Abstract
We provide the first description of the desmosome network in keratinocytes using a systems level approach. The desmo-adhesome consists of 59 proteins connected by 128 direct interactions and forms different functional subnets. Whilst the structure appears to be extremely robust against random perturbations, network fragmentation analysis suggests that the desmo-adhesome is susceptible to targeted attacks. To confirm this prediction, we applied this model to the autoimmune disease Pemphigus Vulgaris (PV), a paradigm of external perturbation of the desmosome. Our analysis showed that the adaptor protein plakophilin (Pkp) 3 was in the highest percentile group for both connectivity rate and gene expression changes in experimental PV. This observation led us to speculate that Pkp3 was crucial in desmosomal remodelling, and therefore we designed the experiments to verify this hypothesis. Our data demonstrate that, whilst Pkp3 is important in conferring adhesive strength to keratinocytes, it also acts as a central molecule mediating cell–cell detachment induced by PV IgG.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0139-7) contains supplementary material, which is available to authorised users.
Keywords: Systems biology, Adhesome, Desmosome, Plakophilin 3, Pemphigus vulgaris, Keratinocytes
Introduction
The study of individual genes and molecules is the focus of the majority of work in the biomedical sciences, but the shift to understanding the nature of molecular networks will be crucial for comprehending disease pathogenesis and developing appropriate molecular tools for therapeutic intervention. Historically, molecular bioscience has examined the function of individual molecules, an approach that may not predict the consequences of biological interactions [1]. The function of living organisms, however, cannot be addressed satisfactorily by examining molecules alone, not even if every molecule is studied individually in this way [2]. Indeed, proteins seldom carry out their function in isolation but rather operate through interactions with other biomolecules. Computational analysis and experimental verification of the complex networks formed by individual protein–protein interactions (PPIs) is one of the major challenges of the post-genomic era [3] and is essential if there is to be an understanding of the collective function of these activities. Many technologies, such as microarrays, two-dimensional SDS-PAGE, two-hybrid assays, co-immunoprecipitation and the chIP–chip approach, have been used to study global changes and molecular interactions. The development of these high throughput techniques, together with the availability of large biological datasets, are the tools by which cellular functions can be analysed at a systems level. Tentative steps are now being taken to use this approach to study human disease [4] and to develop new therapeutic interventions [5, 6].
Cell–cell adhesion is involved in all aspects of tissue behaviour in multi-cellular organisms, from tissue morphogenesis (regulation of cell shape, apoptosis, cell movement and the development of complex structures) to ageing and disease [7]. Organised rearrangements of cell–cell contacts in epithelial cells, for example, are invariably associated with spatio-temporal changes in the expression of cadherins, the main transmembrane proteins of desmosomes and adherens junctions [8]. Desmosomes are the principal intercellular junctions of stratified epithelia and play a central role in epithelial tissue homeostasis [9]. The desmosome consists of transmembrane adhesion molecules, including the cadherins [desmogleins (Dsg), desmocollins] and intracellular plaque proteins belonging to the families of plakins (desmoplakin, periplakin, envoplakin) and Armadillo proteins [plakoglobin and plakophilins (Pkps)]. Because desmosomes also link intracellularly to keratin intermediate filaments (KIF) in the cytoskeleton, termed the desmosome/intermediate filament complex (DIFC), an adhesive bond is established that confers mechanical stability to tissues [10]. Gene targeting has been employed to analyse the hierarchical function of desmosomal components, but by contrast to studies of adherens junctions, only a small number of genes (desmocollin 1, Dsg2, Dsg3, plakoglobin, pkp2, desmoplakin) have been ablated in mice [11]. However, the crucial role of the desmosome in maintaining epidermal integrity is widely demonstrated by the large number of diseases, be they genetic, immunological or infectious in origin, that occur when the function of one or more of desmosomal constituents is impaired [9, 12].
Knowledge of the desmosome has evolved from the notion of a static, punctuate, adhesive barrier structure to one of a multi-component, multifunctional complex involved in the regulation of numerous and diverse aspects of keratinocyte physiology and disease [13]. An insight into the mechanisms that regulate the dynamics of keratinocyte adhesion, therefore, requires not only an understanding of desmosomal structure and function but also an understanding of how the desmosome responds to perturbations and the regulation of component parts. These perturbations can be either genetic (i.e. internal), resulting from one or more mutations or altered expression of desmosomal genes, or environmental (i.e. external), consequent to the processing of specific desmocadherins during infection or to the blocking of molecules such as Dsg in mucocutaneous autoimmune diseases (Electronic Supplementary Material, ESM, Fig. S1). Autoantibodies targeting keratinocyte adhesion molecules, for example, trigger massive changes in the desmosomal network leading to cell–cell detachment, as is known to occur in Pemphigus Vulgaris (PV) [14]. In this disease, autoantibodies cause desmosomal remodelling, a process where PV IgG binds to Dsg1 and/or Dsg3, together with regulatory pathways triggered by non-Dsg3 antibodies, to induce molecular changes in desmosome composition and phosphorylation that lead to a failure of intercellular adhesion and cell detachment [14, 15].
Recent in silico studies have demonstrated the usefulness of a system level analysis to build a roadmap of cell adhesion proteins and signalling [16, 17]. A detailed understanding of the desmosome, however, is still lacking. Furthermore, there have been no attempts to apply such molecular networks of adhesion to human disease. The development of a keratinocyte-centred desmo-adhesome is of paramount importance since most diseases affecting the skin and mucous membranes involve desmosomes.
In the present study, we used a systems level approach to characterise the “desmo-adhesome” and to make predictions, based on the topology and connectivity of major desmosomal proteins, as to the effect of external perturbations on this structure. We postulated that alterations of key molecules in this process would also be involved in the autoimmune disease PV, a paradigm of external perturbation of cell adhesion. We then verified these predictions experimentally using an in vitro model of PV. The results of this study clearly illustrate the benefits of using a systems level approach to understand disease pathogenesis and to identify potential therapeutic targets.
Materials and methods
Criteria for inclusion in the keratinocyte desmosomal interactome (desmo-adhesome)
We distinguished between intrinsic proteins, i.e. structural components that have been reported to physically reside within the desmosome, and accessory proteins which interact directly with the intrinsic components in a junctional context. The search was performed through literature databases (PubMed) and PPI databases (Human Protein Reference Database, HPRD; functional protein association network, STRING; and BioGrid). We combined data from keratinocytes of different types (e.g. normal, immortal, neoplastic) to broaden the search process. Only interactions taking place in a junctional context were taken into account.
The primary literature was accessed for each interaction. The following components were excluded: (1) intrinsic or accessory proteins occurring in desmosomes in cells from tissues other than stratified squamous epithelia (desmin interacts with desmoplakin in muscle cells); and (2) several intrinsic proteins such as Pkp2 localize to adhesion sites and cytoplasm as well as in the nucleus. PPIs of Pkp2 occurring in the nucleus were excluded.
Network clustering
Proteins and their PPIs were combined to form networks and subnets using Cytoscape 2.4 [18]. The resulting network was visualised as a series of graphs in which each node (N) represented a protein and the interactions between proteins were shown by edges (E) in the graph. Only direct interactions between proteins are reported. We distinguished between binding interactions (which are always non-directional) and signalling interactions which have direction, being either activating or inhibitory. Nodes and edges were assigned different shapes and colours according to the function of the respective proteins.
Connectivity and network fragmentation
The average connectivity (k) of a whole network was calculated as k = 2E/N, corresponding to the average number of interactive partners for each protein. More detailed analysis involved a consideration of all node degrees expressed as a global quantity or the connectivity distribution, i.e. P(k) = N k/N, where N k is the number of nodes with k neighbours. This measurement gave the percentage of nodes for each degree k and facilitated computations of the variations in connectivity in the network [19].
The robustness and resilience of the desmo-adhesome was assessed by network fragmentation, as described by Albert et al. [20]. Nodes were removed sequentially in decreasing order of connectivity (k) (targeted attack), and at each step (t), the number of nodes remaining in the fragment (N) was determined. As a control, the nodes were removed from the network randomly (non-targeted attack). The size of the largest fragment (S) was expressed as the ratio between N(t) [node still present at each time point (t)] and the initial number of nodes N(t 0). Then, S was plotted as a function of the fraction (f) of removed nodes.
Antibodies, reagents and sera
Rabbit IgG raised against the epitope corresponding to amino acids 855–999 mapping at the C-terminus of Dsg3, 23E/4 mouse monoclonal IgG2b raised against the amino acids 570–589 of Pkp3, rabbit polyclonal antibodies to human Dsg1, Dsg2, desmocollin 2, desmoplakin, plakoglobin, E-cadherin, anti-tubulin polyclonal IgG, HRP-conjugated anti-rabbit and anti-mouse antibodies and reagents for siRNA transfections were from Santa Cruz Biotecnology (Santa Cruz, CA). FITC-conjugated anti-rabbit IgG were from Dako Denmark. Nitrocellulose filters were purchased from Invitrogen (Carlsbad, CA); the ECL chemiluminescent immunodetection system and Hyperfilms were from Amersham (Buckinghamshire, UK). Protease inhibitors and cell culture reagents were from Sigma (St. Louis, MO).
Sera of two patients with mucocutaneous PV (PV1 and PV2) and a healthy volunteer were used in this study. PV sera, containing both anti-Dsg3 IgG (average ELISA score 127 and 53) and anti-Dsg1 IgG (average ELISA score 75 and 196), have been shown to cause suprabasal epithelial cleft formation in mice [21]. The diagnosis of PV was established using clinical, histological and immunological criteria, as detailed previously [21, 22]. Total IgG were purified with protein A-Sepharose following standard procedures [23].
Cell culture and siRNA transfection experiments
The non-tumorigenic human keratinocyte cell line HaCaT was used as an in vitro model of cell adhesion/detachment; previous studies have demonstrated normal differentiation in vitro and in vivo [24]. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS in a humidified atmosphere containing 5% CO2 at 37°C.
The Pkp3-directed siRNA pool (sc-62826) and the negative control pool (sc-37007) were transfected at a final concentration of 80 nM, according to the manufacturer’s instructions. Cultured HaCaT cells were incubated overnight with the siRNAs in serum-free transfection medium (sc-36868) and then for an additional 24 h in complete medium with 10% FBS. Cells were then treated with IgG, as specified in “Results”. The efficiency of transfection was monitored by western blot analysis of Pkp3 expression.
Cell dissociation assay
To quantify the strength of cell-to-cell adhesion, dispase-based cell dissociation assays were conducted, as reported previously [25]. The ratio of total to single cells is indicative of intercellular adhesive strength.
Immunofluorescence and morphometric analysis of cell–cell detachment
Immunofluorescence was performed according to standard procedures [26]. The extent of cell detachment (acantholysis) in cell monolayers was measured by immunofluorescence according to our previously published protocols [27].
Western blotting
Cells were rinsed (2×) with PBS and then scraped into cold PBS containing 1 mM protease inhibitor phenylmethyl-sulfonylfluoride (PMSF). After centrifugation (800g for 10 min at 4°C), the pellets were re-suspended in Cirillo’s high salt-urea (C-HSU) buffer (50 mM HEPES, 800 mM NaCl, 6 M urea, 0,2% Triton X-100, 1 mM DTT, 1 mM PMSF, 10 μg/ml leupeptin and 10 μg/ml aprotinin). Protein extracts were mixed with 4× SDS sample buffer, separated by 10% SDS-PAGE and transferred overnight onto nitrocellulose membranes. Western blotting was carried out according to standard procedures [28]. Band intensity was quantified by Adobe Photoshop (Adobe System, San Jose, CA).
Statistical analysis
Data relating to densitometric analysis and cell dissociation scores data were expressed as a mean ± SD of independent experiments, as detailed in the figure legends. Differences were assessed by the Student t test for unpaired samples and a P value of <0.05 was considered to be significant. The fold change in gene expression by DNA microarray was reported as Signal Log Ratio (SLR). The percentile was calculated using Microsoft Excel and the genes were grouped as being in the high (≥0.85, i.e. above or equal to the 85 percentile), medium (0.50–0.85) or low (≤0.50) percentile bracket.
Results
The keratinocyte desmo-adhesome
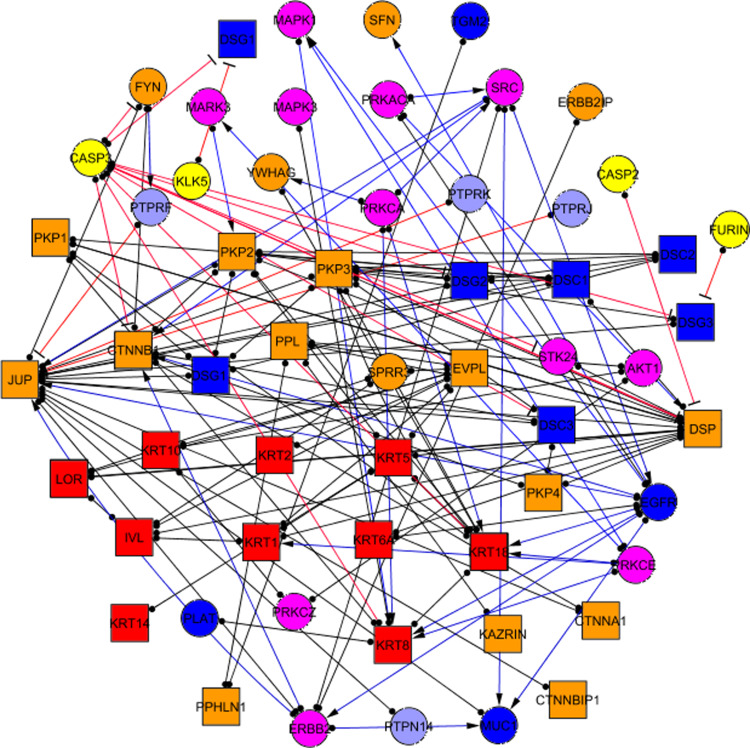
Desmosomes are formed as a scaffold that connects desmosomal cadherins to the keratin cytoskeleton. Additional molecules act as peripheral components and affect desmosome structure and regulation. We identified 59 proteins that were physically located within desmosomal adhesion sites and which were intrinsic components that directly interacted with peripheral components (accessory proteins) (Table 1). The findings were strictly based on specific adhesion–localisation data. For instance, β-catenin—a crucial adaptor protein of adherens junctions —has been reported to bind desmocollin 3, thus including it into the desmosomal interactome as an intrinsic component. However, only the interactions occurring in a desmosomal context were taken into account among the myriad of known interacting partners of β-catenin. Although this approach may have introduced some artificial combinations of connectivity, such limitations were outweighed by the ability to have a general view of the desmosomal adhesome. Of the 59 proteins forming the desmo-adhesome, 30 intrinsic and 29 accessory components were identified (Fig. 1).
Table 1.
Proteins forming the desmosomal interactome
| Gene name | GeneID Entrez | Protein name | ProteinID UniProt | Protein official full name | Functional category | Protein type |
|---|---|---|---|---|---|---|
| CTNNB1 | 1499 | Cnt beta | P35222 | Catenin (cadherin-associated protein), beta 1, 88 kDa | Adaptor | Intr |
| DSC1 | 1823 | Dsc1 | Q08554 | Desmocollin 1 | Membrane | Intr |
| DSC2 | 1824 | Dsc2 | Q02487 | Desmocollin 2 | Membrane | Intr |
| DSC3 | 1825 | Dsc3 | Q14574 | Desmocollin 3 | Membrane | Intr |
| DSG1 | 1828 | Dsg1 | Q02413 | Desmoglein 1 | Membrane | Intr |
| DSG2 | 1829 | Dsg2 | Q14126 | Desmoglein 2 | Membrane | Intr |
| DSG3 | 1830 | Dsg3 | P32926 | Desmoglein 3 | Membrane | Intr |
| DSG4 | 147409 | Dsg4 | Q86SJ6 | Desmoglein 4 | Membrane | Intr |
| DSP | 1832 | DP | P15924 | Desmoplakin | CytoAdapt | Intr |
| EVPL | 2125 | EVPL | Q92817 | Envoplakin | CytoAdapt | Intr |
| IVL | 3713 | IVL | P07476 | Involucrin | Cytoskeleton | Intr |
| JUP | 3728 | PLAK | P14923 | Junction plakoglobin | Adaptor | Intr |
| KRT1 | 3848 | KRT1 | P04264 | Keratin, type II cytoskeletal 1 | Cytoskeleton | Intr |
| KRT10 | 3858 | KRT10 | P13645 | Keratin, type I cytoskeletal 10 | Cytoskeleton | Intr |
| KRT14 | 3861 | KRT14 | P02533 | Keratin, type I cytoskeletal 14 | Cytoskeleton | Intr |
| KRT18 | 3875 | KRT18 | P05783 | Keratin, type I cytoskeletal 18 | Cytoskeleton | Intr |
| KRT2 | 3849 | KRT2 | P35908 | Keratin, type II cytoskeletal 2 epidermal | Cytoskeleton | Intr |
| KRT5 | 3852 | KRT5 | P13647 | Keratin, type II cytoskeletal 5 | Cytoskeleton | Intr |
| KRT6A | 3853 | KRT6A | P02538 | Keratin, type II cytoskeletal 6A | Cytoskeleton | Intr |
| KRT8 | 3856 | KRT8 | Q96J60 | Keratin, type II cytoskeletal 8 | Cytoskeleton | Intr |
| LOR | 4014 | LOR | P23490 | Loricrin | Cytoskeleton | Intr |
| PKP1 | 5317 | PKP1 | Q13835 | Plakophilin-1 | CytoAdapt | Intr |
| PKP2 | 5318 | PKP2 | Q99959 | Plakophilin-2 | CytoAdapt | Intr |
| PKP3 | 11187 | PKP3 | Q9Y446 | Plakophilin-3 | Adaptor | Intr |
| PKP4 | 8502 | PKP4 | Q99569 | Plakophilin-4 | Adaptor | Intr |
| PPHLN1 | 51535 | PPL1 | Q8NEY8 | Periphilin-1 | CytoAdapt | Intr |
| PPL | 5493 | PPK | O60437 | Periplakin | CytoAdapt | Intr |
| CTNNA1 | 1495 | CTNNA1 | P35221 | Catenin (cadherin-associated protein), alpha 1 | Adaptor | Intr |
| CTNNBIP1 | 56998 | CTNNBIP1 | Q9NSA3 | Beta-catenin-interacting protein 1 | Adaptor | Intr |
| KIAA1026 | 23254 | KAZRIN | Q674X7 | Kazrin | Adaptor | Intr |
| AKT1 | 207 | PRKBA | P31749 | RAC-alpha serine/threonine-protein kinase | Kinase | Acc |
| CASP2 | 835 | CASP2 | P42575 | Caspase-2 | Protease | Acc |
| CASP3 | 836 | CASP3 | P42574 | Caspase-3 | Protease | Acc |
| EGFR | 1956 | EGFR | P00533 | Epidermal growth factor receptor precursor | Kinase | Acc |
| ERBB2 | 2064 | ERBB2 | P04626 | Receptor tyrosine-protein kinase erbB-2 precursor | Kinase | Acc |
| ERBB2IP | 55914 | ERBB2IP | Q96RT1 | erbb2 interacting protein | Adaptor | Acc |
| FER | 2241 | FER | P16591 | Proto-oncogene tyrosine-protein kinase FER | Kinase | Acc |
| FURIN | 5045 | FURIN | P09958 | Paired basic amino acid residue cleaving enzyme | Protease | Acc |
| FYN | 2534 | FYN | P06241 | Proto-oncogene tyrosine-protein kinase Fyn | Kinase | Acc |
| KLK5 | 25818 | KLK5 | Q9Y337 | Kallikrein-related peptidase 5 | Protease | Acc |
| MAPK1 | 5594 | MK01 | P28482 | Mitogen-activated protein kinase 1 | Kinase | Acc |
| MAPK3 | 5595 | ERK1 | P27361 | Mitogen-activated protein kinase 3 | Kinase | Acc |
| MARK3 | 4140 | MARK3 | P27448 | MAP/microtubule affinity-regulating kinase 3 | Kinase | Acc |
| MUC1 | 4582 | MUC1 | P15941 | Mucin-1 precursor | Membrane other | Acc |
| PLAT | 5327 | PLAT | Q9BZW1 | Tissue-type plasminogen activator | Membrane other | Acc |
| PRKACA | 5566 | KAPCA | P17612 | cAMP-dependent protein kinase catalytic subunit alpha | Kinase | Acc |
| PRKCA | 5578 | PKCA | P17252 | Protein kinase C alpha type | Kinase | Acc |
| PRKCE | 5581 | PKCE | Q02156 | Protein kinase C epsilon type | Kinase | Acc |
| PRKCZ | 5590 | PKCZ | Q05513 | Protein kinase C zeta type | Kinase | Acc |
| PTPN14 | 5784 | PTN14 | Q15678 | Tyrosine-protein phosphatase non-receptor type 14 | Phosphatase | Acc |
| PTPRF | 5792 | PTPRF | P10586 | Receptor-type tyrosine-protein phosphatase F precursor | Phosphatase | Acc |
| PTPRJ | 5795 | PTPRJ | Q12913 | Receptor-type tyrosine-protein phosphatase eta precursor | Phosphatase | Acc |
| PTPRK | 5796 | PTPRK | Q15262 | Receptor-type tyrosine-protein phosphatase kappa precursor | Phosphatase | Acc |
| SFN | 2810 | 1433S | P31947 | 14-3-3 protein sigma | Adaptor | Acc |
| SPRR3 | 6707 | SPRR3 | Q9UBC9 | Small proline-rich protein 3 | Adaptor | Acc |
| SRC | 6714 | SRC | P12931 | Proto-oncogene tyrosine-protein kinase Src | Kinase | Acc |
| STK24 | 8428 | STK24 | Q9Y6E0 | Serine/threonine-protein kinase 24 | Kinase | Acc |
| TGM2 | 7052 | TGM2 | P21980 | Protein-glutamine gamma-glutamyltransferase 2 | Membrane other | Acc |
| YWHAG | 7532 | 1433G | P61981 | 14-3-3 protein gamma | Adaptor | Acc |
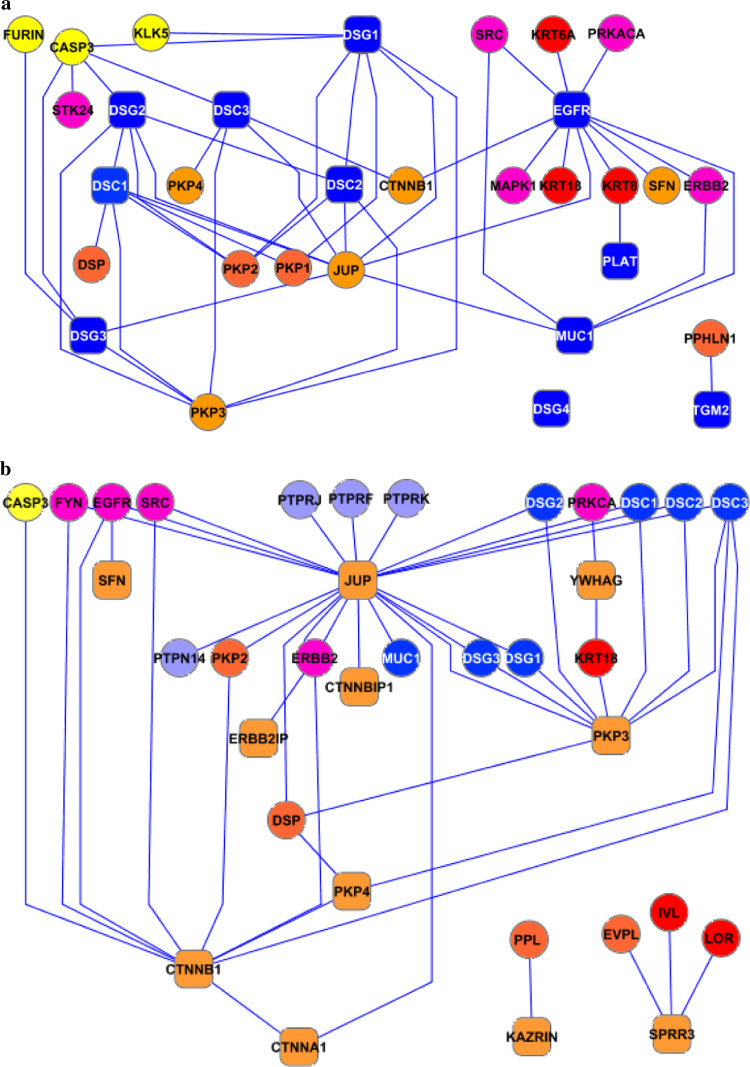
Fig. 1.
The desmo-adhesome network. Intrinsic components are represented as squares whilst accessory nodes are circles. Nodes have been assigned different colours according to the function of the respective proteins, as reported in the figure. Binding interactions (non-directional) are represented as black lines with full circles at their end, whereas signalling interactions have blue lines ending with an arrow (activating) or red line with a stop signal at their end (inhibitory). For the relevant nomenclature, please refer to Table 1
Interactions and connectivity of the network
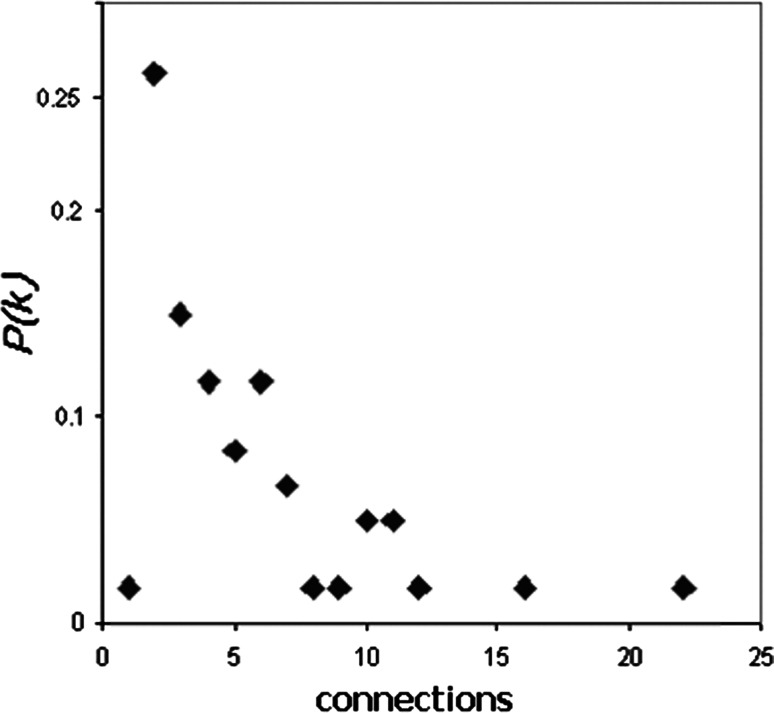
The desmo-adhesome network was connected by 128 PPIs (Fig. 1). Interactions were characterised as non-directional (binding) or directional modifications of a target according to the molecular nature of the modification [16]. The network contained 81 non-directional and 47 directional interactions. The activation events included 31 phosphorylation interactions whilst 3 dephosphorylation and 13 proteolytic degradation processes accounted for the inhibition events (Fig. 1). For some proteins, especially the adaptors, the interactions within the desmo-adhesome constituted the majority of their known activities, whereas with other proteins, notably the protein kinases, there were additional interactions outside of the desmosome network (not shown). The average connectivity was 4.339 edges (interactions) per node. The probability that a node in the network is connected to other nodes is termed the degree of distribution [P(k)] (Fig. 2). The data demonstrate that the desmo-adhesome is an inhomogeneous network, or scale-free network, in which the majority of proteins have a small number of neighbours whilst others act as hubs with large numbers of connections. In general, pure scale-free networks decay as a power-law but the desmo-adhesome shows exponential decay. In fact, there is a slightly faster decaying tail as the network presents fewer and smaller hubs than seen with a pure power law.
Fig. 2.
Distribution of connectivity within the desmo-adhesome. P(k) indicates the fraction of nodes having a given number (x) of connections
Resilience of the desmo-adhesome by network fragmentation
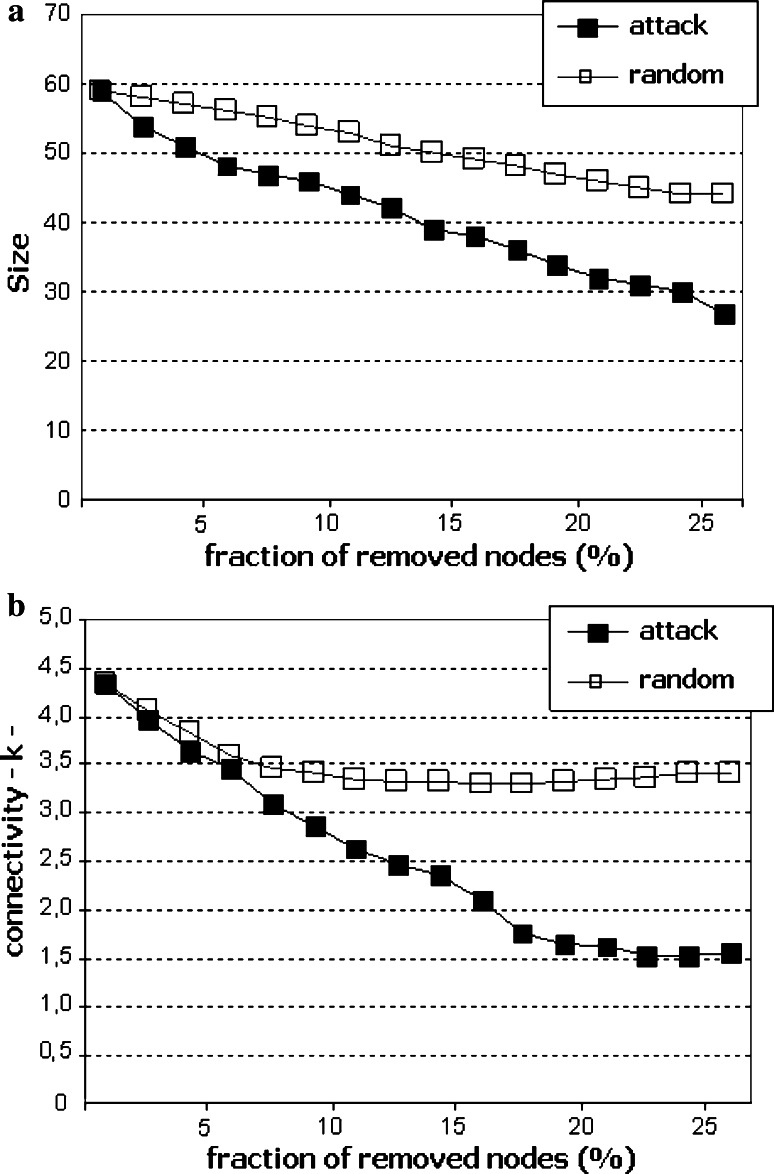
When nodes are removed from a network, clusters of nodes lose their interactions from the main network (fragmentation). The resilience of the network to node removal is determined by monitoring the network while randomly deleting nodes (corresponding to failure or random perturbation) or iteratively removing the nodes with the highest number of connections (corresponding to targeted attack). In the present study, we measured the size of the largest cluster of nodes, and its connectivity, when a fraction of the nodes was removed either randomly or as a targeted attack. Random removal of nodes only marginally affected the size (Fig. 3a) and connectivity (Fig. 3b) of the desmo-adhesome. By contrast, removal of highly connected nodes resulted in a dramatic decrease in the network size and internal connectivity (Fig. 3). For example, removal of 25% of nodes reduced the connectivity of the network by 21.5% (random) or 65% (attack). The data demonstrate that the desmo-adhesome, like most scale-free networks, is extremely robust against random perturbations but highly susceptible to targeted attacks.
Fig. 3.
Size (a) and connectivity k (b) of the network, under random (unfilled squares) or targeted (filled squares) attack, are represented as a function of the percentage of nodes removed
Analysis of the functional categories of desmosome network
An emerging approach to understand the complexity of molecular interactions within networks is to extract functional subnets from the entire network [16]. Unlike previous studies on adhesion structures, we examined only two functional groups, a structural category corresponding to the intrinsic proteins and a regulatory group associated with accessory nodes.
The structural subnet consisted of transmembrane (adhesion) molecules, adaptors, cytoskeleton adaptors and cytoskeleton proteins (mainly keratins) linked through binding interactions. This structural subnet comprised 30 intrinsic proteins connected by 62 internal PPIs (see next paragraph and ESM Fig. S2). There was a group of 7 highly connected proteins in the highest percentile (>85%), 10 in the middle percentile (50–85%) and 13 with a low connection rate (<50%). A hierarchical graphic representation demonstrated constant motifs corresponding to the topological organisation of the desmosome. According to this analysis, structural nodes tended to organise as triads (transmembrane-cyto-adaptor-cytoskeleton; Desmocollin1-Pkp1-Keratin8) or tetrads (transmembrane-adaptor-cyto-adaptor-cytoskeleton; Dsg2-Plakoglobin-Desmoplakin-Keratin6A). The distinction between adaptors and cytoskeleton adaptors, however, was somewhat artificial since many desmosomal proteins bind transmembrane, cytoskeleton and cyto-adaptor proteins at the same time. This concept was demonstrated by Pkp3 which binds keratin 18 and should be included in the cytoskeleton adaptor group. However, a prominent linking function of Pkp3 is via its binding to Dsp, the major cytoskeleton adaptor of the desmosome. Thus, Pkp3 was included as an adaptor protein.
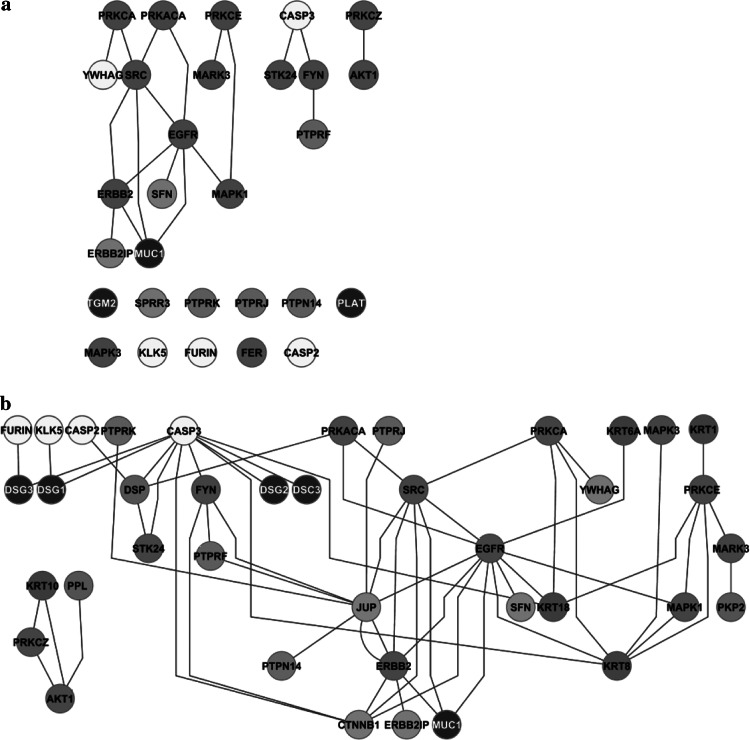
The regulatory subnet was formed by membrane and adaptor proteins together with enzymes. This subnet included 28 accessory proteins linked by only 19 internal connections (Fig. 4a). The disproportion between nodes and edges in the regulatory subnet was due to the fact that the majority of interactions were directed towards the structural subnet. When the whole regulatory network was taken into account, the total number of connections increased to 59 (Fig. 4b).
Fig. 4.
The regulatory subnet (accessory proteins). By considering only internal interactions the subnet is very poorly connected (a) but connectivity increases by inclusion of the interactions with the structural subnet (b)
Dissection of the desmo-adhesome into functional subnets
A useful approach to constructing subnets is to group proteins with similar activities into functional families and then to examine the interactions between these families. Thus, to analyse the desmosome network in further detail, groups of functionally defined proteins were extracted from the desmosomal network and used to construct distinct subnets. According to its known biological activity and topological distribution in keratinocytes, proteins were included in one of the following groups: membrane proteins, adaptor proteins, cytoskeleton-binding (adaptor) proteins, cytoskeleton proteins, kinases, phosphatases and proteases. First neighbours of each protein were included in the graphical representation of the respective subnet, thereby enabling an understanding of how each subnet was intrinsically organised and integrated within the network.
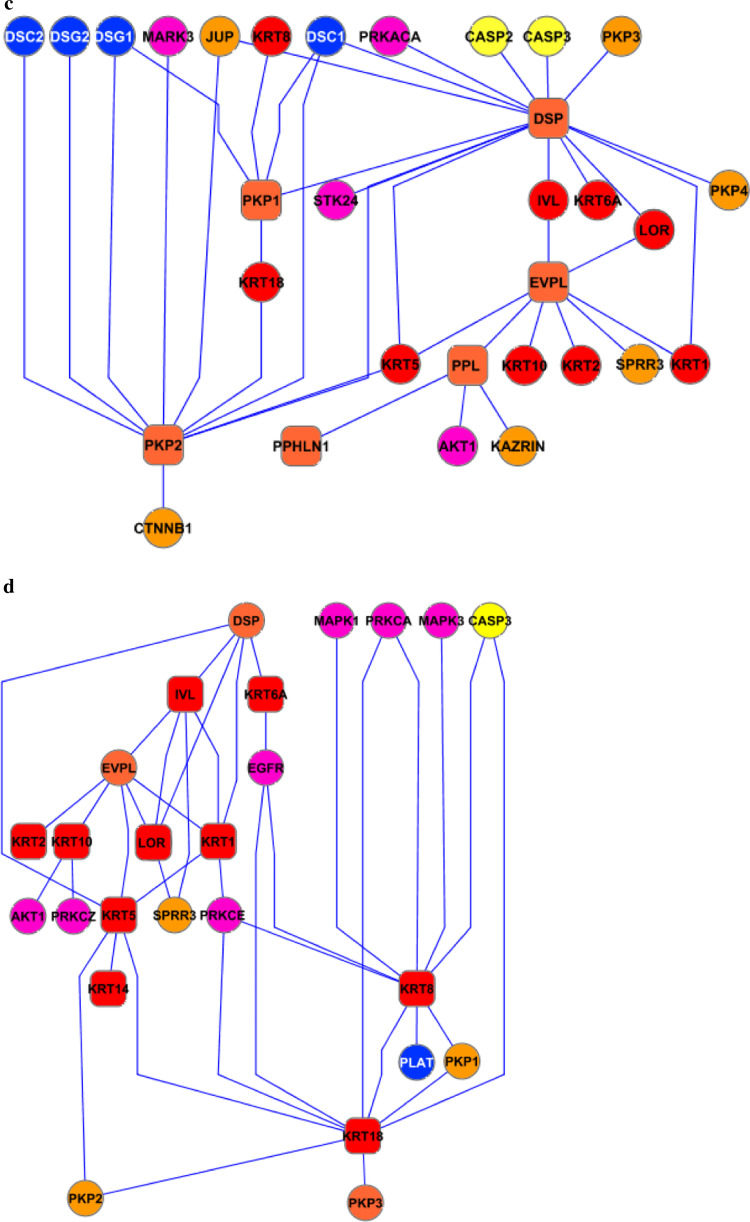
Subnet 1: membrane proteins
This subnet included adhesion molecules such as desmogleins and desmocollins, together with other membrane proteins such as EGFR (which was also categorised under the kinase subnet). Overall, the network consisted of 32 nodes and 47 edges (Fig. 5a). The nine membrane components with at least one neighbour interacted preferentially with adaptor proteins (n = 16), as would have been predicted by their topological organisation. The tendency not to interact also had some biological meaning. Desmosomal cadherins, for example, did not interact directly with the cytoskeleton whereas they were highly connected to adaptor and cytoskeleton-binding proteins. Interestingly, no binding partners of Dsg4 have been identified to date which is likely to reflect the lack of data available for this newly identified protein rather than any atypical biological behaviour. It is also of note that protein kinases did not interact (i.e. phosphorylate) with the desmosomal cadherins suggesting that mechanisms other than phosphorylation may play a role in the regulation of their function and metabolism. Whilst there are some potential phosphorylation sites in the desmosomal cadherins, the lack of interactions with protein kinases may simply reflect a limited literature.
Fig. 5.
Membrane (a), adaptor (b), cytoskeleton adaptor (c) and cytoskeleton (d) subnets. The intrinsic components on each subnet are represented as rounded squares connected to the interacting partners (circles) and are placed in a hierarchical distribution (Cytoscape 2.4)
Subnet 2a: adaptor proteins
Adaptor proteins formed a network of 35 nodes and 48 interactions (Fig. 5b). This group represented some of the most highly connected nodes within the desmo-adhesome. In particular, 2 of 11 adaptor proteins (plakoglobin and Pkp3) made up more then 60% of the total connections. Interestingly, the intrinsic (structural) components tended to interact preferentially with the adhesion molecules rather than with the cytoskeleton. The hierarchical distribution suggested that JUP (plakoglobin, 21 interactions) was the central protein of this network. In this regard, plakoglobin was highly connected to both protein kinases (n = 5) and phosphatases (n = 4), and this raises the possibility that the dynamic regulation of the desmosome may be orchestrated by this protein. By contrast, the other highly connected adaptor protein Pkp3 interacted exclusively with the adhesion molecules, suggesting an important structural role within the desmosomal interactome.
Subnet 2b: cytoskeleton adaptor proteins
Six intrinsic cytoadaptors consisted of 32 nodes and 39 edges (Fig. 5c). The three most connected proteins were desmoplakin, Pkp2 and envoplakin, and together, they made up some 85% of the total interactions. Certain cytoskeleton adaptor proteins, especially the Pkps, bound both membrane adhesion molecules and adaptor proteins. By contrast, desmoplakin (the node with highest connectivity) did not interact with the desmosomal cadherins. Envoplakin did not link any adaptor or membrane proteins, consistent with its role as a stabiliser between the desmosomal core and the cytoskeleton, especially during the formation of the keratinocyte cornified envelop.
Subnet 3: cytoskeleton proteins
The intrinsic components of the cytoskeletal subnet (n = 10) were represented mainly by the keratins, with loricrin and involucrin known to be expressed in the strata granulosum and corneum. Together, they form a network consisting of 26 nodes and 39 edges (Fig. 5d). According to the hierarchical distribution, cytoskeletal proteins are stabilised by envoplakin and connect to the desmosome through desmoplakin and Pkp2. A large number of protein kinases interact with the keratins but the connection can be non-directional suggesting that the keratins may simply be acting as a scaffold for the kinases in addition to being targets for phosphorylation.
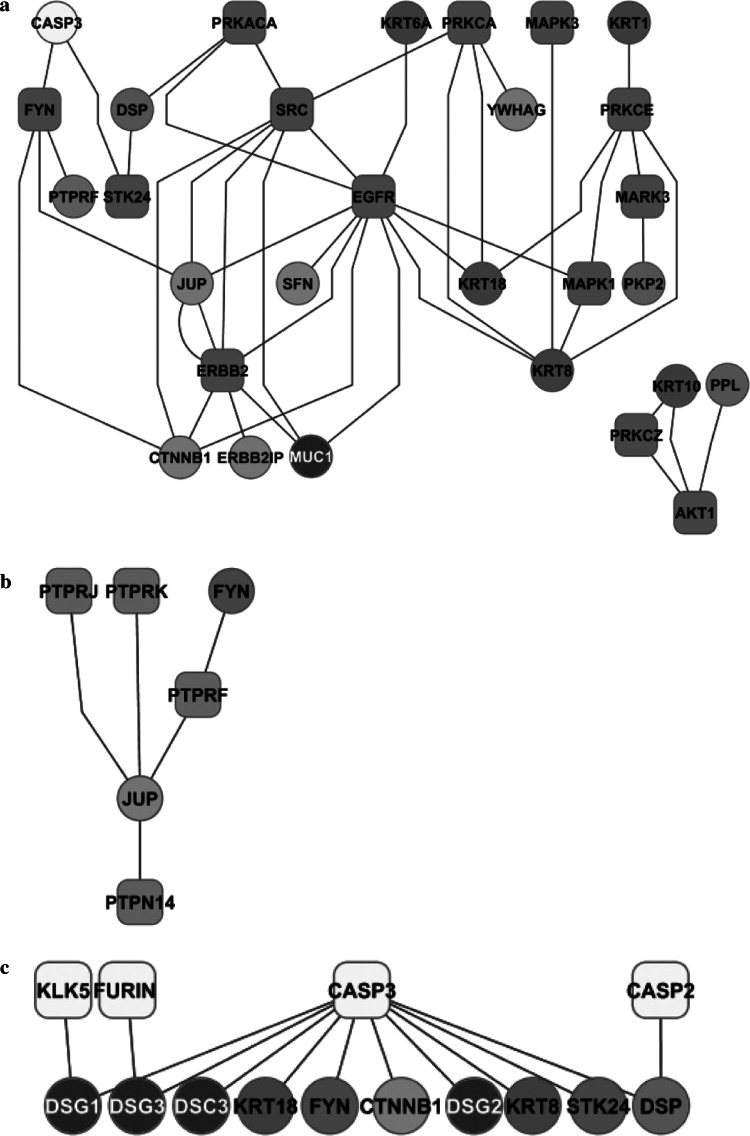
Subnet 4: kinases
The kinase subnet comprises 13 protein kinases. Upstream regulators, downstream substrates and binding partners participated to form a network of 29 nodes linked by 44 connections (Fig. 6a). Whilst the majority of these interactions were directional (activating) and targeted the intrinsic core of the desmosome, they were also associated with kinase–kinase regulation. Unlike adherens junctions, however, no direct interaction was found between adhesion molecules and phosphatases. Both Ser/Thr and Tyr kinases phosphorylate the structural proteins. There is also evidence for regulator-target hierarchies among the kinases. Some Ser/Thr kinases (PRKCA and PRKACA), for example, regulate the Tyr kinase SRC which, in turn, lies upstream of other Tyr kinases (EGFR, ERBB2); all of them regulate the structural proteins.
Fig. 6.
Subnets of kinases (a), phosphatases (b) and proteinases (c). The graphic depicts the enzymes and their substrates/interacting partners within the desmo-adhesome. For information on the nature of the interactions, please refer to Fig. 2
Subnet 5: phosphatases
The phosphatase subnet (Fig. 6b) was small (six nodes and five interactions), a fact that probably reflected the limited literature rather than an absence of function. All of the components were Tyr phosphatases and no Ser/Thr phosphatases were evident. Interestingly, there appeared to be a reciprocal control of plakoglobin by both kinases and phosphatases. The Tyr kinase FYN phosphorylated both plakoglobin and the Tyr phosphatase PTPRF which, in turn, targeted plakoglobin. This is consistent with a pivotal role of plakoglobin in the regulation of desmosomal adhesion.
Subnet 6: proteases
The protease subnet comprised four proteases, with a network of 14 nodes and 13 edges (Fig. 6c). All of the interactions were directional (inhibition) and were associated with proteolytic cleavage. The central enzyme of the protease subnet was caspase 3 which accounted for almost 80% of the network connections. Targets included adhesion molecules, adaptor and cytoskeleton proteins and kinases. Thus, irrespective of whether the adaptor proteins were regulated by phosphorylation events, desmosome remodelling involving adhesion molecules appeared to proceed via caspase 3 activation.
External perturbation on the desmo-adhesome: pemphigus vulgaris
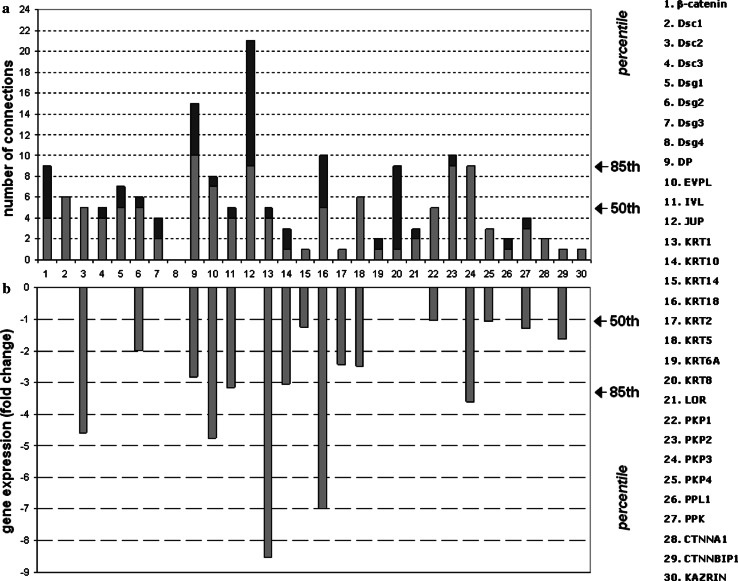
One of the principles of systems biology is the use of high throughput techniques to study the molecular response of a system to perturbation. PV is an autoimmune disease that targets the desmosome and, therefore, represents an ideal model of external perturbation of the desmo-adhesome. Using microarray data, we have shown in a mouse model of PV that a broad spectrum of adhesion molecules, including the highly connected nodes desmoplakin and Pkp3 (Fig. 7a), and many cytokeratins, are down-regulated [21]. In the mouse model of PV, changes in the desmo-adhesome nodes were unidirectional in terms of reduced expression (Fig. 7b). Only a minor decrease in gene expression was found in the accessory signalling nodes such as EGFR and PKC [21]. We classified genes as being in the high (≥0.85; n = 5; FC ≥ 3.44953), medium (0.50–0.85; n = 9; FC 1.05077347–3.44953) or low (<0.50; n = 16; FC ≤ 1.05077347) percentile. Interestingly, the correlation between connectivity rate and down-regulation was consistent for certain proteins such as keratin 18, desmoplakin and Pkp 3 (Fig. 7) suggesting a central role in the regulation of the desmosomal scaffold in PV.
Fig. 7.
Correlation between connectivity rates (a) and gene changes in experimental PV (b) of the 30 intrinsic components forming the desmo-adhesome. The number of connections of the 30 intrinsic nodes reported as a histogram (a). The light colour represents the (binding) interactions with other intrinsic components of the desmosomal interactome, whereas the interactions with nodes of the regulatory subnet are in dark colour. The range of connectivity has been reported as a percentile on the right side of the panel. b Signal log ratio of keratinocyte gene expression changes occurring in the mouse model of PV
Selection of key molecules: Pkp3
The results of this study show that adaptor proteins play a critical role in the regulation of the desmo-adhesome and that their expression is heavily affected during desmosome remodelling by PV antibodies. In particular, Pkp3 showed both a high connection rate and a marked down-regulation in the mouse model of PV, thereby making it a candidate molecule for further study. Further, Pkp3: (1) was one of the most highly connected intrinsic proteins (percentile bracket ≥0.85) but the only protein with 100% connections directed to intrinsic nodes (Fig. 7a); (2) directly binds to transmembrane desmosomal cadherins (including Dsg1 and Dsg3), cytoskeleton adaptor proteins and cytoskeleton keratins; (3) undergoes significant changes by microarray analysis in experimental PV (FC = −3.600083788; >85 percentile) (Fig. 7b); (4) was the only intrinsic protein in the high percentile bracket (≥85) for both number of connections and gene expression changes in PV, with the exception of keratin 18 (Fig. 7); (5) has a cellular location that makes it a candidate signalling molecule, although no data exist at present about its regulation/phosphorylation; and (6) has not been investigated previously. The other highly connected protein undergoing changes in PV, namely keratin 18, was not suitable for further study because it is known to be differentiation-specific and present only marginally in keratinocytes [29]. This apparent incongruence is due to the fact that microarray data have been obtained in mouse skin, whereas the experimental model of PV used in this study was based on human keratinocyte monolayers. Furthermore, keratin 18 executes complex molecular functions that go beyond cell adhesion, since deletion of the gene results in embryonic lethality in mice [30].
Effect of Pkp3 silencing in normal and perturbed desmosomes
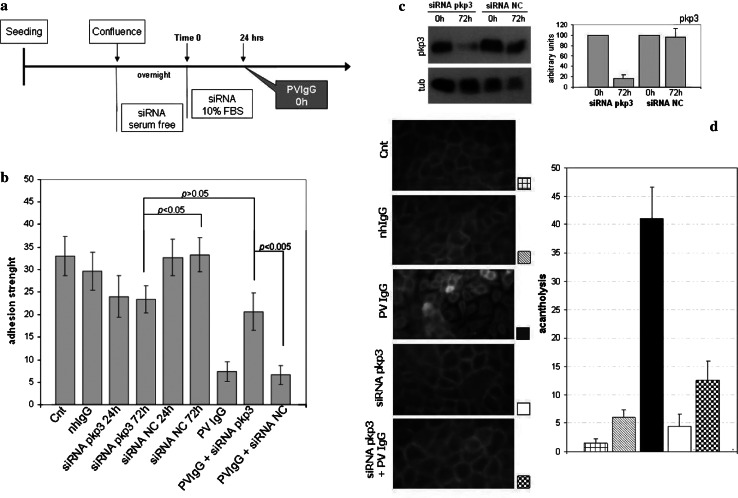
To test the importance of Pkp3 in the desmo-adhesome, we silenced Pkp3 and then evaluated changes in cell adhesion under basal and perturbed conditions. As a model of external perturbation, we used an in vitro model of PV, the best characterised muco-cutaneous disease of desmosomal adhesion. The expression of Pkp3 was decreased in HaCaT cells by siRNA interference and then the cells were exposed to PV autoantibodies (Fig. 8a). In functional assays of adhesion, cells treated with Pkp3 siRNA showed reduced adhesiveness (P < 0.05) 24 and 72 h after transfection compared with untreated controls or cells incubated with random non-targeting siRNAs (NC siRNA, Fig. 8b). In cells incubated with Pkp3 siRNA, the levels of Pkp3 were effectively reduced to approximately 80% after 24 h and did not change significantly at 72 h (Fig. 8c). Despite the reduction in cell–cell adhesion strength, keratinocytes containing Pkp3 siRNA were morphologically normal (Fig. 8d). The expression of other cell adhesion molecules such as Dsg1, Dsg2, Dsg3, desmocollin 2, desmoplakin, plakoglobin or E-cadherin were unchanged in Pkp3-siRNA transfected cells suggesting that the loss of cell adhesion under these conditions was not attributable to decreased expression of these desmosomal proteins (data not shown). Further, control siRNA and nhIgG did not exert any functional or morphologic effect on the HaCaT cells (Fig. 8b, d).
Fig. 8.
Pkp3-deficient cells are resistant to PV acantholysis. a Schematic representation of the experimental protocol. b Dispase-based assays were undertaken on keratinocytes treated as in (a). The ratio of total:single cells was expressed as a histogram. c Cells were treated with Pkp3 siRNAs or control siRNAs and the expression of Pkp3 was assessed by western blotting; tubulin expression was used as a control. d Keratinocytes were grown on glass coverslips and then treated with 0.5 mg nhIgG or PV2IgG for 48 h, with or without Pkp3 siRNAs, or left untreated. For quantification of acantholysis, cells were stained with Dsg3 (H-145 Ab) and processed for immunofluorescence. Five fields were examined with a Leica microscope and analysed for morphometric analysis of cell–cell detachment. Bars indicate standard deviation (SD) of triplicate experiments performed with PV1 and PV2 IgG
The treatment of HaCaT cells with PV IgG for 48 h dramatically disrupted cell–cell adhesion and induced acantholysis (Fig. 8b, d). Morphologically, PV IgG-treated cells were shrunken, rounded and detached (Fig. 8d) and there was a fivefold loss in the adhesive strength of the cell monolayer (Fig. 8b). Interestingly, loss of adhesion (Fig. 8b) and acantholysis (Fig. 8d), the hallmarks of PV in vitro, did not occur in the Pkp3-deficient cells treated with PVIgG for 48 h. Specifically, the adhesion strength of HaCaTs with Pkp3 knockdown remained unchanged (P > 0.05) after incubation with PV IgG (Fig. 8b) such that the adhesion strength was significantly higher than that of wild-type HaCaT exposed to PVIgG. Morphologically, Pkp3-silenced keratinocytes did not show acantholysis and cell–cell detachment after exposure to PV IgG (Fig. 8d). Thus, Pkp3-deficient cells are resistant to PV-induced acantholysis in vitro. The data demonstrate that whilst Pkp3 depletion reduces the desmosomal adhesive strength of keratinocytes, its presence is fundamental to mediate the pathobiological effects of PV IgG.
Discussion
In this study, we present the first characterisation of the desmosome network in keratinocytes at a systems level. We then experimentally challenged this network in the attempt to understand the behaviour of the desmo-adhesome under normal and perturbed conditions. Based on the predictions from an in silico analysis, we demonstrate for the first time that the adaptor protein Pkp3 plays a key role in desmosomal remodelling and, specifically, in the pathogenesis of PV in vitro.
Through data mining, we first used information derived from databases and published studies to address the molecular architecture, structure and signalling of the desmo-adhesome at multiple levels. Then, analysis of the desmosomal interactome was carried out in silico. On the basis of the network fragmentation analysis, we suggest that the desmosome is a heavy-tailed structure resistant to random failures but fragile when an attack occurs at highly connected nodes. Dissection of the network into distinct, yet mutually interacting, families highlighted the functional role of certain components and how they are regulated. Plakoglobin, for example, was the central connector of the desmosome and is the main target of regulation by the kinase machinery. Further, remodelling of desmosomal cadherin function is likely to occur via caspase 3-mediated cleavage. An important challenge for the future will be to define the relative contributions of disease-targeted nodes in the pathogenesis of diseases associated with desmosome adhesion. For example, a lack of desmoplakin might be expected to cause the dramatic changes in skin phenotype that are seen in palmoplantar keratoderma and skin fragility syndrome because of its high connectivity and topological role. Conversely, the distribution of connectivity of Dsg3 (low connectivity rate, above the 50 percentile) suggest that the desmo-adhesome would be unlikely to be sensitive to specific perturbation by anti-Dsg3 antibodies affecting exclusively the adhesion function of Dsg3, as proposed for PV [31]. Further studies are needed to determine the contribution of anti-Dsg3 IgG in disrupting the desmosomal structure and whether these changes proceed via Pkp3.
The desmosome network shows lower connectivity compared to the integrin adhesome (8.66 vs 4.41) and resilience to targeted attack [16]. Similarly, desmo-adhesome connectivity was slightly lower than that found in junctional complexes (tight junctions, adherent junctions and desmosomes) of simple epithelia (4.41 vs 5.85) [17]. Whilst these data are not fully comparable due to differences in the method of analysis and cell type, there is nonetheless some similarity between these systems principally due to the central topological role played by the adaptor proteins. The results of the present study indicate that the following principles apply to the desmo-adhesome. First, adaptors act like major hubs because they have high connectivity; almost all adhesome protein groups have connections with adaptors. Second, protein kinases regulate adaptors plus the protein groups bound to the adaptors; such kinases do not regulate membrane adhesion proteins (desmosomal cadherins). Third, most of the regulation is via kinases, as phosphatases represent only a minor part of the signalling molecules and their regulators have yet to be identified. Fourth, only Tyr phosphatases are present in the network and target, in the main, plakoglobin. And fifth, the majority of proteolytic activity is aimed at transmembrane adhesion molecules. It should be emphasised, however, that the results described herein are biased toward the molecules and pathways so far described in the literature. The advances in molecular techniques are likely to lead to the identification of novel PPIs and mechanisms or regulation of the desmosomal interactome in the near future.
The present analysis of the desmosome network is propaedeutic to the study of human diseases affecting the desmosome. Indeed, the practical contribution of systems biology to human disease is that it advocates new ways of interrogating cellular responses to stimuli and provides a framework for understanding molecular networks. In this study, we demonstrate the usefulness of this approach by predicting and verifying the role of Pkp3 in PV. In accord with our predictions, we show that keratinocytes lacking Pkp3 are less adhesive than wild-type cells. However, in Pkp3 knockdown keratinocytes, the gross architecture of cell–cell contact is not impaired suggesting that this protein may be a stabiliser of the desmosome in a similar way to pkp1 and that alternative partners, such as other Pkp family members or plakoglobin, can substitute Pkp3 in desmosomal adhesion. An important and unexpected finding of the present study was that the cohesion of Pkp3-deficient cells did not decrease in response to PV IgG. We speculate that this phenomenon is due to a signalling role that Pkp3 plays in acantholysis. It is conceivable that binding of PV IgG to Dsg3 induces conformational changes of the target antigen which, in turn, unmasks consensus sequences of Pkp3 causing its phosphorylation and/or detachment from Dsg3. Ongoing experiments in our laboratory suggest that this could be actually the case (unpublished observations).
Pkps are desmosmal plaque proteins which belong to the p120ctn subfamily of Armadillo repeat-containing proteins. Pkp1 is an essential desmosomal plaque molecule of the uppermost differentiated layers of stratified epithelium; skin fragility syndrome is seen in patients lacking PKP1 expression. PKP2 is found in basal cell layers of stratified epithelia and in single-layered epithelium, as well as in non-epithelial desmosomes [9]. Pkp3 and Pkp4 (p0071) are largely expressed in epithelial cells. In addition to being present at desmosomal plaques, Pkp3 is also found in cytoplasmic stress granules. Pkp3 interacts with a broad spectrum of desmosomal proteins including desmoplakin, plakoglobin, three desmogleins and several desmocollins (Fig. 1), thereby implicating Pkp3 in desmosome assembly and physiology. Although Pkp3 knockout mice are viable, they display severe defects in desmosome assembly in the epidermis and are extremely susceptible to skin infections and inflammation [32]. This demonstrates that Pkp3 is involved in control of skin inflammatory responses and suggests a more complex role than simply that of an adaptor protein of the desmosome with structural functions. Using microarray and PCR data, we and others have shown in a mouse model of PV and following treatment of human keratinocyte monolayers with PV IgG that a broad spectrum of adhesion molecules including desmoplakin, Dsgs, Pkps and many cytokeratins, are down-regulated [21, 33, 34]. In the mouse model of PV, changes in the desmo-adhesome nodes were unidirectional in terms of reduced expression. In particular, the desmosomal adaptor proteins were down-regulated with a linear (non-logarithmic) fold change between 2 (periplakin) and 27 (envoplakin) [21]. Significantly, the majority of changes affected the major adaptor proteins such as plakoglobin, desmoplakin and Pkp3 and this supports their central role in the regulation of the desmosomal scaffold. In the present study, we selected Pkp3 for further study and showed that it was required for PV acantholysis. The criterion for selecting Pkp3 as a candidate molecule in PV acantholysis was based on the finding that it falls above the 85 percentile, both in terms of connections and gene changes in experimental PV. This methodology, however, is based exclusively on DNA microarray data and is necessarily limited. Expression of the highly connected protein plakoglobin, for example, is not affected in experimental PV but it is essential for PV pathogenesis since plakoglobin −/− keratinocytes are resistant to cell–cell detachment induced by PV IgG [35]. Subsequent studies have demonstrated that the crucial role of plakoglobin in PV is due to its signalling functions which cannot be properly investigated by DNA microarray technology [36].
The goal of understanding the mechanism associated with PV acantholytic signalling may not be achieved without the use of high throughput techniques such as 2D-SDS-PAGE for the study of protein expression and protein phosphorylation. Changes in gene expression take place relatively slowly and reflect a stable response of the cell to external cues. By contrast, protein phosphorylation is one of the most rapid post-translational modifications that cells use to modulate protein activity, localisation and stability. In this regard, we and others have shown that regulation of protein phosphorylation and signalling molecules are the central mechanisms leading to cell–cell detachment in PV [22, 28, 37]. Since phosphorylation of desmosomal proteins is known to regulate the dynamic of desmosome assembly/disassembly to the sites of intercellular contact, we suggest that the next stage should be to clarify the post-translational modifications of the desmosomal protein network in PV. In this regard, we believe that the limits of the current interactome, built on a concept of direct PPIs, should be overcome by a more functional approach that allows the characterisation of wider regulatory networks. For instance, according to the systems level analysis reported in the literature and used here to develop the desmo-adhesome, a functional link between activation/blockade of keratinocyte cholinergic receptors and phosphorylation desmosomal components could not be demonstrated—yet it is of paramount importance in the homeostasis of cell adhesion and PV pathogenesis [38, 39]. Specifically, a recently characterised pathway suggests that a block of nicotinic receptors can release Src activity which, in turn, phosphorylates desmosomal and/or other cell adhesion proteins [40]. However, because desmosomal proteins directly interact with Src, but not with cholinergic receptors, the latter could not be included in the desmo-adhesome. Therefore, a network considering functional interactions in a desmosomal context should be developed in the future.
In conclusion, we demonstrate that an analysis of the desmosome structure at a systems level can lead to a better understanding of the pathogenesis of human disease. In accord with our predictions, we show for the first time that Pkp3 is required for PV acantholysis. We suggest that the process of combining systems biology with experimental verification is likely to have a broad impact on the study of keratinocyte adhesion in cell physiology and disease.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Schematic representation of the desmosome and skin disease related to failure of desmosomal components (Fig. S1, TIFF 428 kb)
The structural subnet, formed by 30 intrinsic proteins and 62 internal connections (Fig. S2, TIFF 112 kb)
References
- 1.Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends Microbiol. 2007;15:45–50. doi: 10.1016/j.tim.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Hess EL. Origins of molecular biology. Science. 1970;168:664–669. doi: 10.1126/science.168.3932.664. [DOI] [PubMed] [Google Scholar]
- 3.Almaas E. Biological impacts and context of network theory. J Exp Biol. 2007;210:1548–1558. doi: 10.1242/jeb.003731. [DOI] [PubMed] [Google Scholar]
- 4.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 5.Ramaswamy S. Rational design of cancer-drug combinations. N Engl J Med. 2007;357:299–300. doi: 10.1056/NEJMcibr072593. [DOI] [PubMed] [Google Scholar]
- 6.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson WJ. Regulation of cell–cell adhesion by the cadherin–catenin complex. Biochem Soc Trans. 2008;36:149–155. doi: 10.1042/BST0360149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angst BD, Marcozzi C, Magee AI. The cadherin superfamily: diversity in form and function. J Cell Sci. 2001;114:629–641. doi: 10.1242/jcs.114.4.629. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo N. Pathophysiology of the desmosome. Kerala, India: Research Signpost; 2009. [Google Scholar]
- 10.Garrod D, Chidgey M. Desmosome structure, composition and function. Biochim Biophys Acta. 2008;1778:572–587. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Miera A, Foshay K, Stewart A, Gallicano GI. Development of a sticky situation: the role of the desmosome in epidermal morphogenesis. In: Cirillo N, editor. Pathophysiology of the desmosome. Kerala: Research Signpost; 2009. pp. 33–61. [Google Scholar]
- 12.Payne AS, Hanakawa Y, Amagai M, Stanley JR. Desmosomes and disease: pemphigus and bullous impetigo. Curr Opin Cell Biol. 2004;16:536–543. doi: 10.1016/j.ceb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. J Invest Dermatol. 2007;127:2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- 14.Lanza A, Cirillo N, Femiano F, Gombos F. How does acantholysis occur in pemphigus vulgaris: a critical review. J Cutan Pathol. 2006;33:401–412. doi: 10.1111/j.0303-6987.2006.00523.x. [DOI] [PubMed] [Google Scholar]
- 15.Waschke J. The desmosome and pemphigus. Histochem Cell Biol. 2008;130:21–54. doi: 10.1007/s00418-008-0420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paris L, Bazzoni G. The protein interaction network of the epithelial junctional complex: a system-level analysis. Mol Biol Cell. 2008;19:5409–5421. doi: 10.1091/mbc.E08-05-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pieroni E, Bentem S, Mancosu G, Capobianco E, Hirt H, Fuente A. Protein networking: insights into global functional organization of proteomes. Proteomics. 2008;8:799–816. doi: 10.1002/pmic.200700767. [DOI] [PubMed] [Google Scholar]
- 20.Albert R, Jeong H, Barabasi AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 21.Lanza A, Cirillo N, Rossiello R, Rienzo M, Cutillo L, Casamassimi A, de Nigris F, Schiano C, Rossiello L, Femiano F, Gombos F, Napoli C. Evidence of key role of Cdk2 overexpression in pemphigus vulgaris. J Biol Chem. 2008;283:8736–8745. doi: 10.1074/jbc.M702186200. [DOI] [PubMed] [Google Scholar]
- 22.Cirillo N, Lanza M, De Rosa A, Femiano F, Gombos F, Lanza A. At least three phosphorylation events induced by pemphigus vulgaris sera are pathogenically involved in keratinocyte acantholysis. Int J Immunopathol Pharmacol. 2008;21:189–195. doi: 10.1177/039463200802100121. [DOI] [PubMed] [Google Scholar]
- 23.Cirillo N, Gombos F, Lanza A. Pemphigus vulgaris immunoglobulin G can recognize a 130 000 MW antigen other than desmoglein 3 on peripheral blood mononuclear cell surface. Immunology. 2007;121:377–382. doi: 10.1111/j.1365-2567.2007.02585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calautti E, Cabodi S, Stein PL, Hatzfeld M, Kedersha N, Dotto GP. Tyrosine phosphorylation and Src-family kinases control keratinocyte cell–cell adhesion. J Cell Biol. 1998;141:1449–1465. doi: 10.1083/jcb.141.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cirillo N, Lanza M, Femiano F, Gaeta GM, De Rosa A, Gombos F, Lanza A. If pemphigus vulgaris IgG are the cause of acantholysis, new IgG-independent mechanisms are the concause. J Cell Physiol. 2007;212:563–567. doi: 10.1002/jcp.21111. [DOI] [PubMed] [Google Scholar]
- 27.Cirillo N, Lanza M, De Rosa A, Cammarota M, La Gatta A, Gombos F, Lanza A. The most widespread desmosomal cadherin, desmoglein 2, is a novel target of caspase 3-mediated apoptotic machinery. J Cell Biochem. 2008;103:598–606. doi: 10.1002/jcb.21431. [DOI] [PubMed] [Google Scholar]
- 28.Lanza A, Santoro R, De Rosa A, Gaeta GM, Gombos F, Cirillo N. Inhibition of protein phosphorylation, but not synthesis nor lysosomal degradation, prevents keratinocyte adhesion loss induced by pemphigus vulgaris serum. J Stomatol Invest. 2007;1:25–32. [Google Scholar]
- 29.Bragulla HH, Homberger DG. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J Anat. 2009;214:516–559. doi: 10.1111/j.1469-7580.2009.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hesse M, Watson ED, Schwaluk T, Magin TM. Rescue of keratin 18/19 doubly deficient mice using aggregation with tetraploid embryos. Eur J Cell Biol. 2005;84:355–361. doi: 10.1016/j.ejcb.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Waschke J. Direct interference with desmoglein binding in pemphigus. J Stomatol Invest. 2008;2:53–55. [Google Scholar]
- 32.Sklyarova T, Bonné S, D’Hooge P, Denecker G, Goossens S, De Rycke R, Borgonie G, Bösl M, van Roy F, van Hengel J. Plakophilin-3-deficient mice develop hair coat abnormalities and are prone to cutaneous inflammation. J Invest Dermatol. 2008;128:1375–1385. doi: 10.1038/sj.jid.5701189. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen VT, Arredondo J, Chernyavsky AI, Kitajima Y, Pittelkow M, Grando SA. Pemphigus vulgaris IgG and methylprednisolone exhibit reciprocal effects on keratinocytes. J Biol Chem. 2004;279:2135–2146. doi: 10.1074/jbc.M309000200. [DOI] [PubMed] [Google Scholar]
- 34.Stellavato A, Cirillo N. Fate of desmoglein 3 in oral pemphigus vulgaris: an RT-PCR study of cell adhesion molecules. J Stomatol Invest. 2007;1:63–68. [Google Scholar]
- 35.Caldelari R, de Bruin A, Baumann D, Suter MM, Bierkamp C, Balmer V, Müller E. A central role for the armadillo protein plakoglobin in the autoimmune disease pemphigus vulgaris. J Cell Biol. 2001;153:823–834. doi: 10.1083/jcb.153.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson L, Raess NA, Caldelari R, Zakher A, de Bruin A, Posthaus H, Bolli R, Hunziker T, Suter MM, Müller EJ. Pemphigus vulgaris identifies plakoglobin as key suppressor of c-Myc in the skin. EMBO J. 2006;25:3298–3309. doi: 10.1038/sj.emboj.7601224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berkowitz P, Hu P, Warren S, Liu Z, Diaz LA, Rubenstein DS. p38MAPK inhibition prevents disease in pemphigus vulgaris mice. Proc Natl Acad Sci USA. 2006;103:12855–12860. doi: 10.1073/pnas.0602973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen VT, Chernyavsky AI, Arredondo J, Bercovich D, Orr-Urtreger A, Vetter DE, Wess J, Beaudet AL, Kitajima Y, Grando SA. Synergistic control of keratinocyte adhesion through muscarinic and nicotinic acetylcholine receptor subtypes. Exp Cell Res. 2004;294:534–549. doi: 10.1016/j.yexcr.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Chernyavsky AI, Arredondo J, Kitajima Y, Sato-Nagai M, Grando SA. Desmoglein versus non-desmoglein signaling in pemphigus acantholysis: characterization of novel signaling pathways downstream of pemphigus vulgaris antigens. J Biol Chem. 2007;282:13804–13812. doi: 10.1074/jbc.M611365200. [DOI] [PubMed] [Google Scholar]
- 40.Chernyavsky AI, Arredondo J, Piser T, Karlsson E, Grando SA. Differential coupling of M1 muscarinic and alpha7 nicotinic receptors to inhibition of pemphigus acantholysis. J Biol Chem. 2008;283:3401–3408. doi: 10.1074/jbc.M704956200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the desmosome and skin disease related to failure of desmosomal components (Fig. S1, TIFF 428 kb)
The structural subnet, formed by 30 intrinsic proteins and 62 internal connections (Fig. S2, TIFF 112 kb)