Abstract
The cancer cell attractors theory provides a next-generation understanding of carcinogenesis and natural explanation of punctuated clonal expansions of tumor progression. The impressive notion of atavism of cancer is now updated but more evidence is awaited. Besides, the mechanisms that the ectopic expression of some germline genes result in somatic tumors such as melanoma and brain tumors are emerging but are not well understood. Cancer could be triggered by cells undergoing abnormal cell attractor transitions, and may be reversible with “cyto-education”. From mammals to model organisms like Caenorhabditis elegans and Drosophila melanogaster, the versatile Mi-2β/nucleosome remodeling and histone deacetylation complexes along with their functionally related chromatin remodeling complexes (CRCs), i.e., the dREAM/Myb-MuvB complex and Polycomb group complex are likely master regulators of cell attractors. The trajectory that benign cells switch to cancerous could be the reverse of navigation of embryonic cells converging from a series of intermediate transcriptional states to a final adult state, which is supported by gene expression dynamics inspector assays and some cross-species genetic evidence. The involvement of CRCs in locking cancer attractors may help find the recipes of perturbing genes to achieve successful reprogramming such that the reprogrammed cancer cell function in the same way as the normal cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0808-1) contains supplementary material, which is available to authorized users.
Keywords: Caenorhabditis elegans, Drosophila melanogaster, Carcinogenesis, Cell attractors, Chromatin remodeling system, Cellular reprogramming
Introduction
Despite decades of research and billions of dollars invested, cancer, as an ambiguous neoplasm and a “system-in-failing”, still kills millions of people each year. It is of great importance to understand the complex gene regulatory orchestrated by chromatin modification enzymes and transcriptional factors that govern the cell lineage specification, commitment, and differentiation, whose deregulations can cause tumors. Cancer cells bear some immature or embryonic traits and dysregulated developmental genes can act as oncogenes. The Mi-2/nucleosome remodeling and histone deacetylation (NuRD) alongside the dREAM-Myb-MuvB (dREAM/MMB) complex and Polycomb group (PcG) demonstrate that they play essential roles in cancer and normal development in mammals and the model organisms such as the nematode Caenorhabditis elegans and fruit fly Drosophila melanogaster [1–5]. For example, the functions of Mi-2/NuRD in cell differentiation and proliferation link this complex with oncogenesis. Its core subunits are increasingly found in a wide variety of tumors.
The theory of cell attractors, initially proposed by Dr. Kauffman [6], considers cell types as attractors, integrating the original concept of epigenetics from Dr. Waddington [7] in a complex systems approach of a gene expression regulatory network. The cell attractors model can explain the convergence of embryonic and tumorigenic signaling pathways, suggesting how cancers could be considered topologically and functionally linked to an attractor [8]. The notion of cancer attractors might skyrocket as one of most popular theories of carcinogenesis [8–10], but its study with whole animals has just begun.
The cancer attractors hypothesis argues that cancer cells are trapped in abnormal attractors and the “cancer attractors” may provide a natural explanation for “the oncogenesis recapitulates ontogenesis” [8]. Nevertheless, cancer attractors have not yet answered why the stable gene expression profiles of cancer attractors often encode an immature or embryonic program, and the molecular term for such attractors is lacking, and it remains undetermined what its evolution is, why normal somatic cells can be entrapped in and how it couples with cell division [11, 12]. The strategy of cancer prevention and targeting derived from cancer attractors theory remains rather hypothetical and elusive until incorporating the versatile chromatin remodeling systems.
Each NuRD and its functionally related chromatin remodeling complexes (CRCs) may contain different core components, which form distinct multifunctional complexes with specific context-dependent regulators. Particularly, the NuRDs have unique ATP-dependent chromatin remodeling, histone deacetylase and demethylase activities, and higher-order chromatin organization. They can regulate the accessibility of transcription factors or repair DNA proteins. NuRDs may be involved in many, if not most, critical cell signaling pathways during carcinogenesis and development [2, 3, 13]. The NuRDs can inherently integrate deterministic and stochastic biological characteristics. Gene regulatory networks (GRNs) leashed by NuRDs and its functionally related CRCs may provide system-level explanations of development and carcinogenesis [2, 3, 14].
In addition, the cracks of the paradigm of oncogenic mutations and somatic evolution as the driving force of tumorigenesis emerge [15]. Even previous hypothesis emphasizing that six genetic mutations are needed for converting a normal somatic cell into a cancer cell remains active in some corners, however, six mutations is a rare event, at some contexts, it would be predicted that the versatile NuRDs and its functionally related CRCs could help create such mutations or quasi-mutations (i.e., mutation-like state) via some mechanisms such as RNAi mechanism that they get involved [3]. Otherwise, cancer would be viewed as an intrinsic state and a hidden default program, only unleashed by a series of mutations [15]. Consequently, NuRDs and its functionally related CRCs could play diverse roles in response to growth signals, tissue invasion and metastasis, limitless cell replication, and evasion of apoptosis [2, 3]. Finally, NuRDs and its functionally related CRCs can naturally couple non-genetic or environmental and genetic factors as well as developmental processes to evade the tumorigenesis. Nucleosome remodeling and histone deacetylations and its functionally related CRCs could ensure robustness and plasticity during development. Indeed, they can unexpectedly behave as tumor suppressors, but have many more roles than a classic simple tumor suppressor [16]. Thus, the NuRD and its functionally related CRCs linked cancer attractor hypothesis can better explain carcinogenesis.
In this perspectives, we will describe the role of the Mi-2/NuRD complex and its functionally related CRCs within this model alongside computation analysis and some recent genetics evidence.
Global view of NuRDs during the C. elegans embryo development, murine hematopoietic lineage commitment, and differentiation
Cellular differentiation is central to understanding multi-cellular living systems, and why such systems go awry in diseases, such as cancer. Cell fates are multi-dimensional attractor states of the underlying molecular networks.
Similar to initial studies of the molecular terms in cell attractors [17, 18] to obtain experimental evidence for archaic cancer attractors based on the fact that C. elegans postembryonic arrested in the early larva stage can reliably be triggered in vivo, we could assume its endpoint of a stable state of postembryonic development and larva differentiation by a variety of gene RNAi treatment. In general, the animals navigate relatively different state-space trajectories to the differentiated state along with control GFP RNAi and the chromatin factors targeted by let-418/Mi-2β RNAi and mep-1 RNAi, both triggering the same cell fate switch in arrested early larvae. Gene expression profile (GEP) data [19] was applied to monitor genome-wide mRNA steady-state levels at distinct levels of RNAi treatment. These GEPs represent a collective phenotype of hundreds of whole animals subjected to three different target gene RNAi environments. Besides, three replicates of experiments allow the variable further, i.e., more trajectories. The readouts are averaged values. These GEPs could serve as surrogate measures for configuring gene activation states, and hence, for State (S_t.) This allowed us to determine the two trajectories of quantitative variable RNAi and distinct target by special “chemical” agent, i.e., double-stranded RNAi in the two different blocked differentiation processes, State LET-418 (SL_t) and State MEP-1 (SM_t), triggered by let-418/Mi-2β RNAi and mep-1 RNAi as the experimental state space and GFP RNAi as the control, respectively. By utilizing two distinct molecular RNAi stimuli that are likely to target distinct sets of genes, it would be expected that their tracks initially diverge to different regions of the state space in that RNA interference has variable effects. If the final differentiated state is an attractor, it can be approached from different directions within the multi-dimensional state space as the attractor hypothesis predicts that after an initial divergence, the two tracks SL_t and SM_t will converge to their common endpoint. We define these endpoints as “archaic” cell attractors. The gene expression dynamics inspector (GEDI) assay of these GEPs indeed gives a similar pattern for SL_t and SM_t that was dramatically distinct from the control (Fig. 1b). Such convergence of tracks from different directions across different gene dimensions confirms existence of cancer attractor state. The GEDI could also compare the extent of change of individual genes with seeing differences of the individual genes associated to the Kohonen networks represented at the GEDI pixels. For example, both arrested let-418 and mep-1 L1 larvae show ectopic expression of the P granule component PGL-1 in their somatic cells as high as 54.2-fold and 62.7-fold of up-regulation in comparison with the wild-type, suggesting that a LET-418 and MEP-1 containing complex is required to repress pgl-1 in somatic cells. However, these quantitative reverse transcription-PCR data are averaged values of many whole animals [19].
Fig. 1.
Conservation of NuRD-controlled archaic cell attractors represented by characteristic global pattern (GEDI) from RNAi-treated or knockout animals. Top a representative GEDI-maps during the timeline for wild-type C. elegans embryogenesis. Middle left b representative GEDI-maps of four mouse HSC tissue samples. Middle right c representative GEDI-maps of six samples from RNAi C. elegans. Bottom right d representative GEDI-maps of MEF MTA1 wild-type (upper) or knock out (bottom) samples. Bottom left e representative GEDI-maps of rep samples from Drosophila l(3)mbt ts mutant or wild-type. Note: GEDI-maps are for three randomly chosen tissue samples, showing the transcriptome patterns. Each map represents a transcriptome, with pixels representing the same genes in each map and their color is their expression level in the wild-type or experimental sample. Design of each RNAi or knock out treated sample is detailed as following: C. elegans: (1) The timeline for wild-type embryogenesis only. Embryonic cells are developmentally plastic until the 2E stage. At the ~8E stage, cells become committed to a cell fate. Focus from two-cells, 2E (24 cells), 4E (50 cells), and 8E (100 cells, onset of differentiation). (2) GFP is the control, proxy of wild-type animal, MEP-1/KLF4, LET-418/Mi-2β as experimental, proxy of deficient NuRDs. They are arrested at early L1. Mouse: (1) Mouse hematopoietic stem cell (HSC) differentiation. Mi-2β: control or conditional knock out. (2) Mouse embryonic fibroblast (MEF). MTA1: wild-type or knock out. Drosophila: l(3)mbt ts mutant or wild-type
In addition, a total of 1,113 genes changed their expression levels in let-418(RNAi) L1, whereas 1,104 genes were deregulated in mep-1(RNAi) worms. Around 914 (82%) of the deregulated genes were common between let-418(RNAi) and mep-1(RNAi) animals. The majority of them (70%) were up-regulated [19]. Analyses using the statistical software MAGMA with R-scripts revealed a very strong correlation between their deregulation pattern of these genes. A standard correlation factor of R = 0.98 was calculated according to the linear regression (R = 1 means that the relation is linear), demonstrating that gene expression was deregulated very similarly in let-418(RNAi) and mep-1(RNAi) depleted L1 larvae [19]. This can read the difference and similarity by simply clicking GEDI pixels (not shown).
In order to understand the dynamic attractors trajectories, where the order and synchronicity of events is essential, we analyzed the measured temporal evolution of transcriptomes for C. elegans embryogenesis. Different embryos developing from 2-cell stage to onset of differentiation have the convergence of high-dimensional trajectories (across >20,000 genes) (Fig. 1a). This observation focuses on wild-type C. elegans whole animals from 2-cells, 2E (24 cells), 4E (50 cells), and 8E (100 cells, onset of differentiation). The 2-cells to 8E is the timeline for C. elegans embryogenesis. These embryonic blastomeres are developmentally plastic until the 2E stage and this characteristic is lost during gastrulation. At the ~8E stage, cells become committed to a cell fate. There are some variations among the samples, but the tendency from “stem-like” state to “differentiation” state is clear [28] (Fig. 1a). This latter might represent the return of noise-induced deviations of the transcriptome from the border of the basin of attraction back to the attractor state (Fig. 2). They are both dynamical hallmarks of attractor states. Further systematic investigation of NuRD/dREAM/MMB and PcG is ongoing (Zhang et al., unpublished).
Fig. 2.
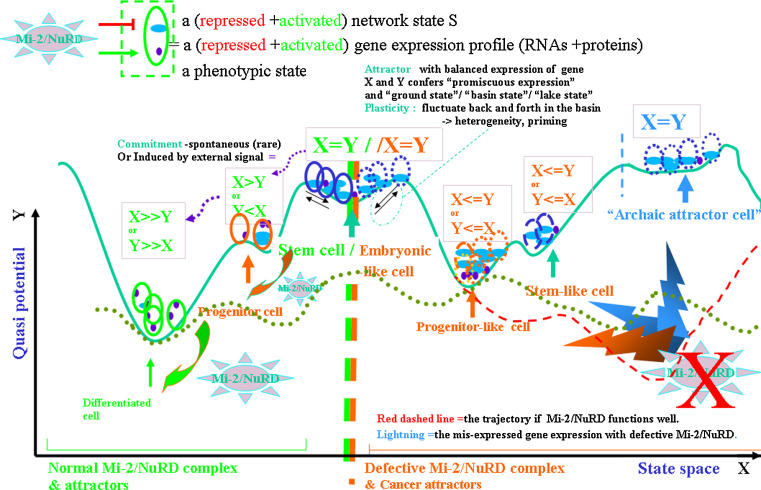
A diagram of the Mi-2/NuRD and multiple-tier cancer attractors and “mountain and valley” metaphor. A hypothetical epigenetic landscape to illustrate the concepts behind the hierarchy of cell type diversification during worm embryo and early larvae development that is regulated by NuRDs, alongside with the notions of “attractor” and basin of attraction by “mountain and valley” analogy. In a two-dimensional representation, the X-axis is a “state space”; the Y-axis (quasi-potential, inverse of probability) represents the relative instability of individual states at each state-space location. The “X” in red means the “loss of function” defect in NuRDs. Basins at top lake and bottom valley correspond to stable equilibrium (equilibrium attractors). Normally, minimal internal fluctuation or external perturbations will move the oval marbles (i.e., cells) away from unstable points (i.e., points at brisk of basins), while the oval marble will generally return to its attractors within its basin of attraction, even under the action of more important perturbations. However, major “potential” energy transfers (e.g., the “lightning”, hereby metaphorically caused by “loss of function” of the major controller of germline stem cell–soma distinction, i.e., NuRD) will be able to induce an oval marbles jumping out of its basin of attraction (i.e., differentiated cell “valley”) and being pushed up and then trapped by another attractor (e.g., the stem-cell “lake” or progenitor cell “basin”). In general, the meta-stable state (e.g., progenitor cell) is a weak attractor from which the oval marble may relatively easily reach a neighboring stable attractor. Note : “=” in figure is “~”, an approximation. This analogy is simplified. Multiple levels of “valleys” indicate the different levels of toti-/pluri-/multi-/mono-/potency, which are likely regulated by NuRD/CRCs. In other exceptional contexts, NuRD might inhibit the potential of differentiation
In mammalian hematopoiesis, the much-studied TF pair comprising GATA1 and PU.1 controls the branch point in the fate map where the erythroid and myeloid lineages separate [18]. However, GATA1 is a proven interactor with NuRD [2, 3], but it remains tempting whether and how NuRDs indeed get involved.
Specialized chromatin environments, which keep stem cell-specific genes active and key differentiation factors repressed but poised for activation, enable somatic stem cells to self-renew and differentiate into downstream lineages. NuRD is responsible for stabilizing the chromatin maintenance factors to keep the HSC pool and lineage differentiation during hematopoiesis [20]. Conditional deletion of Mi-2β expression in bone marrow caused an accumulation of erythroid cells and remained in early stages of development. Significantly, the proerythroblasts in Mi-2β/NuRD-deficient bone marrow and peripheral organs exhibited a neoplastic phenotype. An early increase in HSC numbers was followed by depletion of the pool due to a decrease of quiescence, increased cycling, and apoptosis in the Mi-2β/NuRD-deficient HSC-enriched bone marrow population. These cycling mutant cells readily differentiated into the erythroid lineage but not into the myeloid and lymphoid lineages. Gene expression profiles demonstrated that NuRD permits the repression or expression of different subsets of genes in HSCs [20, 21]. Thus, Mi-2β provides the hematopoietic system with immune cell capability as well as with an extensive regenerative capacity [20].
A similar analysis on Mi-2β in mouse hematopoietic stem cell (HSC) was performed (Fig. 1c). The Mi-2β/NuRD is required for maintenance and multi-lineage differentiation in the early hematopoietic hierarchy. Examination of GEP in the mutant HSC reveals changes in the expression of genes associated with self-renewal and lineage priming, and a pivotal role of Mi-2β in their regulation [20]. Gene expression dynamics inspector shows that the holistic view of the change in the collective genes of Mi-2β knockout mutants in mice is similar to the above-mentioned SL_t and SM_t in C. elegans, and reaches the endpoint attractor states (Fig. 1b). Furthermore, our GEDI analysis performed on mouse embryonic fibroblast (MEF) cell lines with/without MTA1 (wild-type vs. knockout), another core component of NuRD, GEPs showed a similar trend of dramatic change in pattern (Fig. 1d) [22]. So did the GEPs of mouse LSD1 and p66, the other two core subunits of NuRD [23] and Rb [24], a PcG-like protein BMI-1 [25, 26] PcG-like MES-2/E(z) [27], HDA-1/HDAC1 [28] (unpublished). Basically, they (i.e., the collective whole animals’ GEDIs) could possibly represent the common attractors with some extent of variations among similar patterns, taking into account that they were computed from GEPs of cross-species as well as different individuals of whole organisms.
Ectopic expression of germline genes and malignant tumor growth
In C. elegans, the GEPs in MEP-1/LET-418/Mi-2β/NuRD-depleted animals show a change in the expression levels of thousands of common genes [19], one-fourth of which are germline-specific and 49 early embryonic genes. These kinds of up-regulated genes may be ectopically expressed in the soma, particularly encoding the C. elegans germline marker P-granule components. LET-418/Mi-2β and MEP-1 are specifically required during early embryogenesis to down-regulate the early genes with mitotic and meiotic functions. Nucleosome remodeling and histone deacetylation activity is required for the cessation of embryonic cell proliferation and initiation of larval morphogenesis [19].
A number of human tumors, including melanoma and several carcinomas, are known to up-regulate germline-specific genes, such genes may play an important role in some tumor growth. Indeed, ectopic expression of germline genes drives malignant brain tumor growth in Drosophila [4]. Flies with a temperature-sensitive mutation in the lethal (3) malignant brain tumor (l(3)mbt) gene develop large larval brain tumor. This transcriptional repressor acts within dREAM (i.e., the NuRD, MyB, MuvB, retinoblastoma (RB) and HP1-related complex) to remodel the nucleosomes and silence the genes [4]. The human homologues of somatic mutations in L(3)MBT, Rb, and CHD3/Mi-2α exist in some of tumors [29]. It is possible to have more common mutations in the human homologues of L(3)MBT, Rb and its chromatin co-factors in cancer genomes. Interestingly, the C. elegans 32 chromatin factors identified in Rb pathway mutant screen can reverse the developmental defects potentially induced by somatic mis-expression of germline genes. Many of these suppressor genes were in RNA interference and/or small RNA pathways. The somatic cell specification defects in l(3)mbt-type tumors thus may be small and RNA-based [29] (Fig. 1e). In mammals, non-typical RNAs have become topical in tumor biology. It has recently been shown that antisense-to-sense transcript ratios were surprisingly altered in breast cancer [30]. In prostate cancer and colon cancer, the coding and non-coding RNAs can act as trans-regulators by miRNA binding; there might be different miRNA binding sites in pseudogenes of many cancer-related genes. This regulation could be abrogated in Dicer-null carcinoma cells, which are defective in miRNA processing [31]. In fact, one-fourth of the candidate genes that may define tumor cell identity due to l(3)mbt [4], similar to knock-downs of C. elegans LET-418/Mi-2β and MEP-1, encode proteins that are normally expressed specifically in germline cells, including some PIWI machinery. Inactivation of some (but not all) up-regulated genes individually can inhibit the tumor growth, so this suggests a new direction of therapeutic target for such tumors.
Let’s look back a little bit. The volvocine family has been diverging from animals for more than 600 million years. Through further archeological analysis of the mep-1 of C. elegans and Volvox genomes, we consider that mep-1 and regA were probably generated from a progenitor gene in the ancestor of animals and Volvox, which had glutamine (Q)-rich regions ([32, 33]; Zhang, this study). The proteins from these two genes in current C. elegans and Volvox genomes share around 30% identity, spanning over about 100 aa overlap [32, 33]. In humans, our analysis reveals that their functional counterpart could be the stemness factor kruppel-like factor KLF4 [28]. Furthermore, RegA has a SAND domain, which is shared with the CHD1-Like protein in Micromonas sp. RCC299 thus belongs to the same chromo domain (CHD) family as LET-418/Mi-2β/CHD4. It is unclear whether RegA alone in Volvox could perform similar functions to MEP-1-LET-418/Mi-2β complex in C. elegans, but it may be possible to gain some new insights from previous analysis of the somatic regenerator or regA gene. We are currently undergoing further assays of functional domain to obtain new insights. However, it is known that this regA gene is required for maintenance of the somatic cell fate in Volvox; regA mutant somatic cells develop normally at first, but instead of remaining somatic cells their entire lives and then eventually dying, as somatic cells usually do, they enlarge and regenerate as gonidia that eventually divide to produce new spheroids [33]. Therefore, regA somehow prevents somatic cells from growing and dividing, and keeps them from having the stem cell-like potential that gonidia possess. Hence, in different biological systems, they (i.e., functional equivalents) execute germline-specific gene repression. Furthermore, think of regA as a tumor suppressor gene that prevents the sort of uncontrolled growth that cancer cells exhibit [34]. However, Mi-2β/NuRD may evolve and get involved in maintenance of chromatin structure, proper cell replication, cell cycle progression, and many more diversifications. Cancer, superficially the price that we need pay for the evolution from unicellular to multicellular life forms ([15] and Zhang Y, in preparation), at least the germline-specific genes ectopic expression malignant tumor, is leashed by NuRD in advanced organisms. The “normal” GRN, which includes RNAs and proteins, is the current version but with a natural selection pressure and evolution during millions even billions of years. Indeed some RNAs could have more “archaic” characters and probably longer evolutionary times in comparison to the NuRD alongside the evolution of GRN, hence NuRD is unnecessarily secondary to GRN since they could have intertwined with at some stage or NuRD even leashed the GRN as upstream level for prevention of carcinogenesis. Furthermore, its ancestor or functional equivalent might exist before the evolution from unicellular to multi-cellular life forms, assuming that cancer is indeed the above-mentioned tradeoff of multicellular evolution from a unicellular state [15].
LIN-53 (RbAp46/48) and direct cell type conversion
The induced pluripotent stem cell (iPS) has a very similar cellular state transition to that of cancer cells [3, 35, 36]. Understanding this reversibility would allow for precise and directed manipulation, potentially enabling the creation of any cell type from any other. Some examples of reprogramming cultured mammalian cells from one cell type to another have been reported. Researchers are far from having complete control over the process. In C. elegans, through the ectopic expression of the transcription factor che-1, the identity of mitotic germ cells can be directly converted into that of specific neuron types along with the removal of the LIN-53 (RbAp46/48 in humans), a component of NuRD and dREAM. This removal can be mimicked by valproic acid, one inhibitor of histone deacetylases [5]. This direct conversion from germ cells into individual terminally differentiated neuron types demonstrates that the chromatin factor LIN-53 provides a barrier for cellular reprogramming. In dissipative systems far from the equilibrium, even weak forces might break symmetry in stable systems, leading to bifurcation and hysteresis [9, 12]. Tiny perturbations do not always remain tiny [37]. Therefore, further investigations of the functions of these chromatin factors and their coordination could potentially provide fruitful information with which to probe the mechanistic basis for cell-fate conversion and direct reprogramming between any cell types [38–40]. The NuRD and its evolutionally distinct layers of partners like c-myc could contribute much to suppress the “archaic” embryonic attractors, which could be essentially protected by Rb and PcG complexes and antagonized by NuRD at specific stages. The classic iPS reprogramming with defined factors is low efficiency. It would be predicted that a combination of chromatin factor and transcription factor could enhance the reprogramming efficiency, especially after such chromatin factors as master regulators of cancer attractors could be well elucidated in the future. For the time being, we largely rely on a stochastic process at play for the reprogramming. The directly induced lineage conversions, reprogramming of cells that are developmentally closely related, require fewer transcription factors. However, it is possible that NuRD-related transcription factors would have a significant role; for example, two NODE factors, such as Oct-4 and Nanog, could need to be down-regulated in some cases [2, 3, 42]. Thus, some reprogramming could effectively combat cancers.
The NuRDs and the drug-resistance molecules
Previously, by using Ingenuity Program Analysis (IPA), we analyzed the interactors [2, 3] and downstream target genes (not shown) of NuRDs. The results confirm that NuRDs have functions in diverse and distinct pathways. We then compared the NuRDs of mouse hematopoietic stem cells HSC and C. elegans GEPs. Surprisingly, we found the most conserved hub genes shared by both included the drug-resistant molecules, the apoptosis-inducing factor, and metabolism genes. However, it is known that hsf-1 regulates multi-drug-resistant genes in mammalian cell lines [41]. Further investigations are likely to shed light on how the HSF1/NuRD complex relates to the pathological role of MDR1 in cancer. Previously, it was hypothesized that histone deacetylase inhibitors can be used to reverse multiple drug resistance [42].
Genetics of NuRDs, invasion and metastasis
In mammals, the metastasis-associated (MTA) proteins of NuRD components MTA1-3 are critical for epithelial-mesenchymal transition (EMT) [3, 43] and play essential roles in metastasis. Using C. elegans, we and other researchers have demonstrated that NuRD-orchestrated anchor cell (AC) invasion programs could be relevant to metastasis [43, 44]. These works would expand our understanding of the basis of metastasis. Among the critical genes specifically involved in AC invasion through the basement membrane, many are direct targets of the hormonal longevity gene DAF-12/Liver X receptor [44], including HSF1, LIT-1/Nemo-like gene, Hox genes and components of NuRD complex such as HDA1, MEP-1, etc. However, it is known that HSF1 directly regulates multi-drug-resistant genes in mammalian cell lines and Hox genes related to programmed cell death in C. elegans are human oncogene homologs [45].
The extracellular matrix (ECM) and possible reversibility of “archaic” attractors
One of the critical evolutionary changes that brought about multi-cellularity and cellular differentiation is that ECM proteins once peaked at transition from the unicellular life form (e.g., Chlamydomonas) to the multi-cellular (e.g., Volvox). This residual trace exists in the development of immune cells [18]. The difference in number and complexity between the Volvox ECM and the Chlamydomonas cell wall is mirrored by a dramatic increase in Volvox genes for ECMs [2, 46, 47]. In C. elegans, a screen for LET-418/Mi-2β suppression subtractive hybridization (SSH) revealed a number of ECM proteins as target genes, such as sqt-2 and sqt-3 [2]. The microarray profile of HDA-1/HDAC showed that the majority of its target genes are ECM-related [28]. DNA methyltransferase and histone deacetylase (HDAC) inhibitors improve reprogramming efficiency. In particular, valproic acid (VPA), an HDAC inhibitor, improves reprogramming efficiency by more than 100-fold [48].
In general, the ECM proteins function as scaffolding for tissue. Alterations in the ECM could lead to malignancy. However, it was proposed that even after cancer genes have been activated and lesions have formed, the process may still be reversible, and the cells may resume their normal appearance and function. Signaling factors such as ECM and tissue architecture disruption could rewire the gene regulatory network (GRNs) so that unoccupied and inaccessible “archaic” attractors suddenly become accessible from within an adult cell phenotype [18, 40].
The chromatin basis of “archaic” embryonic attractors
The NuRDs may act as a ‘hub’, communicating with a large number of genes to create a network of mutually regulating genes, form circuitry and further leash the cell attractors. Nucleosome remodeling and histone deacetylation can secure robustness by positive transcriptional regulation of loci of downstream target genes, such as CD4 [49] or by repression of other groups of genes, like induced pluripotent stem cell (iPS) cocktail factors, such as c-myc, OCT-4, and LIN-28 [49]. In addition, they can complex with many gene products of their target genes, such as C-MYC, OCT-4 so that such cell-, tissue-, or stage-specific NuRD complexes can modulate the activities of their targets [2, 3]. This can ensure their fine tuning functions. Furthermore, LET-418/Mi-2β/MEP-1/NuRD control at least HDA-1/HDAC1, LET-418/Mi-2β directly [50]. This kind of multiple layer of mutual gene regulations ensures further the different levels of attractors.
Many EMT genes are related to embryonic programs specifically regulated by NuRDs [2, 3]. Most of these embryonic TFs, which are known to play critical roles in orchestrating EMTs during embryogenesis, are also found to be expressed in a variety of human tumor cells; indeed, their expression is often correlated with aggressive tumor cell-associated traits. These TFs, including Slug, Snail, Twist, and SIP-1 are NuRD-regulated genes [3, 27]. They are highly conserved and initially discovered from developmental genetics of frog (Xenopus I) or D. melanogaster, belonging to an initial development early in metazoan evolution and having critical roles in the embryogenesis of these diverse organisms. These various TFs are appropriated in order to enable carcinoma cells to acquire these traits [49, 50].
The histological poorly differentiated tumors show preferential over-expression of genes that are normally enriched in ES cells, combined with preferential repression of PcG-regulated genes. Moreover, activation targets of Nanog and Oct-4 (possibly via NoDE, a NuRD of Nanog and Oct-4) [51] and c-myc (possibly via c-myc/NuRD) are more frequently over-expressed in poorly differentiated tumors compared to well-differentiated tumors. These three subgroups of signatures are all related to NuRD by direct protein–protein interactions, and could thus merge as one big group, i.e., the NuRD group, which has then antagonisms by PcG. This seems conserved at least in C. elegans and mammals. Thus, this link between genes associated with ES cell identity and the tumors raises the possibility of these genes contributing to stem cell-like phenotypes in many tumors [52]. However, key aspects of the ESC gene expression program recapitulated in cancer cells has been argued that this is largely a consequence of c-myc [52–54]. The c-myc amplification is the most frequent somatic copy-number amplification in tumor cells. Tumor cells that over-express c-Myc have enhanced expression of proliferation genes, and this is likely due to the role of c-Myc in recruiting p-TEFb to effect RNA polymerase II pause release at these genes [55, 56]. The recent understandings of dual control of c-myc by NuRD (being targeted and co-targeting) has provided new insights into the molecular pathways affected by mutations in these two key partners [2, 3, 57]. Recruitment of the repressive histone deacetylase activity associated with NuRD is therefore one mechanism by which Myc could act to block expression of specific target genes [58, 59]. It remains mysterious what causes over-expression of c-myc in cancer, but it has been demonstrated that c-myc exists early as hydra.
Mutations that affect the functions or levels of TrxG and PcG chromatin regulators have been implicated in a variety of cancers. In fact, many TrxG-related proteins demonstrate extensive molecular interactions of their counteracting chromatin regulatory protein PcG group [60]. The study of these regulators in ESCs and in cancer cells has revealed how repression of lineage-specific transcription factors and cell-cycle regulators may contribute to cancer phenotypes. Interestingly, the CHD3/Mi-2α proteins PICKLE (PKL) and PICKLE-RELATED2 (PKR2) have TrxG-like functions in plants and are required for the expression of many genes that are repressed by PcG proteins [60]. It is interesting to see if Mi-2α has similar functions in mammals.
The genome of unicellular Chlamydomonas revealed that the Rb and PcG genes are relatively “archaic”, but the genome of multi-cellular Volvox has “novel” SWI/SNF genes [46]. In mammals, PcG is critical for “stemness” [61, 62]. In C. elegans, the PcG-related protein MES-4 is a maternal protein and has a critical function in oogenesis and in the early embryo, especially during the zygote and P1 and/or P2 stages [63–65]. It has been proposed that MES-4 is significant in establishing the germline potential (and pluripotency). Nucleosome remodeling and histone deacetylation is believed to be required for eliminating germline potential (and pluripotency) in the early embryo soma by chromosomal remodeling activity. In later embryo stages, the NuRD-orchestrated remodeling required for germline development, although this is restricted during the P1 to P3 stage by PIE-1 [64]. This raises the exciting possibility that a NuRD-linked “archaic cancer attractor” theory might unify multi-cellularity. Greatly simplified, the zygote stage for germline cells (e.g., in C. elegans) somehow mimics archaic unicellular life forms; two-cell or several cells embryos could mimic the “ground” archaic cancer attractors. Evolutionally, multi-cellularity could have needed to have NuRD activity to drive differentiation; however, “archaic” PcG and Rb complexes or functionally related CRCs could form the reprogramming barrier [52] and GEDI maps of their deficiency are partially overlapped to those of NuRDs (Fig. 1b–f). This could also be the basis for archaic cancer attractor theory and provide a driving force for differentiation with a certain threshold during different stages and in different contexts [64, 65]. Undoubtedly, one could speculate that the “archaic” PcG and Rb complex could have some novel adapted roles. In molecular terms, we may have a holistic view of miRNAs [66], DNA (de)methylation, histone modification, high-order nuclear organization and post-transcription, which are exactly where NuRDs play a critical role [2, 3]. Importantly, as mentioned earlier, MEP-1 directly regulates the transcription of major components of DRM complex [50]. They are LET-418/Mi-2β, GEX-3/Interactor of GEI-11/b-Myb, LIN-35/Rb, EFL-1/E2F1, DPL-1/DP, SSL-1/p400 SWI/SNF ATPase, TRR-1/TRRAP, as well as MES-4/PRC1-like, SOP-2/PcG-like, MES-2/EZH, MES-6/EED, LIN-28, CHD-3/Mi-2α and FZR-1/CDH1, the components of transcriptional machinery, such as AMA-1/Polymerase II, CDK-9, CDK-7. Similar to MEP-1, LIN-15B, a synMuv B pathway gene which possibly cooperates with LET-418/Mi-2β and MEP-1, also directly regulates the transcriptions of major components of DRM complex, LIN-35/Rb, EFL-1/E2F1, DPL-1/DP, MEP-1, EGR-1/MTA1, LIN-15B itself, LIN-61/MBT, as well as MES-4/PRC1-like, P-TEFb/CDK9 [50]. So does heterochromatin protein HP1 functional tightly related synMuv B gene LIN-13 [50]. Such regulatory circuits could ensure fine tuning of GRN. Therefore, NuRD/DRM/dREAM and PcG tightly mutually regulated. However, it remains unclear how these CRCs (i.e., NuRDs, dREAM and PcG) are coordinated well.
How NuRD and its functionally related CRCs are linked to the overall transition dynamics
The GRN in a cell is what it is because over evolutionary time that is what has been selected, i.e., that is what has been best adapted to the environment at the time and survived the transitions necessitated by environmental change. The genomic DNA has been deployed in this system to both act as a “library” for the essential specification of the gene coding sequences (genotype) and as part of the processing that takes place to synthesize the gene products, and possibly contain the program that regulates and organizes the cell—that is what the attractor is assumed to have so it could be intrinsically “instructive”, which needs further confirmation. Fluctuations in environmental conditions and acquisition of genetic mutations during development can have an impact on the phenotypic outcome of a given organism. How biological systems can reduce the impact of these stochastic perturbations to ensure a uniform phenotype is the above-mentioned robustness. Genes that function in part to reduce the effect of environmental or genetic variability are called phenotypic capacitors, and they contribute to robustness and may underlie the stability of biological patterns. We propose that as a result of interactions of the cell/organism with its environment, NuRD (and its GRN) has been selected, though it would be a paradoxical mixture of Darwinism and Lamarckianism [67] (see discussion).
Furthermore, the cell attractors GRN model, particularly one from Dr. Sui Huang [40], emphasizes that the transcriptional factor (TFs) themselves, endowed with sequence-recognition capability, must be in charge of initiating the opening of chromatin at specific sites, and then probably followed by a bidirectional cooperation with the NuRD and its friends that they recruit to their target loci. How about the question “what regulates the regulatory proteins?” Since the expression pattern of TFs is a consequence of the control of the GRN, the sites of chromatin opening and closing must, in principle, mirror the dynamics of the GEP [40].
In fact, it remains unknown whether TFs or CRCs come first at conditionally packed genome context. TFs are sequence-specific, but the chromatin regulators help unpack the nucleosomes and so chromosome.
We could hypothesize that the interaction constitutes a special source of information with variation which is selectable, this is most probably where the much-discussed “missing inheritance” resides that a recent paper in Nat Rev Genet [67] suggests that a source of genetic variation has been missed—epigenetic variation, which might be one coordinator in such contexts. NuRD/CRCs could not serve only as an additional, important layer of stabilization of expression patterns established by the network of transcriptional regulation [40]. If the constraints on gene expression pattern change are dictated by the GRN only, then obviously the epigenetic barriers must be encoded in the particular structure or “architecture” of the GRN [40]. But how do the specific constraints arise from the network interactions? Dr. Sui Huang [40] considers the dynamics of the network, which refers to the temporal behavior of the state space S and NuRD and its friends may play important roles to influence the “systems dynamics”.
The NuRD and its functionally related CRCs emerge as the stabilizers responsible for the robustness, plasticity of life based on their progresses in evolution and developmental biology, through which GRNs in turn likely experience its evolution rather than static or quiescent ones. For example, the HDAC1 and HDAC2, two core components of the NuRD complex, protect the C. elegans genome against mutations [13, 68]. NuRD has important roles in protection of genome integrity, proper DNA replication, cell proliferation (which would correspond to a cycling attractors), and chromatin assembly [13], all of which are crucial to GRN. Mediated by the synMuv B pathway, NuRD can ensure the C. elegans animal survival by antagonizing germline fate in the intestine. Grown at high temperature, a majority of synMuv B mutants irreversibly arrest at the L1 stage. High temperature arrest (HTA) is accompanied by ectopic expression of many genes characteristic of germ line, including genes encoding components of the synaptonemal complex and other meiosis proteins. This high temperature stress plus mutations in such synMuv B genes largely mimics the RNAi treatment on mep-1 and let-418 or their mutant. As a consequence, somatic cells gain a germline program of gene expression in addition to their somatic program, leading to a mixed fate. Somatic expression of germline genes is enhanced at elevated temperature, leading to developmentally compromised somatic cells and arrest of newly hatched larvae. In C. elegans, such a group of synMuv B mutations require further environmental stress to attain the similar functions as LET-418/MEP-1/NuRD, hence NuRD could be dominant in this regulation [69].
In general, chromatin regulators, together with co-repressors or co-activators and/or their receptors, could broadly determine the adaptive value of standing genetic variation and this therefore has influenced the evolution of current genomes and in so doing the evolution of GRNs [70]. At the chromatin level, homologous to human co-repressor SMRT/HDAC1 associated repressor protein (SHARP), C. elegans DIN-1S directly interacts with DAF-12/Liver X receptor as a complex and it serves as a molecular switch that implements slow life history alternatives in response to diminished hormonal signals [71]. We recently demonstrated that daf-12 ensures developmental robustness by committing the animal to adult or dauer developmental programs despite variable internal or external conditions [44].
DAF-21/Hsp90 (heat shock protein 90) was also found to play an important role in buffering genetic and epigenetic variation whose expression led to altered phenotypes. A direct molecular interaction between Hsp90 and chromatin regulator [Trithorax (Trx)] control, together with the members of the PcG, the developmental fate of cells by modulating epigenetic signals. Connecting an epigenetic network controlling major developmental and cellular pathways with a system sensing external cues may explain the rapid fixation and epigenetic inheritance of phenotypic variation as a result of impaired Hsp90 [72, 73]. Both selective inhibition of Hsp90 and temperature stress increase the correlations between genotype and phenotype [74, 75]. Hsp90 also regulates the piRNA pathway through Piwi to mediate the robustness [76].
The new frontiers of carcinogenesis research
Classic incomplete network patterns have failed to provide a description of biological phenomena, since real space (inside the cells and among them) has been usually disregarded. Therefore, one voice is hovering: “cancer research towards cancer therapy may develop faster if cancer is not researched only in terms of molecular biology but rather in terms of systems biology”. Yes or No, see what will happen in the future.
However, in close fields, many researchers make inroads to some “new” aspects, which are aside from mainstream genetic determinism to alternatives that emphasize non-genetic influences, including chronic growth stimulation, extracellular matrix remodeling, alteration of cell mechanics, and disruption of tissue architecture. Importantly, as suggested by experts, the GRN does not work in isolation or in a vacuum: its activity is likely to be considered “permissive” and must be “integrated” by biophysical influences, mediated by the microenvironment (cell-to-cell and cell-to-stroma interactions). Indeed, the adoption of the information paradigm expressed by the central dogma, which is linear, deprived the organism of its physicality resulting in the marginalization of the role of mechanics and electromagnetic forces in morphogenesis [77]. The essays by Saetzler et al. [78] and by Bizzarri et al. [79] focus on the role of mechanics in development and cancer while the others address electromagnetic interactions in cell communication. Saetzler et al. [75] argue for the adoption of the organicist approach, whereby both a bottom-up and top-down causality are taken into consideration when explaining biological phenomena at large, and carcinogenesis in particular. Additionally, they argue that topology, i.e., real space, plays a causal role that should be considered when modeling complex phenomena. Bizzarri et al. [79] propose to concentrate research efforts at a mesoscopic level of organization in their approach to the understanding of cancer, while also highlighting the role of topology and its relationship to metabolism and dynamic processes. Sui Huang [15], while recognizing the role of supracellular phenomena, instead follows Waddington’s epigenetic landscapes to propose a cell-centered approach while focusing it around an integrative concept, namely, that of the attractor states. However, to better understand the carcinogenesis, since TFs function evidently through interactions or communications with NuRD or related CRCs, in our opinion, they could need work together. On the contrary, unexpectedly, chromatin regulators might have both specific and global effects [80] and its mystery needs further clarification. Our hypothesis expands cancer attractors theory to chromatin regulator with a case study of germline-specific ectopic expression in a brain tumor. Finally, it sounds tempting to see if the tissue organization field theory (TOFT) of cancer, which is largely compatible with this NuRD/CRCs involved theory of carcinogenesis, is a testable replacement for the somatic mutation theory (SMT) in the future [81].
Unifying two cell-attractor models
Among a number of carcinogenesis theories (see supporting text for further discussions), in fact, there are even two distinct (cancer) attractor models, but they could now be naturally united by our proposed chromatin remodel system-involved cancer attractor theory.
In one model [the above-mentioned Genetic regulatory network (GRN) model], the attractors are derived from an experimentally determined genetic regulatory network (GRN) for the cell type. It is very popular and has provided fruitful insights since its debut.
In the second model [the independent attractor (IA) model], attractors arising from the interactions between active gene products (mainly proteins) and independent of the genomic sequence, are descended from a pre-cellular state from which life originated. The attractor acts as the interface between the cell and its environment [82, 83]. However, some terms for this model remain elusive and abstract. It is asserted that the evidence from cell and molecular biological research and logic favors the second model, since it is hard to believe that the genome expression profile can solely and exhaustively explain cell differentiation [82, 83].
For some questions, the second model seems relatively easier to explain. For example, the IA model can couple well with cell division and replication. The IA model regards the cell as essentially a self-organized entity, albeit that self-organization extends back into the evolutionary past of the cell, i.e., it is inherited. As such, all functions of the cell, including cell division, are entrained in the dynamics of the processes that sustain the cell. These processes can be represented in the form of a dynamic attractor [82, 83]. The cell is carried though the cell cycle, including cell division at which point the attractor is inherited by two cells. However, the IA model with the introduction of chromatin remodel systems, makes its molecular terms realistic. The chromatin remodeling system indeed has its own specificity grammar and mediates the communication between life organisms and environment as RoE of IA model, although this sounds like Lamarckism. Besides, the rules of interaction between genes and regulators (e.g., CRCs) can also be elaborated in molecular terms. It ensures the relative ease of understanding the evolution of relevant mechanisms and why multi-cellular organisms emerged in the world. Indeed the GEDI map can show that such cell attractors exist in whole animals. It has thus important implications for understanding the evolution of multi-cellularity and hereditary and somatic disease (see below); this is consistent with the IA model. It could be essential that one comprehensive view on carcinogenesis has both protein level and RNAs under consideration [84].
Finally, contradictory to the claim in description of IA model is that MEP-1 has been shown to function in posttranscriptional level [32] (IA model claimed that chromatin marks and nucleosome location cannot influence posttranscriptional processes, lack of stability, and locus specificity) and together with HSP90, NuRD and its functionally related CRCs could get involved in posttranslational regulation [73–75] as well as the RNA processing-related multiple pathways [74–76]. Furthermore, away from the concerns of the description in GRN model, we postulate that NuRD could have both a global and locus-specific role [80]. Hence NuRD could have a comprehensive and profound role in GRN alongside balancing both the plasticity and robustness of animals.
CRCs-linked “archaic” cancer attractors theory
Put together, in Fig. 2, the metaphoric “walking” of cells along the trajectories can be coupled with cell division, since the Mi-2/NuRD complexes couple with cell division via an array of pathways [2, 3]. Considering the metaphorical “potential energy” in Fig. 2 (also see supplementary materials), the non-attractor status or the height of the “mountain” possibly provides a driving force for embryonic/stem/progenitor cell proliferation. This “potential energy” (i.e., an inverse of probability) in a realistic molecular items [12] could be the trend linking inherited genomic program, environmental stresses, the availability of nutrients, and the storage of ATPs for chromatin remodeling. The chemical gradient and/or the quantity of gene products could be the direct “driver” factor. In general, the multi-potency of organisms is lost during development. Stem cells with multi-potency, such as ES cells and HSCs have been proposed to have a variety of weakly activated genes, but a terminally differentiated cell has a smaller number of strongly activated genes [85, 86]. However, this analogy is greatly simplified with the tri-stable switch modified from one initiated by Huang et al. and Kaufmann for its simplicity [40]. It is based on the dynamics of the two-gene regulatory circuit and qualitative explanation to basic principles of dynamical systems for the tri-stable circuits. It almost illustrates the abstract concepts behind the hierarchy of cell type diversification during the development. In reality, or “real space”, the regulatory circuit dynamics need be further formalized as a kind of quasi-potential landscape that captures the global dynamics, typically illustrated by Waddington’s “epigenetic landscape” [7, 40]. However, both dynamical hallmarks of attractor states are represented here [39, 42].
In the pie-1 null mutants of C. elegans, the germline cell P2 differentiates into somatic tissues in response to transcriptional activators that are normally present but inactive in the wild-type germline, so pie-1 plays an important role in protecting germline pluripotency. For example, in the embryonic germ cells, Z2 and Z3 are absent, but excess numbers of differentiated somatic pharyngeal cells are present in pie-1 null mutants [65]. This logically could mimic the “gain of function” of the MEP-1/LET-418/Mi-2/NuRD complex because at this stage; PIE-1 protein biochemically and genetically represses the activities of the MEP-1/LET-418/Mi-2/NuRD complex. In the metaphor of mountains and valleys (Fig. 2), the activities of (i.e., loss or gain of function) in the Mi-2/NuRD complex can manage (i.e., push up or down) the “potential” energy transfer so as to promote differentiation or maintain the germline stem cell state. The somatic cells in animals with loss of function in the Mi-2/NuRD complex retain germ stem cell potential and respond inappropriately to developmental cues that normally regulate germ stem cell-specific GRN. Moreover, the MEP-1/LET-418/Mi-2/NuRD functions at or after the onset of succeeding differentiation events to change the distribution of maintenance factors. This allows the stable specification of new stage-specific chromatin domains sequentially within each cell lineage during development through the concerted action of transcriptional activators and repressors.
Interestingly, for the embryonic somatic cells, the MEP-1/LET-418/Mi-2/NuRD complex stably inactivates germline potential (Figs. 1, 2); however, during larval development for vulval precursor cells, the activities of MEP-1/LET-418/Mi-2/NuRD stably inactivate the potential to undergo vulval differentiation. Therefore, Mi-2/NuRD behaves as a “stabilizer” rather than a simple activator/repressor only, whether cells switch to neighbor “attractors” may be tightly controlled by the powerful Mi-2/NuRD complex and its related CRCs (Fig. 3). A similar mechanism could remain in mammals.
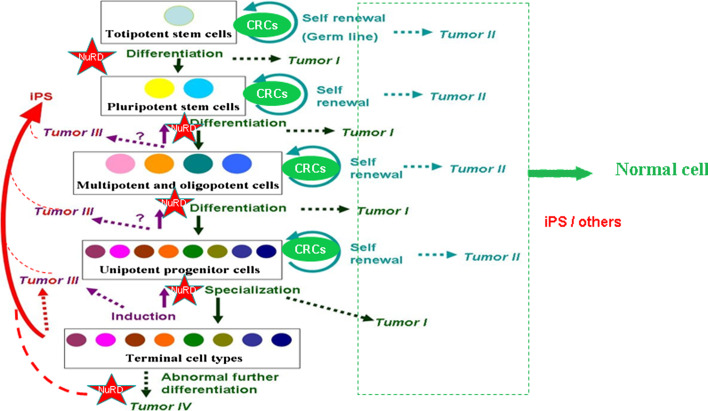
Fig. 3.
Different tiers of tumors based on their origins with respect to development stages of stem cells regulated by NuRD/CRCs. Tumor I and II are derived from so-called ‘stem cells’ in sense of their differentiation potency. Type I occurs when the differentiated cells fail to lose proliferation potential, especially malfunctions of NuRD/CRCs; while type II occurs when abnormal cell types are generated unexpectedly. Tumor III and IV are derived from differentiated cells at each level. Type III occurs when the reverse process of differentiation fails to produce the expected progenitors; type IV occurs when the doomed terminal cells ‘create’ their own descendant cell types, regaining capability of proliferation. NuRD/CRCs could have abnormal functions during such processes. Such stem cell biology could be at part directly tested in models of anchor cell (AC) invasion, vulva development in C. elegans
Conclusions
With GEDI and IPA computation analysis, data-mining, Mi-2/NuRD complexes along with their functionally related dREAM/MMB complex and antagonistic PcG complex emerge as master regulators of such cancer attractors, and the MDR1 gene and some ECM genes as conserved hub molecules for functions of Mi-2/NuRD. The notion of “quasi”—mutation which reflects the nature of Mi-2/NuRD inherent integration of dynamic deterministic and stochastic dynamics as well as non-genetic and genetic heterogeneity, the simplicity of cancer attractors and the versatility of NuRDs are combined, the NuRD—orchestrated “cancer attractors” provides a surprisingly simple, straightforward, and user-friendly explanation for observations of cancer cell behavior and could enlighten the cancer “attractors”-based prevention and therapy. Importantly, this theory (Figs. 2, 3) provides molecular terms for the recent united hypothesis for non-equilibrium dynamics (attractors) and developmental biology (morphogenetic fields), and proposes two big groups (i.e., NuRD and PcG) of embryonic signatures rather than many sets in differentiation cancer (Fig. 3), then emphasizes the “archaic” cancer attractors to own the multiple layers of mutual gene regulation as well as low expression level with more activated genes bearing dynamic variations rather than high expression level but less activated genes [85, 86]. This can give reliable explanatory insights into the complex interactions taking place between cancer cells and embryonic cues. Morphogen-induced network rewiring results in a shift of attractor boundaries, leading to a displacement of the cell population toward a different attractor.
In this perspective, we also presented cross-species evolutional evidence, such as RegA, as a functional equivalent of NuRD in Vovlox Carterei, and summarized the current understanding in protection of genome instability of NuRD, other CRCs together with HSP90 or DAF-12 for cell fate determination, NuRD/CRCs as system sensor of external cues to rapidly fix and epigenetically inherit phenotypic variations as well to ensure developmental robustness during genome evolution. This is just a framework and more detailed work is needed. To some extent, GRN could indeed have the “instructive” as “a Turing machine”, but its evolution could be influenced by NuRD or related CRCs.
Superficially, our work could just add chromatin regulators to cancer attractors theory, however, if it could be proven true, we could propose the strategy: (1) Quantification first of the effects of inhibitors of NuRD/its related for systematically reprogramming and optimizing cell state (i.e., reversing from pathological to normal physiological); (2) “Artificial evolution” by interfering this common frailty (i.e., cancer), which might be the outcome of one evolutional trade-off between the population survival to individual sacrifice/altruism. Such a trade-off is in part proven in the ageing research in C. elegans [87]. It could emerge during the transition from unicellular form to multi-cellular form. Its more advanced form could be Eu-social structure. Hence, cancer is inevitable for human beings during the evolution with the code of natural selection. The active treatments with modern knowledge will make sense for the state/health of individual, but certainly along with a cost of health plan burden since the performance always comes at a price [70, 88].
This hypothesis would also imply reorientation of current treatment principles from cellular destruction therapies to cellular retraining or cyto-education [38]. Obviously, this novel theory integrates well with differentiation theory, i.e., a block of normal differentiation and abnormal reversal of differentiation (i.e., de-differentiation) being the hallmarks of cancer. However, “chemotherapeutic intervention in advanced cancer alone has been an exercise in futility” [12]. One could combine chemotherapeutic intervention selectively inhibiting HSF1 (e.g., quercetin) and proteasome, and targeting this Mi-2/NuRD complex together with differentiation therapy to form “cocktail” therapy, designed to facilitate cancer cells re-entering the differentiation program. Its success depends on appropriate molecular “lever points”, the perturbations of which place the bio-molecular system into states that are poised to differentiate. Indeed, a therapeutic agent or embryonic extracts could allow the system to naturally flow toward an attractor that corresponds to the desired cellular endpoint. Clinical trials performed using zebrafish embryo extracts administered to advanced cancer patients who no longer respond to conventional treatment had marked beneficial effects [12]. Another principally different therapeutic strategy could aim at neutralization of the harmful effects of the cancer cell rather than at its elimination. This approach appeared to be efficient in model systems: antibodies to VEGFR1 and VEGFR2 inhibit formation of metastases in mice in which VEGFR1- and VEGFR2-positive bone marrow cells are involved in formation of a pre-metastatic niche [88]. One alternative interference approach goes to the modified newly developed iPS (induced pluripotent stem) cell technology [35] (Fig. 3). An aneuploid melanoma cell line remains amenable to reprogramming into iPSCs that supported the development of chimeras, suggesting that certain cancer cells are reprogrammable by transcription factors. This observation should be useful for studying the relative contribution of reversible epigenetic and irreversible genetic changes to cancer [89, 90].
Importantly, the simple model organisms such as C. elegans, Drosophila, and zebrafish that have supplied us with many remarkable discoveries have overcome the barriers of resolution not typically achievable in mammalian models, and they could enable us to accelerate the development of a new generation of specific cancer drugs, like co-clinical trials. It would be expected that the small-molecule drug screening for NuRD and other CRCs for cellular reprogramming could be a new class of targets for cancer prevention and therapy. Ideally, a minimal perturbation of “level points” on “archaic” cancer attractors could reverse the diseased states to healthy state attractors, without (or almost without) the short-term or long-term side-effects.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The author is grateful to members of the laboratories of Drs. Mueller, Fisher, and Calderwood for discussions and to Mr. Xiaodong Dang for initially constructing Fig. 3 as well as discussions.
Contributor Information
Yue Zhang, Email: yzhang1@bidmc.harvard.edu, Email: zy1001@yahoo.com.
Hisashi Moriguchi, Email: hisashi.moriguchi@gmail.com.
References
- 1.Ho L, Crabtree GR. Chromatin remodeling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Li Y. The expanding Mi-2/NuRD complexes: a schematic glance. Proteomics Insights. 2010;3:79–109. [Google Scholar]
- 3.Zhang Y. Biology of Mi-2/NuRD in SLAC (stemness, longevity/ageing and cancer) Gene Regul Syst Biol. 2011;5:1–26. doi: 10.4137/GRSB.S6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila . Science. 2010;330(6012):1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- 5.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331(6015):304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kauffman S. Homeostasis and differentiation in random genetic control networks. Nature. 1969;224:177–178. doi: 10.1038/224177a0. [DOI] [PubMed] [Google Scholar]
- 7.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 8.Huang S, Ernberg I, Kauffman S. Cancer attractors: a systems view of tumors from a gene network dynamics and developmental perspective. Semin Cell Dev Biol. 2009;20(7):869–876. doi: 10.1016/j.semcdb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang S, Ingber DE. A non-genetic basis for cancer progression and metastasis: self-organizing attractors in cell regulatory networks. Breast Dis. 2007;26:27–54. doi: 10.3233/bd-2007-26104. [DOI] [PubMed] [Google Scholar]
- 10.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 11.Dinicola S, D’Anselmi F, Pasqualato A, Proietti S, Lisi E, Cucina A, Bizzarri M (2011) A systems biology approach to cancer: fractals, attractors, and nonlinear dynamics. OMICS (Epub ahead of print) [DOI] [PubMed]
- 12.Bizzarri M, Cucina A, Biava PM, Proietti S, D’Anselmi F, Dinicola S, Pasqualato A, Lisi E. Embryonic morphogenetic field induces phenotypic reversion in cancer cells. Curr Pharm Biotechnol. 2011;12(2):243–253. doi: 10.2174/138920111794295701. [DOI] [PubMed] [Google Scholar]
- 13.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11(8):588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demicheli R, Coradini D (2010) Gene regulatory networks: a new conceptual framework to analyse breast cancer behaviour. Ann Oncol (Epub ahead of print) [DOI] [PubMed]
- 15.Huang S (2011) On the intrinsic inevitability of cancer: from foetal to fatal attraction. Semin Cancer Biol (Epub ahead of print) [DOI] [PubMed]
- 16.Liu Y, Clem B, Zuba-Surma EK, El-Naggar S, Telang S, Jenson AB, Wang Y, Shao H, Ratajczak MZ, Chesney J, Dean DC. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell. 2009;4(4):336–347. doi: 10.1016/j.stem.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19(17):2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 18.Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys Rev Lett. 2005;94(12):128701. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- 19.Passannante M, Marti CO, Pfefferli C, Moroni PS, Kaeser-Pebernard S, Puoti A, Hunziker P, Wicky C, Müller F. Different Mi-2 complexes for various developmental functions in Caenorhabditis elegans . PLoS One. 2010;5(10):e13681. doi: 10.1371/journal.pone.0013681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ, Georgopoulos K. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22(9):1174–1189. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramírez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4(8):532–536. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- 22.Ghanta KS, Li DQ, Eswaran J, Kumar R. Gene profiling of mta1 identifies novel gene targets and functions. PLoS One. 2011;6(2):e17135. doi: 10.1371/journal.pone.0017135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marino S, Nusse R. Mutants in the mouse NuRD/Mi2 component P66alpha are embryonic lethal. PLoS One. 2007;2(6):e519. doi: 10.1371/journal.pone.0000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. 2007;179(7):1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagani Z, Wiederschain D, Loo A, He D, Mosher R, Fordjour P, Monahan J, Morrissey M, Yao YM, Lengauer C, Warmuth M, Sellers WR, Dorsch M. The Polycomb group protein Bmi-1 is essential for the growth of multiple myeloma cells. Cancer Res. 2010;70(13):5528–5538. doi: 10.1158/0008-5472.CAN-09-4229. [DOI] [PubMed] [Google Scholar]
- 26.Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, Chang SY, Lee OK, Wu KJ. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12(10):982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 27.Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The Polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell. 2009;16(5):699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whetstine JR, Ceron J, Ladd B, Dufourcq P, Reinke V, Shi Y. Regulation of tissue-specific and extracellular matrix-related genes by a class I histone deacetylase. Mol Cell. 2005;18(4):483–490. doi: 10.1016/j.molcel.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, Ruvkun G. Cancer. Germ cell genes and cancer. Science. 2010;330(6012):1761–1762. doi: 10.1126/science.1200772. [DOI] [PubMed] [Google Scholar]
- 30.Maruyama R, Shipitsin M, Choudhury S, Wu Z, Protopopov A, Yao J, Lo PK, Bessarabova M, Ishkin A, Nikolsky Y, Liu XS, Sukumar S, Polyak K (2010) Breast cancer special feature: altered antisense-to-sense transcript ratios in breast cancer. Proc Natl Acad Sci USA (Epub ahead of print) [DOI] [PMC free article] [PubMed]
- 31.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belfiore M, Mathies LD, Pugnale P, Moulder G, Barstead R, Kimble J, Puoti A. The MEP-1 zinc-finger protein acts with MOG DEAH box proteins to control gene expression via the fem-3 3′ untranslated region in Caenorhabditis elegans . RNA. 2002;8(6):725–739. doi: 10.1017/S1355838202028595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirk MM, Stark K, Miller SM, Müller W, Taillon BE, Gruber H, Schmitt R, Kirk DL. RegA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development. 1999;126(4):639–647. doi: 10.1242/dev.126.4.639. [DOI] [PubMed] [Google Scholar]
- 34.Miller SM. Volvox, Chlamydomonas, and the evolution of multicellularity. Nat Edu. 2010;3(9):65. [Google Scholar]
- 35.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci USA. 2010;107(52):22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janecka IP. Cancer control through principles of systems science, complexity, and chaos theory: a model. Int J Med Sci. 2007;4(3):164–173. doi: 10.7150/ijms.4.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tulloch NL, Pabon L, Murry CE. Get with the (re)program: cardiovascular potential of skin-derived induced pluripotent stem cells. Circulation. 2008;118(5):472–475. doi: 10.1161/CIRCULATIONAHA.108.791442. [DOI] [PubMed] [Google Scholar]
- 40.Huang S. Reprogramming cell fates: reconciling rarity with robustness. BioEssays. 2009;31(5):546–560. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- 41.Vilaboa NE, Galán A, Troyano A, de Blas E, Aller P. Regulation of multidrug resistance 1 (MDR1)/P-glycoprotein gene expression and activity by heat-shock transcription factor 1 (HSF1) J Biol Chem. 2000;275(32):24970–24976. doi: 10.1074/jbc.M909136199. [DOI] [PubMed] [Google Scholar]
- 42.Jiang ZP, Xu P, Wang GP, Zhao XL, Chen FP. Hypothesizing that histone deacetylase inhibitors can be used to reverse multiple drug resistance. Med Hypotheses. 2010;74(1):92–94. doi: 10.1016/j.mehy.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 43.Matus DQ, Li XY, Durbin S, Agarwal D, Chi Q, Weiss SJ, Sherwood DR. In vivo identification of regulators of cell invasion across basement membranes. Sci Signal. 2010;3(120):ra35. doi: 10.1126/scisignal.2000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hochbaum D, Zhang Y, Stuckenholz C, Labhart P, Alexiadis V, Martin R, Knölker H, Fisher AL (2011) DAF-12 regulates a connected network of genes to ensure robust developmental decisions. PLOS Genet 7(7) e1002179 doi:10.1371/journal.pgen.1002179 [DOI] [PMC free article] [PubMed]
- 45.Potts MB, Cameron S. Cell lineage and cell death: Caenorhabditis elegans and cancer research. Nat Rev Cancer. 2011;11(1):50–58. doi: 10.1038/nrc2984. [DOI] [PubMed] [Google Scholar]
- 46.Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, Hellsten U, Chapman J, Simakov O, Rensing SA, Terry A, Pangilinan J, Kapitonov V, Jurka J, Salamov A, Shapiro H, Schmutz J, Grimwood J, Lindquist E, Lucas S, Grigoriev IV, Schmitt R, Kirk D, Rokhsar DS. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329(5988):223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 48.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, De Souza B, Qi X, Keables P, Von Andrian UH, Georgopoulos K. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20(6):719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, Slightha C, Jiang L, Hyman AA, Kim SK, Waterston RH, Gerstein M, Snyder M, Reinke V. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans . Genome Res. 2011;21(2):245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Zn. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 54.Rothenberg ME, Clarke MF, Diehn M. The Myc connection: ES cells and cancer. Cell. 2010;143(2):184–186. doi: 10.1016/j.cell.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 55.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varlakhanova NV, Cotterman RF, de Vries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80(1):9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith KN, Lim JM, Wells L, Dalton S. Myc orchestrates a regulatory network required for the establishment and maintenance of pluripotency. Cell Cycle. 2011;10(4):592–597. doi: 10.4161/cc.10.4.14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strübbe G, Popp C, Schmidt A, Pauli A, Ringrose L, Beisel C, Paro R. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc Natl Acad Sci USA. 2011;108(14):5572–5577. doi: 10.1073/pnas.1007916108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aichinger E, Villar CB, Di Mambro R, Sabatini S, Köhler C (2011) The CHD3 chromatin remodeler PICKLE and Polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell (Epub ahead of print) [DOI] [PMC free article] [PubMed]
- 62.Gieni RS, Ismail IH, Campbell S, Hendzel MJ (2011) Polycomb group proteins in the DNA damage response—A link between radiation resistance and “stemness”. Cell Cycle 10(6) (Epub ahead of print) [DOI] [PubMed]
- 63.Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a homolog of the NuRD complex component Mi-2 act together to maintain germline–soma distinctions in C. elegans . Cell. 2002;111:991–1002. doi: 10.1016/S0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- 64.Shin TH, Mello CC. Chromatin regulation during C. elegans germline development. Curr Opin Genet Dev. 2003;13(5):455–462. doi: 10.1016/S0959-437X(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 65.Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70(1):163–176. doi: 10.1016/0092-8674(92)90542-K. [DOI] [PubMed] [Google Scholar]
- 66.Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D. An integrated approach to uncover drivers of cancer. Cell. 2010;143(6):1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17(4):443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petrella LN, Wang W, Spike CA, Rechtsteiner A, Reinke V, Strome S. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development. 2011;138(6):1069–1079. doi: 10.1242/dev.059501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erren TC. On the origin of cancer: evolution and a mutation paradox. Med Hypotheses. 2099;73(1):124–125. doi: 10.1016/j.mehy.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 71.Ludewig AH, Kober-Eisermann C, Weitzel C, Bethke A, Neubert K, Gerisch B, Hutter H, Antebi A. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 2004;18(17):2120–2133. doi: 10.1101/gad.312604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330(6012):1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tariq M, Nussbaumer U, Chen Y, Beisel C, Paro R. Trithorax requires Hsp90 for maintenance of active chromatin at sites of gene expression. Proc Natl Acad Sci USA. 2009;106(4):1157–1162. doi: 10.1073/pnas.0809669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Specchia V, Piacentini L, Tritto P, Fanti L, D’Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463(7281):662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- 75.Ruden DM, Lu X. Hsp90 affecting chromatin remodeling might explain transgenerational epigenetic inheritance in Drosophila . Curr Genomics. 2008;9(7):500–508. doi: 10.2174/138920208786241207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet. 2011;43(2):153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sonnenschein C, Soto AM (2011) Why systems biology and cancer? Semin Cancer Biol (Epub ahead of print) [DOI] [PubMed]
- 78.Saetzler K, Sonnenschein C, Soto AM (2011) Systems biology beyond networks: generating order from disorder through self-organization. Semin Cancer Biol (Epub ahead of print) [DOI] [PMC free article] [PubMed]
- 79.Bizzarri M, Giuliani A, Cucina A, D’Anselmi F, Soto AM, Sonnenschein C (2011) Fractal analysis in a systems biology approach to cancer. Semin Cancer Biol (Epub ahead of print) [DOI] [PMC free article] [PubMed]
- 80.Lessard JA, Crabtree GR. Regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soto AM, Sonnenschein C. The tissue organization field theory of cancer: a testable replacement for the somatic mutation theory. BioEssays. 2011;33:332–340. doi: 10.1002/bies.201100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee JT, Herlyn M. Embryogenesis meet tumorigenesis. Nat Med. 2006;12:882–883. doi: 10.1038/nm0806-882. [DOI] [PubMed] [Google Scholar]
- 83.Baverstock K (20011) A comparison of two cell regulatory models entailing high dimensional attractors representing phenotype. Prog Biophys Mol Biol (Epub ahead of print) [DOI] [PubMed]
- 84.Gunaratane PH. Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells? Curr Stem Cell Res Ther. 2009;4(3):168–177. doi: 10.2174/157488809789057400. [DOI] [PubMed] [Google Scholar]
- 85.Furusawa C, Kaneko K. Complex organization in multicellularity as a necessity in evolution. Artif Life. 2000;6(4):265–281. doi: 10.1162/106454600300103638. [DOI] [PubMed] [Google Scholar]
- 86.Furusawa C, Kaneko K. Chaotic expression dynamics implies pluripotency: when theory and experiment meet. Biol Direct. 2009;4:17. doi: 10.1186/1745-6150-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hughes SE, Evason K, Xiong C, Kornfeld K. Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 2007;3(2):e25. doi: 10.1371/journal.pgen.0030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lichtenstein AV. Carcinogenesis: evolution of concepts. Biochemistry (Mosc) 2009;74(4):353–361. doi: 10.1134/S0006297909040014. [DOI] [PubMed] [Google Scholar]
- 89.Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122(Pt 19):3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Apostolou E, Hochedlinger K. Stem cells: iPS cells under attack. Nature. 2011;474(7350):165–166. doi: 10.1038/474165a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.