Abstract
Hydrogen sulfide (H2S) plays an important role in inflammation. We showed that macrophages expressed the H2S-forming enzyme cystathionine gamma-lyase (CSE) and produced H2S. Lipopolysaccharide (LPS) stimulated the CSE expression and H2S production rate. l-cysteine reduced LPS-induced nitric oxide (NO) production. CSE inhibitor blocked the inhibitory effect of l-cysteine. CSE knockdown increased, whereas CSE overexpression decreased LPS-induced NO production. Dexamethasone suppressed LPS-induced CSE expression and the H2S production rate as well as NO production. l-arginine increased, whereas NG-nitro-l-arginine methyl ester (l-NAME) decreased LPS-induced CSE expression and H2S production. Dexamethasone plus l-NAME significantly decreased LPS-induced CSE expression and H2S production compared to l-NAME. Our results suggest that macrophages are one of the H2S producing sources. H2S might exert anti-inflammatory effects by inhibiting NO production. Dexamethasone may directly inhibit CSE expression and H2S production, besides the NO-dependent way. Inhibition of H2S and NO production may be a mechanism by which glucocorticoids coordinate the balance between pro- and anti-inflammatory mediators during inflammation.
Keywords: Glucocorticoids, Hydrogen sulfide, Cystathionine γ-lyase, Lipopolysaccharide, Macrophages
Introduction
Hydrogen sulfide (H2S) has recently been suggested to be “the third endogenous gaseous signaling transmitter” in addition to nitric oxide (NO) and carbon monoxide (CO) in mammalian tissues [1, 2]. H2S is naturally synthesized in the body from l-cysteine mainly by the activity of two enzymes, cystathionine γ-lyase (CSE, EC 4.4.1.1) and cystathionine β-synthetase (CBS, EC 4.2.1.22) [3]. In animal models, physiological levels of plasma H2S concentration have been reported to vary from 30 to 300 μM [4–6], and in vivo basal concentrations of H2S are markedly augmented under certain pathological conditions, such as endotoxemia and septic shock [6, 7].
H2S has been indicated to play critical roles in inflammatory processes. It seems that H2S exhibits either pro- or anti-inflammatory effects. It has been shown that exogenous H2S provokes an inflammatory reaction in lung in normal mice [6] and induces the generation of pro-inflammatory cytokine in cultured human monocytes [8]. Inhibition of H2S biosynthesis displays distinct anti-inflammatory activity as evidenced by the attenuation of the organ injury in cases including septic shock, endotoxemia and pancreatitis [6, 9, 10]. On the other hand, H2S has also been reported to play an anti-inflammatory role during inflammation. H2S can scavenge peroxynitrite [11], inhibit leukocyte adherence and infiltration [12, 13], suppress edema formation [13], inhibit formyl-methionyl-leucyl-phenylalanine (fMLP)-induced chemotaxis and degranulation of polymorphonuclear leukocytes [14], induce neutrophil apoptosis [15] and suppress LPS-induced tumor necrosis factor-α (TNF-α) production in microglial cells [16].
Glucocorticoids (GCs) are a class of steroid hormones with pleiotropic effects through interaction with glucocorticoid receptor (GR) in a broad range of cell types. It is well known that GCs have anti-inflammatory effects. They inhibit the expression and production of numerous inflammatory mediators, such as pro-inflammatory cytokines, prostaglandins and reactive oxygen and nitrogen species in various cell types [17]. GCs inhibit leukocyte migration to sites of inflammation and promote lymphocyte apoptosis [18, 19]. However, the role of GCs in the regulation of inflammation is not fully understood. GCs have also been shown to exert pro-inflammatory effects in some cases [20, 21]. It was reported that GCs enhanced migration of leukocytes from the bloodstream to wound sites [22], and low physiological doses of cortisol stimulated the pro-inflammatory cytokine migration inhibitory factor (MIF) production in macrophages [23]. Some studies also showed that GCs could inhibit apoptosis of human neutrophils, thereby contributing to inflammatory processes [24–26].
Macrophages are a heterogeneous population of mononuclear phagocytes found ubiquitously in the body. These cells play a crucial role in innate and adaptive immunity in response to microorganisms and are major contributors to the inflammatory response through generation of numerous pro-inflammatory mediators [27, 28]. Recently, it has been shown that CSE mRNA is induced by lipopolysaccharide (LPS) in cells in the macrophage cell line RAW264.7 [29]. However, the capacity of H2S production in primary macrophages, the possible functions of endogenous H2S and the consequent changes of H2S production in response to inflammatory stimuli remain to be clarified. Moreover, macrophages are the target cells of GCs. GCs have been shown to inhibit the release of pro-inflammatory cytokines including TNF-α, interleukin-1 (IL-1) and interleukin-6 (IL-6) [17], but promote phagocytosis of apoptotic cells by macrophages [30]. In addition, GCs have also been shown to induce changed expression of many pro- and anti-inflammatory genes in human monocytes [31, 32]. Recently, Li et al. [33] reported that dexamethasone decreased the expression of CSE in isolated rat neutrophils. It is of interest to explore the regulation of H2S production by GCs in macrophages.
In this study, we firstly elucidated the capacity of H2S biosynthesis in macrophages by looking at the expression of H2S synthetic enzymes in response to LPS and consequent H2S production. Then, we explored the possible local functions of endogenous H2S by studying the effects of endogenous H2S on NO production. Finally, we investigated the effects of dexamethasone, a potent synthetic glucocorticoid, on the expression of H2S synthetic enzymes as well as H2S biosynthesis in LPS-treated macrophages.
Materials and methods
Materials
Purified LPS (Escherichia coli, O111:B4), 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT), sodium orthovanadate, pyridoxal-5′-phosphate, dl-propargylglycine (PAG),dexamethasone, RU38,486, l-cysteine, l-arginine and N G-nitro-l arginine methyl ester (l-NAME) were obtained from Sigma-Aldrich (St. Louis, MO). TRIzol reagent and superscript reverse transcriptase were purchased from Invitrogen (Grand Island, NY). Antibody to CSE was purchased from Abnova (Taipei, Taiwan). Antibodies to CBS, inducible nitric oxide synthase (iNOS), secondary horseradish peroxidase-conjugated antibody and the enhanced chemiluminescence Western blotting detection system were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture
Peritoneal macrophages were obtained from male C57BL/6 mice, which were purchased from Shanghai SLAC Laboratory Animal Co. (Shanghai, China) and were 8–10 weeks of age when used in the study. The mice were housed in a temperature- and humidity-controlled environment and received standard mouse chow and water ad libitum in compliance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication no. 85-23, revised 1996). Also, all experimental procedures were approved by the Ethics Committee of Experimental Animals of the Second Military Medical University, China. Resident peritoneal exudate cells were washed from the peritoneal cavity with 2 ml ice-cold DMEM medium (Life Technology, Grand Island, NY) as described previously [34]. After centrifugation at 110×g for 10 min, the exudate cells were resuspended in DMEM medium and then plated in six-well tissue culture plates (Corning Glass, Corning, NY) at 1 × 106 cells per well in a final volume of 2 ml. Cells were incubated at 37°C for 2 h to adhere to plastic, and nonadherent cells were rinsed off by vigorously washing with DMEM medium containing 10% FBS. Viability of the adherent cells was >95% as assessed by trypan blue exclusion. Purity was determined by FACScan (Becton–Dickinson, San Jose, CA) with specific monoclonal antibody F4/80 (PharMingen, San Diego, CA), and about 95% of the adherent macrophages were found to express F4/80 marker.
Murine macrophage cell line RAW264.7 was originally obtained from American Type Culture Collection (Rockville, MD) and kindly provided by Shanghai Institute for Biological Sciences. Cells were cultured in DMEM medium containing 10% FBS at 37°C in 5% CO2, 95% air; RAW264.7 cells were also seeded on six-well plates at a density of 1 × 106 cells/well. Both primary and RAW264 macrophages were cultured in DMEM medium containing 10% charcoal-stripped FBS (CD-FBS). Peritoneal macrophages and RAW264.7 cells were then exposed to LPS (10–1,000 ng/ml), dexamethasone (1–1,000 nmol/l), l-cysteine (1 mmol/l), PAG (1 mmol/l), l-arginine (0.1–1 mmol/l), l-NAME (0.25–1 mmol/l) or RU38,486 (1 μmol/l) for the indicated time period. Dexamethasone was first dissolved in absolute ethanol and diluted by the culture media to achieve a final ethanol concentration of less than 0.01%. Control media contained the same final solvent concentrations (0.01%). Each treatment was performed in triplicate for each preparation of cells.
Total RNA extraction, RT-PCR and quantitative real-time RT-PCR
Total RNA of cells was isolated by using TRIzol reagent according to the manufacturer’s instructions; 2 μg RNA was reverse transcribed using superscript reverse transcriptase (Invitrogen) and stored at −20°C. The following sense and antisense primers were used: CSE (accession number NM_145953): 5′-GAGCCTGAGCAATGGAAT-3′ and 5′-GATGGGTAATCGTAATGGTG-3′. CBS (accession number NM_178224): 5′-GAACCAGACGGAGCAAACAG-3′ and 5′-CTTGAACACGCAGACGCCAC-3′.
PCR reaction solution consisted of 2.0 μl diluted RT-PCR product, 0.2 μM of each paired primer, 2.5 mM Mg2+, 250 μM deoxynucleotide triphosphates, 2 U Taq DNA polymerase (Promega, Madison, WI) and 1 × PCR buffer. The PCR reaction was set at 94°C (45 s), 58°C (45 s) and 72°C (1 min) in a total of 40 cycles with a final extension step at 72°C for 10 min. Ten microliters of the reaction mixture were subsequently electrophoresed on a 1.5% agarose gel and visualized by ethidium bromide using a 100-bp DNA ladder (Life Technology) to estimate the band sizes. As a negative control for all of the reactions, distilled water was used in place of cDNA. The identity of the PCR products was confirmed by sequencing. Sequence data were analyzed using Blast nucleic acid database searches from the National Centre for Biotechnology Information (NCBI).
Quantitative real-time PCR analysis was carried out in duplicates using Rotor Gene 3000 (Corbett Research, Sydney, Australia). The real-time PCR solution consisted of 40 ng diluted cDNA product, 0.1–0.3 μM of each paired primer, 2.5 mM Mg2+, 100 μM deoxynucleotide triphosphates, 2 U Taq DNA polymerase and 1 × PCR buffer. SYBR green (BMA, Rockland, ME) was used as detection dye. Quantitative real-time PCR conditions were optimized according to preliminary experiments to achieve linear relationships between the initial RNA concentration and PCR product. The annealing temperature was set at 58°C, and amplification cycles were set at 40 cycles. The temperature range to detect the melting temperature of the PCR product was set from 60–95°C. Amplification of the housekeeping genes β-actin and GAPDH were measured for each sample as an internal control for sample loading and normalization. The melting curve was examined at the end of the amplification to ensure the specificity of PCR products. To determine the relative quantitation of gene expression for both target and housekeeping genes, the comparative Ct (threshold cycle) method with arithmetic formulae (2−ΔΔCt) was used [35]. Because very similar data were obtained by using either β-actin or GAPDH as an internal control, GAPDH was used for calculation of ΔCt in the presentation of results.
Western blot analysis
Cells were scraped off the plate in the presence of lysis buffer consisting of 60 mmol/l Tris-HCl, 2% sodium dodecyl sulfate (SDS), 10% sucrose, 2 mmol/l phenylmethylsulfonyl fluoride (Merck, Darmstadt, Germany), 1 mmol/l sodium orthovanadate and 10 μg/ml aprotinin (Bayer, Leverkusen, Germany). The cell lysates were quickly sonicated and centrifuged at 12,000×g for 5 min at 4°C. The supernatant was collected, and the protein concentration was assayed using a modified Bradford assay. The samples were diluted in sample buffer (250 mmol/l Tris-HCl (pH 6.8), containing 4% SDS, 10% glycerol, 2% β-mercaptoethanol and 0.002% bromophenol blue, and boiled for a further 5 min. Aliquots of proteins were separated by SDS-PAGE (10%) and subsequently transferred to nitrocellulose membranes by electroblotting. The membrane was blocked in 5% skim milk powder in 0.1% Tris-buffered saline/Tween 20 (TBST) at room temperature for 2 h, and then was incubated with antibody raised against iNOS or CBS at a dilution of 1:1,000, and CSE at a dilution of 1:2,000 overnight at 4°C. After three washes with TBST, the membrane was incubated with a secondary horseradish peroxidase-conjugated antibody for 1 h at room temperature. Immunoreactive proteins were visualized using the enhanced chemiluminescence Western blotting detection system. The light-emitting bands were detected with X-ray film. The resulting band intensities were quantified using an image scanning densitometer (Furi Technology, Shanghai, China). To control sampling errors, the ratio of band intensities for iNOS or CSE to β-actin was obtained to quantify the relative protein expression level.
Real-time H2S production measurement
To define the real-time kinetics of H2S production by macrophages, we used a miniaturized H2S micro-respiration sensor (Model H2S-MRCh, Unisense, Aarhus, Denmark) coupled to a Unisense PA2000 amplifier. The 100-μm diameter H2S microsensor is based on the original design of the amperometric microsensor used to measure H2S with high spatial resolution as described before [36, 37]. Since the H2S microsensor was sensitive to pH, temperature and electrical interferences [37, 38], the real-time H2S production measurements were performed in a temperature-controlled micro-respiration chamber (Unisense) containing 1 ml of stirred (160 rpm) culture media (pH 7.2) at 37°C inside a well-grounded Faraday cage. Macrophages were collected and washed in fetal bovine serum-free culture media with 20 mM Hepes substituted for sodium bicarbonate to prevent CO2 bubble formation in the closed respirometer chamber. The final cell pellet was resuspended at a concentration of 1 × 107 cells/ml, and 0.1 ml cell suspension was injected into the respirometer chamber. In addition, to avoid spontaneous H2S oxidation [39], nitrogen was used to deoxygenate the culture media in the respiratory chamber before the addition of macrophages. After the sensor signals stabilized, l-cysteine (1 mmol/l, the substrate of CSE) and pyridoxal-5′-phosphate (PLP, 1 mmol/l, a cofactor of CSE) were added to stimulate H2S production. H2S production rates were determined after the addition of substrate and cofactor at the initial steepest slopes of each trace [37]. Cell viability was assessed by trypan blue exclusion and remained at more than 90% throughout the experiments. The H2S sensor was calibrated after each experiment with freshly prepared anoxic sodium sulfide stock solution (0–100 μmol/l) according to the manufacturer’s manual, using the same buffer and conditions as the experiment.
NO production assay
NO production in supernatants was assessed by measuring nitrite/nitrate, the stable degradation product of NO as described previously [40]. In brief, Griess reagent was prepared by mixing equal volumes of 1% sulfanilamide in 5% phosphoric acid and 0.1% naphtylethylenediamide just before use; 100 μl of Griess reagent was mixed with equal amounts of cell supernatants. After incubation at 37°C for 10 min, the OD value was measured using a Bio-Rad (Hercules, CA) microplate reader at 550 nm. Concentration of nitrite was assessed by reference to the sodium nitrite standard curve.
Establishment of stable CSE-overexpression and CSE-knockdown cell lines
The mouse CSE full-length cDNA was synthesized and cloned (Jinsite Biotechnology Corp., Nanjing, China) into pEGFP-N3 vector (BD Biosciences Clontech, Palo Alto, CA), generating a C-terminal EGFP-tagged fusion protein. The mouse CSE small interfering RNA (siRNA) plasmid was constructed using the pRNAT-U6.1/Neo vector (Jinsite Biotechnology) for knockdown of the CSE gene in vitro. The target sequence of mouse CSE-specific siRNA used in the present study was 5′-TACATGAATGGCCACAGCGAT-3′. Control-siRNA plasmid was supplied by Jinsite Biotechnology, and it expressed a hairpin siRNA with no homology to any known mouse or rat mRNA sequences in the NCBI RefSeq database.
RAW264.7 cells were separately transfected with pEGFP-N3, pEGFP-mCSE, pRNAT-U6.1/Neo-mCSE and pRNAT-U6.1/Neo-Control vector using SofastTM cationic polymer transfection reagent (Sunma Biotech, Xiamen, China) according to the manufacturer’s manual. Forty-eight hours after transfection, stable transfected cells were selected using 800 μg/ml G418 in growth medium for 2 weeks. Surviving clones were then incubated in medium containing 400 μg/ml G418. The stable CSE-overexpression and CSE-knockdown RAW264.7 cell lines were named “RAW-EGFP-mCSE” (“RAW-EGFP” as control) and “RAW-mCSE siRNA” (“RAW-control siRNA” as control), respectively.
Determination of CSE promoter activity
PGL3-luciferase reporter plasmids (Promega Corp, Madison, WI) were used for transient transfections. The pCSE-PGL3 plasmid containing 3.5-kb CSE promoter (−3,498 ~ +18) was a kind gift from Dr. I.Ishii (Gunma University Graduate School of Medicine, Gunma, Japan) [41]. Transient transfections were performed using SofastTM (Sunma Biotech) Cationic Polymer Transfection Reagent according to the manufacturer’s manual. Briefly, 1 day before transfection, RAW264.7 cells were seeded and fed with DMEM containing 10% CD-FBS in 48-well plates. Each well was then transfected with 0.1 μg DNA and 10 ng control DNA (pRL-TK-Renilla-luciferase vector, Promega). Ten hours later, culture media were changed, and cells were treated with various reagents as indicated. Dexamethasone and RU38,486 were added to the treatment media as stock solutions in absolute ethanol. Control media contained the same concentration of vehicle (0.01% vol/vol). Luciferase assays were carried out 12 h later using the dual luciferase assay kit (Promega). Relative luciferase activity is presented as firefly luciferase values normalized to Renilla luciferase activity.
MTT assay
Cell viability was assessed by MTT assay. The assay depends on the reduction of the tetrazolium salt MTT by functional mitochondria to formazan [42]. Cells were seeded, cultured and treated as described above. MTT was added at the last 2 h of treatment. After a 2-h incubation of the cells with MTT at 37°C, cells were lysed with dimethyl sulphoxide and the formazan crystals solubilized. Absorbance was read at 550 nm using a spectrophotometric microplate reader (Bio-Rad).
Statistical analysis
Data were expressed as means ± SEM. Statistical significance was estimated by one-way analysis of variance (ANOVA) followed by the Student-Newman-Keuls test. A P value <0.05 was considered significant.
Results
Expression of H2S synthetic enzymes and H2S synthetic activity in macrophages
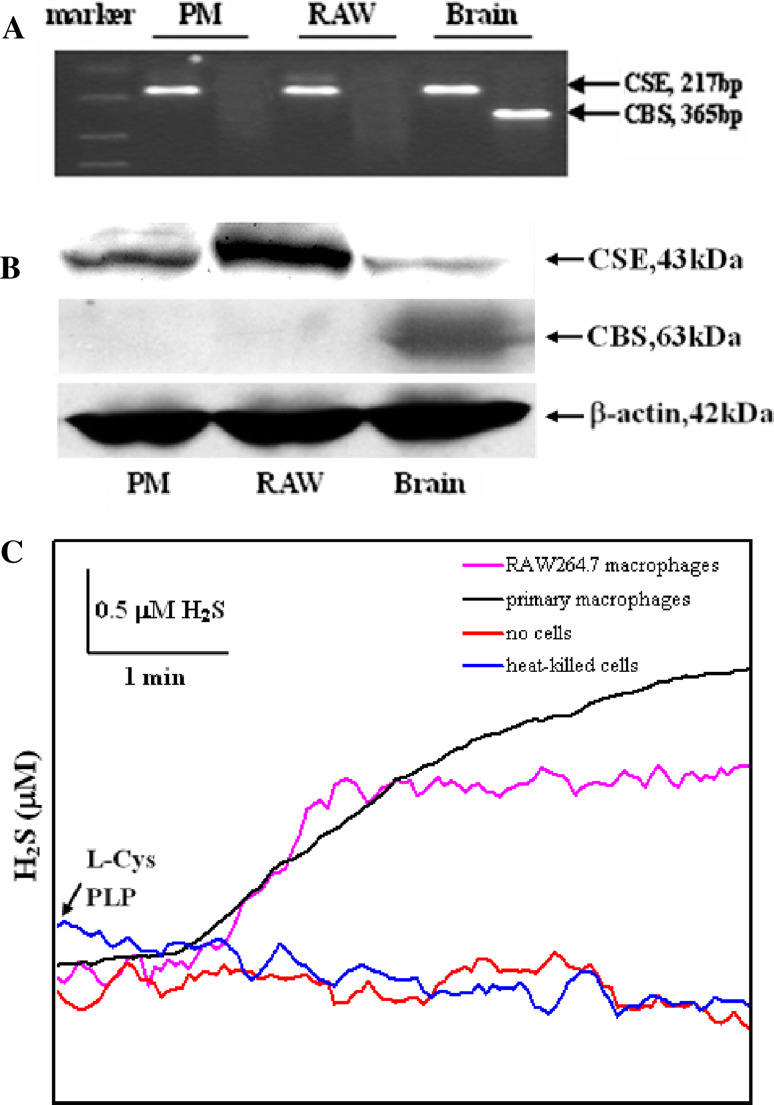
RT-PCR analysis showed that CSE but not CBS mRNA was detected in both primary and RAW 264.7 macrophages (Fig. 1a). Western blot analysis showed that CSE but not CBS protein was detected in both primary and RAW 264.7 macrophages. We detected a single protein band of approximately 43 kDa corresponding to CSE protein in primary macrophages and RAW 264.7 cells (Fig. 1b).
Fig. 1.
Expression of H2S synthetic enzymes and H2S synthetic activity in macrophages. a PCR analysis of CSE and CBS mRNA in macrophages and brain. b Western blot analysis showing protein expression of CSE and CBS in macrophages and brain. c Representative traces of H2S production in suspensions of primary and RAW 264.7 macrophages. H2S production was initiated by the addition of l-cysteine and PLP. There were two control traces of buffered solutions without cells or with heat-killed cells containing l-cysteine and PLP, indicating no spontaneous H2S production. PM primary peritoneal macrophages, RAW RAW 264.7 macrophages, l -Cys l-cysteine
The ability of primary macrophages and RAW 264.7 cells to synthesize H2S from added l-cysteine is demonstrated in Fig. 1c. The initial real-time H2S production rate observed in primary and RAW 264.7 macrophage suspensions after addition of l-cysteine and PLP was 0.04653 ± 0.01023 and 0.04882 ± 0.0998 nmol s−1 mg protein−1 (n = 4), respectively.
LPS enhances CSE expression and H2S synthetic activity in macrophages
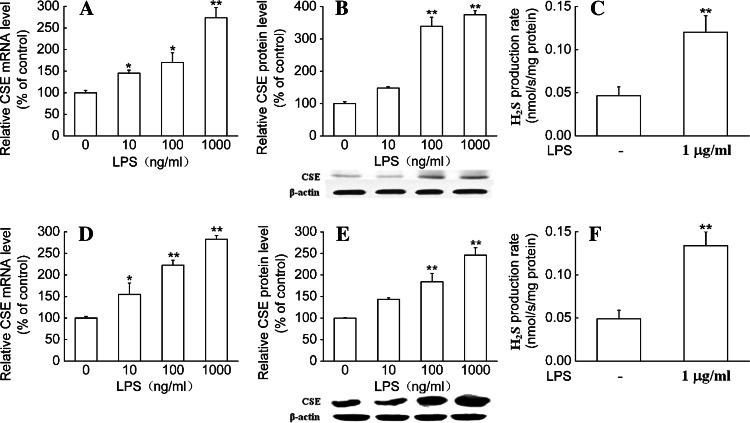
Treatment of primary macrophages with increasing concentrations of LPS (10 ng/ml–1 μg/ml) for 6 h or 24 h resulted in increases in CSE mRNA or protein expression in a dose-dependent manner, respectively. Maximal effect was obtained at a concentration of 1 μg/ml, which caused about 2.5-fold increases in CSE mRNA and protein levels. In the meantime, the initial real-time H2S production rate was also significantly increased in LPS-treated macrophages compared to control (Fig. 2a–c).
Fig. 2.
LPS increased CSE expression and H2S synthetic activity in macrophages. Primary (a–c) and RAW 264.7 (d–f) macrophages were stimulated with LPS at the indicated doses for 6 h (a, d) or 24 h (b, c, e, f), respectively. Quantitative real-time RT-PCR, Western blot analysis and real-time H2S production measurement were used to determine CSE mRNA expression, protein expression and H2S synthetic activity in macrophages. Results of Western blot were quantified by scanning densitometry of blots. H2S production rates were calculated as described in “Materials and Methods.” Data were expressed as mean percentage of control ± SEM for CSE mRNA and protein expression (n = 3), and for H2S production rates (n = 4). *P < 0.05; **P < 0.01 compared with vehicle control
In RAW 264.7 cells, LPS also dose-dependently increased CSE mRNA and protein expression. Maximal effect of LPS was achieved at a concentration of 1 μg/ml. LPS also significantly promoted the initial real-time H2S production rate in RAW264.7 macrophages (Fig. 2d–f).
Endogenous H2S inhibits NO production in LPS-stimulated macrophages
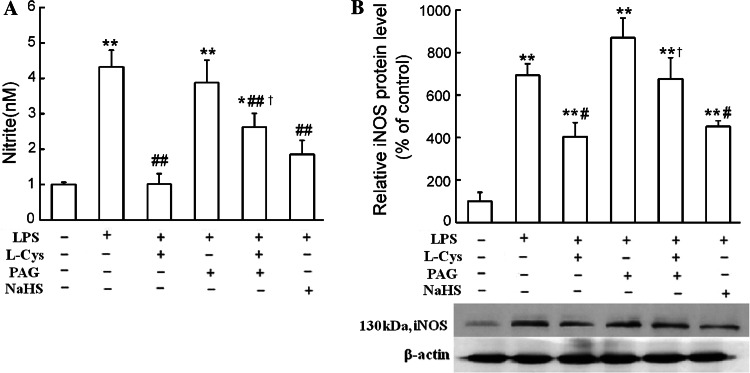
The study of Oh et al. [29] suggested that, in RAW 264.7 cells, one of the attractive functions of H2S was inhibition of NO production. In order to investigate if endogenous H2S has such a function in primary macrophages, we firstly observed the effects of l-cysteine, the precursor of H2S, and PAG, the CSE inhibitor, on NO production in cultured peritoneal macrophages in the presence of LPS. As shown in Fig. 3, treatment with l-cysteine (1 mmol/l) significantly reduced the expression of inducible NO synthase (iNOS), the predominant NOS in macrophages, as well as NO production in a 24-h treatment period. PAG (1 mmol/l) treatment significantly blocked the l-cysteine-induced inhibition of NO production and iNOS expression in LPS-treated macrophages. Treatment of cells with an H2S donor, NaHS (200 μmol/l), for 24 h resulted in significant decreases in iNOS expression and NO production in the presence of LPS.
Fig. 3.
H2S inhibited NO production and iNOS expression in LPS-stimulated macrophages. Primary macrophages were treated with 1 mmol/l l-cysteine, 1 mmol/l PAG, or 200 μmol/l NaHS for 24 h in the absence or presence of LPS (1 μg/ml). NO production (a) and iNOS protein expression (b) were determined as described in “Materials and Methods.” Data were expressed as mean ± SEM for NO production (n = 4) and mean percentage of control ± SEM for iNOS protein expression (n = 3). *P < 0.05; **P < 0.01 compared with vehicle control; #P < 0.05; ##P < 0.01 compared with LPS; †P < 0.05 compared with LPS plus l-Cys. l -Cys l-cysteine
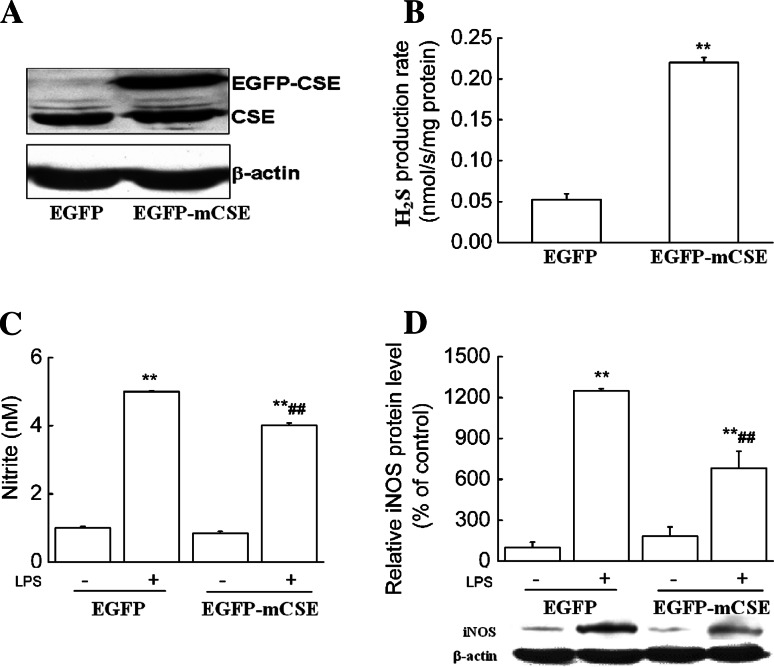
Next, by using genetic approaches including both siRNA and overexpression of CSE, we further confirmed whether endogenous H2S-mediated inhibition of NO production was specifically dependent on CSE. The CSE siRNA and CSE gene were stably transfected into RAW 264.7 macrophages, thus CSE-knockdown (RAW-mCSE siRNA) and CSE-overexpression (RAW-EGFP-mCSE) RAW264.7 cell lines were established, respectively. Cells were then stimulated with LPS (1 μg/ml) for 24 h. As shown Fig. 4, CSE siRNA resulted in approximately 40% decreases in the H2S production rate and significantly increased LPS-induced NO production and iNOS expression compared to control siRNA. In contrast, Fig. 5 demonstrated that CSE overexpression resulted in a more than four-fold increase in the H2S production rate and significantly decreased LPS-induced NO production and iNOS expression compared to the control.
Fig. 4.
CSE siRNA increased LPS-induced NO production and iNOS expression. RAW264.7 macrophages were separately transfected with pRNAT-U6.1/Neo-mCSE and pRNAT-U6.1/Neo-Control vectors. Stable CSE-knockdown RAW264.7 cells were selected and maintained in medium containing G418 and named “RAW-mCSE siRNA” (“RAW-control siRNA” as control). Western blot analysis (a) and real-time H2S production measurement (b) were used to determine CSE protein expression and H2S synthetic activity in CSE-knockdown RAW264.7 cells. After stimulation with LPS (1 μg/ml) for 24 h, NO production (c) and iNOS protein expression (d) were determined as described in “Materials and Methods.” Data were expressed as mean ± SEM for H2S production rates (n = 3) and NO production (n = 4), and mean percentage of control ± SEM for iNOS protein expression (n = 3). *P < 0.05; **P < 0.01 compared with “RAW-control siRNA”; ##P < 0.01 compared with “RAW-control siRNA” stimulated with LPS. Control siRNA: RAW-control siRNA; mCSE siRNA: RAW-mCSE siRNA
Fig. 5.
CSE overexpression decreased LPS-induced NO production and iNOS expression. RAW264.7 macrophages were separately transfected with pEGFP-N3 and pEGFP-mCSE vectors. Stable CSE-overexpression RAW264.7 cells were selected and maintained in medium containing G418 and named “RAW-EGFP-mCSE” (“RAW-EGFP” as control). Western blot analysis (a) and real-time H2S production measurement (b) were used to determine CSE protein expression and H2S synthetic activity in CSE-overexpression RAW264.7 cells. After stimulation with LPS (1 μg/ml) for 24 h, NO production (c) and iNOS protein expression (d) were determined as described in “Materials and Methods.” Data were expressed as mean ± SEM for H2S production rates (n = 3) and NO production (n = 4), and mean percentage of control ± SEM for iNOS protein expression (n = 3). **P < 0.01 compared with “RAW-EGFP”; ##P < 0.01 compared with “RAW-EGFP” stimulated with LPS. EGFP: RAW-EGFP; EGFP-mCSE: RAW-EGFP-mCSE
Dexamethasone suppresses H2S production in LPS-treated macrophages
To determine whether glucocorticoid modulates H2S production in macrophages, the effects of a potent synthetic glucocorticoid, dexamethasone, on the CSE expression and H2S production rate in primary and RAW 264.7 macrophages were examined.
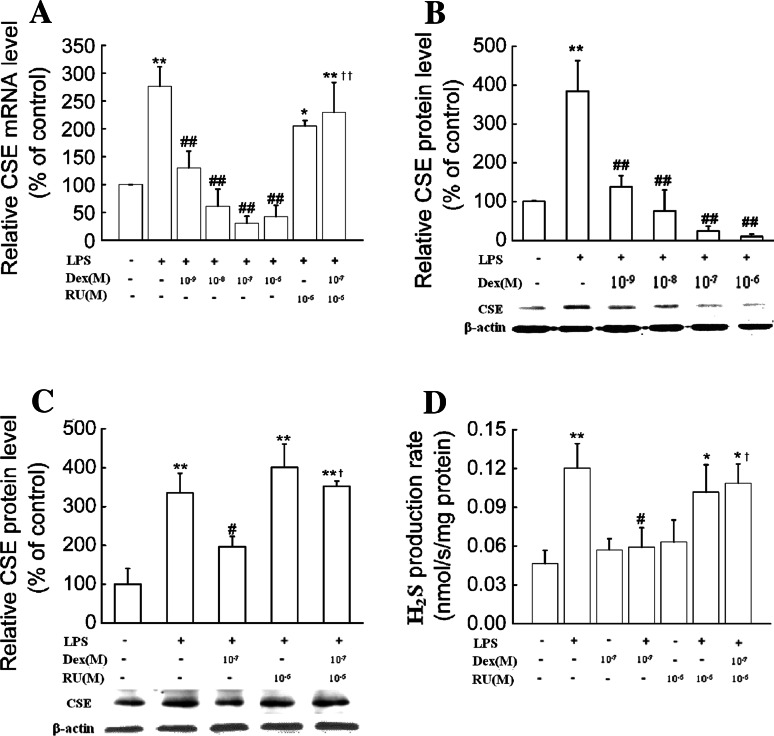
In peritoneal macrophages, dexamethasone (10−9–10−6 mol/l) significantly inhibited LPS (1 μg/ml)-induced CSE expression at both the mRNA and protein levels. Maximal inhibition of dexamethasone occurred at a concentration of 10−6 mol/l, which caused an about 85% decrease in CSE mRNA and 95% decrease in CSE protein levels compared to cells treated with LPS alone (Fig. 6a, b). Consistently, dexamethasone (10−7 mol/l) treatment significantly decreased LPS-induced H2S production rate (Fig.6d). These effects were reversed by RU38,486, a glucocorticoid receptor antagonist (Fig. 6c, d).
Fig. 6.
Dexamethasone suppressed CSE expression and H2S production in primary macrophages. Primary macrophages were treated with dexamethasone at the indicated doses in the absence or presence of RU38,486 (10−6 mol/l) and LPS (1 μg/ml) for 6 h (a) or 24 h (b–d). CSE mRNA (a), protein expression (b, c) as well as H2S production rates (d) were determined as described in “Materials and Methods.” Data were presented as mean percentage of control ± SEM for a–c (n = 3) and mean ± SEM for H2S production rates (n = 4). *P < 0.05, ** P < 0.01 compared with control; #P < 0.05, ##P < 0.01 compared with LPS; †P < 0.05, ††P < 0.01 compared with LPS plus dexamethasone. Dex dexamethasone, RU RU38,486
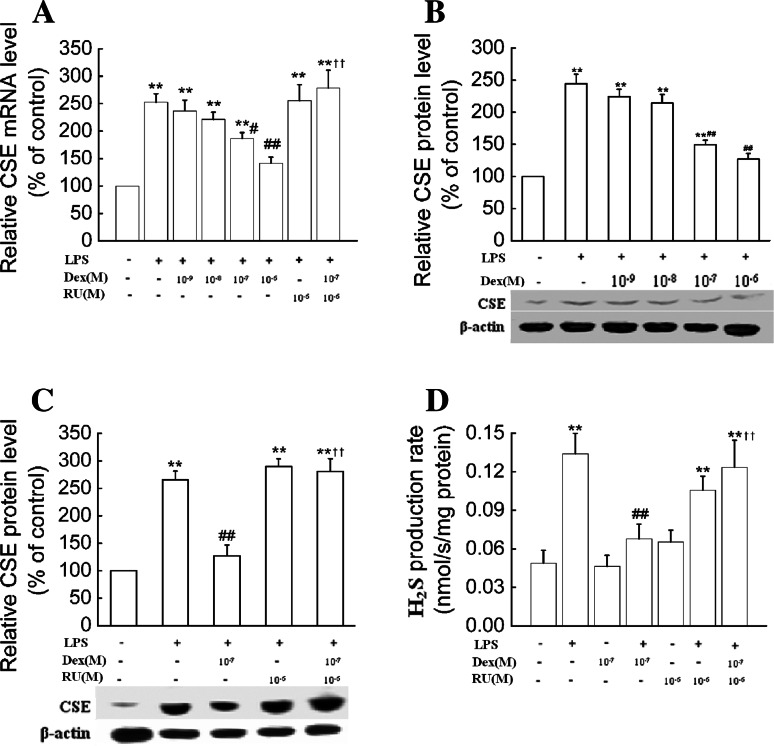
In RAW 264.7 cells, it was also shown that dexamethasone treatment decreased CSE mRNA and protein levels as well as the H2S production rate in the presence of LPS (1 μg/ml) (Fig. 7a–d).
Fig. 7.
Dexamethasone suppressed CSE expression and H2S production in RAW264.7 macrophages. RAW264.7 macrophages were treated with dexamethasone at the indicated doses in the absence or presence of RU38,486 (10−6 mol/l) and LPS (1 μg/ml) for 6 h (a) or 24 h (b–d). CSE mRNA (a), protein expression (b, c) as well as H2S production rates (d) were determined as described in “Materials and Methods.” Data were presented as mean percentage of control ± SEM for a–c (n = 3) and mean ± SEM for H2S production rates (n = 4). **P < 0.01 compared with control; #P < 0.05, ##P < 0.01 compared with LPS; ††P < 0.01 compared with LPS plus dexamethasone. Dex dexamethasone, RU RU38,486
Dexamethasone suppresses NO production in LPS-treated macrophages
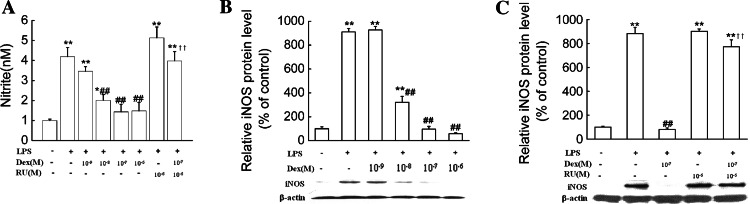
We had shown above that decreased endogenous H2S production resulted in an increase in NO production, and dexamethasone inhibited H2S production in LPS-treated macrophages. Early studies have demonstrated that dexamethasone inhibits NO production and iNOS expression in macrophage cell lines treated with LPS [43]. Thus, it would be of interest to understand iNOS expression and NO production in primary macrophages treated with dexamethasone. Our results showed that NO production and iNOS expression were significantly decreased in primary macrophages treated with LPS plus dexamethasone compared to cells treated with LPS alone. These effects of dexamethasone were also reversed by RU38,486 (Fig. 8).
Fig. 8.
Dexamethasone suppressed NO production and iNOS expression in primary macrophages. Primary macrophages were treated with dexamethasone at the indicated doses in the absence or presence of RU38,486 (10−6 mol/l) and LPS (1 μg/ml) for 24 h. NO production (a) and iNOS expression (b, c) were determined as described in “Materials and Methods.” Data were presented as mean ± SEM for NO production (n = 4) and mean percentage of control ± SEM for iNOS expression (n = 3). *P < 0.05; **P < 0.01 compared with control; ##P < 0.01 compared with LPS; ††P < 0.01 compared with LPS plus dexamethasone. Dex dexamethasone, RU RU38,486
To determine whether dexamethasone treatments were detrimental to cells, cell growth and survival were examined. It was found that treatment of cells with dexamethasone for 24 h had no significant effect on cell growth and viability (data not shown).
The role of NO in dexamethasone inhibition of CSE expression and H2S production in LPS-treated macrophages
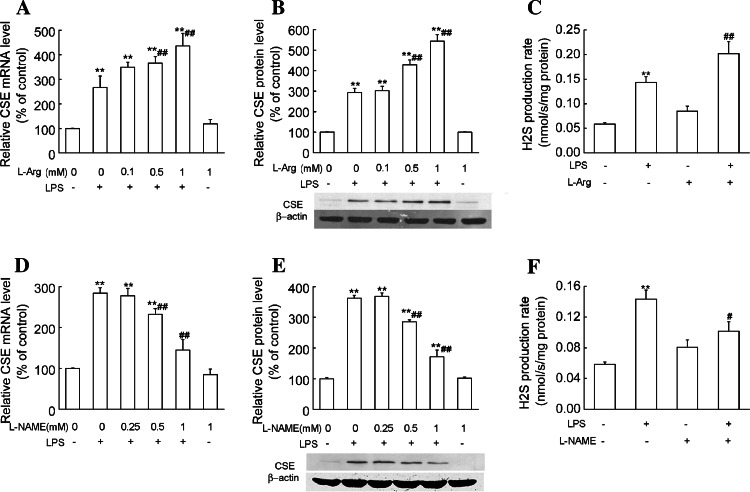
In order to explore if the effect of dexamethasone on H2S production is associated with NO production in LPS-treated macrophages, we firstly investigated the effects of NO precursor and NOS inhibitor on CSE expression and H2S production. Our results showed that l-arginine (0.1–1 mmol/l) dose-dependently enhanced CSE mRNA and protein expression in LPS-treated primary macrophages. Significant effects occurred at concentrations of 0.5 mmol/l and 1 mmol/l. Consistently, l-arginine (1 mmol/l) treatment significantly increased the LPS-induced H2S production rate (Fig. 9a–c). l-NAME (0.25–1 mmol/l), a NOS inhibitor, decreased CSE mRNA and protein levels in a dose-dependent manner. The LPS-induced H2S production rate was significantly decreased by l-NAME (1 mmol/l) treatment (Fig. 9d–f). These results suggest that NO is an important factor for CSE expression and H2S production.
Fig. 9.
Effects of NO precursor and NOS inhibitor on CSE expression and H2S production in primary macrophages stimulated with LPS. Primary macrophages were treated with NO precursor l-arginine (a–c) or NOS inhibitor l-NAME (d–f) at the indicated doses in the absence or presence of LPS (1 μg/ml) for 6 h (a, d) or 24 h (b, c, e, f), respectively. Quantitative real-time RT-PCR, Western blot analysis and real-time H2S production measurement were used to determine CSE mRNA expression, protein expression and H2S synthetic activity in macrophages. Results of Western blot were quantified by scanning densitometry of blots. H2S production rates were calculated as described in “Materials and Methods.” Data were expressed as mean percentage of control ± SEM for CSE mRNA and protein expression (n = 3), and for H2S production rates (n = 4). **P < 0.01 compared with vehicle control. #P < 0.05, ##P < 0.01 compared with LPS. l -arg l-arginine
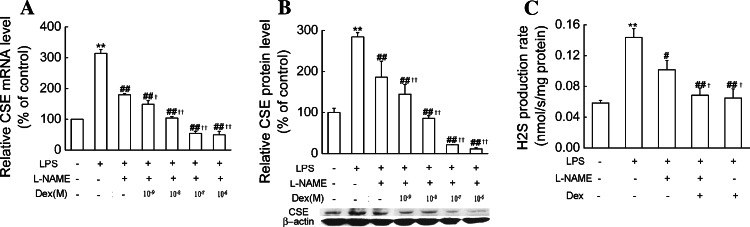
We then observed the effects of dexamethasone on LPS-induced CSE expression and H2S production in the presence of NOS inhibitor. As shown in Fig. 10, dexamethasone (10−9–10−6 mol/l) treatment in the presence of l-NAME (1 mmol/l) significantly decreased LPS-induced expression of CSE mRNA and protein compared with l-NAME. Dexamethasone (10−7 mol/l) plus l-NAME (1 mmol/l) also significantly decreased LPS-induced H2S production rates compared with the cells treated with l-NAME (1 mmol/l). These results suggest that dexamethasone not only indirectly decreases CSE expression and H2S production by inhibiting NO production, but also may directly suppress CSE expression in LPS-treated macrophages.
Fig. 10.
Effects of dexamethasone on LPS-induced CSE expression and H2S production in the presence of NOS inhibitor in primary macrophages. Primary macrophages were treated with dexamethasone at the indicated doses in the absence or presence of l-NAME (1 mmol/l) and LPS (1 μg/ml) for 6 h (a) or 24 h (b–c). CSE mRNA (a), protein expression (b) as well as H2S production rates (c) were determined as described in “Materials and Methods.” Data were presented as mean percentage of control ± SEM for a–b (n = 3) and mean ± SEM for H2S production rates (n = 4). **P < 0.01 compared with control; #P < 0.05, ##P < 0.01 compared with LPS; †P < 0.05, ††P < 0.01 compared with LPS plus l-NAME. Dex dexamethasone
Dexamethasone reduces CSE mRNA stability in primary macrophages
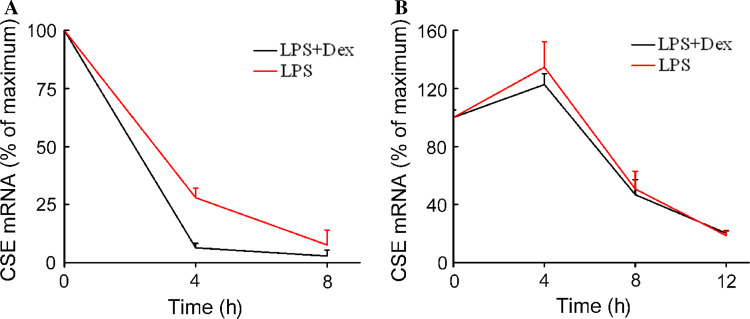
As a first step in elucidating the molecular mechanisms by which dexamethasone decreased CSE mRNA, we measured the rate of CSE mRNA degradation. An inhibitor of mRNA synthesis, actinomycin D (1 μg/ml) was added to the cells 6 h after LPS or LPS plus dexamethasone treatment. Cells were harvested at time points of 0, 4 and 8 h for primary macrophages and 0, 4, 8 and 12 h for RAW 264.7 cells after the addition of actinomycin D.
In primary macrophages, dexamethasone treatment significantly decreased the half-life of CSE mRNA in LPS-treated cells. The half-life of CSE mRNA after LPS treatment was 2.52 ± 0.26 h, and dexamethasone reduced it to 1.67 ± 0.04 h. (***P < 0.05, Fig. 11a). In RAW 264.7 cells (Fig. 11b), the half-life of CSE mRNA was 7.60 ± 0.61 h in LPS-treated cells and 7.59 ± 0.46 h in LPS plus dexmethasone-treated cells. There was no statistical difference between these two groups. These results suggested that dexamethasone reduced CSE mRNA stability in primary macrophages, but not in RAW 264.7 cells.
Fig. 11.
Effects of dexamethasone on CSE mRNA stability in macrophages stimulated with LPS. Primary (a) and RAW 264.7 (b) macrophages were stimulated with LPS (1 μg/ml) in the absence or presence of dexamethasone (10−7 mol/l) as indicated. Actinomycin D (1 μg/ml) was added to the cells after 6 h of incubation. Cells treated with vehicle were used as the maximum point. Cells were harvested for RNA extraction after the incubation time indicated after actinomycin D addition, and CSE and GAPDH mRNA were detected by quantitative real-time RT-PCR. Each data point was expressed as mean percentage of the maximum determined at time zero ± SEM (n = 4). Dex dexamethasone
Dexamethasone reduced CSE transcription in RAW 264.7 macrophages
To determine the effects of dexamethasone on the rate of CSE gene transcription in RAW 264.7 cells, we transfected the pCSE-PGL3 plasmid into RAW 264.7 cells and examined the CSE promoter activity. As shown in Fig. 12, dexamethasone (10−8–10−6 mol/l) treatment significantly decreased the transcriptional activity of CSE genes compared with LPS alone, and these effects of dexamethasone were also reversed by RU38,486, suggesting that dexamethasone suppressed CSE expression by repressing CSE gene transcription in RAW264.7 macrophages.
Fig. 12.
Effects of dexamethasone on CSE promoter activity in RAW264.7 macrophages. RAW264.7 cells were treated with dexamethasone at the indicated doses in the absence or presence of RU38,486 (10−6 mol/l) and LPS (1 μg/ml) for 12 h. CSE promoter activity was assayed as described in “Materials and Methods.” Data were presented as mean percentage of control ± SEM for four independent experiments. **P < 0.01 compared with control; ##P < 0.01 compared with LPS; †P < 0.05 compared with LPS plus dexamethasone. Dex dexamethasone, RU RU38,486
Discussion
Macrophages are shown to produce numerous mediators including the gaseous mediators NO and CO in response to inflammatory stimuli (e.g., LPS) [27, 28]. In this study, we showed that, for the first time, primary macrophages had the capacity to produce H2S, the production rate of which could be further enhanced by LPS treatment; endogenous H2S was capable of inhibiting NO production locally in macrophages.
H2S is formed in mammalian cells mainly by CBS and CSE. CBS and CSE are widely distributed in tissues, although a degree of tissue specificity is apparent. CBS is more abundant in brain, whereas CSE activity is most notable in peripheral tissues such as blood vessels [44]. In liver and kidney, both CBS and CSE have been identified [45]. The present study showed that primary and RAW 264.7 macrophages expressed CSE but not CBS and were capable of producing H2S through CSE. Earlier studies have shown marked increases in the plasma H2S level and H2S production in tissues including liver and kidney during several inflammatory conditions [6, 7, 9, 10]. In the present experiments, the expression of CSE was significantly enhanced by LPS, the major component of the outer membrane of gram-negative bacteria, in both primary and RAW 264.7 macrophages, and consistently H2S synthesis was also elevated. This suggests that macrophages may be one of the H2S-producing sources during gram-negative bacteria-induced inflammation.
There are numerous conflicting data regarding the effects of H2S on inflammation. For example, CSE inhibitor has been reported both to exacerbate [13] and to reduce [46] carrageenan-induced paw edema. Both H2S donors and CSE inhibitors exhibit anti-inflammatory activities such as inhibiting leukocyte/endothelium adhesion [13, 47]. To date, much of our knowledge of the biological effects of H2S comes from the use of inhibitors of H2S synthases, H2S donors or H2S precursor. However, the specificity of these reagents is doubted. For instance, CSE inhibitor PAG targets the pyridoxal 5′-phosphate-binding site of CSE and, as such, may affect other pyridoxal 5′-phosphate-dependent enzymes as well [48]. Thus, interpretations of results from experiments using those above-mentioned reagents must be made with caution. To avoid such limitations in specificity of those reagents, we used both CSE and CSE-siRNA expression vector to specifically increase and decrease CSE expression, respectively. In consistence with the results obtained by using l-cysteine, we demonstrated that CSE siRNA significantly increased, whereas CSE overexpression decreased LPS-induced iNOS expression and NO production. NO is well-recognized as one of the pro-inflammatory mediators, and NO production is markedly increased during various inflammatory conditions such as endoxemia [49]. Our results showed that endogenous H2S produced locally in macrophages has a tonic inhibitory effect on NO production in the presence of LPS. Thus, this suggests that H2S might exert anti-inflammatory effects by inhibiting NO production in macrophages during LPS-induced inflammation.
Several studies have demonstrated the impact of NO on H2S production. Zhao et al. [5] reported that, in cultured smooth muscle cells, NO increases CSE mRNA expression and H2S production. Zhong et al. [50] found that inhibition of NO production caused a decrease in H2S production and CSE activity in artery in vivo. However, a study of Anuar and coworkers suggested that NO can inhibit H2S production by showing that administration of NO donor to LPS-treated rat resulted in a decrease in the formation of H2S and CSE expression in liver [51]. Our findings that NOS inhibitor decreased whereas NO precursor increased H2S production and CSE expression in LPS-treated macrophages suggest that NO is an important endogenous stimulus in the formation of H2S during LPS-induced inflammation.
GCs are known to exert their anti-inflammatory effects by inhibiting the expression and production of various pro-inflammatory mediators, such as TNF-α, IL-1 and NO, as well as stimulating the expression of genes coding for anti-inflammatory proteins, such as lipocortin-1 and interleukin-10 [17]. Our findings that dexamethasone inhibited NO production through iNOS expression are consistent with previous studies [43, 52]. Interestingly, our present study also found that decreased H2S production resulted in increases in NO production and iNOS expression, and dexamethasone reduced H2S production. This would suggest that dexamethasone inhibition of NO production and iNOS expression in LPS-treated macrophages was not related to H2S. As mentioned above, the present study found that NO had a stimulatory effect on CSE expression and H2S production in LPS-treated macrophages. Moreover, when NO production was inhibited by NOS inhibitor, l-NAME, dexamethasone could intensively inhibit CSE expression and H2S production compared to l-NAME. This suggests that dexamethasone may directly inhibit CSE expression and H2S production, besides the NO-dependent way.
More recently, Li et al. proposed that inhibition of H2S production in neutrophils contributes to the anti-inflammatory effect of dexamethasone in endotoxic shock [33]. However, H2S has been indicated to have anti-inflammatory effects in previous investigations [13, 48, 53]. The present study also indicates that H2S might play an anti-inflammation role by inhibiting NO production in macrophages. In this case, our findings that GCs inhibit H2S and NO production provide evidence that GCs coordinate the production of pro- and anti-inflammatory mediators during inflammation. Recently, Van Molle and Libert [20] proposed that GCs could balance between pro- and anti-inflammatory mediators and control their own strength by inducing the pro-inflammatory cytokine MIF.
The classic effects of glucocorticoids are mediated through GR. RU38,486 is a steroid that competitively binds to GR and inhibits the effect of glucocorticoids [54]. In the presence of RU38,486, dexamethasone did not inhibit CSE expression and the H2S production rate in LPS-treated cells, indicating that suppressed H2S production is a GR-mediated process. Glucocorticoid-GR complexes are able to activate or inhibit gene transcription [55]. In the present study, we also found that dexamethasone repressed CSE transcription in LPS-treated RAW 264.7 cells.
In addition to its effects on gene transcription, GCs have been shown to regulate the mRNA stability of some genes. Korhonen et al. [43] showed that dexamethasone decreases LPS-induced iNOS expression by destabilizing iNOS mRNA in J774 macrophages. The study of Walker and coworkers suggested that dexamethasone inhibits iNOS expression by destabilizing mRNA and repressing gene transcription interferon-γ-stimulated macrophages [52]. We found that dexamethasone reduced CSE mRNA stability in primary macrophages, but did not affect that in RAW 264.7 cells. In RAW 264.7 cells, dexamethasone suppressed CSE promoter activity. These findings suggest that dexamethasone inhibition of CSE expression takes place through different mechanisms in primary macrophages and macrophage cell lines. On the other hand, these data also suggest that the mechanism of action of glucocorticoid on CSE expression might be related to cell type.
In conclusion, this study demonstrated that macrophages expressed CSE and had the capacity to produce H2S through CSE. Endogenous H2S had a tonic inhibitory effect on NO production in macrophages. Dexamethasone suppressed the LPS-induced H2S production rate through regulating CSE expression in macrophages. Dexamethasone could also inhibit CSE expression and H2S production by inhibiting NO production because endogenous NO enhanced CSE expression and dexamethasone inhibited NO production in LPS-treated macrophages. Destabilizing mRNA was involved in dexamethasone inhibition of LPS-induced CSE expression and H2S production in primary macrophages. Repression of gene transcription was responsible for dexamethasone inhibition of CSE expression and H2S production in RAW 264.7 cells. These data expanded our knowledge of the mechanisms of LPS-induced inflammatory processes as well as GCs' regulation of inflammation.
Acknowledgments
The authors wish to thank Dr. I.Ishii, Gunma University Graduate School of Medicine, Gunma, Japan, for his gift of pCSE-PGL3 plasmid. This work was supported by the National Natural Science Foundation of China, grant nos. 30670815 and 30770846, and the Science and Technology Commission of Shanghai Municipals (09XD1405600).
Abbreviations
- H2S
Hydrogen sulfide
- NO
Nitric oxide
- CO
Carbon monoxide
- CSE
Cystathionine gamma-lyase
- CBS
Cystathionine β-synthetase
- fMLP
Formyl-methionyl-leucyl-phenylalanine
- TNF-α
Tumor necrosis factor-α
- GCs
Glucocorticoids
- GR
Glucocorticoid receptor
- MIF
Migration inhibitory factor
- LPS
Lipopolysaccharide
- IL-1
Interleukin-1
- IL-6
Interleukin-6
- MTT
3-[4, 5-Dimethylthiazol-2-yl]-2, 5-diphenyl tetrazolium bromide
- PAG
dl-propargylglycine
- l-NAME
NG-nitro-l-arginine methyl ester
- iNOS
Inducible nitric oxide synthase
- CD-FBS
Charcoal-stripped FBS
- PLP
Pyridoxal-5′-phosphate
- siRNA
Interfering RNA
- RAW-EGFP-mCSE
CSE-overexpression RAW264.7 cell lines
- RAW-mCSE siRNA
CSE-knockdown RAW264.7 cell lines
- TBST
Tris-buffered saline/Tween 20
Footnotes
X.-Y. Zhu and S.-J. Liu contributed equally to this manuscript.
References
- 1.Moore PK, Bhatia M, Moochhala S. Hydrogen sulfide: from the smell of the past to the mediator of the future? Trends Pharmacol Sci. 2003;24:609–611. doi: 10.1016/j.tips.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Wang R. Two’s company, three’s a crowd: can H2S be the third endogenous gaseous transmitter? FASEB J. 2002;16:1792–1798. doi: 10.1096/fj.02-0211hyp. [DOI] [PubMed] [Google Scholar]
- 3.Bhatia M. Hydrogen sulfide as a vasodilator. IUBMB Life. 2005;57:603–606. doi: 10.1080/15216540500217875. [DOI] [PubMed] [Google Scholar]
- 4.Qingyou Z, Junbao D, Weijin Z, Hui Y, Chaoshu T, Chunyu Z. Impact of hydrogen sulfide on carbon monoxide/heme oxygenase pathway in the pathogenesis of hypoxic pulmonary hypertension. Biochem Biophys Res Commun. 2004;317:30–37. doi: 10.1016/j.bbrc.2004.02.176. [DOI] [PubMed] [Google Scholar]
- 5.Zhao W, Zhang J, Lu Y, Wang R. The vasorelaxant effect of H2S as a novel endogenous gaseous KATP channel opener. EMBO J. 2001;20:6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Bhatia M, Zhu YZ, Zhu YC, Ramnath RD, Wang ZJ, Anuar FB, Whiteman M, Salto-Tellez M, Moore PK. Hydrogen sulfide is a novel mediator of lipopolysaccharide-induced inflammation in the mouse. Faseb J. 2005;19:1196–1198. doi: 10.1096/fj.04-3583fje. [DOI] [PubMed] [Google Scholar]
- 7.Hui Y, Du J, Tang C, Bin G, Jiang H. Changes in arterial hydrogen sulfide (H(2)S) content during septic shock and endotoxin shock in rats. J Infect. 2003;47:155–160. doi: 10.1016/S0163-4453(03)00043-4. [DOI] [PubMed] [Google Scholar]
- 8.Zhi L, Ang AD, Zhang H, Moore PK, Bhatia M. Hydrogen sulfide induces the synthesis of proinflammatory cytokines in human monocyte cell line U937 via the ERK-NF-kappaB pathway. J Leukoc Biol. 2007;81:1322–1332. doi: 10.1189/jlb.1006599. [DOI] [PubMed] [Google Scholar]
- 9.Collin M, Anuar FB, Murch O, Bhatia M, Moore PK, Thiemermann C. Inhibition of endogenous hydrogen sulfide formation reduces the organ injury caused by endotoxemia. Br J Pharmacol. 2005;146:498–505. doi: 10.1038/sj.bjp.0706367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatia M, Wong FL, Fu D, Lau HY, Moochhala SM, Moore PK. Role of hydrogen sulfide in acute pancreatitis and associated lung injury. Faseb J. 2005;19:623–625. doi: 10.1096/fj.04-3023fje. [DOI] [PubMed] [Google Scholar]
- 11.Whiteman M, Armstrong JS, Chu SH, Jia-Ling S, Wong BS, Cheung NS, Halliwell B, Moore PK. The novel neuromodulator hydrogen sulfide: an endogenous peroxynitrite ‘scavenger’? J Neurochem. 2004;90:765–768. doi: 10.1111/j.1471-4159.2004.02617.x. [DOI] [PubMed] [Google Scholar]
- 12.Fiorucci S, Antonelli E, Distrutti E, Rizzo G, Mencarelli A, Orlandi S, Zanardo R, Renga B, Di Sante M, Morelli A, Cirino G, Wallace JL. Inhibition of hydrogen sulfide generation contributes to gastric injury caused by anti-inflammatory nonsteroidal drugs. Gastroenterology. 2005;129:1210–1224. doi: 10.1053/j.gastro.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 13.Zanardo RC, Brancaleone V, Distrutti E, Fiorucci S, Cirino G, Wallace JL. Hydrogen sulfide is an endogenous modulator of leukocyte-mediated inflammation. FASEB J. 2006;20:2118–2120. doi: 10.1096/fj.06-6270fje. [DOI] [PubMed] [Google Scholar]
- 14.Mariggio MA, Pettini F, Fumarulo R. Sulfide influence onpolymorphonuclear functions: a possible role for Ca2+ involvement. Immunopharmacol Immunotoxicol. 1997;19:393–404. doi: 10.3109/08923979709046984. [DOI] [PubMed] [Google Scholar]
- 15.Mariggio MA, Minunno V, Riccardi S, Santacroce R, De Rinaldis P, Fumarulo R. Sulfide enhancement of PMN apoptosis. Immunopharmacol Immunotoxicol. 1998;20:399–408. doi: 10.3109/08923979809034822. [DOI] [PubMed] [Google Scholar]
- 16.Hu LF, Wong PT, Moore PK, Bian JS. Hydrogen sulfide attenuates lipopolysaccharide-induced inflammation by inhibition of p38 mitogen-activated protein kinase in microglia. J Neurochem. 2007;100:1121–1128. doi: 10.1111/j.1471-4159.2006.04283.x. [DOI] [PubMed] [Google Scholar]
- 17.Barnes PJ. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clin Sci (Lond) 1998;94:557–572. doi: 10.1042/cs0940557. [DOI] [PubMed] [Google Scholar]
- 18.Pitzalis C, Pipitone N, Perretti M. Regulation of leukocyte-endothelial interactions by glucocorticoids. Ann N Y Acad Sci. 2002;966:108–118. doi: 10.1111/j.1749-6632.2002.tb04208.x. [DOI] [PubMed] [Google Scholar]
- 19.Tuckermann JP, Kleiman A, McPherson KG, Reichardt HM. Molecular mechanisms of glucocorticoids in the control of inflammation and lymphocyte apoptosis. Crit Rev Clin Lab Sci. 2005;42:71–104. doi: 10.1080/10408360590888983. [DOI] [PubMed] [Google Scholar]
- 20.Van Molle W, Libert C. How glucocorticoids control their own strength and the balance between pro- and anti-inflammatory mediators. Eur J Immunol. 2005;35:3396–3399. doi: 10.1002/eji.200535556. [DOI] [PubMed] [Google Scholar]
- 21.Wilckens T, De Rijk R. Glucocorticoids and immune function: unknown dimensions and new frontiers. Immunol Today. 1997;18:418–424. doi: 10.1016/S0167-5699(97)01111-0. [DOI] [PubMed] [Google Scholar]
- 22.Dhabhar FS, Miller AH, McEwen BS, Spencer RL. Stress-induced changes in blood leukocyte distribution. Role of adrenal steroid hormones. J Immunol. 1996;157:1638–1644. [PubMed] [Google Scholar]
- 23.Calandra T, Bernhagen J, Metz CN, Spiegel LA, Bacher M, Donnelly T, Cerami A, Bucala R. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377:68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 24.Meagher LC, Cousin JM, Seckl JR, Haslett C. Opposing effects of glucocorticoids on the rate of apoptosis in neutrophilic and eosinophilic granulocytes. J Immunol. 1996;156:4422–4428. [PubMed] [Google Scholar]
- 25.Sivertson KL, Seeds MC, Long DL, Peachman KK, Bass DA. The differential effect of dexamethasone on granulocyte apoptosis involves stabilization of Mcl-1L in neutrophils but not in eosinophils. Cell Immunol. 2007;246:34–45. doi: 10.1016/j.cellimm.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liles WC, Dale DC, Klebanoff SJ. Glucocorticoids inhibit apoptosis of human neutrophils. Blood. 1995;86:3181–3188. [PubMed] [Google Scholar]
- 27.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 28.Ravasi T, Wells C, Forest A, Underhill DM, Wainwright BJ, Aderem A, Grimmond S, Hume DA. Generation of diversity in the innate immune system: macrophage heterogeneity arises from gene-autonomous transcriptional probability of individual inducible genes. J Immunol. 2002;168:44–50. doi: 10.4049/jimmunol.168.1.44. [DOI] [PubMed] [Google Scholar]
- 29.Oh GS, Pae HO, Lee BS, Kim BN, Kim JM, Kim HR, Jeon SB, Jeon WK, Chae HJ, Chung HT. Hydrogen sulfide inhibits nitric oxide production and nuclear factor-kappaB via heme oxygenase-1 expression in RAW264.7 macrophages stimulated with lipopolysaccharide. Free Radic Biol Med. 2006;41:106–119. doi: 10.1016/j.freeradbiomed.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 30.Heasman SJ, Giles KM, Ward C, Rossi AG, Haslett C, Dransfield I. Glucocorticoid-mediated regulation of granulocyte apoptosis and macrophage phagocytosis of apoptotic cells: implications for the resolution of inflammation. J Endocrinol. 2003;178:29–36. doi: 10.1677/joe.0.1780029. [DOI] [PubMed] [Google Scholar]
- 31.Yeager MP, Pioli PA, Wardwell K, Beach ML, Martel P, Lee HK, Rassias AJ, Guyre PM. In vivo exposure to high or low cortisol has biphasic effects on inflammatory response pathways of human monocytes. Anesth Analg. 2008;107:1726–1734. doi: 10.1213/ane.0b013e3181875fb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yona S, Gordon S. Inflammation: glucocorticoids turn the monocyte switch. Immunol Cell Biol. 2007;85:81–82. doi: 10.1038/sj.icb.7100034. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Whiteman M, Moore PK. Dexamethasone inhibits lipopolysacharide-induced hydrogen sulfide biosynthesis in intact cells and in an animal model of endotoxic shock. J Cell Mol Med. 2009;13:2684–2692. doi: 10.1111/j.1582-4934.2008.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei H, Ju DW, Yu Y, Tao Q, Chen G, Gu S, Hamada H, Cao X. Induction of potent antitumor response by vaccination with tumor lysate-pulsed macrophages engineered to secrete macrophage colony-stimulating factor and interferon-gamma. Gene Ther. 2000;7:707–713. doi: 10.1038/sj.gt.3301162. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Jeroschewski P, Steuckart C, Kuhl M. An amperometric micro-sensor for the determination of H2S in aquatic environments. Anal Chem. 1996;68:4351–4357. doi: 10.1021/ac960091b. [DOI] [Google Scholar]
- 37.Doeller JE, Isbell TS, Benavides G, Koenitzer J, Patel H, Patel RP, Lancaster JR, Jr, Darley-Usmar VM, Kraus DW. Polarographic measurement of hydrogen sulfide production and consumption by mammalian tissues. Anal Biochem. 2005;341:40–51. doi: 10.1016/j.ab.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 38.Benavides GA, Squadrito GL, Mills RW, Patel HD, Isbell TS, Patel RP, Darley-Usmar VM, Doeller JE, Kraus DW. Hydrogen sulfide mediates the vasoactivity of garlic. Proc Natl Acad Sci USA. 2007;104:17977–17982. doi: 10.1073/pnas.0705710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tapley D, Buettner G, Shick J. Free radicals and chemiluminescence as products of the spontaneous oxidation of sulfide in seawater, and their biological implications. Biol Bull. 1999;196:52–56. doi: 10.2307/1543166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu DZ, Wang ST, Deng JF, Liu MY. Epinephrine protects against severe acute gastric bleeding in rats: role of nitric oxide and glutathione. Shock. 2005;23:253–257. [PubMed] [Google Scholar]
- 41.Ishii I, Akahoshi N, Yu XN, Kobayashi Y, Namekata K, Komaki G, Kimura H. Murine cystathionine gamma-lyase: complete cDNA and genomic sequences, promoter activity, tissue distribution and developmental expression. Biochem J. 2004;381:113–123. doi: 10.1042/BJ20040243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Twentyman PR, Luscombe M. A study of some variables in a tetrazolium dye (MTT) based assay for cell growth and chemosensitivity. Br J Cancer. 1987;56:279–285. doi: 10.1038/bjc.1987.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korhonen R, Lahti A, Hamalainen M, Kankaanranta H, Moilanen E. Dexamethasone inhibits inducible nitric-oxide synthase expression and nitric oxide production by destabilizing mRNA in lipopolysaccharide-treated macrophages. Mol Pharmacol. 2002;62:698–704. doi: 10.1124/mol.62.3.698. [DOI] [PubMed] [Google Scholar]
- 44.Lowicka E, Beltowski J. Hydrogen sulfide (H2S)—the third gas of interest for pharmacologists. Pharmacol Rep. 2007;59:4–24. [PubMed] [Google Scholar]
- 45.Kimura H. Hydrogen sulfide as a neuromodulator. Mol Neurobiol. 2002;26:13–19. doi: 10.1385/MN:26:1:013. [DOI] [PubMed] [Google Scholar]
- 46.Bhatia M. Hydrogen sulphide is a mediator of carrageenan-induced hindpaw oedema in the rat. Br J Pharmacol. 2005;145:141–144. doi: 10.1038/sj.bjp.0706186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Zhi L, Moochhala SM, Moore PK, Bhatia M. Endogenous hydrogen sulfide regulates leukocyte trafficking in cecal ligation and puncture-induced sepsis. J Leukoc Biol. 2007;82:894–905. doi: 10.1189/jlb.0407237. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Salto-Tellez M, Tan CH, Whiteman M, Moore PK. GYY4137, a novel hydrogen sulfide-releasing molecule, protects against endotoxic shock in the rat. Free Radic Biol Med. 2009;47:103–113. doi: 10.1016/j.freeradbiomed.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 49.Moilanen E, Whittle BJR, Moncada S (1999) Nitric oxide as a factor in inflammation. In: Gallin JI, Snyderman R (ed) Inflammation: principles and clinical correlations. Lippincott Williams & Wilkins, Philadelphia, pp 787–800
- 50.Zhong GZ, Chen FR, Cheng YQ, Tang CS, Du JD. The role of hydrogen sulfide generation in the pathogenesis of hypertension in rats induced by inhibition of nitric oxide synthase. J Hypertens. 2003;21:1879–1885. doi: 10.1097/00004872-200310000-00015. [DOI] [PubMed] [Google Scholar]
- 51.Anuar F, Whiteman M, Siau JL, Kwong SE, Bhatia M, Moore PK. Nitric oxide-releasing flurbiprofen reduces formation of proinflammatory hydrogen sulfide in lipopolysaccharide-treated rat. Br J Pharmacol. 2006;147:966–974. doi: 10.1038/sj.bjp.0706696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker G, Pfeilschifter J, Kunz D. Mechanisms of suppression of inducible nitric-oxide synthase (iNOS) expression in interferon (IFN)-gamma-stimulated RAW 264.7 cells by dexamethasone. Evidence for glucocorticoid-induced degradation of iNOS protein by calpain as a key step in post-transcriptional regulation. J Biol Chem. 1997;272:16679–16687. doi: 10.1074/jbc.272.26.16679. [DOI] [PubMed] [Google Scholar]
- 53.Wallace JL. Hydrogen sulfide-releasing anti-inflammatory drugs. Trends Pharmacol Sci. 2007;28:501–505. doi: 10.1016/j.tips.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Cadepond F, Ulmann A, Baulieu EE. RU486 (mifepristone): mechanisms of action and clinical uses. Annu Rev Med. 1997;48:129–156. doi: 10.1146/annurev.med.48.1.129. [DOI] [PubMed] [Google Scholar]
- 55.Hayashi R, Wada H, Ito K, Adcock IM. Effects of glucocorticoids on gene transcription. Eur J Pharmacol. 2004;500:51–62. doi: 10.1016/j.ejphar.2004.07.011. [DOI] [PubMed] [Google Scholar]