Abstract
In vertebrates, internal organs are positioned asymmetrically across the left–right (LR) axis, placing them in a defined area within the body. This LR asymmetric placement is a conserved feature of the vertebrate body plan. Events determining LR asymmetry occur during embryonic development, and are regulated by the coordinated action of genetic mechanisms that are evolutionarily conserved among vertebrates. Recent studies using zebrafish have provided new insights into how the Kupffer’s vesicle organizer region is generated, and how it relays LR asymmetry information to the lateral plate mesoderm. In this review, we summarize recent advances in zebrafish and describe our current understanding of the mechanisms underlying these processes.
Keywords: Left–right patterning, Kupffer’s vesicle, Cell signaling, Positive feedback loop
Introduction
In zebrafish embryos, the developmental processes that establish left–right (LR) asymmetry are divided into the following four phases (Fig. 1). First, the initial breaking of bilateral symmetry may occur at cleavage stages. Second, an organizer region called Kupffer’s vesicle (KV) forms by early somitogenesis. Third, LR information is transmitted from KV to the lateral plate mesoderm (LPM) during somitogenesis. Finally, LR asymmetric signals are relayed to the organ primordia, eventually leading to the establishment of left- or right-specific morphogenesis. Accumulating evidence points to the participation of multiple signal pathways in KV organization, ciliogenesis and LR information transfer from KV to the LPM, and reveals that positive feedback loops play key roles in generating a robust difference from a small difference during progressive phases of LR asymmetric patterning. In this review, we summarize recent findings about these topics and describe the cellular and molecular mechanisms underlying zebrafish LR asymmetric patterning.
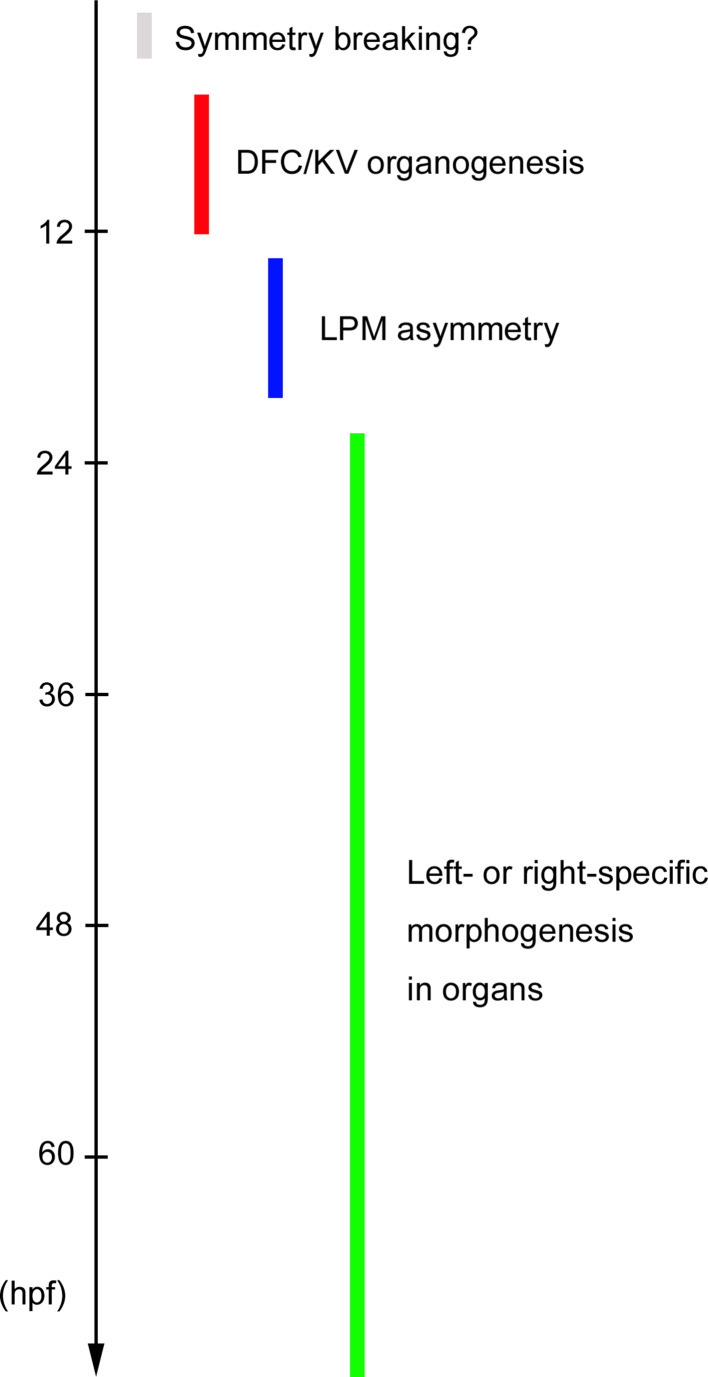
Fig. 1.
Phases of LR asymmetric patterning in zebrafish. A symmetry-breaking process may occur by 3 hpf (cleavage stage). The organizer region named KV is formed by 12 hpf (6-somite stage) and generates fluid flow to create an LR difference around KV. Asymmetric signals then transfer from KV to the LPM. Left- or right-specific morphogenesis finally occurs in organs at much later stages
Onset of left–right axis determination in zebrafish
Embryos treated with H+/K+-ATPase inhibitors at the cleavage stages display randomized expression of the nodal-related gene southpaw (spaw) and its target pitx2 in the LPM, both of which lead to randomization of heart looping and are typical phenotypes of LR patterning defects [23]. H+/K+-ATPase may thus be involved in symmetry breaking in zebrafish, as it is in the frog [27]. Although H+/K+-ATPase transcripts are asymmetrically distributed in frog embryos [27], mRNA and protein of the α isoform of H+/K+-ATPase are distributed broadly in the zebrafish embryo at cleavage stages and show no apparent asymmetry [23]. It is likely that, in zebrafish, H+/K+-ATPase α operates at post-translational level to generate LR differences within the embryo, and/or that other H+/K+-ATPases contribute to the initial breaking of LR symmetry at cleavage stages. However, additional experiments will be required to evaluate the importance of H+/K+-ATPases to zebrafish LR patterning.
Kupffer’s vesicle is an organizer region in zebrafish
Discovered in 1868 [26], KV is a fluid-filled organ that forms transiently at the posterior end of the notochord at the early somite stages in teleosts [14]. In zebrafish, KV is generated from a cluster of 20–30 dorsal forerunner cells (DFCs), which appear adjacent to the embryonic shield at midgastrulation. The DFC cluster then migrates towards the vegetal pole by late gastrulation, and generates KV by the 4- to 6-somite stages [11, 32]. Although the outlines of KV organogenesis had been clarified by 1996 (Fig. 2), how KV contributes to the zebrafish body plan remained unknown until very recently.
Fig. 2.
KV organogenesis from DFCs. KV organogenesis is divided into three steps. First, DFCs are produced from dorsal surface epithelial cells. Second, 20–30 DFCs generate a cluster, and DFCs migrate collectively toward the vegetal pole. Third, the migrated DFCs form a rosette structure at the tailbud, and the lumen and cilia are then generated in the mature vesicle. Several signals including Nodal, FGF, nc-Wnt, Hedgehog and Notch (red) are involved in a specific step(s) of DFC/KV morphogenesis. Right schematic representation of processes from DFC clustering to KV organogenesis
This issue was clarified by considering similarities between diverse vertebrates. In mouse, a transient embryonic organ called the ventral node, which is localized at the anterior tip of the primitive streak, generates a leftward extracellular fluid flow (nodal flow) created by dynein-dependent motility of monocilia, thereby establishing LR asymmetry [19, 42]. Essner et al. [15] discovered the existence of monocilia and the expression of left–right dynein (lrd) in KV in zebrafish as well as in the equivalent organs of chick and frog, suggesting that a similar, conserved mechanism underlies LR asymmetric determination in all vertebrates.
In KV of zebrafish embryos, monocilia (a typical 9+2 microtubule arrangement) form at the apical membrane of the cells facing the lumen, and lrd-dependent cilium rotation creates a counterclockwise fluid flow inside KV [14, 25]. Either ablation of DFCs or surgical removal of KV results in randomization of laterality at later stages, demonstrating that KV is the organizer region that establishes LR asymmetric patterning in zebrafish [14]. Importantly, lrd knockdown disrupts fluid flow inside KV without affecting KV structure or cilium formation; the resulting randomization of spaw expression in the LPM and of cardiac laterality [14, 23] indicates that fluid flow is required for zebrafish LR asymmetric patterning, as in the mouse. Although it is not fully understood how fluid flow breaks LR asymmetry, Ca2+ elevation occurs only in cells on the left side of KV, perhaps in response to morphogen binding or mechanical stress sensing (refs. [16, 22, 40] and for details, see below).
DFC-specific MO/mRNA transfer
A DFC-specific loss-of-function approach, developed by Amack and Yost [5], has contributed greatly to our understanding of how KV is generated from DFCs and whether DFC/KV morphogenesis is required for proper LR asymmetric pattering. It was already known that, although all embryonic cells in embryos at cleavage stages are connected with the yolk by cytoplasmic bridges, almost all such bridges are closed by the 64-cell stage [2 h postfertilization (hpf)], with the exception of DFCs, which retain these bridges until the sphere stage (4 hpf) [11]. Focusing on this difference, Amack and Yost injected fluorescein-tagged MOs into the yolk of embryos at the 256–512-cell stages (2.5–2.75 hpf), and succeeded in delivering MOs into DFCs but not other embryonic cells [4, 5]. Since this method delivers MOs into both the yolk and the yolk syncytial layer (YSL), it is possible that gene function within the yolk/YSL is involved in proper KV formation and LR patterning. To exclude this possibility, MOs have to be delivered only into yolk/YSL by injection of MOs into the yolk of embryos at the sphere–dome stages (4–4.3 hpf), as an important control. We and Esguerra et al. have demonstrated recently that mRNAs injected into the yolk of embryos at 256–512-cell stages yield proteins derived from the injected mRNAs in DFCs, indicating that this strategy is also useful for gain-of-function approaches and rescue experiments [13, 31]. By several strategies, including normal injection of MO/mRNA, DFC-specific MO/mRNA transfer and genetic analysis, it has been revealed that multiple signals including the fibroblast growth factor (FGF), Nodal, Notch, Hedgehog and Wnt pathways play crucial roles in DFC/KV organogenesis.
KV organogenesis
From the appearance of DFCs at midgastrulation to the formation of KV at early somitogenesis, several processes including DFC specification, clustering, collective migration, cluster compaction, lumen formation and ciliogenesis occur in a coordinated fashion. Based on our and others’ findings, a model of KV organogenesis can be proposed as follows (Fig. 2). In response to Nodal signaling, a cluster of 20–30 DFCs appears adjacent to the embryonic shield at midgastrulation [14]. The DFC cluster then migrates towards the vegetal pole during mid- to late gastrulation: this feature is known as collective migration and comprises both directed migration and cell clustering. Collective migration of DFCs is regulated by different types of cell adhesions: directed migration occurs passively through the interaction of DFCs with the overlapping surface ectoderm (OSE) [37], and DFC clustering is maintained by Cadherin1-mediated cell junctions between adjacent DFCs controlled by an FGF positive feedback loop [31]. During late gastrulation, the DFC cluster undergoes progressive compaction and becomes a polarized, bottle-shaped DFC cluster, a process that is regulated by non-canonical Wnt (nc-Wnt) signaling while the cluster remains in contact with the OSE [38]. At the end of gastrulation, the bottle-shaped DFC cluster detaches from the OSE and generates multiple 3D rosette structures, which then rearrange into a single rosette concurrent with the formation of the lumen by the 4- to 6-somite stages. KV formation is completed by the 6-somite stage with the generation of motile monocilia on the apical membranes of KV cells facing the lumen [37, 38].
Defects in either DFC specification, clustering or compaction lead to failures in KV organogenesis, ciliogenesis and LR asymmetric patterning at later stages. For example, in cas mutants of the Nodal-responsive gene sox32, DFC specification, ciliogenesis and KV formation are all abolished [14]. When the positive feedback loop of FGF signaling is disrupted by knockdown of the FGF positive regulator canopy1, the DFC cluster is broken up into multiple groups of cells, leading to defects in lumen formation and control of cilium length and number [31]. Although knockdown of prickle1, a component of nc-Wnt signaling, does not affect earlier processes including DFC specification and clustering, the DFC cluster in prickle1 morphants fails to compact, leading to the appearance of fragmented lumens and shortened cilia [38]. In addition to these three, many more genes have been found to regulate one or more processes of DFC/KV organogenesis, as listed in Table 1. Although the causes and types of KV defects differ from one another, all of them result in randomized body laterality. These findings therefore suggest that earlier events occurring in DFCs are prerequisite for proper KV organogenesis, which is an indispensable step for the establishment of the LR asymmetric body plan. However, it is also known that there is a parallel pathway(s) to generate motile cilia whose disruption does not affect KV organogenesis.
Table 1.
Genes essential for DFC/KV organogenesis
| Loss-of-function | Gene | Protein | DFCs | KV | KV ciliogenesis | References |
|---|---|---|---|---|---|---|
| Mutation | aei/deltaD | Notch ligand | n.d. | Normal | Shortened | [30] |
| Mutation | cas/sox32 | TF induced by Nodal | No DFCs | No KV | No cilia | [14] |
| Mutation | oep | Nodal co-receptor | No DFCs | No KV | No cilia | [14] |
| DFC-KD; KD | Ttrap | Nodal antagonist | Spread | n.d. | n.d | [13] |
| KD | frizzled-2 | nc-Wnt receptor | n.d. | Normal | Shortened/reduced | [36] |
| KD | rock2b | nc-Wnt mediator | n.d. | Small lumen | Abnormal cilium arrangement | [47] |
| KD | prickle1a | nc-Wnt mediator | Failed compaction | Fragmented lumens | Shortened | [38] |
| Mutation | MZvangl2 | nc-Wnt mediator | n.d. | Expanded | Abnormal cilium orientation | [7] |
| Mutation | MZkny | nc-Wnt mediator | n.d. | Expanded | Abnormal cilium orientation | [7] |
| KD | duboraya | nc-Wnt mediator | n.d. | Normal | Shortened/reduced | [36] |
| KD | foxj1a | TF induced by Hedgehog | n.d. | Small | Shortened/reduced | [45] |
| DFC-KD | no tail | TF induced by FGF | n.d. | Disorganized | Shortened/reduced | [5] |
| DFC-KD; KD | tbx16 | TF induced by FGF | Broken-up clusters | Small/disrupted | Reduced | [4, 31] |
| DFC-KD; KD | fgfr1a | FGF receptor | Normal | Normal | Shortened | [35] |
| DFC-KD; KD | canopy1 | FGF positive regulator | Broken-up clusters | Small/disrupted | Shortened/reduced | [31] |
| KD | ire1 | FGF mediator | Broken-up clusters | Small/disrupted | Shortened/reduced | [20] |
| KD | fibp1 | FGF mediator | Broken-up clusters | Small/disrupted | Shortened/reduced | [20] |
| Mutation | ace/fgf8 | FGF ligand | Broken-up clusters | Small/disrupted | Reduced | [3, 31, 35] |
| DFC-KD | cadherin1 | Adhesion molecule induced by FGF | Broken-up clusters | Small/disrupted | Shortened/reduced | [13, 31, 38] |
| KD | CaMK-II | Ca2+ signal mediator | n.d. | Small | Shortened/reduced | [16] |
| DFC-KD; KD | lrd | IFT component | Normal | Normal | Non-motile | [14, 23] |
| KD | ift57 | IFT component | n.d. | n.d. | No/shortened cilia | [25] |
| KD | ift88 | IFT component | n.d. | n.d. | No/shortened cilia | [25] |
| DFC-KD; KD | connexin43.4 | Gap junction component | n.d. | Small | Shortened | [18] |
| KD | miR-92 | Fine-tuner of Gata5 | n.d. | Small/disrupted | Shortened/reduced | [28] |
| DFC-KD; KD | integrin a5 | Adhesion molecule to ECM | Broken-up clusters | Small | Shortened/reduced | [1] |
| KD | nde1 | Centrosomal protein | n.d. | Small | Elongated | [24] |
DFC-KD DFC-specific knockdown, KD knockdown, TF transcription factor, n.d. not determined
KV ciliogenesis
During ciliogenesis, centrioles form basal bodies that anchor cilia to the cell surface and nucleate the synthesis of ciliary axonemes. The cilium is built and maintained by intraflagellar transport (IFT) genes such as ift57 and ift88. Recently, Stubbs et al. [45] and Yu et al. [49] have identified a transcription factor named foxj1a, a target of the Hedgehog pathway, as a master regulator of the production of motile cilia. Knockdown of foxj1a thus down-regulates the expression of cilium/basal body components such as lrd and centrin2, resulting in no or shortened cilia and no fluid flow, and leading in turn to LR patterning defects. Knockdown of either ift57 or ift88 has the same consequences [25]. Furthermore, in mutants of the Notch ligand deltaD (aei) or in knockdown embryos of either a paralog of fgf receptor 1s (fgfr1a) or the nc-Wnt mediator duboraya, cilium components and/or the IFT genes are down-regulated in DFCs, KV cilia become short, and LR asymmetric defects ensue [30, 35, 36]. These findings indicate that ciliogenesis, which is controlled by FGF, Hedgehog, Notch and nc-Wnt signaling, is required for the generation of fluid flow in KV as well as the establishment of LR asymmetric patterning. However, cilium length control is insufficient for determining the proper LR axis, because knockdown of nuclear distribution gene E homologue 1 (nde1), encoding a centrosomal protein, results in increased cilium length in KV but yields LR patterning defects at later stages [24]. In nde1 knockdown embryos, interestingly, KV abnormality accompanies these defects: the KV structure becomes small, perhaps due to a suppression of cell division. This evidence raises the possibility that KV/lumen formation also has a crucial role in relaying LR information.
Signal transfer from KV to the LPM
Nodal, a member of the transforming growth factor-β (TGF-β) family of secreted morphogens, is required to establish proper LR asymmetry in all vertebrates studied to date [41]. In zebrafish, the nodal-related gene spaw is expressed bilaterally in the cells surrounding KV at the 4- to 6-somite stage, whereafter spaw expression becomes restricted to the left LPM beginning at the 10- to 12-somite stage [29]. Because spaw expression is induced by Spaw itself, a positive feedback loop of Spaw is crucial for generating lateral plate asymmetry [41, 42]. While it is not yet fully understood how bilaterally distributed Spaw around KV stimulates spaw expression only in the left LPM, recent findings have pointed out the following possible mechanisms (Fig. 3).
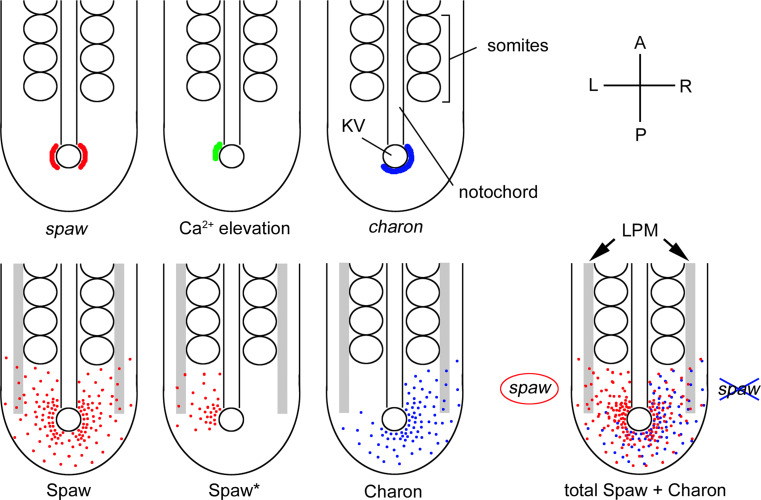
Fig. 3.
Signal transfer from KV to the left LPM. Since spaw is expressed bilaterally (red) around KV at the 10-somite stage, Spaw (red dots) may diffuse symmetrically in the tailbud. In response to fluid flow in the KV, intracellular Ca2+ elevation (green) occurs only on the left side of the KV. The local Ca2+ activates a positive feedback loop of Ca2+ via CaMK-II, leading to enhanced local secretion/processing of Spaw (Spaw* in the lower panel). charon expression becomes asymmetric (blue) in a fluid flow-dependent manner, leading to Charon (blue dots) diffusing toward the right side. Because Charon antagonizes Spaw by binding to it, diffused Spaw cannot activate its own expression in the right LPM. However, Spaw reaches the left LPM without antagonism from Charon, where it stimulates spaw expression. Top right A, anterior; P, posterior; L, left; R, right
Counterclockwise fluid flow promotes intracellular Ca2+ elevation in cells localized on the left side of KV. This induces phosphorylation of Ca2+/CaM-dependent protein kinase II (CaMK-II), to activate both Ca2+ release from the endoplasmic reticulum (ER) and extracellular Ca2+ influx [16, 22, 40]. This positive feedback loop of Ca2+ signals may promote processing and/or secretion of Spaw only in the left side of KV (Spaw* in Fig. 3). Charon, a member of the Cerberus/Dan family, also contributes to generating Spaw asymmetry around KV [17]. charon is expressed bilaterally in KV cells at the 6-somite stage, but its expression switches to a right-sided asymmetric pattern at the 8- to 10-somite stages in a fluid flow-dependent manner [30]. Since Charon binds to Spaw and antagonizes Spaw functions [17], the rightward gradient of Charon around KV tends to inhibit Spaw strongly in the right side. Due to the opposing gradients of activator (Spaw) and inhibitor (Charon), Spaw cannot stimulate expression of spaw at the right side of the LPM. Conversely, Spaw induces its own expression in the left-side LPM by positive feedback regulation. We therefore propose that opposed gradients between Spaw and Charon around KV contribute to initiating spaw expression in the left-side LPM.
Progressive expression of spaw in the LPM
Knockdown of spaw abolishes expression of its target genes, including pitx2, lefty1, and lefty2 in the LPM and lefty1 in the midline (notochord), leading to a loss of left-specific morphogenesis that is consistent with the phenotype seen in mouse Nodal mutants [29]. These findings indicate the presence in zebrafish of a conserved genetic cascade, initiated by Spaw, which is crucial for the transfer of directional LR asymmetric information into the organ primordia.
Recent findings suggest a mechanism by which the Spaw signal expands to the anterior LPM only on the left side. Once spaw expression begins at the posterior end of the left LPM at the 10-somite stage, newly synthesized Spaw in the LPM further stimulates the progressive, posterior-to-anterior expression of spaw, pitx2, lefty1, and lefty2 in the left LPM at a rate of 2.3 somite lengths per somite generation time [29, 48], suggesting that this progression depends on the positive feedback loop of Spaw.
Expression of lefty1, a Nodal inhibitor, is activated by Spaw and another TGF-β family member, BMP, only in the midline between about the 10- to 18-somite stages [10, 44]. In the absence of lefty1, left-sided expression of spaw in the LPM initiates normally, but Spaw then leaks to the right side and stimulates its own expression in the right LPM [44, 48]. Once spaw expression occurs here, the Spaw positive feedback loop expands its own expression in a similar manner to that on the left side, thus leading to bilateral spaw expression in the LPM with the left side leading [48]. These findings indicate that lefty1 knockdown embryos retain a left-side bias, that Lefty1 acts as a midline barrier, and that Spaw can diffuse over a long distance, from the left to the right LPM. Furthermore, in lefty1 morphants, propagation of spaw expression from the posterior to the anterior LPM is faster than that in wild-type embryos [48], suggesting that Lefty1 optimizes spaw expansion in the left LPM. This interpretation is consistent with experimental data showing that Lefty1 is a major component of the midline barrier in the mouse [33], and is supported by a theoretical model in which a diffusible inhibitor (Lefty1) in the midline contributes to the establishment of lateral plate asymmetry by Nodal [34].
The reaction–diffusion system, a theoretical model, has two components: a feedback activator and a feedback inhibitor [46]. Depending on the particular features of the activator/inhibitor pair, multiple patterns such as waves, strips, spirals, and pulses can be generated. A reaction–diffusion system comprising a local activator and a long-range inhibitor can generate a robust asymmetric difference from a small difference between two separated regions. Several studies have thus proposed that LR asymmetry in the LPM is generated by the reaction–diffusion system using the Nodal/Lefty pair [8, 34, 41, 42]. However, our understanding as described above differs from this reaction–diffusion model: it seems possible that Spaw is not a local activator, but rather acts as a long-range activator.
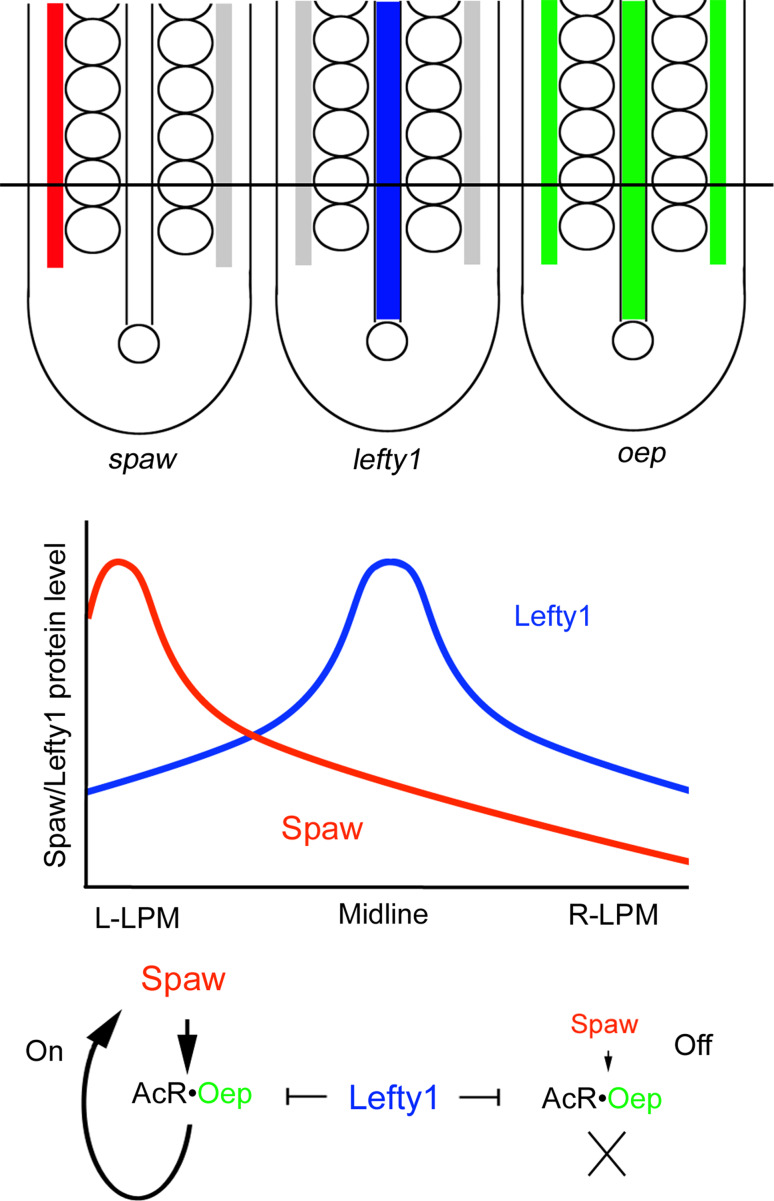
This difference may be explained by the existence of a system that converts a long-range activator into a local one, which we propose here (Fig. 4). Nodal requires epidermal growth factor-Cripto/FRL-1/Cryptic (EGF-CFC) co-receptors to activate a Nodal signal through Activin receptors [9]. Although activin receptors are broadly expressed within embryos, expression of an EGF-CFP co-receptor named one-eyed pinhead (oep) is restricted to the two LPMs and the notochord at early- to mid-somitogenesis. Importantly, Lefty1 reportedly antagonizes Nodal signaling by binding to EGF-CFC co-receptors [9]. Even if Spaw tends to act as a long-range activator, Oep distribution and Lefty1 function can convert Spaw into a local activator. In the left LPM, where Spaw is abundant and Lefty1 is sparse, Spaw stimulates its own expression only in Oep-positive LPM cells. In contrast, the small amounts of Spaw derived from the left LPM are insufficient to activate spaw expression in the right LPM, because Oep expressed in LPM cells is antagonized by Lefty1.
Fig. 4.
Generation of a robust difference between the left and right LPMs. Expression patterns of spaw (red), lefty1 (blue) or oep (green) at the 15- to 18-somite stages. We hypothesize that Spaw and Lefty1 can diffuse at a similar rate, as shown in the central graph. In the left LPM (L-LPM), Spaw binds to an Activin receptor (AcR) and Oep pair, and activates spaw expression even in the presence of Lefty1. In contrast, the small amounts of Spaw in the right LPM (R-LPM) cannot stimulate spaw expression due to the antagonistic effect of Lefty1
However, whether Spaw and Lefty1 can diffuse over a long distance within the embryo remains unknown. Additional experiments measuring the diffusion of Spaw and Lefty1 in live zebrafish embryos will be important to elucidate the exact nature of the midline barrier, and thus to determine how the Nodal signal is restricted to the left side.
Organ laterality
Organs such as the brain, heart, liver, and pancreas display LR asymmetry. It is thought that the left-sided Spaw signals in the LPM are relayed to the organ primordia, eventually leading to left-specific morphogenesis. In fact, knockdown of spaw in Tg[lefty1::GFP], a transgenic line which can monitor Nodal activity in zebrafish, results in the loss of left-sided activation of Nodal signaling in the diencephalon, suggesting that Spaw in the LPM regulates Nodal activity in the brain [29]. The consequent brain asymmetry then leads to expansion of the sub-nucleus in the left habenula by the early larva stage [2]. In abf mutants, which exhibit right-sided activation of Nodal in the diencephalon, asymmetry of the habenula sub-nucleus is reversed with 100 % penetrance, indicating that a Nodal-dependent mechanism is essential for determining habenula laterality. However, in mutants in which Nodal is activated bilaterally (ntl) or is absent (abf or LZoep) in the diencephalon, habenula asymmetry is established but laterality of the habenula sub-nucleus is randomized, indicating that habenula laterality is independent of the Nodal handedness in the diencephalon. These findings suggest that there is a later and unidentified mechanism that specifies habenula laterality.
Aizawa et al. [2] also reported that, in abf mutants, brain laterality does not correlate with laterality of visceral organs such as the heart and gall bladder. abf mutation results in randomization of heart looping: 50 % of abf mutants show normal heart looping, and the rest display reversed looping. In about half of abf mutants having reversed heart looping, the Nodal signal is activated only in the right diencephalon, consistent with the reversed cardiac laterality. However, the other reversed-heart abf mutants display no activation of Nodal on either side of the diencephalon, representing an inconsistency between cardiac and brain laterality. Why differences between heart and brain lateralities occur in these mutants and how such differences are generated remain unknown.
Heart primordia appear bilaterally at the anterior part of the LPM at about the 18- to 20-somite stages, and migrate toward the midline to form a single tube of the heart. The heart tube then undergoes a series of looping events that may be regulated by asymmetric activities of Nodal and BMP signaling. After fusion of the bilateral heart primordia, leftward involution of the right heart field generates the ventral floor, while the noninvoluting left heart field forms the dorsal roof of the primary heart tube [39]. The primary heart tube then rotates a clockwise due to the leftward migration of myocardial cells, resulting in the conversion of the LR axis into the dorsal–ventral axis of the tube and determining the laterality of heart jogging. This rotation is directly influenced by LR information such as asymmetric expression of bmp4, lefty1, lefty2, and pitx2 [6, 12, 43].
Asymmetry of the gut tube is also generated by a looping series, and the looping is mediated by asymmetric migration of the LPM, depending on LR information [21]. In normal embryos, the left and right LPMs migrate dorsally and ventrolaterally, respectively, eventually leading to a leftward shift of the developing intestine. In contrast, spaw knockdown leads to randomized LPM migration and gut looping, suggesting that asymmetries in LPM migration and gut looping are regulated by LR gene expression in the LPM. However, we do not yet know how LR information in the LPM is transduced to organ primordia, or how asymmetric cell behavior and/or tissue migration give rise to left- or right-specific morphogenesis in organs.
Positive feedback regulation for LR asymmetric patterning
As described above, several positive feedback loops are involved in LR asymmetric patterning. During KV organogenesis, the cluster of KV progenitor cells (DFCs) is maintained by a Canopy1-mediated FGF positive feedback loop, which is a prerequisite for proper generation of KV [31]. When this positive loop is disrupted, the DFC cluster is broken up into small groups of cells without affecting the total number of DFCs at midgastrulation. However, at late gastrulation, an abnormal rosette-like structure containing a reduced number of DFCs is formed, surrounded by apparently dead cells. This finding suggests that the FGF positive feedback loop regulates DFC clustering to prevent DFCs from dying and to ensure that they carry out later steps of KV organogenesis properly.
The positive feedback loop of either Ca2+ in the left side of KV or Spaw in the left LPM is utilized to amplify their signals as a pulse at the restricted area. We thus think that positive feedback loops, acting in concert with inhibitors (Charon or Lefty1), generate a robust difference from a small difference in LR patterning [16, 41, 42]. Because such a difference can be seen in the diencephalon and habenula at much later stages, it is possible that positive feedback loops contribute to generating brain laterality.
Concluding remarks
During the 8 years that have passed since the discovery of fluid flow in the KV in zebrafish embryos [14, 25], many studies using zebrafish as an experimental model have provided us with new insights into how zebrafish LR asymmetric patterning is established. We now know that many genes and signals are involved in KV organogenesis, ciliogenesis, and the establishment of LR asymmetric patterning, and regulatory mechanisms underlying these processes have been proposed, especially for KV organogenesis and signal transfer from KV to the LPM. Despite this substantial progress, many questions still remain. We do not know the exact mechanism of the initial breaking of LR symmetry; how fluid flow is sensed; how far Spaw and Lefty1 can diffuse within the embryo; or why there are inconsistencies among lateralities of the heart, diencephalon, and habenula. Filling these gaps will be essential to understanding the entire mechanism underlying the LR asymmetric body plan. Further studies using zebrafish embryos will undoubtedly yield exciting new discoveries in this field in the near future.
Acknowledgments
We are grateful to Ian Smith for advice, helpful discussions and critical reading of the manuscript. We also thank the Ministry of Education, Culture, Sports, Science and Technology (MEXT), the Uehara Memorial Foundation, the Nakajima Foundation and the Mochida Memorial Foundation for past and current support.
References
- 1.Ablooglu AJ, Tkachenko E, Kang J, Shattil SJ. Integrin alphaV is necessary for gastrulation movements that regulate vertebrate body asymmetry. Development. 2010;137:3449–3458. doi: 10.1242/dev.045310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizawa H, Goto M, Sato T, Okamoto H. Temporally regulated asymmetric neurogenesis causes left-right difference in the zebrafish habenular structures. Dev Cell. 2007;12:87–98. doi: 10.1016/j.devcel.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Albertson RC, Yelick PC. Roles for fgf8 signaling in left-right patterning of the visceral organs and craniofacial skeleton. Dev Biol. 2005;283:310–321. doi: 10.1016/j.ydbio.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 4.Amack JD, Wang X, Yost HJ. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Dev Biol. 2007;310:196–210. doi: 10.1016/j.ydbio.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 5.Amack JD, Yost HJ. The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr Biol. 2004;14:685–690. doi: 10.1016/j.cub.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Baker K, Holtzman NG, Burdine RD. Direct and indirect roles for Nodal signaling in two axis conversions during asymmetric morphogenesis of the zebrafish heart. Proc Natl Acad Sci USA. 2008;105:13924–13929. doi: 10.1073/pnas.0802159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borovina A, Superina S, Voskas D, Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat Cell Biol. 2010;12:407–412. doi: 10.1038/ncb2042. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Schier AF. Lefty proteins are long-range inhibitors of squint-mediated nodal signaling. Curr Biol. 2002;12:2124–2128. doi: 10.1016/S0960-9822(02)01362-3. [DOI] [PubMed] [Google Scholar]
- 9.Cheng SK, Olale F, Brivanlou AH, Schier AF. Lefty blocks a subset of TGFbeta signals by antagonizing EGF-CFC coreceptors. PLoS Biol. 2004;2:E30. doi: 10.1371/journal.pbio.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chocron S, Verhoeven MC, Rentzsch F, Hammerschmidt M, Bakkers J. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol. 2007;305:577–588. doi: 10.1016/j.ydbio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Cooper MS, D’Amico LA. A cluster of noninvoluting endocytic cells at the margin of the zebrafish blastoderm marks the site of embryonic shield formation. Dev Biol. 1996;180:184–198. doi: 10.1006/dbio.1996.0294. [DOI] [PubMed] [Google Scholar]
- 12.de Campos-Baptista MI, Holtzman NG, Yelon D, Schier AF. Nodal signaling promotes the speed and directional movement of cardiomyocytes in zebrafish. Dev Dyn. 2008;237:3624–3633. doi: 10.1002/dvdy.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esguerra CV, Nelles L, Vermeire L, Ibrahimi A, Crawford AD, Derua R, Janssens E, Waelkens E, Carmeliet P, Collen D, Huylebroeck D. Ttrap is an essential modulator of Smad3-dependent Nodal signaling during zebrafish gastrulation and left-right axis determination. Development. 2007;134:4381–4393. doi: 10.1242/dev.000026. [DOI] [PubMed] [Google Scholar]
- 14.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 15.Essner JJ, Vogan KJ, Wagner MK, Tabin CJ, Yost HJ, Brueckner M. Conserved function for embryonic nodal cilia. Nature. 2002;418:37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]
- 16.Francescatto L, Rothschild SC, Myers AL, Tombes RM. The activation of membrane targeted CaMK-II in the zebrafish Kupffer’s vesicle is required for left-right asymmetry. Development. 2010;137:2753–2762. doi: 10.1242/dev.049627. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto H, Rebagliati M, Ahmad N, Muraoka O, Kurokawa T, Hibi M, Suzuki T. The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development. 2004;131:1741–1753. doi: 10.1242/dev.01070. [DOI] [PubMed] [Google Scholar]
- 18.Hatler JM, Essner JJ, Johnson RG. A gap junction connexin is required in the vertebrate left-right organizer. Dev Biol. 2009;336:183–191. doi: 10.1016/j.ydbio.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 19.Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left-right asymmetry. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Hong SK, Dawid IB. FGF-dependent left-right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proc Natl Acad Sci USA. 2009;106:2230–2235. doi: 10.1073/pnas.0812880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horne-Badovinac S, Rebagliati M, Stainier DY. A cellular framework for gut-looping morphogenesis in zebrafish. Science. 2003;302:662–665. doi: 10.1126/science.1085397. [DOI] [PubMed] [Google Scholar]
- 22.Jurynec MJ, Xia R, Mackrill JJ, Gunther D, Crawford T, Flanigan KM, Abramson JJ, Howard MT, Grunwald DJ. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc Natl Acad Sci USA. 2008;105:12485–12490. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Izpisua Belmonte JC. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat Cell Biol. 2011;13:351–360. doi: 10.1038/ncb2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 26.Kupffer C. Beobachtungen über die Entwicklung der Knochenfische. Arch Mikrob Anat. 1868;4:209–272. doi: 10.1007/BF02955363. [DOI] [Google Scholar]
- 27.Levin M, Thorlin T, Robinson KR, Nogi T, Mercola M. Asymmetries in H+/K+-ATPase and cell membrane potentials comprise a very early step in left-right patterning. Cell. 2002;111:77–89. doi: 10.1016/S0092-8674(02)00939-X. [DOI] [PubMed] [Google Scholar]
- 28.Li N, Wei C, Olena AF, Patton JG. Regulation of endoderm formation and left-right asymmetry by miR-92 during early zebrafish development. Development. 2011;138:1817–1826. doi: 10.1242/dev.056697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 30.Lopes SS, Lourenco R, Pacheco L, Moreno N, Kreiling J, Saude L. Notch signalling regulates left-right asymmetry through ciliary length control. Development. 2010;137:3625–3632. doi: 10.1242/dev.054452. [DOI] [PubMed] [Google Scholar]
- 31.Matsui T, Thitamadee S, Murata T, Kakinuma H, Nabetani T, Hirabayashi Y, Hirate Y, Okamoto H, Bessho Y. Canopy1, a positive feedback regulator of FGF signaling, controls progenitor cell clustering during Kupffer’s vesicle organogenesis. Proc Natl Acad Sci USA. 2011;108:9881–9886. doi: 10.1073/pnas.1017248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melby AE, Warga RM, Kimmel CB. Specification of cell fates at the dorsal margin of the zebrafish gastrula. Development. 1996;122:2225–2237. doi: 10.1242/dev.122.7.2225. [DOI] [PubMed] [Google Scholar]
- 33.Meno C, Gritsman K, Ohishi S, Ohfuji Y, Heckscher E, Mochida K, Shimono A, Kondoh H, Talbot WS, Robertson EJ, Schier AF, Hamada H. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol Cell. 1999;4:287–298. doi: 10.1016/S1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T, Mine N, Nakaguchi E, Mochizuki A, Yamamoto M, Yashiro K, Meno C, Hamada H. Generation of robust left-right asymmetry in the mouse embryo requires a self-enhancement and lateral-inhibition system. Dev Cell. 2006;11:495–504. doi: 10.1016/j.devcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oishi I, Kawakami Y, Raya A, Callol-Massot C, Izpisua Belmonte JC. Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya. Nat Genet. 2006;38:1316–1322. doi: 10.1038/ng1892. [DOI] [PubMed] [Google Scholar]
- 37.Oteiza P, Koppen M, Concha ML, Heisenberg CP. Origin and shaping of the laterality organ in zebrafish. Development. 2008;135:2807–2813. doi: 10.1242/dev.022228. [DOI] [PubMed] [Google Scholar]
- 38.Oteiza P, Koppen M, Krieg M, Pulgar E, Farias C, Melo C, Preibisch S, Muller D, Tada M, Hartel S, Heisenberg CP, Concha ML. Planar cell polarity signalling regulates cell adhesion properties in progenitors of the zebrafish laterality organ. Development. 2010;137:3459–3468. doi: 10.1242/dev.049981. [DOI] [PubMed] [Google Scholar]
- 39.Rohr S, Otten C, Abdelilah-Seyfried S. Asymmetric involution of the myocardial field drives heart tube formation in zebrafish. Circ Res. 2008;102:e12–e19. doi: 10.1161/CIRCRESAHA.107.165241. [DOI] [PubMed] [Google Scholar]
- 40.Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- 43.Smith KA, Chocron S, von der Hardt S, de Pater E, Soufan A, Bussmann J, Schulte-Merker S, Hammerschmidt M, Bakkers J. Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev Cell. 2008;14:287–297. doi: 10.1016/j.devcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Smith KA, Noel E, Thurlings I, Rehmann H, Chocron S, Bakkers J. Bmp and nodal independently regulate lefty1 expression to maintain unilateral nodal activity during left-right axis specification in zebrafish. PLoS Genet. 2011;7:e1002289. doi: 10.1371/journal.pgen.1002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet. 2008;40:1454–1460. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turing AM. The chemical basis of morphogenesis. Bull Math Biol. 1953;52(153–97):119–152. doi: 10.1007/BF02459572. [DOI] [PubMed] [Google Scholar]
- 47.Wang G, Cadwallader AB, Jang DS, Tsang M, Yost HJ, Amack JD. The Rho kinase Rock2b establishes anteroposterior asymmetry of the ciliated Kupffer’s vesicle in zebrafish. Development. 2011;138:45–54. doi: 10.1242/dev.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Yost HJ. Initiation and propagation of posterior to anterior (PA) waves in zebrafish left-right development. Dev Dyn. 2008;237:3640–3647. doi: 10.1002/dvdy.21771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]