Abstract
The transcription factor Pax6 is crucial for the embryogenesis of multiple organs, including the eyes, parts of the brain and the pancreas. Mutations in one allele of PAX6 lead to eye diseases including Peter's anomaly and aniridia. Here, we use fluorescence recovery after photobleaching to show that Pax6 and also other Pax family proteins display a strikingly low nuclear mobility compared to other transcriptional regulators. For Pax6, the slow mobility is largely due to the presence of two DNA-binding domains, but protein-protein interactions also contribute. Consistently, the subnuclear localization of Pax6 suggests that it interacts preferentially with chromatin-rich territories. Some aniridia-causing missense mutations in Pax6 have impaired DNA-binding affinity. Interestingly, when these mutants were analyzed by FRAP, they displayed a pronounced increased mobility compared to wild-type Pax6. Hence, our results support the conclusion that disease mutations result in proteins with impaired function because of altered DNA- and protein-interaction capabilities.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0429-0) contains supplementary material, which is available to authorized users.
Keywords: Pax6, Transcription factors, FRAP, Nuclear mobility, Aniridia mutants
Introduction
The Pax family of transcription factors separates themselves from other homeodomain (HD) proteins by the presence of a 128-amino acid DNA-binding domain named the paired domain (PD) [1]. The vertebrate Pax family consists of nine members, Pax1–Pax9, which can be divided into four subgroups based on sequence comparisons [2]. All nine Pax proteins contain an N-terminal PD, while the full-length HD is present in Pax3, Pax4, Pax6 and Pax7. Pax2, Pax5 and Pax8 contain a truncated remnant of the HD that cannot bind DNA [3]. All Pax proteins contain a C-terminal transactivation domain (TAD) rich in proline, serine and threonine residues. A conserved octapeptide (O) is located between the PD and the HD in all Pax proteins except for Pax4 and Pax6 [4]. The Pax-specific PD can be divided into two subdomains, PAI and RED. Each of these subdomains bind DNA through their helix-turn-helix motif [5]. The PD and HD of Pax6 are also important for protein-protein interactions [6], and they can modulate each other's properties by binding directly to each other [7]. Protein-protein interactions involving Pax6 occur both dependently and independently of DNA [8].
Similar to the other Pax proteins, Pax6 is evolutionarily conserved. Murine and human Pax6s are 100% identical, while the zebrafish and human proteins are 97% identical [9]. PAX6 encodes predominantly the canonical 422-amino-acid-long protein of 46 kDa, in addition to several variants of varying sizes and properties [10–12].
PAX6 has been named the universal control gene for eye development [13] and is also important for development of other organs such as the nose, the pancreas, the pituitary and the pineal glands (reviewed in [9]). Recently, Pax6 was found to be aberrantly expressed in various tumor cells, acting as a tumor suppressor [14–17], or to actively contribute to cancer progression [18]. Thus, Pax6 is important not only in embryogenesis, but may also have a role in or contribute to the development and maintenance of cancer cells.
The developmental importance of this particular transcription factor is illustrated by the fact that homozygous PAX6 mutants completely lack eyes and nose, and are not viable. There are two reported exceptions to this fact. In 1994, Glaser et al. [19] described an individual with compound heterozygous nonsense mutations in the PD and TAD of PAX6 who died 8 days after birth. A recent publication reported the only known living human with mutations in both PAX6 alleles, namely a 4-year-old boy with microphthalmia, diabetes mellitus, hypopituitarism and complex brain anomalies [20]. This boy has one allele with a missense mutation in the PD, while the other allele contains a nonsense mutation in the HD.
Heterozygous mutations in PAX6 cause a broad range of ocular defects that include human aniridia, Peter’s anomaly and cataracts, the mouse small eye mutant and the Drosophila eyeless mutant (reviewed in [21]). On the other hand, ectopic expression of PAX6 results in the formation of ectopic eyes in flies [22, 23].
Aniridia is a bilateral, panocular disorder characterized by absence of the iris. In most cases, the cornea, the lens, the retina, the optic nerve and the anterior chamber angle are also affected [24]. The human PAX6 Allelic Variant Database [25] contains a record of PAX6 mutations associated with diseases. In 2005, Tzoulaki et al. [26] analyzed the 309 records in the database at that time. Of these 309 entries, 286 referred to pathological mutations in the PAX6 coding region or the splice sites flanking the exons. Of the pathological mutations, 90% were found to be associated with aniridia and 10% with phenotypes like foveal hypoplasia, microphthalmia and optic nerve defects. The aniridia phenotype is predominantly linked to premature translational termination, while the non-aniridia phenotypes primarily are associated with missense mutations. The database also contains details on patients with aniridia or other ocular defects where the normal termination codon is mutated. This causes continued 3′ translation that seemingly, based on patient phenotypes, generates loss-of-function alleles.
Fluorescence recovery after photobleaching (FRAP) makes it possible to monitor the mobility of proteins in vivo (reviewed in [27]). Utilizing this method, fluorescent molecules in a small region of a cell are irreversibly photobleached using a high-powered laser beam. When performed correctly, the photobleaching does not deplete the bleached region of the protein in question, nor does it affect protein-DNA or protein-protein interactions. The measured recovery kinetics of non-bleached fluorescent molecules into the photobleached area represent the dynamic behavior of proteins in their undisturbed state [28]. However, recent studies have shown that phototoxicity and photounbinding can compromise the method, leading to misinterpreted quantitative results [29, 30]. Although proteins with diverse functions move by passive, randomly directed diffusion within the nucleus [31], the nature of a protein's motion can range from full mobility to almost complete immobility. FRAP studies of several transcription factors, including GTFs [32], p53 [33], FOXL2 [34], NF-κB [35, 36] and members of the steroid receptor superfamily [37–42], confirm that most nuclear proteins display high mobility. However, nucleosomal proteins like histone H2B constitute an exception with almost no movement and stable association with chromatin [43].
In this paper we used FRAP to compare the mobility of Pax6 with that of other transcriptional regulators. Surprisingly, we found that Pax6 and also other Pax family members display relatively low nuclear mobility. We attribute the slowed mobility to the presence of two DNA-binding domains in Pax6, the paired domain and the homeodomain. Pax6 is preferentially localized to Hoechst 33258-positive chromatin in what we denote as chromatin-rich territories in the nucleus. Furthermore, aniridia-causing point mutations in Pax6 led to increased mobility compared to the wild-type protein. Our FRAP studies in live cells support and extend previous biochemical studies showing that these mutants have altered DNA- and protein-protein interaction capabilities, which disrupt the normal function of Pax6.
Materials and methods
Plasmid constructs
Plasmids and primers used in this work are listed in Table 1 and supplementary material Table S1, respectively. Details of their construction are available upon request. Oligonucleotides for mutagenesis, PCR and DNA sequencing were purchased from Operon. cDNA constructs were subcloned into Gateway entry vectors and expression clones made as described in the Gateway cloning technology instruction manual (Invitrogen). Point mutations were made using the QuickChange site-directed mutagenesis kit from Stratagene. All constructs made in this study were verified by DNA sequencing (BigDye sequencing kit, Applied Biosystems).
Table 1.
Plasmids used in this work
| Constructs | Description |
|---|---|
| cDNA constructs made by traditional subcloning | |
| pEntr HoxB7 | Human HoxB7 cDNA subcloned into pEntr1A (Invitrogen) |
| pEntr Histone H2B | Human histone H2B cDNA subcloned into pEntr1A |
| pEntr POU5F1 DBD | The two DNA-binding domains of human POU5F1 (aa 136–360) subcloned into pEntr1A |
| pEntr Pax2 | Zebrafish Pax2 cDNA subcloned into pEntr1A |
| pEntr Pax3 | Mouse Pax3 subcloned into pEntr1A |
| pEntr Pax4 | Human Pax4 subcloned into pEntr1A |
| pEntr Pax5 | Human Pax5 (BSAP) subcloned into pEntr1A |
| pEntr Pax6 | Mouse Pax6 cDNA subcloned into pEntr1A |
| pEntr Pax8 | Human Pax8 subcloned into pEntr 1A |
| pEntr Pax9 | Zebrafish Pax9 cDNA subcloned into pEntr1A |
| pEGFP zPax6 | Zebrafish Pax6 [6] |
| pEGFP Pax6 | Mouse Pax6 cDNA subcloned into pEGFP-C1 (Clontech) |
| pPax6 EGFP | Mouse Pax6 cDNA subcloned into pEGFP-N1 (Clontech) |
| cDNA constructs made by LR gateway reactions or site-directed mutagenesis | |
| pDest EGFP Pax6 | Gateway from pEntr Pax6 to pDest EGFP-C1 [75] |
| pDest Cherry Pax6 | Gateway from pEntr Pax6 to pDest Cherry [76] |
| pDest EGFP Pax6(5A) | Mouse Pax6 splice variant (I. Mikkola) |
| pDest EGFP Pax6 PD | The paired domain of mouse Pax6 (E. Thomassen) |
| pDest EGFP Pax6 HD | The homeodomain of mouse Pax6 (E. Thomassen) |
| pDest EGFP Pax6 TAD | The transactivation domain of mouse Pax6 (E. Thomassen) |
| pDest EGFP Pax6 PD-HD | Mouse Pax6 without the transactivation domain (E. Thomassen) |
| pDest EGFP Pax6 HD-TAD | Mouse Pax6 without the paired domain (E. Thomassen) |
| pDest EGFP Pax6 G18R | Amino acid 18 in mouse Pax6 mutated from Gly to Arg |
| pDest EGFP Pax6 I87R | Amino acid 87 in mouse Pax6 mutated from Ile to Arg |
| pDest EGFP Pax6 R262A | Amino acid 262 in mouse Pax6 mutated from Arg to Ala |
| pDest EGFP Pax6 R266A R267A | Amino acids 266 and 267 in mouse Pax6 mutated from Arg to Ala |
| pDest EGFP Pax6 P375Q | Amino acid 375 in mouse Pax6 mutated from Phe to Gln |
| pDest EGFP Pax6 Q422A | Amino acid 422 in mouse Pax6 mutated from Gln to Ala |
| pDest EGFP Pax6 Q422R | Amino acid 422 in mouse Pax6 mutated from Gln to Arg |
| pDest EGFP Pax6 Q422X | Amino acid 422 in mouse Pax6 mutated from Gln to Stop |
| pDest zPax6-YFP | Zebrafish Pax6 with C-terminal EYFP tag [7] |
| pDest EGFP Pax2 | Gateway from pEntr Pax2 to pDest EGFP-C1 |
| pDest EGFP Pax3 | Gateway from pEntr Pax3 to pDest EGFP-C1 |
| pDest EGFP Pax4 | Gateway from pEntr Pax4 to pDest EGFP-C1 |
| pDest EGFP Pax5 | Gateway from pEntr Pax5 to pDest EGFP-C1 |
| pDest EGFP Pax8 | Gateway from pEntr Pax8 to pDest EGFP-C1 |
| pDest EGFP Pax9 | Gateway from pEntr Pax9 to pDest EGFP-C1 |
| pDest EGFP AR | Human androgen receptor (E. Thomassen) |
| pDest EGFP GTFIIH | Human general transcription factor II H (E. Thomassen) |
| pDest EGFP HoxB7 | Gateway from pEntr HoxB7 to pDest EGFP-C1 |
| pDest EGFP Klf4 | Human Klf4 (E. Alemu) |
| pDest EGFP POU5F1 DBD | Gateway from pEntr POU5F1 DBD to pDest EGFP-C1 |
| pDest EGFP Sp1 | Human Sp1 (E. Alemu) |
| pDest EGFP SPBP | Human SPBP |
| pDest EGFP histone H2B | Gateway from pEntr Histone H2B to pDest EGFP-C1 |
| pDest Cherry histone H2B | Gateway from pEntr Histone H2B to pDest Cherry |
| pDest Cherry histone H2A | PCR from pET3-huH2A-ATA (gift from J. A. Goldman) |
| pDest Cherry histone H3.1 | PCR from pIM-H3.1-Venus [77] |
| pDest Cherry HP1α | Gateway from pIRAOp969Bo957D-HP1 purchased from the PlasmidID repository at the DF/HCC DNA Resource core at the Harvard Institute of Proteomics |
| pDest Cherry Pax3 | Gateway from pEntr Pax3 to pDest Cherry |
| pDest Cherry HoxB7 | Gateway from pEntr HoxB7 to pDest Cherry |
| pDest Cherry Sp1 | Human Sp1 (E. Alemu) |
Unless otherwise stated, all Pax6 constructs are of murine origin
Transient transfections
HeLa cells (ATCC CCL2) were cultured in Eagle’s minimum essential medium (MEM), and U2OS cells (ATCC HTB-96) were cultured in Dulbecco’s modified Eagle’s medium (DME). Both MEM and DME were supplemented with 10% fetal calf serum, penicillin (100 U/ml) and streptomycin (100 μg/ml) (Invitrogen). Subconfluent cells were transfected with the different expression constructs using Lipofectamine PLUS (Invitrogen), Metafectene Pro (Biontex) or TransIT-LT1 (Mirus Bio) according to the manufacturer’s protocol.
Reporter gene assays
Subconfluent HeLa or U2OS cells in 24-well tissue culture dishes (Becton-Dickinson) were transiently transfected with 5–150 ng of pEGFP-Pax6 or pPax6-EGFP and 50 ng of the reporter plasmid pP6xCON-Luc [44]. pcDNA3 (Invitrogen) was used to equalize the concentration of DNA in each transfection. To normalize for variations in transfection efficiency, 5 ng of pCMV-β-galactosidase (Stratagene) was included in each transfection. Extracts were prepared 24 h after transfection using a dual-light luciferase and β-galactosidase reporter gene assay system (Tropix) and analyzed in a Labsystems Luminoskan RT dual-injection luminometer. The assays were performed in triplicates and repeated three times.
Cell fractioning and chromatin association assay
HeLa cells were resuspended in cold buffer A [10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 5% glycerol, 0.5 mM DTT, protease (Roche Diagnostics) and phosphatase (Calbiochem) inhibitors], 300 μl/107 cells, and incubated on ice for 30 min. A 10% solution of Triton X-100 was added to a final concentration of 0.2% followed by further incubation on ice for 3 min. The cytoplasmic fraction was collected after centrifugation at 5,000 rpm for 5 min at 4°C in a microcentrifuge. The pellet was washed once in buffer A before resuspension in 300 μl cold buffer B [20 mM HEPES pH 7.9, 450 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, protease (Roche Diagnostics) and phosphatase (Calbiochem) inhibitors]. After incubation on ice for 30 min, the nuclear fraction was collected by centrifugation at maximum speed for 10 min at 4°C in a microcentrifuge. The remaining pellet was resuspended in 300 μl 2× SDS loading buffer with 20 mM DTT, boiled and sonicated. The fractions were subsequently subjected to immunoblotting as described in the following section (protocol adapted from Wu et al. [45]).
Immunoblotting
Transiently transfected HeLa cells in 24-well tissue culture dishes were harvested in 2× SDS loading buffer with 20 mM DTT 24 h after transfection. The lysates were boiled and sonicated before separation on 6, 8 or 15% SDS-PAGE gels in Tris-glycine buffer (25 mM Tris, 250 mM glycine, 0.1% SDS). The Biotinylated Protein Ladder (Cell Signaling) was used as a molecular weight marker. The proteins were subsequently immunoblotted on Hybond-ECL nitrocellulose membranes (Amersham) in Towbin buffer (25 mM Tris, 192 mM glycine, 20% methanol) at 100 V for 1 h. The membranes were incubated in 5% non-fat dried milk for blocking at 1 h at room temperature. Primary antibody was added overnight at 4°C. The primary antibodies used were anti-GFP (1:1,000; Abcam), anti-Pax6 (1:1,000; Chemicon), anti-RNA polymerase II (1:200; Santa Cruz), anti-β-tubulin (1:1,000; Sigma) and anti-histone H3 (1:2,000; AbCam). Secondary antibodies, anti-mouse or -rabbit HRP (1:2,000, Pierce) and anti-biotin HRP (1:2,000, Cell Signaling), were added for 1 h at room temperature. Detection was performed using the Western Blotting Luminol Reagent kit (Santa Cruz Biotechnology) and a LumiAnalyst imager (Roche Applied Sciences).
Immunostaining and confocal microscopy
HeLa cells cultured in eight-chambered cover slides (Nunc) were transiently transfected with 50–100 ng of the different expression constructs. Images of live cells were taken the following day, with the cells kept in 1× HBSS (Invitrogen) supplemented with 10% fetal calf serum. Before staining, the cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Endogenous histone modifications were stained using rabbit polyclonal anti-H3 K4 me3 (Abcam, 1:2,000), rabbit polyclonal anti-acetyl H3 (Millipore, 1:1,000) and rabbit polyclonal anti-H4 K20 me1 (Abcam, 1:1,000) for 60 min at room temperature, followed by the secondary antibody Alexa Fluor 488 goat anti-rabbit IgG (1:500) for 30 min at room temperature. DNA was stained with Hoechst 33258 (Invitrogen, 0.5 μg/ml) for 5 min at room temperature. Pictures were taken with a Zeiss LSM510 confocal microscope using the 488-, 568- and 633-nm laser lines, or a Leica TCS-SP5 confocal microscope using the 561-nm and the UV laser line. Colocalization was studied using the Quantify tool in the Leica software.
Fluorescent recovery after photobleaching (FRAP)
The FRAP experiments were performed 24 h after transient transfection with the cells kept in 1× HBSS (Invitrogen) supplemented with 10% fetal calf serum at 37°C, 5% CO2 during imaging. The 63× H2O objective of a Leica TCS-SP5 confocal microscope was used to perform the experiments. Settings were configured to produce five pre-bleach images followed by ten bleach pulses of 0.113 s each with the 488-nm line of a 25-mW argon laser operating at 100% laser power. These settings resulted in a bleaching efficiency between 90 and 95% for all proteins studied except GFP (50%) and GFP-Pax6 TAD (80%). For all cells selected for bleaching, the pre-bleach green fluorescent intensity of the respective GFP fusion proteins was in the linear range of detection. Three hundred post-bleach images were captured with 0.113-s intervals followed by 50 images with 1-s intervals and 45 images with 5-s intervals. The total time for the experiments conducted was approximately 5 min. Fluorescence recovery was monitored at 5% for the 488-nm laser line. Recovery was measured in a circle with a diameter of 5 μm. A summary of the settings used is listed in supplementary material Table S2. FRAP recovery curves were generated using double normalization as described by Phair et al. [28], correcting for both background and loss of fluorescence during imaging. Each protein was tested in three separate experiments, 8–12 cells each time. The 25–30 experiments were then averaged to generate a single FRAP curve. The t 1/2 value, here defined as the time point after bleaching where the fluorescence has increased to 50% of the initial level, was determined based on the normalized data.
Results
Pax6 is preferentially localized to chromatin-rich nuclear territories
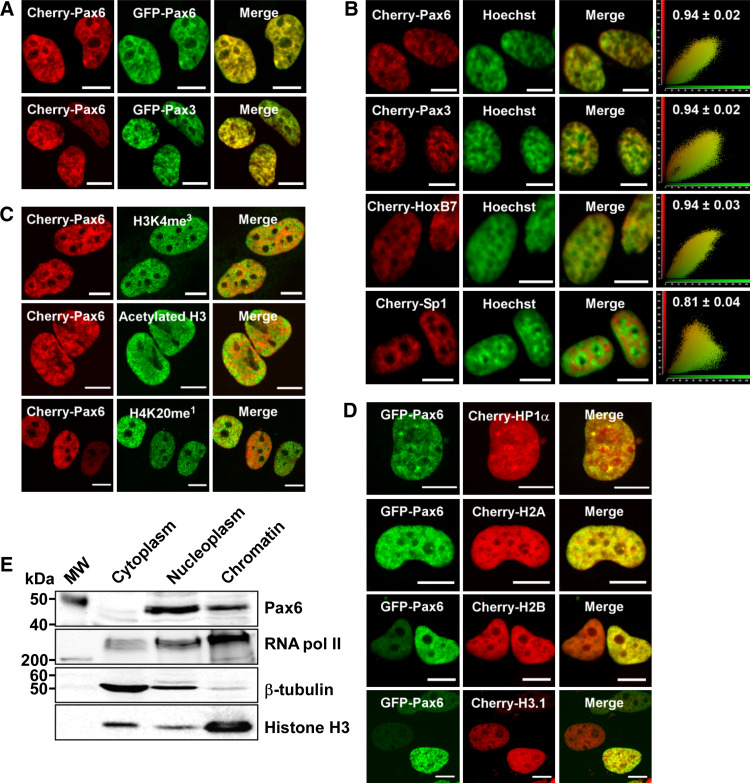
To study the subnuclear localization of Pax6, we first made many attempts to stain endogenous Pax6. However, none of the six different commercially available antibodies we tested gave satisfying results without significant background staining when tried on several different cell lines expressing Pax6. Hence, we analyzed the localization of Pax6 by confocal fluorescence microscopy following transfection of human HeLa cells with GFP- or mCherry-tagged Pax6, taking care to analyze cells with only moderate expression levels. As shown in Fig. 1, Pax6 is located throughout the nucleus, but is excluded from the nucleolus. Pax6 is enriched in speckles in the nucleoplasm, frequently found around the nuclear membrane and the nucleoli. These regions of the nucleus are often associated with heterochromatin (reviewed in [46, 47]). The punctuated localization pattern is similar to that observed for endogenous Pax3 [48]. As shown in Fig. 1a, Pax6 and Pax3 have nearly identical localization patterns in the nucleus. The images in supplementary material Figure S1 show that the localization pattern of Cherry-Pax6 or GFP-Pax6 is not caused by the GFP or Cherry tags themselves.
Fig. 1.
Subnuclear localization of Pax6. The confocal pictures show HeLa cells transiently transfected with different fusion constructs, fixed and stained as described in “Methods.” Scale bars 10 μm. a Cherry-Pax6 and GFP-Pax6 (upper panel) and Cherry-Pax6 and GFP-Pax3 (lower panel) have identical subnuclear localization patterns. b Colocalization between Hoechst 33258 and Cherry-Pax6, Cherry-Pax3, Cherry-HoxB7 and Cherry-Sp1. The scatter plots are made using the Quantify tool in the Leica software. The Pearson’s correlation coefficient is indicated above each scatter plot and is based on the average of 20–30 nuclei for each protein. c Colocalization between Cherry-Pax6 and two markers for active transcription, histone H3 K4 me3 and acetylated histone H3 (upper and middle panel, respectively), and between Cherry Pax6 and a marker for inactive transcription, histone H4 K20 me1 (lower panel). d Colocalization between GFP-Pax6 and Cherry-HP1α, Cherry-histone H2A, Cherry-histone H2B or Cherry-histone H3.1. e Chromatin association assay. Cytoplasmic, nucleoplasmic and chromatin fractions were prepared from HeLa cells as described in “Methods.” The fractions were separated on 8 or 15% SDS polyacrylamide gels and subjected to Western blotting with anti-rabbit Pax6, anti-mouse RNA polymerase II, anti-mouse β-tubulin and anti-rabbit histone H3 antibodies
Hoechst 33258 is a dye with preference for AT-rich DNA regions, which are abundant in centromeric heterochromatin [49]. We compared the Hoechst 33258 colocalization profiles of the transcription factors Pax6, Pax3, HoxB7 and Sp1 (Fig. 1b). Interestingly, Pax6, Pax3 and HoxB7 colocalize strongly (Pearson’s correlation coefficient of 0.94) with the Hoechst staining. All three are homeodomain-containing transcription factors. Sp1, on the other hand, has a very different localization pattern in the nucleus with a rather weak colocalization with Hoechst-positive, AT-rich heterochromatin (Pearson’s correlation coefficient of 0.81). A contributing factor here could be that Sp1 binds to GC-rich target sequences.
Together with acetylated histone H3, trimethylation of lysine 4 in histone H3 is considered a marker for active transcription, while monomethylation of lysine 20 in histone H4 is considered a marker for inactive transcription. Immunostaining of Cherry-Pax6 expressing HeLa cells with antibodies against these three histone modifications indicated that Pax6 is localized to both transcriptionally active and inactive regions (Fig. 1c). The enrichment of GFP-Pax6 on chromatin was further visualized by colocalization with Cherry-H2A, Cherry-H2B and Cherry-H3.1 (Fig. 1d). There was also some colocalization between Pax6 and heterochromatin protein 1α (HP1α) (upper panel Fig. 1d). As anticipated, a cell fractionation assay showed that endogenous Pax6 was found both in the chromatin fraction and in the nucleoplasmic fraction (Fig. 1e). Taken together, the results in Fig. 1 show that Pax6 is enriched in chromatin-rich, nuclear territories.
Pax6 displays a slow nuclear mobility
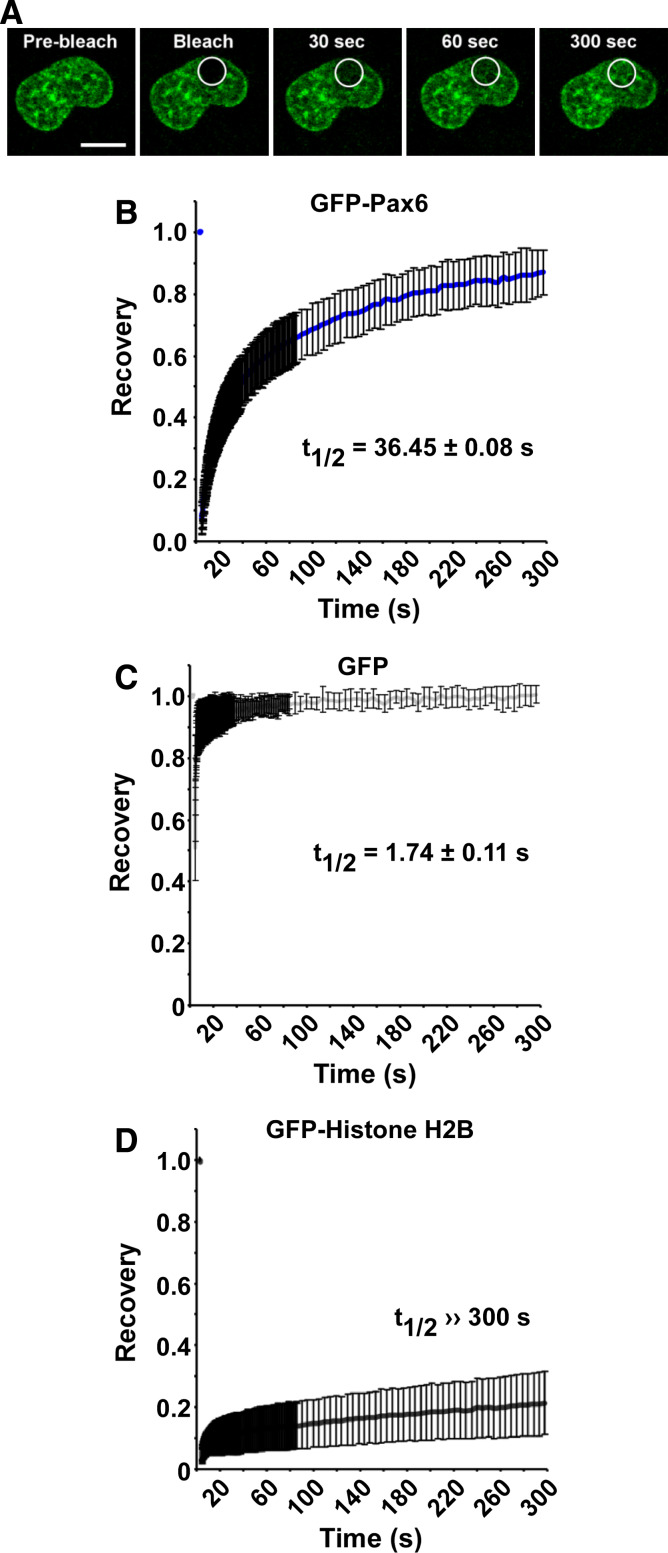
To study the chromatin association of Pax6 further, we investigated the nuclear mobility of Pax6. To this end, FRAP experiments were performed using HeLa cells transiently expressing GFP-Pax6. Figure 2a shows images of a representative nucleus at selected time points before, during and after photobleaching. The conditions and normalization used for the FRAP experiments are described in “Methods.” The recovery kinetics displayed in Fig. 2b are calculated based on the average of 30 individual cells. For Pax6, approximately 90% recovery was obtained after 5 min. This indicates that Pax6 has an immobile fraction. However, since the recovery was still increasing even after 5 min, we could not calculate the exact size of this fraction. Importantly, the FRAP experiments show that the Pax6 distribution pattern is recovered after bleaching, suggesting that Pax6 is specifically recruited to specific chromatin territories.
Fig. 2.
Pax6 displays a slow nuclear mobility. a Time-lapse imaging of HeLa cells transiently expressing N-terminal EGFP-tagged mouse Pax6 (pDest EGFP-Pax6), before and at selected time points after bleaching. The confocal microscope settings used are described in “Methods” and in supplementary material Table S2. The white circles indicate the bleached area of the nucleus. Scale bar 10 μm. b FRAP recovery curve displaying the relative recovery rate into the bleached area over the indicated time period. The curve is based on the average of three independent experiments, each including ~10 individual cells. Standard error bars are included. The recovery kinetics are estimated using double normalization [28], correcting for both background and loss of fluorescence during the imaging. Note that the curve is not corrected for bleaching efficiency, resulting in underestimated recovery. The t 1/2 value, here defined as the time point after bleaching where the fluorescence has increased to 50% of the initial level, was determined based on the normalized data. c, d FRAP recovery curves for GFP and GFP-histone H2B, respectively, with t 1/2 values indicated. These two proteins were included as controls for the FRAP settings used
The FRAP conditions employed in this study were tested using two proteins with known low and high mobility, namely histone H2B and GFP (green fluorescent protein). Histone H2B fused to GFP has been shown to stably incorporate into nucleosomes without affecting cell cycle progression [43]. With the settings used, there was nearly no migration of GFP-histone H2B molecules into the bleached area (Fig. 2d). In fact, only 20% of its initial fluorescence was retained in the bleached area over a 5-min time period. GFP, on the other hand, recovered 50% of its initial fluorescence less than 2 s after bleaching, reflecting its free diffusion in the nucleus (Fig. 2c). The rapid movement of GFP caused an apparent bleaching efficiency of only approximately 50% compared to 95% for GFP-histone H2B. Note that GFP is located both in the cytoplasm and the nucleus. FRAP was conducted on nuclear GFP, but movement of GFP from the cytoplasm to the nucleus and vice versa probably also influences the overall recovery curve.
For GFP-Pax6, it took 36.5 s before the relative fluorescent intensity in the bleached area returned to 50% of the initial intensity. In order to calculate the recovery half-time (t 1/2), the fluorescent recovery must reach a steady-state plateau level [50]. For Pax6 and a few other proteins studied here, such a level was not reached even after 5 min. Hence, we set t 1/2 as the time point after bleaching when the fluorescence had recovered to 50% of the initial level and used it to compare the recovery of different proteins.
The HeLa cells used in this study had a moderate level of endogenous Pax6. FRAP experiments using the same settings were also conducted in U2OS cells, which have a much lower endogenous Pax6 expression level. However, the recovery curves were practically identical in the two cell lines (supplementary material Figure S2A). This is in line with a previous study showing that specific proteins have similar kinetic properties in different cell types [51]. Also, placing the fluorescent tag at the N-terminus or the C-terminus did not alter the functionality of Pax6. N- and C-terminally tagged Pax6 behaved identically in FRAP experiments (supplementary material Figure S2B). Reporter gene assays with a pP6xCON-luc reporter, containing six consensus Pax6 PD-binding sites upstream of the adenovirus ElB minimal promoter [44], showed that GFP-Pax6 and Pax6-GFP transactivated in a similar manner in both HeLa and U2OS cells (supplementary material Figure S2C). Hence, the location of the tag does not seem to disturb the biological activity of Pax6, and subsequent experiments were performed with N-terminal EGFP-tagged fusion proteins.
Pax6 has a lower nuclear mobility than other transcriptional regulators
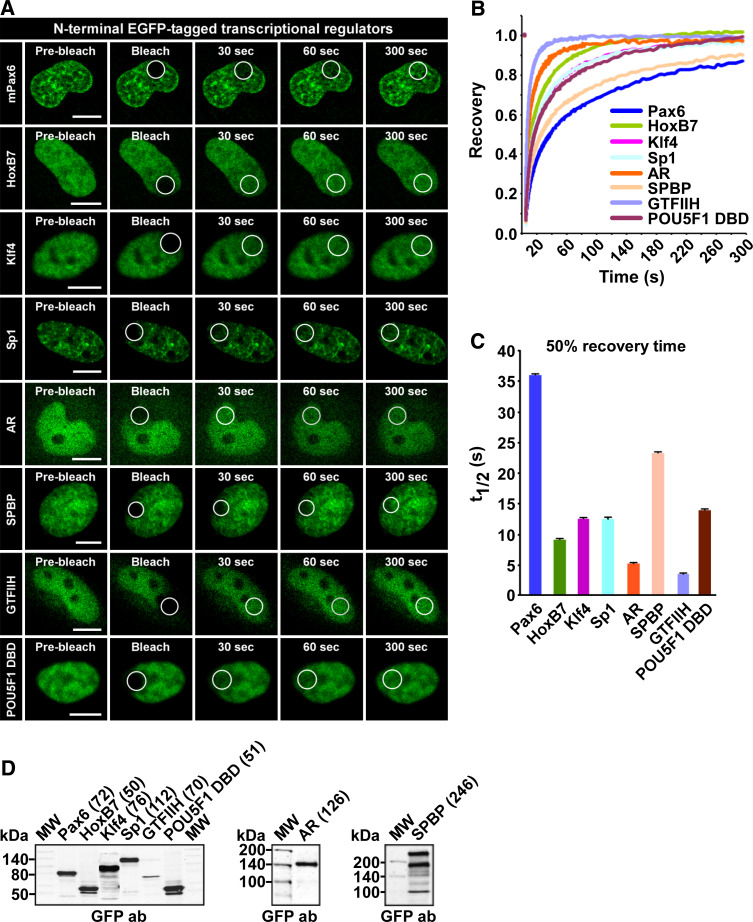
To compare the mobility of Pax6 with that of other nuclear proteins involved in transcriptional regulation, we chose proteins representing various transcription factor families and various sizes, as indicated in the Western blot in Fig. 3d. GTFIIH is one of the many general transcription factors required by RNA polymerase II to initiate transcription. The homeodomain containing protein HoxB7 is a sequence-specific transcription factor belonging to the same superfamily as the Pax proteins. SPBP (stromelysin-1 platelet-derived growth factor-responsive element-binding protein) is an ubiquitously expressed, large nuclear protein able to act both as a transcriptional co-activator and as a co-repressor [52–54]. Klf4 and Sp1 are members of the Krüppel-like family of zinc-finger-containing transcription factors, while the androgen receptor (AR) belongs to the nuclear receptor superfamily. Previously, the latter was extensively studied with FRAP and was shown to behave differently before and after stimulation with androgen [39, 41]. The FRAP experiments conducted in this study were done without androgen stimulation of the cells. Hence, AR is found both in the nucleus and cytoplasm. However, the cells are kept in serum that has not been charcoal-filtered, thus containing some ligands that will stimulate the receptor. Finally, we analyzed POU5F1 (Oct 3/4), a transcription factor containing two DNA-binding domains, the POU domain and a homeodomain. FRAP was conducted with a construct containing the DNA-binding domains of POU5F1 (amino acid 136–360).
Fig. 3.
Pax6 has low mobility compared to other transcriptional regulators. a Time-lapse imaging of HeLa cells transiently expressing the indicated N-terminal EGFP-tagged nuclear proteins, before and at selected time points after bleaching. Scale bars 10 μm. b FRAP recovery curves displaying the relative recovery rate into the bleached area over the indicated time period. See legend to Fig. 2 for details. c Graphical display of t 1/2 values, here defined as the time point after bleaching where the fluorescence has increased to 50% of the initial level, with standard error bars included. d Western blot of extracts from transiently transfected HeLa cells, using an anti-rabbit GFP antibody (AbCam). The numbers in brackets are the theoretical size of each fusion protein in kDa
As shown in Fig. 3, Pax6 has the slowest recovery curve of all the proteins tested (see also supplementary material Figure S3). The results also show that size has a relatively modest influence on the mobility of proteins in the nucleus. AR, SPBP and Sp1, all considerably larger than Pax6, have higher recovery rates than the Pax protein. According to Sprouse et al. [32], the dependence of diffusion rate on molecular volume means that an eight-fold increase in mass would be required for a two-fold decrease in the free diffusion rate. Hence, differences in size cannot explain the observed differences in mobility.
Most Pax family members display low nuclear mobility
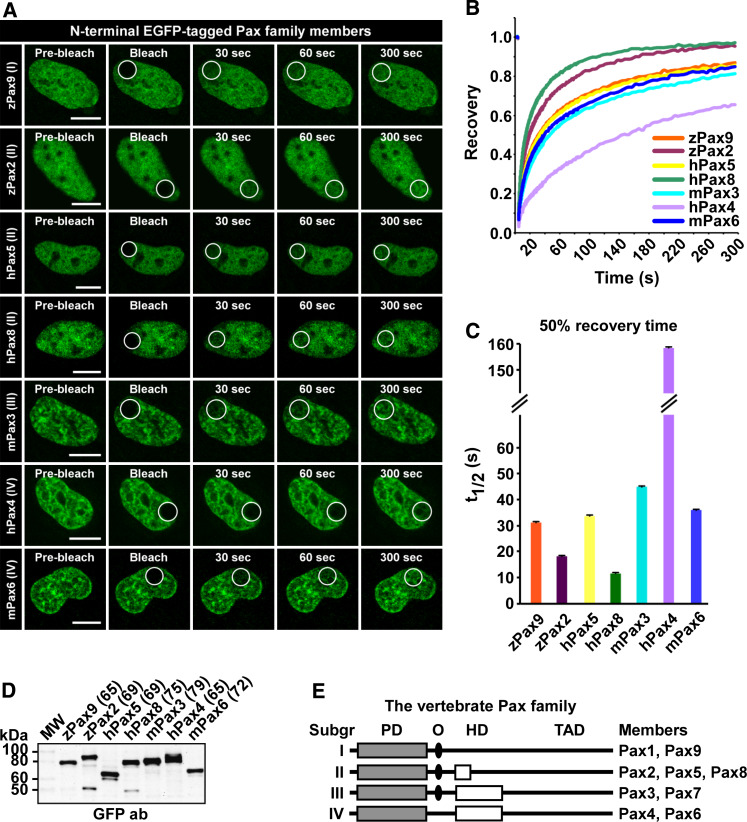
The nine members of the vertebrate Pax family are divided into four groups based on structural similarity and domain architecture (Fig. 4e). FRAP was conducted to compare the mobility of different Pax family members. The results of this comparison are presented in Fig. 4 and supplementary material Figure S4. Pax6 is not the only Pax family member with relatively low mobility in HeLa cells. In fact, Pax4 has a t 1/2 value of 158 s (Fig. 4c) and is by far the most slowly moving transcription factor tested. In addition, Pax3 also has relatively slow recovery, with a t 1/2 value of 47 s. Common for Pax3, Pax4 and Pax6 is that they all contain the PD plus an intact HD, suggesting that the presence of two DNA-binding domains strongly contributes to the slow mobility observed. Also, the Pax proteins with only one DNA-binding domain displayed a more diffuse localization pattern than the ones with two DNA-binding domains (Fig. 4a).
Fig. 4.
The Pax family of transcription factors displays different nuclear mobility. a Time-lapse imaging of HeLa cells transiently expressing N-terminal EGFP-tagged Pax family proteins, before and at selected time points after bleaching. Scale bars 10 μm. b FRAP recovery curves displaying the relative recovery rate into the bleached area over the indicated time period. See legend to Fig. 2 for details. c Graphical display of t 1/2 values, here defined as the time point after bleaching where the fluorescence has increased to 50% of the initial level, with standard error bars included. d Western blot of extracts from transiently transfected HeLa cells, using an anti-rabbit GFP antibody (AbCam). The numbers in brackets are the theoretical size of each fusion protein in kDa. e Schematic illustration of the structural features of the different subgroups of the vertebrate Pax family. PD paired domain, O octapeptide, HD homeodomain, TAD transactivation domain, z zebrafish, h human, m mouse
However, the presence of two DNA-binding domains cannot be the only explanation for the slow recovery curves obtained for these three Pax proteins. Pax9 and Pax5, both having only a single DNA-binding domain, have almost the same 50% recovery time as Pax6 (31.5 and 33.3 vs. 36.5 s, respectively). Members within the different groups, although structurally similar, do not share the same nuclear mobility. Pax6 and Pax4 are one example, Pax2, Pax5 and Pax8 another. For group II, we found that Pax2 and Pax8 diffuse relatively rapidly, with 50% recovery of 17.7 and 11.9 s, respectively. As mentioned above, Pax5, on the other hand, has a 50% recovery of 33.3 s. The differences in nuclear mobility across the groups are also illustrated by their immobile fractions, although the exact size of these fractions cannot be calculated. In this context, the Pax family members segregate into three groups. Pax4 has an immobile fraction of approximately 40%, which is by far the largest of all. The second group consists of Pax3, Pax5, Pax6 and Pax9, with an immobile fraction of about 10%, while Pax8 and Pax2 have reached almost full recovery after 5 min.
Taken together, the presence or absence of two DNA-binding domains is not the sole cause of slow nuclear mobility when looking at different Pax family proteins. However, all Pax proteins with two DNA-binding domains display slower mobility than those with only one.
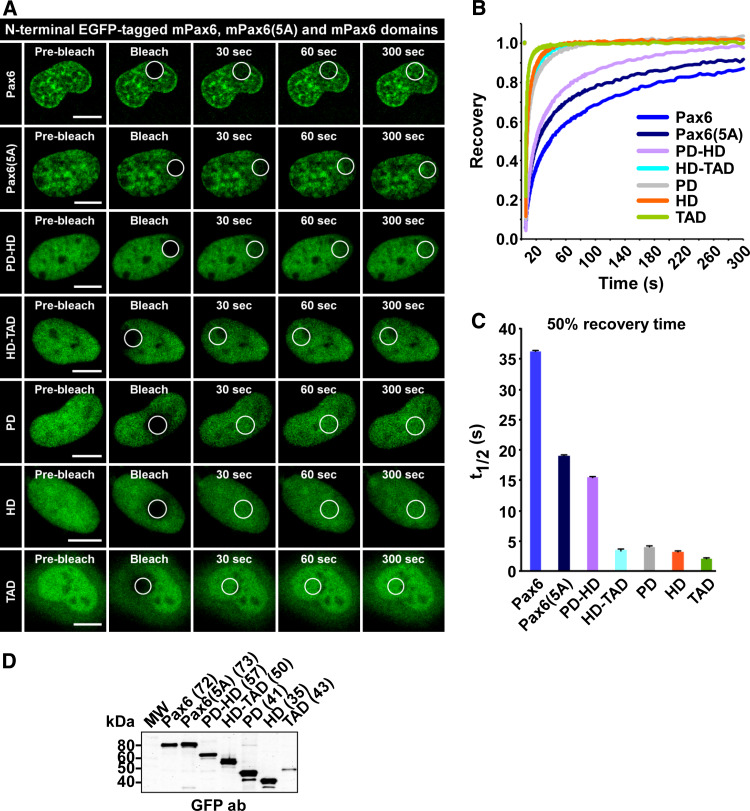
Two DNA-binding domains are required for the low mobility of Pax6
The PD and the HD of Pax6 are important both for DNA-binding and protein-protein interactions [6, 7]. To assess if certain domains of the transcription factor are more important for the mobility of Pax6 than others, the various Pax6 domains alone or in combination were assayed in FRAP experiments. The recovery curves in Fig. 5b and the diagram in 5c show that each of the three domains (PD, HD and TAD) has a 50% recovery time into the bleached area from 2.2 to 4.0 s when assayed separately (GFP alone = 1.7). The recovery curves are also presented separately with standard error bars in supplementary material Figure S5. When both DNA-binding domains (PD and HD) are present, the recovery curve is slowed down considerably. This suggests that these two domains together are important for the low mobility of Pax6. Supporting the notion that the presence of more than one DNA-binding domain reduces nuclear mobility is our finding that the two DNA-binding domains of POU5F1 display a similar 50% recovery time (14 s; Fig. 3) as the isolated PD-HD construct of Pax6 (15.5 s). However, the 50% recovery time for the Pax6 PD-HD fusion protein is more than twice as fast as for full-length Pax6. This indicates that all parts of the protein are necessary for proper interactions with DNA, chromatin, protein complexes or other structures in the nucleus. It is well known that Pax6 interacts with other transcription factors [55] and chromatin remodeling complexes [56, 57]. These interactions most probably influence the mobility of Pax6. The results in Fig. 5 show that there is a correlation between the recovery kinetics for the different Pax6 domains and their subnuclear localization pattern. The PD, HD and TAD all display a relatively diffuse nuclear distribution and a high recovery rate. Combining the PD and the HD gives a nuclear distribution pattern that resembles that of full-length Pax6, with resulting reduced nuclear mobility.
Fig. 5.
The DNA-binding domains of Pax6 are important for the low mobility of the transcription factor. a Time-lapse imaging of HeLa cells transiently expressing N-terminal EGFP-tagged Pax6 domains and the Pax6(5A) splice variant, before and at selected time points after bleaching. Scale bars 10 μm. b FRAP recovery curves displaying the relative recovery rate into the bleached area over the indicated time period. See legend to Fig. 2 for details. c Graphical display of t 1/2 values, here defined as the time point after bleaching where the fluorescence has increased to 50% of the initial level, with standard error bars included. d Western blot of extracts from transiently transfected HeLa cells, using an anti-rabbit GFP antibody (AbCam). The numbers in brackets are the theoretical size of each fusion protein in kDa. PD paired domain, HD homeodomain, TAD transactivation domain
The PD can be further divided in two subdomains, PAI and RED, that both bind DNA independently [5]. The Pax6(5A) splice variant has a 14-amino acid insertion in the PAI subdomain of the PD. This insertion disrupts DNA binding by PAI, resulting in altered DNA-binding specificity [58]. We found that Pax6(5A) has increased mobility compared to wild-type Pax6 (Fig. 5), supporting that the splice variant has lower DNA-binding affinity, in addition to altered protein-protein interaction capabilities.
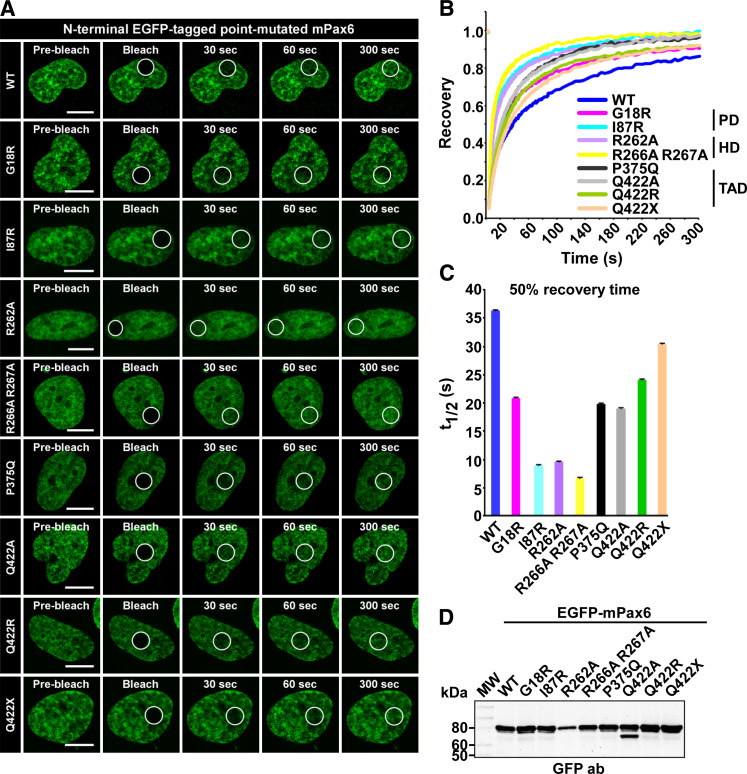
Disease-causing missense mutations influence the mobility of Pax6
Aniridia and other ocular defect-causing mutations of Pax6 are reported to affect DNA-binding and transactivation [59–61]. In order to test if some of the missense mutations associated with eye diseases also affected the nuclear mobility of Pax6, we conducted FRAP experiments using two mutants with mutations in the PD (G18R and I87R) and two in the C-terminal TAD (P375Q and Q422R). In addition to the disease-causing mutations, the HD mutations R262A and R266A R267A were also studied. These were hitherto not known to cause any diseases, but were included in this study because they are reported to affect protein-protein interactions (both mutations) and DNA binding (R262A only) [7].
As the FRAP-recovery curves in Fig. 6b and supplementary material Figure S6 show, all the mutants tested displayed higher recovery rates than the wild-type protein. The mutant Pax6 with the highest recovery rate is the R266A R267A double mutant, previously known to affect protein-protein interactions. This particular mutant has a t 1/2 value of 6.5 s (Fig. 6c) compared to the 36.5 s used by wild-type Pax6. The HD mutant R262A, known to affect both protein-protein interactions and DNA binding, also has a relatively high recovery rate, with a t 1/2 value of approximately 10 s. Of the known disease-causing mutations, I87R has particularly high mobility compared with the wild-type protein. This mutant has been reported to have a reduced DNA-binding ability in vitro [62]. This is consistent with the results of our in vivo analyses using FRAP.
Fig. 6.
Disease-causing point mutations increase the mobility of Pax6. a Time-lapse imaging of HeLa cells transiently expressing N-terminal EGFP-tagged point mutated Pax6, before and at selected time points after bleaching. Scale bars 10 μm. b FRAP recovery curves displaying the relative recovery rate into the bleached area over the indicated time period. See legend to Fig. 2 for details. c Graphical display of t 1/2 values, here defined as the time point after bleaching where the fluorescence has increased to 50% of the initial level, with standard error bars included. d Western blot of extracts from transiently transfected HeLa cells, using an anti-rabbit GFP antibody (AbCam)
The FRAP results further indicate that the last amino acid of Pax6 is important for the mobility of the protein. Changing this amino acid from glutamine to alanine, to arginine or to a stop codon all had different effects on the recovery curves. The alanine mutant (Q422A) had a 50% recovery of 19 s, whereas the aniridia-causing arginine (Q422R) mutant used 24 s. Deleting this last amino acid decreased the 50% recovery time from 36.5 to 30.5 s. The results clearly suggest that the size, properties and presence of the C-terminal amino acid are all important for the mobility of Pax6 in the nucleus. This may be attributed to the finding that this mutation leads to loss of DNA binding by the HD of Pax6 [59]. P375Q, another aniridia-causing mutation in the TAD, is reported to have normal transactivation activity, but decreased DNA binding via the PD compared to the wild-type protein [59]. This decreased DNA binding is most likely the reason why the P375Q mutant has nearly twice as high 50% recovery as wild-type Pax6.
The G18R mutation, thought to behave as a partial loss of function mutation [60], also has increased nuclear mobility compared to wild-type Pax6. This mutation is located in the PAI subdomain of the PD, as is the insertion in the Pax6(5A) splice variant. Interestingly, superimposing the recovery curves for Pax6 G18R and Pax6(5A) shows that the two proteins have near identical recovery into the bleached area. The insertion in Pa6(5A) is known to disrupt DNA binding by the PAI subdomain of the PD. The fact that the G18R mutant behaves in a similar manner as Pax6(5A) in FRAP indicates that this point mutation also disrupts DNA-binding by the PAI subdomain.
The FRAP results correlate well with the subnuclear distribution of the different mutants. As shown in Fig. 6a and supplementary material Figure S7, a large fraction of the cells expressing I87R, R262A and R266A R267A shows a much more diffuse localization than the “territorial pattern” characteristic for wild-type Pax6. The other mutants with wild-type DNA-binding domains have a localization pattern highly similar to the wild-type protein.
Taken together, these FRAP results strongly suggest that DNA binding and protein-protein interactions together are responsible for the slow nuclear mobility of Pax6.
Discussion
In this study we report that Pax6 and also other Pax family proteins have surprisingly low nuclear mobility compared to other transcription factors. Since the AR, SPBP and Sp1, all considerably larger than Pax6, have several fold higher mobility, size alone cannot explain the recovery curves observed. Our results show that the presence of two DNA-binding domains, the PD and the HD, is crucial for the slow mobility. This is supported by the fact that both Pax3 and Pax4, two other members of the Pax family also having an intact PD and HD, actually display even slower nuclear mobility than Pax6. However, the two DNA-binding domains are not solely determinative for the relatively low nuclear mobility, since Pax5 and Pax9 have recovery curves similar to Pax6, although both lack the HD. Furthermore, Pax2, Pax5 and Pax8 all have the same structural features, but do not display similar mobility. This implies that observed differences in recovery curves cannot simply be explained by additive effects of DNA-binding domains. The affinity and strength of both protein-DNA and protein-protein interactions influence the mobility. In addition, the number of available DNA-binding sites is of importance. Pax6 binds to a subset of Alu elements constituting approximately 0.2% of the Alu elements in the human genome. There are about 5,700 such Alu elements containing a Pax6-binding site [63].
Slow nuclear mobility may reflect the presence of multiple binding sites and/or interaction partners. The presence of two DNA-binding domains both increases the number of binding sites and perhaps also the binding affinity, thereby contributing strongly to the observed slow nuclear mobility of Pax6. The results of the chromatin association assay show that a significant portion of Pax6 is bound to chromatin. We further found that Pax6 is highly co-localized with chromatin-rich territories preferentially stained with Hoechst 33258. Since strong Hoechst staining has been correlated with heterochromatin, one could speculate that this colocalization suggests that Pax6 may predominantly act as a transcriptional repressor rather than a transcriptional activator in HeLa cells. The notion that colocalization with Hoechst in chromatin-rich territories can be linked to slow nuclear mobility is supported by the results found for Pax3. Consistently, Sp1, which has a rather high nuclear mobility, shows the lowest correlation with the Hoechst staining. However, since HoxB7, with a Pearson’s correlation coefficient equal to Pax6 and Pax3, has a 50% recovery time below 10 s, colocalization with Hoechst-positive chromatin is not always an indication of slow nuclear mobility. It is striking that the three homeodomain proteins, Pax6, Pax3 and HoxB7, all have a localization pattern that overlaps with the Hoechst dye. We also found the subnuclear distribution pattern of Pax6 to be very similar to what is reported for other transcription factors like Pax3 [48] and Pit-1 [64].
We observed that the most slowly moving proteins gave the highest standard error bars on the recovery curves. This could reflect that there are at least two populations of these particular proteins. In this model, one pool is more dynamic than the other, and the diffusion might also depend on the activity level in the cell. The presence of populations with different nuclear mobility has been shown for both heterochromatin protein 1 [65] and the fibroblast growth factor receptor-1 [66]. In the latter case, FGFR1 exists as three different populations where the slowest moving reflects the chromatin-bound fraction, while the immobile population is bound to the nuclear matrix. Transcriptional activation through the co-activator CBP converted FGFR1 from immobile and fast moving to the slower chromatin-bound form. It would be interesting to test if this applies to Pax6 as well.
A few publications link disease-causing Pax6 mutations to altered biochemical properties. I87R, a paired domain missense mutation from a patient with aniridia, displayed reduced DNA-binding and transactivation capabilities [62]. P375Q and Q422R, two aniridia-causing missense mutations in the transactivation domain, were investigated by Singh et al. [59]. P375Q had normal transactivation activity, but reduced DNA binding through the paired domain, while Q422R was unable to bind DNA through the homeodomain. These biochemical data are supported by the increased mobility observed for these mutations in our FRAP experiments conducted with live cells. The fact that amino acids in the transactivation domain also are found mutated in patients with ocular defects, and that these mutations display increased mobility, underscores that not only the conserved DNA-binding domains are important for the nuclear mobility of Pax6. The importance of an intact, wild-type transactivation domain is further supported by the fact that several aniridia patients have a mutation in the stop codon that is predicted to cause continued translation into the 3’ untranslated region of Pax6, possibly causing a poly-lysine tract in the mutant protein [59, 67–71].
The paired domain missense mutation G18R isolated from a patient with Peter’s anomaly is thought to result in partial loss of function [60], although no biochemical data have been published for this particular mutation. However, the increased mobility observed here supports this suggestion. Further support for the increased nuclear mobility being caused by altered DNA- and/or protein-binding properties comes from the observation that the homeodomain mutations R262A and R266A R267A both have increased mobility. R262 has previously been shown to be important for both DNA- and protein-protein interactions, while R266 and R267 are important for protein-protein interactions [7]. These residues all lie in the third helix of the homeodomain, the helix that mediates specific binding of the HD to the RED subdomain of the PD. This binding can occur both intra- and inter-molecularly, and is not dependent of DNA [7]. Most homeodomains bind DNA as a monomer. The paired class homeodomains, however, bind cooperatively as homodimers [72, 73].
In a recent publication, Corry et al. [48] showed that Pax3 mutations known to cause Waardenburg syndrome displayed increased mobility compared to wild-type Pax3 in accordance with what we found for Pax6. Corry et al. further found that the Pax3 mutants fell into two classes. One class was characterized by diffuse distribution and increased mobility, but retained DNA-binding activity. The other class showed subnuclear compartmentalization similar to wild-type Pax3 and slightly increased mobility, but deficient DNA-binding. It is suggested that the increased Pax3 mobility reflects an impaired ability to form functional transcription complexes. They further suggest that this is a general characteristic of disease-causing missense alleles, something our FRAP results on disease-causing Pax6 mutations support. Several studies of transcription factor mutations show a correlation between increased mobility and decreased DNA-binding affinity [35, 64, 74]. However, the opposite was found for disease-causing mutations in Pax3 where intact DNA binding correlated with increased nuclear mobility [48]. Contrary to this situation, our FRAP results clearly show that mutants of Pax6 adversely affecting DNA binding display an increased mobility. This is also the case for the double mutant in the homeodomain, R266A R267A, which only affects protein-protein interactions but not DNA binding. It actually showed the most pronounced increased mobility of the mutants tested. However, only a few seconds separate this particular mutant from the single mutants I87R and R262A, the latter of which is known to affect both DNA- and protein-protein interactions.
In conclusion, our live cell analyses using FRAP have revealed that Pax6 displays slow nuclear mobility in HeLa cells largely because of the presence of two DNA-binding domains. Importantly, we also show that disease-causing mutations affecting the PD and the TAD of Pax6 alter the nuclear mobility of the protein significantly.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Confocal pictures of HeLa cells transiently expressing GFP, Cherry, GFP and Cherry, GFP and Cherry-Pax6, or GFP-Pax6 and Cherry. The pictures show that the punctuated nuclear pattern observed for Pax6 is not caused by the tags used. Subconfluent HeLa cells were transfected with the different constructs as described in the methods section. The following day, confocal images were taken with the cells kept in 1 × HBSS (Invitrogen) supplemented with 10% fetal calf serum. Scale bars: 10 μm. Supplementary material 2 (TIFF 2.18 mb)
a FRAP recovery curves for GFP-Pax6 in HeLa and U2OS cells. b FRAP recovery curves for zebrafish Pax6 with N-terminal GFP or C-terminal YFP tag. Changing the location of the tag does not alter the mobility significantly. c Transactivation assay with HeLa and U2OS cells transiently transfected with the Pax6 paired domain reporter P6xCON-luc and 50 to 150 ng of GFP-Pax6 and Pax6-GFP. pCMV-βgal was used to normalize the transfection efficiency, and pcDNA3 was used to equal the DNA concentration in each well. The fold activation is determined based on the background level obtained with pcDNA3 alone. The results are the average of three independent experiments, triplicates each time, with standard error bars included. Supplement to figure 2. Supplementary material 3 (TIFF 252 kb)
FRAP recovery curves for different N-terminal GFP-tagged transcriptional regulators compared with GFP-Pax6. Supplement to figure 3. Supplementary material 4 (TIFF 558 kb)
FRAP recovery curves for different N-terminal GFP-tagged Pax family members compared with GFP-Pax6. z, zebrafish; m, mouse; h, human. Supplement to figure 4. Supplementary material 5 (TIFF 551 kb)
FRAP recovery curves for different N-terminal GFP-tagged parts of Pax6 and the splice variant Pax6(5A) compared with full-length GFP-Pax6. PD, paired domain; HD, homeodomain; TAD, transactivation domain. Supplement to figure 5. Supplementary material 6 (TIFF 440 kb)
FRAP recovery curves for different N-terminal GFP-tagged point mutations in Pax6 compared with the wild-type protein. Supplement to figure 6. Supplementary material 7 (TIFF 725 kb)
Subnuclear localization of mutated versus wild-type GFP-tagged Pax6. Subconfluent HeLa cells were transfected with the different constructs as described in the methods section. The following day, confocal images were taken with the cells kept in 1 × HBSS (Invitrogen) supplemented with 10% fetal calf serum. Scale bars: 5 μm. Supplementary material 8 (TIFF 12.4 mb)
Acknowledgments
We are grateful to Ernst Thomassen, Ingvild Mikkola, Endalkachew A. Alemu, J.Y. Thuret and J.A. Goldman for their generous gifts of the plasmid constructs used in this work. We thank the BioImaging FUGE core facility at IMB for use of instrumentation and expert assistance. We are indebted to Kenneth Bowitz Larsen for guidance on the use of confocal microscopy software. This study was supported by grants from the Norwegian Cancer Society and the Blix Foundation to TJ.
Conflict of interest statement
None.
Abbreviations
- FRAP
fluorescence recovery after photobleaching
- PD
paired domain
- HD
homeodomain
- TAD
transactivation domain
- GFP
green fluorescent protein
References
- 1.Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell. 1986;47:1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- 2.Noll M. Evolution and role of Pax genes. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437X(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 3.Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development. 1991;113:1193–1206. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- 4.Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/S0168-9525(01)02594-X. [DOI] [PubMed] [Google Scholar]
- 5.Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 6.Mikkola I, Bruun J-A, Holm T, Johansen T. Superactivation of Pax6-mediated transactivation from paired domain-binding sites by DNA-independent recruitment of different homeodomain proteins. J Biol Chem. 2001;276:4109–4118. doi: 10.1074/jbc.M008882200. [DOI] [PubMed] [Google Scholar]
- 7.Bruun J-A, Thomassen EIS, Kristiansen K, Tylden G, Holm T, Mikkola I, Bjørkøy G, Johansen T. The third helix of the homeodomain of paired class homeodomain proteins acts as a recognition helix both for DNA and protein interactions. Nucleic Acids Res. 2005;33:2661–2675. doi: 10.1093/nar/gki562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Planque N, Leconte L, Coquelle FM, Martin P, Saule S. Specific Pax-6/microphthalmia transcription factor interactions involve their dna-binding domains and inhibit transcriptional properties of both proteins. J Biol Chem. 2001;276:29330–29337. doi: 10.1074/jbc.M101812200. [DOI] [PubMed] [Google Scholar]
- 9.Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- 10.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 11.Carriere C, Plaza S, Martin P, Quatannens B, Bailly M, Stehelin D, Saule S. Characterization of quail Pax-6 (Pax-QNR) proteins expressed in the neuroretina. Mol Cell Biol. 1993;13:7257–7266. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Lauderdale JD. Analysis of Pax6 expression using a BAC transgene reveals the presence of a paired-less isoform of Pax6 in the eye and olfactory bulb. Dev Biol. 2006;292:486–505. doi: 10.1016/j.ydbio.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 13.Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 14.Mishra DK, Chen Z, Wu Y, Sarkissyan M, Koeffler HP, Vadgama JV. Global methylation pattern of genes in androgen-sensitive and androgen-independent prostate cancer cells. Mol Cancer Therapeutics. 2010;9:33–45. doi: 10.1158/1535-7163.MCT-09-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shyr C-R, Tsai M-Y, Yeh S, Kang H-Y, Chang Y-C, Wong P-L, Huang C-C, Huang K-E, Chang C. Tumor suppressor PAX6 functions as androgen receptor co-repressor to inhibit prostate cancer growth. Prostate. 2010;70:190–199. doi: 10.1002/pros.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y-H, Tan F, Hess KR, Yung WKA. The expression of PAX6, PTEN, vascular endothelial growth factor, and epidermal growth factor receptor in gliomas. Clin Cancer Res. 2003;9:3369–3375. [PubMed] [Google Scholar]
- 17.Zhou Y-H, Wu X, Tan F, Shi Y-X, Glass Y-X, Liu TJ, Wathen K, Hess KR, Gumin J, Lang F, Yung WKA. PAX6 suppresses growth of human glioblastoma cells. J Neurooncol. 2005;71:223–229. doi: 10.1007/s11060-004-1720-4. [DOI] [PubMed] [Google Scholar]
- 18.Mascarenhas JB, Young KP, Littlejohn EL, Yoo BK, Salgia R, Lang D. PAX6 is expressed in pancreatic cancer and actively participates in cancer progression through activation of the MET tyrosine kinase receptor gene. J Biol Chem. 2009;284:27524–27532. doi: 10.1074/jbc.M109.047209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 20.Solomon BD, Pineda-Alvarez DE, Balog JZ, Hadley D, Gropman AL, Nandagopal R, Han JC, Hahn JS, Blain D, Brooks B, Muenke M. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am J Med Genet A. 2009;149A:2543–2546. doi: 10.1002/ajmg.a.33081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pichaud F, Desplan C. Pax genes and eye organogenesis. Curr Opin Genet Dev. 2002;12:430–434. doi: 10.1016/S0959-437X(02)00321-0. [DOI] [PubMed] [Google Scholar]
- 22.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 23.Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- 24.Nelson LB, Spaeth GL, Nowinski TS, Margo CE, Jackson L. Aniridia. A review. Surv Ophthalmol. 1984;28:621–642. doi: 10.1016/0039-6257(84)90184-X. [DOI] [PubMed] [Google Scholar]
- 25.Brown A, McKie M, van Heyningen V, Prosser J. The human PAX6 mutation database. Nucleic Acids Res. 1998;26:259–264. doi: 10.1093/nar/26.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzoulaki I, White I, Hanson I. PAX6 mutations: genotype-phenotype correlations. BMC Genet. 2005;6:27. doi: 10.1186/1471-2156-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White J, Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/S0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- 28.Phair RD, Gorski SA, Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 2004;375:393–414. doi: 10.1016/S0076-6879(03)75025-3. [DOI] [PubMed] [Google Scholar]
- 29.Dobrucki JW, Feret D, Noatynska A. Scattering of exciting light by live cells in fluorescence confocal imaging: phototoxic effects and relevance for FRAP studies. Biophys J. 2007;935:1778–1786. doi: 10.1529/biophysj.106.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinze KG, Costantino S, De Koninck P, Wiseman PW. Beyond photobleaching, laser illumination unbinds fluorescent proteins. J Phys Chem B. 2009;113:5225–5233. doi: 10.1021/jp8060152. [DOI] [PubMed] [Google Scholar]
- 31.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 32.Sprouse RO, Karpova TS, Mueller F, Dasgupta A, McNally JG, Auble DT. Regulation of TATA-binding protein dynamics in living yeast cells. Proc Natl Acad Sci USA. 2008;105:13304–13308. doi: 10.1073/pnas.0801901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinow P, Rogers CE, Barbieri CE, Pietenpol JA, Kenworthy AK, DiBenedetto E. The DNA binding activity of p53 displays reaction-diffusion kinetics. Biophys J. 2006;91:330–342. doi: 10.1529/biophysj.105.078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moumne L, Dipietromaria A, Batista F, Kocer A, Fellous M, Pailhoux E, Veitia RA. Differential aggregation and functional impairment induced by polyalanine expansions in FOXL2, a transcription factor involved in cranio-facial and ovarian development. Hum Mol Genet. 2008;17:1010–1019. doi: 10.1093/hmg/ddm373. [DOI] [PubMed] [Google Scholar]
- 35.Schaaf MJM, Willetts L, Hayes BP, Maschera B, Stylianou E, Farrow SN. The relationship between intranuclear mobility of the NF-κB subunit p65 and its dna binding affinity. J Biol Chem. 2006;281:22409–22420. doi: 10.1074/jbc.M511086200. [DOI] [PubMed] [Google Scholar]
- 36.Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-κB-dependent gene activity. EMBO J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNally JG, Müller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 38.Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farla P, Hersmus R, Geverts B, Mari PO, Nigg AL, Dubbink HJ, Trapman J, Houtsmuller AB. The androgen receptor ligand-binding domain stabilizes DNA binding in living cells. J Struct Biol. 2004;147:50–61. doi: 10.1016/j.jsb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Rayasam GV, Elbi C, Walker DA, Wolford R, Fletcher TM, Edwards DP, Hager GL. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol Cell Biol. 2005;25:2406–2418. doi: 10.1128/MCB.25.6.2406-2418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klokk TI, Kurys P, Elbi C, Nagaich AK, Hendarwanto A, Slagsvold T, Chang C-Y, Hager GL, Saatcioglu F. Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol Cell Biol. 2007;27:1823–1843. doi: 10.1128/MCB.01297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuda K, Nishi M, Takaya H, Kaku N, Kawata M. Intranuclear mobility of estrogen receptor alpha and progesterone receptors in association with nuclear matrix dynamics. J Cell Biochem. 2008;103:136–148. doi: 10.1002/jcb.21393. [DOI] [PubMed] [Google Scholar]
- 43.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/S0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 44.Nornes S, Clarkson M, Mikkola I, Pedersen M, Bardsley A, Martinez JP, Krauss S, Johansen T. Zebrafish contains two Pax6 genes involved in eye development. Mech Dev. 1998;77:185–196. doi: 10.1016/S0925-4773(98)00156-7. [DOI] [PubMed] [Google Scholar]
- 45.Wu Z-H, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 46.Fedorova E, Zink D. Nuclear genome organization: common themes and individual patterns. Curr Opin Genet Dev. 2009;19:166–171. doi: 10.1016/j.gde.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Towbin BD, Meister P, Gasser SM. The nuclear envelope—a scaffold for silencing? Curr Opin Genet Dev. 2009;19:180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Corry GN, Hendzel MJ, Underhill DA. Subnuclear localization and mobility are key indicators of PAX3 dysfunction in Waardenburg syndrome. Hum Mol Genet. 2008;17:1825–1837. doi: 10.1093/hmg/ddn076. [DOI] [PubMed] [Google Scholar]
- 49.Singh BK, Gupta JP. Hoechst fluorescence pattern of heterochromatin in three closely related members of Drosophila. Chromosoma. 1982;87:503–506. doi: 10.1007/BF00333471. [DOI] [PubMed] [Google Scholar]
- 50.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Global nature of dynamic protein–chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rekdal C, Sjottem E, Johansen T. The nuclear factor SPBP contains different functional domains and stimulates the activity of various transcriptional activators. J Biol Chem. 2000;275:40288–40300. doi: 10.1074/jbc.M006978200. [DOI] [PubMed] [Google Scholar]
- 53.Gburcik V, Bot N, Maggiolini M, Picard D. SPBP is a phosphoserine-specific repressor of estrogen receptor α. Mol Cell Biol. 2005;25:3421–3430. doi: 10.1128/MCB.25.9.3421-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sjottem E, Rekdal C, Svineng G, Johnsen SS, Klenow H, Uglehus RD, Johansen T. The ePHD protein SPBP interacts with TopBP1 and together they co-operate to stimulate Ets1-mediated transcription. Nucleic Acids Res. 2007;35:6648–6662. doi: 10.1093/nar/gkm739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 56.Hussain MA, Habener JF. Glucagon gene transcription activation mediated by synergistic interactions of pax-6 and cdx-2 with the p300 co-activator. J Biol Chem. 1999;274:28950–28957. doi: 10.1074/jbc.274.41.28950. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Stopka T, Golestaneh N, Wang Y, Wu K, Li A, Chauhan BK, Gao CY, Cveklova K, Duncan MK, Pestell RG, Chepelinsky AB, Skoultchi AI, Cvekl A. Regulation of [alpha]A-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 59.Singh S, Chao LY, Mishra R, Davies J, Saunders GF. Missense mutation at the C-terminus of PAX6 negatively modulates homeodomain function. Hum Mol Genet. 2001;10:911–918. doi: 10.1093/hmg/10.9.911. [DOI] [PubMed] [Google Scholar]
- 60.van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002;11:1161–1167. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- 61.Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional properties of natural human PAX6 and PAX6(5a) mutants. Invest Ophthalmol Vis Sci. 2004;45:385–392. doi: 10.1167/iovs.03-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang HK, Chao LY, Saunders GF. Functional analysis of paired box missense mutations in the PAX6 gene. Hum Mol Genet. 1997;6:381–386. doi: 10.1093/hmg/6.3.381. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Y-H, Zheng JB, Gu X, Saunders GF, Yung WKA. Novel PAX6 binding sites in the human genome and the role of repetitive elements in the evolution of gene regulation. Genome Res. 2002;12:1716–1722. doi: 10.1101/gr.188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharp ZD, David LS, Maureen GM, Ilia IO, Michael AM. Inactivating Pit-1 mutations alter subnuclear dynamics suggesting a protein misfolding and nuclear stress response. J Cell Biochem. 2004;92:664–678. doi: 10.1002/jcb.20028. [DOI] [PubMed] [Google Scholar]
- 65.Schmiedeberg L, Weisshart K, Diekmann S, Meyer zu Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dunham-Ems SM, Lee Y-W, Stachowiak EK, Pudavar H, Claus P, Prasad PN, Stachowiak MK. Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol Biol Cell. 2009;20:2401–2412. doi: 10.1091/mbc.E08-06-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baum L, Pang CP, Fan DS, Poon PM, Leung YF, Chua JK, Lam DS. Run-on mutation and three novel nonsense mutations identified in the PAX6 gene in patients with aniridia. Hum Mutat. 1999;14:272–273. doi: 10.1002/(SICI)1098-1004(1999)14:3<272::AID-HUMU21>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 68.Sisodiya SM, Free SL, Williamson KA, Mitchell TN, Willis C, Stevens JM, Kendall BE, Shorvon SD, Hanson IM, Moore AT, van Heyningen V. PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet. 2001;28:214–216. doi: 10.1038/90042. [DOI] [PubMed] [Google Scholar]
- 69.Mitchell TN, Free SL, Williamson KA, Stevens JM, Churchill AJ, Hanson IM, Shorvon SD, Moore AT, van Heyningen V, Sisodiya SM. Polymicrogyria and absence of pineal gland due to PAX6 mutation. Ann Neurol. 2003;53:658–663. doi: 10.1002/ana.10576. [DOI] [PubMed] [Google Scholar]
- 70.De Becker I, Walter M, Noël LP. Phenotypic variations in patients with a 1630 A>T point mutation in the PAX6 gene. Can J Ophthalmol. 2004;39:272–278. doi: 10.1016/s0008-4182(04)80125-x. [DOI] [PubMed] [Google Scholar]
- 71.Chao L-Y, Mishra R, Strong LC, Saunders GF. Missense mutations in the DNA-binding region and termination codon in PAX6. Hum Mutat. 2003;21:138–145. doi: 10.1002/humu.10163. [DOI] [PubMed] [Google Scholar]
- 72.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 73.Wilson DS, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 74.Schaaf MJM, Cidlowski JA. Molecular determinants of glucocorticoid receptor mobility in living cells: the importance of ligand affinity. Mol Cell Biol. 2003;23:1922–1934. doi: 10.1128/MCB.23.6.1922-1934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 76.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 77.Galvani A, Courbeyrette R, Agez M, Ochsenbein F, Mann C, Thuret JY. In vivo study of the nucleosome assembly functions of ASF1 histone chaperones in human cells. Mol Cell Biol. 2008;28:3672–3685. doi: 10.1128/MCB.00510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confocal pictures of HeLa cells transiently expressing GFP, Cherry, GFP and Cherry, GFP and Cherry-Pax6, or GFP-Pax6 and Cherry. The pictures show that the punctuated nuclear pattern observed for Pax6 is not caused by the tags used. Subconfluent HeLa cells were transfected with the different constructs as described in the methods section. The following day, confocal images were taken with the cells kept in 1 × HBSS (Invitrogen) supplemented with 10% fetal calf serum. Scale bars: 10 μm. Supplementary material 2 (TIFF 2.18 mb)
a FRAP recovery curves for GFP-Pax6 in HeLa and U2OS cells. b FRAP recovery curves for zebrafish Pax6 with N-terminal GFP or C-terminal YFP tag. Changing the location of the tag does not alter the mobility significantly. c Transactivation assay with HeLa and U2OS cells transiently transfected with the Pax6 paired domain reporter P6xCON-luc and 50 to 150 ng of GFP-Pax6 and Pax6-GFP. pCMV-βgal was used to normalize the transfection efficiency, and pcDNA3 was used to equal the DNA concentration in each well. The fold activation is determined based on the background level obtained with pcDNA3 alone. The results are the average of three independent experiments, triplicates each time, with standard error bars included. Supplement to figure 2. Supplementary material 3 (TIFF 252 kb)
FRAP recovery curves for different N-terminal GFP-tagged transcriptional regulators compared with GFP-Pax6. Supplement to figure 3. Supplementary material 4 (TIFF 558 kb)
FRAP recovery curves for different N-terminal GFP-tagged Pax family members compared with GFP-Pax6. z, zebrafish; m, mouse; h, human. Supplement to figure 4. Supplementary material 5 (TIFF 551 kb)
FRAP recovery curves for different N-terminal GFP-tagged parts of Pax6 and the splice variant Pax6(5A) compared with full-length GFP-Pax6. PD, paired domain; HD, homeodomain; TAD, transactivation domain. Supplement to figure 5. Supplementary material 6 (TIFF 440 kb)
FRAP recovery curves for different N-terminal GFP-tagged point mutations in Pax6 compared with the wild-type protein. Supplement to figure 6. Supplementary material 7 (TIFF 725 kb)
Subnuclear localization of mutated versus wild-type GFP-tagged Pax6. Subconfluent HeLa cells were transfected with the different constructs as described in the methods section. The following day, confocal images were taken with the cells kept in 1 × HBSS (Invitrogen) supplemented with 10% fetal calf serum. Scale bars: 5 μm. Supplementary material 8 (TIFF 12.4 mb)