Abstract
Retroelements comprise a considerable fraction of eukaryotic genomes. Since their initial discovery by Barbara McClintock in maize DNA, retroelements have been found in genomes of almost all organisms. First considered as a “junk DNA” or genomic parasites, they were shown to influence genome functioning and to promote genetic innovations. For this reason, they were suggested as an important creative force in the genome evolution and adaptation of an organism to altered environmental conditions. In this review, we summarize the up-to-date knowledge of different ways of retroelement involvement in structural and functional evolution of genes and genomes, as well as the mechanisms generated by cells to control their retrotransposition.
Keywords: Retroelements, Genome rearrangements, Alteration of gene expression, Genome evolution, Cell defense
Introduction
The eukaryotic genome is a complex and dynamic structure. Only about 3% of the human genome are composed of protein-coding sequences in comparison with ~50% constituted by transposable elements (TEs). Transposable or mobile elements are DNA sequences able to jump into new locations within genomes [1]. They can reach very high copy numbers and represent the major fraction of the eukaryotic genomes. Since their initial discovery in 1956 by Barbara McClintock in maize DNA [2], mobile elements have been found in genomes of almost all organisms. They constitute more than 50% of the maize genome [3], 22% of the Drosophila genome [4], and 42% of human DNA [5]. Considered first as a “junk” DNA or genomic parasites, mobile elements are now suggested to be “functional genome reshapers”, able to alter gene expression and promote genome evolution [1, 6–8].
There are two major groups of mobile elements. Class II elements or DNA transposons comprise about 3% of the human genome and move by a so-called “cut and paste” mechanism. No currently active DNA transposons have been identified in mammals [6]. Class I representatives are called retroelements (REs). They move by a "copy and paste” mechanism involving reverse transcription of an RNA intermediate and insertion of its cDNA copy at a new position within the host genome. This review is focused on retroelements: their characteristics and involvement in the eukaryotic genome functioning.
General characteristics of retroelements
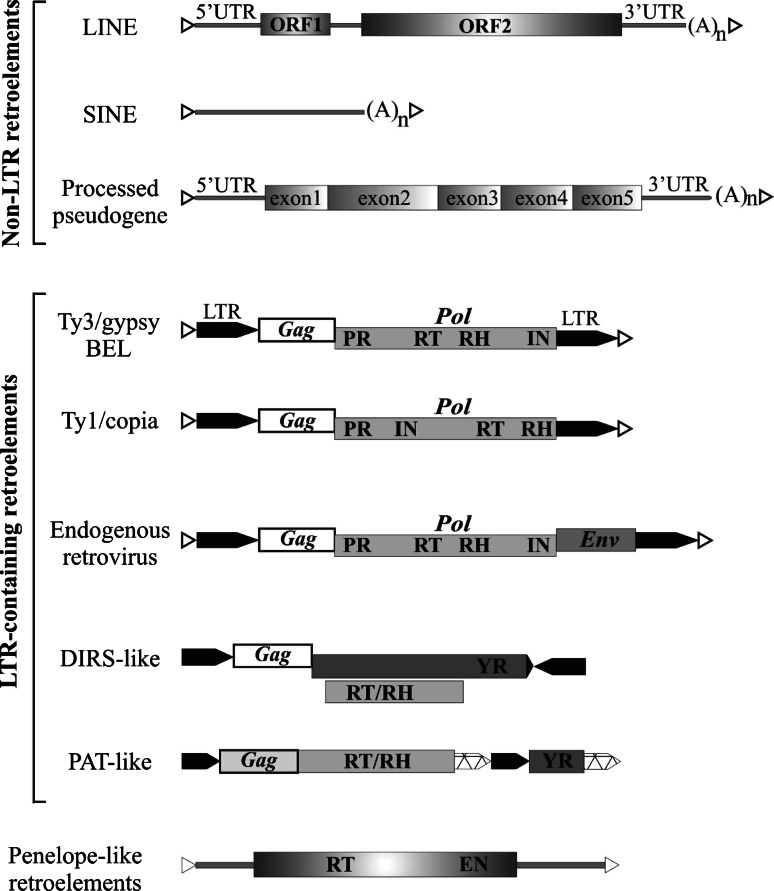
REs constitute about 90% of all transposable elements present in the human genome [9]. The main characteristic feature of REs is that their proliferation in the host genome is dependent on the process of reverse transcription. On the basis of presence or absence of long terminal repeats (LTRs), all retroelements can be divided into two major groups. The first group—LTR-containing retroelements—is represented by LTR retrotransposons, tyrosine recombinase retrotransposons, and endogenous retroviruses. The second group is called non-LTR retroelements, and the main representatives of this group are long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and processed pseudogenes (Fig. 1). Endogenous retroviruses and LINEs are referred to autonomous REs because they encode the proteins necessary for their proliferation and transposition. SINEs and processed pseudogenes are non-autonomous elements. They are suggested to transpose using LINE enzymatic machinery [10]. REs also include recently characterized in the Drosophila genome Penelope-like elements [11].
Fig. 1.
Schematic representation of different retroelements. White triangles short direct repeats (target site duplications), UTR untranslated region, ORF open reading frame, LTR long terminal repeat, PR protease, RT reverse transcriptase, RH ribonuclease H, IN integrase, Env envelope, YR tyrosine recombinase, EN endonuclease
Long interspersed nuclear elements (LINEs)
LINEs are autonomous non-LTR retrotransposons. They are widely distributed in eukaryotes. Nearly 17% of human DNA is occupied by LINEs [5]. However, they are mostly 5′-truncated inactive copies with only about 80–100 elements potentially capable of retrotransposition [12]. LINEs-1 (or L1s) are believed to be the only currently active autonomous transposable elements in human [13, 14].
The full-length human L1 is 6 kb long. It has a 900-nt-long 5′ untranslated region (UTR) that functions as an internal promoter for RNA polymerase II, two open reading frames (ORF1 and ORF2), a short 3′UTR, and a poly(A) tail. ORF1 encodes a nucleic acid binding protein that lacks sequence similarity with any other known protein [8, 15]. The ORF2 protein contains endonuclease (EN) and reverse transcriptase (RT) activities, as well as a Cys-rich domain, which are absolutely required for retrotransposition [16]. L1 integrations are usually flanked by short direct repeats, also called target site duplications.
L1 retrotransposition is thought to occur by target primed reverse transcription (TPRT) [17]. They can integrate at a very large number of sites in the genome, because their endonuclease preferentially cleaves DNA at a short consensus sequence 5′-TTTT/AA-3′ [18, 19]. The enzymatic machinery of a retrotransposition-competent L1 predominantly transposes its own copies; this phenomenon is usually called “cis-preference” of L1 transposition [20]. However, L1s are capable of transposing other sequences, mostly Alu retroposons, but also copies of different cellular RNAs, thus forming pseudogenes [21].
Apart from L1, mammalian genomes contain ancient and extinct LINE elements belonging to LINE2 and CR1/LINE3 families. They constitute about 3% and 0.38% (LINE2), and 0.3% and 0.05% (LINE3), of human and mouse DNA, respectively [22]. In spite of the low-copy number, their presence may be valuable for the host genome. For example, a LINE-2 fragment was shown to be a potent T-cell-specific silencer [23]
Short interspersed nuclear elements (SINEs)
SINEs comprise about 12% of the human genome. These elements are generally quite short (<500 bp) and do not code any proteins. Therefore, they need “exogenous” RT for their transposition. It is generally accepted that LINEs are used as a source of RT for SINE proliferation [24]. SINE sequences generally contain a poly(A) tail or, less frequently, another A-rich stretch on their 3′end. In the human genome, SINEs are represented by two main families: mammalian-wide interspersed repeats (MIR)—tRNA-like SINEs that constitute about 2% of the human genome; and Alu—7SL RNA-derived elements, comprising 10% of human DNA [5]. Alu elements are the most abundant repeats in the human genome. The major burst of Alu retroposition took place 50–60 million years ago and has since dropped to a frequency of one new retroposition for every 20–125 new births [25, 26].
Processed pseudogenes
Pseudogenes are sequences homologous to known genes which appeared due to reverse transcription of different cellular RNAs and subsequent insertion of the cDNA copy into the genome. These elements normally do not contain introns, have a poly(A) tail and are flanked by short direct repeats. Such pseudogenes are referred to as processed pseudogenes [27]. Generally there are 1–10 (in some cases up to 100) pseudogenes for each human gene [28]. It is believed that LINE RT is used for the formation of processed pseudogenes [21].
Long terminal repeat (LTR)-retrotransposons and endogenous retroviruses
This group combines diverse elements that possess LTRs in their sequence. LTRs contain multiple regulatory elements. All LTR-containing elements comprise about 8% of the human genome [9].
The consensus structure of LTR-retrotransposons is similar to that of retroviruses except for the absence of env (envelope) gene in most elements [29]. They contain gag gene, encoding for a structural protein with nucleic acid binding activity, and pol, which encodes polyprotein with protease, reverse transcriptase, RnaseH, and integrase activities. There are three major classes of LTR-retrotransposons in vertebrates: the Ty1/copia, Ty3/gypsy, and BEL families.
Endogenous retroviruses (ERVs) are believed to represent genomic traces of ancient germ-cell retroviral infections. They have been found in all vertebrate genomes and constitute about 1% of the human DNA [30, 31]. The env gene of ERVs confers the potential to spread between cells and individuals. However, most of the ERVs sequences have undergone extensive deletions and mutations, therefore becoming transpositionally deficient and transcriptionally silent [32]. Moreover, the majority of ERVs reside in the genome in the form of solitary LTRs, arisen most probably due to homologous recombination between two LTRs of a full-length element.
Another group of LTR-containing retroelements is represented by the so-called tyrosine-recombinase encoding retrotransposons (or YR-retrotransposons) [33]. These elements have structures quite distinct from the retroelements mentioned above. The most important difference is that YR-retrotransposons do not encode for integrase, but instead encode a tyrosine recombinase (YR). The first element of this group was identified in the slime mold, Dictyostelium discoideum, and was called DIRS [34]. Later on, related elements were found in the genomes of numerous fungi, plants, and animals [33]. All these elements could be divided into two basic groups: DIRS-like elements, flanked by inverted repeats and containing internal complementary region, and elements of the PAT and Ngaro type, having split direct repeats. The unusual structure of the terminal repeats of the YR-retrotransposons was suggested to be necessary for their replication via free circular intermediate [34, 35]. Site-specific recombination is believed to integrate the circle without formation of target-site duplications. The human genome contains a DNA sequence similar to a large fragment of a DIRS1-like recombinase gene. However, no full-length mammalian DIRS-like elements have been found to date.
Penelope-like elements
Penelope-like elements (PLEs) constitute a novel class of eukaryotic retroelements distinct from both non-LTR and LTR retrotransposons [11]. They were first discovered in Drosophila virilis as elements responsible for the hybrid dysgenesis syndrome, characterized by simultaneous mobilization of several unrelated TE families in the progeny of dysgenic crosses. PLEs were further found in genome databases of different eukaryotes. They have a rather complex and highly variable organization. These elements were shown to possess internal promoter [36] and contain one ORF, encoding for reverse transcriptase and endonuclease that differ from the corresponding proteins of LTR-containing and non-LTR retrotransposons [11]. The PLE endonuclease belong to the URI protein family, which includes, inter alia, a catalytic module of the GIY-YIG endonucleases of group I introns, as well as bacterial UvrC DNA repair proteins. The reverse transcriptase of PLEs mostly resembles the RT domain of telomerase. Both the RT and EN domains encoded by D. virilis Penelope are functionally active, but the mechanism of their transposition still remains a mystery.
Structural genomic changes caused by RE activity
Formation of new retrotransposons
RE integrations into the genome can cause multiple effects and, among them, they can lead to the formation of new REs, as in case of SVA elements. SVA is a composite element consisting of four parts: hexamer repeats (CCCTCT)n, Alu, 15-23 tandemly repeated sequences (VNTR), and SINE-R (SVA = SINE-R + VNTR + Alu) [37, 38]. The SVA family originated <25 million years ago and has increased to ~3,000 copies in the human genome [38]. The first SVA probably appeared in the genome due to the integration of several elements into the same genomic locus [37]. SVA elements are flanked by target site duplications, terminate in a poly(A) tail, and they are occasionally truncated and inverted during their integration into the genome. Therefore, they were suggested to represent non-autonomous retrotransposons that are mobilized by L1 encoded proteins in trans. SVA elements remain active in the human genome. To date, at least five diseases have been reported to be the result of SVA insertions [6, 38, 39] (Table 1).
Table 1.
Impact of retroelements on genome structure and functioning
| RE function | Examples |
|---|---|
| Structural genomic changes | |
| Formation of new retrotransposons | Formation of SVA [37], LTR-containing retrotransposons [40] and tRNA-derived SINEs [10] |
| Recombination events | Recombination between retroelements may cause various diseases [6, 44–46]. Human glycophorin gene family evolved through several duplication steps that involved recombination between Alu elements [41] |
| Transduction of 3′-flanking sequences | SVA-mediated transduction duplicated the entire AMAC gene three times in the human genome [54] |
| Formation of processed pseudogenes | Mouse PMSE2b [56] and PHGP [57] pseudogenes, TRIMCyp gene of owl monkey [58] |
| Template switch during reverse transcription | Formation of bipartite and tripartite chimeric elements in eukaryotic genomes [61–63] |
| cis-regulation of gene activity | |
| REs as promoters | LTRs cause placental-specific expression of CYP19 [71] and regulate transcription of NAIP gene [77]; LTR represent the only known promoter for the liver-specific BAAT gene [78] |
| REs as transcriptional enhancers | Expression of salivary amylase in humans is a result of HERV-E integration [82]; ERV9 LTR is an enhancer elements in the β-globin locus control region [83]; Alu sequence is a part of enhancer element of the human CD8 alpha gene [88] |
| REs as providers of novel splice sites | Muscle-specific inclusion of an Alu-derived exon in SEPN1 mRNA in humans [94]; generation of alternative VEGFR-3 transcript due to the use of a non-canonical acceptor splice site within LTR sequence [104] |
| REs as a sourse of new polyadenylation signals | HERV-F LTR may function as an alternative polyadenylation site for gene ZNF195 [110]; HERV-H LTRs are major polyadenylation signals for human HHLA2 and HHLA3 genes [111] |
| REs as transcriptional silencers | A part of Alu element is a transcriptional silencer of the human BRCA gene [117]; endogenous retroviral sequence RTVL-Ia may serve as a silencer of the human Hpr gene [120] |
| REs as antisense regulators of host gene transcription | Human-specific HERV-K LTRs generate antisense transcripts to SLC4A8 and IFT172 mRNAs [128] |
| REs as insulator elements | B2 SINE element located in the murine growth hormone locus serves as a boundary to block the influence of repressive chromatin modifications [134]; Drosophila LTR retrotransposon gypsy in the 5′ region of the gene yellow blocks the action of the upstream located enhancers and is responsible for the pigmentation of cuticula [137] |
| REs as regulators of translation | Alu and L1 segments in the 5′UTR of human ZNF177 gene modify gene expression on the protein level by decreasing translation efficiency [140] |
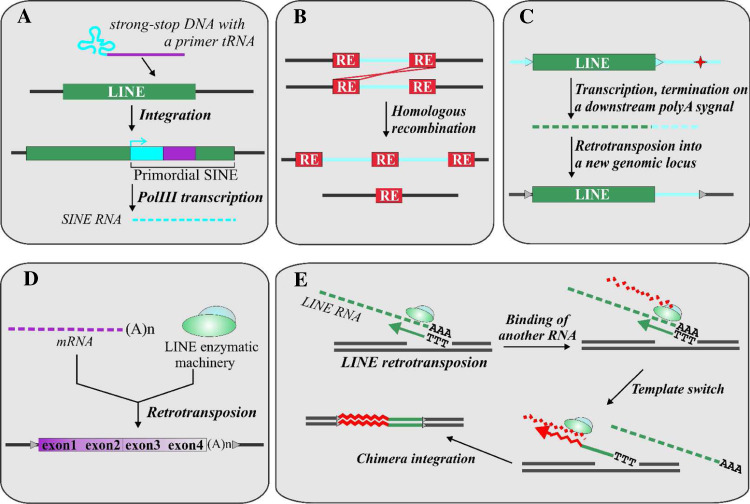
Moreover, it is speculated that LTR-containing retrotransposons and SINEs themselves represent chimeric elements [10, 40]. A phylogenetic analysis of the ribonuclease H domain revealed that LTR-containing retroelements might have been formed as a fusion between DNA transposon and non-LTR retrotransposon [40]. tRNA-derived SINEs likely descended from retroviral strong-stop DNAs [10]. They consist of two regions: a conservative one, including a tRNA promoter and a core domain, and a variable one similar to 3′-terminal sequence of different LINE families. The core domain of tRNA-like SINEs has conservative regions similar to fragments of lysine tRNA-primed retroviral LTRs. On the basis of these structural peculiarities, it was suggested that tRNA-derived SINEs emerged due to the integration of retroviral strong-stop DNA into the LINE 3′-terminal part (Fig. 2a). The RE formed could be transcribed by RNA polymerase III and spread through the genome. Such a mechanism of SINE formation could also explain how these elements can transpose in the genome. Namely, it seems very likely that they recruited the enzymatic machinery from LINEs through a common “tail” sequence [10].
Fig. 2.
a Formation of tRNA-like SINEs as an example of a role of REs in generation of new genetic elements; b retroelements as substrates for recombination events; c transduction of 3′-flanking sequences; d formation of processed pseudogenes by LINE enzymatic machinery; e generation of chimeric elements by template switches during LINE retrotransposition
REs and recombination events
Recombination is a powerful factor of evolution that produces genetic variability by using already existing blocks of biological information [41]. Because of their high copy number and sequence similarity, REs are the substrates for illegitimate homologous recombination, also called ectopic recombination (Fig. 2b). The chance that an ectopic recombination will occur depends on the number of homologous sequences and on the length of the elements [42, 43]. Recombination causes genetic rearrangements that can be deleterious, advantageous, or null.
There are numerous reported cases of human diseases caused by recombination between REs. For example, glycogen storage disease [44], Alport syndrome [45] as a result of recombination between L1 elements, and complete germ cell aplasia due to recombination between HERV-I [46]. Alu elements were implicated in almost 50 disease-causing recombination events [47, 48].
Apart from deleterious effects, recombination between REs can also have positive consequences. For example, human glycophorin gene family evolved through several duplication steps that involved recombination between Alu elements [41]. Furthermore, Alu-derived ectopic recombination generated 492 human-specific deletions, the distribution of which is biased toward gene-rich regions of the genome [49]. About 60% of Alu recombination-mediated deletions were shown to be located in genes and, in at least three cases, exons have been deleted in human genes relative to their chimpanzee orthologs. Finally, L1s were shown to join DNA breaks by inserting into the genome through an endonuclease-independent pathway, thus participating in DNA double-strand breaks repair [50]
Transduction of 3′-flanking sequences
The ability to transduce 3′-flanking DNA to new genomic loci was firstly shown for L1 elements [51–53] (Fig. 2c). L1s have a rather weak polyadenylation signal; therefore, RNA polymerase sometimes gets through it and terminates an RNA synthesis on any polyadenylation site located downstream. It was estimated that ~20% of all L1 inserts contain transduced DNA at the 3′ends. The length of these sequences varies from a small number of bases to over 1 kb. Taken together, such transduced DNA makes up ~0.6–1% of the human genome. Therefore, L1-mediated transductions have the potential to shuffle exons and regulatory sequences to new genomic sites.
Recently, it was shown that SVA elements are also able to transduce downstream sequence, and it was estimated that about 10% of human SVA elements were involved in DNA transduction events [38, 39]. Moreover, SVA-mediated transduction can serve as a previously uncharacterized mechanism for gene duplication and the creation of new gene families [54]. The authors identified one group of SVA elements that duplicated the entire AMAC (acyl-malonyl condensing enzyme 1) gene three times through SVA-mediated transduction events, which happened before the divergence of humans and African great apes. The three transduced AMAC copies contain intact ORFs in the human genome, and at least two of them are transcribed in different human tissues.
Formation of processed pseudogenes
Genomes of all higher eukaryotes contain pseudogenes. These elements normally do not contain introns, end in a poly(A) tail, and are flanked by short direct repeats. Such pseudogenes are referred to as processed pseudogenes [27] and are believed to be produced by the action of LINE retrotransposons [21] (Fig. 2d).
As long as RNA polymerase II-transcribed genes generally lack any promoter sequence in their RNA, processed pseudogenes were classically thought to be transcriptionally silent. Indeed, there were not so many reported cases of active pseudogenes that happened to integrate within an existing transcription unit and give rise to a novel gene or a novel transcriptional pattern of the existing ones. These include jingway element of Drosophila yakuba and D. melanogaster formed due to integration of alcohol dehydrogenase pseudogene into yellow-emperor gene [55], mouse PMSE2b retrogene inserted into the L1 sequence under the control of LINE promoter [56], mouse PHGP pseudogene, which is expressed from its 5′-adjacent sequence in a tissue-specific manner [57], TRIMCyp gene of owl monkey, formed by retrotransposition of cyclophilin A transcript to intron 7 of TRIM5 ubiquitin ligase and shown to confer HIV-1 resistance in owl monkey [58], and several others. However, recent genome-wide analysis of EST databases as well as transcriptional analyses of individual pseudogenes have revealed that up to a third of processed pseudogenes are transcribed, most of them specifically in testes [58, 59]. In humans, >1,000 pseudogene transcripts were detected, and the number of functionally active pseudogenes was estimated to be ~120 [59]. Interestingly, a striking predominance of autosomal retrogenes, which are copies of X-linked parental genes, was shown. These autosomal substitutes probably sustain essential functions during male X chromosome inactivation in the process of spermatogenesis [59].
Chimeric retrogene formation during reverse transcription
Apart from RE retrotransposition and formation of pseudogenes, reverse transcriptase (RT) is also able to change templates during cDNA synthesis. This feature of RT is well known for retroviruses. The RT jumps from one place of the template to another are necessary for the synthesis of retroviral LTRs [60].
Template switches can also occur during LINE-directed reverse transcription. Recently, our group has identified bipartite and tripartite chimeric retrogenes in three mammalian and one fungal genomes. A total of 82, 116, 66, and 31 elements were found in human, mouse, rat, and rice blast fungus Magnaporthe grisea DNAs, respectively [61–64]. These elements are composed of DNA copies of different cellular transcripts either directly fused to each other or more frequently fused to the 3′ part of a LINE retrotransposon. The various cellular transcripts found in these chimeras correspond to messenger RNAs, ribosomal RNAs, small nuclear RNAs, 7SL RNA, and Alu retroposon. The chimeras have the following common features: (1) 5′-parts are full-length copies of cellular RNAs, whereas 3′-parts are 5′-truncated copies of the corresponding RNAs (mostly LINEs); (2) both parts are directly joined with the same transcriptional orientation; (3) chimeras have a poly(A) tail at their 3′-end; and (4) chimeras are flanked by short direct repeats. The last structural feature demonstrates that these elements were transposed as bipartite DNA copies. The simultaneous integration of both parts of these chimeras was further supported by the data obtained from PCR-based multi-species insertion polymorphism assay [64]. We further suggested that the chimeras were formed by a template switch in the process of LINE reverse transcription (Fig. 2e). This suggestion was supported by the results obtained during the analysis of LINE retrotransposition in vitro and in vivo [65, 66]. The presence of structurally similar chimeric elements in evolutionary distinct organisms shows that template switching during LINE reverse transcription represents an evolutionary conserved mechanism of genome rearrangement. Moreover, many of the chimeras can be considered as new genes, as they were shown to be transcribed, some of them in a tissue-specific manner [62, 64].
Apart from generating chimeric retrogenes, template switches during LINE reverse transcription could give rise to chimeric SINE elements [67] and to mosaic rodent L1 structures [68, 69]. Evolution of certain LINE families might also involve change of a template during reverse transcription, resulting in the fusion of the 3′ part of a LINE to a new sequence, as suggested by the observation that the 5′-untranslated regions of human, mouse, rat, and rabbit L1 families are not homologous to each other [70].
cis-regulation of gene activity by retroelements
REs as promoters for a host gene transcription
Recent whole-genome analysis revealed that about 25% of all human promoters contain REs in their sequence [71]. Moreover, 7–10% of experimentally characterized transcription factor binding sites (TFBS) were shown to be derived from repetitive sequences including simple sequence repeats and transposable elements [72]. TFBS that originated from repeats evolve more rapidly than non-repetitive TFBS but still show signs of sequence conservation on functionally critical residues. Such rapidly evolving TFBS are likely to direct species-specific regulation of gene expression, thus participating in evolutionary processes (Fig. 3; Table 1).
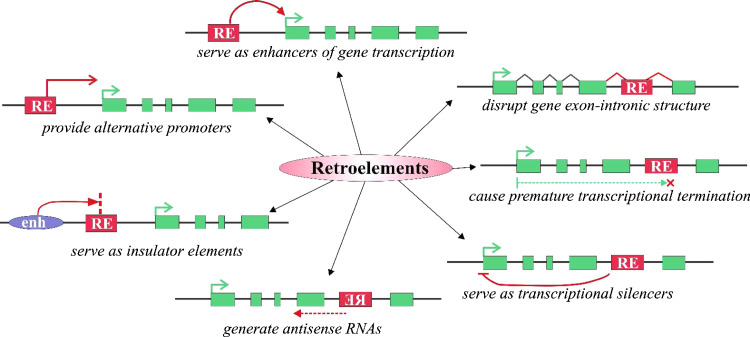
Fig. 3.
Different mechanisms of RE influence on gene transcription. Red boxes retroelements, green boxes gene exons, green arrow gene transcriptional start site, purple oval enhancer element
In the majority of examples reported to date, REs act as alternative promoters. REs can either influence the level of a corresponding RNA transcription or change the tissue-specificity of its expression. For example, LTR integration into CYP19 gene, encoding for aromatase P450, the key enzyme in estrogen biosynthesis, led to the formation of alternative promoter located 100 kb upstream of the coding region [71]. This event resulted in the primate-specific transcription of CYP19 in the syncytiotrophoblast layer of the placenta. Placental-specific expression might play an important role in controlling estrogen levels during pregnancy. Placental-specific transcription driven from endogenous retroviral promoters were also shown for Mid1 gene linked with inheritable Opitz syndrome [73], endothelin B receptor [74], and insulin-like growth factor INSL4 [75].
Solitary ERV-L LTR was shown to promote β3GAL-T5 transcription in various tissues, being especially active in colon, where it is responsible for the majority of gene transcripts [76]. β3GAL-T5 is involved in the synthesis of type 1 carbohydrate chains in gastrointestinal and pancreatic tissues. Interestingly, murine β3GAL-T5 gene is also expressed primarily in colon, despite the absence of an orthologous LTR in the mouse genome. It is likely that in humans the LTR adopted the function of an ancestral mammalian promoter active in colon [76]. An interesting example of gene transcriptional regulation by LTR was shown for NAIP (BIRC1) gene coding for neuronal apoptosis inhibitory protein [77]. Although human and rodent NAIP promoter regions share no similarity, in both cases LTR serve as an alternative promoter. Thus, two different LTR retrotransposons were recruited independently in primate and rodent genomes for the gene transcriptional regulation.
REs may also represent the only known promoter for some human genes. For example, the only apparent promoter of the liver-specific BAAT gene recently implicated in familial hypercholanemia is an ancient LTR in human but not in mouse [78]. Antisense L1 and Alu sequences were shown to act as the only known promoter for HYAL-4 gene, necessary for hyaluronan catabolism [71].
The application of novel high-throughput techniques such as cap analysis of gene expression (CAGE) and paired-end ditag (PET) sequencing revealed 51197 ERV-derived promoter sequences. In 1,743 cases, ERVs were located in gene proximal or 5′ untranslated regions. Also, 114 ERV-derived transcription start sites can be demonstrated to drive transcription of 97 human genes, producing chimeric transcripts initiated within LTR and read-through into known gene sequences [79].
Recently, we developed a technique called genomic repeat expression monitor (GREM) that can be applied to genome-wide isolation and quantitative analysis of any kind of transcriptionally active repetitive elements [80]. With the help of this technique, we have found that at least 50% of human-specific HERV-K LTRs possess promoter activity, and the level of their expression ranges from ~0.001 to ~3% of the beta-actin gene transcriptional level [81]. We have also shown that 5′-proviral LTR is more transcriptionally active than 3′-proviral or solitary LTRs and that the relative content of promoter-active LTRs in gene-rich regions is significantly higher than that in gene-poor loci.
REs as transcriptional enhancers for cellular genes
One of the first striking reports of the involvement of REs in tissue-specific gene transcriptional regulation was for the human amylase locus [82]. In humans, amylase is produced in pancreas and in salivary glands. Human amylase locus includes two genes of pancreatic amylase (AMY2A and AMY2B) and three genes of salivary amylase (AMY1A, AMY1B, AMY1C). The latter three genes are likely products of a recent triplication, because in the chimpanzee genome there is only one gene for AMY1. Exon-intronic structures of these genes are identical, except for an additional untranslated exon at the 5′ terminus of the salivary amylase genes. Moreover, all genes for salivary amylase contain a full-length insert of HERV-E upstream of their transcription start site. It was hypothesized that the insertion of full length endogenous retrovirus activated a cryptic promoter that drives the transcription of amylase in salivary glands. When there is a solitary LTR instead of full-length HERV-E provirus, cryptic promoter cannot be activated and the gene is expressed only in pancreas.
There are several other well-supported examples of LTR involvement in gene regulation. For example, the ERV9 LTR element upstream of the DNase I hypersensitive site 5 (HS5) of the locus control region in the human β-globin cluster might be responsible for controlling expression of this cluster in erythroid cells [83]. It was suggested that the enhancer effect might be caused by LTR-initiated transcription driven in the direction of associated gene promoter [84, 85]. Another example is the mouse Slp (sex-limited protein) gene. Endogenous retrovirus located upstream of the Slp in antisense orientation was shown to direct androgen specific expression of this gene in males [86].
LINEs and SINEs can also serve as transcriptional enhancers. The enhancer of human apoliprotein A was shown to reside within LINE element [87]. Alu sequence is a part of enhancer element located in the last intron of the human CD8 alpha gene [88]. Expression of this gene is restricted to cells of lymphoid lineage and is developmentally regulated during thymopoesis. A CORE-SINE retroelement (ancient tRNA-derived SINE with a conserved core sequence) was found to represent a neuronal enhancer for the POMC (proopiomelanocortin) gene [89]. POMC encodes a prohormone that gives rise to several bioactive peptides that participate in the stress response, skin and hair pigmentation, analgesia, the regulation of food intake, and energy balance. CORE-SINE was shown to be responsible for the expression of POMC in ventral hypothalamic neurons. Recently, AmnSINEs (a new SINE family identified in the genomes of Amniota) have been shown to act as distal transcriptional enhancers for FGF8 (fibroblast growth factor 8) and SATB2 genes in developing mouse forebrain [90].
REs as providers of novel splice sites for the host genes
Apart from the modulation of transcription, retroelements can also regulate splicing of pre-mRNA. An outstanding role here belongs to SINE elements, namely Alu retrotransposons in case of human transcriptome. It is current opinion that it is the exonization of Alu elements that plays a crucial role in birth of new exons in primate genomes [91, 92]. Alu elements have several sequence motifs resembling consensus splice sites in both sense and antisense orientations [93]. Younger Alu-derived exons have weaker splice-sites and lower absolute values for the relative abundance of putative splicing regulators between exonic and adjacent intronic regions. This relative abundance was shown to increase with exon age [91]. There is an excess of Alu-derived internal exons in the 5′-untranslated regions (UTRs) of the genes as compared to the 3′-UTRs. This phenomenon likely reflects stronger purifying selection pressure against exon creation in 3′-UTR because such exons may trigger mRNA nonsense-mediated decay [92]. In addition, there is an ‘exclusion zone’ in intron sequences flanking exons, where insertion of Alu elements is presumably under purifying selection [94].
Overall, Alu-derived exons had significantly weaker splicing signals compared to non-repetitive constitutively and alternatively spliced exons, most probably due to a lower density of exonic splicing regulatory elements. However, at least six Alu-containing exons (in genes FAM55C, NLRP1, ZNF611, ADAL, RPP38, and RSPH10B) are constitutively spliced in human tissues [92, 95]. In addition, Alu sequence provided a donor splice site to one of the constitutive exons of the human gene encoding survivin (a member of the apoptosis inhibitor family that is overexpressed in many malignancies) [96]. Furthermore, using exon array data of 330 Alu-derived exons in 11 human tissues and detailed RT-PCR analyses of 38 exons, it has been demonstrated that some Alu-derived exons are constitutively spliced in a broad range of human tissues, and may display strong tissue-specific switch in their transcript inclusion levels. Most of these latter exons were derived from ancient Alu elements in the genome [92]. This is probably due to the fact that exons derived from older Alu elements had more evolutionary time to accumulate nucleotide substitutions that strengthened exon inclusion in the transcript products.
Interestingly, Alu exonization might have played a role in human speciation. For example, there is a muscle-specific inclusion of an Alu-derived exon in mRNA of gene SEPN1 (gene implicated in a form of congenital muscular dystrophy), which appeared due to a human-specific splicing change after the divergence of humans and chimpanzees [94]. Some novel polymorphic Alu inserts interfere with the normal pre-mRNA splicing by providing additional splicing enhancers, thus causing inheritable diseases [97]. Importantly, it was shown that more Alus flank alternatively spliced exons than constitutively spliced ones. This implies that Alu insertions may change the mode of splicing of the flanking exons [94].
LINE elements may also be involved in the splicing of cellular RNAs, although with relatively lower frequencies. Proportion of L1 elements in gene introns is significantly lower than the one of Alu repeats, although both retrotransposons utilize the same retrotranspositional mechanism [98]. This bias is probably due to a purifying selection acting against accumulation of L1s in genes [99]. Interestingly, an increased ratio of constitutively spliced L1s relatively to alternatively spliced ones has been reported compared to Alu elements. Mammalian L1s contain numerous functional internal splice sites that generate a variety of processed L1 transcripts (most probably useless for L1 retrotransposition) and also contribute to the generation of hybrid transcripts between L1 elements and host genes. Interestingly, L1 splicing is delayed during the course of L1 expression [100]. This delay may serve to protect host genes from the excessive burden of L1 interference with their normal expression via aberrant splicing. In other vertebrate genomes, LINEs have also been reported to generate new chimeric spliced mRNA variants for the host functional genes, e.g., in zebrafish [101] or in pig cells [102].
LTR retrotransposons also may contribute to a diversity of alternatively spliced RNAs [103]. For example, two different isoforms of the human endothelial angeogenesis controlling receptor are encoded by the same gene VEGFR-3/FLT4. Polypeptide encoded by the shorter transcript lacks 65 C-terminal aminoacids. The short VEGFR-3 transcript is formed because of the use of a non-canonical acceptor splice site within the endogenous retroviral sequence located between exons 1 and 2. These different forms of VEGFR-3 gene product probably have different biological functions [104].
Recently, it has been proposed that intronic repeats may also affect gene exon-intronic structure through RNA editing mechanism by adenosine deaminase acting on RNA (ADAR) enzymes (reviewed in [105]). ADARs convert adenosine to inosine in double-stranded RNA substrates. An inosine is interpreted by the splicing machinery as guanosine. A → I RNA editing could therefore create or delete splice donor and acceptor sites. It was discovered that the most frequent targets of A → I RNA editing are dsRNAs that are formed from inverted Alu located in introns and UTRs of mRNAs. Several examples of exclusion and inclusion of the Alu exon due to editing of the Alu dsRNA have been identified through the analysis of human cDNA sequences [106].
REs as a sources of new polyadenylation signals
mRNA polyadenylation is an essential step for the maturation of almost all eukaryotic mRNAs. A polyadenylation signal (AAUAAA) nearby the 3′ end of pre-mRNA is required for poly(A) synthesis. The protein complex involved in the pre-mRNA polyadenylation is coupled with RNA polymerase II during the transcription of a gene, and only RNA polymerase II—products are polyadenylated with the remarkable exception of two polyadenylated polymerase III—transcribed RNAs [107]. Autonomous retrotransposons encode proteins necessary for their transposition and utilize functional poly(A) signals at the 3′-termini of their genes. Therefore, insertions of these elements in genes in the sense orientation can influence the expression of neighboring genes by providing new poly(A) signals. This is probably the right explanation for the clearly seen strong negative selective pressure on such elements oriented in the same transcriptional direction as the enclosing gene [103, 108].
There are numerous reported cases of transcriptional termination on RE polyadenylation signal. For example, in breast cancer cell line T47D, four mRNAs polyadenylated at the sequence of HERV-K retroviral LTR were identified [109]. 5′ LTR of the retrovirus HERV-F may function as an alternative polyadenylation site for gene ZNF195 [110]. Human genes HHLA2 and HHLA3 utilize HERV-H LTRs as the major polyadenylation signals. In the baboon genome, orthologous loci lack retroviral inserts and these genes recruit other polyadenylation sites [111].
Recently, it was shown that, apart from the main polyadenyaltion signal in the 3′UTR, L1s possess an additional transcription termination site located in the 3′ part of ORF2 in antisense orientation [112, 113]. Therefore, regardless of its orientation in a gene, L1s have the potential to produce a truncated RNA and thus a new mRNA isoform.
Of the 1.1 million human Alu retrotransposons, about 10,000 are inserted in the 3′ untranslated regions of protein-coding genes and 1% of these (107 events) are active as poly(A) sites [114]. Alu inserts usually represent weak or cryptic poly(A) signals, but they may as well constitute the major or the unique poly(A) site in a gene. Strikingly, although Alus in 3′ UTRs are equally inserted in the forward or reverse direction, 99% of polyadenylation-active Alu sequences are forward oriented [114].
Recently, it was estimated that ~8% of all poly(A) sites are associated with TEs [115]. Interestingly, human poly(A) sites that are not conserved in mouse were found to be associated with TEs to a much greater extent than the conserved ones. This result suggests the involvement of TEs in creation or modulation of poly(A) sites in evolution.
REs as transcriptional silencers
Some retrotransposons are known to function as transcriptional silencers downregulating transcription of the enclosing genes. For example, 1 out of 44 Alu repeats located in human GH locus, encoding for human growth hormones, harbors a regulatory element that most probably acts by decreasing the rate of promoter-associated histone acetylation resulting in a significant decrease of RNA polymerase II recruitment to the promoter. This silencer likely provides for regulatory control of hGH gene expression in pituitary cells [116].
Expression of the tumor suppressor protein BRCA2 is tightly regulated throughout development. Sharan et al. [117] identified a transcriptional silencer at the distal part of the human BRCA2 gene promoter. This silencer was involved in the tissue-specific negative regulation of BRCA2 expression in breast cell lines. The former mapped 221-bp-long silencer region was also a part of a full-length Alu element.
Another example is the transcriptional regulation of a human gene Hpr for haptoglobin-related protein. Hpr sequence is 92% identical to haptoglobin gene HP [118]. Both genes are transcribed at the highest levels in liver. Hpr promoter is stronger than HP promoter, but the concentration of Hpr liver transcripts is ~17-fold lower than that of HP mRNA [119]. The major distinction between these genes is the endogenous retroviral sequence RTVL-Ia in the intron of Hpr [120]. RTVL-Ia fragment has demonstrated significant silencer activity in a series of luciferase transient transfection experiments [119]. The mechanism of the negative Hpr regulation by the RTVL-Ia endogenous retrovirus is not clear, but the authors propose that this effect is due to an aberrant splicing of the Hpr transcript with the retroviral sequences.
REs as antisense regulators of the host gene transcription
It has been demonstrated that retrotransposons in gene introns are preferentially fixed in antisense to the enclosing gene orientation [103, 121]. Therefore, promoters of intronic retrotransposons may drive transcription of RNAs that are complementary to gene introns and/or exons. Moreover, some retrotransposons are also known to possess bidirectional promoter [122–125], and even downstream insertions of these elements relative to genes may result in production of an antisense RNA. These complementary RNAs may alter functional host gene expression. Retroposition likely accounts for the origin of a significant number of functional sense–antisense pairs in eukaryotic genomes [126]. Moreover, recently applied cap analysis of gene expression (CAGE) technique identified 48,718 human gene antisense transcriptional start sites within transposable elements [127].
Recently, we found the first evidence for the human-specific antisense regulation of gene expression occurring due to promoter activity of HERV-K (HML-2) endogenous retroviral inserts [128]. It was found that human-specific LTRs located in the introns of genes SLC4A8 (for sodium bicarbonate cotransporter) and IFT172 (for intraflagellar transport protein 172) in vivo generate transcripts that are complementary to exons within the corresponding mRNAs in a variety of human tissues. Overexpression of antisense transcripts resulted in 3.9-fold decrease in SLC4A8 and 2.9-fold decrease in IFT172 mRNA level. It is interesting to mention that about 34% of human-specific TE insertions are located within known genes. These insertions represent a form of species-specific genetic variation and may have contributed to the evolutionary process. Similarly to LTRs in SLC4A8 and IFT172 genes, intronically located representatives of an LTR retrotransposon family from rice genome called Dasheng likely regulate tissue-specific expression of several adjacent functional genes via antisense transcripts driven by the LTRs [129].
Smalheiser and Torvik [130] showed that a few mammalian microRNA precursors are derived from intronic insertion of two adjacent LINE retrotransposons in opposite orientation. Some other elements have an intrinsic hairpin structure and/or serve as microRNA precursors when inserted into transcriptionally active genomic regions [131, 132]. In order to catalogue the data on transposable elements that may have an impact on gene regulation and functioning, a comprehensive database termed “TranspoGene database” has been constructed that covers genomes of seven species: human, mouse, chicken, zebrafish, fruit fly, nematode, and sea squirt [133]. A variant of this database termed “microTranspoGene” collects data on human, mouse, zebrafish, and nematode TE-derived microRNAs [133].
REs as insulator elements
The temporal and spatial regulation of gene expression is linked to the establishment of functional chromatin domains. Several lines of evidence have been provided recently that retrotransposons can serve in vivo as insulator sequences that distinguish blocks of active and transcriptionally silent chromatin. For example, a B2 SINE element located in the murine growth hormone locus is required for the correct spatio-temporal activation of that gene. This repeat serves as a boundary to block the influence of repressive chromatin modifications by generating short transcripts, which are necessary and sufficient to enable gene activation [134]. Mammalian LINE elements are frequently found within matrix attachment regions (MARs) [135, 136]. Some Drosophila LTR retrotransposons have insulator activity and may block the activity of transcriptional enhancer elements when located between enhancer and promoter [137–139]. For example, in some fruitfly lineages, there is an insert of LTR retrotransposon gypsy into the 5′ region of the gene yellow that is responsible for the pigmentation of cuticula. Upstream of the gypsy element, there are two enhancer elements that account for the transcription of yellow in different tissues; another enhancer that is responsible for the yellow expression in cilia is located downstream. In the lineage y 2, gypsy insertion between the promoter and two upstream enhancers blocks these enhancers and downregulates yellow in the corresponding tissues, but the yellow expression in cilia remains unaffected [137].
REs as regulators of translation
Although retroelements have been found in UTRs of many functional cellular genes, the effect of their presence on the translational regulation of gene expression is still poorly investigated. Among the few known examples, there is human zinc-finger gene ZNF177, which incorporates Alu and L1 segments into the 5′ UTR of transcripts. The presence of the Alu and L1 segments which form one 5′ UTR exon modifies gene expression on the protein level by decreasing translation efficiency. Interestingly, the same Alu and L1 repeats in the 5′ UTR of ZNF177 exert a positive transcriptional enhancer effect, but repress translation [140]. Approximately 4% of human 5′ UTRs harbor Alu sequences, indicating that the expression of many genes might be influenced by Alu repeats [140]. In the mouse genome, there is a SINE retrotransposon-derived gene for neuronal dendrite-specific BC1 RNA. This small, non-protein coding RNA is thought to somehow regulate translation in dendritic microdomains. However, the mechanism of such a regulation remains a mystery, and further efforts are needed to investigate this phenomenon [141].
Cell defense mechanisms against RE proliferation
Given the various deleterious effects of REs on genome structure and functioning, it is not surprising that the cell has generated multiple mechanisms controlling their proliferation. These mechanisms include methylation of RE sequences, RNAi-based silencing, accumulation of RE’s proteins in stress granules, and nucleic acid editing.
Epigenetic modifications controlling the activity of transposable elements were first reported more than 20 years ago [142]. Since then, many genes involved in epigenetic silencing of TEs (including DNA methyltransferases and demethylases, histone modifying enzymes, chromatin remodeling enzymes, and genes involved in small RNA metabolism) were characterized. Defects in different components of silencing mechanisms were shown to increase transposition events [143].
Most of the methylated cytosines in mammalian genome reside in repetitive elements, and it has been proposed that DNA methylation evolved primarily to suppress the activity of transposable elements and to protect the host cell [144]. Hypomethylation of REs was demonstrated to be associated with genomic instability in cancer [145].
On the one hand, chromatin condensation may suppress the activity of REs. On the other hand, DNA methylation, initiated within RE, may spread to the surrounding regions and, therefore, suppress their functional activity. Methylation spreading from SINE into flanking genomic regions was suggested to create distal epigenetic modifications in plants [146]. Human Alu elements were proposed as potential de novo methylation centers implicated in tumor suppressor gene silencing in neoplasia [147]. Moreover, L1 elements may play a role in X-chromosome inactivation. It was suggested that they are the “boosters” which promote the spread of inactivation [148].
Recent studies have shown the involvement of RNAi-related mechanism in the control of TE activities, in particular in DNA-methylation of TE sequences and in the formation of heterochromatin. Plants, yeasts, and animals use different strategies to detect transposons and to generate small RNAs against them (reviewed in [149, 150]). In Drosophila and vertebrate germ lines, TEs are silenced with the help of Argonaute proteins of the Piwi family and a class of short RNAs called Piwi-interacting RNAs (piRNAs) [151, 152]. piRNAs are 24–30 nt long and, unlike siRNAs or miRNAs, they originate from a single-stranded precursor without the involvement of a Dicer protein. A subset of piRNAs primarily derived from transposons and other repeated sequences is called repeat-associated small interfering RNAs (rasiRNAs) [153]. They are suggested to participate in the formation of heterochromatin enriched with repetitive elements, but the mechanism of their action remains obscure.
Moreover, miRNAs and siRNAs may also participate in TEs silencing. Four miRNAs have been shown to target MIR/LINE-2 elements, and almost 30 human miRNAs exhibit typical short-seed complementarity with a specific region within Alu [130, 154]. Furthermore, it was shown that double-stranded L1 RNA generated in vitro can be converted into functional siRNAs by Dicer and that these siRNAs suppress L1 retrotransposition in cell culture assay [155, 156]
Another possible mechanism of cell defense against REs proliferation is the sequestration of L1 RNPs in stress granules (SG), which are discrete cytoplasmic aggregates formed in response to a range of stress conditions. It was recently demonstrated that under stress conditions ORF1p cosegregates with a large pool of mRNA in SGs [15]. Considering the cis preference of ORF1p for binding its own RNA, the authors suggested that L1 RNA is also present in SG. By targeting L1 RNP to SGs, the cell could reduce the number of L1 or SINE transposition, as well as the number of newly formed processed pseudogenes.
REs activity could also be controlled by nucleic acid editing through the action of APOBEC3 proteins (reviewed in [157]). APOBEC3 proteins belong to a family of cytosine deaminase. In primates, there are seven APOBEC genes compared with only one gene in mouse. In cell culture experiments, it was shown that hA3A, hA3B, hA3C, and hA3F proteins are the major inhibitory factors of L1 retrotransposition; Alu mobilization was inhibited by hA3A, hA3B, and hA3G. It is not known whether the inhibitory effect is caused by DNA-editing, and the mechanism of APOBEC3 action on REs remains to be revealed. The observation that the decline of retrotransposition activity in primates coincided with the expansion of APOBEC3 gene cluster suggests an important role of APOBEC3 in the host genome protection.
Conclusion
As was mentioned at the beginning of this paper, retroelements occupy a large portion of essentially all eukaryotic genomes. However, their regulatory potential has been studied insufficiently even for the sequenced genomes of such important model organisms like mouse, rat, the plant Arabidopsis, the fruitfly Drosophila, and the worm Caenorhabditis elegans, and also for human DNA. Only a few elements have been investigated in detail, whereas hundreds of thousands of other retroelements are awaiting further studies. Of course, the examples that were given in this review are rather fragmentary and cannot give an integral picture of the functional interplay between the host genomes and their numerous selfish inhabitors. However, these examples clearly demonstrate that retroelements may be an important creative force in genome evolution and in the adaptation of an organism to altered environmental conditions. Most probably, the majority of retroelements are silent and do not influence cell functioning. Nevertheless, the authors of this review believe that in few years they will be able to mention many new examples of retrotransposons playing important roles in the regulation of gene expression, and that the number of these examples will be growing exponentially.
Massive sequencing of genomic DNAs opened a new so-called “post-genomic” era in molecular biology, genetics, and biomedicine. Functional characterization of genomes, high-throughput identification of important regulatory regions, will be the next step. It is our belief that the extensive studying of gene-regulatory potential of transposable elements is a prerequisite of solving this difficult problem.
Acknowledgments
The authors were supported by the Molecular and Cellular Biology Program of the Presidium of the Russian Academy of Sciences, by the grant of the President of the Russian Federation and by the grant 08-04-00720-a and 09-04-12302 from the Russian Foundation for Basic Research. We apologize to authors whose primary references have not been cited due to space limitations.
References
- 1.Bohne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008;16:203–215. doi: 10.1007/s10577-007-1202-6. [DOI] [PubMed] [Google Scholar]
- 2.McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 3.Wessler SR. Transposable elements and the evolution of gene expression. Symp Soc Exp Biol. 1998;51:115–122. [PubMed] [Google Scholar]
- 4.Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci USA. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 7.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 9.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101(suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohshima K, Hamada M, Terai Y, Okada N. The 3′ ends of tRNA-derived short interspersed repetitive elements are derived from the 3′ ends of long interspersed repetitive elements. Mol Cell Biol. 1996;16:3756–3764. doi: 10.1128/mcb.16.7.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evgen’ev MB, Arkhipova IR. Penelope-like elements—a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet Genome Res. 2005;110:510–521. doi: 10.1159/000084984. [DOI] [PubMed] [Google Scholar]
- 12.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 14.Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 15.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/S0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 17.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 18.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/S0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 19.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 22.Gentles AJ, Wakefield MJ, Kohany O, Gu W, Batzer MA, Pollock DD, Jurka J. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica . Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnelly SR, Hawkins TE, Moss SE. A conserved nuclear element with a role in mammalian gene regulation. Hum Mol Genet. 1999;8:1723–1728. doi: 10.1093/hmg/8.9.1723. [DOI] [PubMed] [Google Scholar]
- 24.Eickbush TH. Transposing without ends: the non-LTR retrotransposable elements. New Biol. 1992;4:430–440. [PubMed] [Google Scholar]
- 25.Shen MR, Batzer MA, Deininger PL. Evolution of the master Alu gene(s) J Mol Evol. 1991;33:311–320. doi: 10.1007/BF02102862. [DOI] [PubMed] [Google Scholar]
- 26.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 28.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/S0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 29.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leib-Mosch C, Seifarth W. Evolution and biological significance of human retroelements. Virus Genes. 1995;11:133–145. doi: 10.1007/BF01728654. [DOI] [PubMed] [Google Scholar]
- 31.Sverdlov ED. Retroviruses and primate evolution. Bioessays. 2000;22:161–171. doi: 10.1002/(SICI)1521-1878(200002)22:2<161::AID-BIES7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 32.Mager D, Medstrand P (2002) Retroviral repeat sequences. In: Gardiner K (ed) Encyclopedia of the human genome. Nature Publishing, London
- 33.Poulter RT, Goodwin TJ. DIRS-1 and the other tyrosine recombinase retrotransposons. Cytogenet Genome Res. 2005;110:575–588. doi: 10.1159/000084991. [DOI] [PubMed] [Google Scholar]
- 34.Cappello J, Handelsman K, Lodish HF. Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell. 1985;43:105–115. doi: 10.1016/0092-8674(85)90016-9. [DOI] [PubMed] [Google Scholar]
- 35.Goodwin TJ, Poulter RT. A new group of tyrosine recombinase-encoding retrotransposons. Mol Biol Evol. 2004;21:746–759. doi: 10.1093/molbev/msh072. [DOI] [PubMed] [Google Scholar]
- 36.Schostak N, Pyatkov K, Zelentsova E, Arkhipova I, Shagin D, Shagina I, Mudrik E, Blintsov A, Clark I, Finnegan DJ, Evgen’ev M. Molecular dissection of Penelope transposable element regulatory machinery. Nucleic Acids Res. 2008;36:2522–2529. doi: 10.1093/nar/gkm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon–intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- 38.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 39.Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- 41.Makalowski W. Genomic scrap yard: how genomes utilize all that junk. Gene. 2000;259:61–67. doi: 10.1016/S0378-1119(00)00436-4. [DOI] [PubMed] [Google Scholar]
- 42.Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci USA. 2006;103:9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song M, Boissinot S. Selection against LINE-1 retrotransposons results principally from their ability to mediate ectopic recombination. Gene. 2007;390:206–213. doi: 10.1016/j.gene.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 44.Burwinkel B, Kilimann MW. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J Mol Biol. 1998;277:513–517. doi: 10.1006/jmbi.1998.1641. [DOI] [PubMed] [Google Scholar]
- 45.Segal Y, Peissel B, Renieri A, de Marchi M, Ballabio A, Pei Y, Zhou J. LINE-1 elements at the sites of molecular rearrangements in Alport syndrome-diffuse leiomyomatosis. Am J Hum Genet. 1999;64:62–69. doi: 10.1086/302213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–2572. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- 47.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 48.Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, Jorde LB (2009) Mobile elements create structural variation: analysis of a complete human genome. Genome Res (in press) [DOI] [PMC free article] [PubMed]
- 49.Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 51.Goodier JL, Ostertag EM, Kazazian HH., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 52.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 54.Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci USA. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long M, Wang W, Zhang J. Origin of new genes and source for N-terminal domain of the chimerical gene, jingwei, in Drosophila . Gene. 1999;238:135–141. doi: 10.1016/S0378-1119(99)00229-2. [DOI] [PubMed] [Google Scholar]
- 56.Zaiss DM, Kloetzel PM. A second gene encoding the mouse proteasome activator PA28beta subunit is part of a LINE1 element and is driven by a LINE1 promoter. J Mol Biol. 1999;287:829–835. doi: 10.1006/jmbi.1999.2656. [DOI] [PubMed] [Google Scholar]
- 57.Boschan C, Borchert A, Ufer C, Thiele BJ, Kuhn H. Discovery of a functional retrotransposon of the murine phospholipid hydroperoxide glutathione peroxidase: chromosomal localization and tissue-specific expression pattern. Genomics. 2002;79:387–394. doi: 10.1006/geno.2001.6715. [DOI] [PubMed] [Google Scholar]
- 58.Babushok DV, Ostertag EM, Kazazian HH., Jr Current topics in genome evolution: molecular mechanisms of new gene formation. Cell Mol Life Sci. 2007;64:542–554. doi: 10.1007/s00018-006-6453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Temin HM. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buzdin A, Gogvadze E, Lebrun MH. Chimeric retrogenes suggest a role for the nucleolus in LINE amplification. FEBS Lett. 2007;581:2877–2882. doi: 10.1016/j.febslet.2007.05.034. [DOI] [PubMed] [Google Scholar]
- 62.Gogvadze E, Barbisan C, Lebrun MH, Buzdin A. Tripartite chimeric pseudogene from the genome of rice blast fungus Magnaporthe grisea suggests double template jumps during long interspersed nuclear element (LINE) reverse transcription. BMC Genomics. 2007;8:360. doi: 10.1186/1471-2164-8-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fudal I, Bohnert HU, Tharreau D, Lebrun MH. Transposition of MINE, a composite retrotransposon, in the avirulence gene ACE1 of the rice blast fungus Magnaporthe grisea . Fungal Genet Biol. 2005;42:761–772. doi: 10.1016/j.fgb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Buzdin A, Gogvadze E, Kovalskaya E, Volchkov P, Ustyugova S, Illarionova A, Fushan A, Vinogradova T, Sverdlov E. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 2003;31:4385–4390. doi: 10.1093/nar/gkg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hayward BE, Zavanelli M, Furano AV. Recombination creates novel L1 (LINE-1) elements in Rattus norvegicus . Genetics. 1997;146:641–654. doi: 10.1093/genetics/146.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brosius J. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica. 1999;107:209–238. doi: 10.1023/A:1004018519722. [DOI] [PubMed] [Google Scholar]
- 70.Furano AV. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol. 2000;64:255–294. doi: 10.1016/S0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- 71.van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 72.Polavarapu N, Marino-Ramirez L, Landsman D, McDonald JF, Jordan IK. Evolutionary rates and patterns for human transcription factor binding sites derived from repetitive DNA. BMC Genomics. 2008;9:226. doi: 10.1186/1471-2164-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Landry JR, Rouhi A, Medstrand P, Mager DL. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol Biol Evol. 2002;19:1934–1942. doi: 10.1093/oxfordjournals.molbev.a004017. [DOI] [PubMed] [Google Scholar]
- 74.Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J Biol Chem. 2001;276:1896–1903. doi: 10.1074/jbc.M006557200. [DOI] [PubMed] [Google Scholar]
- 75.Bieche I, Laurent A, Laurendeau I, Duret L, Giovangrandi Y, Frendo JL, Olivi M, Fausser JL, Evain-Brion D, Vidaud M. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod. 2003;68:1422–1429. doi: 10.1095/biolreprod.102.010322. [DOI] [PubMed] [Google Scholar]
- 76.Dunn CA, van de Lagemaat LN, Baillie GJ, Mager DL. Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5. Gene. 2005;364:2–12. doi: 10.1016/j.gene.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 77.Romanish MT, Lock WM, de Lagemaat LN, Dunn CA, Mager DL. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007;3:e10. doi: 10.1371/journal.pgen.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carlton VE, Harris BZ, Puffenberger EG, Batta AK, Knisely AS, Robinson DL, Strauss KA, Shneider BL, Lim WA, Salen G, Morton DH, Bull LN. Complex inheritance of familial hypercholanemia with associated mutations in TJP2 and BAAT. Nat Genet. 2003;34:91–96. doi: 10.1038/ng1147. [DOI] [PubMed] [Google Scholar]
- 79.Conley AB, Piriyapongsa J, Jordan IK. Retroviral promoters in the human genome. Bioinformatics. 2008;24:1563–1567. doi: 10.1093/bioinformatics/btn243. [DOI] [PubMed] [Google Scholar]
- 80.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. GREM, a technique for genome-wide isolation and quantitative analysis of promoter active repeats. Nucleic Acids Res. 2006;34:e67. doi: 10.1093/nar/gkl335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J Virol. 2006;80:10752–10762. doi: 10.1128/JVI.00871-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meisler MH, Ting CN. The remarkable evolutionary history of the human amylase genes. Crit Rev Oral Biol Med. 1993;4:503–509. doi: 10.1177/10454411930040033501. [DOI] [PubMed] [Google Scholar]
- 83.Long Q, Bengra C, Li C, Kutlar F, Tuan D. A long terminal repeat of the human endogenous retrovirus ERV-9 is located in the 5′ boundary area of the human beta-globin locus control region. Genomics. 1998;54:542–555. doi: 10.1006/geno.1998.5608. [DOI] [PubMed] [Google Scholar]
- 84.Ling J, Pi W, Bollag R, Zeng S, Keskintepe M, Saliman H, Krantz S, Whitney B, Tuan D. The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol. 2002;76:2410–2423. doi: 10.1128/jvi.76.5.2410-2423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ling J, Pi W, Yu X, Bengra C, Long Q, Jin H, Seyfang A, Tuan D. The ERV-9 LTR enhancer is not blocked by the HS5 insulator and synthesizes through the HS5 site non-coding, long RNAs that regulate LTR enhancer function. Nucleic Acids Res. 2003;31:4582–4596. doi: 10.1093/nar/gkg646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loreni F, Stavenhagen J, Kalff M, Robins DM. A complex androgen-responsive enhancer resides 2 kilobases upstream of the mouse Slp gene. Mol Cell Biol. 1988;8:2350–2360. doi: 10.1128/mcb.8.6.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Z, Boffelli D, Boonmark N, Schwartz K, Lawn R. Apolipoprotein(a) gene enhancer resides within a LINE element. J Biol Chem. 1998;273:891–897. doi: 10.1074/jbc.273.2.891. [DOI] [PubMed] [Google Scholar]
- 88.Hambor JE, Mennone J, Coon ME, Hanke JH, Kavathas P. Identification and characterization of an Alu-containing, T-cell-specific enhancer located in the last intron of the human CD8 alpha gene. Mol Cell Biol. 1993;13:7056–7070. doi: 10.1128/mcb.13.11.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Santangelo AM, de Souza FS, Franchini LF, Bumaschny VF, Low MJ, Rubinstein M. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 2007;3:1813–1826. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sasaki T, Nishihara H, Hirakawa M, Fujimura K, Tanaka M, Kokubo N, Kimura-Yoshida C, Matsuo I, Sumiyama K, Saitou N, Shimogori T, Okada N. Possible involvement of SINEs in mammalian-specific brain formation. Proc Natl Acad Sci USA. 2008;105:4220–4225. doi: 10.1073/pnas.0709398105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Corvelo A, Eyras E. Exon creation and establishment in human genes. Genome Biol. 2008;9:R141. doi: 10.1186/gb-2008-9-9-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin L, Shen S, Tye A, Cai JJ, Jiang P, Davidson BL, Xing Y. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 2008;4:e1000225. doi: 10.1371/journal.pgen.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gotea V, Makalowski W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22:260–267. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 94.Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G. Intronic Alus influence alternative splicing. PLoS Genet. 2008;4:e1000204. doi: 10.1371/journal.pgen.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mola G, Vela E, Fernandez-Figueras MT, Isamat M, Munoz-Marmol AM. Exonization of Alu-generated splice variants in the survivin gene of human and non-human primates. J Mol Biol. 2007;366:1055–1063. doi: 10.1016/j.jmb.2006.11.089. [DOI] [PubMed] [Google Scholar]
- 97.Gu Y, Kodama H, Watanabe S, Kikuchi N, Ishitsuka I, Ozawa H, Fujisawa C, Shiga K. The first reported case of Menkes disease caused by an Alu insertion mutation. Brain Dev. 2007;29:105–108. doi: 10.1016/j.braindev.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 98.Buzdin AA. Retroelements and formation of chimeric retrogenes. Cell Mol Life Sci. 2004;61:2046–2059. doi: 10.1007/s00018-004-4041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pavlicek A, Jabbari K, Paces J, Paces V, Hejnar JV, Bernardi G. Similar integration but different stability of Alus and LINEs in the human genome. Gene. 2001;276:39–45. doi: 10.1016/S0378-1119(01)00645-X. [DOI] [PubMed] [Google Scholar]
- 100.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tamura M, Kajikawa M, Okada N. Functional splice sites in a zebrafish LINE and their influence on zebrafish gene expression. Gene. 2007;390:221–231. doi: 10.1016/j.gene.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 102.Sironen A, Vilkki J, Bendixen C, Thomsen B. Infertile Finnish Yorkshire boars carry a full-length LINE-1 retrotransposon within the KPL2 gene. Mol Genet Genomics. 2007;278:385–391. doi: 10.1007/s00438-007-0256-7. [DOI] [PubMed] [Google Scholar]
- 103.van de Lagemaat LN, Medstrand P, Mager DL. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 2006;7:R86. doi: 10.1186/gb-2006-7-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hughes DC. Alternative splicing of the human VEGFGR-3/FLT4 gene as a consequence of an integrated human endogenous retrovirus. J Mol Evol. 2001;53:77–79. doi: 10.1007/s002390010195. [DOI] [PubMed] [Google Scholar]
- 105.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Borodulina OR, Kramerov DA. Transcripts synthesized by RNA polymerase III can be polyadenylated in an AAUAAA-dependent manner. RNA. 2008;14:1865–1873. doi: 10.1261/rna.1006608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cutter AD, Good JM, Pappas CT, Saunders MA, Starrett DM, Wheeler TJ. Transposable element orientation bias in the Drosophila melanogaster genome. J Mol Evol. 2005;61:733–741. doi: 10.1007/s00239-004-0243-0. [DOI] [PubMed] [Google Scholar]
- 109.Baust C, Seifarth W, Germaier H, Hehlmann R, Leib-Mosch C. HERV-K-T47D-related long terminal repeats mediate polyadenylation of cellular transcripts. Genomics. 2000;66:98–103. doi: 10.1006/geno.2000.6175. [DOI] [PubMed] [Google Scholar]
- 110.Kjellman C, Sjogren HO, Salford LG, Widegren B. HERV-F (XA34) is a full-length human endogenous retrovirus expressed in placental and fetal tissues. Gene. 1999;239:99–107. doi: 10.1016/S0378-1119(99)00372-8. [DOI] [PubMed] [Google Scholar]
- 111.Mager DL, Hunter DG, Schertzer M, Freeman JD. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3) Genomics. 1999;59:255–263. doi: 10.1006/geno.1999.5877. [DOI] [PubMed] [Google Scholar]
- 112.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 113.Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen C, Ara T, Gautheret D. Using Alu elements as polyadenylation sites: a case of retroposon exaptation. Mol Biol Evol. 2009;26:327–334. doi: 10.1093/molbev/msn249. [DOI] [PubMed] [Google Scholar]
- 115.Lee JY, Ji Z, Tian B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res. 2008;36:5581–5590. doi: 10.1093/nar/gkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trujillo MA, Sakagashira M, Eberhardt NL. The human growth hormone gene contains a silencer embedded within an Alu repeat in the 3′-flanking region. Mol Endocrinol. 2006;20:2559–2575. doi: 10.1210/me.2006-0147. [DOI] [PubMed] [Google Scholar]
- 117.Sharan C, Hamilton NM, Parl AK, Singh PK, Chaudhuri G. Identification and characterization of a transcriptional silencer upstream of the human BRCA2 gene. Biochem Biophys Res Commun. 1999;265:285–290. doi: 10.1006/bbrc.1999.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maeda N. Nucleotide sequence of the haptoglobin and haptoglobin-related gene pair. The haptoglobin-related gene contains a retrovirus-like element. J Biol Chem. 1985;260:6698–6709. [PubMed] [Google Scholar]
- 119.Hatada S, Grant DJ, Maeda N. An intronic endogenous retrovirus-like sequence attenuates human haptoglobin-related gene expression in an orientation-dependent manner. Gene. 2003;319:55–63. doi: 10.1016/S0378-1119(03)00791-1. [DOI] [PubMed] [Google Scholar]
- 120.Maeda N, Kim HS. Three independent insertions of retrovirus-like sequences in the haptoglobin gene cluster of primates. Genomics. 1990;8:671–683. doi: 10.1016/0888-7543(90)90254-R. [DOI] [PubMed] [Google Scholar]
- 121.Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Domansky AN, Kopantzev EP, Snezhkov EV, Lebedev YB, Leib-Mosch C, Sverdlov ED. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 2000;472:191–195. doi: 10.1016/S0014-5793(00)01460-5. [DOI] [PubMed] [Google Scholar]
- 123.Dunn CA, Romanish MT, Gutierrez LE, van de Lagemaat LN, Mager DL. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene. 2006;366:335–342. doi: 10.1016/j.gene.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 124.Huh JW, Kim DS, Kang DW, Ha HS, Ahn K, Noh YN, Min DS, Chang KT, Kim HS. Transcriptional regulation of GSDML gene by antisense-oriented HERV-H LTR element. Arch Virol. 2008;153:1201–1205. doi: 10.1007/s00705-008-0105-y. [DOI] [PubMed] [Google Scholar]
- 125.Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006:71753. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Galante PA, Vidal DO, de Souza JE, Camargo AA, de Souza SJ. Sense-antisense pairs in mammals: functional and evolutionary considerations. Genome Biol. 2007;8:R40. doi: 10.1186/gb-2007-8-3-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Conley AB, Miller WJ, Jordan IK. Human cis natural antisense transcripts initiated by transposable elements. Trends Genet. 2008;24:53–56. doi: 10.1016/j.tig.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 128.Gogvadze E, Stukacheva E, Buzdin A, Sverdlov E. Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J Virol. 2009;83:6098–6105. doi: 10.1128/JVI.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kashkush K, Khasdan V. Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics. 2007;177:1975–1985. doi: 10.1534/genetics.107.080234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–326. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 131.Hernandez-Pinzon I, de Jesus E, Santiago N, Casacuberta JM. The frequent transcriptional readthrough of the tobacco tnt1 retrotransposon and its possible implications for the control of resistance genes. J Mol Evol. 2009;68:269–278. doi: 10.1007/s00239-009-9204-y. [DOI] [PubMed] [Google Scholar]
- 132.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Levy A, Sela N, Ast G. TranspoGene and microTranspoGene: transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 2008;36:D47–D52. doi: 10.1093/nar/gkm949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lunyak VV, Prefontaine GG, Nunez E, Cramer T, Ju BG, Ohgi KA, Hutt K, Roy R, Garcia-Diaz A, Zhu X, Yung Y, Montoliu L, Glass CK, Rosenfeld MG. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 135.Akopov SB, Ruda VM, Batrak VV, Vetchinova AS, Chernov IP, Nikolaev LG, Bode J, Sverdlov ED. Identification, genome mapping, and CTCF binding of potential insulators within the FXYD5-COX7A1 locus of human chromosome 19q13.12. Mamm Genome. 2006;17:1042–1049. doi: 10.1007/s00335-006-0037-3. [DOI] [PubMed] [Google Scholar]
- 136.Purbowasito W, Suda C, Yokomine T, Zubair M, Sado T, Tsutsui K, Sasaki H. Large-scale identification and mapping of nuclear matrix-attachment regions in the distal imprinted domain of mouse chromosome 7. DNA Res. 2004;11:391–407. doi: 10.1093/dnares/11.6.391. [DOI] [PubMed] [Google Scholar]
- 137.Dorsett D. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila . Genetics. 1993;134:1135–1144. doi: 10.1093/genetics/134.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Girard L, Freeling M. Regulatory changes as a consequence of transposon insertion. Dev Genet. 1999;25:291–296. doi: 10.1002/(SICI)1520-6408(1999)25:4<291::AID-DVG2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 139.Kostyuchenko MV, Savitskaya EE, Volkov IA, Golovnin AK, Georgiev PG. Study of functional interaction between three copies of the insulator from the MDG4 transposable element in the model system of the miniwhite gene of Drosophila melanogaster . Dokl Biochem Biophys. 2008;421:239–243. doi: 10.1134/s1607672908040194. [DOI] [PubMed] [Google Scholar]
- 140.Landry JR, Medstrand P, Mager DL. Repetitive elements in the 5′ untranslated region of a human zinc-finger gene modulate transcription and translation efficiency. Genomics. 2001;76:110–116. doi: 10.1006/geno.2001.6604. [DOI] [PubMed] [Google Scholar]
- 141.Khanam T, Raabe CA, Kiefmann M, Handel S, Skryabin BV, Brosius J. Can ID repetitive elements serve as cis-acting dendritic targeting elements? An in vivo study. PLoS ONE. 2007;2:e961. doi: 10.1371/journal.pone.0000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chandler VL, Walbot V. DNA modification of a maize transposable element correlates with loss of activity. Proc Natl Acad Sci USA. 1986;83:1767–1771. doi: 10.1073/pnas.83.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Weil C, Martienssen R. Epigenetic interactions between transposons and genes: lessons from plants. Curr Opin Genet Dev. 2008;18:188–192. doi: 10.1016/j.gde.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 144.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/S0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 145.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK, Liloglou T. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer. 2009;124:81–87. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 146.Arnaud P, Goubely C, Pelissier T, Deragon JM. SINE retroposons can be used in vivo as nucleation centers for de novo methylation. Mol Cell Biol. 2000;20:3434–3441. doi: 10.1128/MCB.20.10.3434-3441.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322–22329. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 148.Lyon MF. Do LINEs have a role in X-chromosome inactivation? J Biomed Biotechnol. 2006;2006:59746. doi: 10.1155/JBB/2006/59746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 151.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 152.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 153.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 155.Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 2005;33:846–856. doi: 10.1093/nar/gki223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 157.Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35:637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]