Abstract
The pyridoxal 5′-phosphate-dependent enzyme cystathionine β-lyase (CBL) catalyzes the penultimate step in the de novo biosynthesis of Met in microbes and plants. Absence of CBL in higher organisms makes it an important target for the development of antibiotics and herbicides. The three-dimensional structure of cystathionine β-lyase from Arabidopsis was determined by Patterson search techniques, using the structure of tobacco (Nicotiana tabacum) cystathionine γ-synthase as starting point. At a resolution of 2.3 Å, the model was refined to a final crystallographic R-factor of 24.9%. The overall structure is very similar to other pyridoxal 5′-phosphate-dependent enzymes of the γ-family. Exchange of a few critical residues within the active site causes the different substrate preferences between Escherichia coli and Arabidopsis CBL. Loss of interactions at the α-carboxyl site is the reason for the poorer substrate binding of Arabidopsis CBL. In addition, the binding pocket of Arabidopsis CBL is larger than that of E. coli CBL, explaining the similar binding of l-cystathionine and l-djenkolate in Arabidopsis CBL in contrast to E. coli CBL, where the substrate binding site is optimized for the natural substrate cystathionine.
The whole genome sequence of the flowering plant Arabidopsis has been determined very recently (Arabidopsis Genome Initiative, 2000). This is the first known complete plant genome sequence and it provides the foundation for detailed functional characterization of plant genes. The function of a gene product is closely related to its three-dimensional structure and can be fully understood only when its three-dimensional structure is known. Projects to determine the three-dimensional structures of the gene products of whole genomes with high-throughput techniques have been started and are designated as structural genomics projects (Burley, 2000).
Cystathionine β-lyase (CBL) from Arabidopsis is an enzyme of the Met biosynthesis pathway of this plant. Met is an essential amino acid that can only be synthesized by microorganisms and plants.
In bacteria and higher plants, the synthesis of homo-Cys, the direct precursor of Met, proceeds via a transsulfuration reaction involving two pyridoxal 5′-phosphate (PLP)-dependent enzymes. Cystathionine γ-synthase (CGS) first catalyzes the synthesis of cystathionine from Cys and activated homo-Ser. Cystathionine is then cleaved by CBL to produce homo-Cys, pyruvate, and ammonia. Both enzymes (CGS and CBL) belong to the γ-family of PLP-dependent enzymes (Alexander et al., 1994). CGS is homologous to CBL in its amino acid sequence (Belfaiza et al., 1986) and quaternary structure (Holbrook et al., 1990; Kreft et al., 1994; Kaplan and Flavin, 1996). The native recombinant Arabidopsis CBL is a tetramer composed of four identical subunits of 46 kD, each being associated with one molecule of pyridoxal 5′-phosphate. A Schiff base is formed between the cofactor and the holoenzyme at the ε-amino group of Lys-278 (Ravanel et al., 1996). The amino acid sequence deduced from the sequence of Arabidopsis CBL (aCBL) cDNA showed significant homology with the Escherichia coli CBL (eCBL; 22% identity). Higher identities are obtained with E. coli and Arabidopsis CGS (32% and 28%, respectively; Ravanel et al., 1995) and tobacco (Nicotiana tabacum) CGS (36.8% for a common stretch of 367 amino acid residues). The structures of eCBL (Clausen et al., 1996), E. coli CGS (Clausen et al., 1998), and tobacco CGS (Steegborn et al., 1999) have been solved by x-ray crystallography at a resolution of 1.83, 1.5, and 2.9 Å, respectively.
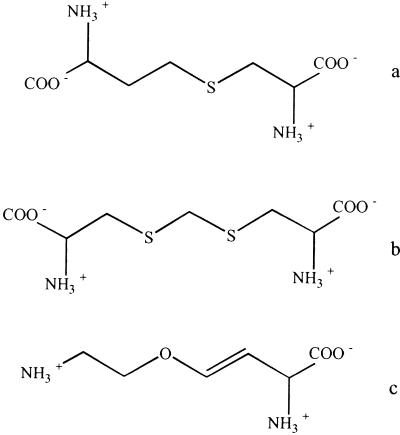
Plant and bacterial CBLs are inhibited by l-amino-ethoxyvinylglycine (AVG), an antimicrobial amino acid (Martel et al., 1987; Droux et al., 1995; Ravanel et al., 1996; Clausen et al., 1997; Turner et al., 1998), and rhizobitoxine, an antibacterial and phytotoxic amino acid (Owens et al., 1968; Giovanelli et al., 1972; Rando, 1975; Minamisawa et al., 1990). The CBL substrates l-cystathionine and l-djenkolate as well as the inhibitor AVG are shown in Figure 1. In addition, plant CBL is sensitive to thiol-blocking inhibitors such as N-ethylmaleimide and iodoacetamide (Giovanelli and Mudd, 1971; Burnell and Whatley, 1977; Gentry-Weeks et al., 1993; Gentry-Weeks et al., 1995; Ravanel et al., 1996). The 50% inhibition of initial activity values of 2.5 μm and 1.5 mm were determined for N-ethylmaleimide and iodacetamide, respectively (Ravanel et al., 1996). Therefore, it was suggested that at least one of the five Arabidopsis CBL Cys residues is located in or near the active site of the enzyme. The Arg-blocking reagent phenylglyoxal has no inhibitory effect on the enzyme, leading the authors to the conclusion that no Arg residue is involved in the catalytic function of CBL from Arabidopsis (Ravanel et al., 1996).
Figure 1.
Structures of CBL substrates. a, l-Cystathione; b, l-djenkolate; c, AVG (inhibitor).
The mature enzyme from Arabidopsis is a poly-peptide of approximately 46 kD. It is originally synthesized as a 50.4-kD precursor that is processed within the chloroplast. The exact location of the maturation site is not known; Gln44 appears to be a good candidate for the cleavage of the precursor, leaving a protein of 45.7 kD (Ravanel et al., 1996).
Here, we report the crystal structure of Arabidopsis CBL at a resolution of 2.3Å. The size of the substrate binding pocket, as well as the exchange of several amino acids in the active site of E. coli and Arabidopsis CBL, underlie the differences in substrate affinity. As enzymes from anabolic pathways are important for plant growth they are attractive targets for the development of inhibitors used as herbicides.
RESULTS
Cloning, Expression, and Protein Purification
The cDNA coding for the putative mature aCBL was cloned into the E. coli expression vector pASK75 (IBA, Institut für Bioanalytik, Göttingen, Germany). The paCBL1 construct had a C-terminal affinity-peptide Strep-I tag (Schmidt et al., 1996), which after overexpression in E. coli strain JM83/paCBL1 could be used during the purification on a Streptavidin Sepharose column (IBA). About 10 mg of purified enzyme could be obtained from 1 L of cell culture.
Structure Solution and Quality of the Final Model
The crystal structure of Arabidopsis CBL was solved by molecular replacement at 2.3 Å resolution. The three-dimensional structure of tobacco CGS was taken as search model because tobacco CGS exhibits the highest amount of identity (36.8%) to aCGL. Patterson search techniques facilitate an x-ray crystal structure determination in such a way that one has to collect an x-ray data set from the native crystal only. The preparation of heavy-metal or selenomethionyl derivatives is not necessary. The phase information is obtained by searching the orientation and translation of the model of the related structure in the unit cell of the other macromolecule by Patterson search methods. In this study the program package AMoRe (Navaza, 1994) was used, and the calculations revealed one prominent solution with a correlation coefficient of 45.3 and a crystallographic R-value of 54.1% for all diffraction data to 3.5 Å resolution.
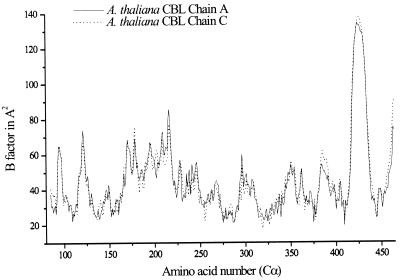
The final model was made up of two identical monomers of 379 amino acid and 253 water molecules. The N-terminal amino acids (Met57-Glu84) did not appear in the electron density, indicating that this part of the enzyme is highly flexible. In addition, residues 418–433 of the C-terminal domain and the C-terminal Strep-tag are not defined (Fig. 2). The final crystallographic R-factor of R = 24.9% and Rfree = 30.9% in a resolution range from 6.0 Å to 2.3 Å was rather high, due to the above-mentioned flexibilities. Properties of the final model are summarized in Table I. A Ramachandran plot (Ramachandran and Sasisekharan, 1968) shows 86.1% of the residues in the most favored regions, 12.5% in the favored regions, 1.2% in the generously allowed regions, and 0.3% in the disallowed region, as indicated by the program Procheck (Laskowski et al., 1993). The residues located in the disallowed region, Ser-258, Lys-278, and Ser-405, are well defined in the electron density map. Lys-278 and Ser-405 are active site residues. The 2Fo-Fc maps contoured at 1.2 ς showed continuous density for all main chain atoms except for the N-terminal residues already mentioned and for residues 418–433, which are localized on the surface of the protein. All active site side chains including the cofactor PLP are well defined; a bicarbonate, which was already shown to be present in E. coli CBL (Clausen et al., 1996), was also found in the active site of Arabidopsis CBL.
Figure 2.
Plot of the B-factors of the aCBL amino acids. Only the B-factors of the Cα-atoms are shown. Note the undefined region 417–436 (see text). The plot was created using the program Origin (Microcal, Northampton, MA).
Table I.
Data collection and refinement statistics
| Structure Refinement | aCBL Data |
|---|---|
| Space group | C2221 (orthorhombic) |
| Unit cell dimensions | a = 107.75 Å, b = 154.33 Å, c = 118.76 Å; α = β = γ = 90° |
| Resolution limits (Å) | 6.0–2.3 |
| No. of reflections | |
| Observed | 102,185 |
| Unique | 37,727 |
| Completeness (%) | |
| Total | 94.3 |
| Last shell | 81.6 |
| Rmergea (%) | |
| Total | 6.8 |
| Last shell | 13.5 |
| No. of atoms | |
| Protein | 5,671 |
| PLP cofactor | 30 |
| CO3 | 8 |
| SO4 | 5 |
| Solvent | 253 |
| R-valueb/Rfree-valuec (%) | 24.9/30.9 |
| r.m.s. Deviation | |
| Bond length (Å) | 0.015 |
| Bond angles (°) | 1.87 |
| B-factors (Å2) | |
| All atoms | 44.3 |
| Protein atoms | 44.5 |
| Main-chain atoms | 45.0 |
| PLP cofactor | 42.0 |
| CO3 | 49.3 |
| SO4 | 48.8 |
| Solvent atoms | 39.7 |
Rmerge = Σ(I − 〈I〉)/ΣI.
R = Σ(IFobs I − IFcalc I)/Σ IFobs I.
Rfree was calculated from 5% of measured reflections omitted from refinement.
Overall Fold
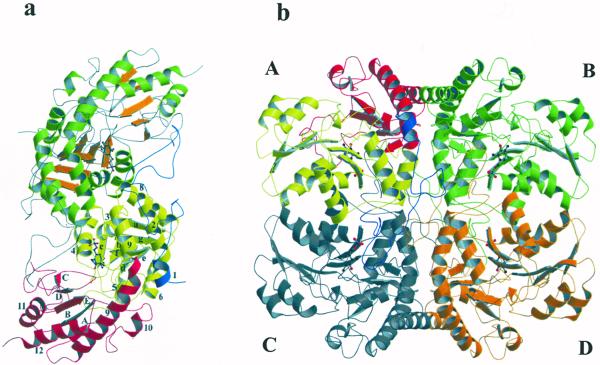
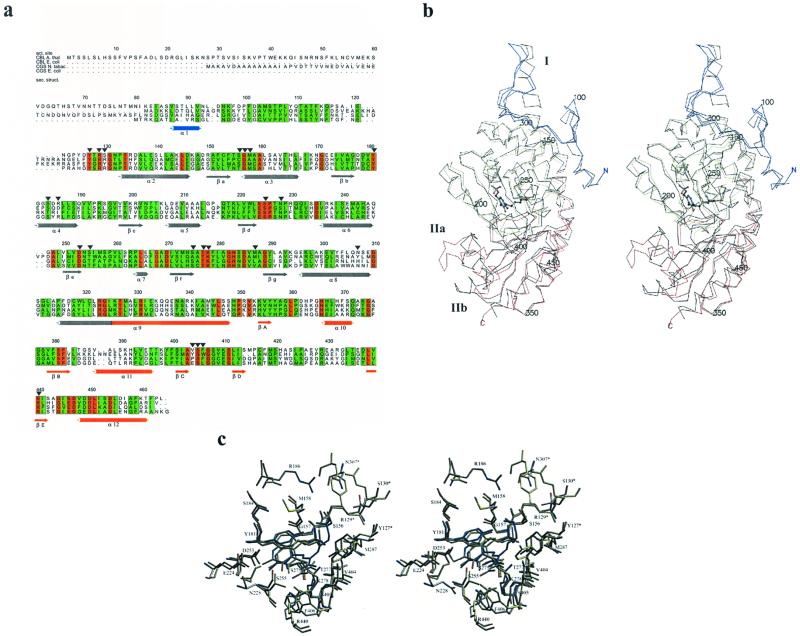
aCBL exists as an α4 tetramer in solution (Ravanel et al., 1996) and crystalline state. Figure 3 shows ribbon plots of the active dimer and the homotetramer. The asymmetric unit of the C2221 crystal form is built up by two monomers that share active site residues, thus forming the active dimer. The two monomers are related by a non-symmetric diad axis. Two active dimers, related by 2-fold crystallographic symmetry, form the tetramer. The dimensions of one monomer and the tetramer are 85 × 50 × 40 Å and 96 × 89 × 64 Å, respectively. Each monomer contains one PLP cofactor and 379 amino acids with 34.3% α-helical, 2.2% 310 helical, 14.1% β-sheet conformation, and 28.9% turns, leaving 20.5% random coil structure. The cofactors of the active dimers are separated by 22 Å. Like most PLP-dependent enzymes of the γ-family, the monomer of aCBL comprises three structurally and functionally distinct regions. The N-terminal domain (residues 57–131) is only partially defined (residues 57–84 are flexible) and interacts with the active site of the neighboring monomer. The large PLP-binding domain (residues 132–325) carries most of the catalytically important residues and is connected to the smaller C-terminal domain (residues 326–464) by the long α-helix 9 with a kink near residue Ala-329. The N-terminal domain consists of one α-helix and an extended loop structure. A seven-stranded mainly parallel β-sheet (a, g, f, e, d, b, c, with g in antiparallel orientation) that is bent around helix 3 is the central part of the PLP-binding domain. It is sandwiched between helices 2, 5, 6, and 7, shielding it from the solvent accessible surface, and helices 3, 4, and 8, which are part of the interdomain surface. PLP, covalently attached to Lys-278 (Ravanel et al., 1996) is located near the C termini of β-sheets d, e, and f. The phosphate group binds near the N terminus of helix 3. The major part of the C-terminal domain is a five-stranded, almost antiparallel β-sheet. Strands A, B, E, D, and C with directions +, −, +, −, and − are shielded from solvent by helices 10–12.
Figure 3.
Overall three-dimensional structure of aCBL. a, Active dimer of aCBL in ribbon representation. Monomer I shows the three domains: N-terminal domain is blue, PLP-binding domain is yellow, and C-terminal domain is red. The α-helices and β-strands are numbered as in the text. Monomer II illustrates the secondary structure elements: α-helices are green and β-strands are orange. PLP is drawn as a ball and stick model. b, The CBL tetramer is built up by two active dimers. The figure was produced using MOLSCRIPT (Kraulis, 1991) and Raster3D (Merrit and Murphy, 1994).
The active dimer-forming monomers associate tightly through hydrogen bonds, hydrophobic interactions and intermolecular active site interactions. The cofactor PLP is involved in several strong hydrogen bonds to amino acid residues of the adjacent subunit (Arg-129* and Tyr-127*). The extended loop of the N-terminal domain additionally stabilizes the dimer formation. Ala-119 and Gln-116 associate with Glu-395* and Ile-402* of the C-terminal domain. The helices 3, 4, 8, and 9 of the PLP-binding domain are in close contact with each other. Several hydrophobic residues of both symmetry mates (Leu-187, Val-191, Val-192, Leu-313, Ala-314, and Phe-316) build a large hydrophobic patch, which is further stabilized by hydrogen bonds between residues Gly-310(O)-Ser-156(OG), Ser-156(OG)-Ala-112(O), Asp-285(O)-Gln-110(NE2), and Arg-194(NH1)-His-166(O).
Upon association of the dimers AC and BD, a tetramer of overall 222 symmetry is formed. The most important elements for tetramer stabilization are the N-terminal half of helix 9 and the extended loop structure of the N-terminal domain.
Active Site Structure
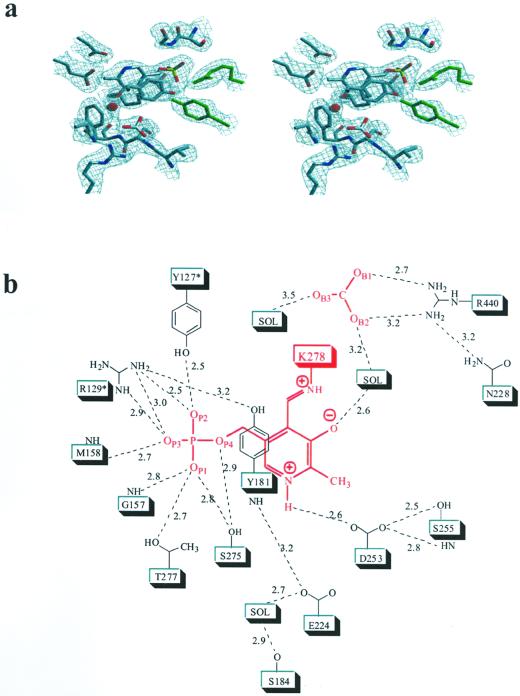
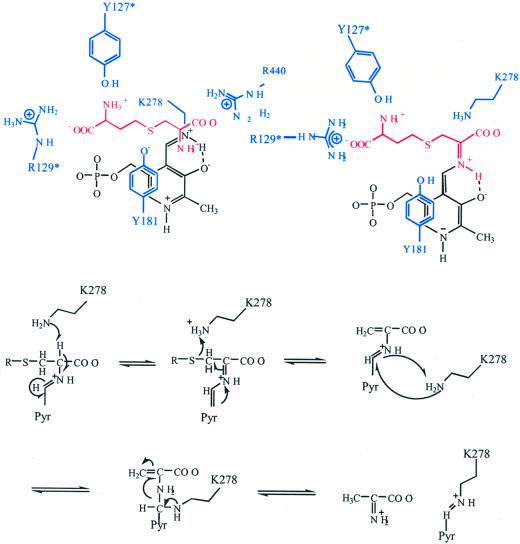
Figure 4a shows the 2Fo-Fc electron density map at 2.3-Å resolution of the aCBL active site region including the PLP cofactor. Interatomic distances are indicated in Figure 4b. In addition to the PLP-binding domain, parts of the C-terminal (C-terminal loop of strand C and strand D) and the neighboring N-terminal domain (loop around Tyr-127*) are involved in building the substrate binding pocket. The PLP cofactor is embedded by the N-terminal part of helix 3, the C termini of β-strands d, e, and f, and their connecting loops. Asp-253 plays a key role in stabilizing the positive charge of the pyridine N1 nitrogen, thereby increasing the electrophilic character of the cofactor. Its carboxylate OD2 forms a hydrogen bond (2.6 Å) with the PLP nitrogen N1; two further interactions between OD1 and the hydroxyl group (2.5 Å) as well as the amide nitrogen (2.8 Å) of Ser-255 tether Asp-253 in an optimal position to interact with the pyridine nitrogen. There are no specific interactions between the methyl group at C2 and the apoprotein. The hydroxyl group at C3 of PLP (presumably ionized) is placed in hydrogen bonding distance to a well-defined water molecule (2.6 Å). The aldehyde function of PLP at position 4 is observed as the internal aldimine resulting from the reaction with the ε-amino group of Lys-278. UV absorbance of the protein near 425 nm indicates protonation of this internal aldimin. The nitrogen NZ is involved in a weak hydrogen bond with the hydroxyl group of Ser-275 (3.1 Å).
Figure 4.
Active site structure of aCBL. a, Final 2Fo-Fc electron density, contoured at 1.2ς and calculated at 2.3 Å resolution. The region containing the cofactor PLP, the bicarbonate, and the immediate protein vicinity is shown. Molecules are colored by atom type, except residues of the neighboring subunit, which are shown in green. b, Schematic representation of the active site around the PLP cofactor (red). Hydrogen bonds and interatomic distances in angstroms are indicated. The figure was produced with SETOR (Evans, 1993). Residues of the neighboring subunit are marked by an asterisk.
The phosphate group is the main anchor of the cofactor within the active site. The four phosphate oxygens are involved in a total of nine interactions with the apoprotein. Gly-157 and Met-158 of the N terminus of α-helix 3 interact with OP1 and OP3, respectively. The hydroxyl groups of Ser-275 and Thr-277 are within hydrogen bonding distance to OP1 (2.8 and 2.7 Å); Ser-275 interacts additionally with OP4 (2.9 Å) and the nitrogen NZ at position 4 (see above). All other interactions originate from the neighboring subunit. Arg-129* and Tyr-127* are involved in a total of four hydrogen bonds to OP2 and OP3, thereby compensating the negative charge of the phosphate group. Arg-129* additionally interacts with the phenolic hydroxyl group of Tyr-181. The PLP pyridine ring is sandwiched between Ser-255 and Ser-275 on the protein facing side and Tyr-181 on the opposite side of the cofactor. The side chains of Ser-255 and Ser-275 lie within van der Waals distance to the pyridine ring atoms, thereby minimizing any movement of the cofactor in this direction. Furthermore, they can form stabilizing H-bonds with the π-electron system of this ring. The phenolic ring of Tyr-181 is almost coplanar to the pyridine ring of PLP. Stacking interactions between an aromatic side chain and the PLP ring system are found in most PLP-dependent enzymes and are thought to increase the electron sink character of the cofactor (Hayashi et al., 1990; John, 1995). The positive charges of the guanidinium group of Arg-129* and the protonated aldimine near Tyr-181 should lower the pKa of the Tyr hydroxylic group, thereby enabling this residue to play an essential role in catalysis by activating the incoming substrate for transaldimation. This is similar to the mechanism earlier proposed for eCBL (Clausen et al., 1996) and E. coli CGS (Clausen et al., 1998).
Comparison of aCBL with Other PLP-Dependent Enzymes
Overall Structure
As mentioned before, aCBL shows 22% identity to eCBL and 32% identity to E. coli CGS. A sequence alignment is shown in Figure 5a. PLP-dependent enzymes of the γ-family fold almost identically, despite moderate amino acid identity. A least square superposition of the aCBL and eCBL dimers as well as a superposition of their active site residues was carried out using the program O (Jones et al., 1991). The resulting overlays of the Cα traces and the active site residues are shown in Figure 5, b and c, respectively.
Figure 5.
Comparison of aCBL and E. coli CBL. a, Sequence alignment; numbering is given for aCBL. Arrows show aCBL active site residues; secondary structure elements (colored by domain) are shown. Conserved and homologous amino acids are boxed red and green, respectively. The figure was produced using the GCG package (GCG, Inc., Madison, Wisconsin) and the program ALSCRIPT (Barton, 1993). b, Stereo plot of the Cα-traces of aCBL (N-terminal domain is blue, PLP-binding domain is yellow, and C-terminal domain is red); the corresponding trace of eCBL (black) is superimposed. PLP is drawn as a ball and stick model. The figure was created using MOLSCRIPT (Kraulis, 1991). c, Stereo plot of the superimposed active sites of aCBL and eCBL. The aCBL residues with the PLP cofactor are colored by atom type, the residues of eCBL and the inhibitor AVG are drawn in yellow and blue, respectively. The figure was produced with SETOR (Evans, 1993).
The N-terminal regions of aCBL and eCBL fold similarly, despite the absence of any sequence similarity. The position of α-helix 3 (Fig. 5b, region I) in eCBL corresponds to a gap in the amino acid sequence of aCBL; the region around this gap is poorly defined in the aCBL structure. The only conserved amino acids, Tyr-127* and Arg-129*, are active site residues of the neighboring subunit. All secondary structure elements of the PLP-binding and most of the C-terminal domain superimpose almost perfectly. The C-terminal domain differs in secondary structure elements and structural folds at two nearby surface regions, marked IIa and IIb in Figure 5b. The aCBL amino acids 421 to 432 of region IIa were exchanged by poly-Ala because they were weakly defined in the electron density map during refinement. Here, the B-factors are approximately 120 Å2 (Fig. 2), indicating a very flexible surface region. The corresponding region in eCBL (residues 349–366) contains α-helix 15 and a following loop structure. Due to the flexibility of the aCBL fold, no information of structural differences in this region was available. Region IIb (382–396 in aCBL, 314–330 in eCBL) is situated near region IIa, forming an extended loop, including α-helix 11. Here, the B-factors are only slightly above the average (around 60 Å2). The extended eCBL loop is exposed to the solvent, in contrast to the corresponding loop structure of aCBL, which is embedded by the C-terminal domain.
The PLP-binding and C-terminal domains contain several conserved residues that are not directly involved in catalysis or binding of cofactor and substrate. The small number of identical residues together with the conservations in hydrophobicity pattern are obviously sufficient to determine the common fold of the proteins. The central β-sheets contain only very few identical amino acids (all residue numbers refer to aCBL amino acid sequence; Asp-253, Gly-289, Ser-381, Phe-382, Ala-403, and Arg-440), whereas hydrophobic regions are strictly conserved (hydrophobic: 152, 174, 175, 202, 220, 221, 222, 252, 271, 293, 355, 380, 383, 412, 413, 439, and 441; hydrophilic: 274, 354, 356, 401, and 442). Similar results are obtained for the α-helices. The loop and turn regions often show conserved Gly (131, 171, 268, 282, 310, 375, 407, and 444) or Pro (226 and 350) residues, which allow tight turns or otherwise unfavorable backbone geometry. There are several other identical amino acids in both eCBL and aCBL which are not involved in active site or secondary structure building. The hydrophilic residues Glu-224, Asp-235, Asp-270, Glu-446, and Asp-447 stabilize the termini of their corresponding secondary structure elements, whereas the conserved amino acids around residue 283 form a loop in the active site region.
Active Site Structure
Detailed descriptions of the eCBL active site and the structure of the CBL inhibitor complex with AVG have been published (Clausen et al., 1996; Clausen et al., 1997). A superposition of the active sites of aCBL and eCBL is shown in Figure 5c. The inhibitor AVG was modeled into the structure by least square superposition of aCBL and the eCBL-AVG inhibitor complex, using the program O. The important catalytic residues Lys-278, Tyr-181, and Asp-253 align almost perfectly. Similarly, the hydrogen bonding network around the PLP-phosphate (Thr-277, Gly-157, Met-158, Arg-129*, and Tyr-127*) is essentially conserved. There are two additional interactions with the PLP-phosphate group in the aCBL structure, namely hydrogen bonds between the hydroxyl group of Ser-275 and OP1 as well as OP4. In addition, there are slight differences in the vicinity of Asp-253. The equivalent Asp-185 from eCBL interacts with the hydroxyl group of Thr-187 and the main chain nitrogen of Asn-186. A water molecule connects Asp-185 and Glu-154, which itself interacts with Ser-158. Thr-187 is exchanged to Ser-255 in aCBL. The interaction with the hydroxyl group is conserved. Glu-224 (Glu-154 in eCBL) interacts with the main chain nitrogen of Tyr-181 instead of Asp-253.
Probably the most important amino acid exchanges involve the eCBL amino acids Trp-340, Asp-116 and Tyr-238*, which are exchanged to Phe-406, Arg-186 and Asn-307* in aCBL. These exchanges result in altered positioning of the amino acid side chains. Furthermore, the quality of aromatic interaction is changed (Trp versus Phe). More importantly, there is an inversion of charges (Asp to Arg) as well as an exchange of an aromatic residue by a polar, uncharged functionality (Tyr to Asn). As a result, substrate binding in Arabidopsis should be altered, as will be discussed below. The anion binding site Arg-440 (Arg-372 in eCBL) and the bicarbonate are located further away from the cofactor PLP, compared to the bacterial structure.
Enzyme Kinetics
The Km values for the substrates cystathionine and djenkolate (structures see Fig. 1) are identical for aCBL (0.22 mm in each case), while they differ by a factor of nine for eCBL (0.04 and 0.36 mm, respectively). eCBL binds the natural substrate cystathione much stronger (0.04 mm, (Dwivedi et al., 1982)) than aCBL (0.22 mm (Ravanel et al., 1996)). Vmax values for l-cystathionine and l-djenkolate are 5-fold higher with eCBL than with aCBL, but each enzyme has similar Vmax values for both substrates. Values of the kinetic constants for both enzymes are summarized in Table II.
Table II.
Enzyme kinetic data
| Substrate |
Km
|
Vmax
|
||
|---|---|---|---|---|
| aCBLa | eCBLb | aCBLa | eCBLb | |
| mm | units mg−1 | |||
| l-Cystathionine | 0.22 | 0.04 | 51.9 | 249 |
| l-Djenkolate | 0.22 | 0.36 | 42.4 | 198 |
30°C, pH 8.5, 5 mm substrate (Ravanel et al., 1996).
37°C, pH 9.0, 2 mm substrate (Dwivedi et al., 1982).
DISCUSSION
Substrate Binding and Inhibitory Studies
The substrate binding pocket of eCBL is smaller than that of aCBL. This is due to the replacement of residues with larger side chains lining the substrate binding site pocket by residues with smaller side chains in the aCBL structure (Trp-340 to Phe-406, Tyr-338 to Val-404, Phe-55* to Asp-126*, Glu-112 to Gly-182 and Tyr-238* to Asn-307*). Several interactions which are present in the active site of eCBL are lost or weakened in aCBL due to active site residue exchanges. Trp-340 of eCBL, which is positioned at the α-carboxyl site of the substrate, forms a hydrogen bond to OB2 of the bicarbonate in the native structure and to O2B of l-aminoethoxyvinylglycine in the complex, whereas the hydrophobic amino acid Phe-406 in aCBL lacks this function. In addition, on the distal site of the pocket Tyr-238*, which interacts indirectly with the amino group of the inhibitor, is replaced by Asn-307*. The shorter side chain of Asn-307* is too far away from the substrate binding pocket to interact directly or via an active site water molecule with inhibitor or substrate. These interactions that are involved in eCBL substrate binding are not present in the aCBL active site.
However, several substrate binding interactions are conserved. The anion binding sites aCBL Arg-129* and eCBL Arg-58*, as well as aCBL Tyr-181 and eCBL Tyr-111, are positioned almost identically in both enzymes. Furthermore, an additional interaction, which is only relevant in the Arabidopsis active site, results from the exchange of eCBL Asp-116 to aCBL Arg-186. Asp-116 with a rather small side chain is positioned too far from the binding pocket to interact with the substrate, whereas the significantly longer side chain of Arg-186 in aCBL, pointing toward the distal part of the substrate, further interacts with the distal carboxyl group. The dimension of the eCBL substrate binding pocket is optimized for the substrate cystathionine, whereas the larger binding site of aCBL should accomodate cystathionine and the longer substrate djenkolate with equal affinity. Without considering the size of the active site cavity, substrate binding to eCBL seems to occur with higher affinity, which may be due to the stronger interaction at the α-carboxyl site.
The influence of Arg- and thiol-blocking agents was examined earlier (Ravanel et al., 1996). Although the Arg blocker phenylglyoxal had no inhibitory effect on the enzyme, plant CBL was found to be sensitive to the thiol-blocking inhibitors N-ethylmaleimide and iodoacetamide, which in turn had no effect on the bacterial enzymes. It was concluded that no Arg residue was involved in the catalysis of aCBL, whereas at least one of the five aCBL Cys residues was postulated to be located near the active site of the plant enzyme (Ravanel et al., 1996). In contrast, our structure clearly showed several Arg, but no Cys in the active site of the enzyme. The lack of inhibition by the Arg blocker phenylglyoxal indicates that the interactions involving the three active site Arg are strong enough to prevent the blocker from binding. The guanidinium group of Arg-440 interacts with OB1 and OB2 of the bicarbonate; the side chain of Arg-129* forms several hydrogen bonds to the PLP phosphate OP2, OP3, and the hydroxyl group of Tyr-181. One of the guanidino nitrogens of Arg-186 lies within hydrogen bond distance (3.2 Å) to the ND2 nitrogen of Asn-309*. The shape of the active site in E. coli CBL may provide additional interactions favoring the binding of phenylglyoxal. The blocker alternatively could bind to an Arg outside the active site, thereby acting as a noncompetitive inhibitor.
Similarly, the inhibitory effects of N-ethylmaleimide can only be explained by allosteric effects because no Cys residue was identified in or near the active site.
Postulated Catalytic Mechanism
The catalytic mechanism of Arabidopsis CBL is similar to that proposed for eCBL (Clausen et al., 1996). Clausen et al. already postulated an induced fit mechanism analogous to that of AAT (Jansonius and Vincent, 1987). Further experiments to examine the mechanism included the crystallization and structure determination of ApoCBL from E. coli (Breitinger, 2000).
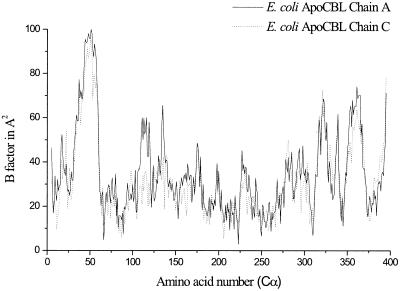
Attempts were made to solve the structure of the eCBL substrate complex by soaking the crystals with cystathionine. Under this treatment, the crystals cracked after several hours, indicating a conformational change of the protein upon substrate binding. The apo-eCBL crystals were unstable, but the structure could be solved to a resolution of 2.5 Å. It showed flexible regions in the N-terminal and PLP-binding domain. The loop, including residues Tyr56* and Arg58*, which interacts through several hydrogen bonds with the PLP phosphate in the native structure, is no longer defined in the apoprotein (Fig. 6). The region around Tyr-111 is also highly flexible. These flexibilities indicate that parts of the protein change their conformation during substrate binding, thereby shielding the active site region. Because Tyr-56* and Arg-58* are the only conserved amino acids in the specific region of the N-terminal domain, interaction directed to these residues seems to be the main driving force for this transition. The native structures of CBL, therefore, show the open conformation of the enzyme, where the active site is accessible to substrate.
Figure 6.
B-factors of the E. coli ApoCBL amino acids. Only the B-factors of the Cα-atoms are shown. Note the undefined region (see text). The plot was created using the program Origin (Microcal).
As shown in Figure 7, we propose that the substrate enters the active site cleft from a position above the conserved amino acids Arg-129* and Tyr-181. The substrate will be oriented in proper position for transaldimation by interaction with the general α-carboxylate docking site Arg-440, the ionized PLP hydroxyl group, Tyr-181, and Arg-129*. In the next step Tyr-181, a conserved residue in all cystathionine-utilizing enzymes, abstracts a proton from the α-amino group of the bound substrate. After formation of the external aldimine, the α-carboxylate and the sulfur atom of the substrate are fixed in a position, which allows the amino group of Lys-278 to abstract the proton from Cα. The PLP cofactor delocalizes the negative charge of the developing carbanion over its entire conjugated π-system. Subsequently, Lys-278 transfers the proton directly from Cα to Sγ of the substrate. During this process, the positively charged ε-amino group of Lys-278 is attracted by the negative charge of the PLP phosphate group, directing it to hydrogen distance position to Sγ, thereby inducing Cβ-S bond cleavage by protonation of Sγ, followed by the release of homo-Cys. The remaining PLP derivative of aminoacrylate is converted to iminopropionate by proton addition on Cβ. After release of homo-Cys, the transaldimation equilibrium between external and internal aldimine should be shifted toward the latter, resulting in release of iminopropionate, which is hydrolyzed to pyruvate and ammonia outside the enzyme.
Figure 7.
Proposed reaction mechanism for aCBL. The colors used are: apoprotein, blue; PLP cofactor, black; and substrate, red.
Knowledge of the substrate binding site of CBL, coupled with detailed information of the differences in function, as well as three-dimensional structures of plant and bacterial CBLs should serve as a starting point for the development of specific enzyme inhibitors, herbicides, and antibiotics.
MATERIALS AND METHODS
Cloning Expression and Purification
The cDNA coding for the putative mature aCBL polypeptide, starting with Met-57 of the respective coding sequence region of the GenBank/EMBL accession number L40511, was amplified from an Arabidopsis cDNA library (Minet et al., 1992) by PCR with Pfu DNA polymerase (Stratagene, La Jolla, CA) according to the manufacturer's recommendations using phosphorothioate-protected primers. The aCBL gene was amplified using the oligonucleotides 5′-GCACTATCTAGATAACGAGGGCAAAACATGGAGAAAAGTGT-3′ and 5′-GCTGATCCCGGGGAGAGGGAAGGTTTTGAAGG-3′ as primers. The PCR product was digested by XbaI/SmaI and cloned into the XbaI/Eco47III-digested Escherichia coli expression vector pASK75 (IBA) yielding the plasmid paCBL1, which codes for an aCBL polypeptide that is tagged at its C terminus by the affinity peptide Strep-tag I (Schmidt et al., 1996). The paCBL1 construct was used to transform E. coli JM83 (Yanisch-Perron et al., 1985). For overexpression, the E. coli strain JM83/paCBL1 was grown at 26°C in Luria-Bertani medium containing 100 μg mL−1 ampicillin to a cell density of A550 = 0.5 and induced with 200 μg L−1 anhydrotetracycline (Skerra, 1994) for 16 h. Cells were harvested by centrifugation, resuspended in CP buffer (100 mm Tris/HCl, pH 8.0, 1 mm EDTA, 10 μm pyridoxalphosphate) and disrupted by two passages through a French pressure cell at 15,000 psi. The cell debris was removed by centrifugation (18,000g for 30 min), and the supernatant was subjected to 0% to 33% ammonium sulfate fractionation. The precipitate was resuspended in CP buffer and affinity purified on a Streptavidin Sepharose column (IBA). This protocol yielded about 10 mg of purified enzyme per liter cell culture.
Crystallization and Data Collection
Crystals were grown by sitting drop vapor diffusion against a reservoir containing 100 mm MES[2-(N-morpholino)-ethanesulfonic acid]·NaOH, pH 6.0, and 300 mm (NH4)2SO4. Droplets were mixed from 4 μL of protein solution in 10 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.0, 10 μm PLP, and 2 μL of reservoir. The crystals were grown at room temperature to an approximate size of 200 μm × 100 μm × 40 μm within 1 week. The crystals belong to the orthorhombic space group C2221 with cell dimensions of a = 107.8 Å, b = 154.3 Å, c = 118.8 Å, and one homodimer per asymmetric unit. Diffraction data were collected at the Deutsche Elektronen Synchrotron (beamline BW7B, Hamburg, Germany) at 100 K (λ = 0.83 Å) on a 345-mm detector (MAR Research, Norderstedt, Germany). Before freezing, the crystals were incubated for a few seconds in 30% (v/v) PEG400, 50 mm HEPES (pH 7.0), 300 mm (NH4)2SO4. A single crystal yielded a data set complete to 2.3 Å resolution in a 90° φ-scan with a 1.0° frame width. Diffraction data were processed using the Denzo program package (Otwinowski and Minor, 1996). The intensities were scaled, reduced, and truncated to structure factors by the programs of the CCP4 program suite (Collaborative Computational Project, 1994). The data collection statistics are summarized in Table I.
Molecular Replacement and Structure Refinement
Structure solution by Patterson search techniques was carried out using the program package AMoRe (Navaza, 1994). The coordinates of the tobacco (Nicotiana tabacum) CGS dimer (Steegborn et al., 1999) were used as a search model. Residues were mutated to Ala or Gly, and the PLP cofactor was omitted. The AMoRe calculations revealed one prominent solution with a correlation coefficient of 45.3 and a crystallographic R-factor of 54.1% using diffraction data to a resolution of 3.5 Å.
The quality of the electron density maps for model building was improved by 2-fold real space averaging using the program AVE (Jones, 1992). The non-crystallographic symmetry (NCS) operators were determined using program LSQMAN (Kleywegt and Jones, 1994). Model building was done with program O (Jones et al., 1991) and refinement with the CNS software (Brünger et al., 1998) using the parameters of Engh and Huber (1991). No NCS restraints were used due to the sufficient good resolution. The minimize protocol with an overall anisotropic B-factor was applied. B-factors were refined isotropically, yielding a final root mean square deviation value for bonded Bs of 3.45. The crystallographic R-factor was used for monitoring the stage of the refinement procedure, omitting 5% of the structure factors in order to calculate Rfree values. The initial averaged 2Fo-Fc electron density map showed well-defined electron density for the PLP cofactor. At the end of the refinement model, building was done in unaveraged electron density maps independently in both monomers. Finally, 253 water and two bicarbonate molecules were incorporated.
Rebuilding and refinement cycles converged at Rcryst and Rfree values of 24.9% and 30.9%, respectively. Three stretches of the polypeptide chain were invisible in the electron density maps; the N-terminal part, i.e. residues 57 to 84; the region between residues 418 to 433; and the 11 amino acids of the C-terminal Strep-tag (Fig. 2). These parts are invisible most probably due to their high flexibility. This feature of the structure accounts for the relatively high final R values. The final model contained 5,671 protein atoms, 30 cofactor atoms, and 266 solvent atoms. Further details are given in Table I. The stereochemistry of the model was analyzed with programs CNS and PROCHECK (Laskowski et al., 1993). For the comparison of NCS-related molecules, the program LSQMAN was used.
Coordinates
The coordinates of the refined model of aCBL have been deposited in the Research Collaboratory for Structural Bioinformatics Protein data bank (accession code 1IBJ).
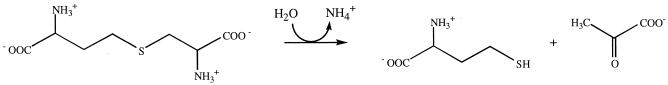
Figure.
Cleavage of cystathionine by CBL.
LITERATURE CITED
- Alexander FW, Sandmeier E, Mehta PK, Christen P. Evolutionary relationships among pyridoxal-5′-phosphate-dependent enzymes: regio-specific α, β and γ-family. Eur J Biochem. 1994;219:953–960. doi: 10.1111/j.1432-1033.1994.tb18577.x. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Belfaiza J, Parsot C, Martel A, Bouthier de la Tour C, Margerita D, Cohen GN, Saint-Girons I. Evolution in biosynthetic pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci USA. 1986;83:867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitinger U. Biochemische und kristallographische Charakterisierung der Cystathionine β-Lyase. Diploma thesis. Erlangen, Germany: University of Erlangen; 2000. [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burley SK. An overview of structural genomics. Nat Struct Biol Suppl. 2000;7:932–934. doi: 10.1038/80697. [DOI] [PubMed] [Google Scholar]
- Burnell JN, Whatley FR. Sulphur metabolism in Paracoccus denitrificans: purification, properties and regulation of cysteinyl- and methionyl-tRNA synthetase. Biochim Biophys Acta. 1977;481:266–278. doi: 10.1016/0005-2744(77)90158-9. [DOI] [PubMed] [Google Scholar]
- Clausen T, Huber R, Laber B, Pohlenz HD, Messerschmidt A. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine β-lyase from Escherichia coli at 1.83 angstrom. J Mol Biol. 1996;262:202–224. doi: 10.1006/jmbi.1996.0508. [DOI] [PubMed] [Google Scholar]
- Clausen T, Huber R, Messerschmidt A, Pohlenz HD, Laber B. Slow-binding inhibition of Escherichia coli cystathionine β-lyase by L-aminoethoxyvinylglycine: a kinetic and X-ray study. Biochemistry. 1997;36:12633–12643. doi: 10.1021/bi970630m. [DOI] [PubMed] [Google Scholar]
- Clausen T, Huber R, Prade L, Wahl MC, Messerschmidt A. Crystal structure of Escherichia coli cystathionine γ-synthase at 1.5 angstrom resolution. EMBO J. 1998;17:6827–6838. doi: 10.1093/emboj/17.23.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droux M, Ravanel S, Douce R. Methionine biosynthesis in higher plants: II. Purification and characterization of cystathionine β-lyase from spinach chloroplasts. Arch Biochem Biophys. 1995;316:585–595. doi: 10.1006/abbi.1995.1078. [DOI] [PubMed] [Google Scholar]
- Dwivedi CM, Ragin RC, Uren JR. Cloning, purification and characterization of β-cystathionase from Escherichia coli. Biochemistry. 1982;21:3064–3069. doi: 10.1021/bi00256a005. [DOI] [PubMed] [Google Scholar]
- Engh RA, Huber R. Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- Evans SV. SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Gentry-Weeks CR, Keith JM, Thompson J. Toxicity of Bordetella avium β-cystathionase toward MC3T3–E1 osteogenic cells. J Biol Chem. 1993;268:7298–7314. [PubMed] [Google Scholar]
- Gentry-Weeks CR, Spokes J, Thompson J. β-Cyst-athionase from Bordetella avium: role(s) of lysine 214 and cysteine residues in activity and cytotoxicity. J Biol Chem. 1995;270:7695–7702. doi: 10.1074/jbc.270.13.7695. [DOI] [PubMed] [Google Scholar]
- Giovanelli J, Mudd SH. Transsulfuration in higher plants: partial purification and properties of β-cyst-athionase of spinach. Biochim Biophys Acta. 1971;227:654–670. doi: 10.1016/0005-2744(71)90015-5. [DOI] [PubMed] [Google Scholar]
- Giovanelli J, Owens LD, Mudd SH. In vivo inactivation by rhizobitoxine and role of the enzyme in methionine biosynthesis in corn seedlings. Plant Physiol. 1972;51:492–503. doi: 10.1104/pp.51.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Inoue Y, Kuramitsu S, Morino Y, Kagamiyama H. Effects of replacement of tryptophan-140 by phenylalanine or glycine on the function of Escherichia coli aspartate aminotransferase. Biochem Biophys Res Commun. 1990;167:407–412. doi: 10.1016/0006-291x(90)92037-z. [DOI] [PubMed] [Google Scholar]
- Holbrook EL, Greene RC, Krueger JH. Purification and properties of cystathionine γ-synthase from overproducing strains of Escherichia coli. Biochemistry. 1990;29:435–442. doi: 10.1021/bi00454a019. [DOI] [PubMed] [Google Scholar]
- Jansonius JN, Vincent MG. Structural basis for catalysis by aspartate aminotransferase. In: Jurnak FA, McPherson A, editors. Biological Macromolecules and Assemblies. Vol. 3. New York: Wiley & Sons; 1987. pp. 187–288. [Google Scholar]
- John RA. Pyridoxal phosphate-dependent enzymes. Biochim Biophys Acta. 1995;1248:81–96. doi: 10.1016/0167-4838(95)00025-p. [DOI] [PubMed] [Google Scholar]
- Jones TA. a, yaap, asap, @**?: a set of averaging programs. In: Dodson EJ, Gover S, Wolf W, editors. Molecular Replacement (CCP4). Warrington, UK: Daresbury Laboratory; 1992. pp. 99–105. [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjieldgaard M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kaplan MM, Flavin M. Cystathionine γ-synthase of Salmonella: structural properties of a new enzyme in bacterial methionine biosynthesis. J Biol Chem. 1996;241:5781–5789. [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. A super position. ESF/CCP4 Newsletter. 1994;31:9–14. [Google Scholar]
- Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Kreft BD, Townsend A, Pohlenz HD, Laber B. Purification and properties of cystathionine γ-synthase from wheat (Triticum aestivum) Plant Physiol. 1994;104:1215–1220. doi: 10.1104/pp.104.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, McArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Martel A, Bouthier de la Tour C, Le Goffic F. Cystathionine γ-synthase and β-cystathionase have common peptide sequences of the active site and are inhibited similarly by aminoethoxyvinylglycine. In: Korpela TK, Christen P, editors. Biochemistry of Vitamin B6. Basel: Birkhäuser Verlag; 1987. pp. 341–344. [Google Scholar]
- Merrit EA, Murphy MEP. Raster 3d, version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Minamisawa K, Fukai K, Asami T. Rhizobitoxine inhibition of hydrogenase synthesis in free-living Bradyrhizobium japonicum. J Bacteriol. 1990;172:4505–4509. doi: 10.1128/jb.172.8.4505-4509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Owens LD, Guggenheim S, Hilton JL. Rhizobium synthesized phytotoxin: an inhibitor of β-cystathionase in Salmonella typhimurium. Biochim Biophys Acta. 1968;158:219–225. doi: 10.1016/0304-4165(68)90134-7. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Rando RR. Mechanisms of action of naturally occurring irreversible enzyme inhibitors. Accounts Chem Res. 1975;8:281–287. [Google Scholar]
- Ravanel S, Job D, Douce R. Purification and properties of cystathionine β-lyase from Arabidopsis thaliana overexpressed in Escherichia coli. Biochem J. 1996;320:383–392. doi: 10.1042/bj3200383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Ruffet M, Douce R. Cloning of an Arabidopsis thaliana cDNA encoding cystathionine β-lyase by functional complementation in Escherichia coli. Plant Mol Biol. 1995;29:875–882. doi: 10.1007/BF00041177. [DOI] [PubMed] [Google Scholar]
- Schmidt TGM, Koepke J, Frank R, Skerra A. Molecular interaction between the Strp-tag affinity peptide and its cognate target streptavidin. J Mol Biol. 1996;255:753–766. doi: 10.1006/jmbi.1996.0061. [DOI] [PubMed] [Google Scholar]
- Skerra A. Use of the tetracyclin promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene. 1994;151:131–135. doi: 10.1016/0378-1119(94)90643-2. [DOI] [PubMed] [Google Scholar]
- Steegborn C, Messerschmidt A, Laber B, Streber W, Huber R, Clausen T. The crystal structure of cystathionine γ-synthase from Nicotiana tabacum reveals its substrate and reaction specificity. J Mol Biol. 1999;290:983–996. doi: 10.1006/jmbi.1999.2935. [DOI] [PubMed] [Google Scholar]
- Turner WL, Pallett KE, Lea PJ. Cystathionine β-lyase from Echinochloa colonum tissue culture. Phytochem. 1998;47:189–196. [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]