Abstract
Dps-like proteins are key factors involved in the protection of prokaryotic cells from oxidative damage. They act by either oxidizing iron to prevent the formation of oxidative radicals or by forming Dps-DNA complexes to physically protect DNA. All Dps-like proteins are characterized by a common three-dimensional architecture and are found as spherical dodecamers with a hollow central cavity. Despite their structural similarities, recent biochemical and structural data have suggested different functions among members of the family that range from protection inside the cells in response to various stress signals to adhesion and virulence during bacterial infections. Moreover, the Dps-like proteins have lately attracted considerable interest in the field of nanotechnology owing to their ability to act as protein cages for iron and various other metals. A better understanding of their function and mechanism could therefore lead to novel applications in biotechnology and nanotechnology.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0168-2) contains supplementary material, which is available to authorized users.
Keywords: Oxidative stress, Mini-ferritins, Metal binding, Ferroxidase, Fenton reaction, Iron binding, Zinc
Introduction
Iron is an abundant chemical element on earth and an essential co-factor for several enzymes found in living organisms. Although it is widely distributed in cells, its tendency to oxidize to insoluble Fe3+ compounds can disrupt normal cellular functions. Consequently, most of the cytosolic iron is complexed with different proteins with only a low concentration of free iron present inside the cells (typical concentrations 10−18 and 10−8 M of free Fe3+ and Fe2+, respectively) [1]. Most importantly, the presence of Fe2+ can lead to the generation of toxic hydroxyl radicals according to the Fenton reaction:
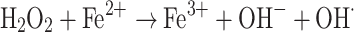 |
Hydroxyl radicals, in turn, are reactive oxygen species (ROS) able to induce DNA breakage, lipid peroxidation and degradation of biomolecules [2, 3].
DNA-binding protein from starved cells (Dps)-like proteins comprise a well-conserved family of prokaryotic proteins found in bacteria and archaea. These proteins are able to store iron in a bioavailable form and protect cells against oxidative stress [4–11]. The oxidative protection is achieved in two ways: either by binding Fe2+ ions and preventing the Fenton reaction-catalyzed formation of toxic hydroxyl radicals or by binding DNA and thus shielding it from oxidative radicals.
The first Dps protein was discovered in Escherichia coli and found to protect cells from hydrogen peroxide and to bind DNA in a sequence-independent manner [4]. The protection from hydrogen peroxide damage was suggested to be achieved by DNA-binding. The crystal structure of Dps revealed structural homology to ferritins, a large family of iron storage proteins [12]. It was proposed that protection from oxidative damage could also be achieved by metal sequestering. Subsequently, an iron-binding capability was shown for Listeria innocua Dps. Interestingly, it was found that L. innocua Dps does not bind DNA, in contrast to E. coli Dps [5]. Furthermore, the crystal structure of L. innocua Dps revealed a novel iron-binding site inside the protein analogous to that of E. coli Dps [13].
To date, over a thousand Dps-like proteins have been identified (http://www.uniprot.org). Of them, approximately 97% were found in bacteria and the remaining 3% in archaea. The structural information of these proteins has significantly grown [currently over 40 entries related to Dps-like proteins are available in the Protein Data Bank (http://www.rcsb.org)], revealing a highly conserved fold shared by all family members [14–21]. Despite the fold conservation, Dps-like proteins exhibit a variety of activities in addition to protection from oxidative stress and DNA-binding. Among their additional functions, members of the Dps family act as important virulence factors conferring cell protection against multiple stresses, including metal stress, and heat and cold shock [22–24]. Moreover, they have been involved in inflammatory processes. Dps proteins have also been suggested to be useful in many applications in biotechnology and nanotechnology.
Biological regulation and function
Regulation of Dps proteins is largely related to oxidative stress, metal regulation and general stress responses under starvation (Table 1). E. coli Dps is the prototype of the protein family (Fig. 1), and its regulation, together with that of B. subtilis Dps-like proteins, has been extensively studied. The importance of E. coli Dps has been shown in stationary-phase E. coli cells, where it was found to be the most abundant protein in the cell with as many as ~180,000 Dps molecules present [25]. In E. coli, dps mRNA levels are controlled by transcriptional activator OxyR, transcription initiation factor σs and histone-like IHF protein. OxyR-dependent induction is induced by H2O2, while σs and IHF-dependent expression of dps occurs during the stationary phase [25]. OxyR is a common regulator of expression of dps in other bacteria as well [26, 27]. EcDps levels are also controlled by proteolysis. Indeed, EcDps is proteolytically degraded in carbon-abundant conditions, but the degradation stops upon carbon starvation [28]. Intriguingly, EcDps was recently reported to act as a checkpoint by interacting with DnaA to delay initiations of new rounds of DNA replication until the damaged DNA has been repaired [29].
Table 1.
Modes of oxidative stress protection and other functions of some Dps-like proteins
| Protein | Oxidative stress protection | Other functions | |
|---|---|---|---|
| Ferroxidase activity | DNA-binding | ||
| E. coli Dps | Yes [50] | Yes [4] | Protection from copper stress [70] and acid stress [71]. Checkpoint during initiation [29] |
| B. brevis Dps | Yes [14] | Yes [14] | n.k. |
| B. anthracis Dps1 | Yes [9] | No [9] | n.k. |
| B. anthracis Dps2 | Yes [9] | No [9] | n.k. |
| C. jejuni Dps | Yes [33] | No [33] | n.k. |
| L. innocua Dps | Yes [5] | No [5] | n.k. |
| H. pylori NAP | Yes [72] | Yes [73] | Involved in virulence [38–42] |
| D. radiodurans Dps | Yes [74] | Yes [74] | n.k. |
| M. smegmatis Dps | Yes [7] | Yes [7] | n.k. |
| L. lactis DpsA | No [45] | Yes [45] | n.k. |
| L. lactis DpsB | No [45] | Yes [45] | n.k. |
| S. solfataricus Dps | Yes [75] | n.k. | n.k. |
| P. furiosus Dps | Yes [10] | n.k. | n.k. |
| A. tumefaciens Dps | Yes [76] | No [76] | n.k. |
| S. suis Dpr | Yes [77] | No [77] | n.k. |
| S. mutans Dpr | Yes [11] | No [11] | n.k. |
| S. pyogenes Dpr | Yes [64] | No [64] | Protection from pH and metal stress [64] |
| S. enterica serovar Typhimurium Dps | Yes [8] | n.k. | Involved in virulence [8] |
| P. gingivalis Dps | Yes [27] | n.k. | Involved in virulence [27] |
| B. burgdorferi Dps | No [35] | n.k. | Involved in virulence [35] |
n.k. Not known
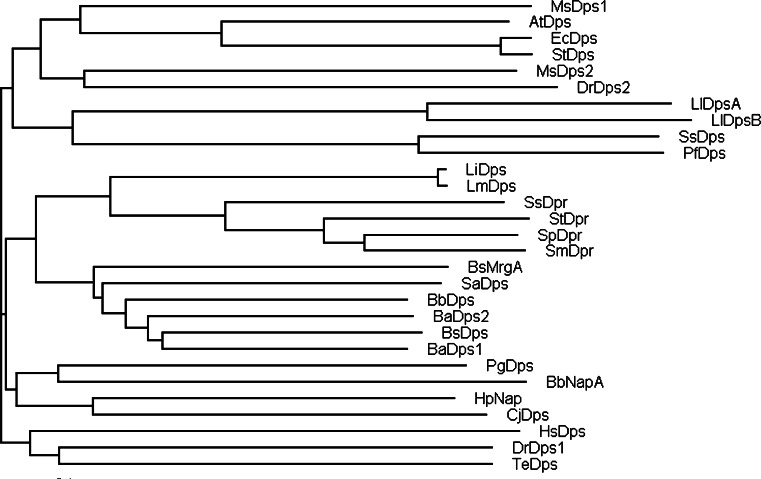
Fig. 1.
Phylogenetic tree of Dps-family representatives. The figure was created with TreeView [78]. The sequence alignment (supplementary material, Fig. S1) was performed using CLUSTALW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and default settings. AtDps, A. tumefaciens Dps; BaDps1, B. anthracis Dps1; BaDps2, B. anthracis Dps2; BbNapA, B. burgdorferi NapA; BbDps, B. brevis Dps; BsDps, B. subtilis Dps; BsMrgA, B. subtilis MrgA; CjDps, C. jejuni Dps; EcDps, E. coli Dps; HpNap, H. pylori Nap; HsDps, H. salinarium Dps; LiDps, L. innocua Dps; LlDpsA, L. lactis DpsA; LlDpsB, L. lactis DpsB; LmDps, L. monocytogenes Dps; MsDps1, M. smegmatis Dps1; MsDps2, M. smegmatis Dps2; PfDps, P. furiosus Dps; PgDps, P. gingivalis Dps; SaDps, S. aureus Dps; SmDpr, S. mutans Dpr; SpDpr, S. pyogenes Dpr; SsDps, S. solfataricus Dps; SsDpr, S. suis Dpr; StDpr, S. thermophilus Dpr; StDps, S. typhimurium Dps; TeDps, T. elongatus Dps
Regulation of B. subtilis Dps-like proteins has attracted significant interest because of the existence of two Dps-like proteins known as DpsA and MrgA. Both are functional Dps-like proteins, but their expression is differentially regulated. The expression of DpsA, for instance, is induced by heat, glucose starvation, salt and ethanol stress. However, it is not induced by H2O2, and requires the general stress and starvation sigma factor σB. In contrast, the expression of MrgA is induced by oxidative stress. MrgA is also expressed at the early stationary phase provoked by the limitation of iron and manganese [28]. The expression of MrgA is regulated by PerR [30], which also functions as a regulator for Dps proteins in other bacteria [31, 32]. An interesting exception to the general expression pattern of dps comes from C. jejuni Dps, which is constitutively expressed during exponential and stationary phases and no induction is observed after treatment with H2O2 [33]. Taken together, Dps-like proteins exhibit a variety of regulatory mechanisms and function. As they are present in different types of organisms, their regulatory properties may be dependent on special needs of each organism.
Role of Dps proteins in virulence
During infection, pathogenic bacteria face increased amounts of H2O2 produced by respiratory burst within phagocytes. Oxidative stress resistance through Dps-like proteins is a key factor in the virulence of several bacterial species. Indeed, bacteria with loss-of-function dps mutations have lower viability than wild-type cells in infected hosts [8, 27, 34]. A recent study in Borrelia burgdorferi, an agent of Lyme disease, showed that Dps enhances the infection levels of the bacterium and aids its survival in nature [35].
The virulence of the neutrophil-activating protein from Helicobacter pylori (HP-NAP) is the best-studied example among Dps-like proteins. H. pylori infects more than 50% of the world’s population, resulting in chronic gastritis, peptic ulcer disease and gastric carcinoma [36, 37]. HP-NAP is a major pro-inflammatory factor of H. pylori and is able to stimulate human neutrophils to produce ROS [38, 39]. Recently, it was demonstrated that HP-NAP promotes leukocyte adhesion to endothelium in vivo [38, 40, 41]. It was also shown that HP-NAP promotes helper-T-cell immune responses and stimulates human neutrophils to produce chemokines [42].
Structural characteristics
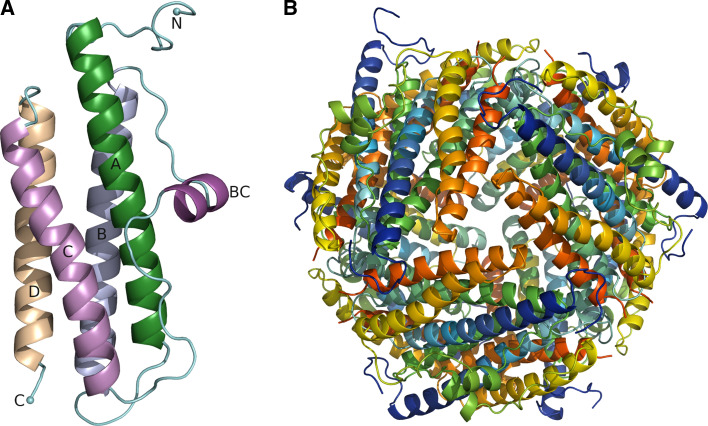
Dps monomers are small proteins with a MW of approximately 20 kDa that fold to a compact four-helix bundle (Fig. 2a). The fold is essentially similar to that of ferritins and bacterioferritins, suggesting a common evolutionary ancestor [43]. However, in contrast to ferritins that form 24-mers with 432-point-group symmetry, Dps monomers pack together to form a native dodecameric structure with 23-point-group symmetry (Fig. 2b). The lack in Dps proteins of a fifth helix (helix E) that is found in ferritins and involved in fourfold symmetry interactions has been suggested to be responsible for the formation of 12-mers rather than 24-mers in Dps proteins [12]. Accordingly, it has been speculated that the 24-mers evolved from a common dodecameric protein, which subsequently became adapted to a variety of functions giving rise to the Dps family. The dodecamer has a spherical architecture with a hollow cavity in the middle that serves as an iron storage compartment able to accommodate up to 500 iron atoms. The external and internal diameter of the dodecamer is 80–90 and 40–50 Å, respectively. The surface as well as the interior of the dodecamer is highly negatively charged. The only exception is HP-NAP, which has a positively charged exterior that may be related to HP-NAP’s function as an adhesin [18]. Recent studies, however, have implicated the C-terminal region of HP-NAP in human neutrophil stimulation and their adhesion to endothelial cells [44].
Fig. 2.
Monomer fold and the quaternary structure of Dps-like proteins. a The monomer of EcDps. b EcDps dodecamer viewed along the threefold axis. Figure was created with PyMol (DeLano Scientific LLC)
Even though the dodecameric structure is very compact, the Dps monomers have a highly flexible N-terminus that usually protrudes out of the dodecamer. This flexibility may be important for the DNA-binding ability for some of the Dps-like proteins. Two Dps proteins from L. lactis have shown an ordered N-terminus that adopts an α-helical (αN) conformation linked to DNA-binding ability [45]. Notably, the N-terminal tail has also been suggested to stabilize the formation of the dodecamer, and deletion of nine of its residues leads to the formation of trimers instead of dodecamers in Mycobacterium smegmatis Dps [46].
Mechanism of iron entry and oxidation
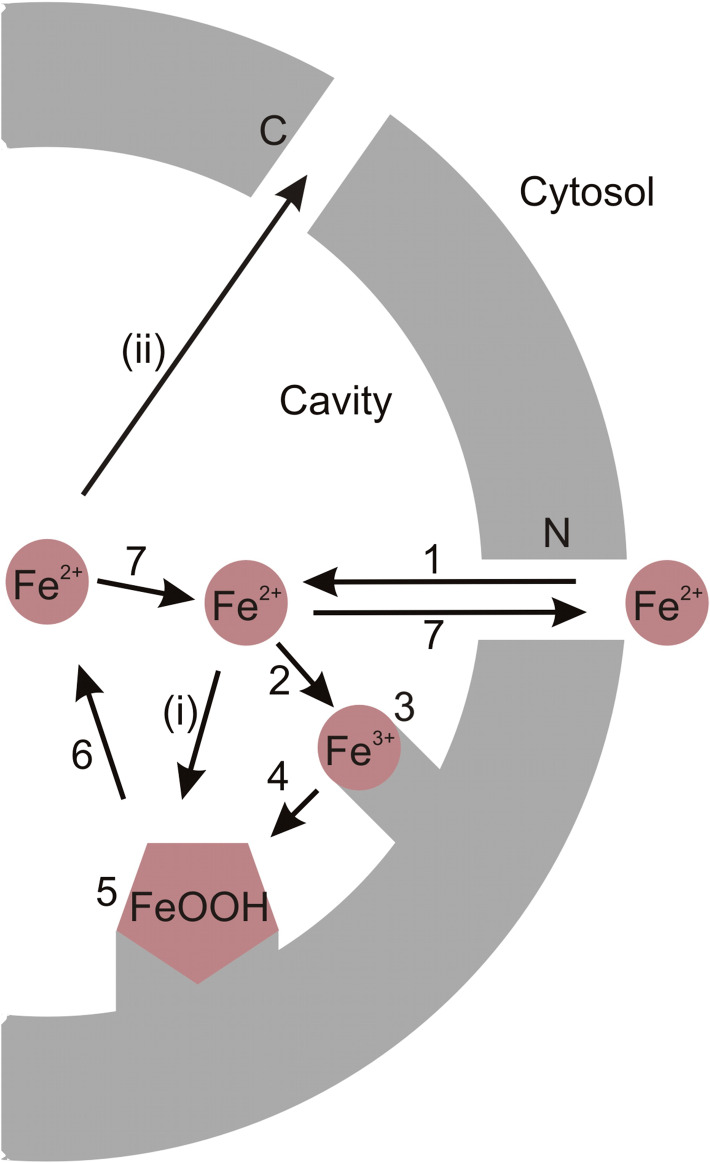
The reaction path for the iron oxidation has been determined for L. innocua Dps and E. coli Dps [47–51]. Briefly, it involves five phases: (1) iron entry inside the dodecamer, (2) binding of iron to the ferroxidase center, (3) oxidation of iron at the ferroxidase center, (4) nucleation and (5) mineralization (Fig. 3).
Fig. 3.
Iron entry, oxidation, mineralization and reutilization. 1 Fe2+ enters the dodecamer via the N-terminal pores. 2 Fe2+ binds to the ferroxidase site and 3 is oxidized to Fe3+. 4 Fe3+ moves to the nucleation site. 5 Mineralization starts with the formation of FeOOH at the nucleation site. 6 Iron is reutilized by reducing FeOOH back to Fe2+ and 7 transported back to cytosol through N-terminal pores in response to cellular needs. i Fe2+ flowing to the cavity might also get oxidized on the surface of the growing mineral. ii Iron transport out of the protein might also occur via the C-terminal pores
The ferroxidase reaction with H2O2 as an oxidant proceeds as follows in E. coli and L. innocua Dps:
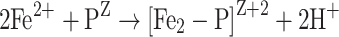 |
1 |
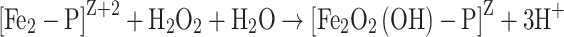 |
2 |
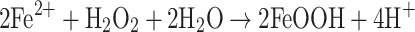 |
3 |
PZ represents the apo-form of the protein and [Fe2-P]Z+2 the dinuclear iron complexed with the ferroxidase center. Iron is oxidized to Fe3+ in the second phase of the reaction path, where [Fe2O2(OH)-P]Z is an oxidized iron complex at the ferroxidase site. In the third phase, the iron mineralizes as hydrous ferric oxide (FeOOH). In contrast to ferritins, core formation is faster than ferroxidase reaction [50, 51].
Also O2 can be utilized in the ferroxidase reaction, but it is not as potent as H2O2. In L. innocua, the ferroxidation reaction with dioxygen as an oxidant follows the following scheme [48]:
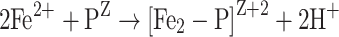 |
4 |
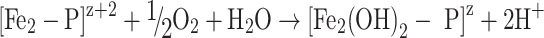 |
5 |
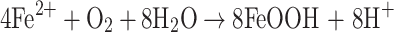 |
6 |
Entry channels
The 23-symmetry of the Dps dodecamer creates four threefold axes that pass through the protein leading to eight trimeric interfaces around the protein. These interfaces are formed at the end of either the C- or N-termini of the monomers creating channels that connect the exterior of the dodecamer to the internal cavity. The pores formed at the N-terminus (Fig. 4a) are usually hydrophilic and have a length of about 10 Å with outer and inner openings of 9–17 and 7–11 Å, respectively, based on Cα–Cα distances (supplementary, Table SI). These negatively charged pores are similar to the iron-uptake pores in ferritins and have been proposed as the main route for iron entry in Dps proteins as well [13, 14]. Moreover, the negative charge of the outer surface of the protein is thought to guide metal ions to the negatively charged pores, from where they are transported to the ferroxidase center through an electrostatic gradient [52]. The same pores have also been found to be utilized as an exit route for iron [52].
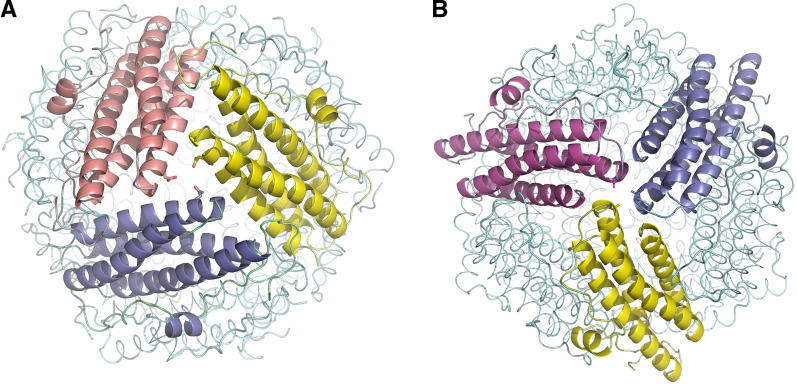
Fig. 4.
The threefold interfaces of Dps-like proteins. Monomers are depicted in different colors. a N-terminal pore at the threefold interface of EcDps. b C-terminal pore at the threefold interface of EcDps. Asp146 and Ala61 from the N- and C-terminal pore, respectively, are shown in stick representation. Figure was created with PyMol
The C-terminal pores are usually highly hydrophobic (Fig. 4b). They show more sequence variability among Dps proteins than the N-terminal pores. They have a length of 7–21 Å with outer and inner openings of approximately 6–14 and 8–10 Å, respectively, measured between Cα–Cα distances, and are generally blocked by hydrophobic residues. Because of their smaller size and hydrophobic nature, the C-terminal pores are usually not considered as the main path for cations. However, they might function as an auxiliary route for cation passage after some structural rearrangements [15, 17].
Crystal structures of Deinococcus radiodurans Dps have provided some evidence for the function of the pores. In the iron-loaded structure, an iron atom was observed in the N-terminal pores. In the native structure, no iron was found in this position, thus supporting the idea that the N-terminal pores are responsible for iron entry. Similarly, the iron-loaded structure showed an iron atom in the C-terminal pore near the cavity. Because of the hydrophobic character of the pore, it was proposed that it could function as a channel for iron release rather than as an auxiliary route for iron entry [15]. An exception to the iron entry via threefold channels was found in the archaeal Halobacterium salinarium Dps where the threefold pores are blocked. Instead, 12 twofold channels appear to be used for iron entry [19].
Ferroxidase site
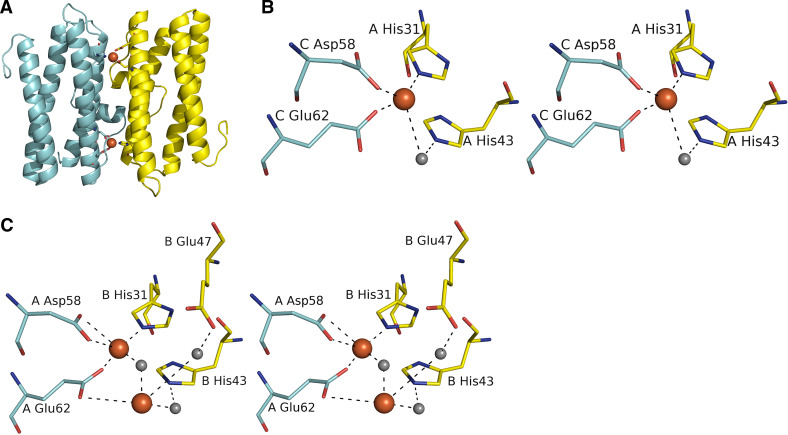
Twelve ferroxidase sites exist in the dodecamer, two between each dimer (Fig. 5a). The binding site is composed of conserved histidine and carboxylate residues. In several structures iron has been found bound in this site. In L. innocua Dps one iron is bound to the ferroxidase center complexed to Glu62 and Asp58 from one monomer and His31 from another (Fig. 5b). In addition, His43 from the same subunit binds to iron via a water molecule. In B. brevis Dps, two irons were found to bind to the active site with a relative distance of 3.3 Å, which is a typical distance of two irons in proteins with di-iron site (Fig. 5c). The first iron was coordinated to Asp58 and Glu62 from one subunit and His31 from another subunit. The second iron was coordinated to Glu62 and Glu47 via a water molecule from one subunit and to His43 from another. Strong evidence also pointed to the presence of an μ-oxo bridge between the two irons, a feature only observed in functional di-iron proteins [19].
Fig. 5.
The conserved ferroxidase site of Dps-like proteins. a The dimer interface of LiDps showing two ferroxidase sites. b The mono-iron ferroxidase site of LiDps. c The di-iron ferroxidase site of BbDps
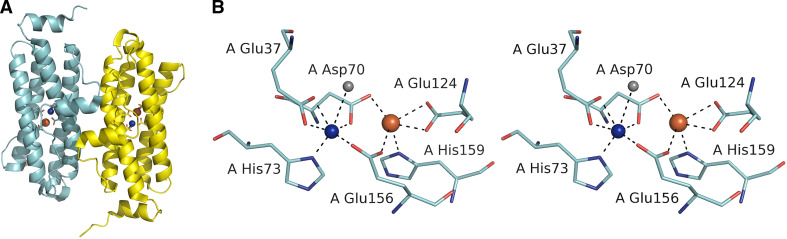
With the exception of L. lactis Dps [45], all bacterial Dps proteins possess a ferroxidase site between two symmetry-related monomers. However, a Dps from the thermoacidophilic archeon Sulfolobus solfataricus revealed a bacterioferritin-like ferroxidase center located inside the four-helix bundle monomer (Fig. 6a). Two metal ions identified in the ferroxidase site were found coordinated by two histidines and four acidic residues, forming a metal-binding motif that resembled that of bacterioferritins (Fig. 6b). The two metals were 3.7 Å apart from each other. Although Sulfolobus solfataricus Dps has greater structural similarity with Dps proteins, the superposition of the active sites shows a close similarity to E. coli bacterioferritin and Lactobacillus plantarum manganese catalase [20].
Fig. 6.
The bacterioferritin-like ferroxidase site of Dps-like protein from S. solfataricus. a The dimer interface of SsDps showing two intersubunit ferroxidase sites. b The bacterioferritin-like intersubunit ferroxidase site of SsDps
A conserved water molecule is modeled instead of iron in the ferroxidase center of some Dps-like proteins. This water has an atypical distance to the iron at the ferroxidase center (about 1 Å longer than expected), which actually corresponds well with a typical 3.1 Å distance of μ-oxo bridged irons. Due to the nature of this second iron as a catalytical intermediate, it is possible that water modeled in some Dps structures is actually a low occupancy iron atom. In accordance with this, in the comparison of low and high iron-stated structures of H. salinarum DpsA, a water molecule in the ferroxidase center of the low-state iron structure was found replaced by an iron atom in the high-state iron structure. The distance between the two irons was 3.2 Å, which also points towards μ-oxo bridged species. Furthermore, a third iron atom was found in the ferroxidase center ligated by two Glu residues in the same monomer. Because two of the iron-binding sites are only transiently occupied, it is possible that they are occupied only by Fe2+, which is rapidly oxidized to Fe3+, and therefore not present in crystals under long exposure to oxygen. The third iron site might also stabilize the ferroxidase site, slowing down iron incorporation and thus offering better bioavailability of iron [19].
Nucleation
The mineralization of oxidized iron to ferrihydrite begins at nucleation sites. Dps nucleation sites share common residues with those found in mammalian ferritin l-chain proteins. In L. innocua Dps, residues Glu44 and Asp47 at the twofold symmetry axis have been proposed to play a role in core nucleation based on structural similarities with the l-chain of horse spleen ferritin [13]. However, these residues are not strictly conserved, and different residues may be employed for core nucleation in other Dps proteins. Indeed, there are several negatively charged residues around the ferroxidase center that might also facilitate nucleation. Once the nucleated Fe3+ mineral has grown to a sufficient size, it is possible that Fe2+ ions flowing into the core also get oxidized at the surface of the growing mineral as observed in ferritins [47].
An alternative nucleation site was proposed for H. salinarium DpsA since Glu44 and Asp47 are replaced by leucine and glycine, respectively. In this structure, three irons were found trapped between four glutamic acids (Glu72 and Glu75 from the dimeric interface). Also, another nucleation site was found in the protein, where iron was coordinated to three symmetry-related Glu154 residues. Because H. salinarium Glu72 and Glu75 are not conserved in Dps proteins, archaeal Dps proteins may have different mechanisms for iron nucleation [19].
Iron core formation
The oxidized iron is deposited in the hollow core as ferrihydrite. The iron cores of ferritins and Dps proteins have been studied with a variety of techniques, including X-ray absorption spectroscopy (XAS), Mössbauer spectroscopy and polarized single-crystal absorption microspectrophotometry. Two different core types have been described: (1) a native core, which is present in the ‘as purified’ protein with no additional iron introduced to the protein, and (2) an in vitro core, which is formed after addition of iron to the protein.
The native iron core of Dps-like proteins contains only tens of iron atoms, with different ratios of phosphate [11, 53]. The core can also contain other metals, such as Zn, Cu, Cr, Mn, Co, Ni and Mo [11]. According to XAS measurements, the native iron core of S. suis Dps-like peroxide resistance protein (Dpr) consists of ~16 iron atoms with a P/Fe ratio of 0.7. Each iron atom is coordinated with six ligands in an octahedral manner. The core consists of small FeO6 iron clusters linked by corner shearing interactions and is the smallest ferritin-type core analyzed so far.
The in vitro loaded iron core of Dps-like proteins is widely characterized [53–55]. Typically, the in vitro loaded core contains up to 500 iron atoms. This core has more irregular structure than the native core and appears to exist as a mixture of octahedrally and tetrahedrally coordinated ferrihydrite [53, 56]. The different sizes of the two cores may contribute to their diverse geometries. The disordered structure of the in vitro loaded iron core makes iron more readily usable than the well-ordered, crystalline state. It has also been proposed that a small amount of iron is continuously being released from and re-deposited to the core, making iron highly bioavailable for various needs [57]. The amount of phosphate in the in vitro loaded cores is higher than in the native cores. It has been reported that phosphate can affect the crystallinity and chemical reactivity of ferritin cores, but the physiological significance of this is still unknown [58]. Further studies on the in vitro loaded iron core could provide insights into the use of Dps to create tailor-made iron cores for various applications [59].
DNA-binding
A number of Dps-like proteins are able to bind DNA. When viewed with electron microscopy, the Dps-DNA complexes appear to form two-dimensional, highly ordered honeycomb-like structures. The complexes are extremely stable and do not dissociate even after treatment with detergents or solvents, or when heated up to 100°C [4]. Because the surfaces of both Dps and DNA are negatively charged, the positively charged residues at the N-terminus have been suggested to bind DNA [6, 12, 45, 46, 60]. In support of that, the deletion of the N-terminus in L. innocua Dps resulted in the lack of DNA-binding [13]. Moreover, in L. lactis Dps, a novel N-terminal helix was found to be crucial for DNA-binding [45].
Further studies suggested that the N-terminal residues are not directly responsible for the DNA-binding, but the interaction is mediated by divalent cations that form multiple ion bridges between Dps and DNA [45, 61]. These ion bridges form only under a particular cation concentration range. Interestingly, below or above this cation concentration, the surfaces of both Dps and DNA become similarly charged, and thus electrostatically repel each other. Even though the N-terminus, the N-terminal Lys residues and divalent cations have all been found important for DNA-binding, the exact binding mechanism is still not fully understood.
The binding of Dps-like proteins to DNA results in the formation of Dps–DNA biocrystals. Based on the crystal structures of Dps proteins and electron microscopy studies, two models of DNA-Dps complexes have been proposed. The first model stemmed from the crystal structure of E. coli Dps and proposed that the DNA helix binds to channels between three adjacent dodecamers in a pseudo-hexagonal lattice. The DNA-binding was suggested to be mediated by three lysines at the disordered N-terminus of the each monomer, so that each channel at the hexagonal lattice would be lined by nine lysine residues, three from each monomer [12].
The second model was derived from electron microscopic studies of E. coli Dps. These studies revealed that the addition of DNA to Dps does not disturb the intra-planar lattice spacing of the crystals. This indicated that DNA is packed within the Dps–DNA crystals and that DNA and Dps are stacked in alternating layers [62]. This model was further supported by the crystal structure of Bacillus brevis Dps [14]. The structure showed that the hexagonal packing of Dps creates grooves that lead to channels. These channels could accommodate DNA in such a way that Dps and DNA would form alternating layers in the crystal structure.
Zinc-binding sites and potential implications
Zinc is an essential trace metal, but becomes toxic in elevated amounts. It usually has a catalytic or structural function in proteins and is often found in DNA-binding proteins. A distinct zinc-binding site consisting of His40 and His44 was identified in S. suis Dpr approximately 8.8 Å from the ferroxidase center [63]. These residues are not conserved in Dps proteins, but the most closely related streptococcal Dpr proteins have either histidine or other residue that can bind zinc at this site. Interestingly, Streptococcus pyogenes Dpr has been suggested to play a role in zinc stress, although the second histidine responsible for zinc binding in S. suis Dpr is replaced by alanine in S. pyogenes Dpr [64]. Ferritins have been suggested to participate in the removal of zinc from the cytoplasm. This might also be the function for the zinc site of S. suis Dpr.
A novel intrasubunit metal binding site was also found in L. lactis Dps in the loop connecting the αN-helix with the rest of the monomer. The site was found to bind zinc, iron and manganese by employing two His residues. It is worth mentioning that L. lactis Dps lacks a conventional ferroxidase center, and this site is probably not related with ferroxidase activity [45].
Deinococcus radiodurans Dps also contains a novel zinc-binding site near the N-terminus. This site is located on the surface of the dodecamer and is solvent-accessible. The zinc is coordinated by two histidines, an aspartate and a glutamate. Interestingly, the coordination geometry of the center represents the coordination of a zinc site in prokaryotic metalloregulatory transcriptional repressors where it functions as a metal sensor. It is also possible that the zinc-binding site stabilizes the N-terminal extension of the protein, thus facilitating DNA-binding [15]. In L. lactis Dps, however, zinc binding was not found to considerably alter the conformation of the N-terminal extension [45].
Biotechnological applications
Several potential applications that could also be exploited with Dps proteins have been described for ferritins in the field of biotechnology and drug design. For instance, ferritins have been used as platforms for antigen presentation and vaccine development [65]. Recently, the use of ferritins in the production of nanoparticle conjugates for cancer immunotherapy was described [66]. Additional applications of ferritins have been reported in the fields of drug delivery and molecular imaging [67].
The potential of iron core-forming proteins, including ferritins, as nanoreactors in the field of nanotechnology has also been put forward [68]. Because Dps-like proteins are capable of forming small metal particles with different metals, they can be used, for instance, in the production of nanoparticles with different compositions, sizes and properties. More recently, the use of Dps proteins to produce iron particles able to catalyze the formation of carbon nanotubes with lower size distribution was reported [69]. The benefit of using protein cages is that the cores can be selectively loaded to control the size of the particle. The protein cage also prevents the aggregation of particles in the preparation and growth of carbon nanotubes. In addition, different nanoparticles with specific magnetic properties have been created using ferritin cores, and similar approaches could be used for Dps-like proteins as well.
Future perspectives
Dps-like proteins have mainly evolved to protect bacteria from ROS. Besides, they have also developed a wide array of other roles in response to a multitude of different stresses and survival mechanisms of bacteria. However, many different aspects of Dps-like proteins still remain unclear. Although the basic mechanisms of iron entry and oxidation are known, many important questions about iron core formation and the mechanism of iron release in response to cellular needs still require elucidation. Furthermore, the exact site and mechanism of DNA-binding are still unknown. Some Dps-like proteins also function as adhesins. Although this topic is not very widely studied, the adhesive function probably plays a significant role in virulence. The recent identification of an interacting partner for E. coli Dps opens new research avenues and calls for similar studies in other family members. Future studies on Dps-like proteins may provide a better understanding of their protective role against various forms of stress and offer new leads to various applications in biotechnology and nanotechnology.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1. Sequence alignment of Dps-family representatives. The secondary structures are presented according to E. coli Dps. The blue triangles indicate the iron-binding residues at the mono-iron ferroxidase center. The blue boxes show residues that share at least 70% similarity in physico-chemical properties. Figure was created with ESPript (http://www.espript.ibcp.fr/). (TIF 3391 kb)
References
- 1.Williams RJ. Free manganese (II) and iron (II) cations can act as intracellular cell controls. FEBS Lett. 1982;140:3–10. doi: 10.1016/0014-5793(82)80508-5. [DOI] [PubMed] [Google Scholar]
- 2.Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/S1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 3.Imlay JA. Cellular defences against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almirón M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli . Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 5.Bozzi M, Mignogna G, Stefanini S, Barra D, Longhi C, Valenti P, Chiancone E. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua . J Biol Chem. 1997;272:3259–3265. doi: 10.1074/jbc.272.6.3259. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Helmann JD. Bacillus subtilis MrgA is a Dps(PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol Microbiol. 1995;18:295–300. doi: 10.1111/j.1365-2958.1995.mmi_18020295.x. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S, Chatterji D. Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. J Biol Chem. 2003;278:5235–5241. doi: 10.1074/jbc.M208825200. [DOI] [PubMed] [Google Scholar]
- 8.Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect Immun. 2004;72:1155–1158. doi: 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papinutto E, Dundon WG, Pitulis N, Battistutta R, Montecucco C, Zanotti G. Structure of two iron-binding proteins from Bacillus anthracis . J Biol Chem. 2002;277:15093–15098. doi: 10.1074/jbc.M112378200. [DOI] [PubMed] [Google Scholar]
- 10.Ramsay B, Wiedenheft B, Allen M, Gauss GH, Lawrence CM, Young M, Douglas T. Dps-like protein from the hyperthermophilic archaeon Pyrococcus furiosus . J Inorg Biochem. 2006;100:1061–1068. doi: 10.1016/j.jinorgbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto Y, Poole LB, Hantgan RR, Kamio Y. An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J Bacteriol. 2002;184:2931–2939. doi: 10.1128/JB.184.11.2931-2939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 13.Ilari A, Stefanini S, Chiancone E, Tsernoglou D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat Struct Biol. 2000;7:38–43. doi: 10.1038/71236. [DOI] [PubMed] [Google Scholar]
- 14.Ren B, Tibbelin G, Kajino T, Asami O, Ladenstein R. The multi-layered structure of Dps with a novel di-nuclear ferroxidase center. J Mol Biol. 2003;329:467–477. doi: 10.1016/S0022-2836(03)00466-2. [DOI] [PubMed] [Google Scholar]
- 15.Romao CV, Mitchell EP, McSweeney S. The crystal structure of Deinococcus radiodurans Dps protein (DR2263) reveals the presence of a novel metal centre in the N terminus. J Biol Inorg Chem. 2006;11:891–902. doi: 10.1007/s00775-006-0142-5. [DOI] [PubMed] [Google Scholar]
- 16.Kauko A, Haataja S, Pulliainen AT, Finne J, Papageorgiou AC. Crystal structure of Streptococcus suis Dps-like peroxide resistance protein Dpr: implications for iron incorporation. J Mol Biol. 2004;338:547–558. doi: 10.1016/j.jmb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Franceschini S, Ceci P, Alaleona F, Chiancone E, Ilari A. Antioxidant Dps protein from the thermophilic cyanobacterium Thermosynechococcus elongatus . FEBS J. 2006;273:4913–4928. doi: 10.1111/j.1742-4658.2006.05490.x. [DOI] [PubMed] [Google Scholar]
- 18.Zanotti G, Papinutto E, Dundon W, Battistutta R, Seveso M, Giudice G, Rappuoli R, Montecucco C. Structure of the neutrophil-activating protein from Helicobacter pylori . J Mol Biol. 2002;323:125–130. doi: 10.1016/S0022-2836(02)00879-3. [DOI] [PubMed] [Google Scholar]
- 19.Zeth K, Offermann S, Essen LO, Oesterhelt D. Iron-oxo clusters biomineralizing on protein surfaces: structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states. Proc Natl Acad Sci USA. 2004;101:13780–13785. doi: 10.1073/pnas.0401821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauss GH, Benas P, Wiedenheft B, Young M, Douglas T, Lawrence CM. Structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like dimetal binding site within a DPS-like dodecameric assembly. Biochemistry. 2006;45:10815–10827. doi: 10.1021/bi060782u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thumiger A, Polenghi A, Papinutto E, Battistutta R, Montecucco C, Zanotti G. Crystal structure of antigen TpF1 from Treponema pallidum . Proteins. 2006;62:827–830. doi: 10.1002/prot.20828. [DOI] [PubMed] [Google Scholar]
- 22.Perrin C, Guimont C, Bracquart P, Gaillard JL. Expression of a new cold shock protein of 21.5 kDa and of the major cold shock protein by Streptococcus thermophilus after cold shock. Curr Microbiol. 1999;39:342–347. doi: 10.1007/s002849900469. [DOI] [PubMed] [Google Scholar]
- 23.Hébraud M, Guzzo J. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol Lett. 2000;190:29–34. doi: 10.1016/S0378-1097(00)00310-4. [DOI] [PubMed] [Google Scholar]
- 24.Nicodeme M, Perrin C, Hols P, Bracquart P, Gaillard JL. Identification of an iron-binding protein of the Dps family expressed by Streptococcus thermophilus . Curr Microbiol. 2004;48:51–56. doi: 10.1007/s00284-003-4116-3. [DOI] [PubMed] [Google Scholar]
- 25.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha ER, Owens GJ, Smith CJ. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis . J Bacteriol. 2000;182:5059–5069. doi: 10.1128/JB.182.18.5059-5069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueshima J, Shoji M, Ratnayake DB, Abe K, Yoshida S, Yamamoto K, Nakayama K. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis . Infect Immun. 2003;71:1170–1178. doi: 10.1128/IAI.71.3.1170-1178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephani K, Weichart D, Hengge R. Dynamic control of Dps protein levels by ClpXP and ClpAP proteases in Escherichia coli . Mol Microbiol. 2003;49:1605–1614. doi: 10.1046/j.1365-2958.2003.03644.x. [DOI] [PubMed] [Google Scholar]
- 29.Chodavarapu S, Gomez R, Vicente M, Kaguni JM. Escherichia coli Dps interacts with DnaA protein to impede initiation: a model of adaptive mutation. Mol Microbiol. 2008;67:1331–1346. doi: 10.1111/j.1365-2958.2008.06127.x. [DOI] [PubMed] [Google Scholar]
- 30.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 31.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus . Infect Immun. 2001;69:3744–3754. doi: 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brenot A, King KY, Caparon MG. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes . Mol Microbiol. 2005;55:221–234. doi: 10.1111/j.1365-2958.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa T, Mizunoe Y, Kawabata S, Takade A, Harada M, Wai SN, Yoshida S. The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni . J Bacteriol. 2003;185:1010–1017. doi: 10.1128/JB.185.3.1010-1017.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen KN, Larsen MH, Gahan CG, Kallipolitis B, Wolf XA, Rea R, Hill C, Ingmer H. The Dps-like protein Fri of Listeria monocytogenes promotes stress tolerance and intracellular multiplication in macrophage-like cells. Microbiology. 2005;151:925–933. doi: 10.1099/mic.0.27552-0. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, Anderson JF, Radolf JD, Fikrig E. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol Microbiol. 2007;63:694–710. doi: 10.1111/j.1365-2958.2006.05550.x. [DOI] [PubMed] [Google Scholar]
- 36.Montemurro P, Barbuti G, Dundon WG, Del Giudice G, Rappuoli R, Colucci M, De Rinaldis P, Montecucco C, Semeraro N, Papini E. Helicobacter pylori neutrophil-activating protein stimulates tissue factor and plasminogen activator inhibitor-2 production by human blood mononuclear cells. J Infect Dis. 2001;183:1055–1062. doi: 10.1086/319280. [DOI] [PubMed] [Google Scholar]
- 37.D’Elios MM, Appelmelk BJ, Amedei A, Bergman MP, Del Prete G. Gastric autoimmunity: the role of Helicobacter pylori and molecular mimicry. Trends Mol Med. 2004;10:316–323. doi: 10.1016/j.molmed.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Evans DJ, Jr, Evans DG, Takemura T, Nakano H, Lampert HC, Graham DY, Granger DN, Kvietys PR. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect Immun. 1995;63:2213–2220. doi: 10.1128/iai.63.6.2213-2220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montemurro P, Nishioka H, Dundon WG, de Bernard M, Del Giudice G, Rappuoli R, Montecucco C. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a potent stimulant of mast cells. Eur J Immunol. 2002;32:671–676. doi: 10.1002/1521-4141(200203)32:3<671::AID-IMMU671>3.3.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 40.Brisslert M, Enarsson K, Lundin S, Karlsson A, Kusters JG, Svennerholm AM, Backert S, Quiding-Jarbrink M. Helicobacter pylori induce neutrophil transendothelial migration: role of the bacterial HP-NAP. FEMS Microbiol Lett. 2005;249:95–103. doi: 10.1016/j.femsle.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Polenghi A, Bossi F, Fischetti F, Durigutto P, Cabrelle A, Tamassia N, Cassatella MA, Montecucco C, Tedesco F, de Bernard M. The neutrophil-activating protein of Helicobacter pylori crosses endothelia to promote neutrophil adhesion in vivo. J Immunol. 2007;178:1312–1320. doi: 10.4049/jimmunol.178.3.1312. [DOI] [PubMed] [Google Scholar]
- 42.Amedei A, Cappon A, Codolo G, Cabrelle A, Polenghi A, Benagiano M, Tasca E, Azzurri A, D’Elios MM, Del Prete G, de Bernard M. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J Clin Invest. 2006;116:1092–1101. doi: 10.1172/JCI27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peña MM, Bullerjahn GS. The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. Linkage of the Dps and bacterioferritin protein families. J Biol Chem. 1995;270:22478–22482. doi: 10.1074/jbc.270.38.22478. [DOI] [PubMed] [Google Scholar]
- 44.Kottakis F, Befani C, Asiminas A, Kontou M, Koliakos G, Choli-Papadopoulou T. The C-terminal region of HPNAP activates neutrophils and promotes their adhesion to endothelial cells. Helicobacter. 2009;14:177–179. doi: 10.1111/j.1523-5378.2009.00678.x. [DOI] [PubMed] [Google Scholar]
- 45.Stillman TJ, Upadhyay M, Norte VA, Sedelnikova SE, Carradus M, Tzokov S, Bullough PA, Shearman CA, Gasson MJ, Williams CH, Artymiuk PJ, Green J. The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding. Mol Microbiol. 2005;57:1101–1112. doi: 10.1111/j.1365-2958.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- 46.Roy S, Saraswathi R, Gupta S, Sekar K, Chatterji D, Vijayan M. Role of N and C-terminal tails in DNA binding and assembly in Dps: structural studies of Mycobacterium smegmatis Dps deletion mutants. J Mol Biol. 2007;370:752–767. doi: 10.1016/j.jmb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Yang X, Chen-Barrett Y, Arosio P, Chasteen ND. Reaction paths of iron oxidation and hydrolysis in horse spleen and recombinant human ferritins. Biochemistry. 1998;37:9743–9750. doi: 10.1021/bi973128a. [DOI] [PubMed] [Google Scholar]
- 48.Yang X, Chiancone E, Stefanini S, Ilari A, Chasteen ND. Iron oxidation and hydrolysis reactions of a novel ferritin from Listeria innocua . Biochem J. 2000;349(3):783–786. doi: 10.1042/bj3490783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Le Brun NE, Thomson AJ, Moore GR, Chasteen ND. The iron oxidation and hydrolysis chemistry of Escherichia coli bacterioferritin. Biochemistry. 2000;39:4915–4923. doi: 10.1021/bi992631f. [DOI] [PubMed] [Google Scholar]
- 50.Zhao G, Ceci P, Ilari A, Giangiacomo L, Laue TM, Chiancone E, Chasteen ND. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli . J Biol Chem. 2002;277:27689–27696. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- 51.Su M, Cavallo S, Stefanini S, Chiancone E, Chasteen ND. The so-called Listeria innocua ferritin is a Dps protein. Iron incorporation, detoxification, and DNA protection properties. Biochemistry. 2005;44:5572–5578. doi: 10.1021/bi0472705. [DOI] [PubMed] [Google Scholar]
- 52.Bellapadrona G, Stefanini S, Zamparelli C, Theil EC, Chiancone E. Iron translocation into and out of Listeria innocua Dps and size distribution of the protein-enclosed nanomineral are modulated by the electrostatic gradient at the 3-fold “ferritin-like” pores. J Biol Chem. 2009;284:19101–19109. doi: 10.1074/jbc.M109.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauko A, Pulliainen AT, Haataja S, Meyer-Klaucke W, Finne J, Papageorgiou AC. Iron incorporation in Streptococcus suis Dps-like peroxide resistance protein Dpr requires mobility in the ferroxidase center and leads to the formation of a ferrihydrite-like core. J Mol Biol. 2006;364:97–109. doi: 10.1016/j.jmb.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 54.Ilari A, Ceci P, Ferrari D, Rossi GL, Chiancone E. Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J Biol Chem. 2002;277:37619–37623. doi: 10.1074/jbc.M206186200. [DOI] [PubMed] [Google Scholar]
- 55.Castruita M, Saito M, Schottel PC, Elmegreen LA, Myneni S, Stiefel EI, Morel FM. Overexpression and characterization of an iron storage and DNA-binding Dps protein from Trichodesmium erythraeum . Appl Environ Microbiol. 2006;72:2918–2924. doi: 10.1128/AEM.72.4.2918-2924.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eggleton RA, Fitzpatrick RW. New data and a revised structural model for ferrihydrite. Clays Clay Miner. 1988;36:111–124. doi: 10.1346/CCMN.1988.0360203. [DOI] [Google Scholar]
- 57.Castruita M, Elmegreen LA, Shaked Y, Stiefel EI, Morel FM. Comparison of the kinetics of iron release from a marine (Trichodesmium erythraeum) Dps protein and mammalian ferritin in the presence and absence of ligands. J Inorg Biochem. 2007;101:1686–1691. doi: 10.1016/j.jinorgbio.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 58.Johnson JL, Cannon M, Watt RK, Frankel RB, Watt GD. Forming the phosphate layer in reconstituted horse spleen ferritin and the role of phosphate in promoting core surface redox reactions. Biochemistry. 1999;38:6706–6713. doi: 10.1021/bi982727u. [DOI] [PubMed] [Google Scholar]
- 59.Flenniken ML, Uchida M, Liepold LO, Kang S, Young MJ, Douglas T. A library of protein cage architectures as nanomaterials. Curr Top Microbiol Immunol. 2009;327:71–93. doi: 10.1007/978-3-540-69379-6_4. [DOI] [PubMed] [Google Scholar]
- 60.Roy S, Gupta S, Das S, Sekar K, Chatterji D, Vijayan M. X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules. J Mol Biol. 2004;339:1103–1113. doi: 10.1016/j.jmb.2004.04.042. [DOI] [PubMed] [Google Scholar]
- 61.Frenkiel-Krispin D, Levin-Zaidman S, Shimoni E, Wolf SG, Wachtel EJ, Arad T, Finkel SE, Kolter R, Minsky A. Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J. 2001;20:1184–1191. doi: 10.1093/emboj/20.5.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A. DNA protection by stress-induced biocrystallization. Nature. 1999;400:83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- 63.Havukainen H, Haataja S, Kauko A, Pulliainen AT, Salminen A, Haikarainen T, Finne J, Papageorgiou AC. Structural basis of the zinc- and terbium-mediated inhibition of ferroxidase activity in Dps ferritin-like proteins. Protein Sci. 2008;17:1513–1521. doi: 10.1110/ps.036236.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsou CC, Chiang-Ni C, Lin YS, Chuang WJ, Lin MT, Liu CC, Wu JJ. An iron-binding protein, Dpr, decreases hydrogen peroxide stress and protects Streptococcus pyogenes against multiple stresses. Infect Immun. 2008;76:4038–4045. doi: 10.1128/IAI.00477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li CQ, Soistman E, Carter DC. Ferritin nanoparticle technology. A new platform for antigen presentation and vaccine development. Ind Biotechnol. 2006;2:143. [Google Scholar]
- 66.Wu H, Wang J, Wang ZM, Fisher DR, Lin YH. Apoferritin-templated yttrium phosphate nanoparticle conjugates for radioimmunotherapy of cancers. J Nanosci Nanotechnol. 2008;8:2316–2322. doi: 10.1166/jnn.2008.177. [DOI] [PubMed] [Google Scholar]
- 67.Uchida M, Terashima M, Cunningham CH, Suzuki Y, Willits DA, Willis AF, Yang PC, Tsao PS, McConnell MV, Young MJ, Douglas T. A human ferritin iron oxide nano-composite magnetic resonance contrast agent. Magn Reson Med. 2008;60:1073–1081. doi: 10.1002/mrm.21761. [DOI] [PubMed] [Google Scholar]
- 68.Galvez N, Sanchez P, Dominguez-Vera JM. Preparation of Cu and CuFe Prussian Blue derivative nanoparticles using the apoferritin cavity as nanoreactor. Dalton. 2005;Trans7(15):2492–2494. doi: 10.1039/b506290j. [DOI] [PubMed] [Google Scholar]
- 69.Kramer RM, Sowards LA, Pender MJ, Stone MO, Naik RR. Constrained iron catalysts for single-walled carbon nanotube growth. Langmuir. 2005;21:8466–8470. doi: 10.1021/la0506729. [DOI] [PubMed] [Google Scholar]
- 70.Thieme D, Grass G (2009) The Dps protein of Escherichia coli is involved in copper homeostasis. Microbiol Res. doi:10.1016/j.micres.2008.12.003 [DOI] [PubMed]
- 71.Jeong K, Hung K, Baumler D, Byrd J, Kaspar C. Acid stress damage of DNA is prevented by Dps binding in Escherichia coli O157:H7. BMC Microbiol. 2008;8:181. doi: 10.1186/1471-2180-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tonello F, Dundon WG, Satin B, Molinari M, Tognon G, Grandi G, Del Giudice G, Rappuoli R, Montecucco C. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol Microbiol. 1999;34:238–246. doi: 10.1046/j.1365-2958.1999.01584.x. [DOI] [PubMed] [Google Scholar]
- 73.Ceci P, Mangiarotti L, Rivetti C, Chiancone E. The neutrophil-activating Dps protein of Helicobacter pylori, HP-NAP, adopts a mechanism different from Escherichia coli Dps to bind and condense DNA. Nucleic Acids Res. 2007;35:2247–2256. doi: 10.1093/nar/gkm077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grove A, Wilkinson S. Differential DNA binding and protection by dimeric and dodecameric forms of the ferritin homolog Dps from Deinococcus radiodurans . J Mol Biol. 2005;347:495–508. doi: 10.1016/j.jmb.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 75.Wiedenheft B, Mosolf J, Willits D, Yeager M, Dryden KA, Young M, Douglas T. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus . Proc Natl Acad Sci USA. 2005;102:10551–10556. doi: 10.1073/pnas.0501497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ceci P, Ilari A, Falvo E, Chiancone E. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: X-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J Biol Chem. 2003;278:20319–20326. doi: 10.1074/jbc.M302114200. [DOI] [PubMed] [Google Scholar]
- 77.Pulliainen AT, Haataja S, Kahkonen S, Finne J. Molecular basis of H2O2 resistance mediated by Streptococcal Dpr. Demonstration of the functional involvement of the putative ferroxidase center by site-directed mutagenesis in Streptococcus suis . J Biol Chem. 2003;278:7996–8005. doi: 10.1074/jbc.M210174200. [DOI] [PubMed] [Google Scholar]
- 78.Page RD (2002) Visualizing phylogenetic trees using TreeView. Curr Protoc Bioinformatics, chap 6, unit 6 2 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence alignment of Dps-family representatives. The secondary structures are presented according to E. coli Dps. The blue triangles indicate the iron-binding residues at the mono-iron ferroxidase center. The blue boxes show residues that share at least 70% similarity in physico-chemical properties. Figure was created with ESPript (http://www.espript.ibcp.fr/). (TIF 3391 kb)