Abstract
Aryl hydrocarbon receptor nuclear translocator (ARNT) binds to other basic helix-loop-helix Per/ARNT/Sim (bHLH-PAS) proteins to form functional transcriptional complexes in order to regulate specific biological pathways. Here, we report a novel mechanism that upon EGF treatment, ARNT associated with non-bHLH-PAS transcription factors, c-Jun/Sp1, and regulated gene expression, through forming a c-Jun/ARNT/Sp1 complex and binding to the Sp1 site of the gene promoter. EGF-induced promoter activity and the mRNA level of 12(S)-lipoxygenase as well as the association between c-Jun and Sp1 were reduced by ARNT knockdown. Notably, dominant negative c-Jun mutant, TAM-67, blocked ARNT-mediated 12(S)-lipoxygenase expression, demonstrating that c-Jun was responsible for the transcriptional activation. Moreover, ARNT knockdown also inhibited other EGF-induced c-Jun/Sp1 mediated gene expression, such as p21WAF1/CIP1. Our results reveal a novel mechanism by which ARNT acts as a modulator to bridge the c-Jun/Sp1 interaction and plays a role in EGF-mediated gene expression under normoxic conditions.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0392-9) contains supplementary material, which is available to authorized users.
Keywords: Epidermal growth factor (EGF), Aryl hydrocarbon receptor nuclear translocator (ARNT), Gene expression, c-Jun/Sp1, Protein–DNA interaction
Introduction
Aryl hydrocarbon receptor nuclear translocator (ARNT), also known as hypoxia-inducible factor-1β (HIF-1β), is a member of the basic helix-loop-helix Per/ARNT/Sim (bHLH-PAS) family of transcription factors, which can form homo- or heterodimers with other bHLH-PAS domain-containing proteins, such as aryl hydrocarbon receptor (AhR), HIF-1α, and single-minded (SIM) proteins, in regulating downstream gene expression upon various environmental stresses [1]. For instance, ARNT forms heterodimers with HIF-1α and regulates the expression of numerous genes in tumorigenesis and hypoxia [2, 3]. In response to xenobiotics, the heterodimers of ARNT/AhR can recognize the xenobiotic response element (XRE) of the target gene thus promoting gene transcription, most notably of the CYP1A1 gene, which is important in detoxification [4]. In addition, SIM-1, SIM-2, and ARNT have been reported to be involved in the neural development of normal embryos [5–7]. Despite the above-mentioned functions, ARNT has recently been shown to be involved in the regulation of CD30-mediated nuclear factor (NF)-κB activity [8]. Our previous report also showed that ARNT could associate with c-Jun in response to EGF and bind to the CRE site of the COX-2 promoter, resulting in the transcriptional activation of the COX-2 gene and squamous cell carcinoma formation [9]. These findings imply a possible role of ARNT in tumorigenesis under normoxic conditions.
Growth factors such as epidermal growth factor (EGF) and vascular endothelial growth factor (VEGF) play important roles in the regulation of cell growth and tumorigenesis, by mediating gene expression [10]. EGF receptor (EGFR) is expressed at high levels in a variety of human tumors and has been associated with poor prognosis and low survival rate [11]. Numerous effects of the activation of EGFR signaling pathways have been reported; they include increased proliferation and angiogenesis and decreased apoptosis [12]. The activation of EGFR signaling leads to increased mitogen-activated protein kinase (MAPK) activity, which results in c-Jun/Sp1-mediated induction of transcription of genes such as 12(S)-lipoxygenase [13, 14]. Furthermore, overexpressed 12(S)-lipoxygenase has been reported to mediate an increase in VEGF promoter activity and to regulate angiogenesis in prostate cancer cells [15].
ARNT has long been regarded as a general partner to HIF-1α in hypoxia and in mediating tumor growth. Unlike HIF-1α, which would be degraded in normoxic conditions, ARNT is expressed ubiquitously and constitutively in cells. Although its functions in hypoxic conditions and in the stimulation of xenobiotics have been largely discussed, little is known about its role in response to growth factor activation. Growth factors play important roles in a variety of physiological and pathophysiological functions, including cell proliferation, differentiation, and tumorigenesis. Therefore, we proposed that ARNT might play some role in growth factor-induced gene expression. Thus, from a DNA affinity precipitation assay (DAPA) carried out previously in our work, that pulled down Sp1-binding proteins after treatment with EGF, ARNT was also precipitated. Genes that are regulated by c-Jun/Sp1 interactions, such as 12(S)-lipoxygenase and p21WAF1/CIP1 [13, 16], were then studied.
In this study, the immunoprecipitation assay showed that ARNT formed a complex with c-Jun and Sp1 and participated in c-Jun/Sp1-regulated gene expression. Knocking down ARNT inhibited the interaction between c-Jun and Sp1; moreover, the binding of the c-Jun/ARNT/Sp1 complex to the promoter regions was also attenuated with the knockdown of Sp1. Our results revealed a novel mechanism by which ARNT acts as a modulator to bridge the c-Jun/Sp1 interaction and plays a role in EGF-mediated gene expression under normoxic conditions.
Materials and methods
Cell culture
The cell line of human squamous cell carcinoma (A431), c4 and vT2 were grown at 37°C under 5% CO2 in 10-cm plastic dishes containing 10 ml of Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin, and 100 U/ml penicillin. In this series of experiments, cells were treated with 50 ng/ml EGF (Pepro Technology, Rocky Hill, NJ, USA) in culture medium supplemented with 10% fetal bovine serum, unless stated otherwise. C4 and vT2 cells origin from the same cell type, where c4, a 3,4-benzopyrene-resistant mutant clone (c4) of the mouse hepatoma Hepa-1c1c7 cell line, was examined for a single point mutation, leading to replacement of Gly326 with Asp between two internal repeats in the highly conserved PAS domain that causes the defective function of ARNT; the vT2 cell line was derived from c4 cell line, possesses a complete transfected ARNT cDNA, and expresses the ARNT gene [17].
Plasmid construction
ARNT cDNA fragments were generated by polymerase chain reaction (PCR) and were subcloned into pcDNA3.1- myc/His vector (Invitrogen, Grand Island, NY, USA) at the BamH I and EcoR V sites. The forward and reverse primers were 5′-CGGGATCCAT GGCGGCGACTACTG-3′ and 5′-CGGATATCTTCTGAAAAGGGGGGAAAC-3′, respectively. C-Jun cDNA was amplified by PCR and inserted into the BamHI and Bgl II sites in pcDNA3.1/myc-His to generate myc-c-Jun. The forward and reverse primers were 5′-CGGGATCCATGACTGCAAAGATG-3′ and 5′- GGAAGATCTTCAAAATGTTTGC-3′, respectively. The vector sequences were confirmed by DNA sequencing. The construct of N-terminal-truncated c-Jun mutant (TAM-67) was kindly provided by Michael J Barrer of the National Institutes of Health in USA.
Transfection of cells with plasmids and luciferase assay
Luciferase vectors bearing wild-type (pXLO-7-1) or Sp1 mutants (SPM7) of 12(S)-lipoxygenase gene promoter and pPLA599 of cPLA 2 gene promoter were used [18, 19]. Transient transfection of cells with plasmids was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions but with slight modifications. A431 cells were replated for 36 h before transfection at a density of 3 × 105 cells in 2 ml of fresh culture medium in a 3.5-cm plastic dish. For use in transfection, 2 μl of Lipofectamine 2000 were incubated with 0.5 μg of pXLO-7-1 and SPM7 luciferase plasmids, or plasmids where indicated such as myc-ARNT, pRSVjun and myc-c-Jun in 1 ml of Opti-MEM medium for 30 min at room temperature. Cells were transfected by changing medium with 1 ml of Opti-MEM containing plasmids and Lipofectamine 2000, and then incubated at 37°C in a humidified atmosphere of 5% CO2 for 24 h. Following the change of Opti-MEM medium to 2 ml of fresh culture medium, cells were incubated for an additional 24 h, unless stated otherwise. The luciferase activity in cell lysate was determined as described previously [18].
Reverse transcription–PCR
Total RNA was isolated using the TRIzol RNA extraction kit (Invitrogen), and 2.5 μg of RNA were subjected to reverse transcription PCR with SuperScriptTMII (Invitrogen). The 12(S)-lipoxygenase-specific primers (sense, 5′-CTGGGCCACCTGGAAG-3′; antisense, 5′-GAGAGGTCGAGCGTCT-3′), ARNT-specific primers (sense, 5′-TGGGTCCAGCCATTGCCTCT-3′; antisense, 5′-CG AGCCAGGGCACTACAGGT-3′) and GAPDH primers were used. The PCR products were separated by 1% agarose-gel electrophoresis and visualized with ethidium bromide staining.
Quantitative real-time PCR
Primers specifically used for quantitative real-time PCR were as follows: (F) 5′-CTCAGATGGAGGAATTTTTG-3′ and (R) 5′-TATGTCATCCCTTTGGTAGAAG-3′. For each reaction the PCR mixture consisted of 5 μl of 2× SYBR® Advantage® qPCR premix (Clontech, Mountain view, CA, USA), 4 μl of reverse transcription reaction product, 500 nM of each forward and reverse primers and sterile distilled water was added to a final volume of 10 μl. Amplification and detection of specific products were carried out on a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA).
Western blotting
The nuclear extracts of cells were prepared for western blot analysis according to the method described [20]. An analytical 10% sodium dodecyl sulfate polyacrylamide slab gel electrophoresis (SDS–PAGE) was performed, and 30 μg of protein were analyzed, unless stated otherwise. For immunoblotting, proteins in the SDS gels were transferred onto a polyvinylidene difluoride membrane by an electroblot apparatus. Antibodies against human Sp1 (Upstate, Lake Placid, NY, USA), c-Jun (Santa Cruz Biotechology, Santa Cruz, CA, USA), phospho-c-Jun (Santa Cruz), ARNT (Santa Cruz) and β-actin (Santa Cruz) were used as the primary antibodies. Mouse or rabbit IgG antibodies coupled to horseradish peroxidase were used as secondary antibodies. An enhanced chemiluminescence kit (Pierce, Rockford, IL, USA) was used for detection.
Coimmunoprecipitation
For this, 200 μg protein of nuclear extracts were incubated under gentle shaking at 4°C overnight with a mixture of anti-c-Jun, anti-Sp1 or anti-ARNT antibodies and protein A agarose in 300 μl of immunoprecipitation buffer (20 mM HEPES, pH 7.9, 420 mM NaCl, 1.5 mM MgCl2 and 25% glycerol (v/v), 0.5 mM phenylmethylsulfonyl fluoride, 1 mM orthovanadate, 2 μg/ml pepstatin A, and 2 μg/ml leupeptin). Beads were pelleted at 7,500g for 2 min and washed three times with RIPA buffer (50 mM Tris–HCl, pH 7.5, 1% IGEPAL CA-630 (v/v), 150 mM NaCl, and 0.5% sodium deoxycholate). Protein was removed from the beads by boiling in sample buffer (120 mM Tris–HCl, pH 6.8, 10% glycerol, 3% SDS, 20 mM DTT, and 0.4% bromophenol blue) for 5 min and subjected to SDS–PAGE on a 10% gel. Western blot analysis was carried out as described above.
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation assay was performed as previously reported [21] with minor modifications. Briefly, A431 cells were treated with 1% formaldehyde for 15 min. The crosslinked chromatin was sonicated to 400- to 500-bp fragments. Lysates were precleaned with protein A beads and incubated overnight at 4°C with antibodies specific to ARNT (Santa Cruz), c-Jun (Santa Cruz), Sp1 (Upstate) or control rabbit IgG. Immune complexes were precipitated with protein A beads pre-absorbed with sonicated ssDNA and BSA. After reversal of cross-linking, levels of precipitated Sp1 promoter DNA were determined by PCR. Oligonucleotides spanning the Sp1 binding sites of 12(S)-lipoxygenase and p21WAF1/CIP1 were as follows: 12(S)-lipoxygenase: sense, 5′-GGGAAGTGTT CTCATCTATG-3′; antisense, 5′-GGCCACTTCCAACCTTTAAA-3′ and p21WAF1/CIP1: sense, 5′-ACCAACGCAGGCGAGGGACT-3′; antisense, 5′-CCGGCTCCACAAGGAACTGA-3′. The PCR products were separated by 1% agarose-gel electrophoresis and visualized with ethidium bromide staining.
DNA affinity precipitation assay (DAPA)
Quantitation of the change of c-Jun, Sp1 and ARNT binding to 12(S)-lipoxygenase promoter element was achieved by DNA affinity precipitation assay according to the method reported previously [22]. In brief, 5′-biotinylated oligonucleotides corresponding to the promoter region of 12(S)-lipoxygenase and p21WAF1/CIP1 promoters, which sense −170 to −110 and −64 to −84 bp, respectively, and antisense strands elements were annealed. The DNA affinity precipitation assay was performed by incubating 2 μg of biotinylated DNA probe with 200 μg of nuclear extract and 20 μl of streptavidin-agarose beads in phosphate-buffered saline at room temperature for 1 h with rotation. Beads were collected and washed with ice-cold phosphate-buffered saline for three times. The bound proteins were eluted by loading buffer and separated by SDS–PAGE, followed by western blot analysis probed with specific antibodies.
Results
EGF induces formation of the c-Jun/ARNT/Sp1 complex and its binding to the Sp1 sites of the gene promoter
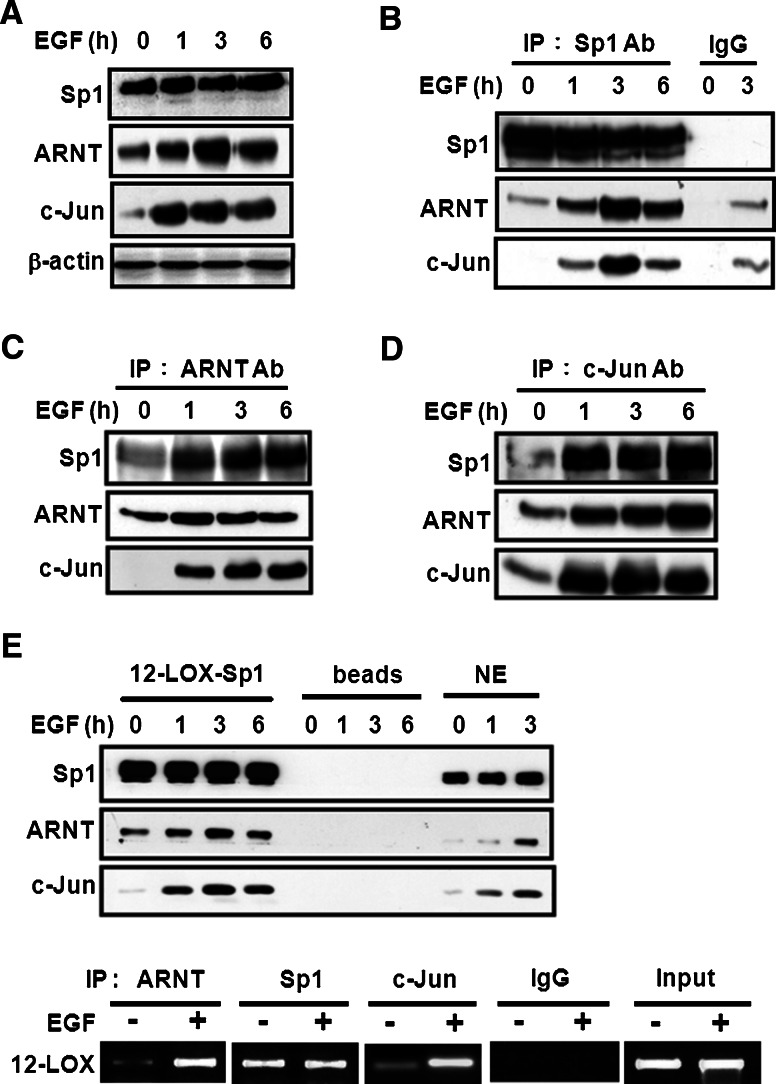
In order to investigate the factors associated with c-Jun/Sp1 in the regulation of EGF-induced gene expression, nuclear-extracted proteins from EGF-treated A431 cells were immunoprecipitated by using Sp1 antibodies. The immunoprecipitated proteins were subsequently analyzed by two-dimensional SDS–PAGE and were then identified by using matrix-associated laser desorption ionization time-of flight (MALDI-TOF) mass spectrometry. ARNT, one of the Sp1-associated proteins, was identified (data not shown). As shown in our previous report, after treatment with EGF, ARNT could bind with c-Jun and induce gene expression [9]. Therefore, it was then assumed that, after treatment with EGF, ARNT may possibly play a role in c-Jun/Sp1-mediated gene expression. To this end, western blotting was carried out and the nuclear proteins were extracted after being treated with EGF for various time periods. As shown in Fig. 1a, EGF dramatically induced the nuclear accumulation of c-Jun and ARNT in a time-dependent manner, whereas the expression of Sp1 did not change. The formation of c-Jun/ARNT/Sp1 complex induced by EGF treatment was also confirmed by the immunoprecipitation assay that used Sp1 antibodies (Fig. 1b), ARNT antibodies (Fig. 1c), and c-Jun antibodies (Fig. 1d), independently. Since EGF induced the interaction among Sp1, c-Jun, and ARNT, we then studied whether the complex could bind to the Sp1 site of the gene promoter, using Sp1 binding sequences as probes and performing DAPA. As shown in Fig. 1e (upper panel), EGF enhanced the binding of c-Jun and ARNT to DNA. The same results were obtained in ChIP assay (Fig. 1e, lower panel). Thus, we confirmed that these proteins did form a complex and that the c-Jun/ARNT/Sp1 complex induced by EGF could bind to the Sp1 sites of 12(S)-lipoxygenase gene promoter. These results indicated that ARNT may be involved in EGF-regulated gene expression through formation of the c-Jun/ARNT/Sp1 complex.
Fig. 1.
EGF induces formation of the c-Jun/ARNT/Sp1 complex and its binding to the Sp1 sites of the gene promoter. Cells were starved for 18 h in serum-free culture medium and then treated with 50 ng/ml EGF for a different time period as indicated. Nuclear extracts were used for the following experiments. a The Sp1, ARNT and c-Jun proteins were detected by anti-Sp1, anti-ARNT and anti-c-Jun antibodies, respectively. b–d Nuclear extracts were immunoprecipitated (IP) with antibodies (Ab) against Sp1, ARNT and c-Jun. The proteins were subjected to SDS–PAGE and analyzed by western blotting with antibodies against c-Jun, ARNT and Sp1. IgG was for the negative control of antibodies. e After EGF treatment for different time period as indicated, nuclear extracts were prepared, chromatin immunoprecipitation (ChIP, lower panel) and DNA affinity precipitation assay (upper panel) were performed. Binding of Sp1, ARNT and c-Jun to Sp1 binding sequence of 12(S)-lipoxygenase promoter (12-LOX-Sp1) was analyzed by western blot. The streptavidin-agarose beads were used as a nonspecific binding control. NE protein from nuclear extracts
ARNT regulates EGF-induced 12(S)-lipoxygenase gene expression through binding of Sp1 protein to the Sp1 binding sites
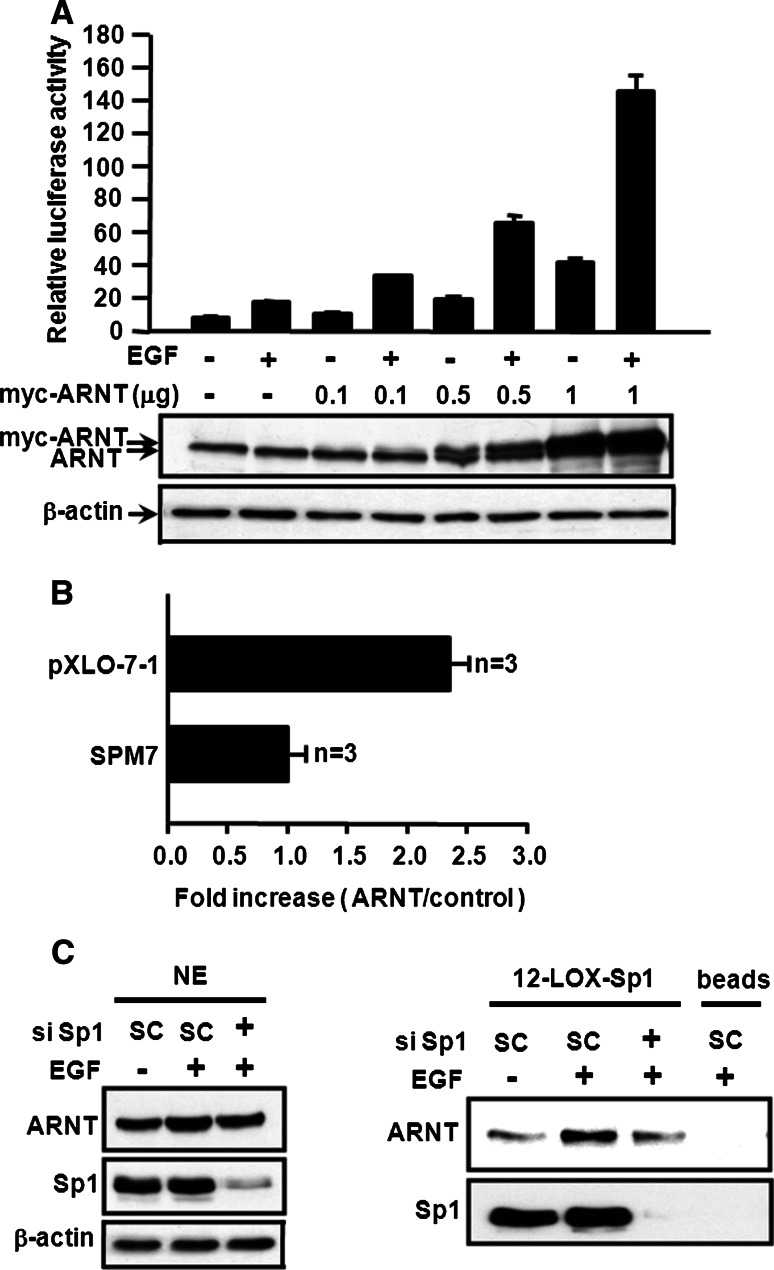
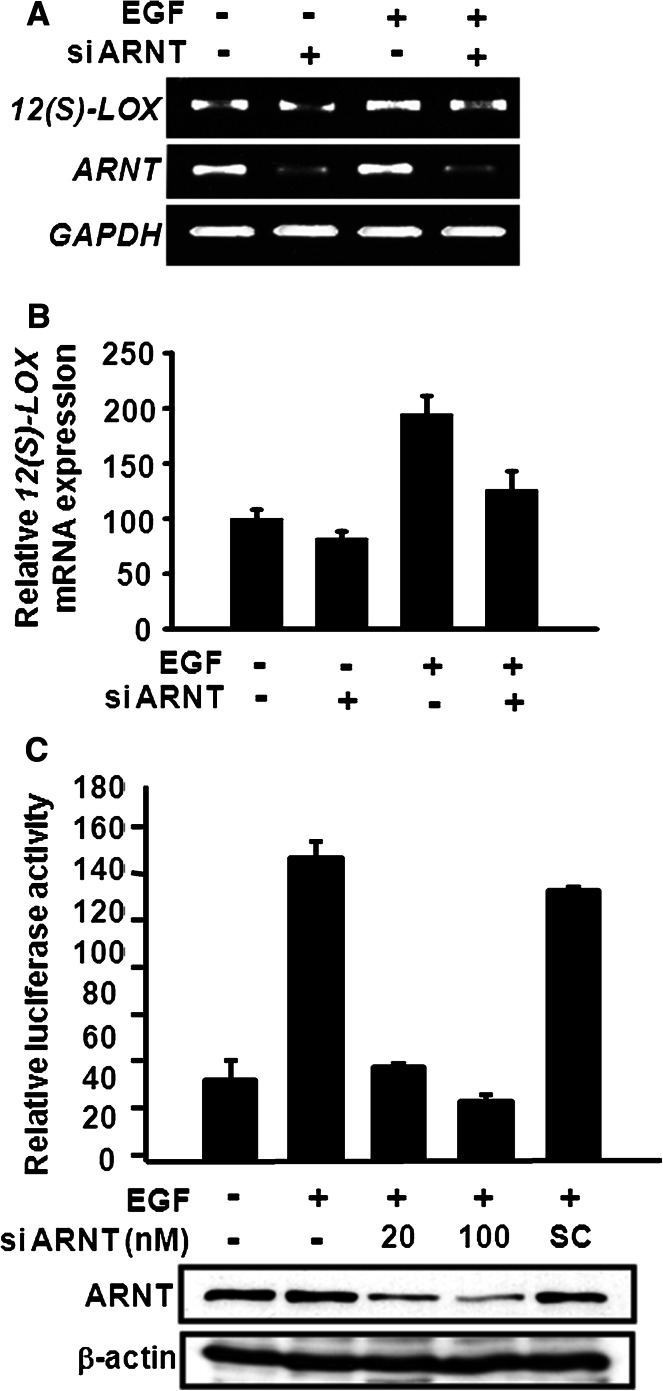
Since the EGF-induced expression of certain genes, such as 12(S)-lipoxygenase, is mediated through c-Jun/Sp1 [13], we investigated whether ARNT of the c-Jun/ARNT/Sp1 complex was involved in the EGF-induced gene regulation. A reporter assay was then carried out to study the effect of ARNT on EGF-induced 12(S)-lipoxygenase expression. In Fig. 2a, overexpression of ARNT alone induced 12(S)-lipoxygenase gene expression in a dose-dependent manner. ARNT also enhanced the effect of EGF on the transcriptional activation of 12(S)-lipoxygenase gene (Fig. 2a). Since Sp1 binding sites of 12(S)-lipoxygenase promoter are required for EGF-induced gene expression [18], we further clarified whether ARNT-mediated 12(S)-lipoxygenase gene expression was specific through Sp1 binding sites. Luciferase vectors, pXLO-7-1, which bears wild-type 12(S)-lipoxygenase promoter, and SPM7, which contains mutants on the Sp1 binding sequence of 12(S)-lipoxygenase promoter region, were then used. As shown in Fig. 2b, ARNT increased the pXLO-7-1 promoter activity of 12(S)-lipoxygenase, but it had no effect on SPM7, indicating that a Sp1 binding site is essential in ARNT-mediated 12(S)-lipoxygenase gene expression. ARNT also cooperated with Sp1 to enhance 12(S)-lipoxygenase promoter activity (Electronic supplementary material, ESM, Fig. 1). Furthermore, as shown in the DAPA assay (Fig. 2c, right panel), the EGF-induced binding of ARNT to Sp1 site was decreased in the Sp1 knockdown cells, while the protein level of ARNT was not affected (Fig. 2c, left panel). These results demonstrated that Sp1 protein was required for the binding of ARNT to the DNA. To further confirm the functional role of endogenous ARNT in regulating the EGF-induced 12(S)-lipoxygenase expression, ARNT small interfering RNA (siRNA) oligonucleotides were transfected into A431 cells. The mRNA expression (Fig. 3a, b) and promoter activity (Fig. 3c) of 12(S)-lipoxygenase induced by EGF were attenuated in ARNT-knockdown cells. These results revealed that ARNT was involved in the regulation of EGF-induced 12(S)-lipoxygenase expression through binding of Sp1 protein to the Sp1 binding sequence.
Fig. 2.
ARNT transactivates the promoter activity of 12(S)-lipoxygenase. a Cells were transfected with wild-type 12(S)-lipoxygenase promoter (pXLO-7-1), and myc-ARNT expression vector by lipofection. Cells were treated with 50 ng/ml EGF and further cultured in fresh medium up to 18 h. The luciferase activities and protein concentrations were then determined and normalized. Values represent means ± SEM of three determinations. b Cells were transfected with ARNT expression vector, wild-type (pXLO-7-1) and Sp1 sites mutant (SPM7) of 12(S)-lipoxygenase promoter by lipofection. The luciferase activities and protein concentrations were then determined and normalized. Values represent means ± SEM of three determinations. c Cells were transfected with 20 nM Sp1 siRNA by lipofection. After EGF treatment for 3 h, nuclear extracts were prepared, DNA affinity precipitation assay (DAPA, right panel) was performed. Binding of Sp1 and ARNT to Sp1 binding sequence of 12(S)-lipoxygenase promoter (12-LOX-Sp1) was analyzed by western blot. The streptavidin-agarose beads were used as a nonspecific binding control. NE protein from nuclear extracts
Fig. 3.
Effect of ARNT on EGF-induced 12(S)-lipoxygenase gene expression. a Cells were transfected with 20 nM ARNT siRNA by lipofection. After EGF treatment for 18 h, total RNA was extracted for reverse transcription PCR with 12(S)-lipoxygenase (12(S)-LOX), ARNT and GAPDH primers. b Quantitative real-time PCR analysis of 12(S)-lipoxygenase was performed as described under “Materials and methods”. Each column represents the mean ± SEM (n = 3). c Cells were transfected with pXLO-7-1 and various amounts of ARNT siRNA or scramble oligonucleotides (SC) by lipofection. After EGF treatment for 18 h, the luciferase activities and protein concentrations were then determined and normalized. Values represent means ± SEM of three determinations. Expressions of ARNT and β-actin proteins were analyzed by western blot analysis using anti-ARNT and anti-β-actin antibodies, respectively
ARNT cooperates with c-Jun in the regulation of 12(S)-lipoxygenase gene transcription
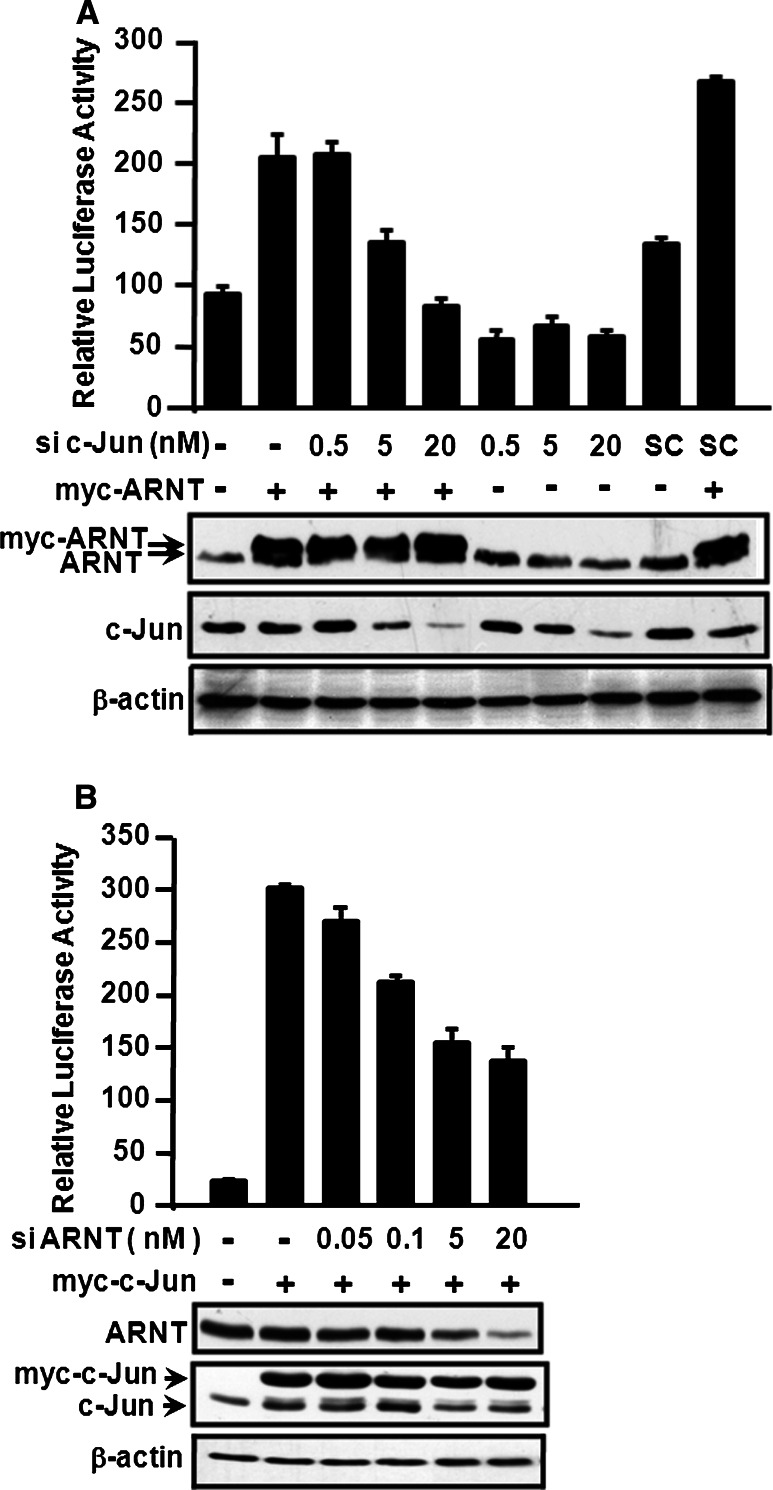
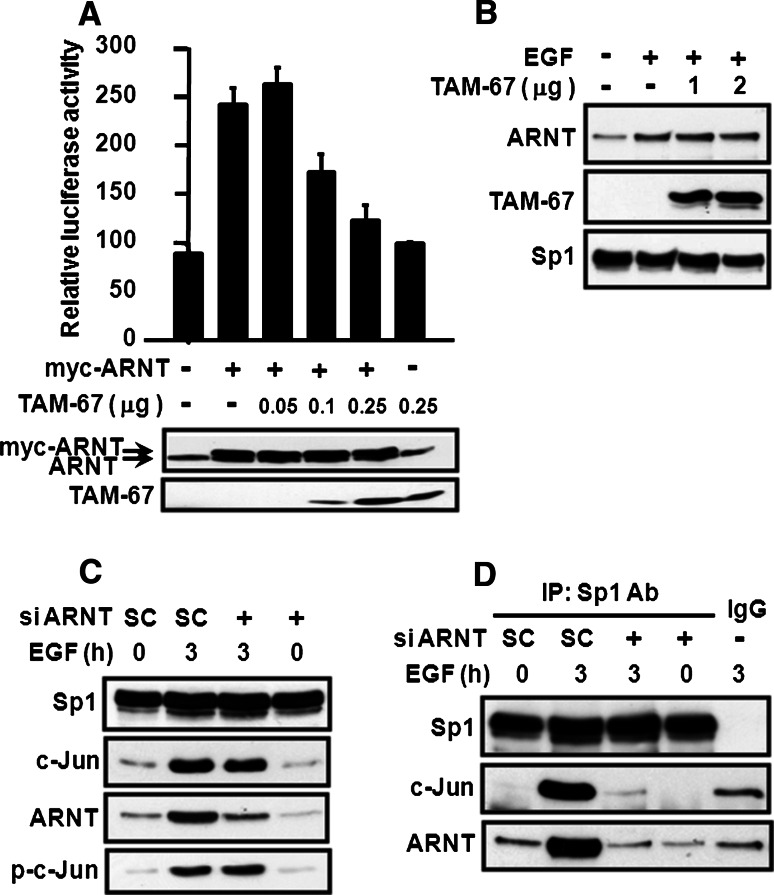
The results of our previous studies indicated that the transcription factor c-Jun is essential for EGF-induced 12(S)-lipoxygenase gene expression [13]. Therefore, we tested whether the interaction between c-Jun and ARNT was required in the regulation of EGF-induced 12(S)-lipoxygenase transcriptional expression. To determine this, c-Jun siRNA was used in the 12(S)-lipoxygenase promoter assay. Notably, the ARNT-induced 12(S)-lipoxygenase promoter activity decreased with the knockdown of c-Jun, in a dose-dependent manner (Fig. 4a). Consistently, ARNT siRNA also decreased the c-Jun-induced 12(S)-lipoxygenase promoter activity (Fig. 4b). These results indicated that the cooperation between c-Jun and ARNT was essential for 12(S)-lipoxygenase gene expression. To further assess whether the transcriptional activity of c-Jun is involved in ARNT-activated gene expression, the dominant negative c-Jun TAM-67, which lacks the transactivation domain but retains the DNA-binding and dimerization domains [13, 23], was then used. As shown in Fig. 5a, TAM-67 dramatically decreased ARNT-induced 12(S)-lipoxygenase promoter activity in a dose-dependent manner, indicating the involvement of the transcriptional activity of c-Jun in ARNT-mediated gene expression. However, the possible contribution of the transcriptional activity of ARNT could not be ruled out, since the inhibitory effect of TAM-67 may also result from less interaction between ARNT and c-Jun, thus reducing the binding of ARNT to DNA. Therefore, we studied the binding of TAM-67 and ARNT to the promoter of 12(S)-lipoxygenase. As shown in Fig. 5b, from DAPA results, we found that ARNT, TAM-67, and Sp1 were able to form a protein complex and bound to Sp1 site upon EGF treatment. Notably, TAM-67 had no effect on EGF-induced binding of ARNT to DNA (Fig. 5b). In addition, the transfected dominant negative mutant (TAM-67) also competed with endogenous c-Jun for interacting with Sp1 [13]. These results indicate that c-Jun, and not ARNT, provides the transcriptional activity in the regulation of 12(S)-lipoxygenase gene expression. Binding of c-Jun to Sp1 is necessary for EGF-induced 12(S)-lipoxygenase gene expression because there are only Sp1 binding sites present in the 12(S)-lipoxygenase promoter region, and not c-Jun binding sites [13]. It was therefore interesting to evaluate the effect of ARNT on the interaction between c-Jun and Sp1 under EGF treatment. As shown in Fig. 5c, knockdown of ARNT did not decrease phospho-c-Jun and c-Jun expression upon EGF stimulation, but it did affect the association between c-Jun and Sp1 (Fig. 5d). These results demonstrated that ARNT mediated EGF-induced gene expression by regulating the interaction between c-Jun and Sp1 but not through regulating the transcriptional activity or expression of c-Jun.
Fig. 4.
Cooperation between ARNT and c-Jun on promoter activation of 12(S)-lipoxygenase gene. a Cells were transfected with pXLO-7-1, myc-ARNT, c-Jun siRNA and scramble oligonucleotides (SC) by lipofection. The luciferase activities and protein concentrations were then determined and normalized. Values represent means ± SEM of three determinations. Expressions of myc-ARNT and c-Jun proteins were analyzed by western blot analysis using anti-ARNT and anti-c-Jun antibodies. b Cells were transfected with various amounts of ARNT siRNA oligonucleotides, pXLO-7-1 and myc-c-Jun by lipofection. The luciferase activities and protein concentrations were then determined and normalized. Values represent means ± SEM of three determinations. Expressions of myc-c-Jun, ARNT and β-actin proteins were analyzed by western blot analysis using anti-c-Jun and anti-ARNT antibodies
Fig. 5.
ARNT mediates transcriptional effect of c-Jun on promoter activation of 12(S)-lipoxygenase gene. a Cells were transfected with pXLO-7-1, myc-ARNT and TAM-67 by lipofection. The luciferase activities and protein concentrations were then determined and normalized. Values represent means ± SEM of three determinations. Expression of myc-ARNT and TAM-67 proteins were analyzed by western blot analysis using anti-ARNT and anti-c-Jun antibodies. b Cells were transfected with TAM-67 and then treated with 50 ng/ml EGF for 3 h. Nuclear extracts were prepared and DNA affinity precipitation assay was performed. Binding of Sp1, ARNT and TAM-67 to Sp1 probes was analyzed by western blot. c Cells were transfected with 20 nM ARNT siRNA and scramble (SC) oligonucleotides and then treated with 50 ng/ml EGF for 3 h. Nuclear extracts were prepared and the Sp1, ARNT and c-Jun proteins were detected by anti-Sp1, anti-ARNT, anti-phospho-c-Jun (Ser 73) and anti-c-Jun antibodies, respectively. d Nuclear extracts were immunoprecipitated (IP) with antibodies (Ab) against Sp1. The proteins were subjected to SDS–PAGE and analyzed by western blotting with antibodies against c-Jun, ARNT and Sp1. IgG indicates the negative control of antibodies
ARNT mediates other c-Jun/Sp1-regulated gene expressions
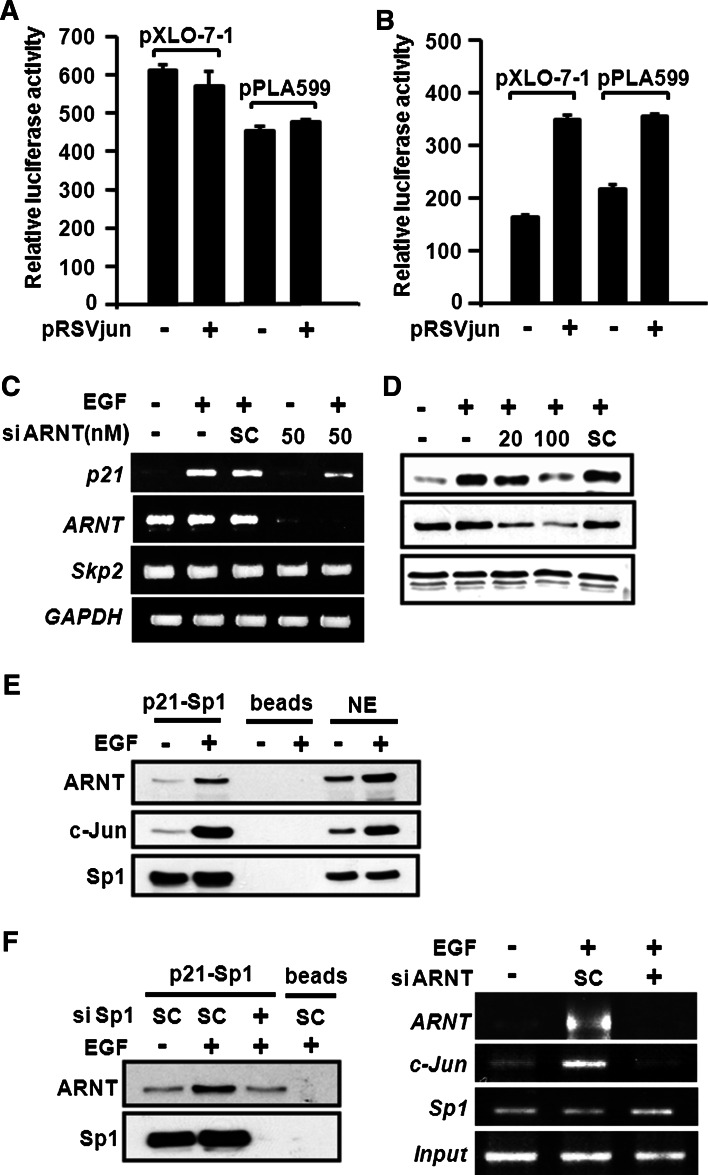
Since ARNT mediated the interaction between c-Jun and Sp1 (Fig. 5d), we then checked whether ARNT was essential for other c-Jun/Sp1-regulated gene expressions [24, 25]. The c4 cell line, which lacks functional ARNT owing to a point mutation in the ARNT gene, and the vT2 cell line, which possesses completely transfected ARNT cDNA, were examined in the c-Jun-induced promoter activation. In addition, the mutated ARNT in c4 is degraded more rapidly than the wild-type protein in vT2 [17]. As shown in Fig. 6a, we found that the promoter activity of 12(S)-lipoxygenase (pXLO-7-1) and cytosolic phospholipase A2 (cPLA599) were not activated in pRSVjun-transfected c4 cells (the cells lacking ARNT). However, pRSVjun significantly induced transcriptional activity of 12(S)-lipoxygenase and cPLA2 in vT2 cells, which possess ARNT expression (Fig. 6b). These results demonstrate that the presence of ARNT is essential in c-Jun mediated gene expression. To further confirm the importance of ARNT in c-Jun/Sp1-dependent gene expression, we knocked down ARNT expression by siRNA and evaluated EGF-induced p21WAF1/CIP1 expression. As shown in Fig. 6c, d, ARNT knockdown inhibited EGF-induced p21WAF1/CIP1 expression both at the mRNA and protein levels. Furthermore, the EGF-induced c-Jun/ARNT/Sp1 complex could bind to the Sp1 sites of the p21WAF1/CIP1 promoter (Fig. 6e). In addition, the EGF-induced binding of c-Jun to Sp1 site was also inhibited in the ARNT knockdown cells (Fig. 6f, right panel). Sp1 knockdown dramatically inhibited EGF-induced interaction between ARNT and p21WAF1/CIP1 promoter (Fig. 6f, left panel), indicating that EGF-induced gene expression was mediated by the c-Jun/ARNT/Sp1 complex through the binding of Sp1 to the Sp1 binding site. Taken together, ARNT played an essential role in the regulation of c-Jun/Sp1-mediated gene expression by regulating the interaction between c-Jun and Sp1. In summary (Fig. 7), ARNT is a key component to bridge the c-Jun/Sp1 interaction, resulting in the formation of c-Jun/ARNT/Sp1 complex to regulate EGF-induced gene expression.
Fig. 6.
ARNT is essential for c-Jun/Sp1-regulated gene expression. c4 cells (a) and vT2 cells (b) were transfected with pXLO-7-1 of 12(S)-lipoxygenase gene promoter, pPLA599 of cPLA 2 gene promoter and pRSVjun by lipofection. The luciferase activities and protein concentrations were then determined and normalized. Values represent means ± SEM of three determinations. c A431 cells were transfected with 50 nM ARNT siRNA or scramble(SC ) oligonucleotides by lipofection. After EGF treatment for 4 h, total RNA was extracted for reverse transcription PCR with p21WAF1/CIP1, Skp2, ARNT and GAPDH primers. d A431 cells were transfected with 20 or 100 nM ARNT siRNA or scramble (SC) oligonucleotides by lipofection. After EGF treatment for 4 h, the p21WAF1/CIP1, ARNT and Skp2 proteins were detected by anti-p21WAF1/CIP1, anti-ARNT and anti-Skp2 antibodies, respectively. e Cells were starved for 18 h in serum-free culture medium and then treated with 50 ng/ml EGF for 3 h. Nuclear extracts were prepared and DNA affinity precipitation assay was performed. Binding of Sp1, ARNT and c-Jun to Sp1 binding site of p21WAF1/CIP1 promoter (p21-Sp1) was analyzed by western blot. The streptavidin-agarose beads were used as a nonspecific binding control. f Cells were transfected with 20 nM ARNT siRNA by lipofection. After EGF treatment for 3 h, nuclear extracts were prepared, chromatin immunoprecipitation (ChIP, right panel), and DNA affinity precipitation assay (DAPA, left panel) was performed as described under “Materials and methods.” Binding of Sp1 and ARNT to Sp1 site of p21WAF1/CIP1 promoter region (p21-Sp1) was analyzed by western blot. The streptavidin-agarose beads were used as a nonspecific binding control. NE protein from nuclear extracts
Fig. 7.
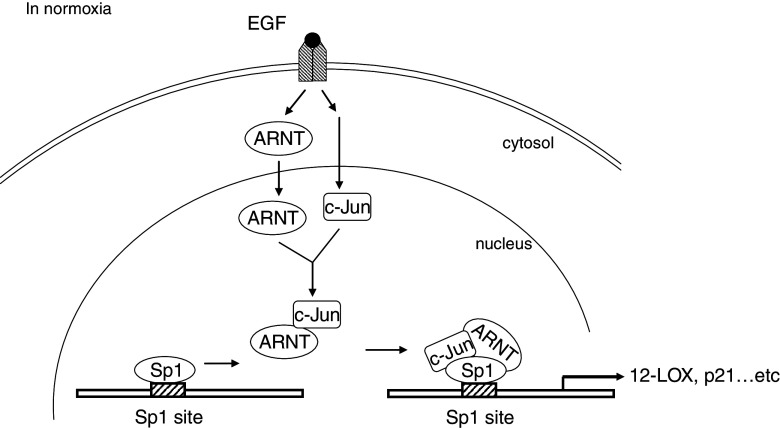
Putative transcriptional mechanism of ARNT involvement in EGF-induced gene expression. After the stimulation of EGF in normoxia, ARNT accumulates in the nucleus where it interacts with c-Jun and Sp1 to form c-Jun/ARNT/Sp1 complex. These complexes then bind to the Sp1 site of the gene promoter through Sp1 protein and turn on relative gene expression, in which c-Jun transcriptional activity was required. 12-LOX 12(S)-lipoxygenase
Discussion
In this study, we demonstrated that ARNT, a general partner factor for bHLH-PAS proteins, is essential in mediating c-Jun/Sp1-regulated gene expression upon EGF treatment. Although the protein amount of c-Jun expression induced by EGF did not decrease in ARNT knockdown cells (Fig. 5c), the EGF-induced association between c-Jun and Sp1 was inhibited, resulting in the attenuation of c-Jun/Sp1-mediated gene expression (Figs. 3, 5d and 6). The requirement of ARNT for the interaction between c-Jun and Sp1 indicates that ARNT may play a pivotal role in the regulation of c-Jun/Sp1-dependent gene expression. In addition to the genes mentioned in this study, such as 12(S)-lipoxygenase, cPLA2, and p21WAF1/CIP1, it is assumed that ARNT may participate in the regulation of expression of more genes through the regulation of c-Jun/Sp1 interaction. Functions of these c-Jun/Sp1-regulated target genes have been well documented. For example, 12(S)-lipoxygenase plays important physiological roles in tumor metastasis [26, 27]. Nicotinic acetylcholine receptors (nAChRs) are important for synaptic transmission in the nervous system [28]. cPLA2 hydrolyzes membrane phospholipids to release arachidonic acid and mediates cell injury [29]. Keratin 16 is the marker of keratinocyte hyperproliferation in psoriasis [30], and p21WAF1/CIP1 modulates cyclin-dependent kinase activity resulting in cell growth arrest or progression [31]. These imply that ARNT in response to different stimuli may play diverse roles in the regulation of a variety of biological functions under normoxic conditions.
In hypoxic conditions, HIF-1α requires the cooperation of ARNT to form a heterodimer and regulates several genes involved in tumorigenesis [32]. Contrary to the hypoxic conditions and consistent with our findings that HIF-1α was barely detectable either in the control or in the EGF-treated cells (data not shown), ARNT/HIF-1β, and not HIF-1α plays a more important role in mediating cellular functions under normoxic conditions. Functions of ARNT in glucose metabolism under normoxic conditions have been reported. Reducing ARNT levels results in markedly impaired glucose-stimulated insulin release and changes in gene expression which are similar to those in human type 2 diabetes [33].
ARNT is essential for the normal functions of HIF-1α, HIF-2α, and AhR. These heterodimeric complexes are required for cellular responses to hypoxia (HIF proteins) and environmental toxins (AhR) [1, 6]. ARNT-containing dimers have been reported to regulate the expression of many genes. Many of these promoters have multiple potential ARNT binding sites. Under hypoxic conditions, HIF-1α/ARNT dimers activate the transcription of a number of target genes whose promoters contain the binding motif termed as the hypoxia-responsive element (HRE) [34]. In response to environmental pollutants, such as dioxin, AhR translocates to the nucleus and binds with ARNT to form heterodimers, which then specifically bind to the XRE of the CYP1A1 gene promoter region. Interestingly, ARNT also regulates the transcriptional activation of genes that do not contain ARNT binding sites, such as 12(S)-lipoxygenase and p21 WAF1/CIP1 in this study. This indicates that gene expression may be regulated by ARNT either directly through the binding of the heterodimers to the DNA or indirectly through the interaction with other transcriptional factors. It is intriguing that, since ARNT is a partner for other transcription factors, cross-talks between the ARNT mediated genes regulation might take place. A recent report from Sutter has demonstrated that EGFR signaling competes binding of p300 and interferes with AhR-mediated differentiation in keratinocytes [35]. In order to clarify whether ARNT plays a role in the cross-talk between TCDD- and EGF-induced gene expression. A431 cells were treated with TCDD and EGF separately to study whether TCDD could turn on 12(S)-lipoxygenase gene expression or whether EGF has any effects on the CYP1A1 gene expression. In ESM Fig. 2, the RT–PCR result demonstrated that EGF was not able to induce CYP1A1 gene expression. In addition, the promoter assay revealed that TCDD did not significantly induce 12(S)-lipoxygenase promoter activity even in higher dose of TCDD such as 10 nM (Supplementary Fig. 3). It is therefore quite clear that although ARNT can form transcriptional complex with other transcription factors, upon specific stimuli, ARNT associates with different partners to turn on specific gene expression. In addition, EGF induces c-Jun/ARNT protein complex formation and binds to cAMP response element (CRE) on the COX-2 promoter region, thus resulting in an increase in gene expression [9]. Since the Sp1 site is not required for EGF-induced COX-2 promoter activity, the activation of COX-2 gene was mediated through c-Jun/ARNT complex rather than c-Jun/ARNT/Sp1 mediation. It is interesting, however, that in this study EGF induced the formation of c-Jun/ARNT/Sp1 complex formation and turned on 12(S)-lipoxygenase and p21WAF1/CIP1 gene expression through binding to Sp1 site. Thus, the mechanism of ARNT-associated protein complex in regulating gene expression is also dependent on the responsive element presenting on the gene promoter region.
ARNT has transactivation domains in the C-terminus and could drive gene expression by its transcriptional activity [1]. In this study, we found that the function of ARNT in the regulation of gene expression was through regulating the interaction between c-Jun and Sp1. Because c-Jun in the c-Jun/ARNT complex possesses the transcriptional activity that regulated the transcriptional activation of the gene (Fig. 5), we concluded that ARNT acts as a bridge, mediating the binding between c-Jun and Sp1. Association between c-Jun and Sp1 was identified by in vitro and in vivo experiments [13, 16]. However, in the current study, we observed no interaction between c-Jun and Sp1 in ARNT knockdown cells (Figs. 5d, 6f). These results implied that c-Jun might not directly bind to Sp1 upon growth factor stimulation and needs the mediation of other cofactors such as ARNT. Therefore, we concluded that, without the presence of the DNA binding sequence, ARNT could still mediate gene expression by recruiting transcription factors c-Jun and Sp1 upon growth factor stimulation and thus activate specific gene expression.
It is well documented that the post-translational modifications of proteins play an important role in the regulation of protein–protein interactions. Recently, we found that protein phosphotase 2B (PP2B)-mediated dephosphorylation of c-Jun at the C-terminus regulates c-Jun/Sp1 interaction [36]. Since both ARNT and PP2B regulate the interaction between c-Jun and Sp1, we raised the hypothesis that the dephosphorylation of c-Jun by PP2B might be essential for c-Jun and ARNT association. It has been reported that the sumoylation of ARNT inhibits its ability to interact with cooperative transcriptional proteins [37]. In addition, EGF also stimulates the phosphorylation of ARNT in the nucleus [9], implying that a posttranslational modification may be involved in the nuclear localization of ARNT as well as the c-Jun/ARNT/Sp1 complex formation, resulting in an increase in 12(S)-lipoxygenase and p21WAF1/CIP1 gene expression. Thus, whether the posttranslational modification of ARNT is the key factor in regulating its binding or association to various transcription factors upon different stimuli remains to be elucidated.
Although HIF-1α is overexpressed in most tumors, some tumors do not stain positive for it [38]. Alternatively, other bHLH-PAS transcription factors that may have biological properties similar to that of HIF-1α, such as HIF-2α or HIF-3α, may also mediate hypoxic adaptation. Consistent with this hypothesis, in this study, we revealed that ARNT, in place of HIF-1α, mediated c-Jun/Sp1-dependent gene expression in normoxic conditions. Since tumorigenesis usually starts in normoxic conditions, we concluded that ARNT may regulate tumorigenesis under normoxic conditions. In our recent study, we found that ARNT also regulates EGF-induced COX-2 gene expression, resulting in cell migration, which is highly correlated with tumorigenesis [9]. In conclusion, our results revealed a novel regulatory mechanism, apart from forming dimers with other bHLH-PAS members, in which ARNT acts a modulator to bridge the c-Jun/Sp1 interaction and plays a role in EGF-mediated gene expression under normoxic conditions. This broadens the role that ARNT plays in physiological and pathophysiological functions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by grant NSC 98-2320-B-006-014 from the National Science Council of the Republic of China and by the National Cheng Kung University Project of Promoting Academic Excellence and Developing World-Class Research Centers.
Footnotes
W.-C. Huang and S.-T. Chen contributed equally to this work.
Contributor Information
Wen-Chang Chang, Email: wcchang@mail.ncku.edu.tw.
Ben-Kuen Chen, Phone: +886-6-2757575, FAX: +886-6-2083663, Email: bkchen58@mail.ncku.edu.tw.
References
- 1.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36:189–204. doi: 10.1016/S1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 2.Maltepe E, Schmidt JV, Baunoch D, Bradfield CA, Simon MC. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature. 1997;386:403–407. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P, Dor Y, Herbert J-M, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshet E. Role of HIF-1[alpha] in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 4.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 5.Woods SL, Whitelaw ML. Differential activities of murine single minded 1 (SIM1) and SIM2 on a hypoxic response element. Cross-talk between basic helix-loop-helix/per-Arnt-Sim homology transcription factors. J Biol Chem. 2002;277:10236–10243. doi: 10.1074/jbc.M110752200. [DOI] [PubMed] [Google Scholar]
- 6.Kozak KR, Abbott B, Hankinson O. ARNT-deficient mice and placental differentiation. Dev Biol. 1997;191:297–305. doi: 10.1006/dbio.1997.8758. [DOI] [PubMed] [Google Scholar]
- 7.Moffett P, Pelletier J. Different transcriptional properties of mSim-1 and mSim-2. FEBS Lett. 2000;466:80–86. doi: 10.1016/S0014-5793(99)01750-0. [DOI] [PubMed] [Google Scholar]
- 8.Wright CW, Duckett CS. The aryl hydrocarbon nuclear translocator alters CD30-mediated NF-kappaB-dependent transcription. Science. 2009;323:251–255. doi: 10.1126/science.1162818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang KY, Shen MR, Lee MY, Wang WL, Su WC, Chang WC, Chen BK. Epidermal growth factor-activated aryl hydrocarbon receptor nuclear translocator/HIF-1{beta} signal pathway up-regulates cyclooxygenase-2 gene expression associated with squamous cell carcinoma. J Biol Chem. 2009;284:9908–9916. doi: 10.1074/jbc.M806210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Cruijsen H, Giaccone G, Hoekman K. Epidermal growth factor receptor and angiogenesis: opportunities for combined anticancer strategies. Int J Cancer. 2005;117:883–888. doi: 10.1002/ijc.21479. [DOI] [PubMed] [Google Scholar]
- 11.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-I. [DOI] [PubMed] [Google Scholar]
- 12.Yarden Y, Sliwkowski MX. Untangling the ErbB signaling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 13.Chen BK, Chang WC. Functional interaction between c-Jun and promoter factor Sp1 in epidermal growth factor-induced gene expression of human 12(S)-lipoxygenase. Proc Natl Acad Sci USA. 2000;97:10406–10411. doi: 10.1073/pnas.180321497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BK, Kung HC, Tsai TY, Chang WC. Essential role of mitogen-activated protein kinase pathway and c-Jun induction in epidermal growth factor-induced gene expression of human 12-lipoxygenase. Mol Pharmacol. 2000;l57:153–161. [PubMed] [Google Scholar]
- 15.Nie D, Krishnamoorthy S, Jin R, Tang K, Chen Y, Qiao Y, Zacharek A, Guo Y, Milanini J, Pages G, Honn KV. Mechanisms regulating tumor angiogenesis by 12-lipoxygenase in prostate cancer cells. J Biol Chem. 2006;281:18601–18609. doi: 10.1074/jbc.M601887200. [DOI] [PubMed] [Google Scholar]
- 16.Kardassis D, Papakosta P, Pardali K, Moustakas A. c-Jun transactivates the promoter of the human p21(WAF1/Cip1) gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J Biol Chem. 1999;274:29572–29581. doi: 10.1074/jbc.274.41.29572. [DOI] [PubMed] [Google Scholar]
- 17.Numayama-Tsuruta K, Kobayashi A, Sogawa K, Fujii-Kuriyama Y. A point mutation responsible for defective function of the aryl-hydrocarbon-receptor nuclear translocator in mutant Hepa-1c1c7 cells. Eur J Biochem. 1997;246:486–495. doi: 10.1111/j.1432-1033.1997.00486.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu YW, Arakawa T, Yamamoto S, Chang WC. Transcriptional activation of human 12-lipoxygenase gene promoter is mediated through Sp1 consensus sites in A431 cells. Biochem J. 1997;324:133–140. doi: 10.1042/bj3240133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsou JH, Chang KY, Wang WC, Tseng JT, Su WC, Hung LY, Chang WC, Chen BK. Nucleolin regulates c-Jun/Sp1-dependent transcriptional activation of cPLA2alpha in phorbol ester-treated non-small cell lung cancer A549 cells. Nucleic Acids Res. 2008;36:217–227. doi: 10.1093/nar/gkm1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor {kappa}B recruitment to target promoters. J Exp Med. 2001;193:1351–1360. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Saunders MA, Yeh H, Deng W-g, Wu KK. Dynamic regulation of cyclooxygenase-2 promoter activity by isoforms of CCAAT/enhancer-binding proteins. J Biol Chem. 2002;277:6923–6928. doi: 10.1074/jbc.M108075200. [DOI] [PubMed] [Google Scholar]
- 23.Dong Z, Crawford HC, Lavrovsky V, Taub D, Watts R, Matrisian LM, Colburn NH. A dominant negative mutant of jun blocking 12-O-tetradecanoylphorbol-13-acetate-induced invasion in mouse keratinocytes. Mol Carcinog. 1997;19:204–212. doi: 10.1002/(SICI)1098-2744(199707)19:3<204::AID-MC8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 24.Blaine SA, Wick M, Dessev C, Nemenoff RA. Induction of cPLA2 in lung epithelial cells and non-small cell lung cancer is mediated by Sp1 and c-Jun. J Biol Chem. 2001;276:42737–42743. doi: 10.1074/jbc.M107773200. [DOI] [PubMed] [Google Scholar]
- 25.Kardassis D, Papakosta P, Pardali K, Moustakas A. c-Jun transactivates the promoter of the human p21WAF1/Cip1 gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J Biol Chem. 1999;274:29572–29581. doi: 10.1074/jbc.274.41.29572. [DOI] [PubMed] [Google Scholar]
- 26.Timar J, Raso E, Dome B, Li L, Grignon D, Nie D, Honn KV, Hagmann W. Expression, subcellular localization and putative function of platelet-type 12-lipoxygenase in human prostate cancer cell lines of different metastatic potential. Int J Cancer. 2000;87:37–43. doi: 10.1002/1097-0215(20000701)87:1<37::AID-IJC6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 27.Pidgeon GP, Lysaght J, Krishnamoorthy S, Reynolds JV, O’Byrne K, Nie D, Honn KV. Lipoxygenase metabolism: roles in tumor progression and survival. Cancer Metastasis Rev. 2007;26:503–524. doi: 10.1007/s10555-007-9098-3. [DOI] [PubMed] [Google Scholar]
- 28.Cordero-Erausquin M, Marubio LM, Klink R, Changeux JP. Nicotinic receptor function: new perspectives from knockout mice. Trends Pharmacol Sci. 2000;21:211–217. doi: 10.1016/S0165-6147(00)01489-9. [DOI] [PubMed] [Google Scholar]
- 29.Leslie CC. Properties and regulation of cytosolic phospholipase A2. J Biol Chem. 1997;272:16709–16712. doi: 10.1074/jbc.272.27.16709. [DOI] [PubMed] [Google Scholar]
- 30.Leigh IM, Navsaria H, Purkis PE, McKay IA, Bowden PE, Riddle PN. Keratins (K16 and K17) as markers of keratinocyte hyperproliferation in psoriasis in vivo and in vitro. Br J Dermatol. 1995;133:501–511. doi: 10.1111/j.1365-2133.1995.tb02696.x. [DOI] [PubMed] [Google Scholar]
- 31.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giaccia A, Siim BG, Johnson RS. HIF-1 as a target for drug development. Nat Rev Drug Discov. 2003;2:803–811. doi: 10.1038/nrd1199. [DOI] [PubMed] [Google Scholar]
- 33.Gunton JE, Kulkarni RN, Yim S, Okada T, Hawthorne WJ, Tseng Y-H, Roberson RS, Ricordi C, O’Connell PJ, Gonzalez FJ, Kahn CR. Loss of ARNT/HIF1[beta] mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 34.Semenza GL. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med. 2001;7:345–350. doi: 10.1016/S1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- 35.Sutter CH, Yin H, Li Y, Mammen JS, Bodreddigari S, Stevens G, Cole JA, Sutter TR. EGF receptor signaling blocks aryl hydrocarbon receptor-mediated transcription and cell differentiation in human epidermal keratinocytes. Proc Natl Acad Sci USA. 2009;106:4266–4271. doi: 10.1073/pnas.0900874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen BK, Huang CC, Chang WC, Chen YJ, Kikkawa U, Nakahama K-i, Morita I, Chang WC. PP2B-mediated dephosphorylation of c-Jun C terminus regulates phorbol ester-induced c-Jun/Sp1 interaction in A431 cells. Mol Biol Cell. 2007;18:1118–1127. doi: 10.1091/mbc.E06-09-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tojo M, Matsuzaki K, Minami T, Honda Y, Yasuda H, Chiba T, Saya H, Fujii-Kuriyama Y, Nakao M. The aryl hydrocarbon receptor nuclear transporter is modulated by the SUMO-1 conjugation system. J Biol Chem. 2002;277:46576–46585. doi: 10.1074/jbc.M205987200. [DOI] [PubMed] [Google Scholar]
- 38.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.