Abstract
Class I Histone deacetylases (HDACs) play a central role in controlling cell cycle regulation, cell differentiation, and tissue development. These enzymes exert their function by deacetylating histones and a growing number of non-histone proteins, thereby regulating gene expression and several other cellular processes. Class I HDACs comprise four members: HDAC1, 2, 3, and 8. Deletion and/or overexpression of these enzymes in mammalian systems has provided important insights about their functions and mechanisms of action which are reviewed here. In particular, unique as well as redundant functions have been identified in several paradigms. Studies with small molecule inhibitors of HDACs have demonstrated the medical relevance of these enzymes and their potential as therapeutic targets in cancer and other pathological conditions. Going forward, better understanding the specific role of individual HDACs in normal physiology as well as in pathological settings will be crucial to exploit this protein family as a useful therapeutic target in a range of diseases. Further dissection of the pathways they impinge on and of their targets, in chromatin or otherwise, will form important avenues of research for the future.
Keywords: HDAC, Cell cycle, Differentiation, Development, Chromatin, HDAC inhibitors, Deacetylation, Transcription factors
Introduction
Acetylation of histones is a dynamic process controlled by two families of enzymes: histone acetyl transferases (HATs) and histone deacetylases (HDACs). HATs catalyze the transfer of an acetyl group to lysine residues of histone tails, thereby neutralizing the positive charge of histones. This change decreases the affinity between DNA and histones and relaxes chromatin structure, making it more accessible for transcription factors. Acetylated histones also act as docking sites for transcription factors carrying a bromodomain [1]. Therefore, HATs are considered as transcription co-activators. The activity of HATs is balanced by the HDACs that deacetylate histone tails, promote chromatin compaction, and act as co-repressors. In addition to histones, HDACs can deacetylate non-histone proteins such as transcription factors and a growing number of other proteins [2, 3]. As a result, HDACs are now also called KDACs, or lysine deacetylases [4]. Recently, mass spectrometric analysis identified several thousand acetylation sites in HeLa cells, a large fraction of which was altered upon treatment of cells with HDAC inhibitors (HDACis) [3]. The large number of potential HDACs substrates suggests that these enzymes may play diverse functions in many cellular processes beyond the regulation of gene expression. Many of the early investigations on HDACs function have been based on HDACis and their effect on tumor cell proliferation, in particular induction of cell cycle arrest and apoptosis. Although these initial studies demonstrated the important role of HDACs in cell cycle regulation, the specific role of individual members of the HDAC family has only recently been addressed genetically. Several recent studies based on deletion and/or overexpression of specific HDACs have highlighted the complex network of cellular processes that involve HDACs action.
In this review, we will focus on the role of class I HDACs in cell cycle regulation, cell differentiation, and tissue development in mammalian systems. We first briefly describe the properties of the class I members and their multisubunit complexes. Then, we describe the phenotypes linked to the deletion or overexpression of these enzymes in mammalian systems and the molecular mechanisms involved in their function. Finally, we discuss the effects of HDACis as promising anti-cancer drugs.
Protein deacetylases
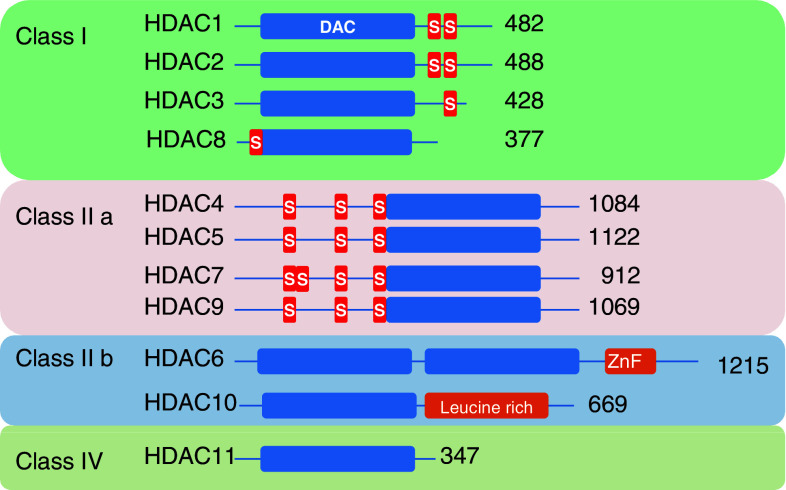
The mammalian genomes encode 18 enzymes with deacetylase activity which comprise two distinct groups: classical HDACs, whose enzymatic activity requires Zn2+, and sirtuins, which are NAD+-dependent. The classical HDAC family contains 11 members divided into four classes. Class I contains HDAC1, 2, 3, and 8, class IIa comprises HDAC4, 5, 7, and 9, class IIb includes HDAC6 and 10, and class IV comprises the unique member HDAC11 (Fig. 1).
Fig. 1.
HDAC classification. The HDAC classes are indicated (class III HDACs—sirtuins—are not depicted). Blue rectangles depict the Deacetylase Domain (DAC), red rectangles indicate phosphorylation sites; Zinc finger (ZnF) and leucine rich motifs are also indicated. The numbers specify the number of amino acids
The sirtuin enzymes are generally classified as class III HDACs. This family contains seven members, SIRT1–SIRT7 which are organized into four classes. Sirtuin class I contains SIRT1, 2, and 3. SIRT4 and SIRT5 constitute class II and III, respectively, and class IV comprises SIRT6 and SIRT7. Sirtuins are involved in many biological processes including genome stability, cell cycle regulation and play critical roles in several metabolic pathways (for review, see [5]). The reader is referred to several excellent reviews discussing the classification and regulation of the different HDACs [6, 7]. Here, we focus on summarizing what is known about class I HDACs, and, in particular, what has been learned from their genetic analysis in higher eukaryotes.
Class I HDACs complexes and their function in mammals
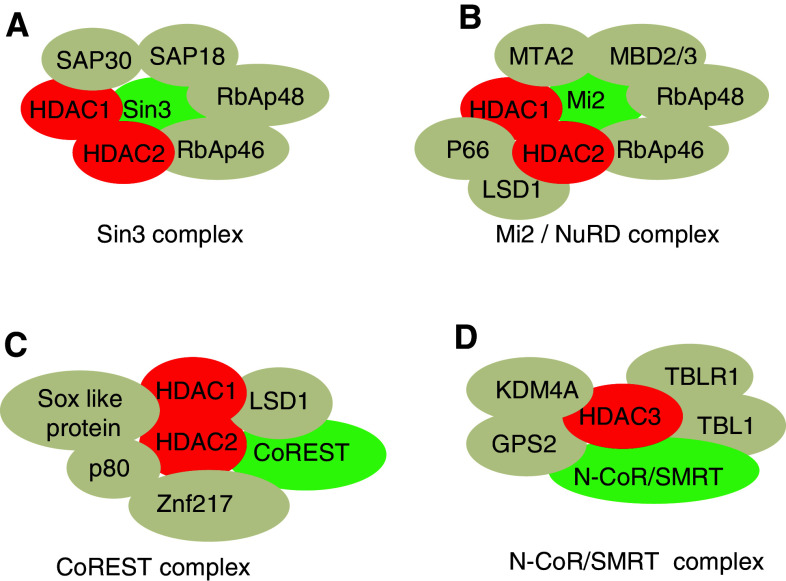
HDACs lack a DNA-binding motif and require interaction with other proteins to be recruited to their chromatin targets. Class I HDACs, except HDAC8, are found as subunits of several multisubunit complexes and interact with various transcription factors. Mammalian HDAC1 and 2 originated from a common ancestor via gene duplication [8]. They exhibit high sequence homology, having 87% amino acid identity in mice (Fig. 1), and their C-terminal tail contains tandem casein kinase-2 (CK2) phosphorylation sites [9]. HDAC3 shares the homologous catalytic domain with HDAC1 and 2, but has only one CK2 phosphorylation site [10]. HDAC8 has a conserved motif for protein kinase A phosphorylation [8, 11]. HDAC1 and 2 form a heterodimer and constitute the catalytic core of the Sin3, NuRD, and CoREST complexes, while HDAC3 is a subunit of the N-CoR/SMRT complex (Fig. 2). Class I HDAC-containing complexes may have different subunits in different cell types, at specific developmental stages or depending on the purification methods (for reviews, see [4, 12, 13]).
Fig. 2.
Class I HDACs complexes and their components. a The Sin3 complex contains six subunits: the transcriptional co-repressor Sin3, two Sin3 associated proteins (SAP18 and 30) and two Rb associated proteins (RbAp46 and 48). HDAC1 and 2 form the catalytic core of this complex [115, 116]. b The Mi2/NuRD complex shares the dimers HDAC1 and 2 and RbAP46 and 48 with the Sin3 complex. In addition this complex contains the chromatin remodeler Mi2, the methyl CpG binding domain protein MBD2 or MBD3, the lysine-specific demethylase LSD1 [117], the transcriptional repressor p66 and MTA2 (a member of metastasis associated family) [118–120]. c The CoREST complex contains the co-repressor CoREST, the lysine-specific demethylase LSD1, the Kruppel-like zinc-finger protein Znf217, the Sox-like protein and p80 [121]. d The N-CoR/SMRT complex contains N-CoR (Nuclear receptor CoRepressor) or SMRT (Silencing Mediator for Retinoid and Thyroid receptor), TBL1 (transducin β-like 1), TBLR1 (TBL related 1), GPS2 (G-protein pathway suppressor 2), the lysine-specific demethylase KDM4A [122] and HDAC3 ([123]; for review, see [12]). HDAC complexes are reviewed in [4, 13]
In mammals, class I HDACs are ubiquitously expressed suggesting a general role in transcription repression. However, deletion of these enzymes individually leads to deregulation of a limited set of genes, indicating a specific role of class I HDACs in transcription regulation [14, 15]. Furthermore, class I HDACs interact with many lineage-specific transcription factors suggesting that these enzymes may play an important role in controlling specific transcriptional programs. The majority of recent findings about the role of class I HDACs come from studies based on deletion or overexpression of these enzymes in mammalian systems. In the following section, we will discuss the phenotypes linked to the deregulation of class I HDACs expression with emphasis on knockout (KO) mouse models, either globally or in a tissue-specific manner. We will also discuss the molecular mechanisms involved in HDACs action.
Germline deletion of class I HDACs
Every class I HDAC has been deleted either globally or in a tissue-specific manner in the mouse (see Table 1). Germline deletion of HDAC1 in mice leads to early embryonic lethality before E9.5 due to severe proliferation defects and retardation in development [16]. In contrast to the clear effect of HDAC1 global deletion, the impact of HDAC2 ablation in mice varies depending on the genetic approach used. Indeed, independent studies investigating the effect of HDAC2 ablation in mice reported somewhat divergent conclusions. The various strategies used to delete HDAC2 might explain—at least in part—these different findings. The Hdac2 KO was performed by Montgomery and colleagues [17] using a conditionally targeted Hdac2 locus which leads to deletion of exons 2, 3, and 4, thus compromising part of the (oligo) hetero-/homodimerization domain and the catalytic domain required for enzymatic activity. This strategy generated a mutant Hdac2 allele with two splice variants, an all-in-frame transcript and an out-of-frame transcript. In mice carrying this modified allele, no HDAC2 protein was detectable. Although born at the expected Mendelian ratio, these Hdac2-deficient mice died within 24 h after birth [17]. Detailed histological analysis of these Hdac2-deficient neonates revealed morphological changes of the heart [17]. The complete lethality of these HADC2-null mice contrasts with the findings in several other studies reporting that the majority of Hdac2-deficient mice were partially viable with a transient growth retardation phenotype. Deletion of HDAC2 using a gene-trap insertion led to partial embryonic lethality, with about 50% death within the first months [18, 19]. In this case, the random insertion of LacZ into the Hdac2 locus exon 9 generated an enzymatically inactive, in-frame HDAC2–LacZ fusion protein which lacks the C-terminus [19]. Finally, similar results were obtained with a conditional gene targeting of Hdac2 exons 5 and 6 by Guan and colleagues and of Hdac2 exon 6 in our laboratory; these alleles affected the histone deacetylase domain and also led to partial perinatal lethality and reduced body size [15, 20]. In the detailed study from Trivedi and colleagues [18], the postnatal mutant Hdac2 mice had smaller hearts than wild-type mice and a thickened myocardium at day 8 after birth. In Hdac2-deficient hearts, the gene encoding inositol polyphosphate-5-phosphatase f (Inpp5f) was upregulated, resulting in constitutive activation of glycogen synthase kinase 3β (GSK3β) and protection from hypertrophy [18]. Chemical inhibition of activated Gsk3β caused these Hdac2-deficient mice to become sensitive to hypertrophic stimulation. In contrast, transgenic mice overexpressing HDAC2 displayed increased hypertrophy coupled to inactivated Gsk3β [18]. This suggested that HDAC2 and GSK3β are components of a regulatory pathway involved in cardiac hypertrophic response. The KO studies of HDAC1 and 2 demonstrated specific and distinct roles of these two enzymes in mouse embryogenesis and development. Global Hdac3 KO led to gastrulation defects and death of the embryo prior to E9.5 [21, 22]. In contrast, HDAC8 deletion led to perinatal lethality due to skull instability [23].
Table 1.
List of phenotypes observed in mice
| Target organ/tissue | Cre driver | Observation/phenotype | Reference | |
|---|---|---|---|---|
| HDAC1 and 2 | Germline | Global | H1 KO lethal E9.5–10.5 due to proliferation defect and dev. retardation | [16, 17]; Yamaguchi, p.c. |
| Germline | Global | H1 KO combined with p21 KO no rescue possible | [14] | |
| Germline | Global | H2 KO neonates die shortly after birth (24 h), due to cardiac defects | [17] | |
| Germline | Global | H2 KO 50% perinatal lethal (1 mo), smaller in size, hypertrophy in the heart, 50% survive until adulthood | [15, 18–20] | |
| T cell dev. | Cd4-cre | H1 KO leads to increased proliferation and increased inflammation response in an allergic airway model | [54] | |
| Hippocampal/Neurons | Transfection | H1 OE protects neurons from DNA damage and neurotoxicity | [49] | |
| Synapse dev. | Lentiviral infection | H2 KO leads to reduced synaptic activity; DKO increased synaptic activity and synapse numbers, involved in synaptic maturation | [51] | |
| Neurons | Nestin-cre | H2 KO leads to enhanced learning and memory formation; H2 OE leads to impairment, rescue with HDACis possible | [20] | |
| Brain | Global and GLAST::CreERT2/Z/EG | H2 KO leads to differentiation block and apoptosis during adult neurogenesis | [47] | |
| CNS | GFAP-cre | DKO in neuronal progenitors increases proliferation and block in diff., alters lineage-specificity | [45] | |
| Oligodendrocyte diff. | Olig1-cre | DKO mice die postnatal 2 w, required for oligodendrocyte specification and diff. | [46] | |
| PNS | Dhh-cre | DKO in PNS leads to apoptosis of Schwann cells and abrogated myelination | [42, 48] | |
| Heart | MHC-cre | DKO is neonatal lethal due to cardiac defects | [17] | |
| Epidermis | KRT14-cre | DKO leads to impaired epidermal and hair follicle development, perinatal lethal, block in cell cycle | [25] | |
| Liver | Mx-cre | DKO in hematoptiesis alters erythrocyte–megakaryocyte diff., increase in apoptosis | [24] | |
| B-cell dev. | Mb1-cre or CD23-cre | DKO impairs B-cell diff., cell cyle G1 blocks and increased apoptosis (p21 not altered, own observation) | [15] | |
| HDAC3 | Germline | Global | lethal prior to E9.5 due to gastrulation defects | [22, 58] |
| Liver | Albumin cre | Long term study leads to genomic instability and to hepatocellular carcinoma (mean survival 10 mos) | [58] | |
| Liver | Albumin cre | Leads to increased liver size, metabolic changes carbohydrate and lipid imbalance | [59] | |
| Heart | MHC-cre | Cardiac-specific loss leads to lethality by 3–4 mos, due to cardiac defects, hypertrophy and deregulated metabolism | [22] | |
| Heart | Myocyte-specific OE | Heart defects without hypertrophy | [61] | |
| Skeletal dev. | Osterix-cre | Causes bone formation defects, reduced volume and cell number | [62] | |
| Brain | Genetic and pharmacological approach | Enhanced long-term memory formation, | [64] | |
| HDAC8 | Germline | Global | Perinatal lethal, due to skull instability (craniofacial defects) | [23] |
| Neural crest | Wnt1-cre | Phenocopied the global deletion, skull dysmorphism | [23] |
Knockout (KO) and overexpression (OE) of class I HDACs
H1 HDAC1, H2 HDAC2, DKO double knockout HDAC1 and 2, h hour, w week, mo month, dev. development, diff. differentiation, p.c. personal communication, PNS peripheral nervous system, CNS central nervous system
Tissue-specific analysis of class I HDACs
To overcome the early embryonic lethality associated with class I HDAC ablation, conditional alleles in combination with various cre-expressing strains were generated. These strategies allowed the investigation of the function of class I HDACs in specific tissues. In addition to KO experiments, several ex vivo studies based on cultured cell lines were conducted to address the mechanisms involved in class I HDACs functions.
HDAC1 and 2
Role of HDAC1 and 2 in cell cycle regulation and DNA damage control
HDAC1 and 2 are usually co-expressed and often show redundancy in their function in many adult tissues and cultured cell lines [15, 24, 25]. However, several studies reported specific functions of these two homologues during development. It is important to mention that, in several cellular systems, deletion of HDAC1 leads to an increased level of HDAC2 protein and vice versa, without corresponding change of the mRNA level [15, 16, 24, 26]. This compensatory effect suggests common mechanisms regulating HDAC1 and 2 protein levels and reinforces the idea that these two proteins are implicated in shared regulatory pathways. Several studies have demonstrated that HDAC1 and 2 play a crucial role in cell cycle regulation. Hdac1-deficient embryonic stem cells show reduced proliferation, which correlates with decreased cyclin A and cyclin E-associated kinase activities and elevated levels of the cyclin-dependent kinase inhibitors p21 and p27; this is correlated to hyperacetylation of histone H3 and H4 on the promoters of these genes, suggesting that these promoters may be direct targets of HDAC1 in ES cells [16]. Indeed, chromatin immunoprecipitation (ChIP) experiments showed that HDAC1 binds to the p21 promoter in ES cells. Importantly, ablation of p21 in Hdac1-deficient ES cells rescues the proliferation defect; however, this does not overcome the embryonic developmental block, indicating that this phenotype is not only due to a proliferation defect [14].
In primary mouse embryonic fibroblasts (MEFs), deletion of HDAC1 reduces proliferation and causes a partial G1/S arrest, while deletion of HDAC2 has little or no effect on proliferation [15]. By contrast, combined deletion of both enzymes led to strong cell cycle arrest at the G1 phase accompanied by concomitant upregulation of p21 and p57, followed by apoptosis. This upregulation is direct, since HDAC1 and 2 bind to the p21 and p57 promoters and can repress their transcription. Importantly, siRNA-mediated knockdown of these two genes partially rescued the proliferation defect [15]. In another study, it was also shown that deletion of HDAC1 and 2 or inactivation of their deacetylase activity in primary or oncogenic-transformed fibroblasts led to senescence-like G1 cell cycle arrest accompanied by upregulation of p21 and p53. Strikingly, in this case, ablation by siRNA of these two proteins did not rescue the proliferation defect, indicating that HDAC1 and 2 also regulate p53–p21 independent pathways critical for maintaining cell cycle progression [24].
SiRNA-mediated knockdown of HDAC1 and 2 in various human tumor cells indicated that the requirement for HDAC1 and/or HDAC2 for cell cycle progression may be cell type-dependent [26]. Indeed, knockdown of HDAC1 only or HDAC1 plus HDAC2 in U2OS (osteosarcoma cell line) and MCF7 (mammary carcinoma cell line) cells led to cell cycle arrest in the G1 phase or apoptosis during the G2/M transition. Conversely, this phenotype was not observed in the non-tumorigenic epithelial mammary gland cell line MCF10A. Strikingly, deletion of HDAC1 alone or HDAC1 plus HDAC2 led to p21 upregulation only in U2OS cells. Unlike the phenotype due to HDAC1 deletion, HDAC2 knockdown showed no effect on cell proliferation in the three analyzed cell lines [26]. Although the knockdown efficiency can vary between cell lines and this variability can influence the outcome of this study, these results suggest a cell-specific role of HDAC1 and 2 in cell cycle regulation.
HDAC1 and 2 also play a role in the DNA damage response. Depletion of HDAC1 and 2 causes hypersensitivity to the double-strand breaks (DSB)-inducing agents ionizing radiation and phleomycin [27]. HDAC1 and 2 were found to localize to the sites of DNA damage and are responsible for deacetylation of H3K56ac, which is believed to play a role in DNA damage response [28]. As repair of DNA lesions constitutes an important cell cycle check point, this suggests that HDAC1 and 2 can also affect cell cycle progression via their action on DNA damage control.
Recruitment of HDAC1 and 2 to cell cycle regulator genes by various transcription factors
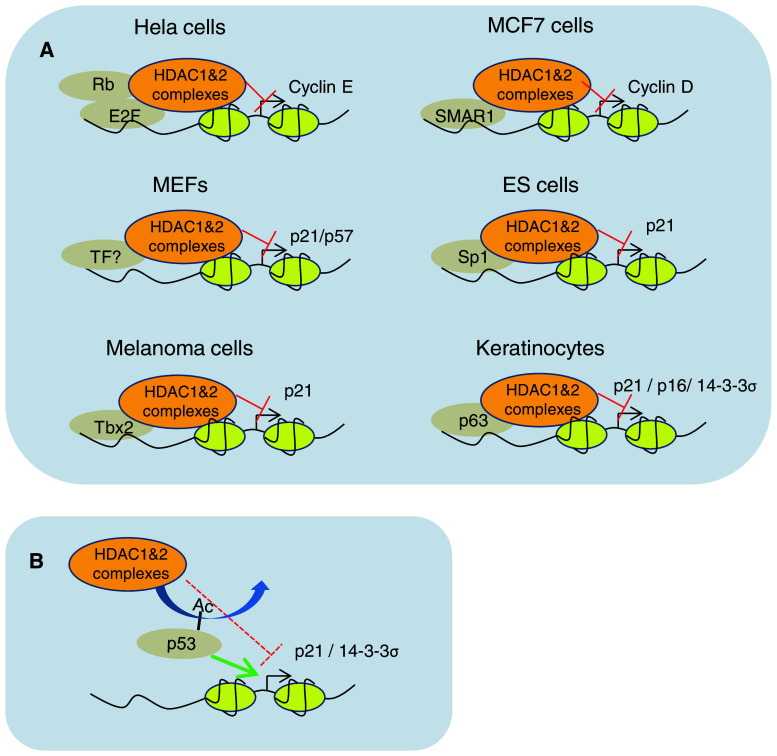
The majority of studies that addressed the role of HDAC1 and 2 in cell cycle regulation demonstrated that this regulation occurs at least partially via direct transcriptional repression of cell cycle regulators such as p21, p27, and p57. Since HDACs lack a DNA binding domain, a central question is how are HDACs complexes recruited to the promoters/genes they regulate? The transcription factor SP1 has been found to be required for HDAC1-mediated transcriptional repression of the thymidine kinase promoter [29]. HDAC1 binds directly to the C-terminus of Sp1, which is also involved in the interaction with p53 [29, 30]. It was shown that p53 and HDAC1 are antagonistic regulators of p21: p53 cooperates with Sp1 to activate the p21 promoter, whereas HDAC1 counteracts p53 action by repressing p21 transcription via its interaction with Sp1 (Fig. 3). Induction of p53 in response to DNA-damaging agents resulted in the formation of a p53–Sp1 complex and dissociation of the HDAC1–Sp1 complex. Chromatin immunoprecipitation (ChIP) experiments showed that HDAC1 is recruited to the p21 promoter in proliferating cells, while genotoxic stress leads to recruitment of p53, reduced binding of HDAC1, and hyperacetylation of core histones at the p21 promoter [30]. These data indicate that Sp1 may recruit HDAC1 to the p21 promoter to repress its activity in proliferating cells. Other critical transcription factors were shown to require HDAC activity to exert their functions: The retinoblastoma protein (pRb) regulates the G1/S transition by repressing a subset of genes that are controlled by the E2F family of transcription factors [31, 32]. The complex formed by HDAC1, pRb, and E2F represses transcription of E2F target genes such as cyclin E [33–35]. It was also shown that the tumor suppressor and chromatin modulator matrix attachment region (MAR) binding protein (SMAR1) recruits the Sin3/HDAC1 complex to the cyclin D1 promoter, leading to deacetylation of histone residues H3K9 and H4K8 at that locus and reduced expression [36].
Fig. 3.
HDAC1- and 2-mediated repression of cell cycle regulators. a Recruitment of HDAC1 and 2 complexes to their target genes by various transcription factors in different cellular systems. Transcription factors (TF) are schematized as gray circles and nucleosomes as yellow circles, cellular systems and target promoters are indicated; red inhibition symbols indicate direct repression by deacetylation of histones at the indicated loci. b Schematic representation of indirect repression of the p21 promoter by HADCs via p53 deacetylation
In melanoma cells, the T-box transcription factor Tbx2 plays an important role in maintaining proliferation and suppressing senescence. Tbx2 represses p21 expression by recruiting HDAC1 to its promoter (Fig. 3). Expression of a dominant-negative Tbx2 leads to displacement of HDAC1 and upregulation of p21 expression which correlate with the induction of replicative senescence in melanoma cells [37]. In mice, combined deletion of Hdac1 and 2 in the ectoderm led to perinatal lethality due to defects in epidermal proliferation and differentiation accompanied by repression of p21, p16, and 14-3-3σ. The repression of these genes is mediated by the key ectodermal transcription factor p63 and requires HDAC1 and 2. This suggests that p63 recruits HDAC1 and 2 to these genes to repress their transcription (Fig. 3) [25]. In rat smooth muscle cells (SMC), knockdown of either Hdac1, 2, or 3 prevents mitogen-induced SMC proliferation. Reduction of SMC proliferation by HDACs depletion involves a growth arrest in the G1 phase of the cell cycle which correlates with inhibition of pRB phosphorylation, as well as transcriptional and post-transcriptional regulation of p21 and p27 [38].
In addition to the role of HDACs in regulating gene expression via histones deacetylation, it has been reported that they can also impact on gene expression by deacetylating other non-histone proteins such as transcription factors. In vitro studies suggest that p53 activity is modulated in an HDAC1-dependent manner through removal of acetyl groups from its C-terminal lysine residues and ensuing modulation of its DNA-binding affinity [39–41]. It has also been shown that HDAC1 and 2 suppress p53 hyperacetylation in embryonic epidermis (Fig. 3) [25]. HDAC1 and 2 also target other transcription factors such as NFκB [42] (see below), and the myogenic differentiation factors MyoD [43], E2F1 [44], and several others (for review, see [2]).
HDAC1 and 2 function in the nervous system
Mice lacking both HDAC1 and 2 in the central nervous system (CNS) displayed major aberration of cortical, hippocampal, and cerebellar development, and die postnatally by day 7 [45]. In addition, differentiation of neuronal precursors into neurons requires the presence of either HDAC1 or HDAC2, strongly suggesting functional redundancy [45]. HDAC1 and 2 are also required for oligodendrocyte specification and differentiation, where they play a role in stabilizing β-catenin and thereby activating Wnt signaling [46]. Mice lacking HDAC1 and 2 in oligodendrocyte lineage developed severe tremors and died postnatally within 2 weeks [46]. Interestingly, comparing neurogenesis during development and adulthood, HDAC2 is found to be essential for appropriate differentiation and survival of adult generated neurons, however unnecessary during development [47]. Loss of HDAC2 in adult neurons leads to an aberrant maintenance of Sox2 expression and an increased rate of progenitor proliferation [47].
In the peripheral nervous system (PNS), ablation of HDAC1 and 2 in the Schwann cell lineage results in severe myelination defects in sciatic nerves accompanied by altered development of Schwann cells [42, 48]. Two distinct mechanisms have been proposed to explain this phenotype. First, it was observed that, in the absence of both HDAC1 and 2 in Schwann cells, the NF-κB p65 subunit becomes hyperacetylated, leading to the deregulation of NFκB-dependent genes controlling Schwann cell differentiation. This indicates that HDAC1 and 2 regulate NF-κB function by deacetylating its p65 subunit [42]. Second, it was demonstrated that, in Schwann cells, HDAC1 and 2 also exert specific primary functions: HDAC2 activates the transcriptional program of myelination in synergy with Sox10, whereas HDAC1 controls Schwann cell survival by regulating the level of β-catenin expression [48]. Other specific functions for HDAC1 and 2 have been observed in the neuronal system. HDAC1 activity plays a clear role in neuroprotection, while the loss of HDAC2 enhances memory formation [20, 49]. CK-p25 is a mouse model of rapid neurodegeneration, mimicking an Alzheimer’s-like disease. p25 is a truncated form of p35, a regulatory subunit associated with cyclin-dependent kinase 5 (Cdk5) [50]. CK-p25 mice show aberrant cell-cycle and double-strand DNA breaks leading to neurotoxicity, due to the inhibition of HDAC1 activity directly mediated by p25/Cdk5 induction [49]. Importantly, in these mice, the neurotoxicity and DNA damage could be rescued by HDAC1 overexpression [49]. Likewise, HDAC1 gain-of-function protected neurons against ischemia-induced DNA damage and neurotoxicity in a stroke model in vivo [49]. In contrast, mice with a neuron-specific HDAC2 deletion exhibited enhanced learning and memory formation, similar to chronic treatment with HDAC inhibitors (HDACis) [20]. HDAC2 interacts with CoREST which in turn negatively regulates neurogenesis and memory. In addition, HDAC2 was found to bind to the promoter regions of genes involved in synaptic plasticity and neuronal activity [20]. It has also been shown that neuron-specific overexpression of HDAC2, but not of HDAC1, resulted in memory impairment and in decreased synaptic plasticity, which could be rescued by HDACis [20]. Using primary dissociated hippocampal neurons from newborns as an ex vivo model of synapse development, Akhtar and colleagues [51] observed complementary functions of HDAC1 and 2 in synapse development. This suggests that HDAC1 and 2 act as a developmental switch to control maturation of synapses and function.
HDAC1 and 2 function in other systems
Cardiac-specific KO of either Hdac1 or Hdac2 had no apparent effect on cardiac function, indicating functional redundancy. By contrast, combined loss of both genes leads to cardiomyopathy, arrhythmias, and postnatal lethality by day 14, accompanied by deregulation of genes encoding skeletal muscle-specific proteins and calcium channels [17]. Recently, GATA binding protein 4 (GATA4) was found as a novel non-histone target of HDAC2 [52]. HDAC2 directly interacts with GATA4, and this interaction is stabilized by a small HOP homeobox protein (Hopx) [52]. Together, they regulate cardiac myocyte proliferation during embryonic development. Global ablation of HDAC2 and Hopx leads to cardiac developmental defects, increased cardiac myocyte proliferation and perinatal lethality. Moreover, the dual loss leads to GATA4 hyperacetylation and repression of GATA4-dependent cell cycle genes [52].
HDAC1 and 2 also play an important role in epidermis development. In this system, loss of either Hdac1 or Hdac2 showed no apparent phenotype [25]. In contrast, in the absence of both enzymes, embryos died perinatally with multiple ectodermal defects. In addition to having an undifferentiated epidermis, these embryos also failed to develop hair follicles, tongue papillae, eyelids, and teeth. This epidermal phenotype is similar to the loss of the epidermal master regulator p63. Genes that are normally repressed by p63 were upregulated in Hdac-deficient undifferentiated keratinocytes, in particular genes involved in cell cycle regulation and epidermal development [25]. These results suggest that p63 exert its repressive function via HDAC1 and 2.
It has also been reported recently that HDAC1 and 2 act redundantly to control adipogenesis: simultaneous deletion of these two enzymes in MEFs leads to reduced lipid accumulation and blocks the in vitro differentiation of MEFs to adipocytes. By contrast, deletion of HDAC1 or 2 only does not affect adipogenesis [53].
In the hematopoietic system, deletion of either HDAC1 or HDAC2 has no striking effect, while dual inactivation of both enzymes leads to apoptosis of megakaryocytes and thrombocytopenia (decrease of platelets in blood) [24]. Similar results were obtained in the B cell lineage. It was found that, in the absence of HDAC1 or 2, B cell development proceeds normally, indicating functional redundancy between HDAC1 and 2. By contrast, dual inactivation of Hdac1 and 2 in early B cell progenitors led to a dramatic block in B cell development at the pre-B cell stage, accompanied by G1 cell cycle arrest and apoptosis [15]. Additionally, while one allele of Hdac2 in the absence of HDAC1 is not sufficient to rescue proper B cell development, one Hdac1 allele in the absence of HDAC2 is able to restore B cell differentiation. This haplo-insufficiency of HDAC2 indicates that HDAC1 and 2 have some non-redundant functions (Yamaguchi et al., unpublished data). In contrast to other systems, conditional deletion of only HDAC1 in T cells results in increased proliferation [54]. In an in vivo allergic airway inflammation model, the T cell-specific loss of HDAC1 leads to an increased inflammatory response which correlates with enhanced Th2 cytokine production and cellular proliferation [54]. This indicates that the influence of HDACs on proliferation is cell type-specific and depends on the physiological environment.
HDAC3
Role of HDAC3 in cell cycle regulation and DNA damage control
HDAC3 is an attractive target for cancer research due to increasing evidence supporting its role in cell cycle progression and DNA damage control [21, 55]. HDAC3 has been found to be required for normal mitotic progression and maximal phosphorylation of H3S10 (phosphorylation of H3S10 is an evolutionary conserved mitotic event [56]) [55]. HDAC3 forms a complex with A-Kinase-Anchoring Proteins AKAP95 and HA95, which are targeted to mitotic chromosomes [55]. Deacetylation of H3 in mitosis requires AKAP95/HA95 and HDAC3 and provides a hypoacetylated H3 tail that is the preferred substrate for Aurora B kinase that phosphorylates H3S10 [55]. These observations were supported by conditional inactivation of HDAC3 in MEFs [21]. The loss of HDAC3 also led to a delay in cell cycle progression during S phase accompanied by increasing DNA damage and apoptosis [21]. However, no mitotic alterations and phosphorylation changes of H3Ser10 were observed in these Hdac3-deficient cells [21]. In addition, DNA damage observed in interphase MEFs correlated with impaired DNA double-strand break repair. Interestingly, arrested or quiescent MEFs lacking HDAC3 were protected from DNA damage. These results might explain the findings that HDACis preferentially influence cancer cells or highly proliferating cells rather than primary or quiescent cells [57]. In an additional study, Bhaskara and colleagues showed that deletion of HDAC3 in MEFs led to impaired DNA repair, reduced chromatin compaction and heterochromatin content. This correlates with an increase in histone H3K9, H3K14, H4K5, and H4K12 acetylation in late S phase of the cell cycle. Importantly, histone marks such as H4K5ac and H4K12ac, which are commonly associated with histone deposition onto newly synthesized DNA, were maintained in quiescent Hdac3-deficient cells [58]. In the same study, it was shown that liver-specific deletion of Hdac3 led to hepatocellular carcinoma accompanied by downregulation of NCOR1 [58]. These data indicate the importance of the HDAC3/NCOR axis in liver function; however, the exact role of these factors remains to be further elucidated [58].
HDAC3 function in other systems
Liver-specific loss of HDAC3 results in aberrant lipid and cholesterol biosynthesis shortly after birth. Later, the liver displays disturbed lipid and cholesterol homeostasis, leading to an accumulation of lipids and a decrease in glycogen storage [59]. This phenotype is due to the derepression of a gene program that is usually under the control of nuclear hormone receptors [59]. A recent study unraveled a link between HDAC3 and the circadian rhythm in the mouse liver [60]. Importantly, the circadian nuclear receptor Rev-erbα recruits HDAC3 to genes regulating lipid metabolism [60]. Deletion of HDAC3 or Rev-erbα in mouse liver led to hepatic steatosis and deregulated gene expression, correlating with changes in histone acetylation. These findings indicate that genomic recruitment of HDAC3 by Rev-erbα directs a circadian rhythm of histone acetylation and gene expression that is required for normal hepatic lipid homeostasis [60].
In the cardiac system, loss of HDAC3 in cardiomyocytes was lethal within 3–4 months after birth [22]. Massive cardiac hypertrophy and derepression of genes that control fatty-acid uptake and metabolism were observed in mice lacking HDAC3, and expression of p21 was upregulated in Hdac3-deficient hearts [22]. Furthermore, myocyte-specific overexpression of HDAC3 also resulted in heart defects and cardiomyocyte hyperplasia, leading to increased thickness of the myocardium [22]. In contrast to the HDAC2 overexpression model mentioned above, the increased thickness of the myocardium was not associated with hypertrophy [61]. Therefore, HDAC3 is another important regulator of cardiac myocyte proliferation during cardiac development. Hdac3-deficient mice were smaller and had a reduced life span, as compared to wild-type animals. HDAC3 ablation caused severe defects in bone formation and increased bone marrow adipogenesis [62]. HDAC3 was shown to be associated to an orphan nuclear receptor TLX and to mediate neural stem cell proliferation and self-renewal by transcriptional repression [63]. Ablation of HDAC3 leads to increased expression of the p21 and phosphatase and tensin homologue (pten) genes and inhibition of neuronal stem cell proliferation [63]. Similar to HDAC2, specific HDAC3 inactivation in the dorsal hippocampus leads to enhanced long-term memory and elevated expression of nuclear receptor subfamily 4, group A, member 2 (Nr4a2), a gene associated with long-term memory formation [64]. Recently, the loss of HDAC3 was analyzed during embryonic cardiac and neural crest development [65]. Ablation of HDAC3 led to a block in smooth muscle differentiation and downregulation of the Notch ligand Jagged1 [65]. These reports demonstrate the critical role of HDAC3 in the regulation of specific factors that are important for lineage differentiation during development. Recently, HDAC3 was found to also be required for gene induction by transforming growth factor beta (TGFβ) via activation (phosphorylation) of PI3K (phosphoinositide 3-kinase) and ERK (extracellular signal regulated kinase). The exact mechanism by which HDAC3 affects these pathways needs to be further investigated [66].
HDAC8
HDAC8 has been described as playing an important role in proliferation of sympathetic neuroblastoma cells [67]. High HDAC8 expression levels correlate with advanced stage childhood neuroblastoma, clinical and genetic risk factors, and poor long-term survival. HDAC8 ablation in neuroblastoma cells induces morphologic changes, inhibits proliferation and colony formation, and causes cell cycle arrest [67]. Despite these facts, little is known about the role of HDAC8 in the CNS. Additional insights come from the role of HDAC8 as a regulator of cAMP response element-binding (CREB)-dependent gene expression [68]. CREB target genes are essential in diverse processes such as glucose metabolism, cell survival, and neuronal plasticity (for review, see [69]). It has been reported that HDAC8, like HDAC1, promotes the dephosphorylation of CREB at serine 133 via a stable interaction with protein phosphatase 1 (PP1), and therefore inhibits CREBs transcriptional activity [55, 57].
HDACs in the clinic: use of HDACs inhibitors to treat disease
As discussed above, recent genetic studies on HDACs have begun to identify specific and redundant functions for many of these enzymes, but further analysis will be needed to uncover the full spectrum of their physiological roles. Initial studies on HDACs relied heavily on small molecules inhibitors (HDACis), and described early on that in vitro proliferation of tumor cell lines is impaired by HDAC inhibition [70]. In addition, several HDACs were found to be deregulated (overexpressed) in various kinds of tumors or cancer cell lines, suggesting that epigenetic (de)regulation could play a role in cancer [71–74]. Together, these findings sparked a very intense research into the possible use of HDACis for therapeutic purposes. Although the research on the therapeutic potential of HDACis was initially concentrated in oncology, other areas such as neurodegeneration or autoimmunity have recently been investigated. Because of the high medical relevance of these observations, we will review here some of the important advances in HDACis and their use.
HDACis can be grouped into four main structural classes, with a growing list of novel compounds: first, the hydroxamic acids [e.g. trichostatin A (TSA), Suberoylanilide Hydroxamic Acid (SAHA, Vorinostat, Zolinza®), and LBH589 (Panobinostat)]; second, the short chain carboxylic acids [Phenylbutyrate and Valproic acid (VPA)]; third, the benzamides [MS-275 (Entinostat)]; and fourth, the cyclic tetrapeptides [Romidepsin (FK-228 2, Istodax®)]. They all target the deacetylase enzymatic activity of HDACs by binding the active site and competing with the Zn2+ ion in the active pocket [75, 76]. As a result, the substrate pocket is blocked and the enzyme inactivated, thereby causing an increase in histones acetylation (for review, see [77]). HDACis also affect other biological processes than chromatin dynamics and increase the acetylation level of non-histone proteins, such as tubulin and others [78–80]. A common effect of HDACis in cancer cells is the induction of the cell cycle regulator p21 [81]. This is often conducted in a p53-independent manner and is at the core of the proliferation-inhibiting effects of HDACis in cancer cells (for review, see [82]). Many pan-HDACis induce cell cycle arrest and inhibit proliferation, leading to induction of differentiation, growth arrest, and apoptosis of transformed cells, by downregulating a common set of genes [83, 84]. These effects are dose- and inhibitor-dependent. Several HDACis have been investigated in clinical trials for their potential as anticancer drugs [82, 85]. The first identified and most prominent HDACi is the naturally occurring antifungal antibiotic TSA [70]. TSA is one of the most potent HDACis, exhibiting EC50 values in low nanomolar range against HDAC1, 2, 4, 6, 7, and 9, but less efficient against HDAC8 [86]. Although TSA appeared very promising in preclinical investigations, it was not suitable for clinical use due to unfavorable pharmacological side effects (for review, see [87]).
To date, two HDACis have been approved as anticancer drug in humans: Vorinostat (SAHA, Zolinza®; Merck Research Laboratories) and Romidepsin (FK-228 2, Istodax®; Gloucester Pharmaceuticals). SAHA has been approved since 2006 as a therapeutic approach in patients with progressive, persistent, or recurrent cutaneous T cell lymphoma (CTCL) after one or more lines of chemotherapy [88–92]. SAHA efficiently inhibits HDAC1, 2, 3, 4, 6, 7, and 9, similar to TSA, but 5- to 30-fold less potent [86]. This classic pan-HDACi can induce apoptosis as well as autophagy in cancer cells. Romidepsin (FK-228 2), one of the more selective inhibitors, has also been approved since 2009 to treat CTCL [93–96]. It belongs to the cyclic peptides that strongly inhibit HDAC1 and 2, and inhibits HDAC class II enzymes at higher concentrations [97–99]. The fact that Romidepsin, which preferentially inhibits HDAC1 and 2, is also effective in CTCL may suggest that this disease is particularly dependent on the function of these two enzymes.
The major side effects associated with HDACis are gastrointestinal symptoms, bone marrow suppression [100], cardiac toxicity [101], thrombocytopenia, neutropenia, diarrhea, nausea, vomiting, and fatigue [6, 102]. Although these side effects should be taken into consideration, it is important to mention that HDACis are well tolerated in the majority of patients compared to other anticancer treatments. Several HDACis have already demonstrated preclinical efficacy as monotherapy as well as in combination with other anticancer agents (for review, see [87]). Moreover, an exciting finding is the observation that HDACis administered at low concentrations are efficacious for treating a range of diseases that are not related to cancer. For example, HDACis exhibit anti-inflammatory properties due to a reduction of cytokine production as well as inhibition of cytokine effects (for reviews, see [103], [104]). Furthermore, experiments in mouse models have shown beneficial effects of HDACis in neurodegenerative conditions and autoimmunity and several clinical trials are currently underway in these areas (for reviews, see [104, 105]).
It is likely that a specific HDACi will have clinical benefits only in a specific tumor type and/or in a subset of patients. For future treatment and effective selection of the “right” HDACis for a given therapy, the availability of predictive and prognostic biomarkers will be crucial. Recently, the ubiquitin-like fusion protein HR23B (RAD23 homolog B of S. cerevisiae) has been described as a possible candidate cancer biomarker in CTCL patients [106, 107]. Yet, the few identified HDACis biomarkers known so far do not reflect tumor response or correlate to the dosage of the HDACis needed [108], and more work is required to make progress in this important area.
Recently, while modeling the acute response to various anti-cancer agents in drug-sensitive human tumor cell lines, a small drug-tolerant cancer cell subpopulation has been described [109]. This cell subpopulation emerges de novo and is reversibly drug-tolerant, indicating that its resistance to drug treatment is not due to mutation but rather to changes in gene expression [109]. Treatment of these cells with HDACis, such as TSA, selectively ablates them, presumably by influencing the chromatin. This correlates with an induction of DNA damage response via γ-H2AX [109]. Moreover, in these in vitro experiments, it was shown that constant treatment with HDACis prevents the development of drug tolerance in cancer cells [109]. These observations indicate that HDACis, or other chromatin modifying agents, may have a general impact on the emergence of drug-tolerant cells and, thus, may be useful in combination with other drug treatments.
Pan versus specific HDAC inhibitors and their benefits
Class I selective HDACis display similar toxicity as pan-HDACis. This suggests that the inhibition of HDAC1, 2, and 3 are the main reasons for most of the observed side effects. Fatigue syndrome, gastrointestinal difficulties, and cardiac toxicity are all observed using class I selective inhibitors (MS-275 or Depsipeptide) [110, 111] or pan-HDACis (Vorinostat) [89]. This is perhaps not surprising since HDAC1, 2, and 3 have been reported to hold key functions in various pathways evidenced in knockout studies, for example cell proliferation arrest and apoptosis, and metabolic and cardiac abnormalities [16, 17, 59]. Importantly, it has been shown that class I and class II or individual HDACs are often differentially expressed in human cancers; for example, HDAC1 or 2 expression is increased in gastric cancer [71, 72]. In contrast, in prostate cancer, only HDAC1 is upregulated [73], and in lung cancer, only HDAC3 [74]. Recently, HDAC8 was identified to be a crucial regulator of tumorigenesis in neuroblastoma cells [67]. APCmin (Min, multiple intestinal neoplasia) mice carry a point mutation in the Adenomatous polyposis coli (APC) gene and are highly susceptible to spontaneous intestinal adenoma formation [112, 113]. Elevated HDAC2 levels were observed in tumors induced by APC mutations [114], and it was reported that the loss of HDAC2 in APCmin mice leads to 10% decreased tumor rates in the gut [19]. Moreover these genetic approaches gave comparable results to studies conducted with HDACis. Recent studies of tumor cell survival in vitro indicate that dual loss of HDAC1 and 2 mimics the reported effects of HDACis in cancer cells [7].
Conclusion and perspectives
The recent advances in the study of HDACs have shown that these enzymes exert a large variety of functions by deacetylating histones as well as non-histone proteins. Class I HDACs were found to be important for specific functions in several tissues where they are involved in the regulation of differentiation, proliferation, cell cycle progression, and apoptosis. It was demonstrated that HDAC1 and 2 directly control the expression of genes involved in cell cycle regulation such as p21. It has also been shown that HDAC1, 2, and 3 play a role in DNA damage response, and subsequently can be important for DNA replication and cell cycle progression. Some of the important pathways have been identified, but many of the chromatin targets of HDACs remain to be identified in different cellular settings. The transcription factors or mechanisms recruiting HDAC complexes have so far only been described in few cases and further analysis is needed. Genome-wide mapping of HDACs and their partners will address this issue by defining direct target genes and will allow the distinguishing of common from specific targets.
The increasing number of acetylated non-histone proteins suggests an important role for HDACs in the regulation of cellular processes beyond chromatin and gene expression. Acetylome analyses in cells lacking specific HDACs will be a powerful tool to identify specific substrates and define the pathways in which these enzymes are involved. As highlighted above, class I HDACs are excellent therapeutic candidates for anticancer treatment. A better understanding of the role of individual HDACs, in normal physiology as well as in various pathological settings, will give a framework for the development of more specific inhibitors. While having selective inhibitors should be useful to reduce side effects, in particular for non-cancer applications, it may be necessary to target more than one HDAC to achieve clinical benefits in some cancer settings. Further genetic studies in model systems will be needed to address this issue.
Acknowledgments
We wish to thank Camille Du Roure (Phocus, Basel), Benjamin Herquel (IGBMC, Strasbourg), Arnaud Krebs, and Oliver Truee for useful comments and suggestions on the manuscript. This work was supported by the Novartis Research Foundation and the SystemsX RTD Cellplasticity project.
Footnotes
N. Reichert and M.-A. Choukrallah contributed equally to this work.
References
- 1.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 2.Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5(10):981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 4.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Natl Rev Mol Cell Biol. 2008;9(3):206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao J, Sack MN. Protein deacetylation by sirtuins: delineating a post-translational regulatory program responsive to nutrient and redox stressors. Cell Mol Life Sci. 2010;67(18):3073–3087. doi: 10.1007/s00018-010-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walkinshaw DR, Tahmasebi S, Bertos NR, Yang XJ. Histone deacetylases as transducers and targets of nuclear signaling. J Cell Biochem. 2008;104(5):1541–1552. doi: 10.1002/jcb.21746. [DOI] [PubMed] [Google Scholar]
- 7.Haberland M, Johnson A, Mokalled MH, Montgomery RL, Olson EN. Genetic dissection of histone deacetylase requirement in tumor cells. Proc Natl Acad Sci USA. 2009;106(19):7751–7755. doi: 10.1073/pnas.0903139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Pflum MK, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem. 2001;276(50):47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 2005;19(7):827–839. doi: 10.1101/gad.1286005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco S. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci USA. 2004;101(42):15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11(2):109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 13.Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011;2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, Jurkin J, Rembold M, Meunier D, Egger G, Lagger S, Chiocca S, Propst F, Weitzer G, Seiser C. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol. 2010;30(5):1171–1181. doi: 10.1128/MCB.01500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi T, Cubizolles F, Zhang Y, Reichert N, Kohler H, Seiser C, Matthias P. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24(5):455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21(11):2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21(14):1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13(3):324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann S, Kiefer F, Prudenziati M, Spiller C, Hansen J, Floss T, Wurst W, Minucci S, Gottlicher M. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67(19):9047–9054. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]
- 20.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459(7243):55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, Hiebert SW. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30(1):61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118(11):3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23(14):1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilting RH, Yanover E, Heideman MR, Jacobs H, Horner J, van der Torre J, DePinho RA, Dannenberg JH. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29(15):2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LeBoeuf M, Terrell A, Trivedi S, Sinha S, Epstein JA, Olson EN, Morrisey EE, Millar SE. Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev Cell. 2010;19(6):807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, Bernard L, Draetta GF, Alcalay M, Seiser C, Chiocca S. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27(13):4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17(9):1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28(13):1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol. 1999;19(8):5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagger G, Doetzlhofer A, Schuettengruber B, Haidweger E, Simboeck E, Tischler J, Chiocca S, Suske G, Rotheneder H, Wintersberger E, Seiser C. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol. 2003;23(8):2669–2679. doi: 10.1128/MCB.23.8.2669-2679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nevins JR. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258(5081):424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 32.Helin K, Lees JA, Vidal M, Dyson N, Harlow E, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70(2):337–350. doi: 10.1016/0092-8674(92)90107-N. [DOI] [PubMed] [Google Scholar]
- 33.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391(6667):601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 34.Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391(6667):597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 35.Morrison AJ, Sardet C, Herrera RE. Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol Cell Biol. 2002;22(3):856–865. doi: 10.1128/MCB.22.3.856-865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rampalli S, Pavithra L, Bhatt A, Kundu TK, Chattopadhyay S. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol Cell Biol. 2005;25(19):8415–8429. doi: 10.1128/MCB.25.19.8415-8429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 2005;65(6):2260–2268. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 38.Findeisen HM, Gizard F, Zhao Y, Qing H, Heywood EB, Jones KL, Cohn D, Bruemmer D. Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition. Arterioscler Thromb Vasc Biol. 2011;31(4):851–860. doi: 10.1161/ATVBAHA.110.221952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higashitsuji H, Masuda T, Liu Y, Itoh K, Fujita J. Enhanced deacetylation of p53 by the anti-apoptotic protein HSCO in association with histone deacetylase 1. J Biol Chem. 2007;282(18):13716–13725. doi: 10.1074/jbc.M609751200. [DOI] [PubMed] [Google Scholar]
- 40.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21(22):6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, Wang H, Yoon SO, Xu X, Hottiger MO, Svaren J, Nave KA, Kim HA, Olson EN, Lu QR. HDAC-mediated deacetylation of NF-kappaB is critical for Schwann cell myelination. Nat Neurosci. 2011;14(4):437–441. doi: 10.1038/nn.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 2001;20(7):1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19(4):662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci USA. 2009;106(19):7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin–TCF interaction. Nat Neurosci. 2009;12(7):829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jawerka M, Colak D, Dimou L, Spiller C, Lagger S, Montgomery RL, Olson EN, Wurst W, Gottlicher M, Gotz M. The specific role of histone deacetylase 2 in adult neurogenesis. Neuron Glia Biol. 2010;6(2):93–107. doi: 10.1017/S1740925X10000049. [DOI] [PubMed] [Google Scholar]
- 48.Jacob C, Christen CN, Pereira JA, Somandin C, Baggiolini A, Lotscher P, Ozcelik M, Tricaud N, Meijer D, Yamaguchi T, Matthias P, Suter U. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci. 2011;14(4):429–436. doi: 10.1038/nn.2762. [DOI] [PubMed] [Google Scholar]
- 49.Kim D, Frank CL, Dobbin MM, Tsunemoto RK, Tu W, Peng PL, Guan JS, Lee BH, Moy LY, Giusti P, Broodie N, Mazitschek R, Delalle I, Haggarty SJ, Neve RL, Lu Y, Tsai LH. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60(5):803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, Tsai LH. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402(6762):615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- 51.Akhtar MW, Raingo J, Nelson ED, Montgomery RL, Olson EN, Kavalali ET, Monteggia LM. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J Neurosci. 2009;29(25):8288–8297. doi: 10.1523/JNEUROSCI.0097-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, Hannenhalli S, Epstein JA. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell. 2010;19(3):450–459. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haberland M, Carrer M, Mokalled MH, Montgomery RL, Olson EN. Redundant control of adipogenesis by histone deacetylases 1 and 2. J Biol Chem. 2010;285(19):14663–14670. doi: 10.1074/jbc.M109.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grausenburger R, Bilic I, Boucheron N, Zupkovitz G, El-Housseiny L, Tschismarov R, Zhang Y, Rembold M, Gaisberger M, Hartl A, Epstein MM, Matthias P, Seiser C, Ellmeier W. Conditional deletion of histone deacetylase 1 in T cells leads to enhanced airway inflammation and increased Th2 cytokine production. J Immunol. 2010;185(6):3489–3497. doi: 10.4049/jimmunol.0903610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006;20(18):2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cobb J, Miyaike M, Kikuchi A, Handel MA. Meiotic events at the centromeric heterochromatin: histone H3 phosphorylation, topoisomerase II alpha localization and chromosome condensation. Chromosoma. 1999;108(7):412–425. doi: 10.1007/s004120050393. [DOI] [PubMed] [Google Scholar]
- 57.Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, Aladjem MI, Pommier Y. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70(11):4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, Wells CE, Kaiser JF, Washington MK, Zhao Z, Wagner FF, Sun ZW, Xia F, Holson EB, Khabele D, Hiebert SW. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18(5):436–447. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27(7):1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trivedi CM, Lu MM, Wang Q, Epstein JA. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem. 2008;283(39):26484–26489. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Razidlo DF, Whitney TJ, Casper ME, McGee-Lawrence ME, Stensgard BA, Li X, Secreto FJ, Knutson SK, Hiebert SW, Westendorf JJ. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLoS One. 2010;5(7):e11492. doi: 10.1371/journal.pone.0011492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci USA. 2007;104(39):15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, Wood MA. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31(2):764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh N, Trivedi CM, Lu M, Mullican SE, Lazar MA, Epstein JA. Histone deacetylase 3 regulates smooth muscle differentiation in neural crest cells and development of the cardiac outflow tract. Circ Res. 2011 doi: 10.1161/CIRCRESAHA.111.255067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barter MJ, Pybus L, Litherland GJ, Rowan AD, Clark IM, Edwards DR, Cawston TE, Young DA. HDAC-mediated control of ERK- and PI3K-dependent TGF-beta-induced extracellular matrix-regulating genes. Matrix Biol. 2010;29(7):602–612. doi: 10.1016/j.matbio.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 67.Oehme I, Deubzer HE, Wegener D, Pickert D, Linke JP, Hero B, Kopp-Schneider A, Westermann F, Ulrich SM, von Deimling A, Fischer M, Witt O. Histone deacetylase 8 in neuroblastoma tumorigenesis. Clin Cancer Res. 2009;15(1):91–99. doi: 10.1158/1078-0432.CCR-08-0684. [DOI] [PubMed] [Google Scholar]
- 68.Gao J, Siddoway B, Huang Q, Xia H. Inactivation of CREB mediated gene transcription by HDAC8 bound protein phosphatase. Biochem Biophys Res Commun. 2009;379(1):1–5. doi: 10.1016/j.bbrc.2008.11.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barco A, Marie H. Genetic approaches to investigate the role of CREB in neuronal plasticity and memory. Mol Neurobiol. 2011 doi: 10.1007/s12035-011-8209-x. [DOI] [PubMed] [Google Scholar]
- 70.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265(28):17174–17179. [PubMed] [Google Scholar]
- 71.Choi JH, Kwon HJ, Yoon BI, Kim JH, Han SU, Joo HJ, Kim DY. Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn J Cancer Res. 2001;92(12):1300–1304. doi: 10.1111/j.1349-7006.2001.tb02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song J, Noh JH, Lee JH, Eun JW, Ahn YM, Kim SY, Lee SH, Park WS, Yoo NJ, Lee JY, Nam SW. Increased expression of histone deacetylase 2 is found in human gastric cancer. Apmis. 2005;113(4):264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- 73.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59(2):177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 74.Bartling B, Hofmann HS, Boettger T, Hansen G, Burdach S, Silber RE, Simm A. Comparative application of antibody and gene array for expression profiling in human squamous cell lung carcinoma. Lung Cancer. 2005;49(2):145–154. doi: 10.1016/j.lungcan.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 75.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401(6749):188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 76.Sternson SM, Wong JC, Grozinger CM, Schreiber SL. Synthesis of 7200 small molecules based on a substructural analysis of the histone deacetylase inhibitors trichostatin and trapoxin. Org Lett. 2001;3(26):4239–4242. doi: 10.1021/ol016915f. [DOI] [PubMed] [Google Scholar]
- 77.Miller TA, Witter DJ, Belvedere S. Histone deacetylase inhibitors. J Med Chem. 2003;46(24):5097–5116. doi: 10.1021/jm0303094. [DOI] [PubMed] [Google Scholar]
- 78.Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, Seto E, Bhalla K. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280(29):26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 79.Sun C, Zhang M, Shan X, Zhou X, Yang J, Wang Y, Li-Ling J, Deng Y. Inhibitory effect of cucurbitacin E on pancreatic cancer cells growth via STAT3 signaling. J Cancer Res Clin Oncol. 2010;136(4):603–610. doi: 10.1007/s00432-009-0698-x. [DOI] [PubMed] [Google Scholar]
- 80.Blagosklonny MV, Robey R, Sackett DL, Du L, Traganos F, Darzynkiewicz Z, Fojo T, Bates SE. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol Cancer Ther. 2002;1(11):937–941. [PubMed] [Google Scholar]
- 81.Rocchi P, Tonelli R, Camerin C, Purgato S, Fronza R, Bianucci F, Guerra F, Pession A, Ferreri AM. p21Waf1/Cip1 is a common target induced by short-chain fatty acid HDAC inhibitors (valproic acid, tributyrin and sodium butyrate) in neuroblastoma cells. Oncol Rep. 2005;13(6):1139–1144. [PubMed] [Google Scholar]
- 82.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 83.Fang JY. Histone deacetylase inhibitors, anticancerous mechanism and therapy for gastrointestinal cancers. J Gastroenterol Hepatol. 2005;20(7):988–994. doi: 10.1111/j.1440-1746.2005.03807.x. [DOI] [PubMed] [Google Scholar]
- 84.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2(2):151–163. doi: 10.4161/cbt.2.2.349. [DOI] [PubMed] [Google Scholar]
- 85.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 86.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, Qian X, Mills E, Berghs SC, Carey N, Finn PW, Collins LS, Tumber A, Ritchie JW, Jensen PB, Lichenstein HS, Sehested M. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409(2):581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 87.Thurn KT, Thomas S, Moore A, Munster PN. Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer. Future Oncol. 2011;7(2):263–283. doi: 10.2217/fon.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kelly WK, O’Connor OA, Marks PA. Histone deacetylase inhibitors: from target to clinical trials. Expert Opin Investig Drugs. 2002;11(12):1695–1713. doi: 10.1517/13543784.11.12.1695. [DOI] [PubMed] [Google Scholar]
- 89.Kelly WK, O’Connor OA, Krug LM, Chiao JH, Heaney M, Curley T, MacGregore-Cortelli B, Tong W, Secrist JP, Schwartz L, Richardson S, Chu E, Olgac S, Marks PA, Scher H, Richon VM. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol. 2005;23(17):3923–3931. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12(10):1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 91.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25(1):84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 92.Grant S, Easley C, Kirkpatrick P. Vorinostat. Nat Rev Drug Discov. 2007;6(1):21–22. doi: 10.1038/nrd2227. [DOI] [PubMed] [Google Scholar]
- 93.Klimek VM, Fircanis S, Maslak P, Guernah I, Baum M, Wu N, Panageas K, Wright JJ, Pandolfi PP, Nimer SD. Tolerability, pharmacodynamics, and pharmacokinetics studies of depsipeptide (romidepsin) in patients with acute myelogenous leukemia or advanced myelodysplastic syndromes. Clin Cancer Res. 2008;14(3):826–832. doi: 10.1158/1078-0432.CCR-07-0318. [DOI] [PubMed] [Google Scholar]
- 94.Schrump DS, Fischette MR, Nguyen DM, Zhao M, Li X, Kunst TF, Hancox A, Hong JA, Chen GA, Kruchin E, Wright JJ, Rosing DR, Sparreboom A, Figg WD, Steinberg SM. Clinical and molecular responses in lung cancer patients receiving Romidepsin. Clin Cancer Res. 2008;14(1):188–198. doi: 10.1158/1078-0432.CCR-07-0135. [DOI] [PubMed] [Google Scholar]
- 95.Piekarz RL, Frye R, Turner M, Wright JJ, Allen SL, Kirschbaum MH, Zain J, Prince HM, Leonard JP, Geskin LJ, Reeder C, Joske D, Figg WD, Gardner ER, Steinberg SM, Jaffe ES, Stetler-Stevenson M, Lade S, Fojo AT, Bates SE. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27(32):5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Whittaker SJ, Demierre MF, Kim EJ, Rook AH, Lerner A, Duvic M, Scarisbrick J, Reddy S, Robak T, Becker JC, Samtsov A, McCulloch W, Kim YH. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin Oncol. 2010;28(29):4485–4491. doi: 10.1200/JCO.2010.28.9066. [DOI] [PubMed] [Google Scholar]
- 97.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6(3):238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, Horinouchi S. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62(17):4916–4921. [PubMed] [Google Scholar]
- 99.Grant C, Rahman F, Piekarz R, Peer C, Frye R, Robey RW, Gardner ER, Figg WD, Bates SE. Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther. 2010;10(7):997–1008. doi: 10.1586/era.10.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Q, Xu W. Novel anticancer targets and drug discovery in post genomic age. Curr Med Chem Anticancer Agents. 2005;5(1):53–63. doi: 10.2174/1568011053352631. [DOI] [PubMed] [Google Scholar]
- 101.Karagiannis TC, El-Osta A. Will broad-spectrum histone deacetylase inhibitors be superseded by more specific compounds? Leukemia. 2007;21(1):61–65. doi: 10.1038/sj.leu.2404464. [DOI] [PubMed] [Google Scholar]
- 102.Piekarz RL, Frye AR, Wright JJ, Steinberg SM, Liewehr DJ, Rosing DR, Sachdev V, Fojo T, Bates SE. Cardiac studies in patients treated with depsipeptide, FK228, in a phase II trial for T-cell lymphoma. Clin Cancer Res. 2006;12(12):3762–3773. doi: 10.1158/1078-0432.CCR-05-2095. [DOI] [PubMed] [Google Scholar]
- 103.Adcock IM. HDAC inhibitors as anti-inflammatory agents. Br J Pharmacol. 2007;150(7):829–831. doi: 10.1038/sj.bjp.0707166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dinarello CA, Fossati G, Mascagni P. Histone deacetylase inhibitors for treating a spectrum of diseases not related to cancer. Mol Med. 2011;17(5–6):333–352. doi: 10.2119/molmed.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31(12):605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 106.Khan O, Fotheringham S, Wood V, Stimson L, Zhang C, Pezzella F, Duvic M, Kerr DJ, La Thangue NB. HR23B is a biomarker for tumor sensitivity to HDAC inhibitor-based therapy. Proc Natl Acad Sci USA. 2010;107(14):6532–6537. doi: 10.1073/pnas.0913912107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fotheringham S, Epping MT, Stimson L, Khan O, Wood V, Pezzella F, Bernards R, La Thangue NB. Genome-wide loss-of-function screen reveals an important role for the proteasome in HDAC inhibitor-induced apoptosis. Cancer Cell. 2009;15(1):57–66. doi: 10.1016/j.ccr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 108.Steele NL, Plumb JA, Vidal L, Tjornelund J, Knoblauch P, Rasmussen A, Ooi CE, Buhl-Jensen P, Brown R, Evans TR, DeBono JS. A phase 1 pharmacokinetic and pharmacodynamic study of the histone deacetylase inhibitor belinostat in patients with advanced solid tumors. Clin Cancer Res. 2008;14(3):804–810. doi: 10.1158/1078-0432.CCR-07-1786. [DOI] [PubMed] [Google Scholar]
- 109.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ryan QC, Headlee D, Acharya M, Sparreboom A, Trepel JB, Ye J, Figg WD, Hwang K, Chung EJ, Murgo A, Melillo G, Elsayed Y, Monga M, Kalnitskiy M, Zwiebel J, Sausville EA. Phase I and pharmacokinetic study of MS-275, a histone deacetylase inhibitor, in patients with advanced and refractory solid tumors or lymphoma. J Clin Oncol. 2005;23(17):3912–3922. doi: 10.1200/JCO.2005.02.188. [DOI] [PubMed] [Google Scholar]
- 111.Sandor V, Bakke S, Robey RW, Kang MH, Blagosklonny MV, Bender J, Brooks R, Piekarz RL, Tucker E, Figg WD, Chan KK, Goldspiel B, Fojo AT, Balcerzak SP, Bates SE. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res. 2002;8(3):718–728. [PubMed] [Google Scholar]
- 112.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247(4940):322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 113.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256(5057):668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 114.Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP, Gottlicher M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5(5):455–463. doi: 10.1016/S1535-6108(04)00114-X. [DOI] [PubMed] [Google Scholar]
- 115.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89(3):357–364. doi: 10.1016/S0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 116.Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1(7):1021–1031. doi: 10.1016/S1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 117.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138(4):660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 118.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95(2):279–289. doi: 10.1016/S0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 119.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13(15):1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26(3):843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST—human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98(4):1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12(3):723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 123.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9(3):611–623. doi: 10.1016/S1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]