Abstract
The Thioredoxin (Trx) fold is a versatile protein scaffold consisting of a four-stranded β-sheet surrounded by three α-helices. Various insertions are possible on this structural theme originating different proteins, which show a variety of functions and specificities. During evolution, the assembly of different Trx fold domains has been used many times to build new multi-domain proteins able to perform a large number of catalytic functions. To clarify the interaction mode of the different Trx domains within a multi-domain structure and how their combination can affect catalytic performances, in this review, we report on a structural and functional analysis of the most representative proteins containing more than one catalytically active Trx domain: the eukaryotic protein disulfide isomerases (PDIs), the thermophilic protein disulfide oxidoreductases (PDOs) and the hybrid peroxiredoxins (Prxs).
Keywords: Trx fold, Disulfide bond, Redox sites, Protein disulfide oxidoreductase, Protein disulfide isomerase, Hybrid peroxiredoxin
Introduction
The thioredoxin (Trx) fold consists of a four-stranded β-sheet surrounded by three α-helices (Fig. 1) [1]. The Trx superfamily comprises proteins with different functions, characterized by the presence of at least one Trx fold [2–4]. The main components of the superfamily are the ubiquitous Trxs and glutaredoxins (Grxs) [5], the bacterial disulfide bond forming (Dsb) proteins [6], the eukaryotic protein disulfide isomerase (PDI) and its homologs [7], the thermophilic protein disulfide oxidoreductases (PDOs) [8], and the eukaryotic flavoprotein quiescin-sulfhydryl oxidase (QSOX) [9, 10], all involved in the thiol-disulfide exchange reactions. The superfamily (SCOP: http://scop.mrc-lmb.cam.ac.uk/scop/) also includes glutathione S-transferase (GSH-transferase), which catalyzes the conjugation of GSH to electrophilic substrates, the bacterial Escherichia coli arsenate reductase (ArsC) [11], catalyzing the reduction of arsenate, the glutathione peroxidase (GPX) and the peroxiredoxins (Prxs), which are involved in the reduction of hydroperoxides [12], and finally the chloride intracellular channel (CLIC) proteins and the copper-ion binding protein Sco1 [13, 14].
Fig. 1.
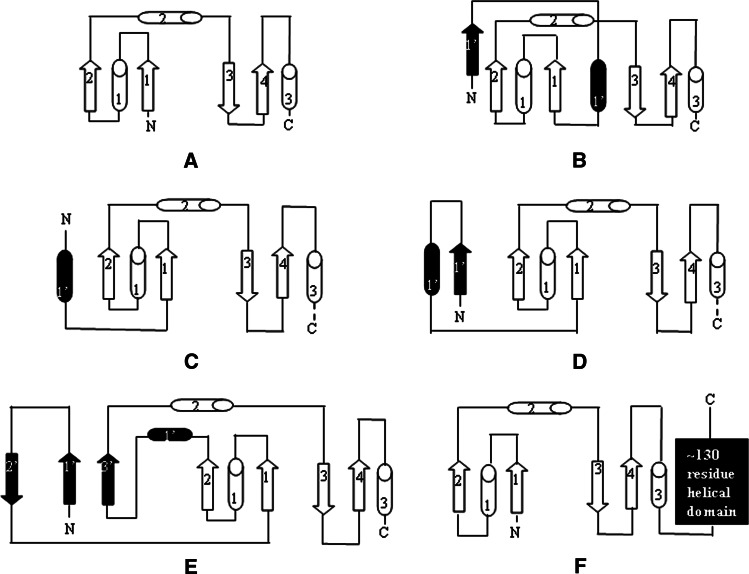
The Trx fold and its variations in different components of the Trx superfamily. The basic Trx fold of Grxs is reported in a and b–f show modifications of Trx fold in other thiol oxidoreductases: b Trx, c A. pernix PDO-N unit, d yeast PDI a domain, e S. solfataricus Bcp1 and f GSH-transferase. Additional secondary structural elements with respect to the basic Trx fold are shown in black. β-sheets are drawn as arrows and α-helices as cylinders
These proteins do not present a high level of sequence similarity, although they show considerable structural similarity thanks to the presence of the common Trx fold. However, multiple variations of this fold can be observed (Fig. 1). Grx is the only member of the family with the basic Trx fold; an additional β-strand and α-helix are inserted at the N-terminus in Trxs [1, 3]. An insertion of secondary structure elements between the second β-strand and the second α-helix is very common in several proteins, such as Prxs, GPXs, ArsC, and DsbA-like enzymes. In the latter two cases, this insertion consists of four or five α-helices. Further modifications as insertions at the C-terminus can be observed in GSH-transferase [15].
Members of the Trx superfamily involved in the thiol-disulfide exchange reaction all have an active site containing the CXXC motif and share the same catalytic mechanism. In the CXXC motif, always located at the N-terminus of helix α1, the N-terminal cysteine, referred to as the nucleophilic cysteine [13], is deprotonated at physiological pH, largely exposed and consequently hyper-reactive. By contrast, the C-terminal cysteine is buried and is usually protonated [13]. Other hallmarks of this subgroup are the following conserved residues: (1) a cis-proline located in the loop region before strand β3 and juxtaposed to the active site, which is implicated in substrate binding [16, 17]; (2) a conserved proline in the middle of helix α1, which introduces a kink in the helix; (3) charged residues in the vicinity of the active site, which are implicated in proton-transfer reactions required for the redox mechanism [18, 19]. The nature of the amino acids between the two cysteines varies considerably among the members of the superfamily, influencing the pKa of the nucleophilic cysteine and consequently the redox properties of these proteins [20]. Indeed, the redox potential ranges from −270 mV of the reductant Trx to −95 mV of the oxidant DsbL [21, 22], leading to several functional differences [23]. A recent work by Ren et al. [23] showed that protein functions are also influenced by the loop containing the cis-proline: modifications in the residue preceding the cis-proline play a crucial role both in the redox properties and in regulating the ability to interact with substrates [23].
The canonical CXXC motif can be modified, as regards the number and position of the cysteines involved in the catalysis with consequent variations in catalytic functions. For example, the C-terminal cysteine can be substituted by other residues, such as in monothiol Grxs [1], the two cysteines can be separated by three/four residues, such as in Sco1 [24], both cysteines can be substituted by other residues, with consequent loss of redox activity, such as in calsequestrin or some PDI homologs [2, 9, 10, 25, 26].
Taken together, these observations highlight the fact that the Trx fold is very versatile. Such versatility makes the proteins of the Trx superfamily suitable to perform several catalytic functions in numerous processes, ranging from protein folding to detoxification and metabolite synthesis [27].
During evolution, the assembly of different Trx fold domains has often been used to build new proteins. In particular, several examples are available where the duplication of the Trx fold has occurred generating multi-domain proteins [14]. However, although the Trx superfamily has received widespread attention in the literature, few studies have made a comparative structural and functional analysis of the multi-domain proteins. Thus, to provide insights into the mutual interaction of different Trx domains within a multi-domain structure and how their combination can influence physiological functions, this review will focus on the most representative proteins of the Trx superfamily containing at least two catalytically active Trx domains: the eukaryotic PDIs [28], the thermophilic PDOs [29] and the hybrid Prxs [30].
An overview of the PDI family
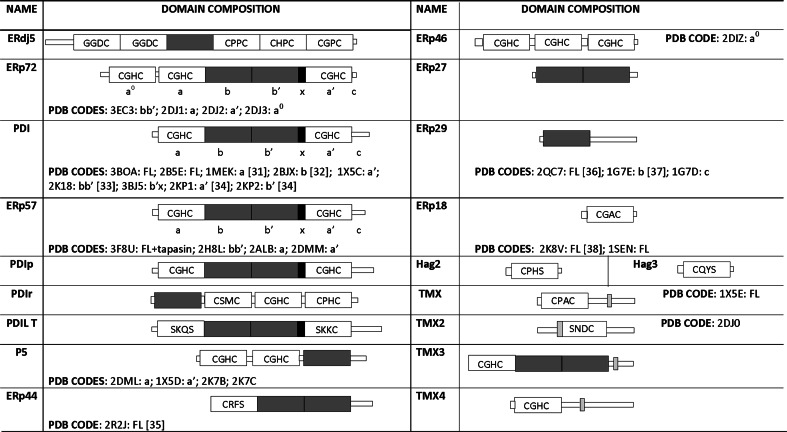
To fold in the oxidative environment of the endoplasmic reticulum (ER) proteins need to form the correct disulfide bonds. For this purpose, the cell produces a large number of proteins, belonging to the protein disulfide isomerase (PDI) family, which catalyze the oxidation and isomerization of disulfide bonds [7, 28]. This protein family comprises PDI and PDI-like proteins, with four members in Saccharomyces cerevisiae and 19 members in mammals so far identified [7] (Fig. 2). The reason for such a large number of PDI family members and how they function together to cooperate to the protein folding is still a mystery. One hypothesis is that each family member has the ability to act on a specific set of substrates and/or that each enzyme catalyzes a precise type of reaction [39, 40].
Fig. 2.
The human PDI family. Catalytic domains are shown in white, while non-catalytic ones in dark grey. The x and transmembrane regions are also shown in black and light grey, respectively. PDB codes are given only for mammalian and fungal members of the family
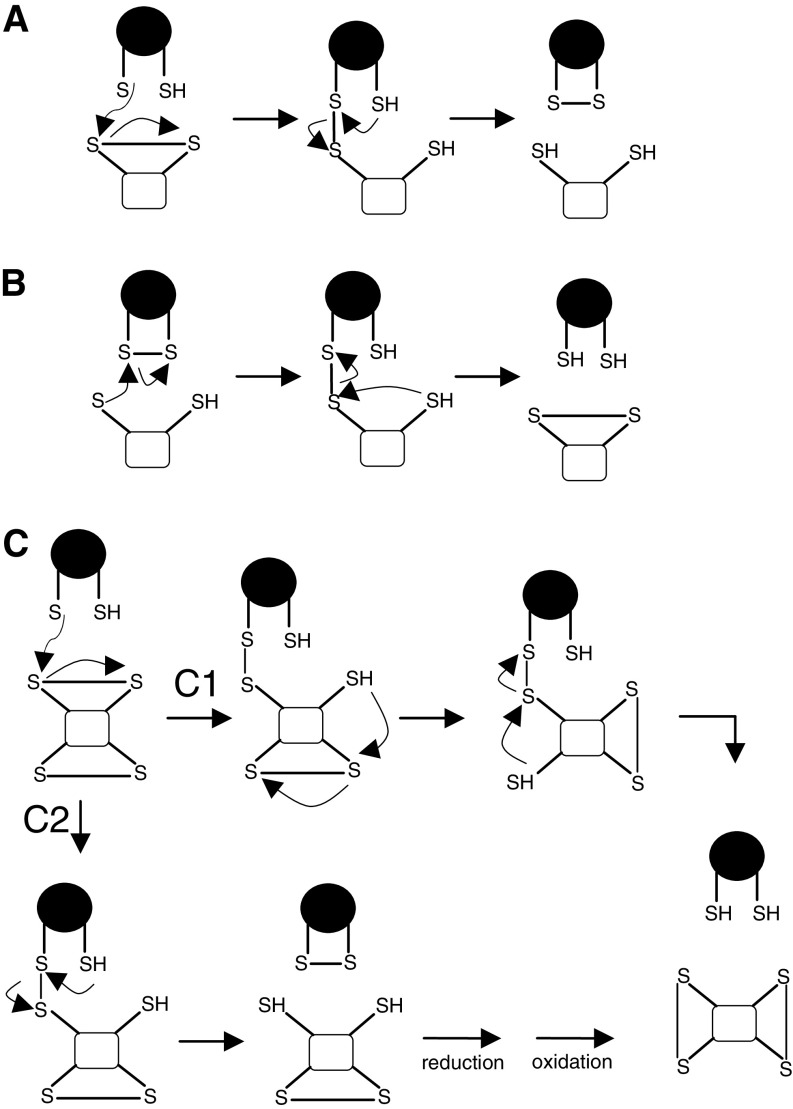
PDI family members differ in tissue distribution, substrate specificity, and ability to catalyze dithiol-disulfide exchange reactions [41], but all contain an ER-localization motif and at least one Trx-like domain, which can be either catalytic (hereafter indicated as domain a) or non-catalytic (hereafter indicated as domain b) (Fig. 2). The CXXC motif is a hallmark of the catalytic domains. If it is in the reduced state, substrate disulfide can be reduced (Fig. 3a); if it is in the oxidized state, the disulfide can be transferred to the substrate protein and the PDI active site released in its reduced state (Fig. 3b). The isomerization of wrongly formed disulfides (Fig. 3c) can occur directly or indirectly, via several reduction–oxidation cycles, until the substrate disulfide becomes sufficiently stable to resist reduction by PDIs [7].
Fig. 3.
Schematic representation of reactions catalyzed by PDI (in white substrates and in black PDI): a reduction, b oxidation, c isomerization. The last reaction can occur directly (C1) or indirectly, via several reduction–oxidation cycles (C2)
Many research efforts have been devoted in recent years to understanding the structure and enzymatic properties of PDIs. Below, the most abundant and best characterized members of the family, namely PDI, Erp57, and Erp72, will be reviewed both from a structural and functional point of view.
PDI
PDI is the best characterized member of the PDI family and is highly conserved in all eukaryotic cells [28]. Besides its role in protein-folding catalysis, PDI operates as a chaperone by preventing aggregation of proteins that do not contain cysteine residues [42], is the β-subunit of propyl-4-hydrolase [43], is a subunit of microsomal triglyceride transfer protein [44] and is involved in the regulation of NAD(P)H oxidase [45].
In vitro PDI catalyzes the reduction, oxidation, and isomerization of disulfide bonds in a large range of substrates [46–48], while in vivo it is more likely to act only as an isomerase and as an oxidase [28]. Since the latter activity leaves PDI in the reduced form, a further source of oxidizing equivalents is necessary to complete the catalytic cycle. The proteins responsible for this oxidation in vivo are Ero1-Lα and Ero1-Lβ in mammalian cells [49, 50], and Ero1p [51–53] or Erv2p [54–56] in yeast. These proteins combine disulfide bond formation to reduction of molecular oxygen, with the help of the cofactor flavin adenine dinucleotide (FAD) [52, 53]. However, it cannot be excluded, as was thought for several years, that GSSG may also provide oxidizing equivalents to PDI. Indeed, GSSG together with GSH forms a redox buffer that helps to maintain the ER redox homoeostasis and can influence the in vivo redox state of PDI [57].
Few PDI natural substrates have so far been identified [58–62], even if the promiscuity of the enzyme in vitro is expected to be reflected also in vivo [28].
PDI is a multi-domain protein with two catalytic domains, a and a′, each containing the WCGHC sequence, separated by two non-catalytic domains b and b′. In addition, between b′ and a′ there is a linker region named x, and at the C-terminus a highly acidic extension is present, termed c, which contains the ER-localization motif (Fig. 2).
Catalytic assay experiments on PDI mutant forms and on individual domains or their combinations have provided insights into the contribution of the catalytic domains to the different functions of the enzyme [63–69]. These studies demonstrated that domains a and a′ operate independently, in that each catalytic center alone imparts activity to the enzyme [63, 69], even though mammalian and yeast PDI differ in the contribution of each domain to overall catalytic activity. Specifically, in yeast PDI in vitro experiments, where the Ero1p pathway was reconstituted, the a domain functioned mainly as an isomerase, while a′ presented mostly an oxidative activity [70]. These results were also in agreement with previously reported in vivo experiments [69, 71]. More ambiguous results were obtained for human PDI. In this case, in vitro experiments showed that at substrate concentrations near the KM the two active sites presented comparable isomerase activity [63], while at saturating concentration of substrate the isomerase activity was mainly carried out by the a domain [65]. Subsequent experiments which reconstituted the Ero1Lα/PDI oxidative folding system demonstrated surprisingly that both oxidase and isomerase activity were mainly performed by the a′ domain, and that this domain was preferentially oxidized by Ero1Lα [72]. In the same study, the authors showed also that the functional asymmetry of the active sites was not to be ascribed to their intrinsic catalytic properties, but rather to the specific order of the two domains in the full-length PDI [72]; indeed if the position of the two catalytic domains was swapped, domain a became more active than a′ [72].
Insights into molecular determinants of substrate recognition were also obtained, showing that the b′ domain was sufficient for binding small peptides and domains a and a′ were necessary for binding the larger proteins [73]. Within the b′ domain, the proposed ligand-binding site is a small hydrophobic pocket, defined by residues Leu242, Leu244, Phe258, and Ile272 (numbering refers to human PDI). They are all implicated in substrate binding with a major role for Ile272 [74].
These studies were in line with other experiments showing that simple oxidation reactions required only a and a′, simple isomerization reactions required a linear combination of b′ with either a or a′, while complex isomerization reactions were only catalyzed by full length PDI (excluding the c region) [75].
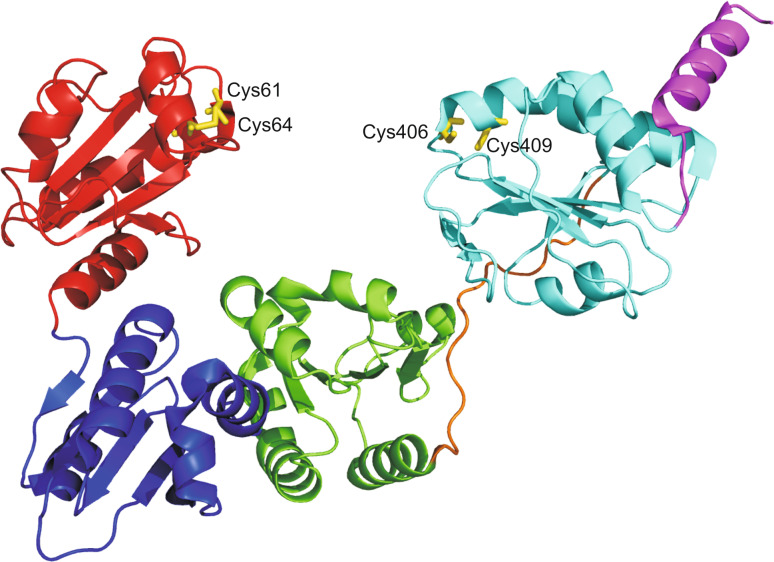
Although several structures of individual PDI domains have been determined (Fig. 2), only the three-dimensional structure of the full-length yeast PDI (PDB code: 2b5e) showed how the individual domains were arranged with respect to one another [76]. Analysis of this structure revealed a monomeric enzyme in which the four domains were organized in a twisted ‘U’ shape, with a and a′ active sites facing each other and domains b and b′ forming a rigid base [76] (Fig. 4). The inner surface of both domains b and b′ comprises the large hydrophobic region involved in substrate interaction [73]. The c region is located at the top of domain a′ pointing toward the external part of the U-shaped molecule [76]. The a and a′ catalytic domains present a structure very similar to Trx, while domains b and b′ adopt some variations with respect to it. Domains a and a′ share with other Trx family members all the structural features and residues important for catalysis. However, one of their unique features is the presence of an arginine residue (corresponding to Arg126 in domain a and Arg471 in domain a′), which is critical for the catalytic function [77, 78]. Indeed, this residue resolves the paradox of the different requirement for a high pKa (when the oxidation of the substrate has to take place) and a low pKa (when the reoxidation of the PDI occurs to complete the catalytic cycle) of the C-terminal cysteine. Specifically, this arginine undergoes a conformational change, from an ‘out’ to an ‘in’ conformation, to destabilize and stabilize the thiolate form of the C-terminal cysteine [77, 78].
Fig. 4.
Overall fold of yeast PDI. a domain is shown in red, b domain in blue, b′ domain in green, x region in orange, a′ domain in cyan and C-terminal region in magenta. Cysteine residues of catalytic sites are colored in yellow and shown in stick representation
Interestingly, the two active sites were found in the structure in two different redox states: a oxidized and a′ reduced. This finding was in agreement with redox potentials of two active sites (−188 and −152 mV for domains a and a′, respectively) and with theoretical pKa calculations which indicated that the pKa of the N-terminal cysteine of the a domain was slightly higher than that of a′ [79].
More recently, the full-length yeast PDI has been crystallized (PDB code: 3boa) at a different temperature and analysis of this structure has revealed a dimeric arrangement of the enzyme, with the extensive intermolecular contacts between the two monomers primarily mediated by their b′ domains [80]. Comparison with the previous structure shows that in this case a drastic rotation and translation of the a domain takes place, with the result that the two active sites no longer face each other and the substrate-binding site in the b′ domain becomes buried. This conformational change is allowed principally by the presence of a flexible loop connecting the a and b domains [80]. These data suggest that PDI can exist in two different states: the monomeric form represents the ‘active’ state, because both active sites and the substrate binding site are accessible, while the dimeric form represents the ‘inactive’ state of the enzyme, with the two active sites far apart and the substrate binding site buried [80]. Further support for this hypothesis comes from the observation that the recombinant human PDI b′x domain can assume two conformations corresponding to different oligomeric states. In the first conformation, corresponding to a monomeric form, the x region interacts with domain b′, ‘capping’ its hydrophobic site, while in the second conformation, corresponding to a dimeric form, the x region is far removed from the b′ domain, which becomes less buried and can promote the formation of the dimer [81–83].
Taken together, these data suggest that PDI is a flexible molecule and this flexibility is important to perform enzymatic activity. In line with this observation, recently reported small-angle X-ray scattering (SAXS) and spectroscopic experiments on PDI from Humicola insolens demonstrated that oxidation of the a′ domain causes a change in the relative orientation of a′ and b′ domains, with a consequent modification in the exposure of the hydrophobic surface responsible for substrate binding [34]. On the basis of these data, it has been proposed that when the a′ domain transfers its own disulfide bond to the substrate, positioned on the hydrophobic surface of the binding site, the enzyme adopts a ‘closed’ conformation, liberating the oxidized substrate [34].
Erp57
Erp57 is another member of the PDI family characterized in depth in recent years, which was shown to efficiently catalyze both in vivo [59, 60, 84, 85] and in vitro [78, 86, 87] disulfide reduction, disulfide isomerization, and dithiol oxidation in substrate proteins [7]. As already observed for PDI, recent experiments suggest that the oxidizing equivalent required for oxidation could be provided in vivo by Ero1Lα [70, 85].
Erp57 is the closest homolog of PDI, sharing with it 27% sequence identity and showing the same domain composition and the same active site motif in both catalytic domains (Fig. 2). Major differences between these two enzymes are located in the c region, which is extremely acidic in PDI, while it contains several lysine residues in Erp57 [28].
Several biochemical studies have allowed the substrate specificity of Erp57 to be identified: it is specific to catalyze the isomerization of non-native disulfide bonds in glycoproteins with unstructured disulfide-rich domains [85, 88]. To meet this challenge, Erp57 interacts with two ER resident lectin-like chaperons, calnexin (CNX) and calreticulin (CRT). CNX is membrane-bound while CRT is a soluble luminal protein [89] and both have been demonstrated to be directly associated with glycoproteins [39, 86, 88, 90, 91]. Erp57 forms with these two lectins distinct 1:1 complexes with dissociation constants in the micromolar range [86, 92, 93]. NMR and mutagenesis studies have identified the b′ domain of Erp57 and the P-domain of both lectins, a proline-rich arm-like domain, as the main players in the formation of these complexes [92, 94–96].
Erp57 also plays an important role in the immune system, forming part of the major histocompatibility complex (MHC) class I peptide loading complex, where it stabilizes the complex and facilitates the proper assembly of class I molecules, through the formation of a disulfide-linked adduct with tapasin, a MCH class I-specific chaperone [97, 98].
Mutagenesis studies provided insights into the role of different catalytic domains to enzymatic activity. In particular, the observation that full-length Erp57, with the C-terminal cysteines of both active sites mutated in serines, partially retains reductase activity, while the recombinant a′c fragment, with the same mutation, was inactive, suggested that domains a and a′ were not functionally equivalent [99–101]. Redox properties of the catalytic sites of Erp57 were also characterized, showing redox potentials of −167 and −156 mV for domains a and a′, respectively [86]. These values were comparable with redox potentials determined for domains a and a′ of PDI [102], suggesting that these two proteins could possess similar catalytic properties. However, a quantitative comparative analysis of the activity of full-length Erp57 with human PDI revealed that Erp57 reduces and isomerizes less efficiently than PDI [86], probably due to a more efficient interaction of PDI with its substrates [86].
The first structural studies on Erp57 were performed by NMR experiments on the isolated a and a′ domains (PDB codes: 2alb; 2dmm) [103, 104], showing that they present a structure very similar to that of Trx. Subsequently, the crystal structure of the Erp57 bb′ fragment was solved at 2.0 Å resolution (PDB code: 2h8l) [93], and together with NMR experiments, provided important information on the CNX/CRT binding site. Specifically, it was shown that this site was mainly localized on the b′ domain and was characterized by a large patch of positively charged residues, which interacted with the negatively charged residues of the lectin P-domain [93]. These experiments confuted previous studies which indicated a role for the positively charged C-terminus of Erp57 in CNX binding [105]. Residues R282 and K274 of the b′ domain were identified as being mainly involved in this binding, while a minor contribution of K214 of the b domain was also proved [93]. Interestingly, the corresponding surface in the b′ domain of yeast PDI is rich in negatively charged residues, in agreement with the lack of interaction with CNX [93]. These data together with the observation that the b′ domain of PDI is directly involved in substrate binding [73], indicate that the b′ domain of these proteins has an important role in determining enzyme specificity.
Preliminary structural information on full-length Erp57 was derived from SAXS experiments, which allowed a model of the enzyme to be obtained [93]. Superposition of this model with the yeast PDI crystal structure [76] showed that the shape of Erp57 was strikingly similar to the U-shaped molecule of PDI, with the relative position of the domains conserved [93]. Surprisingly, in this model the CNX-binding site was placed far from catalytic thiols, on the outer surface of the base of the U formed by domains b and b′. Thus, it has been suggested that glycoproteins could be properly positioned close to the catalytic sites of the a and a′ domains thanks to the peculiar elongated arm-like shape of the P-domain, which curves around Erp57 [93]. Altogether these studies suggest a mechanism following which the binding of Erp57 to CNX is necessary not only to bind but also to properly position substrates next to the redox active site.
More recently, the crystal structure of the full-length Erp57 in complex with tapasin was solved (PDB code: 3f8u) [106], confirming that the overall arrangement of the four Trx-like domains of Erp57 resembles that observed in yeast PDI [76]. However, small differences in the relative orientations of these domains are observed, which result in a different distance between the two active sites (~34 Å for Erp57 and ~26 Å for PDI). In the complex structure, a disulfide bond is present between the N-terminal cysteine of the active site of the Erp57 a domain and tapasin Cys95 residue. Beyond this covalent bond, a large number of protein–protein interactions, involving both Erp57 catalytic domains, account for the high stability of this heterodimer [106]. It is worth noting that, despite the specificity of Erp57 for tapasin, interacting residues of Erp57 are conserved also in PDI. It has thus been suggested that the lack of interaction of PDI with tapasin could be attributed to the lower distance between its a and a′ active site with respect to that observed in Erp57. Although the aforementioned domain flexibility could allow PDI to have a range of distances and orientation of the catalytic domains, it may well not be enough to reach the right distance for proper interaction with tapasin [106].
Erp72
Erp72, unlike PDI and Erp57, contains five Trx-like domains (Fig. 2), three of which, namely a°, a and a′, present the WCGHC catalytic motif [107]. The a° domain is a rather unique feature of this enzyme and presents a higher sequence identity with the a domain (54%) compared to a′ (39%), suggesting that it could derive from duplication of the a domain during evolution [108]. At the N-terminus of the a° domain, a highly negatively charged sequence is present, which resembles the c region of PDI and has been reported to bind Ca2+ ions and interact with positively charged substrates [109].
Erp72 has been shown to catalyze in vitro the reduction, formation, and isomerization of disulfide bonds [110–112] and is supposed to have the same function in vivo [28, 48]. It has been detected in complex with various substrate proteins [62, 113–115] and often targets misfolded proteins with their consequent retention in the ER [114–116].
Erp72 is more similar to Erp57 (41% sequence identity, excluding a° domain from the alignment) than PDI (30% sequence identity, excluding a° domain from the alignment). Even if many of the residues that are implicated in ERp57–CRT interactions are conserved in ERp72 [18], no experimental evidence has been found to suggest a direct interaction between ERp72 and CNX or CRT [28, 84]. However, recent studies demonstrated that Erp72 can functionally substitute in part for Erp57 in knockout cell lines [84].
Although several functional studies have been carried out on this protein, the participation of each catalytic domain in disulfide bond formation and isomerization remains to be determined. However, using site-directed mutagenesis experiments the function of individual domains in the insulin reduction assay system was recently identified [117]. These studies demonstrated that all three catalytic domains participate in catalyzing insulin reduction and, rather than simply having additive effects, act synergistically [117]. However, while the active site of the a° domain contributes mainly to catalytic activity, the active sites of the a and a′ domains mainly enhance the recognition and binding of substrate in the steady state [117].
To date, few structural studies have been performed on Erp72. In 2006, the NMR structure of the individual catalytic domains of this protein was deposited in the PDB (Fig. 2), although no paper has yet been associated with these structures. Only recently the crystal structure of the central non-catalytic domains (bb′) was solved at 1.92 Å resolution (PDB code: 3ec3) [108]. Analysis of the structure revealed as expected the presence of two Trx domains connected by a short linker. Observation of two perfectly superimposable bb′ fragments in the asymmetric unit together with NMR experiments strongly suggested that the b and b′ domains of Erp72 form a rigid pair and the inter-domain linker is not flexible. Structural comparison of the bb′ fragments of yeast PDI [76] and Erp57 [93] shows that the main differences are observed in the charge distribution on the protein surface. In particular, Erp72 lacks the exposed hydrophobic patch of yeast PDI involved in substrate interaction, thus suggesting that it is unlikely to bind unfolded substrates in the same manner as PDI. Moreover, of the three Erp57 basic residues fundamental for CNX binding, namely Lys214, Arg282, and Lys274, the first two are conserved and correspond to Lys364 and Lys435, while the third is substituted by a threonine. This substitution explains why Erp72 does not bind CNX and CRT and acts independently of the association with these two lectins [28, 84].
SAXS experiments were also performed to determine the overall shape and domain organization of the protein [108]. These studies suggested that the shape of the protein region containing domains a, b, b′ and a′ is very similar to the U-shaped structure of yeast PDI [76] and Erp57 [93, 106], while the additional a° domain was located above the a domain, resulting in a structure where the three catalytic domains were close to each other on the same side of the protein. However, alternative conformations characterized by different orientations of the a and a° domains were also possible [108].
PDOs: the discovery of a dual Trx fold in thermophilic microorganisms
While in the past it was largely accepted that disulfide bonds were rarely found in intracellular proteins, recent studies have strongly disputed this belief, demonstrating that intracellular proteins of some thermophilic bacteria and archaea are rich in disulfide bonds [118]. Moreover, a clear correlation between disulfide abundance and maximal growth temperature was also observed [119, 120], suggesting a critical role for disulfide bonds in thermal stabilization [118].
A potential role in disulfide bond formation in the intracellular proteins of thermophilic organisms, and consequently in adaptation to high temperature, has been recently attributed to a new protein family, termed PDO. Members of this protein family are characterized by a molecular mass of about 26 kDa and two Trx folds, one at the N-terminus and the other at the C-terminus, each containing the CXXC active site motif [121]. Some proteins of the family present additional cysteines involved in a structural disulfide bridge [122].
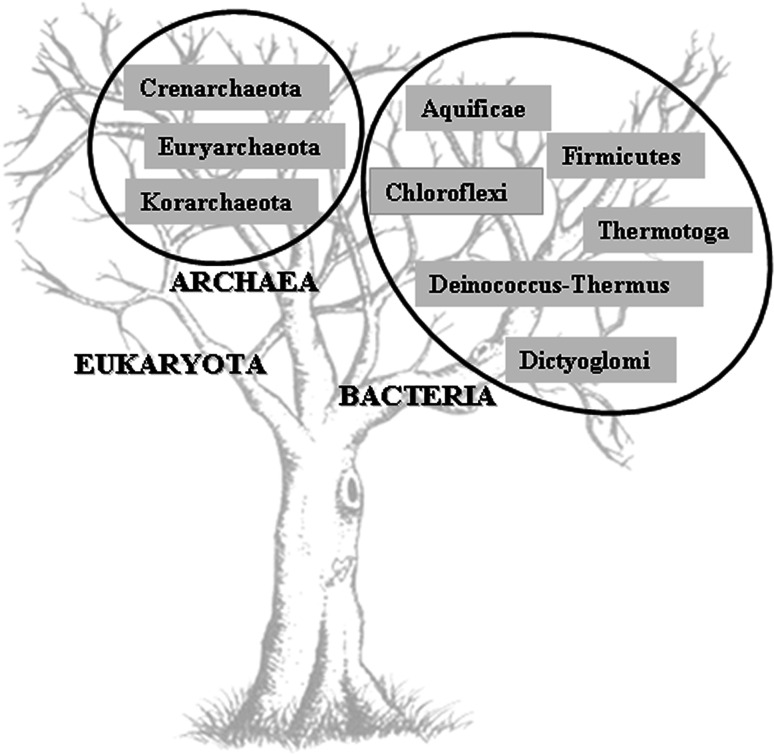
PDOs have so far been identified both in archaea such as Pyrococcus furiosus and Aeropyrum pernix and in thermophilic bacteria, such as Aquifex aeolicus, Dictyoglomus turgidum DSM 6724, and Roseiflexus sp. RS-1 (Fig. 5). Phylogenetic analysis has suggested that they first evolved in the Crenarchaeota, dispersing later into bacteria by horizontal gene transfer events (HGT) via the Euryarchaeota [123]. The preferential HGT generally observed between archaea and hyperthermophilic bacteria, such as Aquifex and Thermotoga [121, 124], further supports this evolutionary hypothesis.
Fig. 5.
A representation of phyla in the kingdoms of Bacteria and Archaea based on PDOs sequences annotated in the genomes
The first member of the PDO family was isolated in 1995 from the crude extracts of the hyperthermophilic archaeon P. furiosus [125]. This protein, termed PfPDO, showed two active sites, CQYC and a typical Grx sequence CPYC, and thioltransferase activity. It was thus considered a Grx-like protein [125, 126]. The resolution of its crystal structure (PDB code: 1a8l) opened up a completely new scenario, revealing structural details that correlated PfPDO with the multidomain PDI [127, 128]. So far several members of this family have been characterized in detail both at structural and functional level: one from a hyperthermophilic bacterium A. aeolicus [129], and four from the archaea A. pernix [79], P. furiosus [121, 127, 128], P. horikoshii [130], and Sulfolobus solfataricus [122, 131].
Insight into PDO functions
Although the characterized PDOs present a variety of sequences within the redox active site (Table 1), they are all active as dithiol oxidants of synthetic peptides, reductants of insulin disulfides and isomerases of disulfide bond in scrambled ribonuclease [79, 122, 125, 129]. Mutagenesis studies performed on PDOs from P. furiosus (PfPDO), P. horikoshii (PhPDO), and S. solfataricus (SsPDO) have clarified the contribution of each active site to overall catalytic activity, showing that the C-terminal site has a fundamental role in the thiol-transferase activity, whereas both sites are indispensable for isomerase activity, where the two units are presumed to function synergistically [121, 122, 126, 130]. By contrast, mutagenesis studies performed on PDO from A. aeolicus (AaPDO) showed a different behavior; indeed in this case each redox site was able to perform all catalytic activities operating independently, but their contribution was not equivalent. In particular, the C-terminal site was able to perform an activity comparable to the wild-type, while the N-terminal site presented a slightly lower activity [129].
Table 1.
Comparison of active site sequences, estimated pKas, solvent accessibility and disulfide conformations for the cysteine residues of PDOs from different sources
| Protein unit | Active site sequence | Cys1 | S-S | Cys2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pKa [66] | SEA* [116] | χ1(°) | χ2(°) | χ3(°) | χ1′(°) | χ2′(°) | SEA* [116] | pKa [66] | ||
| AaPDO-N | CESC | 9.5 | 2.42 | 171 | −148 | 88 | −79 | 78 | 0 | 23.0 |
| AaPDO-C | CGYC | 6.0 | 1.99 | 168 | −150 | 86 | −72 | 79 | 0 | 26.9 |
| PfPDO-N | CQYC | 12.0 | 0.11 | −135 | −138 | 47 | −116 | 99 | 0 | 26.5 |
| PfPDO-C | CPYC | 8.0 | 5.63 | 174 | −153 | 80 | −81 | 80 | 0 | 25.3 |
| ApPDO-N | CETC | 9.2 | 1.73** | 176 | −144 | 87 | −84 | 75 | 0** | 25.4 |
| ApPDO-C | CPYC | 7.3 | 1.40** | 172 | −149 | 89 | −72 | 76 | 0** | >30 |
* Sγ atom solvent exposed area measured in Å2, ** Data unpublished
A chaperone activity in the presence of ATP was determined for SsPDO [131], following the refolding of an alcohol dehydrogenase isolated from S. solfataricus. This activity, reported also for PDI [132], further supports the correlation between PDO and PDI [132]. In agreement with this property, an ATPase activity was identified both for SsPDO (Pedone E. pers. comm.) and PfPDO [121]. In the latter case, a more detailed characterization showed for this activity a maximum pH around basic values and an optimum temperature of 90°C [121, 133].
FT-IR spectroscopy and molecular dynamic simulation studies, performed at different pHs and temperatures, were used to evaluate the effect of pH and temperature on the stability of PfPDO [134]. These studies demonstrated that at pH 10.0, and at a temperature between 90.0 and 99.5°C, optimal conditions for ATPase activity, the protein undergoes partial unfolding with a concomitant relaxation of the tertiary structure, followed by a reorganization of part of the structure into a new β-conformation. The α-helix regions proved more affected by the increase in pH and temperature than the β-sheets [134].
Very little information is currently available on PDO substrates, while in several cases Prxs have been identified as Trx target proteins [135–137]. In correlation with these findings, recently two Prxs of S. solfataricus, bacterioferritin comigratory protein (Bcp)1 and Bcp4 [138], were reported to utilize the reconstituted SsPDO/SsTrx reductase (SsTr) (SSO2416) system for recycling, in place of the common Trx/Tr system [131], thus suggesting their potential role as PDO substrates. A similar recycling system had been previously characterized also in P. horikoshii (PhPDO/PhTr) [130].
Many reports have highlighted the peculiar role of sulfhydryl groups in the oxidative stress response and mainly of the Grx/Trx system in the maintenance of cell redox homeostasis [131, 139]. The in vivo role of PDOs has also been investigated, analyzing the expression of SsPDO under oxidative stress [131]. However, only a very small increase in mRNA and protein levels has been observed in such conditions, suggesting no direct role of SsPDO in the oxidative stress response [131]. In agreement with these data, a recent transcriptomic, proteomic, and chemical reactivity analysis, performed in oxidative stress conditions, failed to show any up-regulation of SsPDO, unlike other antioxidant enzymes [140–143]. By contrast, similar studies conducted on P. furiosus showed that PfPDO is one of the most strongly up-regulated proteins in response to cold adaptation from 95 to 72°C [144]. Microarray experiments performed independently by adding elemental sulfur to growing P. furiosus cells also indicated the up-regulation of PfPDO [145].
Recently, in vitro transcription experiments allowed the identification in P. furiosus of a transcriptional repressor of pdo, namely Sulfur Response Regulator (SurR). The surR gene is positioned 132 bp downstream of pdo and is divergently oriented [146]. SurR seems to effect its transcriptional regulation in the absence of elemental sulfur, activating genes that are down-regulated during the primary elemental sulfur response and repressing genes like pdo, that are up-regulated under the same conditions [146].
Altogether, these results reveal a complex scenario for PDOs, demonstrating that they can be finely regulated as shown in the anaerobic P. furiosus or constitutively expressed, as observed in S. solfataricus [131].
Insights into structural features of the PDO family
The resolution of the structure of PfPDO (PDB code: 1a8l; 1.9 Å resolution), AaPDO (PDB code: 2ayt; 2.4 Å resolution), and ApPDO (PDB code: 2hls; 1.9 Å resolution), revealed the presence of two structural units, one at the N-terminus and another at the C-terminus (Fig. 6), each consisting of a typical Trx fold with an additional α-helix inserted at the N-terminus [79, 127, 129]. For all the structures the two structural units superimpose well, despite a rather low sequence identity (about 20%). A homology model of SsPDO has also been built, confirming the general fold observed for the other PDOs [122]. In AaPDO and ApPDO the two active sites [79, 129] were accessible to the solvent and presented similar structural features, with dihedral angles comparable and in agreement with stable disulfide bonds (Table 1) [129]. By contrast, in PfPDO the two active sites showed strikingly different geometrical parameters and solvent accessibilities. Indeed, while the C-terminal site had a stable and exposed disulfide bond [127, 129], as shown in Table 1, the N-terminal site was completely buried and showed unusual dihedral angles, indicating the existence of a strong conformational strain [127]. Surprisingly, crystallographic data showed a greater mobility of the N-terminal segment with respect to the C-terminal one, in agreement with simulation studies conducted at different temperatures [124, 127, 134].
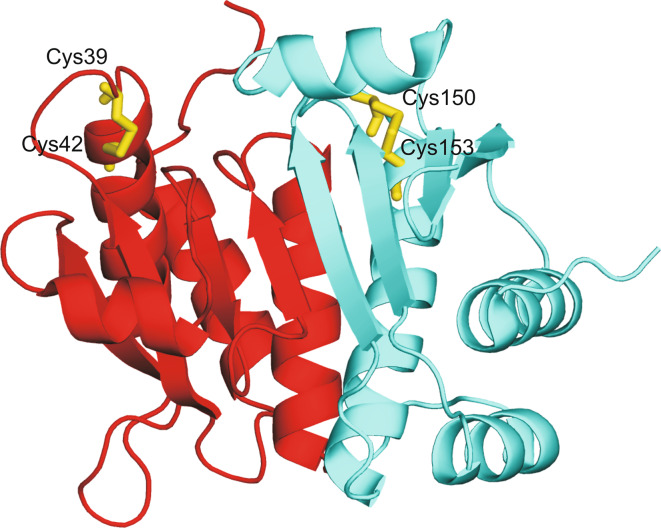
Fig. 6.
Overall fold of ApPDO [79]. The N-terminal unit is shown in red, while the C-terminal one in cyan. Cysteine residues of catalytic sites are colored in yellow and shown in stick representation
In the Trx superfamily, the pKa values of the nucleophilic cysteines have been demonstrated to be related to the redox potential, hence to the distinct reactivities of the CXXC motif [78, 147, 148]. Thus, a theoretical pKa study for the active site cysteines of the three different members of the PDO family with known crystallographic structure, ApPDO, AaPDO, PfPDO, was carried out [79, 127, 129] (Table 1). In all cases, the pKa values obtained for the N-terminal cysteines for both PDO active sites were found to be lower than those determined for the C-terminal cysteines highlighting the higher reactivity of the N-terminal cysteines. Moreover, while for ApPDO and AaPDO pKa values of the first cysteine range between 9.2 and 9.5 for the N-terminal active site and between 6 and 7.3 for the C-terminal one, in PfPDO the first cysteine of the N-terminal active site shows a significantly higher pKa (Table 1). This result can be attributed to its low solvent exposure and is in agreement with above reported mutagenesis and kinetic studies, which indicated that this site does not perform any reductive or oxidative activity.
Very little is known about the substrate binding site in PDOs [124], unlike PDI and the bacterial Dsbs. In 2006, Ladenstein and Ren built a model for the putative peptide binding site in PfPDO, comparing the crystal packing contacts, which can generally mimic substrate binding interactions [124], and the interaction region between human Trx and peptides from Ref-1 and NFkB [149, 150]. The authors observed that residues around the two PfPDO active sites form two grooves, denominated N and C, which could constitute two substrate binding sites [127]. The N groove is very narrow and deep and is delimited by residues belonging to both Trx units, while the C groove, whose shape is very similar to that of Trx, is much larger and is delimited only by residues of the C-terminal domain [127]. These residues are mainly hydrophobic and strictly conserved in all PDOs structurally characterized to date (Fig. 7). Interestingly, the hydrophobic nature of the C groove is a distinctive feature also of the PDI b′ domain [74].
Fig. 7.
Sequence alignment of the characterized PDOs performed with ClustalW2. PhPDO: Pyrococcus horykoshii PDO; PfPDO: Pyrococcus furiosus PDO; AaPDO: Aquifex aeolicus PDO; ApPDO: Aeropyrum pernix PDO; SsPDO: Sulfolobus solfataricus PDO. Residues of the CXXC motif are boxed, residues constituting the N groove are bold, while residues forming the C groove are underlined
Similarities and differences between PDI and PDO: is PDO an ancestor?
In light of the findings reported above, PDOs and PDI clearly present many similarities. Indeed, all these enzymes are able to reduce, oxidize, and isomerize disulfide bridges with high catalytic efficiency. Moreover, they show two catalytically active Trx-like domains and present a hydrophobic region for substrate recognition. However, a detailed comparison also highlights the presence of major differences. First of all, although in PDI it is clearly recognized that each active site has a different role in catalytic activity, the data available on PDOs do not define the same role for each active site of every member belonging to the family. Furthermore, these enzymes present a different structural organization; in particular, in PDOs the two Trx units are packed together, whereas in PDI they are structurally separated [8], although the distance between the two active sites is comparable (about 20 Å). Finally, in PDOs the substrate binding site is located in the same domain as the active site, whereas in PDI this region is situated in an additional non-active Trx domain. Altogether these data suggest that PDOs could be considered precursors of PDI, representing a simpler version of the eukaryotic enzyme, even though the observed differences in these two biological systems indicate very different mechanisms of action.
Heterodimerization of redox-active Trx fold: hybrid Prxs
The variability of Trx-like proteins can also be obtained by the fusion of genes encoding different Trx folds [30]. The hybrid Prxs are a peculiar subgroup of the Prx family [151], in which the Prx domain is fused with a Grx domain. These proteins represent a solution adopted by different bacteria to optimize hydroperoxide reduction [152–156].
Prxs are ubiquitous enzymes that degrade hydroperoxides and alkyl hydroperoxides to defend the cells from oxidative stress, and in eukaryotic cells they also play a key role in regulating H2O2-mediated cell signaling [157, 158]. To date, a large number of crystal structures of Prxs have been solved, showing that they all contain a canonical Trx fold with additional secondary structure elements, such as an extension of the N-terminal region, an insertion between β2 and α2, and sometimes an extension of the C-terminal region [12]. Prxs can be divided into 2-Cys and 1-Cys type based on the number of cysteine residues involved in the catalysis [157]. The first step in the catalytic mechanism, common to 2-Cys and 1-Cys Prxs, is the nucleophilic attack of a conserved cysteine located in the N-terminal region, called the peroxidatic cysteine (CPSH), on the peroxide and its consequent oxidation to cysteine sulphenic acid (CPSOH). In 2-Cys Prxs the CPSOH resolution depends on a second redox-active cysteine, termed the resolving cysteine (CRSH). Through a conformational change, from a fully folded to a locally unfolded form, a disulfide bond is formed connecting the two redox-active cysteines located in the same monomer (atypical 2-Cys Prxs) or in two different subunits (typical 2-Cys Prxs). In 1-Cys Prxs the CRSH is absent and CPSOH is regenerated with a mechanism not yet well elucidated. In this case, the CPSH is stabilized by hydrogen bond formation with a conserved histidine, which prevents its overoxidation to sulfinic and sulfonic acids [157].
The structure of the active site is highly conserved among Prxs. Indeed, the CPSH is always located in a loop-helix region exposed to the solvent and surrounded by three conserved residues: Pro38, Thr42, and Arg112 (the numbering refers to Bcp1 from S. solfataricus) [151]. The first residue restricts the solvent accessibility and protects the reactive sulfenic acid intermediate from further oxidation, Thr42 facilitates the proper position of the CPSH, allowing an unidentified catalytic base to extract a proton and finally Arg112 lowers the pKa of CPSH.
Different thiol redox systems are used to regenerate Prxs, the main one being the flavoprotein Tr coupled to Trx [157]. A variety of other recycling systems [159, 160] have also been described, such as the Alkyl hydroperoxide reductase (AhpF), in which AhpF represents the fusion between Tr and Trx domains and regenerates the Prx AhpC [161, 162], the previously described Tr/PDO system associated to the reduction of Bcps in S. solfataricus [138, 151], and the GSH reductase/GSH/Grx involved in the regeneration of Prx from Poplar [163].
Grxs are small (9–15 kDa) thioltransferases characterized by a Trx fold and an active site containing a CXXC motif in dithiol Grxs or a CXXS motif in monothiol Grxs [1, 164]. They specifically use GSH for their regeneration and catalyze the reduction of proteins with disulfide bridges and GSH-containing mixed disulfide bonds. Their role spans a great number of functions related to defense against oxidative stress, but also to protein folding, sulphur assimilation, coordination of [2Fe–2S] cluster, regulation of cellular differentiation, and transcription in eukaryotic cells [164].
In the hybrid Prxs, the Prx domain is always localized at the N-terminus, while the Grx domain is located at the C-terminus and is involved in the regeneration of the Prx domain. These proteins have so far been characterized in the ancestral microorganism Chromatium gracile [152] and in pathogenic bacteria such as Vibrio cholerae [155], Haemophilus influenzae [153, 154], and Neisseria meningitidis [156] (Fig. 8). In addition, genomic analysis of other pathogenic bacteria and cyanobacteria highlights both the diffusion of these types of proteins among pathogens and their ancestral role in the detoxification from hydroperoxides (Fig. 8). The multiple sequence alignment, reported in Fig. 8, shows that in all these proteins the Prx domain resembles the Type II Prx [151], while the Grx domain resembles E. coli Grx3 [166]. Hybrid Prxs can be divided into two subgroups on the basis of the number of conserved cysteines in the Prx domain: members of the first subgroup possess two conserved cysteines, while members of the second have only one conserved cysteine (Fig. 8).
Fig. 8.
ClustalW2 multiple alignments of hybrid Prxs from C. gracile [165], V. cholerae AAF95778, N. meningitidis NP_273984.1, and H. influenzae AAC22230.1 with homologous hybrid Prxs from pathogens (Yersinia aldovae ZP_04621281.1; Serratia proteamaculans YP_001480997.1, Erwinia pyrifoliae YP_002647197.1) and cyanobacteria (Anabaena variabilis Q3M358; Synechoccus sp YP_002370726.1), with Type II Prx YP_353982.1 from R. sphaeroides and Grx3 NP_418067.1 from E. coli. Boxed regions indicate the Prx and Grx domains; the two regions were separated by a linker. The active site cysteines of Prx domains are in bold, while the conserved CXXC motif of Grx domains is underlined
The only proteins of the first subgroup to be characterized are from C. gracile (CgPrx), V. cholerae (VcPrx), and N. menigitidis (NmPrx). CgPrx was demonstrated to be involved, together with the NADH, a glutathione amide reductase (GAR) and a glutathione amide (GASH) molecule, in the detoxification of H2O2 and small alkyl hydroperoxides of C. gracile, an anaerobic prototroph sulphur-oxidizing bacterium [152]. This protein contains in the Prx domain two cysteines in position 50 and 75, but no direct information is available on their catalytic role.
The first information on the catalytic mechanism of this subgroup comes from a kinetic study performed on VcPrx [155]. This protein reduces lipid hydroperoxides such as linoleic hydroperoxide, suggesting its action in vivo as a lipid hydroperoxide peroxidase, but unlike CgPrx, the recycling system is GSH supported. VcPrx has been shown to exist as a monomer and to function as a Type II 2-Cys atypical Prx, with Cys51 and Cys77 acting as CPSH and CRSH, respectively. A direct exchange of reducing equivalents between the Prx active site and that of Grx is thought to perform the complete catalytic mechanism.
More detailed information is available on the catalytic mechanism of the second subgroup, thanks to the crystal structure resolution of the hybrid Prx from H. influenzae (HiPrx) (PDB code: 1nm3) [153] (Fig. 9). HiPrx has two redox active sites: one in the Prx domain, which contains a unique cysteine (Cys49), corresponding to CPSH, and the other in the Grx domain containing two cysteines, Cys180, and Cys183 in the CXXC motif [154]. Analysis of the crystal structure reveals that the Prx domain (residues 3–162) presents the canonical Trx fold with the addition of several insertions common to other Prxs, while the Grx domain (residues 171–214), joined to the Prx domain through a linker loop, is characterized by the basic Trx fold (Fig. 9). The protein presents a tetrameric organization, which is achieved mainly by two strong subunit contacts: the first is between two Prx domains and the second between two Grx domains. In this structural organization the active sites of the Prx and Grx domains of two different monomers come close to each other, allowing the regeneration of the peroxidase activity of the enzyme. Indeed, after the oxidation of the CPSH, a direct electron transfer may occur from the CXXC redox site of the Grx domain of one monomer to the CPSOH of the Prx domain of another monomer.
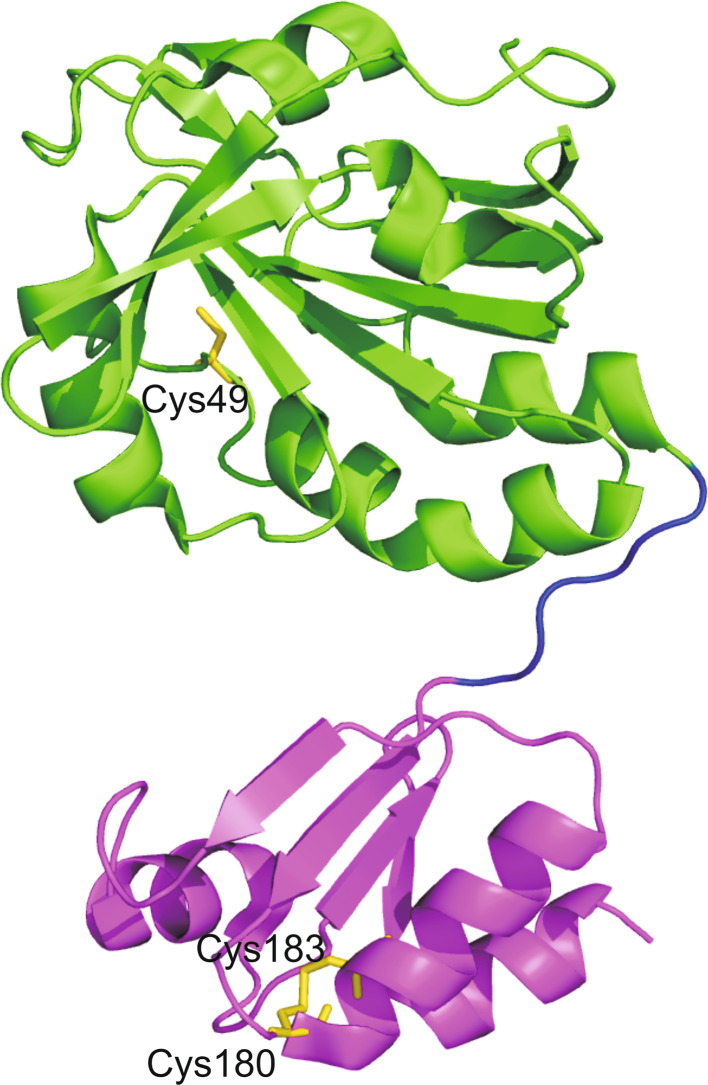
Fig. 9.
Overall fold of HiPrx. The Prx domain is shown in green, the linker region in blue and the Grx domain in magenta. The cysteine residues of catalytic sites are colored in yellow and shown in stick representation
The same quaternary organization has also been hypothesized for NmPrx [156] belonging to the first group of hybrid Prxs. The catalytic activity of the full length enzyme and of the two separated domains was tested, showing the better efficiency of the fused enzyme with respect to the reconstituted system. These results indicate that the hybrid enzyme can optimize both peroxidase activity and the reduction reaction to recycle the Prx domain [156].
Taken together, these data show that the aforementioned pathogens have acquired a key enzyme to defend themselves against both the attack of the human antimicrobial system and the oxidant environment, such as the nasopharynx, where H. influenzae and N. meningitidis can localize.
Concluding remarks
Trx fold is a widespread and versatile protein scaffold. Various insertions are possible on this structural theme originating different proteins, which span from the simple Grxs to the more complex GPXs or PDIs. The different structural complexity of these proteins reflects a variety of catalytic functions which range from dithiol-disulfide exchange reactions to hydroperoxide reduction. Analysis of the sequences of Trx-like proteins in all the kingdoms of life indicate that while most members contain just one copy of the Trx fold, some classes contain multiple copies. PDI represents the most interesting example of the presence of multiple copies of the same Trx fold in a single protein. Within the overall multi-domain organization, each catalytic domain specializes in a different dithiol-disulfide exchange reaction, while the non-catalytic domains are simply involved in substrate recognition. A simpler form of PDI is represented by the prokaryotic PDOs, in which there are only two Trx units packed together, and such a specific division of the functions is not possible. Finally, within the same multi-domain structure, combinations of different Trx domains with different functions may also be found. The hybrid Prxs are an example in which a Prx, involved in hydroperoxide reduction, is fused with a Grx, responsible for its regeneration.
Taken together these examples show that generally the presence of two or more catalytically active Trx fold in the same protein results either in the possibility to perform different catalytic functions or to fine tune the substrate specificity, thus representing a very useful strategy to improve enzyme catalytic performances.
References
- 1.Berndt C, Lillig CH, Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim Biophys Acta. 2008;1783:641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Kinch LN, Baker D, Grishin NV. Deciphering a novel thioredoxin-like fold family. Proteins. 2003;52:323–331. doi: 10.1002/prot.10425. [DOI] [PubMed] [Google Scholar]
- 3.Martin JL. Thioredoxin—a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/S0969-2126(01)00154-X. [DOI] [PubMed] [Google Scholar]
- 4.Eklund H, Cambillau C, Sjöberg BM, Holmgren A, Jörnvall H, Höög JO, Brändén CI. Conformational and functional similarities between glutaredoxin and thioredoxins. EMBO J. 1984;3:1443–1449. doi: 10.1002/j.1460-2075.1984.tb01994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 6.Inaba K. Disulfide bond formation system in Escherichia coli . J Biochem. 2009;146:591–597. doi: 10.1093/jb/mvp102. [DOI] [PubMed] [Google Scholar]
- 7.Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Pedone E, Limauro D, Bartolucci S. The machinery for oxidative protein folding in thermophiles. Antioxid Redox Signal. 2008;10:157–169. doi: 10.1089/ars.2007.1855. [DOI] [PubMed] [Google Scholar]
- 9.Heckler EJ, Rancy PC, Kodali VK, Thorpe C. Generating disulfides with the Quiescin-sulfhydryl oxidases. Biochim Biophys Acta. 2008;1783:567–577. doi: 10.1016/j.bbamcr.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorpe C, Coppock DL. Generating disulfides in multicellular organisms: emerging roles for a new flavoprotein family. J Biol Chem. 2007;282:13929–13933. doi: 10.1074/jbc.R600037200. [DOI] [PubMed] [Google Scholar]
- 11.Messens J, Martins JC, Van Belle K, Brosens E, Desmyter A, De Gieter M, Wieruszeski J, Willem R, Wyns L, Zegers I. All intermediates of the arsenate reductase mechanism, including an intramolecular dynamic disulfide cascade. Proc Natl Acad Sci USA. 2002;99:8506–8511. doi: 10.1073/pnas.132142799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copley SD, Novak WR, Babbitt PC. Divergence of function in the thioredoxin fold suprafamily: evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry. 2004;43:13981–13995. doi: 10.1021/bi048947r. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho AP, Fernandes PA, Ramos MJ. Similarities and differences in the thioredoxin superfamily. Prog Biophys Mol Biol. 2006;91:229–248. doi: 10.1016/j.pbiomolbio.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Atkinson HJ, Babbitt PC. An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput Biol. 2009;5:e1000541. doi: 10.1371/journal.pcbi.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson HJ, Babbitt PC. Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry. 2009;48:11108–11116. doi: 10.1021/bi901180v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 17.Guddat LW, Bardwell JC, Zander T, Martin JL. The uncharged surface features surrounding the active site of Escherichia coli DsbA are conserved and are implicated in peptide binding. Protein Sci. 1997;6:1148–1156. doi: 10.1002/pro.5560060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyson HJ, Jeng MF, Tennant LL, Slaby I, Lindell M, Cui DS, Kuprin S, Holmgren A. Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: structural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry. 1997;36:2622–2636. doi: 10.1021/bi961801a. [DOI] [PubMed] [Google Scholar]
- 20.Fomenko DE, Gladyshev VN. CxxS: fold-independent redox motif revealed by genome-wide searches for thiol/disulfide oxidoreductase function. Protein Sci. 2002;11:2285–2296. doi: 10.1110/ps.0218302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito K, Inaba K. The disulfide bond formation (Dsb) system. Curr Opin Struct Biol. 2008;18:450–458. doi: 10.1016/j.sbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Grimshaw JPA, Stirnimann CU, Brozzo MS, Malojcic G, Grütter MG, Capitani G, Glockshuber R. DsbL and DsbI form a specific dithiol oxidase system for periplasmic arylsulfate sulfotransferase in uropathogenic Escherichia coli . J Mol Biol. 2008;380:667–680. doi: 10.1016/j.jmb.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 23.Ren G, Stephan D, Xu Z, Zheng Y, Tang D, Harrison RS, Kurz M, Jarrott R, Shouldice SR, Hiniker A, Martin JL, Heras B, Bardwell JC. Properties of the thioredoxin fold superfamily are modulated by a single amino acid residue. J Biol Chem. 2009;284:10150–10159. doi: 10.1074/jbc.M809509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banci L, Bertini I, Ciofi-Baffoni S, Hadjiloi T, Martinelli M, Palumaa P. Mitochondrial copper(I) transfer from Cox17 to Sco1 is coupled to electron transfer. Proc Natl Acad Sci USA. 2008;105:6803–6808. doi: 10.1073/pnas.0800019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, Trumble WR, Liao H, Wesson CR, Dunker AK, Kang CH. Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat Struct Biol. 1998;5:476–483. doi: 10.1038/nsb0698-476. [DOI] [PubMed] [Google Scholar]
- 26.Alanen HI, Williamson RA, Howard MJ, Hatahet FS, Salo KE, Kauppila A, Kellokumpu S, Ruddock LW. ERp27, a new non-catalytic endoplasmic reticulum-located human protein disulfide isomerase family member, interacts with ERp57. J Biol Chem. 2006;281:33727–33738. doi: 10.1074/jbc.M604314200. [DOI] [PubMed] [Google Scholar]
- 27.Pan JL, Bardwell JC. The origami of thioredoxin-like folds. Protein Sci. 2006;15:2217–2227. doi: 10.1110/ps.062268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 29.Freedman RB. Novel disulfide oxidoreductase in search of a function. Nat Struct Biol. 1998;5:531–532. doi: 10.1038/783. [DOI] [PubMed] [Google Scholar]
- 30.Rouhier N, Gama F, Wingsle G, Gelhaye E, Gans P, Jacquot JP. Engineering functional artificial hybrid proteins between poplar peroxiredoxin II and glutaredoxin or thioredoxin. Biochem Biophys Res Commun. 2006;341:1300–1308. doi: 10.1016/j.bbrc.2006.01.099. [DOI] [PubMed] [Google Scholar]
- 31.Kemmink J, Darby N, Dijkstra K, Nilges M, Creighton TE. Structure determination of the N-terminal thioredoxin-like domain of protein disulfide isomerase using multidimensional heteronuclear 13C/15N NMR spectroscopy. Biochemistry. 1996;35:7684–7691. doi: 10.1021/bi960335m. [DOI] [PubMed] [Google Scholar]
- 32.Kemmink J, Dijkstra K, Mariani M, Scheek RM, Penka E, Nilges M, Darby NJ. The structure in solution of the b domain of protein disulfide isomerase. J Biomol NMR. 1999;13:357–368. doi: 10.1023/A:1008341820489. [DOI] [PubMed] [Google Scholar]
- 33.Denisov AY, Maattanen P, Dabrowski C, Kozlov G, Thomas DY, Gehring K. Solution structure of the bb′ domains of human protein disulfide isomerase. FEBS J. 2009;276:1440–1449. doi: 10.1111/j.1742-4658.2009.06884.x. [DOI] [PubMed] [Google Scholar]
- 34.Serve O, Kamiya Y, Maeno A, Nakano M, Murakami C, Sasakawa H, Yamaguchi Y, Harada T, Kurimoto E, Yagi-Utsumi M, Iguchi T, Inaba K, Kikuchi J, Asami O, Kajino T, Oka T, Nakasako M, Kato K. Redox-dependent domain rearrangement of protein disulfide isomerase coupled with exposure of its substrate-binding hydrophobic surface. J Mol Biol. 2010;396:361–374. doi: 10.1016/j.jmb.2009.11.049. [DOI] [PubMed] [Google Scholar]
- 35.Wang LK, Wang L, Vavassori S, Li SJ, Ke H, Anelli T, Degano M, Ronzoni R, Sitia R, Sun F, Wang CC. Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail. Embo Rep. 2008;9:642–647. doi: 10.1038/embor.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barak NN, Neumann P, Sevvana M, Schutkowski M, Naumann K, Malesevic M, Reichardt H, Fischer G, Stubbs MT, Ferrari DM. Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29. J Mol Biol. 2009;385:1630–1642. doi: 10.1016/j.jmb.2008.11.052. [DOI] [PubMed] [Google Scholar]
- 37.Liepinsh E, Baryshev M, Sharipo A, Ingelman-Sundberg M, Otting G, Mkrtchian S. Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51-kDa dimer. Structure. 2001;9:457–471. doi: 10.1016/S0969-2126(01)00607-4. [DOI] [PubMed] [Google Scholar]
- 38.Rowe ML, Ruddock LW, Kelly G, Schmidt JM, Williamson RA, Howard MJ. Solution structure and dynamics of ERp18, a small endoplasmic reticulum resident oxidoreductase. Biochemistry. 2009;48:4596–4606. doi: 10.1021/bi9003342. [DOI] [PubMed] [Google Scholar]
- 39.Jessop CE, Tavender TJ, Watkins RH, Chambers JE, Bulleid NJ. Substrate specificity of the oxidoreductase ERp57 is determined primarily by its interaction with calnexin and calreticulin. J Biol Chem. 2009;284:2194–2202. doi: 10.1074/jbc.M808054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maattanen P, Kozlov G, Gehring K, Thomas DY. ERp57 and PDI: multifunctional protein disulfide isomerases with similar domain architectures but differing substrate-partner associations. Biochem Cell Biol. 2006;84:881–889. doi: 10.1139/O06-186. [DOI] [PubMed] [Google Scholar]
- 41.Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci. 2006;31:455–464. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Wang CC, Tsou CL. Protein disulfide isomerase is both an enzyme and a chaperone. FASEB J. 1993;7:1515–1517. doi: 10.1096/fasebj.7.15.7903263. [DOI] [PubMed] [Google Scholar]
- 43.Koivu J, Myllyla R, Helaakoski T, Pihlajaniemi T, Tasanen K, Kivirikko KI. A single polypeptide acts both as the β subunit of prolyl 4-hydroxylase and as a protein disulfide-isomerase. J Biol Chem. 1987;262:6447–6449. [PubMed] [Google Scholar]
- 44.Wetterau JR, Combs KA, McLean LR, Spinner SN, Aggerbeck LP. Protein disulfide isomerase appears necessary to maintain the catalytically active structure of the microsomal triglyceride transfer protein. Biochemistry. 1991;30:9728–97235. doi: 10.1021/bi00104a023. [DOI] [PubMed] [Google Scholar]
- 45.Janiszewski M, Lopes LR, Carmo AO, Pedro MA, Brandes RP, Santos CXC, Laurindo FRM. Regulation of NAD(P)H oxidase by associated protein disufide isomerase in vascular smooth muscle cells. J Biol Chem. 2005;280:40813–40819. doi: 10.1074/jbc.M509255200. [DOI] [PubMed] [Google Scholar]
- 46.Creighton TE. Protein folding coupled to disulphide bond formation. Biol Chem. 1997;378:731–744. doi: 10.1515/bchm.1997.378.8.731. [DOI] [PubMed] [Google Scholar]
- 47.Ruddon RW, Bedows E. Assisted protein folding. J Biol Chem. 1997;272:3125–3128. doi: 10.1074/jbc.272.6.3125. [DOI] [PubMed] [Google Scholar]
- 48.Satoh M, Shimada A, Kashiwai A, Saga S, Hosokawa M. Differential cooperative enzymatic activities of protein disulfide isomerase family in protein folding. Cell Stress Chaperones. 2005;10:211–220. doi: 10.1379/CSC-109R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R. Endoplasmic reticulum oxidoreductin 1-lβ (ERO1-Lβ), a human gene induced in the course of the unfolded protein response. J Biol Chem. 2000;275:23685–236292. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- 50.Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 51.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/S0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 52.Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci USA. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sevier CS, Kaiser CA. Disulfide transfer between two conserved cysteine pairs imparts selectivity to protein oxidation by Ero1. Mol Biol Cell. 2006;17:2256–22566. doi: 10.1091/mbc.E05-05-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sevier CS, Cuozzo JW, Vala A, Aslund F, Kaiser CA. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- 55.Vala A, Sevier CS, Kaiser CA. Structural determinants of substrate access to the disulfide oxidase Erv2p. J Mol Biol. 2005;354:952–966. doi: 10.1016/j.jmb.2005.09.076. [DOI] [PubMed] [Google Scholar]
- 56.Gross E, Sevier CS, Vala A, Kaiser CA, Fass D. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat Struct Biol. 2002;9:61–67. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
- 57.Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashworth JL, Kelly V, Wilson R, Shuttleworth CA, Kielty CM. Fibrillin assembly: dimer formation mediated by amino-terminal sequences. J Cell Sci. 1999;112:3549–3558. doi: 10.1242/jcs.112.20.3549. [DOI] [PubMed] [Google Scholar]
- 59.Di Jeso B, Park YN, Ulianich L, Treglia AS, Urbanas ML, High S, Arvan P. Mixed-disulfide folding intermediates between thyroglobulin and endoplasmic reticulum resident oxidoreductases ERp57 and protein disulfide isomerase. Mol Cell Biol. 2005;25:9793–9805. doi: 10.1128/MCB.25.22.9793-9805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molinari M, Helenius A. Glycoproteins form mixed disulfides with oxidoreductases during folding in living cells. Nature. 1999;402:90–93. doi: 10.1038/47062. [DOI] [PubMed] [Google Scholar]
- 61.Roth RA, Pierce SB. In vivo cross-linking of protein disulfide isomerase to immunoglobulins. Biochemistry. 1987;26:4179–4182. doi: 10.1021/bi00388a001. [DOI] [PubMed] [Google Scholar]
- 62.Vandenbroeck K, Martens E, Alloza I. Multi-chaperone complexes regulate the folding of interferon-gamma in the endoplasmic reticulum. Cytokine. 2006;33:264–273. doi: 10.1016/j.cyto.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 63.Vuori K, Myllylä R, Pihlajaniemi T, Kivirikko KI. Expression and site-directed mutagenesis of human protein disulfide isomerase in Escherichia coli. This multifunctional polypeptide has two independently acting catalytic sites for the isomerase activity. J Biol Chem. 1992;267:7211–7214. [PubMed] [Google Scholar]
- 64.LaMantia ML, Lennarz WJ. The essential function of yeast protein disulfide isomerase does not reside in its isomerase activity. Cell. 1993;74:899–908. doi: 10.1016/0092-8674(93)90469-7. [DOI] [PubMed] [Google Scholar]
- 65.Lyles MM, Gilbert HF. Mutations in the thioredoxin sites of protein disulfide isomerase reveal functional nonequivalence of the N- and C-terminal domains. J Biol Chem. 1994;269:30946–30952. [PubMed] [Google Scholar]
- 66.Laboissiere MC, Sturley SL, Raines RT. The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. J Biol Chem. 1995;270:28006–28009. doi: 10.1074/jbc.270.47.28006. [DOI] [PubMed] [Google Scholar]
- 67.Darby NJ, Creighton TE. Characterization of the active site cysteine residues of the thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:16770–16780. doi: 10.1021/bi00051a027. [DOI] [PubMed] [Google Scholar]
- 68.Walker KW, Lyles MM, Gilbert HF. Catalysis of oxidative protein folding by mutants of protein disulfide isomerase with a single active-site cysteine. Biochemistry. 1996;35:1972–1980. doi: 10.1021/bi952157n. [DOI] [PubMed] [Google Scholar]
- 69.Westphal V, Darby NJ, Winther JR. Functional properties of the two redox-active sites in yeast protein disulphide isomerase in vitro and in vivo. J Mol Biol. 1999;286:1229–1239. doi: 10.1006/jmbi.1999.2560. [DOI] [PubMed] [Google Scholar]
- 70.Kulp MS, Frickel EM, Ellgaard L, Weissman JS. Domain architecture of protein-disulfide isomerase facilitates its dual role as an oxidase and an isomerase in Ero1p-mediated disulfide formation. J Biol Chem. 2006;281:876–884. doi: 10.1074/jbc.M511764200. [DOI] [PubMed] [Google Scholar]
- 71.Holst B, Tachibana C, Winther JR. Active site mutations in yeast protein disulfide isomerase cause dithiothreitol sensitivity and a reduced rate of protein folding in the endoplasmic reticulum. J Cell Biol. 1997;138:1229–1238. doi: 10.1083/jcb.138.6.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L, Li SJ, Sidhu A, Zhu L, Liang Y, Freedman RB, Wang CC. Reconstitution of human Ero1-Lα/protein-disulfide isomerase oxidative folding pathway in vitro. Position-dependent differences in role between the a and a′ domains of protein-disulfide isomerase. J Biol Chem. 2009;284:199–206. doi: 10.1074/jbc.M806645200. [DOI] [PubMed] [Google Scholar]
- 73.Klappa P, Ruddock LW, Darby NJ, Freedman RB. The b′ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 1998;17:927–935. doi: 10.1093/emboj/17.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pirneskoski A, Klappa P, Lobell M, Williamson RA, Byrne L, Alanen HI, Salo KE, Kivirikko KI, Freedman RB, Ruddock LW. Molecular characterization of the principal substrate binding site of the ubiquitous folding catalyst protein disulfide isomerase. J Biol Chem. 2004;279:10374–10381. doi: 10.1074/jbc.M312193200. [DOI] [PubMed] [Google Scholar]
- 75.Darby NJ, Penka E, Vincentelli R. The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J Mol Biol. 1998;276:239–247. doi: 10.1006/jmbi.1997.1504. [DOI] [PubMed] [Google Scholar]
- 76.Tian G, Xiang S, Noiva R, Lennarz WJ, Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell. 2006;124:61–73. doi: 10.1016/j.cell.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 77.Karala AR, Lappi AK, Ruddock LW. Modulation of an active-site cysteine pKa allows PDI to act as a catalyst of both disulfide bond formation and isomerization. J Mol Biol. 2010;396:883–892. doi: 10.1016/j.jmb.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 78.Lappi AK, Lensink MF, Alanen HI, Salo KE, Lobell M, Juffer AH, Ruddock LW. A conserved arginine plays a role in the catalytic cycle of the protein disulphide isomerases. J Mol Biol. 2004;335:283–295. doi: 10.1016/j.jmb.2003.10.051. [DOI] [PubMed] [Google Scholar]
- 79.D’Ambrosio K, Pedone E, Langella E, De Simone G, Rossi M, Pedone C, Bartolucci S. A novel member of the protein disulfide oxidoreductase family from Aeropyrum pernix K1: structure, function and electrostatics. J Mol Biol. 2006;362:743–752. doi: 10.1016/j.jmb.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 80.Tian G, Kober FX, Lewandrowski U, Sickmann A, Lennarz WJ, Schindelin H. The catalytic activity of protein-disulfide isomerase requires a conformationally flexible molecule. J Biol Chem. 2008;283:33630–33640. doi: 10.1074/jbc.M806026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen VD, Wallis K, Howard MJ, Haapalainen AM, Salo KE, Saaranen MJ, Sidhu A, Wierenga RK, Freedman RB, Ruddock LW, Williamson RA. Alternative conformations of the x region of human protein disulphide-isomerase modulate exposure of the substrate binding b′ domain. J Mol Biol. 2008;383:1144–1155. doi: 10.1016/j.jmb.2008.08.085. [DOI] [PubMed] [Google Scholar]
- 82.Byrne LJ, Sidhu A, Wallis AK, Ruddock LW, Freedman RB, Howard MJ, Williamson RA. Mapping of the ligand-binding site on the b′ domain of human PDI: interaction with peptide ligands and the x-linker region. Biochem J. 2009;423:209–217. doi: 10.1042/BJ20090565. [DOI] [PubMed] [Google Scholar]
- 83.Wallis AK, Sidhu A, Byrne LJ, Howard MJ, Ruddock LW, Williamson RA, Freedman RB. The ligand-binding b′ domain of human protein disulphide-isomerase mediates homodimerization. Protein Sci. 2009;18:2569–2577. doi: 10.1002/pro.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soldà T, Garbi N, Hämmerling GJ, Molinari M. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem. 2006;281:6219–6226. doi: 10.1074/jbc.M513595200. [DOI] [PubMed] [Google Scholar]
- 85.Jessop CE, Chakravarthi S, Garbi N, Hämmerling GJ, Lovell S, Bulleid NJ. ERp57 is essential for efficient folding of glycoproteins sharing common structural domains. EMBO J. 2007;26:28–40. doi: 10.1038/sj.emboj.7601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frickel EM, Frei P, Bouvier M, Stafford WF, Helenius A, Glockshuber R, Ellgaard L. ERp57 is a multifunctional thiol-disulfide oxidoreductase. J Biol Chem. 2004;279:18277–18287. doi: 10.1074/jbc.M314089200. [DOI] [PubMed] [Google Scholar]
- 87.Zapun A, Darby NJ, Tessier DC, Michalak M, Bergeron JJ, Thomas DY. Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J Biol Chem. 1998;273:6009–6012. doi: 10.1074/jbc.273.11.6009. [DOI] [PubMed] [Google Scholar]
- 88.Oliver JD, van der Wal FJ, Bulleid NJ, High S. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–88. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- 89.Ellgaard L, Helenius A. ER quality control: towards an understanding at the molecular level. Curr Opin Cell Biol. 2001;13:431–437. doi: 10.1016/S0955-0674(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 90.Oliver JD, Roderick HL, Llewellyn DH, High S. ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin. Mol Biol Cell. 1999;10:2573–2582. doi: 10.1091/mbc.10.8.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ellgaard L, Frickel EM. Calnexin, calreticulin, and ERp57: teammates in glycoprotein folding. Cell Biochem Biophys. 2003;39:223–247. doi: 10.1385/CBB:39:3:223. [DOI] [PubMed] [Google Scholar]
- 92.Frickel EM, Riek R, Jelesarov I, Helenius A, Wuthrich K, Ellgaard L. TROSY-NMR reveals interaction between ERp57 and the tip of the calreticulin P-domain. Proc Natl Acad Sci USA. 2002;99:1954–1959. doi: 10.1073/pnas.042699099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kozlov G, Maattanen P, Schrag JD, Pollock S, Cygler M, Nagar B, Thomas DY, Gehring K. Crystal structure of the bb′ domains of the protein disulfide isomerase ERp57. Structure. 2006;14:1331–1339. doi: 10.1016/j.str.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 94.Ellgaard L, Bettendorff P, Braun D, Herrmann T, Fiorito F, Jelesarov I, Güntert P, Helenius A, Wüthrich K. NMR structures of 36 and 73-residue fragments of the calreticulin P-domain. J Mol Biol. 2002;322:773–784. doi: 10.1016/S0022-2836(02)00812-4. [DOI] [PubMed] [Google Scholar]
- 95.Leach MR, Cohen-Doyle MF, Thomas DY, Williams DB. Localization of the lectin, ERp57 binding, and polypeptide binding sites of calnexin and calreticulin. J Biol Chem. 2002;277:29686–29697. doi: 10.1074/jbc.M202405200. [DOI] [PubMed] [Google Scholar]
- 96.Russell SJ, Ruddock LW, Salo KE, Oliver JD, Roebuck QP, Llewellyn DH, Roderick HL, Koivunen P, Myllyharju J, High S. The primary substrate binding site in the b′ domain of ERp57 is adapted for endoplasmic reticulum lectin association. J Biol Chem. 2004;279:18861–18869. doi: 10.1074/jbc.M400575200. [DOI] [PubMed] [Google Scholar]
- 97.Dick TP, Bangia N, Peaper DR, Cresswell P. Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity. 2002;16:87–98. doi: 10.1016/S1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 98.Peaper DR, Wearsch PA, Cresswell P. Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J. 2005;24:3613–3623. doi: 10.1038/sj.emboj.7600814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hirano N, Shibasaki F, Sakai R, Tanaka T, Nishida J, Yazaki Y, Takenawa T, Hirai H. Molecular cloning of the human glucose-regulated protein ERp57/GRP58, a thiol-dependent reductase. Identification of its secretory form and inducible expression by the oncogenic transformation. Eur J Biochem. 1995;234:336–342. doi: 10.1111/j.1432-1033.1995.336_c.x. [DOI] [PubMed] [Google Scholar]
- 100.Antoniou AN, Ford S, Alphey M, Osborne A, Elliott T, Powis SJ. The oxidoreductase ERp57 efficiently reduces partially folded in preference to fully folded MHC class I molecules. EMBO J. 2002;21:2655–2663. doi: 10.1093/emboj/21.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beynon-Jones SM, Antoniou AN, Powis SJ. Mutational analysis of the oxidoreductase ERp57 reveals the importance of the two central residues in the redox motif. FEBS Lett. 2006;580:1897–1902. doi: 10.1016/j.febslet.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 102.Lundström J, Holmgren A. Determination of the reduction-oxidation potential of the thioredoxin-like domains of protein disulfide-isomerase from the equilibrium with glutathione and thioredoxin. Biochemistry. 1993;32:6649–6655. doi: 10.1021/bi00077a018. [DOI] [PubMed] [Google Scholar]
- 103.Silvennoinen L, Karvonen P, Koivunen P, Myllyharju J, Kivirikko K, Kilpeläinen I. Assignment of 1H, 13C and 15N resonances of the a′ domain of ERp57. J Biomol NMR. 2001;20:385–386. doi: 10.1023/A:1011291316810. [DOI] [PubMed] [Google Scholar]
- 104.Silvennoinen L, Koivunen P, Myllyharju J, Kilpeläinen I, Permi P. NMR assignment of the N-terminal domain a of the glycoprotein chaperone ERp57. J Biomol NMR. 2005;33:136. doi: 10.1007/s10858-005-2720-1. [DOI] [PubMed] [Google Scholar]
- 105.Pollock S, Kozlov G, Pelletier MF, Trempe JF, Jansen G, Sitnikov D, Bergeron JJ, Gehring K, Ekiel I, Thomas DY. Specific interaction of ERp57 and calnexin determined by NMR spectroscopy and an ER two-hybrid system. EMBO J. 2004;23:1020–1029. doi: 10.1038/sj.emboj.7600119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dong G, Wearsch PA, Peaper DR, Cresswell P, Reinisch KM. Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity. 2009;30:21–32. doi: 10.1016/j.immuni.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mazzarella RA, Srinivasan M, Haugejorden SM, Green M. ERp72, an abundant luminal endoplasmic reticulum protein, contains three copies of the active site sequences of protein disulfide isomerase. J Biol Chem. 1990;265:1094–1101. [PubMed] [Google Scholar]
- 108.Kozlov G, Määttänen P, Schrag JD, Hura GL, Gabrielli L, Cygler M, Thomas DY, Gehring K. Structure of the noncatalytic domains and global fold of the protein disulfide isomerase ERp72. Structure. 2009;17:651–659. doi: 10.1016/j.str.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 109.Van PN, Rupp K, Lampen A, Söling HD. CaBP2 is a rat homolog of ERp72 with protein disulfide isomerase activity. Eur J Biochem. 1993;213:789–795. doi: 10.1111/j.1432-1033.1993.tb17821.x. [DOI] [PubMed] [Google Scholar]
- 110.Kramer B, Ferrari DM, Klappa P, Pöhlmann N, Söling HD. Functional roles and efficiencies of the thioredoxin boxes of calcium-binding proteins 1 and 2 in protein folding. Biochem J. 2001;357:83–95. doi: 10.1042/0264-6021:3570083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lundström-Ljung J, Birnbach U, Rupp K, Söling HD, Holmgren A. Two resident ER-proteins, CaBP1 and CaBP2, with thioredoxin domains, are substrates for thioredoxin reductase: comparison with protein disulfide isomerase. FEBS Lett. 1995;357:305–358. doi: 10.1016/0014-5793(94)01386-F. [DOI] [PubMed] [Google Scholar]
- 112.Rupp K, Birnbach U, Lundström J, Van PN, Söling HD. Effects of CaBP2, the rat analog of ERp72, and of CaBP1 on the refolding of denatured reduced proteins. Comparison with protein disulfide isomerase. J Biol Chem. 1994;269:2501–2507. [PubMed] [Google Scholar]
- 113.Miyaishi O, Kozaki K, Iida K, Isobe K, Hashizume Y, Saga S. Elevated expression of PDI family proteins during differentiation of mouse F9 teratocarcinoma cells. J Cell Biochem. 1998;68:436–445. doi: 10.1002/(SICI)1097-4644(19980315)68:4<436::AID-JCB4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 114.Cotterill SL, Jackson GC, Leighton MP, Wagener R, Mäkitie O, Cole WG, Briggs MD. Multiple epiphyseal dysplasia mutations in MATN3 cause misfolding of the A-domain and prevent secretion of mutant matrilin-3. Hum Mutat. 2005;26:557–565. doi: 10.1002/humu.20263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sørensen S, Ranheim T, Bakken KS, Leren TP, Kulseth MA. Retention of mutant low density lipoprotein receptor in endoplasmic reticulum (ER) leads to ER stress. J Biol Chem. 2006;281:468–476. doi: 10.1074/jbc.M507071200. [DOI] [PubMed] [Google Scholar]
- 116.Menon S, Lee J, Abplanalp WA, Yoo SE, Agui T, Furudate S, Kim PS, Arvan P. Oxidoreductase interactions include a role for ERp72 engagement with mutant thyroglobulin from the rdw/rdw rat dwarf. J Biol Chem. 2007;282:6183–6191. doi: 10.1074/jbc.M608863200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Satoh M, Shimada A, Keino H, Kashiwai A, Nagai N, Saga S, Hosokawa M. Functional characterization of 3 thioredoxin homology domains of ERp72. Cell Stress Chaperones. 2005;10:278–284. doi: 10.1379/CSC-116R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mallick P, Boutz DR, Eisenberg D, Yeates TO. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc Natl Acad Sci USA. 2002;99:9679–9684. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.O’Connor B, Yeates TO. GDAP: a web tool for structural disulfide bond prediction. Nucleic Acids Res. 2004;32:360–364. doi: 10.1093/nar/gkh376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Beeby M, O’Connor BD, Ryttersgaard C, Boutz DR, Perry LJ, Yeates TO. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 2005;3:1549–1558. doi: 10.1371/journal.pbio.0030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pedone E, Ren B, Ladenstein R, Rossi M, Bartolucci S. Functional properties of the protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus . Eur J Biochem. 2004;271:3437–3448. doi: 10.1111/j.0014-2956.2004.04282.x. [DOI] [PubMed] [Google Scholar]
- 122.Limauro D, Saviano M, Galdi I, Rossi M, Bartolucci S, Pedone E. Sulfolobus solfataricus protein disulphide oxidoreductase: insight into the roles of its redox sites. Protein Eng Des Sel. 2009;22:19–26. doi: 10.1093/protein/gzn061. [DOI] [PubMed] [Google Scholar]
- 123.Becerra A, Delaye L, Lazcano A, Orgel LE. Protein disulfide oxidoreductases and the evolution of thermophily: was the last common ancestor a heat-loving microbe? J Mol Evol. 2007;65:296–303. doi: 10.1007/s00239-007-9005-0. [DOI] [PubMed] [Google Scholar]
- 124.Ladenstein R, Ren B. Protein disulfides and protein disulfide oxidoreductases in hyperthermophiles. FEBS J. 2006;273:4170–4185. doi: 10.1111/j.1742-4658.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 125.Guagliardi A, de Pascale D, Cannio R, Nobile V, Bartolucci S, Rossi M. The purification, cloning, and high level expression of a glutaredoxin-like protein from the hyperthermophilic archaeon Pyrococcus furiosus . J Biol Chem. 1995;270:5748–5755. doi: 10.1074/jbc.270.11.5748. [DOI] [PubMed] [Google Scholar]
- 126.Bartolucci S, de Pascale D, Rossi M. Protein disulfide oxidoreductase from Pyrococcus furiosus: biochemical properties. Methods Enzymol. 2001;334:62–73. doi: 10.1016/S0076-6879(01)34459-2. [DOI] [PubMed] [Google Scholar]
- 127.Ren B, Tibbelin G, de Pascale D, Rossi M, Bartolucci S, Ladenstein R. A protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus contains two thioredoxin fold units. Nat Struct Biol. 1998;5:602–611. doi: 10.1038/862. [DOI] [PubMed] [Google Scholar]
- 128.Ren B, Ladenstein R. Protein disulfide oxidoreductase from Pyrococcus furiosus: structural properties. Methods Enzymol. 2001;334:74–88. doi: 10.1016/S0076-6879(01)34460-9. [DOI] [PubMed] [Google Scholar]
- 129.Pedone E, D’Ambrosio K, De Simone G, Rossi M, Pedone C, Bartolucci S. Insights on a new PDI-like family: structural and functional analysis of a protein disulfide oxidoreductase from the bacterium Aquifex aeolicus . J Mol Biol. 2006;356:155–164. doi: 10.1016/j.jmb.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 130.Kashima Y, Ishikawa K. A hyperthermostable novel protein-disulfide oxidoreductase is reduced by thioredoxin reductase from hyperthermophilic archaeon Pyrococcus horikoshii . Arch Biochem Biophys. 2003;418:179–185. doi: 10.1016/j.abb.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 131.Pedone E, Limauro D, D’Alterio R, Rossi M, Bartolucci S. Characterization of a multifunctional protein disulfide oxidoreductase from Sulfolobus solfataricus . FEBS J. 2006;273:5407–5420. doi: 10.1111/j.1742-4658.2006.05533.x. [DOI] [PubMed] [Google Scholar]
- 132.Quemeneur E, Guthapfel R, Gueguen P. A major phosphoprotein of the endoplasmic reticulum is protein disulfide isomerase. J Biol Chem. 1994;269:5485–5488. [PubMed] [Google Scholar]
- 133.Zapun A, Creighton TE, Rowling PJ, Freedman RB. Folding in vitro of bovine pancreatic trypsin inhibitor in the presence of proteins of the endoplasmic reticulum. Proteins. 1992;14:10–15. doi: 10.1002/prot.340140104. [DOI] [PubMed] [Google Scholar]
- 134.Pedone E, Saviano M, Bartolucci S, Rossi M, Ausili A, Scirè A, Bertoli E, Tanfani F. Temperature-, SDS-, and pH-induced conformational changes in protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus: a dynamic simulation and Fourier transform infrared spectroscopic study. J Proteome Res. 2005;4:1972–1980. doi: 10.1021/pr050152z. [DOI] [PubMed] [Google Scholar]
- 135.Hisabori T, Hara S, Fujii T, Yamazaki D, Hosoya-Matsuda N, Motohashi K. Thioredoxin affinity chromatography: a useful method for further understanding the thioredoxin network. J Exp Bot. 2005;56:1463–1468. doi: 10.1093/jxb/eri170. [DOI] [PubMed] [Google Scholar]
- 136.Goyer A, Haslekås C, Miginiac-Maslow M, Klein U, Le Marechal P, Jacquot JP, Decottignies P. Isolation and characterization of a thioredoxin-dependent peroxidase from Chlamydomonas reinhardtii . Eur J Biochem. 2002;269:272–282. doi: 10.1046/j.0014-2956.2001.02648.x. [DOI] [PubMed] [Google Scholar]
- 137.Broin M, Cuiné S, Eymery F, Rey P. The plastidic 2-cysteine peroxiredoxin is a target for a thioredoxin involved in the protection of the photosynthetic apparatus against oxidative damage. Plant Cell. 2002;14:1417–1432. doi: 10.1105/tpc.001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Limauro D, Pedone E, Galdi I, Bartolucci S. Peroxiredoxins as cellular guardians in Sulfolobus solfataricus—characterization of Bcp1, Bcp3 and Bcp4. FEBS J. 2008;275:2067–2077. doi: 10.1111/j.1742-4658.2008.06361.x. [DOI] [PubMed] [Google Scholar]
- 139.Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST20051375. [DOI] [PubMed] [Google Scholar]
- 140.Maaty WS, Wiedenheft B, Tarlykov P, Schaff N, Heinemann J, Robison-Cox J, Valenzuela J, Dougherty A, Blum P, Lawrence CM, Douglas T, Young MJ, Bothner B. Something old, something new, something borrowed; how the thermoacidophilic archaeon Sulfolobus solfataricus responds to oxidative stress. PLoS One. 2009;4:e6964. doi: 10.1371/journal.pone.0006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vivancos AP, Jara M, Zuin A, Sanso M, Hidalgo E. Oxidative stress in Schizosaccharomyces pombe: different H2O2 levels, different response pathways. Mol Genet Genomics. 2006;276:495–502. doi: 10.1007/s00438-006-0175-z. [DOI] [PubMed] [Google Scholar]
- 142.Rabilloud T, Chevallet M, Luche S, Leize-Wagner E. Oxidative stress response: a proteomic view. Expert Rev Proteomics. 2005;2:949–956. doi: 10.1586/14789450.2.6.949. [DOI] [PubMed] [Google Scholar]
- 143.Vandenbroucke K, Robbens S, Vandepoele K, Inze D, de Peer YV, Van Breusegem F. Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol. 2008;25:507–516. doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- 144.Trauger SA, Kalisak E, Kalisiak J, Morita H, Weinberg MV, Menon AL, Poole FL, 2nd, Adams MW, Siuzdak G. Correlating the transcriptome, proteome, and metabolome in the environmental adaptation of a hyperthermophile. J Proteome Res. 2008;7:1027–1035. doi: 10.1021/pr700609j. [DOI] [PubMed] [Google Scholar]
- 145.Schut GJ, Bridger SL, Adams MW. Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme A-dependent NAD(P)H sulfur oxidoreductase. J Bacteriol. 2007;189:4431–4441. doi: 10.1128/JB.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lipscomb GL, Keese AM, Cowart DM, Schut GJ, Thomm M, Adams MW, Scott RA. SurR: a transcriptional activator and repressor controlling hydrogen and elemental sulphur metabolism in Pyrococcus furiosus . Mol Microbiol. 2009;71:332–349. doi: 10.1111/j.1365-2958.2008.06525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 148.Kortemme T, Darby NJ, Creighton TE. Electrostatic interactions in the active site of the N-terminal thioredoxin-like domain of protein disulfide isomerase. Biochemistry. 1996;35:14503–14511. doi: 10.1021/bi9617724. [DOI] [PubMed] [Google Scholar]
- 149.Qin J, Clore GM, Kennedy WMP, Kuszewski J, Gronenborn AM. The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. Structure. 1996;4:613–620. doi: 10.1016/S0969-2126(96)00065-2. [DOI] [PubMed] [Google Scholar]
- 150.Qin J, Clore GM, Kennedy WMP, Huth JR, Gronenborn AM. Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NFκB. Structure. 1995;3:289–297. doi: 10.1016/S0969-2126(01)00159-9. [DOI] [PubMed] [Google Scholar]
- 151.D’Ambrosio K, Limauro D, Pedone E, Galdi I, Pedone C, Bartolucci S, De Simone G. Insights into the catalytic mechanism of the Bcp family: functional and structural analysis of Bcp1 from Sulfolobus solfataricus . Proteins. 2009;76:995–1006. doi: 10.1002/prot.22408. [DOI] [PubMed] [Google Scholar]
- 152.Vergauwen B, Pauwels F, Jacquemotte F, Meyer TE, Cusanovich MA, Bartsch RG, Van Beeumen JJ. Characterization of glutathione amide reductase from Chromatium gracile. Identification of a novel thiol peroxidase (Prx/Grx) fueled by glutathione amide redox cycling. J Biol Chem. 2001;276:20890–20897. doi: 10.1074/jbc.M102026200. [DOI] [PubMed] [Google Scholar]
- 153.Kim SJ, Woo JR, Hwang YS, Jeong DG, Shin DH, Kim K, Ryu SE. The tetrameric structure of Haemophilus influenza hybrid Prx5 reveals interactions between electron donor and acceptor proteins. J Biol Chem. 2003;278:10790–10798. doi: 10.1074/jbc.M209553200. [DOI] [PubMed] [Google Scholar]
- 154.Pauwels F, Vergauwen B, Vanrobaeys F, Devreese B, Van Beeumen JJ. Purification and characterization of a chimeric enzyme from Haemophilus influenzae Rd that exhibits glutathione-dependent peroxidase activity. J Biol Chem. 2003;278:16658–16666. doi: 10.1074/jbc.M300157200. [DOI] [PubMed] [Google Scholar]
- 155.Cha MK, Hong SK, Lee DS, Kim IH. Vibrio cholerae thiol peroxidase-glutaredoxin fusion is a 2-Cys TSA/AhpC subfamily acting as a lipid hydroperoxide reductase. J Biol Chem. 2004;279:11035–11041. doi: 10.1074/jbc.M312657200. [DOI] [PubMed] [Google Scholar]
- 156.Rouhier N, Jacquot JP. Molecular and catalytic properties of a peroxiredoxin-glutaredoxin hybrid from Neisseria meningitidis . FEBS Lett. 2003;554:149–153. doi: 10.1016/S0014-5793(03)01156-6. [DOI] [PubMed] [Google Scholar]
- 157.Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sc. 2003;28:32–40. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 158.Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins—structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Castro H, Tomás AM. Peroxidases of trypanosomatids. Antioxid Redox Signal. 2008;10:1593–1606. doi: 10.1089/ars.2008.2050. [DOI] [PubMed] [Google Scholar]
- 160.Lee SP, Hwang YS, Kim YJ, Kwon KS, Kim HJ, Kim K, Chae HZ. Cyclophilin a binds to peroxiredoxins and activates its peroxidase activity. J Biol Chem. 2001;276:29826–29832. doi: 10.1074/jbc.M101822200. [DOI] [PubMed] [Google Scholar]
- 161.Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 162.Hall A, Parsonage D, Horita D, Karplus PA, Poole LB, Barbar E. Redox-dependent dynamics of a dual thioredoxin fold protein: evolution of specialized folds. Biochemistry. 2009;48:5984–5993. doi: 10.1021/bi900270w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rouhier N, Gelhaye E, Jacquot JP. Glutaredoxin-dependent peroxiredoxin from poplar: protein–protein interaction and catalytic mechanism. J Biol Chem. 2002;277:13609–13614. doi: 10.1074/jbc.M111489200. [DOI] [PubMed] [Google Scholar]
- 164.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 165.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 166.Aslund F, Nordstrand K, Berndt KD, Nikkola M, Bergman T, Ponstingl H, Jörnvall H, Otting G, Holmgren A. Glutaredoxin-3 from Escherichia coli. Amino acid sequence, 1H AND 15N NMR assignments, and structural analysis. J Biol Chem. 1996;271:6736–6745. doi: 10.1074/jbc.271.12.6736. [DOI] [PubMed] [Google Scholar]