Abstract
MicroRNAs (miRNAs) are a family of small, non-coding RNAs that control gene expression at the post-transcriptional level by destabilizing and inhibiting translation of their target messenger RNAs. MiRNAs are involved in the regulation of a number of fundamental biological processes, and their dysregulation is thought to contribute to several disease processes. Emerging evidence suggests that miRNAs also play a critical role in protecting the heritable genome by contributing to the regulation of the DNA damage response. Consequently, much recent investigative effort has been directed towards an improved understanding of how miRNAs are regulated in response to DNA damage. In this review, we discuss the most recent findings regarding the regulation of miRNA expression and the functional roles of miRNAs in the DNA damage response.
Keywords: DNA damage response, MicroRNA, DNA repair, Transcriptional regulation, Post-transcriptional regulation, MicroRNA biogenesis
Introduction
Maintenance of genomic integrity is an important challenge that allows the faithful transmission of genetic information to our offspring, thereby ensuring our own survival [1]. One can appreciate the magnitude of this challenge by taking into consideration the fact that each cell in the human body is the recipient of tens of thousands of DNA lesions each day [2]. DNA damage may arise from genomic infidelities that take place during DNA replication or from endogenously produced reactive oxygen species that are a byproduct of normal metabolic processes. In addition, DNA may be damaged by several extrinsic genotoxic stresses, such as ultraviolet (UV) light, ionizing radiation (IR), and a variety of industrial and environmental chemical compounds [1]. In order to preserve the integrity of the genome, eukaryotic cells rely on a sophisticated cellular system referred to as the DNA damage response (DDR) that is designed to detect DNA damage and then activate the most appropriate signaling pathway to mediate its repair.
The DDR consists of various processes, including DNA repair, cell-cycle checkpoints, and apoptosis. This DDR is a highly regulated process that involves a variety of proteins that function as sensors, transducers, and effectors. The information collected and transmitted by these proteins will ultimately be used in making a cell fate determination—either arrest the cell cycle to allow repair of damaged DNA or, if the damage is beyond repair, initiate programs that instruct the cell to undergo apoptosis [3, 4].
Defects in DNA damage signaling or repair may have profound implications for the well-being of an individual in that a diminished capacity for DNA repair has been associated with several human disease processes, including cancer [5]. Perhaps one of the most widely recognized genetic syndromes involving the DDR is ataxia telangiectasia (A-T), which is caused by mutations in the ATM (Ataxia Telangiectasia Mutated) gene. ATM is a serine/threonine protein kinase that functions as a transducing protein in the DNA damage response. ATM is recruited to DNA double-strand breaks where it functions to reduce cyclin-dependent kinase (CDK) activity [5]. Covalent modifications (phosphorylation, ubiquitination, SUMOylation, and acetylation) of proteins often promote the assembly/disassembly of checkpoint proteins and the DNA repair machinery at DNA break points [6, 7]. While the DDR activation leads to a dramatic change in transcription programs in the cell, expression of DDR-associated genes is also highly regulated. Results from recent studies suggest that microRNAs play a key role in mediating the expression of genes involved in the initiation, activation, and maintenance of the DDR. MiRNAs are small (18–25 nucleotides) endogenous noncoding RNAs that regulate gene expression by repressing translation or promoting the degradation of their target mRNAs [8]. To date, miRNAs have been assigned a regulatory role in virtually every biological process, including development, differentiation, cell-cycle control, and apoptosis [9]. MiRNAs suppress gene expression by binding to the 3′ untranslated region (UTR) of their target mRNAs and mediating mRNA degradation or translational inhibition [8]. Similar to protein-coding genes, expression of miRNA genes following DNA damage appears to be modulated at the transcriptional and post-transcriptional levels [10, 11]. In this review, we describe the miRNAs involved in the regulation of the DDR and discuss recent findings regarding the functional relationship between the DDR and miRNA biogenesis.
Canonical microRNA biogenesis: miRNA expression and maturation
MiRNAs are produced through a series of endonucleolytic cleavage steps that are mediated by two evolutionarily conserved RNase III enzymes, referred to as Drosha and Dicer [12]. MiRNAs can be transcribed from two different pathways. Approximately half of miRNAs (intergenic miRNAs) are derived from non-protein-coding RNA transcripts, whereas intronic or exonic miRNAs are located within protein coding genes and share a common promoter with their host genes [13, 14]. The majority of miRNAs are transcribed by RNA Pol II as primary miRNAs (pri-miRNAs) [15]. Following transcription, the pri-miRNA is recognized and processed in the nucleus by the Drosha-DGCR8 microprocessor. This first cleavage step generates an approximately 70-nucleotide hairpin structure called precursor miRNA (pre-miRNA) that contains 25- to 30-base pair stems and relatively small loops with 3′ overhangs [16]. In the next step, the pre-miRNA is translocated from the nucleus to the cytoplasm through an interaction with exportin-5, a RanGTP-binding nuclear transporter [17]. The pre-miRNA is then subjected to additional processing by Dicer, which yields a 20- to 25- nucleotide double-stranded mature miRNA consisting of a functional guide strand and passenger strand. The mature miRNA is then loaded into the RNA-induced silencing complex (RISC) where it directs Argonaute 2 (Ago2) to target mRNAs and repress protein expression [18, 19]. Although our understanding of miRNA biogenesis has increased dramatically over recent years, the precise cellular and molecular mechanisms that coordinate the degradation and turnover of miRNAs have remained elusive [20]. However, recent evidence suggests the steady-state levels of miRNAs may be regulated by ‘microRNases’ in plants [21] and animals [22].
MiRNAs modulate the DNA damage response
The DDR is initiated at damaged DNA sites by DDR sensors/mediators. Upon recognition of DNA damage, transducers, such as ATM, ATR (ataxia telangiectasia and Rad3 related), and DNA-PKcs (DNA-dependent protein kinase catalytic subunit) relay and amplify the original damage signal to effectors in downstream pathways, including the DNA repair, cell cycle–checkpoint, and apoptosis pathways [23]. In silico prediction of miRNA target sites and experimental validation have shown that miRNAs regulate the DDR by modulating key components of DDR pathways. An examination of the 3′-UTR of 142 DDR genes that were analyzed using two independent target prediction algorithms (Miranda and Targetscan), determined that more than half of the DNA repair and DNA damage checkpoint genes contained conserved miRNA target sites [24]. Several review articles have described miRNAs that regulate the DDR as well as their predicted target binding sites of DDR genes [10, 24, 25]. In the following section, we describe some of the key proteins in the DDR pathway that are regulated by miRNAs (Fig. 1).
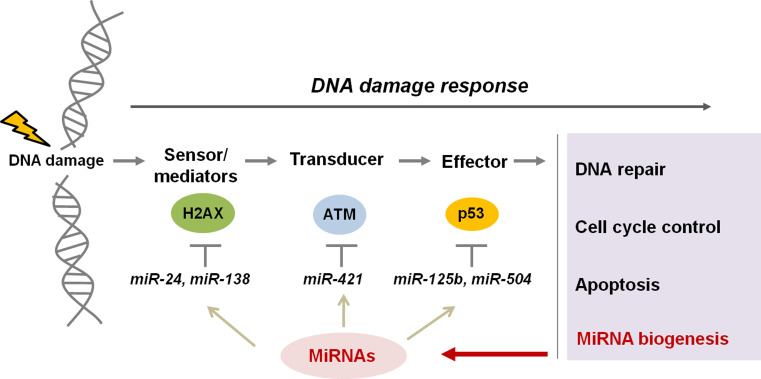
Fig. 1.
Crosstalk between the DNA damage response pathway and miRNAs. The DNA damage response (DDR) pathway is comprised of a number of different proteins that function to coordinate the repair of DNA damage and preserve genomic integrity. These proteins are classified as either sensors, transducers, or effectors, depending on their specific function. Recent studies have shown that miRNAs may play a regulatory role in the DDR by targeting and modulating the expression of genes involved in the DDR. For example, H2AX is targeted by miR-24 and miR-138, and ATM is controlled by miR-421. P53, a critical effector protein in the DDR pathway, is targeted by miR-125b and miR-504. There is also data to suggest that the DDR is also involved in the regulation of miRNA biogenesis
MiRNA-mediated regulation of DDR genes
Sensors/mediators of the DNA damage response: H2AX-miR-24/miR-138
Immediately following DNA damage, the histone variant H2AX is extensively phosphorylated at DNA break sites by the ATM and ATR kinases. Phosphorylation of H2AX represents one of the earliest DNA damage-induced signals and is essential for the sustained recruitment of various checkpoint and DNA repair proteins to the damage site [6]. Lal and colleagues [26] were the first to show that upregulated miR-24 inhibited H2AX expression and DNA repair in terminally differentiated blood cells and rendered the cells hypersensitive to the effects of γ-irradiation and genotoxic drugs. In other studies, both miR-138 and miR-542-3p were shown to reduce γH2AX (phosphorylated H2AX at serine 139) foci formation after DNA damage in human U2OS cells [27]. Overexpression of miR-138 was found to inhibit homologous recombination and enhance cellular sensitivity to multiple DNA-damaging agents. However, miR-24 was not identified in this screening using human U2OS cells, indicating that the functions and targets of miRNAs might be cell-specific and context-dependent.
Transducers of the DNA damage response: ATM-miR-421
ATM is a major PI3 kinase-related kinase that is responsible for initiating the activation of a number of signaling pathways following DNA damage. It is estimated that ATM may phosphorylate as many as 700 proteins [28], underscoring its manifold interfaces with various biological processes in the DDR. In response to DNA damage, ATM becomes activated by autophosphorylation on serine residues. Homeostatic regulation of ATM activity in the DDR is primarily mediated by the Wip1 phosphatase [29]. A recent examination of miRNA-binding motifs in the 3′-UTR region of the ATM gene led to the identification of miR-421 as a new regulator of ATM. Additional experiments revealed that overexpression of miR-421 was sufficient to attenuate the S-phase cell-cycle checkpoint and sensitize HeLa cells to ionizing radiation [30]. N-myc was also shown to transcriptionally activate miR-421. Gene amplification of N-myc aberrantly upregulated miR-421 expression in neuroblastoma. This study identified a novel regulatory mechanism for ATM and, moreover, a potential target for neuroblastoma treatment [30].
Effectors of DNA repair: BRCA1-miR-182
BRCA1, a tumor suppressor gene, encodes a nuclear phosphoprotein that plays a key role in maintaining genomic stability. Mutations in BRCA1 are associated with a substantially increased risk of developing breast and ovarian cancer [31]. BRCA1, along with other tumor suppressors, DNA-damage sensors, and signal transducers forms a large multisubunit protein complex known as the BRCA1-associated genome surveillance complex (BASC) that is recruited to DNA lesions where it facilitates DNA repair and cell-cycle regulation [31]. A recent report demonstrated that miR-182 targeted BRCA1 mRNA and inhibited its expression [32]. Overexpression of miR-182 impaired homologous recombination repair and rendered cells hypersensitive to IR. Furthermore, miR-182-overexpressing breast tumor cells were hypersensitive to inhibitors of poly (ADP-ribose) polymerase 1 (PARP1), showing a BRCA1-deficiency phenotype (BRCAness). In that study, antagonizing miR-182 enhanced BRCA1 levels and induced resistance to a PARP1 inhibitor [32].
A genome-wide screen of single-nucleotide polymorphisms (SNP) was recently performed to identify SNPs that are associated with breast cancer risk, from which Nicoloso et al. further conducted a case–control population study and observed that germline occurrence of rs799917-BRCA1 varies significantly among populations with different risks of developing breast cancer. Differential regulation of BRCA1 in cancer cells with different genotypes appeared to be associated with the BRCA1-interacting miR-638. This study not only identified a clinically relevant miRNA that targets BRCA1 but also discovered a novel mechanism of gene regulation in which transcribed target SNPs alter miRNA gene regulation and protein expression, contributing to the likelihood of cancer susceptibility [33].
Effectors of cell-cycle checkpoints and apoptosis: p53-miR-125b/miR-504 and MDM2-miR-605
The tumor suppressor p53 plays a central role in the cellular stress response. P53 protein level and activity are tightly controlled to execute its critical functions in the cell [34]. While previous studies have concentrated on the stabilization and activation of p53 proteins following DNA damage, recent examinations have sought to determine the potential relationship between p53 and miRNAs. By searching conserved miRNA-binding sites in the 3′-UTR of the p53 gene, miR-125b was identified as a potential highly conserved p53-targeting miRNA. Functional studies in human neuroblastoma cells and lung fibroblasts revealed that overexpression of miR-125b reduces endogenous p53 level and suppresses apoptosis [35]. Loss of miR-125b increased p53-dependent apoptosis in zebrafish, leading to severe defects in embryonic development. Using a similar approach, Hu et al. [36] identified miR-504 as another p53-targeting miRNA in human colon HCT116 p53 +/+, lung H460, and breast MCF7 cells. These investigators reported that MiR-504 inhibited p53-mediated activities in human cells expressing wild-type p53 and significantly increased the growth of human colorectal xenografts tumors in nude mice [36]. Sachdeva et al. [37] found an inverse correlation between miR-145 and c-Myc. Interestingly, miR-145 is also transcriptionally activated by p53, suggesting p53 represses c-Myc through induction of the tumor suppressor miR-145 [37]. As a putative tumor suppressor, miR-145 levels were found to be underexpressed in human breast and colon cancer [37].
Mdm2 is an E3 ubiquitin ligase and a known negative regulator of p53. Following DNA damage, the p53/Mdm2 interaction is disassociated, which leads to the rapid activation of p53 [38, 39]. Xiao et al. [40] recently identified miR-605 as a key cofactor in the p53 regulatory network. Specifically, the p53 protein was found to bind to the promoter region of the miR-605 gene and transcriptionally activate miR-605. Transactivated miR-605 directly decreases the expression of Mdm2 and thus indirectly enhances the transcriptional activity of p53. These results reveal a novel p53/miR-605/Mdm2 positive feedback loop that ensures rapid accumulation of p53 after DNA damage [40].
Regulators of the DNA damage response: Wip1-miR-16/miR-29, PTEN-miR-22
Protein phosphorylation is a major event in the DNA damage signaling transduction pathway. The ATM/ATR-initiated kinase cascade activates cell-cycle checkpoints and DNA repair pathways [1]. Once DNA damage is repaired, the cell needs to shut off the DNA damage response pathways and resume its normal activity. Wip1 is a serine/threonine phosphatase that serves as a master inhibitor in the ATM-p53 DNA damage signaling pathway [41]. Recent studies identified a number of key proteins in the DDR as Wip1 dephosphorylation substrates, including p38 MAPK, CHK1, CHK2, p53, MDM2, and ATM [29, 41–45]. Wip1 has been shown to be an oncogene and is amplified and overexpressed in several human tumor types [46–50]. In vivo studies showed that mice lacking Wip1 are resistant to spontaneous and oncogene-induced tumors due to enhanced DNA damage and p53 responses [44, 51–54]. Wip1 inhibitors have been shown to significantly reduce tumor cell proliferation, suggesting that therapeutic agents that inhibit Wip1 may have potent anticancer activity. Although the Wip1 gene is initially transactivated by p53 at the early stage of the DDR, the increase in Wip1 protein level is significantly delayed when compared to that of its mRNA level, thereby preventing premature inactivation of ATM/ATR signaling and ensuring the completion of the early DDR. We [55] and others [56] have shown that MiR-16 and miR-29 are the two principal miRNAs that target Wip1. Rapid induction of these two miRNAs after DNA damage may facilitate the proper DNA damage signaling through inhibition of Wip1 induction. Overexpression of miR-16 markedly inhibits Wip1 expression and sensitizes MCF-7 human breast cancer cells to the chemotherapeutic drug doxorubicin. Both miR-16 and miR-29 also have important physiological roles in vivo. Overexpression of miR-16 appears to suppress self-renewal and growth of mouse mammary tumor stem cells, and miR-29 is significantly upregulated during aging in the normal mouse [55, 56].
Phosphatase and tensin homolog (PTEN) is one of the most important tumor suppressor genes that controls cell growth by inhibiting the PI3 K/AKT pathway [57]. It is frequently mutated, deleted, or silenced in many types of human cancers [58, 59]. Recent findings suggest that PTEN also contributes to the DNA damage response and DNA repair pathways. PTEN ablation in mouse embryonic fibroblasts results in genomic instability, and PTEN-deficient cells have defective double-stranded DNA repair due to lack of or downregulation of Rad51 and lack of PTEN at centromeres [59]. Ming et al. [60, 61] showed that PTEN is essential for the activity of nucleotide excision repair in UV radiation cells. A recent study demonstrated that UV-induced miR-22 suppressed PTEN expression in HEK293T and HaCaT cells, which may promote cell survival after UV radiation [62].
Regulation of miRNA biogenesis in the DDR
An expanding body of evidence suggests that there are bidirectional communication signals between miRNAs and the DDR. For example, MiRNA-mediated gene silencing has been shown to modulate the activity of the DDR, and the DDR is a known regulator of miRNA expression at the transcriptional and post-transcriptional levels [10]. In addition, several studies have shown that genotoxic agents, such as UV light, hydrogen peroxide, IR, and radiomimetic drugs all have a dramatic effect on miRNA expression profiles in a variety of cell types [24]. These studies utilized miRNA microarrays and quantitative real-time PCR to document global changes in miRNA expression levels after the induction of DNA damage [63–71]. IR induces a substantial change in miRNA expression in different cell lines, including human lymphoblastic cells, non-small cell lung cancer cells, and prostate cancer cells [63, 64, 66, 68–71]. Other DNA damaging agents, such as UV light, hydrogen peroxide, and etoposide also resulted in the alteration of unique, as well as shared sets of miRNAs within the same cell type, suggesting that miRNAs respond differently to DNA damage depending on the type and/or intensity of DNA damage [64, 65, 69, 71]. While there are clearly some variations among DNA damage–responsive miRNAs, they are all predicted to target genes in the DDR pathway [63, 64, 66, 68–71]. Whether there exists a common core miRNA signature that is characteristic of DNA damage in all cells requires additional investigation.
DNA damage–responsive miRNAs
Persengiev and colleagues were the first to show that expression levels of a large subset of miRNAs are rapidly modulated after UV damage [67]. An examination of the kinetics of miRNA expression following UV damage suggests that measurable alterations in expression levels occur within a few hours after the injury, and that miRNA expression returns to basal levels after a 24-h period. These data suggest that the onset of miRNA responses following DNA damage precedes the transcriptional events modulated by p53, but is slower than the events involving post-translational protein modifications (phosphorylation and ubiquitination) on downstream effectors of p53. That is, the miRNAs appear to act in the time between the transcriptional and post-transcriptional responses to modulate the DDR [24]. Recently, our laboratory examined DNA damage–responsive miRNAs using the radiomimetic drug neocarzinostatin (NCS) to generate double-strand DNA breaks in Atm +/+ and Atm −/− mouse embryonic fibroblasts (MEFs). Compared to untreated MEFs, we observed that the expression of 71 miRNAs, approximately one-quarter of those examined, was increased more than twofold in an ATM-dependent manner upon DNA damage. Interestingly, the induction of these miRNAs resulted from increased levels of mature miRNAs, but not their primary transcripts (pri-miRNAs) suggesting global post-transcriptional regulation of miRNAs upon DNA damage [71]. Accumulating evidence suggests that there are critical connections between the DDR and miRNA biogenesis and that miRNA expression is regulated in response to DNA damage at both the transcriptional and post-transcriptional levels. In the next section, we discuss miRNA biogenesis and its regulation in the DDR.
Transcriptional regulation of miRNAs after DNA damage
Many miRNA promoters have characteristics that are similar to those of normal protein-coding genes, suggesting that miRNA transcription can be controlled by common transcription factors. In response to DNA damage, p53 family members transactivate multiple target genes that instruct cells for growth arrest or programmed death, serving as a good example of how DNA damage can modulate miRNA transcription (Fig. 2) [72]. Transcription factors, such as p53, can regulate miRNA expression by directly binding to miRNA promoters and modulating their transcription or by manipulating the expression of components of the miRNA processing machinery. Su et al. [73] reported that TAp63, a member of the p53 family, directly binds to the Dicer promoter and activates its transcription as well as that of miR-130b. Furthermore, TAp63 knockout mice and human cells deficient in TAp63 express Dicer at very low levels. TAp63 may also play an important role in suppressing tumor progression and it has been suggested that TAp63 may suppress metastasis formation by regulating the expression of Dicer and miRNAs (Fig. 2) [73]. While these preliminary studies support the notion that DNA damage leads to transcriptional regulation of miRNAs, a more comprehensive understanding of miRNA transcriptional regulation in the DDR requires additional investigation.
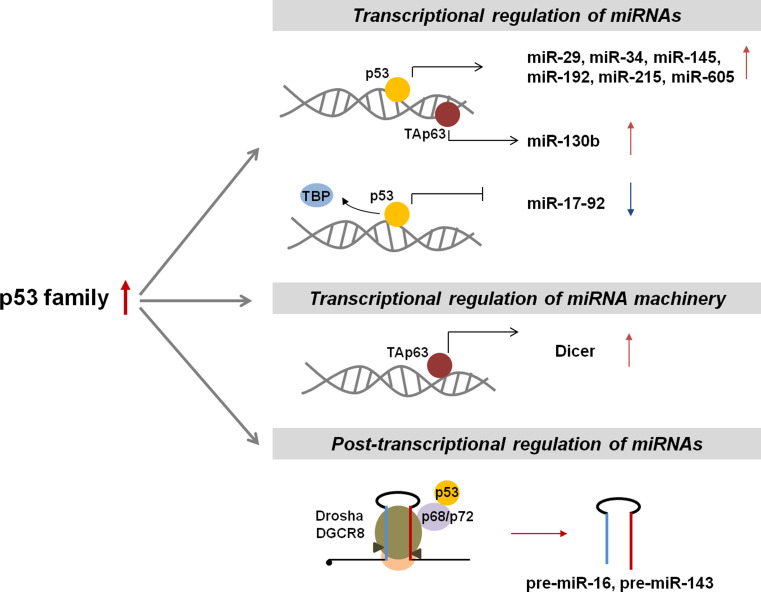
Fig. 2.
Regulation of miRNA expression by the p53 family. P53 tumor-suppressor family members are activated after DNA damage and participate in the regulation of miRNA expression at both the transcriptional and post-transcriptional levels. As a transcription factor, p53 increases or decreases the transcription of several miRNAs. In addition, p53 is functionally linked to Drosha/DGCR8 through direct interaction with p68/p72, which enhances processing of pri-miRNAs to pre-miRNAs. Another p53 family member, TAp63, induces transcription of Dicer, a critical component of the miRNA biogenesis machinery, and miR-130b
P53, a positive and negative transcriptional regulator of miRNAs
The level and activity of p53 are induced by a number of diverse stress signals. In response to DNA damage, the ATM kinase activates p53, which in turn transactivates genes in multiple pathways, including cell-cycle regulation, tumor suppression, and apoptosis [74, 75]. The miR-34 family was first reported to be a direct p53 transcriptional target [76, 77]. DNA damage induces the level of miR-34 transcripts by increasing the activity and stability of p53. Similar to p53-induced proteins, miR-34 functions as a downstream effector to amplify the p53 signal, which heightens the cells sensitivity to external signals. Increased levels of miR-34a mediate G1 cell-cycle arrest by downregulating multiple cell cycle-related transcripts [76]. Ectopic miR-34 expression induces apoptosis, cell-cycle arrest (G1-arrest), or senescence by interfering with the mRNA transcripts of several genes, including Bcl-2, Cyclin D1, c-MYC, and SIRT1 [78–82]. Interestingly, miR-34a-targeted SIRT1 may form a positive feedback loop by increasing the acetylation of p53, which induces the expression of p21 and PUMA, both of which are transcriptional targets of p53 that regulate the cell cycle and apoptosis, respectively [83]. Thus, miR-34 fine-tunes the p53 signaling pathway through its regulation of a number of p53 targets (Fig. 3) [81].
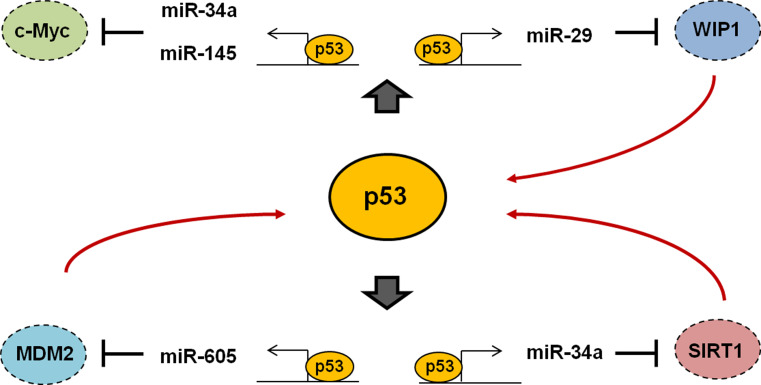
Fig. 3.
Positive feedback loop of p53 through transactivation of miRNAs. P53 transcriptionally activates miR-29, miR-34a, miR-145, and miR-605. Each of the induced miRNAs represses their targets such as Wip1, c-Myc, SIRT1, and MDM2 mRNAs, which in turn regulates p53 expression and activity. MiR-605 inhibition of Mdm2 and miR-29 inhibition of Wip1 lead to an activation of p53, and miR-34 inhibition of SIRT1 results in increased p53 acetylation and activation. P53 induces miR-34 and miR-145 transcription, and they directly target the c-Myc mRNA, suggesting that p53 represses c-Myc functions through regulation of miRNA expression
P53 also positively regulates the expression of miR-145, miR-192, and miR-215. These miRNAs induce cell-cycle arrest by inhibiting the transcripts of several genes that regulate the G1 and G2 checkpoints [84, 85]. In p53+/+ HCT116 colorectal carcinoma cells, but not p53−/− HCT116 cells, miR-192 was found to increase the level of p21, indicating that this p53-transactivated miRNA may in turn regulate the activity of p53 [85]. P53 induces miR-145 transcription by directly binding to its promoter, and miR-145 directly targets the oncogene c-Myc, suggesting that p53 may repress c-Myc through induction of miR-145 [37, 86] (Fig. 3). Ugalde et al. [56] recently reported that expression of the miR-29 family is increased in response to DNA damage in a p53-dependent manner. MiR-29 targets and represses Wip1 phosphatase, a master inhibitor in the DDR that inhibits the activation and stabilization of p53, leading to p53 induction [56].
P53 can also function as a transcriptional suppressor to silence miRNA expression by binding to miRNA promoters and preventing the recruitment of transcriptional activators. For example, p53 suppress the transcription of the miR-17–92 cluster gene by preventing recruitment of the TATA-binding protein to the TAATA site in the promoter (Fig. 2). It appears that the miR-17–92 cluster is repressed under hypoxic conditions through a p53-dependent and c-Myc-independent mechanism and that this leads to the sensitization of cells to hypoxia-induced apoptosis [87].
Post-transcriptional regulation of miRNAs after DNA damage
Current studies suggest that post-transcriptional controls for miRNA expression are mediated by two types of proteins: proteins associated with the Drosha or Dicer microprocessor complex and proteins that bind to specific miRNAs [88]. Here, we discuss two recent findings on how DNA damage signals are transmitted to the miRNA pathway. First, DNA damage-induced p53 is directly associated with the Drosha microprocessor machinery through interactions with p68/p72. Binding of p53 to this complex promotes the processing of some pri-miRNAs to pre-miRNAs. Second, ATM mediates the phosphorylation of the KH-type splicing regulatory protein (KSRP), which binds to the terminal loop of a subset of pri- and pre-miRNAs and enhances processing by the Drosha and Dicer complex (Fig. 4).
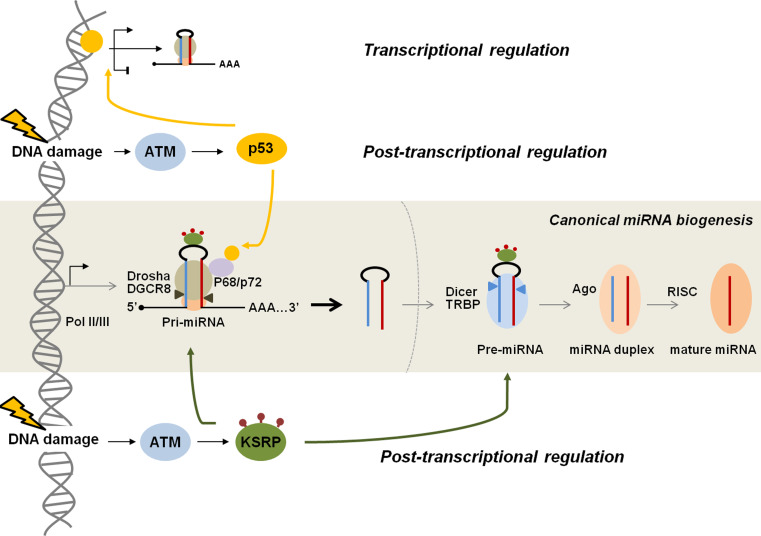
Fig. 4.
Regulation of miRNA biogenesis by the DNA damage response. Once the cell recognizes DNA damage, ATM, a major transducer of the DNA damage response, transmits DNA damage signals to the miRNA biosynthesis pathway through DDR-specific proteins, such as p53 and the RNA-binding protein KSRP. Primary regulation of miRNAs in the DDR may be post-transcriptionally regulated in the nucleus. ATM-mediated phosphorylation of KSRP promotes its interaction with the terminal loop of a subset of miRNA precursors and enhances the processing of target pri-miRNAs to pre-miRNAs by the Drosha/DGCR8 microprocessor complex
The p53-p68/p72-Drosha/DGCR8 complex enhances pri-miRNA processing
In addition to transcriptional regulation, the DDR is also involved in the post-transcriptional regulation of miRNAs. As discussed above, p53 has been shown to activate the transcription of several miRNA genes (miR-34, miR-192, and miR-215) [76, 84, 85]. Subsequent studies have demonstrated that p53 also promotes pri-miRNA processing. Suzuki and colleagues [89] observed that miR-16 and miR-143 were induced in irradiated human HCT116 cells in a p53-depentent and p68/p72-dependent manner. The DNA damage increased the levels of pre-miR-16, pre-miR-143, and other precursor miRNAs, but not their corresponding pri-miRNAs [89]. DNA damage-induced miR-16 and miR-143 reduced cell proliferation and suppressed important regulators of the cell cycle and cell proliferation, such as K-Ras and CDK6 [90]. Consistent with a previous report on the interaction between p53 and p68/p72 [91], it was shown that p53 interacts with the Drosha/DGCR8 complex through its association with p68 [89]. The DEAD-box RNA helicases p68 (DDX5) and p72 (DDX17) were initially identified as components of the Drosha microprocessor complex by mass spectrometry [92]. In p72- or p68-knockout MEFs, the levels of a subset of pre-miRNAs were reduced, whereas levels of their corresponding pri-miRNAs remained unchanged [93], suggesting that p68/p72 are required for the maturation of miRNAs in Drosha-mediated processing [92, 93]. Thus, a direct interaction between p53 and p68/p72 facilitates the processing of pri-miRNAs to pre-miRNAs in response to DNA damage. Indeed, inactive p53 mutants interfere with the functional interaction between p68 and the Drosha complex, resulting in the attenuation of miRNAs processing [89].
ATM-dependent phosphorylation of KSRP enhances pri-miRNA processing
Results from recent reports indicate that ATM, a key signaling component in the DDR, is also directly involved in miRNA processing. We [71] noted that following the induction of DNA double-strand breaks, approximately one-quarter of the miRNAs that we examined were significantly induced in an ATM-dependent manner whereas a small group of miRNAs (19 miRNAs) were significantly reduced [71]. These results suggest that ATM may play a central role in regulating DNA damage-mediated miRNA expression. In addition, we noted that previously reported KSRP-dependent miRNAs were also induced in an ATM-dependent manner upon DNA damage [71]. KSRP is a multifunctional single-strand RNA-binding protein that regulates several aspects of RNA metabolism, including RNA splicing, localization, and degradation [94]. Trabucchi et al. [95] reported that KSRP interacts with 5′ guanosine-rich regions, including GGG triplets in the terminal loop (TL) of a class of pre-miRNA precursors, and serves as a component of both the Drosha and Dicer complexes. Thus, KSRP positively regulates the maturation of a cohort of miRNA precursors by promoting the maturation of their target pri-miRNAs, including pri-miR-1, pri-miR-15, pri-miR-21, and pri-let-7 [95].
A genome-wide screen identified KSRP as a potential phosphorylation target of the ATM and ATR kinases [28], and we [71] were able to confirm this in human U2OS and GM0637 cells. Phosphorylation of KSRP significantly enhanced the recruitment of target pri-miRNAs to the Drosha complex and increased processing of pri-miRNAs, whereas mutating the ATM phosphorylation sites on KSRP impaired its miRNA-regulating activity [71]. These findings suggest a critical link between the DDR and miRNA-processing pathway in which ATM stimulates the activity of modulators of miRNA biogenesis. In our study, the levels of some miRNAs were reduced after DNA damage in an ATM-dependent manner, indicating that ATM may also be involved in inhibitory pathways that downregulates miRNA expression [11]. However, because the KSRP-dependent miRNAs only represent a portion of the DNA damage-induced miRNAs, there could be alternative KSRP-independent mechanisms that mediate DNA damage-induction of miRNAs. Indeed, other RNA-binding proteins, including hnRNP A1 and Lin-28, have been shown to bind to the terminal loop of a subset of precursor miRNAs and facilitate or attenuate their processing [96, 97]. Thus, it is likely that these RNA-binding proteins can be regulated in the DNA damage signaling pathways. In addition, ATM-independent regulation may also connect DNA damage signaling to the miRNA pathway.
Conclusions and future perspectives
When considered collectively, the available evidence suggests that DNA damage signaling participates in miRNA biogenesis by regulating both transcriptional and post-transcriptional machineries. However, these studies require confirmation and several other possibilities need to be considered. For example, it is likely that transcription factors other than p53 may be involved in the regulation of miRNA gene expression in response to DNA damage. It is also conceivable that other components in the miRNA processing machinery may be direct or indirect targets of DNA damage signaling. In addition, the export of pre-miRNAs from the nucleus to the cytoplasm may also be altered after DNA damage. Whereas the majority of studies examining miRNA regulation in response to DNA damage have focused on events that take place in the nucleus, it will be important to expand investigations to determine the contribution of cytoplasmic regulation of miRNA maturation following DNA damage. For example, it will be interesting to determine whether DNA damage signals can modulate the modification, turnover, stabilization, and degradation of miRNAs. The stability of Ago2 and the Dicer-TRBP complex have been shown to be regulated through phosphorylation by the p38 mitogen-activated protein kinase (MAPK) pathway and MAPK/Erk signaling, respectively [98, 99]. Phosphorylated TRBP stabilizes the Dicer-TRBP complex and increases mature miRNA production [99]. Interestingly, Erk and other MAPKs are phosphorylated and activated after DNA damage [100], suggesting a potential connection between DDR and the activity of the Dicer complex. In addition to canonical miRNA biogenesis, various alternative mechanisms, such as Drosha-independent and Dicer-independent miRNA biogenesis, have emerged over the past few years [101], expanding the number of regulatory pathways that may be targeted by the DDR.
Defects in the DDR and global repression of miRNAs are now considered hallmarks of many types of human cancer [102–105]. As we described earlier, several core proteins in the DDR pathway are regulated by miRNAs. DDR-regulated miRNAs and miRNAs that target the DDR are involved in the initiation and progression of tumorigenesis and also modulate the sensitivity of cells to DNA damaging agents [24]. Thus, further identification and characterization of new miRNA targets in the DDR are required and an improved understanding of the roles of these miRNAs may provide new insight into the sensitivity or resistance of cancer cells to genotoxic drugs and possibly lead to the development of novel therapeutic strategies. In conclusion, although many questions still remain, it is now clear that DNA damage signals communicate with miRNAs by regulating multiple steps of miRNA biogenesis. Ongoing and further efforts to answer remaining questions will eventually lead to the development of novel diagnostic and therapeutic strategies for many human diseases with DNA damage processing defects, including cancer.
Acknowledgments
X.L. is supported by a National Institutes of Health grant (CA136549) and a research grant from the American Cancer Society (119135-RSG-10-185-01-TBE).
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SP. The DNA-damage response: new molecular insights and new approaches to cancer therapy. Biochem Soc Trans. 2009;37(Pt 3):483–494. doi: 10.1042/BST0370483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiloh Y. ATM and ATR: networking cellular responses to DNA damage. Curr Opin Genet Dev. 2001;11(1):71–77. doi: 10.1016/S0959-437X(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 5.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huen MS, Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci. 2010;35(2):101–108. doi: 10.1016/j.tibs.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25(5):409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 10.Wan G, Mathur R, Hu X, Zhang X, Lu X. miRNA response to DNA damage. Trends Biochem Sci. 2011;36(9):478–484. doi: 10.1016/j.tibs.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Lu X. Posttranscriptional regulation of miRNAs in the DNA damage response. RNA Biol. 2011;8(6):960–963. doi: 10.4161/rna.8.6.17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 13.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26(3):775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci USA. 2007;104(45):17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 17.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17(1):5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321(5895):1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans . Nature. 2009;461(7263):546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 23.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28(5):739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Wouters MD, van Gent DC, Hoeijmakers JH, Pothof J. MicroRNAs, the DNA damage response and cancer. Mutat Res. 2011;717(1–2):54–66. doi: 10.1016/j.mrfmmm.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Hu H, Gatti RA. MicroRNAs: new players in the DNA damage response. J Mol Cell Biol. 2011;3(3):151–158. doi: 10.1093/jmcb/mjq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, Bentwich Z, Lieberman J, Chowdhury D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. 2009;16(5):492–498. doi: 10.1038/nsmb.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Huang JW, Li M, Cavenee WK, Mitchell PS, Zhou X, Tewari M, Furnari FB, Taniguchi T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol Cancer Res. 2011;9(8):1100–1111. doi: 10.1158/1541-7786.MCR-11-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19(10):1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc Natl Acad Sci USA. 2010;107(4):1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jhanwar-Uniyal M. BRCA1 in cancer, cell cycle and genomic stability. Frontiers Biosci J Virtual Library. 2003;8:s1107–s1117. doi: 10.2741/1131. [DOI] [PubMed] [Google Scholar]
- 32.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, Chowdhury D. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41(2):210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L, Manoukian S, Secreto G, Ravagnani F, Wang X, Radice P, Croce CM, Davuluri RV, Calin GA. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70(7):2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23(7):862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, Tang LH, Levine AJ, Feng Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell. 2010;38(5):689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Nat Acad Sci USA. 2009;106(9):3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 39.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 40.Xiao J, Lin H, Luo X, Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 2011;30(3):524–532. doi: 10.1038/emboj.2010.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu X, Nguyen TA, Donehower LA. Reversal of the ATM/ATR-mediated DNA damage response by the oncogenic phosphatase PPM1D. Cell Cycle. 2005;4(8):1060–1064. doi: 10.4161/cc.4.8.1876. [DOI] [PubMed] [Google Scholar]
- 42.Fujimoto H, Onishi N, Kato N, Takekawa M, Xu XZ, Kosugi A, Kondo T, Imamura M, Oishi I, Yoda A, Minami Y. Regulation of the antioncogenic Chk2 kinase by the oncogenic Wip1 phosphatase. Cell Death Differ. 2006;13(7):1170–1180. doi: 10.1038/sj.cdd.4401801. [DOI] [PubMed] [Google Scholar]
- 43.Lu X, Nguyen TA, Zhang X, Donehower LA. The Wip1 phosphatase and Mdm2: cracking the “Wip” on p53 stability. Cell Cycle. 2008;7(2):164–168. doi: 10.4161/cc.7.2.5299. [DOI] [PubMed] [Google Scholar]
- 44.Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, Minami Y, Appella E, Bulavin DV. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell. 2006;23(5):757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 45.Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J. 2000;19(23):6517–6526. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castellino RC, De Bortoli M, Lu X, Moon SH, Nguyen TA, Shepard MA, Rao PH, Donehower LA, Kim JY. Medulloblastomas overexpress the p53-inactivating oncogene WIP1/PPM1D. J Neurooncol. 2008;86(3):245–256. doi: 10.1007/s11060-007-9470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuku T, Semba S, Yutori H, Yokozaki H. Increased wild-type p53-induced phosphatase 1 (Wip1 or PPM1D) expression correlated with downregulation of checkpoint kinase 2 in human gastric carcinoma. Pathol Int. 2007;57(9):566–571. doi: 10.1111/j.1440-1827.2007.02140.x. [DOI] [PubMed] [Google Scholar]
- 48.Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, Yokoyama T, Nozawa S, Inazawa J, Imoto I. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res. 2003;9(6):1995–2004. [PubMed] [Google Scholar]
- 49.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, Gabriele T, McCurrach ME, Marks JR, Hoey T, Lowe SW, Powers S. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31(2):133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 50.Saito-Ohara F, Imoto I, Inoue J, Hosoi H, Nakagawara A, Sugimoto T, Inazawa J. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003;63(8):1876–1883. [PubMed] [Google Scholar]
- 51.Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, Fornace AJ., Jr Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet. 2004;36(4):343–350. doi: 10.1038/ng1317. [DOI] [PubMed] [Google Scholar]
- 52.Choi J, Nannenga B, Demidov ON, Bulavin DV, Cooney A, Brayton C, Zhang Y, Mbawuike IN, Bradley A, Appella E, Donehower LA. Mice deficient for the wild-type p53-induced phosphatase gene (Wip1) exhibit defects in reproductive organs, immune function, and cell cycle control. Mol Cell Biol. 2002;22(4):1094–1105. doi: 10.1128/MCB.22.4.1094-1105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrison M, Li J, Degenhardt Y, Hoey T, Powers S. Wip1-deficient mice are resistant to common cancer genes. Trends Mol Med. 2004;10(8):359–361. doi: 10.1016/j.molmed.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 54.Nannenga B, Lu X, Dumble M, Van Maanen M, Nguyen TA, Sutton R, Kumar TR, Donehower LA. Augmented cancer resistance and DNA damage response phenotypes in PPM1D null mice. Mol Carcinog. 2006;45(8):594–604. doi: 10.1002/mc.20195. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, Lu X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70(18):7176–7186. doi: 10.1158/0008-5472.CAN-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ugalde AP, Ramsay AJ, de la Rosa J, Varela I, Marino G, Cadinanos J, Lu J, Freije JM, Lopez-Otin C. Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J. 2011;30(11):2219–2232. doi: 10.1038/emboj.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3 K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6(3):184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 58.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100(4):387–390. doi: 10.1016/S0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 59.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128(1):157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 60.Ming M, Feng L, Shea CR, Soltani K, Zhao B, Han W, Smart RC, Trempus CS, He YY. PTEN positively regulates UVB-induced DNA damage repair. Cancer Res. 2011;71(15):5287–5295. doi: 10.1158/0008-5472.CAN-10-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ming M, Shea CR, Feng L, Soltani K, He YY. UVA induces lesions resembling seborrheic keratoses in mice with keratinocyte-specific PTEN downregulation. J Invest Dermatol. 2011;131(7):1583–1586. doi: 10.1038/jid.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan G, Shi Y, Wu ZH. MicroRNA-22 promotes cell survival upon UV radiation by repressing PTEN. Biochem Biophys Res Commun. 2012;417(1):546–551. doi: 10.1016/j.bbrc.2011.11.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cha HJ, Shin S, Yoo H, Lee EM, Bae S, Yang KH, Lee SJ, Park IC, Jin YW, An S. Identification of ionizing radiation-responsive microRNAs in the IM9 human B lymphoblastic cell line. Int J Oncol. 2009;34(6):1661–1668. doi: 10.3892/ijo_00000297. [DOI] [PubMed] [Google Scholar]
- 64.Faraonio R, Salerno P, Passaro F, Sedia C, Iaccio A, Bellelli R, Nappi TC, Comegna M, Romano S, Salvatore G, Santoro M, Cimino F. A set of miRNAs participates in the cellular senescence program in human diploid fibroblasts. Cell Death Differ. 2011;19(4):713–721. doi: 10.1038/cdd.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupe P, Robert T, Ripoche H, Lazar V, Harel-Bellan A, Dessen P, Barillot E, Kroemer G. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70(5):1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 66.Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68(15):1599–1606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pothof J, Verkaik NS, van Ijcken W, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van Gent DC, Hoeijmakers JH, Persengiev SP. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28(14):2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK, Jin HO, Park IC, Jin YW, An S. Alteration of miRNA profiles by ionizing radiation in A549 human non-small cell lung cancer cells. Int J Oncol. 2009;35(1):81–86. [PubMed] [Google Scholar]
- 69.Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Gius D, Mitchell JB. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS ONE. 2009;4(7):e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X, Wan G, Berger FG, He X, Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41(4):371–383. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin YL, Sengupta S, Gurdziel K, Bell GW, Jacks T, Flores ER. p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet. 2009;5(10):e1000680. doi: 10.1371/journal.pgen.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, Wistuba I, Flores ER. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467(7318):986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51(23 Pt 1):6304–6311. [PubMed] [Google Scholar]
- 75.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 76.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67(18):8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 78.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17(15):1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 79.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67(18):8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 80.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 82.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 83.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105(36):13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Orntoft TF, Andersen CL, Dobbelstein M. p53-Responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68(24):10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68(24):10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 86.Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, Tanaka Y, Dahiya R. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis. 2011;32(5):772–778. doi: 10.1093/carcin/bgr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, Sun SH. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 2009;28(18):2719–2732. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24(11):1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 90.Suzuki HI, Miyazono K. Emerging complexity of microRNA generation cascades. J Biochem. 2011;149(1):15–25. doi: 10.1093/jb/mvq113. [DOI] [PubMed] [Google Scholar]
- 91.Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, Gregory DJ, Lane DP, Perkins ND, Fuller-Pace FV. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 2005;24(3):543–553. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 93.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, Akimoto C, Yamamoto Y, Katagiri T, Foulds C, Takezawa S, Kitagawa H, Takeyama K, O’Malley BW, Kato S. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9(5):604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 94.Briata P, Chen CY, Giovarelli M, Pasero M, Trabucchi M, Ramos A, Gherzi R. KSRP, many functions for a single protein. Front Biosci. 2011;16:1787–1796. doi: 10.2741/3821. [DOI] [PubMed] [Google Scholar]
- 95.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459(7249):1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14(7):591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 97.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 98.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139(1):112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413(3):429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 100.Dent P, Yacoub A, Fisher PB, Hagan MP, Grant S. MAPK pathways in radiation responses. Oncogene. 2003;22(37):5885–5896. doi: 10.1038/sj.onc.1206701. [DOI] [PubMed] [Google Scholar]
- 101.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics. 2010;284(2):95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 102.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 103.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 104.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 105.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]