Abstract
In mammals, the matrix extracellular phosphoglycoprotein (MEPE) is known to activate osteogenesis and mineralization via a particular region called dentonin, and to inhibit mineralization via its ASARM (acidic serine-aspartate rich MEPE-associated motif) peptide that also plays a role in phosphatemia regulation. In order to understand MEPE evolution in mammals, and particularly that of its functional regions, we conducted an evolutionary analysis based on the study of selective pressures. Using 37 mammalian sequences we: (1) confirmed the presence of an additional coding exon in most placentals; (2) highlighted several conserved residues and regions that could have important functions; (3) found that dentonin function was recruited in a placental ancestor; and (4) revealed that ASARM function was present earlier, pushing the recruitment of MEPE deep into amniote origins. Our data indicate that MEPE was involved in various functions (bone and eggshell mineralization) prior to acquiring those currently known in placental mammals.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0185-1) contains supplementary material, which is available to authorized users.
Keywords: MEPE, OC116, Mammals, Evolution, ASARM, SIBLINGs, Purifying selection, Positive selection, Dentonin
Introduction
The matrix extracellular phosphoglycoprotein (MEPE; also called OF45) was discovered the same year in humans [1] and the rat [2], followed shortly thereafter by its identification in the mouse [3]. MEPE was shown to belong to the small integrin-binding ligand N-linked glycoprotein (SIBLING) family, comprising five proteins principally expressed in the extracellular matrix of bone and dentin [4]: dentin sialophosphoprotein (DSPP); dentin matrix protein 1 (DMP1); bone sialoprotein (IBSP); MEPE; and osteopontin (OPN/SPP1). The SIBLINGs are characterized by similar molecular properties including binding to the cell membrane via integrins, and linking calcium through phosphorylation sites. These proteins play a role, albeit poorly understood, in the mineralization of the collagen matrix, and some of them are overexpressed in various forms of cancer.
In humans, the MEPE gene was first identified in a patient suffering from oncogenic hypophosphatemic osteomalacia (OHO) tumors [1], a disease in which the gene is overexpressed in osteoblasts [2, 5]. This disorder is characterized by hypophosphatemia caused by renal phosphate wasting. A similar overexpression of MEPE in osteoblasts occurs in patients suffering from X-linked hypophosphatemic (XLH) rickets [6]. In normal conditions, MEPE is mainly expressed in osteoblasts, osteocytes and odontoblastes [6].
MEPE inactivation in the mouse leads to an increase in bone mass and mineralization, indicating that the encoded protein plays an inhibitor role in bone formation and mineralization [3]. In humans, this role has been confirmed along with a function in renal phosphate regulation [7]. These two functions are ensured by a small peptide called ASARM (acidic serine-aspartate rich MEPE-associated motif), located at the extremity of the C-terminus of the protein. This peptide is not degraded by proteases and is active when released in blood circulation after cleavage by cathepsin-B. When phosphorylated, ASARM can bind to hydroxyapatite crystals and inhibit mineralization [8]. Its function is modulated through the interaction of MEPE with an endopeptidase known as PHEX (phosphate-regulating endopeptidase homolog, X-linked) [9]. This interaction protects MEPE from cathepsin-B and prevents ASARM from being released into the blood circulation. The PHEX gene is also expressed in osteoblasts, osteocytes and odontoblasts [6]. Most physiological disorders involving MEPE result from PHEX mutations, leading to a malfunction of PHEX–MEPE interaction and a large release of ASARM. The increased ASARM level in blood circulation has various pathological effects including disruption of renal function, loss of bone mass, cardiovascular problems, tumors, and defects in the regulation of cell proliferation.
ASARM is not the only important region identified in MEPE. Several phosphorylation sites are known, and a region containing a RGD and a SGDG motif has attracted attention in the search for a bioactive peptide. The end result is a 23 amino acid synthetic peptide containing these two motifs called AC-100, or more commonly dentonin [10, 11]. Due to their respective interactions with integrins and glycosaminoglycans, RGD and SGDG motifs have two functions: (1) regulation of bone homeostasis by facilitating cell–matrix interactions and differentiation of osteoblasts; and (2) activation of cell proliferation [10]. Dentonin was successfully used to activate osteogenesis in vitro and in vivo [10], and to stimulate dental pulp stem cell proliferation in order to repair dentin damages in rodents [11, 12].
To date, MEPE is known only in human, macaque, rat, and mouse. It is widely acknowledged that comparative data, acquired from many species and analyzed in an evolutionary context, may shed new light on protein functions. Notably, such studies tend to reveal regions and residues that are potentially functionally important (i.e., conserved vs variable sequences). The functions of these regions/residues are then eligible to be tested experimentally. The utility of such evolutionary molecular analysis has been well demonstrated with amelogenin, the major enamel matrix protein [13–16]. One of the key results expected from such analyses is to predict in humans the medical risks associated with possible changes in those important regions or residues, and to validate substitutions identified in clinical diagnostics [17–19].
We conducted an evolutionary analysis of MEPE in mammals with two principal objectives. First, we wanted to identify regions and/or residues that may have important functions (conserved sequences) as possible targets for further functional studies and to predict disease-associated mutations in humans. Second, we sought to better understand the evolutionary patterns of MEPE in mammals and when and how its functional regions were acquired during evolution.
Materials and methods
Sequences in databases
Four published sequences of MEPE (human, macaque, rat and mouse) were extracted from the NCBI database (http://www.ncbi.nlm.nih.gov). Unpublished mammalian MEPE sequences (computer predicted from assembled genomes) were searched for in Ensembl (http://www.ensembl.org). Some partial sequences were completed and new sequences were found using BLAST (basic local alignment search tool) in NCBI Trace Archives repository. A total of 37 sequences of mammalian MEPE, either partial or complete, were obtained with the dataset representative of most therian (marsupials + placentals) lineages (Table 1; Fig. 1). These methods were unable to find the MEPE sequence in the currently available platypus (Ornithorhynchus anatinus: Monotremata) genome. Ultimately, platypus MEPE was searched in the Trace Archives using the putative ancestral sequence calculated from the 37 therian MEPE.
Table 1.
Preferred common names, scientific names, family and orders of the 37 species of mammals used for the evolutionary analysis of MEPE, with accession number in Genbank
| Common name | Genus and species | Family | Order | Source |
|---|---|---|---|---|
| Armadilloa | Dasypus novemcinctus | Dasypodidae | Xenarthra | FJ999672 |
| Baboona | Papio hamadryas | Cercopithecidae | Primates | FJ999673 |
| Bushbabya | Otolemur garnettii | Galagidae | Primates | FJ999674 |
| Cat | Felis catus | Felidae | Carnivora | FJ999675 |
| Chimpanzeea | Pan troglodytes | Hominidae | Primates | FJ999676 |
| Cowa | Bos taurus | Bovidae | Cetartiodactyla | FJ999677 |
| Doga | Canis familiaris | Canidae | Carnivora | FJ999678 |
| Dolphina | Tursiops truncatus | Delphinidae | Cetartiodactyla | FJ999679 |
| Elephanta | Loxodonta africana | Elephantidae | Proboscidea | FJ999680 |
| Gibbona | Nomascus leucogenys | Hylobatidae | Primates | FJ999681 |
| Gorillaa | Gorilla gorilla | Hominidae | Primates | FJ999682 |
| Guinea piga | Cavia porcellus | Caviidae | Rodentia | FJ999683 |
| Hedgehog | Erinaceus europaeus | Erinaceidae | Eulipotypla | FJ999684 |
| Horse | Equus caballus | Equidae | Perissodactyla | FJ999685 |
| Humana | Homo sapiens | Hominidae | Primates | FJ999686 |
| Hyraxa | Procavia capensis | Hyracoidea | Hyracoidea | FJ999687 |
| Kangaroo rata | Dipodomys ordii | Heteromyidae | Rodentia | FJ999688 |
| Macaquea | Macaca fascicularis | Cercopithecidae | Primates | FJ999689 |
| Macaquea | Macaca mulatta | Cercopithecidae | Primates | FJ999690 |
| Marmoseta | Callithrix jacchus | Cebidae | Primates | FJ999691 |
| Megabat | Pteropus vampyrus | Pteropodidae | (Mega)Chiroptera | FJ999692 |
| Microbata | Myotis lucifugus | Vespertilionidae | (Micro)Chiroptera | FJ999693 |
| Mouse lemura | Microcebus murinus | Cheirogaleidae | Primates | FJ999694 |
| Mousea | Mus musculus | Muridae | Rodentia | FJ999695 |
| Opossuma | Monodelphis domestica | Didelphidae | Didelphimorphia | FJ999696 |
| Orangutana | Pongo pygmaeus | Hominidae | Primates | FJ999697 |
| Pig | Sus scrofa | Suidae | Cetartiodactyla | FJ999698 |
| Pika | Ochotona princeps | Ochotonidae | Lagomorpha | FJ999699 |
| Rabbita | Oryctolagus cuniculus | Leporidae | Lagomorpha | FJ999700 |
| Rata | Rattus norvegicus | Muridae | Rodentia | FJ999701 |
| Shrew | Sorex araneus | Soricidae | Eulipotypla | FJ999702 |
| Slotha | Choloepus hoffmanni | Megalonychidae | Xenarthra | FJ999703 |
| Squirrela | Spermophilus tridecemlineatus | Sciuridae | Rodentia | FJ999704 |
| Tarsier | Tarsius syrichta | Tarsiidae | Primates | FJ999705 |
| Tenreca | Echinops telfairi | Tenrecidae | Afrosoricida | FJ999706 |
| Tree shrewa | Tupaia belangeri | Tupaiidae | Scandentia | FJ999707 |
| Vicugnaa | Vicugna vicugna | Camelidae | Cetartiodactyla | FJ999708 |
| Wallabya | Macropus eugenii | Macropodidae | Diprotodontia | FJ999709 |
A total of 33 new sequences were obtained in this work
aThe 29 species for which the coding MEPE sequences are complete
Fig. 1.
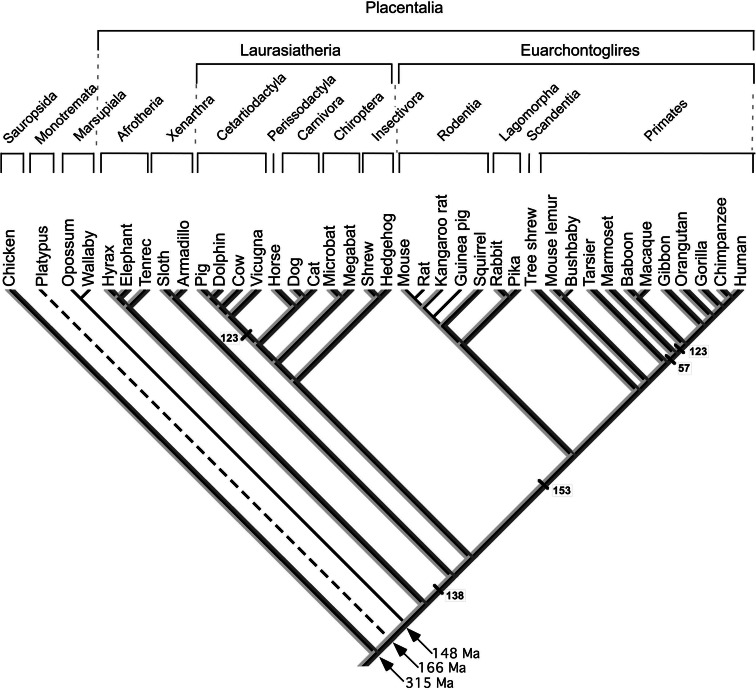
Location of the 37 species studied in the phylogenetic tree of mammals [19]. The lineage of Monotremata (platypus) is indicated with a dashed line as no MEPE sequence has been identified yet. However, an orthologue of MEPE (OC-116) is present in the chicken (Sauropsida). The five positions (amino acids) identified through analysis as positively selected are located in the tree (numbers refer to our alignment). The black line indicates the presence of MEPE. The light and dark gray lines represent exons 3 and 4, respectively; exon 3 and/or exon 4 were lost in several lineages during evolution
The sequence of the MEPE ortholog in chicken, OC-116, was also used for comparison (accession number: AF148716).
Sequence alignment and calculation of the ancestral therian sequence
The 37 MEPE sequences were aligned using Clustal X 2.0 and checked by hand using Se-Al v2.0a11 software (http://tree.bio.ed.ac.uk/software/seal) [20], taking the human sequence as a reference. The alignment, hereafter referred to as “our alignment”, resulted in a 632 amino acid sequence (including the numerous indels). The putative ancestral sequence of therian MEPE was calculated using the HyPhy program (http://www.hyphy.org [21]). First, a matrix was established taking into account the phylogenetic relationships of the various species [19] using MacClade 4.08 software (http://macclade.org [22]). Then, the matrix was used to establish the character state at each evolutionary node (HyPhy). The ancestral sequence was calculated with the following parameters: maximum likelihood (ML) method and Dayhoff’s protein substitution model with local parameter estimates from the dataset using ML.
The putative ancestral MEPE sequence, specifically the most conserved regions identified in our alignment, was unsuccessfully used to search by BLAST in the platypus genome. Therefore, our evolutionary analysis of MEPE concerned only the Theria.
Purifying selection analysis using the Selecton method
The sites of MEPE under purifying selection (i.e., slow evolving positions: unchanged and conservative residues) were identified using the Selecton Server 2.2 (http://selecton.tau.oc.il), in which ML allows to estimate dN/dS ratio at each position [23, 24]. dN/dS was calculated as follows: dN/dS = (NN/EN)/(NS/ES), in which NN and NS represent, respectively, the number of non-synonymous (N) and synonymous (S) substitutions observed, and EN and ES represent the expected number of normalized non-synonymous and synonymous substitutions. The values that are significantly smaller than 1 are indicative of purifying selection. The results were displayed on the human MEPE sequence.
Selective pressure analysis using the Hyphy method
Sliding window analysis (mean substitution rate)
In order to identify strong functional constraints, a sliding window analysis of nucleotide sequence variability [25] was conducted on MEPE alignment using HyPhy. The mean substitution rate was calculated using the ML method based on HKY 85 model [26]. Contrary to other methods, Hyphy uses the Ln likelihood in order to measure the selective pressure. At each position the probability for the observed data is calculated by the likelihood algorithm taking into account the phylogenetic relationships. Then, the logarithm (Ln) of the product of the probabilities is calculated for a window of 15 base pairs with an overlap of 5 bp between windows. Indeed, when applying the Hyphy method, it is not necessary to use large sliding windows and it is even recommended to avoid a “smoothing” effect, i.e., a loss of evolutionary information. In addition, when using the Ln likelihood, the evolution rate in a given sequence is not represented by a rate of change but by a probability. This value is more interesting as it does not necessitate to take into account numerous parameters and does not need to identify non-synonymous and synonymous mutations.
Non-synonymous changes
To get a clear idea of the selective constraints acting on MEPE at each site, we used the codon-based SLAC method (Single Likelihood Ancestor Counting: http://www.datamonkey.org) to study all non-synonymous substitutions (dN) in the alignment. These non-synonymous changes were compared to the number of non-synonymous changes expected at random in absence of selective pressure.
Positive selection
We used the SLAC method in order to determine the positions subjected to positive selection [27, 28]. This method estimates the selection in a phylogenetic context, inferring the ancestral condition at each site. By using a phylogeny, SLAC calculates the number of synonymous and non-synonymous substitutions expected at random, and compares them to the number of synonymous and non-synonymous substitutions observed. The dN/dS value is estimated at each site of the alignment. When dN > dS, the codon is considered positively selected. The results were statistically tested (binomial distribution) at each site. The test assumes that, under neutral hypothesis, the probability for a random substitution to be synonymous is P = ES/(ES + EN). A minimum value of P = 0.1 was chosen to consider the result significant. Indeed, with our dataset of 37 sequences a P value of 0.1 is considered more appropriate in order to detect true positive positions using SLAC, which is the highest conservative method [28]. We also tested our results with P = 0.05 and P = 0.01.
These evolutionary events were placed on a mammalian phylogeny in MacClade 4.08 using the “trace character” option. We only considered mutations that occurred during the evolution of different lineages of mammals, and then became unchanged. Mutations in terminal branches were not considered informative.
Distance matrix
A distance matrix was used to calculate the evolutionary distance between the 37 nucleotide sequences of MEPE, with the aim of testing their relevance for the analysis. This calculation was made using HyPhy program and the Tamura and Nei [29] distance algorithm. A distance tree was obtained using the neighbor-joining method [30] to illustrate the evolutionary distances between taxa. These distances were estimated from the 37 amino acid sequences using ML estimate method, with TN93 algorithm [29].
Results
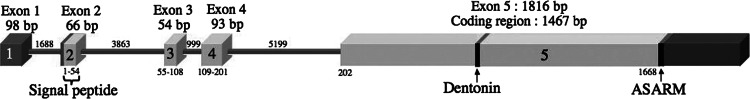
When first published in humans and rats, MEPE transcripts were known to have four exons, three of which were coding exons [1, 2]. That same year, Osada and colleagues submitted online (NCBI, No. accession AB046056) a MEPE sequence containing an additional coding exon. This transcript (93 bp) was identified in brain mRNAs from the crab macaque (Macaca fascicularis). Fisher and Fedarko [4] mentioned this additional exon in human brain mRNAs along with another extra exon, but these sequences remain unpublished (L. Fisher, personal communication, Dec 2008). Therefore, it appears that MEPE is composed of five exons, at least in humans and macaques: exon 1 (98 bp) is untranslated; exon 2 (66 bp) is composed of 12 untranslated bp and encodes the signal peptide (48 bp) and the two-first residues of the protein (6 bp); exon 3 (54 bp), exon 4 (93 bp) and the large exon 5 (1,467 bp), encode for the protein; exon 5 ends with the 3′ untranslated region (Fig. 2). The two regions known to play an important role, dentonin and ASARM peptide, are encoded by exon 5.
Fig. 2.
Structure of human MEPE as deduced from our study, i.e., including exon 4. The size of the exons and introns (gray lines) is indicated (bp). Light gray coding regions, dark gray untranslated regions. The signal peptide (bp 1–48) is located in exon 2 and the dentonin region and ASARM peptide are located in exon 5
Sequence comparison
A set of well representative sequences
A total of 33 new sequences (25 complete) of mammalian MEPE were obtained from the databases. Combined with the four complete sequences already known, our evolutionary analysis was carried out using a dataset of 37 sequences, representing 32 families distributed in 15 orders (Fig. 1; Table 1). The monotreme lineage (platypus and echidnas) is not represented in these sequences, which means that our MEPE analysis covers an evolutionary period of approximately 150 million years (Ma), during which time therians diversified [31].
Our alignment confirmed that four exons code for MEPE in most species, including humans (Fig. 3; electronic supplementary material, ESM, 1). This is accurately established when a gDNA sequence aligns clearly with published sequences (same length and conserved residues), and when the donor and acceptor intron splice sites are present. In contrast, an invalidated (i.e., no longer functional) exon is easily recognizable when (1) one or both splice sites are mutated, (2) there is a stop codon in the correct reading frame, or (3) the sequence is different (i.e., not evolutionarily constrained) when compared to the ortholog sequence in a closely related species. These three conditions were often met when the exon invalidation occurred early in the lineage and/or when the species displayed a rapid evolution rate (e.g., rodents). Our interpretation is that the exon was coding (i.e., functional) in an ancestor before becoming invalidated. However, this exon is still recognizable in the targeted gDNA region as a pseudo-exon.
Fig. 3.
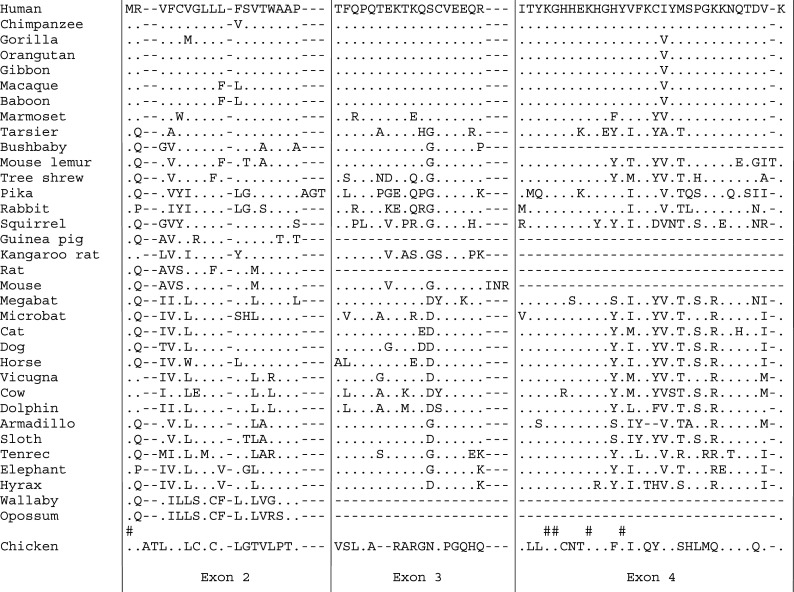
Alignment of the amino acid sequence encoded by exon 2, exon 3 and exon 4 of 34 mammalian MEPE and chicken OC-116. The latter was not included in the evolutionary analysis. The human sequence is used as the reference sequence for the alignement, which takes into consideration mammalian relationships (see Fig. 1). In these regions the residues are well conserved. Exon 3 and/or exon 4 are invalidated in some species while they are present in the chicken. (.) residue identical to that in human sequence; (-) indels; (#) residue identical in all sequences
Our alignment reveals variable and conserved regions and/or residues during therian evolution. Conserved positions include unchanged residues and residues that can be replaced with residues possessing the same characteristics (conservative positions). Unchanged and conservative regions and residues indicate a strong selective pressure, which we interpret as related to their important function. Among these conserved regions are included the signal peptide, the dentonin region, the ASARM peptide, and the regions encoded by exons 3 and 4. In addition, our evolutionary analysis identified five other important regions encoded by exon 5 and numerous important residues distributed along the MEPE sequence.
Exons 2, 3 and 4
The structure of MEPE is generally conserved in Theria, with the exception of exons 3 and 4 which are sometimes lacking in some species (e.g., marsupials and rodents). In contrast, exons 2 and 5 are present in all species. The length of the exon 2 protein sequence is different in only two species: a single amino acid insertion in the guinea pig and the addition of three amino acids in the pika (ESM 1). The cleavage site of the signal peptide is unchanged.
Exon 3 was lacking (no sequence similarity in the expected gDNA region) in the two marsupials (opossum and kangaroo), and it is invalidated in the rat (stop codon) and guinea pig (uncorrect splice site) (Fig. 3). Exon 3 is present and probably coding in the other rodents investigated (mouse, kangaroo rat and squirrel). Its invalidation in rat and guinea pig while probably coding in the mouse and kangaroo rat suggests that it was lost independently in these two rodent lineages. In the other species, the comparison of coding exon 3 sequences reveals that most residues are well conserved, which indicates that (1) these amino acids have an important function and (2) this region of the protein has an ancient origin (Fig. 3). The length of the protein sequence encoded by exon 3 is different in the mouse only, with the addition of three residues: INR. In the near future, it would be interesting to look for additional MEPE transcripts in mouse cDNA that would include this exon.
Exon 4 is absent in marsupials and it is not coding in some rodents (mouse, rat, kangaroo rat, and guinea pig). In these latter, a pseudo-exon 4 is still recognizable in the gDNA, exhibiting a non-functional splice site and a highly variable sequence. In the squirrel, exon 4 sequence shows several substitutions but there is no evidence for this exon as being no longer coding (conserved splice sites). These data suggest that exon 4 was probably invalidated in these lineages after their separation from the sciurognath lineage (squirrel). Finally, in primates, exon 4 was found invalidated (stop codon and splice site mutation) in the bushbaby only; this event has occurred recently in this lineage because the residues encoded by the no longer functional exon 4 are well conserved compared to those in related species.
We compared the mammalian MEPE sequences with the chicken orthologue ovocleidin 116 (OC-116). Exons 3 and 4 are present in the chicken, but exon 4 only shows a high sequence similarity between chicken and mammalian MEPE (Fig. 3). This strongly suggests that the ancestral sequence of MEPE/OC-116 comprised at least five exons. In addition, the long-lasting conservation of exon 4 sequence through geological times (310 Ma) indicates that the amino acids it encodes play probably an important role in both lineages.
Exon 5 and functional regions
Most of the sequence encoded by MEPE exon 5 is characterized by high variability, as indicated by the numerous substitutions observed in our alignment (Fig. 4; ESM 1). This exon is more variable in the two marsupials than in placentals. In addition, at its 5′ end of rat and mouse MEPE, exon 5 is 174 nucleotides shorter than the other MEPE.
Fig. 4.
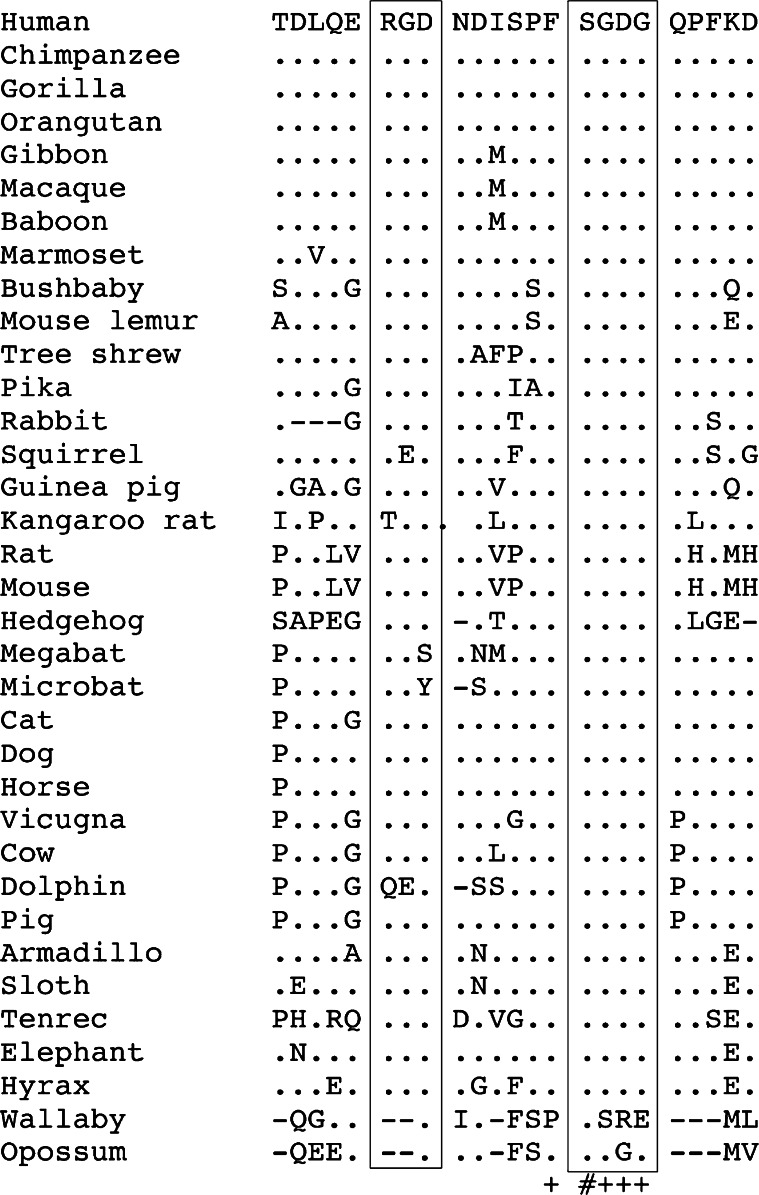
Alignment of the amino acid sequence encoding the dentonin region of 35 mammalian MEPE. This 23 amino acid-long region (synthetical peptide AC-100) is located in the central region of MEPE encoded by exon 5 (see Fig. 2). It contains two important motives, RGD and SGDG (boxed), which are preserved in most species. However, RGD is absent in several sequences and the two motives are missing in marsupials. (+) residue identical in all therian sequences. See Fig. 3 for the other symbols
Our comparative analysis indicates that the dentonin region is well conserved in placentals, the RGD and SGDG motifs being present in most MEPE sequences (Fig. 4). However, five sequences (squirrel, kangaroo rat, the two bats, and dolphin) show a substitution of at least one residue of the RGD, which means that this short peptide probably cannot play its function. In addition, there is no RGD motif elsewhere in these five sequences. In contrast, the SGDG motif is conserved in all placentals (Fig. 4).
It is worth noting that the two marsupial MEPE lack both the RGD and SGDG motifs.
The ASARM peptide is well conserved in all placentals. It is also present in marsupials, in which it is capped at its C-terminal by a dozen amino acids (Fig. 5).
Fig. 5.
Alignment of the amino acid sequence encoding the ASARM peptide of 35 mammalian MEPE and chicken OC-116. The latter was not included in the evolutionary analysis. The residues are well conserved in all the sequences studied, but in the two marsupials and the chicken the ASARM sequence is capped by several residues. (=) unknown residue. See Fig. 3 and 4 for the other symbols
The two N-glycosylation sites in exon 5 (N477 and N478 = positions N569 and N570 in our alignment) described in humans by Rowe et al. [1] are not conserved in all therian MEPE (ESM 1). However, in the sequences that do not possess these sites, several potential N-glycosylation sites were identified elsewhere with computer-prediction programs.
The comparison of MEPE and chicken OC-116 reveals the presence of a short sequence resembling the ASARM peptide, but capped by several amino-acids as described in marsupials (Fig. 5). This suggests strongly that the origin of this region was present in the ancestral sequence of MEPE/OC-116.
Purifying selection
The analysis of the 37 MEPE sequences using Selecton allowed us to identify 154 positions (out of 489) subjected to purifying selection during 150 Ma of therian evolution, i.e., either unchanged or conservative positions. These constraints indicate that the concerned positions have important functions (Fig. 6; ESM 1).
Fig. 6.
Human MEPE sequence on which are displayed the results of evolutionary analysis. Purifying selection: the residue on a black background have not changed during mammalian evolution; residues on a gray background are those positions that have kept their properties (conservative positions). In humans, we predict that mutations on these residues could lead to a genetic disease. The residues positively selected are indicated with an asterisk. The signal peptide is underlined. The already known functional regions (RGD and SGDG motifs of dentonin region (dashed line), and the ASARM peptide) are indicated. The new important motifs identified by our analysis are boxed. Arrows putative N-glycosylated residues in humans
In the region encoded by exon 2, five amino acids are under purifying selection, three of them being functionally related to the signal peptide. Eight residues (out of 18) were identified in the region corresponding to exon 3. Of the 31 residues encoded by exon 4, 16 are under purifying selection. Among them, a potential site of N-glycosylation, N69 in our alignment, has previously been identified in the chick (ESM 1, and arrow in Fig. 6). The only MEPE sequences of pika and tenrec lack this asparagin. In the region encoded by exon 5, 125 positions (out of 489) are subjected to purifying selection. Among these are the RGD and SGDG residues of the dentonin region and 13 residues (out of 23) of the ASARM peptide. In addition to these interesting residues and regions, our evolutionary analysis revealed the presence of five conserved regions whose functions are unknown: they are amino acids 266–271, 333–341, 368–387, 395–406, and 428–436 in human MEPE (Fig. 6; ESM 1). Among these conserved regions, the first one (DFEGSG) is located close to the RGD/SGDG motif, and its sequence similarity with SGDG would mean a possible similar function. Interestingly, this motif is present in marsupials and in species lacking the RGD motif.
Sliding window analysis
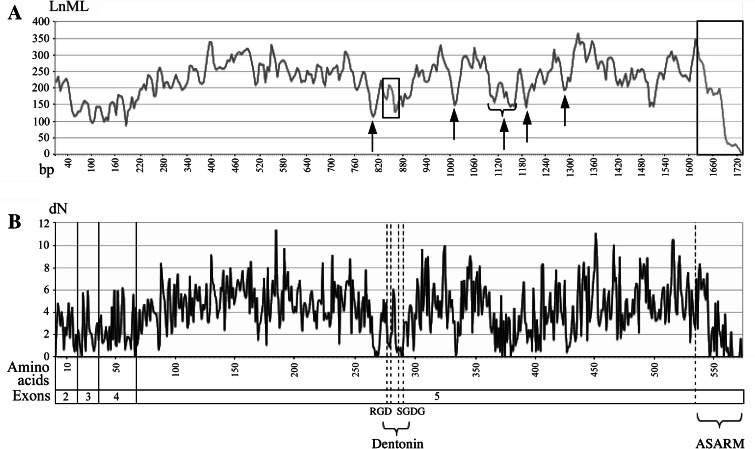
The functional constraints acting on MEPE were inferred by means of a sliding window analysis (dN/dS ratio) (Fig. 7a). The amino acid sequence of MEPE is characterized by selection pressures, alternatively low and high, resulting in high and low values of maximum likelihood (ML), respectively. Exons 2, 3 and 4 protein regions are under strong functional constraints as indicated by the relatively low ML values in the analysis. Exon 5 protein region exhibits seven regions under strong functional pressure, characterized by low ML values (Fig. 7). The analysis of non-synonymous substitution rates allowed us to localize precisely these regions on the MEPE sequence (Fig. 7b).
Fig. 7.
Substitution rate analysis of the 37 mammalian MEPE. a Overview of functional regions inferred by the sliding window analysis (dN/dS ratio) of the nucleotide sequences (pb). The dentonin region and the ASARM peptide, whose functions are known, are boxed. The arrows indicate areas under strong functional constraints, but whose functions are not established yet. b Analysis of non-synonymous substitution rate (dN) in amino acid sequences. The regions identified with arrows in a correspond to regions with the lowest rate of non-synonymous substitutions. LnML log of maximum likelihood. Note that the regions encoded by exons 2, 3 and 4 have a low substitution rate
Substitution rates
In MEPE, the number of amino acid substitutions is highly variable from one species to the next. These variations were quantified using a distance matrix. Among primates, the substitution rate in MEPE is low (0.01 when comparing MEPE in humans and chimpanzee; 0.08 between marmoset and rhesus monkey). These results were confirmed on the distance tree through short branches, indicative of low substitution levels (ESM 2). When comparing species from other mammalian lineages, the values are higher (i.e., comprised between 0.1 and 0.6), and are reflected as long branches on the distance tree. The lower values of substitution rates in primates are statistically significant when compared to those in other mammals (Student’s t test, P < 0.001). In some species, such as tenrec (Afrotheria), rodents and insectivores (hedgehog and shrew), which are known to have rapid evolution rates, MEPE displays a high substitution rate leading to long branches on the distance tree. Therefore, the discrepancy between the MEPE tree and mammalian phylogeny is a result of long-branch attraction, linked in particular to the rapid evolution of exon 5 for species that have a high substitution rate. This result indicates that the MEPE sequence as a whole cannot be used as phylogenetical marker, except perhaps in primates.
Positive selection
The evolutionary analysis suggested that nine positions in our alignment are subjected to positive selection. These sites are located in the region encoded by exon 3 (1 position), exon 4 (1) and exon 5 (7) (Fig. 6). Although varying in different lineages, five of them, were kept unchanged in terminal lineages (positions 32, 94, 223, 390, and 502 in our alignment, ESM 1). The other four positions, more informative, were kept unchanged at various steps of mammalian evolution (Fig. 1): position 138 in the common ancestor to Euarchontoglires and Laurasiatheria, position 123 in the common ancestor of Cetartiodactyla, position 153 in the common ancestor of Euarchontoglires, and positions 57 and 123 in Primates.
We tested the positive selection with a value of P = 0.01 and obtained no significant results. With a significance level of P = 0.05, we found two positively-selected sites: positions 123 and 153. Therefore, these positions are the more informative.
Discussion
A set of well-representative mammalian MEPE sequences, exclusive of monotremes
While only four MEPE sequences have been published to date, our study provides 33 new sequences of mammalian MEPE. With the addition of the four already known sequences, our analysis was conducted with a dataset of 37 sequences, representing most major mammalian lineages, with the exception of Monotremata. Despite our efforts, including the use of the putative ancestral therian sequence calculated with the 37 sequences, MEPE was not found in the currently sequenced platypus genome. It is possible that this gene was invalidated after the divergence of this lineage, or that the genomic region in which it should be located was not correctly assembled in the current release (OA 5.0, Dec 2005). Indeed, the exploration of the targeted intergenic DNA region, i.e., between IBSP and OPN/SPP1, revealed the probable presence of a reverse transcriptase gene. This is the first report of such a gene in this region among currently annotated mammalian genomes. Moreover, many DNA regions were lacking in the targeted region. In addition, the only MEPE sequence that could be identified when blasting DNA is the large exon 5, which is unfortunately highly variable. All these reasons could explain why we failed to find MEPE sequences in the current genome assembly (Ensembl) and in the trace archives (NCBI) of platypus.
In birds, the MEPE orthologue ovocleidin 116 (OC-116) encodes a protein involved in eggshell mineralization [32–34]. However, recent studies in the chick revealed that MEPE/OC-116 was also expressed in osteoblasts and osteocytes of cortical bone [35, Bardet et al., submitted]. Taken together, these data indicate (1) a dual function for MEPE/OC-116 in bone and eggshell mineralization in the chick, and (2) the presence of MEPE/OC-116 in the genome of the common amniote ancestor (mammals and sauropsids). The platypus, whose lineage ancestor is supposed to have diverged from an ancestral Theria approximately 160 Ma [36], possesses both mammalian (hair, milk) and sauropsid features (cleidoic eggs). Platypus eggs are encased within a parchment-like shell, which has a crystalline component [37]. In fact, all parchment-like shells of amniotes, as in most turtles, lizards, snakes, and monotremes, contain calcium carbonate crystals [38]. Therefore, in addition to a role in bone mineralization, we believe that MEPE was also involved in eggshell formation in the common amniote ancestor. Both functions support the probable presence of MEPE in the platypus genome. However, the difficulties associated with (1) finding the variable region of exon 5 in platypus DNA, and (2) working with a poor sequence assembly in the target region prevent any possibility of obtaining MEPE using synteny. We hope that this will be corrected in the next release of platypus genome assembly.
We know, therefore, that MEPE/OC-116 has been recruited (1) after the duplication of a member of the SIBLING family [34, 39], and (2) before the divergence of the two amniote lineages. Was it present earlier during evolution, i.e., in the common tetrapod ancestor or even earlier? This would mean that MEPE/OC-116 could be present at least in lissamphibians. Unfortunately, until now, we have failed to find this gene in the only currently available genome in a lissamphibian, the clawed toad, Xenopus tropicalis. Again, this failure could be explained because either the assembly of the xenopus genome is not complete in the target region, or the MEPE/OC-116 xenopus sequence is too much divergent, or the gene is really lacking. On the other hand, we know that MEPE is not present in teleost fish (e.g., fugu, zebrafish, stickleback, and medaka). Indeed, several genes belonging to the same family of SCPPs (for secretory calcium-binding phosphoproteins), but not a MEPE orthologue, have been characterized in teleosts [34, 39]. Therefore, to our current knowledge, the origin of MEPE/OC-116 is to be found somewhere in the sarcopterygian lineage but before the divergence of amniote lineages.
Four exons encode MEPE
Beginning with the studies by Rowe et al. [1] and Petersen et al. [2], MEPE was considered to be composed of four exons. Our evolutionary analysis confirmed the presence of a fifth exon, located between exons 3 and 4. This additional exon has already been reported in isoforms of MEPE transcripts expressed in monkey brain [40], but this sequence was deposited only in GenBank and remained long underestimated. This exon was also identified in isoforms of MEPE transcripts from human brain by Fisher and Fedarko [4], but remains unpublished. This exon is also annotated in Ensembl for the human MEPE, but this is a computer-predicted data (ENST00000395102). The presence of a fifth exon supports the relationships of MEPE with the other members of the SIBLING gene cluster (i.e., DSPP, DMP1, IBSP and OPN/SPP1). Indeed, as the SIBLING cluster was created by tandem duplication from an ancestral SIBLING, one would expect to see a similar gene structure (six exons) for each SIBLING gene, with five small exons (the first being untranslated) followed by a large exon [4, 34, 41]. Our BLAST search in the MEPE sequences available in databases allowed the identification of an additional (fifth) exon in many mammals, but failed to reveal a sixth, small exon, as expected from the SIBLING gene structure. This indicates that the MEPE organization consisting in five exons existed in the common mammalian ancestor. The additional exon is named exon 4 and the former exon 4 is now exon 5.
In chicken, OC-116 is also composed of five exons, and no isoforms have been reported to date. It is worth noting that OC-116 exon 4 sequence is similar to that of MEPE exon 4. This implies that (1) this exon was present in MEPE/OC-116 in the common ancestor of amniotes, and (2) the amino acids it encodes probably play an important structural and/or functional role. Therefore, in contrast to most SIBLINGs, MEPE/OC-116 possesses only five exons. This indicates that an exon was lost during the evolutionary period running from the creation of MEPE/OC-116 after duplication of an ancestral SIBLING to its functional recruitment in the common ancestor of amniotes [4, 34, 41].
The inherited gene structure from the ancestral SIBLING has also been modified during evolution in various members of the family, as currently observed in mammals. In DSPP, the last intron was lost, leading to the current large exon 5. Conversely, in SPP1 and IBSP, an intron was created in the last exon, splitting it into the current exons 6 and 7. In IBSP, the border between exon 6 and 7 might shift to the upstream region by reorganization of the exon–intron boundaries in the amniote lineage after the divergence from amphibians [42]. In chicken IBSP, two exons are absent compared to mammals: an exon at the 5′UTR and one exon through the fusion of the two-first exons that encode the signal peptide and N-terminal end of the mature IBSP peptide [43]. Moreover, isoforms have been identified that miss either exon 4 or exon 5 in SPP1, and exon 5 in DMP1 [4]. Therefore, the exon–intron structure of the SIBLINGs has been subjected to several variations during tetrapod evolution. This plasticity is probably related to the recruitment of the encoded proteins into distinct functions, not only for each SIBLING but also for the various isoforms.
The evolutionary analysis reveals selection pressure acting on various MEPE regions
As the other members of the SCPPs’—and nearly 40% of the proteins’—MEPE belongs to a category of proteins known as intrinsically unstructured/natively disordered proteins, there is ample evidence that the unstructured state, common to all living organisms, is essential for basic cellular functions [44]. The disordered regions accumulate numerous substitution, and this is the case for a large part of the region encoded by MEPE exon 5. Therefore, one could deduce that there is no selection pressure except to maintain the main biochemical properties. Such an interpretation is not correct as the disorder does not apply to the whole sequence. Many regions of the protein are structured and subjected to selection pressure. It is known that disordered proteins contain functional motifs that are conserved across nearly all family members [45].
Invalidation and alternative splicing of exons 3 and 4
In the two marsupials investigated, MEPE does not possess coding exons 3 and 4, although these regions are present in a number of placentals. Possibly, these exons were invalidated in a marsupialan ancestor prior to the divergence of this lineage. In the mouse, exon 3 was not previously identified in MEPE transcripts sequenced [5], although the DNA sequence we have obtained indicates that this exon is likely coding (conserved residues and correct splice sites). This suggests that murine MEPE exon 3 is alternatively spliced and that two MEPE isoforms are translated. The transcripts lacking exon 3 are the only reported to date, which suggests that the transcript containing exon 3 could be present in other locations as reported below for primate exon 4. This is not the case in rats and guinea pigs, in which this exon is no longer functional, leading us to the conclusion that MEPE exon 3 was independently invalidated in these two lineages.
Among placentals, exon 4 was invalidated in rats, mice, kangaroo rat, guinea pig, and bushbaby, a lemurian primate. In humans, our comparative analysis clearly indicates that exon 4 is coding, as in most mammalian lineages (conserved residues and correct splice sites). However, this exon was not identified in human MEPE transcripts sequenced from bone marrow and bone tumor cells [1]. This suggests that MEPE exon 4 is alternatively spliced and that there are two isoforms of human MEPE. The isoform that includes exon 4 could be restricted to particular tissues (e.g., the primate brain). MEPE isoforms that possess regions encoded by exon 4 and/or exon 3 are interpreted to have an important function because selective forces are acting to conserve most of their residues.
Among therians, the current organization of MEPE exons was inherited from an ancestral amniote (or tetrapod). Ancestrally, MEPE encoded a protein that probably had other/additional functions besides those currently known for therians. This interpretation is supported by: (1) the high divergence of exon 5 sequences when comparing the two orthologs, OC-116 and MEPE; and (2) the expression of OC-116 during eggshell mineralization in chicken. In the latter, the region encoded by exon 4 possesses a site of N-glycosylation at position N62 [46]. This amino acid is present in most therian MEPE (N69 in our alignment), with the exception of pika (Lagomorpha) and tenrec (Afrotheria). Unlike MEPE in humans and rhesus monkey, OC-116 seems not to be subjected to alternatively splicing as exon 4 was found in all transcripts in hen oviduct cells [32]. Whether or not exon 4 is present in non-primate therian MEPE transcripts is currently uncertain, and isoform regulation is complex and still poorly understood [47]. In order to understand the function of the MEPE region encoded by exon 4, and why this region can be lost without any consequences in some species, it is critical to study non-primate species, in which we have identified this exon as functional. This includes possible transcripts expressed in the brain, the only locus in which MEPE isoforms including exon 4 were found in primates [4, 40]. We speculate that isoforms either containing or lacking exon 4, which contains an N-glycosylation site, would modulate cell adhesion of MEPE in connection with the presence of the dentonin region, which also possess cell adhesion properties through its RGD and SGDG motifs.
A contradiction about the importance of MEPE regions encoded by exons 3 and 4
MEPE exons 3 and 4 are coding in most mammals [4], and our evolutionary analysis reveals that some amino acids they encode are under strong selective pressure, which indicates that they play important (albeit presently unknown) structural and/or functional roles. However, in contrast to this interpretation, our analysis also shows that some species lack one or two of these exons without any obvious detriment. This suggests that the regions encoded by exons 3 and 4 are not important, contradicting our previous conclusion.
The conservation of the first and second codon and the saturation in mutations of the third codon (high dS values) confirm that a strong selective pressure acts on these residues. However, in mammals, the expression pattern of MEPE in bone and dentin cells reveals that exon 3 (mice) and exon 4 (primates) are not present in the transcripts [1, 2, 6]. Interestingly, in macaque, exon 4 was found expressed in brain tissues, and alternative splicing of this exon was reported in genomic databases. In addition, as discussed above, several SIBLINGs possess isoforms that result from alternative splicing occurring in this N-terminal region, a finding that indicates a regulation of gene expression [4]. It is known that some isoforms can play other roles, as reported for amelogenin, an enamel matrix protein belonging to the SCPP family, and DMP1 [48, 49]. The only hypothesis that could explain the contradiction revealed by our evolutionary analysis is that MEPE exons 3 and 4 encode peptides that are required to fulfil a function elsewhere than in dentin and bone (may be in the brain), but such a role is still unknown. For an unknown reason, the constraint on these residues was relaxed in some mammals (rat, mouse, and a primate) and the encoding exons were subsequently invalidated through mutation of the splice acceptor site.
It is noteworthy that there are many examples of proteins showing a combination of intrinsically disordered regions and alternative splicing: they are interpreted as providing mechanisms enabling evolution [50]. In most species that lack exon 3 and/or exon 4, the pseudo-exon is easy to identify, which means that the invalidation of these exons could have occurred relatively recently (in the context of geological times) in the mammalian lineages concerned.
Exon 5: variable and functional regions
The length and nucleotide sequence of the large exon 5 vary depending on the species. This explains why the encoded region was not easy to align. Moreover, in rats and mice, the MEPE region encoded by exon 5 is shorter than in the other therians due to the loss of circa 60 amino acids at its N-ter extremity. Such a large deletion can be explained by the invalidation of the primary 5′ splice acceptor site that was simultaneously compensated by the presence of a cryptic splice acceptor site located farther in exon 5 sequences. This deletion being not present in the other rodent MEPE (guinea pig, kangaroo rat, and squirrel), we conclude that it occurred in the common ancestor of mice and rats. It appears that the loss of 60 residues in this MEPE region was not a negative event for the subsequent evolution of this lineage, which suggests that this region was not under high functional constraint when this mutation occurred.
Most of the protein encoded by MEPE exon 5 is variable, with the exception of eight short, non-contiguous motifs, among which are the RGD and SGDG motifs of the dentonin region and the ASARM peptide, whose biological functions are well known. Dentonin and ASARM are conserved in all placentals. However, in the two marsupials, a dentonin region was not identified, and the ASARM peptide is different. In addition, our study demonstrates five other conserved motifs in the region encoded by exon 5.
Dentonin region
The dentonin region contains both the RGD integrin-binding motif (cell–matrix adhesion) and the SGDG glycosaminoglycan-binding motif, which was shown to support bone formation and osteoblast proliferation [10, 11].
The presence of an RGD motif is a feature characterizing all members of the SIBLING family, i.e., DSPP, DMP1, IBSP, MEPE, and SPP1 [4]. This motif was probably inherited from a common ancestral SIBLING and transmitted to all members through tandem duplication events. Then, it has been either conserved or not in various lineages probably in connection with a slight advantage offered by its presence. Indeed, RGD is present in many placentalian MEPE, but it is lacking in kangaroo rat, squirrel, dolphin, and bats, and in marsupials. In all these species, no RGD was found elsewhere in MEPE, although the sequence encoded by exon 5 is rich in R (Arg), G (Gly), and D (Asp). There is no RGD motif in chicken OC-116. Therefore, it appears that this motif is not under a strong selective pressure in amniotes and would not be a crucial feature for MEPE/OC-116 function. This finding seems to contradict the result of our analysis using Selecton, which indicated that the RGD motif was under purifying selection. It is important to keep in mind that Selecton takes into account the physicochemical properties of the residues, but does not consider the presence of a motif. Therefore, when a residue of the RGD is substituted with an amino acid possessing the same properties, Selecton considers the position conserved, but this is not functionally correct.
The RGD motif alone is not sufficient for an optimal settlement of biomaterial surfaces by osteoblasts [51]. Nevertheless, there is evidence that the presence of an RGD close to the SGDG motif improves the mitogenic activity of dentonin, while the only presence of SGDG increases cell proliferation [11].
The main feature of the dentonin region is the presence of a SGDG motif and, for many placentals, its combination with a RGD motif. A SGDG motif is not found in the other SIBLINGs although their sequence is rich in G, D, and S (Ser). In MEPE, SGDG is found in all placentals, while it is lacking in marsupials. In chicken OC-116, a dentonin region is lacking, but a SGDG motif exists. Although MEPE/OC-116 probably inherited a SGDG motif from their amniote ancestor, the event creating the dentonin region seems to have occurred in the common ancestor of placentals, after the divergence of marsupials, some 148 Ma [36]. This typical feature was secondary kept unchanged in placentals as probably improving MEPE function.
Both RGD and SGDG motifs are connected with cell membranes, respectively via integrins and glycosaminoglycans. This cell-MEPE interaction allows (1) the regulation of bone homeostasis, facilitating cell–matrix adhesion and osteoblast differentiation, and (2) the activation of cell proliferation [10]. We infer that the function of the RGD motif could be compensated, in its absence, by SGDG. In marsupials that lack the two motifs, the dentonin function could be compensated by the newly identified conserved motif (DFEGSG) located close to the dentonin region. This hypothesis remains to be tested, as should also be tested the bioactivity of a synthetic peptide including the three motifs: DFEGSG, RGD and SGDG. It is also worth noting that an EGSG motif is present in chicken OC-116, but located at distance from the SGDG motif.
ASARM peptide
Our evolutionary analysis indicates that the region known as ASARM peptide [1] is present at the C-terminus of all placental MEPE. In humans, rats and mice, ASARM peptides are known as an inhibitor of mineralization (the so-called minhibins) when released [7, 8, 52]. The high sequence conservation of the ASARM peptide suggests that it plays a similar function in all other placentals. In the two marsupial sequences, however, the C-terminal ASARM of MEPE is capped by a dozen amino acids. In chicken OC-116, a serine-rich sequence similar to the ASARM peptide in mammals was identified in the C-ter region. However, it is not located at the extremity of the protein and is capped with several residues. Accordingly, this sequence resembles the ASARM peptide condition of marsupialian MEPE. We postulate that this ASARM-like sequence in chicken was inherited from the common ancestor of MEPE/OC-116. This particular motif probably originated in the ancestral SIBLING as ASARM-like sequences were identified in virtually all members of the family [4].
Therefore, we question whether this region could be processed differently in marsupials (see below). MEPE possess many potentially phosphorylated sites. The exact number of these sites that are present in vivo has yet to been accurately determined. However, other SIBLINGs possessing an ASARM peptide have also various phosphorylated forms [53], especially osteopontin (SPP1). In osteoblasts, most computer-predicted phosphorylated sites of SPP1 were demonstrated to be phosphorylated in vivo [54]. Phosphorylations are known to (1) modify the extracellular binding properties of proteins [55], (2) modulate cell–matrix interactions, (3) facilitate cell adhesion [56], and (4) promote cell migration [57]. The ASARM peptide of MEPE possesses at least one potentially phosphorylated serine that is conserved in all therian MEPE. When it is phosphorylated, this serine inhibits mineralization through an increasing affinity of the phosphorylated peptide with the surface of hydroxyapatite crystals. Phosphorylation also potentially regulates the sensitivity of ASARM to degradation by PHEX, a Zn metalloendopeptidase family member [8]. Indeed, the degradation of ASARM by PHEX facilitates extracellular matrix mineralization. When phosphorylated, the ASARM peptide binds to PHEX with stronger affinity and is protected from cathepsin B proteolysis, hence modulating osteogenesis inhibition and phosphatemia regulation. When PHEX is defective, MEPE-PHEX interaction is lost and MEPE is exposed to increased proteolysis, leading to genetic diseases [6]. For instance, there is a five-fold increase in concentration of ASARM peptide in the serum of HYP patients [58].
Does the presence of a C-terminal sequence cap in marsupials modify ASARM function? If additional amino acids change the conformation of ASARM peptide, this MEPE region will no longer be able to interact with PHEX, and/or its affinity with hydroxyapatite crystals when phosphorylated would be lost or reduced (see above). However, because (1) a large part of the ASARM sequence is similar to that in placental MEPE, and (2) this sequence was retained during million years of mammalian evolution, we speculate that it has an important function. The chicken ASARM-like peptide, the function of which is still unknown, is also capped by several amino acids. This leads us to assume that in placental MEPE the ASARM peptide is derived from an ASARM-like sequence capped by several amino acids as observed in marsupials and birds. The amino acids capping the C-terminus were probably present in the common therian ancestor then lost in the ancestor of placentals. It is worth noting that such a C-ter cap sequence is somewhat reminiscent of DMP1, a related SIBLING protein, which has a conserved C-ter sequence that caps a region with ASARM-motif similarities [52]. Could this ASARM sequence reveals a potential relationship of MEPE and DMP1? It is interesting to note here that several SIBLINGs (e.g., DMP1 and DSPP) possess an ASARM peptide. In DMP1, ASARM is similarly processed as in MEPE although separated by approximately 30 residues from the C-ter extremity. When released in the blood circulation, it is responsible, along with the ASARM peptide of MEPE, for mineralization defects [52]. This indicates that these protease-resistant peptides can be cleaved, and remain active, even when located at distance from the C-terminus of the protein.
Conserved motifs and adaptive selection: some new functions?
The evolutionary analysis of mammalian MEPE allowed us to identify five motifs, whose residues are well conserved, in addition to RGD and SGDG in the dentonin region and to the ASARM peptide. This conservation of amino acids indicates that these motifs are subjected to strong selective pressures and, therefore, certainly play an important role. However, to date, none of these conserved motifs were identified in protein databases and they are not recognized as bearing a particular function. In the future, to complete our knowledge of MEPE evolution, it would be interesting to test the possible functions of these five motifs, and in particular of the EGSG motif located close to the dentonin region, as they probably bear important properties.
Nine positions were predicted as being subjected to positive selection. Positive selection is defined as the evolutionary mechanism whereby newly produced mutants have higher fitnesses than the average in the population, and the frequencies of the mutants increase in the following generations [59]. All these positions are located in MEPE regions that are not associated with a known biological function. Substitutions occurring at random in regions with weak selective constraints could have led to residues that are structurally and/or functionally advantageous. Hence, these amino acids were positively selected in this ancestor and remained unchanged during evolution of this lineage, as suggested by our analysis. Although their function still remains to be defined, we speculate that the mutation of one of these residues could lead to some disorders in the considered lineage.
Finally, our evolutionary analysis has demonstrated that many amino acid positions were kept unchanged or were conservative during placental evolution. This allows us to predict that when a mutation of one of these positions will occur in humans this will result in a genetic disease. This provides additional support to the importance of such an evolutionary analysis of MEPE because it will allow to validate any point mutation (amino acid substitution) suspected to be responsible for a genetic disease.
Conclusion
Together with evidence of the presence of an additional exon in most placental MEPE, we have shown that the dentonin region of MEPE, with its two cell–matrix adhesion motifs (RGD and SGDG), is undoubtedly an innovation of the placental lineage. Its recruitment as an advantageous adaptation probably occurred after the placental–marsupial split, some 148 Ma. In contrast, the function of the ASARM peptide was likely selected earlier in evolution, at least in the common ancestor of amniotes (315 Ma), if one considers the presence of an ASARM-like sequence in the C-terminal region of chicken OC-116. It is possible that this event occurred even earlier as the ASARM peptide of MEPE was inherited from the duplication of an ancestral SIBLING that possessed a functional ASARM peptide.
The ancestral function of MEPE/OC-116 might have been linked to bone and eggshell mineralization [35, Bardet et al., submitted]. MEPE involvment in bone mineralization might explain why MEPE was not invalidated after eggshell was lost in therian ancestors. The functional constraints related to eggshell mineralization were relaxed and MEPE accumulated many substitutions, in particular in the large exon 5, as highlighted in our evolutionary analysis. At this time, MEPE function was restricted to bone and dentin matrix mineralization, and the relaxed functional pressure on exon 5 probably allowed the recruitment of new functions.
The evolutionary rates among the various placentalian MEPE are high, with the exception of primate sequences, which could indicate a stronger selection of MEPE residues and/or regions in this lineage. This rapid evolution, characterized by long branches in the distance tree, does not reflect current mammalian phylogenies [31], and it is obvious that MEPE is not a good phylogenetic marker, at least at the ordinal level and above (Fig. 1).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Alignment of the 37 amino acid sequences of mammalian MEPE. The human sequence was used as a reference sequence for this alignment, which takes into consideration mammalian relationships (see Fig. 1). Note that exon 3 and/or exon 4 are invalidated in some species. The 23 amino acid-long dentonin region (synthetical peptide AC-100) is located in the central region encoded by exon 5. It contains two important motifs, RGD and SGDG (boxed), which are preserved in most species. However, RGD is absent in several sequences and the two motifs are missing in marsupials. The amino acids of the ASARM peptide (Cterminus, boxed) are well conserved in all sequences studied, but in the two marsupials the ASARM sequence is followed by a dozen of residues. Newly identified important motifs are boxed in gray background. [ ] exon limits; (.) residue identical to human sequence; (-) Indel; (=) Unknown residue; (#) Residue identical in all sequences; (+) Residue identical in all therian sequences. (PDF 111 kb)
Tree distance obtained using the distance matrix of the 37 mammalian MEPE. The relationships between species do not refelct current mammalian phylogeny (compare with Fig. 1). The species in which MEPE shows a high rate of evolution are artefactually attracted to the base of the tree (e.g., rodents and insectivores). (JPG 450 kb)
Acknowledgments
We express our sincere thanks to IFRO (Institut Français pour la Recherche Odontologique) for financial support to CB and to Dr Matthew Vickarious (Ontario Veterinary College, University of Guelph, Canada) for English corrections. We also are grateful to Professor Jorge Cubo (UPMC) for his help in performing statistical analyses and to Dr Guillaume Achaz (UPMC) for helpful discussion. This work was supported by CNRS and UPMC (UMR 7138) grants.
References
- 1.Rowe PS, de Zoysa PA, Dong R, Wang HR, White KE, Econs MJ, Oudet CL. MEPE, a new gene expressed in bone marrow and tumors causing osteomalacia. Genomics. 2000;67:54–68. doi: 10.1006/geno.2000.6235. [DOI] [PubMed] [Google Scholar]
- 2.Petersen DN, Tkalcevic GT, Mansolf AL, Rivera-Gonzalez R, Brown TA. Identification of osteoblast/osteocyte factor 45 (OF45), a bone-specific cDNA encoding an RGD-containing protein that is highly expressed in osteoblasts and osteocytes. J Biol Chem. 2000;275:36172–36180. doi: 10.1074/jbc.M003622200. [DOI] [PubMed] [Google Scholar]
- 3.Gowen LC, Petersen DN, Mansolf AL, Qi H, Stock JL, Tkalcevic GT, Simmons HA, Crawford DT, Chidsey-Frink KL, Ke HZ, McNeish JD, Brown TA. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J Biol Chem. 2003;278:1998–2007. doi: 10.1074/jbc.M203250200. [DOI] [PubMed] [Google Scholar]
- 4.Fisher LW, Fedarko NS. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res. 2003;44:33–40. [PubMed] [Google Scholar]
- 5.Argiro L, Desbarats M, Glorieux FH, Ecarot B. Mepe, the gene encoding a tumor-secreted protein in oncogenic hypophosphatemic osteomalacia, is expressed in bone. Genomics. 2001;74:342–351. doi: 10.1006/geno.2001.6553. [DOI] [PubMed] [Google Scholar]
- 6.Rowe PS. The wrickkened pathways of FGF23, MEPE and PHEX. Crit Rev Oral Biol Med. 2004;15:264–281. doi: 10.1177/154411130401500503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowe PS, Kumagai Y, Gutierrez G, Garrett IR, Blacher R, Rosen D, Cundy J, Navvab S, Chen D, Drezner MK, Quarles LD, Mundy GR. MEPE has the properties of an osteoblastic phosphatonin and minhibin. Bone. 2004;34:303–319. doi: 10.1016/j.bone.2003.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Addison WN, Nakano Y, Loisel T, Crine P, McKee MD. MEPE-ASARM peptides control extracellular matrix mineralization by binding to hydroxyapatite: an inhibition regulated by PHEX cleavage of ASARM. J Bone Miner Res. 2008;23:1638–1649. doi: 10.1359/jbmr.080601. [DOI] [PubMed] [Google Scholar]
- 9.Guo R, Rowe PS, Liu S, Simpson LG, Xiao ZS, Quarles LD. Inhibition of MEPE cleavage by Phex. Biochem Biophys Res Commun. 2002;297:38–45. doi: 10.1016/S0006-291X(02)02125-3. [DOI] [PubMed] [Google Scholar]
- 10.Hayashibara T, Hiraga T, Yi B, Nomizu M, Kumagai Y, Nishimura R, Yoneda T. A synthetic peptide fragment of human MEPE stimulates new bone formation in vitro and in vivo. J Bone Miner Res. 2004;19:455–462. doi: 10.1359/JBMR.0301263. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Li W, Gao C, Kumagai Y, Blacher RW, DenBesten PK. Dentonin, a fragment of MEPE, enhanced dental pulp stem cell proliferation. J Dent Res. 2004;83:496–499. doi: 10.1177/154405910408300612. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg M, Lacerda-Pinheiro S, Jegat N, Six N, Septier D, Priam F, Bonnefoix M, Tompkins K, Chardin H, Denbesten P, Veis A, Poliard A. The impact of bioactive molecules to stimulate tooth repair and regeneration as part of restorative dentistry. Dent Clin North Am. 2006;50:277–298. doi: 10.1016/j.cden.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Delgado S, Casane D, Bonnaud L, Laurin M, Sire JY, Girondot M. Molecular evidence for precambrian origin of amelogenin, the major protein of vertebrate enamel. Mol Biol Evol. 2001;18:2146–2153. doi: 10.1093/oxfordjournals.molbev.a003760. [DOI] [PubMed] [Google Scholar]
- 14.Delgado S, Girondot M, Sire JY. Molecular evolution of amelogenin in mammals. J Mol Evol. 2005;60:12–30. doi: 10.1007/s00239-003-0070-8. [DOI] [PubMed] [Google Scholar]
- 15.Delgado S, Couble ML, Magloire H, Sire JY. Cloning, sequencing, and expression of the amelogenin gene in two scincid lizards. J Dent Res. 2006;85:138–143. doi: 10.1177/154405910608500205. [DOI] [PubMed] [Google Scholar]
- 16.Sire JY, Delgado S, Girondot M. The amelogenin story: origin and evolution. Eur J Oral Sci. 2006;114(Suppl 1):64–77. doi: 10.1111/j.1600-0722.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian S, Kumar S. Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics. 2006;7:306. doi: 10.1186/1471-2164-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delgado S, Ishiyama M, Sire JY. Validation of amelogenesis imperfecta inferred from amelogenin evolution. J Dent Res. 2007;86:326–330. doi: 10.1177/154405910708600405. [DOI] [PubMed] [Google Scholar]
- 19.Springer MS, Murphy WJ. Mammalian evolution and biomedicine: new views from phylogeny. Biol Rev Camb Philos Soc. 2007;82:375–392. doi: 10.1111/j.1469-185X.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- 20.Rambaut A (1996) Se-Al sequence alignment editor. Zoology Department, University of Oxford, Oxford
- 21.Kosakovsky Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 22.Maddison DR, Maddison WP (2005) MacClade4 Version 4.08. Sinauer, Sunderland, Mass.
- 23.Doron-Faigenboim A, Stern A, Mayrose I, Bacharach E, Pupko T. Selecton: a server for detecting evolutionary forces at a single amino-acid site. Bioinformatics. 2005;21:2101–2103. doi: 10.1093/bioinformatics/bti259. [DOI] [PubMed] [Google Scholar]
- 24.Stern A, Doron-Faigenboim A, Erez E, Martz E, Bacharach E, Pupko T. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007;35:506–511. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsunoyama K, Gojobori T. Evolution of nicotinic acetylcholine receptor subunits. Mol Biol Evol. 1998;15:518–527. doi: 10.1093/oxfordjournals.molbev.a025951. [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 27.Kosakovsky Pond SL, Frost SD. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 28.Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hincke MT, Gautron J, Tsang CP, McKee MD, Nys Y. Molecular cloning and ultrastructural localization of the core protein of an eggshell matrix proteoglycan, ovocleidin-116. J Biol Chem. 1999;274:32915–32923. doi: 10.1074/jbc.274.46.32915. [DOI] [PubMed] [Google Scholar]
- 33.Consortium ICS. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 34.Kawasaki K, Weiss KM. Evolutionary genetics of vertebrate tissue mineralization: the origin and evolution of the secretory calcium-binding phosphoprotein family. J Exp Zool B Mol Dev Evol. 2006;306:295–316. doi: 10.1002/jez.b.21088. [DOI] [PubMed] [Google Scholar]
- 35.Horvat-Gordon M, Yu F, Burns D, Leach RM., Jr Ovocleidin (OC 116) is present in avian skeletal tissues. Poult Sci. 2008;87:1618–1623. doi: 10.3382/ps.2008-00031. [DOI] [PubMed] [Google Scholar]
- 36.Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT, Yang SP, Heger A, Locke DP, Miethke P, Waters PD, Veyrunes F, Fulton L, Fulton B, Graves T, Wallis J, Puente XS, López-Otín C, Ordóñez GR, Eichler EE, Chen L, Cheng Z, Deakin JE, Alsop A, Thompson K, Kirby P, Papenfuss AT, Wakefield MJ, Olender T, Lancet D, Huttley GA, Smit AF, Pask A, Temple-Smith P, Batzer MA, Walker JA, Konkel MK, Harris RS, Whittington CM, Wong ES, Gemmell NJ, Buschiazzo E, Vargas Jentzsch IM, Merkel A, Schmitz J, Zemann A, Churakov G, Kriegs JO, Brosius J, Murchison EP, Sachidanandam R, Smith C, Hannon GJ, Tsend-Ayush E, McMillan D, Attenborough R, Rens W, Ferguson-Smith M, Lefèvre CM, Sharp JA, Nicholas KR, Ray DA, Kube M, Reinhardt R, Pringle TH, Taylor J, Jones RC, Nixon B, Dacheux JL, Niwa H, Sekita Y, Huang X, Stark A, Kheradpour P, Kellis M, Flicek P, Chen Y, Webber C, Hardison R, Nelson J, Hallsworth-Pepin K, Delehaunty K, Markovic C, Minx P, Feng Y, Kremitzki C, Mitreva M, Glasscock J, Wylie T, Wohldmann P, Thiru P, Nhan MN, Pohl CS, Smith SM, Hou S, Nefedov M, de Jong PJ, Renfree MB, Mardis ER, Wilson RK. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hughes RL, Carrick FN. Reproduction in female monotremes. Aust Zool. 1978;20:233–253. [Google Scholar]
- 38.Palmer BD, Guillette LJ. Oviductal proteins and their influence on embryonic development in birds and reptiles. In: Deeming DC, Ferguson MWJ, editors. Egg incubation: its effects on embryonic development in birds and reptiles. Cambridge: Cambridge University Press; 2004. pp. 29–46. [Google Scholar]
- 39.Kawasaki K. The SCPP gene repertoire in bony vertebrates and graded differences in mineralized tissues. Dev Genes Evol. 2009;219:147–157. doi: 10.1007/s00427-009-0276-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osada N, Hida M, Kusuda J, Tanuma R, Iseki K, Hirai M, Terao K, Suzuki Y, Sugano S, Hashimoto K (2000) Isolation of full-length cDNA clones from macaque brain cDNA libraries. [http://www.ncbi.nlm.nih.gov/nuccore/13874559]
- 41.Kawasaki K, Weiss KM. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc Natl Acad Sci USA. 2003;100:4060–4065. doi: 10.1073/pnas.0638023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shintani S, Kamakura N, Kobata M, Toyosawa S, Onishi T, Sato A, Kawasaki K, Weiss KM, Ooshima T. Identification and characterization of integrin-binding sialoprotein (IBSP) genes in reptile and amphibian. Gene. 2008;424:11–17. doi: 10.1016/j.gene.2008.07.035. [DOI] [PubMed] [Google Scholar]
- 43.Yang R, Gerstenfeld LC. Structural analysis and characterization of tissue and hormonal responsive expression of the avian bone sialoprotein (BSP) gene. J Cell Biochem. 1997;64:77–93. doi: 10.1002/(SICI)1097-4644(199701)64:1<77::AID-JCB11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 44.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/S0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 45.Chen JW, Romero P, Uversky VN, Dunker AK. Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J Proteome Res. 2006;5:888–898. doi: 10.1021/pr060049p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mann K, Hincke MT, Nys Y. Isolation of ovocleidin-116 from chicken eggshells, correction of its amino acid sequence and identification of disulfide bonds and glycosylated Asn. Matrix Biol. 2002;21:383–387. doi: 10.1016/S0945-053X(02)00031-8. [DOI] [PubMed] [Google Scholar]
- 47.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veis A. Amelogenin gene splice products: potential signaling molecules. Cell Mol Life Sci. 2003;60:38–55. doi: 10.1007/s000180300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayanan K, Gajjeraman S, Ramachandran A, Hao J, George A. Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J Biol Chem. 2006;281:19064–19071. doi: 10.1074/jbc.M600714200. [DOI] [PubMed] [Google Scholar]
- 50.Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dee KC, Andersen TT, Bizios R. Design and function of novel osteoblast-adhesive peptides for chemical modification of biomaterials. J Biomed Mater Res. 1998;40:371–377. doi: 10.1002/(SICI)1097-4636(19980605)40:3<371::AID-JBM5>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 52.Martin A, David V, Laurence JS, Schwarz PM, Lafer EM, Hedge AM, Rowe PS. Degradation of MEPE, DMP1 and release of SIBLING ASARM-peptides (minhibins): ASARM-peptide(s) are directly responsible for defective mineralization in HYP. Endocrinology. 2008;149:1757–1772. doi: 10.1210/en.2007-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasugai S, Zhang Q, Overall CM, Wrana JL, Butler WT, Sodek J. Differential regulation of the 55 and 44 kDa forms of secreted phosphoprotein 1 (SPP-1, osteopontin) in normal and transformed rat bone cells by osteotropic hormones, growth factors and a tumor promoter. Bone Miner. 1991;13:235–250. doi: 10.1016/0169-6009(91)90071-7. [DOI] [PubMed] [Google Scholar]
- 54.Christensen B, Kazanecki CC, Petersen TE, Rittling SR, Denhardt DT, Sorensen ES. Cell type-specific post-translational modifications of mouse osteopontin are associated with different adhesive properties. J Biol Chem. 2007;282:19463–19472. doi: 10.1074/jbc.M703055200. [DOI] [PubMed] [Google Scholar]
- 55.Boskey AL. Osteopontin and related phosphorylated sialoproteins: effects on mineralization. Ann N Y Acad Sci. 1995;760:249–256. doi: 10.1111/j.1749-6632.1995.tb44635.x. [DOI] [PubMed] [Google Scholar]
- 56.Ek-Rylander B, Flores M, Wendel M, Heinegard D, Andersson G. Dephosphorylation of osteopontin and bone sialoprotein by osteoclastic tartrate-resistant acid phosphatase. Modulation of osteoclast adhesion in vitro. J Biol Chem. 1994;269:14853–14856. [PubMed] [Google Scholar]
- 57.Al-Shami R, Sorensen ES, Ek-Rylander B, Andersson G, Carson DD, Farach-Carson MC. Phosphorylated osteopontin promotes migration of human choriocarcinoma cells via a p70 S6 kinase-dependent pathway. J Cell Biochem. 2005;94:1218–1233. doi: 10.1002/jcb.20379. [DOI] [PubMed] [Google Scholar]
- 58.Bresler D, Bruder J, Mohnike K, Fraser WD, Rowe PS. Serum MEPE-ASARM-peptides are elevated in X-linked rickets (HYP): implications for phosphaturia and rickets. J Endocrinol. 2004;183:R1–R9. doi: 10.1677/joe.1.05989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki Y, Gojobori T. A Method for detecting positive selection at single amino acid sites. Mol Biol Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the 37 amino acid sequences of mammalian MEPE. The human sequence was used as a reference sequence for this alignment, which takes into consideration mammalian relationships (see Fig. 1). Note that exon 3 and/or exon 4 are invalidated in some species. The 23 amino acid-long dentonin region (synthetical peptide AC-100) is located in the central region encoded by exon 5. It contains two important motifs, RGD and SGDG (boxed), which are preserved in most species. However, RGD is absent in several sequences and the two motifs are missing in marsupials. The amino acids of the ASARM peptide (Cterminus, boxed) are well conserved in all sequences studied, but in the two marsupials the ASARM sequence is followed by a dozen of residues. Newly identified important motifs are boxed in gray background. [ ] exon limits; (.) residue identical to human sequence; (-) Indel; (=) Unknown residue; (#) Residue identical in all sequences; (+) Residue identical in all therian sequences. (PDF 111 kb)
Tree distance obtained using the distance matrix of the 37 mammalian MEPE. The relationships between species do not refelct current mammalian phylogeny (compare with Fig. 1). The species in which MEPE shows a high rate of evolution are artefactually attracted to the base of the tree (e.g., rodents and insectivores). (JPG 450 kb)