Abstract
How mutations in the protein emerin lead to the cardiomyopathy associated with X-linked Emery-Dreifuss muscular dystrophy (X-EDMD) is unclear. We identified emerin at the adherens junction of the intercalated disc, where it co-localised with the catenin family of proteins. Emerin bound to wild type β-catenin both in vivo and in vitro. Mutating the GSK3β phosphorylation sites on β-catenin abolished this binding. Wild type but not mutant forms of emerin associated with X-EDMD were able to reduce β-catenin protein levels. Cardiomyocytes from emerin-null mice hearts exhibited erroneous β-catenin distribution and intercalated disc architecture. Treatment of wild type cardiomyocytes with phenylephrine, which inactivates GSK3β, redistributed emerin and β-catenin. Emerin was identified as a direct target of GSK3β activity since exogenous expression of GSK3β reduced emerin levels at the nuclear envelope. We propose that perturbation to or total loss of the emerin–β-catenin complex compromises both intercalated disc function and β-catenin signalling in cardiomyocytes.
Keywords: Emerin, β-catenin, Intercalated disc, Emery-Dreifuss muscular dystrophy, GSK3β
Introduction
X-linked Emery-Dreifuss muscular dystrophy (X-EDMD) is a neuromuscular condition characterised by progressive skeletal muscle wasting, contractures and a dilated cardiomyopathy (DCM) with an associated conduction defect [1]. X-EDMD arises due to mutations in the nuclear envelope protein emerin, which exhibits highest expression in skeletal and cardiac muscle [2]. Emerin has a large cohort of binding partners including lamin A/C, barrier to autointegration factor (BAF), LUMA, nesprin-1 and -2, the transcriptional repressors germ cell-less (GCL) and Btf, Lim-domain only 7 (Lmo7), β-catenin and actin [3–5]. Many of these binding partners possess over-lapping binding sites, e.g. β-catenin and nesprin-1/-2, which bind to emerin residues 169–180 and 140–176 respectively [6–8]. Thus, it is likely that different emerin-containing protein complexes confer cell-specific functions. Disruption of these emerin-containing complexes results in compromised nuclear envelope integrity and deregulation of downstream tissue-specific gene transcription [9–11]. In hearts from emerin-null mice, the extracellular signal-regulated kinase (ERK1/2) branch of the mitogen-activated protein kinase (MAPK) pathway is upregulated leading to alterations in the expression of downstream genes associated with cardiomyopathy [12]. Emerin-null mouse embryonic fibroblasts (MEFs) exhibit impaired response to mechanical strain, displaying abnormalities in mechanosensitive gene transcription and increased sensitivity to apoptosis [13].
Emerin locates to the inner nuclear membrane in all tissues studied, but additionally targets to other subcellular locations in a cell type-specific manner [14–19]. In the heart, emerin co-localises with vinculin and N-cadherin at the adherens junction (AJ) of the intercalated disc (ID) of cardiomyocytes [6, 14]. The ID is a specialised plasma membrane region that mediates electrical and mechanical coupling between adjacent cardiomyocytes. Disruption to ID integrity arising from mutations in ID proteins generates a cardiopathic phenotype, e.g. plakophilin [20], desmoplakin [21] and N-cadherin [22]. β-catenin, a component of the AJ, is also important for maintaining ID integrity. In muscle-LIM protein (MLP) null mice, a model for DCM, β-catenin expression is increased at the ID [23] whilst deletion of the ID protein mXinalpha, a binding partner of β-catenin, produces a cardiomyopathy [24]. Since β-catenin is a binding partner of emerin, we can speculate that in the absence of emerin, β-catenin function may be compromised, which in the heart may lead to the development of a DCM. It is already known that emerin interacts with β-catenin through the adenomatous polyposis coli-like domain on emerin and regulates β-catenin’s nuclear accumulation through a CRM1-dependent export pathway in HEK293 cells [7]. Recent evidence suggests emerin- and lamin A/C-containing nuclear envelope complexes affect adipogenesis by regulating the nucleocytoplasmic location of β-catenin [25]. We speculate that the emerin–β-catenin complex has a functional role in maintaining the differentiated state.
Here, we identified an emerin–β-catenin complex in the heart and confirmed emerin localises to the AJs of the IDs. In cardiomyocytes from emerin–null hearts, β-catenin distribution, ID architecture and cell shape were perturbed consistent with a DCM phenotype. Both the interaction and the distribution of emerin and β-catenin were regulated by GSK3β activity. This is a novel function for emerin demonstrating yet again the diverse functions associated with nuclear envelope proteins.
Materials and methods
Cell culture
Neonatal rat cardiomyocytes (NRCs) were isolated from P1 Sprague–Dawley rat pups according to [26]. NRCs were cultured in maintenance medium [DMEM, 4% (w/v) horse serum, 2 mM glutamine, 10 U/ml penicillin/streptomycin] containing phenylephrine (PE, 100 μM; Sigma) and cytosine-β-D-arabino-furanoside hydrochloride (AraC, 10 μM; Sigma). HEK293 and C2C12 myoblasts were cultured according to [7, 27].
Antibodies
We generated a new affinity purified sheep antibody (APS20) against rat emerin residues 114–183 as described in [27]. Affinity purified rabbit polyclonal antibody, AP8, against human emerin residues 1–70 was described previously [28]. APS20 and AP8 antibodies were used at 1:1,000 and 1:3,000 dilution on immunoblots respectively and 1:100 for immunoprecipitation and immunofluorescence experiments. The following antibodies were also used: α-catenin (Sigma), β-catenin (Sigma), desmoplakin (Serotec), connexin 43 (Zymed), laminin (Sigma), plakoglobin (BD Transduction Labs), FLAG tag (Sigma), myomesin [29], sarcomeric α-actinin (Clone EA53; Sigma), anti-HA tag (Roche) and anti-αII-spectrin [30]. Primary antibodies used were visualised with fluorochrome-conjugated secondary antibodies from either Molecular Probes or Chemicon.
Emd null mice
Emd null mice were a kind gift from Colin Stewart [31]. Wild type litter mates were used as a control.
Emerin, β-catenin and GSK3β cDNA constructs
cDNA encoding pET29b-1-254 (wild type, full length) emerin and emerin 1-221 (lacks transmembrane and C-terminal tail), GFP-1-254 emerin, GFP-emerin 1-221 and GFP-emerin mutants S54F, Δ95-99, Q133H, 1-169(208) [frameshift at residue 169 followed by a premature stop codon at residue 208] and P183T have been described previously [8, 27, 28]. cDNA encoding pET15-emerin 1-221 with a clustered missense mutation (SSL-to-AAA) at residues 175–177 (emerin m175) was provided by Kathy Wilson [32]. pCMV-Tag2A-emerin 1-254 was subcloned from pcDNA-emerin [28]. GFP-emerinΔ168–186 [deletion of β-catenin binding site and surrounding residues] was a kind gift from Christopher Hutchison (Durham, UK), while GFP-β-catenin wild type and GFP-β-catenin stabilised, in which amino acids Ser33, Ser37, Thr41 and Ser45 are substituted with Ala, thus removing all four GSK3β phosphorylation sites, were a gift from Peter Zammit (King’s College London) [33]. HA-tagged GSK3β wild type and constitutively active S9A GSK3β were gifts from Britta Eickholt (King’s College London).
Transfections
For transfection of NRCs, cDNA encoding GFP-emerin 1-254, GFP-emerin 1-221, GFP-β-catenin wild type and GFP-β-catenin stabilised (1 μg) was mixed with 4 μl of Escort III transfection reagent (Sigma), and transfections were carried out according to manufacturer’s instructions. HA-tagged GSK3β wild type together with HA-tagged GSK3β S9A (1 μg) was mixed with lipofectamine 2000 (Invitrogen) at a ratio of 1 μg DNA to 2.5 μl lipofectamine, and transfections were performed according to manufacturer’s instructions. Following an incubation of 5–6 h at 37°C, cells were switched to maintenance medium containing PE and further incubated for 12–86 h. Cells were fixed in 4% (w/v) paraformaldehyde (PFA). For transient transfections of HEK293 cells, cDNA encoding GFP-emerin 1-254, GFP-emerin mutants (S54F, Δ95–99, Q133H, 1-169(208) and P183T), GFP-β-catenin wild type and stabilised and GFP-emerin Δ168–186 (2 μg) was mixed with lipofectamine (Invitrogen) at a ratio of 1 μg DNA to 2.5 μl lipofectamine. After 4–6 h, the medium was replaced and the cells further incubated for 24 h.
Immunofluorescence and immunohistochemistry
For microscopic visualisation cells were fixed and stained according to Fairley et al. [27]. Sections of rat, mouse and human heart were similarly stained except that sections were fixed in acetone at −20°C for 5 min and blocked with blocking buffer containing 5% (v/v) horse serum for 30 min at room temperature. Images were captured on a Zeiss Axiovert 200 M inverted microscope fitted with a AxioCam HR camera using a 40×/0.75NA or 63×/1.4NA oil objective and analysed using AxioVision LE version 4.4 software. Confocal images were obtained using a Leica TCS SP5 microscope with Argon HeNe lasers using a 63×/1.4NA oil objective. All figures were assembled in Adobe Illustrator (version CS2).
Cryosectioning, immunogold labelling and electron microscopy
Papillary muscles from adult rat hearts were prepared, fixed and cryosectioned according to [34, 35]. Primary antibodies were all used at 1:50 and secondary at 1:100 with both 10 nm and 5 nm colloidal gold (BB International). Each double-labelling reaction was performed so that each antigen was separately labelled with either 5 or 10 nm gold, and the 10 nm gold label was always added first. This was to counter any steric hindrance effects from the larger gold size. Five grids were used per antibody pair and the experiment was repeated three times. Stained sections were examined on a FEI Tecnai T12 BioTWIN transmission electron microscope and images captured on a Gatan BioScan Model 792 MSC SI003 1 camera and analysed using Digital Micrograph 3.7.1 software. Files were exported as TIFF files at a minimum of 1024 × 1024 pixel resolution, into Adobe Illustrator CS2.
Immunoprecipitation of emerin and immunoblotting
Immunoprecipitation and immunoblotting was performed as described previously [8, 27].
In vitro glycogen synthase kinase 3β assay
His6 tagged emerin 1–221 wild type and m175 [32] were expressed and purified as described previously [8]. A 5 μg sample of each purified fusion protein was incubated in the absence or presence of 250 units GSK3β (NEB) in 1 × GSK3β reaction buffer (20 mM Tris–HCl, 10 mM MgCl2, 5 mM DTT, 200 mM ATP, 100 μCi [32P] γ-ATP) for 30 min at 30°C. The GSK3β inhibitor SB216763 (20 μM) was incubated with emerin fusion proteins for 30 min at 4°C prior to the addition of GSK3β. Reactions were stopped by the addition of SDS PAGE sample buffer and samples heated at 70°C for 10 min. Proteins were resolved by 4–20% SDS PAGE, dried and autoradiographed.
RNA interference
Scrambled rat Emd siRNA (sense nucleotide: 5′ ACGAGACCCAGAGAAGGAGACUCUU-3′) and rat Emd siRNA (sense nucleotide: 5′-GCAAGGACUAUAAUGAUGACUACUA-3′) together with control siRNA duplexes were obtained from Invitrogen. siRNA was diluted into OPTI-MEM (Invitrogen) to 20 nM and added to OPTI-MEM containing lipofectamine (4 μl). Following an incubation of 20 min at room temperature, the siRNA:lipofectamine was added to NRCs and incubated for 4 h at 37°C. Cells were transferred to maintenance medium and incubated for 24 h, after which the transfection was repeated. Cells were further incubated for 48 h and then fixed.
Phenylephrine stimulation of NRCs
NRCs were incubated in maintenance medium lacking PE for 24 h at 37°C. Cells were treated with maintenance medium containing DMSO (0.1% v/v), PE (100 μM), SB216763 (20 μM, diluted in DMSO) or PE together with SB216763 for 30 min, 4 h and 24 h. Cells were fixed and immunostained with antibodies against emerin (APS20) and β-catenin.
Statistical analysis and quantification
All measurements are given as the mean ± SEM. Statistical significance was calculated using the Student’s t test (two sample assuming equal variance). Cell shape was calculated by importing 80 cells of cardiomyocytes each taken from three wild type and Emd −/y mice stained for laminin and β-catenin into Image J and measuring the length and width of each cell. Only cardiomyocytes that were cut in a longitudinal section and that had clearly defined boundaries were included. The mean band intensities of exogenously expressed GFP-labelled constructs were measured using the image analysis programme Image J (http://rsb.info.nih.gov/ij/). To calculate nuclear region intensities, images of NRCs stained for emerin and β-catenin in the presence of either 0.1% DMSO (v/v) 100 μM PE or 100 μM PE +20 μM SB216763 were imported into Image J. The nuclear intensity for emerin and β-catenin was calculated by making a mask of the nucleus followed by thresholding the image and then subtracting this new binary image from the original red (emerin) and green (β-catenin) channels respectively. A total of 75 cells were analysed per treatment and per time point.
Results
Characterisation of the subcellular localisation of emerin in the heart
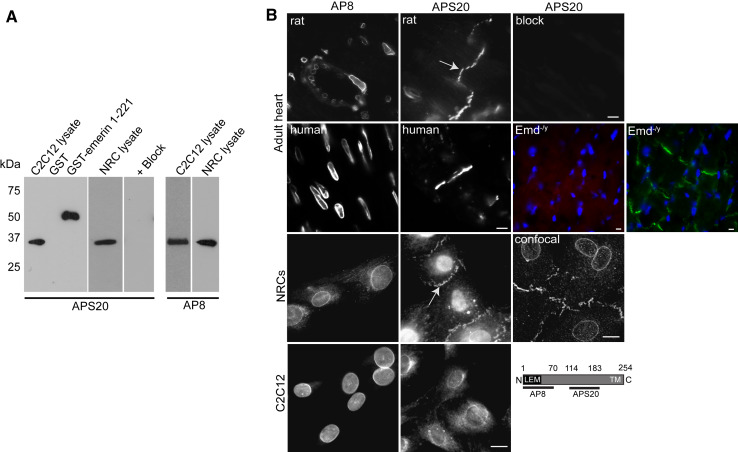
To clarify the localisation of emerin in the rat heart we used two antibodies which recognise different epitopes in emerin. These are the previously described affinity purified rabbit polyclonal AP8 antibody [27, 28] against the N-terminus of emerin (residues 1–70) and a new affinity purified sheep polyclonal antibody against residues 114–183 which we term APS20. Both antibodies recognised their respective fusion proteins and the same molecular weight band in both neonatal rat cardiomyocytes (NRCs) and C2C12 (mouse myoblasts) cell lysate by immunoblotting (Fig. 1a). Pre-incubation of the new APS20 antibody with its fusion protein abrogated all immunoreactivity. We then performed immunofluorescence studies on wild type adult rat, human and Emd −/y mouse hearts, NRCs and C2C12s (Fig. 1b). Both AP8 and APS20 produced an emerin staining pattern in C2C12s, consistent with its previously described location [16, 17, 27]. However, in NRCs and adult heart tissue, APS20 additionally labelled the intercalated discs (IDs) (arrows, Fig. 1b), confirming previous reports [6, 14]. In adult heart sections, the APS20 antibody recognised NE emerin more weakly than AP8. A likely explanation for this is that we optimised for ID staining by cutting longitudinal sections. To exclude the possibility that APS20 cross-reacts with a non-specific protein located at the ID, APS20 was pre-incubated with its antigen before immunostaining of the adult rat heart sections. No staining of either the NE or ID was observed (Fig. 1b). Similarly, heart sections from 5-week-old Emd −/y mice showed no emerin staining at either the NE or the ID, with the ID clearly labelled for β-catenin (Fig. 1b). Since AP8 and APS20 recognise different epitopes on emerin, these data raise the possibility that either another isoform or a stable pool of differentially regulated emerin exists at the ID.
Fig. 1.
Characterisation of APS20 emerin antibody. a Lysates (20 μg) of C2C12s and NRCs, GST (5 μg) and GST-emerin 1-221 (5 μg) fusion proteins were resolved by SDS-PAGE and immunoblotted with AP8 and APS20. Blocking of APS20 was performed by pre-incubating the antibody with GST-emerin 114–183 fusion protein (40 μg) overnight. b Immunostaining of cryosectioned adult wild type and Emd −/y hearts (top two panels), NRCs (middle panel) and C2C12s (lower panel) with emerin antibodies. APS20 specificity was confirmed by pre-incubating antibody with emerin GST-114-183 fusion protein (APS20 + block). IDs are shown by arrows while the schematic depicts the region against which AP8 and APS20 were raised. Emd −/y hearts are stained with APS20 (red), β-catenin (green) and DAPI (blue). All images were captured on a Zeiss Axiovert inverted microscope, except the image labelled as ‘confocal’ which was captured on a Leica TCS SP5. Scale bars 10 μm
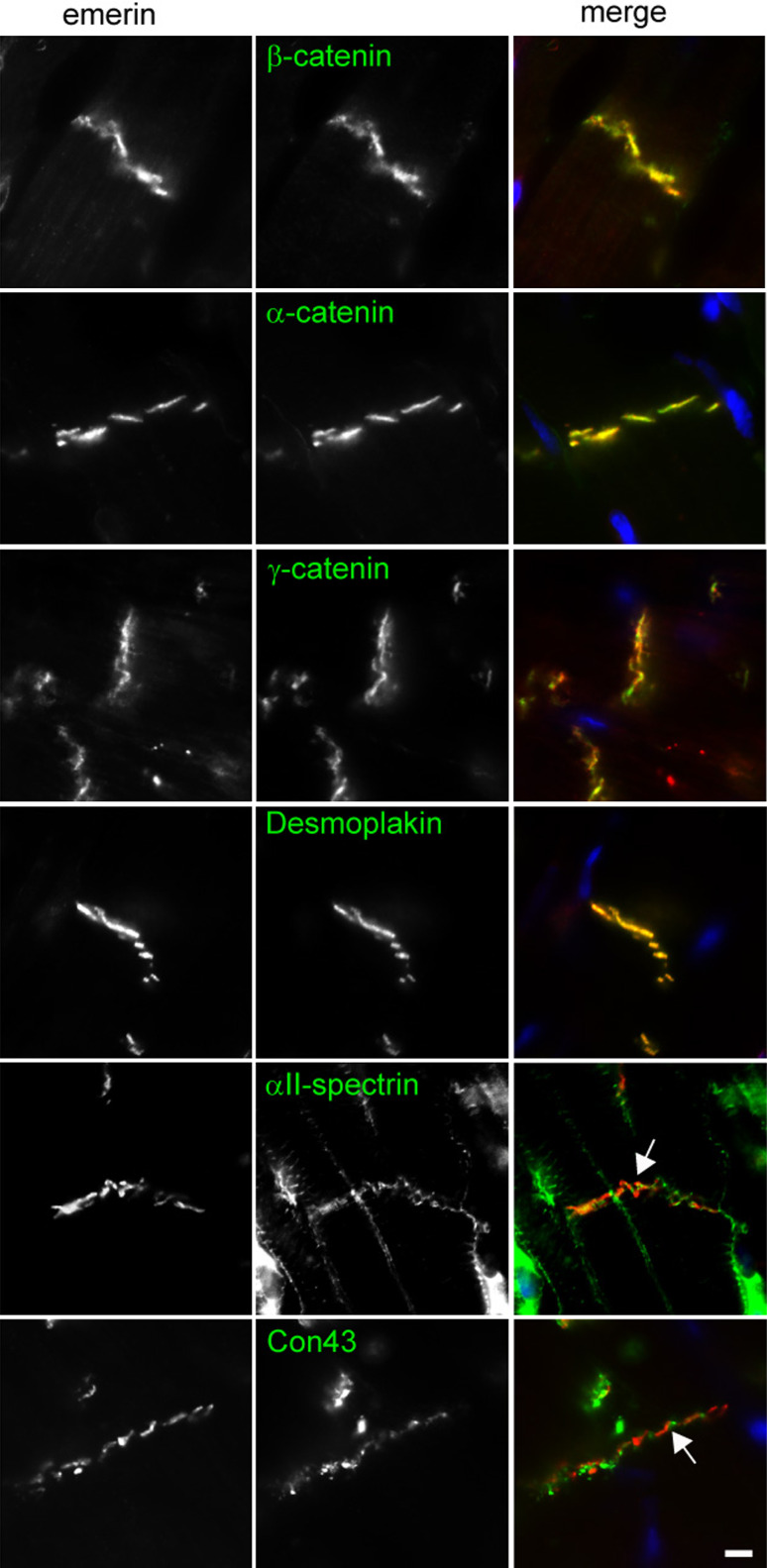
To further characterise the ID localisation of emerin, we co-stained sectioned adult rat heart for emerin and a number of ID proteins. A strong co-localisation with α-, β- and γ-catenin (plakoglobin) and desmoplakin was observed suggesting emerin is a component of the adherens junction (AJ) and possibly of the desmosome too (Fig. 2). We observed no co-localisation with either connexin 43 or αII-spectrin (arrows in Fig. 2), suggesting it is unlikely that emerin is a component of either the gap or transitional junction [30].
Fig. 2.
Emerin co-localises with specific junctional components of the intercalated disc. Immunostaining adult rat heart sections with APS20 (red) and for a range of ID (green) proteins illustrates emerin co-localises with α-catenin, β-catenin, plakoglobin and desmoplakin and not with αII-spectrin (arrow) and connexin 43 (arrow). Scale bar 10 μm
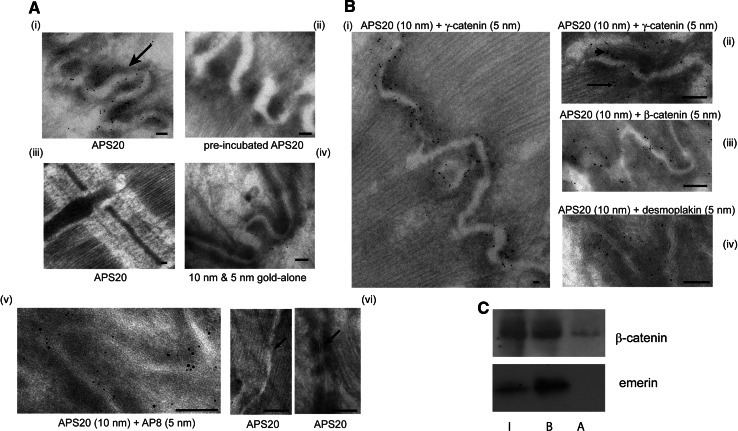
We next used immunogold labelling to determine whether emerin was a component of the AJs and/or of the desmosomes in the papillary muscles of adult rat heart. Both emerin antibodies labelled the ID and nuclear envelope but differed in their affinity for each. APS20 labelled the NE less strongly than AP8 (data not shown) whereas APS20 labelled membranous structures at the ID (Fig. 3a, panel i), consistent with our immunohistochemistry results. No immunogold emerin labelling was observed at any other subcellular localisations (Fig. 3a, panel iii). Pre-incubation of APS20 with its antigen reduced immunogold labelling to background levels (Fig. 3a, compare panels ii with iv). Double-labelling emerin with APS20 (10 nm) and AP8 (5 nm) demonstrated AP8 labelled the ID less strongly (Fig. 3a, panel v). Similar immunogold distribution patterns were observed in our double-labelled samples regardless of which emerin antibody was detected by the larger immunogold. Furthermore, emerin only located to junctional structures within the ID complex folds and not to ID riser regions which contain gap junctions (Fig. 3a, panel vi, left) or to desmosomes (Fig. 3a, panel vi, right).
Fig. 3.
Emerin co-localises with the catenins at the adherens junction of intercalated discs (ID) and binds to β-catenin in vivo. a Electron micrographs of adult rat heart ID regions (arrow in i) in cryosections immunogold labelled for i APS20 (10 nm gold), ii APS20 pre-incubated with its antigen (10 nm gold), iii APS20 (10 nm gold) on Z-disc region of sarcomere, iv 10 and 5 nm gold-alone on ID, v APS20 (10 nm gold) and AP8 (5 nm gold), and vi APS20 (10 nm gold) on ID riser region (arrow; left hand panel) and desmosomes (arrow; right hand panel). Scale bars 0.1 μm. b Electron micrographs of adult rat ID regions in cryosections immunogold labelled for emerin and AJ proteins: i APS20 (10 nm gold) and γ-catenin (5 nm gold), ii APS20 (10 nm gold) and γ-catenin (5 nm gold), with arrowhead labelling the cytoplasmic plaque and arrow pointing to the terminal ends of the myofibrils at the AJ, iii APS20 (10 nm gold) and β-catenin (5 nm gold), iv APS20 (10 nm gold) and desmoplakin (5 nm gold). Scale bars i 0.1 μm, ii–iii 0.05 μm. c AP8 (1:100) was added to NRC lysates to immunoprecipitate emerin and immunoblotted for β-catenin. I 5% total cell lysates input, B immunoprecipitated protein, A no primary antibody
Immunogold double-labelling also confirmed emerin co-localised strongly with β-and γ-catenin, but less so with desmoplakin at the AJs (Fig. 3b, panels i–iv). The AJs are found at the end of the myofibrils, and under EM appear as dense, slightly fuzzy extra mass regions (known as the cytoplasmic plaque; [30]) adjacent to the ID membranes (arrowhead on Fig. 3b, panel ii). Emerin was found spread throughout the width of the cytoplasmic plaque, and in a few places to the edge of the cytoplasmic plaque where it juxtaposes the myofibrillar network (arrow on Fig. 3b, panel ii). This may be because emerin is interacting with myofibrillar components since it has previously been shown to bind in vitro to the pointed end of F-actin and to nuclear myosin I [27, 36, 37].
We then quantified the immunogold co-localisation between emerin and the other ID components examined. The distance of the specific C-terminal epitopes of β- and γ-catenin and desmoplakin from the ID membranes midline used in this study has been previously calculated as <20.0, 52.0 and 10.8 nm respectively [30]. For each of these epitopes and for APS20, we analysed five digital micrographs at 127,000× magnification, with an average of 100 gold particles/figure. Greater than 80% of the gold particles counted were calculated to be at an average of 18.3 ± 0.75 nm for β-catenin, 55.3 ± 0.42 nm for desmoplakin and 12.1 ± 0.62 nm for γ-catenin, consistent with the published values. Emerin exhibited quite a different distribution. Nearly 90% of the gold particles evenly spanned 10–23 nm from the midline, which covers most of the width of the cytoplasmic plaque, with the remaining 10% labelling the cytoplasmic plaque edge, 60 nm from the ID membrane midline. Thus ultra-structural analysis demonstrates emerin co-localises with both β- and γ-catenin with equal merit.
The EM data confirm emerin as an ID component of the AJ and not of the desmosomes and it additionally localises with the catenin-family of proteins. This is particularly interesting given that an emerin–β-catenin interaction has been previously described in HEK293 cells [7]. We next investigated whether a similar complex exists in heart. We confirmed an interaction between emerin and β-catenin in cardiomyocytes by immunoprecipitating endogenous emerin from lysates of NRCs and immunoblotting for β-catenin (Fig. 3c).
Loss of emerin redistributes β-catenin in cardiomyocytes
In most clinical cases of X-EDMD, emerin is totally absent. We next examined whether reduced emerin expression had any effect on either cardiomyocyte morphology or β-catenin localisation by knocking-down emerin in cultures of NRCs with siRNA. Compared to cells treated with either control or scrambled emerin siRNA, in cells transfected with emerin siRNA, emerin expression was still detectable but reduced at both the NE and the plasma membrane (Fig. 4a). Cardiomyocytes expressing reduced levels of emerin failed to form a continuous monolayer. Instead extended filopodial extensions were observed between cell colonies (Fig. 4b; arrows). Interestingly, β-catenin immunostaining at the plasma membrane was less regular, being more intense at regions not involved in cell–cell contacts (arrowheads in Fig. 4a). In addition, the F-actin network needed to anchor the AJs was slightly perturbed (Fig. 4c). Immunoblotting demonstrated that total β-catenin levels remained unchanged in emerin siRNA treated cardiomyocytes (Fig. 4d). Quantification of cardiomyocyte cell size confirmed no significant change of cardiomyocyte size (data not shown). These findings suggest that knocking down emerin prevents or delays the maturation of cell–cell junctions in NRCs.
Fig. 4.
Analysing NRCs treated with siRNA emerin (a–d) and emerin null hearts for changes in morphology and catenin localisation (e–h). NRCs were transfected with control siRNA, emerin siRNA or scrambled emerin siRNA for 72 h in medium containing PE. a Cells were fixed and immunostained with APS20 (red) to demonstrate reduced emerin at both the NE and the ID in the presence of emerin siRNA. Counterstaining for β-catenin (green) and DAPI (blue) reveals mislocalisation of β-catenin at the plasma membrane (arrowheads) in emerin siRNA treated cardiomyocytes compared with control cells. Scale bars 10 μm. b DIC images of the conditions shown in a. Arrows label the filopodial extensions. Scale bar 50 μm. c NRCs treated with control or emerin siRNA for 72 h were immunostained for emerin (red) and F-actin (green) and counterstained with DAPI (blue). d Immunoblot of control, scrambled and emerin siRNA treated NRCs demonstrates that reduced emerin expression does not alter total β-catenin levels. e Immunoblot of wild type and Emd −/y hearts shows that total β-catenin levels are unaffected by the loss of emerin. f Cryosections of 5-week-old wild type and Emd −/y mice hearts immunostained for either β- or γ-catenin. Insets represent higher magnification to illustrate increased convolution of the IDs in emerin-null mice. g Immunostaining of sections from 5-week-old wild type and Emd −/y mice hearts for β-catenin (green) and laminin (red). Insets represent higher magnification of IDs to illustrate β-catenin lateral staining. h The ratio of length to width of 80 cardiomyocytes from wild type and Emd −/y mice stained as for g were calculated using Image J. Data are shown from three individual mice as well as a mean value for each experimental group. **p < 0.001. Images were colleted on a Leica TCS SP5 confocal microscope for g only. c–e Scale bars 10 μm
We next went on to determine whether the integrity of the ID was also compromised by analysing the emerin-null mouse heart for both catenin and laminin localisation and expression levels. As found with cardiomyocytes treated with emerin siRNA, total β-catenin protein levels were similar between Emd−/y hearts and wild type hearts (Fig. 4e). Co-staining Emd −/y hearts for catenins and laminin confirmed that mild changes in ID integrity can arise from a loss of emerin function. Both β- and γ-catenin staining gave a broader fluorescent signal at the ID in hearts from emerin-null mice, compared with wild type litter mates, indicative of a higher degree of convolution of the ID (Fig. 4f and inset for β-catenin). We also observed brighter immunofluorescence of β- and γ-catenin at the lateral plasma membranes of Emd −/y mice compared to wild type litter mates (Fig. 4g and inset for β-catenin). Individual cardiomyocytes from Emd −/y hearts appeared more elongated (Fig. 4g). Quantification of cell shape confirmed emerin null cardiomyocytes exhibit an increase in the cell length-to-width ratio consistent with a DCM phenotype (Fig. 4h). Since this feature was not observed in emerin-knock-down NRCs, it suggests it arises from a developmental abnormality. Overall heart sarcomeric arrangement, ascertained by α-actinin and myomesin staining, was unaffected (data not shown). Our findings are consistent with the previously described mild cardiomyocyte pathology observed in the Emd −/y mouse [31, 38].
Expression of emerin and β-catenin in cultured cardiomyocytes
We next determined whether exogenous expression of either GFP-tagged emerin or β-catenin (constructs shown schematically in Fig. 5a) altered the localisation of endogenous β-catenin and emerin respectively in NRCs (Fig. 5b). In contrast to endogenous emerin, full-length emerin 1–254 tagged with either GFP or FLAG localised predominately to the NE, with only minor localisation at the ID (Fig. 5b, panel i; shown for GFP only). Emerin appeared juxtaposed to β-catenin on the cytoplasmic side of the ID (Fig. 5b, panel i inset; note yellow under the red at ID), consistent with our EM data where emerin is present at the cytoplasmic plaque of the AJs and not in the ID membrane. We then expressed GFP-emerin 1–221 in NRCs. This construct lacks the transmembrane domain and C-terminal tail and has previously been shown to localise exclusively to the nucleoplasm of HeLa, C2C12 or COS7 cells [6, 17, 39] respectively. In NRCs, this GFP-emerin construct localised to the sarcomere and the ID, where it co-localised with β-catenin. This emerin construct also appeared juxtaposed to β-catenin on the cytoplasmic side of the ID (Fig. 5b panel ii, yellow under the red). The targeting of other exogenously expressed proteins to the ID does not appear to be effected by GFP [40]. However, it is possible that the exogenous expression of full length emerin causes it to fold in such a way as to mask or prevent the addition of signals required for efficient targeting to the ID. Endogenous β-catenin mainly localised to the ID with a little in the cytoplasm, and expression of any GFP-emerin construct did not alter its intracellular distribution. This contrasts with HEK293 cells where, in cells transfected with GFP-emerin, nuclear β-catenin is reduced while staining at the AJ is increased [7]. This apparent discrepancy may reflect cell type-specific β-catenin localisation.
Fig. 5.
Emerin and β-catenin expression in NRCs. a Schematic diagram showing cDNA constructs used. Blue box represents the transmembrane, white-shadowed box the LEM domain, GFP-emerin full length 1-254, GFP-emerin 1-221, GFP-emerin Δ168-186 (β-catenin binding site [169-180] and surrounding residues deleted) and stabilised GFP-β-catenin (residues Ser33, Ser37, Thr41, Ser45 replaced with Ala). b NRCs in medium containing PE were transiently transfected with cDNA encoding i GFP-emerin 1-254, ii GFP-emerin 1-221, iii wild type GFP-β-catenin and iv stabilised GFP-β-catenin. Counterstaining for anti-β-catenin and anti-emerin is shown in red. Inset in panel i represents the captured image at a longer exposure and higher magnification of the ID region demonstrating low levels of GFP-emerin 1-254 at the ID. Scale bar 10 μm. Images were collected on a Leica TCS SP5 confocal microscope
After 48 h of expression, wild type GFP-β-catenin mainly targeted to the IDs with some in the cytoplasm of cells expressing higher GFP-β-catenin levels (Fig. 5b, panel iii). Similarly, stabilised GFP-β-catenin (residues Ser33, Ser37, Thr41 and Ser45 replaced with Ala so it cannot be phosphorylated by GSK3β; [33]) targeted mainly to the ID (Fig. 5b, panel iv). Both GFP-β-catenin proteins co-localised with endogenous emerin at the IDs. Expression of neither wild type nor stabilised β-catenin dramatically altered the localisation of endogenous ID emerin. Under the experimental conditions used here, we suggest stabilised β-catenin has a preponderance for targeting to the ID over the nucleus in cardiomyocytes.
Emerin is unable to interact with a stabilised form of β-catenin
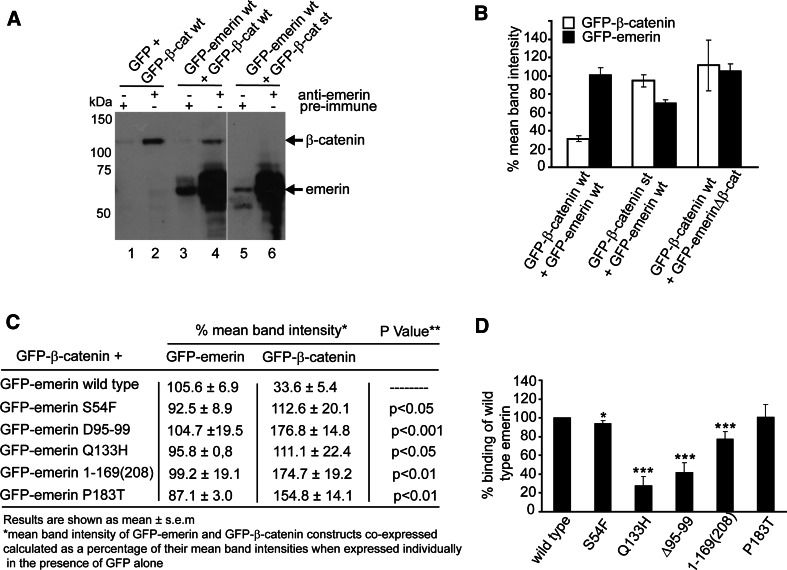
GSK3β is an important kinase in regulating β-catenin function. We next investigated whether the formation of the emerin–β-catenin complex required GSK3β-dependent phosphorylation of β-catenin, by co-expressing GFP-emerin and GFP-β-catenin constructs in HEK 293 cells. We first confirmed that wild type GFP-β-catenin could be immunoprecipitated from cell lysates with the anti-emerin AP8 antibody (Fig. 6a, lane 2). GFP-emerin was co-immunoprecipitated with GFP-wild type but not stabilised β-catenin (Fig. 6a; compare lanes 4 with 6). To see if emerin could regulate β-catenin cellular levels, we expressed GFP-tagged emerin and β-catenin constructs both individually and together in HEK293 cells and determined protein levels by immunoblotting with an anti-GFP antibody (data not shown). The mean band intensity of GFP-emerin and GFP-β-catenin constructs co-expressed was calculated as a percentage of their mean band intensities when expressed individually in the presence of GFP-alone (Fig. 6b). Wild type GFP-emerin was able to reduce the levels of wild type but not stabilised GFP-β-catenin. Emerin’s ability to do this appeared to be determined by its ability to directly bind to β-catenin, since GFP-emerinΔ168-186 (β-catenin binding site deleted) was unable to reduce the levels of wild type GFP-β-catenin (Fig. 6b). We conclude that emerin regulates wild type but not stabilised β-catenin levels through an interaction with β-catenin and that GSK3β phosphorylation on β-catenin may be important in regulating this interaction.
Fig. 6.
Characterisation of the interaction between emerin and β-catenin. a Emerin binds to wild type but not stabilised β-catenin. Lysates of HEK293 transfected with GFP-emerin and GFP-β-catenin constructs were immunoprecipitated with AP8 and immunoblotted with anti-GFP antibody to visualise binding between emerin and β-catenin. GFP-β-cat st represents stabilised GFP-β-catenin construct. A representative blot from three independent experiments is shown. b Emerin regulates wild type but not stabilised β-catenin cellular levels through its interaction with β-catenin. Lysates of HEK293 cells were transfected with cDNA encoding GFP-emerin and GFP-β-catenin constructs and immunoblotted with anti-GFP. The mean band intensity of GFP-emerin and GFP-β-catenin constructs co-expressed was calculated as a percentage of their mean band intensities when expressed individually in the presence of GFP alone. c Emerin mutations disrupt its ability to regulate cellular β-catenin levels. HEK293 cells were co-transfected with wild type GFP-β-catenin and GFP-emerin constructs expressing X-EDMD mutations. Lysates were immunoblotted with anti-GFP antibody and mean band intensities for each GFP-construct calculated and tabulated as described for b above. d Emerin mutations disrupt the emerin–β-catenin affinity. Each GFP-emerin cDNA was expressed in HEK293 and normalised for protein levels and mixed with lysates containing an equal amount of GFP-β-catenin. Lysates were immunoprecipitated for β-catenin and immunoblotted for emerin. Binding is shown as a percentage of binding between wild type GFP-β-catenin and wild type GFP-emerin. For b–d, results are shown as the mean of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001
To determine the relevance of this interaction for X-EDMD, wild type GFP-β-catenin was expressed in HEK293 cells as above, but in the presence of GFP-emerin constructs carrying in-frame mutations known to cause X-EDMD. These are a suitable model because X-EDMD patients exhibit a fairly consistent clinical phenotype, regardless of whether they are null or expressing in-frame mutations [41]. The emerin mutants expressed at similar levels compared with wild type, and in all cases β-catenin levels were significantly elevated in a mutation-dependent manner (Fig. 6c). Thus mutant emerin is unable to regulate β-catenin’s protein levels. Next, we examined whether this effect could be attributed to the mutations reducing the affinity of the emerin–β-catenin interaction. Each GFP-emerin cDNA was expressed in HEK293 cells and quantified for the amount produced by immunoblotting. An equal amount of each mutant emerin protein was mixed with lysates containing an equal amount of GFP-β-catenin, immunoprecipitated for β-catenin and immunoblotted for emerin. The binding affinity between β-catenin and each emerin mutant was calculated as a percentage of the binding between β-catenin and wild type emerin (Fig. 6d). β-catenin binding to several of the GFP-emerin mutants was significantly reduced. Binding of β-catenin to the emerin mutants S54F and P183T was similar to the wild type. Given that X-EDMD mutations are known to disrupt emerin-protein interactions [8, 36, 42, 43], our findings are not surprising. Although the 1–168(208) mutation spans the nesprin and β-catenin binding regions, it affected β-catenin binding less than the Q133H mutation which lies outside this region. This suggests that a secondary mechanism, such as emerin phosphorylation, additionally influences the emerin–β-catenin interaction or that a second binding site to β-catenin exists. Interestingly, mutations at residue P183 are associated with both a milder clinical phenotype and normal emerin-protein interactions [36, 41, 44], consistent with our finding that the P183T mutation had little effect on the emerin–β-catenin interaction. Moreover, these findings provide further evidence that the emerin–β-catenin complex is likely to be functionally relevant for the aetiology of X-EDMD as first proposed by Markiewicz et al. [7].
Emerin is phosphorylated by GSK3β
The results shown in Fig. 6a and b suggest GSK3β may regulate the emerin–β-catenin interaction. Emerin has 12 identified phosphorylation sites (reviewed in [5]). Two of these sites, Ser66 and Ser175 (which is in the β-catenin binding site), are predicted to be phosphorylated by GSK3β [45]. To determine whether emerin is also a substrate for GSK3β, we performed an in vitro kinase assay. Purified recombinant his6-tagged emerin 1–221 [28] was incubated with GSK3β in the absence and presence of the GSK3β inhibitor SB216763. Under these conditions GSK3β is auto-phosphorylated and is represented by a band that migrates at approximately 50 kDa (Fig. 7a). Several phosphorylated emerin bands were identified migrating at 34 to 37 kDa suggesting the presence of more than one GSK3β phosphorylation site. Phosphorylation of both GSK3β and emerin was inhibited by the addition of SB216763. To see if Ser175 is a site for GSK3β phosphorylation, we repeated the GSK3β kinase assay on purified recombinant emerin 1–221 containing clustered Ala substitutions such that residues 175–177 (SSL) are mutated to AAA (m175) [32]. Phosphorylation of emerin m175 was similar to that of the wild type protein, although an additional band at approximately 40 kDa was present. This likely reflects differences in folding between wild type and mutant emerin. From the above, we conclude that emerin is a substrate for GSK3β, but residue S175 is not a site of phosphorylation by this kinase. Nevertheless, this experiment demonstrates the emerin–β-catenin interaction may also be regulated by GSK3β phosphorylation on emerin, as well as on β-catenin.
Fig. 7.
GSK3β phosphorylates emerin and regulates its intracellular location. a Emerin 1-221His6 (5 μg) and emerin 1-221His6 m175 (5 μg) were incubated in the absence (−) or presence (+) of 250U recombinant GSK3β in 1× reaction buffer for 30 min at 30°C. Several phospho-emerin bands are evident (M r 34–37 kDa) as is an auto-phosphorylated GSK3β (M r ~ 50 kDa). Inclusion of the GSK3β inhibitor SB216763 in the reaction completely abolished emerin phosphorylation. b Cultures of HEK293s and NRCs were transfected with cDNA encoding HA-tagged wild type GSK3β or HA-tagged GSK3β S9A. After 24 h cells were fixed. Immunostaining with anti-HA demonstrates exogenous expression of GSK3β whilst counterstaining with either APS20 (NRCs, red) or AP8 (HEK293s, green) demonstrates the loss of emerin from the NE in GSK3β expressing cells. Scale bars 10 μm
We then investigated whether GSK3β activity regulates emerin’s intracellular location, which may indirectly influence the availability of emerin to form complexes with β-catenin. HEK293s and NRCs transiently transfected with either HA-tagged wild type GSK3β or constitutively active GSK3β S9A were immunostained for emerin and HA (Fig. 7b). Endogenous GSK3β locates diffusely to both the cytoplasm and nucleus and is catalytically active [46]. As expected, both HA-GSK3β and HA-GSK3β S9A targeted similarly to endogenous GSK3β. In all cases, endogenous emerin at the NE was reduced, but with no discernible increase at either the cell surface or cytoplasm. This effect did not appear to be more severe with constitutively active GSK3β. Since no mis-targeting of emerin was observed in either cell type, it raises the possibility that the NE population of emerin is targeted for degradation when phosphorylated by GSK3β. This also implies that in cardiomyocytes the NE and ID populations of emerin are functionally different. GSK3β activity could thus have a functional role on both components of the emerin–β-catenin complex and differentially regulate the complex at different intracellular locations.
Inactivation of GSK3β re-locates both emerin and β-catenin in cardiomyocytes
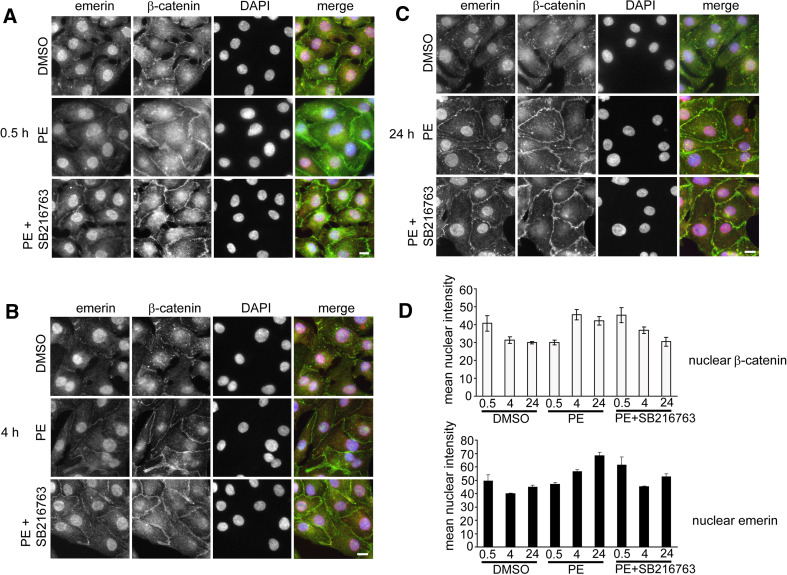
Given the emerging relationship between emerin, β-catenin and GSK3β, we sought to identify whether stabilising β-catenin co-ordinately affected the intracellular re-localisation of emerin and β-catenin in vivo. β-catenin stabilisation in NRCs can be achieved by the addition of hypertrophic stimuli such as phenylephrine (PE). Under these conditions, GSK3β is inhibited by the phosphorylation on Ser9 by protein kinase B (PKB/Akt) [47]. NRCs were first incubated in maintenance medium lacking PE for 24 h and then treated with DMSO, PE, the GSK3β inhibitor SB216763 or PE together with SB216763. Cells were fixed at 30 min, 4 h and 24 h and stained with antibodies against β-catenin and emerin (Fig. 8a–c). Quantification of the amount of emerin and β-catenin in the nucleus at each time point was performed by importing images into Image J and calculating the mean intensity of the nuclear region (Fig. 8d). Since PE promotes proper myofibril anchorage at the IDs, it was not unexpected that in its absence, emerin appeared predominantly nuclear and β-catenin mainly cytoplasmic (Fig. 8a–c; DMSO panels). When cells were treated with PE, an increase in nuclear β-catenin was evident by 4 h (Fig. 8b, panel PE; d). Although this was less by 24 h, it did not return to basal levels (Fig. 8b, PE panel; d). Nuclear and ID emerin levels and ID β-catenin levels increased steadily over 24 h (Fig. 8b and c; PE panels; d). As expected, cells treated with PE and SB216763 exhibited a faster response, so that the level of nuclear emerin and β-catenin were maximum at 30 min after stimulation, compared with PE alone (Fig. 8a–c, panels PE + SB216763; d). Treatment of cells with SB216763 alone demonstrated a similar but milder effect of PE (data not shown). From these results we can conclude that both emerin and β-catenin’s subcellular localisation and expression levels in cardiomyocytes can be regulated by GSK3β. In addition, the increase in endogenous emerin levels observed in the presence of a GSK3β inhibitor is consistent with a decrease in endogenous emerin at the NE in the presence of exogenously expressed GSK3β (Fig. 7b).
Fig. 8.
Stimulation of NRCs with PE relocates both β-catenin and emerin. a–c NRCs pre-incubated in maintenance medium lacking PE for 24 h were then treated with 0.1% (v/v) DMSO, 100 μM PE or 100 μM PE + SB216763 for 30 min, 4 h and 24 h. Cardiomyocytes were fixed with 4% PFA and immunostained with APS20 (red), β-catenin (green) and DAPI (blue). Scale bar 10 μm. d Images collected from a–c were imported into Image J, and the mean intensity of the nuclear region was calculated for both emerin and β-catenin staining in the presence of DMSO, PE or PE + SB216762. Images were taken from three independent experiments; n = 75. Images were captured on a Zeiss Axiovert inverted microscope
Discussion
Mutations in emerin lead to both skeletal and cardiac muscle defects. Although many downstream gene targets have been identified, primary mechanism involvement, particularly at the ID, remains to be elucidated. We discuss our findings in the context of the aetiology of the cardiomyopathy associated with X-EDMD.
Emerin can occur in non-nuclear membranes
The initial report of emerin being a component of the ID was shown by Cartegni et al. [6] who used an affinity purified rabbit polyclonal antisera against mouse emerin residues 1–168, which encompasses most of the nucleoplasmic domain. However, this was subsequently revoked as an artefact of rabbit antibody specificity in heart tissue by Manilal et al. [14]. This was despite reporting that 7 monoclonal antibodies from a panel of 16 supported an ID localisation for emerin. These monoclonal antibodies also mapped across the entire nucleoplasmic domain of emerin. Furthermore, Manilal et al. performed no EM analysis. We believe our antibody data conclusively demonstrate emerin is a component of the ID. From our EM experiments, both APS20 (affinity-purified sheep polyclonal raised against rat emerin 114–183) and AP8 (affinity-purified rabbit polyclonal against human emerin residues 1–70) recognised emerin at the ID. Thus, we show that two emerin antibodies against different epitopes and animal species, raised in two different animals both located emerin to the ID. In addition, since our APS20 antibody (sheep) exhibited a greater accessibility than AP8 (rabbit) for emerin at the ID, this rules out non-specific immunological effects from innate rabbit antigens. The inability of AP8 to recognise emerin at the ID by immunohistochemistry may be attributed to epitope masking, a known feature of NE proteins, such as lamin B, which can influence their detection by immunofluorescence [48].
Emerin was originally identified as being an inner nuclear membrane protein [2]. However, it is now clear that it can locate to alternative subcellular locations, with the exact distribution being cell type-specific. These include the ER, sarcoplasmic reticulum, nucleoplasm, outer nuclear membrane and plasma membrane [6, 15, 17–19, 49]. How can a protein with nuclear targeting motifs target to non-nuclear membranes? Emerin, like many tail-anchored membrane proteins, is post-translationally inserted into the ER, en route to its final destination at the NE [17, 28, 50]. This class of membrane proteins are also commonly associated with secretory pathway components [2]. Indeed, COS 7 cells over-expressing GFP-emerin are able to re-direct emerin from the ER to the Golgi-plasma membrane trafficking pathway [17]. Emerin is a heavily phosphorylated protein. To date, only a few functions have been associated with emerin’s phosphorylation including cell cycle-mediated events [28, 51], dissociation from BAF [45] and its interaction with F-actin [52]. We propose that cell type-specific signalling events, such as phosphorylation, on emerin in either the ER or cytoplasm prior to ER entry are responsible for targeting emerin to alternative cellular localisations. In the heart this could mean emerin would additionally target to the ID. We have evidence to support this idea. Protein kinase A phosphorylation on emerin residue S49 has no effect on targeting emerin in myoblasts [51], but relocates emerin to the sarcoplasmic reticulum of cardiomyocytes (M. A. Wheeler, unpublished). We propose that in cardiomyocytes a sub-population of emerin may be phosphorylated to target it to the ID.
Are there other components of the emerin–β-catenin complex at the ID?
The junctional complexes at the ID contain many proteins, and emerin is known to have a large cohort of binding partners and so it is more than likely that other proteins may be involved in interacting with the emerin–β-catenin complex at the ID. Indeed our immunoEM data suggest other catenin members are likely contenders. An even stronger candidate is Lmo7, a LIM-only protein which was recently shown to be a binding partner of emerin and is expressed at high levels in the heart [53, 54]. It also exhibits dual localisation to the nucleus and AJs of epithelial cells and like β-catenin is able to shuttle between the nucleus and plasma membrane [53, 55]. Lmo7 is proposed to regulate emerin, Rb1, MyoD and their co-activator transcripts such as CREBBP, which are known to be up-regulated in skeletal muscle from X-EDMD patients and emerin-null mice [31, 53, 56]. In this light, it will be interesting to see if Lmo7 is a component of the emerin–β-catenin complex at the ID and if so whether its location is disrupted in emerin null mice.
The significance of an emerin–β-catenin complex in contributing to the cardiomyopathy observed in X-EDMD
Analysis of the two available mouse models for X-EDMD [31, 38] offers little evidence of cardiac pathology until the mice are aged 21 weeks, when in both the atria and ventricles, vacuoles bordering the myonuclei are observed [38]. However, a mild prolongation of the atrioventricular conduction time is not seen in these mice until they are older than 40 weeks. This is in contrast to the lamin A/C-null mice, which exhibit a rapidly progressive DCM, with death commonly occurring by 8 weeks [57]. Despite this more severe phenotype, normal ID morphology is observed in 4- to 6-week-old lamin A/C-null mice, but no extensive molecular analysis has been reported. Similarly, in 21-week-old emerin-null mice only connexin 40 and 43 immunostaining has been performed, which were both normal [38], consistent with our finding that emerin does not localise to this complex. Indeed the only molecular change published which may affect ID function is in the AD-EDMD-mouse model, where disruptions to the desmin network, particularly at the nuclear attachment points, are observed. Desmin is also a component of the ID, where it maintains the lateral stability of the myocytes and so in this scenario ID function could be indirectly compromised. We report here for the first time that the protein composition of the ID disc is altered for at least β-catenin in 5- to 6-week-old emerin-null mice. In addition, we observed architectural changes to both the cell shape and to the ID. Such changes are likely to affect force transmission from the contracting myofibrils, which are anchored by AJs. Collectively these findings are indicative of a DCM [58]. However, since mice of this age are sub-clinical, it suggests up-regulation of functionally compensating proteins occurs, at least in the young animal. Contenders for this role include α-catenin, α-II-spectrin and filamin, all of which exhibit elevated mRNAs in X-EDMD patient fibroblasts [59]. We can speculate that this ‘rescue package’ fails over the longer term, possibly as part of the normal ageing process, at which point clinical symptoms develop. This is consistent with the human EDMD phenotype, since an age of 4 weeks in a mouse equates to the human age of 12–14 years, which in the majority of cases is prior to any clinical cardiac involvement, normally not seen until the third decade of life [1].
β-catenin’s nuclear function in non-cycling cells such as cardiomyocytes appears to differ from that in proliferating cells. In cycling cells, activation of the canonical Wnt signalling pathway results in the inhibition of GSK3β, which stabilises and thus activates β-catenin, which translocates to the nucleus and promotes gene expression through TCF/LEF transcriptional activation [60]. Stabilisation of β-catenin in this manner favours proliferation over differentiation in cycling cells [61]. The role of β-catenin signalling in fully differentiated cells, such as cardiomyocytes, is less clear, although current data suggest it is Wnt-independent. In this cell type GSK3β is inactivated by PKB/Akt, which in turn leads to the stabilisation of β-catenin and subsequent increase in TCF/LEF reporter gene expression [47, 62]. Our results also link the emerin–β-catenin complex to GSK3β activity on both components, although further studies are required to ascertain its precise role. The phosphorylation status of β-catenin at the ID is not well characterised. Recently it has been shown that N-terminally phosphorylated forms of β-catenin can exist at cell–cell contacts [63]. This is consistent with our findings that emerin interacts predominately with phosphorylated β-catenin. We can speculate that in non-cycling cardiomyocytes, emerin, as well as playing a role in regulating the nuclear accumulation of β-catenin, can additionally regulate the amount of β-catenin at the AJs. Therefore, at least in cardiomyocytes, the absence of emerin is likely to affect both β-catenin-dependent gene expression and cell–cell contact formation.
In conclusion, our study shows that β-catenin is indeed a binding partner of emerin in the heart and that the formation and intracellular location of this complex is important for ID structure. Changes to ID architecture and protein composition are classic features of DCM and thus likely to be a strong factor contributing to the cardiomyopathy associated with X-EDMD. Therefore, in addition to emerin’s acclaimed role in nuclear architecture, it can also act as a ‘gate-keeper’ for β-catenin function in cells of cardiac lineage.
Acknowledgements
Matthew Wheeler was supported by a BHF grant awarded to Juliet Ellis (PG/06/062/20926). We thank Pauline Bennett for help in analysing our EM images, Colin Stewart for the Emd −/y mice and Peter Zammit for critically reading the manuscript.
References
- 1.Emery AE. Emery-Dreifuss muscular dystrophy—a 40 year retrospective. Neuromuscul Disord. 2000;10:228–232. doi: 10.1016/S0960-8966(00)00105-X. [DOI] [PubMed] [Google Scholar]
- 2.Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- 3.Bengtsson L, Wilson KL. Multiple and surprising new functions for emerin, a nuclear membrane protein. Curr Opin Cell Biol. 2004;16:73–79. doi: 10.1016/j.ceb.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Ellis JA. Emery-Dreifuss muscular dystrophy at the nuclear envelope: 10 years on. Cell Mol Life Sci. 2006;63:2702–2709. doi: 10.1007/s00018-006-6247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holaska JM. Emerin and the nuclear lamina in muscle and cardiac disease. Circ Res. 2008;103:16–23. doi: 10.1161/CIRCRESAHA.108.172197. [DOI] [PubMed] [Google Scholar]
- 6.Cartegni L, di Barletta MR, Barresi R, Squarzoni S, Sabatelli P, Maraldi N, Mora M, Di Blasi C, Cornelio F, Merlini L, Villa A, Cobianchi F, Toniolo D. Heart-specific localization of emerin: new insights into Emery-Dreifuss muscular dystrophy. Hum Mol Genet. 1997;6:2257–2264. doi: 10.1093/hmg/6.13.2257. [DOI] [PubMed] [Google Scholar]
- 7.Markiewicz E, Tilgner K, Barker N, van de Wetering M, Clevers H, Dorobek M, Hausmanowa-Petrusewicz I, Ramaekers FC, Broers JL, Blankesteijn WM, Salpingidou G, Wilson RG, Ellis JA, Hutchison CJ. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J. 2006;25:3275–3285. doi: 10.1038/sj.emboj.7601230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler MA, Davies JD, Zhang Q, Emerson LJ, Hunt J, Shanahan CM, Ellis JA. Distinct functional domains in nesprin-1alpha and nesprin-2beta bind directly to emerin and both interactions are disrupted in X-linked Emery-Dreifuss muscular dystrophy. Exp Cell Res. 2007;313:2845–2857. doi: 10.1016/j.yexcr.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 9.Cohen M, Lee KK, Wilson KL, Gruenbaum Y. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem Sci. 2001;26:41–47. doi: 10.1016/S0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- 10.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson KL. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/S0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]
- 12.Muchir A, Pavlidis P, Bonne G, Hayashi YK, Worman HJ. Activation of MAPK in hearts of EMD null mice: similarities between mouse models of X-linked and autosomal dominant Emery Dreifuss muscular dystrophy. Hum Mol Genet. 2007;16:1884–1895. doi: 10.1093/hmg/ddm137. [DOI] [PubMed] [Google Scholar]
- 13.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manilal S, Sewry CA, Pereboev A, Man N, Gobbi P, Hawkes S, Love DR, Morris GE. Distribution of emerin and lamins in the heart and implications for Emery-Dreifuss muscular dystrophy. Hum Mol Genet. 1999;8:353–359. doi: 10.1093/hmg/8.2.353. [DOI] [PubMed] [Google Scholar]
- 15.Manta P, Terzis G, Papadimitriou C, Kontou C, Vassilopoulos D. Emerin expression in tubular aggregates. Acta Neuropathol. 2004;107:546–552. doi: 10.1007/s00401-004-0851-1. [DOI] [PubMed] [Google Scholar]
- 16.Nagano A, Koga R, Ogawa M, Kurano Y, Kawada J, Okada R, Hayashi YK, Tsukahara T, Arahata K. Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat Genet. 1996;12:254–259. doi: 10.1038/ng0396-254. [DOI] [PubMed] [Google Scholar]
- 17.Ostlund C, Ellenberg J, Hallberg E, Lippincott-Schwartz J, Worman HJ. Intracellular trafficking of emerin, the Emery-Dreifuss muscular dystrophy protein. J Cell Sci. 1999;112(Pt 11):1709–1719. doi: 10.1242/jcs.112.11.1709. [DOI] [PubMed] [Google Scholar]
- 18.Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Squarzoni S, Sabatelli P, Capanni C, Petrini S, Ognibene A, Toniolo D, Cobianchi F, Zauli G, Bassini A, Baracca A, Guarnieri C, Merlini L, Maraldi NM. Emerin presence in platelets. Acta Neuropathol. 2000;100:291–298. doi: 10.1007/s004019900169. [DOI] [PubMed] [Google Scholar]
- 20.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Drenckhahn J, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E, Thierfelder L. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36:1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 21.Sen-Chowdhry S, Syrris P, McKenna WJ. Desmoplakin disease in arrhythmogenic right ventricular cardiomyopathy: early genotype-phenotype studies. Eur Heart J. 2005;26:1582–1584. doi: 10.1093/eurheartj/ehi343. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Patel VV, Kostetskii I, Xiong Y, Chu AF, Jacobson JT, Yu C, Morley GE, Molkentin JD, Radice GL. Cardiac-specific loss of N-cadherin leads to alteration in connexins with conduction slowing and arrhythmogenesis. Circ Res. 2005;97:474–481. doi: 10.1161/01.RES.0000181132.11393.18. [DOI] [PubMed] [Google Scholar]
- 23.Ehler E, Horowits R, Zuppinger C, Price RL, Perriard E, Leu M, Caroni P, Sussman M, Eppenberger HM, Perriard JC. Alterations at the intercalated disk associated with the absence of muscle LIM protein. J Cell Biol. 2001;153:763–772. doi: 10.1083/jcb.153.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustafson-Wagner EA, Sinn HW, Chen YL, Wang DZ, Reiter RS, Lin JL, Yang B, Williamson RA, Chen J, Lin CI, Lin JJ. Loss of mXinalpha, an intercalated disk protein, results in cardiac hypertrophy and cardiomyopathy with conduction defects. Am J Physiol Heart Circ Physiol. 2007;293:H2680–H2692. doi: 10.1152/ajpheart.00806.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tilgner K, Wojciechowicz K, Jahoda C, Hutchison C, Markiewicz E. Dynamic complexes of A-type lamins and emerin influence adipogenic capacity of the cell via nucleocytoplasmic distribution of {beta}-catenin. J Cell Sci. 2009;122:401–413. doi: 10.1242/jcs.026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange S, Auerbach D, McLoughlin P, Perriard E, Schafer BW, Perriard JC, Ehler E. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J Cell Sci. 2002;115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- 27.Fairley EA, Kendrick-Jones J, Ellis JA. The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J Cell Sci. 1999;112(Pt 15):2571–2582. doi: 10.1242/jcs.112.15.2571. [DOI] [PubMed] [Google Scholar]
- 28.Ellis JA, Craxton M, Yates JR, Kendrick-Jones J. Aberrant intracellular targeting and cell cycle-dependent phosphorylation of emerin contribute to the Emery-Dreifuss muscular dystrophy phenotype. J Cell Sci. 1998;111(Pt 6):781–792. doi: 10.1242/jcs.111.6.781. [DOI] [PubMed] [Google Scholar]
- 29.Grove BK, Kurer V, Lehner C, Doetschman TC, Perriard JC, Eppenberger HM. A new 185,000-dalton skeletal muscle protein detected by monoclonal antibodies. J Cell Biol. 1984;98:518–524. doi: 10.1083/jcb.98.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett PM, Maggs AM, Baines AJ, Pinder JC. The transitional junction: a new functional subcellular domain at the intercalated disc. Mol Biol Cell. 2006;17:2091–2100. doi: 10.1091/mbc.E05-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melcon G, Kozlov S, Cutler DA, Sullivan T, Hernandez L, Zhao P, Mitchell S, Nader G, Bakay M, Rottman JN, Hoffman EP, Stewart CL. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum Mol Genet. 2006;15:637–651. doi: 10.1093/hmg/ddi479. [DOI] [PubMed] [Google Scholar]
- 32.Holaska JM, Lee KK, Kowalski AK, Wilson KL. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J Biol Chem. 2003;278:6969–6975. doi: 10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Ruiz A, Ono Y, Gnocchi VF, Zammit PS. beta-Catenin promotes self-renewal of skeletal-muscle satellite cells. J Cell Sci. 2008;121:1373–1382. doi: 10.1242/jcs.024885. [DOI] [PubMed] [Google Scholar]
- 34.Felts PA, Woolston AM, Fernando HB, Asquith S, Gregson NA, Mizzi OJ, Smith KJ. Inflammation and primary demyelination induced by the intraspinal injection of lipopolysaccharide. Brain. 2005;128:1649–1666. doi: 10.1093/brain/awh516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davey KA, Garlick PB, Warley A, Southworth R. Immunogold labeling study of the distribution of GLUT-1 and GLUT-4 in cardiac tissue following stimulation by insulin or ischemia. Am J Physiol Heart Circ Physiol. 2007;292:H2009–H2019. doi: 10.1152/ajpheart.00663.2006. [DOI] [PubMed] [Google Scholar]
- 36.Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:E231. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holaska JM, Wilson KL. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozawa R, Hayashi YK, Ogawa M, Kurokawa R, Matsumoto H, Noguchi S, Nonaka I, Nishino I. Emerin-lacking mice show minimal motor and cardiac dysfunctions with nuclear-associated vacuoles. Am J Pathol. 2006;168:907–917. doi: 10.2353/ajpath.2006.050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchiya Y, Hase A, Ogawa M, Yorifuji H, Arahata K. Distinct regions specify the nuclear membrane targeting of emerin, the responsible protein for Emery-Dreifuss muscular dystrophy. Eur J Biochem. 1999;259:859–865. doi: 10.1046/j.1432-1327.1999.00112.x. [DOI] [PubMed] [Google Scholar]
- 40.Zuppinger C, Schaub MC, Eppenberger HM. Dynamics of early contact formation in cultured adult rat cardiomyocytes studied by N-cadherin fused to green fluorescent protein. J Mol Cell Cardiol. 2000;32:539–555. doi: 10.1006/jmcc.1999.1086. [DOI] [PubMed] [Google Scholar]
- 41.Yates JR, Bagshaw J, Aksmanovic VM, Coomber E, McMahon R, Whittaker JL, Morrison PJ, Kendrick-Jones J, Ellis JA. Genotype-phenotype analysis in X-linked Emery-Dreifuss muscular dystrophy and identification of a missense mutation associated with a milder phenotype. Neuromuscul Disord. 1999;9:159–165. doi: 10.1016/S0960-8966(98)00121-7. [DOI] [PubMed] [Google Scholar]
- 42.Lee KK, Haraguchi T, Lee RS, Koujin T, Hiraoka Y, Wilson KL. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J Cell Sci. 2001;114:4567–4573. doi: 10.1242/jcs.114.24.4567. [DOI] [PubMed] [Google Scholar]
- 43.Haraguchi T, Holaska JM, Yamane M, Koujin T, Hashiguchi N, Mori C, Wilson KL, Hiraoka Y. Emerin binding to Btf, a death-promoting transcriptional repressor, is disrupted by a missense mutation that causes Emery-Dreifuss muscular dystrophy. Eur J Biochem. 2004;271:1035–1045. doi: 10.1111/j.1432-1033.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 44.Ellis JA, Yates JR, Kendrick-Jones J, Brown CA. Changes at P183 of emerin weaken its protein-protein interactions resulting in X-linked Emery-Dreifuss muscular dystrophy. Hum Genet. 1999;104:262–268. doi: 10.1007/s004390050946. [DOI] [PubMed] [Google Scholar]
- 45.Hirano Y, Segawa M, Ouchi FS, Yamakawa Y, Furukawa K, Takeyasu K, Horigome T. Dissociation of emerin from barrier-to-autointegration factor is regulated through mitotic phosphorylation of emerin in a xenopus egg cell-free system. J Biol Chem. 2005;280:39925–39933. doi: 10.1074/jbc.M503214200. [DOI] [PubMed] [Google Scholar]
- 46.Matsuda T, Zhai P, Maejima Y, Hong C, Gao S, Tian B, Goto K, Takagi H, Tamamori-Adachi M, Kitajima S, Sadoshima J. Distinct roles of GSK-3alpha and GSK-3beta phosphorylation in the heart under pressure overload. Proc Natl Acad Sci USA. 2008;105:20900–20905. doi: 10.1073/pnas.0808315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haq S, Michael A, Andreucci M, Bhattacharya K, Dotto P, Walters B, Woodgett J, Kilter H, Force T. Stabilization of beta-catenin by a Wnt-independent mechanism regulates cardiomyocyte growth. Proc Natl Acad Sci USA. 2003;100:4610–4615. doi: 10.1073/pnas.0835895100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tunnah D, Sewry CA, Vaux D, Schirmer EC, Morris GE. The apparent absence of lamin B1 and emerin in many tissue nuclei is due to epitope masking. J Mol Histol. 2005;36:337–344. doi: 10.1007/s10735-005-9004-7. [DOI] [PubMed] [Google Scholar]
- 49.Lattanzi G, Ognibene A, Sabatelli P, Capanni C, Toniolo D, Columbaro M, Santi S, Riccio M, Merlini L, Maraldi NM, Squarzoni S. Emerin expression at the early stages of myogenic differentiation. Differentiation. 2000;66:208–217. doi: 10.1046/j.1432-0436.2000.660407.x. [DOI] [PubMed] [Google Scholar]
- 50.Soullam B, Worman HJ. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts RC, Sutherland-Smith AJ, Wheeler MA, Jensen ON, Emerson LJ, Spiliotis II, Tate CG, Kendrick-Jones J, Ellis JA. The Emery-Dreifuss muscular dystrophy associated-protein emerin is phosphorylated on serine 49 by protein kinase A. FEBS J. 2006;273:4562–4575. doi: 10.1111/j.1742-4658.2006.05464.x. [DOI] [PubMed] [Google Scholar]
- 52.Lattanzi G, Cenni V, Marmiroli S, Capanni C, Mattioli E, Merlini L, Squarzoni S, Maraldi NM. Association of emerin with nuclear and cytoplasmic actin is regulated in differentiating myoblasts. Biochem Biophys Res Commun. 2003;303:764–770. doi: 10.1016/S0006-291X(03)00415-7. [DOI] [PubMed] [Google Scholar]
- 53.Holaska JM, Rais-Bahrami S, Wilson KL. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum Mol Genet. 2006;15:3459–3472. doi: 10.1093/hmg/ddl423. [DOI] [PubMed] [Google Scholar]
- 54.Rozenblum E, Vahteristo P, Sandberg T, Bergthorsson JT, Syrjakoski K, Weaver D, Haraldsson K, Johannsdottir HK, Vehmanen P, Nigam S, Golberger N, Robbins C, Pak E, Dutra A, Gillander E, Stephan DA, Bailey-Wilson J, Juo SH, Kainu T, Arason A, Barkardottir RB, Nevanlinna H, Borg A, Kallioniemi OP. A genomic map of a 6-Mb region at 13q21–q22 implicated in cancer development: identification and characterization of candidate genes. Hum Genet. 2002;110:111–121. doi: 10.1007/s00439-001-0646-6. [DOI] [PubMed] [Google Scholar]
- 55.Ooshio T, Irie K, Morimoto K, Fukuhara A, Imai T, Takai Y. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and alpha-actinin in epithelial cells. J Biol Chem. 2004;279:31365–31373. doi: 10.1074/jbc.M401957200. [DOI] [PubMed] [Google Scholar]
- 56.Bakay M, Wang Z, Melcon G, Schiltz L, Xuan J, Zhao P, Sartorelli V, Seo J, Pegoraro E, Angelini C, Shneiderman B, Escolar D, Chen YW, Winokur ST, Pachman LM, Fan C, Mandler R, Nevo Y, Gordon E, Zhu Y, Dong Y, Wang Y, Hoffman EP. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- 57.Nikolova V, Leimena C, McMahon AC, Tan JC, Chandar S, Jogia D, Kesteven SH, Michalicek J, Otway R, Verheyen F, Rainer S, Stewart CL, Martin D, Feneley MP, Fatkin D. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perriard JC, Hirschy A, Ehler E. Dilated cardiomyopathy: a disease of the intercalated disc? Trends Cardiovasc Med. 2003;13:30–38. doi: 10.1016/S1050-1738(02)00209-8. [DOI] [PubMed] [Google Scholar]
- 59.Tsukahara T, Tsujino S, Arahata K. CDNA microarray analysis of gene expression in fibroblasts of patients with X-linked Emery-Dreifuss muscular dystrophy. Muscle Nerve. 2002;25:898–901. doi: 10.1002/mus.10085. [DOI] [PubMed] [Google Scholar]
- 60.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 61.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/S0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 62.Shevtsov SP, Haq S, Force T. Activation of beta-catenin signaling pathways by classical G-protein-coupled receptors: mechanisms and consequences in cycling and non-cycling cells. Cell Cycle. 2006;5:2295–2300. doi: 10.4161/cc.5.20.3357. [DOI] [PubMed] [Google Scholar]
- 63.Maher MT, Flozak AS, Stocker AM, Chenn A, Gottardi CJ. Activity of the beta-catenin phosphodestruction complex at cell-cell contacts is enhanced by cadherin-based adhesion. J Cell Biol. 2009;186:219–228. doi: 10.1083/jcb.200811108. [DOI] [PMC free article] [PubMed] [Google Scholar]