Abstract
Aging—defined as the progressive impairment of an organism’s functional capacity, resulting from deleterious changes in cells, organs, and biological systems—is one of the most fundamental features of Eukaryotes, from humans to the unicellular budding yeast Saccharomyces cerevisiae. It has recently been reported that this may also be the case for certain (if not all) types of bacteria. In this paper, the current view on the mechanistic background and evolutionary significance of bacterial kind of aging is presented, with particular emphasis on the role of asymmetric cell division, the characteristics of stationary growth phase, and the role of oxidative protein damage.
Keywords: Asymmetric division, Bacterial aging, Protein aggregates, Conditional senescence, Stationary phase
Defining aging
In terms of biology, aging is a progressive phenomenon characterized by a set of morphological and functional changes that disturb an organism’s ability to maintain homeostasis and which increase the probability of death. Most extensively, aging was studied in various types of human somatic cells, where this process manifests as a terminal exhaustion of the cell’s capacity to divide, which is termed cellular/replicative senescence. The maximal number of population doublings a cell may undergo before entering senescence (the so-called Hayflick limit) depends on its type and embryological origin, and ranges from over a dozen divisions in epithelial cells to several dozen divisions in the case of fibroblasts. Cell senescence has been shown to proceed independently from the lapse of chronological time and stems from a combination of pre-determined internal events (shortening and/or uncapping of telomeric DNA) and environmental variables of a stochastic character (oxidative stress). In addition, cell senescence may be triggered artificially in young proliferating or quiescent cells by their exposure to various stresses, including ionizing radiation, oxidants (e.g., H2O2), DNA-damaging agents (e.g., mitomycin C), chromatin perturbations (e.g., histone acetylation/methylation), oncogene activation (RAS) and culture shock (e.g., inadequate medium composition). Under these conditions, cell-growth arrest proceeds rapidly and is known as the stress-induced premature senescence (SIPS) [1, 2].
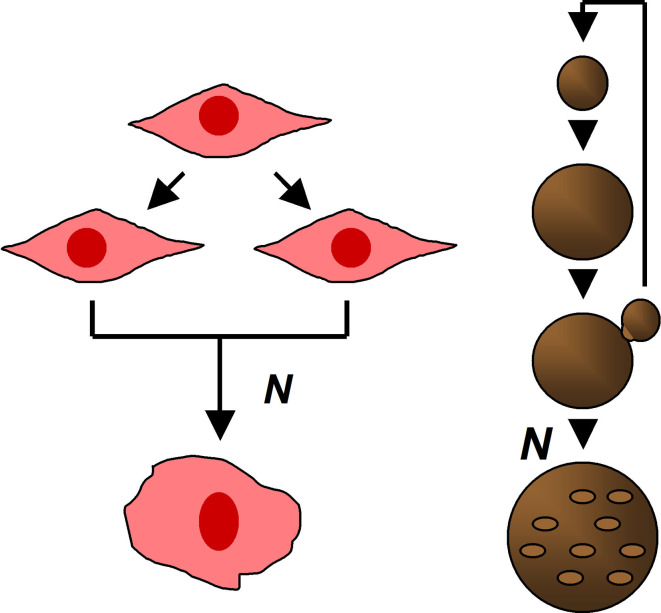
The existence of a fixed number of divisions that a cell can endure is not only a feature of cells derived from higher Eukaryotes. A similar phenomenon has been described in the unicellular budding yeast Saccharomyces cerevisiae, in which the cell replicative lifespan can be measured as the number of new buds which a mother cell can produce throughout its lifespan [3]. Generally, S. cerevisiae are able to endure between 10 and 30 divisions, a limit which is believed to be species- and strain-specific. Significantly, however, because of the asymmetric nature of yeast replication and the so-called ‘replicative reset phenomenon’ (in which the mother cell undergoes successive divisions and ultimately senesces, while a newly formed bud does not inherit the mother’s replicative history and starts its lifespan from zero), when one considers yeast colonies growing in a permissive milieu, one may describe them as having an infinite capacity to replicate. The mechanism by which S. cerevisiae reaches its Hayflick limit is not a function of telomere disruption, since these cells express active telomerase—an enzyme that restores the telomeric structure. On the other hand, there is strong evidence that yeast aging may be associated with the accumulation of ‘senescence factors’ in the cytoplasm, of which the extrachromosomal rDNA circles (ERCs) seem to play a key role. The increased level of protein carbonylation suggests that this process may also be, at least in part, related to oxidative stress. Moreover, as in human cells, several genes associated with cell-cycle control, genome stability, transcriptional silencing, and DNA repair, have been recognized to be involved [4] (Fig. 1).
Fig. 1.
Cell replication pattern resulting in senescence in multi- and unicellular Eukaryotes. Left panel human somatic cells, right panel budding yeast S. cerevisiae. N values indicate the maximal number of divisions a cell can undergo before entering senescence state (Hayflick limit). In the case of human cells, N ranges from 10 to 100 divisions (depending on cell type), while in yeast, ranges from 10 to 30 divisions
Bacteria also get old
Although bacteria may reproduce in several, often widely disparate ways (e.g., budding, endospore and multiple spore formation, multiple fission), the most common mechanism by which a bacterial cell divides is binary fission [5]. This kind of division, controlled by the FtsZ protein—a structural homologue of the eukaryotic cytoskeletal constituent tubulin [6]—results in two identical (morphologically and genetically) and equally young cells [7]. This symmetricity, in turn, has become the main source of the dogma that bacteria do not undergo any events which could be considered equivalent to aging. In other words, under optimal growth conditions (of nutrients, temperature, oxygen level, pH, osmolarity, etc.), bacteria have been considered to be immortal [8].
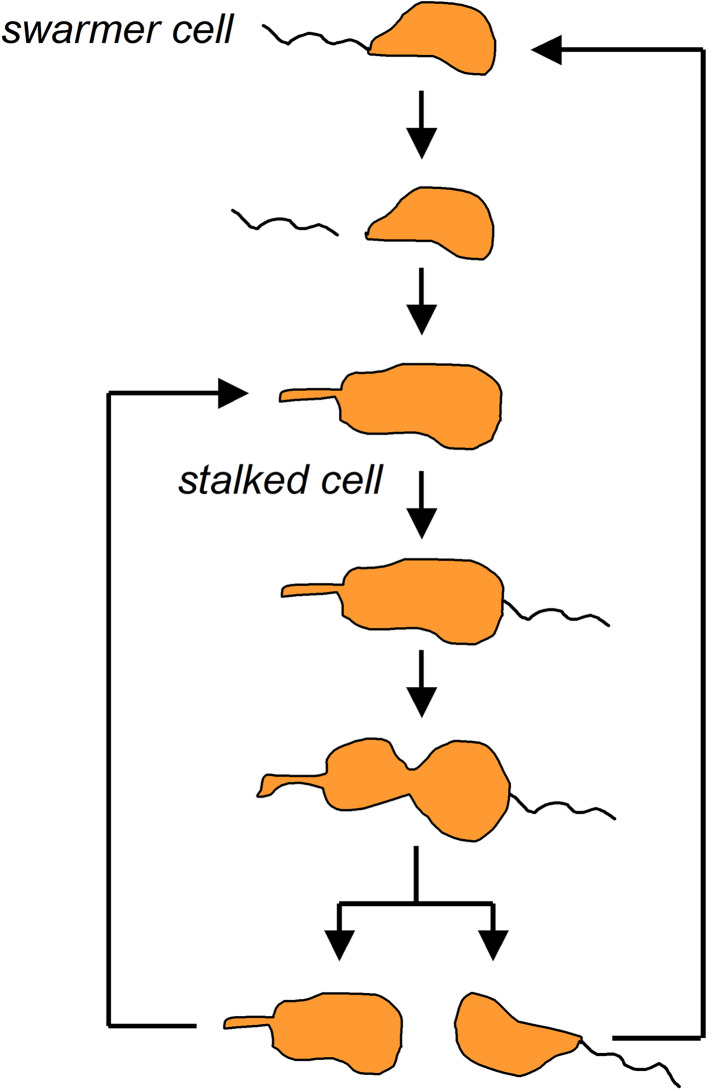
For the first time, this classic view has been blurred in studies conducted on Caulobacter crescentus, an aquatic Gram-negative bacterium displaying a unique asymmetric reproduction and a dimorphic life cycle. During division, the parent C. crescentus cell produces two types of daughter cells that markedly differ from one another in both structure and function. When the bacterium divides, first arises a mobile swarmer cell, which is equipped with a flagellum for movement. After a dozen minutes of swimming, the swarmer cell sheds the flagellum and transforms into a sessile form in which a stalked tubule-like structure protruding from one end allows the cell to adhere to surfaces with a holdfast. Importantly, of these two progeny, only the stalked cell is capable of DNA replication and entry into the next round of the division cycle [9] (Fig. 2). As shown by Ackermann et al. [10], the time a stalked cell requires to generate the next swarmer counterpart gradually increases, which has been postulated to constitute a manifestation of bacterial aging.
Fig. 2.
The asymmetric division and dimorphic life cycle in Caulobacter crescentus. As a result of this type of cell division, two morphologically distinct cells (swarmer cell and stalked cell) arise. Only stalked cells enter a new division cycle
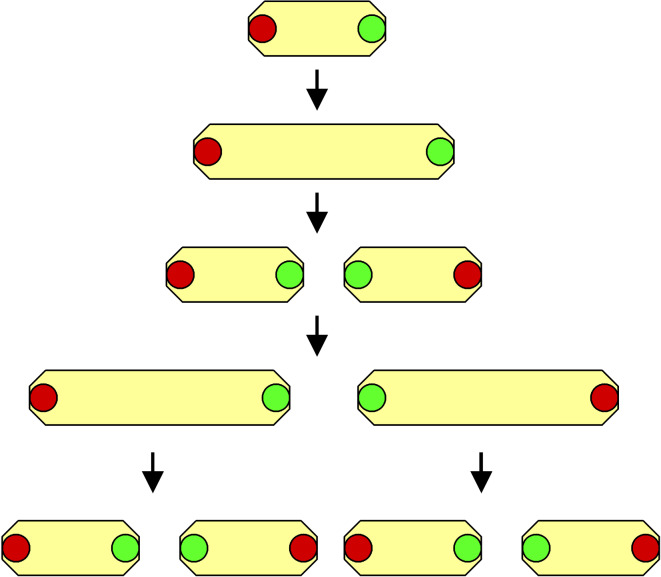
The asymmetrical cell division reflected by morphological diversity of the offspring is not a limiting condition for the appearance of aging in bacterial populations. As shown in the studies on the rod-shaped gut bacterium Escherichia coli, a model organism characterized by symmetrical reproduction, despite the morphological identity of the daughter cells, they may significantly differ at the level of cellular organization (functional asymmetry), which may be attributed to an uneven segregation of certain mothers cell’s constituents during binary fission. It has been observed using an elegant fluorescence-based individual cell tracking system that one of the E. coli progeny cells inherits some pre-existing elements of the mothers cell’s (‘old pole’), while the second descendant synthesizes these elements de novo (‘new pole’). The experiments, in which the fates of more than 35,000 cells were carefully traced, revealed that those lineages that vertically succeeded old pole from the mother cell exhibited features that could be equated with the aging process, including decelerated growth rate, decreased offspring production, and increased probability of death (Fig. 3). It has been recorded that the reproductive lifespan of the old pole-bearing cells has been ultimately terminated upon reaching about 100 divisions. Although the exact nature of the old-pole elements is still uncertain, it is likely that they may include fragments of cell wall, harmful/damaged DNA molecules, and modified proteins [11].
Fig. 3.
The functionally asymmetric division in Escherichia coli. Red dots indicate the old-pole elements, which are inherited by only one offspring. Green dots indicate newly formed elements of new poles, which subsequently become ‘old’ as a result of cell aging. These elements accumulate in a group of cells leading to their aging and death
Bacterial aging in the stationary phase
One of the most important features of bacteria is their highly developed ability to cope with unfavorable environmental conditions. In fact, the conditions that favor vigorous growth of bacteria populations in nature are rather seldom [12]. Bacteria usually live in competitive relationships when the limited resources of nutrients force them to launch a number of adaptative reactions that are necessary to allow them to survive until the external conditions improve. The best recognized mode by which bacteria confront adverse, oligotrophic environments, is an entry into stationary (stasis) phase [13].
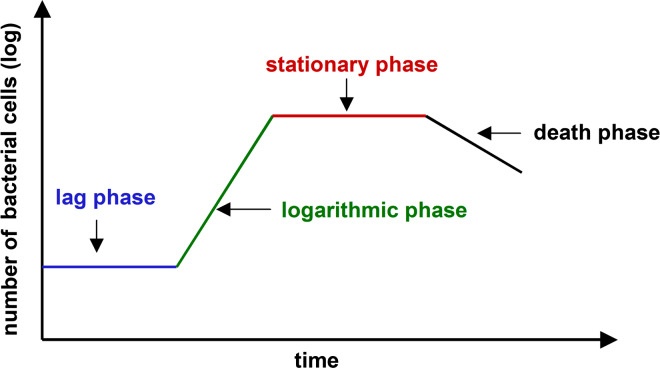
The stationary phase can also be reproduced in laboratory conditions, when experimental purposes do not require the collection of large numbers of cells. In this case, bacteria cultures are maintained in a closed system (batch cultures), in which cells are deprived of a supply of fresh nutrients (especially a carbon source) and an outflow of waste. When plotted as the logarithm of cell number versus incubation time, growth of such a culture can be depicted as a four-phase curve, which encompasses a lag phase, a logarithmic growth phase, a stationary phase, and a death phase (Fig. 4). In the initial period (lag phase), inoculated bacteria do not replicate and the population adapts to the fresh culture environment; cells grow in size, store nutrients, and generate vital structural constituents to prepare for binary fission. The lag phase is followed by logarithmic growth phase, in which cells divide at a very high rate and the number of bacteria increases exponentially (it doubles with each generation time). As a result of vigorous cell growth, the availability of growth-supporting substrates dramatically declines, which is accompanied by a reduction of booming growth, the accumulation of toxic metabolism by-products, and the development of a steady-state equilibrium between cell growth and die-off. These are the major features of the stationary growth phase. In the last, death phase, the number of cells dying, markedly exceeds the number of new cells formed [14].
Fig. 4.
The typical pattern of bacteria growth curve in batch culture
It has been observed that E. coli cells entering the stationary phase undergo various degenerative changes, of which the decline in proliferative capacity is the leading one. In addition, starved bacteria are characterized by condensed chromosome, which is attributed to an induction of Dps protein with an ability to compact DNA in nucleoprotein complex [15], and by dimerized (translationally inactive) ribosomes, which is a response to guanosine pentaphosphate (ppGpp)-dependent activation of ribosome modulation factor (rmf) [16]. These changes are also accompanied by the drift in cell morphology and size. Namely, starved E. coli cells become smaller and more spherical or ovoid than exponentially growing ones, which is primarily attributed to the activity of sigma factor σ S/RpoS (a major element of stress response) and its control over the morphogene bolA [17]. Factor σ S/RpoS also exerts a similar effect also through the ftsQAZ operon, which by regulating reductive cell division, partly contributes to changes in bacterial appearance during starvation [18]. Some abnormalities have also been reported within the structure of cell membranes. In the outer membrane, one may find decreased efficiency of peptidoglycan synthesis, which is probably associated with an incorporation of certain d-amino acids into the polymer [19]. In turn, the fluidity of the inner membrane markedly goes down, which may be causatively linked with conversion of unsaturated fatty acids into cyclopropyl derivatives and an increase in phosphatidylglycerol to phosphatidylethanolamine ratio [20]. At the same time, which is worthy to note, stationary E. coli cultures are still viable and capable of continue protein synthesis (mainly of those proteins allowing prolonged survival in the low-nutrient environment) [21]. The other issue that should be stressed is that the changes adopted by bacteria in the stationary phase, especially growth cessation, have a permanent character, namely they persist even when starved culture will be transferred to a nutrient-rich environment [22]. Although it is very hard (and risky) to compare the phenotypic features of starved bacteria with senescence phenotype of eukaryotic (e.g., human) cells, the starvation-induced senescence in E. coli cultures (currently termed as ‘conditional senescence’ [23]) can be considered as the nearest equivalent of SIPS described in eukaryotic cells, e.g., in human fibroblasts in which concentration of serum (source of nutrients) in culture medium has been reduced to an extreme value of 0.25% [24, 25].
Molecular causes of conditional senescence
Research into the molecular causes responsible for cell entry into conditional senescence using quantitative microarray analysis has revealed that E. coli cells that have been rendered senescent by their movement into the stationary phase for extended periods of time (16 days) are characterized by down-regulated expression of genes involved in crucial processes regulating bacterial growth, such as carbohydrate catabolism and energy generation (e.g., sdhA gene encoding a component of succinate dehydrogenase; cydA and cydB expressing the subunits of cytochrome bd-I terminal oxidase) and macromolecules synthesis (e.g., murI gene that encodes glutamate racemase, an enzyme engaged in peptidoglycan production) [26].
Researchers’ attention has also been focused on the accumulation of aggregated, aberrant proteins. It must be stressed, however, that the beginning of this process has been found to occur in the earlier, exponential phase of cell growth. As shown by Maisonneuve et al., exponentially growing E. coli cells tend to generate aggregated and structurally abnormal (e.g., cross-linked) proteins. These structures, which are regarded as the waste-storing organelles in the intracellular protein degradation pathway, included chaperone DnaK/HSP70 and protease ClpXP, substrates known to be very sensitive to unfolding and subsequent aggregation [27]. These observations have been strengthened by Lindner et al., who, tracking E. coli cultures growing under permissive conditions, found the spontaneous generation of inclusion bodies rich in aggregated proteins within cell old poles. Moreover, they demonstrated the anti-proliferative activity of these proteins by showing a correlation between their storage and a significant decrease in old-pole-inheriting cells’ ability to reproduce [28]. Given that these old pole-localized aggregated proteins are actively or passively (the mechanism is still speculative) sorted to only one of sibling cells, the causative role of these structures in the progression of cell aging seems to be reasonable. Moreover, given that factors causatively involved in aging should also increase the probability of cell/organism death, it is worthy to note that this assumption is in keeping with the effect of aggregated proteins on bacteria survival in the stationary phase. Namely, Maisonneuve et al. [29] observed the positive correlation between the relative amount of aggregated proteins and the incidence of cell death, which was attributed to several-fold higher content of these abnormal constituents in the non-viable bacteria.
The role of oxidative stress
Oxidative stress is one of the major culprits of aging considered at both the cellular and organismal levels [30, 31], and this may also be the case in bacteria [32]. In this context, it has been observed that senescent E. coli cultures contain increased amounts of carbonylated proteins—a key indicator of oxidative stress-mediated protein damage [33]. Moreover, as found in studies employing E. coli mutants defective in genes responsible for oxidative stress defense (superoxide dismutase and catalase), the level of modified proteins in cells undergoing starvation-mediated aging was considerably enhanced [27]. Conversely, in bacteria overexpressing Mn-superoxide dismutase, the magnitude of protein modifications was lower compared to the wild-type strain, and this effect was evident in both the exponential and stationary stages of growth [29]. Cell protection against oxidative stress is also associated with activity of factor σ S/RpoS, as mutants devoid of σ S senesce at an accelerated rate [34], with a remarkably higher load of oxidized proteins [33, 35].
Although bacteria are able to release significant amounts of reactive oxygen species (ROS) from their membrane-associated electron-transport machinery (mostly at the NADH dehydrogenase and ubiquinone sites) [36], increased oxidative deterioration of proteins in cells experiencing conditional senescence is puzzling, since steady-state ROS level in starved cultures seems to be constant, and the activity of major antioxidative enzymes is even increased [35]. This apparent contradiction has been exposed by Dukan et al. [37], who, using an antibiotic that promotes increased mistranslation (streptomycin) at a concentration not affecting cell growth and not inducing oxidative stress (either by stimulation of ROS release or by reducing antioxidant activities), found that under these conditions, the magnitude of protein carbonylation may rise several times over. Since the protein modification observed was a generalized reaction and concerned virtually all available proteins, the authors concluded that such a modification must proceed differently than in eukaryotic cells; namely, not as a response to classically expressed oxidative stress (understood as an imbalance between ROS production and neutralization [38]), but as a result of some kind of biochemical abnormality in the oxidized substrates themselves. Detailed analysis using proteomic techniques confirmed this view, and showed that certain classes of aggregated/aberrant proteins abundant in senescent cells display an increased susceptibility to oxidative modifications, as compared to their plain counterparts [37]. This frailty is probably attributable to defects arising during the translation process, which may include, e.g., framing and missense errors and stop codon read-through [39–41]. This scenario agrees with studies on E. coli mutants equipped with faultless ribosomes in which the content of oxidized proteins was remarkably lower [34, 41].
The above-presented findings indicate that oxidative stress-dependent increase in protein damage in fast growing E. coli cultures over time and their subsequent vertical transfer to successive generations of daughter cells may contribute to individual cell aging and death. It is worthy to note that the conclusive piece of evidence for the leading role of oxidative stress in bacterial aging is the observation that the loss of culturability in cells maintained in the stationary phase for extended period of time may be forcibly reversed by reducing oxygen pressure [35]. This result also implies the broad similarity between the triggers of aging in eukaryotic and prokaryotic cells, as with eukaryotic cells, the reduction of oxygen pressure in the environment also resulted in an extended replicative cell lifespan and delayed the development of the senescent phenotype [42].
Evolutionary significance of aging, or benefits from asymmetry
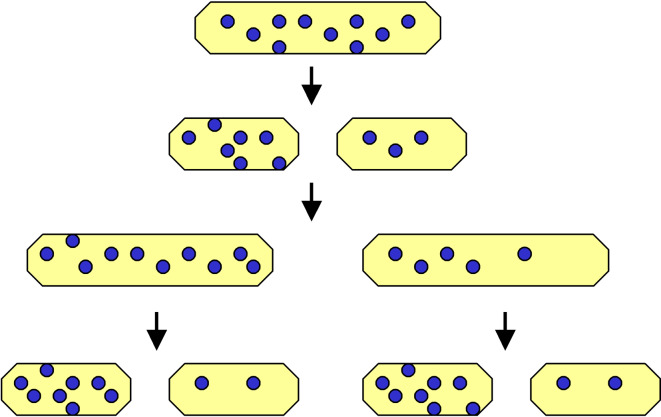
‘Why has aging evolved?’ is still one of the fundamental questions in biogerontology. Therefore, having shown that aging is also encountered in bacteria, it is of interest to ask whether its evolution is subject to the same rules as in Eukaryotes. According to Kirkwood’s [43] disposable soma theory, the finite lifespan of organisms, ending in aging and death, is elicited by the limited energy resources it has available to allocate to body maintenance/repair, in order to ensure adequate fecundity. As classic examples of this trade-off strategy, two classes of human cells are often referenced, i.e., the energetically favored germline cells, which are responsible for reproductive success (priority of evolution), and the body-forming somatic cells, which receive inadequate energy resources (a side-effect of evolution). In other words, from an evolutionary point of view, the benefits of the whole population supersede the particular welfare of individuals which constitute it [44]. The same argument can be adduced with respect to aging yeast, in which the mother cell collects damaged and/or toxic compounds (e.g., ERCs), ages, and ultimately dies in order to allow successive generations of young buds—and thus the whole culture—to rejuvenate and continue to grow. The asymmetrical (morphologically and/or functionally) bacterial cell division may constitute another example of such a strategy (Fig. 5).
Fig. 5.
The asymmetrical distribution of cellular damage in dividing bacteria. This pattern of distribution, based on the trade-off strategy, results in dilution of damaged molecules (blue dots) and rejuvenation in one offspring at the expense of an accumulation of damage, senescence, and death in the second one
The E. coli mother cell’s old pole probably contains damaged and possibly irreparable molecules (e.g., proteins) which steadily compromise reproductive performance, but at the same time, the new-pole-bearing offspring (see the analogy with stalked cells in C. crescentus) have a chance to completely renew their structures and multiply at the optimal rate [11]. Such a concept fits the results of studies by Watve et al. [45], who used a modified Leslie matrix framework to estimate that bacteria dividing in an asymmetrical fashion (resulting in aging and death) are characterized by intensified growth rate with concomitant reduction of biomass yield, which may be favored by evolution under certain pressures, e.g., in competitive environments.
Conclusions
Although bacterial cells have joined the narrow canon of cellular aging models, our knowledge about the triggers and molecular mechanisms of aging in Prokaryotes still remains elusive. Among factors that limit the scientific progress in this field, the small number of species investigated seems to be the major one. On the other hand, one must be aware that a unique organizational simplicity of bacteria (a single cell is at the same time a whole organism) combined with a great potential for genetic manipulations provide an excellent opportunity to transfer the ideas/observations/results arising during bacteria studies on the ground of aging research in higher Eukaryotes. The clear parallels in many aspects of aging in bacteria and eukaryotic cells (e.g., the causative role of oxidative stress, the involvement of stress response mechanisms, and the evolutionary context of this process based on a trade-off strategy) indicate that the issue of bacterial aging is worth a thorough examination.
References
- 1.Campisi J, d`Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 2.Toussaint O, Remacle J, Dierick JF, Pascal T, Frippiat C, Zdanov S, Magalhaes JP, Royer V, Chainiaux F. From the Hayflick mosaic to the mosaics of ageing role of stress-induced premature senescence in human ageing. Int J Biochem Cell Biol. 2002;34:1415–1429. doi: 10.1016/S1357-2725(02)00034-1. [DOI] [PubMed] [Google Scholar]
- 3.Powell CD, Van Zandycke SM, Quain DE, Smart KA. Replicative ageing and senescence in Saccharomyces cerevisiae and the impact on brewing fermentations. Microbiology. 2000;146:1023–1034. doi: 10.1099/00221287-146-5-1023. [DOI] [PubMed] [Google Scholar]
- 4.Roux AE, Chartrand P, Ferbeyre G, Rokeach LA. Fission yeast and other yeasts as emergent models to unravel cellular aging in eukaryotes. J Gerontol A Biol Sci Med Sci. 2010;65:1–8. doi: 10.1093/gerona/glp152. [DOI] [PubMed] [Google Scholar]
- 5.Angert ER. Alternatives to binary fission in bacteria. Nat Rev Microbiol. 2005;3:214–224. doi: 10.1038/nrmicro1096. [DOI] [PubMed] [Google Scholar]
- 6.Lowe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 7.Harry E, Monahan L, Thompson L. Bacterial cell division: the mechanism and its precision. Int Rev Cytol. 2006;253:27–94. doi: 10.1016/S0074-7696(06)53002-5. [DOI] [PubMed] [Google Scholar]
- 8.Rose M. Evolutionary biology of aging. New York: Oxford University Press; 1991. [Google Scholar]
- 9.Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus . Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ackermann M, Stearns SC, Jenal U. Senescence in a bacterium with asymmetric division. Science. 2003;300:1920. doi: 10.1126/science.1083532. [DOI] [PubMed] [Google Scholar]
- 11.Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:0295–0300. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morita R. Bacteria in oligotrophic environments: starvation survival lifestyle. Berlin Heidelberg New York: Springer; 1997. [Google Scholar]
- 13.Kolter R, Siegele DA, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 14.Pommerville JC (2010) Microbial growth and nutrition. In: Alcamo’s fundamentals of microbiology: body systems. Jones and Bartlett Publishers International, London
- 15.Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A. DNA protection by stress-induced biocrystallization. Nature. 1999;400:83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida H, Yamamoto H, Uchiumi T, Wada A. RMF inactivates ribosomes by covering the peptidyl transferase centre and entrance of peptide exit tunnel. Genes Cells. 2004;9:271–278. doi: 10.1111/j.1356-9597.2004.00723.x. [DOI] [PubMed] [Google Scholar]
- 17.Lange R, Hengge-Aronis R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J Bacteriol. 1991;173:4474–4481. doi: 10.1128/jb.173.14.4474-4481.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballesteros M, Kusano S, Ishihama A, Vicente M. The ftsQ1p gearbox promoter of Escherichia coli is a major sigma S-dependent promoter in the ddlB-ftsA region. Mol Microbiol. 1998;30:419–430. doi: 10.1046/j.1365-2958.1998.01077.x. [DOI] [PubMed] [Google Scholar]
- 19.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cronan JE., Jr Phospholipid alterations during growth of Escherichia coli . J Bacteriol. 1968;95:2054–2061. doi: 10.1128/jb.95.6.2054-2061.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli . Mol Microbiol. 1991;5:3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 22.Ericsson M, Hanstorp D, Hagberg P, Enger J, Nystrom T. Sorting out bacterial viability with optical tweezers. J Bacteriol. 2000;182:5551–5555. doi: 10.1128/JB.182.19.5551-5555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nystrom T. Conditional senescence in bacteria: death of the immortals. Mol Microbiol. 2003;48:17–23. doi: 10.1046/j.1365-2958.2003.03385.x. [DOI] [PubMed] [Google Scholar]
- 24.Wright WE, Shay JW. Cellular senescence as a tumor-protection mechanism: the essential role of counting. Curr Opin Genet Dev. 2001;11:98–103. doi: 10.1016/S0959-437X(00)00163-5. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez RD, Morales CP, Herbert BS, Rohde JM, Passons C, Shay JW, Wright WE. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pin C, Rolfe MD, Munoz-Cuevas M, Hinton JC, Peck MW, Walton NJ, Baranyi J. Network analysis of the transcriptional pattern of young and old cells of Escherichia coli during lag phase. BMC Syst Biol. 2009;3:108. doi: 10.1186/1752-0509-3-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maisonneuve E, Fraysse L, Moinier D, Dukan S. Existence of abnormal protein aggregates in healthy Escherichia coli cells. J Bacteriol. 2008;190:887–893. doi: 10.1128/JB.01603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maisonneuve E, Ezraty B, Dukan S. Protein aggregates: an aging factor involved in cell death. J Bacteriol. 2008;190:6070–6075. doi: 10.1128/JB.00736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucleic Acids Res. 2007;35:7545–7556. doi: 10.1093/nar/gkm1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Passos JF, Von Zglinicki T, Saretzki G. Mitochondrial dysfunction and cell senescence: cause or consequence? Rejuvenation Res. 2006;9:64–68. doi: 10.1089/rej.2006.9.64. [DOI] [PubMed] [Google Scholar]
- 32.Nystrom T. The free-radical hypothesis of aging goes prokaryotic. Cell Mol Life Sci. 2003;60:1333–1341. doi: 10.1007/s00018-003-2310-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dukan S, Nystrom T. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 1998;12:3431–3441. doi: 10.1101/gad.12.21.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli . Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 35.Dukan S, Nystrom T. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J Biol Chem. 1999;274:26027–26032. doi: 10.1074/jbc.274.37.26027. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli . J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 37.Dukan S, Farewell A, Ballesteros M, Taddei F, Radman M, Nystrom T. Protein oxidation in response to increased transcriptional or translational errors. Proc Natl Acad Sci USA. 2000;97:5746–5749. doi: 10.1073/pnas.100422497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerschman R, Gilbert DL, Nye SW, Dwyer P, Fenn WO. Oxygen poisoning and x-irradiation: a mechanism in common. Science. 1954;119:623–626. doi: 10.1126/science.119.3097.623. [DOI] [PubMed] [Google Scholar]
- 39.O’Farrell PH. The suppression of defective translation by ppGpp and its role in the stringent response. Cell. 1978;14:545–557. doi: 10.1016/0092-8674(78)90241-6. [DOI] [PubMed] [Google Scholar]
- 40.Wenthzel AM, Stancek M, Isaksson LA. Growth phase dependent stop codon readthrough and shift of translation reading frame in Escherichia coli . FEBS Lett. 1998;421:237–242. doi: 10.1016/S0014-5793(97)01570-6. [DOI] [PubMed] [Google Scholar]
- 41.Ballesteros M, Fredriksson A, Henriksson J, Nystrom T. Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001;20:5280–5289. doi: 10.1093/emboj/20.18.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q, Fischer A, Reagan JD, Yan LJ, Ames BN. Oxidative DNA damage and senescence of human diploid fibroblast cells. Proc Natl Acad Sci USA. 1995;92:4337–4341. doi: 10.1073/pnas.92.10.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirkwood TB, Holliday R. The evolution of ageing and longevity. Proc R Soc Lond B Biol Sci. 1979;205:531–546. doi: 10.1098/rspb.1979.0083. [DOI] [PubMed] [Google Scholar]
- 44.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 45.Watve M, Parab S, Jogdand P, Keni S. Aging may be a conditional strategic choice and not an inevitable outcome for bacteria. Proc Natl Acad Sci USA. 2006;103:14831–14835. doi: 10.1073/pnas.0606499103. [DOI] [PMC free article] [PubMed] [Google Scholar]