Abstract
DNA aneuploidy has been identified as a prognostic factor in the majority of epithelial malignancies. We aimed at identifying ploidy-associated protein expression in endometrial cancer of different prognostic subgroups. Comparison of gel electrophoresis-based protein expression patterns between normal endometrium (n = 5), diploid (n = 7), and aneuploid (n = 7) endometrial carcinoma detected 121 ploidy-associated protein forms, 42 differentially expressed between normal endometrium and diploid endometrioid carcinomas, 37 between diploid and aneuploid endometrioid carcinomas, and 41 between diploid endometrioid and aneuploid uterine papillary serous cancer. Proteins were identified by mass spectrometry and evaluated by Ingenuity Pathway Analysis. Targets were confirmed by liquid chromatography/mass spectrometry. Mass spectrometry identified 41 distinct polypeptides and pathway analysis resulted in high-ranked networks with vimentin and Nf-κB as central nodes. These results identify ploidy-associated protein expression differences that overrule histopathology-associated expression differences and emphasize particular protein networks in genomic stability of endometrial cancer.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0752-0) contains supplementary material, which is available to authorized users.
Keywords: Aneuploidy, Endometrial carcinoma, Genomic instability, Mass spectrometry, Pathway analysis, Two-dimensional gel electrophoresis
Introduction
Endometrial cancer (EnCa) is a common malignancy and accounts for about 6% of all female malignancies in the Western world. Due to early bleeding symptoms, about 70% of affected women are detected at tumor stage I when the tumor is limited to the corpus uteri and mean survival is close to 87%. Next to histopathology, tumor stage, tumor grade, and DNA ploidy are used as prognostic factors with aneuploidy being significantly related to an increased risk of nodal metastases, recurrence, and disease-related death [1, 2]. For example, uterine papillary serous carcinoma (UPSC) has an inferior prognosis compared to that of endometrioid carcinomas [3]. The prognostic potential of DNA ploidy was shown in a study involving 376 endometrial carcinoma patients with a more favorable 5-year survival rate of 94% for patients with diploid cell populations as opposed to those with aneuploid malignancies (83%) [4].
DNA ploidy can be assessed by flow or image cytometry [5], the latter case assessing cells on tissue sections [6]. We demonstrated for breast carcinomas that not only the ploidy status per se but also the grade of genomic instability has an impact on patient prognosis [7]. Instability can be assessed by detection of gains and losses of chromosomal regions. For instance, in breast carcinomas, a gain of 1q is a common alteration, whereas for vaginal carcinomas, the most frequent aberration is a gain of 3q [8, 9]. In endometrial carcinomas, significant allelic gains have been reported for many chromosome arms, such as 1q, 3q, 8q, and 10q [10–12]. The predominance of these tumor-specific chromosomal aneuploidies is associated with increased expression of genes located on particular chromosomes in a manner that seems to be independent of tissue and cell type [13]. The multitude of chromosomal aberrations seems to lead to an irreversible disturbance of transcriptional regulation and a loss of genomic integrity in aneuploid tumor cells [14]. Furthermore, we found that aneuploidy-associated genomic instability also translates into protein expression alterations in colorectal cancer cell lines [15]. However, acquiring knowledge of the underlying molecular network mechanisms characterized by chromosomal abnormalities remains a major challenge in the EnCa to elucidate.
In the present study, we identified genomic instability-associated protein expression patterns of interest for risk evaluation and therapy in endometrial carcinomas.
Materials and methods
Patients, sample collection, and preparation
Endometrial tumor material and normal endometrium were collected after hysterectomies for EnCa or benign affections. Collection was performed between 1997 and 2003 after patient consent and according to ethical review board approvals (#00-385 and #03-034) at the Karolinska Hospital, Stockholm, Sweden. Clinical data of 14 malignancies and five normal samples are provided in S1 and S2. Cells were immediately after collection harvested from the surface of the tumor and the normal epithelium by scalpel scraping [16]. This technique supplies a high percentage of representative cells for analysis. After preparation, each sample was quality checked by comparison of Giemsa-stained smears with histological slides. Only samples with more than 95% tumor cells were used. The samples were stored at −80°C until two-dimensional gel electrophoresis (2-DE).
Histopathology and image cytometry
Histopathological subtype and degree of differentiation were evaluated according to the FIGO grading system [17]. Ten tumors were of endometrioid subtype and four of the less common UPSC type. DNA ploidy determination was performed on paraffin-embedded tumor specimens of Feulgen-stained histopathological sections (8 μm) using image cytometry as described [5]. At least 100 nuclei per sample were selected interactively and their DNA contents measured using the ACAS image system (Ahrens ACAS, Hamburg, Germany). In addition, small lymphocytes were measured as internal controls. The classification of the DNA profiles was performed according to Auer et al. [5].
Two-dimensional gel electrophoresis
Protein concentrations of samples were determined in quadruplicates by addition of 25 μl of concentrated assay reagent (Bio-Rad) to 1 μl solubilized sample diluted in 100 μl Milli-Q water using 96-well microplates [18] followed by isoelectric focusing and SDS-PAGE of 75-μg samples as described [16] (S3). Gels were stained with silver nitrate, scanned with a flatbed densitometer (GS 710, Bio-Rad), and analyzed with PDQuest software (Bio-Rad, Hercules, CA, USA, version 8.0.1).
Statistical analysis
Only protein spots that showed expression in at least 50% of the samples within each ploidy group were used for statistical analyses by Welch t tests for unequal variances to compare protein expression in four analytical sets, i.e., those of (1) normal endometrium versus diploid endometrioid cancer, (2) diploid versus aneuploid endometrioid cancer, (3) aneuploid endometrioid cancer versus aneuploid UPSC, and (4) diploid endometrioid cancer versus aneuploid UPSC [19]. For the pairwise comparison, the significance level was set to 0.001, whereas 0.01 was used for the remaining tests due to low sample size. To detect proteins with increasing or decreasing expression from normal endometrial versus the tumor tissues, an exact version of the Jonckheere–Terpstra test was applied at a significance level of 0.001. Fold change was calculated as logarithmized means of each group. Principal component analysis (PCA) was used to discriminate normal mucosa and carcinomas with the software package R version 2.10.1 for statistical analyses with R packages clinfun version 0.8.10 (Jonckheere–Terpstra test) and FactoMineR version 1.14 (PCA).
In-gel digestion and mass spectrometry (MALDI-TOF)
2-DE gel spots were manually cut with a scalpel in a laminar air-flow bench. After destaining and digestion with trypsin as described [20] (S4), the fragments were analyzed by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) in an Ultraflex III TOF/TOF instrument (Bruker Daltonics, Bremen, Germany).
Network and functional pathway analysis
Differentially expressed and identified proteins were analyzed by the Ingenuity Pathway Analysis software (Ingenuity, Mountain View, CA, USA; http:\\www.ingenuity.com, version 8.5-2803). The IPA-generated networks are ranked using a score that is calculated as the negative logarithm of the p value. This p value indicates the likelihood of the proteins of interest being found together within a network. ANXA4, ANXA5, HSP90AB1, KRT9, KRT10, and PDIA3 were identified from at least two protein spots and showed non-homogenous up- or down-regulation between groups because of possible post-translational modification and were excluded from pathway analysis. For the evaluation of aneuploid endometrioid cancer versus aneuploid UPSC, only one differentially expressed protein was detected and no relevant network analysis was performed.
Confirmation by liquid chromatography and tandem mass spectrometry (LC–MS/MS)
Picked protein spots were processed and digested with trypsin using a robotic protein handling system (MassPREP, Waters). Peptides were extracted with 30 μl of 5% formic acid/2% acetonitrile and subsequently with 24 μl 2.5% formic acid/50% acetonitrile. Tryptic fragments were analyzed using a quadrupole time-of-flight mass spectrometer (Q-TOF Premier API, Waters, Milford, MA, USA) with a standard Z-spray source coupled to a Waters nanoAcquity system (S5).
Results
Genomic instability
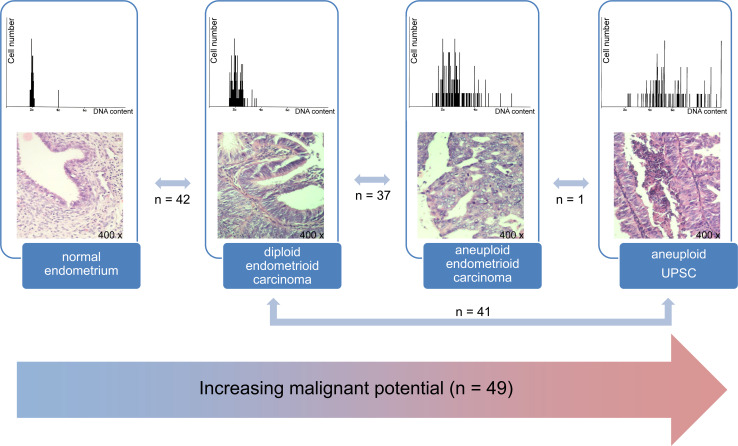
DNA image cytometry showed seven of the 14 carcinomas to be of diploid endometrioid histopathology and the remaining seven to be aneuploid with endometrioid (n = 3) and UPSC (n = 4) histopathology. Based on clinical routine assessment of more than 25 normal endometrial samples, diploidy was presumed for the normal endometrium being part of this study.
Two-dimensional gel electrophoresis and pathway analysis
PCA results showed an overall clustering of each of the normal endometrial mucosa (n = 5), diploid endometrioid cancer (n = 7), and aneuploid EnCa samples, including the endometrioid (n = 3) and UPSC (n = 4) subtypes. However, two samples, CP 27 (aneuploid UPSC) and CP 35 (diploid endometrioid cancer), showed aberrant locations in the PCA plot and were therefore excluded from analysis, resulting in a perfect separation of all three groups (S2, S6).
Normal endometrium versus diploid endometrioid carcinomas
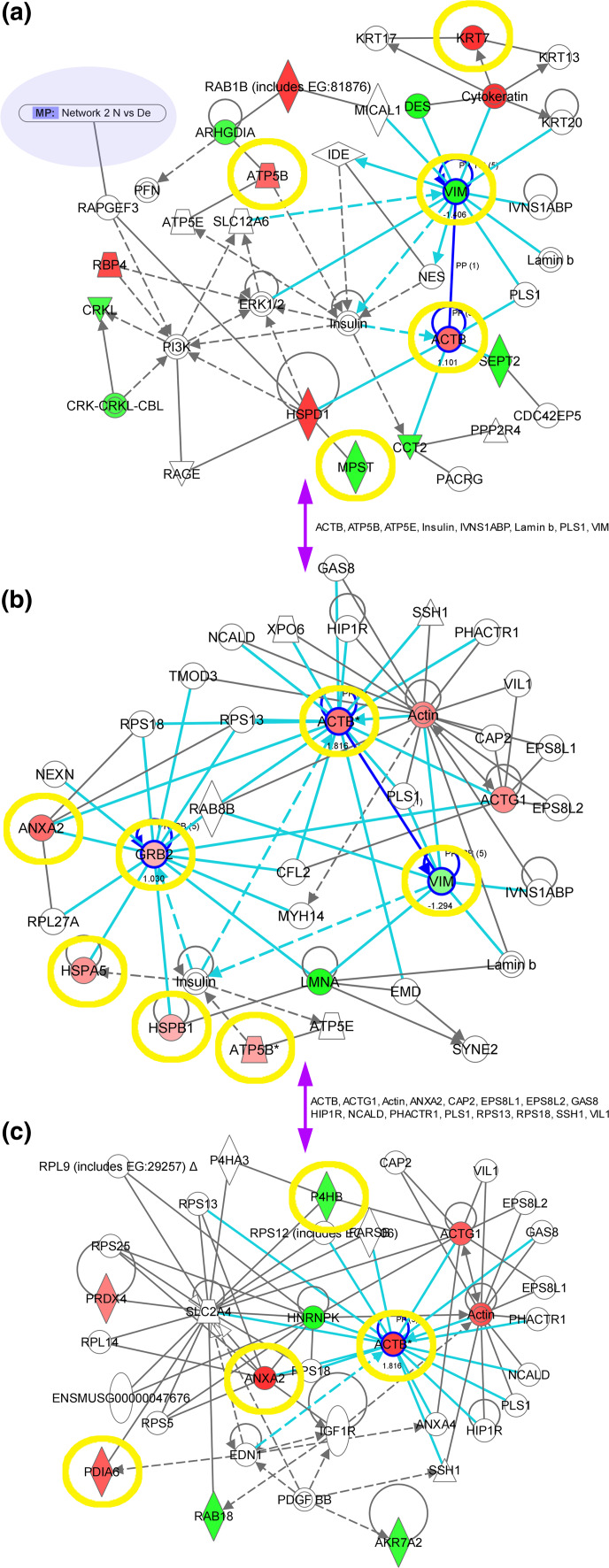
A total of 42 differentially expressed proteins were detected (p ≤ 0.001); 19 (45%) were identified by MALDI-TOF-MS: seven over- and 12 under-expressed in the diploid group (Fig. 1, S7). These up- and down-regulated proteins were further analyzed using the Ingenuity Pathway Knowledge Base (IPA) generating networks of interaction. The first network with a score of 33 is associated with Cellular Assembly and Organization and Small Molecule Biochemistry, the second with a score of 10 with Development and Function, Cell Morphology, Skeletal and Muscular System Development and Function. Vimentin (VIM), β-actin (ACTB), and nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) are central nodes of these networks. These three proteins are associated with diseases and functions related to cancer, neurological disease, and genetic disorders (p < 0.00001 to p < 0.0464; Fig. 2a, S8). Interestingly, both networks are connected via RAPGEF3 (Fig. 2a).
Fig. 1.
Histopathological subtypes and ploidy groups. Numbers show differentially expressed protein spots in the statistical pairwise (top) and trend (bottom) tests. DNA histograms show DNA content on the x-axis and the total number of cell on the y-axis
Fig. 2.
Networks of differentially expressed proteins. Normal mucosa and diploid endometrioid carcinoma are compared in a, diploid endometrioid and aneuploid endometrioid carcinoma in b, and diploid endometrioid carcinoma and aneuploid UPSC in c. Red and green designations indicate over- and under-expressed proteins, blue arrows and circulations central nodes of the networks, encircling in yellow LC–MS/MS-verified evaluations, and arrows in purple those proteins that connect different networks
Diploid versus aneuploid endometrioid carcinomas
A total of 37 differentially expressed protein spots were detected, 20 (54%) identified by MALDI-TOF: 17 higher- and three lower-expressed in the aneuploid group (Fig. 1, S9), and ten eligible for IPA analysis interacting in a network (score 25) associated with Cellular Assembly and Organization, Nucleic Acid Metabolism, and Small Molecule Biochemistry. Vimentin, growth factor receptor-bound protein 2 (GRB2) and β-actin are central nodes of this network and associated with diseases and functions regarding cancer, neurological disease, and genetic disorders (p < 0.00001 to p < 0.0486) and cellular growth and proliferation and cellular assembly and organization (p < 0.00001 to p < 0.0382; Fig. 2b, S8).
Diploid endometrioid cancer versus aneuploid UPSC
A total of 41 spots were differentially expressed (p ≤ 0.01), 15 (37%) identified by peptide mass fingerprinting: ten higher- and five lower-expressed in the aneuploid group (Fig. 1, S10), and involved in a network associated with Lipid Metabolism, Small Molecule Biochemistry, and Cell Morphology (score 25). The differentially expressed β-actin, annexin A2 (ANXA2), and the heterogeneous nuclear ribonucleoprotein K isoform A (HNRNPK) are identified as major central nodes of this network and associated with cellular functions of cancer, gastrointestinal disease, and inflammatory response (p < 0.00001 to p < 0.0479; Fig. 2c, S8).
A network comparison between all significant networks mentioned above showed eight proteins in common (β-actin, ATP synthase 5B (ATP5B), ATP5E, insulin, IVNS1ABP, lamin b, PLS1, and vimentin) that connect the network of the normal endometrium versus diploid endometrioid carcinomas with the network of the diploid versus aneuploid endometrioid carcinomas. A total of 16 proteins (β-actin, γ-actin (ACTG1), actin, annexin A2, CAP2, EPS8L1, EPS8L2, GAS8, HIP1R, NCALD, PHACTR1, PLS1, PRS13, PRS18, SSH1, VIL1) associate the diploid versus aneuploid endometrioid carcinomas network with the diploid endometrioid cancer versus aneuploid UPSC network.
Trend analysis of normal endometrium versus diploid endometrioid carcinomas, aneuploid endometrioid carcinomas and aneuploid UPSC
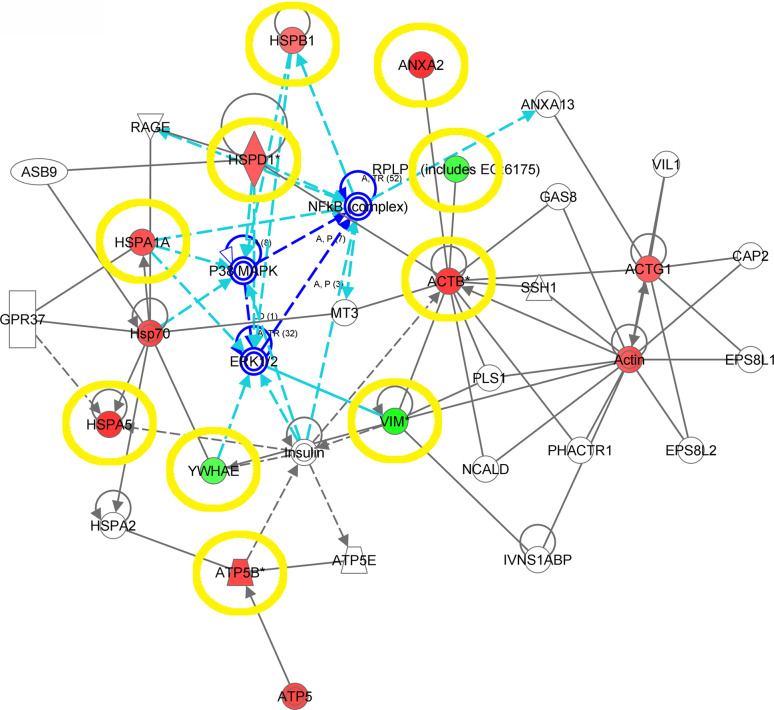
A total of 49 proteins were detected with an increase or decrease over all four groups (p ≤ 0.001), 31 spots (63%) identified, ten down-regulated and 21 up-regulated towards the aneuploid groups (Fig. 1; Table 1). Two overlapping networks interact with Cellular Function and Maintenance, Cellular Compromise and Nucleic Acid Metabolism (score 29); and Lipid Metabolism, Molecular Transport and Small Molecule Biochemistry (score 18). NF-κB, mitogen-activated protein kinase 1/2 (ERK1/2) and p38 mitogen-activated protein kinase (P38MAPK) act as central nodes and were found to be associated with cellular functions regarding cancer, gastrointestinal disease, inflammatory response as well as with cellular function and maintenance (p < 0.00001 to p < 0.0476; Fig. 3, S8).
Table 1.
Identifications of the trend analysis
| Protein name | Accession number | Sequence coverage (%) | Matched/searched peptides | Score | Trend (N-De-Ae-Au) | Fold change (logarithmized) |
|---|---|---|---|---|---|---|
| Lower expression in aneuploid UPSC | ||||||
| Keratin 10a | AAH34697 | 15 | 6/47 | 0.81 | ↓ | 0.73 |
| 14-3-3 ε | NP_006752 | 35 | 11/61 | 0.011 | ↓ | 0.82 |
| Cytokeratin 9a | CAA82315 | 18 | 6/73 | 0.83 | ↓ | 0.51 |
| Vimentin | AAA61279 | 68 | 39/96 | 2.8 × 10−5 | ↓ | 0.85 |
| Annexin A5a | NP_001145 | 56 | 19/96 | 5.3 × 10−4 | ↓ | 0.84 |
| Thioredoxin-like 1 | NP_004777 | 43 | 8/93 | 0.034 | ↓ | 0.82 |
| Vimentin | NP_003371 | 45 | 22/68 | 1.6 × 10−4 | ↓ | 0.64 |
| HSP90 alpha (cytosolic), class Ba | NP_031381 | 28 | 17/57 | 2.7 × 10−4 | ↓ | 0.35 |
| Ribosomal protein P0 | NP_000993 | 48 | 10/34 | 1.7 × 10−5 | ↓ | 0.82 |
| Annexin A4a | NP_001144 | 19 | 5/34 | 0.41 | ↓ | 0.77 |
| Higher expression in aneuploid UPSC | ||||||
| Annexin A5a | NP_001145 | 26 | 8/33 | 0.015 | ↑ | 1.34 |
| ATP-Synthase 5B | NP_001677 | 15 | 5/43 | 0.33 | ↑ | 1.82 |
| HSP70 | CAI18465 | 10 | 3/40 | 0.73 | ↑ | 1.66 |
| β-actin | AAH12854 | 28 | 7/33 | 1.8 × 10−3 | ↑ | 1.61 |
| β-actin | AAH12854 | 28 | 7/33 | 3.7 × 10−4 | ↑ | 1.56 |
| β-actin | AAH12854 | 28 | 7/33 | 9.4 × 10−4 | ↑ | 1.85 |
| HSP90 alpha (cytosolic), class Ba | NP_031381 | 28 | 17/57 | 0.16 | ↑ | 1.44 |
| HSP60 | AAA36022 | 21 | 8/75 | 0.28 | ↑ | 1.64 |
| Cytokeratin 9a | CAA82315 | 18 | 6/73 | 1.3 × 10−3 | ↑ | 1.87 |
| Keratin 10a | AAH34697 | 15 | 6/47 | 0.018 | ↑ | 1.51 |
| ATP-synthase 5B | NP_001677 | 15 | 5/43 | 0.047 | ↑ | 1.45 |
| γ-actin | AAH10417 | 31 | 4/46 | 0.21 | ↑ | 1.51 |
| Prot disulfide-isomerase A6 | NP_005733 | 31 | 9/52 | 7.3 × 10−4 | ↑ | 1.58 |
| HSP60 | AAA36022 | 21 | 8/75 | 0.65 | ↑ | 1.49 |
| HSP70 protein 5 | NP_005338 | 26 | 14/100 | 2.8 × 10−5 | ↑ | 1.90 |
| β-actin | AAH12854 | 28 | 7/33 | 2.0 × 10−7 | ↑ | 1.39 |
| Annexin A2 | AAH09564 | 43 | 15/53 | 2.9 × 10−4 | ↑ | 1.94 |
| Phospholipase C beta 4 isoform | NP_000924 | 10 | 4/39 | 0.64 | ↑ | 1.50 |
| HSP27 | NP_001531 | 25 | 5/43 | 0.74 | ↑ | 1.35 |
| Protein disulfide-isomerase family A member 3a | NP_005304 | 23 | 10/48 | 3.4 × 10−4 | ↑ | 1.71 |
| Annexin A4a | NP_001144 | 52 | 21/64 | 5.9 × 10−6 | ↑ | 1.33 |
aSix proteins (ANXA4, ANXA5, KRT9, KRT10, PDIA3 and HSP90AB1) were identified out of at least two protein spots and showed non-homogenous up- or down regulation between groups due to possible post-translational modification and were thus excluded from all pathway analysis
N normal, De diploid endometrioid, Ae aneuploid endometrioid, Au aneuploid UPSC
Fig. 3.
Network for continuous trends in expression of differentially expressed proteins. Compared are proteins from normal endometrium over diploid endometrioid carcinomas and aneuploid endometrioid carcinomas to aneuploid UPSC. Proteins denoted in red are overexpressed and those in green underexpressed in the UPSC group. Blue arrows and encirclings indicate central nodes of the networks, yellow ovals LC–MS/MS-verified proteins
Confirmation by liquid chromatography/tandem mass spectrometry (LC–MS/MS)
Based on fold changes, pathway analysis and molecular functions, 28 protein identities were selected for confirmation by LC–MS/MS, 14 with a higher and 14 with a lower expression in the aneuploid carcinomas relative to the diploid carcinomas. Nineteen protein identities were then confirmed by LC–MS/MS, while spectra of nine proteins (RAB1B, ARHGDIA, RBP4, CRKL, MPST, ACTG1, PRDX4, HNRNPK, RAB18) showed no identification.
In summary, we could confirm our initial protein identifications by LC-MS/MS for 19 of 28 (68%) proteins. Repeated IPA pathway analysis after exclusion of the non-validated nine protein identities did not change results regarding molecular functions in different ploidy and histology subtypes of tumors.
Discussion
In the present study, we focused on differences between protein profiles in diploid and aneuploid EnCa and normal endometrium. The results provide molecular relationships with network differences in relation to polyploidization.
Evaluation with a 2-DE approach showed 121 distinct protein spots with significant expression differences within all defined subgroups. We observed that aneuploid EnCas are distinct in their 2-DE protein expression profiles from diploid tumors and normal endometrium. We identified 69 polypeptides (57%), representing 41 distinct proteins, the majority connected in networks as determined by pathway analysis. The functions extracted from these networks concerned cellular function and maintenance, cancer, gastrointestinal disease, cell morphology, and small molecule biochemistry, with NF-κB, vimentin, and β-actin as central nodes. The identification of differentially expressed proteins in top pathways in connection of networks with cell functions strongly suggests that these proteins should be submitted to extensive downstream assessment.
Interestingly, we detected only one protein spot that distinguishes aneuploid endometrioid cancer from aneuploid UPSC. Along with the fact of more significant spots between samples of the same histopathology but different ploidy status, we suggest that aneuploidy could be of greater impact regarding poor prognosis than the histological subtype. Interpretations from other data also highlight the importance of aneuploidy for the prognosis [21].
All 69 proteins identified with altered expression changes belong to several functional groups. For instance, the LC–MS/MS-identified β-actin, γ-actin, annexin A2, and ATP synthase 5B concern functions of cellular growth and proliferation and thus reflect different rates of cell division between diploid and aneuploid carcinomas. Another group includes proteins involved in cell-to-cell signaling and interaction, e.g., the serine peptidase inhibitors. A third group, with increased expression levels in the diploid and aneuploid samples was identified as a member of the heat shock protein (HSP) family. HSPs are abundant intracellular polypeptides up-regulated by stress (e.g., temperature, cancer, infection). Their release is considered as a putative danger signal and maintains the structural and metabolic integrity of the cells. Several reports using therapeutic cancer vaccines based on HSPs have been published [22, 23] showing HSP27 overexpression to be a good independent prognostic indicator (p < 0.005) in patients with endometrial adenocarcinoma [24]. Up-regulation of HSP27 has also been described as a potential marker for primary vaginal carcinoma [25] and as immunohistochemical tumor marker in the endometrium [26]. This member of the heat shock family is up-regulated in our comparison between diploid and aneuploid endometrioid tumor samples and was confirmed by LC–MS/MS. It is considered as a putative biomarker for polyploidization, regardless of the histopathological background, and reflects structural and metabolic disintegrity of cells. In addition, up-regulation of HSP27 is associated with aggressive tumor behavior, as shown by the fact that only poorly differentiated tumors overexpress HSP27, and that the level of up-regulation in Barrett’s esophagus is higher in samples presenting lymph node metastasis [27]. This suggests that the higher the level of expression, the more aggressive the tumor. This conclusion is in agreement with results from analyses of also gastric and esophageal squamous cell carcinomas [28, 29] and with our present finding of aneuploid EnCas with increased HSP27 expression.
Yet, another group consists of proteins involved in mechanisms of genetic disorders and maintenance of genomic stability. We found tyrosine 3-monooxygenase/tryptophan-5-monooxygenase activation protein, epsilon (14-3-3-ε) and vimentin to be less expressed in aneuploid samples. Both protein identifications were confirmed by LC–MS/MS. The 14-3-3-ε protein encoded by the YWHAE gene, binds to phosphoserine-containing proteins and mediates signal transduction. For example, 14-3-3 binding is required for the stabilization of active RAF-1 [30] and CDC25-mediated cell cycle control [31], whereas its interaction with BAD (BCL2-associated agonist of cell death) and BAX (BCL2-associated X protein) prevents their proapoptotic release to the mitochondrial membrane [32, 33]. Furthermore, we found NF-κB as a central node in our pathway analysis. This is consistent with other findings linking NF-κB with EnCa [34]. The NF-κB signal transduction pathway plays important roles in the establishment and maintenance of cell phenotypes, through regulation of the expression of many genes. NF-κB has also been shown to regulate the epithelial-mesenchymal transition marker vimentin, which is down-regulated in aneuploid samples [35, 36] and required to maintain cellular integrity [37, 38]. Hence, our finding of higher expression of vimentin in diploid endometrioid carcinomas is in agreement with all data, and down-regulation of vimentin is concluded to be of importance for genomic instability and polyploidization.
Particular attention should be drawn to one protein, thioredoxin-like 1 (TXNL1), now verified by LC–MS/MS. This protein cannot be classified into the groups mentioned above. It has been reported that TXNL1 overexpression can increase the transcriptional repressor function through its binding to the transcription factor B-Myb [39] and that TXNL1 is down-regulated in aneuploid colorectal cancer cell lines and primary colorectal cancer [15]. We therefore now conclude that higher TXNL1 expression might be important to maintain genomic stability.
In conclusion, using a proteomic approach to identify protein signatures of genomic stability/instability irrespective of histological subgroups we have identified 41 distinct polypeptides. Although 2-DE represents a subset analysis of the overall proteome, we were able to deliver a complex map of intact proteins, in relation to molecular networks, that reflect aneuploidy-associated protein expression alterations available for clinical testing. The agreements with the data for other tumors suggest that the differentially expressed proteins follow mechanisms and pathways in common, independent of histopathology or tumor entity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
S2: Clinical parameters of patients included in the study (PDF 109 kb)
S3: Two-dimensional gel electrophoresis (PDF 111 kb)
S4: In-gel digestion and mass spectrometry (MALDI-TOF-MS) (PDF 147 kb)
S5: Confirmation by LC–MS/MS (PDF 107 kb)
S6: PCA map of the protein expression data (PDF 117 kb)
S7: Identifications of the normal endometrium versus diploid endometrioid carcinoma analysis (PDF 164 kb)
S8: Overview of top networks (PDF 110 kb)
S9: Identifications of the diploid versus aneuploid endometrioid carcinoma analysis (PDF 152 kb)
S10: Identifications of the diploid endometrioid cancer versus aneuploid UPSC analysis (PDF 150 kb)
S11: Mass spectrometry peak lists (MALDI-TOF) (PDF 152 kb)
S13: Statistical data for expression analysis (PDF 146 kb)
Acknowledgments
We thank Gunvor Alvelius, Marie Ståhlberg, and Carina Palmberg for mass spectrometric analysis and the Surgical Center for Translational Oncology—Lübeck (SCTO-L) for cooperation. Grants from the Swedish Cancer Society, the Cancer Society Stockholm, the Swedish Research Council, King Gustav V Jubilee Fund, Wallenberg Consortium North, Knut and Alice Wallenberg Foundation, Werner and Clara Kreitz Foundation and the Ad Infinitum Foundation are gratefully acknowledged.
Conflict of interest
All authors declare that they have no competing financial interests.
Abbreviations
- 2-DE
Two-dimensional gel electrophoresis
- Ae
Aneuploid endometrioid
- Ambic
Ammoniumbicarbonate
- Au
Aneuploid uterine papillary serous cancer
- CGH
Comparative genomic hybridization
- De
Diploid endometrioid
- EnCa
Endometrial cancer
- FIGO
Fédération Internationale de Gynécologie et d’Obstétrique
- IPA
Ingenuity Pathway Analysis
- LC–MS/MS
Liquid chromatography and tandem mass spectrometry
- MALDI-TOF-MS
Matrix-assisted laser desorption/ionization time of flight mass spectrometry
- PCA
Principal component analysis
- UPSC
Uterine papillary serous cancer
References
- 1.Creasman WT, Odicino F, Maisonneuve P, Beller U, Benedet JL, Heintz AP, Ngan HY, Sideri M, Pecorelli S. Carcinoma of the corpus uteri. J Epidemiol Biostat. 2001;6:47–86. [PubMed] [Google Scholar]
- 2.Fox H. Ploidy in gynaecological cancers. Histopathology. 2005;46:121–129. doi: 10.1111/j.1365-2559.2005.02087.x. [DOI] [PubMed] [Google Scholar]
- 3.Zaino RJ, Kurman RJ, Diana KL, Morrow CP. The utility of the revised International Federation of Gynecology and Obstetrics histologic grading of endometrial adenocarcinoma using a defined nuclear grading system. A Gynecologic Oncology Group study. Cancer. 1995;75:81–86. doi: 10.1002/1097-0142(19950101)75:1<81::AID-CNCR2820750114>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Lundgren C, Auer G, Frankendal B, Moberger B, Nilsson B, Nordstrom B. Nuclear DNA content, proliferative activity, and p53 expression related to clinical and histopathologic features in endometrial carcinoma. Int J Gynecol Cancer. 2002;12:110–118. doi: 10.1046/j.1525-1438.2002.01079.x. [DOI] [PubMed] [Google Scholar]
- 5.Auer GU, Caspersson TO, Wallgren AS. DNA content and survival in mammary carcinoma. Anal quant Cytol. 1980;2:161–165. [PubMed] [Google Scholar]
- 6.Gerling M, Meyer KF, Fuchs K, Igl BW, Fritzsche B, Ziegler A, Bader F, Kujath P, Schimmelpenning H, Bruch HP, Roblick UJ, Habermann JK (2010) High frequency of aneuploidy defines ulcerative colitis-associated carcinomas: a comparative prognostic study to sporadic colorectal carcinomas. Ann Surg [Epub ahead of print] [DOI] [PubMed]
- 7.Kronenwett U, Huwendiek S, Ostring C, Portwood N, Roblick UJ, Pawitan Y, Alaiya A, Sennerstam R, Zetterberg A, Auer G. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64:904–909. doi: 10.1158/0008-5472.CAN-03-2451. [DOI] [PubMed] [Google Scholar]
- 8.Habermann JK, Doering J, Hautaniemi S, Roblick UJ, Bundgen NK, Nicorici D, Kronenwett U, Rathnagiriswaran S, Mettu RK, Ma Y, Kruger S, Bruch HP, Auer G, Guo NL, Ried T. The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int J Cancer. 2009;124:1552–1564. doi: 10.1002/ijc.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habermann JK, Hellman K, Freitag S, Heselmeyer-Haddad K, Hellstrom AC, Shah K, Auer G, Ried T. A recurrent gain of chromosome arm 3q in primary squamous carcinoma of the vagina. Cancer Genet Cytogenet. 2004;148:7–13. doi: 10.1016/S0165-4608(03)00245-0. [DOI] [PubMed] [Google Scholar]
- 10.Pere H, Tapper J, Wahlstrom T, Knuutila S, Butzow R. Distinct chromosomal imbalances in uterine serous and endometrioid carcinomas. Cancer Res. 1998;58:892–895. [PubMed] [Google Scholar]
- 11.Muresu R, Sini MC, Cossu A, Tore S, Baldinu P, Manca A, Pisano M, Loddo C, Dessole S, Pintus A, Tanda F, Palmieri G. Chromosomal abnormalities and microsatellite instability in sporadic endometrial cancer. Eur J Cancer. 2002;38:1802–1809. doi: 10.1016/S0959-8049(02)00152-1. [DOI] [PubMed] [Google Scholar]
- 12.Muslumanoglu HM, Oner U, Ozalp S, Acikalin MF, Yalcin OT, Ozdemir M, Artan S. Genetic imbalances in endometrial hyperplasia and endometrioid carcinoma detected by comparative genomic hybridization. Eur J Obstet Gynecol Reprod Biol. 2005;120:107–114. doi: 10.1016/j.ejogrb.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 13.Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habermann JK, Paulsen U, Roblick UJ, Upender MB, McShane LM, Korn EL, Wangsa D, Kruger S, Duchrow M, Bruch HP, Auer G, Ried T. Stage-specific alterations of the genome, transcriptome, and proteome during colorectal carcinogenesis. Genes Chromosom Cancer. 2007;46:10–26. doi: 10.1002/gcc.20382. [DOI] [PubMed] [Google Scholar]
- 15.Gemoll T, Roblick UJ, Szymczak S, Braunschweig T, Becker S, Igl BW, Bruch HP, Ziegler A, Hellman U, Difilippantonio MJ, Ried T, Jornvall H, Auer G, Habermann JK (2011) HDAC2 and TXNL1 distinguish aneuploid from diploid colorectal cancers. Cell Mol Life Sci [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 16.Roblick UJ, Hirschberg D, Habermann JK, Palmberg C, Becker S, Kruger S, Gustafsson M, Bruch HP, Franzen B, Ried T, Bergmann T, Auer G, Jornvall H. Sequential proteome alterations during genesis and progression of colon cancer. Cell Mol Life Sci. 2004;61:1246–1255. doi: 10.1007/s00018-004-4049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shepherd JH. Revised FIGO staging for gynaecological cancer. Br J Obstet Gynaecol. 1989;96:889–892. doi: 10.1111/j.1471-0528.1989.tb03341.x. [DOI] [PubMed] [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Welch BL. On the comparison of several mean values: an alternative approach. Biometrika. 1951;38:330–336. [Google Scholar]
- 20.Hellman U. Sample preparation by SDS/PAGE and in-gel digestion. EXS. 2000;88:43–54. doi: 10.1007/978-3-0348-8458-7_3. [DOI] [PubMed] [Google Scholar]
- 21.Lundgren C, Auer G, Frankendal B, Nilsson B, Nordstrom B. Prognostic factors in surgical stage I endometrial carcinoma. Acta Oncol. 2004;43:49–56. doi: 10.1080/02841860310018990. [DOI] [PubMed] [Google Scholar]
- 22.Vanaja DK, Grossmann ME, Celis E, Young CY. Tumor prevention and antitumor immunity with heat shock protein 70 induced by 15-deoxy-delta12, 14-prostaglandin J2 in transgenic adenocarcinoma of mouse prostate cells. Cancer Res. 2000;60:4714–4718. [PubMed] [Google Scholar]
- 23.Murshid A, Gong J, Calderwood SK. Heat-shock proteins in cancer vaccines: agents of antigen cross-presentation. Expert Rev Vaccines. 2008;7:1019–1030. doi: 10.1586/14760584.7.7.1019. [DOI] [PubMed] [Google Scholar]
- 24.Geisler JP, Geisler HE, Tammela J, Miller GA, Wiemann MC, Zhou Z. A study of heat shock protein 27 in endometrial carcinoma. Gynecol Oncol. 1999;72:347–350. doi: 10.1006/gyno.1998.5283. [DOI] [PubMed] [Google Scholar]
- 25.Hellman K, Silfversward C, Nilsson B, Hellstrom AC, Frankendal B, Pettersson F. Primary carcinoma of the vagina: factors influencing the age at diagnosis. The Radiumhemmet series 1956–96. Int J Gynecol Cancer. 2004;14:491–501. doi: 10.1111/j.1048-891x.2004.014310.x. [DOI] [PubMed] [Google Scholar]
- 26.Ioachin E. Immunohistochemical tumour markers in endometrial carcinoma. Eur J Gynaecol Oncol. 2005;26:363–371. [PubMed] [Google Scholar]
- 27.Lemieux P, Oesterreich S, Lawrence JA, Steeg PS, Hilsenbeck SG, Harvey JM, Fuqua SA. The small heat shock protein hsp27 increases invasiveness but decreases motility of breast cancer cells. Invasion Metastasis. 1997;17:113–123. [PubMed] [Google Scholar]
- 28.Doak SH, Jenkins GJ, Parry EM, Griffiths AP, Baxter JN, Parry JM. Differential expression of the MAD2, BUB1 and HSP27 genes in Barrett’s oesophagus-their association with aneuploidy and neoplastic progression. Mutat Res. 2004;547:133–144. doi: 10.1016/j.mrfmmm.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Kapranos N, Kominea A, Konstantinopoulos PA, Savva S, Artelaris S, Vandoros G, Sotiropoulou-Bonikou G, Papavassiliou AG. Expression of the 27-kDa heat shock protein (HSP27) in gastric carcinomas and adjacent normal, metaplastic, and dysplastic gastric mucosa, and its prognostic significance. J Cancer Res Clin Oncol. 2002;128:426–432. doi: 10.1007/s00432-002-0357-y. [DOI] [PubMed] [Google Scholar]
- 30.Roy S, McPherson RA, Apolloni A, Yan J, Lane A, Clyde-Smith J, Hancock JF. 14-3-3 facilitates Ras-dependent Raf-1 activation in vitro and in vivo. Mol Cell Biol. 1998;18:3947–3955. doi: 10.1128/mcb.18.7.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorson JA, Yu LW, Hsu AL, Shih NY, Graves PR, Tanner JW, Allen PM, Piwnica-Worms H, Shaw AS. 14-3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol Cell Biol. 1998;18:5229–5238. doi: 10.1128/mcb.18.9.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Won J, Kim DY, La M, Kim D, Meadows GG, Joe CO. Cleavage of 14-3-3 protein by caspase-3 facilitates bad interaction with Bcl-x(L) during apoptosis. J Biol Chem. 2003;278:19347–19351. doi: 10.1074/jbc.M213098200. [DOI] [PubMed] [Google Scholar]
- 33.Nomura M, Shimizu S, Sugiyama T, Narita M, Ito T, Matsuda H, Tsujimoto Y. 14-3-3 Interacts directly with and negatively regulates pro-apoptotic Bax. J Biol Chem. 2003;278:2058–2065. doi: 10.1074/jbc.M207880200. [DOI] [PubMed] [Google Scholar]
- 34.Pallares J, Martinez-Guitarte JL, Dolcet X, Llobet D, Rue M, Palacios J, Prat J, Matias-Guiu X. Abnormalities in the NF-kappaB family and related proteins in endometrial carcinoma. J Pathol. 2004;204:569–577. doi: 10.1002/path.1666. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/S0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 36.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nisolle M, Casanas-Roux F, Donnez J. Coexpression of cytokeratin and vimentin in eutopic endometrium and endometriosis throughout the menstrual cycle: evaluation by a computerized method. Fertil Steril. 1995;64:69–75. [PubMed] [Google Scholar]
- 38.Goldman RD, Khuon S, Chou YH, Opal P, Steinert PM. The function of intermediate filaments in cell shape and cytoskeletal integrity. J Cell Biol. 1996;134:971–983. doi: 10.1083/jcb.134.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim KY, Lee JW, Park MS, Jung MH, Jeon GA, Nam MJ. Expression of a thioredoxin-related protein-1 is induced by prostaglandin E(2) Int J Cancer. 2006;118:1670–1679. doi: 10.1002/ijc.21572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S2: Clinical parameters of patients included in the study (PDF 109 kb)
S3: Two-dimensional gel electrophoresis (PDF 111 kb)
S4: In-gel digestion and mass spectrometry (MALDI-TOF-MS) (PDF 147 kb)
S5: Confirmation by LC–MS/MS (PDF 107 kb)
S6: PCA map of the protein expression data (PDF 117 kb)
S7: Identifications of the normal endometrium versus diploid endometrioid carcinoma analysis (PDF 164 kb)
S8: Overview of top networks (PDF 110 kb)
S9: Identifications of the diploid versus aneuploid endometrioid carcinoma analysis (PDF 152 kb)
S10: Identifications of the diploid endometrioid cancer versus aneuploid UPSC analysis (PDF 150 kb)
S11: Mass spectrometry peak lists (MALDI-TOF) (PDF 152 kb)
S13: Statistical data for expression analysis (PDF 146 kb)