Abstract
Heparan sulfate proteoglycans are a remarkably diverse family of glycosaminoglycan-bearing protein cores that include the syndecans, the glypicans, perlecan, agrin, and collagen XVIII. Members of this protein class play key roles during normal processes that occur during development, tissue morphogenesis, and wound healing. As key components of basement membranes in organs and tissues, they also participate in selective filtration of biological fluids, in establishing cellular barriers, and in modulation of angiogenesis. The ability to perform these functions is provided both by the features of the protein cores as well as by the unique properties of heparan sulfate, which is assembled as a polymer of N-acetylglucosamine and glucuronic acid and modified by specific enzymes to generate specialized biologically active structures. This article discusses the structures and functions of this amazing family of proteoglycans and provides a platform for further study of the individual members.
Keywords: Heparan sulfate, Proteoglycan, Glycosaminoglycan, Perlecan, Syndecan, Glypican, Agrin, Collagen XVIII
Introduction
Proteoglycans are remarkably complex molecules with functions associated not only with their protein cores but also with their constituent carbohydrate chains. These functions range from structural roles in the extracellular matrix to control of growth factor gradients, cell migration and behavior. Proteoglycans are classified based on the types of carbohydrate chains they contain. In this regard, the signature carbohydrate chains are large, linear, highly negatively charged polymers called glycosaminoglycans. Glycosaminoglycans are further classified based on their characteristic disaccharide repeating structures. Heparan sulfate (HS) is one class of glycosaminoglycans initially assembled as a polymer of N-acetylglucosamine and glucuronic acid and subsequently modified in several ways to generate more complex, biologically active structures. HS proteoglycans (HSPGs) are found in all animals from worms and insects to higher mammals, and mutations associated with them usually have large phenotypic consequences.
While much has been learned about the diverse roles these molecules play, we still do not completely understand the full extent of HSPG function. This review will focus on several well-described HSPGs of the extracellular matrix. We will discuss their structures, the variety of interactions they support, both via their protein cores and constituent HS chains, and the impact mutations in HSPG core proteins and HS biosynthetic enzymes play in various developmental and biological processes. For convenience, all structural information provided is for the RefSeq human version of these proteins. Information on alternate forms can be found on the Swiss-Prot database (http://www.uniprot.org/) pages using the ID provided.
Core proteins
Perlecan
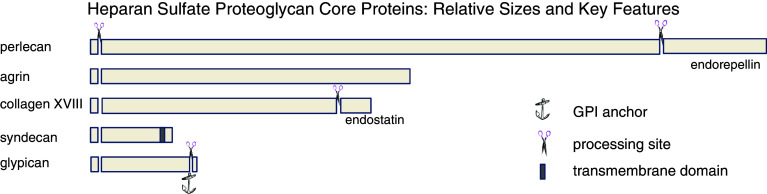
Perlecan, also called heparan sulfate proteoglycan 2 (HSPG2), is an ancient, structurally conserved HS bearing proteoglycan that is wholly secreted into the extracellular space, where it is incorporated into the basement membranes separating epithelial and stromal compartments in tissues or into the territorial matrices surrounding mesenchymal cells [1, 2]. The core protein of human pre-perlecan consists of 4,391 amino acids of which the first 21 are removed by signal peptidase (Fig. 1). The mature core protein is modular, containing 1 sperm protein–enterokinase–agrin (SEA) module in unique domain I, 4 EGF-like domains, 11 laminin epidermal growth factor (EGF)-like domains, 22 Ig-like C2 domains, 3 laminin G-like domains, 3 laminin IV type A domains, and 4 low density lipoprotein (LDL) receptor class A domains (UniProtKB/Swiss-Prot entry P98160). The core protein is by nature functionally complex, offering a very large number of functional surfaces for interactions with other proteins which can be HS-dependent, HS-influenced, or HS-independent. Many important functions have been ascribed to perlecan including modulation of angiogenesis [3], solute filtration [4, 5], growth factor delivery [6], initiation of chondrogenesis [2], regulation of cell adhesion [7, 8], and fibrillogenesis [9]. For instance, the C-terminus of perlecan can be processed by proteases into a fragment (Fig. 1) with anti-angiogenic activity called endorepellin [10, 11].
Fig. 1.
Comparison of relative sizes of HSPG core proteins and their processed fragments. The relative sizes of five major HSPG core proteins are illustrated by the length of the horizontal bars. For those with multiple family members, such as the syndecans and glypicans, the relative size of the largest family member is shown. All lose an approximately 20 amino acid N-terminal signal peptide in the secretory route. Only syndecan is retained as a transmembrane protein (darker box), although glypicans receive a lipid anchor (shown as anchor symbol) after removal of a C-terminal fragment which retains them on the cell surface. Gaps in the sequences of the core proteins with scissors show demonstrated sites of protein processing. For endostatin (collagen XVIII) and endorepellin (perlecan), the released fragment has potent biological activity
Agrin
Agrin (AGRN), once known as neural aggregating factor, is best known for its ability to cluster synaptic proteins including acetylcholine receptors at neuromuscular junctions, although there are recent challenges to the mechanistic paradigm [12, 13]. The mature agrin core protein is produced by removal of a 29 amino acid signal sequence from a 2,045 amino acid precursor form prior to secretion (Fig. 1). The agrin core protein has a unique N-terminus or NtA domain followed by eight Kazal-like domains. The central portion of the agrin core protein contains an SEA domain flanked by two Ser/Thr-rich regions to which multiple heparan sulfate glycosaminoglycan chains are attached. Chondroitin sulfate also may be present in this region [14]. C-terminal to the central region are alternating EGF-like and laminin G domains, four of the first and three of the second (UniProtKB/Swiss-Prot entry O00468). Agrin, primarily known as a modulator of synaptogenesis, is increasingly recognized for a broader range of functions in the nervous system [15].
Collagen XVIII
This multifunctional protein, once called MFP, is characterized as a “multiplexin”, a class of proteins with multiple triple helix domains interspaced with non-triple helical domains [16]. The core protein (UniProtKB/Swiss-Prot entry P39060) contains 1,754 amino acids, the first 23 of which comprise the signal sequence (Fig. 1). A unique feature of collagen XVIII (COL18A1) is that the C-terminus is cleaved post-translationally to produce endostatin (amino acids 1572–1754), an effective blocker of both endothelial cell proliferation and angiogenesis [17]. The core protein also contains a frizzled (FZ) domain (amino acids 329–446), a cysteine rich domain (CRD) that contains ten conserved cysteines that form five disulfide bridges. FZ domains are present in diverse group of proteins including several receptor tyrosine kinases [18]. The CRD in the Drosophila gene product Frizzled is part of the extracellular region implicated as the Wnt binding domain [19]. Collagen XVIII also contains a thrombospondin-type laminin G domain (TspN) also found in various laminins, the N-terminus of thrombospondin and pentraxins. TspN-containing proteins typically are multi-domain adhesive matrix proteins that function as molecular connectors between cells and matrix, and participate in cellular communication [20]. An RGD cell adhesion motif is also present on the core protein (amino acids 1330–1332) just upstream of the cleavage site for generation of endostatin. Multiple sites for both N-linked and O-linked glycosylation are also present.
Syndecans
There are four syndecans (SDC1–4), all of which are single pass type I transmembrane proteins with a large glycosaminoglycan-bearing ectodomain, an approximately 20 amino acid transmembrane region, and a comparatively short cytoplasmic tail (Fig. 1). Syndecan 3 (UniProtKB/Swiss-Prot entry O75056) is the longest at 442 amino acids, Syndecan 1 (UniProtKB/Swiss-Prot entry P18827) is significantly shorter at 310 amino acids, Syndecan 2 (UniProtKB/Swiss-Prot entry P34741) is shorter at 201, and Syndecan 4 (UniProtKB/Swiss-Prot entry P31431) shorter still at 198 amino acids. All four mature syndecans are produced by cleavage of an 18–22 amino acid signal peptide present in the pre-protein. Syndecan 1, also designated CD138, is present in both stromal and epithelial cells. The mature protein core contains a unique 232 amino acid extracellular domain that is modified by carbohydrate chains that include both O-linked chondroitin sulfate and N-linked structures in addition to HS. A 21 amino acid transmembrane segment links the extracellular domain to the 35 amino acid long cytoplasmic tail on the cell’s interior. A site for phosphorylation is conserved at tyrosine 309. Syndecan 2, common name fibroglycan, is typically made by fibroblasts. It has a 126 amino acid long ectodomain, followed by short transmembrane and cytoplasmic domains. There are three conserved sites of HS modification at amino acids 41, 55, and 57. Syndecan 3, or N-syndecan, is expressed in the nervous system, the adrenal gland, and the spleen. It has a large ectodomain of some 387 amino acids that contains six potential sites for glycosaminoglycan attachment. Isoform 1 of this protein is considered the reference sequence, but several isoforms have been reported, one of which (isoform 3) is missing amino acids 112–125. Syndecan 4, also called amphiglycan or ryudocan, has a 127 amino acid extracellular domain with three sites for addition of HS, at amino acids 39, 61 and 63. The transmembrane and cytoplasmic domains are of approximately equal lengths, 25 and 28 amino acids, respectively. A conserved site for tyrosine phosphorylation is at amino acid 197 on the cytoplasmic tail.
Glypicans
The hallmark quality of the six glypicans is their attachment to the cell membrane by a glycosyl phosphatidylinositol linkage (Fig. 1). For this feature, they have been dubbed the “glypican-related integral membrane proteoglycan family” (GRIPS) [21]. All are proteins of some 550–580 amino acids that are made as precursors with a signal peptide destined for cleavage during biosynthesis. Additionally, all glypicans have a large globular cysteine-rich domain with 14 invariant cysteines residues followed by the HS modified “stalk”. Glypican 1 (UniProtKB/Swiss-Prot entry P35052) is lipid modified at serine 530, and has four potential sites for gagosylation along with two sites for N-linked modification. The mature protein is generated after removal of amino acids 531–558 at the C-terminus. Glypican 2 (UniProtKB/Swiss-Prot entry Q8N158) is also called cerebroglycan. It is lipid modified at amino acid 554 which becomes the C-terminus after processing removes amino acids 555–579. Glypican 3 (UniProtKB/Swiss-Prot entry P51654) is also called intestinal protein OCI-5. Its 580 amino acid long precursor protein core is processed similarly to the other glypicans. Glypican 4 (UniProtKB/Swiss-Prot entry O75487) has been termed K-glypican for its abundance in the kidney. It is derived from a precursor of 556 amino acids, with a mature form ending at amino acid 529 after addition of the lipid anchor and removal of the C-terminus. It contains both HS and N-linked carbohydrate additions. Glypican 5 (UniProtKB/Swiss-Prot entry P78333) has multiple sites for modification by N-linked carbohydrate chains and for addition of HS. Between amino acids 446 and 509, there are five sites for O-gagosylation; thus, a high negative charge density can be achieved in this portion of the molecule near the cell surface. There are several potential sites for addition of the lipid modification near the C-terminus of glypican 5, and it is not certain which of these is used. Glypican 6 (UniProtKB/Swiss-Prot entry Q9Y625) is lipid modified on amino acid 529 and is processed at both ends from a 555 amino acid precursor like the other glypican family members.
Core protein interactions
Perlecan
Perlecan is a giant modular protein with domains for interacting with many binding partners. A domain-by-domain list of those potential binding interactions, including those with many basement membrane proteins such as laminin and collagen IV, was recently published by our group and will not be repeated here [1]. Perlecan has a strong tendency to aggregate, and has been found in a variety of extracellular deposits including those containing β-amyloid [22], β2-microglobulin [23], and collagen [24]. Perlecan is processed by a variety of proteases, with proteolytic processing by BMP-1/Tolloid-like metalloproteases responsible for the production of the C-terminal fragment, endorepellin [10], which has potent anti-angiogenic properties [11]. Domain IV of perlecan contains a tandem of immunoglobulin repeats with the capacity to interact with other proteins containing similar repeats in a “zipper-like” fashion. Cell surface proteins with Ig modules include ectodomains of ion channels [25], cadherins [26], and other immunoglobulin superfamily members [27]. A novel peptide derived from one of these Ig modules in domain IV of perlecan interacts with cell surface receptors and alters cell adhesion [7]. Fibroblast growth factor-18 (FGF-18) has been reported to bind to the core protein of perlecan in domain III [28]. Interactions of perlecan core protein with WARP, a member of the von Willebrand factor A domain superfamily found in cartilage matrix, have also been reported [29]. Mapping studies also showed interaction of perlecan domains I and II with a central region of fibrillin-1 [30].
Agrin
Known as “neural aggregating factor”, agrin made by nerve cells clusters acetylcholine receptors at neuromuscular junctions by virtue of its interactions with the signaling complex located at the synapse. The so-called “laminin/integrin–agrin–α-dystroglycan network” [31] links the signaling complex to the basement membrane and is key to synaptic transmission and prevention of muscular dystrophy [32]. The binding of agrin to laminin occurs through the NtA domain (amino acids 30–157) and has been analyzed in detail [33]. The interaction with α-dystroglycan is glysosylation-dependent [34]. Agrin has also been found along with glypican 1 in amyloid fibrils [35]. Agrin can also interact with α-synuclein and by doing so increase its insolubility and tendency to form fibrils, an occurrence which has been implicated in the development of Parkinson’s disease [36].
Collagen XVIII
Collagen XVIII and its derivative, endostatin, interact with a wide variety of protein components of the basement membrane [37]. Endostatins interactions with a host of cell surface receptors and extracellular matrix proteins responsible for its potent anti-angiogenic activities have been well reviewed [38, 39]. Interestingly, endostatin was recently reported to interact with fibrinogen [40].
Syndecans
Syndecan family members classically link the functions of the cell surface heparin-binding growth factor receptors to the cytoskeleton, a function facilitated by their uniqueness as transmembrane HSPGs. Acting as high capacity, low affinity co-receptors able to deliver bound heparin-binding growth factors to their cognate low capacity, high affinity receptors, syndecans play key roles in signal transduction [41]. Syndecan-1 was recently shown to interact with CD147, an association proposed to be essential for cyclophilin B-induced activation of p44/42 mitogen-activated protein kinases and promotion of cell adhesion and chemotaxis [42]. Interactions with proteases lead to ectodomain shedding [43]. Muramatsu et al. [44] recently described an approach for identifying proteins that interact with HSPGs and discriminating between those which interact with protein cores and those which require intact proteoglycan. Syndecan 4 was used as the binding partner for this demonstration. The conserved site for tyrosine phosphorylation on the cytoplasmic tail of syndecans is thought to be important in regulating interactions with the cytoskeleton. Syntenin-1 recruitment, for example, depends on the dephosphorylation of Tyr-309 located within the syndecan-1 PDZ binding domain EFYA [45].
Glypicans
The core protein of the glypicans is unique in that the C-terminus is processed prior to addition of the lipid anchor. Glypicans are modified by proprotein convertases, including furin, but remain together as disulfide-linked complexes [46–48]. Glypican interactions with slit proteins are important in axonal guidance and related developmental processes in nervous tissue [49]. Glypican-1 has also been reported to be present in amyloid plaques of Alzheimer’s disease, perhaps by binding directly to beta-amyloid protein [50]. Altogether, the molecular processing and interactions of glypican family members remain relatively poorly understood.
Heparan sulfate
Synthesis and structure
As mentioned above, heparan sulfate (HS) chains are linear polysaccharides consisting of alternating glucosamine and uronic acid residues >50 disaccharide units in length. These polymers are initially assembled as a simple polymer of d-N-acetylglucosamine and d-glucuronic acid. Subsequently, the polymer is variously N-deacetylated and N-sulfated, glucuronic acid epimerized to iduronic acid and O-sulfated. These modifications occur through an ordered series of reactions which appear to be controlled primarily by changes in the levels of expression of the various enzymes that act at each step. In multiple cases, more than one distinct gene encodes isozymes of HS biosynthetic activities providing redundancy, tissue specificity and opportunity for differential gene regulation (reviewed in [51]).
Interactions
Heparan sulfate polymer modification tends to occur in block regions along the large HS chains resulting in highly modified regions and regions with little or no modifications [52]. As a general rule, HS-binding proteins preferentially bind to the more modified, and therefore more highly charged, regions. Examples of proteins that preferentially bind to particular HS structures and the structures they prefer are listed in Table 1. Many of the preferred structures have similar or lower charge densities than less preferred or poorly interacting structures indicating that these interactions involve more than simple charge interactions. Nonetheless, it should be kept in mind that HS is the most highly negatively charged extracellular molecule in higher species, and therefore interactions of many proteins with a net positive charge with HS have little specificity beyond charge interactions.
Table 1.
Preference of growth factor binding to HS structures
| HS-binding growth factor | Preferred HS binding features | Refs. |
|---|---|---|
| FGF2 | [IdoA(2-O-SO3) → GlcNSO3]2-4 | [149–151] |
| FGF4 | [GlcNSO4(6-O-SO3) → IdoA (2-O-SO3)]3 | [150, 151] |
| VEGF165 | Sequences >8 saccharides containing both 2- and 6-O-SO3 modifications | [150] |
| HGF | IdoA → GlcNSO3(6-O-SO3) and [IdoA(2-O-SO3) → GlcNSO3(6-O-SO3)]2 | [150, 152–154] |
Heparanases and Sulfs
Just as there are extracellular activities that hydrolyze specific sites of HSPG protein cores, i.e., metalloproteases, there are extracellular activities that hydrolyze specific motifs in HS chains, namely heparanase-1 and Sulfs (reviewed in [53, 54]). Only one enzymatically active heparanase has been identified, heparanase-1. Although transcripts that appear to encode isozymes of heparanase-1 have been detected, no protein product or enzymatic activity has been reported for these species. Heparanase-1 is a endo-beta-d-glucuronidase cleaving between glucuronosyl and N-sulfated acetylglucosaminyl residues in HS chains. Although the substrate specificity of this enzyme is not completely elucidated, the saccharide motifs recognized by heparanase-1 are found in restricted, less modified, low-sulfated regions of HS chains and, therefore, tend to be inbetween regions active in protein binding [55]. While minimum sulfation is required for heparanase-1 activity, specific sulfation patterns such as 3-O-sulfation of the glucosamine residue on the reducing side of the target glucuronic acid residue were found to be inhibitory when present in highly sulfated domains [56]. Consequently, in addition to breaking down the extracellular matrix, heparanase-1 can release complexes of HS fragments bound to proteins. In the case of HS-binding growth factors, this has particular biological significance since various receptors for HS-binding growth factors also bind HS and require the formation of a ternary growth factor:HS:growth factor receptor complex for efficient signal transduction [57–59].
In most cases, it appears that the bulk of heparanase-1 produced by cells remains intracellular where it presumably participates in lysosomal HS degradation. Consistent with this is the observation that heparanase-1 displays an acidic pH optimum [60, 61]. In fact, at neutral pH, heparanase-1 only displays 10–20% of its maximal activity. In this regard, it has been suggested that at the cell surface heparanase-1 may function more as a HS-binding protein [62, 63]. In agreement with this notion is the finding that heparanase-1 mutants that are catalytically inactive can still reach the cell surface and participate in HS-dependent cell adhesion processes [63].
Heparanase-1 is highly expressed by endothelial cells [64], various tumor cells [65], trophoblast [66] and decidual cells of mouse embryo implantation sites [67]. Its patterns of expression are consistent with a role in extracellular matrix remodeling as well as growth factor release. Two proteins have been identified that are naturally occurring inhibitors of heparanase activity, namely eosinophil major basic protein [68] and heparin/HS-binding protein/RPL29 [69]. In addition to inhibiting heparanase-1 activity, HIP/RPL29 also displaces HS-binding growth factors from HS-bearing substrates [69]. Thus, a complex scenario exists where HIP/RPL29 displays seemingly antagonistic actions in terms of HS-binding growth factor release. It is possible that HIP/RPL29, and possibly eosinophil basic protein, provide a means of releasing and increasing HS-binding growth factor bioavailability while simultaneously promoting stability of HS-bearing extracellular matrices. In addition to these natural inhibitors, several synthetic heparanase-1 inhibitors have been identified [70]. One of these, PI-88, has been used in vivo as an antagonist of tumor progression [71] and has potential in clinical applications.
Sulfs are secreted enzymes that remove 6-O-sulfates from glucosaminyl residues in HS chains. Two proteins have been identified, Sulf1 and Sulf2, that are products of two distinct genes [54]. Unlike heparanase-1, sulfs are maximally active at neutral pH and, therefore, are expected to be fully active upon secretion [72]. Release of 6-O-sulfate residues is not expected to alter the physical structure of the extracellular matrix to a significant extent; however, this modification would be expected to modify the biological properties of the extracellular matrix substantially. Many HS-binding proteins recognize 6-O-sulfates as part of the high affinity binding motif in HS (see Table 1 for examples). Thus, Sulf action may either destroy binding sites in HS and prevent subsequent HS-binding protein interaction or cause release of HS-bound growth factors. In the latter case, it would be expected that at least transient HS-binding growth factor dissociation must occur to provide access to the 6-O-sulfate residues. Destruction of HS-binding growth factor binding sites would be expected to increase growth factor diffusion and reduce local concentrations of these proteins. A wide variety of cell types express Sulfs in developing and adult tissues as well as various tumors [73–77]. In some cases, Sulfs enhance tumor progression while in others tumor progression is inhibited [75, 76]. These seemingly contradictory observations may reflect differences in tumor types or responses to individual HS-binding growth factors. For example, growth factors that can activate their cognate receptors in the absence of HS, e.g., BMPs, may be more actively released from HS-bearing substrata by Sulfs and enhance tumor progression. On the other hand, activity of growth factors that require formation of HS-containing complexes with their cognate receptors, e.g., FGFs, would be reduced by Sulf action. No Sulf inhibitors, either naturally occurring or synthetic, have been reported although these would be very useful tools to study the biology of these enzymes and also in therapeutic applications.
Genetics
Perlecan
Null mutations of the Pln gene in mice (also known as Hspg2) are not compatible with life and result in severe developmental abnormalities affecting primarily heart, brain, and cartilage functions [78, 79]. Between embryonic stages E10 and E12, absence of the Pln protein was found to compromise basement membrane integrity and induced either cell–cell detachment in the contracting heart muscle wall or ectopic brain tissue invasions of the cephalic mesenchyme [78, 80]. Although death caused by hemorrhages and exencephalies was the main consequence of these anatomical defects, a small proportion of Pln-null mouse embryos developed further and died perinatally because of narrow thorax and respiratory distress. These mice displayed skeletal defects including craniofacial malformations (flat face, cleft palate) and disproportionate dwarfism due to severe chondrodysplasia in the cartilage growth plate of long bones.
Patients diagnosed with dysegmental dysplasia of the Silverman–Handmaker syndrome (DDSH, MIM 224410) exhibit PLN loss-of-function mutations and present life-threatening symptoms equivalent to the defects observed in Pln-deficient mice. In contrast to loss-of-function studies, over-expression of Pln core protein in multiple tissues including brain and heart did not result in obvious developmental defects [81]. Recently, knock-in mouse models for the non-lethal autosomal recessive Schwartz–Jampel type I syndrome (SJS, MIM 255800) in which reduced amounts of perlecan protein are expressed have been created [82, 83]. Like in these hypomorphic mutants, the majority of the HSPG2 mutations (deletions/insertions, missense, nonsense, splice site mutations) found in SJS patients cause aberrant transcription and/or instability of Pln mRNAs. Thus, most of these mutations result in a common disease phenotype consisting of short stature associated with enhanced joint dysplasia and neuromyotonia associated with deficiency in acetylcholinesterase at the neuromuscular junction [80, 83–85].
Pln mouse mutants, lacking the three glycoaminoglycan attachment sites in the N-terminal domain I of Pln, display severe structural defects of the lens capsule leading to lens degeneration [86]. Also, this mutation promoted smooth muscle cell proliferation and intimal hyperplasia of artery walls indicative of a role for Pln HS chains in the modulation of atherosclerosis [87, 88]. Interestingly, reduced perlecan expression was associated with the presence of atherosclerotic lesions in human arteries [89]. In addition, although no significant abnormalities were observed in the glomerular basement membrane of the kidney, susceptibility to protein overload was observed in these mouse mutants suggesting a function for Pln HS chains in glomerular filtration [5].
Agrin
The targeted disruption of the Agrin gene at exons encoding for the neuron-specific isoform (z-agrin) produced null mutants for z-agrin as well as severe hypomorphs for all other forms of agrin, and provoked death at birth due to respiratory distress. This mutation resulted in defective synapse organization at the neuromuscular junction by impairing both pre- and post-synaptic differentiation. Anomalies included severe defects in acetylcholine receptors clustering and nerve branching [90]. The generation of isoform specific mutants for nerve-derived and muscle-derived agrin demonstrated that only neural z-agrin is required to induce post-synaptic differentiation and that its absence can also indirectly affect pre-synaptic differentiation through inhibition of proper retrograde signals [91].
Recently, rescue of perinatal death in agrin-deficient mice was achieved by transgenic expression of neural isoform of chick agrin and uncovered a reduction in skeletal growth [92]. In another recent study, the conditional disruption of agrin in kidney cells did not compromise the glomerular basement membrane architecture and function even when challenged with a protein excess. Thus, unlike Pln, agrin is thought to be dispensable for proper glomerular filtration [93].
Collagen (XVIII)
Absence of collagen XVIII in Col18a1-null mice resulted in severe ocular defects affecting vasculogenesis of the retina and integrity of basement membranes located in both the anterior and posterior portion of the eye [94, 95]. After derivation onto a defined genetic background, this mutation was also linked to the increase in width of various type of basement membranes such as those found in brain choroid plexuses and kidney proximal tubules. Such defects were shown to be responsible for accumulation of cerebrospinal fluid in the brain and altered filtration capacity in the kidney, respectively [96]. The phenotypes observed in Col18a1-null mice are consistent with symptoms observed in patients with mutations in the COL18A1 gene and diagnosed with the recessive Knobloch type I syndrome (MIM 267750) [97, 98]. In addition, high serum levels of the endostatin fragment in Down syndrome patients carrying an additional copy of the COL18A1 gene was associated with prevention of solid tumors, and nucleotide polymorphisms in this fragment were linked to the development of different type of cancers [99–101].
Although collagen XV and XVIII are similar molecules, compound Col15a1 and Col18a1 knock-out mice did not display any new phenotype, suggesting that no compensation mechanism takes place in single mutants and that they exert their scaffolding activity between fibrillar collagens and basement membranes at distinct anatomical sites [102].
Membrane spanning HSPGs
Syndecan-1
Syndecan-1 (Sdc1) knock-out mice do not display major developmental anomalies. Nonetheless, Bernfield’s group demonstrated that hyperplasia occurring in the Wnt-1 transgenic model of mammary tumorigenesis was greatly inhibited on a Sdc1-null background [103]. These experiments established that Sdc1 is a modulator of Wnt signaling. Recently, the tumor protective effect of Sdc1 deficiency was confirmed by evaluating the potential of multiple organs including liver and lung to develop carcinogen-induced tumors [104]. Another group demonstrated that considerable resistance to microbial lung infection was observed in Sdc1−/− newborn mice and was attributed to the lack of Sdc1 shedding and the absence of HS chains on epithelial cell surfaces [105]. Interestingly, both knock-out and over-expression of Sdc1 induced defects in wound healing processes [106, 107].
Other syndecans
Although syndecan-2 (Sdc2) null mice have not been reported, recent siRNA studies revealed that Sdc2 plays an important role in angiogenesis [108]. Mice lacking the syndecan-3 (Sdc3) protein have mild defects in synaptogenesis as revealed by impaired hippocampal functioning and locomotion [109, 110]. In addition to neural migration defects in the brain, Sdc3-null mice show partial resistance to obsesity under high-fat diet [111]. Furthermore, Sdc3−/− skeletal muscles are dystrophic and display hyperplastic nuclei in both myoblasts and satellite cells [110]. Syndecan-4 (sdc4) null mice exhibit fewer muscular defects that are limited to satellite cell function. On the other hand, absence of Sdc4 triggered degeneration of placental vessels in Sdc4−/− embryos and delayed repair after wounding [112, 113]. Nonetheless, except for some deficiencies exacerbated by stress/injury, syndecan knock-outs have been shown to be fertile and viable. This implies that overlapping functions exist between members of the syndecan family (reviewed in [114]).
Glypican-3
The Glypican-3 gene (GPC3) is mutated in patients diagnosed with Simpson-Golabi-Behmel type 1 syndrome (SGBS, MIM# 312870), a rare X-linked disease characterized by skeletal anomalies and overgrowth of various tissues and organs including bone during both pre- and postnatal life [115]. Gpc3-deficient mice exhibit most of the clinical features of SGBS patients [116]. More recently, altered hematopoiesis that result in decreased osteoclast differentiation was found to be responsible for delayed endochondral ossification in Gpc3-null mice [117]. In contrast to Gpc-3, absence of glypican-2 did not result in any obvious phenotypic defect [118]. To date, no mouse models are available for the other four glypican family members.
Heparan sulfate biosynthesis and processing enzymes
EXT1 and 2
Patients diagnosed with the hereditary multiple exostoses (HME) disease carry a mutation in one of the EXT genes (EXT1–3), develop benign cartilage-capped bony outgrowth from long bones, and often display short stature. Although disruption of one allele of the Ext1 gene only induces reduction of HS chain production without exostoses formation, Ext1-deficient embryos completely lack HS chains and die at gastrulation (E8.5) before cartilage formation takes place [119]. The cause of embryonic lethality involves regulation of HS-binding growth factor activity and is not primarily due to alteration in the structure of basement membrane as seen for knock-outs of core proteins such as agrin and perlecan. Conditional genetic disruption of EXT1 specifically in brain resulted in defects in brain development associated with abnormal midline axon guidance [120].
In contrast to Ext1 heterozygotes, disruption of one allele of Ext2 gene resulted in a significant development of exostoses as well as various defects in cartilage differentiation at distinct histological sites [121]. Over-expression of Ext 2 resulted in up-regulation of HS production, enhanced expression of Ext1, and increased formation of bone trabeculae [122]. Together, Ext2 transgenic and halploinsufficiency studies indicate that Ext2 is an essential component of the Golgi-located glycosyltransferase Ext1/Ext2 complex [123]. A recent study demonstrated that levels of Ext1 and Ext2 can affect the expression of a specific N-sulfotransferase isoform (NDST1) and global HS sulfation levels, possibly through competitive interactions in a HS biosynthetic complex called GAGosome [124].
Glucosaminyl N-deacetylase/N-sulfotransferases and C5-epimerase
Out of the four genes encoding glucosaminyl N-deacetylase/N-sulfotransferases (NDSTs) only three knock-outs have been reported. Mouse embryos carrying null mutations in the Ndst1 gene exhibited dramatic reduction in N-sulfation of HS in a wide range of tissues and died at birth from respiratory failure [125]. In addition, these newborn displayed severe developmental defects of forebrain-derived structures and developed cerebral hypoplasia, eye and facial defects [126]. Interestingly, absence of Ndst1 negatively regulated O-sulfation and epimerization reactions indicating a regulatory role in subsequent HS modifications and specific binding of HS-binding growth factors. Interestingly, C5-epimerase (Hsepi−/−) knock-out resulted in HS structural changes similar to the alterations seen in Ndst1-null mice [127]. Like for NDST1 knock-out, C5-epimerase targeted disruption resulted in perinatal lethality due to lung defects [128]. In addition, phenotypic analysis of Hsepi-null newborns revealed renal agenesis and skeletal malformations [128].
Targeted inactivation of the Ndst2 gene did not result in HS structure changes in most tissues, but induced a severe lack of granules in connective tissue-type mast cells leading to reduction in histamine and to the absence of sulfated heparin and proteases production [129]. This mast cell phenotype showed some similarities with the phenotype induced by the absence of the SG core protein in mast cells. However, targeting of the Ndst2 gene in mice remained limited to connective-tissue-type mast cells that primarily synthesize highly sulfated HS chains whereas SG deficiency induced additional defects in other cell types. These differences are likely due to the presence of both HS and chondroitin sulfate chains on SG core protein. Recent studies demonstrated that only subtle hematological and behavioral abnormalities were observed in Ndst3-null mice [130].
Lastly, compound knock-out mutations in Ndst1 and Ndst2 genes result in early embryonic lethality, and double null embryonic stem cells lacked N-sulfate but synthesized HS with 6-O-sulfate and N-unsubstituted glucosamine residues [131, 132]. These data indicated that, in the absence of Ndst1 and Ndst2, the N-deacetylating activity can be accomplished by other Ndst isozymes such as Ndst3 and that 6-O-sulfation can be partially carried out without prior N-sulfation. Further studies showed that, in fact, only one copy of the Ndst1 allele is required, as Ndst1+/−/Ndst2−/− mice developed normally to term [133]. Studies using these mouse models determined that biosynthesis of N-sulfated HS chains is crucial for the generation of binding sites for key ligands (e.g., FGF, hedgehog, Wnt) involved in important regulatory pathways during development [134, 135].
O-Sulfotransferase
Mice deficient in HS 2-O-sulfotransferase (Hs2st) display severe bilateral kidney morphogenetic defects and died as neonates due to a complete failure of kidney function [136]. Additionally, Hs2st−/− mice showed eye and skeleton abnormalities but did not exhibit lung defects as seen in Ndst1−/− neonates. Thus, distinct HS sulfation patterns are important for the development of specific organs. Although HS chains of Hs2st−/− mice lack 2-O-sulfate groups, compensatory increases in N- and 6-O-sulfation are believed to be responsible for normal affinity and signaling response to HS-binding growth factors [137].
Out of the three genes encoding HS 6-O-sulfotransferases, Hs6st1 and Hs6st2 have been knocked-out in mice. Absence of Hs6st1 has been associated with bilateral abnormal retinal innervation [138]. In addition, the majority of Hs6st1-deficient mice died between gestational stage E15.5 and birth due to severe growth retardation linked to aberrant placental angiogenesis [139]. In contrast, Hs6st2−/− develop normally, survive, and are fertile [140]. Double knock-out studies demonstrated that Hs6st1−/−/Hs6st2−/− mice displayed very little 6-O-sulfates on their HS chains and died earlier than Hs6st1−/− embryos. In these mutants, compensatory mechanisms also led to a significant increase in Hs2st activity [140]. Differential affinity and signaling properties of HS-binding growth factors in 2-O- and 6-O-sulfated depleted backgrounds when compared to wild-type indicated that both 2-O- and 6-O-sulfates are likely to regulate signaling by inducing different interactions between ligands and their receptors.
Genetic disruption of the HS 3-O-sulfotransferase-1 (Hs3st1) gene encoding for the isozyme controlling the synthesis of anticoagulant heparin species did not result in a coagulopathic phenotype possibly because 3-O-sulfates modification are rare and can be accomplished by seven distinct 3-O-sulfotransferase isoforms [141].
Secreted sulfatases
Due to redundancy between the two sulfatase (Sulf) proteins that remove 6-O-sulfate residues from HS, neither Sulf 1 nor Sulf2 genes resulted in major developmental defects [74, 142]. In contrast, 50% of Sulf1−/−/Sulf2−/− double mutants died around birth, and among the survivors only 20% became adults and displayed a short stature phenotype in conjunction with renal and skeletal defects [142]. This double mutant phenotype has been proposed to be the result of the hyper-activation of the FGF signaling pathway. Interestingly, double Sulf1−/−/Sulf2−/− mice displayed a similar, but less severe, phenotype than that observed in Hs2st-deficient mice (see above) and exhibited HS chains with increased 6-O-sulfates and decreased 2-O-sulfates levels [137, 143].
Because only double Sulf1−/−/Sulf2−/− mutants exhibit significant postnatal defects, Sulf 1 and Sulf 2 are believed to cooperate functionally [143]. However, the relatively mild developmental defect observed in Sulf1−/−/Sulf2−/− mice is in contradiction with previous studies reporting that over-expression of Sulfs in cell culture can modulate the activity of HS-binding growth factors important for embryonic patterning, and suggests that these two enzymes are rather involved in the fine-tuning of HS selectivity [144, 145].
Heparanase
Homozygous transgenic mice over-expressing human heparanase (HPSE, an endo-β-d-glucuronisade cleaving HS chains at only a few sites) under the control of the chicken β-actin promoter resulted in accelerated tissue remodeling exemplified by a significant increase in embryonic implantation sites, overbranching and widening of ducts in mammary glands, and accelerated hair regrowth and vascularization [146]. Additionally, over-expression of HPSE in osteoblasts and osteocytes induced a significant increase in bone apposition rates indicating that HS chains of proteoglycans are inhibitors of osteoblast activity and bone formation [147]. More recently, transgenic over-expression of HPSE was also shown to correlate with increased HS sulfation and increased interaction of HS-binding growth factors with their cognate receptor [148].
Summary
Molecular biology and molecular genetics have revealed a wealth of information on the structures of HSPGs, the enzymes that assemble HS chains and the essential functions these gene products play in a diverse array of biological processes. HSPG core proteins are generally large, modular and encoded by complex genes. We know little about whether these genes are alternatively spliced or to what extent these proteins are proteolytically modified in ways that generate novel functions, although a few provocative examples of this are available. We also have much to learn as to how HSPGs function in large extracellular complexes which may change in composition under different physiological states. Consequently, significantly more needs to be done to understand the biology of this important class of molecules.
References
- 1.Farach-Carson MC, Carson DD. Perlecan—a multifunctional extracellular proteoglycan scaffold. Glycobiology. 2007;17:897–905. doi: 10.1093/glycob/cwm043. [DOI] [PubMed] [Google Scholar]
- 2.Farach-Carson MC, Hecht JT, Carson DD. Heparan sulfate proteoglycans: key players in cartilage biology. Crit Rev Eukaryot Gene Expr. 2005;15:29–48. doi: 10.1615/critreveukaryotgeneexpr.v15.i1.30. [DOI] [PubMed] [Google Scholar]
- 3.Bix G, Iozzo RV. Novel interactions of perlecan: unraveling perlecan’s role in angiogenesis. Microsc Res Tech. 2008;71:339–348. doi: 10.1002/jemt.20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvey SJ, Miner JH. Revisiting the glomerular charge barrier in the molecular era. Curr Opin Nephrol Hypertens. 2008;17:393–398. doi: 10.1097/MNH.0b013e32830464de. [DOI] [PubMed] [Google Scholar]
- 5.Morita H, Yoshimura A, Inui K, Ideura T, Watanabe H, Wang L, Soininen R, Tryggvason K. Heparan sulfate of perlecan is involved in glomerular filtration. J Am Soc Nephrol. 2005;16:1703–1710. doi: 10.1681/ASN.2004050387. [DOI] [PubMed] [Google Scholar]
- 6.Whitelock JM, Melrose J, Iozzo RV. Diverse cell signaling events modulated by perlecan. Biochemistry. 2008;47:11174–11183. doi: 10.1021/bi8013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farach-Carson MC, Brown AJ, Lynam M, Safran JB, Carson DD. A novel peptide sequence in perlecan domain IV supports cell adhesion, spreading and FAK activation. Matrix Biol. 2008;27:150–160. doi: 10.1016/j.matbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwiatkowska D, Kwiatkowska-Korczak J. Adhesive glycoproteins of the extracellular matrix. Postepy Hig Med Dosw. 1999;53:55–74. [PubMed] [Google Scholar]
- 9.Castillo GM, Ngo C, Cummings J, Wight TN, Snow AD. Perlecan binds to the beta-amyloid proteins (A beta) of Alzheimer’s disease, accelerates A beta fibril formation, and maintains A beta fibril stability. J Neurochem. 1997;69:2452–2465. doi: 10.1046/j.1471-4159.1997.69062452.x. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez EM, Reed CC, Bix G, Fu J, Zhang Y, Gopalakrishnan B, Greenspan DS, Iozzo RV. BMP-1/tolloid-like metalloproteases process endorepellin, the angiostatic C-terminal fragment of perlecan. J Biol Chem. 2005;280:7080–7087. doi: 10.1074/jbc.M409841200. [DOI] [PubMed] [Google Scholar]
- 11.Bix G, Castello R, Burrows M, Zoeller JJ, Weech M, Iozzo RA, Cardi C, Thakur ML, Barker CA, Camphausen K, Iozzo RV. Endorepellin in vivo: targeting the tumor vasculature and retarding cancer growth and metabolism. J Natl Cancer Inst. 2006;98:1634–1646. doi: 10.1093/jnci/djj441. [DOI] [PubMed] [Google Scholar]
- 12.Kummer TT, Misgeld T, Sanes JR. Assembly of the postsynaptic membrane at the neuromuscular junction: paradigm lost. Curr Opin Neurobiol. 2006;16:74–82. doi: 10.1016/j.conb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Phillips WD. Acetylcholine receptors and the cytoskeletal connection. Clin Exp Pharmacol Physiol. 1995;22:961–965. doi: 10.1111/j.1440-1681.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 14.Winzen U, Cole GJ, Halfter W. Agrin is a chimeric proteoglycan with the attachment sites for heparan sulfate/chondroitin sulfate located in two multiple serine-glycine clusters. J Biol Chem. 2003;278:30106–30114. doi: 10.1074/jbc.M212676200. [DOI] [PubMed] [Google Scholar]
- 15.Williams S, Ryan C, Jacobson C. Agrin and neuregulin, expanding roles and implications for therapeutics. Biotechnol Adv. 2008;26:187–201. doi: 10.1016/j.biotechadv.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Oh SP, Warman ML, Seldin MF, Cheng SD, Knoll JH, Timmons S, Olsen BR. Cloning of cDNA and genomic DNA encoding human type XVIII collagen and localization of the alpha 1 (XVIII) collagen gene to mouse chromosome 10 and human chromosome 21. Genomics. 1994;19:494–499. doi: 10.1006/geno.1994.1098. [DOI] [PubMed] [Google Scholar]
- 17.Bix G, Iozzo RV. Matrix revolutions: “tails” of basement-membrane components with angiostatic functions. Trends Cell Biol. 2005;15:52–60. doi: 10.1016/j.tcb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Xu YK, Nusse R. The Frizzled CRD domain is conserved in diverse proteins including several receptor tyrosine kinases. Curr Biol. 1998;8:R405–R406. doi: 10.1016/s0960-9822(98)70262-3. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Mlodzik M. The frizzled extracellular domain is a ligand for Van Gogh/Stbm during nonautonomous planar cell polarity signaling. Dev Cell. 2008;15:462–469. doi: 10.1016/j.devcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckmann G, Hanke J, Bork P, Reich JG. Merging extracellular domains: fold prediction for laminin G-like and amino-terminal thrombospondin-like modules based on homology to pentraxins. J Mol Biol. 1998;275:725–730. doi: 10.1006/jmbi.1997.1510. [DOI] [PubMed] [Google Scholar]
- 21.David G. Integral membrane heparan sulfate proteoglycans. Faseb J. 1993;7:1023–1030. doi: 10.1096/fasebj.7.11.8370471. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu H, Ghazizadeh M, Sato S, Oguro T, Kawanami O. Interaction between beta-amyloid protein and heparan sulfate proteoglycans from the cerebral capillary basement membrane in Alzheimer’s disease. J Clin Neurosci. 2009;16:277–282. doi: 10.1016/j.jocn.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 23.Ohashi K. Pathogenesis of beta2-microglobulin amyloidosis. Pathol Int. 2001;51:1–10. doi: 10.1046/j.1440-1827.2001.01156.x. [DOI] [PubMed] [Google Scholar]
- 24.Leung EW, Rife L, Smith RE, Kay EP. Extracellular matrix components in retrocorneal fibrous membrane in comparison to corneal endothelium and Descemet’s membrane. Mol Vis. 2000;6:15–23. [PubMed] [Google Scholar]
- 25.Davis TH, Chen C, Isom LL. Sodium channel beta1 subunits promote neurite outgrowth in cerebellar granule neurons. J Biol Chem. 2004;279:51424–51432. doi: 10.1074/jbc.M410830200. [DOI] [PubMed] [Google Scholar]
- 26.Kuma K, Iwabe N, Miyata T. Motifs of cadherin- and fibronectin type III-related sequences and evolution of the receptor-type-protein tyrosine kinases: sequence similarity between proto-oncogene ret and cadherin family. Mol Biol Evol. 1993;10:539–551. doi: 10.1093/oxfordjournals.molbev.a040033. [DOI] [PubMed] [Google Scholar]
- 27.Hirata K, Ishida T, Penta K, Rezaee M, Yang E, Wohlgemuth J, Quertermous T. Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. J Biol Chem. 2001;276:16223–16231. doi: 10.1074/jbc.M100630200. [DOI] [PubMed] [Google Scholar]
- 28.Smith SM, West LA, Hassell JR. The core protein of growth plate perlecan binds FGF-18 and alters its mitogenic effect on chondrocytes. Arch Biochem Biophys. 2007;468:244–251. doi: 10.1016/j.abb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allen JM, Bateman JF, Hansen U, Wilson R, Bruckner P, Owens RT, Sasaki T, Timpl R, Fitzgerald J. WARP is a novel multimeric component of the chondrocyte pericellular matrix that interacts with perlecan. J Biol Chem. 2006;281:7341–7349. doi: 10.1074/jbc.M513746200. [DOI] [PubMed] [Google Scholar]
- 30.Tiedemann K, Sasaki T, Gustafsson E, Gohring W, Batge B, Notbohm H, Timpl R, Wedel T, Schlotzer-Schrehardt U, Reinhardt DP. Microfibrils at basement membrane zones interact with perlecan via fibrillin-1. J Biol Chem. 2005;280:11404–11412. doi: 10.1074/jbc.M409882200. [DOI] [PubMed] [Google Scholar]
- 31.Bezakova G, Ruegg MA. New insights into the roles of agrin. Nat Rev Mol Cell Biol. 2003;4:295–308. doi: 10.1038/nrm1074. [DOI] [PubMed] [Google Scholar]
- 32.Meinen S, Barzaghi P, Lin S, Lochmuller H, Ruegg MA. Linker molecules between laminins and dystroglycan ameliorate laminin-alpha2-deficient muscular dystrophy at all disease stages. J Cell Biol. 2007;176:979–993. doi: 10.1083/jcb.200611152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mascarenhas JB, Ruegg MA, Sasaki T, Eble JA, Engel J, Stetefeld J. Structure and laminin-binding specificity of the NtA domain expressed in eukaryotic cells. Matrix Biol. 2005;23:507–513. doi: 10.1016/j.matbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Muntoni F, Torelli S, Brockington M. Muscular dystrophies due to glycosylation defects. Neurotherapeutics. 2008;5:627–632. doi: 10.1016/j.nurt.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmer NM, van Horssen J, Otte-Holler I, Wilhelmus MM, David G, van Beers J, de Waal RM, Verbeek MM. Amyloid beta induces cellular relocalization and production of agrin and glypican-1. Brain Res. 2009;1260:38–46. doi: 10.1016/j.brainres.2008.12.063. [DOI] [PubMed] [Google Scholar]
- 36.Liu IH, Uversky VN, Munishkina LA, Fink AL, Halfter W, Cole GJ. Agrin binds alpha-synuclein and modulates alpha-synuclein fibrillation. Glycobiology. 2005;15:1320–1331. doi: 10.1093/glycob/cwj014. [DOI] [PubMed] [Google Scholar]
- 37.Marneros AG, Keene DR, Hansen U, Fukai N, Moulton K, Goletz PL, Moiseyev G, Pawlyk BS, Halfter W, Dong S, Shibata M, Li T, Crouch RK, Bruckner P, Olsen BR. Collagen XVIII/endostatin is essential for vision and retinal pigment epithelial function. EMBO J. 2004;23:89–99. doi: 10.1038/sj.emboj.7600014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickstrom SA, Alitalo K, Keski-Oja J. Endostatin signaling and regulation of endothelial cell–matrix interactions. Adv Cancer Res. 2005;94:197–229. doi: 10.1016/S0065-230X(05)94005-0. [DOI] [PubMed] [Google Scholar]
- 39.Marneros AG, Olsen BR. Physiological role of collagen XVIII and endostatin. FASEB J. 2005;19:716–728. doi: 10.1096/fj.04-2134rev. [DOI] [PubMed] [Google Scholar]
- 40.Tang H, Fu Y, Lei Q, Han Q, Ploplis VA, Castellino FJ, Li L, Luo Y. Fibrinogen facilitates the anti-tumor effect of nonnative endostatin. Biochem Biophys Res Commun. 2009;380:249–253. doi: 10.1016/j.bbrc.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delehedde M, Lyon M, Sergeant N, Rahmoune H, Fernig DG. Proteoglycans: pericellular and cell surface multireceptors that integrate external stimuli in the mammary gland. J Mammary Gland Biol Neoplasia. 2001;6:253–273. doi: 10.1023/a:1011367423085. [DOI] [PubMed] [Google Scholar]
- 42.Pakula R, Melchior A, Denys A, Vanpouille C, Mazurier J, Allain F. Syndecan-1/CD147 association is essential for cyclophilin B-induced activation of p44/42 mitogen-activated protein kinases and promotion of cell adhesion and chemotaxis. Glycobiology. 2007;17:492–503. doi: 10.1093/glycob/cwm009. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Gotte M, Bernfield M, Reizes O. Constitutive and accelerated shedding of murine syndecan-1 is mediated by cleavage of its core protein at a specific juxtamembrane site. Biochemistry. 2005;44:12355–12361. doi: 10.1021/bi050620i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muramatsu T, Muramatsu H, Kojima T. Identification of proteoglycan-binding proteins. Methods Enzymol. 2006;416:263–278. doi: 10.1016/S0076-6879(06)16017-6. [DOI] [PubMed] [Google Scholar]
- 45.Sulka B, Lortat-Jacob H, Terreux R, Letourneur F, Rousselle P. Tyrosine dephosphorylation of the syndecan-1 PDZ binding domain regulates syntenin-1 recruitment. J Biol Chem. 2009;284:10659–10671. doi: 10.1074/jbc.M807643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Capurro MI, Shi W, Sandal S, Filmus J. Processing by convertases is not required for glypican-3-induced stimulation of hepatocellular carcinoma growth. J Biol Chem. 2005;280:41201–41206. doi: 10.1074/jbc.M507004200. [DOI] [PubMed] [Google Scholar]
- 47.De Cat B, Muyldermans SY, Coomans C, Degeest G, Vanderschueren B, Creemers J, Biemar F, Peers B, David G. Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J Cell Biol. 2003;163:625–635. doi: 10.1083/jcb.200302152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronca F, Andersen JS, Paech V, Margolis RU. Characterization of slit protein interactions with glypican-1. J Biol Chem. 2001;276:29141–29147. doi: 10.1074/jbc.M100240200. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe N, Araki W, Chui DH, Makifuchi T, Ihara Y, Tabira T. Glypican-1 as an A beta binding HSPG in the human brain: its localization in DIG domains and possible roles in the pathogenesis of Alzheimer’s disease. FASEB J. 2004;18:1013–1015. doi: 10.1096/fj.03-1040fje. [DOI] [PubMed] [Google Scholar]
- 51.Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 52.Vives RR, Pye DA, Salmivirta M, Hopwood JJ, Lindahl U, Gallagher JT. Sequence analysis of heparan sulphate and heparin oligosaccharides. Biochem J. 1999;339(Pt 3):767–773. [PMC free article] [PubMed] [Google Scholar]
- 53.Vlodavsky I, Elkin M, Abboud-Jarrous G, Levi-Adam F, Fuks L, Shafat I, Ilan N. Heparanase: one molecule with multiple functions in cancer progression. Connect Tissue Res. 2008;49:207–210. doi: 10.1080/03008200802143281. [DOI] [PubMed] [Google Scholar]
- 54.Lamanna WC, Kalus I, Padva M, Baldwin RJ, Merry CL, Dierks T. The heparanome—the enigma of encoding and decoding heparan sulfate sulfation. J Biotechnol. 2007;129:290–307. doi: 10.1016/j.jbiotec.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 55.Gong F, Jemth P, Escobar Galvis ML, Vlodavsky I, Horner A, Lindahl U, Li JP. Processing of macromolecular heparin by heparanase. J Biol Chem. 2003;278:35152–35158. doi: 10.1074/jbc.M300925200. [DOI] [PubMed] [Google Scholar]
- 56.Okada Y, Yamada S, Toyoshima M, Dong J, Nakajima M, Sugahara K. Structural recognition by recombinant human heparanase that plays critical roles in tumor metastasis. Hierarchical sulfate groups with different effects and the essential target disulfated trisaccharide sequence. J Biol Chem. 2002;277:42488–42495. doi: 10.1074/jbc.M206510200. [DOI] [PubMed] [Google Scholar]
- 57.Luo Y, Ye S, Kan M, McKeehan WL. Structural specificity in a FGF7-affinity purified heparin octasaccharide required for formation of a complex with FGF7 and FGFR2IIIb. J Cell Biochem. 2006;97:1241–1258. doi: 10.1002/jcb.20724. [DOI] [PubMed] [Google Scholar]
- 58.Kemp LE, Mulloy B, Gherardi E. Signalling by HGF/SF and Met: the role of heparan sulphate co-receptors. Biochem Soc Trans. 2006;34:414–417. doi: 10.1042/BST0340414. [DOI] [PubMed] [Google Scholar]
- 59.Robinson CJ, Stringer SE. The splice variants of vascular endothelial growth factor (VEGF) and their receptors. J Cell Sci. 2001;114:853–865. doi: 10.1242/jcs.114.5.853. [DOI] [PubMed] [Google Scholar]
- 60.Miao HQ, Navarro E, Patel S, Sargent D, Koo H, Wan H, Plata A, Zhou Q, Ludwig D, Bohlen P, Kussie P. Cloning, expression, and purification of mouse heparanase. Protein Expr Purif. 2002;26:425–431. doi: 10.1016/s1046-5928(02)00558-2. [DOI] [PubMed] [Google Scholar]
- 61.Podyma-Inoue KA, Yokote H, Sakaguchi K, Ikuta M, Yanagishita M. Characterization of heparanase from a rat parathyroid cell line. J Biol Chem. 2002;277:32459–32465. doi: 10.1074/jbc.M203282200. [DOI] [PubMed] [Google Scholar]
- 62.Gilat D, Hershkoviz R, Goldkorn I, Cahalon L, Korner G, Vlodavsky I, Lider O. Molecular behavior adapts to context: heparanase functions as an extracellular matrix-degrading enzyme or as a T cell adhesion molecule, depending on the local pH. J Exp Med. 1995;181:1929–1934. doi: 10.1084/jem.181.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldshmidt O, Zcharia E, Cohen M, Aingorn H, Cohen I, Nadav L, Katz BZ, Geiger B, Vlodavsky I. Heparanase mediates cell adhesion independent of its enzymatic activity. FASEB J. 2003;17:1015–1025. doi: 10.1096/fj.02-0773com. [DOI] [PubMed] [Google Scholar]
- 64.Elkin M, Ilan N, Ishai-Michaeli R, Friedmann Y, Papo O, Pecker I, Vlodavsky I. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15:1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 65.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goshen R, Hochberg AA, Korner G, Levy E, Ishai-Michaeli R, Elkin M, de Groot N, Vlodavsky I. Purification and characterization of placental heparanase and its expression by cultured cytotrophoblasts. Mol Hum Reprod. 1996;2:679–684. doi: 10.1093/molehr/2.9.679. [DOI] [PubMed] [Google Scholar]
- 67.D’Souza SS, Daikoku T, Farach-Carson MC, Carson DD. Heparanase expression and function during early pregnancy in mice. Biol Reprod. 2007;77:433–441. doi: 10.1095/biolreprod.107.061317. [DOI] [PubMed] [Google Scholar]
- 68.Temkin V, Aingorn H, Puxeddu I, Goldshmidt O, Zcharia E, Gleich GJ, Vlodavsky I, Levi-Schaffer F. Eosinophil major basic protein: first identified natural heparanase-inhibiting protein. J Allergy Clin Immunol. 2004;113:703–709. doi: 10.1016/j.jaci.2003.11.038. [DOI] [PubMed] [Google Scholar]
- 69.D’Souza S, Yang W, Marchetti D, Muir C, Farach-Carson MC, Carson DD. HIP/RPL29 antagonizes VEGF and FGF2 stimulated angiogenesis by interfering with HS-dependent responses. J Cell Biochem. 2008;105:1183–1193. doi: 10.1002/jcb.21899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferro V, Dredge K, Liu L, Hammond E, Bytheway I, Li C, Johnstone K, Karoli T, Davis K, Copeman E, Gautam A. PI-88 and novel heparan sulfate mimetics inhibit angiogenesis. Semin Thromb Hemost. 2007;33:557–568. doi: 10.1055/s-2007-982088. [DOI] [PubMed] [Google Scholar]
- 71.Kudchadkar R, Gonzalez R, Lewis KD. PI-88: a novel inhibitor of angiogenesis. Expert Opin Investig Drugs. 2008;17:1769–1776. doi: 10.1517/13543784.17.11.1769. [DOI] [PubMed] [Google Scholar]
- 72.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao W, Sala-Newby GB, Dhoot GK. Sulf1 expression pattern and its role in cartilage and joint development. Dev Dyn. 2006;235:3327–3335. doi: 10.1002/dvdy.20987. [DOI] [PubMed] [Google Scholar]
- 74.Lum DH, Tan J, Rosen SD, Werb Z. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol Cell Biol. 2007;27:678–688. doi: 10.1128/MCB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narita K, Chien J, Mullany SA, Staub J, Qian X, Lingle WL, Shridhar V. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282:14413–14420. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- 76.Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, Zou H, Shire AM, Nagorney DM, Sanderson SO, Adjei AA, Lee JS, Thorgeirsson SS, Roberts LR. Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology. 2008;47:1211–1222. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Backen AC, Cole CL, Lau SC, Clamp AR, McVey R, Gallagher JT, Jayson GC. Heparan sulphate synthetic and editing enzymes in ovarian cancer. Br J Cancer. 2007;96:1544–1548. doi: 10.1038/sj.bjc.6603747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Costell M, Gustafsson E, Aszodi A, Morgelin M, Bloch W, Hunziker E, Addicks K, Timpl R, Fassler R. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol. 1999;147:1109–1122. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arikawa-Hirasawa E, Watanabe H, Takami H, Hassell JR, Yamada Y. Perlecan is essential for cartilage and cephalic development. Nat Genet. 1999;23:354–358. doi: 10.1038/15537. [DOI] [PubMed] [Google Scholar]
- 80.Sasse P, Malan D, Fleischmann M, Roell W, Gustafsson E, Bostani T, Fan Y, Kolbe T, Breitbach M, Addicks K, Welz A, Brem G, Hescheler J, Aszodi A, Costell M, Bloch W, Fleischmann BK. Perlecan is critical for heart stability. Cardiovasc Res. 2008;80:435–444. doi: 10.1093/cvr/cvn225. [DOI] [PubMed] [Google Scholar]
- 81.Hart M, Li L, Tokunaga T, Lindsey JR, Hassell JR, Snow AD, Fukuchi K. Overproduction of perlecan core protein in cultured cells and transgenic mice. J Pathol. 2001;194:262–269. doi: 10.1002/1096-9896(200106)194:2<262::AID-PATH882>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 82.Rodgers KD, Sasaki T, Aszodi A, Jacenko O. Reduced perlecan in mice results in chondrodysplasia resembling Schwartz–Jampel syndrome. Hum Mol Genet. 2007;16:515–528. doi: 10.1093/hmg/ddl484. [DOI] [PubMed] [Google Scholar]
- 83.Stum M, Girard E, Bangratz M, Bernard V, Herbin M, Vignaud A, Ferry A, Davoine CS, Echaniz-Laguna A, Rene F, Marcel C, Molgo J, Fontaine B, Krejci E, Nicole S. Evidence of a dosage effect and a physiological endplate acetylcholinesterase deficiency in the first mouse models mimicking Schwartz–Jampel syndrome neuromyotonia. Hum Mol Genet. 2008;17:3166–3179. doi: 10.1093/hmg/ddn213. [DOI] [PubMed] [Google Scholar]
- 84.Nicole S, Davoine CS, Topaloglu H, Cattolico L, Barral D, Beighton P, Hamida CB, Hammouda H, Cruaud C, White PS, Samson D, Urtizberea JA, Lehmann-Horn F, Weissenbach J, Hentati F, Fontaine B. Perlecan, the major proteoglycan of basement membranes, is altered in patients with Schwartz–Jampel syndrome (chondrodystrophic myotonia) Nat Genet. 2000;26:480–483. doi: 10.1038/82638. [DOI] [PubMed] [Google Scholar]
- 85.Stum M, Davoine CS, Vicart S, Guillot-Noel L, Topaloglu H, Carod-Artal FJ, Kayserili H, Hentati F, Merlini L, Urtizberea JA, Hammouda el H, Quan PC, Fontaine B, Nicole S. Spectrum of HSPG2 (Perlecan) mutations in patients with Schwartz–Jampel syndrome. Hum Mutat. 2006;27:1082–1091. doi: 10.1002/humu.20388. [DOI] [PubMed] [Google Scholar]
- 86.Rossi M, Morita H, Sormunen R, Airenne S, Kreivi M, Wang L, Fukai N, Olsen BR, Tryggvason K, Soininen R. Heparan sulfate chains of perlecan are indispensable in the lens capsule but not in the kidney. EMBO J. 2003;22:236–245. doi: 10.1093/emboj/cdg019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tran PK, Tran-Lundmark K, Soininen R, Tryggvason K, Thyberg J, Hedin U. Increased intimal hyperplasia and smooth muscle cell proliferation in transgenic mice with heparan sulfate-deficient perlecan. Circ Res. 2004;94:550–558. doi: 10.1161/01.RES.0000117772.86853.34. [DOI] [PubMed] [Google Scholar]
- 88.Tran-Lundmark K, Tran PK, Paulsson-Berne G, Friden V, Soininen R, Tryggvason K, Wight TN, Kinsella MG, Boren J, Hedin U. Heparan sulfate in perlecan promotes mouse atherosclerosis: roles in lipid permeability, lipid retention, and smooth muscle cell proliferation. Circ Res. 2008;103:43–52. doi: 10.1161/CIRCRESAHA.108.172833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tran PK, Agardh HE, Tran-Lundmark K, Ekstrand J, Roy J, Henderson B, Gabrielsen A, Hansson GK, Swedenborg J, Paulsson-Berne G, Hedin U. Reduced perlecan expression and accumulation in human carotid atherosclerotic lesions. Atherosclerosis. 2007;190:264–270. doi: 10.1016/j.atherosclerosis.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 90.Gautam M, Noakes PG, Moscoso L, Rupp F, Scheller RH, Merlie JP, Sanes JR. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- 91.Burgess RW, Nguyen QT, Son YJ, Lichtman JW, Sanes JR. Alternatively spliced isoforms of nerve- and muscle-derived agrin: their roles at the neuromuscular junction. Neuron. 1999;23:33–44. doi: 10.1016/s0896-6273(00)80751-5. [DOI] [PubMed] [Google Scholar]
- 92.Hausser HJ, Ruegg MA, Brenner RE, Ksiazek I. Agrin is highly expressed by chondrocytes and is required for normal growth. Histochem Cell Biol. 2007;127:363–374. doi: 10.1007/s00418-006-0258-2. [DOI] [PubMed] [Google Scholar]
- 93.Harvey SJ, Jarad G, Cunningham J, Rops AL, van der Vlag J, Berden JH, Moeller MJ, Holzman LB, Burgess RW, Miner JH. Disruption of glomerular basement membrane charge through podocyte-specific mutation of agrin does not alter glomerular permselectivity. Am J Pathol. 2007;171:139–152. doi: 10.2353/ajpath.2007.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fukai N, Eklund L, Marneros AG, Oh SP, Keene DR, Tamarkin L, Niemela M, Ilves M, Li E, Pihlajaniemi T, Olsen BR. Lack of collagen XVIII/endostatin results in eye abnormalities. EMBO J. 2002;21:1535–1544. doi: 10.1093/emboj/21.7.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ylikarppa R, Eklund L, Sormunen R, Kontiola AI, Utriainen A, Maatta M, Fukai N, Olsen BR, Pihlajaniemi T. Lack of type XVIII collagen results in anterior ocular defects. FASEB J. 2003;17:2257–2259. doi: 10.1096/fj.02-1001fje. [DOI] [PubMed] [Google Scholar]
- 96.Utriainen A, Sormunen R, Kettunen M, Carvalhaes LS, Sajanti E, Eklund L, Kauppinen R, Kitten GT, Pihlajaniemi T. Structurally altered basement membranes and hydrocephalus in a type XVIII collagen deficient mouse line. Hum Mol Genet. 2004;13:2089–2099. doi: 10.1093/hmg/ddh213. [DOI] [PubMed] [Google Scholar]
- 97.Kliemann SE, Waetge RT, Suzuki OT, Passos-Bueno MR, Rosemberg S. Evidence of neuronal migration disorders in Knobloch syndrome: clinical and molecular analysis of two novel families. Am J Med Genet A. 2003;119A:15–19. doi: 10.1002/ajmg.a.20070. [DOI] [PubMed] [Google Scholar]
- 98.Sertie AL, Sossi V, Camargo AA, Zatz M, Brahe C, Passos-Bueno MR. Collagen XVIII, containing an endogenous inhibitor of angiogenesis and tumor growth, plays a critical role in the maintenance of retinal structure and in neural tube closure (Knobloch syndrome) Hum Mol Genet. 2000;9:2051–2058. doi: 10.1093/hmg/9.13.2051. [DOI] [PubMed] [Google Scholar]
- 99.Iughetti P, Suzuki O, Godoi PH, Alves VA, Sertie AL, Zorick T, Soares F, Camargo A, Moreira ES, di Loreto C, Moreira-Filho CA, Simpson A, Oliva G, Passos-Bueno MR. A polymorphism in endostatin, an angiogenesis inhibitor, predisposes for the development of prostatic adenocarcinoma. Cancer Res. 2001;61:7375–7378. [PubMed] [Google Scholar]
- 100.Zorick TS, Mustacchi Z, Bando SY, Zatz M, Moreira-Filho CA, Olsen B, Passos-Bueno MR. High serum endostatin levels in Down syndrome: implications for improved treatment and prevention of solid tumours. Eur J Hum Genet. 2001;9:811–814. doi: 10.1038/sj.ejhg.5200721. [DOI] [PubMed] [Google Scholar]
- 101.Zambon L, Honma HN, Lourenco GJ, Saad IA, Mussi RK, Lima CS. A polymorphism in the angiogenesis inhibitor, endostatin, in lung cancer susceptibility. Lung Cancer. 2008;59:276–278. doi: 10.1016/j.lungcan.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 102.Ylikarppa R, Eklund L, Sormunen R, Muona A, Fukai N, Olsen BR, Pihlajaniemi T. Double knockout mice reveal a lack of major functional compensation between collagens XV and XVIII. Matrix Biol. 2003;22:443–448. doi: 10.1016/s0945-053x(03)00074-x. [DOI] [PubMed] [Google Scholar]
- 103.Alexander CM, Reichsman F, Hinkes MT, Lincecum J, Becker KA, Cumberledge S, Bernfield M. Syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice. Nat Genet. 2000;25:329–332. doi: 10.1038/77108. [DOI] [PubMed] [Google Scholar]
- 104.McDermott SP, Ranheim EA, Leatherberry VS, Khwaja SS, Klos KS, Alexander CM. Juvenile syndecan-1 null mice are protected from carcinogen-induced tumor development. Oncogene. 2007;26:1407–1416. doi: 10.1038/sj.onc.1209930. [DOI] [PubMed] [Google Scholar]
- 105.Park PW, Pier GB, Hinkes MT, Bernfield M. Exploitation of syndecan-1 shedding by Pseudomonas aeruginosa enhances virulence. Nature. 2001;411:98–102. doi: 10.1038/35075100. [DOI] [PubMed] [Google Scholar]
- 106.Elenius V, Gotte M, Reizes O, Elenius K, Bernfield M. Inhibition by the soluble syndecan-1 ectodomains delays wound repair in mice overexpressing syndecan-1. J Biol Chem. 2004;279:41928–41935. doi: 10.1074/jbc.M404506200. [DOI] [PubMed] [Google Scholar]
- 107.Stepp MA, Gibson HE, Gala PH, Iglesia DD, Pajoohesh-Ganji A, Pal-Ghosh S, Brown M, Aquino C, Schwartz AM, Goldberger O, Hinkes MT, Bernfield M. Defects in keratinocyte activation during wound healing in the syndecan-1-deficient mouse. J Cell Sci. 2002;115:4517–4531. doi: 10.1242/jcs.00128. [DOI] [PubMed] [Google Scholar]
- 108.Fears CY, Gladson CL, Woods A. Syndecan-2 is expressed in the microvasculature of gliomas and regulates angiogenic processes in microvascular endothelial cells. J Biol Chem. 2006;281:14533–14536. doi: 10.1074/jbc.C600075200. [DOI] [PubMed] [Google Scholar]
- 109.Kaksonen M, Pavlov I, Voikar V, Lauri SE, Hienola A, Riekki R, Lakso M, Taira T, Rauvala H. Syndecan-3-deficient mice exhibit enhanced LTP and impaired hippocampus-dependent memory. Mol Cell Neurosci. 2002;21:158–172. doi: 10.1006/mcne.2002.1167. [DOI] [PubMed] [Google Scholar]
- 110.Cornelison DD, Wilcox-Adelman SA, Goetinck PF, Rauvala H, Rapraeger AC, Olwin BB. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004;18:2231–2236. doi: 10.1101/gad.1214204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Strader AD, Reizes O, Woods SC, Benoit SC, Seeley RJ. Mice lacking the syndecan-3 gene are resistant to diet-induced obesity. J Clin Invest. 2004;114:1354–1360. doi: 10.1172/JCI20631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ishiguro K, Kadomatsu K, Kojima T, Muramatsu H, Nakamura E, Ito M, Nagasaka T, Kobayashi H, Kusugami K, Saito H, Muramatsu T. Syndecan-4 deficiency impairs the fetal vessels in the placental labyrinth. Dev Dyn. 2000;219:539–544. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1081>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 113.Echtermeyer F, Streit M, Wilcox-Adelman S, Saoncella S, Denhez F, Detmar M, Goetinck P. Delayed wound repair and impaired angiogenesis in mice lacking syndecan-4. J Clin Invest. 2001;107:R9–R14. doi: 10.1172/JCI10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alexopoulou AN, Multhaupt HA, Couchman JR. Syndecans in wound healing, inflammation and vascular biology. Int J Biochem Cell Biol. 2007;39:505–528. doi: 10.1016/j.biocel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 115.Pilia G, Hughes-Benzie RM, MacKenzie A, Baybayan P, Chen EY, Huber R, Neri G, Cao A, Forabosco A, Schlessinger D. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet. 1996;12:241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 116.Cano-Gauci DF, Song HH, Yang H, McKerlie C, Choo B, Shi W, Pullano R, Piscione TD, Grisaru S, Soon S, Sedlackova L, Tanswell AK, Mak TW, Yeger H, Lockwood GA, Rosenblum ND, Filmus J. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146:255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Viviano BL, Silverstein L, Pflederer C, Paine-Saunders S, Mills K, Saunders S. Altered hematopoiesis in glypican-3-deficient mice results in decreased osteoclast differentiation and a delay in endochondral ossification. Dev Biol. 2005;282:152–162. doi: 10.1016/j.ydbio.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 118.Lander AD, Selleck SB. The elusive functions of proteoglycans: in vivo veritas. J Cell Biol. 2000;148:227–232. doi: 10.1083/jcb.148.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin X, Wei G, Shi Z, Dryer L, Esko JD, Wells DE, Matzuk MM. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 2000;224:299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- 120.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 121.Stickens D, Zak BM, Rougier N, Esko JD, Werb Z. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development. 2005;132:5055–5068. doi: 10.1242/dev.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morimoto K, Shimizu T, Furukawa K, Morio H, Kurosawa H, Shirasawa T. Transgenic expression of the EXT2 gene in developing chondrocytes enhances the synthesis of heparan sulfate and bone formation in mice. Biochem Biophys Res Commun. 2002;292:999–1009. doi: 10.1006/bbrc.2002.6770. [DOI] [PubMed] [Google Scholar]
- 123.McCormick C, Duncan G, Goutsos KT, Tufaro F. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA. 2000;97:668–673. doi: 10.1073/pnas.97.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Presto J, Thuveson M, Carlsson P, Busse M, Wilen M, Eriksson I, Kusche-Gullberg M, Kjellen L. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc Natl Acad Sci USA. 2008;105:4751–4756. doi: 10.1073/pnas.0705807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ringvall M, Ledin J, Holmborn K, van Kuppevelt T, Ellin F, Eriksson I, Olofsson AM, Kjellen L, Forsberg E. Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J Biol Chem. 2000;275:25926–25930. doi: 10.1074/jbc.C000359200. [DOI] [PubMed] [Google Scholar]
- 126.Grobe K, Inatani M, Pallerla SR, Castagnola J, Yamaguchi Y, Esko JD. Cerebral hypoplasia and craniofacial defects in mice lacking heparan sulfate Ndst1 gene function. Development. 2005;132:3777–3786. doi: 10.1242/dev.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L, Spillmann D. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J Biol Chem. 2004;279:42732–42741. doi: 10.1074/jbc.M405382200. [DOI] [PubMed] [Google Scholar]
- 128.Li JP, Gong F, Hagner-McWhirter A, Forsberg E, Abrink M, Kisilevsky R, Zhang X, Lindahl U. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking l-iduronic acid and in neonatal lethality. J Biol Chem. 2003;278:28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- 129.Forsberg E, Pejler G, Ringvall M, Lunderius C, Tomasini-Johansson B, Kusche-Gullberg M, Eriksson I, Ledin J, Hellman L, Kjellen L. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 1999;400:773–776. doi: 10.1038/23488. [DOI] [PubMed] [Google Scholar]
- 130.Pallerla SR, Lawrence R, Lewejohann L, Pan Y, Fischer T, Schlomann U, Zhang X, Esko JD, Grobe K. Altered heparan sulfate structure in mice with deleted NDST3 gene function. J Biol Chem. 2008;283:16885–16894. doi: 10.1074/jbc.M709774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Forsberg E, Kjellen L. Heparan sulfate: lessons from knockout mice. J Clin Invest. 2001;108:175–180. doi: 10.1172/JCI13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Holmborn K, Ledin J, Smeds E, Eriksson I, Kusche-Gullberg M, Kjellen L. Heparan sulfate synthesized by mouse embryonic stem cells deficient in NDST1 and NDST2 is 6-O-sulfated but contains no N-sulfate groups. J Biol Chem. 2004;279:42355–42358. doi: 10.1074/jbc.C400373200. [DOI] [PubMed] [Google Scholar]
- 133.Grobe K, Ledin J, Ringvall M, Holmborn K, Forsberg E, Esko JD, Kjellen L. Heparan sulfate and development: differential roles of the N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes. Biochim Biophys Acta. 2002;1573:209–215. doi: 10.1016/s0304-4165(02)00386-0. [DOI] [PubMed] [Google Scholar]
- 134.Pallerla SR, Pan Y, Zhang X, Esko JD, Grobe K. Heparan sulfate Ndst1 gene function variably regulates multiple signaling pathways during mouse development. Dev Dyn. 2007;236:556–563. doi: 10.1002/dvdy.21038. [DOI] [PubMed] [Google Scholar]
- 135.Pan Y, Woodbury A, Esko JD, Grobe K, Zhang X. Heparan sulfate biosynthetic gene Ndst1 is required for FGF signaling in early lens development. Development. 2006;133:4933–4944. doi: 10.1242/dev.02679. [DOI] [PubMed] [Google Scholar]
- 136.Bullock SL, Fletcher JM, Beddington RS, Wilson VA. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 1998;12:1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Merry CL, Bullock SL, Swan DC, Backen AC, Lyon M, Beddington RS, Wilson VA, Gallagher JT. The molecular phenotype of heparan sulfate in the Hs2st−/− mutant mouse. J Biol Chem. 2001;276:35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- 138.Pratt T, Conway CD, Tian NM, Price DJ, Mason JO. Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm. J Neurosci. 2006;26:6911–6923. doi: 10.1523/JNEUROSCI.0505-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007;282:15578–15588. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- 140.Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J Biol Chem. 2008;283:10366–10376. doi: 10.1074/jbc.M705948200. [DOI] [PubMed] [Google Scholar]
- 141.Shworak NW, HajMohammadi S, de Agostini AI, Rosenberg RD. Mice deficient in heparan sulfate 3-O-sulfotransferase-1: normal hemostasis with unexpected perinatal phenotypes. Glycoconj J. 2002;19:355–361. doi: 10.1023/A:1025377206600. [DOI] [PubMed] [Google Scholar]
- 142.Holst CR, Bou-Reslan H, Gore BB, Wong K, Grant D, Chalasani S, Carano RA, Frantz GD, Tessier-Lavigne M, Bolon B, French DM, Ashkenazi A. Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS ONE. 2007;2:e575. doi: 10.1371/journal.pone.0000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lamanna WC, Baldwin RJ, Padva M, Kalus I, Ten Dam G, van Kuppevelt TH, Gallagher JT, von Figura K, Dierks T, Merry CL. Heparan sulfate 6-O-endosulfatases: discrete in vivo activities and functional co-operativity. Biochem J. 2006;400:63–73. doi: 10.1042/BJ20060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang S, Ai X, Freeman SD, Pownall ME, Lu Q, Kessler DS, Emerson CP., Jr QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc Natl Acad Sci USA. 2004;101:4833–4838. doi: 10.1073/pnas.0401028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP., Jr QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zcharia E, Metzger S, Chajek-Shaul T, Aingorn H, Elkin M, Friedmann Y, Weinstein T, Li JP, Lindahl U, Vlodavsky I. Transgenic expression of mammalian heparanase uncovers physiological functions of heparan sulfate in tissue morphogenesis, vascularization, and feeding behavior. FASEB J. 2004;18:252–263. doi: 10.1096/fj.03-0572com. [DOI] [PubMed] [Google Scholar]
- 147.Kram V, Zcharia E, Yacoby-Zeevi O, Metzger S, Chajek-Shaul T, Gabet Y, Muller R, Vlodavsky I, Bab I. Heparanase is expressed in osteoblastic cells and stimulates bone formation and bone mass. J Cell Physiol. 2006;207:784–792. doi: 10.1002/jcp.20625. [DOI] [PubMed] [Google Scholar]
- 148.Escobar Galvis ML, Jia J, Zhang X, Jastrebova N, Spillmann D, Gottfridsson E, van Kuppevelt TH, Zcharia E, Vlodavsky I, Lindahl U, Li JP. Transgenic or tumor-induced expression of heparanase upregulates sulfation of heparan sulfate. Nat Chem Biol. 2007;3:773–778. doi: 10.1038/nchembio.2007.41. [DOI] [PubMed] [Google Scholar]
- 149.Habuchi H, Suzuki S, Saito T, Tamura T, Harada T, Yoshida K, Kimata K. Structure of a heparan sulphate oligosaccharide that binds to basic fibroblast growth factor. Biochem J. 1992;285(Pt 3):805–813. doi: 10.1042/bj2850805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ashikari-Hada S, Habuchi H, Kariya Y, Itoh N, Reddi AH, Kimata K. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J Biol Chem. 2004;279:12346–12354. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- 151.Noti C, de Paz JL, Polito L, Seeberger PH. Preparation and use of microarrays containing synthetic heparin oligosaccharides for the rapid analysis of heparin–protein interactions. Chemistry. 2006;12:8664–8686. doi: 10.1002/chem.200601103. [DOI] [PubMed] [Google Scholar]
- 152.Deakin JA, Lyon M. Differential regulation of hepatocyte growth factor/scatter factor by cell surface proteoglycans and free glycosaminoglycan chains. J Cell Sci. 1999;112(Pt 12):1999–2009. doi: 10.1242/jcs.112.12.1999. [DOI] [PubMed] [Google Scholar]
- 153.Lyon M, Deakin JA, Mizuno K, Nakamura T, Gallagher JT. Interaction of hepatocyte growth factor with heparan sulfate. Elucidation of the major heparan sulfate structural determinants. J Biol Chem. 1994;269:11216–11223. [PubMed] [Google Scholar]
- 154.Ashikari S, Habuchi H, Kimata K. Characterization of heparan sulfate oligosaccharides that bind to hepatocyte growth factor. J Biol Chem. 1995;270:29586–29593. doi: 10.1074/jbc.270.49.29586. [DOI] [PubMed] [Google Scholar]