. Protein-DNA binding can be measured by the signal intensity of fluorescence after probing protein microarray with DNA samples. It should be noted that for a similar reason that the PBM assay requires extensive washing to avoid nonspecific binding, protein microarrays can only detect interactions with high affinity. Because this approach provides simultaneous profiles of PDIs for thousands of proteins and because multiple DNA probes can be tested in parallel, it enables a rapid mapping of PDIs on a proteome-wide scale.
Applications
To identify novel DNA-binding activity in yeast, Hall et al. [44] probed a yeast proteome microarray composed of 5,800 yeast proteins with labeled total genomic DNA. They identified >200 proteins, half of which were not previously known to encode any DNA-binding activity. To validate this surprising result, eight of them were chosen and four were confirmed to interact with DNA in vivo by using ChIP-chip assay. They then focused on a yeast metabolic enzyme Arg5,6, which is involved in arginine biosynthesis, and found that it indeed bound to a specific DNA motif in the promoter of a mitochondrial gene, COX1, and regulated its expression in vivo. This study is the first to demonstrate the power of functional protein microarray in identifying unconventional DNA-binding proteins (uDBPs).
Later, the Snyder and Johnston groups collaborated to identify yeast TFs that could bind to 75 evolutionarily conserved DNA motifs using a yeast TF microarray containing 287 known and predicted yeast TF proteins [45]. Besides a number of previously known PDIs they recovered, they also identified more than 100 novel PDIs. One of the previously uncharacterized DNA-binding proteins, Yjl103, was validated both in vitro and in vivo.
More recently, protein microarrays containing 802 Arabidopsis TFs were constructed for identification of protein-DNA and protein-protein interactions. To evaluate the efficacy of detecting protein–DNA-binding activity, the authors focused on the AP2/ERF family by designing dsDNAs based on the known binding sites for four representative members [49]. Subsequent analysis showed that in addition to the eight previously reported TFs, the microarray analysis also detected sequence-specific DNA binding by 49 previously uncharacterized members of the AP2/ERF family.
To date, the largest PDI study using protein microarray was undertaken by our team using protein microarrays composed of 4,191 unique human proteins in full length probed with 460 individual DNA motifs [41]. The unique feature of this human protein microarray is that it not only contains ~90% of known and predicted human TFs, but also proteins that were not previously known to specifically interact with DNA (i.e., uDBPs), including protein kinases, RNA binding proteins, transcription coregulators, and many metabolic enzymes. Of the 460 DNA motifs, 400 were transcriptional regulatory motifs predicted using various bioinformatic algorithms based on comparative genomics, tissue specificity, and gene expression profiles [50–54]. However, the proteins that bound these sites were not known. The other 60 motifs were known ones retrieved from the TRANSFAC database and used as positive controls [18].
To identify proteins that specifically recognize these DNA motifs, protein microarrays were individually probed with 460 labeled DNA motifs. A total of 17,718 PDIs were determined using a stringent identification threshold. Many known PDIs were recovered for TFs, illustrating the high quality of our PDI dataset. More importantly, many new DNA consensus sites were determined for over 200 TFs, which doubled the number of previously reported DNA motifs for human TFs [41, 55]. EMSA analysis was used to confirm 17 of 21 (82%) new PDIs between a TF and its predicted consensus sequence, suggesting that these newly identified PDIs were also of high fidelity.
Surprisingly, over 300 proteins previously not known to specifically bind to DNA showed sequence-specific PDIs. Forty-one of the 45 randomly chosen PDIs between a uDBP and its predicted motif were validated by EMSA. These uDBPs include RNA-binding proteins, chromatin-associated proteins, nucleotide-binding proteins, mitochondrial proteins, protein kinases, and metabolic enzymes, which raises an intriguing hypothesis that many uDBPs may regulate transcription as a moonlighting function. To further investigate the physiological relevance of the DNA-binding activities of these uDBPs, we decided to focus on a well studied MAP kinase, ERK2. Using a series of in vitro and in vivo approaches, including EMSA, luciferase reporter assay, mutagenesis, and ChIP, a novel DBD was identified in ERK2 and showed that its DNA-binding activity is independent of ERK2’s kinase activity. Using a combination of mutants detected in DNA binding or kinase activity, it was further demonstrated that ERK2 directly regulates expression of IRF9 and OAS1, two interferon-induced genes, which led us to determine that ERK2 acts as a transcription repressor during the course of the interferon-gamma signaling in cells. The study illustrates the power of protein microarrays used for an unbiased identification of PDIs on a proteome-wide scale.
A complex landscape of DNA-binding activities
DNA-binding specificities for TFs
Although it was known that some homeodomain TFs recognize substantially different DNA sequences using a common DNA-binding structure [56, 57], the large-scale PDI studies as described above provided dramatic confirmation that recognition of multiple different DNA sequences is quite common among members of this protein family [24, 36, 58]. In general, overall amino acid sequence similarity in the homeodomain correlates overall with DNA-binding specificity. There are also many cases that two homeodomains with very similar protein sequences bind to diverse DNA sequences. This is not completely unanticipated, as it has long been known that single amino acid changes are sufficient to drastically alter DNA-binding specificity for some TFs, both by directly altering DNA-contacting residues and by changing the secondary structure of the DBD [59, 60]. Some later studies examining the DNA sequence preferences for many members of other DBD family extended this observation, reporting that diverse DNA-binding specificities exist for similarly related proteins in some other DBD families, such as nuclear hormone receptors [27].
A more recent study by Badis et al. makes the concept of diverse DNA-binding specificities even more complex [27]. Secondary binding sites were found for a large fraction of TFs tested using PBM (nearly half of 104 mouse TFs). Based on the different types of secondary sites, the authors categorized the binding activities into four classes: (1) variable spacer lengths, (2) positional interdependence of base preference, (3) alternate recognition interfaces, and (4) multiple combinations of the first three classes. To find out the possible physiological relevance of these secondary binding sites, the authors analyzed in vivo usage of secondary motifs by considering their TF occupancy. Interestingly, the secondary motifs are enriched among sequences bound by the TF that lack primary motifs for this TF, suggesting the secondary motif may recruit the TFs to genomic loci independently of the primary motif.
Unconventional DNA-binding proteins
Perhaps one of the most striking findings among the large-scale PDI studies is the notion that a large number of proteins that are not currently annotated as TFs do in fact function as TFs [34, 41, 61, 62]. These studies add yet another dimension to this increasingly complex model of transcriptional regulation by identifying more players from a variety of different protein families. They also broaden our functional understanding of a large number of proteins. Recent data, primarily from microorganisms, have begun to indicate that many proteins belonging to well-annotated gene families are actually associated with unconventional or moonlighting functions, among them regulation of transcription [63, 64]. We envision that such moonlighting functions may also be widespread in higher eukaryotes, given the fact that the study only examined around 20% of human proteins.
In vitro versus in vivo DNA-binding specificities
Except for the ChIP-based approaches, all the techniques we discussed in this review interrogate PDIs either in vitro—as is the case for SELEX, PBMs, and protein microarrays—or, in the case of one-hybrid approaches, in heterologous cell-based systems. The direct physiological relevance of data obtained using these approaches is thus not necessarily clear. However, for many domain classes, and in organisms ranging from yeast to human, in vivo binding sites detected by ChIP-based approaches typically contain sequences that reflect those preferred in vitro [15, 24, 65]. For four Drosophila homeodomain-containing proteins, a good correlation was found between monomer binding in vitro and in vivo on many actively transcribed genes [66]. Berger et al. examined six ChIP-chip or ChIP-seq data sets in the literature that involved immunoprecipitation of homeodomain proteins analyzed by PBM [24]. They observed a strong enrichment for binding sites identified by PBM in immunoprecipitated genomic DNA, suggesting a good agreement between ChIP and PBM data. A similar observation was also found for ETS proteins between the in vitro data studied by PBM and in vivo data studied by ChIP-seq [67]. Jolma et al. analyzed whether the binding profiles obtained via SELEX matched those detected using ChIP-seq for 14 different human TF classes. They tested two cases and found that TF binding sequences identified using ChIP-seq closely matched those obtained using SELEX [20]. More interestingly, good agreement between in vitro and in vivo data was also seen for uDBPs, where protein microarray was used for in vitro PDI identification [41].
On the other hand, there is also ample evidence that DNA-binding sequences of a TF retrieved from in vitro experiments may not necessarily match those seen in vivo [68]. For example, the majority of sequences bound in vivo by the TF HNF1 do not contain the highest affinity binding sequences identified in vitro by PBM analysis [24]. There are several reasons that may explain the disparity of in vitro and in vivo DNA-binding motifs: (1) A TF may interact with other proteins in vivo (coregulators), which may induce its conformational change. Therefore, the TF may bind to a different DNA sequence that does not correspond to its in vitro DNA-binding sequences [69]. (2) The accessibility of cis-elements in vivo is greatly influenced by the packaging of genomic DNA in chromatin [70]. (3) ChIP-based approaches only measure PDI occurring in a particular cell or at a particular time point. (4) PDIs identified by ChIP-based approaches may reflect both direct interactions between genomic DNA and the TF of interest, and indirect interactions between DNA and protein complex containing the TF of interest. So the observed binding sequences may in fact be mediated by other DNA proteins than the target TF [65].
Given the complementary strengths and weaknesses of the various technologies available for analyzing PDIs, an integrated approach that brings biochemical, genetic, and ChIP-based approaches to bear on the problem may ultimately prove the best approach to obtain a clear picture of the myriad PDIs that underlie the cellular transcriptional regulatory network.
Conclusion
Experimental technologies have significantly advanced in the last decade, allowing the high-throughput analysis of PDIs. A number of complementary approaches are now available to characterize PDIs with substantially lower cost and higher resolution than were previously possible. Each of these approaches has unique strengths and limitations, and investigators should carefully choose or combine the approaches before designing experiments.
Many recent studies using these technologies have provided large-scale, high quality sets of PDI data from multiple different organisms. These resources will greatly facilitate analysis of the complex DNA-binding mechanisms for proteins. Furthermore, many findings also challenged some conventional concepts of PDI, such as multiple distinct sequence preferences observed for some TFs and the recent detection of large numbers of different unconventional DNA-binding proteins.
Although we have addressed six high-throughput approaches for charactering PDIs here, this is by no means an exhaustive review describing all the recent advances in this field. Some other important technologies include the luciferase-based PDI mapping technique [71] and the microfluidics-based PBM approach [72], where both techniques can not only discover DNA-binding sites for a given TF, but also measure relative binding affinity.
Finally, it should be noted that analysis of PDIs is just one step in understanding complex transcriptional regulatory networks. In many cases, monomeric protein binding sites are not sufficient for the functions of a protein, as we have discussed in the previous section. There are also many examples in which DNA-binding proteins often bind DNA cooperatively to create protein complexes that precisely control gene expression [73]. Furthermore, the recent findings suggest that the sequence context of a binding site may also influence binding energetics [74]. In addition, there are now more than 1,500 structures of protein-DNA complexes available in the Protein Data Bank [75] that are critical in understanding biophysical roles of molecules in the protein-DNA structures [76]. In the future, understanding of complex DNA-binding specificities of proteins will benefit from an integrated analysis of multiple data, including in vitro DNA-binding sites, in vivo PDIs, protein-protein interactions, protein-DNA structures, and other biochemical and biophysical data.
Acknowledgments
We are grateful to NIH for funding support.
References
- 1.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 2.Lane D, Prentki P, Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992;56:509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. Footprinting: a method for determining the sequence selectivity, affinity and kinetics of DNA-binding ligands. Methods. 2007;42:128–140. doi: 10.1016/j.ymeth.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 5.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walhout AJ. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- 8.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 9.Massie CE, Mills IG. ChIPping away at gene regulation. EMBO Rep. 2008;9:337–343. doi: 10.1038/embor.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 11.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 12.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 14.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 15.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae . Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 17.Gordan R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roulet E, Busso S, Camargo AA, Simpson AJ, Mermod N, Bucher P. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- 20.Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpaa MJ, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci USA. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc Natl Acad Sci USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- 29.Yokoe H, Anholt RR. Molecular cloning of olfactomedin, an extracellular matrix protein specific to olfactory neuroepithelium. Proc Natl Acad Sci USA. 1993;90:4655–4659. doi: 10.1073/pnas.90.10.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 31.Reece-Hoyes JS, Deplancke B, Barrasa MI, Hatzold J, Smit RB, Arda HE, Pope PA, Gaudet J, Conradt B, Walhout AJ. The C. elegans Snail homolog CES-1 can activate gene expression in vivo and share targets with bHLH transcription factors. Nucleic Acids Res. 2009;37:3689–3698. doi: 10.1093/nar/gkp232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng J, Yan J, Wang T, Mosbrook-Davis D, Dolan KT, Christensen R, Stormo GD, Haussler D, Lathrop RH, Brachmann RK, et al. Genome wide screens in yeast to identify potential binding sites and target genes of DNA-binding proteins. Nucleic Acids Res. 2008;36:e8. doi: 10.1093/nar/gkm1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deplancke B, Dupuy D, Vidal M, Walhout AJ. A gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stamm L, Reece-Hoyes JS, Hope IA, et al. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 35.Meng X, Brodsky MH, Wolfe SA. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat Biotechnol. 2005;23:988–994. doi: 10.1038/nbt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CS, Zhu H (2006) Protein microarrays. Biotechniques 40:423, 425, 427 passim [DOI] [PubMed]
- 38.Tao SC, Chen CS, Zhu H. Applications of protein microarray technology. Comb Chem High Throughput Screen. 2007;10:706–718. doi: 10.2174/138620707782507386. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J, Gopinath K, Murali A, Yi G, Hayward SD, Zhu H, Kao C. RNA-binding proteins that inhibit RNA virus infection. Proc Natl Acad Sci USA. 2007;104:3129–3134. doi: 10.1073/pnas.0611617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao SC, Li Y, Zhou J, Qian J, Schnaar RL, Zhang Y, Goldstein IJ, Zhu H, Schneck JP. Lectin microarrays identify cell-specific and functionally significant cell surface glycan markers. Glycobiology. 2008;18:761–769. doi: 10.1093/glycob/cwn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu J, Liao G, Shan L, Zhang J, Chen MR, Hayward GS, Hayward SD, Desai P, Zhu H. Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J Virol. 2009;83:5219–5231. doi: 10.1128/JVI.02378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, Hewison M, Adams JS. Functional characterization of heterogeneous nuclear ribonuclear protein C1/C2 in vitamin D resistance: a novel response element-binding protein. J Biol Chem. 2006;281:39114–39120. doi: 10.1074/jbc.M608006200. [DOI] [PubMed] [Google Scholar]
- 44.Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- 45.Ho SW, Jona G, Chen CT, Johnston M, Snyder M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc Natl Acad Sci USA. 2006;103:9940–9945. doi: 10.1073/pnas.0509185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 47.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 48.Lin J, Xie Z, Zhu H, Qian J. Understanding protein phosphorylation on a systems level. Brief Funct Genomics. 2010;9:32–42. doi: 10.1093/bfgp/elp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong W, He K, Covington M, Dinesh-Kumar SP, Snyder M, Harmer SL, Zhu YX, Deng XW. The development of protein microarrays and their applications in DNA-protein and protein-protein interaction analyses of Arabidopsis transcription factors. Mol Plant. 2008;1:27–41. doi: 10.1093/mp/ssm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elemento O, Tavazoie S. Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol. 2005;6:R18. doi: 10.1186/gb-2005-6-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci USA. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu X, Lin J, Zack DJ, Qian J. Computational analysis of tissue-specific combinatorial gene regulation: predicting interaction between transcription factors in human tissues. Nucleic Acids Res. 2006;34:4925–4936. doi: 10.1093/nar/gkl595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Z, Hu S, Blackshaw S, Zhu H, Qian J. hPDI: a database of experimental human protein–DNA interactions. Bioinformatics. 2010;26(2):287–289. doi: 10.1093/bioinformatics/btp631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-L. [DOI] [PubMed] [Google Scholar]
- 57.Wolberger C, Vershon AK, Liu B, Johnson AD, Pabo CO. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 58.Affolter M, Slattery M, Mann RS. A lexicon for homeodomain-DNA recognition. Cell. 2008;133:1133–1135. doi: 10.1016/j.cell.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 59.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-X. [DOI] [PubMed] [Google Scholar]
- 60.Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: structures of two Cys2His2 zinc finger/TATA box complexes. Structure. 2001;9:717–723. doi: 10.1016/S0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 61.Dioum EM, Wauson EM, Cobb MH. MAP-ping unconventional protein-DNA interactions. Cell. 2009;139:462–463. doi: 10.1016/j.cell.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 62.Casci T. Gene expression: regulators hidden in human proteome. Nat Rev Genet. 2009;10:820. [Google Scholar]
- 63.Gancedo C, Flores CL. Moonlighting proteins in yeasts. Microbiol Mol Biol Rev. 2008;72:197–210. doi: 10.1128/MMBR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huberts DHEW, van der Klei IJ. Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta Mol Cell Res. 2010;1803:520–525. doi: 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 65.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 66.Carr A, Biggin MD. A comparison of in vivo and in vitro DNA-binding specificities suggests a new model for homeoprotein DNA binding in Drosophila embryos. Embo J. 1999;18:1598–1608. doi: 10.1093/emboj/18.6.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. Embo J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horie-Inoue K, Takayama K, Bono HU, Ouchi Y, Okazaki Y, Inoue S. Identification of novel steroid target genes through the combination of bioinformatics and functional analysis of hormone response elements. Biochem Biophys Res Commun. 2006;339:99–106. doi: 10.1016/j.bbrc.2005.10.188. [DOI] [PubMed] [Google Scholar]
- 69.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 70.Simpson RT. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 71.Hallikas OK, Aaltonen JM, von Koskull H, Lindberg LA, Valmu L, Kalkkinen N, Wahlstrom T, Kataoka H, Andersson L, Lindholm D, et al. Identification of antibodies against HAI-1 and integrin alpha6beta4 as immunohistochemical markers of human villous cytotrophoblast. J Histochem Cytochem. 2006;54:745–752. doi: 10.1369/jhc.5A6816.2006. [DOI] [PubMed] [Google Scholar]
- 72.Fordyce PM, Gerber D, Tran D, Zheng J, Li H, DeRisi JL, Quake SR. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat Biotechnol. 2010;28:970–975. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ptashne M, Gann A. Genes and signals. New York: Cold Spring Harbor Laboratory; 2002. [Google Scholar]
- 74.Carlson CD, Warren CL, Hauschild KE, Ozers MS, Qadir N, Bhimsaria D, Lee Y, Cerrina F, Ansari AZ. Specificity landscapes of DNA binding molecules elucidate biological function. Proc Natl Acad Sci USA. 2010;107:4544–4549. doi: 10.1073/pnas.0914023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]