Abstract
Host defense peptides and proteins are important components of the innate host defense against pathogenic microorganisms. They target negatively charged bacterial surfaces and disrupt microbial cytoplasmic membranes, which ultimately leads to bacterial destruction. Throughout evolution, pathogens devised several mechanisms to protect themselves from deleterious damage of host defense peptides. These strategies include (a) inactivation and cleavage of host defense peptides by production of host defense binding proteins and proteases, (b) repulsion of the peptides by alteration of pathogen’s surface charge employing modifications by amino acids or amino sugars of anionic molecules (e.g., teichoic acids, lipid A and phospholipids), (c) alteration of bacterial membrane fluidity, and (d) expulsion of the peptides using multi drug pumps. Together with bacterial regulatory network(s) that regulate expression and activity of these mechanisms, they represent attractive targets for development of novel antibacterials.
Keywords: Pathogens, Cationic peptides, Teichoic acids, Lipid A, D-alanylation, Aminoarabinose, Ethanolamine, Multidrug efflux pumps, Proteases
Introduction
In order to survive on and within the host, bacterial pathogens have evolved numerous mechanisms to combat host immune system. Among these are secreted molecules that interfere with recognition of bacterial pathogens by host immune system and pathogen-associated components that make the invader more resistant to the arsenal of host antimicrobial molecules such as lysoyzme, group IIA phospholipase A2, and small cationic antimicrobial peptides (CAMPs) [1]. Bacterial resistance mechanisms to the latter will be the focus of this brief.
Infection is typically initiated when a breach of the skin or mucosal barriers allows bacterial pathogen access to adjoining tissues or the bloodstream. Whether an infection is contained or spreads depends on a complex interplay between the pathogen’s virulence determinants and host defense mechanisms. The effective host immune response is based on quick recognition, isolation, and elimination of the pathogen. Innate immunity is the first line of defense against the invading microbial pathogens [1, 2]. The swift action of cellular (e.g., phagocytes) and secreted compounds (e.g., CAMPs) of innate immunity is in part due to the ability of innate immunity to recognize invariant structures on pathogens, so-called pathogen-associated molecular patterns or PAMPs [3]. While the killing by phagocytes occurs by combined action of reactive oxygen species and CAMPs, CAMPs are also secreted by various types of epithelial cells and therefore represent an important component of host defense on their own [1, 4, 5]. Many CAMPs have additional immunomodulatory roles that contribute to antimicrobial host defense [6].
Host defense peptides and their role in innate immune defense
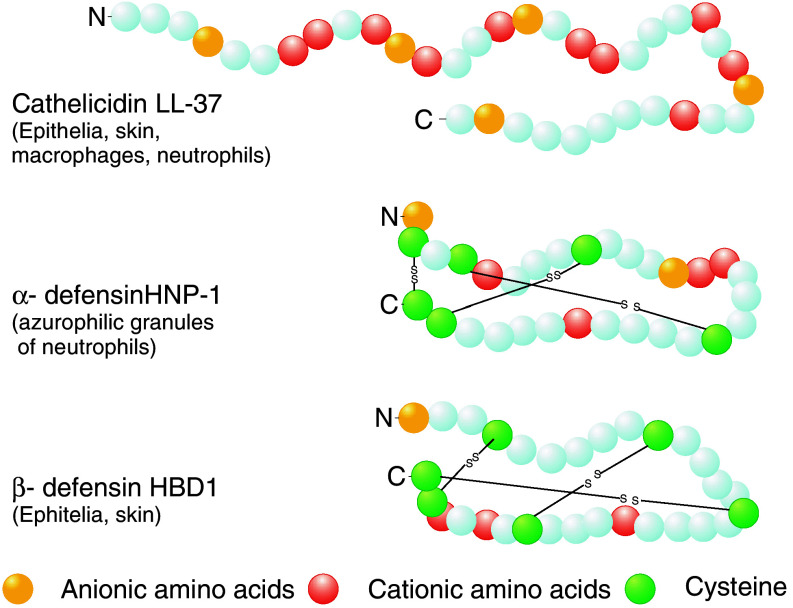
CAMPs represent an evolutionary ancient part of immune response that is found in all kingdoms of life, from bacteria to plants and animals [5, 7]. These are small (12–50 amino acids) hydrophobic molecules that usually have a net positive charge of +2 or more and display a broad spectrum antimicrobial activity against bacteria as well as fungi [5, 8]. In humans, CAMPs can be placed in three distinct groups based on their peptide structure, amino acid composition, and number of disulfide bonds [5, 9]. Cathelicidins are CAMPs with α-helical structure, which do not contain cysteine residues. The only human representative of cathelicidin is LL-37 (see Fig. 1) [9]. Defensins have a β-sheet structure that is stabilized by three disulphide bridges. The pattern of disulphide bridges determines whether defensins belong to the group of α- or β-defensins. Defensins belonging to the two groups also differ by which cell type they are produced. While α-defensins [e.g., human neutrophil peptides (HNPs)] are mainly produced by neutrophil granulocytes and the intestinal Paneth cells, β-defensins are produced by epithelial tissues such as skin and epithelia of gastrointestinal, respiratory, and genitourinary tracts (Fig. 1) [5, 8, 10–12]. Kinocidins are the third group of CAMPs related to chemokines that are released from platelets upon contact with bacteria (thrombocidins) or other cells [13, 14]. Thrombocidins have been shown to participate in endovascular infections, e.g., in elimination of bacteria in endocardial vegetations [15–17]. In addition to the three above-mentioned major groups of CAMPs, further human antimicrobial peptides have been described including anionic dermcidins produced in human sweat, the iron-regulatory hormone hepcidin [18], and split products form human proteins such as α-melanocyte-stimulating hormone [19].
Fig. 1.
Structure of selected human cationic antimicrobial peptides. The only representative of cathelicidins in humans is α-helical LL-37. S S Disulphide bonds between cysteines. HNP-1 human neutrophil peptide 1. HBD-1 human β-defensin 1. Modified from [121]
There are several proposed mechanisms of how CAMPs kill bacteria. Whether CAMPs induce pore formation or disrupt the membrane in any other way is still a matter of debate and may be different for different CAMPs. The modes of action may include cytoplasmic leakage and additional antibacterial mechanisms such as inhibition of membrane-bound cell wall biosynthetic steps (e.g., interaction of human-β defensin-3 (HBD3) and plectasin with lipidII, or binding of friulimicin B to bactoprenol phosphate carrier C55–P) [20–22], all ultimately leading to death of bacteria [7, 23, 24]. CAMPs mechanism of action can be described in at least three steps [5, 25, 26]. In order to reach the bacterial membrane, CAMPs need to initially bind to the bacterial cell surface and traverse the bacterial cell wall. The third step is insertion of CAMPs in the bacterial membrane that ultimately leads to cell membrane disruption and cell death [5, 8, 23]. The cationic nature of CAMP allows the peptides to distinguish between largely neutrally charged host membranes and mostly anionic bacterial membranes. Besides bacterial membrane damage, CAMPs can assist in killing of microorganisms by modulating host immune responses. To this end, CAMPs can bind lipopolysaccharides (LPS), the outer leaflet of the outer membrane of all Gram-negative bacteria with high proinflammatory capacity, to dampen host inflammatory response [27, 28]. In addition, many CAMPs can act as chemokines, can induce apoptosis, or play a role in activation of autoimmune response [7, 28, 29]. CAMP-derived synthetic peptides lacking antimicrobial activity have been shown to modulate immune responses and contribute to the clearance of infection [30], which underscores the importance of immunomodulatory CAMP activities.
How do bacteria defend against host defense peptides?
Extracellular mechanisms
Bacteria can defend themselves against host defense peptides using several different strategies. Some pathogens produce extracellular molecules that bind and trap CAMPs and thereby prevent their antimicrobial action. An example of such a protein is Staphylococcus aureus (S. aureus) staphylokinase. Besides binding of plasminogen that may facilitate S. aureus invasion of tissues, staphylokinase also forms complexes with α-defensins (HNP 1-3) resulting in over 80% reduced activity of these CAMPs against S. aureus [31]. Streptococcal inhibitor of complement (SIC) closely resembles the protective action of staphylokinase against the CAMPs. Even though SIC has been initially discovered as a complement-binding protein, Pence et al. [32] recently found that SIC-deficient Streptococcus pyogenes is more sensitive to LL-37. It has been proposed that SIC protects S. pyogenes by direct binding and inactivation of CAMPs [33].
Alternatively, bacteria can intercept host defense peptides before they reach their target—the bacterial cytoplasmic membrane—using cell envelope-associated or secreted proteases. Several pathogenic microbes use proteases that cleave CAMPs, thereby abolishing their antibacterial activity. Examples of such secreted proteases can be found in S. aureus (e.g., V8 and aureolysin proteases [34]), Proteus mirabilis (ZapA [35]), S. pyogenes (streptopain SpeB [35]), or Pseudomonas aeruginosa (elastase [35]). Alternatively, CAMP-degrading proteases can also be associated with the bacterial cell envelope either anchored in the cell wall or in the outer membrane such as PgtE of Salmonella enterica [36] and E. coli OmpT [37]. It has been suggested that CAMP evolution led to introduction of multiple cysteine bridges, resulting in a form that is substantially more resistant to proteolysis [7].
Modifications of cell surface
Synthesis of capsular polysaccharides
To protect themselves from host immune response including host defense peptides, bacteria can encase themselves by producing elaborate extracellular matrices. S. epidermidis, for example, produces cationic exopolymer polysaccharide intercellular adhesin (PIA) and anionic poly-γ-glutamic acid (PGA), which were both shown to play a role in resistance to cationic LL-37 and HBD3 as well as anionic defense peptide dermcidin [38–40]. Because these polysaccharide matrices protect bacteria from both cationic and anionic host defense peptides, the resistance mechanism probably involves both electrostatic and mechanical (charge-independent) sequestration of host defense peptides, far from their ultimate target—bacterial cytoplasmic membrane.
Role of teichoic acids (TA) in resistance to host defense peptides
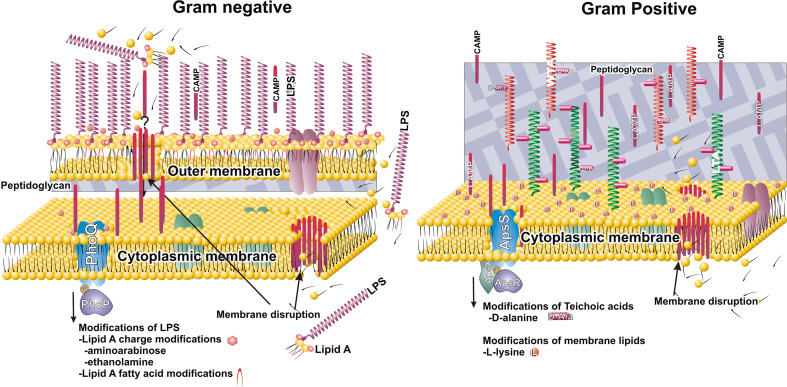
Host evolution of defense peptides led to selection of mostly positively charged compounds. For measuring bactericidal effects of defense peptides, it seems that the rule of ‘the more cationic the better’ may be applied [5, 41, 42]. Net positive charge may be beneficial in targeting of bacterial surfaces that are generally negatively charged. Molecules that contribute to the net negativity of bacterial surface include peptidoglycan, anionic polymers such as TA, teichuronic acid, lipopolysaccharide (LPS) and anionic phospholipids in the bacterial cytoplasmic membrane. In the last decade, substantial advances in uncovering novel bacterial resistance mechanism that reduce the net negative charge of bacterial surfaces have been shown to confer resistance to CAMPs. These mechanisms differ between Gram-positive and Gram-negative bacteria due to the different composition of their cell surfaces (see Fig. 2). The envelope of Gram-negative bacteria consists of a thin layer of peptidoglycan and an extra outer membrane with inner phospholipid layer and outer LPS layer [43]. In contrast, Gram-positive bacteria lack the outer membrane, but have a thick, multiple-peptidoglycan layer cell wall. Analogous to LPS, the cell envelope of Gram-positive bacteria also contains various glyco- or glycolipid polymers, namely TA, which are attached to either peptidoglycan (wall teichoic acid, WTA) or cell membrane (lipoteichoic acid, LTA) [44, 45]. Both LPS and teichoic acids are negatively charged and contribute to the net negativity of bacterial surface [43–46].
Fig. 2.
Sensing and responding to CAMPs in Gram-negative and Gram-positive bacteria. Initial binding of CAMPs is dependent on electrostatic interactions between cationic surface of CAMPs and negatively charged moieties in bacterial cell envelope (lipid A and teichoic acids: WTA, wall teichoic acid, LTA, lipoteichoic acid). Before disruption of the cytoplasmic membrane, CAMPs need to penetrate the outer membrane (Gram-negative bacteria) or cell wall composed of thick layers of peptidoglycan (Gram-positive bacteria). PhoQ and ApsS are sensor histidine kinases that respond to CAMPs or CAMPs-mediated membrane perturbation. PhoP and ApsR are transcriptional regulators that upregulate expression of CAMPs resistance genes. Substitutions of lipid A by aminoarabinose and ethanolamine, modification of phosphatidylglycerol by l-lysine, and d-alanine modification of WTA and LTA, act to neutralize the net negative charge of bacterial cell envelope. In addition, increased resistance is achieved by changing membrane fluidity (e.g., by changes of fatty acid acylation pattern of lipid A)
Structurally, WTA and LTA differ depending on bacterial species. For example in S. aureus, they consist of repeating units of ribitol and glycerol phosphates, respectively [47, 48]. The length of the glycerol phosphate polymers of LTA can exceed 40 U [49], extending through the peptidoglycan layers. Together, LTA and WTA form a negatively charged lattice that bridges bacterial cell membrane and cell wall. The biochemical role of TA in bacterial physiology has been a focus of many studies. TA have been implicated in maintaining proton gradient, cationic homeostasis in bacterial cell wall, regulating enzymatic activity of autolysins and assembly of division site [50]. The biochemical properties of TA have been tightly linked to the substitutions of TA by the amino acid d-alanine. d-alanine’s positively charged free amino group partially influences the net negativity of TA [44, 50, 51]. In turn, d-alanyl esters of TA modulate functions of TA that significantly increase resistance of Gram-positive bacteria to CAMPs and proteins [52–58].
d-Alanylation of TA
At least four proteins, encoded by a single operon dltABCD, are necessary for d-alanylation of both WTA and LTA. d-alanyl carrier protein ligase (Dcl; dltA) activates d-alanine using ATP. With assistance of DltD (dltD), this activated complex is delivered to the d-alanine carrier protein (Dcp) encoded by dltC. DltB (dltB) is predicted to be a transmembrane protein and is thought to be involved in passage of the d-alanyl-Dcp complex across the cytoplasmic membrane, where d-alanine is transferred to the glycerol phosphate backbone of LTA and WTA [44]. The glycerol phosphate backbone is derived from phosphatidyl-glycerol (PG), the major phospholipid on growing S. aureus. Transfer of d-alanine most likely occurs at the base of the growing chain of LTA and WTA [59]. An additional open reading frame (“dltX”) present within the 5′ end of the dlt operon of S. aureus [60] and other Gram-positive bacteria has been recently identified, however, its function is not yet fully understood.
Inactivation of any of dltA–D genes results in complete loss of d-alanine esters from TA [52, 61–63]. Such dlt mutants have been created in numerous Gram-positive organisms including S. aureus, Streptococcal species, Listeria, Bacillus, and Enterococcus [52–58, 64]. In vitro studies have shown that dltA S. aureus is more sensitive to host cationic peptides (defensins) and proteins, especially group IIA PLA2 [41, 52] and is defective in adherence to artificial surfaces and formation of biofilms [65]. In addition, this mutant strain is virulence-attenuated in several animal models [53, 54, 66, 67]. Similar phenotypes have been observed in other bacterial species (see Table 1 for details). Of note, d-alanylation may also regulate pro-inflammatory activity of LTA [68], which may be an important determinant in establishing persistent infections (e.g., biofilms) and colonization of special niches. d-alanylation of TA in Lactobacillus have been show to be crucial for establishing successful colonization of the gastrointestinal tract [69].
Table 1.
Summary of bacterial resistance mechanisms to CAMPs
| Mechanism of resistance | Product name/gene | Organism | Virulence of the mutant | Reference |
|---|---|---|---|---|
| CAMP binding and inactivation | Staphylokinase | S. aureus | ↑ systemic infection | [122, 123] |
| SIC | S. pyogenes | ↓ systemic infection | [32] | |
| CAMP proteolytic cleavage | V8 protease | S. aureus | ND | [34] |
| Aureolysin | S. aureus | ND | [34] | |
| ZapA | P. mirabilis | ↓ colonization of urinary tract | [35, 124] | |
| Streptopain SpeB | S. pyogenes | ND | [35, 125] | |
| Elastase LasA | P. aeruginosa | ↓ chronic infection in rat | [35, 126, 127] | |
| Capsular polysaccharides | PIA, PGA | S. epidermidis | ↓virulence | [38–40, 128, 129] |
| Capsule synthesis gene cluster | N. meningitidis | ↓virulence | [130–132] | |
| K. pneumoniae | ↓ virulence in murine model of pneumonia | [133–135] | ||
| Surface charge neutralization | S. aureus |
↓ virulence in mouse, ↓ virulence in rabbit model of endocarditis |
[53, 54, 136] | |
| d-alanine modification of teichoic acids | dltABCD | S. pyogenes | ND | [58, 137] |
| L. monocytogenes | ↓ mouse model of infection | [55] | ||
| L. reuteri | ↓ colonization of mouse | [69] | ||
| l-Lysine modification of PG | mprF | S. aureus | ↓ virulence in mouse model of infection | [53, 85, 138] |
| ↓ virulence rabbit model of endocarditis | ||||
| Lipid A modifications Aminoarabinose Ethanolamine | pmrAB, pmrE, pmrFHIJKL pmrC, | S. enterica ser. typhimurium | ↓ virulence in mice | [98, 100, 139, 140] |
| Decrease in membrane fluidity | ||||
| Modification of lipid A acylation | pagP, pagL, | S. enterica ser. typhimurium | No phenotype | [141, 142] |
| Production of carotenoids | crtOPQMN | S. aureus | ↓virulence in subcutaneous abscess model | [143–145] |
| Presence of efflux pumps | qacA | S. aureus | ND | [16, 119, 146] |
| CAMP expulsion | mtrCDE | N. gonorrhoeae | ↓ virulence in murine model of infection | [117, 147, 148] |
↑ increased virulence, ↓ decreased virulence or virulence attenuated, ND not determined
The mechanism of d-alanyl-TA resistance to cationic peptides
CAMPs killing action is accomplished in three steps: (1) binding of CAMPs to the cell wall that depends on electrostatic interaction between cationic surfaces of defense peptides and negatively charged bacterial surfaces, (2) penetration of the cell wall, and, (3) insertion of CAMPs in the cell membrane leading to membrane disruption [4, 5]. In the absence of d-alanine esters, increased binding of CAMPs has been observed, suggesting that initial binding of CAMPs may be the critical step controlled by d-alanine esters [52]. However, additional binding sites for CAMPs may be revealed secondary to CAMPs’ inflicted cell damage that may mask the additional effect of d-alanine esters on CAMP action. Additional insights on CAMPs action may be gained from studies of another cationic protein that is part of mammalian innate immune response, human group IIA phospholipase A2 (gIIA PLA2). Antibacterial action of gIIA PLA2 is besides enzymatic degradation of phospholipids closely similar to the smaller CAMPs, but each of the steps can be controlled by manipulation of the presence of Ca2+ in the medium [41, 70, 71]. The results of these studies suggest that lack of d-alanine esters in TA may promote the penetration and release of autolytic enzymes that enhance the activity of cell wall bound gIIA PLA2 [41].
Regulation of d-alanylation of TA
Environmental factors such as pH, temperature, and salt (e.g., NaCl) concentration [72–74] are known to affect the degree to which TA are substituted by d-alanines. For example, the degree of d-alanylation of LTA is 0.77 mol d-alanine/mol glycerol phosphate when bacteria are grown under low-salt conditions (0.2% NaCl), but decreased to only 0.3 mol d-alanine/mol glycerol phosphate when bacteria are grown in medium with much higher salt (7.5% NaCl) concentration [75]. d-alanylation of TA could be further modulated by regulating the activity of the proteins encoded by dlt operon [76] or their abundance by regulation of transcription of dlt genes.
In Bacillus subtilis, the dlt operon is part of the σx regulon and is regulated by the global transcriptional regulators AbrB and Spo0A [61]. In Streptococcus agalactiae, a two-component system, dltRS, that is part of the dlt operon, is presumably involved in transcriptional regulation of dlt expression [77]. In S. aureus, transcription profiling studies have demonstrated increased dlt mRNA in an accessory gene regulator (agr) mutant and decreased dlt mRNA in a rot (repressor of toxins) mutant suggesting a role for the agr and rot global regulators in negative and positive regulation of dlt, respectively [78, 79]. In addition, S. aureus represses dlt transcription in response to increased concentration of mono and divalent cations that is in part dependent on ArlRS two-component system [60]. Most recently, studies aimed to identify genome-wide responses of S. epidermidis to HBD3 revealed a novel three-component system ApsRSX also termed GraRSX in S. aureus [80–84]. ApsRSX is an unusual three components system, composed of the classical combination of sensor histidine kinase and response regulator plus an additional protein, the function of which has yet to be determined. Most significantly, ApsS sensor kinase has been shown to be activated in the presence of CAMPs, in turn upregulating expression of resistance genes against CAMPs: dlt operon, mprF for modification of phospholipids involved in resistance to CAMPs [85] (see Fig. 2 and below), and vraFG operon encoding a transporter conferring resistance to glycopeptide vancomycin [86].
Modification of bacterial cell membrane
Charge neutralization
The ultimate step in CAMPs is integration into bacterial membrane leading to bacterial destruction [5]. This final step has been shown to be driven by electrostatic interactions between cationic defense peptides and anionic lipids in bacterial cell membrane [24, 87]. Neutralization mechanisms similar to charge compensation of anionic TA by d-alanine described above have also been described for bacterial membrane lipids [53, 82, 85]. In many Gram-positive and Gram-negative pathogens, phosphatidylglycerol (PG), a major component of bacterial cell membrane, is modified by positively charged amino acid lysine. The synthesis of lysyl-PG is dependent on the MprF protein [85]. MprF is highly conserved in Gram-positive and Gram-negative pathogens and is a 97-kDa integral membrane protein composed of two functional domains. A C-terminal domain is responsible for synthesis of lysyl-PG at the inner leaflet of cytoplasmic membrane using PG and lysyl-tRNA as substrates [85, 88, 89], while the N-terminal—flippase domain—is responsible for translocation of lysyl-PG to the outer leaflet of cytoplasmic membrane [89]. In addition to increased sensitivity to CAMPs, mprF mutants are more sensitive to killing by neutrophils and virulence-attenuated in multiple animal models (Table 1) [53, 85]. MprF has also been shown to be under control of ApsRSX regulator [81, 83]. Increased lysyl-PG content and point mutation resulting in MprF gain of function have been recently reported to increase resistance to daptomycin in vitro and in vivo [90–92]. Daptomycin is an anionic antibiotic but has CAMP-like properties in the presence of calcium ions [93] and is used as one of the last resort drugs in treatment of multiple drug resistant S. aureus (MRSA). In addition, mprF affects sensitivity to several other antibiotics such as gentamicin, vancomycin, and moenomycin [94–96].
Gram-negative bacteria can repel CAMPs by regulating anionic surface charge. Lipid A, the anionic component of LPS, consists of two glucosamine units with free phosphate groups linked to four or more acyl chains [43]. Salmonella can modify lipid A and LPS core sugars by incorporation of aminoarabinose or phosphoethanol amine (Fig. 2), leading to reduction in the net negative charge of lipid A and increased resistance to the CAMP polymyxin B [97–99]. The genes responsible for decoration of lipid A by aminoarabinose (pmrEHFIJKL) and ethanolamine (pmrC) are under control of PmrAB two-component system. The activity of PmrA transcriptional regulator is controlled by two different systems: (a) a sensor kinase PmrB that senses iron, zinc and mild acidic conditions, and (b) PhoPQ two-component system that regulates PmrA activity through PmrD protein [97, 100].
Changes in membrane fluidity
Salmonella typhimurium PhoPQ is one of the most studied bacterial two-component systems that controls several groups of genes important for bacterial survival in the host [101–103]. PhoPQ is a Mg2+ sensor, responds to changes in pH [104–106] and to the presence of CAMPs [107]. Crystal structure of PhoQ sensor kinase has revealed a patch of acidic amino acids that are responsible for binding of divalent cations and CAMPs (Fig. 2) [107]. PhoP has been shown to directly or indirectly influence expression >100 genes. Among them are Mg2+ transporters (MgtA and MgtCB), SlyA, which regulates genes important for survival inside macrophages, RpoS that regulates genes important for resistance to oxidative stress, and PmrAB, a two-component system that regulates genes for modifications of LPS by amino arabinose [104, 108, 109]. One of the PhoPQ-mediated responses is also upregulation of pagP that is responsible for additional acylation of the lipid A and pagL that is involved in deacylation of lipid A (Fig. 2). Such modified acylation of lipid A in turn reduces fluidity and permeability of the bacterial outer membrane and renders bacteria more resistant to CAMPs [36, 110, 111]. PhoPQ homologs can be found in many Gram-negative pathogens including Shigella flexneri, Yersinia pestis, and P. aeruginosa, suggesting this two-component system and its function may be one of the evolutionary adaptations to pathogen life within the host [112].
Changes in membrane fluidity have also been shown to influence CAMP resistance of Gram-positive organisms. For example, incorporation of longer chain unsaturated fatty acids in membrane lipids results in an increased membrane fluidity and resistance of S. aureus to platelet-derived CAMP [113]. Several studies also suggested that pigment production in S. aureus may influence sensitivity to CAMPs. Just recently, Mishra et al. [114] discovered that an S. aureus carotenoid staphyloxanthin increases S. aureus resistance to human neutrophil defensin 1, platelet-derived CAMPs, and polymyxin B. However, the mechanism of resistance proposed in this study is that staphyloxanthin increases membrane rigidity [114]. Why extreme increase and extreme decrease in membrane fluidity lead to increased resistance to CAMPs is still subject of investigation, however, it appears that the mechanism of resistance may be specific for different CAMPs.
Expulsion of host defense peptides
CAMPs that after damaging the membrane end up inside the cytoplasm, can also be actively exported from the cell by certain multi drug resistance exporters (MDR). Such examples have been found in several bacterial species including Neisseria gonorrhoeae [115–117]. N. gonorrhoeae, utilizes the MtrCDE MDR exporter to expel diverse antibiotics and confer resistance to the CAMPs protegrin PG1 and LL-37. However, in a recent study using E. coli overexpressing certain MDR exporters s of S. aureus, P. aeruginosa, and E. coli failed to show increased resistance to several CAMPs, indicating that most MDR exporters do not mediate broad CAMP resistance and that only some MDR exporters can expel certain CAMPs [118]. Further more, resistance mediated by S. aureus MDR exporter QacA to rabbit tPMP has been suggested not to be associated with the ability of QacA active transport of CAMP but rather attributed to the secondary QacA mediated changes on membrane fluidity [119]
Conclusions
Several advances in bacterial host defenses against host antimicrobial peptides have been recently reported. They include secreted proteins or cell surface-associated proteins that irreversibly bind or cleave CAMPs or glycopolymeric matrices that trap CAMPs to prevent their access to bacterial cytoplasmic membrane (see Table 1). Perhaps the biggest advances were made in identification of cell wall-associated mechanisms, the primary action of which appears to be electrostatic repulsion of CAMPs (Fig. 2). Whether it is modification of lipid A of Gram-negative LPS by aminoarabinose or ethanolamine, modification of phosphatidylglycerol by lysine, or esterification of TA by d-alanine, the common denominator to all is a reduction of bacterial net negative surface charge. In the era of increasing numbers of bacterial infections resistant to multiple antibiotics, novel antibiotics are in dire need. Even though CAMPs can be designed in such a way that they are more resistant to proteolytic degradation, high concentrations are needed for their effectiveness and their potentially harmful immunomodulatory effect may not make them suitable for further considerations [7]. In contrast, drugs that target bacterial responses to CAMPs that appear to be conserved among pathogens may be much more suitable. Inhibition of d-alanylation of TA offers a premium target due to the multiple effects that lack of d-alanine esters have on pathogenicity of several Gram-positive pathogens (see Table 1). To this end, inhibitors of DltA, a protein involved in the first step of d-alanylation have shown remarkable success as potential therapeutics in vitro and in vivo, especially when used in combination with other antibiotics [120]. MprF may be an even better target, since it is present in both Gram-positive as well as Gram-negative pathogens. Furthermore, because of involvement of MprF in resistance to daptomycin, inhibitors of MprF could be of even greater value.
Perhaps the newest notion to exploit is a discovery that bacteria can sense and respond to the presence of CAMPs by upregulation of genes responsible for their resistance. These bacterial sensing systems are well conserved among pathogens and may present attractive targets for developing new antimicrobials.
Acknowledgments
Our work is supported in part by Grants from Marie Curie International Grant 249285 to TK, the German Research Foundation (SFB766, TRR34) and the German Ministry of Education and Research (SkinStaph, MENAGE) to AP.
Abbreviations
- CAMPs
Cationic antimicrobial peptides
- gIIA PLA2
group IIA phospholipase A2
- HBD3
Human β-defensin 3
- HNPs
Human neutrophil peptides
- LPS
Lipopolysaccharide
- LTA
Lipoteichoic acid
- MDR
Multidrug resistance
- PAMPs
Pathogen-associated molecular patterns
- PGA
Anionic poly-γ -glutamic acid
- TA
Teichoic acids
- PG
Phosphatidylglycerol
- PIA
Polysaccharide intercellular adhesin
- SIC
Streptococcal inhibitor of complement
- WTA
Wall teichoic acid
References
- 1.Weiss J, Bayer AS, Yeaman M. Cellular and extracellular defenses against Staphylococcal infections. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-positive pathogens. Washington, DC: ASM Press; 2006. [Google Scholar]
- 2.DeLeo FR, Diep BA, Otto M. Host defense and pathogenesis in Staphylococcus aureus infections. Infect Dis Clin North Am. 2009;23:17–34. doi: 10.1016/j.idc.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 5.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 6.Easton DM, Nijnik A, Mayer ML, Hancock REW. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 2009;27:582–590. doi: 10.1016/j.tibtech.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peschel A, Sahl H-G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Micro. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 8.Izadpanah A, Gallo RL. Antimicrobial peptides. J Am Acad Dermatol. 2005;52:381–390. doi: 10.1016/j.jaad.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 9.Lehrer RI, Ganz T. Cathelicidins: a family of endogenous antimicrobial peptides. Curr Opin Hematol. 2002;9:18–22. doi: 10.1097/00062752-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Ayabe T. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nature Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 11.Bals R. Mouse [beta]-defensin 3 is an inducible antimicrobial peptide expressed in the epithelia of multiple organs. Infect Immun. 1999;67:3542–3547. doi: 10.1128/iai.67.7.3542-3547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bensch KW, Raida M, Magert HJ, Schulz-Knappe P, Forssmann WG. hBD-1: a novel [beta]-defensin from human plasma. FEBS Lett. 1995;368:331–335. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 13.Yeaman MR. Platelets in defense against bacterial pathogens. Cell Mol Life Sci. 2010;67:525–544. doi: 10.1007/s00018-009-0210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald JR, Foster TJ, Cox D. The interaction of bacterial pathogens with platelets. Nat Rev Micro. 2006;4:445–457. doi: 10.1038/nrmicro1425. [DOI] [PubMed] [Google Scholar]
- 15.Bayer AS, Cheng D, Yeaman MR, Corey GR, McClelland RS, Harrel LJ, Fowler VG., Jr In vitro resistance to thrombin-induced platelet microbicidal protein among clinical bacteremic isolates of Staphylococcus aureus correlates with an endovascular infectious source. Antimicrob Agents Chemother. 1998;42:3169–3172. doi: 10.1128/aac.42.12.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kupferwasser LI, Yeaman MR, Shapiro SM, Nast CC, Bayer AS. In vitro susceptibility to thrombin-induced platelet microbicidal protein is associated with reduced disease progression and complication rates in experimental Staphylococcus aureus endocarditis: microbiological, histopathologic, and echocardiographic analyses. Circulation. 2002;105:746–752. doi: 10.1161/hc0602.103721. [DOI] [PubMed] [Google Scholar]
- 17.Dhawan VK, Bayer AS, Yeaman MR. In vitro resistance to thrombin-induced platelet microbicidal protein is associated with enhanced progression and hematogenous dissemination in experimental Staphylococcus aureus infective endocarditis. Infect Immun. 1998;66:3476–3479. doi: 10.1128/iai.66.7.3476-3479.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganz T. Hepcidin–a peptide hormone at the interface of innate immunity and iron metabolism. Curr Top Microbiol Immunol. 2006;306:183–198. doi: 10.1007/3-540-29916-5_7. [DOI] [PubMed] [Google Scholar]
- 19.Madhuri ST, Venugopal SK, Ghosh D, Gadepalli R, Dhawan B, Mukhopadhyay K. In vitro antimicrobial activity of alpha-melanocyte stimulating hormone against major human pathogen Staphylococcus aureus . Peptides. 2009;30:1627–1635. doi: 10.1016/j.peptides.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Schneider T, Gries K, Josten M, Wiedemann I, Pelzer S, Labischinski H, Sahl HG. The lipopeptide antibiotic friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob Agents Chemother. 2009;53:1610–1618. doi: 10.1128/AAC.01040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sass V, Schneider T, Wilmes M, Korner C, Tossi A, Novikova N, Shamova O, Sahl H-G. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun. 2010;78:2793–2800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventos DS, Neve S, Ravn B, Bonvin AMJJ, De Maria L, Andersen AS, Gammelgaard LK, Sahl H-G, Kristensen H-H. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science. 2010;328:1168–1172. doi: 10.1126/science.1185723. [DOI] [PubMed] [Google Scholar]
- 23.Lai Y, Gallo RL. AMPed Up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 25.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462:55–70. doi: 10.1016/S0005-2736(99)00200-X. [DOI] [PubMed] [Google Scholar]
- 26.Wimley WC, Selsted ME, White SH. Interactions between human defensins and lipid bilayers: evidence for formation of multimeric pores. Protein Sci. 1994;3:1362–1373. doi: 10.1002/pro.5560030902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott MG, Vreugdenhil AC, Buurman WA, Hancock RE, Gold MR. Cutting edge: cationic antimicrobial peptides block the binding of lipopolysaccharide (LPS) to LPS binding protein. J Immunol (Baltimore, MD, 1950) 2000;164:549–553. doi: 10.4049/jimmunol.164.2.549. [DOI] [PubMed] [Google Scholar]
- 28.Durr M, Peschel A. Chemokines meet defensins: the merging concepts of chemoattractants and antimicrobial peptides in host defense. Infect Immun. 2002;70:6515–6517. doi: 10.1128/IAI.70.12.6515-6517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mader JS, Marcet-Palacios M, Hancock REW, Bleackley RC. The human cathelicidin, LL-37, induces granzyme-mediated apoptosis in cytotoxic T lymphocytes. Exp Cell Res. 2011;317:531–538. doi: 10.1016/j.yexcr.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Nijnik A, Madera L, Ma S, Waldbrook M, Elliott MR, Easton DM, Mayer ML, Mullaly SC, Kindrachuk J, Jenssen Hv, Hancock REW. Synthetic cationic peptide IDR-1002 provides protection against bacterial infections through chemokine induction and enhanced leukocyte recruitment. J Immunol. 2010;184:2539–2550. doi: 10.4049/jimmunol.0901813. [DOI] [PubMed] [Google Scholar]
- 31.Jin T, Bokarewa M, Foster T, Mitchell J, Higgins J, Tarkowski A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J Immunol. 2004;172:1169–1176. doi: 10.4049/jimmunol.172.2.1169. [DOI] [PubMed] [Google Scholar]
- 32.Pence MA, Rooijakkers SHM, Cogen AL, Cole JN, Hollands A, Gallo RL, Nizet V. Streptococcal inhibitor of complement promotes innate immune resistance phenotypes of invasive M1T1 Group A Streptococcus . J Innate Immun. 2010;2:587–595. doi: 10.1159/000317672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frick I-M, Akesson P, Rasmussen M, Schmidtchen A, Bjorck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- 34.Sieprawska-Lupa M, Mydel P, Krawczyk K, Wojcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, McAleese F, Foster T, Travis J, Potempa J. Degradation of human antimicrobial p LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother. 2004;48:4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidtchen A, Frick I-M, Andersson E, Tapper H, Björck L. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol Microbiol. 2002;46:157–168. doi: 10.1046/j.1365-2958.2002.03146.x. [DOI] [PubMed] [Google Scholar]
- 36.Guina T, Yi EC, Wang H, Hackett M, Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to [alpha]-helical antimicrobial peptides. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/JB.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hui C-Y, Guo Y, He Q-S, Peng L, Wu S-C, Cao H, Huang S-H. Escherichia coli outer membrane protease OmpT confers resistance to urinary cationic peptides. Microbiol Immunol. 2010;54:452–459. doi: 10.1111/j.1348-0421.2010.00238.x. [DOI] [PubMed] [Google Scholar]
- 38.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 39.Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-γ-dl-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis . J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, De Leo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 41.Koprivnjak T, Peschel A, Gelb MH, Liang NS, Weiss JP. Role of charge properties of bacterial envelope in bactericidal action of human group IIA phospholipase A2 against Staphylococcus aureus . J Biol Chem. 2002;277:47636–47644. doi: 10.1074/jbc.M205104200. [DOI] [PubMed] [Google Scholar]
- 42.Koprivnjak T, Weidenmaier C, Peschel A, Weiss JP. Wall teichoic acid deficiency in Staphylococcus aureus confers selective resistance to mammalian group IIA phospholipase A2 and human beta-defensin -3. Infect Immun. 2008;76:2169–2176. doi: 10.1128/IAI.01705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neuhaus FC, Baddiley J. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 46.Trent MS, Stead CM, Tran AX, Hankins JV. Invited review: diversity of endotoxin and its impact on pathogenesis. J Endotxin Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- 47.Weidenmaier C, Kokai-Kun JF, Kulauzovic E, Kohler T, Thumm G, Stoll H, Gotz F, Peschel A. Differential roles of sortase-anchored surface proteins and wall teichoic acid in Staphylococcus aureus nasal colonization. Int J Med Microbiol. 2008;298:505–513. doi: 10.1016/j.ijmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Baddiley J, Buchanan JG, Hardy FE, Martin RO, Rajbhandary UL, Sanderson AR. The structure of the ribitol teichoic acid of Staphylococcus aureus H. Biochim Biophys Acta. 1961;52:406–407. doi: 10.1016/0006-3002(61)90699-0. [DOI] [PubMed] [Google Scholar]
- 49.Morath S, Geyer A, Hartung T. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus . J Exp Med. 2001;193:393–397. doi: 10.1084/jem.193.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer W. Lipoteichoic acid and lipids in the membrane of Staphylococcus aureus . Med Microbiol Immunol (Berl) 1994;183:61–76. doi: 10.1007/BF00277157. [DOI] [PubMed] [Google Scholar]
- 51.Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus . Int J Med Microbiol. 2009;300:148–154. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 52.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 53.Weidenmaier C, Peschel A, Kempf VAJ, Lucindo N, Yeaman MR, Bayer AS. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect Immun. 2005;73:8033–8038. doi: 10.1128/IAI.73.12.8033-8038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins LV, Kristian SA, Weidenmaier C, Faigle M, Van Kessel KP, Van Strijp JA, Gotz F, Neumeister B, Peschel A. Staphylococcus aureus strains lacking d-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis. 2002;186:214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 55.Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes . Mol Microbiol. 2002;43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- 56.Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R. A Functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in Gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae . J Bacteriol. 2006;188:5797–5805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abi Khattar Z, Rejasse A, Destoumieux-Garzon D, Escoubas JM, Sanchis V, Lereclus D, Givaudan A, Kallassy M, Nielsen-Leroux C, Gaudriault S. The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J Bacteriol. 2009;191:7063–7073. doi: 10.1128/JB.00892-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cox KH, Ruiz-Bustos E, Courtney HS, Dale JB, Pence MA, Nizet V, Aziz RK, Gerling I, Price SM, Hasty DL. Inactivation of DltA modulates virulence factor expression in Streptococcus pyogenes . PLoS ONE. 2009;4:e5366. doi: 10.1371/journal.pone.0005366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haas R, Koch HU, Fischer W. Alanyl turnover from lipoteichoic acid to teichoic acid in Staphylococcus aureus . FEMS Microbiol Lett. 1984;21:27–31. doi: 10.1111/j.1574-6968.1984.tb00180.x. [DOI] [Google Scholar]
- 60.Koprivnjak T, Mlakar V, Swanson L, Fournier B, Peschel A, Weiss JP. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus . J Bacteriol. 2006;188:3622–3630. doi: 10.1128/JB.188.10.3622-3630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. Incorporation of d-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem. 1995;270:15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- 62.Grundling A, Missiakas DM, Schneewind O. Staphylococcus aureus mutants with increased lysostaphin resistance. J Bacteriol. 2006;188:6286–6297. doi: 10.1128/JB.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grundling A, Schneewind O. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus . J Bacteriol. 2006;188:2463–2472. doi: 10.1128/JB.188.7.2463-2472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fabretti F, Theilacker C, Baldassarri L, Kaczynski Z, Kropec A, Holst O, Huebner J. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect Immun. 2006;74:4164–4171. doi: 10.1128/IAI.00111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gross M, Cramton SE, Gotz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun. 2001;69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weidenmaier C, Peschel A, Xiong YQ, Kristian SA, Dietz K, Yeaman MR, Bayer AS. Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J Infect Dis. 2005;191:1771–1777. doi: 10.1086/429692. [DOI] [PubMed] [Google Scholar]
- 67.Kristian SA, Lauth X, Nizet V, Goetz F, Neumeister B, Peschel A, Landmann R. Alanylation of teichoic acids protects Staphylococcus aureus against Toll-like receptor 2-dependent host defense in a mouse tissue cage infection model. J Infect Dis. 2003;188:414–423. doi: 10.1086/376533. [DOI] [PubMed] [Google Scholar]
- 68.Deininger S, Stadelmaier A, von Aulock S, Morath S, Schmidt RR, Hartung T. Definition of structural prerequisites for lipoteichoic acid-inducible cytokine induction by synthetic derivatives. J Immunol. 2003;170:4134–4138. doi: 10.4049/jimmunol.170.8.4134. [DOI] [PubMed] [Google Scholar]
- 69.Walter J, Loach DM, Alqumber M, Rockel C, Hermann C, Pfitzenmaier M, Tannock GW. D-alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100–23 results in impaired colonization of the mouse gastrointestinal tract. Environ Microbiol. 2007;9:1750–1760. doi: 10.1111/j.1462-2920.2007.01292.x. [DOI] [PubMed] [Google Scholar]
- 70.Foreman-Wykert AK, Weinrauch Y, Elsbach P, Weiss J. Cell-wall determinants of the bactericidal action of group IIA phospholipase A2 against Gram-positive bacteria. J Clin Invest. 1999;103:715–721. doi: 10.1172/JCI5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weiss J, Inada M, Elsbach P, Crowl RM. Structural determinants of the action against Escherichia coli of a human inflammatory fluid phospholipase A2 in concert with polymorphonuclear leukocytes. J Biol Chem. 1994;269:26331–26337. [PubMed] [Google Scholar]
- 72.Heptinstall S, Archibald AR, Baddiley J. Teichoic acids and membrane function in bacteria. Nature. 1970;225:519–521. doi: 10.1038/225519a0. [DOI] [PubMed] [Google Scholar]
- 73.MacArthur AE, Archibald AR. Effect of culture pH on the d-alanine ester content of lipoteichoic acid in Staphylococcus aureus . J Bacteriol. 1984;160:792–793. doi: 10.1128/jb.160.2.792-793.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hurst A, Hughes A, Duckworth M, Baddiley J. Loss of d-alanine during sublethal heating of Staphylococcus aureus S6 and magnesium binding during repair. J Gen Microbiol. 1975;89:277–284. doi: 10.1099/00221287-89-2-277. [DOI] [PubMed] [Google Scholar]
- 75.Koch HU, Doker R, Fischer W. Maintenance of d-alanine ester substitution of lipoteichoic acid by reesterification in Staphylococcus aureus . J Bacteriol. 1985;164:1211–1217. doi: 10.1128/jb.164.3.1211-1217.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kiriukhin MY, Neuhaus FC. d-alanylation of lipoteichoic acid: role of the d-alanyl carrier protein in acylation. J Bacteriol. 2001;183:2051–2058. doi: 10.1128/JB.183.6.2051-2058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poyart C, Lamy MC, Boumaila C, Fiedler F, Trieu-Cuot P. Regulation of d-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J Bacteriol. 2001;183:6324–6334. doi: 10.1128/JB.183.21.6324-6334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Said-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol. 2003;185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Herbert S, Bera A, Nerz C, Kraus D, Peschel A, Goerke C, Meehl M, Cheung A, Gotz F. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in Staphylococci. PLoS Pathogens. 2007;3:e102. doi: 10.1371/journal.ppat.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meehl M, Herbert S, Gotz F, Cheung A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus . Antimicrob Agents Chemother. 2007;51:2679–2689. doi: 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kraus D, Herbert S, Kristian SA, Khosravi A, Nizet V, Gotz F, Peschel A. The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol. 2008;8:85. doi: 10.1186/1471-2180-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus . Mol Microbiol. 2007;66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 84.Otto M. Bacterial sensing of antimicrobial peptides. Contrib Microbiol. 2009;16:136–149. doi: 10.1159/000219377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peschel A. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuroda M, Kuwahara-Arai K, Hiramatsu K. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun. 2000;269:485–490. doi: 10.1006/bbrc.2000.2277. [DOI] [PubMed] [Google Scholar]
- 87.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim Biophys Acta Biomembranes. 2009;1788:1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 88.Staubitz P, Neumann H, Schneider T, Wiedemann I, Peschel A. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol Lett. 2004;231:67–71. doi: 10.1016/S0378-1097(03)00921-2. [DOI] [PubMed] [Google Scholar]
- 89.Ernst CM, Staubitz P, Mishra NN, Yang S-J, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. The Bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus . Antimicrob Agents Chemother. 2006;50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rubio A, Conrad M, Haselbeck R, Kedar GC, Driver V, Finn J, Silverman J. Regulation of mprF by antisense restores daptomycin susceptibility to daptomycin-resistant isolates of Staphylococcus aureus . Antimicrob Agents Chemother. 2011;55:364–367. doi: 10.1128/AAC.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang S-J, Xiong YQ, Dunman PM, Schrenzel J, Francois P, Peschel A, Bayer AS. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob Agents Chemother. 2009;53:2636–2637. doi: 10.1128/AAC.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Straus SK, Hancock REW. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biophys Biochim Acta Biomembranes. 2006;1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 94.Camargo ILBdC, Neoh H-M, Cui L, Hiramatsu K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother. 2008;52:4289–4299. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruzin A, Severin A, Moghazeh SL, Etienne J, Bradford PA, Projan SJ, Shlaes DM. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus . Biochim Biophys Acta. 2003;1621:117–121. doi: 10.1016/s0304-4165(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 96.Nishi H, Komatsuzawa H, Fujiwara T, McCallum N, Sugai M. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus . Antimicrob Agents Chemother. 2004;48:4800–4807. doi: 10.1128/AAC.48.12.4800-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gunn JS, Miller SI. PhoP–PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 99.Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotxin Res. 2001;7:57–62. [PubMed] [Google Scholar]
- 100.Lee H, Hsu F-F, Turk J, Groisman EA. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica . J Bacteriol. 2004;186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Groisman EA, Heffron F, Solomon F. Molecular genetic analysis of the Escherichia coli phoP locus. J Bacteriol. 1992;174:486–491. doi: 10.1128/jb.174.2.486-491.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Groisman EA, Chiao E, Lipps CJ, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci USA. 1989;86:7077–7081. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garcia Vescovi E, Soncini FC, Groisman EA, Vescovi EG, Ayala YM, Di Cera E. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 104.Chamnongpol S, Cromie M, Groisman EA. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica . J Mol Biol. 2003;325:795–807. doi: 10.1016/S0022-2836(02)01268-8. [DOI] [PubMed] [Google Scholar]
- 105.Vescovi EG, Ayala YM, Di Cera E, Groisman EA. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+ . J Biol Chem. 1997;272:1440–1443. doi: 10.1074/jbc.272.3.1440. [DOI] [PubMed] [Google Scholar]
- 106.Alpuche Aranda CM, Swanson JA, Loomis WP, Miller SI. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci USA. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 108.Charles RC, Harris JB, Chase MR, Lebrun LM, Sheikh A, LaRocque RC, Logvinenko T, Rollins SM, Tarique A, Hohmann EL, Rosenberg I, Krastins B, Sarracino DA, Qadri F, Calderwood SB, Ryan ET. Comparative proteomic analysis of the phoP regulon in Salmonella enterica serovar typhi versus typhimurium . PLoS ONE. 2009;4:e6994. doi: 10.1371/journal.pone.0006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Prost LR, Sanowar S, Miller SI. Salmonella sensing of anti-microbial mechanisms to promote survival within macrophages. Immunol Rev. 2007;219:55–65. doi: 10.1111/j.1600-065X.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 110.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP–phoQ . Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 111.Miller SI, Pulkkinen WS, Selsted ME, Mekalanos JJ. Characterization of defensin resistance phenotypes associated with mutations in the phoP virulence regulon of Salmonella typhimurium . Infect Immun. 1990;58:3706–3710. doi: 10.1128/iai.58.11.3706-3710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Prost LR, Daley ME, Bader MW, Klevit RE, Miller SI. The PhoQ histidine kinases of Salmonella and Pseudomonas spp. are structurally and functionally different: evidence that pH and antimicrobial peptide sensing contribute to mammalian pathogenesis. Mol Microbiol. 2008;69:503–519. doi: 10.1111/j.1365-2958.2008.06303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bayer AS, Prasad R, Chandra J, Koul A, Smriti M, Varma A, Skurray RA, Firth N, Brown MH, Koo S-P, Yeaman MR. In Vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein is associated with alterations in cytoplasmic membrane fluidity. Infect Immun. 2000;68:3548–3553. doi: 10.1128/IAI.68.6.3548-3553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mishra NN, Liu GY, Yeaman MR, Nast CC, Proctor RA, McKinnell J, Bayer AS. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob Agents Chemother. 2011;55:526–531. doi: 10.1128/AAC.00680-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bengoechea JA, Skurnik M. Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia . Mol Microbiol. 2000;37:67–80. doi: 10.1046/j.1365-2958.2000.01956.x. [DOI] [PubMed] [Google Scholar]
- 116.Tzeng Y-L, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. Cationic antimicrobial peptide resistance in Neisseria meningitidis . J Bacteriol. 2005;187:5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shafer WM, Qu X, Waring AJ, Lehrer RI. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rieg S, Huth A, Kalbacher H, Kern WV. Resistance against antimicrobial peptides is independent of Escherichia coli AcrAB, Pseudomonas aeruginosa MexAB and Staphylococcus aureus NorA efflux pumps. Int J Antimicrob Ag. 2008;33:174–176. doi: 10.1016/j.ijantimicag.2008.07.032. [DOI] [PubMed] [Google Scholar]
- 119.Bayer AS, Kupferwasser LI, Brown MH, Skurray RA, Grkovic S, Jones T, Mukhopadhay K, Yeaman MR. Low-level resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein 1 In Vitro associated with qacA gene carriage is independent of multidrug efflux pump activity. Antimicrob Agents Chemother. 2006;50:2448–2454. doi: 10.1128/AAC.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.May JJ, Finking R, Wiegeshoff F, Weber TT, Bandur N, Koert U, Marahiel MA. Inhibition of the d-alanine:d-alanyl carrier protein ligase from Bacillus subtilis increases the bacterium’s susceptibility to antibiotics that target the cell wall. FEBS. 2005;272:2993–3003. doi: 10.1111/j.1742-4658.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- 121.Peschel A. How do bacteria resist human antimicrobial peptides? Trend Microbiol. 2002;10:179–186. doi: 10.1016/S0966-842X(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 122.Jin T, Bokarewa M, McIntyre L, Tarkowski A, Corey GR, Reller LB, Fowler VG., Jr Fatal outcome of bacteraemic patients caused by infection with staphylokinase-deficient Staphylococcus aureus strains. J Med Microbiol. 2003;52:919–923. doi: 10.1099/jmm.0.05145-0. [DOI] [PubMed] [Google Scholar]
- 123.Kwiecinski J, Josefsson E, Mitchell J, Higgins J, Magnusson M, Foster T, Jin T, Bokarewa M. Activation of plasminogen by staphylokinase reduces the severity of Staphylococcus aureus systemic infection. J Infect Dis. 2010;202:1041–1049. doi: 10.1086/656140. [DOI] [PubMed] [Google Scholar]
- 124.Belas R, Manos J, Suvanasuthi R. Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect Immun. 2004;72:5159–5167. doi: 10.1128/IAI.72.9.5159-5167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Johansson L, Thulin P, Sendi P, Hertzen E, Linder A, Akesson P, Low DE, Agerberth B, Norrby-Teglund A. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect Immun. 2008;76:3399–3404. doi: 10.1128/IAI.01392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Woods DE, Cryz SJ, Friedman RL, Iglewski BH. Contribution of toxin A and elastase to virulence of Pseudomonas aeruginosa in chronic lung infections of rats. Infect Immun. 1982;36:1223–1228. doi: 10.1128/iai.36.3.1223-1228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blackwood LL, Stone RM, Iglewski BH, Pennington JE. Evaluation of Pseudomonas aeruginosa exotoxin A and elastase as virulence factors in acute lung infection. Infect Immun. 1983;39:198–201. doi: 10.1128/iai.39.1.198-201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/Hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rupp ME, Fey PD, Heilmann C, Gotz F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183:1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- 130.Jones A, Georg M, Maudsdotter L, Jonsson A-B. Endotoxin, capsule, and bacterial attachment contribute to Neisseria meningitidis resistance to the human antimicrobial peptide LL-37. J Bacteriol. 2009;191:3861–3868. doi: 10.1128/JB.01313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yi K, Stephens DS, Stojiljkovic I. Development and evaluation of an improved mouse model of meningococcal colonization. Infect Immun. 2003;71:1849–1855. doi: 10.1128/IAI.71.4.1849-1855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Spinosa MR, Progida C, Tala A, Cogli L, Alifano P, Bucci C. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect Immun. 2007;75:3594–3603. doi: 10.1128/IAI.01945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cortes G, Borrell N, de Astorza B, Gomez C, Sauleda J, Alberti S. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect Immun. 2002;70:2583–2590. doi: 10.1128/IAI.70.5.2583-2590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Campos MA, Vargas MA, Regueiro V, Llompart CM, Alberti S, Bengoechea JA. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun. 2004;72:7107–7114. doi: 10.1128/IAI.72.12.7107-7114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology. 2008;154:3877–3886. doi: 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- 136.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10:243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 137.Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. d-alanylation of teichoic acids promotes group a streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol. 2005;187:6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kristian SA, Durr M, Van Strijp JAG, Neumeister B, Peschel A. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect Immun. 2003;71:546–549. doi: 10.1128/IAI.71.1.546-549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA–PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and Oral virulence of Salmonella enterica serovar typhimurium . Infect Immun. 2000;68:6139–6146. doi: 10.1128/IAI.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhou Z, Ribeiro AA, Lin S, Cotter RJ, Miller SI, Raetz CRH. Lipid A modifications in polymyxin-resistant Salmonella typhimurium . J Biol Chem. 2001;276:43111–43121. doi: 10.1074/jbc.M106960200. [DOI] [PubMed] [Google Scholar]
- 141.Belden WJ, Miller SI. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect Immun. 1994;62:5095–5101. doi: 10.1128/iai.62.11.5095-5101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guo L. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/S0092-8674(00)81750-X. [DOI] [PubMed] [Google Scholar]
- 143.Pelz A, Wieland K-P, Putzbach K, Hentschel P, Albert K, Gotz F. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus . J Biol Chem. 2005;280:32493–32498. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- 144.Clauditz A, Resch A, Wieland K-P, Peschel A, Gotz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kupferwasser LI, Skurray RA, Brown MH, Firth N, Yeaman MR, Bayer AS. Plasmid-mediated resistance to thrombin-induced platelet microbicidal protein in Staphylococci: role of the qacA locus. Antimicrob Agents Chemother. 1999;43:2395–2399. doi: 10.1128/aac.43.10.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Warner DM, Folster JP, Shafer WM, Jerse AE. Regulation of the MtrC–MtrD–MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae . J Infect Dis. 2007;196:1804–1812. doi: 10.1086/522964. [DOI] [PubMed] [Google Scholar]
- 148.Jerse AE, Sharma ND, Simms AN, Crow ET, Snyder LA, Shafer WM. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect Immun. 2003;71:5576–5582. doi: 10.1128/IAI.71.10.5576-5582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]