Abstract
Phage display, the presentation of (poly)peptides as fusions to capsid proteins on the surface of bacterial viruses, celebrates its 25th birthday in 2010. The technique, coupled with in vitro selection, enables rapid identification and optimization of proteins based on their structural or functional properties. In the last two decades, it has advanced tremendously and has become widely accepted by the scientific community. This by no means exhaustive review aims to inform the reader of the key modifications in phage display. Novel display formats, innovative library designs and screening strategies are discussed. I will also briefly review some recent uses of the technology to illustrate its incredible versatility.
Keywords: Phage display, Molecular evolution, Library screening, Library design, Selection strategy
The technology
Bacteriophage (also simply called phage) are a diverse group of viruses that infect prokaryotic cells. Due to their genetic and structural simplicity (as compared with their more complex eukaryotic “cousins”), as well as the ability to simply grow them on bacterial hosts in the laboratory, phage have been extensively used in basic and applied life sciences ever since their discovery in the early twentieth century (reviewed in [1, 2]). With the advent of genetic engineering in the late 1970s, phage-based vectors were also among the first cloning vehicles [3–6].
Phage display is a term describing display of foreign (poly)peptides on the surface of phage particle. This is achieved by splicing a gene encoding such a peptide into a gene encoding a capsid structural protein. The capsid protein should ideally serve only as an anchor for the displayed peptide and not interfere with its structure; the idea is to have a peptide tethered to a replicating particle via a peptide linker, yet retaining its structure/function as it would be free in solution. Using recombinant DNA technology, collections of billions of peptides, protein variants, gene fragment- or cDNA-encoded proteins presented on phage (so-called phage-displayed libraries) can be constructed and surveyed for specific affinity or activity (Fig. 1). Several excellent books dealing with design, construction, and applications of phage display libraries have been published [7–11]. Related technologies include ribosome display [12, 13], mRNA display [13], display on the surface of bacterial [14] or eukaryotic cells [15–17], and on capsids of eukaryotic viruses [17, 18]. These have been extensively reviewed elsewhere and are beyond the scope of this review.
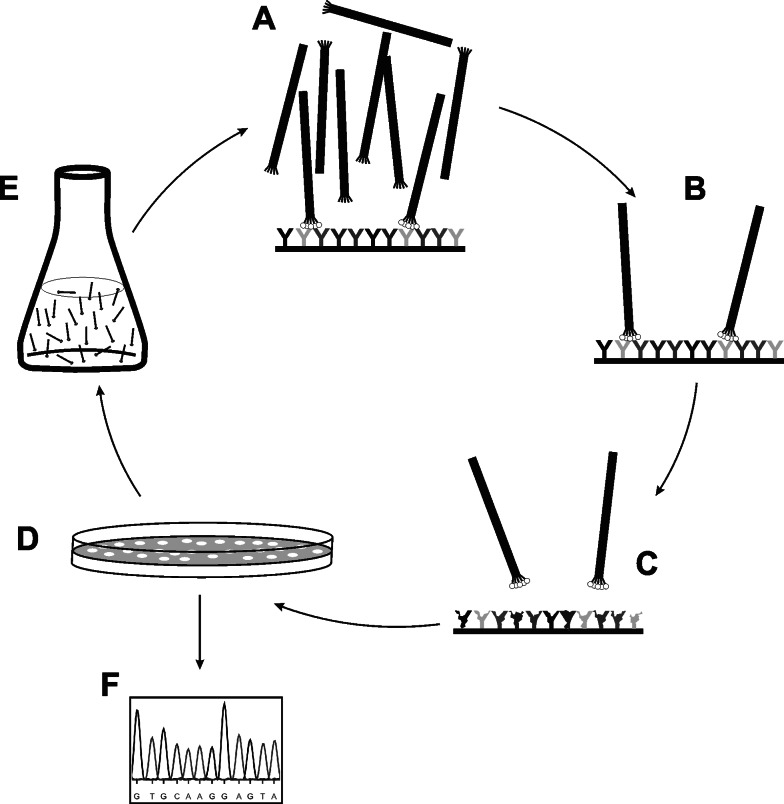
Fig. 1.
Basic principle of phage library screening. Library phage are incubated over an immobilized target (a). Non-binding phage are washed away (b) whereas target-binders are eluted (c). Eluted phage are plated on host bacteria (d), amplified in liquid growth medium (e) and subjected to further rounds of in vitro selection and in vivo amplification. Individual phage clones can be isolated from eluted subpopulation at any stage. Finally, primary structure of displayed ligand is easily identified via determination of insert’s nucleotide sequence (f). Usually, selected ligands (either on-phage or soluble synthetic/recombinant) are further analyzed to assess binding specificity and affinity (not shown)
Conventional phage display formats
The first report describing display of a foreign polypeptide on the surface of a bacteriophage particle dates from 1985 [19]. Smith demonstrated that fusions to the minor capsid protein p3 (product of gene III) of the non-lytic filamentous phage f1 (the overall architecture of filamentous phage is depicted in Fig. 2a and schematic structure of p3 is given in Fig. 2b) were fairly well tolerated by cloning a fragment of the EcoRI restrictase gene in the middle section of the gene III. Moreover, he was able to effectively enrich the recombinant phage population from a mixture of dominating wild-type phage by affinity capture with polyclonal antibodies against EcoRI. The insert, however, interfered with virion assembly by reducing expression of p3 relative to other phage structural proteins, as well as compromised phage infectivity (see caption of Fig. 2b).
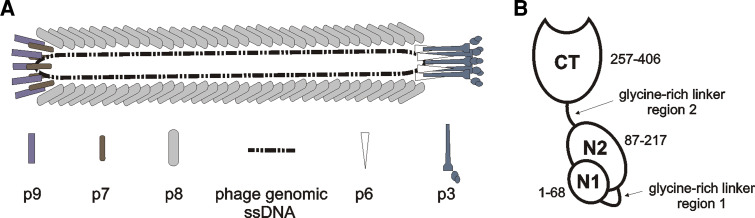
Fig. 2.
a Filamentous phage architecture (adapted from [21]). Filamentous phage are rod-like viruses with a circular ssDNA genome. The tube-like capsid is composed of several thousand copies of tightly packed major coat protein [gene VIII product (p8)], capped by five copies of p3 and p6 on one end and five copies of p7 and p9 on the opposite end. b Schematic representation of minor coat protein p3 structure (adapted from [21]). Two N-terminal domains (N2 and N1) are essential for phage infectivity. They are implicated in binding the F-pilus and TolA membrane protein of bacterial host, respectively, which act as phage receptors. The N-terminal part of p3 also renders infected cell immune to superinfection by another filamentous phage. C-terminal domain (CT) takes part in the final stages of virion assembly on inner bacterial membrane, capping the capsid end together with p6. Residue numbering is according to the mature p3
Phage vectors readily support presentation of short peptides on all five copies of p3, but are reluctant to display large proteins. An ingenious solution to this serious drawback came in the form of a special display vector called a phagemid [20]. Phagemids combine features of plasmids (i.e., carry antibiotic resistance and enable replication of dsDNA) with features of phage vectors (i.e., allow for production and packing of ssDNA into virions). Phagemids are engineered to express recombinant p3 fusions under controlled conditions, but do not encode any viral structural or replication proteins. Only upon superinfection of a bacterial host by a helper phage which contributes the missing genes can the phagemid ssDNA replication and packing take place. Helper phage are defective in their origin of replication or packaging signal thereby ensuring preferential packing of phagemids. Progeny phagemid virions carry recombinant as well as wild-type p3 (contributed by helper phage), so their infectivity is not crucially affected. Phagemid display on p3 is regarded as monovalent, although there is typically on average less then one copy of the fusion protein present per phage particle [21].
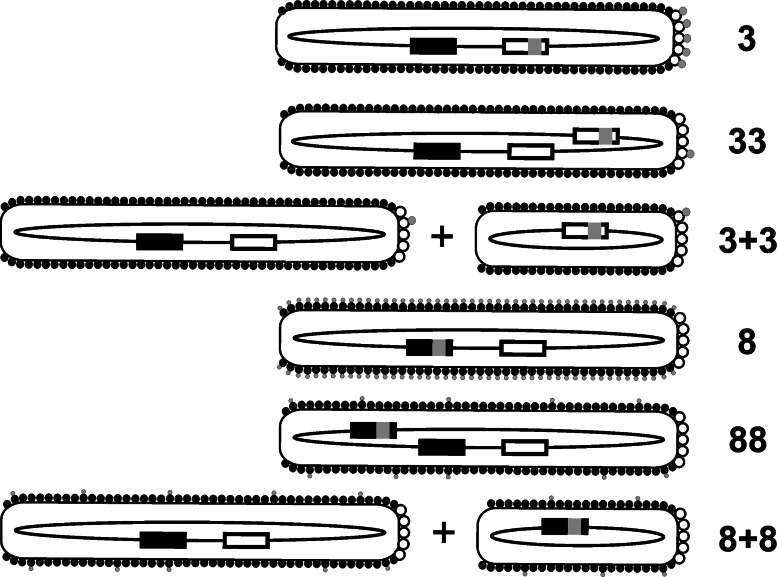
According to Smith’s classification [22, 23] (Fig. 3), the p3 phagemid display is denoted as a 3+3 type system (implying there are two different forms of p3). Type 3 system stands for p3 phage display where all five copies of p3 are transcribed from a single fusion gene on a phage vector. If a single phage vector carries both recombinant and wild-type gIII genes, this is referred to as a type 33 system.
Fig. 3.
Classification of major phage display formats (adapted from [23]). White and black boxes represent wild-type gIII and gVIII, respectively. Foreign DNA inserts encoding displayed peptides (gray circles) are depicted by gray squares. White and black circles symbolize wild-type p3 and p8, respectively. In 3+3 and 8+8 format depictions, smaller phage represent phagemid virions, whereas larger ones depict helper phage. See text for details
All the basic principles of p3 display are also true for display on p8. Perhaps the most striking difference between using either of the two capsid proteins as the display platform is display valency: whereas (poly)peptides fused to p3 can be displayed in up to five copies per virion, p8 phage display enables presentation of hundreds (type 88) or thousands of peptides (i.e., on all copies of p8; type 8) on a single phage. Again, the use of a different display format (i.e. 8+8 phagemid system) allows for oligomeric display of larger proteins. It is important to stress that affinity selections from high-valency display libraries (e.g., 88 or 8+8) as a rule lead to low affinity ligands due to avidity effects. With the use of type 8 display format, an extreme of such phenomena can be observed. Dense packing of peptide-p8 fusions is responsible for forcing the displayed peptides into a particular conformation. Thus, when synthesized as soluble entities, selected peptides are unlikely to retain binding activity. Peptide libraries based on type 8 vectors are meant for special applications (where whole phage particles are to be used as ligands [24]) and are thus seldom used.
Alternative phage display formats
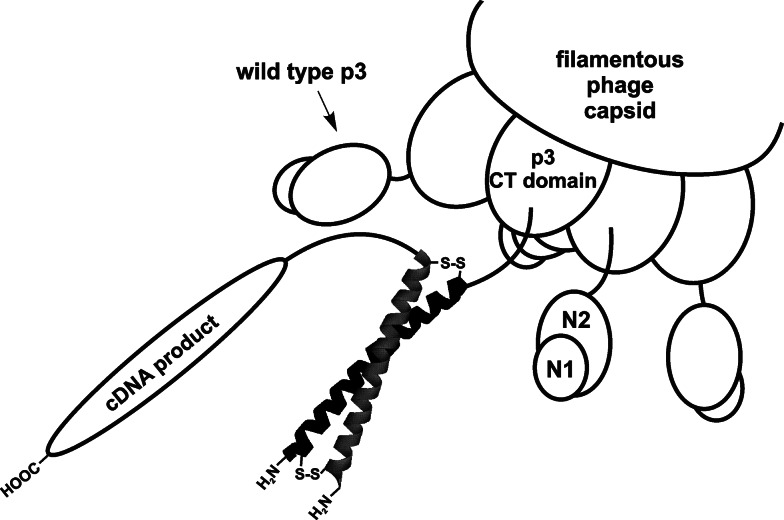
In all display formats discussed so far, polypeptides are fused to the surface exposed N-terminus of either p3 or p8. However, conventional phage display does have certain limitations. It is incompatible with presentation of cDNA protein products with inherent stop codons being present at the 3′ portion of the gene transcript. Capsid proteins p3 and p8 require a leader peptide sequence at the N-terminus for trafficking to the periplasm. Therefore, any fusions have to be inserted in the correct reading frame between the leader sequence and mature coat protein, which makes construction of gene fragment libraries rather inefficient. Also, the free C-terminus of a protein interacting with a specific target might be essential in some cases. To this end, Crameri and Suter [25] devised a specialized 3+3 display system with two fusion proteins encoded in the phagemid vector pJuFo. pJuFo enables co-expression of (1) c-Jun protein leucine zipper domain, N-terminally displayed on p3 which is integrated into mature phage particles, and (2) a soluble peptide to be displayed fused to the C-terminus of the c-Fos protein leucine zipper domain. Jun and Fos domains associate to form a super secondary structural motif called the leucine zipper (Fig. 4). To avoid interphage exchange of Fos-fusions, cysteins were engineered into Fos and Jun domains at the interacting surfaces thus enabling formation of covalent bonds. The method was used in cloning and affinity selection of the fungus Aspergillus fumigates protein products from a cDNA library [26]. It is noteworthy to emphasize that even with the use of pJuFo display only 1 in 6 cloned cDNA will have the correct orientation and correct reading frame (compared to 1 in 18 with the use of conventional 3+3 display). Alternatively, one can subject inserts to open reading frame selection (i.e., cloning of cDNA fragments upstream of gene encoding β lactamase and plating transformed bacteria on selective media) prior to construction of conventional phage library [27]. Although this approach solves the problem of poor cDNA display in the initial library, the required procedures are somewhat more laborious and the library still encompasses only a fraction of all potential genes.
Fig. 4.
Depiction of p3-displayed cDNA products on a bicomponent Fos-Jun complex. Jun domain (black helix) is fused to minor capsid protein p3 while cDNA proteins are coupled to the C-terminus of Fos-domain (gray helix). Fos and Jun form a coiled coil complex (leucine zipper) reinforced with disulphide bonds
Jespers et al. [28] demonstrated successful presentation of proteins attached to the C-terminus of p6 in a phagemid format (i.e., 6+6), although display efficiency was reported to be two orders of magnitude lower than with a comparable 3+3 format. They constructed a series of three phagemids which provide cloning sites in all three reading frames; the inserted cDNA, however, still oriented randomly.
More recently, direct C-terminal display has also been achieved with p3 [29] and p8 [30] in the phagemid format. Although the p3 and p8 C-termini are believed to be buried in the phage capsid, Sidhu’s group managed to empirically optimize amino acid sequences of the linker connecting them to displayed peptides to reach display levels comparable to conventional N-terminal display.
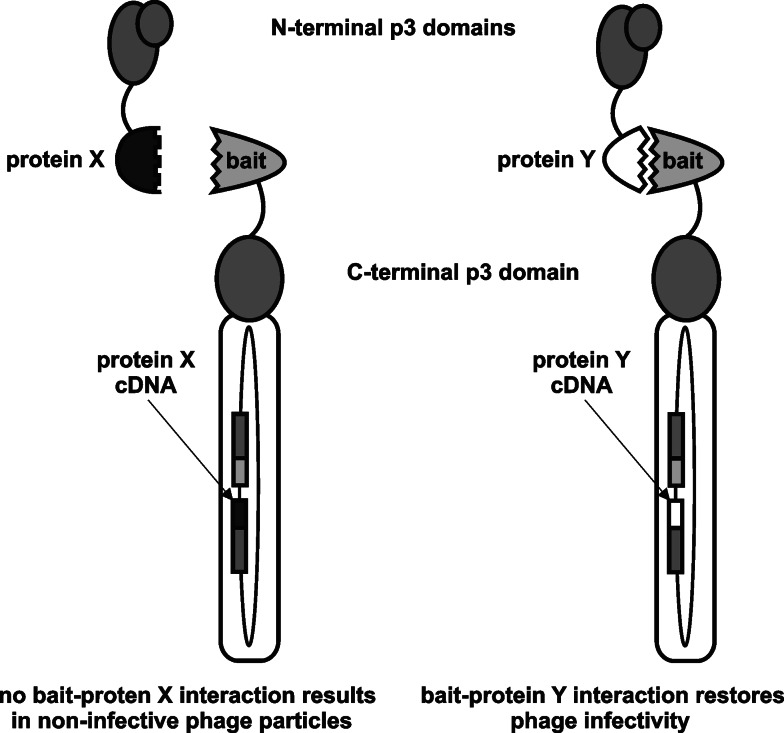
A clever display system for identification of interacting receptor-ligand pairs in vivo, termed direct interaction rescue, was proposed by Schaffner and co-workers [31]. Phagemid-encoded recombinant p3 was separated into two parts with respect to their function (infectivity or structural role). In a reported model system, authors replaced the N-terminal domains of p3 with the c-Jun protein leucine zipper domain, making the phage non-infective, whereas the N-terminal part of p3 was fused to the c-Fos protein leucine zipper domain. Phage were rescued by infecting cells harboring the phagemid with a special p3-defective helper phage. Infectivity was restored upon receptor–ligand (i.e., Jun–Fos) interaction to form a fully functional p3 (Fig. 5). This technology circumvents affinity capture-based screening of phage libraries, vastly eliminating background phage resulting from nonspecific adsorption. Although the utility of direct interaction rescue was later demonstrated by successful co-selection of single-chain variable fragment (scFv)-antigen pairs [32], the method has not gained much popularity. The reasons for that might be low expression of some protein fusions, improper folding in prokaryotic host, and/or incapability of the whole complex or the individual fusion proteins to cross the bacterial membrane during phage assembly, thus inefficiently restoring phage infectivity. Instead, the yeast two-hybrid system [33] is better suited for identification of protein–protein interactions from cDNA libraries, i.e., on the transcriptome-wide scale [34, 35].
Fig. 5.
The principle of direct interaction rescue technique (adapted from [31]). Separate N- and C-terminal halves of p3 are rejoined through interaction of heterologous interacting protein domains, restoring phage infectivity
Minor capsid proteins p7 and p9, located proximately to each other at the distal capsid tip with respect to p3 and p6, have also been employed as display platforms. Gao et al. [36] developed a kind of “7/9+7/9” phagemid display format with heavy- (VH) and light-chain variable regions (VL) fused to the N-termini of p7 and p9, respectively. Heterodimers of VH and VL formed on the phage apex, resulting in functional FV antibody domain. The system is a valuable addition to display formats and should in theory facilitate construction of combinatorial libraries of heterodimeric proteins. Successful N-terminal display of scFv on p9 (in a 9+9 format [37]) and p7 (in a 7 format [38]) alone have also been reported.
An exciting research area in the field of phage display is design of mutated or artificial (i.e., nonessential) coat proteins which get incorporated into phage particles together with non-replaceable wild-type structural proteins. Sidhu’s group devised several new capsid proteins to serve as improved anchoring proteins for more efficient polypeptide display or to serve specific needs in protein presentation [39–44].
In recent years, display on bacteriophage types other than filamentous phage has gained popularity. A major drawback to filamentous phage as the display platform is that certain fusion proteins are not capable of crossing the cell membrane in the course of viral assembly and will therefore not be expressed on the capsid. With lytic species (e.g., T7, T4, or lambda), phage assembly takes place inside the infected cell and progeny phage are released by cell lysis. Indeed, Krumpe et al. [45] demonstrated that T7 phage-displayed random peptide libraries exhibit less sequence bias than filamentous phage-displayed libraries of comparable complexity.
A series of phage vectors for display of peptides and proteins on the surface of T7 phage were developed by Rosenberg et al. [46] and are now commercialized by Novagen, Merck Biosciences. The T7 capsid head has an icosahedral shape and is composed of 415 copies of capsid protein 10. This protein exists in two forms: the shorter 10A and the longer 10B, the latter resulting from a translational frameshift. It was observed that a functional capsid can be made entirely of either 10A or 10B or of various ratios thereof, which led Rosenberg and co-workers to introduce restriction sites at the 3′ end of gene 10, accomplishing display of (poly)peptides as fusions to the C-terminus of the truncated protein 10B. With the upstream cloning of promoters of different strength and the presence of modified translation initiation sites, diverse display valency was achieved. Only relatively short peptides (<50 amino acids) can be displayed at high valency. Display of extremely large proteins (up to 1,200 amino acids) is feasible, but is limited to a few copies per phage. The native protein 10A that complements the capsid is expressed from a plasmid harbored by the host bacteria.
The T4 phage capsid head is also icosahedral, but has a more complex architecture [47]. Faces of the shell are composed of the hexagonally arranged major capsid protein gp23 (product of gene 23) with pentamers of gp24 corner protein at the vertices. Two dispensable proteins symmetrically decorate the head: Soc (small outer capsid protein) forms a continuous intertwined network on the surface of gp23 hexameres, and Hoc (highly antigenic outer capsid protein) is situated in the middle of each gp23 hexagon. Both accessory proteins have been developed as display platforms. Ren and co-workers [48] have developed two T4 Soc display formats. In one display system, fusion to the C-terminus of Soc was expressed separately and combined with the prohead (capsid lacking the wild-type Soc) in vitro to form artificial phage particles. The second display format was based on the in vivo integration of the recombinant soc gene, carried by a positive selection plasmid pRH, into the T4 phage genome. In the pRH vector, soc gene was flanked by specific T4 gene fragments to allow for homologous recombination with T4 genome. Cells housing recombination plasmids were infected by a specially designed recombination-integration phage deleted for wild-type soc to achieve production of fusion viral particles. In a proof-of-principle experiment a short peptide, a small protein domain and a large protein have all been successfully displayed on phage capsid, although display efficiency decreased with polypeptide size [48, 49]. In a similar manner polypeptides fused to the N- or the C-terminus of Hoc can be displayed [49–51]. Of particular interest to structural biochemists is the in vitro T4 display system exploiting both Soc and Hoc proteins as anchor proteins. Recently, display of a macromolecular complex (the anthrax toxin with Mr 710 kDa) on T4 was reported [52]: two bipartite fusion proteins were constructed, the anthrax lethal factor (LF) fused to Hoc, and the N-terminal domain of the LF (LFn) coupled to Soc. Proheads were decorated with Hoc and Soc fusions in vitro. Subsequently, the protective antigen fragment (PA63) was assembled into heptamers via interactions with LFn domain. Finally, the edema factor (EF) was bound to unoccupied sites of PA63 heptamers to yield the complete anthrax toxin. Over 200 of such macromolecular complexes were attached to a single phage particle, increasing the capsid mass by 2.7-fold. Such robust display systems could potentially be used for production of multicomponent vaccines.
Another platform in T4 display is the fibrous α-helical protein fibritin [53]. This product of wac gene (whisker antigen control) forms a collar (so-called “whiskers”) in the head–tail joining region of the virion. It is present in 6 copies per phage particle, enabling low valency display in contrast to Soc and Hoc (810 and 155 copies per T4 capsid head, respectively). A polypeptide of 53 amino acids was reported effectively displayed fused to the C-terminus of fibritin [53].
Bacteriophage lambda is yet another alternative to filamentous display vectors. Its icosahedral capsid head is composed of 415 copies of the gpE (product of gene E). 135 to 140 trimers of gpD bind the prohead to stabilize the capsid. A tubular tail made from gpV hexamer rings is attached to the head of the virion [54]. Polypeptides have been displayed on the surface of lambda phage attached to gpD and gpV. Sternberg and Hoess [55] reported a site-specific recombination technique to construct phage lambda libraries of polypeptides fused to the N-terminus of gpD. gpD fusions with specific recombination sites at the 5′ and 3′ gene ends were encoded on a plasmid. Bacterial hosts expressing recombinase and containing the plasmids were exposed to the recombination prone phage lacking the wild-type gD. After in vivo recombination, phage carrying the fusion gD genes were produced. Later, Mikawa et al. [56] demonstrated that displayed proteins can also be fused to the C-terminus, even though both gpD termini are buried in the phage capsid. As could be expected, display efficiency decreased with protein size, probably as a consequence of coat protein proteolysis. Large proteins (e.g., β-galactosidase and agglutinin from the tree Bauhinia purpurea whose sizes are 465 and 120 kDa, respectively) have been displayed fused to the C-terminus of gpV [57]. To enable functional tail formation (i.e., prevent detrimental steric hindrance of gpV fusions), the amber stop codon was introduced between the gV and the gene encoding the protein to be displayed. This allowed for expression of conditional fusions in an amber suppression bacterial strain, resulting in a mosaic phage tail. Display of shorter peptides, on the other hand, was well tolerated at all 192 copies of gpV [58].
Further technical improvements
Creating libraries of high complexity
The fate of affinity selection is determined by numerous factors, making systematic empirical optimization of selection conditions virtually impossible. Perhaps most importantly, one should always start with a highly diverse library and check that the cloned genes are efficiently displayed on the phage capsid. In general, the higher the diversity (i.e., the repertoire of phage-displayed polypeptides), the higher the success rate of attaining high affinity ligands. Creating complex libraries at the nucleic acid level using (synthetic) fragment ligation, error-prone PCR, or overlap extension PCR is relatively straightforward. However, it is the inefficiency of transformation of the genetic information into the bacterial host that severely limits phage-displayed (poly)peptide diversity. This section provides a short overview of the attempts to enhance the complexities of phage display libraries using innovative cloning strategies and taking advantage of recombination techniques.
In some cases, e.g., with antibody fragments (Fig. 6), the modular structure of the displayed proteins facilitates generation of highly complex libraries. Antibody libraries are constructed by reverse-transcribing and PCR amplifying genes encoding antigen-binding domains of heavy (VH) and light (VL) chains from lymphocyte total RNA. By combining different VH and VL, a process termed chain-shuffling, unique antibody fragments are formed. Traditionally, this was achieved by randomly combining the V-genes through fragment PCR assembly and subsequent subcloning of the scFv gene constructs [59], or by sequential cloning of the genes encoding the light and heavy chains into display vectors [60]. To raise a reasonably complex library, many parallel ligation reactions and electroporation steps were necessary. Hoogenboom et al. [61], on the other hand, cloned heavy chain repertoires into a phagemid (for soluble protein expression), while inserting the light chain genes into a phage vector as fusions to gIII. An E. coli host strain was transfected with the phagemid library and subsequently infected with the phage library (functioning as a helper phage), giving rise to functional Fabs displayed on phage particles. Since infection is extremely efficient, combinatorial diversity of Fab fragments was only limited by the number of bacterial cells in the culture. A downside to this approach is that the infected bacteria produced both packaged phage and phagemids. Although both types of virions displayed light and heavy chains, they only carried genetic information for one of the chains. The genes for both antibody chains thus had to be recovered separately by plating the bacteria harboring them on selective media.
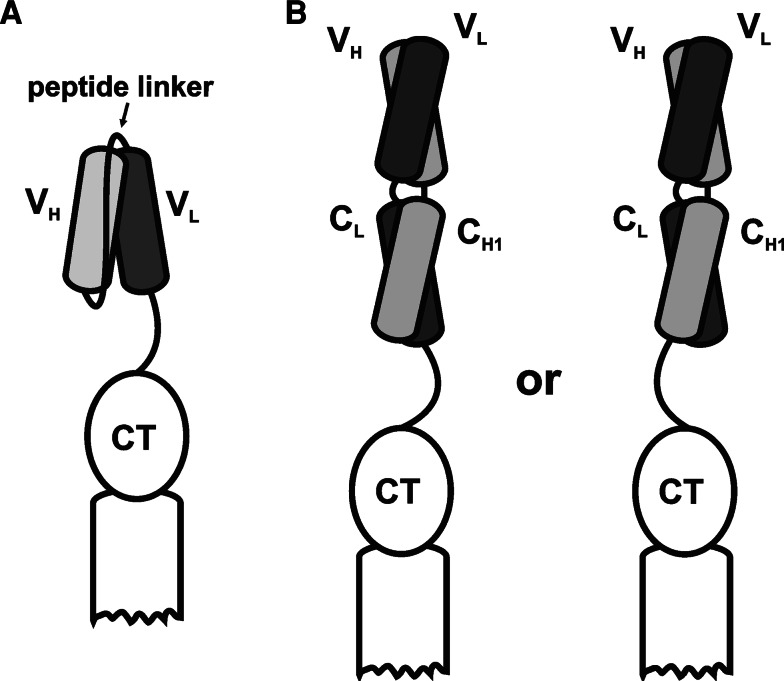
Fig. 6.
Schematic representation of antibody fragment types displayed as fusions to p3: a single-chain variable fragment (scFv); b antigen-binding fragment (Fab) with light chain-p3 fusion (left) or heavy chain-p3 fusion (right)
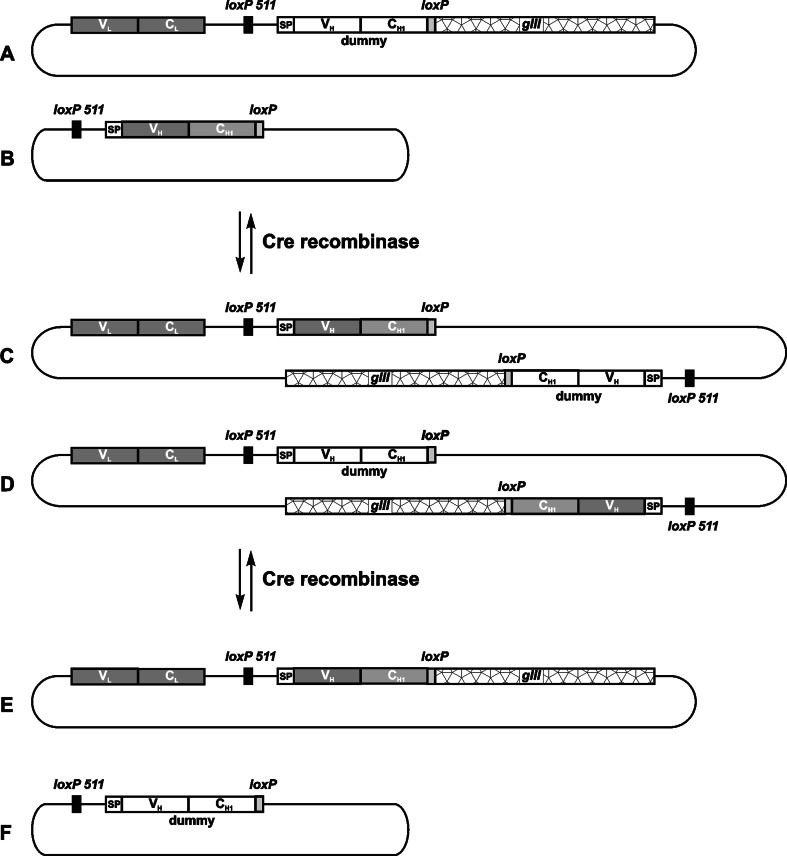
Waterhouse et al. [62] proposed in vivo recombination for enhancing the diversity of the phage-displayed antibody libraries. In a proof-of-concept experiment, the Cre-lox site-specific recombination system of phage P1 [63] was used to lock together the heavy and light chains encoded on a plasmid and a phage vector, respectively (Fig. 7). On both vectors, the heavy chain genes were floxed (i.e., flanked by heterologous loxP sites). Bacteria were transfected with the plasmid and infected with the recombinant phage. Upon Cre recombinase expression (provided by superinfection with P1 phage), two new vectors were formed, one of which encoded the functional Fab fragment (i.e., the combination of the relevant light and heavy chains). A similar study was reported by Tsurushita et al. [64] who constructed a functional phagemid-displayed scFv via the Cre-lox recombination system. Here, the loxP sequence was used to encode a part of the peptide linker connecting the VH and VL domains. The same principle was later applied to create large phage-displayed antibody libraries [65–67]. Further improvement of the method came from including a selection marker (i.e., an antibiotic resistance gene), the expression of which is only activated after the correct recombination event takes place, thereby enabling elimination of non-relevant background phage particles from libraries [66, 67].
Fig. 7.
The principle of chain shuffling by in vivo recombination using the Cre-lox recombination system as reported by Waterhouse et al. [62]. Two replicons enter recombination: the acceptor fd phage vector (a), carrying the relevant light chain gene (VL–CL) and a dummy heavy chain gene (VH–CH1_dummy) fused to gIII, and the pUC19 donor plasmid (b), encoding the relevant heavy chain gene (VH-CH1). On both vectors, heavy chain genes are flanked by heterologous recombination sites (loxP and loxP 511). In the presence of Cre recombinase, chimaeric vectors are formed (c,d), which can undergo further recombination to yield either the original replicons (a,b) or two new vectors (e,f). Newly formed fd vector (e) now carries genetic information for a functional antigen-binding fragment
Sblattero and Bradbury [68] devised a more efficient in vivo recombination method to produce large scFv libraries. Their technique relies on a single phagemid vector. In this way, all the recombination products are functional. First, a primary library of scFv is constructed with floxed VH genes. An easily attainable small library (<108 members) is sufficient. V genes are then recombined in the host bacterial cells expressing Cre recombinase. However, there is a problem with infecting multiple phagemids (each encoding a different combination of VH and VL) into a single bacterial cell, a prerequisite for recombination to take place. This is solved by using a high excess of phage particles over bacteria while repressing expression of p3. Phage particles rescued after recombination encapsulate a single phagemid, but their capsid is not necessarily composed only of structural proteins encoded by that phagemid. Therefore, phage have to be isolated and used to infect bacteria not expressing Cre recombinase at a multiplicity of infection of less than 1 to couple phenotype and genotype. In a report by Cen et al. [69], a slightly different approach to increase scFv library complexity is described. The authors used the Cre-lox system for in vivo recombination of variable domain genes whose CDR3 sequences were previously fully randomized by cassette replacement. Multiple phagemids (presumably 9 per cell) were infected into bacterial host cells as concatemers. For that, phagemids had to be modified to carry λ packaging signal that was used for in vitro packaging of concatemers into λ phage capsids. Concatemers were constructed by cutting phagemids with a specific restrictase and re-ligation at high concentration.
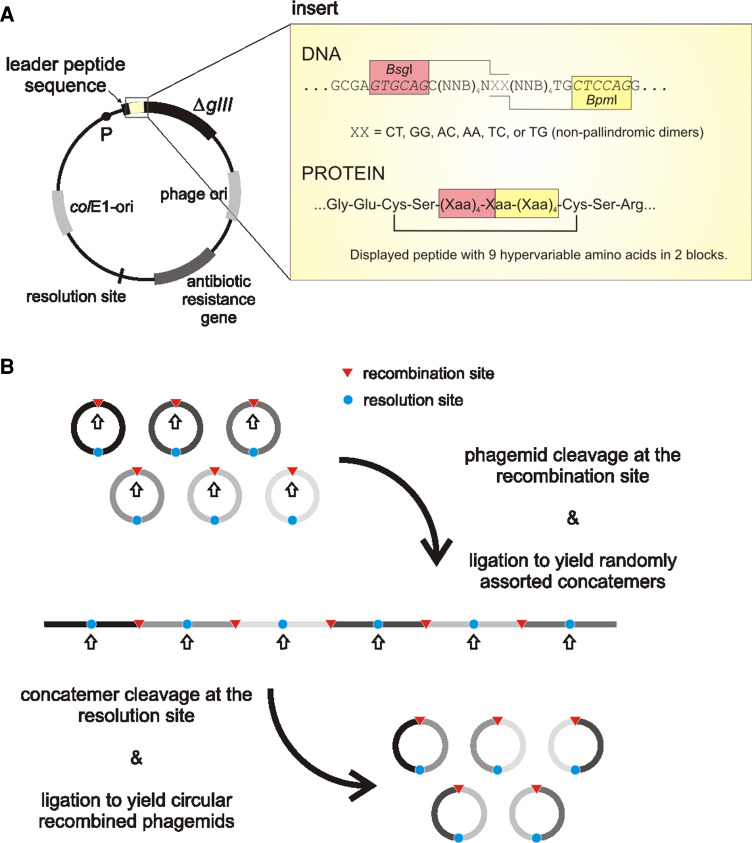
An elegant method of accessing extremely high diversity of displayed (poly)peptides termed cosmix-plexing was developed by Collins and Röttgen [70] (see also the review by Collins et al. [71]). A primary library is first constructed using a phagemid vector (to ensure monovalent display) in such a way that the insert coding for the displayed randomized polypeptides is divided into several fragments (i.e., hypervariable DNA cassettes), separated by restriction sites. The library is actually composed of a number of subset libraries, differing in two base-pairs at non-pallindromic restriction sites (termed recombination points) (Fig. 8a). Phagemids also contain a second restriction site (called the resolution site). One or two rounds of affinity selection are initially performed with the primary library to enrich for ligands of weak/intermediate affinity. It is assumed that such ligands share sequence motifs, which will merge upon recombination to form unique ligands exhibiting stronger binding to the target. Therefore, a limited pool of genes encoding (poly)peptides enriched for target binders is recombined in vitro (i.e., cut at recombination points, concatenated, and then recirculized through cleavage at the resolution sites and ligation; Fig. 8b). Finally, the recombined phagemid library is subjected to further rounds of affinity selection. Since, in the primary library, each hypervariable cassette contains all possible permutations of amino acids, longer stretches of polypeptides can be optimized as compared to selections from naïve libraries. Indeed, peptide ligands selected through cosmix-plexing exhibit binding affinities comparable to those of antibodies [71].
Fig. 8.
a General features of a cosmix-plexing phagemid vector. The insert of CPS1 library (a set of six sub-libraries) displaying cyclic (disulphide constrained) decapeptides is depicted as an example. The two hypervariable regions are separated by restriction sites BsgI/BpmI. In vitro recombination is induced by cleavage with either of the restrictases: in any case, a dinucleotide cohesive-end 3′-extension containing the sequence XX is produced. Limiting the sequence XX to one of the six shown, ensures that subsequent re-ligation only produces concatemers with the correct fragment orientation. P Promoter, ΔgIII gene encoding the C-terminal domain of p3, colE1-ori plasmid’s origin of replication, N any nucleotide (A, G, T, or C), B nucleotides C, G or T, Xaa any amino acid. b In vitro recombination procedure (adapted from [71])
Fisch et al. [72] devised an in vivo recombination system (called exon shuffling) for creating highly diverse peptide phage libraries. The Cre-lox P system was used to shuffle hypervariable DNA cassettes (exons), encoding random decapeptides, from a donor plasmid sub-library to an acceptor phage sub-library vector. A self-splicing intron from Tetrahymena thermophila 26S rRNA with an engineered lox P site was inserted between the two exon regions. In this way, the relatively large recombination site (34 nucleotides encoding at least 12 amino acids) was removed from the mRNA upon transcription, thus bringing the two exon regions closer together. At the protein level, the spliced peptide repertoires were separated by a spacer of only five residues.
Addressing the problem of low display efficiency
Phagemid vectors permit surface display of medium and large size proteins on filamentous phage. Superinfection of bacteria harboring phagemids with helper phage initiates extrusion of mosaic phage particles, preferentially packed with phagemid ssDNA. In contrast, type 33 or 88 vectors supply both versions (native and recombinant p3 and p8, respectively) in cis (Fig. 3), which makes amplification of protein-displaying virions more convenient, yet they are rarely used. The main reason for this is their genetic instability (the tendency to delete any unnecessary DNA) and difficulty of manipulation (preparing DNA, cloning, and transfection efficiency) [73]. On the other hand, with phagemid display (especially type 3+3) low display efficiency can often be observed. The majority of progeny phage particles can be of wild-type phenotype, i.e., composed entirely from helper phage-contributed structural proteins. This causes high background of non-displaying phage that cannot participate in affinity selection.
Theoretically, one way to increase display of phagemid-encoded fusion coat proteins is to place them under the control of strong transcriptional promoters. However, such an approach is likely to lead to bacterial stress [74]. Beekwilder et al. [75] reported on an attempt to select trypsin inhibitors from a phagemid library of variants of potato protease inhibitor 2 (PI2). PI2-p3 fusion was transcriptionally controlled by the lac promoter. Although native PI2 is a natural trypsin inhibitor, the authors noted that, instead of true target binders, only mutants carrying deletions or stop codons were recovered. Severe host cell stress resulted in growth advantage of clones encoding truncated fusion proteins. The problem was solved by replacing the promoter with the weaker psp (phage shock protein) promoter, but only at the cost of low display efficiency. Krebber et al. [74] showed that library diversity can be maintained by inserting a strong terminator upstream of the lac promoter governing transcription of p3-fusions from a phagemid. This prevents background expression resulting from independent transcriptional starting points upstream of the lac promoter region and only slightly affects expression upon IPTG induction.
Alternatively, display levels can be improved by using more efficient translation regulators and optimized leader peptide sequences. Jestin et al. [76] partially randomized the 92 base region located upstream of the encoded signal peptide pelB (pectate lyase B) cleavage site of p3. This region is known to affect translational regulation and translocation. Phagemids were rescued by a trypsin sensitive helper phage KM13 [77] which has a trypsin cleavage site engineered between the second and the third domain of p3. Cleavage of p3 destroys the infectivity of those phage particles whose minor coat proteins are encoded solely from the helper phage. Phagemid virions were exposed to multiple rounds of trypsin digestion and reinfection to select for clones with high p3-fusion display efficiency. A clone enabling 50-fold higher display levels of a model protein (Stoffel fragment of Taq polymerase) than the original phagemid was identified.
Phagemid rescue with a p3-deficient helper phage imposes incorporation of recombinant p3-fusions at the capsid tip in up to five copies. In reality, some fusions undergo proteolysis so display efficiency is never 100%. A helper phage called hyperphage (now available from Progen and RD, Fitzgerald Industries Int.) was developed by Dübel and co-workers [78] by completely deleting gIII from the standard helper phage M13K07 (ATTC). A specifically designed bacterial host strain with wild-type gIII inserted in the chromosomal DNA was prepared to allow amplification of hyperphage. With hyperphage, display level of single chain antibody fragments (scFv) fused to intact p3 was demonstrated to be more than two orders of magnitude higher than with the parent helper phage. Enrichment factors in affinity selections were considerably higher leading to fewer selection rounds required to identify antigen binders. Similarly, Kramer et al. [79] constructed a helper phage (called CT helper phage, a derivative of the VCSM13; Stratagene, Agilent Technologies) by deleting the 5′ section of gIII encoding the two N-terminal domains of p3. Two other derivatives of M13K07 with defective p3 have also been reported. Ex-phage [80] and phaberge [81] have point mutation introduced into gIII so that one and two sense codons, respectively, are replaced by amber stop codons. These helper phage can be amplified in amber suppressive host strains. A comparative study confirmed similar display levels of Fab fragments achieved by using hyperphage, ex-phage, or phaberge [82]. However, virion production and percentage of infective phagemid virions were highly dependent not only on the helper phage but also on the host strain and the phagemid used.
While cells housing phagemids have to be complemented with vectors expressing phage proteins in trans, these vectors need not necessarily be helper phage. Chasteen et al. [73] constructed a series of “helper plasmids”, carrying all the M13 genes. The primary origin of replication/packaging signal was replaced by a low copy ori and a chloramphenicol resistance gene was added into the phage nonessential intergenic region. Plasmids differed in gIII to allow production of either intact or truncated p3, or were completely incapable of producing p3. Thereby, packaging cell lines harboring the different plasmid constructs could be established that enabled a simple switch from monovalent to multivalent display on phage with a single library. An additional benefit of relying on described helper plasmids rather than helper phage is that helper phage ssDNA is not produced at all. Although crucial for helper phage amplification in creating viral stocks, partially functional packaging can cause problems in affinity selections. In situations of helper phage overgrowth, high background of mosaic capsid virions (encapsulating helper phage ssDNA; see Fig. 3) competes with phagemid virions for target binding leading to failed selection.
Phage display of recalcitrant proteins
Intuitively, one would expect only the soluble proteins to be effectively displayed on phage. However, there are scarce reports demonstrating the opposite. Olszewski et al. [83] successfully displayed a hydrophobic HIV-1 virulence factor Nef (negative factor) in an 8+8 format. A series of in vitro binding assays with Nef-specific antibodies as well as Nef’s known cellular ligands was conducted to verify that the displayed protein was indeed fully functional. A recent publication [84] coming from the same laboratory describes effective on-phage presentation of an integral monotopic protein (human caveolin-1) and a transmembrane protein (gp41 from HIV) with an 8+8 phagemid system. Even though attempts to produce recombinant eukaryotic membrane proteins in bacteria usually fail, the authors explain their success by the fact that a large phage body should significantly improve the solubility of otherwise extremely hydrophobic proteins. It is worth mentioning that a special helper phage M13K07+ was used for both caveolin and gp41 display. M13K07+ is a derivative of M13K07 with a tetrapeptide (amino acid sequence Ala-Lys-Ala-Ser [85]) inserted near the N-terminus of surface exposed p8. This modification, primarily exploited for disrupting non-specific binding between the negatively charged capsid and high pI target proteins, further augments phage particle solubility.
Zanghi et al. [86] noted that certain proteins were not well tolerated when trying to display them at high copy number as fusions to gpD major coat protein of lambda phage. To this end, they designed a dual plasmid-based expression system (encoding the wild-type and recombinant gpD, respectively) to co-complement lambda lysogens devoid of gD. Successful display of a previously problematic protein (a mutant of the tenth fibronectin type III domain) was achieved upon lysogen induction. Due to lower display levels, no interference of the protein with capsid assembly was observed that would manifest itself in reduced phage titers or loss of infectivity.
Widening the chemical space of phage-displayed libraries
In phage libraries each displayed peptide is specified by the corresponding nucleotide insert. This provides the means for construction of large pooled libraries with all the components (i.e., phage clones) mixed together, and greatly simplifies identification of individual screening hits. However, dealing with biological systems, chemical diversity of polypeptides is restricted to only 20 naturally occurring amino acids. In contrast, synthetic peptide libraries are typically far smaller than their biologic counterparts and the process of deconvolution is more tedious as these libraries are usually not encoded. Importantly, however, synthetic libraries can encompass a more diverse set of amino acids and their surrogates, thereby precisely defining peptide conformations which can lead to increased target–ligand affinity. Additionally, novel structural features can protect peptides from proteolysis or provide reactive sites for irreversible binding to receptor active sites.
Two proof-of-principle studies have been conducted to show that integration of genetically encoded unusual amino acids in phage-displayed peptides is feasible [87, 88]. For a bacterial host to recognize a certain codon and incorporate an unnatural amino acid in its respective place, it has to express the specific mutant tRNA and aminoacyl-tRNA synthetase as well as grow on medium supplemented with the corresponding synthetic amino acid (or a suitable precursor). Using this approach, Tian et al. [87] demonstrated successful incorporation of o-methyltyrosine, p-azidophenylalanine, p-acetylphenylalanine, p-benzophenylalanine, and 3-(2-naphtyl)alanine into a defined position of a decapeptide on p3 of M13 filamentous phage. Sandman et al. [88] managed to display selenopeptides (containing selenocysteine) fused to p3 of M13 phage using the TGA codon and selenocysteine insertion sequence.
Chemical space can also be expanded by covalent attachment of synthetic molecules to native coat proteins or displayed peptides. Meyer et al. [89] constructed a peptide library attached to the N-terminus of Fos domain which was in turn fused to p3. A synthetic Jun domain with a conjugated small molecule (a known inhibitor of protein kinase A) was prepared and added to the library phage. The small molecule was functionally tethered to the library phage via non-covalent self-assembly of a leucine-zipper heterodimer and used as a warhead to guide displayed peptides to the very proximity of the enzyme’s active site with the goal of bisubstrate analogue inhibitor discovery.
A hybrid system of phage display and combinatorial synthetic chemistry was reported by Cwirla and co-workers [90]. Here, a random nucleotide sequence was introduced into non-expressed regions of filamentous phage and T7 genomes. Individual clones of a “phenotypically silent” phage library were arrayed and grown in microtiter plate wells. A fraction of the phage array was spotted on a nylon membrane whereas the rest was coupled to small molecules via polyethylenglycol linkers (on a one compound–one clone principle). Unique nucleotide sequences were used as codes for small molecules attached to the major coat proteins (i.e., p8 and product of gene 10). Finally, clones were pooled. In one application, a T7 library with 980 folic acid analogues was selected against KB carcinoma cells over-expressing folate receptors at the surface. Endocytosed phage particles were released by cell lysis and amplified in bacterial hosts. Next, radiolabeled probes were synthesized based on the DNA-insert sequences from the extracted virion population and hybridized to archive nylon arrays to deduce the identities of displayed folic acid mimetics.
Dwyer et al. [91] developed a method termed “biosynthetic phage display” in which one part of the protein is displayed fused to the p3 or p8 coat protein, whereas the other part is produced synthetically. Both parts are chemically ligated before subjecting the library to affinity selection. The authors ligated the N-terminal half of eglin (a serine protease inhibitor) with the non-natural amino acids kynurenine or norvaline replacing the buried Tyr25, to the C-terminal half with two randomized positions known to be in close proximity of Tyr25 in the properly folded protein. The library was screened against the immobilized protease genenase to sort out viable eglin variants. Thereby, some surprising residues allowing accommodation of the two nonisosteric amino acids were identified. The biosynthetic display combining peptide synthesis and phage display mutagenesis adds a whole new dimension to protein engineering.
Innovative screening strategies
Two inversely related parameters are typically manipulated in the course of affinity selection—stringency and yield. Stringency, as defined by Smith and Petrenko [23], is “the degree to which peptides with higher fitness are favored over peptides with lower fitness”. As stringency is inversely proportional to yield (“the fraction of particles with a given fitness that survive selection” [23]), selection conditions should be tailored towards lower stringency in the first selection round [where there are typically only ~100 phage clones representing each library (poly)peptide] and then gradually intensified towards higher stringency with ongoing selection cycles (performed with amplified phage subpopulations). Most common parameters to manipulate are the duration of incubation of library phage with target (to steer selections on the basis of interaction kinetics [92]), conditions of washing steps (e.g., detergent concentration, pH value of washing buffer [93–95], or incubation time [96]), and target concentration/density (less target available for phage binding imposes higher stringency [97]). Elution conditions as a critical parameter affecting the outcome of affinity selection are often overlooked by researchers. Ideally, one would use high-affinity, highly specific ligands to the employed target as eluents, which would, together with the optimized washing conditions, guarantee selective binders. Unfortunately, in most cases, such eluents are not available and a non-specific elution step has to be attempted. In a model experiment, our group has recently demonstrated that the widely accepted non-specific elution buffer 0.2 M Gly·HCl, pH 2.2 used with filamentous phage libraries was unable to break the interaction between streptavidin and a cyclic peptide previously selected through biotin elution [98]. However, when the streptavidin-coated microwell was treated in the same buffer with ultrasound, the phage clone was efficiently detached (most likely due to target protein desorption). Moreover, following rigorous washing under acidic conditions, a specific streptavidin binder was isolated from phage library after a single selection round with the help of sonication.
Optimization of screening conditions, which have to be empirically determined for each target individually, and characterization of individual clones after completed selection can be a laborious process. Recently, biosensors such as Biacore (based on surface plasmon resonance) [99, 100] and quartz crystal microbalance apparatus (based on piezoelectric effect) [101, 102] have emerged as screening devices. These biosensors have extremely high sensitivity and permit monitoring of phage-target interactions in real time, thereby greatly simplifying optimization of washing and elution conditions. In addition, specific binders can be acquired in a single round of selection, significantly reducing experimental procedures.
A number of publications deal with reducing hands-on and total experimental time with phage library selections and characterization of the individual selected clones. Vanhercke et al. [103] developed a procedure that they termed RISE (rescue and in situ selection and evaluation). Their method eliminates the need to purify the amplified phage pool before each panning round. Instead, phagemids are rescued, selected, and evaluated for binding in a single microtiter well. For that, host cells harboring phagemids have to be grown in a microwell precoated with the target of interest. After the phagemids are rescued via helper phage infection, the progeny phage particles that are released into the medium are directly captured by the target. Washing steps remove non-binding phage as well as host bacterial cells, and the bound phage can be immediately detected in an ELISA assay and/or eluted to enter a new selection cycle. The procedure allows for simple monitoring of selection progression as well as omits some of the steps traditionally performed, such as growing phage in large culture volumes and precipitating and isolating phage particles. It is also amenable to evaluation of single phage clones and has the advantage of being automated. Similarly, Ladner and co-workers [104] devised a protocol called URSA (ultra rapid selection of antibodies) that allows up to five selection rounds to be performed in a single day. Their method is based on shortening the filamentous phage amplification steps, as only progeny phage produced from one or two extrusions are used as input for the next selection cycle. Input phage are extruded in the presence of the target, therefore the standard overnight propagation and phage purification steps are avoided. Morohashi et al. [105] used real-time PCR to quantify phage instead of performing traditional microbiological titration that tends to be imprecise and time-consuming. Combining this approach with a single-step affinity selection on an avidin agarose gel column, they successfully enriched peptide binders to a biotinylated small molecule (NK109, a benzo[c]phenanthridine derivative) from a T7 phage library. More recently, Mattiason and co-workers [106, 107] introduced cryogel monolith chromatography to phage affinity selections, coining the phrase chromato-panning. Cryogel monolith columns are produced by gelation of polymers at subzero temperatures. Resulting hydrogels are macroporous and can accommodate large biological particles, such as phage and bacteria (or even eukaryotic cells), while having an immense active surface. The authors report selection of strong peptide binders to human lactoferrin covalently coupled to the column after a single panning cycle involving elution with on-column cell infection. In addition to reducing experimental time, affinity selection in a chromatographic mode also has the advantage of avoiding phage amplification between successive panning rounds, which inherently introduces bias into the phage population due to the variability of growth characteristics of different phage clones.
Sufficient target protein production can also be the bottleneck of the otherwise high-throughput ligand selection from phage libraries. Target biotinylation and subsequent coating on streptavidin-covered surfaces can be of benefit when trying to limit target protein consumption. Castillo et al. [108], on the other hand, displayed target peptides on the T7 capsid (by using high display valency vector T7Select415) and screened a filamentous phage library of scFv directly against the adsorbed whole T7 phage clones. Thus, target expression time was cut down to merely 2 h, and its isolation was confined to simple phage precipitation with polyethylene glycol. Subtractive panning steps with T7 phage displaying no foreign peptide prior to incubation of library M13 phage with T7 fusion phage (the actual target) were performed to avoid selection of anti-T7 scFv. As T7 phage are incapable of infecting male (F pilus containing) host cells [109] required for filamentous phage propagation, no cross contamination was observed. Bowley et al. [110] took a step further to develop a dual display system for identification of antibody–antigen pairs by library-against-library screening. In a proof-of-concept study, they panned a mocked library of antigen-displaying phage against a mocked library of scFv-displaying yeast cells. The phage library was enriched for library yeast cell binders in two conventional in vitro selection rounds before separating the yeast–phage clonal pairs by flow cytometry. The authors further envision a reversal of libraries, i.e., display of antigens on yeast cells and antibody fragments on phage, as yeast display systems support the presentation of extremely large and glycolsylated proteins. Such an approach should permit the saturation of whole proteomes with antibodies.
Often, a phage display project is launched with the ultimate goal of drug development. Selected (poly)peptides typically bind to biologically relevant sites on the target protein, thus acting as modulators of protein function [111]. However, peptide-based drugs suffer from metabolic instability and potential immunogenicity. Attempts to design non-peptide drugs (i.e., peptidomimetics) based on the active peptide structure are arduous and only rarely fruitful. To this end, Schumacher et al. [112] ingeniously panned a phage library expressing L-amino acid peptides against a synthetic SH3 domain of c-Src, composed entirely of D-amino acids. The selected peptide was then synthesized in the D-amino acid configuration and was demonstrated to interact with the target protein of the natural handedness.
In contrast to traditional panning procedures where a subset of the phage population is captured from a library based on the affinity of the displayed (poly)peptides towards a target, Hawinkels et al. [113] when screening for protease inhibitors relied on the selection pressure exerted by proteases to inactivate or degrade phage particles. They incubated peptide libraries of type 3+3 and type 8+8 with either proteinase K or cathepsin S. A protease inhibitor was added to the mixture to inactivate the protease activity and stop the selection procedure, and intact virions were amplified as an input for the next selection cycle. Surprisingly, low valency display on p3 was sufficient to effectively prevent certain phage clones of being inactivated and yielded peptide inhibitors with somewhat lower IC50 values.
Recent exciting applications of phage display technology
Applications of phage display have been extensively reviewed elsewhere [23, 114–132] and will not be discussed here in detail. The technology, often requiring creative modifications, was used for epitope mapping of antibodies [114], screening for receptor agonists and antagonists [115, 133, 134], in vitro evolution of antibodies [116, 117] and antibody surrogates in the form of randomized fragments on diverse scaffold proteins [118, 119], discovery of enzyme substrates [120, 121] and inhibitors [113, 122, 135], identifying functionally coupled proteins and analysis of protein–protein interactions [123, 124], designing catalytic antibodies (abzymes) and enzymes with novel specificities [125], improving proteolytic and folding stability of muteins [126], vaccine design [131, 132], and construction of gene delivery vehicles [127]. Protein ligands selected from phage libraries as well as whole recombinant phage have been used as affinity matrices in chromatographic techniques [128, 129] and biosensing [130]. This last section briefly discusses selected exciting applications of the technology, complementing previous reviews.
Discovery of biopharmaceuticals
According to the latest report by Visiongain [136], therapeutic monoclonal antibodies (MAbs) total sales reached almost US$32 billion in 2008, representing more than 30% of the global biopharmaceutical drug market. Traditionally, therapeutic antibodies were developed from murine IgGs (produced by hybridoma technology [137]) which needed to be modified in order to reduce immunogenicity in man. Using genetic engineering, antigen-binding variable regions from the immunized animal were fused to human constant region domains to produce chimeric antibodies, or the complementarity determining loops (CDRs) of murine IgG were grafted onto a human antibody framework to yield humanized antibodies. However, immunogenicity of therapeutic antibodies remained a serious problem until methods for developing fully human recombinant MAbs had emerged. There are now two well-established platforms for direct discovery of human sequence antibodies. Antibodies can be retrieved from phage libraries, constructed from human peripheral blood mononuclear cells-isolated antibody genes. These libraries are usually naïve (i.e., nonimmune), as immunization of human subjects in unethical, but can also be made (semi)synthetically by grafting random peptide sequences in place of CDRs [138]. The second approach relies on immunization of transgenic mice that express human antibody repertoires [139].
Adalimumab (Humira), the first fully human MAb to enter the clinic, targets tumor necrosis factor alpha (TNFα), thereby blocking its inflammatory effects. It is indicated for the treatment of moderate and severe forms of rheumatoid arthritis, polyarticular juvenile idiopathic arthritis, psoriatic arthritis, ankylosing spondylitis, Crohn’s disease, and plaque psoriasis. Adalimumab was developed using an innovative phage display screening technique guiding the selection of human antibody light and heavy chains, successively, from a rodent monoclonal antibody as a template [140]. First, the heavy chain of a rodent antibody directed against TNFα was cloned in pair with a repertoire of human light chains to display combined human–mouse Fab fragments on filamentous phage and select for optimal TNFα binders with respect to the human light chain. Next, selected human light chains were displayed on phage in pairs with a repertoire of human heavy chains and the Fab fragments were again subjected to affinity selection against TNFα, this time yielding fully human TNFα-binding Fabs. One of the optimized antibody fragments was subcloned for expression in Chinese hamster ovary cell culture as a glycosylated human antibody. An excellent recent review by Lonberg [141] discusses additional 16 phage-derived human antibodies (as well as 47 MAbs from transgenic mouse platform, including a registered anticancer drug panitumumab, Vectibix, an anti-human epidermal growth factor receptor IgG2) that are at the moment being evaluated at various stages of clinical trials.
Phage display has also had a significant impact on direct development of biopharmaceuticals other than MAbs. A prominent example is ecallantide (DX-88), a small-protein inhibitor of plasma protease kallikrein based on the first Kunitz domain of the human tissue factor pathway inhibitor [142, 143]. Ecallantide is currently undergoing clinical trials for two indications: hereditary angioedema [144, 145] (a rare genetic disorder characterized by spontaneous periodic attacks of oedema in various parts of the body as a result of C1 esterase inhibitor deficiency) and for reduction of blood loss during on-pump cardiothoracic surgery [145].
Discovery of conformation- and superstructure-specific ligands
It has well been recognized that ligands selected from phage-displayed libraries can interact specifically with target macromolecules of distinct conformations. Although this feature can pose a problem when trying to screen for functionally active modulators of target protein function (as some conformers might act as decoys), it can be of benefit if one is able to stabilize the target molecule to a particular form. Nizak et al. [146] used a GTP-locked form of inactive mutant of Rab6, a small GTPase, as a target antigen to screen for conformation-specific scFv from a phage library. One of the selected antibody fragments interacted preferentially with the form of Rab6 that was preactivated by GTP-γ-S, a slowly hydrolysable analog of GTP, indicating specificity for the GTP-bound conformation in vitro. The recombinant scFv also specifically detected Rab6·GTP by immunofluorescence in HeLa cells in vivo. Ultimately, the scFv was fused to enhanced yellow fluorescent protein and expressed in HeLa cells as an intrabody, which allowed for spatiotemporal monitoring of endogenous Rab6 activation by GTP binding in live cells. More recently, Wells and co-workers [147] reported a strategy for identifying conformation-specific antibodies by using small molecule ligands to lock target proteins into the conformation of interest. They demonstrated the applicability of the approach by identifying two scFvs, each recognizing a distinct conformation of caspase-1, induced by an active-site and an allosteric inhibitor, respectively. Corresponding Fab and IgG derivatives were used for specific immunocytochemical detection of catalytically competent and inactive forms of the enzyme. The authors proposed this method as a general strategy for generating highly specific inhibitors (and possibly activators) of proteins.
While trying to isolate ligands of human IgG-Fc region from T7-displayed library of random peptides, Sakamoto et al. [148] accidentally stumbled upon a series of peptides selectively recognizing a conformation that was induced by acidic treatment of IgG during the isolation process. This non-native conformer, which maintained the ability to bind antigen and the Fc receptor, but was shown to be prone to aggregation, was effectively depleted from IgG preparations using one of the synthetic peptides as affinity matrix in column chromatography.
Phage library-derived antibody fragments specific for different oligomeric forms of the same protein have also been reported. Sierks and co-workers [149, 150] identified two scFvs against aggregates of α-synuclein, a major component of Lewy bodies found in brains of patients affected by Parkinson’s disease. The antibodies were found to recognize two distinct oligomeric forms of α-synuclein prepared in vitro, the morphology of which was verified by atomic force microscopy. Both scFvs inhibited α-synuclein aggregation in vitro and blocked cytotoxicity of α-synuclein aggregates in human neuroblastoma cell lines. The antibody against the larger later stage oligomeric form [150] also bound selectively to naturally occurring aggregates in Parkinson’s patients’ brains. Such agents hold promise in diagnosis and therapy of Parkinson’s disease.
Development of artificial transcription factors
Transcription factors bind to specific stretches of nucleotide sequence on regulatory segments of DNA to induce RNA polymerase attachment and subsequent mRNA transcription. The Cys2/His2 zinc finger motif (i.e., domain of ~30 amino acids with a characteristic ββα fold stabilized by hydrophobic interactions and chelation of a zinc ion) is one of the most common structural motifs of DNA binding proteins found in eukaryotes. Its α-helix protrudes to the major groove of the DNA double helix and is accountable for recognizing three consecutive DNA bases. Transcription factors are typically composed of a string of DNA binding domains, allowing for highly specific recognition of longer nucleotide sequences.
The modular structure of zinc finger transcription factors has inspired Barbas and co-workers to design a series of elegant experiments in which zinc finger domains capable of distinguishing among different nucleotide triplets were identified [151–154]. Libraries of phage-displayed zinc finger muteins were screened against synthetic hairpin oligonucleotides with specific target sequence (i.e. individual members of the 5′-GNN-3′, the 5′-ANN-3′, the 5′-CNN-3′ and the 5′-TNN-3′ families of triplets). To prevent enrichment of non-specific DNA binders and guide selection towards the relevant target triplet, sheared DNA and competitor hairpin oligonucleotides containing the decoy sequence NNN, respectively, were added to the phage–target mixture. Recombinant zinc finger domains selectively recognizing all or most 5′-GNN-3′ [151], 5′-ANN-3′ [152], 5′-CNN-3′ [153], and 5′-TNN-3′ triplets [154] were identified. Polydactyl proteins composed of six zinc-finger domains were then fused to regulatory domains. Such artificial proteins targeted specific regulatory sites on chromosomes and were capable of transcriptional regulation of reporter as well as selected endogenous genes [152–154].
Phage display at the crossroads with inorganic surface chemistry
Everyone working with phage display has probably faced failed selections due to enrichment of clones adhering to supports used for target immobilization. The observation that certain peptides preferentially adsorb to polymeric materials has led researchers to screen random peptide libraries against a range of organic and inorganic materials (reviewed in [155]). There are at least two major utilizations of peptides with affinity to inorganic materials [155]: one is to functionalize materials which are not modified easily (i.e., introduction of moieties such as amino-, carboxy- or hydroxyl-groups is difficult), and the other is to trigger nucleation and stimulate growth of nanocrystals.
Khoo et al. [156] panned numerous phage-displayed combinatorial peptide libraries against a titanium alloy used for manufacturing of orthopedic devices. The tripeptide motif KHK was identified as the main contributor to titanium binding. A synthetic peptide containing repeats of the KHK motif separated by diglycine linkers was prepared and demonstrated to selectively adsorb to the titanium surface from dilute aqueous solutions. The PEGylated analogue of the peptide spontaneously formed a hydrophilic film on the metal surface, thereby effectively preventing nonspecific protein adsorption and bacterial colonization, two common problems accompanying orthopedic implantation procedures. Phage library-derived peptides have also been developed with the goal of promoting cell adhesion to inorganic materials. Segvich et al. [157] identified three diverse peptides that preferentially adsorb to apatite-based substrates, bone-like minerals used as scaffolds in bone tissue engineering. These peptides, when fused to specific cell-binding recognition sequences, provide the means to promote cell attachment, proliferation, and ultimately differentiation into true bony tissue.
Inorganic nanocrystals are an important component of modern electronic devices. The observation that macromolecules in biological systems can direct growth of inorganic crystals has stimulated researchers into developing protein tools for controlled assembly of nanostructures from solutions. By phage library screening Tomczak et al. [158] identified a peptide that binds to and directs the growth of ZnO hexagonal nanocrystals. Moreover, by altering the peptide’s concentration, the nanocrystal morphology could be tailored. A recent publication from the Belcher laboratory [159] describes the use of genetically engineered filamentous phage for self assembly into microelectrodes. Tetraglutamate was displayed in the type 8 format and viruses were left to adhere onto multiple bilayers of linear-polyethylenimine/polyacrylic acid deposited on polydimethylsiloxane surface in micrometer islands. The assembled phage film was dipped into solution of cobalt chloride for cobalt nucleation on viral templates. Spontaneous oxidation of cobalt ions resulted in deposition of cobalt oxide nanocrystals on the phage capsid, yielding plannary arranged cobalt oxide nanowires. Viral nanowires served as the active anode material in the battery anode, whereas the polyelectrolyte multilayers functioned as the battery electrolyte. The two battery components were finally stamped onto a platinum microband current collector, generating fully functional electrode arrays. This exciting approach paves the way towards fabrication of high-performance nanostructured batteries.
Conclusions
Over the course of evolution, Nature has developed an amazing repertoire of finely-tuned protein instruments, capable of performing highly specialized tasks. However, even with modern structural techniques, advanced genetic engineering and bioinformatic tools, proteins are not easily amenable to design. Combining rational approach (as in identification of residues that form binding “hot-spots” on the parent protein’s surface) with the power of combinatorial biology and in vitro selection has made the task of engineering proteins for novel functions and specificities a great deal more tangible.
Phage display is the oldest biological display technique developed. It is easy to implement and inexpensive, making it also the most popular. Typically, phage are only used as display vehicles, allowing replication and straightforward identification of the displayed (poly)peptide. Using simple recombinant DNA techniques, vast libraries of protein variants tethered to phage coat proteins can be prepared and screened for specific affinity or activity by mimicking natural evolutionary concepts. In certain cases, however, decorated whole phage can be exploited as structural or functional entities for downstream applications. These include phage biosensors, phage-based vaccines (wherein antigens are either displayed on phage directly, or a recombinant capsid with specific cell-targeting ligands encapsulates a DNA vaccine), affinity chromatographic matrices, and nanoscale templates for bioassembly of inorganic materials.
Phage display as a method for focused molecular evolution has itself tremendously evolved in the two and a half decades since its introduction. Numerous academic and industrial laboratories have adopted the technique and contributed to its advancement. Versatile display vectors, library designs, and innovative screening strategies have been developed to meet specific needs. Most importantly, applications of phage display seem limited only by the imagination of the one practising it. Taken together, the unexhausted flexibility and applicability of this technique assert that a wide range of life and material sciences will continue to benefit from phage display and all of the many related technologies it has inspired in years to come.
Acknowledgments
I am grateful to Miha Kosmač, Mojca Lunder and Matjaž Ravnikar for helpful discussions and critical reading of the manuscript. I apologize to those authors whose work is not mentioned due to space restrictions.
References
- 1.Pennazio S. The origin of phage virology. Riv Biol. 2006;99:103–129. [PubMed] [Google Scholar]
- 2.Sulakvelidze A, Alavidze Z, Morris JG., Jr Bacteriophage therapy. Antimicrob Agents Chemother. 2001;45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maniatis T, Hardison RC, Lacy E, Lauer J, O’Connell C, Quon D, Sim GK, Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- 4.Sargent TD, Wu JR, Sala-Trepat JM, Wallace RB, Reyes AA, Bonner J. The rat serum albumin gene: analysis of cloned sequences. Proc Natl Acad Sci USA. 1979;76:3256–3260. doi: 10.1073/pnas.76.7.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zacher AN, 3rd, Stock CA, Golden JW, 2nd, Smith GP. A new filamentous phage cloning vector: fd-tet. Gene. 1980;9:127–140. doi: 10.1016/0378-1119(80)90171-7. [DOI] [PubMed] [Google Scholar]
- 6.Hines JC, Ray DS. Construction and characterization of new coliphage M13 cloning vectors. Gene. 1980;11:207–218. doi: 10.1016/0378-1119(80)90061-X. [DOI] [PubMed] [Google Scholar]
- 7.Kay BK, Winter J, McCafferty J, editors. Phage display of peptides and proteins. A laboratory manual. San Diego: Academic; 1996. [Google Scholar]
- 8.Barbas CFI, Burton DR, Scott JK, Silverman GJ (eds) (2001) Phage display. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- 9.Clackson T, Lowman HB, editors. Phage display. A practical approach. New York: Oxford University Press; 2004. [Google Scholar]
- 10.Sidhu SS (ed) (2005) Phage display in biotechnology and drug discovery. CRC, Boca Raton
- 11.Aitken R, editor. Antibody Phage Display. Methods and Protocols. New York: Humana; 2009. [Google Scholar]
- 12.Yan X, Xu Z. Ribosome-display technology: applications for directed evolution of functional proteins. Drug Discov Today. 2006;11:911–916. doi: 10.1016/j.drudis.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Lipovsek D, Pluckthun A. In vitro protein evolution by ribosome display and mRNA display. J Immunol Methods. 2004;290:51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Daugherty PS. Protein engineering with bacterial display. Curr Opin Struct Biol. 2007;17:474–480. doi: 10.1016/j.sbi.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Pepper LR, Cho YK, Boder ET, Shusta EV. A decade of yeast surface display technology: where are we now? Comb Chem High Throughput Screen. 2008;11:127–134. doi: 10.2174/138620708783744516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho M, Pastan I. Mammalian cell display for antibody engineering. Methods Mol Biol. 2009;525:337–352. doi: 10.1007/978-1-59745-554-1_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makela AR, Oker-Blom C. The baculovirus display technology–an evolving instrument for molecular screening and drug delivery. Comb Chem High Throughput Screen. 2008;11:86–98. doi: 10.2174/138620708783744525. [DOI] [PubMed] [Google Scholar]
- 18.Michelfelder S, Lee MK, de Lima-Hahn E, Wilmes T, Kaul F, Muller O, Kleinschmidt JA, Trepel M. Vectors selected from adeno-associated viral display peptide libraries for leukemia cell-targeted cytotoxic gene therapy. Exp Hematol. 2007;35:1766–1776. doi: 10.1016/j.exphem.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 20.Bass S, Greene R, Wells JA. Hormone phage: an enrichment method for variant proteins with altered binding properties. Proteins. 1990;8:309–314. doi: 10.1002/prot.340080405. [DOI] [PubMed] [Google Scholar]
- 21.Russel M, Lowman HB, Clackson T. Introduction to phage biology and phage display. In: Clackson T, Lowman HB, editors. Phage display. A practical approach. New York: Oxford University Press; 2004. pp. 1–26. [Google Scholar]
- 22.Smith GP. Preface. Surface display and peptide libraries. Gene. 1993;128:1–2. doi: 10.1016/0378-1119(93)90145-S. [DOI] [PubMed] [Google Scholar]
- 23.Smith GP, Petrenko VA. Phage Display. Chem Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 24.Nanduri V, Sorokulova IB, Samoylov AM, Simonian AL, Petrenko VA, Vodyanoy V. Phage as a molecular recognition element in biosensors immobilized by physical adsorption. Biosens Bioelectron. 2007;22:986–992. doi: 10.1016/j.bios.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 25.Crameri R, Suter M. Display of biologically active proteins on the surface of filamentous phages: a cDNA cloning system for selection of functional gene products linked to the genetic information responsible for their production. Gene. 1993;137:69–75. doi: 10.1016/0378-1119(93)90253-Y. [DOI] [PubMed] [Google Scholar]
- 26.Crameri R, Jaussi R, Menz G, Blaser K. Display of expression products of cDNA libraries on phage surfaces. A versatile screening system for selective isolation of genes by specific gene-product/ligand interaction. Eur J Biochem. 1994;226:53–58. doi: 10.1111/j.1432-1033.1994.tb20025.x. [DOI] [PubMed] [Google Scholar]
- 27.Faix PH, Burg MA, Gonzales M, Ravey EP, Baird A, Larocca D. Phage display of cDNA libraries: enrichment of cDNA expression using open reading frame selection. Biotechniques. 2004;36:1018–1029. doi: 10.2144/04366RR03. [DOI] [PubMed] [Google Scholar]
- 28.Jespers LS, Messens JH, De Keyser A, Eeckhout D, Van den Brande I, Gansemans YG, Lauwereys MJ, Vlasuk GP, Stanssens PE. Surface expression and ligand-based selection of cDNAs fused to filamentous phage gene VI. Biotechnology (N Y) 1995;13:378–382. doi: 10.1038/nbt0495-378. [DOI] [PubMed] [Google Scholar]
- 29.Fuh G, Sidhu SS. Efficient phage display of polypeptides fused to the carboxy-terminus of the M13 gene-3 minor coat protein. FEBS Lett. 2000;480:231–234. doi: 10.1016/S0014-5793(00)01946-3. [DOI] [PubMed] [Google Scholar]
- 30.Fuh G, Pisabarro MT, Li Y, Quan C, Lasky LA, Sidhu SS. Analysis of PDZ domain-ligand interactions using carboxyl-terminal phage display. J Biol Chem. 2000;275:21486–21491. doi: 10.1074/jbc.275.28.21486. [DOI] [PubMed] [Google Scholar]
- 31.Gramatikoff K, Georgiev O, Schaffner W. Direct interaction rescue, a novel filamentous phage technique to study protein-protein interactions. Nucleic Acids Res. 1994;22:5761–5762. doi: 10.1093/nar/22.25.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krebber C, Spada S, Desplancq D, Pluckthun A. Co-selection of cognate antibody-antigen pairs by selectively-infective phages. FEBS Lett. 1995;377:227–231. doi: 10.1016/0014-5793(95)01348-2. [DOI] [PubMed] [Google Scholar]
- 33.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 34.Legrain P, Selig L. Genome-wide protein interaction maps using two-hybrid systems. FEBS Lett. 2000;480:32–36. doi: 10.1016/S0014-5793(00)01774-9. [DOI] [PubMed] [Google Scholar]
- 35.Miller J, Stagljar I. Using the yeast two-hybrid system to identify interacting proteins. Methods Mol Biol. 2004;261:247–262. doi: 10.1385/1-59259-762-9:247. [DOI] [PubMed] [Google Scholar]
- 36.Gao C, Mao S, Lo CH, Wirsching P, Lerner RA, Janda KD. Making artificial antibodies: a format for phage display of combinatorial heterodimeric arrays. Proc Natl Acad Sci USA. 1999;96:6025–6030. doi: 10.1073/pnas.96.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao C, Mao S, Kaufmann G, Wirsching P, Lerner RA, Janda KD. A method for the generation of combinatorial antibody libraries using pIX phage display. Proc Natl Acad Sci USA. 2002;99:12612–12616. doi: 10.1073/pnas.192467999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwasnikowski P, Kristensen P, Markiewicz WT. Multivalent display system on filamentous bacteriophage pVII minor coat protein. J Immunol Methods. 2005;307:135–143. doi: 10.1016/j.jim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Weiss GA, Wells JA, Sidhu SS. Mutational analysis of the major coat protein of M13 identifies residues that control protein display. Protein Sci. 2000;9:647–654. doi: 10.1110/ps.9.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss GA, Sidhu SS. Design and evolution of artificial M13 coat proteins. J Mol Biol. 2000;300:213–219. doi: 10.1006/jmbi.2000.3845. [DOI] [PubMed] [Google Scholar]
- 41.Sidhu SS. Engineering M13 for phage display. Biomol Eng. 2001;18:57–63. doi: 10.1016/S1389-0344(01)00087-9. [DOI] [PubMed] [Google Scholar]
- 42.Roth TA, Weiss GA, Eigenbrot C, Sidhu SS. A minimized M13 coat protein defines the requirements for assembly into the bacteriophage particle. J Mol Biol. 2002;322:357–367. doi: 10.1016/S0022-2836(02)00769-6. [DOI] [PubMed] [Google Scholar]
- 43.Held HA, Sidhu SS. Comprehensive mutational analysis of the M13 major coat protein: improved scaffolds for C-terminal phage display. J Mol Biol. 2004;340:587–597. doi: 10.1016/j.jmb.2004.04.060. [DOI] [PubMed] [Google Scholar]
- 44.Sidhu SS, Feld BK, Weiss GA. M13 bacteriophage coat proteins engineered for improved phage display. Methods Mol Biol. 2007;352:205–219. doi: 10.1385/1-59745-187-8:205. [DOI] [PubMed] [Google Scholar]
- 45.Krumpe LR, Atkinson AJ, Smythers GW, Kandel A, Schumacher KM, McMahon JB, Makowski L, Mori T. T7 lytic phage-displayed peptide libraries exhibit less sequence bias than M13 filamentous phage-displayed peptide libraries. Proteomics. 2006;6:4210–4222. doi: 10.1002/pmic.200500606. [DOI] [PubMed] [Google Scholar]
- 46.Rosenberg A, Griffin K, Studier FW, McCormick M, Berg J, Novy R, Mierendorf R. T7Select phage display system: a powerful new protein display system based on bacteriophage T7. Innovations. 1996;6:1–6. [Google Scholar]
- 47.Iwasaki K, Trus BL, Wingfield PT, Cheng N, Campusano G, Rao VB, Steven AC. Molecular architecture of bacteriophage T4 capsid: vertex structure and bimodal binding of the stabilizing accessory protein. Soc Virol. 2000;271:321–333. doi: 10.1006/viro.2000.0321. [DOI] [PubMed] [Google Scholar]
- 48.Ren ZJ, Lewis GK, Wingfield PT, Locke EG, Steven AC, Black LW. Phage display of intact domains at high copy number: a system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 1996;5:1833–1843. doi: 10.1002/pro.5560050909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren Z, Black LW. Phage T4 SOC and HOC display of biologically active, full-length proteins on the viral capsid. Gene. 1998;215:439–444. doi: 10.1016/S0378-1119(98)00298-4. [DOI] [PubMed] [Google Scholar]
- 50.Ren ZJ, Baumann RG, Black LW. Cloning of linear DNAs in vivo by overexpressed T4 DNA ligase: construction of a T4 phage hoc gene display vector. Gene. 1997;195:303–311. doi: 10.1016/S0378-1119(97)00186-8. [DOI] [PubMed] [Google Scholar]
- 51.Shivachandra SB, Li Q, Peachman KK, Matyas GR, Leppla SH, Alving CR, Rao M, Rao VB. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine. 2007;25:1225–1235. doi: 10.1016/j.vaccine.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Q, Shivachandra SB, Leppla SH, Rao VB. Bacteriophage T4 capsid: a unique platform for efficient surface assembly of macromolecular complexes. J Mol Biol. 2006;363:577–588. doi: 10.1016/j.jmb.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 53.Efimov VP, Nepluev IV, Mesyanzhinov VV. Bacteriophage T4 as a surface display vector. Virus Genes. 1995;10:173–177. doi: 10.1007/BF01702598. [DOI] [PubMed] [Google Scholar]
- 54.Hoess RH. Bacteriophage lambda as a vehicle for peptide and protein display. Curr Pharm Biotechnol. 2002;3:23–28. doi: 10.2174/1389201023378481. [DOI] [PubMed] [Google Scholar]
- 55.Sternberg N, Hoess RH. Display of peptides and proteins on the surface of bacteriophage lambda. Proc Natl Acad Sci USA. 1995;92:1609–1613. doi: 10.1073/pnas.92.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mikawa YG, Maruyama IN, Brenner S. Surface display of proteins on bacteriophage lambda heads. J Mol Biol. 1996;262:21–30. doi: 10.1006/jmbi.1996.0495. [DOI] [PubMed] [Google Scholar]
- 57.Maruyama IN, Maruyama HI, Brenner S. Lambda foo: a lambda phage vector for the expression of foreign proteins. Proc Natl Acad Sci USA. 1994;91:8273–8277. doi: 10.1073/pnas.91.17.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunn IS. Total modification of the bacteriophage lambda tail tube major subunit protein with foreign peptides. Gene. 1996;183:15–21. doi: 10.1016/S0378-1119(96)00400-3. [DOI] [PubMed] [Google Scholar]
- 59.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-U. [DOI] [PubMed] [Google Scholar]
- 60.Barbas CF, 3rd, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterhouse P, Griffiths AD, Johnson KS, Winter G. Combinatorial infection and in vivo recombination: a strategy for making large phage antibody repertoires. Nucleic Acids Res. 1993;21:2265–2266. doi: 10.1093/nar/21.9.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 64.Tsurushita N, Fu H, Warren C. Phage display vectors for in vivo recombination of immunoglobulin heavy and light chain genes to make large combinatorial libraries. Gene. 1996;172:59–63. doi: 10.1016/0378-1119(96)00170-9. [DOI] [PubMed] [Google Scholar]
- 65.Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, Prospero TD, Hoogenboom HR, Nissim A, Cox JPL, Harrison JL, Zaccolo M, Gherardi E, Winter G. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13:3245–3260. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zahra DG, Vancov T, Dunn JM, Hawkins NJ, Ward RL. Selectable in vivo recombination to increase antibody library size—an improved phage display vector system. Gene. 1999;227:49–54. doi: 10.1016/S0378-1119(98)00593-9. [DOI] [PubMed] [Google Scholar]
- 67.Geoffroy F, Sodoyer R, Aujame L. A new phage display system to construct multicombinatorial libraries of very large antibody repertoires. Gene. 1994;151:109–113. doi: 10.1016/0378-1119(94)90639-4. [DOI] [PubMed] [Google Scholar]
- 68.Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotechnol. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- 69.Cen X, Bi Q, Zhu S. Construction of a large phage display antibody library by in vitro package and in vivo recombination. Appl Microbiol Biotechnol. 2006;71:767–772. doi: 10.1007/s00253-006-0334-5. [DOI] [PubMed] [Google Scholar]
- 70.Collins J, Rottgen P (1997) Generation of diversity in combinatorial libraries. EP 9710539.1 (WO 98/33901)
- 71.Collins J, Horn N, Wadenback J, Szardenings M. Cosmix-plexing: a novel recombinatorial approach for evolutionary selection from combinatorial libraries. J Biotechnol. 2001;74:317–338. doi: 10.1016/s1389-0352(01)00019-8. [DOI] [PubMed] [Google Scholar]
- 72.Fisch I, Kontermann RE, Finnern R, Hartley O, Soler-Gonzalez AS, Griffiths AD, Winter G. A strategy of exon shuffling for making large peptide repertoires displayed on filamentous bacteriophage. Proc Natl Acad Sci USA. 1996;93:7761–7766. doi: 10.1073/pnas.93.15.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chasteen L, Ayriss J, Pavlik P, Bradbury AR. Eliminating helper phage from phage display. Nucleic Acids Res. 2006;34:e145. doi: 10.1093/nar/gkl772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krebber A, Burmester J, Pluckthun A. Inclusion of an upstream transcriptional terminator in phage display vectors abolishes background expression of toxic fusions with coat protein g3p. Gene. 1996;178:71–74. doi: 10.1016/0378-1119(96)00337-X. [DOI] [PubMed] [Google Scholar]
- 75.Beekwilder J, Rakonjac J, Jongsma M, Bosch D. A phagemid vector using the E. coli phage shock promoter facilitates phage display of toxic proteins. Gene. 1999;228:23–31. doi: 10.1016/S0378-1119(99)00013-X. [DOI] [PubMed] [Google Scholar]
- 76.Jestin JL, Volioti G, Winter G. Improving the display of proteins on filamentous phage. Res Microbiol. 2001;152:187–191. doi: 10.1016/S0923-2508(01)01191-3. [DOI] [PubMed] [Google Scholar]
- 77.Kristensen P, Winter G. Proteolytic selection for protein folding using filamentous bacteriophages. Fold Des. 1998;3:321–328. doi: 10.1016/S1359-0278(98)00044-3. [DOI] [PubMed] [Google Scholar]
- 78.Rondot S, Koch J, Breitling F, Dubel S. A helper phage to improve single-chain antibody presentation in phage display. Nat Biotechnol. 2001;19:75–78. doi: 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- 79.Kramer RA, Cox F, van der Horst M, van der Oudenrijn S, Res PC, Bia J, Logtenberg T, de Kruif J. A novel helper phage that improves phage display selection efficiency by preventing the amplification of phages without recombinant protein. Nucleic Acids Res. 2003;31:e59. doi: 10.1093/nar/gng058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baek H, Suk KH, Kim YH, Cha S. An improved helper phage system for efficient isolation of specific antibody molecules in phage display. Nucleic Acids Res. 2002;30:e18. doi: 10.1093/nar/30.5.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soltes G, Barker H, Marmai K, Pun E, Yuen A, Wiersma EJ. A new helper phage and phagemid vector system improves viral display of antibody Fab fragments and avoids propagation of insert-less virions. J Immunol Methods. 2003;274:233–244. doi: 10.1016/S0022-1759(02)00294-6. [DOI] [PubMed] [Google Scholar]
- 82.Soltes G, Hust M, Ng KK, Bansal A, Field J, Stewart DI, Dubel S, Cha S, Wiersma EJ. On the influence of vector design on antibody phage display. J Biotechnol. 2007;127:626–637. doi: 10.1016/j.jbiotec.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olszewski A, Sato K, Aron ZD, Cohen F, Harris A, McDougall BR, Robinson WE, Jr, Overman LE, Weiss GA. Guanidine alkaloid analogs as inhibitors of HIV-1 Nef interactions with p53, actin, and p56lck. Proc Natl Acad Sci USA. 2004;101:14079–14084. doi: 10.1073/pnas.0406040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Majumdar S, Hajduczki A, Mendez AS, Weiss GA. Phage display of functional, full-length human and viral membrane proteins. Bioorg Med Chem Lett. 2008;18:5937–5940. doi: 10.1016/j.bmcl.2008.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Houten NE, Zwick MB, Menendez A, Scott JK. Filamentous phage as an immunogenic carrier to elicit focused antibody responses against a synthetic peptide. Vaccine. 2006;24:4188–4200. doi: 10.1016/j.vaccine.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zanghi CN, Lankes HA, Bradel-Tretheway B, Wegman J, Dewhurst S. A simple method for displaying recalcitrant proteins on the surface of bacteriophage lambda. Nucleic Acids Res. 2005;33:e160. doi: 10.1093/nar/gni158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tian F, Tsao ML, Schultz PG. A phage display system with unnatural amino acids. J Am Chem Soc. 2004;126:15962–15963. doi: 10.1021/ja045673m. [DOI] [PubMed] [Google Scholar]
- 88.Sandman KE, Benner JS, Noren CJ. Phage display of selenopeptides. J Am Chem Soc. 2000;122:960–961. doi: 10.1021/ja992462m. [DOI] [Google Scholar]
- 89.Meyer SC, Shomin CD, Gaj T, Ghosh I. Tethering small molecules to a phage display library: discovery of a selective bivalent inhibitor of protein kinase A. J Am Chem Soc. 2007;129:13812–13813. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
- 90.Woiwode TF, Haggerty JE, Katz R, Gallop MA, Barrett RW, Dower WJ, Cwirla SE. Synthetic compound libraries displayed on the surface of encoded bacteriophage. Chem Biol. 2003;10:847–858. doi: 10.1016/j.chembiol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 91.Dwyer MA, Lu W, Dwyer JJ, Kossiakoff AA. Biosynthetic phage display: a novel protein engineering tool combining chemical and genetic diversity. Chem Biol. 2000;7:263–274. doi: 10.1016/S1074-5521(00)00102-2. [DOI] [PubMed] [Google Scholar]
- 92.El Zoeiby A, Sanschagrin F, Darveau A, Brisson JR, Levesque RC. Identification of novel inhibitors of Pseudomonas aeruginosa MurC enzyme derived from phage-displayed peptide libraries. J Antimicrob Chemother. 2003;51:531–543. doi: 10.1093/jac/dkg010. [DOI] [PubMed] [Google Scholar]
- 93.D’Mello F, Howard CR. An improved selection procedure for the screening of phage display peptide libraries. J Immunol Methods. 2001;247:191–203. doi: 10.1016/S0022-1759(00)00318-5. [DOI] [PubMed] [Google Scholar]
- 94.Lunder M, Bratkovic T, Kreft S, Strukelj B. Peptide inhibitor of pancreatic lipase selected by phage display using different elution strategies. J Lipid Res. 2005;46:1512–1516. doi: 10.1194/jlr.M500048-JLR200. [DOI] [PubMed] [Google Scholar]
- 95.Yu H, Dong X, Sun Y. An alternating elution strategy for screening high affinity peptides from a phage display library. Biochem Eng J. 2004;18:169–175. doi: 10.1016/j.bej.2003.08.006. [DOI] [Google Scholar]
- 96.Zhuang G, Katakura Y, Furuta T, Omasa T, Kishimoto M, Suga K. A kinetic model for a biopanning process considering antigen desorption and effective antigen concentration on a solid phase. J Biosci Bioeng. 2001;91:474–481. doi: 10.1263/jbb.91.474. [DOI] [PubMed] [Google Scholar]
- 97.Kretzschmar T, Zimmermann C, Geiser M. Selection procedures for nonmatured phage antibodies: a quantitative comparison and optimization strategies. Anal Biochem. 1995;224:413–419. doi: 10.1006/abio.1995.1059. [DOI] [PubMed] [Google Scholar]
- 98.Lunder M, Bratkovic T, Urleb U, Kreft S, Strukelj B. Ultrasound in phage display: a new approach to nonspecific elution. Biotechniques. 2008;44:893–900. doi: 10.2144/000112759. [DOI] [PubMed] [Google Scholar]
- 99.Malmborg AC, Duenas M, Ohlin M, Soderlind E, Borrebaeck CA. Selection of binders from phage displayed antibody libraries using the BIAcore biosensor. J Immunol Methods. 1996;198:51–57. doi: 10.1016/0022-1759(96)00159-7. [DOI] [PubMed] [Google Scholar]
- 100.Yuan B, Schulz P, Sierks M-R. Improved affinity selection using phage display technology and off-rate based selection. Electron J Biotechnol. 2006;9:171–175. [Google Scholar]
- 101.Matsubara T, Ishikawa D, Taki T, Okahata Y, Sato T. Selection of ganglioside GM1-binding peptides by using a phage library. FEBS Lett. 1999;456:253–256. doi: 10.1016/S0014-5793(99)00962-X. [DOI] [PubMed] [Google Scholar]
- 102.Takakusagi Y, Takakusagi K, Kuramochi K, Kobayashi S, Sugawara F, Sakaguchi K. Identification of C10 biotinylated camptothecin (CPT-10-B) binding peptides using T7 phage display screen on a QCM device. Bioorg Med Chem. 2007;15:7590–7598. doi: 10.1016/j.bmc.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 103.Vanhercke T, Ampe C, Tirry L, Denolf P. Rescue and in situ selection and evaluation (RISE): a method for high-throughput panning of phage display libraries. J Biomol Screen. 2005;10:108–117. doi: 10.1177/1087057104271956. [DOI] [PubMed] [Google Scholar]
- 104.Hogan S, Rookey K, Ladner R. URSA: ultra rapid selection of antibodies from an antibody phage display library. Biotechniques. 2005;38:536–538. doi: 10.2144/05384BM03. [DOI] [PubMed] [Google Scholar]
- 105.Morohashi K, Arai T, Saito S, Watanabe M, Sakaguchi K, Sugawara F. A high-throughput phage display screening method using a combination of real-time PCR and affinity chromatography. Comb Chem High Throughput Screen. 2006;9:55–61. doi: 10.2174/138620706775213840. [DOI] [PubMed] [Google Scholar]
- 106.Plieva FM, Galaev IY, Noppe W, Mattiasson B. Cryogel applications in microbiology. Trends Microbiol. 2008;16:543–551. doi: 10.1016/j.tim.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 107.Noppe W, Plieva F, Galaev IY, Pottel H, Deckmyn H, Mattiasson B. Chromato-panning: an efficient new mode of identifying suitable ligands from phage display libraries. BMC Biotechnol. 2009;9:21. doi: 10.1186/1472-6750-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Castillo J, Goodson B, Winter J. T7 displayed peptides as targets for selecting peptide specific scFvs from M13 scFv display libraries. J Immunol Methods. 2001;257:117–122. doi: 10.1016/S0022-1759(01)00454-9. [DOI] [PubMed] [Google Scholar]
- 109.Wang WF, Cheng X, Molineux IJ. Isolation and identification of fxsA, an Escherichia coli gene that can suppress F exclusion of bacteriophage T7. J Mol Biol. 1999;292:485–499. doi: 10.1006/jmbi.1999.3087. [DOI] [PubMed] [Google Scholar]
- 110.Bowley DR, Jones TM, Burton DR, Lerner RA. Libraries against libraries for combinatorial selection of replicating antigen-antibody pairs. Proc Natl Acad Sci USA. 2009;106:1380–1385. doi: 10.1073/pnas.0812291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Grøn H, Hyde-DeRuyscher R. Peptides as tools in drug discovery. Curr Opin Drug Discov Dev. 2000;3:636–645. [PubMed] [Google Scholar]
- 112.Schumacher TN, Mayr LM, Minor DL, Jr, Milhollen MA, Burgess MW, Kim PS. Identification of D-peptide ligands through mirror-image phage display. Science. 1996;271:1854–1857. doi: 10.1126/science.271.5257.1854. [DOI] [PubMed] [Google Scholar]
- 113.Hawinkels LJ, van Rossenberg SM, de Jonge-Muller ES, Molenaar TJ, Appeldoorn CC, van Berkel TJ, Sier CF, Biessen EA. Efficient degradation-aided selection of protease inhibitors by phage display. Biochem Biophys Res Commun. 2007;364:549–555. doi: 10.1016/j.bbrc.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 114.Rowley MJ, O’Connor K, Wijeyewickrema L. Phage display for epitope determination: a paradigm for identifying receptor-ligand interactions. Biotechnol Annu Rev. 2004;10:151–188. doi: 10.1016/S1387-2656(04)10006-9. [DOI] [PubMed] [Google Scholar]
- 115.Schooltink H, Rose-John S. Designing cytokine variants by phage-display. Comb Chem High Throughput Screen. 2005;8:173–179. doi: 10.2174/1386207053258550. [DOI] [PubMed] [Google Scholar]
- 116.Hoogenboom HR. Overview of antibody phage-display technology and its applications. Methods Mol Biol. 2002;178:1–37. doi: 10.1385/1-59259-240-6:001. [DOI] [PubMed] [Google Scholar]
- 117.Conrad U, Scheller J. Considerations on antibody-phage display methodology. Comb Chem High Throughput Screen. 2005;8:117–126. doi: 10.2174/1386207053258532. [DOI] [PubMed] [Google Scholar]
- 118.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18:295–304. doi: 10.1016/j.copbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 119.Gronwall C, Stahl S. Engineered affinity proteins–generation and applications. J Biotechnol. 2009;140:254–269. doi: 10.1016/j.jbiotec.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 120.Deperthes D. Phage display substrate: a blind method for determining protease specificity. Biol Chem. 2002;383:1107–1112. doi: 10.1515/BC.2002.119. [DOI] [PubMed] [Google Scholar]
- 121.Sedlacek R, Chen E. Screening for protease substrate by polyvalent phage display. Comb Chem High Throughput Screen. 2005;8:197–203. doi: 10.2174/1386207053258541. [DOI] [PubMed] [Google Scholar]
- 122.Kay BK, Hamilton PT. Identification of enzyme inhibitors from phage-displayed combinatorial peptide libraries. Comb Chem High Throughput Screen. 2001;4:535–543. doi: 10.2174/1386207013330760. [DOI] [PubMed] [Google Scholar]
- 123.Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol. 2007;17:481–487. doi: 10.1016/j.sbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 124.Hertveldt K, Belien T, Volckaert G. General M13 phage display: M13 phage display in identification and characterization of protein-protein interactions. Methods Mol Biol. 2009;502:321–339. doi: 10.1007/978-1-60327-565-1_19. [DOI] [PubMed] [Google Scholar]
- 125.Fernandez-Gacio A, Uguen M, Fastrez J. Phage display as a tool for the directed evolution of enzymes. Trends Biotechnol. 2003;21:408–414. doi: 10.1016/S0167-7799(03)00194-X. [DOI] [PubMed] [Google Scholar]
- 126.Jung S, Honegger A, Pluckthun A. Selection for improved protein stability by phage display. J Mol Biol. 1999;294:163–180. doi: 10.1006/jmbi.1999.3196. [DOI] [PubMed] [Google Scholar]
- 127.Sergeeva A, Kolonin MG, Molldrem JJ, Pasqualini R, Arap W. Display technologies: application for the discovery of drug and gene delivery agents. Adv Drug Deliv Rev. 2006;58:1622–1654. doi: 10.1016/j.addr.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Clonis YD. Affinity chromatography matures as bioinformatic and combinatorial tools develop. J Chromatogr A. 2006;1101:1–24. doi: 10.1016/j.chroma.2005.09.073. [DOI] [PubMed] [Google Scholar]
- 129.Noppe W, Plieva FM, Vanhoorelbeke K, Deckmyn H, Tuncel M, Tuncel A, Galaev IY, Mattiasson B. Macroporous monolithic gels, cryogels, with immobilized phages from phage-display library as a new platform for fast development of affinity adsorbent capable of target capture from crude feeds. J Biotechnol. 2007;131:293–299. doi: 10.1016/j.jbiotec.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 130.Mao C., Liu A., Cao B. Virus-based chemical and biological sensing. Angew Chem Int Ed Engl. 2009;48:6790–6810. doi: 10.1002/anie.200900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.De Berardinis P, Haigwood NL. New recombinant vaccines based on the use of prokaryotic antigen-display systems. Expert Rev Vaccines. 2004;3:673–679. doi: 10.1586/14760584.3.6.673. [DOI] [PubMed] [Google Scholar]
- 132.Manoutcharian K, Gevorkian G, Cano A, Almagro JC. Phage displayed biomolecules as preventive and therapeutic agents. Curr Pharm Biotechnol. 2001;2:217–223. doi: 10.2174/1389201013378671. [DOI] [PubMed] [Google Scholar]
- 133.Lee SC, Ibdah R, Van Valkenburgh C, Rowold E, Abegg A, Donnelly A, Klover J, Merlin S, McKearn JP. Phage display mutagenesis of the chimeric dual cytokine receptor agonist myelopoietin. Leukemia. 2001;15:1277–1285. doi: 10.1038/sj.leu.2402163. [DOI] [PubMed] [Google Scholar]
- 134.McConnell SJ, Dinh T, Le MH, Brown SJ, Becherer K, Blumeyer K, Kautzer C, Axelrod F, Spinella DG. Isolation of erythropoietin receptor agonist peptides using evolved phage libraries. Biol Chem. 1998;379:1279–1286. doi: 10.1515/bchm.1998.379.10.1279. [DOI] [PubMed] [Google Scholar]
- 135.Hyde-DeRuyscher R, Paige LA, Christensen DJ, Hyde-DeRuyscher N, Lim A, Fredericks ZL, Kranz J, Gallant P, Zhang J, Rocklage SM, Fowlkes DM, Wendler PA, Hamilton PT. Detection of small-molecule enzyme inhibitors with peptides isolated from phage-displayed combinatorial peptide libraries. Chem Biol. 2000;7:17–25. doi: 10.1016/S1074-5521(00)00062-4. [DOI] [PubMed] [Google Scholar]
- 136.Monoclonal antibody therapeutics 2009-2024. Market report. Visiongain. 06/08/2009
- 137.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 138.Bostrom J, Fuh G. Design and construction of synthetic phage-displayed fab libraries. In: Aitken R, editor. Antibody phage display: methods and protocols. New York: Humana; 2009. pp. 17–35. [DOI] [PubMed] [Google Scholar]
- 139.Lonberg N (2008) Human monoclonal antibodies from transgenic mice. Handb Exp Pharmacol 69–97 [DOI] [PMC free article] [PubMed]
- 140.Jespers LS, Roberts A, Mahler SM, Winter G, Hoogenboom HR. Guiding the selection of human antibodies from phage display repertoires to a single epitope of an antigen. Biotechnology (N Y) 1994;12:899–903. doi: 10.1038/nbt0994-899. [DOI] [PubMed] [Google Scholar]
- 141.Lonberg N. Fully human antibodies from transgenic mouse and phage display platforms. Curr Opin Immunol. 2008;20:450–459. doi: 10.1016/j.coi.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 142.Williams A, Baird LG. DX-88 and HAE: a developmental perspective. Transfus Apher Sci. 2003;29:255–258. doi: 10.1016/S1473-0502(03)00170-8. [DOI] [PubMed] [Google Scholar]
- 143.Levy JH, O’Donnell PS. The therapeutic potential of a kallikrein inhibitor for treating hereditary angioedema. Expert Opin Investig Drugs. 2006;15:1077–1090. doi: 10.1517/13543784.15.9.1077. [DOI] [PubMed] [Google Scholar]
- 144.Epstein TG, Bernstein JA. Current and emerging management options for hereditary angioedema in the US. Drugs. 2008;68:2561–2573. doi: 10.2165/0003495-200868180-00003. [DOI] [PubMed] [Google Scholar]
- 145.Lehmann A. Ecallantide (DX-88), a plasma kallikrein inhibitor for the treatment of hereditary angioedema and the prevention of blood loss in on-pump cardiothoracic surgery. Expert Opin Biol Ther. 2008;8:1187–1199. doi: 10.1517/14712598.8.8.1187. [DOI] [PubMed] [Google Scholar]
- 146.Nizak C, Monier S, del Nery E, Moutel S, Goud B, Perez F. Recombinant antibodies to the small GTPase Rab6 as conformation sensors. Science. 2003;300:984–987. doi: 10.1126/science.1083911. [DOI] [PubMed] [Google Scholar]
- 147.Gao J, Sidhu SS, Wells JA. Two-state selection of conformation-specific antibodies. Proc Natl Acad Sci USA. 2009;106:3071–3076. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sakamoto K, Ito Y, Hatanaka T, Soni PB, Mori T, Sugimura K. Discovery and characterization of a peptide motif that specifically recognizes a non-native conformation of human IgG induced by acidic pH conditions. J Biol Chem. 2009;284:9986–9993. doi: 10.1074/jbc.M807618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Emadi S, Barkhordarian H, Wang MS, Schulz P, Sierks MR. Isolation of a human single chain antibody fragment against oligomeric alpha-synuclein that inhibits aggregation and prevents alpha-synuclein-induced toxicity. J Mol Biol. 2007;368:1132–1144. doi: 10.1016/j.jmb.2007.02.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Emadi S, Kasturirangan S, Wang MS, Schulz P, Sierks MR. Detecting morphologically distinct oligomeric forms of alpha-synuclein. J Biol Chem. 2009;284:11048–11058. doi: 10.1074/jbc.M806559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 153.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 154.Blancafort P, Magnenat L, Barbas CF., 3rd Scanning the human genome with combinatorial transcription factor libraries. Nat Biotechnol. 2003;21:269–274. doi: 10.1038/nbt794. [DOI] [PubMed] [Google Scholar]
- 155.Kriplani U, Kay BK. Selecting peptides for use in nanoscale materials using phage-displayed combinatorial peptide libraries. Curr Opin Biotechnol. 2005;16:470–475. doi: 10.1016/j.copbio.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 156.Khoo X, Hamilton P, O’Toole GA, Snyder BD, Kenan DJ, Grinstaff MW. Directed assembly of PEGylated-peptide coatings for infection-resistant titanium metal. J Am Chem Soc. 2009;131:10992–10997. doi: 10.1021/ja9020827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Segvich SJ, Smith HC, Kohn DH. The adsorption of preferential binding peptides to apatite-based materials. Biomaterials. 2009;30:1287–1298. doi: 10.1016/j.biomaterials.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tomczak MM, Gupta MK, Drummy LF, Rozenzhak SM, Naik RR. Morphological control and assembly of zinc oxide using a biotemplate. Acta Biomater. 2009;5:876–882. doi: 10.1016/j.actbio.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 159.Nam KT, Wartena R, Yoo PJ, Liau FW, Lee YJ, Chiang YM, Hammond PT, Belcher AM. Stamped microbattery electrodes based on self-assembled M13 viruses. Proc Natl Acad Sci USA. 2008;105:17227–17231. doi: 10.1073/pnas.0711620105. [DOI] [PMC free article] [PubMed] [Google Scholar]