Abstract
The influence of chromatin on immediate-early gene expression has been studied in a model of Egr1 induction in intact mouse cells. ChIP analysis of factor and RNA polymerase binding reveals that the gene is constitutively poised for transcription in nonstimulated cells, but a repressing chromatin structure hampers productive transcription. Stimulation with phorbol esters results in a transient activation, which starts at 5 min and peaks at 30 min. Quantitative mapping of promoter occupancy by the different factors shows for the first time that no direct competition between SP1 and EGR1 occurs. The phosphorylation of ELK1 and CREB, which involves both the cascades of MEK1/2 and p38 kinases, is required for gene expression, which ceases following the binding of NAB1 and NAB2 to the promoter. The changes in histone acetylation and the differential recruitment of histone-modifying complexes further show the role of chromatin in the activation of this immediate-early gene.
Keywords: Immediate-early genes, Chromatin structure, Factor binding, Histone deacetylases, Histone acetyltransferases
Introduction
Activation of cellular immediate-early (IE) genes occurs within a few minutes following the reception of an appropriate stimulus, which reaches the nucleus usually via signalling cascades. The roles played by IE gene products are multiple, ranging from transcription factors to secreted proteins to cellular enzymes (see [1] for an early review). The protooncogenes c-Myc, c-Fos, and c-Jun were the founder members of the IE gene class, to which a large number of genes have been added.
The activation of IE genes usually occurs in a transient fashion. For instance, in the course of a study on liver regeneration, we have described that the expression of c-Myc, Id2, c-Fos, and Mat2A genes, as measured by RNAPol-ChIP [2], is detected within 30 min after partial hepatectomy [3, 4] and declines afterwards, dropping to the basal values by 6 h. The mechanisms by which expression of IE genes occurs, including the cascades involved in signal transduction to nuclei, the nuclear targets of kinases, and the nature of the events that eventually result in gene transcription, differ from one gene to another. Some epigenetic modifications, such as the phosphorylation of H3 serine 10, followed by the acetylation of its lysine 14 (reviewed in [5]), are common hallmarks of the induction of IE genes, but the nature of trans factors required for the induction, their specific modifications, and the order in which they are recruited to the cis sequences depend on the particular gene considered. In the same manner, the mechanisms responsible for the repression of a given IE gene after its transient activation are largely gene-dependent. Finally, many of the alterations of chromatin structure that allow transcription to take place are also gene-specific. Moreover, some of the IE genes required to start a proliferating event may be expressed again at later stages of the cell cycle, and this second wave of expression often implies a mechanism different from that followed to achieve the early response. For instance, we have described that the early expression of c-Myc, concomitant with hepatocyte priming, follows a different mechanism from that occurring at the second wave of c-Myc expression, at the G1-S interface [3].
All the above considerations make it interesting to study, at a chromatin level, the mechanisms of the transient activation of IE genes. In the present study, we have focused our attention on the mechanisms involved in the induction of the Egr1 (early growth response) gene. This gene codes for a zinc-finger transcription factor preferentially binding G-rich sequences, which largely colocalize with CpG islands [6]. The Egr1 gene itself has a binding site for its product, which eventually causes its own transcriptional down-regulation (reviewed in [7]) by a mechanism not yet fully understood, which probably involves the EGR1-induced expression of the corepressor NGFI-A binding (NAB) protein 2 as a negative feedback loop [8].
Several factors are needed for full, regulated transactivation of the Egr1 gene. Its promoter contains two clusters of serum-responsive elements (SRE), which require not only the binding of the serum response factor (SRF), but also that of a ternary complex factor, which may be any of the Ets-like-1 (ELK1) or SRF accessory proteins. The role of ELK1 has been studied in detail; the phosphorylation of the protein results in the activation of the ternary complex [9]. Moreover, ELK1 can recruit histone acetyltransferases (HATs) and coactivators, tethered by its phosphorylated C-terminal domain, but phosphorylation also enhances the activity of the previously associated HATs [10]. Nevertheless, the function of ELK1 is complex, because it also can recruit histone deacetylases (HDACs) through its N-terminal domain, resulting in the repression of the target genes [11].
Another type of cis element in Egr1 promoter is the cyclic AMP response element. Although it does not function as a cAMP-inducible enhancer element [7], it is capable of binding CREB, which also becomes phosphorylated in response to the signalling events. This phosphorylation has been postulated to be an essential requirement for the CREB transactivating capacity in IE genes [12]. The phosphorylation-linked transactivating capacity of CREB has been related to the recruitment of the CREB binding protein (CBP), which, besides acting as a HAT, may also play an adaptor role allowing, for instance, the interaction of CREB with SRE-bound SRF [13] and even with EGR1 itself [14].
The specificity protein 1 (SP1), a zinc-finger-containing protein, also plays a role in regulating Egr1 expression. SP1 is a ubiquitous factor that may activate or repress the transcription of its target genes. In many instances SP1 and EGR1 play opposing roles, and it has been suggested that this behavior may result from a direct competition for their partially overlapping binding sites [15–21], albeit other reports conclude that such a competition does not occur [22]. Finally, NAB1, a NAB2 related, constitutively expressed corepressor, also regulates Egr1 transcription. Both NAB1 [23] and NAB2 [24] bind EGR1 in a yeast two-hybrid system, and this might account for the above-mentioned negative feedback loop [8], although experimental evidence is still missing.
Many of the experimental approaches to the transcriptional regulation of the Egr1 gene have been carried out thus far in systems that do not ensure the maintenance of a physiologically functional chromatin structure, so that re-evaluating many of the known data by using intact cells will give a closer physiological approach.
Only a few studies on Egr1 induction have been conducted in intact cells. These studies agree that phosphorylation of ELK1 [25, 26] and of CREB [26, 27] is required for Egr1 expression in different cells. In every case tested, the phosphorylation of these factors depends on the extracellular signal-regulated kinase (ERK) pathway [26, 27] and is required for the mediator complex recruitment [25]. In other genes, the phosphorylation of CREB results in the recruitment and/or stimulation of HAT activities [28], and this may be the cause of the enhanced histone acetylation observed in the Egr1 promoter upon stimulation of murine embryo stem cells [25]. Indirect evidence points to the importance of histone acetylation for the expression of Egr1 gene [29]. Nevertheless, several questions are still waiting for a definite answer to understand the chromatin-related mechanisms that allow transcription of the Egr1 gene.
First, with the above-mentioned exceptions, nothing is known about the binding of factors and of chromatin-modifying complexes to Egr1 promoter in intact cells. The binding of these different factors to chromatin should be studied in a time-dependent manner to establish the temporal coincidences between binding and transcription of the gene. Second, additional experiments are necessary to check whether there is a direct competition between EGR1 and SP1 for the Egr1 promoter. Moreover, it remains to be determined whether NAB1/2 bind EGR1 in vivo as they do in vitro. These questions, among others, are addressed in the present paper by using a combination of chromatin immunoprecipitation (ChIP), mapping of factor binding by quantitative ChIP, and a kinetic analysis of the binding and/or modification of factors correlated with the real-time detection of gene transcription.
Materials and methods
Cell culture, induction, and inhibition
Cells from the mouse progenitor hepatocyte cell line MLP29 were grown in Dulbecco modified Eagle’s medium (Sigma), supplemented with 10% fetal bovine serum (Gibco), 200 Mm l-glutamine (Gibco), 100 U/ml penicillin (Sigma), 100 μg/ml streptomycin (Sigma), and 2.5 μg/ml fungizone (Gibco). Cells were plated in parallel, and they were grown to a 70–80% confluence. They were then serum-starved (0.5% fetal bovine serum) for 16–24 h, and to stimulate Egr1 transcription, 12-O-tetradecanoylphorbol-13-acetate (TPA) (Sigma) was added to a final concentration of 50 nM. When required, cells were pre-treated with the following protein kinase inhibitors at the indicated final concentrations: 100 μM STAT3 inhibitor, 10 μM hypericin, 100 μM PD98059, 100 μM SB203580, 100 μM JNK inhibitor, 20 μM ERK inhibitor, 1 μM wortmanin, 10 μM H89, and 25 μM NFκB inhibitor. All the inhibitors were from Calbiochem, except hyperycin and cycloheximide, which were purchased from Sigma. Controls were carried out by treating the cells with the correspondent solvent only.
ChIP analysis
Cells were fixed in 1% formaldehyde in PBS buffer for 4 min at room temperature, with constant shaking. To obtain chromatin fragments, chromatin was subjected to 12 cycles, 10 s each, of sonication at a 37% setting in a Vibra-Cell VCX-500 sonicator. The samples were allowed to stand for 1 min on ice between two consecutive cycles. Under these conditions, the average size of chromatin fragments was of 500 ± 200 bp. ChIP and RNApol-ChIP analyses, as well as the PCR amplification of the DNA recovered from the immunoprecipitates, were carried out essentially as described by Sandoval et al. [2], with the primers described in Table 1. The antibodies used for ChIP assays were α-SRF (sc-335), α-SP1 (sc-59), α-ELK1 (sc-355), α-CREB (sc-186), α-EGR-1 (sc-110), α-CREB-P (sc-7978), α-ELK-1-P (sc-8406), α-NAB1 (sc-22813), α-NAB2 (sc-22815), α-RNAPol II (sc-899), α-mSIN3A (sc-994), α-HDAC3 (sc-11417), α-NCoR (sc-8994), α-GCN5 (sc-6304), and α-CBP (sc-369) from Santa Cruz; α-H3K4Me2 (ab 7766), α- H3K4Me3 (ab 8580), α-H3K9Ac (ab 4441), α-H3K18Ac (ab 1191), α-H4K5Ac (ab 1758), and α-H4K8Ac (ab 1760) from Abcam; α-H3K14Ac (07–353), α-H3K27Ac (07–360), α-H3S10PhK14Ac (07–081), α-H4hyperAc (06–598), α-H4K16Ac (07–329), and αμ-H3S10PhK14Ac (07–081) from Upstate/Millipore. The specificity of the most relevant antibodies against the different factors was checked by Western blot of MLP29 cell extracts.
Table 1.
Primers used for PCR
| Amplicon | Size (bp) | From | To | Primer sequence: (5′–3′); (5′–3′) |
|---|---|---|---|---|
| P (Egr1 promoter) | 352 | −579 | −228 | TTCCCTAGCCCAGCTCGCAC; GAACAGCAGCAGTGGCCTCC |
| C (Egr1 2nd exon) | 263 | +1764 | +2027 | CAGAAGCCCTTCCAGTGTCG; GATGGGTAGGAGGTAGCCAC |
| a (Egr1 promoter) | 102 | −1250 | −1148 | AACGTGGAGGCGACGGAAGA; CAGTGCGGAGGACCCGTTTA |
| b (Egr1 promoter) | 97 | −1133 | −1036 | CAGACTGCCCAAAGCCAAGT; ATGGTTAGACACTTCCTGAC |
| c (Egr1 promoter) | 97 | −987 | −890 | TCAGGGTTCCGCAATCCAGG; CTGGGGCTTCCAATGCACCA |
| d (Egr1 promoter) | 112 | −960 | −848 | AGGTGGGATCCTCAACCGCA; TGCATACTCGGCCACCAGTC |
| f (Egr1 promoter) | 77 | −685 | −608 | TCTCCGCAGGCTCGCTCCCA; AGTCAGGCCGGGGTTCTACAGCC |
| g (Egr1 promoter) | 72 | −633 | −561 | GACGGCTGTAGAACCCCGGCCTGA; TGCGAGCTGGGCTAGGGAAGCC |
| h (Egr1 promoter) | 112 | −569 | −458 | TTCCCTAGCCCAGCTCGCAC; TCCCAACTGGGAGCCTGAGC |
| i (Egr1 promoter) | 141 | −465 | −325 | AGTTGGGAACCAAGGAGGGG; AGGGCCATATTGGGCCACTC |
| k (Egr1 promoter) | 113 | −334 | −222 | ATATGGCCCTGCCGCTTCCGGCTC; GTATTGGAACAGCAGCAGTGGCCTC |
| m (Egr1 promoter) | 103 | −286 | −184 | GGGGCAAGCTGGGAACTCCA; AGAGTGGTGGGCACCTCCAC |
| n (Egr1 promoter) | 101 | −247 | −147 | GGAGGCCACTGCTGCTGTTC; CATCCAAGAGTGGTGGGCAC |
| o (Egr1 promoter) | 107 | −135 | −29 | CGTCACTCCGGGTCCTCCCG; AGGGTCTGGAACAGCACGGGCC |
| s (Egr1 promoter-coding) | 50 | −33 | +17 | ACCCTTGAAATAGAGGCCG; GCGCGCTGGGATCTCTC |
| t (Egr1 coding) | 91 | +243 | +333 | CCAACCACCCAACATCAGTT; AGAGATCTGCAGCGGAGACATC |
| u (Egr1 coding) | 91 | +547 | +637 | CCGAGCGAACAACCCTATGA; CTGGACGTTTGGGTCGTCAC |
| α-actin (promoter) | 227 | −484 | −257 | CACCTGACCACAGGGCATCC; AACTGGCTCCAAGGCTCACG |
| α-actin (coding) | 169 | +2647 | +2816 | ATCCTGGCCTCGCTGTCCAC; CACCCTGCAACCACAGCACG |
| β-actin (promoter) | 306 | −541 | −235 | TCTGGCTTTCCGGCTATTGC; AGTTTTGGCGATGGGTGCTG |
| 18S rRNA (coding) | 433 | +4 | +437 | TGGTTGATCCTGCCAGTAGC; CTCTCCGGAATCGAA CCCTG |
| 18S rRNA (coding) | 71 | +67 | +137 | CACGGCCGGTACAGTGAAA; AGAGGAGCGAGCGACCAA |
Quantitative mapping of factor binding was carried out by ChIP analysis using the appropriate antibodies. Immunoprecipitated DNA was quantified by real-time PCR carried out with an ABI GeneAmp 7000 Sequence Detection System (Perkin-Elmer, Applied Biosystems) with the appropriate primers to analyze short amplicons (around 100 bp) over the Egr1 promoter and proximal coding region (Table 1). The resulting values for DNA concentration were normalized relative to the values obtained for the input fractions, which were quantified in the same manner. In this way, the deviations due to the distinct efficiency of the different sets of primers were corrected, making the data obtained for the binding of a given factor at different amplicons fully comparable. Of course, the comparison between the data obtained for different factors can only be done at a semiquantitative scale because, when different antibodies have to be used for the immunoprecipitation, and hence, different input fractions have to be quantified, the accuracy of the method relies on the estimation of the amount of chromatin used as the input fraction. At any rate, the method provides a convenient approach to study the occupancy of the different target sites for a given factor.
RNA isolation and RT-PCR analysis
Total RNA was obtained by the TRIZOL method [30]. To obtain cDNA, 1 μg RNA was retrotranscribed using Superscript II RNase H− (Life Technologies, Invitrogen) following the manufacturer’s instructions, although random hexamers (Sigma) were used instead of oligo dT. PCR analysis of cDNA was done as above, with the appropriate primers (Table 1).
Results
Analysis of TPA-induced Egr1 expression
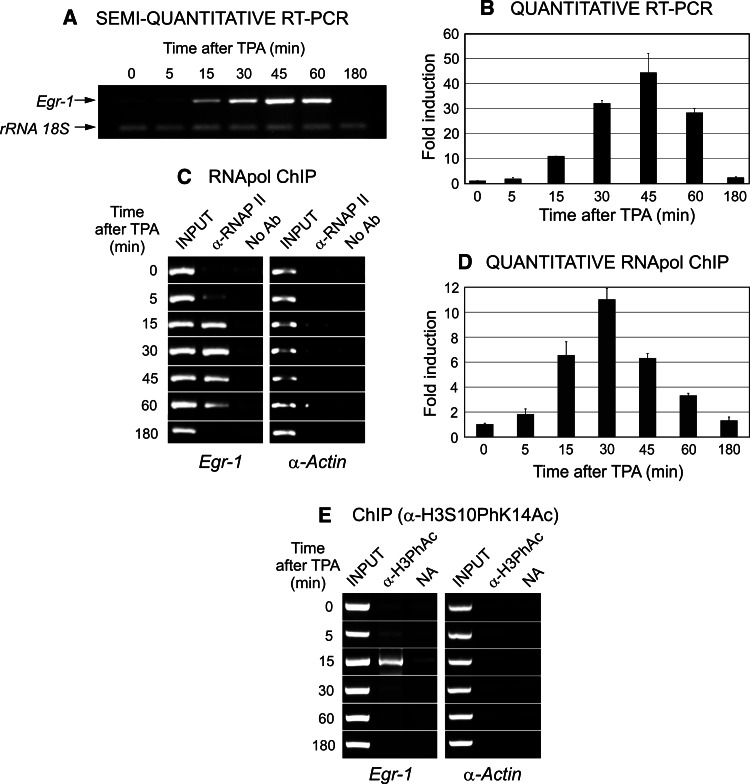
To induce Egr1, we use in the present work TPA, a known inductor of IE and other genes [31]. The expression of Egr1 gene in the MLP29 mouse progenitor hepatocyte cell line was first analyzed by either semi- or quantitative RT-PCR, starting from total RNA samples prepared from nonstimulated cells at different times after adding TPA, as described in the section “Materials and methods.” Results of a representative experiment are given in Fig. 1a, b, which shows that the maximum level of mRNA is obtained 45 min after adding TPA. Nevertheless, the steady state level of mRNA does not allow us to know the actual transcriptional rate of a gene, especially when the half-life of mRNA is comparable with the expanse of the period in which the experimental data are obtained. We have previously described an experimental approach to circumvent this inconvenience, namely the RNApol-ChIP assay, which consists of detecting, by ChIP analysis, the presence of RNA polymerase II in coding regions well apart from the promoter [2]. This presence, which can be quantified, offers a convenient measurement of the actual transcriptional rate. It is obvious from the results obtained by this method, presented in Fig. 1c, d, that Egr1 expression can be detected as early as 5 min after adding TPA and that the peak of its expression occurs at 30 min. The shift of 15 min between this peak and that of Fig. 1b is clearly due to the fact that the balance between mRNA transcription and degradation favors its accumulation. Therefore, RNApol-ChIP offers a more accurate quantification of the transcriptional rate than the measure of the steady state levels of mRNA.
Fig. 1a–e.
TPA-induced expression of Egr1 gene in murine MLP29 progenitor cells. The steady state level of Egr1 mRNA was analyzed by RT-PCR (a, b) and the real-time transcription rate was determined by RNAPol-ChIP (c, d). Results of representative semiquantitative analyses (a, c) and of quantitative (b, d) PCRs are given. The latter represent the average ± SE of three (b) or six (d) determinations. In the quantitative analyses, Egr1 expression is normalized to the value in the absence of TPA and plotted as fold induction. e Results of a ChIP analysis of phosphoacetylation of histone H3
The fast response of Egr1 transcription to the proliferating signal is a property of the IE genes. As the phosphorylation of H3 serine 10 followed by the acetylation of Lys 14 of the same histone is a common hallmark of IE genes [5], we have checked the acquisition of these epigenetic marks in the Egr1 promoter (Fig. 1e). The simultaneous presence of both modifications appears 5 min after TPA treatment, just when the transcription begins to be detected (Fig. 1c, d) and passes a narrow peak at 15 min, prior to the peak of transcriptional rate.
Factor binding to Egr1 promoter
Most of the previously available data on the control of Egr1 induction have been obtained by using electrophoretic mobility shift assays or transient expression systems which, as mentioned above, do not necessarily reveal the actual influence of the different factors in a regulated chromatin context. The experiments of Wang et al. [25], carried out with intact murine embryonic stem cells, pointed towards the importance of phosphorylated ELK1 in the serum-induced expression of Egr1. To test whether this is also valid in the induction by TPA, as well as to check the possible influence of other factors in intact cell systems, we planned a series of ChIP analyses of factor binding to Egr1 promoter. We conducted these experiments in a time-dependent manner to correlate the binding of the different factors with the real-time expression analysis.
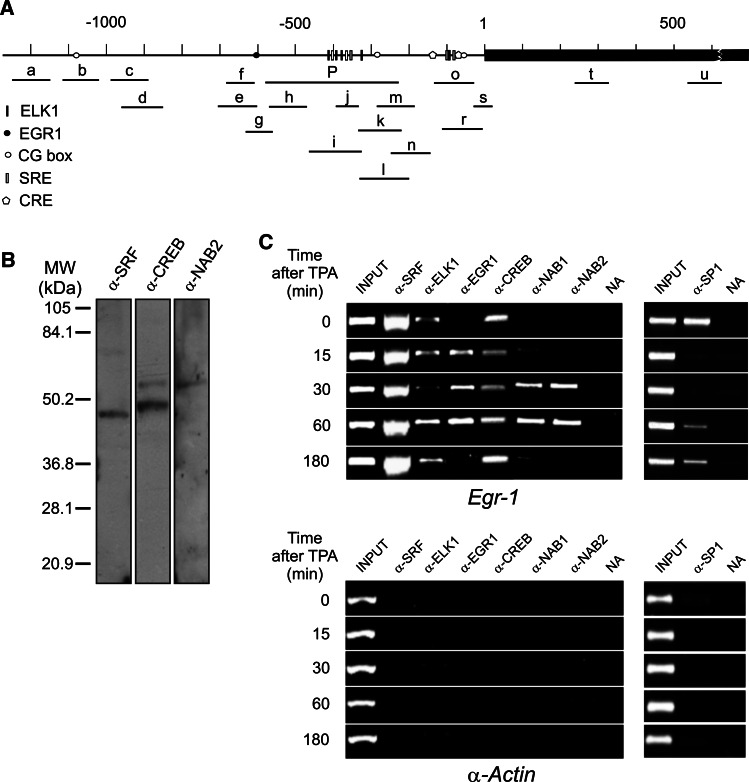
To facilitate the interpretation of all the experiments described in this paper, a map showing the cis elements present in the Egr1 promoter, as well as the different amplicons covering the promoter and part of the coding region used in the PCR analyses, is given in Fig. 2a. Figure 2c shows the results of a representative ChIP experiment to study time-dependent factor binding to the whole promoter. The oligonucleotides used (Table 1) amplify a region of 352 bp from −579 to −227 relative to the transcription start site (TSS). Taking into account that the estimated average size of the chromatin fragments obtained by sonication is about 500 bp, the ChIP experiment covers the proximal regions of the promoter (Fig. 2a). The results of ChIP analysis (Fig. 2c) show that SRF, CREB, ELK1, and SP1 are bound to the nonstimulated Egr1 promoter. The extent of factor binding seems to change when the gene is being actively transcribed, i e., between 15 and 30 min after adding TPA. Particularly, ELK1 binding displays a complex pattern, and it seems to increase over the basal values when Egr1 transcription declines. Further data in this sense will be presented later. All these results of ChIP analysis were consistently repeated in at least three independent experiments.
Fig. 2a–c.
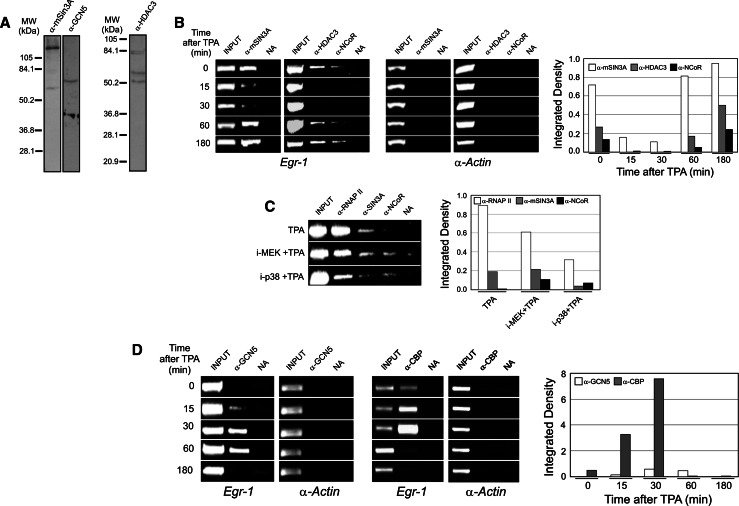
ChIP analysis of factor binding to Egr1 promoter after TPA stimulation. To facilitate the interpretation of the experiments, a map of the Egr1 promoter is depicted (a) in which the relevant elements of the promoter are represented. The bars below the map correspond to the different amplicons used in the PCR analyses (see Table 1 for details). Western blots of extracts from MLP29 progenitor cells, showing the specificity of antibodies used, are given in b. The results of a representative ChIP assay with amplicon P are given in c. As a negative control, c shows the results obtained with the same antibodies in the promoter of α-actin gene. NA Control with no antibody added
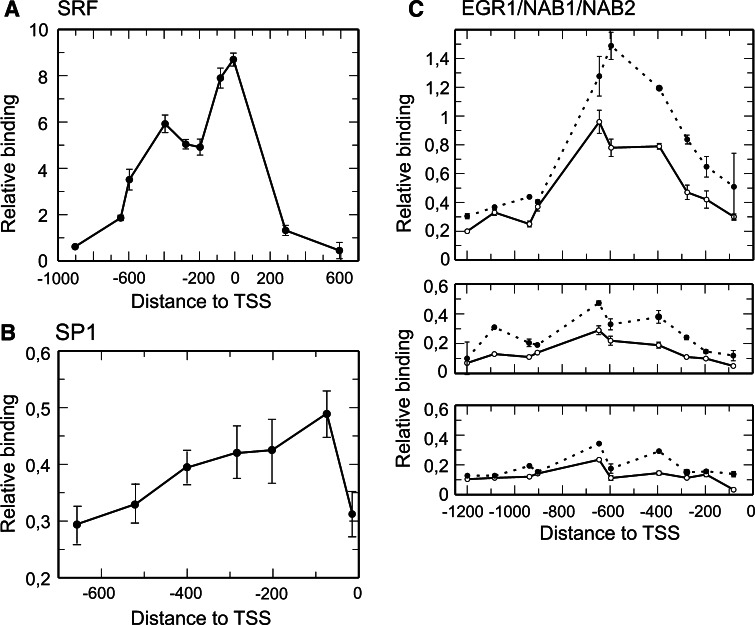
The signal obtained in the ChIP experiments with the SRF antibody is especially intense (Fig. 2c). Apart from the possible differences in antibody effectiveness, it has to be noted that murine Egr1 promoter contains two clusters of SREs, one between −50 and −100, which includes two binding sites, and the other one, comprising four additional sites, located between −300 and −400 relative to the TSS. Bauer et al. [32] reported that in transient expression experiments, both clusters of SREs are operative and one can wonder if they are also operative in a stably chromatin context. To address this issue we mapped the occupancy of Egr1 promoter by SRF. We had previously used a similar ChIP analysis to follow the redistribution of Tup1p over the yeast SUC2 promoter upon induction [33], but the method described here allowed us to obtain quantitative data. Cross-linked chromatin fragments of MLP29 control cells were immunoprecipitated with the antibody against SRF. The purified DNA was amplified by real-time PCR using the appropriate oligonucleotides to analyze several of the short amplicons depicted in the map of Fig. 2a (see Table 1). The plot given in Fig. 3a shows that the relative binding of SRF follows a bimodal distribution with peaks at about −400 and around the TSS. This means that both clusters of SREs are also functional in SRF binding in a chromatin context.
Fig. 3a–c.
Quantitative mapping of the binding of several factors to the Egr1 promoter. a Plot of the relative binding of SRF in nonstimulated cells, determined as detailed in the experimental procedures, against the position of the centers of the analyzed amplicons. The bimodal distribution shows that both clusters of SREs (see Fig. 2a) are being used. b Relative binding of SP1 in nonstimulated cells. c Relative binding of EGR1 (upper panel), NAB1 (middle panel), and NAB2 (lower panel), plotted for 30 min (open symbols and solid line) and 60 min (filled symbols and dashed line) of TPA treatment. To estimate the relative binding of the factors, the DNA concentration obtained for each amplicon was normalized to the value obtained for the input (see the section “Materials and methods”)
Apart from a putative SP1 site at −1089 bp, there are two CG boxes in the Egr1 proximal promoter to which SP1 can bind. The first one, which does not possess a canonical consensus sequence, is located at −49, close to the proximal cluster of SREs, while the second one maps at −277 bp. To determine whether both proximal CG boxes are operative in intact cells, we mapped the SP1 occupancy along the Egr1 proximal promoter in nonstimulated cells by a quantitative ChIP analysis. The results obtained between −600 and the TSS (Fig. 3b) show that the site at −49 is the main target site for SP1. To explore whether competition between SP1 and EGR1 occurs in the Egr1 promoter in intact murine cells, we also determined the time-dependent binding of EGR1 to the promoter of its own gene (Fig. 2c). The results show that SP1 is bound to the promoter in nonstimulated cells (t = 0), and that the binding of EGR1 at 15 min coincides with SP1 leaving. Furthermore, generally speaking, SP1 and EGR1 do not simultaneously occupy Egr1 promoter. Nevertheless, these results do not necessarily mean that a direct competition between both factors occurs, as they might bind different sites. Actually, in addition to the CG box at −49, there exists a second one at −277 and a canonical EGR1 site at −595. To further explore whether SP1 and EGR1 use the same target sites, we carried out a time-dependent mapping of the occupancy of Egr1 promoter by EGR1 after TPA stimulation (Fig. 3c). The binding of EGR1 at 30 min after TPA induction peaks at the amplicon centered at −597, showing that EGR1 mainly uses the site at −595 and, therefore, it can be concluded that SP1 is not displaced by a direct competition with EGR1. After 60 min induction, the presence of EGR1 on its own promoter increases (Fig. 3c), in accordance with the semiquantitative data of Fig. 2b.
For the reasons given in the “Introduction,” we next analyzed the presence of NAB1 and NAB2 at the Egr1 promoter. The results given in Fig. 2c show that both NAB1 and NAB2 are recruited to Egr1 promoter at 30 min after starting TPA treatment, when transcription has passed its maximum (see Fig. 1) and begins to decline. At 180 min, once the Egr1 transcription has again ceased, NAB1 and NAB2 are no longer found at Egr1 promoter. The analysis of the EGR1 binding profile shows a shoulder at about −395, which is especially obvious at 30 min (Fig. 3c). This might be compatible with the binding of EGR1 to an additional site close to that position. The profiles of NAB1 and NAB2 binding, anchored to chromatin by the interaction with EGR1, also agree with this possibility. The analysis of the Egr1 promoter shows the presence of a weak (70% consensus matching) putative EGR1 site at −427 (not shown in Fig. 2a). Our data, however, do not allow us to reach a definite conclusion on the possible usage of this putative site.
Finally, we also analyzed the presence of RNA polymerase II in the promoter. ChIP experiments revealed that the polymerase is present in the promoter even in the absence of TPA induction, although its presence is increased in coincidence with the peak of gene expression. The presence of paused RNA polymerase either at the promoter or at the beginning of the coding region is a common behavior in immediate-early genes [34, 35].
In summary, the results described in the present section show that all the activating factors tested are present in the promoter in nonstimulated cells, and yet the gene is not expressed. It seems obvious that an additional element is required for gene induction. Apart from the factors that contribute to the releasing of elongating polymerase, phosphorylation of CREB [12] and ELK1 [36] is required for these factors to manifest their transactivating capacity in different systems. On the other hand, changes in chromatin structure may play a definite role in controlling gene expression. Both activating effects, namely factor phosphorylation and chromatin modification, need not be mutually excluding, and they will be examined in the next sections.
TPA-induced factor phosphorylation
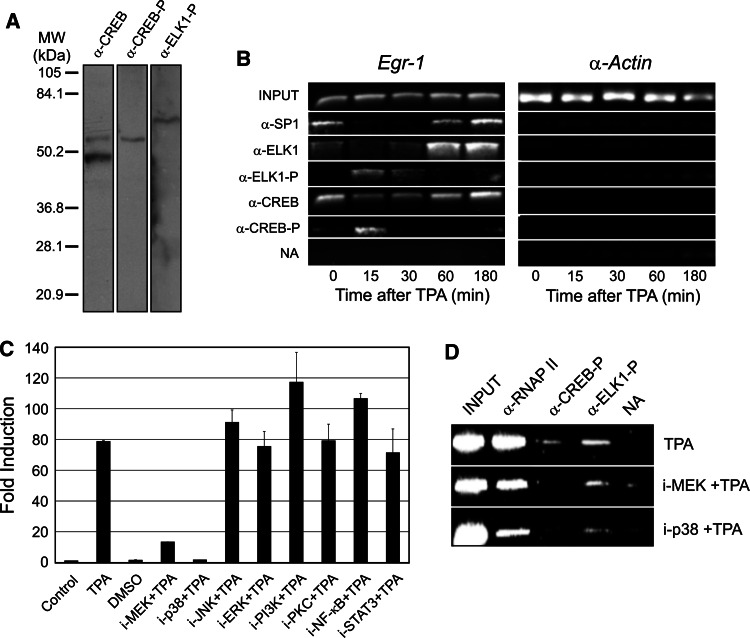
To explore the time-dependent relation between factor phosphorylation and transcriptional activity in intact cells, we conducted a ChIP experiment similar to that of Fig. 2, but using antibodies specific for phosphorylated ELK1 and CREB. The maximum phosphorylation of both ELK1 and CREB in the Egr1 promoter is reached at 15 min, just before the peak of gene expression, and the factors are dephosphorylated when gene transcription declines (Fig. 4b). It is interesting to note that the enhancement of ELK1 binding at 60 and 180 min, which is appreciated in this figure more clearly than in Fig. 2c, corresponds only to the nonphosphorylated form of the factor. On the other hand, the use of kinase inhibitors allows us to determine that both MEK1/2 and p38 kinase cascades are involved in the signal transduction from TPA to Egr1 promoter (Fig. 4c). Moreover, the data of Fig. 4d indicate that the inhibition of either MEK1/2 or p38 kinases reduces the TPA-induced phosphorylation of ELK1 and abolishes that of CREB. At the same time, the recruitment of RNA polymerase II to the Egr1 promoter is clearly diminished after kinase inhibitor treatment, in accordance with the concomitant inhibition of transcription. The specificity of the antibody to phosphorylated CREB is critical to interpret these results. In our hands, although the antibody against CREB gives a faint signal with the phosphorylated factor in an extract of MLP29 cells, the antibody against phosphorylated CREB is absolutely specific (Fig. 4a).
Fig. 4a–d.
Factor phosphorylation and Egr1 activity. a Western blots of extracts from MLP29 progenitor cells, showing the specificity of antibodies used. b Time-dependent phosphorylation of ELK1 and CREB in Egr1 promoter. ChIP analyses, similar to those of Fig. 2, were carried out with antibodies specific for the phosphorylated forms of ELK1 and CREB. c Effects of several kinase inhibitors on TPA-induced Egr1 expression. The average ± SE of three independent quantitative RT-PCR determinations at amplicon C (normalized relative to rRNA 18S gene) is represented for cells treated or not with the inhibitors (i) of the indicated kinases. A control determination (from untreated cells) was arbitrarily set to 1. d ChIP analysis of the effects of kinase inhibitors on the phosphorylation of CREB and ELK1, and on the binding of RNA polymerase II to the Egr1 promoter. The results of the PCR analyses of a and c were integrated and plotted
Recruitment of chromatin-modifying complexes to Egr1 promoter
The above results confirm that phosphorylation events are required for the TPA-induced transcriptional activation of Egr1 promoter in a chromatin context. We next examine the changes that occur in chromatin upon Egr1 induction.
Figure 5b shows that three components of HDAC complexes, namely mSin3A, HDAC3, and N-CoR, are present in the Egr1 promoter in untreated, serum-starved cells, but all these components leave the promoter when the gene is being expressed, to return again after the transient period of gene activation.
Fig. 5a–d.
Binding of HDACs and HATs to Egr1 promoter. ChIP analysis (amplicon P) of some components of HDAC (b) and HAT (d) complexes to Egr1 promoter. The effects of kinase inhibitors on the presence of histone-modifying complexes at the Egr1 promoter of cells stimulated 15 min are shown in c. The results of the PCR analyses were integrated as in Fig. 4. Western blots of extracts from MLP29 progenitor cells, showing the specificity of antibodies used, are given in a
We next asked whether the presence of HDAC complexes was affected by the inhibitors of the cascades responsible for Egr1 induction. To address this issue we carried out ChIP assays with cells previously treated with the corresponding kinase inhibitors. The results shown in Fig. 5c indicate that N-CoR needs some phosphorylation event to be released from the promoter. Moreover, our data are compatible with the possibility that both MEK and p38 kinases are involved in this phosphorylation.
The analysis of HAT binding to Egr1 promoter revealed that a GCN5-containing HAT complex is recruited in response to TPA treatment and that this HAT is present throughout the interval in which the gene is being expressed. CBP is clearly detectable in the nonstimulated promoter, but its presence evidently increases in parallel with TPA-induced transcription of the gene (Fig. 5d). All these results prompted us to examine the acetylation marks in the histones at the Egr1 promoter, as reported in the next section.
Histone acetylation at the Egr1 promoter
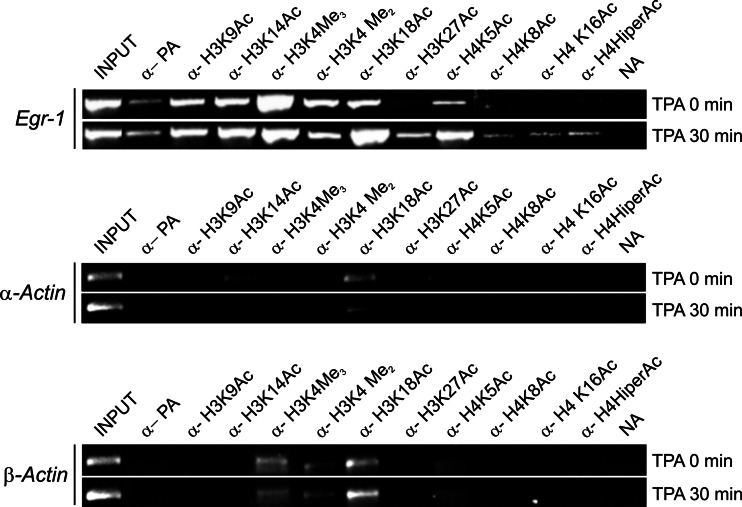
Figure 6 shows the results of a ChIP analysis of some histone modifications at the Egr1 promoter in nonstimulated MLP29 cells and after 30 min of TPA treatment. According to the results of Fig. 1c, d, under the first set of conditions the gene is repressed, whereas in the second case the gene is actively expressed. As a negative control, a similar analysis of the α-actin promoter is presented. This gene is not expressed under the present conditions and the transcription-related epigenetic marks are missing. It can be seen that four of the sites analyzed, namely H3K9, H3K14, H3K18, and H4K5 are acetylated in nonstimulated cells. The last three lysyl residues are substrates of p300/CBP [37] and thus they may become acetylated provided that this HAT is actually present under basal conditions (see above). Acetylation of H3K9 and H3K14 is maintained or even increased under inducing conditions but that of H3K18 and H4K5 is clearly enhanced upon induction. The first of these residues may also be acetylated by GCN5 [37], and the recruitment of HAT complexes containing this activity (Fig. 5c) may account for the observed increase. As to the enhancement of H4K5 acetylation, it may be caused by some non-tested HAT or by the enhanced recruitment of CBP (Fig. 5c).
Fig. 6.
Histone modifications at the promoter of the repressed and active Egr1 gene. The figure shows the results of a ChIP experiment carried out in nonstimulated (TPA 0 min) and TPA-treated cells for 30 min, in which the Egr1 gene is fully active (TPA 30 min) with the indicated antibodies. The immunoprecipitated DNA was PCR-amplified with the primers corresponding to amplicon P (see Table 1). The results obtained at the promoter of the α-actin gene were included as negative control. The appropriate primers (see Table 1) were used
Finally three of the potentially acetylatable lysines tested, namely H3K27, H4K8, and H4K16, are not modified at all in the repressed promoter and become clearly modified after 30 min of TPA treatment (Fig. 6). Taking into account that these lysyl residues may be target sites for GCN5-driven acetylation [37], the increase in their acetylation might be related to the recruitment of GCN5 to the promoter (see Fig. 5c).
Discussion
In the present paper we describe an analysis of the changes that take place upon induction at the promoter of an IE gene, namely the murine Egr1 gene. To do this we have taken advantage of several circumstances. First, the research was conducted in intact murine cells, to maintain the chromatin structure preserved. Secondly, we have used the RNApol-ChIP method [2] to follow transcription in real-time, so the recruiting of factors and the chromatin modifications can be strictly correlated to changes in transcription. Moreover, in many instances we have used a quantitative mapping of factor occupancy, which allowed us to decide which of the potential target sites are actually being used by the different factors in a chromatin context.
It was already known that the activation of Egr1 followed a transient pattern [32, 38]. These considerations are also valid for MLP29 cells. Figure 1 shows that although the level of Egr1 mRNA peaks at 45 min, the actual transcriptional rate of the TPA-induced mouse Egr1 peaks at 30 min. The evaluation of transcriptional rate in real-time is therefore decisive to analyze the relationships between factor binding and/or modification and gene activity. In this sense, it is important to note the transient character of the phosphoacetylation of H3 (Fig. 1e), which may play a signalling role among the other epigenetic marks.
The extent of CREB, SP1, and EGR1 binding to Egr1 promoter changes with gene expression (Fig. 2). The interplay of these three factors is essential for the expression of several genes [17, 22, 32, 39], but there are conflicting reports in the literature concerning the exact nature of that interplay. For instance, we have already commented on the possibility of a binding competition between EGR1 and SP1. In fact, our data indicate that SP1 leaves the promoter when EGR1 enters it (Fig. 2c). A similar inverse correlation has been recently described in several genes [6], but the data of Fig. 3 clearly show that there is no direct competition between EGR1 and SP1 in the Egr1 promoter. Actually, the basal binding of SP1 peaks close to the site at −49, which does not possess a canonical sequence, although it cannot be rejected that some binding occurs at the canonical site at −277. It can be assumed that, in some way, either the chromatin organization or the presence of other factors makes the binding of SP1 to the canonical CG box difficult. On the contrary, EGR1 mainly binds the site at −593 (Fig. 3c), so the displacement of SP1 is not due to a direct competition.
CREB is constitutively bound to the Egr1 promoter (Fig. 2c). As there are two cAMP-response elements (CREs) in close vicinity (Fig. 2a), we cannot decide whether both sites are occupied or just one is. Anyway, both CREs are in close vicinity of SREs (Fig. 2a), so the described interaction between CREB and SRE-bound SRF [13] may be established. On the other hand, a transient phosphorylation of CREB is detected 15 min after adding TPA (Fig. 4b), which can be attributed to the action of both MEK1/2 and p38 kinases (Fig. 4d). Fig. 4c shows that the inhibition of MEK1/2, upstream of the mitogen- and stress-activated protein kinase 1 in the MAPK pathway, results in the failure of TPA to induce Egr1. Our results are, therefore, in agreement with previously reported data [32, 40], but they add two interesting details: the participation of p38 pathway in CREB phosphorylation and a possible cross-talk between MEK and other signalling pathways, as the sole inhibition of ERK, allows the induction of Egr1 (Fig. 4c). It has been recently reported that p38 kinase does not mediate the histamine-induced accumulation of EGR1 in human aortic endothelial cells [28], but the data of Fig. 4c are conclusive in that p38 activity is required for TPA-induced Egr1 expression. Moreover, it seems clear that phosphorylation of CREB, which is essential for this factor to display its transactivating activity in IE genes [12], depends on p38 activity (Fig. 4d). It is possible that histamine and TPA use somewhat different signalling cascades upstream of ELK1 and/or CREB phosphorylation.
Moreover, the data of Fig. 4 point toward the idea that the phosphorylation of CREB is required for Egr1 induction by TPA in intact cells. It is interesting to note that CRE only plays a limited role, if any, when the TPA-induced activation of the gene is studied by transient expression experiments [32]. Probably the reasons for this discrepancy are to be found in the mechanistic differences between the transcriptional regulation in nonintegrated and integrated chromatin templates. In this context it should be mentioned that the ChIP experiments of Cha-Molstad et al. [28], working with PC12 cells, indicated that CREB, constitutively bound to the CRE, regulates their target genes through the phosphorylation-dependent recruitment of CBP. Mayer et al. [26] have recently found that ELK1 links the phosphorylation cascade initiated by the proliferating signals and the expression of Egr1. On the other hand, it has been described that the phosphorylation of ELK1, which physically interacts with the HATs CBP and p300 [41, 42], increases the HAT activity [10], and our results also show that ELK1 becomes phosphorylated 15 min after adding TPA (Fig. 4b). We have already mentioned that some of the changes in histone acetylation occurring at the Egr1 promoter (Fig. 6) may be explained by a CBP/p300-driven modification. The observed recruitment of GCN5-containing complexes, which takes place early during the induction process (Fig. 5d), may account for most of the remaining changes in histone acetylation. The complex role of ELK1 is further illustrated in our case by the increased binding of its nonphosphorylated form to the Egr1 promoter as a prelude to gene inactivation. The function of ELK1 in this context might be the recruitment of HDACs [11], which bind the Egr1 promoter concomitantly with ELK1 (Fig. 5).
As mentioned earlier, the transient activation observed for Egr1 is a common feature in many IE genes. This is surely related to the huge number of often antagonistic roles played by the products of these genes, which may require a nonsustained expression. The early results of Cao et al. [15] suggested that EGR1 acts as a repressor of its own expression. Our time-dependent ChIP analyses (Fig 2b) and the quantitative data of Fig. 3c indicate that EGR1 binding is maintained at 60 min of TPA treatment, when transcription has dropped again almost to its basal level (Fig. 1d). On the other hand, it has been described that NAB1/2 interact with EGR1 in two-hybrid systems [23, 24]. In the present instance, the data of Fig. 3c show that the binding profiles of both NAB1 and NAB2 roughly parallel that of EGR1. It is therefore highly probable that these corepressors actually interact with EGR1 in chromatin and that the recruitment of NAB2 (and perhaps of NAB1 as well) serves as a signal for repression. Actually, NAB2 acts as a repressor by interacting with a component of the nucleosome remodelling and deacetylase (NuRD) complex [43]. We are currently studying the chromatin structure along the Egr1 promoter to check whether the repression observed after 60 min of TPA treatment is accompanied by changes in nucleosome positioning.
The results presented in this paper, therefore, confirm the importance of studying the regulation of eukaryotic transcription in a chromatin context. The Egr1 gene is apparently ready for transcription in the nonstimulated cells, as RNA polymerase II is already poised for transcription. This is a characteristic feature of the immediate-early genes, also observed in many other genes (see [44] and references therein). Thus, a repressing chromatin structure is required to avoid either initiation or elongation of transcription. The interest in the regulation of transcription elongation is growing but, in the present case, changes in the chromatin structure at the promoter are decisive to initiate transcription. In fact, Fig. 5a shows that, in nonstimulated cells, the Egr1 promoter is occupied by N-CoR and mSin3A-containing repressor complexes, and Fig. 5c indicates that the release of N-CoR is phosphorylation-dependent. However, a caveat must be mentioned, because the release of N-CoR may influence transcription not only by changing the chromatin structure at promoter, but also at an elongation level [45]. Finally, the recruitment of NAB1 and NAB2 represents an additional repressing mechanism, which ensures a transient activation of the Egr1 gene. It seems obvious that this expression must be accompanied by changes in the Egr1 promoter chromatin, which we are currently studying.
Acknowledgments
We are very indebted to Dr. J. Sandoval for useful comments and to Drs. Medico and Boccacio for the gift of the MLP29 cell line. This work was supported by grants No. BFU2007-63120 and CSD2006-49 from the Ministry of Science and Innovation (Spain), and ACOMP07/085 from the Generalitat Valenciana to G.L.R. G.T. was a recipient of a fellowship (BMC 2001-2868) from the Ministry of Education (Spain).
Abbreviations
- CBP
CREB binding protein
- ChIP
Chromatin immunoprecipitation
- CRE
cAMP-response element
- CREB
cAMP-response element-binding protein
- ELK
Ets-like
- ERK
Extracellular signal-regulated kinase
- HAT
Histone acetyltransferase
- HDAC
Histone deacetylase
- IE
Immediate-early
- NAB1/2
NGFI-A binding protein 1/2
- S.E
Standard error
- SP1
Specificity protein 1
- SRE
Serum response element
- SRF
Serum response factor
- TPA
12-O-Tetradecanoylphorbol-13-acetate
- TSS
Transcription start site
References
- 1.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 2.Sandoval J, Rodríguez JL, Tur G, Serviddio G, Pereda J, Boukaba A, Sastre J, Torres L, Franco L, López-Rodas G. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004;32:e88. doi: 10.1093/nar/gnh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodríguez JL, Sandoval J, Serviddio G, Sastre J, Morante M, Perrelli MG, Martínez-Chantar ML, Viña J, Viña JR, Mato JM, Ávila MA, Franco L, López-Rodas G, Torres L. Id2 leaves the chromatin of the E2F4–p130-controlled c-myc promoter during hepatocyte priming for liver regeneration. Biochem J. 2006;398:431–437. doi: 10.1042/BJ20060380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez JL, Boukaba A, Sandoval J, Georgieva EI, Latasa MU, García-Trevijano ER, Serviddio G, Nakamura T, Avila MA, Sastre J, Torres L, Mato JM, López-Rodas G. Transcription of the MAT2A gene, coding for methionine adenosyltransferase, is up-regulated by E2F and Sp1 at a chromatin level during proliferation of liver cells. Int J Biochem Cell Biol. 2007;39:842–850. doi: 10.1016/j.biocel.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Kubosaki A, Tomaru Y, Tagami M, Arner E, Miura H, Suzuki T, Suzuki M, Suzuki H, Hayashizaki Y. Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation. Genome Biol. 2009;10:R41. doi: 10.1186/gb-2009-10-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 8.Kumbrink J, Gerlinger M, Johnson JP. Egr-1 induces the expression of its corepressor Nab2 by activation of the Nab2 promoter thereby establishing a negative feedback loop. J Biol Chem. 2005;280:42785–42793. doi: 10.1074/jbc.M511079200. [DOI] [PubMed] [Google Scholar]
- 9.Whitmarsh AJ, Shore P, Sharrocks AD, Davis RJ. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 10.Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green M. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 2003;22:281–291. doi: 10.1093/emboj/cdg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang SH, Vickers E, Brehm A, Kouzarides T, Sharrocks AD. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol Cell Biol. 2001;21:2802–2814. doi: 10.1128/MCB.21.8.2802-2814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiggin GR, Soloaga A, Foster JM, Murray-Tait V, Cohen P, Arthur JSC. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol Cell Biol. 2002;22:2871–2881. doi: 10.1128/MCB.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 14.Silverman ES, Du J, Williams AJ, Wadgaonkar R, Drazen JM, Collins T. cAMP-response-element-binding-protein-binding protein (CBP) and p300 are transcriptional co-activators of early growth response factor-1 (Egr-1) Biochem J. 1998;336:183–189. doi: 10.1042/bj3360183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao X, Mahendran R, Guy GR, Tan YH. Detection and characterization of cellular EGR-1 binding to its recognition site. J Biol Chem. 1993;268:16949–16957. [PubMed] [Google Scholar]
- 16.Fukada T, Tonks NK. The reciprocal role of Egr-1 and Sp family proteins in regulation of the PTP1B promoter in response to the p210 Bcr-AbI oncoprotein-tyrosine kinase. J Biol Chem. 2001;276:25512–25519. doi: 10.1074/jbc.M101354200. [DOI] [PubMed] [Google Scholar]
- 17.Barroso I, Santisteban P. Insulin-induced early growth response gene (Egr-1) mediates a short term repression of rat malic enzyme gene transcription. J Biol Chem. 1999;274:17997–18004. doi: 10.1074/jbc.274.25.17997. [DOI] [PubMed] [Google Scholar]
- 18.Khachigian LM, Williams AJ, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J Biol Chem. 1995;270:27279–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 19.Huang RP, Fan Y, Ni Z, Mercola D, Adamson ED. Reciprocal modulation between Sp1 and Egr-1. J Cell Biochem. 1997;66:489–499. doi: 10.1002/(SICI)1097-4644(19970915)66:4<489::AID-JCB8>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 20.Thottassery JV, Sun D, Zambetti GP, Troutman A, Sukhatme VP, Schuetz JD. Sp1 and egr-1 have opposing effects in the regulation of the rat Pgp2/mdr1b gene. J Biol Chem. 1999;274:3199–3206. doi: 10.1074/jbc.274.5.3199. [DOI] [PubMed] [Google Scholar]
- 21.Malakooti J, Sandoval R, Memark VC, Dudeja PK, Ramaswamy K. Zinc finger transcription factor Egr-1 is involved in stimulation of NHE2 gene expression by phorbol 12-myristate 13-acetate. Am J Physiol Gastrointest Liver Physiol. 2005;289:G653–G663. doi: 10.1152/ajpgi.00010.2005. [DOI] [PubMed] [Google Scholar]
- 22.Russell DL, Doyle KMH, Gonzales-Robayna I, Pipaon C, Richards JS. Egr-1 induction in rat granulosa cells by follicle-stimulating hormone and luteinizing hormone: combinatorial regulation by transcription factors cyclic adenosine 3,5-monophosphate regulatory element binding protein, serum response factor, Sp1, and early growth response factor-1. Mol Endocrinol. 2003;17:520–533. doi: 10.1210/me.2002-0066. [DOI] [PubMed] [Google Scholar]
- 23.Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Mayer SI, Willars GB, Nishida E, Thiel G. Elk-1, CREB, and MKP-1 regulate Egr-1 expression in gonadotropin-releasing hormone stimulated gonadotrophs. J Cell Biochem. 2008;105:1267–1278. doi: 10.1002/jcb.21927. [DOI] [PubMed] [Google Scholar]
- 27.Hao F, Tan M, Xu X, Cui M-Z. Histamine induces Egr-1 expression in human aortic endothelial cells via the H1 receptor-mediated protein kinase Cd-dependent ERK activation pathway. J Biol Chem. 2008;283:26928–26936. doi: 10.1074/jbc.M803071200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cha-Molstad H, Keller DM, Yochum GS, Impey S, Goodman RH. Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc Natl Acad Sci USA. 2004;101:13572–13577. doi: 10.1073/pnas.0405587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubieniecka JM, de Bruijn DRH, Su L, van Dijk AHA, Subramanian S, van de Rijn M, Poulin N, van Kessel AG, Nielsen TO. Histone deacetylase inhibitors reverse SS18–SSX-mediated polycomb silencing of the tumor suppressor early growth response 1 in synovial sarcoma. Cancer Res. 2008;68:4303–4310. doi: 10.1158/0008-5472.CAN-08-0092. [DOI] [PubMed] [Google Scholar]
- 30.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 31.Cochran BH. Regulation of immediate early gene expression. NIDA Res Monogr. 1993;125:3–24. [PubMed] [Google Scholar]
- 32.Bauer I, Hohl M, Al-Sarraj A, Vinson Ch, Thiel G. Transcriptional activation of the Egr-1 gene mediated by tetradecanoylphorbol acetate and extracellular signal-regulated protein kinase. Arch Biochem Biophys. 2005;438:36–52. doi: 10.1016/j.abb.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Boukaba A, Georgieva EI, Myers FA, Thorne AW, López-Rodas G, Crane-Robinson A, Franco L. A short range gradient of histone H3 acetylation and Tup1p redistribution at the promoter of the Saccharomyces cerevisiae SUC2 gene. J Biol Chem. 2004;279:7678–7684. doi: 10.1074/jbc.M310849200. [DOI] [PubMed] [Google Scholar]
- 34.Plet A, Eick D, Blanchard JM. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene. 1995;10:319–328. [PubMed] [Google Scholar]
- 35.Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian J, Karin M. Stimulation of Elk1 transcriptional activity by mitogen-activated protein kinases is negatively regulated by protein phosphatase 2B (calcineurin) J Biol Chem. 1999;274:15173–15180. doi: 10.1074/jbc.274.21.15173. [DOI] [PubMed] [Google Scholar]
- 37.Peterson CL, Laniel M-A. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Lim RW, Varnum BC, O’Brien TG, Herschman HR. Induction of tumor promotor-inducible genes in murine 3T3 cell lines and tetradecanoyl phorbol acetate-nonproliferative 3T3 variants can occur through protein kinase C-dependent and -independent pathways. Mol Cell Biol. 1989;9:1790–1793. doi: 10.1128/mcb.9.4.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raychowdhury R, Schäfer G, Fleming J, Rosewicz S, Wiedenmann B, Wang TC, Höcker M. Interaction of early growth response protein 1 (Egr-1), specificity protein 1 (Sp1), and cyclic adenosine 3′5′-monophosphate response element binding protein (CREB) at a proximal response element is critical for gastrin-dependent activation of the chromogranin A promoter. Mol Endocrinol. 2002;16:2802–2818. doi: 10.1210/me.2001-0292. [DOI] [PubMed] [Google Scholar]
- 40.Arthur JSC, Cohen P. MSK1 is required for CREB phosphorylation in response to mitogens in mouse embryonic stem cells. FEBS Lett. 2000;482:44–48. doi: 10.1016/S0014-5793(00)02031-7. [DOI] [PubMed] [Google Scholar]
- 41.Janknecht R, Nordheim A. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem Biophys Res Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 42.Nissen LJ, Gelly JC, Hipskind RA. Induction-independent recruitment of CREB-binding protein to the c-fos serum response element through interactions between the bromodomain and Elk-1. J Biol Chem. 2001;276:5213–5221. doi: 10.1074/jbc.M007824200. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan R, Mager GM, Ward RM, Mayer J, Svaren J. NAB2 represses transcription by interacting with the CHD4 subunit of the nucleosome remodeling and deacetylase (NuRD) complex. J Biol Chem. 2006;281:15129–15137. doi: 10.1074/jbc.M600775200. [DOI] [PubMed] [Google Scholar]
- 44.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu J, Yoon HG, Qin J, Wong J. Regulation of P-TEFb elongation complex by CDK9 acetylation. Mol Cell Biol. 2007;27:4641–4652. doi: 10.1128/MCB.00857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]