Abstract
The pharmacological effects of morphine and morphine-like drugs are mediated primarily through the µ opioid receptor. Here we show that differential use of an in-frame translational start codon in the 5′-untranslated region of the OPRM1 generates different translational products in vivo and in vitro. The 5′-end of the OPRM1 gene is necessary for initiating the alternate form and for subsequent degradation of the protein. Initiation of OPRM1 at the upstream site decreases the initiation at the main AUG site. However, alternative initiation of the long form of OPRM1 produces a protein with a short half-life, resulting from degradation mediated by the ubiquitin–proteasome pathway. Reporter and degradation assays showed that mutations of this long form at the second and third lysines reduce ubiquitin-dependent proteasome degradation, stabilizing the protein. The data suggest that MOP expression is controlled in part by initiation of the long form of MOP at the alternate site.
Keywords: Human mu opioid receptor, Translational regulation, Protein degradation
Introduction
The pharmacological effects of opioid drugs and the physiological effects of endogenous opioid peptides are initiated through the binding and activation of opioid receptors [1]. The opioid receptors are classified into three major types (μ, δ, and κ) and have been studied both pharmacologically and by molecular cloning [2]. All three types of opioid receptors belong to the superfamily of G-protein-coupled receptors (GPCR). The μ opioid receptor (MOR) plays roles in morphine-induced analgesia, tolerance, and dependence, as indicated by pharmacological studies and analyses of MOR gene knockout mice [3, 4]. MOR expression is regulated by multiple mechanisms, including transcriptional [5–12] and posttranscriptional events [13–15]. However, the translational control of the MOR gene has not been well characterized.
Generation of protein variants by alternative pre-mRNA splicing or alternative initiation of mRNA translation enlarges the coding capacity and might explain the relatively low number of genes actually found in the human genome [16]. However, recent studies showed that the amino terminus of a protein substrate can also serve as the site of ubiquitination [17, 18]. Degradation of many short-lived cellular proteins, such as transcription factors, tumor suppressors, and cell cycle regulators, occurs via the ubiquitin–proteasome pathway [19–21]. The mechanism for GPCR proteolysis generally has been assumed to involve fusion of endosomes with lysosomes [22]. Although trafficking and degradation of several membrane proteins are regulated by ubiquitination catalyzed by E3 ubiquitin ligases, there is little evidence connecting ubiquitination with regulation of mammalian GPCR function [23]. Additionally, lysine residues provide the site of formation for multiubiquitin chains, which comprise several covalently linked ubiquitin moieties [24].
In this study, we demonstrate that an isoform of the human mu-opioid peptide (MOP) is generated by initiation at an alternative in-frame upstream AUG site (uAUG) and that initiation from this site can downregulate MOP expression from the main initiation site. This amino-terminal-extended product of MOP (long-form MOP) contains four lysine residues and therefore is sensitive to proteasome-dependent degradation. These data provide evidence for regulation of MOP expression and function at the translational level.
Materials and methods
Plasmid construction
All site-directed mutagenic PCR were performed as described previously [14]. The plasmid encoding the wild-type construct [huAUG (+)], the SP6 promoter-controlled 5′-UTR/luciferase (LUC)-fused and Human MOR 5′-UTR and an Exon fused to Flag (hMUEF) constructs have all been described previously [15]. The long-form-initiated and main AUGs in the OPRM1 5′-untranslated region (5′-UTR; −291 to +1) were inactivated by introducing point mutations into the start codons via oligonucleotide-directed mutagenesis (Stratagene) according to the manufacturer’s recommendation, using the following oligonucleotides: huAUG_Long: 5′-CGCAGAGGAGAAcTGCAGATGCTC-3′ (forward) and 5′-GAGCATCTGACgTTCTCCTCTGCG′ (reverse); huAUG_Main: 5′-AGTACCAcGGAAGACGCCAAA-3′ (forward) and 5′-TTTGGCGTCTTCCgTGGTACT-3′ (reverse). The lysine-deficient constructs (M_K1, M_K2, M_K3, M_K4, and M_Kall) were prepared via site-directed mutagenesis using the huAUG_Main construct as the template. Both charge-conservative (e.g., substitution of arginine for K1, K2, or K4) and charge-nonconservative (substitution of asparagine for K3) mutant constructs were generated. All mutated constructs were verified by sequencing analysis and then subcloned back to the parental vector.
Cell culture, DNA transfection and reporter gene assay
Human neuroblastoma NMB cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum. Transfection and reporter gene assays were performed as described previously [14]. Briefly, cells were plated in six-well plates at a concentration of 1 × 106 cells/well and cultured overnight before transfection. For luciferase reporter analyses of each promoter construct, 1 μg of the reporter plasmid was mixed with the Effectene transfection reagent (QIAGEN) for 10 min before being added to the well. Forty-eight hours posttransfection, cells were washed once with phosphate-buffered saline (PBS) and lysed with lysis buffer (Promega). To correct for the differences in transfection efficiency, a one-fifth molar ratio of a pCH110 plasmid (Amersham) containing the β-galactosidase gene under the SV40 promoter was included in each transfection for normalization. The luciferase and galactosidase activities of each lysate were determined according to the manufacturer’s instructions (Promega and Tropix, respectively).
Immunoprecipitation and immunoblot analysis
Cells were lysed in RIPA lysis buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.25% sodium deoxycholate, 0.1% Nonidet P-40, 0.1% Triton X-100, 50 mM NaF, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 50 mM sodium pyrophosphate, 10 mM sodium vanadate and 1× protease inhibitor cocktail (Roche)]. Immunoprecipitation of LUC-tagged MOP was performed using the luciferase antibody. After incubation for 2 h at 4°C protein–antibody complexes were captured on protein agarose-G-Sepharose beads (Sigma) by mixing for 18 h at 4°C. Beads were collected by centrifugation and washed three times in lysis buffer. After elution by incubation of beads for 5 min at 90°C in protein gel loading buffer, immunoprecipitated proteins were separated by Novex 3–8% Tris–Acetate precasting gels (Invitrogen).
Immunoblots were performed with approximately 30 μg of protein from each lysate resolved by SDS-PAGE using a 10% polyacrylamide gel. Gels were electroblotted onto polyvinylidene difluoride membranes (Amersham Bioscience) in transfer buffer (48 mM Tris–HCl, 39 mM glycine, and 20% methanol). Membranes were blocked in blocking solution (5% dry milk and 0.1% Tween 20 in Tris–buffered saline) overnight at 4°C. Immunoblotting with anti-LUC (Chemicon), anti-β-actin (Cell Signaling Technology), and anti-Ubiquitin (Millipore) antibodies were performed according to the manufacturer’s instructions. Signals were detected using a Storm 860 PhosphorImager system (Amersham Biosciences).
In vitro transcription/translation and autoradiography
Capped mRNAs were synthesized in vitro with the MAXIscript in vitro Transcription Kit (Ambion) according to the manufacturer’s instructions. Briefly, after linearization by SalI digestion, DNA was gel-purified (QIAGEN). The resulting DNA was transcribed in vitro (i.e., capped) by SP6 RNA polymerase in the presence of 1 mM of the methylated cap analog m7GpppG (Ambion). After a 1-h incubation at 37°C, samples were treated with 2 units of DNase for 15 min at 37°C. After ethanol precipitation and a 70% ethanol wash, RNA was resuspended in DEPC-treated water. RNA integrity was confirmed by gel electrophoresis. The amounts of RNA were analyzed by spectrophotometry and ethidium bromide visualization.
Equal amounts (0.1 μg) of RNA were added to the TnT Quick Coupled Transcription/Translation System (Promega) for translation under conditions recommended by the manufacturer. In vitro-translated proteins were labeled with l-[35S]-methionine (Amersham). Reactions were incubated for 60 min at 30°C and analyzed by SDS-PAGE on 10% polyacrylamide gels. The gels were dried and exposed to a PhosphorImager screen overnight at room temperature. The translated peptides were detected using a Molecular Dynamic Storm 860 PhosphorImager system.
Quantification of LUC and LacZ transcripts by real-time PCR and reverse transcription (RT)-PCR
Total RNA was isolated using TRI reagent (Molecular Research Center, Inc.) according to the supplier’s protocol. After quantification of total RNA by measuring OD at 260 nm, 1 μg of RNA was treated with one unit of DNase I (Invitrogen). Reverse transcription using an oligo-dT primer was performed with the Transcription First strand cDNA synthesis kit (Roche) according to the manufacturer’s protocol. The first strand obtained was quantified by real-time quantitative PCR using a SYBR Green assay on the iCycler Optical System (Bio-Rad). The following oligonucleotides were used to amplify the 159- and 105-bp fragments of the cDNAs (corresponding to LUC or LacZ, respectively): LUC primers, 5′-CCAGGACTGGTTTCTGTAAG-3′ (forward) and 5′-CTTTATGTTTTTGGCGTCTTCC-3′ (reverse); LacZ primers, 5′-GCTGCATAAACCGACTACACAAA-3′ (forward) and 5′-GCCGCACATCTGAACTTCAG-3′ (reverse). After first-strand cDNA synthesis, the samples were amplified by real-time PCR at 95°C for 30 s, 60°C for 30 s, and 70°C for 30 s. Relative mRNA levels were reported as the ratio of LUC mRNA to LacZ mRNA. For RT-PCR, total RNA isolated from human neuroblastoma NMB cells and human brain total RNA (Ambion) were prepared as previously described [7]. Primers specific to MOR mRNA were: primer hrtM (5′-CCAGGACTGGTTTCTGTAAG-3′, located at position −133 in 5′-UTR), P1 (5′-GATCATGGCCCTCTACTCCA-3′, located at position 216 in exon 1), and primer P2 (5′-GCATTTCGGG GAGTACGGAA-3′, located at position 557 in exon 2 to avoid amplification of genomic DNA). The PCR cycle conditions for human MOR consisted of 95°C for 60 s, 60°C for 60 s, and 72°C for 60 s followed by a 10-min extension at 72°C (34 cycles).
Radioligand binding assay
To determine the effects of various protease inhibitors on receptor degradation, NMB cells were transfected transiently with the indicated constructs. After a 24-h transfection, cells were incubated in the absence and presence of different protease inhibitors for 0–12 h. Protein degradation experiments were performed with 25 μM of MG132 (proteasomal inhibitor), leupeptin (calpain and trypsin-like protease inhibitor), or E64 (calpain inhibitor). All protease inhibitors were dissolved in dimethyl sulfoxide (DMSO); control cells were treated with DMSO alone. MOR gene expression was determined by a whole-cell-binding assay using [3H]diprenorphine in 25 mM HEPES buffer, 5 mM MgCl2 (pH 7.6). Specific binding was defined as the difference between the radioactivity bound to the cells in the presence and absence of 100 μM of the μ-opioid specific antagonist, CTOP.
Determination of protein degradation half-life
Protein degradation half-lives were determined as described [25]. Briefly, after transient transfection with the indicated constructs, cells were incubated in serum-free media and harvested at 0–18 h, and then assayed for remaining activity using the luciferase reporter assay.
Results
The 5′-UTR of OPRM1 presents an alternative site for translation initiation
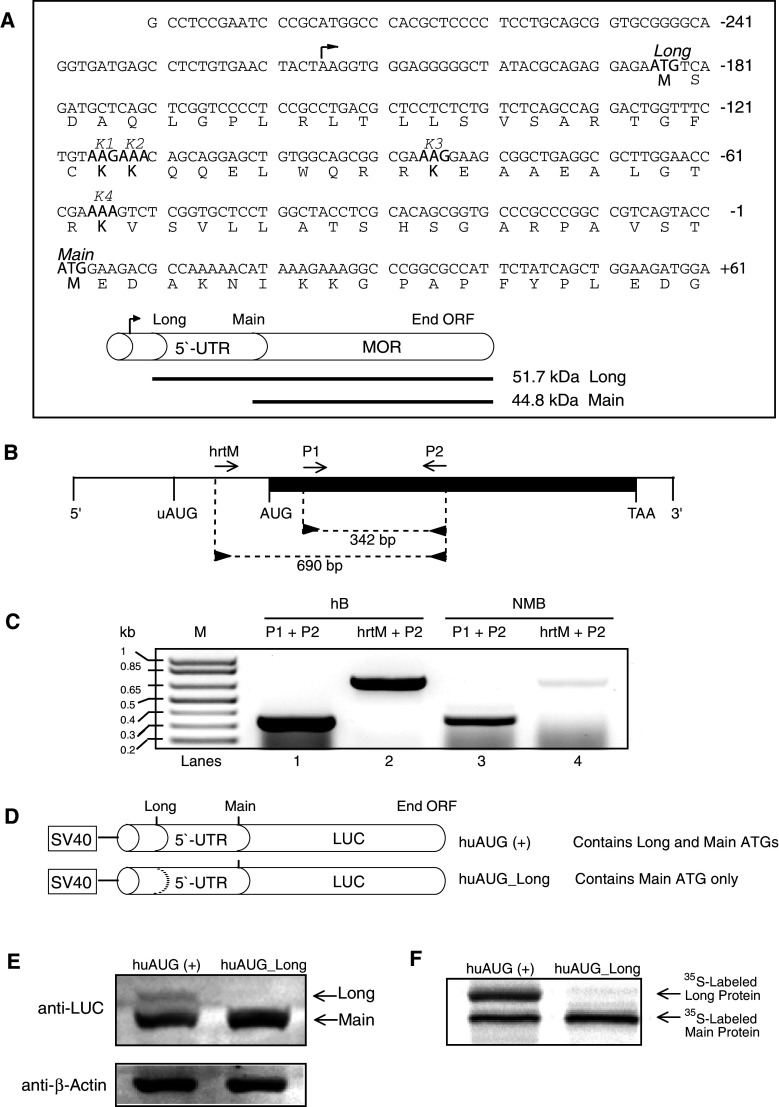
It is well established that many genes encode several variants of proteins by using alternative promoters and alternative splicing, greatly increasing the number of proteins produced. Alternative translation provides another molecular mechanism by which a single mRNA can produce several proteins [26]. The OPRM1 gene contains an in-frame start codon 186 bp upstream of the canonical ATG. Translation from this new open reading frame (ORF) should produce a 51.7-kDa protein similar to the 44.8-kDa mu-opioid peptide (MOP), but bearing an amino-terminal extension of an additional 62 residues (Fig. 1a). RT-PCR of human and NMB RNA using the primers hrtM, P1, and P2, which correspond to nucleotide sequences within the 5′-untranslated region and exon region (Fig. 1b), respectively, of the OPRM1 mRNA, exhibits two amplicons at 690 bp and at 342 bp (Fig. 1c). Our results confirm the existence of an alternative initiated OPRM1 mRNA that encodes a novel 5′-end extended isoform, long form. The functionality of this in-frame AUG codon was investigated by analyzing the proteins produced by constructs of the OPRM1 5′-UTR portion fused to the LUC gene (Fig. 1d). Immunoblotting (Fig. 1e) and in vitro transcription/translation (Fig. 1f) showed that both the regular (Main) and extended (Long) forms of the protein (initiated from the main AUG and uAUG, respectively) were expressed in wild-type cells. However, the long form of the MOP apparently downregulated expression of the main MOP. These results are similar to the previously reported uAUG-mediated downregulation of OPRM1 expression [15].
Fig. 1.
Schematic representation of the human μ opioid receptor (OPRM1) and putative 5′-UTR sequences. a Sequence of the OPRM1 5′-UTR used in this study. The in-frame uAUG (Long) and main AUG (Main) codons are indicated, as are the peptide sequences for the lysine (K1, K2, K3, and K4) codons. The arrowhead above the sequence indicates the transcription initiation site. The upstream and main AUGs encode the Long and Main forms of OPRM1, respectively. b Reverse transcription (RT)-PCR was carried out using oligonucleotide primers (“Materials and methods”). The diagram shows the relative position of sense (hrtM and P1; forward arrows) and antisense (P2; reverse arrows) primers along the OPRM1 mRNA. The darkened area represents the exon region (1,403 bp). Vertical lines represent the relative positions of the common 5′ end, initiation codon for the Long form of OPRM1 (uAUG), initiation codon for the Main form of OPRM1 (AUG), termination codon for both OPRM1s (TAA), and the 3′ end. c RT-PCR analysis of the OPRM1. Total RNA prepared from human brain (hB, lanes 1 and 2; Ambion) and NMB cells were subjected to RT-PCR with the oligonucleotide primer pairs indicated above the lanes followed by agarose gel electrophoresis. Sizes of amplification products obtained with the indicated primer pairs are shown. M denotes DNA markers, the sizes of which are shown on the left. d Luciferase fusion constructs containing portions of OPRM1 extension sequences. The huAUG (+) construct retains both the 5′ in-frame and main AUGs; the huAUG_Long construct retains only the main AUG. The dotted line represents ATGs converted to ACGs by point mutations. e NMB cells were transfected transiently with the constructs shown in d, and protein levels were analyzed by Western blotting for luciferase and β-actin. f Representative autoradiogram of proteins translated in vitro in the presence of [35S]-methionine using a coupled transcription/translation system
The uAUG-initiated long form of MOP contains no MOR activity
Both MOP products were examined for MOR activity. Luciferase reporter assays using the 5′-UTR of OPRM1 fused to a luciferase reporter plasmid (Fig. 2a) showed that inactivation of the long-form OPRM1 initiation site increased luciferase activity, whereas mutation of the main site decreased activity. Subsequently, the 5′-UTR and the coding regions (exons 1–4) of OPRM1 were fused in-frame to FLAG tags. Receptor-binding assays (Fig. 2b) paralleled the reporter assay results. These data support the hypothesis that the long-form MOP shows little, if any, receptor activity, whereas it downregulates expression of MOP produced by initiation at the main site.
Fig. 2.
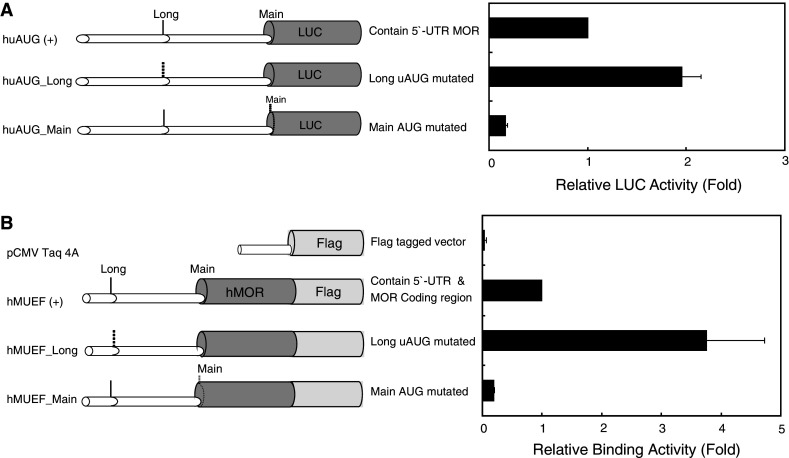
Translation of the OPRM1 gene is controlled by upstream AUG initiation. a Schematic representations of reporter constructs with wild-type and mutant human 5′-UTRs. Dotted lines represent ATGs converted to ACGs by point mutations. Relative LUC activity in NMB cells transfected transiently with the constructs (right panel). Bars indicate the standard errors of triplicate LUC assays. b Schematic representations of constructs containing the OPRM1 promoter and coding regions. The coding regions were fused in-frame to the FLAG tag, except for the negative control vector (pCMV-Taq4A). [3H]diprenorphine-binding assays performed on opioid receptor-expressing NMB cells transfected transiently with the constructs (right panel). Data represent the mean binding ± standard errors from representative triplicate assays
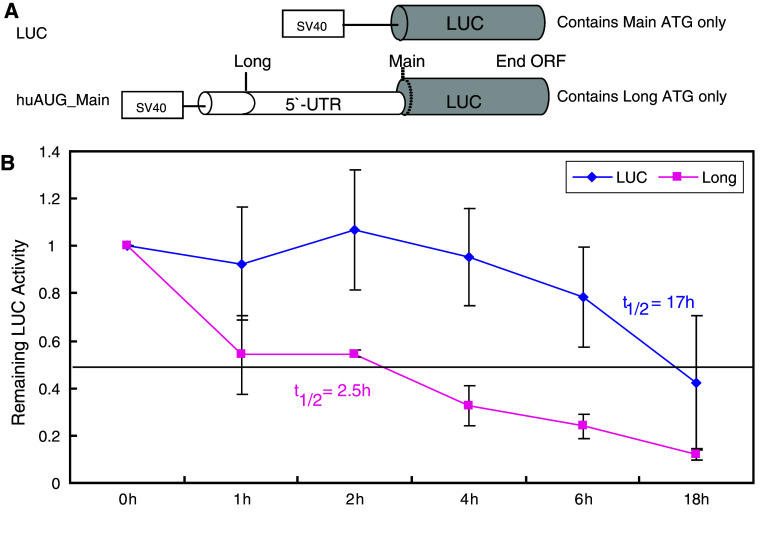
Main-AUG-initiated MOP is more stable than long-form MOP in NMB cells
The stabilities of the main and long forms of MOP were assessed by measuring luciferase activity over time in NMB cells transfected transiently with constructs in which the reporter protein was initiated from the main or alternative sites (Fig. 3a). When incubated in serum-free medium, the half-life of luciferase protein produced in NMB cells expressing the main AUG-initiated protein (LUC) was approximately 17 h. In contrast, the protein degraded much more rapidly in cells expressing the long form of the MOP, with an apparent half-life of 2.5 h (Fig. 3b).
Fig. 3.
The 5′-extended long form of MOP is a short-lived protein. a Schematics of the constructs used. The initiation codon (dotted line) indicates a point mutation of ATG to ACG. b Time course of protein degradation. The relative remaining LUC activity in NMB cells transfected transiently with the constructs shown in a was determined by incubating the cells in serum-free medium at 37°C for 0–18 h. Cell lysates were assayed for luciferase reporter activity, expressed as the ratio LUC/β-gal. Data represent the means and standard errors of three independent assays conducted in duplicate. t 1/2 = half-life
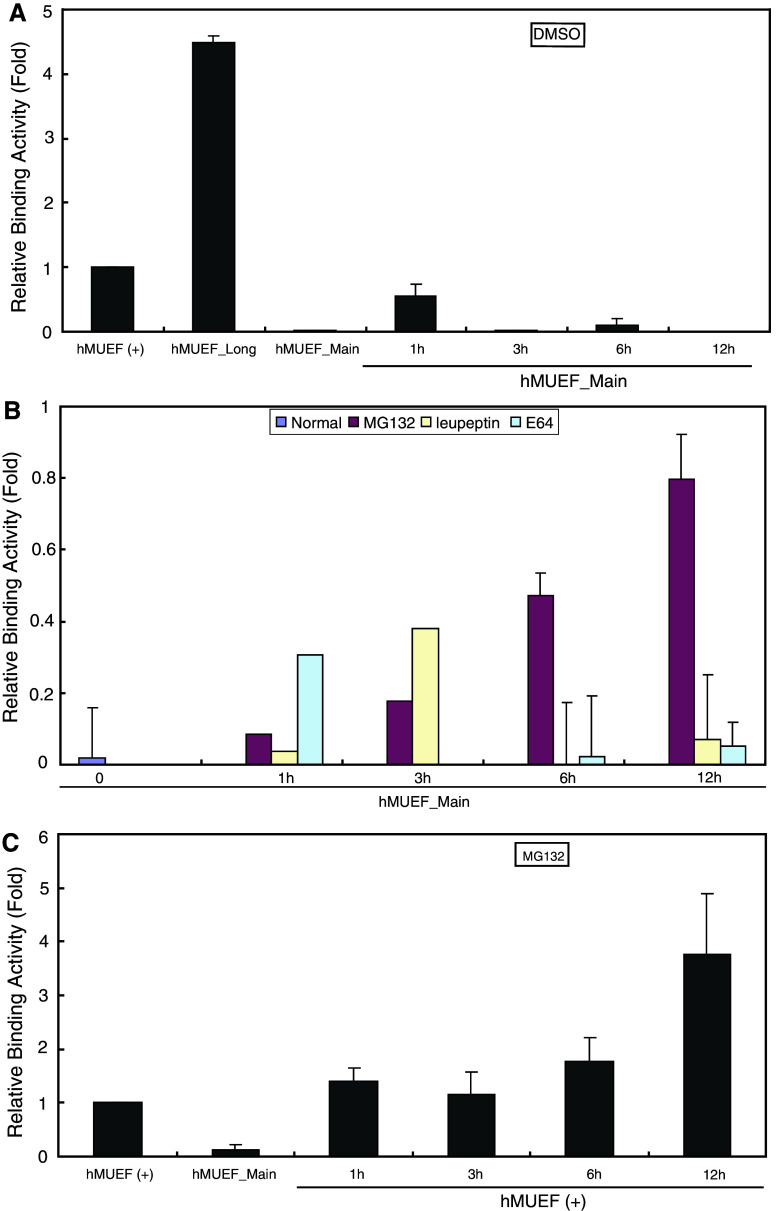
To determine the pathway responsible for the selective degradation of the long form of the protein, we performed receptor-binding experiments in cells transiently transfected with the in-frame-fused, FLAG-tagged constructs and subsequently exposed to several classes of protease inhibitors (Fig. 4). DMSO vehicle alone was unable to prevent the degradation of the long-form protein (Fig. 4a). However, the specific receptor-binding activity of long-form MOP increased significantly when incubated with the 26S proteasome inhibitor MG132. In contrast, two different cysteine protease inhibitors of lysosomal degradation, leupeptin and E64, did not prevent degradation over the course of the 12-h incubation (Fig. 4b). In cells expressing both the long and main forms of MOP, treatment with MG132 also increased specific receptor-binding activity (Fig 4c). Overall, these findings indicate that degradation of the long form of MOP is dependent on 26S proteasome.
Fig. 4.
Effects of protease inhibitors on the degradation of the long form of MOP. NMB cells were transfected for 24 h with the constructs shown in Fig. 2b. Cells were then incubated with or without inhibitor at 37°C for 0–12 h, as indicated. The time course of receptor binding was determined as described in Fig. 2. Mean values ± standard errors from representative triplicate assays are shown. a Receptor binding in cells expressing the long form of MOP (hMUEF_Main) incubated with DMSO alone (i.e., no protease inhibitors). b Receptor binding in cells expressing the long form of MOP incubated with 25 μM MG132, leupeptin, or E64. c Receptor binding in cells expressing both the long and main forms of MOP [hMUEF (+)], incubated with 25 μM MG132
26S proteasome-dependent degradation of MOP is mediated by different lysines, with lysine-3 being the most efficient
Ubiquitin can link covalently to lysine (Lys) side chains as a single monomer or as polyubiquitin chains, in which the internal branching of the residues can vary. Therefore, we used site-directed mutagenesis (Fig. 5a) to examine the roles of the four Lys residues in the 5′-extended region of long-form MOP (Fig. 1a) in proteasome-dependent degradation. Cells were assayed 48 h after transient transfection with the mutant constructs. Mutation of the lysine residues had no effect on the transcript levels produced by the various constructs (Fig. 5b, lower panel). However, mutations of Lys-1 and Lys-4 decreased luciferase activity (fivefold and twofold, respectively). In contrast, mutation of Lys-2 increased activity fivefold, and mutation of Lys-3 had the most pronounced effect, producing a nearly ninefold increase in luciferase activity. When all four lysine residues in the long-form protein were mutated, luciferase activity increased up to sixfold (Fig. 5b, upper panel).
Fig. 5.
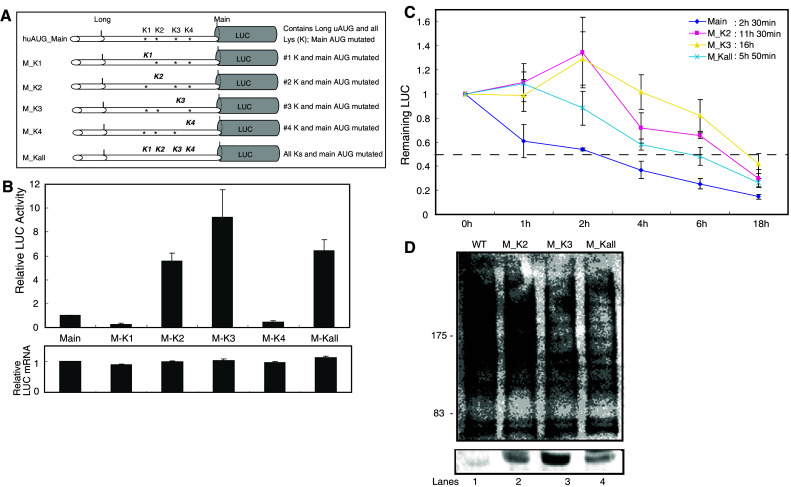
Mutation of OPRM1 lysines significantly affects the protein’s stability in NMB cells. a Schematic representations of OPRM1 constructs with 5′ lysines (K). Mutated lysines are shown in bold and italicized. b Relative LUC activity and mRNA levels (expressed as the ratios LUC/β-gal and LUC/LacZ, respectively) in NMB cells transfected transiently with each mutant construct. Bars indicate the standard errors of triplicate LUC assays. c Time course of protein degradation. The relative remaining LUC activity in NMB cells transfected transiently with the constructs shown in a was determined as described in Fig. 3b. d NMB cells cultured in six-well plates were transfected with 2 μg of wild-type (lane 1) or mutated (lanes 2–4) constructs. Forty-eight hours after transfection, immunoprecipitation was performed with anti-luciferase and analyzed by immunoblot using either the antibody to ubiquitin (upper panel) or the antibody to luciferase (lower panel). Numbers on the left (kDa) indicate position of molecular weight marker
The effects of lysine deletion on the kinetics of receptor downregulation were also examined (Fig. 5c). Transiently transfected NMB cells were incubated in serum-free medium at 37°C for up to 18 h. The long form of MOP was degraded with a short half-life of approximately 2.5 h, whereas mutations of Lys-2, Lys-3, or all four lysine residues were more stable, with half-lives of 11.5, 16, and 5.8 h, respectively.
To further examine this hypothesis, we tested ubiquitination of wild-type (WT), Lys-2, Lys-3, or all four lysine mutated plasmids (Fig. 5a) using NMB cells. Ubiquitination of WT protein was demonstrated by immunoblotting with anti-ubiquitin antibody (Fig. 5d, lane 1). Mutation of Lys-2 had little affect on the intensity of this smear (Fig. 5d, lane 2); by contrast, mutation of Lys-3 and all four lysines combined markedly reduced it (Fig. 5d, lanes 3 and 4). These observations confirmed that MOP is modified by polyubiquitination. They also establish that Lys-3 is the main site of ubiquitination. These results indicate that the lysine-dependent ubiquitination pathway plays a significant role in the degradation of long-form MOP in NMB cells.
Discussion
Upstream ORFs (uORFs) are common genomic features, occurring in up to 25% of mammalian genes [27] and 10–22% of fungal genes [28]. uORFs with canonical start codons are important in mRNAs whose main products are involved in controlling cell growth, such as receptors, growth factors, and other proto-oncogenes [29]. However, many eukaryotic mRNAs contain alternative translation initiation sites (TIS) either upstream of or downstream from the annotated TIS, and the proteins encoded from these alternative sites can be synthesized as isoforms with different amino-terminal segments [30]. In most cases, ribosomes recognize the alternative TIS by leaky scanning. The reinitiation of translation is likely to represent a special situation [26]. It also should be noted that the translation efficiency of many eukaryotic mRNAs must be low to prevent the harmful overproduction of proteins with regulatory functions, and the uAUG/uORFs are frequently used to inhibit the translation efficiency of such mRNAs [26]. In the case of OPRM1 mRNAs, reinitiation of translation can be used as a mechanism for selection of alternative start sites and synthesis of alternative protein isoforms [15]. The present study demonstrates that a 5′-extended long form of MOP is initiated from an in-frame uAUG site, producing a long form of the MOP, and that the uORF peptide encoded by the 5′-UTR of this protein participates in repression of OPRM1 translation.
In eukaryotic cells, a wide variety of proteins with roles in cell cycle progression, transcriptional control, signal transduction and metabolic regulation are degraded by the ubiquitination/proteasome pathway [31, 32]. This study demonstrates that a ubiquitin–proteasome-dependent pathway is involved in the degradation of the long form of MOP. Furthermore, ubiquitination of the long-form MOP protein is apparently dependent on the presence of the lysine residues in the amino-extended region, with Lys-3 exerting the most significant effect on protein stability.
Two major pathways operate in cells to degrade intracellularly retained GPCRs: the ubiquitin-dependent proteasomal pathway and the lysosomal pathway [31, 33]. Integral membrane proteins expressed at the cell surface are internalized and degraded by lysosomes. In contrast, in a degradation pathway known as endoplasmic reticulum quality control (ERQC), membrane proteins retained in the ER by the quality control system are retrotranslocated into the cytosol and degraded by the ubiquitin–proteasome system. Proteins that pass ERQC criteria traffic to their final destinations through the secretory pathway, whereas nonnative and unassembled subunits of multimeric proteins are degraded by the ER-associated degradation (ERAD) pathway [34]. The ERQC system is a cellular process designed to protect the cell from the accumulation of toxic unfolded proteins. Sometimes this system is overzealous and prevents mutants, which could still be biologically active, from leaving the ER (or to escape ERAD), thus leading to disease [35, 36]. It is therefore possible that the long form of MOP is also regulated by the ERAD pathway. The results of the present study are similar to the mechanism observed in the Cys-27 variant of the δ opioid receptor [37], as well as the V2 vasopressin receptor [38].
In summary, translation of OPRM1 mRNA leads to synthesis of an MOP isoform through the use of an alternative in-frame initiation codon. Furthermore, synthesis of this 5′-extended isoform regulates expression of the MOP expressed from the main TIS. However, the product of this isoform is degraded rapidly by a ubiquitin-dependent proteasomal pathway.
Acknowledgments
This work was supported by National Institutes of Health research grants DA000564, DA001583, DA011806, K05-DA070554, DA011190, and DA013926, and by the A&F Stark Fund of the Minnesota Medical Foundation.
References
- 1.Chaturvedi K, Christoffers KH, Singh K, Howells RD. Structure and regulation of opioid receptors. Biopolymers. 2001;55:334–346. doi: 10.1002/1097-0282(2000)55:4<334::AID-BIP1006>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Min BH, Augustin LB, Felsheim RF, Fuchs JA, Loh HH. Genomic structure analysis of promoter sequence of a mouse μ opioid receptor gene. Proc Natl Acad Sci USA. 1994;91:9081–9085. doi: 10.1073/pnas.91.19.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kieffer B. Recent advances in molecular recognition and signal transduction of active peptides: receptors for opioid peptides. Cell Mol Neurobiol. 1995;15:615–635. doi: 10.1007/BF02071128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kieffer B. Opioids: first lessons from knockout mice. Trends Pharmacol Sci. 1999;20:19–26. doi: 10.1016/S0165-6147(98)01279-6. [DOI] [PubMed] [Google Scholar]
- 5.Kim CS, Hwang CK, Choi HS, Song KY, Law PY, Wei LN, Loh HH. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J Biol Chem. 2004;279:46464–46473. doi: 10.1074/jbc.M403633200. [DOI] [PubMed] [Google Scholar]
- 6.Choi HS, Hwang CK, Kim CS, Song KY, Law PY, Wei LN, Loh HH. Transcriptional regulation of mouse mu opioid receptor gene: Sp3 isoforms (M1, M2) function as repressors in neuronal cells to regulate the mu opioid receptor gene. Mol Pharmacol. 2005;67:1674–1683. doi: 10.1124/mol.104.008284. [DOI] [PubMed] [Google Scholar]
- 7.Choi HS, Kim CS, Hwang CK, Song KY, Wang W, Qiu Y, Law PY, Wei LN, Loh HH. The opioid ligand binding of human mu-opioid receptor is modulated by novel splice variants of the receptor. Biochem Biophys Res Commun. 2006;343:1132–1140. doi: 10.1016/j.bbrc.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 8.Kim CS, Choi HS, Hwang CK, Song KY, Lee BK, Law PY, Wei LN, Loh HH. Evidence of the neuron-restrictive silencer factor (NRSF) interaction with Sp3 and its synergic repression to the mu opioid receptor (MOR) gene. Nucleic Acids Res. 2006;34:6392–6403. doi: 10.1093/nar/gkl724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang CK, Song KY, Kim CS, Choi HS, Guo XH, Law PY, Wei LN, Loh HH. Evidence of endogenous mu opioid receptor regulation by epigenetic control of the promoters. Mol Cell Biol. 2007;27:4720–4736. doi: 10.1128/MCB.00073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi HS, Kim CS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Novel function of the poly(C)-binding protein alpha CP3 as a transcriptional repressor of the mu opioid receptor gene. FASEB J. 2007;21:3963–3973. doi: 10.1096/fj.07-8561com. [DOI] [PubMed] [Google Scholar]
- 11.Choi HS, Hwang CK, Kim CS, Song KY, Law PY, Loh HH, Wei LN. Transcriptional regulation of mouse mu opioid receptor gene in neuronal cells by poly(ADP-ribose) polymerase-1. J Cell Mol Med. 2008;12:2319–2333. doi: 10.1111/j.1582-4934.2008.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi HS, Song KY, Hwang CK, Kim CS, Law PY, Wei LN, Loh HH. A proteomics approach for identification of single strand DNA-binding proteins involved in transcriptional regulation of mouse mu opioid receptor gene. Mol Cell Proteomics. 2008;7:1517–1529. doi: 10.1074/mcp.M800052-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei LN, Law PY, Loh HH. Post-transcriptional regulation of opioid receptors in the nervous system. Front Biosci. 2004;9:1665–1679. doi: 10.2741/1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song KY, Hwang CK, Kim CS, Choi HS, Law PY, Wei LN, Loh HH. Translational repression of mouse mu opioid receptor expression via leaky scanning. Nucleic Acids Res. 2007;35:1501–1513. doi: 10.1093/nar/gkm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song KY, Kim CS, Hwang CK, Choi HS, Law PY, Wei LN, Loh HH (2009) uAUG-mediated translational initiations are responsible for human mu opioid receptor gene expression. J Cell Mol Med (in press) [DOI] [PMC free article] [PubMed]
- 16.Uhlmann-Schiffler H, Rössler OG, Stahl H. The mRNA of DEAD box protein p72 is alternatively translated into an 82-kDa RNA helicase. J Biol Chem. 2002;277:1066–1075. doi: 10.1074/jbc.M107535200. [DOI] [PubMed] [Google Scholar]
- 17.Aviel S, Winberg G, Massucci M, Ciechanover A. Degradation of the Epstein–Barr virus latent membrane protein 1 (LMP1) by the ubiquitin–proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J Biol Chem. 2000;275:23491–23499. doi: 10.1074/jbc.M002052200. [DOI] [PubMed] [Google Scholar]
- 18.Reinstein E, Scheffner M, Oren M, Ciechanover A, Schwartz A. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin–proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene. 2000;19:5944–5950. doi: 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- 19.Glickman MH, Ciechanover A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 20.Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/S0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 21.Ciechanover A, Schwartz AL. The ubiquitin–proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci USA. 1998;95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornilova E, Sorkina T, Beguinot L, Sorkin A. Lysosomal targeting of epidermal growth factor receptors via a kinase-dependent pathway is mediated by the receptor carboxyl-terminal residues 1022–1123. J Biol Chem. 1996;271:30340–30346. doi: 10.1074/jbc.271.48.30340. [DOI] [PubMed] [Google Scholar]
- 23.Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 24.Madura K, Varshavsky A. Degradation of G alpha by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 25.Chaturvedi K, Bandari P, Chinen N, Howells RD. Proteasome involvement in agonist-induced down-regulation of mu and delta opioid receptors. J Biol Chem. 2001;276:12345–12355. doi: 10.1074/jbc.M008054200. [DOI] [PubMed] [Google Scholar]
- 26.Kochetov AV. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays. 2008;30:683–691. doi: 10.1002/bies.20771. [DOI] [PubMed] [Google Scholar]
- 27.Crowe ML, Wang XQ, Rothnagel JA. Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides. BMC Genomics. 2006;7:16. doi: 10.1186/1471-2164-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Batürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae . Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov IP, Loughran G, Atkins JF. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Natl Acad Sci USA. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kochetov AV, Ahmad S, Ivanisenko V, Volkova OA, Kolchanov NA, Sarai A. uORFs, reinitiation and alternative translation start sites in human mRNAs. FEBS Lett. 2008;582:1293–1297. doi: 10.1016/j.febslet.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 32.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 33.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin–proteasome system and onto human diseases and drug targeting. Exp Biol Med (Maywood) 2006;231:1197–1211. doi: 10.1177/153537020623100705. [DOI] [PubMed] [Google Scholar]
- 34.Vembar S, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;12:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- 36.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leskelä T, Markkanen PM, Alahuhta IA, Tuusa JT, Petäjä-Repo UE. Phe27Cys polymorphism alters the maturation and subcellular localization of the human delta opioid receptor. Traffic. 2009;10:116–129. doi: 10.1111/j.1600-0854.2008.00846.x. [DOI] [PubMed] [Google Scholar]
- 38.Schwieger I, Lautz K, Krause E, Rosenthal W, Wiesner B, Hermosilla R. Derlin-1 and p97/valosin-containing protein mediate the endoplasmic reticulum-associated degradation of human V2 vasopressin receptors. Mol Pharmacol. 2008;73:697–708. doi: 10.1124/mol.107.040931. [DOI] [PubMed] [Google Scholar]